Bacterial cold-water disease, caused by F. psychrophilum, is a major problem for salmonid aquaculture. Little is known regarding the virulence factors involved in this disease, and control measures are inadequate. A targeted gene deletion method was adapted to F. psychrophilum and used to demonstrate the importance of the T9SS in virulence. Proteins secreted by this system are likely virulence factors and targets for the development of control measures.
KEYWORDS: Flavobacterium, fish pathogen, protein secretion, rainbow trout, virulence
ABSTRACT
Flavobacterium psychrophilum causes bacterial cold-water disease in wild and aquaculture-reared fish and is a major problem for salmonid aquaculture. The mechanisms responsible for cold-water disease are not known. It was recently demonstrated that the related fish pathogen, Flavobacterium columnare, requires a functional type IX protein secretion system (T9SS) to cause disease. T9SSs secrete cell surface adhesins, gliding motility proteins, peptidases, and other enzymes, any of which may be virulence factors. The F. psychrophilum genome has genes predicted to encode components of a T9SS. Here, we used a SacB-mediated gene deletion technique recently adapted for use in the Bacteroidetes to delete a core F. psychrophilum T9SS gene, gldN. The ΔgldN mutant cells were deficient for secretion of many proteins in comparison to wild-type cells. Complementation of the mutant with wild-type gldN on a plasmid restored secretion. Compared to wild-type and complemented strains, the ΔgldN mutant was deficient in adhesion, gliding motility, and extracellular proteolytic and hemolytic activities. The ΔgldN mutant exhibited reduced virulence in rainbow trout and complementation restored virulence, suggesting that the T9SS plays an important role in the disease.
IMPORTANCE Bacterial cold-water disease, caused by F. psychrophilum, is a major problem for salmonid aquaculture. Little is known regarding the virulence factors involved in this disease, and control measures are inadequate. A targeted gene deletion method was adapted to F. psychrophilum and used to demonstrate the importance of the T9SS in virulence. Proteins secreted by this system are likely virulence factors and targets for the development of control measures.
INTRODUCTION
The fish pathogen Flavobacterium psychrophilum is a major cause of disease and mortality for aquaculture-reared salmonids, such as rainbow trout (Oncorhynchus mykiss) (1–3). Ayu (Plecoglossus altivelis), European eel (Anguilla anguilla), and other fish are also impacted by this pathogen (2–5). F. psychrophilum infections cause bacterial cold-water disease (BCWD), which often results in erosion of the tail, destruction of tissues near the dorsal fin (saddleback lesion), and systemic spread to internal organs, such as the spleen and kidney. In young fish, mortality may occur rapidly and without obvious surface lesions, a condition sometimes referred to as rainbow trout fry syndrome. F. psychrophilum infections are a challenge to sustainable aquaculture of salmonids and result in large economic losses (6). The mechanisms used by F. psychrophilum to cause disease are poorly understood, and methods to prevent or control outbreaks are inadequate.
Antibiotic treatments have been the primary method to control outbreaks of BCWD (2, 7). The expense of such treatments, the danger of development of resistant strains, and the potential spread of antibiotic resistance genes to other bacteria make this a poor solution. Vaccine development has been complicated by the need to protect young fry that may not have fully developed immune systems, uncertainties regarding the most protective antigens, and the need to efficiently administer the vaccine to large numbers of fish (6). Bacteriophages are promising tools to control outbreaks, but additional studies are needed to determine if this approach will be broadly successful (8). Rainbow trout lines resistant to F. psychrophilum have also been developed and used to control the disease (9–11). Improved understanding of the mechanisms used by F. psychrophilum to cause disease could aid the continued development of these and other measures to prevent or control BCWD outbreaks.
Previous studies identified F. psychrophilum genes and proteins that may be involved in disease. Peptidases and other secreted enzymes and cell surface adhesins are suspected to be involved (1, 2, 12, 13). F. psychrophilum gliding motility, which allows cells to crawl over surfaces, has also been suggested as a potential virulence factor (7, 14). Genome analyses suggest that F. psychrophilum uses the type IX secretion system (T9SS) to secrete many proteins to the cell surface and beyond (15, 16). T9SSs are common in but apparently confined to members of the phylum Bacteroidetes (16–20). T9SSs secrete cell surface adhesins and soluble or cell-associated peptidases, nucleases, and other hydrolytic enzymes. T9SSs also secrete motility adhesins to the cell surface and are thus required for gliding motility (21–23). These adhesins are propelled along the cell surface by the rest of the gliding motility machinery, resulting in cell movement (24). Core components of the T9SS include the cytoplasmic membrane proteins GldL and GldM, the periplasmic protein GldN, the lipoprotein GldK, and the outer membrane protein SprA (25–29). These proteins are thought to form an envelope-spanning complex that secretes proteins through the outer membrane SprA channel (22, 25, 27). GldL and GldM have been proposed to harvest the proton gradient across the cytoplasmic membrane to power both secretion and gliding (22, 27). Additional proteins involved in secretion are associated with these core components (18–20).
Proteins secreted by the T9SS have N-terminal signal peptides that facilitate their export across the cytoplasmic membrane by the Sec system and C-terminal domains (CTDs) that target them to the T9SS for secretion across the outer membrane (18–20). Most characterized T9SS CTDs belong to either the TIGRFAM protein domain family TIGR04183 (type A CTDs) or to TIGR04131/pfam13585 (type B CTDs) (30, 31). Type A CTDs are typically removed during or after secretion (32), but removal of type B CTDs has not been carefully examined. Some proteins secreted by T9SS are secreted in soluble form, whereas others become covalently attached to the cell surface (17, 33–35). Genome analyses can be used to identify genes predicted to encode proteins with type A CTDs or type B CTDs, thus facilitating discovery of proteins secreted by T9SSs (30, 34). Many Bacteroidetes are predicted to secrete dozens to hundreds of proteins using this system, and T9SS-mediated secretion of many of these proteins has been biochemically verified (34, 36, 37).
Recent experiments demonstrate that T9SSs are required for virulence of the human pathogens Porphyromonas gingivalis and Prevotella melanogenica, the fish pathogen Flavobacterium columnare, and the bird pathogen Riemerella anatipestifer (17, 21, 38–41). In each case, disruption of a gene encoding a core component of the T9SS resulted in secretion defects and loss of virulence. Proteins secreted by the T9SSs of each of these organisms are thus potential virulence factors. The role of core components of the F. psychrophilum T9SS in virulence has not been directly explored, but mutations in gldD and gldG that disrupted gliding motility and virulence also impacted the T9SS (14).
Methods to genetically manipulate F. psychrophilum have been developed (42, 43). These include replicative plasmids, transposons, and methods to make site-directed insertions or deletions by homologous recombination. Genetic manipulations remain challenging, however, at least in part because of the low frequencies of gene transfer into F. psychrophilum (42). This is especially challenging for the gene deletion approach described above, which requires multiple rounds of gene transfer (43). Another method to construct gene deletions was recently developed for F. columnare (44) and modified for use in many other Bacteroidetes (45). This method involves cloning regions upstream and downstream from the target gene into a plasmid that cannot replicate in members of the Bacteroidetes. The plasmid is introduced into the bacterium, and gene deletion occurs as a result of two recombination events between the regions on the plasmid and the identical sequences on the chromosome. The plasmid carries an antibiotic resistance gene that allows selection for the first recombination (integrating the plasmid into the chromosome) and sacB, which confers sensitivity to sucrose, allowing selection for the second recombination (45, 46). Here, we describe the use of this system to construct an F. psychrophilum gldN deletion mutant and demonstrate the importance of the T9SS for F. psychrophilum virulence.
RESULTS
The F. psychrophilum T9SS and predicted T9SS-secreted proteins.
Analysis of the F. psychrophilum strain OSU THCO2-90 genome (47) revealed each of the T9SS components (Table S1 in the supplemental material), as previously reported for other F. psychrophilum strains (15–17). Most known proteins that are secreted by T9SSs have CTDs that belong to either TIGRFAM protein family TIGR04183 (type A CTDs) or TIGR04131/pfam13585 (type B CTDs). We examined the 2,344 predicted F. psychrophilum strain OSU THCO2-90 proteins and identified 49 proteins predicted to be secreted by the T9SS. Thirty-nine of these had type A CTDs (Table 1), and 10 had type B CTDs (Table 2). This probably underestimates the number of T9SS-secreted proteins, because some proteins secreted by T9SSs have novel CTDs that are not easily assigned to either the type A or type B CTD families (33). Included in the 49 predicted secreted proteins are 9 predicted peptidases, 2 predicted nucleases, 1 predicted lipase, 1 predicted glycoside hydrolase, and 28 potential adhesins, any of which may contribute to virulence. The 28 potential adhesins include 17 that contain leucine-rich repeats (LRRs) and are encoded by tandemly organized genes (Table 1). These LRR proteins are similar to Bacteroides forsythus BspA and Treponema denticola LrrA, which are proposed to be involved in attachment to host cells (48, 49). One of the non-LRR adhesins, SprB, is involved in gliding motility in related bacteria (50). Flavobacterium johnsoniae SprB is propelled along the cell surface, resulting in cell movement (24, 50). SprB-mediated gliding of F. psychrophilum cells over and through fish mucus and tissues may be important for virulence. The 49 predicted T9SS-secreted proteins are highly conserved in other F. psychrophilum strains. Orthologs of each protein were present in the three additional F. psychrophilum strains that we examined, JIP 02/86, CSF-259-93, and the type strain ATCC 49418T (12, 13, 51).
TABLE 1.
Predicted T9SS type A CTD-containing proteins of F. psychrophilum strain OSU THCO2-90a
| Locus tag | Protein | Length (aa)b | Molecular mass (kDa)c | Domain(s)d | Description/known or predicted function |
|---|---|---|---|---|---|
| THC0290_0086 | 642 | 70.9 | M1_APN_like (cd09603), peptidase_M1 (pfam01433) | Probable peptidase precursor (M1 family metalloprotease) | |
| THC0290_0091 | 1,139 | 122.8 | Sortilin-Vps10 superfamily (cl25791), peptidase_M6 superfamily (cl29544) | Probable glycoside hydrolase precursor | |
| THC0290_0128 | 113 | 12.8 | |||
| THC0290_0129 | 324 | 33.6 | Putative outer membrane protein precursor | ||
| THC0290_0170 | 398 | 41.7 | LRR_5 (pfam13306) | Probable cell surface protein (leucine-rich repeat protein) precursor | |
| THC0290_0171 | 421 | 44.5 | LRR_5 (pfam13306) | Probable cell surface protein precursor (leucine-rich repeat protein) precursor | |
| THC0290_0172 | 329 | 35.2 | LRR_5 (pfam13306) | Probable cell surface protein (leucine-rich repeat protein) | |
| THC0290_0173 | 444 | 47.2 | LRR_5 (pfam13306) | Probable cell surface protein (leucine-rich repeat protein) precursor | |
| THC0290_0174 | 422 | 44.5 | LRR_5 (pfam13306) | Probable cell surface protein (leucine-rich repeat protein) precursor | |
| THC0290_0175 | 330 | 35.3 | LRR_5 (pfam13306) | Probable cell surface protein (leucine-rich repeat protein) precursor | |
| THC0290_0176 | 375 | 40.1 | LRR_5 (pfam13306) | Probable cell surface protein (leucine-rich repeat protein) precursor | |
| THC0290_0177 | 536 | 56.0 | LRR_5 (pfam13306) | Probable cell surface protein (leucine-rich repeat protein) precursor | |
| THC0290_0178 | 307 | 33.1 | LRR_5 (pfam13306) | Probable cell surface protein (leucine-rich repeat protein) precursor | |
| THC0290_0179 | 421 | 44.3 | LRR_5 (pfam13306) | Probable cell surface protein (leucine-rich repeat protein) precursor | |
| THC0290_0180 | 398 | 42.1 | LRR_5 (pfam13306) | Probable cell surface protein (leucine-rich repeat protein) precursor | |
| THC0290_0181 | 467 | 50.1 | LRR_5 (pfam13306) | Probable cell surface protein (leucine-rich repeat protein) precursor | |
| THC0290_0182 | 329 | 35.0 | LRR_5 (pfam13306) | Probable cell surface protein (leucine-rich repeat protein) precursor | |
| THC0290_0183 | 306 | 33.2 | LRR_5 (pfam13306) | Probable cell surface protein (leucine-rich repeat protein) precursor | |
| THC0290_0184 | 351 | 37.0 | LRR_5 (pfam13306) | Probable cell surface protein (leucine-rich repeat protein) precursor | |
| THC0290_0185 | 329 | 35.3 | LRR_5 (pfam13306) | Probable cell surface protein (leucine-rich repeat protein) precursor | |
| THC0290_0186 | 328 | 35.3 | LRR_5 (pfam13306) | Probable cell surface protein (leucine-rich repeat protein) precursor | |
| THC0290_0237 | Fpp1 | 1,138 | 119.6 | ZnMc superfamily (cl00064), FN3 superfamily (cl21522), P_proprotein superfamily (cl27557) | Peptidase precursor (psychrophilic metalloprotease Fpp1) |
| THC0290_0238 | Fpp2 | 942 | 100.3 | ZnMc_pappalysin_like (cd04275), cleaved_adhesin superfamily (cl06636) | Peptidase precursor (psychrophilic metalloprotease Fpp2) |
| THC0290_0299 | 914 | 100.1 | Peptidase_M36 (pfam02128), PA_subtilisin_1 (cd04818) | Probable peptidase precursor (M36 fungalysin family metalloprotease) | |
| THC0290_0300 | 919 | 99.8 | Peptidase_M36 (pfam02128), PA_subtilisin_1 (cd04818) | Probable peptidase precursor (M36 fungalysin family metalloprotease) | |
| THC0290_0878 | 747 | 78.2 | FN3 (cd00063) | Putative adhesin precursor | |
| THC0290_0931 | 959 | 103.2 | ZnMc superfamily (cl00064) | Peptidase precursor (collagenase; Zn-dependent metalloprotease) | |
| THC0290_0944 | 544 | 58.5 | Peptidases_S8_9 (cd07493) | Probable peptidase precursor (S8 subtilisin family serine endopeptidase) | |
| THC0290_0977 | 585 | 64.9 | Abhydrolase superfamily (cl21494) | Esterase/lipase/thioesterase family protein precursor | |
| THC0290_1054 | 408 | 46.0 | SGNH_hydrolase superfamily (cl01053), PKD repeat (COG3291) | ||
| THC0290_1343 | 1,218 | 134.1 | ZnMc superfamily (cl00064), PKD (COG3291, 3 copies), PCC superfamily (cl28216) | Probable cell surface protein precursor; putative metallopeptidase, metzincin clan | |
| THC0290_1494 | 657 | 71.5 | HNHc superfamily (cl00083) | Probable ribonuclease | |
| THC0290_1520 | 613 | 66.2 | Endonuclease_1 (pfam04231), FN3 (cd00063), COG2374 superfamily (cl28586) | Probable endonuclease precursor | |
| THC0290_1595 | 628 | 67.9 | Putative adhesin precursor | ||
| THC0290_1797 | 263 | 29.4 | DOMON_DOH (cd09631) | ||
| THC0290_2029 | 301 | 32.9 | |||
| THC0290_2146 | PorU | 1,278 | 142.1 | Peptidase_C25_N (cd02258) | Peptidase precursor (T9SS CTD signal peptidase, PorU) |
| THC0290_2157 | 166 | 18.7 | |||
| THC0290_2385 | 456 | 49.2 | ATS1 superfamily (cl34932), RCC1 (pfam00415) | RCC1 (regulator of chromosome condensation) repeat domain protein precursor | |
| Avg | 550 | 59.3 | |||
| Median | 422 | 46.0 | |||
| Range | 113 to 1,278 | 12.8 to 142.1 | |||
The T9SS type A C-terminal domain (CTD) (TIGR04183) is a C-terminal domain that targets a protein for secretion by the T9SS. TIGR04183 is described as “Por secretion system C-terminal sorting domain” on the J. Craig Venter Institute TIGR website (http://www.jcvi.org/cgi-bin/tigrfams/index.cgi). Each of these proteins contains an N-terminal signal peptide for export across the cytoplasmic membrane and a T9SS type A CTD for secretion across the outer membrane by the T9SS, as determined using SignalP 5.0 with a cutoff of 0.8 and the Integrated Microbial Genomes (IMG version 4.0.1 [https://img.jgi.doe.gov/]) tools and trusted cutoffs, respectively.
Length in amino acids (aa) of full-length protein before removal of signal peptide or CTD.
Molecular mass (kDa) of amino acids in full-length protein before removal of signal peptide or CTD.
Domains other than signal peptide and CTDs, with pfam, cl, cd, or COG numbers indicated.
TABLE 2.
Predicted T9SS type B CTD-containing proteins of F. psychrophilum strain OSU THCO2-90a
| Locus tag | Name | Length (aa)b | Molecular mass (kDa)c | Domain(s)d | Description/function or predicted function |
|---|---|---|---|---|---|
| THC0290_0023 | SprC | 477 | 51.2 | FlgD_ig superfamily (cl21544) | Gliding motility protein precursor, SprC |
| THC0290_0025 | SprB | 3,521 | 366.8 | SprB repeat (pfam13573, 3 copies), FlgD Ig-like domain (cl21544) | Gliding motility adhesin precursor, SprB |
| THC0290_0332 | 2,653 | 269.2 | ATS1 superfamily (cl34932), Calx-beta domain (cl02522), herpes_BLLF1 superfamily (cl37540) | Putative adhesin precursor | |
| THC0290_1047 | 798 | 87.8 | Putative adhesin precursor | ||
| THC0290_1048 | 757 | 82.6 | Putative adhesin precursor | ||
| THC0290_1527 | 2,229 | 233.6 | Ig_2 (pfam13895) | Putative adhesin precursor | |
| THC0290_1616 | 1,253 | 129.6 | YjdB (COG5492, 2 copies) Big_2 (cl02708) | Putative adhesin precursor | |
| THC0290_1818 | 1,452 | 158.9 | Putative adhesin precursor | ||
| THC0290_1932 | 2,001 | 200.0 | DUF11 (pfam01345), PRK08026 superfamily (cl35635) | Putative adhesin precursor | |
| THC0290_2338 | 2,008 | 208.3 | fn3 (pfam00041, 2 copies), FN3 (cd00063, 2 copies), CUB (cd00041) | Putative adhesin precursor | |
| Avg | 1,715 | 178.8 | |||
| Median | 1,727 | 179.5 | |||
| Range | 477 to 3,521 | 51.2 to 366.8 | |||
aThe T9SS type B CTD (TIGR04131/pfam13585) is a C-terminal domain that targets a protein for secretion by the T9SS. TIGR04131 is described as “gliding motility-associated C-terminal domain” on the J. Craig Venter Institute TIGR website (http://www.jcvi.org/cgi-bin/tigrfams/index.cgi). Each of these proteins contains an N-terminal signal peptide for export across the cytoplasmic membrane and a T9SS type B CTD for secretion across the outer membrane by the T9SS, as determined using SignalP 5.0 with a cutoff of 0.7 and the IMG tools and trusted cutoffs, respectively. THC0290_0023 (SprC) matched only pfam13585, whereas each of the other proteins matched both TIGR04131 and pfam13585.
bLength in amino acids (aa) of full-length protein before removal of signal peptide or CTD.
cMolecular mass (kDa) of amino acids in full-length protein before removal of signal peptide or CTD.
dDomains other than signal peptide and CTDs, with pfam, cl, cd, or COG numbers indicated.
Previous studies of F. johnsoniae revealed general features of T9SS-secreted proteins (30) that we examined here for F. psychrophilum. As with F. johnsoniae, F. psychrophilum proteins with type B CTDs were typically much larger than those with type A CTDs (Tables 1 and 2). The median sizes of proteins with type A CTDs and type B CTDs were 46.0 kDa and 179.5 kDa, respectively, and the largest proteins with type A CTDs and type B CTDs were 142.1 kDa and 366.8 kDa, respectively. As for F. johnsoniae, many F. psychrophilum proteins with type A CTDs are predicted or known to have enzymatic functions, whereas this was not the case for any proteins with type B CTDs (Tables 1 and 2). Instead, many of the type B CTD proteins are suspected to be cell surface adhesins, at least one of which (SprB) is likely involved in gliding motility.
F. johnsoniae proteins with type B CTDs require the assistance of additional outer membrane proteins that belong to the PorP/SprF protein domain family TIGR03519 for secretion by the T9SS (30, 52). In many cases, these PorP/SprF-like proteins are specific for individual secreted proteins. F. johnsoniae SprB, for example, requires coexpression with SprF for its secretion. PorP/SprF-like proteins are often encoded by the genes immediately downstream from those encoding their cognate type B CTD-containing secreted proteins. F. psychrophilum strain OSU THCO2-90 has seven porP/sprF-like genes (Tables S1 and S2), and six of these lie immediately downstream from genes encoding type B CTD proteins. This synteny suggests that, as for F. johnsoniae, these six F. psychrophilum type B CTD proteins may each require their cognate PorP/SprF-like proteins for secretion. The remaining four type B CTD proteins may use the “orphan” PorP/SprF protein, THC0290_0614, which does not have an obvious cognate type B CTD protein, based on gene arrangement. Three of the F. psychrophilum porP/sprF genes lie immediately upstream from and are transcribed in the same direction as genes encoding predicted peptidoglycan-binding proteins related to P. gingivalis PG1058, which is required for T9SS function (53). These proteins share similarity to the C-terminal peptidoglycan-binding domain of Escherichia coli OmpA (54, 55). The secreted type B CTD proteins, PorP/SprF-like proteins, and PG1058-like proteins may form cell surface complexes, as predicted for similar F. johnsoniae proteins (30).
sacB-mediated deletion of the F. psychrophilum T9SS gene gldN.
To investigate the role of the T9SS in F. psychrophilum virulence, we deleted gldN, which is essential for T9SS function in related bacteria (17, 21, 56). Regions upstream and downstream from gldN of approximately 3 kbp in size were amplified by PCR, inserted into the sacB-containing suicide vector pYT313 (45), and introduced into F. psychrophilum strain OSU THCO2-90 by conjugation. Selection for erythromycin resistance resulted in 123 colonies. These had the plasmid inserted in the genome by homologous recombination either upstream or downstream from gldN. Three colonies were streaked for isolation and then grown in broth culture without selection to allow a second recombination event, resulting in loss of the inserted plasmid. These cultures were selected for sucrose resistance, and hundreds of colonies lacking the plasmid were obtained. Plasmid loss was expected to result in approximately equal numbers of wild-type colonies and gldN deletion mutant colonies, depending on whether the second recombination occurred on the same side of gldN as the first recombination or on the opposite side, respectively. PCR analyses using primers flanking gldN were used to identify deletion mutants. Of 11 independent sucrose-resistant colonies examined, 7 had gldN deleted, and the remaining 4 had gldN intact and were thus wild type. One representative mutant colony (FpT13), which we refer to hereinafter as the ΔgldN mutant, was selected for further analysis.
GldN is required for gliding motility and the formation of spreading colonies.
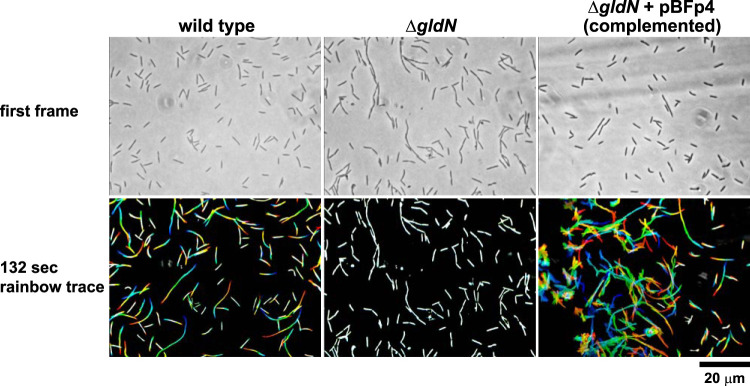
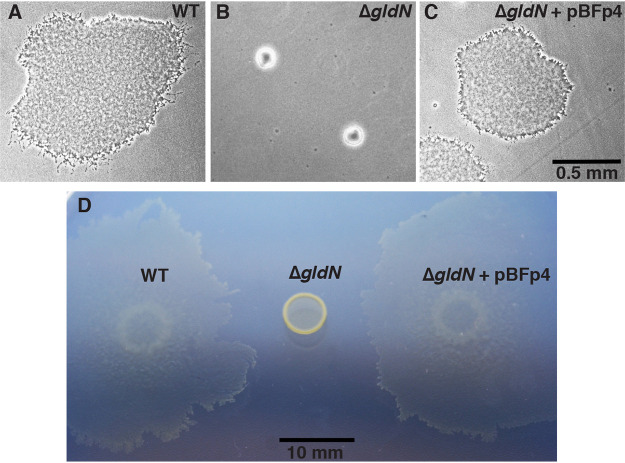
GldN is required for gliding motility in related bacteria (21, 56–58). Cells of wild-type F. psychrophilum moved over agar by gliding, whereas cells of the ΔgldN mutant did not (Fig. 1 and Movie S1). Complementation of the ΔgldN mutant by introduction of pBFp4, which carries gldN, restored gliding motility. As a result of the gliding movements, cells of the wild-type and complemented strains formed thin spreading colonies on 5% tryptone yeast extract salts (TYES) medium (TYES diluted 20-fold) that was solidified with agar (10 g/liter), whereas cells of the ΔgldN mutant formed nonspreading colonies (Fig. 2). In our experiments, the wild type formed spreading colonies on TYES agar media in which all components except the agar had been diluted at least 2-fold, and all of the strains formed nonspreading colonies on full-strength TYES agar (data not shown). This is consistent with previous results that demonstrated that spreading F. psychrophilum colonies were only observed on nutrient-poor media or media prepared with decreased amounts of agar (59).
FIG 1.
Gliding of wild-type and ΔgldN mutant cells on agar. Cells were grown in TYES at 18°C for 24 h to late exponential phase (OD of approximately 1.3). Ten microliters of cultures were spotted on TYES solidified with agar (10 g/liter) and observed for motility using an Olympus BH-2 phase-contrast microscope. Results for wild-type (WT) F. psychrophilum, the ΔgldN mutant, and the ΔgldN mutant complemented with wild-type gldN on pBFp4 are shown. In each case, a series of images were taken using a Photometrics Cool-SNAPcf2 camera. Individual frames were colored from red (time zero) to yellow, green, cyan, and finally blue (132 s) and integrated into one image, resulting in “rainbow traces” of gliding cells. The top row shows the first frame for each strain, and the bottom row shows the corresponding 132-s rainbow trace. White cells in the bottom panel correspond to cells that exhibited little if any net movement. Scale bar at lower right applies to all panels. The rainbow traces correspond to the sequences in Movie S1 in the supplemental material.
FIG 2.
Colonies of wild-type, ΔgldN mutant, and complemented strains of F. psychrophilum. (A to C) Microcolonies of wild type (WT), ΔgldN mutant, and ΔgldN mutant complemented with wild-type gldN on pBFp4. Colonies arose from cells incubated for 32 h at 18°C on 5% TYES solidified with agar (10 g/liter). Photomicrographs were taken with a Photometrics Cool-SNAPcf2 camera mounted on an Olympus IMT-2 phase-contrast microscope. Scale bar in panel C applies to panels A, B, and C. (D) Macrocolonies of wild type, ΔgldN mutant, and complemented mutant. Cells were spotted on 5% TYES solidified with agar (10 g/liter), incubated at 18°C for 12 days, and photographed using a Nikon D7200 camera.
The gldN deletion mutant appears to be deficient in protein secretion.
Cell-free culture fluid samples from wild-type, mutant, and complemented strains were examined by SDS-PAGE for soluble secreted proteins. The ΔgldN mutant cells released much less protein than did wild-type or complemented cells (Fig. 3), suggesting a protein secretion defect. Samples of cell-free culture fluid were examined by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The numbers of spectral counts of predicted secreted proteins detected are shown in Table 3. In most cases, the ΔgldN mutant had far fewer spectral counts than did the wild-type or complemented strains, corroborating a secretion defect in the mutant. The exceptions were primarily the secreted LRR proteins. All predicted LRR proteins had type A T9SS CTDs, and requirement of GldN for their secretion was expected. However, LRR proteins were detected in the cell-free spent culture fluid of both wild-type and mutant cells, and in some cases, larger amounts were detected in the culture fluid of the ΔgldN mutant. We do not know the reason for this observation, but the ΔgldN mutant did not appear to have a general permeability defect, as revealed by the results shown in Fig. 3 and by the full LC-MS/MS data set that includes all proteins detected in the cell-free spent medium (Data Set S1), which had very few spectra corresponding to ribosomal proteins or other abundant cytoplasmic proteins.
FIG 3.
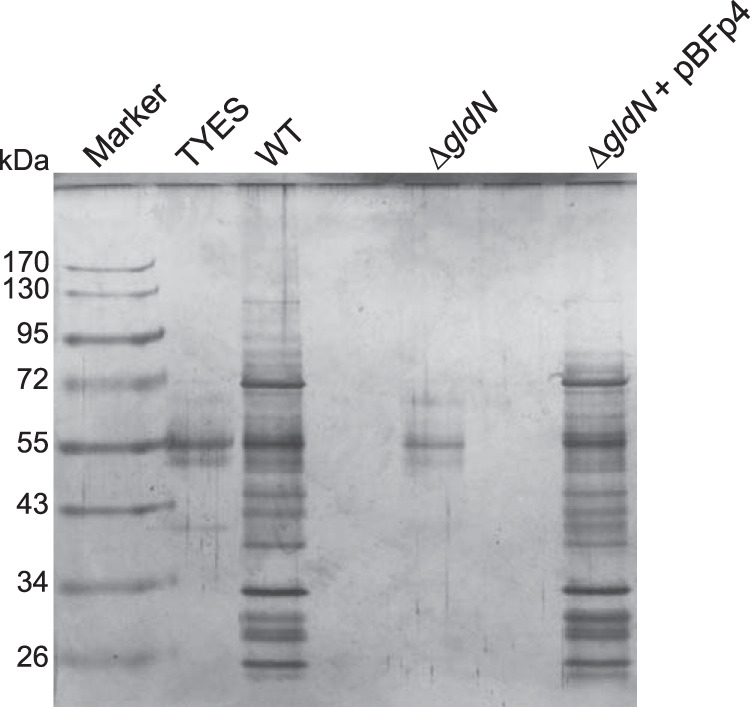
Soluble extracellular proteins of wild-type and ΔgldN mutant cells. Cells of wild-type F. psychrophilum, the ΔgldN mutant, and the ΔgldN mutant complemented with wild-type gldN on pBFp4 were grown in TYES medium at 18°C with shaking until cells reached early stationary phase of growth (Klett readings of 160). Cells were removed by centrifugation followed by filtration (0.45 μm). Proteins from equal amounts of cell-free spent media of wild-type, mutant, and complemented cells were precipitated with TCA, solubilized in loading buffer, and separated by SDS-PAGE. Proteins were detected by silver staining. Lane labeled “TYES” contained an equivalent amount of TYES growth medium that had not been inoculated with F. psychrophilum, indicating bands that correspond to components of the growth medium.
TABLE 3.
F. psychrophilum T9SS type A CTD and type B CTD proteins identified by LC-MS/MS analysis of cell-free spent culture mediuma
| Locus tag/protein | Molecular mass (kDa)b | T9SS CTD typec | Predicted protein function or description | Spectral count of culture fluid from: |
||
|---|---|---|---|---|---|---|
| Wild type | ΔgldN mutant | ΔgldN mutant + pBFp4 | ||||
| Non-LRR T9SS CTD proteins | ||||||
| THC0290_0238/Fpp2 | 100.3 | A | Peptidase | 813 | 72 | 903 |
| THC0290_0025/SprB | 366.8 | B | Gliding motility adhesin SprB | 268 | 59 | 315 |
| THC0290_0931 | 103.2 | A | Peptidase (collagenase) | 170 | 22 | 143 |
| THC0290_2338 | 208.3 | B | Adhesin | 84 | 32 | 94 |
| THC0290_1932 | 200.0 | B | Adhesin | 48 | 17 | 36 |
| THC0290_1520 | 66.2 | A | Endonuclease | 37 | 0 | 40 |
| THC0290_2029 | 32.9 | A | 34 | 19 | 45 | |
| THC0290_1797 | 29.4 | A | 32 | 1 | 23 | |
| THC0290_1616 | 129.6 | B | Adhesin | 27 | 8 | 53 |
| THC0290_1595 | 67.9 | A | Adhesin | 24 | 8 | 16 |
| THC0290_0944 | 58.5 | A | Peptidase | 21 | 0 | 23 |
| THC0290_2146/PorU | 142.1 | A | T9SS-specific CTD peptidase | 20 | 17 | 32 |
| THC0290_1494 | 71.5 | A | Ribonuclease | 19 | 0 | 24 |
| THC0290_0332 | 269.2 | B | Adhesin | 15 | 4 | 16 |
| THC0290_1527 | 233.6 | B | Adhesin | 13 | 5 | 20 |
| THC0290_0023/SprC | 51.2 | B | Gliding motility protein SprC | 9 | 4 | 9 |
| THC0290_1047 | 87.8 | B | Adhesin | 8 | 1 | 15 |
| THC0290_1343 | 134.1 | A | Peptidase | 6 | 0 | 6 |
| THC0290_0086 | 70.9 | A | Peptidase | 6 | 0 | 9 |
| THC0290_0237/Fpp1 | 119.6 | A | Peptidase | 6 | 0 | 5 |
| THC0290_0129 | 33.6 | A | 6 | 1 | 9 | |
| THC0290_1818 | 158.9 | B | Adhesin | 6 | 0 | 6 |
| THC0290_0091 | 122.8 | A | Glycoside hydrolase | 5 | 2 | 4 |
| THC0290_1048 | 82.6 | B | Adhesin | 3 | 0 | 7 |
| THC0290_0299 | 100.1 | A | Peptidase | 1 | 16 | 6 |
| THC0290_1054 | 46.0 | A | 0 | 0 | 3 | |
| THC0290_0128 | 12.8 | A | 0 | 2 | 0 | |
| LRR proteinsd | ||||||
| THC0290_0183 | 33.2 | A | Leucine-rich repeat protein | 51 | 22 | 46 |
| THC0290_0174 | 44.5 | A | Leucine-rich repeat protein | 23 | 13 | 22 |
| THC0290_0171 | 44.5 | A | Leucine-rich repeat protein | 22 | 18 | 32 |
| THC0290_0170 | 41.7 | A | Leucine-rich repeat protein | 20 | 40 | 24 |
| THC0290_0181 | 50.1 | A | Leucine-rich repeat protein | 17 | 18 | 18 |
| THC0290_0184 | 37.0 | A | Leucine-rich repeat protein | 15 | 25 | 23 |
| THC0290_0186 | 35.3 | A | Leucine-rich repeat protein | 12 | 15 | 13 |
| THC0290_0182 | 35.0 | A | Leucine-rich repeat protein | 9 | 17 | 10 |
| THC0290_0175 | 35.3 | A | Leucine-rich repeat protein | 7 | 16 | 12 |
| THC0290_0185 | 35.3 | A | Leucine-rich repeat protein | 7 | 9 | 10 |
| THC0290_0172 | 35.2 | A | Leucine-rich repeat protein | 7 | 19 | 11 |
| THC0290_0180 | 42.1 | A | Leucine-rich repeat protein | 0 | 20 | 0 |
| THC0290_0177 | 56.0 | A | Leucine-rich repeat protein | 0 | 15 | 0 |
Proteins in cell-free culture fluid from wild-type F. psychrophilum, ΔgldN mutant, and ΔgldN mutant complemented with pBFp4 were analyzed by LC-MS/MS. Total/unweighted spectrum count corresponding to total number of spectra associated with a single protein and indicative of relative abundance of that protein is indicated for each strain. Only proteins with predicted T9SS CTDs are shown. For the full data set, which includes additional proteins, see Data Set S1 in the supplemental material. The following type A CTD proteins were not detected: THC0290_0173, THC0290_0176, THC0290_0178, THC0290_0179, THC0290_0300, THC0290_0878, THC0290_0977, THC0290_2157, and THC0290_2385. However, the LRR proteins THC0290_0173, THC0290_0176, THC0290_0178, and THC0290_0179 may have been present, because they are similar in sequence to the other LRR proteins, resulting in ambiguity regarding protein assignment for some peptides (Data Set S1). Similarly, THC0290_300 may have been missed because of similarity in sequence to THC0290_0299.
Molecular mass as calculated for full-length protein before removal of signal peptide.
Type A CTDs belong to TIGRFAM protein domain family TIGR04183. Type B CTDs belong to TIGRFAM protein domain family TIGR04131 and/or pfam13585.
These proteins have paralogs in the genome that complicate assignment of peptides.
Deletion of gldN results in decreased extracellular proteolytic activity.
F. psychrophilum secretes many peptidases (14). We examined wild-type, ΔgldN mutant, and complemented strains of F. psychrophilum for extracellular proteolytic activity on TYES-plus-glucose (TYESG) agar supplemented with casein (Fig. 4A). Wild-type and complemented strains formed clearing zones around the cell growth, indicating digestion of casein, whereas the ΔgldN mutant did not. Extracellular proteolytic activity in cell-free spent culture medium from each strain was quantified using an azocasein-based assay (Fig. 4B). For wild-type and complemented strains, high levels of soluble extracellular proteolytic activities that increased over time through late exponential and early stationary phases of growth were detected. In contrast, extracellular proteolytic activities for the ΔgldN mutant were low and failed to increase over time. In vitro cell growth kinetics in TYESG broth cultures were not affected by gldN deletion (Fig. 4B). TYESG contains tryptone (casein that has been digested by trypsin) and yeast extract, which may explain the lack of requirement for high levels of secreted peptidases for growth in this medium. In contrast, the lack of peptidase secretion may impair the growth of ΔgldN mutant cells within the host. Wild-type F. psychrophilum produces peptidases (some apparently strain dependent) that digest collagen, fibrinogen, elastin, and fish muscle tissue, and fish with BCWD often present signs of tissue erosion (60, 61).
FIG 4.
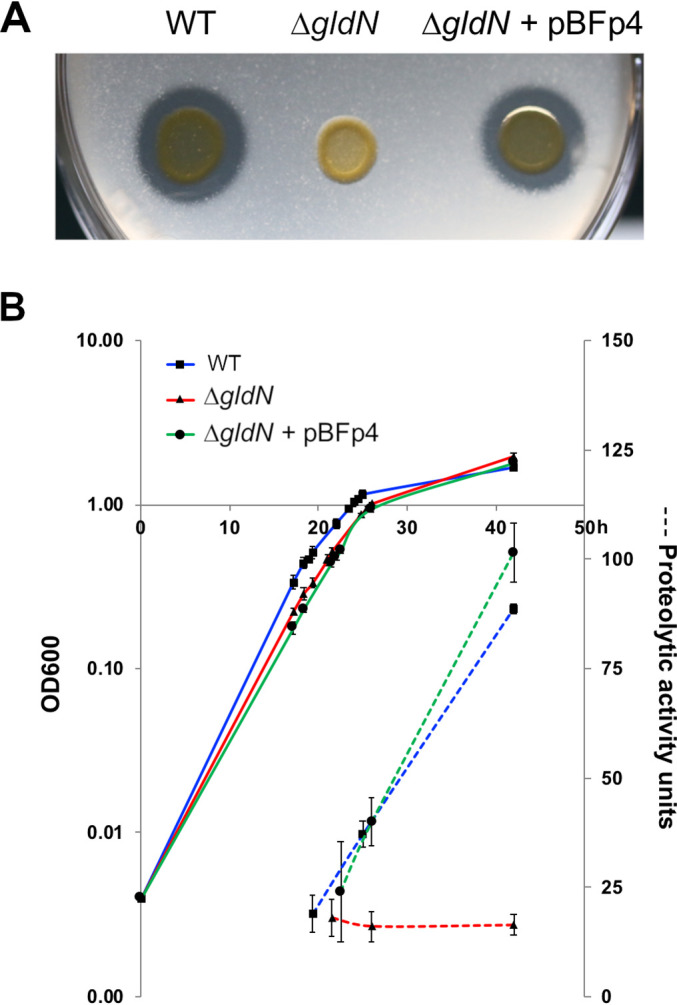
Proteolytic activities of wild-type and ΔgldN mutant cells. Exoproteolytic activities of F. psychrophilum wild type (WT), ΔgldN mutant, and ΔgldN mutant complemented with pBFp4. (A) Stationary-phase cultures were spotted on TYESG agar supplemented with casein (7.5 g/liter), and proteolytic activity was visualized as clearing zones around the bacterial growth after incubation at 18°C for 4 days. (B) Proteolytic activity measured using azocasein as a substrate. Cells were grown in TYESG broth at 18°C with shaking. Proteolytic activity was measured in cell-free supernatants from each strain at different stages of growth. The results of three experiments are presented (values and error bars are means and standard deviations, respectively). Solid lines, OD600, indicating growth; dashed lines, proteolytic activity units.
The gldN deletion mutant is deficient in hemolytic activity and Hb utilization.
Hemoglobin (Hb) is the most abundant reservoir of iron in vertebrates and represents an attractive nutrient source for pathogenic bacteria (62). F. psychrophilum infections in young rainbow trout result in hemorrhagic septicemia. This bacterium lyses rainbow trout erythrocytes and uses hemoglobin as an iron source for growth (63, 64). Wild-type cells grown on TYESG agar supplemented with rainbow trout blood exhibited hemolytic activity, whereas cells of the ΔgldN mutant did not (Fig. 5). Hemolysis by the wild-type strain was accompanied by brown coloration of the colonies, which was not observed for the ΔgldN mutant. The introduction of gldN on pBFp4 into the ΔgldN mutant restored the phenotypic properties associated with the wild-type strain, indicating that hemolysis and the brown coloration are dependent on a functional T9SS. Dark colony pigmentation on blood agar occurs for other pathogenic bacteria and may result from binding of hemoglobin or heme to the surface of bacterial cells. In P. gingivalis, hemoglobin released following lysis of erythrocytes is digested by gingipain proteases that are secreted by the T9SS. The released heme accounts for the black pigmentation of P. gingivalis colonies and contributes to oxidative stress resistance (65). To evaluate the role of the F. psychrophilum T9SS in hemoglobin utilization, wild-type and mutant strains were grown to stationary phase in TYESG broth supplemented with 0.5 μM hemoglobin. Residual free heme and hemoglobin were measured using a porphyrin detection method that relies on growth stimulation of a heme-auxotrophic bacterial reporter strain (66). Stationary-phase wild-type cultures resulted in minimal growth stimulation of the reporter, indicating little if any residual porphyrin in the medium (Fig. 6). In contrast, uninoculated medium or stationary-phase cultures of the ΔgldN mutant stimulated growth of the reporter, indicating that little if any heme porphyrin was used by the ΔgldN mutant cells. Hemoglobin consumption was restored to the ΔgldN mutant by the introduction of gldN on pBFp4. The results indicate that a functional T9SS is required for efficient hemoglobin utilization by F. psychrophilum. This may in part involve secretion of peptidases that cleave hemoglobin, releasing heme. The brown color of wild-type colonies (Fig. 5) suggests that released heme was captured by the cells. Heme was likely imported into wild-type F. psychrophilum cells, because it was absent in the medium after growth.
FIG 5.
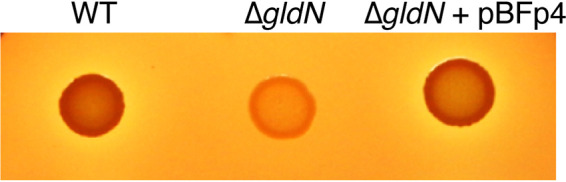
Hemolytic activities of wild-type and ΔgldN mutant cells. Hemolytic activities of cells of F. psychrophilum wild type (WT), ΔgldN mutant, and ΔgldN mutant complemented with pBFp4 incubated for 4 days at 18°C on TYESG agar supplemented with rainbow trout blood (50 ml/liter). The results are representative of three independent experiments.
FIG 6.
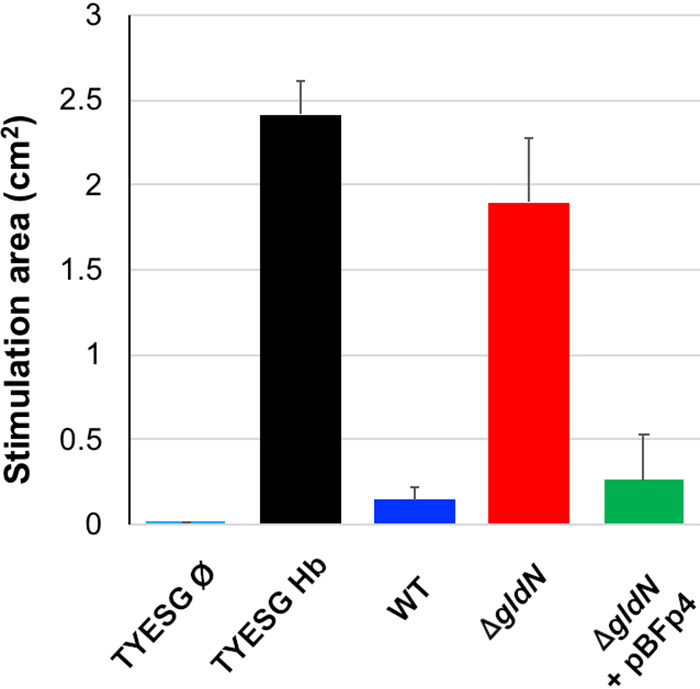
Hemoglobin utilization by wild-type and ΔgldN mutant cells. Detection of residual heme after growth of F. psychrophilum wild type (WT), ΔgldN mutant, and ΔgldN mutant complemented with pBFp4 to stationary phase in TYESG broth supplemented with 0.5 μM hemoglobin. The heme-protoporphyrin IX screen is a bacterial-growth-based assay that exploits Bacteroides thetaiotaomicron heme auxotrophy. The stimulation area (cm2) is proportional to the amount of porphyrin (heme, hemoglobin, or protoporphyrin IX) in the sample. TYESG Hb and TYESG Ø, sterile medium with (Hb) or without (Ø) addition of hemoglobin to 0.5 μM. The values correspond to the results from three independent bacterial cultures (means and standard deviations are presented).
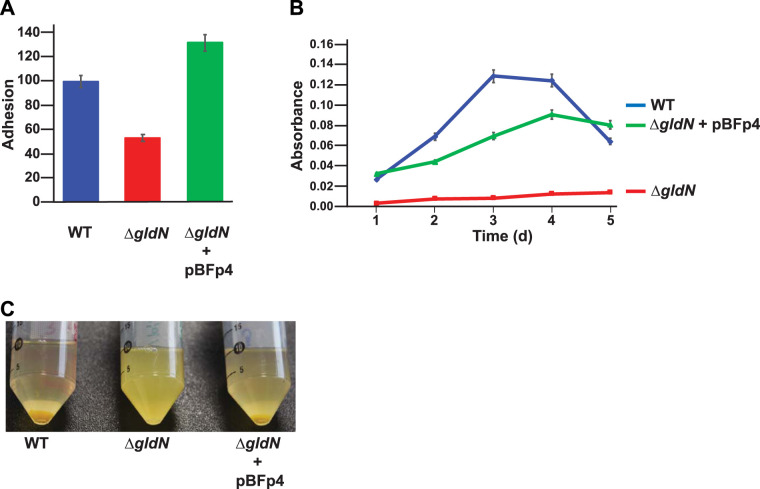
The gldN deletion mutant is deficient in adhesion, biofilm formation, and cell sedimentation.
Wild-type F. psychrophilum cells adhere readily to polystyrene (67). Cells of the ΔgldN mutant were partially deficient in adhesion to polystyrene compared to wild-type cells or to cells of the complemented mutant (Fig. 7A). Adhesion of cells to a surface is the first step in biofilm formation, and F. psychrophilum is known to form biofilms on polystyrene (68, 69). The ΔgldN mutant was deficient in biofilm formation compared to the wild-type and complemented strains (Fig. 7B). F. psychrophilum cells also interact with each other, causing them to sediment from suspension (14). Wild-type and complemented ΔgldN mutant cells sedimented from suspension readily, whereas the ΔgldN mutant cells did not (Fig. 7C). The results indicate that the T9SS is involved in efficient adhesion, biofilm formation, and sedimentation, as was also suggested by analysis of F. psychrophilum gldD and gldG mutants (14). The many predicted adhesins demonstrated to be secreted by the T9SS (Tables 1 to 3) may explain some of the defects in these processes, which each require adhesion, in the ΔgldN mutant cells.
FIG 7.
Adhesion, biofilm formation, and sedimentation of wild-type and ΔgldN mutant cells. (A) Adhesion of wild-type F. psychrophilum (blue), ΔgldN mutant (red), and ΔgldN mutant complemented with pBFp4 (green) to polystyrene after 3 h of incubation at 18°C without shaking as determined by staining with crystal violet and measuring absorbance at 595 nm. Adhesion is shown in relation to the wild-type strain, which was set as 100. (B) Biofilm formation on polystyrene by the same strains grown in half-strength TYES broth for 120 h at 18°C without shaking. (C) Cell sedimentation of the same strains grown in half-strength TYES broth for 96 h at 18°C with shaking at 200 rpm.
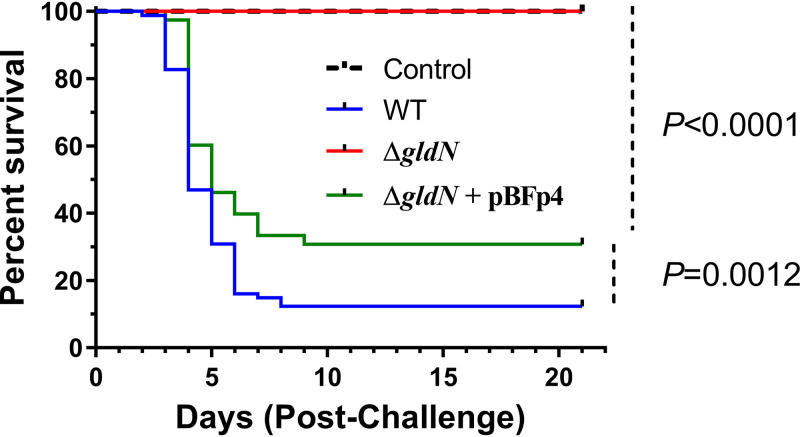
GldN is required for virulence in rainbow trout.
Wild-type, ΔgldN mutant, and complemented strains were examined for virulence using a rainbow trout injection challenge (70). At the dose examined (2.8 × 106 CFU/fish), intramuscular injection of the ΔgldN mutant caused no mortality, whereas exposure to similar doses of the wild-type and complemented strains resulted in more than 60% mortality within 10 days of challenge (Fig. 8). The complemented strain was less virulent than the wild type. F. psychrophilum was isolated from all cases of mortality examined (for the wild type, n = 13/13 fish, and for the ΔgldN mutant complemented with pBFp4, n = 11/11 fish).
FIG 8.
Analysis of virulence of wild-type and ΔgldN mutant cells toward rainbow trout following challenge by injection. Kaplan-Meier survival curves of rainbow trout after intramuscular injection challenge. Four groups of 20 fish (n = 80) were challenged with each F. psychrophilum strain at doses (CFU/fish) of 2.6 × 106 for the wild type, 2.7 × 106 for the ΔgldN mutant, and 2.8 × 106 for the ΔgldN mutant complemented with pBFp4. Survival curves for fish inoculated with PBS (Control) and for fish inoculated with the ΔgldN mutant were identical. Survival curves for fish inoculated with either the wild type or the ΔgldN mutant complemented with pBFp4 are significantly different from survival curves for the ΔgldN mutant (Mantel-Cox log rank test, P < 0.0001). The survival curve of the ΔgldN mutant complemented with pBFp4 was modestly higher than that of the wild type (P = 0.0012).
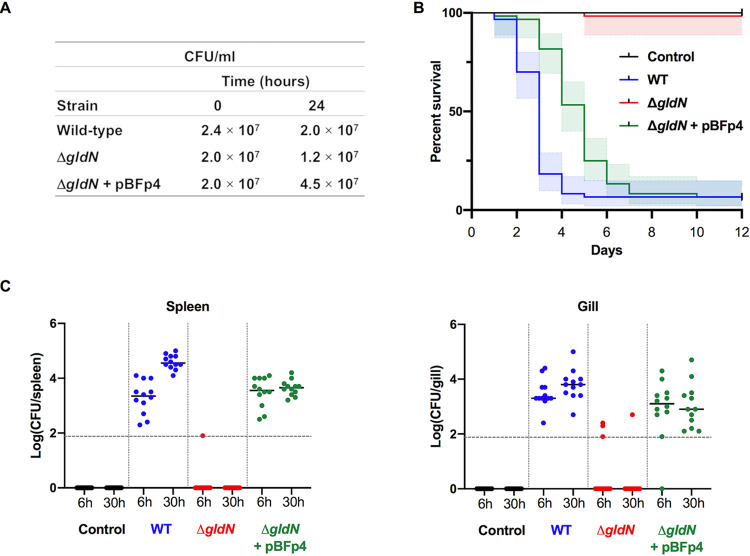
The effect of gldN deletion on the virulence of F. psychrophilum was also investigated using an immersion model that more closely mimics the natural route of infection. Fish were bathed for 24 h in water containing the wild-type, ΔgldN mutant, or complemented strain at a final concentration of ∼2 × 107 CFU/ml (Fig. 9A). Virulence was compared between the three strains by recording the mortality rates and by determining the F. psychrophilum bacterial loads in external (gills) and internal (spleen) organs. Five days after the immersion challenge, 93% of fish infected with the wild-type strain had died (Fig. 9B). In contrast, the cumulative mortality of fish exposed to the ΔgldN mutant was less than 2% and was similar to that of the noninfected control group. Complementation with pBFp4 restored virulence to the ΔgldN mutant, resulting in cumulative mortality similar to that caused by the wild type. The increased time-to-death observed for fish exposed to the complemented strain compared to that of the wild-type strain may be a result of the increased copy number of gldN or of an effect of the gldN deletion on the expression of nearby genes. Six of each group of 42 fish were arbitrarily sampled for the presence of F. psychrophilum 6 h and 30 h after the end of the immersion challenge. At 30 h, F. psychrophilum was detected at high levels in the gills (average, 6 × 103 CFU per gill arch) and spleen (4 × 104 CFU per spleen) of all fish exposed to the wild-type strain (Fig. 9C). Strikingly, the bacterium was detected in the organs of only 5 of 24 fish exposed to the ΔgldN mutant, and bacterial loads were significantly lower than in fish exposed to the wild type. Bacterial loads in the spleen and gills of fish exposed to the complemented mutant were similar to those of fish exposed to the wild type. Together, the results indicate that a functional T9SS is required for full virulence of F. psychrophilum in rainbow trout.
FIG 9.
Analysis of virulence of wild-type and ΔgldN mutant cells toward rainbow trout following immersion challenge. Groups of 42 fish were infected by immersion with F. psychrophilum wild type (WT), ΔgldN mutant, and ΔgldN mutant complemented with pBFp4. The results of two independent experiments are presented. (A) F. psychrophilum bacterial loads in aquarium water during fish infection by immersion. Average results for bacterial quantification determined at the beginning (0 h) and end (24 h) of fish infection challenge. (B) Kaplan-Meier survival curves of rainbow trout after immersion challenge (each group composed of 30 fish). Survival curves for fish challenged with the ΔgldN mutant or with either the wild type or the ΔgldN mutant complemented with pBFp4 are significantly different (Mantel-Cox log rank test, P < 0.0001). Colored shaded areas indicate 95% confidence intervals. (C) Bacterial loads in organs of rainbow trout after immersion challenge. Six fish were sacrificed at 6 h and 30 h postinfection for each group. Serial dilutions of homogenized organs were incubated on TYESG agar supplemented with fetal calf serum (50 ml/liter) to determine the CFU. Bacterial loads of spleen (left) and gills (right) are shown. Horizontal dashed line indicates the detection threshold. Values are significantly lower for the ΔgldN mutant than for wild-type and complemented strains (Mann-Whitney P < 0.0001).
DISCUSSION
F. psychrophilum is an important pathogen of aquaculture-reared salmonids and ayu and results in large economic losses for the aquaculture industry worldwide. Genetic manipulation to identify F. psychrophilum virulence factors has been hampered by the lack of efficient genetic tools. Here, we demonstrate the usefulness of a sacB-mediated gene deletion system to construct mutations in F. psychrophilum, and we use this approach to demonstrate that the T9SS is required for virulence.
A gene deletion procedure was previously developed for F. psychrophilum strain OSU THCO2-90 (43). This double recombination procedure was similar to the one described here, but it relied on an introduced restriction enzyme site for counterselection against strains carrying the integrated plasmid. While the procedure allowed mutant construction, it was inefficient, presumably because of the multiple rounds of gene transfer required to introduce the integrative plasmid and later introduce the restriction enzyme-encoding plasmid. Here, we used a procedure that was demonstrated to allow efficient targeted mutagenesis of other members of the genus Flavobacterium and phylum Bacteroidetes (44, 45). This procedure, adapted to F. psychrophilum, required a single gene transfer event, relied on sacB-mediated sucrose sensitivity for counterselection, and resulted in many F. psychrophilum gene deletion colonies. This approach allows efficient targeted mutagenesis of F. psychrophilum strain OSU THCO2-90. However, many other F. psychrophilum strains have resisted gene transfer (42), and additional research is needed before similar studies will be possible for a diverse collection of F. psychrophilum strains.
Genome analysis revealed F. psychrophilum OSU THCO2-90 genes encoding T9SS components. Forty-nine proteins predicted to be secreted by this system were also identified, including predicted peptidases, nucleases, adhesins, and motility proteins, any of which could contribute to virulence. Each of the predicted secreted proteins was conserved across the genomes of the four F. psychrophilum strains analyzed (OSU THCO2-90, ATCC 49418T, JIP 02/86, and CSF-259-93), and many of them were identified as secreted proteins by proteomic analyses (Table 3). Many of the proteins secreted by the F. psychrophilum T9SS were previously identified as predicted virulence factors in the initial genome analysis of strain JIP 02/86 (12). Of the 13 peptidases identified as potential virulence factors in that study, 7 have type A CTDs (FP0082, FP0086, FP0231, FP0232, FP0280, FP0281, and FP1763). Two additional peptidase-encoding genes appear disrupted in strain JIP 02/86 (FP1776/FP1777 and FP1024/FP1023), whereas strain OSU THCO2-90 has full-length orthologous genes (THC0290_0931 and THC0290_1343, respectively) encoding predicted peptidases with type A CTDs (Tables 1 and 3). Duchaud et al. (12) also identified genes encoding 27 predicted adhesins, including 15 tandemly arrayed genes that encode proteins with LRRs that were similar to B. forsythus BspA and T. denticola LrrA (48, 49). F. psychrophilum OSU THCO2-90 has 17 tandemly arrayed genes encoding LRR proteins that are similar to those of strain JIP 02/86 (Table 1). Each of these has type A CTDs (as do the strain JIP 02/86 LRR proteins) and is thus predicted to be secreted by the T9SS. Other previously predicted T9SS-secreted adhesins of strain JIP 02/86 (12) (with strain OSU THCO2-90 orthologs indicated in parentheses) include the type A CTD protein FP1830 (THC0290_0878) and the type B CTD proteins FP2413 (THC0290_2338), FP0016 (THC0290_0025; SprB), and FP0595 (THC0290_1616).
GldN was originally identified as an F. johnsoniae gliding motility protein (57) and was later shown to be a core component of the F. johnsoniae and P. gingivalis T9SSs (17, 56). gldN is required for T9SS-mediated secretion in F. johnsoniae, F. columnare, and P. gingivalis (17, 21, 56). Similarly, as shown here, F. psychrophilum gldN was required for both gliding motility and T9SS function. The gldN mutant was nonmotile and appeared to be deficient in the secretion of many proteins, including predicted peptidases, hemolysins, and adhesins. Deletion of gldN greatly reduced the extracellular proteolytic and hemolytic activities of cells and also reduced their adhesiveness and ability to form biofilms. Motility, adhesion, proteolysis, hemolysis, and ability to form biofilms have all been suggested to play roles in BCWD (1, 2, 7, 12, 13, 71). The only correlation observed between virulence (estimated by 50% lethal dose [LD50] using intramuscular injection challenge) and these phenotypic traits in a recent study of 26 F. psychrophilum isolates was that the avirulent strain studied was deficient in colony spreading and exoproteolytic activity (7), suggesting a probable T9SS deficiency. Adhesins may allow initial attachment, and motility could facilitate penetration of tissues and spread of the infection. Secreted peptidases and hemolysins may digest host tissues and proteins, providing nutrients to the bacterium and/or destroying host defenses. The ΔgldN mutant was completely deficient in virulence in rainbow trout infected by injection or immersion. An in vivo adhesion defect of the ΔgldN mutant is suggested by the reduced recovery of F. psychrophilum cells from gill tissue 6 h after exposure by immersion (Fig. 9C). Loss of secretion of enzymes, adhesins, and motility proteins may together result in the avirulent phenotype of the mutant. The T9SS was also recently demonstrated to be critical for F. columnare to cause columnaris disease (21), suggesting similarities between at least some aspects of the virulence strategies employed by these related fish pathogens.
The results reported here are consistent with those obtained by Pérez-Pascual et al. (14), who examined transposon-induced motility mutants of F. psychrophilum for virulence. In that study, mutations in the motility genes gldD and gldG were linked to T9SS defects, resulting in reductions in extracellular proteolytic activity, adhesiveness, and virulence (14). GldD and GldG are not core T9SS components and are absent from nonmotile bacteria that have T9SSs, such as P. gingivalis (16). However, recent studies of F. johnsoniae indicate that the gliding motility and T9SS machineries are intertwined (23) and suggest an explanation for the phenotypes of the F. psychrophilum gldD and gldG mutants. In F. johnsoniae, absence of the motility protein GldJ results in apparent instability of the T9SS protein GldK and, thus, in loss of protein secretion (23). Further, mutations in some other motility genes (including gldD and gldG) result in apparent instability of the GldJ protein (72) and, thus, in loss of GldK and of T9SS function (23). The apparent interconnections between the motility and T9SS machineries may explain how mutations in the F. psychrophilum motility genes gldD and gldG perturb T9SS function.
Our results may also help to explain the lack of virulence of rifampin-resistant F. psychrophilum mutants that have been isolated and examined as vaccine candidates (73). The most attenuated of these strains were deficient in motility (74). Given the link between gliding motility and the T9SS described above, the mutants may have also been compromised for protein secretion, which could have contributed to the attenuated phenotype.
The proteomic analysis of spent cell-free culture fluid confirmed our predictions regarding secretion of many proteins that carried type A or type B CTDs, but it also revealed some unexpected results. Numerous LRR proteins, each of which had type A CTDs, were predicted to be secreted by the T9SS, but surprisingly, many of these were found at similar or, for some, higher levels in the cell-free culture fluid of the ΔgldN mutant. The LRR proteins were predicted to be cell surface proteins (12). We do not know why similar amounts of LRR proteins were found in the cell-free spent culture fluids of wild-type and ΔgldN mutant cells. Perhaps the LRR proteins were not actually secreted in soluble form but, instead, were present on the surface of or inside extracellular outer membrane vesicles released by wild-type and mutant cells, respectively. Our proteomic results for secretion of LRR proteins by wild-type and ΔgldN mutant cells differ from those obtained in a similar study of wild-type, gldD, and gldG mutant cells (14). The gldD and gldG mutants secreted much smaller amounts of LRR proteins than did wild-type cells. We do not know the reasons for this difference. As discussed above, GldD and GldG are not thought to be components of the T9SS, but they impact its function. Mutations in gldD and gldG may thus affect the functioning of the T9SS differently than mutations in gldN, which encodes a core component of the system.
The 49 proteins with type A and type B CTDs that are predicted to be secreted by the F. psychrophilum T9SS probably underestimate the actual number of proteins secreted by this system, since some T9SS-secreted proteins from related bacteria have novel CTDs (33). Supporting this, deletion of gldN appeared to disrupt the secretion of many proteins that lacked obvious T9SS CTDs (Data Set S1 in the supplemental material). Included among these proteins are 14 predicted peptidases that may contribute to virulence. Analysis of the C-terminal regions of seven proteins that lacked obvious type A or type B CTDs but that appeared to be secreted by the T9SS, as indicated by Data Set S1 and by the results of a previous study (14), revealed two putative adhesins (THC0290_0754 and THC0290_2201) and one probable peptidase (THC0290_0500) that exhibit some similarity to T9SS CTDs (Fig. S1). These may represent a novel T9SS CTD type or may indicate that the sequence constraints delineating CTD domains may need to be relaxed to avoid false negatives. Identification of additional proteins secreted by the T9SS will not be surprising given our current limited understanding of the functioning of this system. An alternative potential explanation for the apparent T9SS-mediated secretion of some non-CTD proteins is that deletion of gldN, which is expected to result in the failure of numerous cell surface proteins to reach their destination, could have pleotropic effects on the cell surface that perturb other protein secretion systems. F. psychrophilum has multiple predicted type I secretion systems (12) that could have been affected in this way.
Our analyses of F. psychrophilum motility proteins SprC and SprD identified some unexpected features that were not immediately obvious from earlier studies. F. johnsoniae SprC was not previously recognized as a potential T9SS-secreted protein (30, 52). F. psychrophilum SprC also fell below the trusted cutoff for inclusion in type B CTD family TIGR04131, but it had a conserved region (pfam13585) (Table 2) that is found in all type B CTDs. The regions recognized by TIGR04131 and pfam13585 are nearly identical (19). Further, sprD, which is immediately downstream from sprC, encodes a protein with a PorP/SprF-like region (TIGR03519) (Table S2), suggesting that it may interact with SprC and facilitate its secretion by the T9SS. Similar conserved regions are found in SprC (pfam13585) and SprD (TIGR03519) proteins of F. columnare and other related bacteria. sprC, sprD, sprB, and sprF appear to form an operon in F. psychrophilum and in many other Bacteroidetes, and SprC and SprD support SprB-mediated gliding motility (30, 52). As mentioned above, SprB also requires its cognate PorP/SprF-like protein (SprF) for its secretion (30). The results presented here enhance our understanding of the possible interactions between these four common Bacteroidetes gliding motility proteins and their relationships to the T9SS.
The F. psychrophilum T9SS is important for virulence, but which of the many secreted proteins contribute to BCWD remains unclear. The roles of the individual secreted proteins can now be examined systematically using the gene deletion approach described above. Since this deletion technique does not leave antibiotic resistance genes or other foreign DNA in the mutant, it can be used iteratively to generate strains lacking many genes. This is important given the many potential secreted virulence factors and should allow questions of redundancy to be addressed. Construction of mutants lacking one or more critical secreted virulence factors may reveal the most important mechanisms employed by F. psychrophilum to cause disease and could suggest control measures. Such mutants may function as attenuated vaccine strains that interact with fish and generate a protective immune response but fail to cause disease. Since T9SSs are involved in diseases caused by many related animal and human pathogens (17, 21, 38–41), improved understanding of F. psychrophilum virulence may have impacts beyond fish health and sustainable aquaculture.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
F. psychrophilum strain OSU THCO2-90 (47, 75) was the wild-type strain used in this study. F. psychrophilum cultures were grown at 18°C in tryptone yeast extract salts (TYES) medium (76, 77), which contains, per liter, 4 g tryptone, 0.4 g yeast extract, 0.5 g MgSO4·7H2O, and 0.5 g CaCl2·2H2O, with pH adjusted to 7.2. For some experiments, TYESG, which contains, per liter, 4 g tryptone, 0.4 g yeast extract, 0.5 g MgSO4·7H2O, 0.2 g CaCl2·2H2O, and 0.5 g d-glucose, with pH adjusted to 7.2, was used. TYESG differs from TYES only by the addition of glucose and by the reduced amount of CaCl2. For solid media, agar was used at 15 g/liter unless indicated otherwise. For most experiments, F. psychrophilum was recovered from −80°C freezer tubes onto TYES agar and incubated for 72 h at 18°C. Strains were streaked on fresh agar, incubated for 72 h at 18°C, and then used to inoculate 20 ml TYES broth cultures, which were incubated for 24 to 48 h at 18°C with shaking at 200 rpm. Extracellular proteolytic activity on solid medium was visualized using TYESG agar supplemented with 7.5 g casein (product number C8654; Sigma-Aldrich) per liter or 10 g hemoglobin (Hb) (product number 08449; Sigma-Aldrich) per liter. Hemolysis and colony pigmentation were visualized on TYESG agar supplemented with 50 ml rainbow trout blood per liter. Blood was collected by caudal venipuncture of anesthetized rainbow trout and immediately mixed with heparin at a final concentration of 250 IU/ml (25,000 IU/5 ml heparine choay injectable solution; Sanofi Aventis) at 4°C. Colony spreading was examined on 5% TYES (TYES with all components diluted 20-fold) solidified with 10 g agar per liter.
Bacteroides thetaiotaomicron VPI-5482T (78) was grown in Difco M17 medium (catalog number 218561; Becton, Dickinson, Sparks, MD) supplemented with (per liter) 5 g glucose, 0.5 g cysteine, and 6 g agar and is referred to herein as M17-glu. E. coli strains were grown in lysogeny broth (LB) at 37°C (79). Strains and plasmids used in this study are listed in Table 4, and primers are listed in Table 5. Antibiotics were used at the following concentrations when needed: ampicillin, 100 μg/ml, and erythromycin, 10 μg/ml.
TABLE 4.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference(s) |
|---|---|---|
| Strains | ||
| Bacteroides thetaiotaomicron VPI-5482T | Used to measure porphyrin (heme, hemoglobin or protoporphyrin IX) | 78 |
| E. coli DH5α(MCR) | Strain used for general cloning | Life Technologies (Grand Island, NY) |
| E. coli S17-1(λ-pir) | Strain used for conjugation | 87 |
| F. psychrophilum OSU THCO2-90 | Wild type | 47, 75 |
| F. psychrophilum FpT13 | ΔgldN mutation in strain OSU THCO2-90 | This study |
| Plasmids | ||
| pCP11 | E. coli-Flavobacterium shuttle plasmid; Apr (Emr) | 80 |
| pYT313 | Suicide vector carrying sacB; Apr (Emr) | 45 |
| pBFp0 | 2,858-bp region upstream from gldN amplified with primers 2060 and 2061 and cloned into BamHI and SalI sites of pYT313; Apr (Emr) | This study |
| pBFp1 | Construct used to delete gldN; 3,132-bp region downstream from gldN amplified with primers 2062 and 2063 and cloned into SalI and SphI sites of pBFp0; Apr (Emr) | This study |
| pBFp4 | Plasmid for complementation of gldN; gldN was amplified with primers 2076 and 2077 and cloned into KpnI and SphI sites of pCP11; Apr (Emr) | This study |
Antibiotic resistance phenotypes: ampicillin, Apr; erythromycin, Emr. Unless indicated otherwise, the antibiotic resistance phenotypes are those expressed in E. coli. The antibiotic resistance phenotypes given in parentheses are those expressed in F. psychrophilum but not in E. coli.
TABLE 5.
Primers used in this study
| Primer | Sequencea |
|---|---|
| 2060 | 5′-GCTAGGGATCCGCTAGAAATGATAGAGGTTGTTATT-3′ |
| 2061 | 5′-GCTAGGTCGACGTCATTATCTTTTAGTTGTTGTGCT-3′ |
| 2062 | 5′-GCTAGGTCGACCGTTTTAATGCACTGACTTATAAAG-3′ |
| 2063 | 5′-GCTAGGCATGCGTAATTTCGCCATCTAGATATTCT-3′ |
| 2076 | 5′-GCTAGGGTACCAGCTAGCTTCTCTGGAATAG-3′ |
| 2077 | 5′-GCTAGGCATGCATTTGGTAAGAGTTTTTTAA-3′ |
Underlined sequences indicate introduced restriction enzyme sites.
Conjugative transfer of plasmids into F. psychrophilum.
Plasmids were transferred from E. coli strain S17-1(λ-pir) into F. psychrophilum strains by conjugation. Briefly, E. coli strains were incubated overnight with shaking in LB at 37°C. F. psychrophilum strains were incubated for 30 h with shaking in TYES broth at 18°C. Cells were collected by centrifugation at 3,220 × g for 25 min, washed twice with 1 ml TYES, and suspended in 500 μl TYES. The suspensions of E. coli and F. psychrophilum cells were mixed, spotted on TYES agar, and incubated at 18°C for 48 h. The cells were removed from the plate with a scraper and suspended in 2 ml TYES. One hundred-microliter aliquots were spread on TYES agar containing 10 μg erythromycin per ml and incubated at 18°C for 5 to 7 days.
Construction of the gldN deletion mutant.
To delete F. psychrophilum gldN (THC0290_0743), a 2,858-bp fragment spanning the last 387 bp of gldK (THC0290_0740), gldL (THC0290_0741) and gldM (THC0290_0742), and including the first 141 bp of gldN was amplified using Phusion DNA polymerase (New England Biolabs, Ipswich, MA) and primers 2060 (introducing a BamHI site) and 2061 (introducing a SalI site). The fragment was digested with BamHI and SalI and ligated into the suicide vector pYT313, which had been digested with the same enzymes, to generate pBFp0. A 3,132-bp fragment spanning the final 153 bp of gldN, THC0290_0744 and THC0290_0745, and the first 1,220 bp of THC0290_0746 was amplified with primers 2062 (introducing a SalI site) and 2063 (introducing an SphI site). The fragment was digested with SalI and SphI and inserted into pBFp0 that had been digested with the same enzymes to generate the deletion construct pBFp1. pBFp1 was transferred by conjugation into F. psychrophilum OSU THCO2-90, and colonies that had the plasmid integrated into the chromosome by recombination were obtained by selecting for erythromycin resistance. Resistant colonies were streaked for isolation on TYES agar containing erythromycin. An isolated colony was used to inoculate 10 ml TYES broth without erythromycin, which was incubated for 30 h at 18°C to allow loss of the integrated plasmid. Recombinants that had lost the plasmid were selected by plating on TYES containing 50 g/liter sucrose and incubating at 18°C. Isolated sucrose-resistant colonies were picked and streaked for isolation on TYES agar containing sucrose to eliminate background cells that had not lost the plasmid. Colonies were screened by PCR using primers 2076 and 2077, which flank gldN, to identify the gldN deletion mutant. Streaking colonies for isolation on selective media at both the plasmid integration (erythromycin selection) and plasmid loss (sucrose selection) steps was critical to eliminate nonselected cells.
Complementation of the gldN deletion mutant.
A plasmid carrying F. psychrophilum gldN was constructed using shuttle vector pCP11 (80). Primers 2076 (introducing a KpnI site) and 2077 (introducing an SphI site) were used to amplify a 1,161-bp product spanning gldN. The product was digested with KpnI and SphI and ligated into pCP11, which had been digested with the same enzymes, to generate pBFp4, which was transferred into F. psychrophilum by conjugation.
Analysis of colony spreading and cell motility.
F. psychrophilum wild-type, ΔgldN mutant, and complemented strains were serially diluted and plated on 5% TYES (TYES diluted 20-fold) solidified with 10 g agar per liter. The plates were incubated for 32 h at 18°C to obtain isolated colonies. Colonies were examined using a Photometrics Cool-SNAPcf2 camera mounted on an Olympus IMT-2 phase-contrast microscope. Colonies were also obtained by spotting 5 μl of cells at an optical density at 600 nm (OD600) of 1.5 on 5% TYES medium solidified with 10 g agar per liter and incubating for 12 days at 18°C, and these were examined macroscopically using a Nikon D7200 camera. Motility of individual cells on agar was examined by spotting cells on a pad of full-strength TYES solidified with 10 g agar per liter on a glass slide, allowing the spot to dry briefly, and covering it with an O2-permeable Teflon membrane (Yellow Springs Instrument Co., Yellow Springs, OH) that prevented dehydration and served as a coverslip. Cell movements over agar were observed using an Olympus BH-2 phase-contrast microscope. Images were recorded using a Photometrics Cool-SNAPcf2 camera and analyzed using MetaMorph software (Molecular Devices, Downingtown, PA). Rainbow traces of cell movements were made using ImageJ version 1.45s (http://rsb.info.nih.gov/ij/) and the macro Color FootPrint (24).
Analysis of secreted proteins by SDS-PAGE and LC-MS/MS.
F. psychrophilum wild-type, ΔgldN mutant, and complemented strains were grown in TYES broth at 18°C for approximately 48 h. Growth was monitored with a Klett-Summerson colorimeter (Klett Manufacturing Co., Inc., Long Island City, NY), and cultures were harvested in early stationary phase when the Klett readings reached 160. Cultures were centrifuged at 16,873 × g for 10 min at 4°C. The fluid was filtered with 0.45-μm-pore-size HT Tuffryn filters (PALL Life Sciences). Proteins were precipitated by mixing 1 volume of trichloroacetic acid (TCA) with 9 volumes of cell-free spent culture fluid, incubating at −20°C for 1 h, and centrifuging for 45 min at 12,210 × g at 4°C. The pellet was washed with acetone (0°C) and centrifuged for 10 min at 12,210 × g to collect the pellet, which was dried for 10 min at room temperature. Proteins were suspended in SDS-PAGE loading buffer, boiled for 10 min, separated by SDS-PAGE (81) using a 12% polyacrylamide gel, and detected using the Bio-Rad (Hercules, CA) silver stain kit. Protein pellets (TCA precipitated from cell-free spent culture fluid and acetone washed as described above) were also analyzed directly by enzymatic digestion and nanoscale LC-MS/MS at the University of Wisconsin—Madison Mass Spectrometry Facility as outlined on the website (https://www.biotech.wisc.edu/services/massspec) and as described previously (21), except that LC-MS/MS data were searched against proteins encoded by the F. psychrophilum strain OSU THCO2-90 genome (47).
Analysis of proteolytic activity.
Proteolytic activity was quantified using azocasein as a substrate as previously described (82). Briefly, F. psychrophilum strains were grown in 25 ml TYESG broth at 18°C and 200 rpm for 2 days. Two-milliliter volumes of cultures at OD600s of 0.5, 1.0 and 2.0 were centrifuged and the supernatants filtered using 0.22-μm Millipore filters and stored at 4°C for 24 h. For protease activity assay, a solution of 20 g azocasein per liter (Sigma-Aldrich) was prepared in 0.1 M Tris-HCl, pH 7.4. The cell-free supernatant (150 μl) was mixed with 150 μl azocasein solution and incubated for 4 h at 25°C. The reaction was stopped by adding 750 μl of TCA (50 g/liter). The reaction mixture was centrifuged at 20,000 × g for 30 min, and 150 μl of the supernatant was mixed in a 96-well microplate well containing 150 μl of 1 M NaCl. The OD440 was measured using a Tecan microplate reader (Infinite 200 pro), and the negative-control OD440 value was subtracted from this. One unit of proteolytic activity was defined as an increase in absorbance of 0.001 under the conditions of the assay. All cultures were performed in triplicate, and the average results of the three experiments were used as the measure of proteolytic activity over the growth curve.
Hemoglobin utilization.
The ability to use hemoglobin as a heme source was investigated by measuring the residual porphyrin (heme, hemoglobin, or protoporphyrin IX) remaining after growth of F. psychrophilum to stationary phase in TYESG broth supplemented with 0.5 μM hemoglobin (product number H2500; Sigma-Aldrich), using the heme-protoporphyrin IX screening method (66). This method is a bacterial-growth-based assay that exploits the heme auxotrophy of B. thetaiotaomicron. Briefly, a vertical gel set-up with 0.8-cm spacers was filled with M17-glu medium containing 8 × 105 B. thetaiotaomicron CFU/ml. Stationary-phase F. psychrophilum cultures in TYESG broth (with or without 0.5 μM hemoglobin) were heat inactivated by incubation at 37°C for 3 h. The cultures (100-μl volumes) were loaded in wells of the B. thetaiotaomicron-containing gel, and sterile TYESG broth containing 0.5 μM hemoglobin was used as a positive control. The gel was overlaid with a 5-ml agar plug (12-g/liter). B. thetaiotaomicron growth stimulation was visualized as dense growth around wells after overnight incubation at 37°C. The surface area of growth stimulation, which correlates with heme concentration in the sample, was determined using ImageJ (version 1.45s; Wayne Rasband, National Institutes of Health, Bethesda, MD, USA). The results correspond to three independent F. psychrophilum cultures.
Adhesion, biofilm, and sedimentation assays.
Adhesion to polystyrene, biofilm formation on polystyrene, and cell sedimentation were determined as previously described (14) except that cells were grown in TYES instead of TYESG for the adhesion assay and in half-strength TYES instead of half-strength TYESG for the biofilm and sedimentation assays, as outlined briefly below.
For the adhesion assay, cells were grown in TYES broth to an OD600 of 0.7. One milliliter of each culture was centrifuged at 11,000 × g for 5 min, the supernatant was removed, and cell pellets were suspended in 1 ml sterile distilled water. One hundred microliters of each strain suspension was added to a 96-well flat-bottom polystyrene microtiter plate (Nunclon delta surface; ThermoFisher Scientific, Waltham, MA), and sterile water was used as a negative control in noninoculated wells. The plate was incubated at 18°C for 3 h without shaking. Subsequently, wells were washed twice with sterile distilled water and the adherent cells were stained with 100 μl of crystal violet (10 g/liter) for 30 min at room temperature. The wells were washed four times with sterile distilled water, and 100 μl of absolute ethanol was added to each well to solubilize the remaining crystal violet. Cell adhesion was determined by measuring the OD595 using a CLARIOstar microplate reader (BMG Labtech, Ortenberg, Germany). The level of adhesion observed for each strain was compared with the adhesion of the wild-type strain, which was set as 100. All assays were performed in quadruplicate and repeated at least two times. The absorbance of the negative control was subtracted from the absorbance of each strain.
For biofilm formation, cells were grown in half-strength TYES broth to the mid-exponential phase. The cultures were diluted 1:100 in half-strength TYES broth and 150 μl of each diluted bacterial culture was deposited in wells of 96-well flat-bottom polystyrene microtiter plates. The plates were covered with aluminum foil and incubated in a humid environment at 18°C under static conditions for 120 h. Biofilm development was evaluated in four wells per strain, and wells containing sterile noninoculated medium were used as negative controls. The culture fluid was discarded, and the wells were washed twice with 200 μl of sterile distilled water. One hundred fifty microliters of crystal violet (10 g/liter) was added to each well, and plates incubated at room temperature for 30 min. Unbound stain was removed by washing the wells four times with 200 μl of sterile distilled water. Stain bound to biofilm cells was solubilized in 100 μl of ethanol, and the absorbance (OD595) was determined.
To measure bacterial cell sedimentation, cells were grown in half-strength TYES broth at 18°C at 200 rpm for 96 h. Tubes were allowed to stand statically for 10 min before being photographed.
Rainbow trout challenges.
Two experimental infection models differing by the infection route (injection or immersion), size of fish, and line of fish were used. For the intramuscular injection model, rainbow trout (27.5 g average mass) of the ARS-Fp-C line were used, which are intermediate in resistance (83). Four groups of 20 fish (n = 80) were anesthetized and challenged with each F. psychrophilum strain by intramuscular injection at a point midway between the insertion of the dorsal fin and the lateral line. Injection of 25 μl was performed using a 26-gauge needle attached to an Eppendorf repeating syringe. All challenge fish were maintained using flowthrough spring water. Two groups of 20 fish were injected with sterile phosphate-buffered saline (PBS) as a negative control. CFU counts per fish (triplicate dilution series) were 2.6 (± 0.8) × 106 for the wild type, 2.7 (± 1.3) × 106 for the ΔgldN mutant, and 2.8 (± 1.5) × 106 for the ΔgldN complemented mutant CFU. The water temperature during challenge was 13.2°C. Mortality was recorded daily, and dead fish examined for clinical signs of disease and for the presence of F. psychrophilum.
For the bath immersion challenge, the rainbow trout (O. mykiss) homozygous line A36 was used as previously described (84, 85). The uniformity of genetic background in isogenic lines and the high susceptibility of line A36 to F. psychrophilum infection makes this line useful to examine bacterial virulence (14). Fish were reared at 10°C in dechlorinated recirculated water until they reached 1.4 g and were then transferred to continuous-flow aquaria for infection experiments. Bacteria used for infections were prepared as follows: the wild-type strain, ΔgldN mutant, and ΔgldN mutant complemented with pBFp4 were grown in TYESG broth at 200 rpm and 18°C to late-exponential phase (OD600 of 1.3). The bacterial cultures were directly diluted (200-fold) into 15 liters of aquarium water. Bacteria were maintained in contact with fish (n = 42 per group) for 24 h by stopping the water flow and were subsequently removed by restoring flow. Sterile TYESG broth was used for the control group. F. psychrophilum bacterial counts were determined at the beginning and at the end of the immersion challenge by plating serial dilutions of water samples on TYESG agar. Water was maintained at 10°C under continuous aeration during the experiment. Virulence was evaluated according to (i) fish mortality during 12 days postinfection and (ii) bacterial loads in organs. Six of each group of 42 fish were randomly chosen and sacrificed 6 h and 30 h after the end of the immersion challenge to evaluate the F. psychrophilum bacterial loads in the spleen (whole organ) and gills (one gill arch). Organs were mechanically disrupted in lysing matrix tubes containing 400 μl of peptone (10 g/liter water) and 1-mm ceramic beads (Mineralex SAS, Lyon, France). Samples were homogenized at 6.0 m s−1 for two cycles of 20 s on a FastPrep-24 instrument (ThermoFisher Scientific, Waltham, MA). Serial dilutions of the homogenized solution were plated on TYESG agar supplemented with fetal calf serum (50 ml/liter) and incubated for 3 days at 18°C. Two independent experiments were performed. Statistical differences of bacterial loads between groups were analyzed using the Mann-Whitney test; the Kaplan-Meier method was used to draw survival curves that were compared using the Gehan-Breslow-Wilcoxon test with GraphPad Prism 8.1.2 (GraphPad Software, San Diego, CA, USA).
Bioinformatic analyses.
Genome sequences were analyzed for T9SS genes encoding proteins that belong to appropriate TIGRFAM multiple-sequence-alignment families (86). This was accomplished using the Integrated Microbial Genomes (IMG version 4.0.1 [https://img.jgi.doe.gov/]) Function Profile tool to examine the genomes for sequences predicted to encode orthologs of GldK (TIGR03525), GldL (TIGR03513), GldM (TIGR03517), GldN (TIGR03523), and SprA (TIGR04189). The genomes were also examined for genes encoding proteins with type A CTDs (TIGR04183) and type B CTDs (TIGR04131 and pfam13585) (19) and for genes encoding PorP/SprF-like proteins (TIGR03519) in the same way. In each case, the trusted cutoffs assigned by The J. Craig Venter Institute (JCVI) that allow identification of the vast majority of family members with vanishingly few false positives (86) were used. Other potential F. psychrophilum T9SS proteins listed in Table S1 (PorU, PorV, SprE, SprT, Plug, PorQ, PorZ, PorX, PorY, and PG1058) were identified by BLASTP analysis using the appropriate F. johnsoniae or P. gingivalis protein as query.
Ethics statements.
The immersion challenges were performed in accordance with the European Directive 2010/2063/UE regarding animal experiments and were approved by the institutional review ethics committee, COMETHEA, of the INRAE Center in Jouy-en-Josas, France. Authorizations were approved by the Direction of the Veterinary Services of Versailles (authorization number 15-58). Injection challenge was performed under the guidelines of NCCCWA Institutional Animal Care and Use Committee, protocol number 132.
Data availability.
All data associated with this work are included either in the manuscript or in the online supplemental materials.
ACKNOWLEDGMENTS
We are grateful to the staff of the fish facilities (INRAE IERP and PEIMA, France) and to Edwige Quillet and Nicolas Dechamp (INRAE GABI, France) for supplying fish and advice. We thank Victor Folcher for technical help. For challenge experiments conducted at the NCCCWA, we thank Timothy Leeds for providing fish and Travis Moreland and Keira Osbourn for technical help.
This work was financially supported by the Agence Nationale de la Recherche (grant ANR-17-CE20-0020-01 FlavoPatho), by grant number MCB-1516990 from the National Science Foundation to M.J.M., and by U.S. Department of Agriculture-ARS CRIS projects number 5090-31320-004-00D and 8082-32000-007-00-D and cooperative agreement number 5090-31320-004-03S. The views contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the U.S. Government. Mention of trade name, proprietary product, or specific equipment does not constitute a guarantee or warranty by the USDA and does not imply its approval to the exclusion of other products that may be suitable.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Dalsgaard I. 1993. Virulence mechanisms in Cytophaga psychrophila and other Cytophaga-like bacteria pathogenic for fish. Annu Rev Fish Dis 3:127–144. doi: 10.1016/0959-8030(93)90032-7. [DOI] [Google Scholar]
- 2.Nematollahi A, Decostere A, Pasmans F, Haesebrouck F. 2003. Flavobacterium psychrophilum infections in salmonid fish. J Fish Dis 26:563–574. doi: 10.1046/j.1365-2761.2003.00488.x. [DOI] [PubMed] [Google Scholar]
- 3.Starliper CE. 2011. Bacterial coldwater disease of fishes caused by Flavobacterium psychrophilum. J Adv Res 2:97–108. doi: 10.1016/j.jare.2010.04.001. [DOI] [Google Scholar]
- 4.Miwa S, Nakayasu C. 2005. Pathogenesis of experimentally induced bacterial cold water disease in ayu Plecoglossus altivelis. Dis Aquat Organ 67:93–104. doi: 10.3354/dao067093. [DOI] [PubMed] [Google Scholar]
- 5.Soares SMC, Walker A, Elwenn SA, Bayliss S, Garden A, Stagg HEB, Munro ES. 2019. First isolation of Flavobacterium psychrophilum associated with reports of moribund wild European eel (Anguilla anguilla) in Scotland. J Fish Dis 42:1509–1521. doi: 10.1111/jfd.13069. [DOI] [PubMed] [Google Scholar]
- 6.Gomez E, Mendez J, Cascales D, Guijarro JA. 2014. Flavobacterium psychrophilum vaccine development: a difficult task. Microb Biotechnol 7:414–423. doi: 10.1111/1751-7915.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sundell K, Landor L, Nicolas P, Jorgensen J, Castillo D, Middelboe M, Dalsgaard I, Donati VL, Madsen L, Wiklund T. 2019. Phenotypic and genetic predictors of pathogenicity and virulence in Flavobacterium psychrophilum. Front Microbiol 10:1711. doi: 10.3389/fmicb.2019.01711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christiansen RH, Madsen L, Dalsgaard I, Castillo D, Kalatzis PG, Middelboe M. 2016. Effect of bacteriophages on the growth of Flavobacterium psychrophilum and development of phage-resistant strains. Microb Ecol 71:845–859. doi: 10.1007/s00248-016-0737-5. [DOI] [PubMed] [Google Scholar]
- 9.Evenhuis JP, Leeds TD, Marancik DP, LaPatra SE, Wiens GD. 2015. Rainbow trout (Oncorhynchus mykiss) resistance to columnaris disease is heritable and favorably correlated with bacterial cold water disease resistance. J Anim Sci 93:1546–1554. doi: 10.2527/jas.2014-8566. [DOI] [PubMed] [Google Scholar]
- 10.Wiens GD, Palti Y, Leeds TD. 2018. Three generations of selective breeding improved rainbow trout (Oncorhynchus mykiss) disease resistance against natural challenge with Flavobacterium psychrophilum during early life-stage rearing. Aquaculture 497:414–421. doi: 10.1016/j.aquaculture.2018.07.064. [DOI] [Google Scholar]
- 11.Silva RMO, Evenhuis JP, Vallejo RL, Tsuruta S, Wiens GD, Martin KE, Parsons JE, Palti Y, Lourenco DAL, Leeds TD. 2019. Variance and covariance estimates for resistance to bacterial cold water disease and columnaris disease in two rainbow trout breeding populations. J Anim Sci 97:1124–1132. doi: 10.1093/jas/sky478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duchaud E, Boussaha M, Loux V, Bernardet JF, Michel C, Kerouault B, Mondot S, Nicolas P, Bossy R, Caron C, Bessières P, Gibrat JF, Claverol S, Dumetz F, Hénaff ML, Benmansour A. 2007. Complete genome sequence of the fish pathogen Flavobacterium psychrophilum. Nat Biotechnol 25:763–769. doi: 10.1038/nbt1313. [DOI] [PubMed] [Google Scholar]
- 13.Wu AK, Kropinski AM, Lumsden JS, Dixon B, MacInnes JI. 2015. Complete genome sequence of the fish pathogen Flavobacterium psychrophilum ATCC 49418(T.). Stand Genomic Sci 10:3. doi: 10.1186/1944-3277-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pérez-Pascual D, Rochat T, Kerouault B, Gomez E, Neulat-Ripoll F, Henry C, Quillet E, Guijarro JA, Bernardet JF, Duchaud E. 2017. More than gliding: involvement of GldD and GldG in the virulence of Flavobacterium psychrophilum. Front Microbiol 8:2168. doi: 10.3389/fmicb.2017.02168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castillo D, Christiansen RH, Dalsgaard I, Madsen L, Espejo R, Middelboe M. 2016. Comparative genome analysis provides insights into the pathogenicity of Flavobacterium psychrophilum. PLoS One 11:e0152515. doi: 10.1371/journal.pone.0152515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McBride MJ, Zhu Y. 2013. Gliding motility and Por secretion system genes are widespread among members of the phylum Bacteroidetes. J Bacteriol 195:270–278. doi: 10.1128/JB.01962-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato K, Naito M, Yukitake H, Hirakawa H, Shoji M, McBride MJ, Rhodes RG, Nakayama K. 2010. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc Natl Acad Sci U S A 107:276–281. doi: 10.1073/pnas.0912010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McBride MJ. 2019. Bacteroidetes gliding motility and the type IX secretion system. Microbiol Spectr 7:PSIB-0002-2018. doi: 10.1128/microbiolspec.PSIB-0002-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veith PD, Glew MD, Gorasia DG, Reynolds EC. 2017. Type IX secretion: the generation of bacterial cell surface coatings involved in virulence, gliding motility and the degradation of complex biopolymers. Mol Microbiol 106:35–53. doi: 10.1111/mmi.13752. [DOI] [PubMed] [Google Scholar]
- 20.Lasica AM, Ksiazek M, Madej M, Potempa J. 2017. The type IX secretion system (T9SS): highlights and recent insights into its structure and function. Front Cell Infect Microbiol 7:215. doi: 10.3389/fcimb.2017.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li N, Zhu Y, LaFrentz BR, Evenhuis JP, Hunnicutt DW, Conrad RA, Barbier P, Gullstrand CW, Roets JE, Powers JL, Kulkarni SS, Erbes DH, Garcia JC, Nie P, McBride MJ. 2017. The type IX secretion system is required for virulence of the fish pathogen Flavobacterium columnare. Appl Environ Microbiol 83:e01769-17. doi: 10.1128/AEM.01769-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shrivastava A, Johnston JJ, van Baaren JM, McBride MJ. 2013. Flavobacterium johnsoniae GldK, GldL, GldM, and SprA are required for secretion of the cell surface gliding motility adhesins SprB and RemA. J Bacteriol 195:3201–3212. doi: 10.1128/JB.00333-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnston JJ, Shrivastava A, McBride MJ. 2018. Untangling Flavobacterium johnsoniae gliding motility and protein secretion. J Bacteriol 200:e00362-17. doi: 10.1128/JB.00362-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakane D, Sato K, Wada H, McBride MJ, Nakayama K. 2013. Helical flow of surface protein required for bacterial gliding motility. Proc Natl Acad Sci U S A 110:11145–11150. doi: 10.1073/pnas.1219753110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lauber F, Deme JC, Lea SM, Berks BC. 2018. Type 9 secretion system structures reveal a new protein transport mechanism. Nature 564:77–82. doi: 10.1038/s41586-018-0693-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorasia DG, Veith PD, Hanssen EG, Glew MD, Sato K, Yukitake H, Nakayama K, Reynolds EC. 2016. Structural insights into the PorK and PorN components of the Porphyromonas gingivalis type IX secretion system. PLoS Pathog 12:e1005820. doi: 10.1371/journal.ppat.1005820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vincent MS, Canestrari MJ, Leone P, Stathopulos J, Ize B, Zoued A, Cambillau C, Kellenberger C, Roussel A, Cascales E. 2017. Characterization of the Porphyromonas gingivalis type IX secretion trans-envelope PorKLMNP core complex. J Biol Chem 292:3252–3261. doi: 10.1074/jbc.M116.765081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leone P, Roche J, Vincent MS, Tran QH, Desmyter A, Cascales E, Kellenberger C, Cambillau C, Roussel A. 2018. Type IX secretion system PorM and gliding machinery GldM form arches spanning the periplasmic space. Nat Commun 9:429. doi: 10.1038/s41467-017-02784-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glew MD, Veith PD, Chen D, Seers CA, Chen YY, Reynolds EC. 2014. Blue native-PAGE analysis of membrane protein complexes in Porphyromonas gingivalis. J Proteomics 110:72–92. doi: 10.1016/j.jprot.2014.07.033. [DOI] [PubMed] [Google Scholar]
- 30.Kulkarni SS, Johnston JJ, Zhu Y, Hying ZT, McBride MJ. 2019. The carboxy-terminal region of Flavobacterium johnsoniae SprB facilitates its secretion by the type IX secretion system and propulsion by the gliding motility machinery. J Bacteriol 201:e00218-19. doi: 10.1128/JB.00218-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kulkarni SS, Zhu Y, Brendel CJ, McBride MJ. 2017. Diverse C-terminal sequences involved in Flavobacterium johnsoniae protein secretion. J Bacteriol 199:e00884-16. doi: 10.1128/JB.00884-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glew MD, Veith PD, Peng B, Chen YY, Gorasia DG, Yang Q, Slakeski N, Chen D, Moore C, Crawford S, Reynolds E. 2012. PG0026 is the C-terminal signal peptidase of a novel secretion system of Porphyromonas gingivalis. J Biol Chem 287:24605–24617. doi: 10.1074/jbc.M112.369223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kharade SS, McBride MJ. 2014. Flavobacterium johnsoniae chitinase ChiA is required for chitin utilization and is secreted by the type IX secretion system. J Bacteriol 196:961–970. doi: 10.1128/JB.01170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veith PD, Muhammad NAN, Dashper SG, Likic VA, Gorasia DG, Chen D, Byrne SJ, Catmull DV, Reynolds EC. 2013. Protein substrates of a novel secretion system are numerous in the Bacteroidetes phylum and have in common a cleavable C-terminal secretion signal, extensive post-translational modification, and cell-surface attachment. J Proteome Res 12:4449–4461. doi: 10.1021/pr400487b. [DOI] [PubMed] [Google Scholar]
- 35.Gorasia DG, Veith PD, Chen D, Seers CA, Mitchell HA, Chen YY, Glew MD, Dashper SG, Reynolds EC. 2015. Porphyromonas gingivalis type IX secretion substrates are cleaved and modified by a sortase-like mechanism. PLoS Pathog 11:e1005152. doi: 10.1371/journal.ppat.1005152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kharade SS, McBride MJ. 2015. Flavobacterium johnsoniae PorV is required for secretion of a subset of proteins targeted to the type IX secretion system. J Bacteriol 197:147–158. doi: 10.1128/JB.02085-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sato K, Yukitake H, Narita Y, Shoji M, Naito M, Nakayama K. 2013. Identification of Porphyromonas gingivalis proteins secreted by the Por secretion system. FEMS Microbiol Lett 338:68–76. doi: 10.1111/1574-6968.12028. [DOI] [PubMed] [Google Scholar]
- 38.Kondo Y, Sato K, Nagano K, Nishiguchi M, Hoshino T, Fujiwara T, Nakayama K. 2018. Involvement of PorK, a component of the type IX secretion system, in Prevotella melaninogenica pathogenicity. Microbiol Immunol 62:554–566. doi: 10.1111/1348-0421.12638. [DOI] [PubMed] [Google Scholar]
- 39.Chen Z, Wang X, Ren X, Han W, Malhi KK, Ding C, Yu S. 2019. Riemerella anatipestifer GldM is required for bacterial gliding motility, protein secretion, and virulence. Vet Res 50:43. doi: 10.1186/s13567-019-0660-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malhi KK, Wang X, Chen Z, Ding C, Yu S. 2019. Riemerella anatipestifer gene AS87_08785 encodes a functional component, GldK, of the type IX secretion system. Vet Microbiol 231:93–99. doi: 10.1016/j.vetmic.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 41.Guo Y, Hu D, Guo J, Wang T, Xiao Y, Wang X, Li S, Liu M, Li Z, Bi D, Zhou Z. 2017. Riemerella anatipestifer type IX secretion system is required for virulence and gelatinase secretion. Front Microbiol 8:2553. doi: 10.3389/fmicb.2017.02553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alvarez B, Secades P, McBride MJ, Guijarro JA. 2004. Development of genetic techniques for the psychrotrophic fish pathogen Flavobacterium psychrophilum. Appl Environ Microbiol 70:581–587. doi: 10.1128/aem.70.1.581-587.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gomez E, Alvarez B, Duchaud E, Guijarro JA. 2015. Development of a markerless deletion system for the fish-pathogenic bacterium Flavobacterium psychrophilum. PLoS One 10:e0117969. doi: 10.1371/journal.pone.0117969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li N, Qin T, Zhang XL, Huang B, Liu ZX, Xie HX, Zhang J, McBride MJ, Nie P. 2015. Gene deletion strategy to examine the involvement of the two chondroitin lyases in Flavobacterium columnare virulence. Appl Environ Microbiol 81:7394–7402. doi: 10.1128/AEM.01586-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu Y, Thomas F, Larocque R, Li N, Duffieux D, Cladiere L, Souchaud F, Michel G, McBride MJ. 2017. Genetic analyses unravel the crucial role of a horizontally acquired alginate lyase for brown algal biomass degradation by Zobellia galactanivorans. Environ Microbiol 19:2164–2181. doi: 10.1111/1462-2920.13699. [DOI] [PubMed] [Google Scholar]
- 46.Link AJ, Phillips D, Church GM. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol 179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rochat T, Barbier P, Nicolas P, Loux V, Pérez-Pascual D, Guijarro JA, Bernardet J-F, Duchaud E. 2017. Complete genome sequence of Flavobacterium psychrophilum strain OSU THCO2-90. Genome Announc 5:e01665-16. doi: 10.1128/genomeA.01665-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ikegami A, Honma K, Sharma A, Kuramitsu HK. 2004. Multiple functions of the leucine-rich repeat protein LrrA of Treponema denticola. Infect Immun 72:4619–4627. doi: 10.1128/IAI.72.8.4619-4627.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma A, Sojar HT, Glurich I, Honma K, Kuramitsu HK, Genco RJ. 1998. Cloning, expression, and sequencing of a cell surface antigen containing a leucine-rich repeat motif from Bacteroides forsythus ATCC 43037. Infect Immun 66:5703–5710. doi: 10.1128/IAI.66.12.5703-5710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nelson SS, Bollampalli S, McBride MJ. 2008. SprB is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery. J Bacteriol 190:2851–2857. doi: 10.1128/JB.01904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiens GD, LaPatra SE, Welch TJ, Rexroad C III, Call DR, Cain KD, LaFrentz BR, Vaisvil B, Schmitt DP, Kapatral V. 2014. Complete genome sequence of Flavobacterium psychrophilum strain CSF259-93, used to select rainbow trout for increased genetic resistance against bacterial cold water disease. Genome Announc 2:e00889-14. doi: 10.1128/genomeA.00889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rhodes RG, Nelson SS, Pochiraju S, McBride MJ. 2011. Flavobacterium johnsoniae sprB is part of an operon spanning the additional gliding motility genes sprC, sprD, and sprF. J Bacteriol 193:599–610. doi: 10.1128/JB.01203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heath JE, Seers CA, Veith PD, Butler CA, Nor Muhammad NA, Chen YY, Slakeski N, Peng B, Zhang L, Dashper SG, Cross KJ, Cleal SM, Moore C, Reynolds EC. 2016. PG1058 is a novel multidomain protein component of the bacterial type IX secretion system. PLoS One 11:e0164313. doi: 10.1371/journal.pone.0164313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ortiz-Suarez ML, Samsudin F, Piggot TJ, Bond PJ, Khalid S. 2016. Full-length OmpA: structure, function, and membrane interactions predicted by molecular dynamics simulations. Biophys J 111:1692–1702. doi: 10.1016/j.bpj.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Samsudin F, Ortiz-Suarez ML, Piggot TJ, Bond PJ, Khalid S. 2016. OmpA: a flexible clamp for bacterial cell wall attachment. Structure 24:2227–2235. doi: 10.1016/j.str.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 56.Rhodes RG, Samarasam MN, Shrivastava A, van Baaren JM, Pochiraju S, Bollampalli S, McBride MJ. 2010. Flavobacterium johnsoniae gldN and gldO are partially redundant genes required for gliding motility and surface localization of SprB. J Bacteriol 192:1201–1211. doi: 10.1128/JB.01495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Braun TF, Khubbar MK, Saffarini DA, McBride MJ. 2005. Flavobacterium johnsoniae gliding motility genes identified by mariner mutagenesis. J Bacteriol 187:6943–6952. doi: 10.1128/JB.187.20.6943-6952.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu Y, McBride MJ. 2016. Comparative analysis of Cellulophaga algicola and Flavobacterium johnsoniae gliding motility. J Bacteriol 198:1743–1754. doi: 10.1128/JB.01020-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pérez-Pascual D, Menendez A, Fernandez L, Mendez J, Reimundo P, Navais R, Guijarro JA. 2009. Spreading versus biomass production by colonies of the fish pathogen Flavobacterium psychrophilum: role of the nutrient concentration. Int Microbiol 12:207–214. [PubMed] [Google Scholar]
- 60.Bernardet JF, Kerouault B. 1989. Phenotypic and genomic studies of “Cytophaga psychrophila” isolated from diseased rainbow trout (Oncorhynchus mykiss) in France. Appl Environ Microbiol 55:1796–1800. doi: 10.1128/AEM.55.7.1796-1800.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Otis EJ. 1984. Lesions of coldwater disease in steelhead trout (Salmo gairdneri): the role of Cytophaga psychrophila extracellular products. MS thesis. University of Rhode Island, Kingston, RI. [Google Scholar]
- 62.Pishchany G, Skaar EP. 2012. Taste for blood: hemoglobin as a nutrient source for pathogens. PLoS Pathog 8:e1002535. doi: 10.1371/journal.ppat.1002535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Högfors-Rönnholm E, Wiklund T. 2010. Hemolytic activity in Flavobacterium psychrophilum is a contact-dependent, two-step mechanism and differently expressed in smooth and rough phenotypes. Microb Pathog 49:369–375. doi: 10.1016/j.micpath.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 64.Moller JD, Ellis AE, Barnes AC, Dalsgaard I. 2005. Iron acquisition mechanisms of Flavobacterium psychrophilum. J Fish Dis 28:391–398. doi: 10.1111/j.1365-2761.2005.00639.x. [DOI] [PubMed] [Google Scholar]
- 65.Nakayama K. 2015. Porphyromonas gingivalis and related bacteria: from colonial pigmentation to the type IX secretion system and gliding motility. J Periodont Res 50:1–8. doi: 10.1111/jre.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Halpern D, Gruss A. 2015. A sensitive bacterial-growth-based test reveals how intestinal Bacteroides meet their porphyrin requirement. BMC Microbiol 15:282. doi: 10.1186/s12866-015-0616-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Högfors-Rönnholm E, Norrgård J, Wiklund T. 2015. Adhesion of smooth and rough phenotypes of Flavobacterium psychrophilum to polystyrene surfaces. J Fish Dis 38:429–437. doi: 10.1111/jfd.12250. [DOI] [PubMed] [Google Scholar]
- 68.Alvarez B, Secades P, Prieto M, McBride MJ, Guijarro JA. 2006. A mutation in Flavobacterium psychrophilum tlpB inhibits gliding motility and induces biofilm formation. Appl Environ Microbiol 72:4044–4053. doi: 10.1128/AEM.00128-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sundell K, Wiklund T. 2011. Effect of biofilm formation on antimicrobial tolerance of Flavobacterium psychrophilum. J Fish Dis 34:373–383. doi: 10.1111/j.1365-2761.2011.01250.x. [DOI] [PubMed] [Google Scholar]
- 70.Hadidi S, Glenney GW, Welch TJ, Silverstein JT, Wiens GD. 2008. Spleen size predicts resistance of rainbow trout to Flavobacterium psychrophilum challenge. J Immunol 180:4156–4165. doi: 10.4049/jimmunol.180.6.4156. [DOI] [PubMed] [Google Scholar]
- 71.Levipan HA, Avendano-Herrera R. 2017. Different phenotypes of mature biofilm in Flavobacterium psychrophilum share a potential for virulence that differs from planktonic state. Front Cell Infect Microbiol 7:76. doi: 10.3389/fcimb.2017.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Braun TF, McBride MJ. 2005. Flavobacterium johnsoniae GldJ is a lipoprotein that is required for gliding motility. J Bacteriol 187:2628–2637. doi: 10.1128/JB.187.8.2628-2637.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.LaFrentz BR, LaPatra SE, Call DR, Cain KD. 2008. Isolation of rifampicin resistant Flavobacterium psychrophilum strains and their potential as live attenuated vaccine candidates. Vaccine 26:5582–5589. doi: 10.1016/j.vaccine.2008.07.083. [DOI] [PubMed] [Google Scholar]
- 74.Gliniewicz K, Wildung M, Orfe LH, Wiens GD, Cain KD, Lahmers KK, Snekvik KR, Call DR. 2015. Potential mechanisms of attenuation for rifampicin-passaged strains of Flavobacterium psychrophilum. BMC Microbiol 15:179. doi: 10.1186/s12866-015-0518-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bertolini JM, Wakabayashi H, Watral VG, Whipple MJ, Rohovec JS. 1994. Electrophoretic detection of proteases from selected strains of Flexibacter psychrophilus and assessment of their variability. J Aquat Anim Health 6:224–233. doi:. [DOI] [Google Scholar]
- 76.Cain KD, LaFrentz BR. 2007. Laboratory maintenance of Flavobacterium psychrophilum and Flavobacterium columnare. Curr Protoc Microbiol 6:13B.1.1–13B.1.12. doi: 10.1002/9780471729259.mc13b01s6. [DOI] [PubMed] [Google Scholar]
- 77.Holt RA, Rohovec JS, Fryer JL. 1993. Bacterial coldwater disease, p 3–23. In Inglis V, Roberts RJ, Bromage NR (ed), Bacterial diseases of fish. Blackwell Scientific Publications, Oxford, United Kingdom. [Google Scholar]
- 78.Cato EP, Johnson JL. 1976. Reinstatement of species rank for Bacteroides fragilis, B. ovatus, B. distasonis, B. thetaiotaomicron, and B. vulgatus: designation of neotype strains for Bacteroides fragilis (Veillon and Zuber) Castellani and Chalmers and Bacteroides thetaiotaomicron (Distaso) Castellani and Chalmers. Int J Syst Bacteriol 26:230–237. doi: 10.1099/00207713-26-2-230. [DOI] [Google Scholar]
- 79.Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300. doi: 10.1128/JB.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McBride MJ, Kempf MJ. 1996. Development of techniques for the genetic manipulation of the gliding bacterium Cytophaga johnsonae. J Bacteriol 178:583–590. doi: 10.1128/jb.178.3.583-590.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 82.Newton JC, Wood TM, Hartley MM. 1997. Isolation and partial characterization of extracellular proteases produced by isolates of Flavobacterium columnare derived from catfish. J Aquat Anim Health 9:75–85. doi:. [DOI] [Google Scholar]
- 83.Wiens GD, LaPatra SE, Welch TJ, Evenhuis JP, Rexroad CE, Leeds TD. 2013. On-farm performance of rainbow trout (Oncorhynchus mykiss) selectively bred for resistance to bacterial cold water disease: effect of rearing environment on survival phenotype. Aquaculture 388-391:128–136. doi: 10.1016/j.aquaculture.2013.01.018. [DOI] [Google Scholar]
- 84.Quillet E, Dorson M, Le Guillou S, Benmansour A, Boudinot P. 2007. Wide range of susceptibility to rhabdoviruses in homozygous clones of rainbow trout. Fish Shellfish Immunol 22:510–519. doi: 10.1016/j.fsi.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 85.Fraslin C, Dechamp N, Bernard M, Krieg F, Hervet C, Guyomard R, Esquerre D, Barbieri J, Kuchly C, Duchaud E, Boudinot P, Rochat T, Bernardet JF, Quillet E. 2018. Quantitative trait loci for resistance to Flavobacterium psychrophilum in rainbow trout: effect of the mode of infection and evidence of epistatic interactions. Genet Sel Evol 50:60. doi: 10.1186/s12711-018-0431-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Haft DH, Selengut JD, Richter RA, Harkins D, Basu MK, Beck E. 2013. TIGRFAMs and genome properties in 2013. Nucleic Acids Res 41:D387–D395. doi: 10.1093/nar/gks1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.de Lorenzo V, Timmis KN. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol 235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data associated with this work are included either in the manuscript or in the online supplemental materials.