In order to obtain high-level production of antibiotics, oils have been used as the main carbon source for some Streptomyces strains. Based on multiomics analysis, this study provides insight into the relationship between triacylglycerol (TAG) metabolism and antibiotic biosynthesis in S. albus ZD11, an oil-preferring industrial Streptomyces strain. Our investigation into TAG hydrolysis yielded further evidence that this strain utilizes complicated strategies enabling an efficient TAG metabolism. In addition, a novel secreted lipase was identified that exhibited highly hydrolytic activity for medium- and long-chain TAGs. Our findings represent a good start toward clarifying the complicated relationship between TAG catabolism and high-level antibiotic production in the industrial strains.
KEYWORDS: antibiotics, fatty acid, lipase, metabolism, triacylglycerol
ABSTRACT
Streptomyces is well known for biosynthesis of secondary metabolites with diverse bioactivities. Although oils have been employed as carbon sources to produce polyketide antibiotics for several industrial Streptomyces strains, the intrinsic correlation between oil utilization and high production of antibiotics still remains unclear. In this study, we investigated the correlation between oil metabolism and salinomycin biosynthesis in Streptomyces albus ZD11, which employs soybean oil as the main carbon source. Comparative genomic analysis revealed the enrichment of genes related to triacylglycerol (TAG) metabolism in S. albus ZD11. Transcriptomic profiling further confirmed the enhancement of TAG metabolism and acyl coenzyme A biosynthesis in S. albus ZD11. Multiple secreted lipases, which catalyze TAG hydrolysis, were seen to be working in a synergistic and complementary manner in aiding the efficient and stable hydrolyzation of TAGs. Together, our results suggest that enhanced TAG hydrolysis and fatty acid degradation contribute to the high efficiency of oil utilization in S. albus ZD11 in order to provide abundant carbon precursors for cell growth and salinomycin biosynthesis.
IMPORTANCE In order to obtain high-level production of antibiotics, oils have been used as the main carbon source for some Streptomyces strains. Based on multiomics analysis, this study provides insight into the relationship between triacylglycerol (TAG) metabolism and antibiotic biosynthesis in S. albus ZD11, an oil-preferring industrial Streptomyces strain. Our investigation into TAG hydrolysis yielded further evidence that this strain utilizes complicated strategies enabling an efficient TAG metabolism. In addition, a novel secreted lipase was identified that exhibited highly hydrolytic activity for medium- and long-chain TAGs. Our findings represent a good start toward clarifying the complicated relationship between TAG catabolism and high-level antibiotic production in the industrial strains.
INTRODUCTION
Streptomyces is one of the most important genera in the class Actinomycetes due to its ability to produce varieties of secondary metabolites with diverse bioactivities. These include antibiotics, antitumor agents, immunosuppressive agents, and enzymes (1). The composition of the nutrients, such as carbon, nitrogen, and inorganic ions, affects the production of secondary metabolites in the genus Streptomyces (2). Although sugars are the preferred carbon source for most microorganisms (2), several species of Streptomyces, such as Streptomyces clavuligerus (clavulanic acid producer) (3), Streptomyces hygroscopicus (rapamycin producer) (4), and Streptomyces tsukubaensis (FK506 producer) (5), have been found to preferentially use oils as their carbon sources, leading to higher production of antibiotics. Since most of the oil-preferring Streptomyces strains are industrial strains (6, 7), the correlation between an efficient oil metabolism and the high production of antibiotics still remains unclear due to a relative lack of genomic information and the fairly infrequent application of genetic manipulation techniques.
Triacylglycerols (TAGs) are the main ingredients of oils. The process of TAG catabolism normally comprises three steps in Streptomyces. Initially, extracellular TAGs are hydrolyzed to free fatty acids (FFAs) and glycerol by secreted lipases and esterases. FFAs are then absorbed by the cells through passive diffusion and/or protein-facilitated transport. This absorption process has been well characterized in Escherichia coli but remains unclear in Streptomyces (8). Finally, intracellular FFAs are degraded, mainly via the beta-oxidation pathway, resulting in the production of acetyl coenzyme A (acetyl-CoA), which can be easily transformed to acyl building blocks for antibiotic biosynthesis, especially polyketides (9). Acetyl-CoA can also be concurrently utilized in the biosynthesis of other lipids, such as phospholipids and neutral lipids, through the fatty acid (FA) biosynthetic pathway, which can then be used in cell growth or storage (10). For oil-utilizing Streptomyces and some other microorganisms, the activities of metabolic pathways for TAGs, such as TAG hydrolysis and beta-oxidation pathways, are considered the main factors affecting the efficiency of oil utilization (11–14). The hydrolysis of TAGs is the first rate-determining step for the oil-utilizing Streptomyces strains to utilize extracellular oils (13, 15). This process is mainly catalyzed by secreted lipases at the lipid-water interface (16). Lipase (EC 3.1.1.3) is a class of hydrolases that is widely found in bacteria, fungi, plants, and mammalian cells. Although several lipases from Streptomyces strains have been reported, the relationship between lipase and secondary metabolism in the oil-utilizing Streptomyces strains remains obscure (17).
Streptomyces albus ZD11 is a derivative of an industrial salinomycin-producing strain which can produce up to 40 g/liter of salinomycin in a medium containing soybean oil (18). To investigate the mechanism of efficient conversion of TAGs to salinomycin in S. albus ZD11, whole-genome sequencing and transcriptome sequencing were both performed in this study. The metabolic pathways for TAGs, including TAG hydrolysis, beta-oxidation, and FA biosynthesis, were found to be enhanced in S. albus ZD11. Overall, 22 lipases were identified, of which 15 contained predicted secretory signal peptides. Among these, two secreted-lipase genes, lip1 and lip2, showed increased expression levels upon soybean oil addition to the medium. The influence of these two lipases on salinomycin biosynthesis was evaluated, and Lip1 was carefully characterized in vitro. Our results provide evidence that the first step of TAG catabolism, as catalyzed by lipase, is a complicated and complementary process in S. albus ZD11.
RESULTS
Streptomyces albus ZD11 prefers soybean oil as the carbon source in salinomycin biosynthesis.
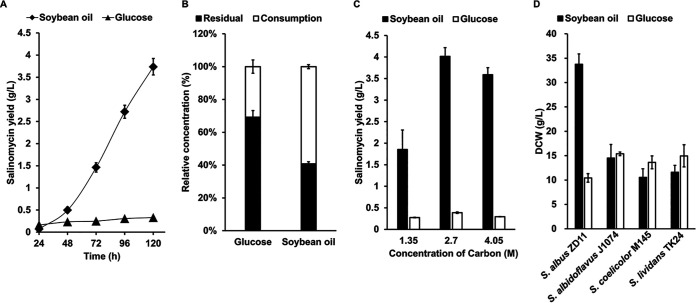
Oils are often employed as primary or auxiliary carbon sources in the production of salinomycin and other antibiotics (19, 20). With its high salinomycin production and ability to use soybean oil, S. albus ZD11 was chosen as an ideal subject to explore the correlation between TAG metabolism and antibiotic biosynthesis. In this study, glucose and soybean oil with equal carbon contents were each provided as the sole carbon source to evaluate salinomycin production in S. albus ZD11. As shown in Fig. 1A, the salinomycin yield in the medium containing soybean oil was up to 3.74 g/liter at 120 h. This is 11-fold higher than the yield in the medium containing glucose (0.33 g/liter). Quantitative analysis of carbon source consumption showed that more soybean oil (59.23%) than glucose (30.80%) had been consumed (Fig. 1B). The conversion ratio of soybean oil carbon to salinomycin carbon was 12.92% (3.08 mmol/mol), while that of glucose was 2.18% (0.52 mmol/mol). Comparison of salinomycin yield in media containing different concentrations of soybean oil or glucose further confirmed that S. albus ZD11 is more inclined to utilize soybean oil than glucose (Fig. 1C).
FIG 1.
S. albus ZD11 prefers soybean oil to glucose as a carbon source. (A) Salinomycin yield in the liquid medium with soybean oil or glucose containing equimolar amounts of carbon (2.7 M). (B) Consumption of soybean oil or glucose in the liquid medium at 120 h. (C) Salinomycin yield in the liquid medium with different concentrations of carbon sources at 120 h. (D) Comparison of cell growth among different Streptomyces strains in the oil medium containing 2.7 M carbon. Error bars indicate the SD for samples tested in triplicate.
To verify whether the oil-preferring property of S. albus ZD11 is distinctive, three Streptomyces type strains, Streptomyces coelicolor M145 [a Streptomyces coelicolor A3(2) derivative], Streptomyces albidoflavus J1074, and Streptomyces lividans TK24, were chosen to compare growth difference in the medium containing glucose or soybean oil as the sole carbon source. According to previous reports, S. coelicolor M145 and S. lividans TK24 are able to utilize long-chain FAs or triolein (11, 13), but we know of no prior report stating that S. albidoflavus J1074 can utilize oils. As shown in Fig. 1D, in the oil medium, the biomass of S. albus ZD11 was more than 2-fold those of the other three strains. In comparison, growth of S. albus ZD11 was a little worse than the other strains in the glucose medium. Among these four strains, only S. albus ZD11 grew much better in the oil medium than in the glucose medium, suggesting that S. albus ZD11 might possess a distinct mechanism of highly efficient oil utilization leading to an increased supply of acyl coenzyme A (acyl-CoA) precursors, not only for its secondary metabolism but also for its primary metabolism.
Comparative genomic analysis reveals an abundant repertoire of genes related to TAG metabolism in S. albus ZD11.
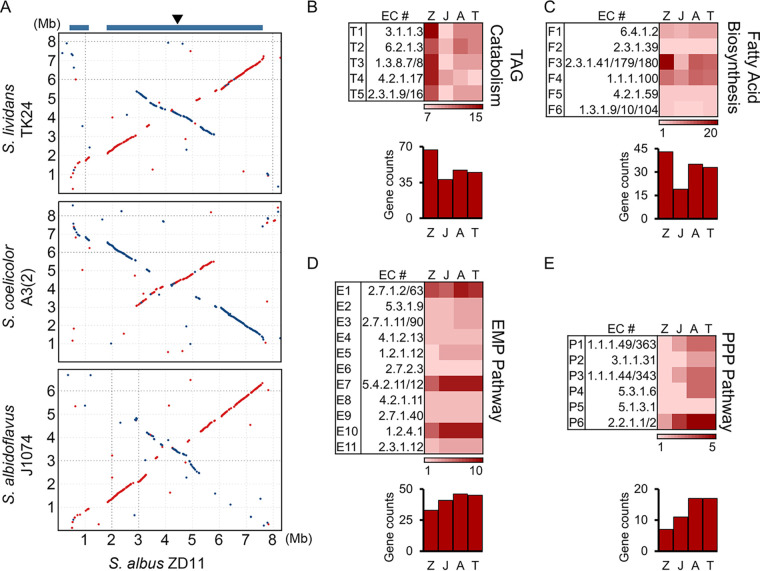
In order to investigate why S. albus ZD11 can utilize oil more efficiently than the other three strains, whole-genome sequencing was performed. S. albus ZD11 has a single linear chromosome of 8,317,371 bp with 7,159 predicted open reading frames (ORFs) and an average GC content of 72.64%. This linear chromosome is nonsymmetrical due to the replication origin (oriC) and dnaA being located at about 207 kb to the right of the chromosome midpoint. Whole-genome alignments between S. albus ZD11 and the other three strains mentioned above identified a 6.4-Mb core genome separated by a 0.7-Mb nonconserved region in S. albus ZD11. Two chromosomal ends and the 0.7-Mb nonconserved region constitute a 1.9-Mb noncore genome for S. albus ZD11 (Fig. 2A).
FIG 2.
Genomic landscape of S. albus ZD11. (A) Whole-genome alignments between S. albus ZD11 and S. albidoflavus J1074, S. coelicolor A3(2), and S. lividans TK24. Matches on the same strand are in red, and those on the opposite strand are in blue. The blue bars at the top represent the conserved core regions on the S. albus ZD11 chromosome. The arrowhead indicates the position of oriC. (B through E) Quantities of genes involved in the catabolic pathways for TAGs (B), the FA biosynthetic pathway (C), the EMP pathway (D), and the PPP (E). The quantity variance of genes corresponding to the KEGG pathway as determined for each biochemical reaction (k number) is color coded, as shown at the bottom of each pathway chart. Z, J, A, and T represent S. albus ZD11, S. albidoflavus J1074, S. coelicolor A3(2), and S. lividans TK24, respectively.
To look further into the relationship between functional genomics and highly efficient oil utilization, the lipid metabolism-related genes were analyzed according to the COG (Clusters of Orthologous Groups) database (21). In S. albus ZD11, more candidate proteins (257 predicted proteins) were clustered into lipid transport and metabolism category (category I) (see Fig. S1 in the supplemental material). Further metabolic pathway analysis based on the KEGG (Kyoto Encyclopedia of Genes and Genomes) database (22) revealed that the number of genes involved in the TAG metabolism, especially the TAG catabolism, including TAG hydrolysis and beta-oxidation, is significantly higher in S. albus ZD11 than in the other strains (Fig. 2B; Data Set S1). Also, the FA biosynthetic pathway is enhanced in the third step, which is catalyzed by 3-oxoacyl-ACP (acyl carrier protein) synthase (EC 2.3.1.41/179/180) (Fig. 2C; Data Set S1). In addition, more than half of these TAG metabolism-related genes are present only in S. albus ZD11 and have no homolog identified in the other three strains, implying that they may play important roles in promoting the efficiency of TAG metabolism in S. albus ZD11.
Conversely, for the metabolic pathways for glucose, such as the Embden-Meyerhof-Parnas (EMP) pathway and the pentose phosphate pathway (PPP), the quantities of related genes present in S. albus ZD11 were generally lower, to various degrees, than those found in the other three strains. In the EMP pathway, the gene counts for main reaction steps in S. albus ZD11 were similar to or less than those in the other three strains, resulting in a lower total number of these genes (Fig. 2D; Data Set S1). In addition, a notably lower gene count for the PPP was observed compared to the other strains (Fig. 2E; Data Set S1). This is consistent with the results of cell growth and salinomycin yield.
Pan-genome analysis was further employed to explore how S. albus ZD11 is distinct from other strains of this species. All 61,202 protein-coding genes from nine S. albus strains are grouped into 24,858 clusters, including 1,541 core gene clusters shared by the nine strains and 15,994 unique gene clusters present in only one strain (Fig. S2A). Compared with the other eight strains, more lipid transport- and metabolism-related genes are identified in S. albus ZD11, and the unique genes account for 52.14% of them (Fig. S2B). In addition, S. albus ZD11 harbors more unique genes involved in the metabolic pathways for TAGs than the other eight strains. However, in the metabolic pathways for glucose, the quantities of total genes and unique genes are similar to or less than those in the other eight strains (Fig. S2C; Data Set S2). This further suggests the enhancement and specificity of TAG metabolism in S. albus ZD11.
Transcriptomic profiling confirms the enhancement of TAG metabolism in S. albus ZD11.
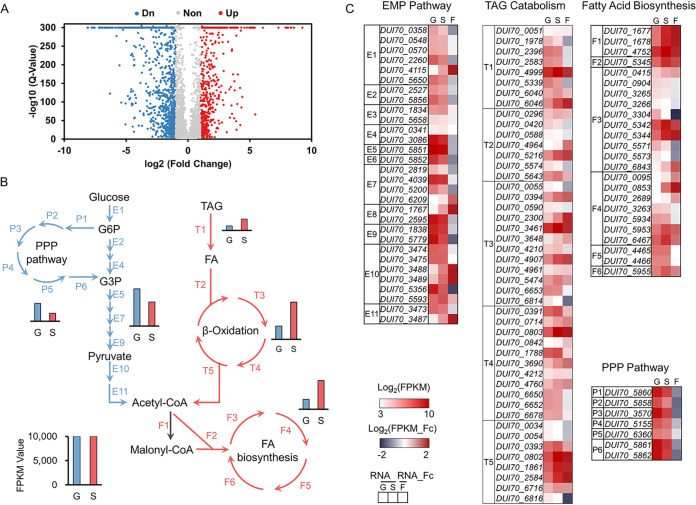
Although more TAG metabolism-related genes are found in the genome of S. albus ZD11, the transcriptional expression behavior of these genes remains unclear. Therefore, transcriptome sequencing (RNA-seq) was performed to profile the transcriptomic changes in S. albus ZD11 when this strain was cultured in the presence of glucose or soybean oil. Comparative transcriptomic analysis results showed that up to 2,093 genes had significant changes in their expression levels between glucose and oil media (fold change > 2; P < 0.001). After low-expression (<10 fragments per kilobase per million [FPKM] in both media) genes were filtered out, the remaining 1,860 genes exhibited differential expression levels (Fig. 3A). Among these genes, the expression levels of 655 were upregulated and 1,205 were downregulated when S. albus ZD11 was cultured in the oil medium compared to the glucose medium.
FIG 3.
Comparative transcriptomic profiling of S. albus ZD11 based on cultivation with different carbon sources. (A) Volcano map of differentially expressed genes. Blue points represent downregulated genes, red points represent upregulated genes, and gray points represent nonregulated genes. (B) Overall expression levels of the metabolic pathways for TAGs or glucose. Red and blue arrows represent the reaction steps involved in the metabolic pathways for TAGs and glucose, respectively. T, F, E, and P represent the reaction steps involved in the TAG hydrolysis and FA beta-oxidation pathways, the FA biosynthetic pathway, the EMP pathway, and the PPP, respectively. The sum of FPKM values of genes in one pathway represents the total expression level of this pathway. (C) The expression levels and fold changes of genes involved in above metabolic pathways. The RNA value is the normalized log2 FPKM value, and RNA fold change (RNA_Fc) is the normalized log2 ratio of FPKM values (FPKMoil/FPKMglucose). G and S indicate samples from glucose and oil medium, respectively. F represents the fold change in gene expression level in the oil medium relative to that in the glucose medium.
As expected, the total expression levels of the metabolic pathways for TAGs in S. albus ZD11 were upregulated and those of the metabolic pathways for glucose were downregulated in the oil medium compared to the glucose medium (Fig. 3B). The total expression level of all the predicted secreted-lipase genes in the oil medium was 2.44-fold that in the glucose medium, and the total expression level of the beta-oxidation-related genes was 2.81-fold higher as well. In the oil medium, the total expression level of genes involved in the FA biosynthetic pathway, of which only the third step is enhanced significantly according to comparative genomic analysis, was also 2.92-fold that in the glucose medium. The activation of FA biosynthetic pathway might be due to the accumulation of intracellular acetyl-CoA resulting from the high efficiency of FA oxidation process in S. albus ZD11. This could also explain why S. albus ZD11 grows better in the oil medium, as reflected by biomass. Conversely, the total expression levels of the EMP pathway and the PPP showed obvious decreases in the oil medium. As shown in Fig. 3C, among the TAG hydrolysis, FA beta-oxidation, and FA biosynthesis pathways, the expression levels of 52.86% of genes increased (FPKMoil/FPKMglucose > 1.5) to various degrees in the oil medium, whereas in the EMP and PPP pathways, 48.65% of genes had reduced expression levels (FPKMoil/FPKMglucose < 0.67) (Data Set S1).
The yield of salinomycin, per liter of culture, is much higher in the oil medium than in the glucose medium. This indicates that a large portion of acetyl-CoA generated by TAG catabolism is converted to various acyl-CoA precursors for salinomycin biosynthesis. Therefore, the expression levels of genes participating in acyl-CoA biosynthesis were also analyzed. As shown in Fig. S3A, acetyl-CoA could be transformed to malonyl-CoA, methylmalonyl-CoA, or ethylmalonyl-CoA, either directly or through some simple reaction steps. The expression levels of genes encoding the acetyl-CoA carboxylase complex were upregulated dramatically in the oil medium, implying an increase in malonyl-CoA supply to support salinomycin or FA biosynthesis. A notable increase in the expression levels of genes involved in the methylmalonyl-CoA or ethylmalonyl-CoA biosynthetic process was also observed (Fig. S3B; Data Set S3). The above results gave further evidence that the metabolic pathways for TAGs are enhanced in S. albus ZD11 and that acetyl-CoA generated by the beta-oxidation pathway could be directed to cell growth or secondary metabolic biosynthesis through the efficient acyl-CoA biosynthetic pathways.
S. albus ZD11 harbors at least 15 predicted secreted lipases that contribute to highly lipolytic activity.
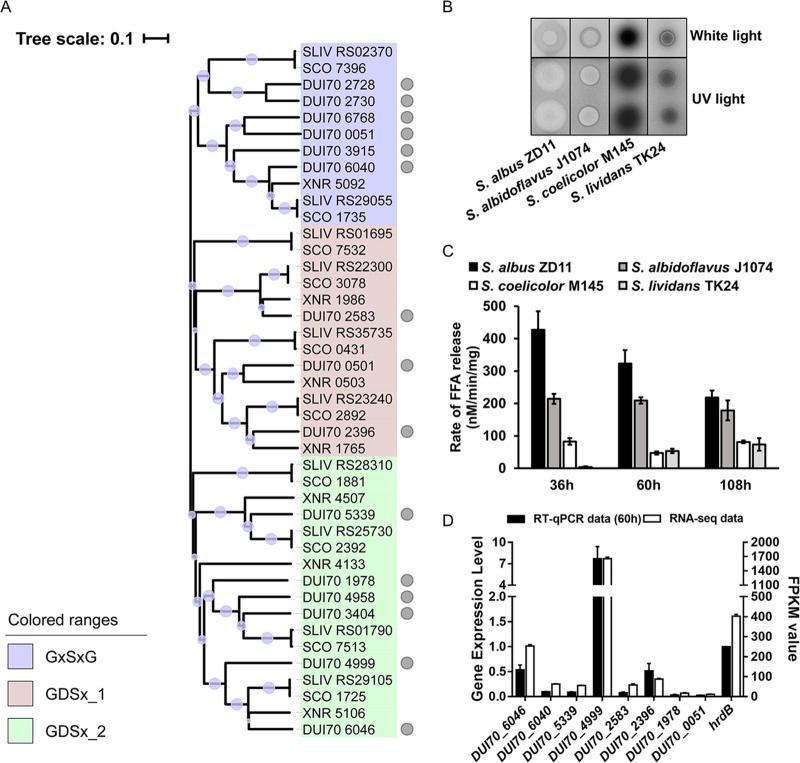
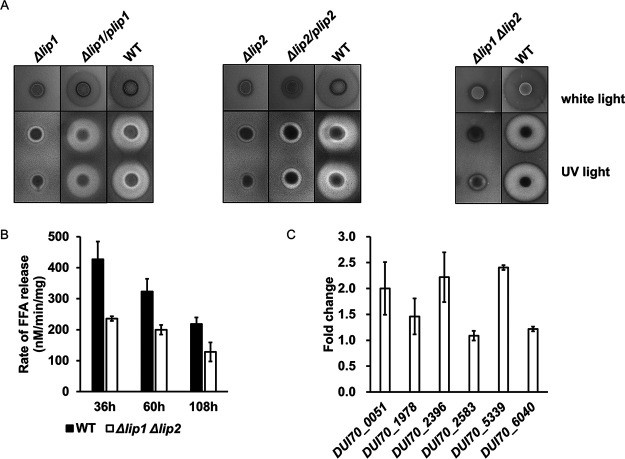
In order to verify the hypothesis that enhanced TAG metabolism at the genomic and transcriptomic levels results in high efficiency of oil utilization in S. albus ZD11, the TAG hydrolysis process was chosen as the focus for investigation. Based on functional annotations in the PFAM database (23), 22 candidate lipases, belonging to the α/β hydrolase family or the SGNH hydrolase family, were predicted in S. albus ZD11. Among them, 15 secreted lipases were screened out according to analysis of the signal peptide sequence (Data Set S1). In addition, the secreted lipases present in the other three strains were also extracted, of which 7, 10, and 10 were extracted from S. albidoflavus J1074, S. coelicolor A3(2), and S. lividans TK24, respectively. Phylogenetic analysis showed that all secreted lipases identified from the four strains could be classified into three clades (Fig. 4A). Clade I contained 11 lipases which shared a canonical GXSXG pentapeptide around a catalytic serine and belonged to the α/β hydrolase family. Six of these lipases were from S. albus ZD11, and the remaining five were from the other three strains. Clades II and III contained 14 and 17 lipases, respectively. The lipases from these two clades belong to the GDS(L) lipase family, a subfamily of the SGNH hydrolase family, and share a GDSX catalytic site motif. The low similarities of amino acid sequences among GDS(L) lipases probably resulted in the distribution into two clades on the phylogenetic tree (24). A rhodamine B (RhB) plate assay was performed to evaluate total lipolytic activity. As shown in Fig. 4B, the fluorescent halos formed by TAG hydrolysis around the aerial mycelial lawns of S. albus ZD11 were much larger than those around S. albidoflavus J1074. The other two strains failed to exhibit any discernible fluorescent halo. The quantitative results further confirmed that the total lipolytic activity of S. albus ZD11 was higher than those of the other three strains, especially at the early stage of fermentation (Fig. 4C).
FIG 4.
Secreted lipase scanning in S. albus ZD11. (A) Phylogenetic relationship of the secreted lipases from the four Streptomyces strains. Gray circles represent the lipases from S. albus ZD11. Diameters of the light purple bubbles on the branches represent the bootstrap values of the N-J tree. Gene IDs with the prefixes SCO, XNR, and SLIV represent lipase genes from S. coelicolor A3(2), S. albidoflavus J1074, and S. lividans TK24, respectively. (B) Determination of total lipolytic activity. Diameters of the fluorescent halos under UV represent lipolytic activities. (C) Quantification of the total lipolytic activity in the oil medium containing 2.7 M carbon. (D) Expression level analysis of secreted-lipase genes based on RNA-seq and RT-qPCR data. Error bars indicate the SD of samples performed in triplicate.
According to our transcriptomic data, eight secreted-lipase genes from S. albus ZD11 could be expressed effectively in both growth media (FPKM value ≥ 10 in either oil or glucose medium). The expression levels of these genes were confirmed by real-time quantitative PCR (RT-qPCR) (Fig. 4D). Two of these lipase genes, DUI70_6046 (named lip1) and DUI70_4999 (named lip2), showed significant increases in their expression levels (fold change > 2; P < 0.001) when S. albus ZD11 was cultured in the oil medium compared to the glucose medium (Fig. S4). This implies that these two lipases may play functional roles in the hydrolytic process of TAGs.
Lip1 and Lip2 play functional roles in the hydrolytic process of TAGs.
To further evaluate the functions of Lip1 and Lip2 in the hydrolytic process of TAGs, Δlip1 and Δlip2 mutants were constructed. The result of the RhB plate assay showed that the fluorescent halos formed by the Δlip1 and Δlip2 mutants were much smaller than those formed by the wild-type (WT) strain, indicating that the total lipolytic activities of the mutants had decreased (Fig. 5A). When each lipase gene was used to complement its deletion mutant, the total lipolytic activity showed a certain degree of recovery in both complemented mutant strains (Δlip1/plip1 and Δlip2/plip2). In the Δlip1/plip1 mutant, the total lipolytic activity recovered dramatically, with fluorescent halos almost as large as those of the WT strain. However, lipolytic activity recovered only a little in the Δlip2/plip2 mutant. The result of expression level measurement showed that the incomplete recovery of the Δlip2/plip2 mutant for total lipolytic activity was due to the lower expression level of complemented lip2 (Fig. S5A), indicating that the genomic context of complemented lip2 was different from that of its WT. In contrast, complemented lip1 showed higher expression level in the Δlip1/plip1 mutant (Fig. S5B). When both lipase genes were disrupted in S. albus ZD11 (Δlip1 Δlip2 mutant), the total lipolytic activity decreased notably on the RhB plates, as expected (Fig. 5A). Further quantitative analysis revealed that the total lipolytic activity of the Δlip1 Δlip2 mutant was about 40% lower than that of the WT strain during the fermentation time course (Fig. 5B). Unexpectedly, the salinomycin yield and oil utilization efficiency of the Δlip1 Δlip2 mutant in flask fermentation did not change significantly (data not shown). Since there are still other secreted lipases in addition to Lip1 and Lip2 present in S. albus ZD11, the remaining lipases might play complementary roles in TAG hydrolysis in the Δlip1 Δlip2 mutant. Among these remaining lipase genes, the expression levels of DUI70_0051, DUI70_2396, and DUI70_5339 in the Δlip1 Δlip2 mutant were more than 2-fold those in the WT strain, and that of DUI70_1978 was about 1.5-fold (Fig. 5C). This indicates that the secreted lipases in S. albus ZD11 could function in a synergistic way in order to maintain efficiently catalytic activity for hydrolyzing TAGs.
FIG 5.
Multiple lipases work in a synergistic way to support efficient and stable TAG hydrolysis in S. albus ZD11. (A) Total lipolytic activity, as determined by the RhB plate assay. (B) Comparison of total lipolytic activities between the Δlip1 Δlip2 mutant and WT strain in a liquid fermentation time course. (C) Expression level analysis of the remaining lipase genes in the Δlip1 Δlip2 mutant. RNA samples were isolated from 60-h cultures. hrdB transcription was monitored and used as the internal control. Values are relative to those of the WT strain. Error bars indicate the SD for samples tested in triplicate.
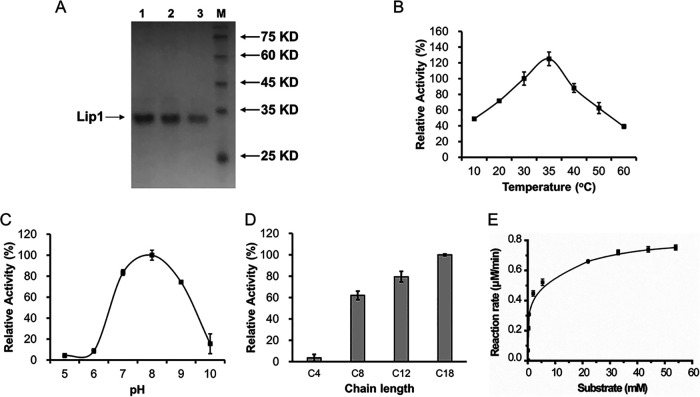
As Lip1 and Lip2 play functional roles in S. albus ZD11, their catalytic activities were investigated in vitro. Both lip1 and lip2 were cloned from S. albus ZD11 and heterologously expressed in E. coli BL21(DE3). The signal peptides of these two lipases were truncated for intracellular soluble expression. The purified proteins of Lip1 and Lip2 were obtained with the aid of the chaperones DnaJ, DnaK, and GrpE (25) (Fig. 6A and Fig. S6).
FIG 6.
Determination of the hydrolytic activity of Lip1. (A) SDS-PAGE of purified truncated Lip1 with a double 6×His tag located at both ends of the peptide. Lane 1, 1.0 μg protein; lane 2, 0.8 μg protein; lane 3, 0.5 μg protein. (B through D) Effects of temperature (B), pH (C), and substrate (D) on the activity of Lip1. C4, C8, C12, and C18 represent tributyrin, tricaprylin, trilaurin, and triolein, respectively. (E) Kinetic curve for Lip1 activity. Error bars indicate the SD for samples tested in triplicate.
The effects of temperature, pH, and substrate on the activities of Lip1 and Lip2 were evaluated. As shown in Fig. 6B, Lip1 was active from 10°C to 60°C and the activity reached a peak at 35°C with para-nitrophenyl palmitate (p-NPP) as the substrate. This temperature is consistent with the fermentation temperature of S. albus ZD11 (33 ± 1°C). To determine the influence of pH on the activity of Lip1, triolein was selected as a substrate, because the p-NPP method is affected by pH variation (26). As shown in Fig. 6C, Lip1 was active from pH 6 to pH 9, with the optimum pH at 8.0. Furthermore, the substrate selectivity of Lip1 was detected by using TAGs with different carbon chain lengths. Our results showed that Lip1 preferred to hydrolyze medium-chain (C8 to C12) and long-chain (>C12) TAGs (Fig. 6D). As oleic acid is one of the major FAs in soybean oil, Lip1 could play an important role in the process of soybean oil hydrolysis. Under the optimum reaction conditions, i.e., 35°C and pH 8.0, Vmax and Km of Lip1 were detected and calculated as 245.90 ± 5.34 U/mg and 0.96 ± 0.08 mM (means ± standard deviations [SD] of three replicates), respectively (Fig. 6E). Compared to the previous reports (27–29), Lip1 exhibited the highest activity for triolein among the GDS(L) family members from the Streptomyces strains studied (27, 30). This further supports the presence of an efficient oil metabolism in S. albus ZD11.
DISCUSSION
Because the industrial strain S. albus ZD11 has an impressive ability to utilize oils efficiently, leading to a high production of salinomycin, the relationship between efficient TAG metabolism and high salinomycin production in S. albus ZD11 was investigated in this study. Multiomics analysis results demonstrate that TAG hydrolysis, beta-oxidation, and FA biosynthesis are all enhanced in S. albus ZD11. With the investigation of the first step of TAG catabolism as our main focus, we found that multiple secreted lipases could work in a synergistic and complementary manner, contributing to highly efficient hydrolysis of extracellular TAGs. Among these lipases, Lip1 exhibited a highly lipolytic activity and was found to play an important role in the hydrolysis of soybean oil.
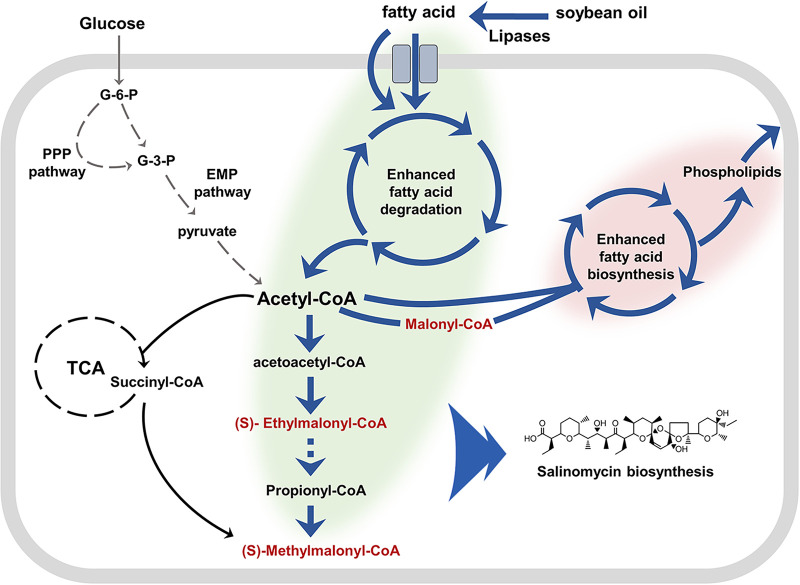
As shown in Fig. 7, we speculate that efficient hydrolysis of extracellular TAGs generates a large number of FFAs which are absorbed later by S. albus ZD11. These FFAs could be degraded by an enhanced beta-oxidation pathway to produce abundant acetyl-CoA, most of which is directly converted to various acyl-CoA precursors for salinomycin biosynthesis. This is consistent with the result in a recent report, which confirmed that efficient beta-oxidation inhibited the activity of tricarboxylic acid (TCA) cycle and directed the acetyl-CoA to polyketide biosynthesis (14). Simultaneously, with the aid of an enhanced FA biosynthetic pathway, S. albus ZD11 was able to utilize acetyl-CoA efficiently to synthesize other lipids, such as phospholipids and neutral lipids. These phospholipids could support fast cell growth, and the neutral lipids could be used for polyketide biosynthesis during the stationary phase (14). Conversely, glucose metabolism is weaker in S. albus ZD11 than in the other three Streptomyces strains tested, implying that S. albus ZD11 has enhanced its ability to utilize oil by reducing its glucose metabolism in order to maintain its competitiveness under special environmental conditions.
FIG 7.
Proposed schematic diagram of enhanced TAG metabolism in S. albus ZD11. Green shading represents the enhanced TAG catabolism and precursor biosynthesis. Pink shading represents the enhanced FA biosynthesis. Dark blue arrows represent the enhanced reaction steps, with gray arrows indicating weakened reaction steps and dashed arrows indicating multiple reaction steps. Intermediates (in red) are the direct precursors of salinomycin biosynthesis.
Microorganisms provide the most abundant source of lipases for biotechnological applications due to their stability, selectivity, and broad substrate specificity (31). Lipases from Streptomyces species have been reported over the last 30 years (27–30). Bielen and colleagues characterized a GDS(L) lipase, SCO1725 from S. coelicolor A3(2), which shows 63.94% sequence similarity to Lip1 (27). However, Lip1 exhibited a 2.5-fold increase in hydrolysis activity for long-chain TAGs and a different optimum temperature compared to SCO1725. The expression level of lip2 was upregulated significantly when S. albus ZD11 was cultured in the oil medium, and the total lipolytic activity of the Δlip2 mutant decreased dramatically. This implies that Lip2 plays an important role in TAG hydrolysis. However, the activity of Lip2 was not detected in vitro. Although most bacterial lipases do not require cofactors (32), some lipases have been reported to require lipase-specific foldases that aid them in correct folding (33). As Lip2 is a unique protein from S. albus ZD11 according to pan-genome analysis, there may therefore be some uncharacterized lipase-specific foldases that participate in Lip2 folding.
As mentioned above, the salinomycin yield showed no obvious decrease in the Δlip1 Δlip2 mutant under our liquid growth conditions. Three reasons might lead to the appearance of this phenotype. First, as multiple secreted lipases work in a synergistic and complementary way in S. albus ZD11, the remaining lipases might restore the lipolytic activity in the Δlip1 Δlip2 mutant to some extent. Second, TAG metabolism is a complicated process, and a decrease in just one step may not significantly impair salinomycin production. Third, given the much lower production of salinomycin in the lab (about 4 g/liter) than that in the industrial setting (40 g/liter), the precursor supply might be still sufficient after deletion of these two lipase genes.
As the beta-oxidation pathway is very important for TAG utilization, a preliminary study on 3-hydroxyacyl-CoA dehydrogenase, which catalyzes the third step of the beta-oxidation pathway, was also carried out by our group. Two highly expressed enzymes (DUI70_0391 and DUI70_0803), predicted to harbor both 3-hydroxyacyl-CoA dehydrogenase and enoyl-CoA hydratase domains, were screened out according to the result of genome-wide mining. Homologs of these two enzymes in S. coelicolor A3(2) have been reported to participate in the degradation of FAs (34). Double deletion of these two genes (DUI70_0391 and DUI70_0803) in S. albus ZD11 led to only about a 50% decrease in salinomycin yield (data not shown). This indicates that although these two enzymes are very important for β-oxidation in S. albus ZD11, there are still some uncharacterized enzymes that play complementary roles. The above results suggest that identifying and characterizing the novel functional proteins will be very helpful for understanding the intrinsic connection between efficient TAG metabolism and high antibiotic production in S. albus ZD11 and other oil-preferring industrial strains.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All experiments were performed with the strains and plasmids listed in Table 1. The mutants were derived from wild-type S. albus ZD11.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| S. albus ZD11 | Derivative obtained by the streak plate method from an industrial salinomycin-producing strain | CGMCC 4.7658 |
| S. albus ZD11 Δlip1 | ZD11 with disruption of lip1; Spr | This study |
| S. albus ZD11 Δlip2 | ZD11 with disruption of lip2; Spr | This study |
| S. albus ZD11 Δlip1/plip1 | Δlip1 harboring plip1 | This study |
| S. albus ZD11 Δlip2/plip2 | Δlip2 harboring plip2 | This study |
| S. albus ZD11 Δlip1 Δlip2 | ZD11 with disruptions of lip1 and lip2; Spr and Apr | This study |
| E. coli TG1 | Host strain for DNA clone | Stratagene |
| E. coli BL21(DE3) | Host strain for protein expression | Stratagene |
| E. coli ET12567(pUZ8002) | dam dcm strain containing helper plasmid pUZ8002 | 44 |
| Plasmids | ||
| pTA2 | TA clone vector; Amr | Toyobo |
| pET28a | Protein expression vector; Kmr | Novagen |
| pKJE7 | Plasmid with para-DnaK-DnaJ-GrpE | Takara |
| pIJ773 | Plasmid with FRT-aac(3)IV-FRT for disruption cassette amplification | 44 |
| pIJ779 | Plasmid with FRT-aadA-FRT for disruption cassette amplification | 44 |
| pSOK804 | Streptomyces-E. coli shuttle vector; Apr | 48 |
| pSOK804H | Streptomyces-E. coli shuttle vector with 6×His tag derived from pSOK804; Apr | This study |
| plip1 | Derived from pSOK804H carrying lip1 with native promoter and terminator | This study |
| plip2 | Derived from pSOK804H carrying lip2 with native promoter and terminator | This study |
| Fosmid SAF16H7 | Fosmid pCC2FOS carrying lip1; Cmr | This study |
| Fosmid SAF5A3 | Fosmid pCC2FOS carrying lip2; Cmr | This study |
| pET-lip1 | lip1 cloned into pET28a; Kmr | This study |
| pET-lip2 | lip2 cloned into pET28a; Kmr | This study |
Amr, ampicillin resistance; Apr, apramycin resistance; Spr, spectinomycin resistance; Kmr, kanamycin resistance; Cmr, chloramphenicol resistance; FRT, Flp recombination target.
Media and growth conditions.
S. albus ZD11 and its derivative strains were grown on ISP4 agar medium (BD, USA) for 7 to 8 days at 30°C for sporulation. Rhodamine B (RhB) agar medium [K2HPO4, 1 g; MgSO4, 1 g; NaCl, 1 g; (NH4)2SO4, 2 g; CaCO3, 2 g; FeSO4, 0.001 g; MgCl2, 0.001 g; ZnSO4, 0.001 g; agar, 20 g; soybean oil, 15 ml; RhB, 0.1 mg per liter] was used to determine lipolytic activity. The culture in tryptic soy broth (TSB) medium (TSB, 30 g per liter) for 24 h was used to obtain high-quality genomic DNA for whole-genome sequencing and genotype confirmation. The industrial seed medium (soybean powder, 30 g; glucose, 40 g; yeast extract, 10 g; CaCO3, 2 g per liter) was used for seed cultivation for 29 h. Ionic medium [NaCl, 2 g; KCl, 2 g; (NH4)2SO4, 5 g; MgSO4, 0.1 g; K2HPO4, 0.2 g; CaCl2, 0.1 g; CaCO3, 5 g per liter] with soybean oil (oil medium) or glucose (glucose medium) containing an equimolar amount of carbon (2.7 M) was used for fermentation at 32°C for 5 days. All E. coli strains were grown at 30°C or 37°C in Luria-Bertani (LB) medium (35). E. coli ET12567(pUZ8002) was used for transferring DNA into S. albus by intergeneric conjugation (18). DNA manipulations were carried out according to standard procedures (36). When appropriate, apramycin, spectinomycin, kanamycin, or chloramphenicol (Sangon, China) was added to the medium at a final concentration of 50, 50, 50, or 25 μg/ml, respectively.
Quantification of carbon source consumption.
To determine the glucose concentration in medium, the mycelia in fermented culture from the glucose medium were removed by centrifugation at 12,000 × g for 2 min. The supernatant was diluted 100 times with water, and the glucose concentration was measured using an SBA-40D biosensor analyzer (Institute of Biology, Shandong Academy of Sciences, China). The soybean oil concentration in medium was measured using a method described previously (6).
Whole-genome sequencing and alignment.
The whole genome of S. albus ZD11 was sequenced using a PacBio RS II platform at the Beijing Genomics Institute (BGI, Shenzhen, China). Gene prediction was performed on a genome assembly using glimmer3 (http://ccb.jhu.edu/software/glimmer/index.shtml) with hidden Markov models. The KEGG and COG databases were used for general function annotation. Whole-genome alignment was performed using MUMmer (version 4.0 beta) with mincluster of 65 and minmatch of 35 (37). Chromosomal sequences of S. albidoflavus J1074 (CP004370.1), S. coelicolor A3(2) (NC_003888.3), and S. lividans TK24 (NZ_CP009124.1) were obtained from GenBank in GenBank format.
RNA-seq analysis.
The total RNA of S. albus ZD11 was prepared from mycelia grown in ionic medium with soybean oil or glucose (0.45 M) as the sole carbon source. Two replicates of 40 μg (wet weight) mycelia were taken at 60 h for both media. Total RNA isolation and RNA-seq were performed according to a previous report (38).
Genome-wide mining and phylogenetic analysis of lipases.
To excavate the lipases in the Streptomyces strains, genome-wide mining of lipase was performed based on functional annotations in the PFAM database (version 32.0). Two clans, CL0264 (SGNH_ hydrolase) and CL0028 (AB_hydrolase), and five lipase families, LIP, Lipase_2, Lipase_GDSL, Lipase_GDSL_2 and Lipase_GDSL_3, were used to screen the putative lipases. Signal peptide analysis was performed on the SignalP web server (http://www.cbs.dtu.dk/services/SignalP/). All sequences of the lipase genes mentioned above were obtained from GenBank in FASTA format. Multiple-sequence alignment (MSA) and phylogenetic tree construction were carried out using MAFFT on the EMBL web server (https://www.ebi.ac.uk/Tools/msa/mafft/) with default parameter settings (39, 40). A bootstrap neighbor-joining (N-J) tree was calculated using MEGA X (41), and the iTOL (Interactive Tree of Life) online software (https://itol.embl.de/) was used to landscape the phylogenetic tree (42).
Determination of total lipolytic activity and biomass.
To determine total lipolytic activity, all Streptomyces strains were cultured in the oil medium containing 2.7 M carbon. The fermented culture was sequentially filtered using a weighted filter cloth (300 mesh) and a 0.45-μm filter. Then, the mycelium wrapped in the filter cloth was dried to a constant weight for biomass determination. Eight milliliters of filtered liquid was concentrated to 200 μl using an ultrafiltration centrifugal tube (10 kDa; Millipore, USA). The concentrated liquid was employed for lipolytic activity determination using the endpoint titration method with soybean oil (5% [vol/vol]) as the substrate (43).
Construction of gene disruption mutants.
A fosmid library was previously generated for the construction of S. albus ZD11 mutants (18). Fosmids SAF16H7, containing lip1, and SAF5A3, containing lip2, were used to construct the gene disruption mutants using the PCR targeting system (44). Part of the Lip1 coding region (amino acids 10 to 254) and part of the Lip2 coding region (amino acids 16 to 237) were displaced by the apramycin and spectinomycin disruption cassette as described previously (45). All mutants were confirmed using PCR and DNA-sequencing analysis. Primer pairs for gene disruption and mutant confirmation are listed in Table S2.
Complementation of the lipase genes in the Δlip1 mutant and the Δlip2 mutant.
The coding region of Lip1 or Lip2 with its native promoter and terminator was amplified and cloned into the EcoRV site of plasmid pSOK804H to generate the plasmid plip1 or plip2. For lip1, 305 bp upward from the start codon and 300 bp downward from the stop codon were selected as its promoter and terminator, respectively. For lip2, 392 bp upward from the start codon and 385 bp downward from the stop codon were selected as its promoter and terminator, respectively. The complementary plasmid was transferred into the Δlip1 or the Δlip2 mutant, respectively, by intergeneric conjugation. The positive transformants were identified by apramycin overlay and confirmed by PCR.
Expression and purification of recombinant proteins.
Signal peptide of either Lip1 (residues 1 to 29) or Lip2 (residues 1 to 30) was truncated when expressed in E. coli BL21(DE3). These two truncated sequences were amplified and cloned into pET28a to generate the recombinant plasmid pET-lip1 or pET-lip2, respectively. E. coli BL21(DE3)/pKJE7 was chosen for heterologous expression of the recombinant lipases. The overnight culture of recombinant E. coli BL21(DE3)/pKJE7 was inoculated into 200 ml liquid LB medium at a final optical density at 600 nm (OD600) of 0.05 with appropriate antibiotics and 1 mM l-arabinose. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added at a final concentration of 0.1 mM when the culture was grown to OD600 of 0.5 to 0.6 at 25°C. After a further 12 h of incubation at 16°C, the cells were harvested by centrifugation and disrupted by sonication on ice. The supernatant was then recovered by centrifugation (4°C, 13,000 × g for 15 min). Soluble Lip1 or Lip2 was purified using nickel-nitrilotriacetic acid (Ni-NTA) His·Bind resin (Novagen, USA) as described by the manufacturer.
Activity assays.
Lipase activities at different temperatures were measured with para-nitrophenyl palmitate (p-NPP; Aladdin, China) as the substrate as described by Rashid et al. (46). One enzyme unit was defined as the amount of enzyme that produced 1 μmol of p-nitrophenyl per min. A modified colorimetric method was used to assess the effect of pH on the lipase activity (47). The experiment was performed at 35°C in different buffers at 50 mM (citrate [pH 5.0 to 6.0], sodium phosphate [pH 6.0 to 8.0], Tris-HCl [pH 8.0 to 9.0], glycine-NaOH [pH 9.0 to 10.0]) containing 1% (wt/vol) gum arabic and 50 mM triolein. The mixture was emulsified by sonification for 4 min and cooled on ice for 1 min; this procedure was repeated three times. The emulsified mixture (500 μl) was dispensed into an Eppendorf tube and preincubated for 5 min. Ten microliters of purified lipase was then added to the tube, followed by another incubation (30 min). The reaction was ended with 500 μl of ethanol. The released FFAs were extracted with 1 ml of isooctane followed by centrifugation at 12,000 × g for 10 min. Eight hundred microliters of the supernatant was mixed with 80 μl of copper reagent (47), followed by another centrifugation at 12,000 × g for 5 min. The optical density of supernatant was measured at 715 nm using a UV spectrophotometer (UV-2450; Shimadzu, Japan). The endpoint titration method was used to determine the substrate specificity (43). Tributyrin, tricaprylin, trilaurin, and triolein were selected as the substrates and hydrolyzed at 35°C for 30 min. A kinetic curve was also drawn using the endpoint titration method at 35°C and pH 8.0, with triolein as the substrate.
Analysis of the yield of salinomycin.
A colorimetric assay based on the vanillin-sulfuric acid reagent was used to determine the production of salinomycin. At first, the fermentation broth (1 ml) was extracted using 9 ml of methanol for 12 h and centrifuged at 12,000 × g for 10 min. Four hundred microliters of the supernatant was mixed with 400 μl of methanol. To this mixture, 200 μl of vanillin-sulfuric acid reagent (vanillin, 3 g; methanol, 900 ml; concentrated sulfuric acid, 100 ml) was added, and the mixture was then incubated at 60°C for 30 min. The absorbance of the reaction product was measured at 520 nm using a UV spectrophotometer. The OD value was used to represent the yield of salinomycin according to the standard curve of the standard salinomycin sample. High-performance liquid chromatography (HPLC) analysis was used for precise determination as described previously (18).
Data availability.
The genome of Streptomyces albus ZD11 was deposited in the GenBank database under accession number CP033071. The sequence reads obtained by RNA-seq were deposited in the SRA database under accession numbers SAMN12251778, SAMN12251779, SAMN12251780, and SAMN12251781.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Key R&D Program of China (2018YFA0903200) and the Zhejiang Provincial Natural Science Foundation of China (no. LZ15C010001).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Hwang KS, Kim HU, Charusanti P, Palsson BO, Lee SY. 2014. Systems biology and biotechnology of Streptomyces species for the production of secondary metabolites. Biotechnol Adv 32:255–268. doi: 10.1016/j.biotechadv.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 2.Sánchez S, Chávez A, Forero A, García-Huante Y, Romero A, Sánchez M, Rocha D, Sánchez B, Avalos M, Guzmán-Trampe S, Rodríguez-Sanoja R, Langley E, Ruiz B. 2010. Carbon source regulation of antibiotic production. J Antibiot (Tokyo) 63:442–459. doi: 10.1038/ja.2010.78. [DOI] [PubMed] [Google Scholar]
- 3.Efthimiou G, Thumser AE, Avignone-Rossa CA. 2008. A novel finding that Streptomyces clavuligerus can produce the antibiotic clavulanic acid using olive oil as a sole carbon source. J Appl Microbiol 105:2058–2064. doi: 10.1111/j.1365-2672.2008.03975.x. [DOI] [PubMed] [Google Scholar]
- 4.Zhao SM, Huang D, Qi HS, Wen JP, Jia XQ. 2013. Comparative metabolic profiling-based improvement of rapamycin production by Streptomyces hygroscopicus. Appl Microbiol Biotechnol 97:5329–5341. doi: 10.1007/s00253-013-4852-7. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, Liu HH, Huang D, Jin LN, Wang C, Wen JP. 2017. Comparative proteomic and metabolomic analysis of Streptomyces tsukubaensis reveals the metabolic mechanism of FK506 overproduction by feeding soybean oil. Appl Microbiol Biotechnol 101:2447–2465. doi: 10.1007/s00253-017-8136-5. [DOI] [PubMed] [Google Scholar]
- 6.Li CX, Hazzard C, Florova G, Reynolds KA. 2009. High titer production of tetracenomycins by heterologous expression of the pathway in a Streptomyces cinnamonensis industrial monensin producer strain. Metab Eng 11:319–327. doi: 10.1016/j.ymben.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Choi DB, Tamura S, Park YS, Okabe M, Seriu Y, Takeda S. 1996. Efficient tylosin production from Streptomyces fradiae using rapeseed oil. J Ferment Bioeng 82:183–186. doi: 10.1016/0922-338X(96)85047-1. [DOI] [Google Scholar]
- 8.van den Berg B. 2010. Going forward laterally: transmembrane passage of hydrophobic molecules through protein channel walls. Chembiochem 11:1339–1343. doi: 10.1002/cbic.201000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan YA, Podevels AM, Kevany BM, Thomas MG. 2009. Biosynthesis of polyketide synthase extender units. Nat Prod Rep 26:90–114. doi: 10.1039/b801658p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gago G, Diacovich L, Arabolaza A, Tsai SC, Gramajo H. 2011. Fatty acid biosynthesis in actinomycetes. FEMS Microbiol Rev 35:475–497. doi: 10.1111/j.1574-6976.2010.00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banchio C, Gramajo HC. 1997. Medium- and long-chain fatty acid uptake and utilization by Streptomyces coelicolor A3(2): first characterization of a Gram-positive bacterial system. Microbiology 143:2439–2447. doi: 10.1099/00221287-143-7-2439. [DOI] [PubMed] [Google Scholar]
- 12.Brigham CJ, Budde CF, Holder JW, Zeng QD, Mahan AE, Rha C, Sinskey AJ. 2010. Elucidation of beta-oxidation pathways in Ralstonia eutropha H16 by examination of global gene expression. J Bacteriol 192:5454–5464. doi: 10.1128/JB.00493-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peacock L, Ward J, Ratledge C, Dickinson FM, Ison A. 2003. How Streptomyces lividans uses oils and sugars as mixed substrates. Enzyme Microb Technol 32:157–166. doi: 10.1016/S0141-0229(02)00278-8. [DOI] [Google Scholar]
- 14.Wang W, Li S, Li Z, Zhang J, Fan K, Tan G, Ai G, Lam SM, Shui G, Yang Z, Lu H, Jin P, Li Y, Chen X, Xia X, Liu X, Dannelly HK, Yang C, Yang Y, Zhang S, Alterovitz G, Xiang W, Zhang L. 2020. Harnessing the intracellular triacylglycerols for titer improvement of polyketides in Streptomyces. Nat Biotechnol 38:76–83. doi: 10.1038/s41587-019-0335-4. [DOI] [PubMed] [Google Scholar]
- 15.Shim MS, Kim WS, Kim JH. 1997. Neutral lipids and lipase activity for actinorhodin biosynthesis of Streptomyces coelicolor A3(2). Biotechnol Lett 19:221–224. doi: 10.1023/A:1018345305250. [DOI] [Google Scholar]
- 16.Javed S, Azeem F, Hussain S, Rasul I, Siddique MH, Riaz M, Afzal M, Kouser A, Nadeem H. 2018. Bacterial lipases: a review on purification and characterization. Prog Biophys Mol Biol 132:23–34. doi: 10.1016/j.pbiomolbio.2017.07.014. [DOI] [PubMed] [Google Scholar]
- 17.Large KP, Mirjalili N, Osborne M, Peacock LM, Zormpaidis V, Walsh M, Cavanagh ME, Leadlay PF, Ison AP. 1999. Lipase activity in Streptomycetes. Enzyme Microb Technol 25:569–575. doi: 10.1016/S0141-0229(99)00080-0. [DOI] [Google Scholar]
- 18.Zhu ZH, Li H, Yu P, Guo YY, Luo S, Chen ZB, Mao XM, Guan WJ, Li YQ. 2017. SlnR is a positive pathway-specific regulator for salinomycin biosynthesis in Streptomyces albus. Appl Microbiol Biotechnol 101:1547–1557. doi: 10.1007/s00253-016-7918-5. [DOI] [PubMed] [Google Scholar]
- 19.Miyazaki Y, Shibata A, Yahagi T, Hara M, Hara K, Yoneda S, Kasahara H, Nakamura Y. July 1980. Method of producing salinomycin antibiotics. US patent US4212942A.
- 20.Zhang XJ, Lu CY, Bai LQ. 2017. Conversion of the high-yield salinomycin producer Streptomyces albus BK3-25 into a surrogate host for polyketide production. Sci China Life Sci 60:1000–1009. doi: 10.1007/s11427-017-9122-8. [DOI] [PubMed] [Google Scholar]
- 21.Huerta-Cepas J, Szklarczyk D, Heller D, Hernandez-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei T, Jensen LJ, von Mering C, Bork P. 2019. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res 47:D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE, Finn RD. 2019. The Pfam protein families database in 2019. Nucleic Acids Res 47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akoh CC, Lee GC, Liaw YC, Huang TH, Shaw JF. 2004. GDSL family of serine esterases/lipases. Prog Lipid Res 43:534–552. doi: 10.1016/j.plipres.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Cui SS, Lin XZ, Shen JH. 2011. Effects of co-expression of molecular chaperones on heterologous soluble expression of the cold-active lipase Lip-948. Protein Expres Purif 77:166–172. [DOI] [PubMed] [Google Scholar]
- 26.Stoytcheva M, Montero G, Zlatev R, Leon JA, Gochev V. 2012. Analytical methods for lipases activity determination: a review. Curr Anal Chem 8:400–407. doi: 10.2174/157341112801264879. [DOI] [Google Scholar]
- 27.Bielen A, Cetkovic H, Long PF, Schwab H, Abramic M, Vujaklija D. 2009. The SGNH-hydrolase of Streptomyces coelicolor has (aryl)esterase and a true lipase activity. Biochimie 91:390–400. doi: 10.1016/j.biochi.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 28.Cote A, Shareck F. 2008. Cloning, purification and characterization of two lipases from Streptomyces coelicolor A3(2). Enzyme Microb Technol 42:381–388. doi: 10.1016/j.enzmictec.2008.01.009. [DOI] [Google Scholar]
- 29.Vujaklija D, Schroder W, Abramic M, Zou PJ, Lescic I, Franke P, Pigac J. 2002. A novel streptomycete lipase: cloning, sequencing and high-level expression of the Streptomyces rimosus GDS(L)-lipase gene. Arch Microbiol 178:124–130. doi: 10.1007/s00203-002-0430-6. [DOI] [PubMed] [Google Scholar]
- 30.Abramic M, Lescic I, Korica T, Vitale L, Saenger W, Pigac J. 1999. Purification and properties of extracellular lipase from Streptomyces rimosus. Enzyme Microb Technol 25:522–529. doi: 10.1016/S0141-0229(99)00077-0. [DOI] [Google Scholar]
- 31.Priyanka P, Tan YQ, Kinsella GK, Henehan GT, Ryan BJ. 2019. Solvent stable microbial lipases: current understanding and biotechnological applications. Biotechnol Lett 41:203–220. doi: 10.1007/s10529-018-02633-7. [DOI] [PubMed] [Google Scholar]
- 32.Liu X, Kokare C. 2017. Microbial enzymes of use in industry, p 267–298. In Branmachari G, Demain AL, Adrio JL (ed), Biotechnology of Microbial Enzymes. Academic Press, London, United Kingdom. [Google Scholar]
- 33.Rosenau F, Tommassen J, Jaeger KE. 2004. Lipase-specific foldases. Chembiochem 5:152–161. doi: 10.1002/cbic.200300761. [DOI] [PubMed] [Google Scholar]
- 34.Menendez-Bravo S, Paganini J, Avignone-Rossa C, Gramajo H, Arabolaza A. 2017. Identification of FadAB complexes involved in fatty acid beta-oxidation in Streptomyces coelicolor and construction of a triacylglycerol overproducing strain. Front Microbiol 8:1428. doi: 10.3389/fmicb.2017.01428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sezonov G, Joseleau-Petit D, D'Ari R. 2007. Escherichia coli physiology in Luria-Bertani broth. J Bacteriol 189:8746–8749. doi: 10.1128/JB.01368-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 37.Marcais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A. 2018. MUMmer4: a fast and versatile genome alignment system. PLoS Comput Biol 14:e1005944. doi: 10.1371/journal.pcbi.1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shan Y, Guo D, Gu Q, Li Y, Li Y, Chen Y, Guan W. 2020. Genome mining and homologous comparison strategy for digging exporters contributing self-resistance in natamycin-producing Streptomyces strains. Appl Microbiol Biotechnol 104:817–831. doi: 10.1007/s00253-019-10131-7. [DOI] [PubMed] [Google Scholar]
- 39.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Letunic I, Bork P. 2016. Interactive Tree Of Life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J, Wang D, Wang B, Mei ZH, Liu J, Yu HW. 2012. Enhanced activity of Rhizomucor miehei lipase by directed evolution with simultaneous evolution of the propeptide. Appl Microbiol Biotechnol 96:443–450. doi: 10.1007/s00253-012-4049-5. [DOI] [PubMed] [Google Scholar]
- 44.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A 100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang TJ, Shan YM, Li H, Dou WW, Jiang XH, Mao XM, Liu SP, Guan WJ, Li YQ. 2017. Multiple transporters are involved in natamycin efflux in Streptomyces chattanoogensis L10. Mol Microbiol 103:713–728. doi: 10.1111/mmi.13583. [DOI] [PubMed] [Google Scholar]
- 46.Rashid N, Shimada Y, Ezaki S, Atomi H, Imanaka T. 2001. Low-temperature lipase from psychrotrophic Pseudomonas sp strain KB700A. Appl Environ Microbiol 67:4064–4069. doi: 10.1128/aem.67.9.4064-4069.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwon DY, Rhee JS. 1986. A simple and rapid colorimetric method for determination of free fatty acids for lipase assay. J Am Oil Chem Soc 63:89–92. doi: 10.1007/BF02676129. [DOI] [Google Scholar]
- 48.Van Mellaert L, Mei LJ, Lammertyn E, Schacht S, Anne J. 1998. Site-specific integration of bacteriophage VWB genome into Streptomyces venezuelae and construction of a VWB-based integrative vector. Microbiology 144:3351–3358. doi: 10.1099/00221287-144-12-3351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome of Streptomyces albus ZD11 was deposited in the GenBank database under accession number CP033071. The sequence reads obtained by RNA-seq were deposited in the SRA database under accession numbers SAMN12251778, SAMN12251779, SAMN12251780, and SAMN12251781.