HMFO is the only enzyme described to date that can catalyze by itself the three consecutive oxidation steps to produce FDCA from HMF. Unfortunately, only one HMFO enzyme is currently available for biotechnological application. This availability is enlarged here by the identification, heterologous production, purification, and characterization of two new HMFOs, one from Pseudomonas nitroreducens and one from an unidentified Pseudomonas species. Compared to the previously known Methylovorus HMFO, the new enzyme from P. nitroreducens exhibits better performance for FDCA production in wider pH and temperature ranges, with higher tolerance for the hydrogen peroxide formed, longer half-life during oxidation, and higher yield and total turnover numbers in long-term conversions under optimized conditions. All these features are relevant properties for the industrial production of FDCA. In summary, gene screening and heterologous expression can facilitate the selection and improvement of HMFO enzymes as biocatalysts for the enzymatic synthesis of renewable building blocks in the production of bioplastics.
KEYWORDS: hydroxymethylfurfural, furandicarboxylic acid, database screening, Pseudomonas genes, Escherichia coli expression, flavooxidases, enzyme kinetics, enzyme stability, biotransformation, bioplastics
ABSTRACT
The enzymatic production of 2,5-furandicarboxylic acid (FDCA) from 5-hydroxymethylfurfural (HMF) has gained interest in recent years, as FDCA is a renewable precursor of poly(ethylene-2,5-furandicarboxylate) (PEF). 5-Hydroxymethylfurfural oxidases (HMFOs) form a flavoenzyme family with genes annotated in a dozen bacterial species but only one enzyme purified and characterized to date (after heterologous expression of a Methylovorus sp. HMFO gene). This oxidase acts on both furfuryl alcohols and aldehydes and, therefore, is able to catalyze the conversion of HMF into FDCA through 2,5-diformylfuran (DFF) and 2,5-formylfurancarboxylic acid (FFCA), with only the need of oxygen as a cosubstrate. To enlarge the repertoire of HMFO enzymes available, genetic databases were screened for putative HMFO genes, followed by heterologous expression in Escherichia coli. After unsuccessful trials with other bacterial HMFO genes, HMFOs from two Pseudomonas species were produced as active soluble enzymes, purified, and characterized. The Methylovorus sp. enzyme was also produced and purified in parallel for comparison. Enzyme stability against temperature, pH, and hydrogen peroxide, three key aspects for application, were evaluated (together with optimal conditions for activity), revealing differences between the three HMFOs. Also, the kinetic parameters for HMF, DFF, and FFCA oxidation were determined, the new HMFOs having higher efficiencies for the oxidation of FFCA, which constitutes the bottleneck in the enzymatic route for FDCA production. These results were used to set up the best conditions for FDCA production by each enzyme, attaining a compromise between optimal activity and half-life under different conditions of operation.
IMPORTANCE HMFO is the only enzyme described to date that can catalyze by itself the three consecutive oxidation steps to produce FDCA from HMF. Unfortunately, only one HMFO enzyme is currently available for biotechnological application. This availability is enlarged here by the identification, heterologous production, purification, and characterization of two new HMFOs, one from Pseudomonas nitroreducens and one from an unidentified Pseudomonas species. Compared to the previously known Methylovorus HMFO, the new enzyme from P. nitroreducens exhibits better performance for FDCA production in wider pH and temperature ranges, with higher tolerance for the hydrogen peroxide formed, longer half-life during oxidation, and higher yield and total turnover numbers in long-term conversions under optimized conditions. All these features are relevant properties for the industrial production of FDCA. In summary, gene screening and heterologous expression can facilitate the selection and improvement of HMFO enzymes as biocatalysts for the enzymatic synthesis of renewable building blocks in the production of bioplastics.
INTRODUCTION
5-Hydroxymethylfurfural oxidases (HMFOs; EC 1.1.3.47) are flavoenzymes of biotechnological interest, classified in the superfamily of glucose-methanol-choline oxidases and dehydrogenases (GMC) (1). The importance of these enzymes resides in their ability to catalyze the three consecutive oxidation steps for the production of 2,5-furandicarboxylic acid (FDCA) from 5-hydroxymethylfurfural (HMF) with only the need of molecular oxygen as a cosubstrate. The final product, FDCA, is reported as a building block for the production of renewable and biodegradable plastics through its polymerization into poly(ethylene-2,5-furandicarboxylate) (PEF) (2), which is expected to substitute for petroleum-derived poly(ethylene-terephthalate) (PET) plastics in the near future.
HMF-oxidizing activity was first identified in Cupriavidus basilensis strain HMF14 (3, 4) and more recently in Pseudomonas putida strain ALS1267 (5), with their metabolic pathways being characterized. The hmfH gene of C. basilensis encodes an HMFO (UniProt D5KB61) that contributes to the conversion of HMF into FDCA, while the lack of this gene in P. putida suggests that, in this species, the conversion of HMF is catalyzed by different enzymes. The only HMFO characterized to date is that from Methylovorus sp. strain MP688 (MetspHMFO), a homologue of the enzyme encoded by the hmfH gene of C. basilensis, which was produced in Escherichia coli (6). This flavoenzyme, with a flavin adenine dinucleotide (FAD) molecule as a prosthetic group, is active on primary alcohols, primary thiols, and hydrated aldehydes (6, 7). Its catalytic mechanism, similar to that of other GMC oxidases (8–11), involves a proton transfer from the hydroxyl (or thiol) group to a conserved catalytic base, MetspHMFO His467 (12), and hydride abstraction from the substrate α-carbon by the oxidized flavin. The reduced flavin is then reoxidized by molecular oxygen, yielding hydrogen peroxide as a by-product (13).
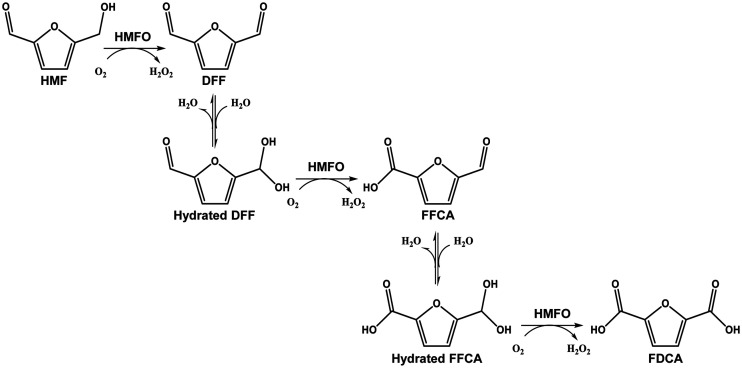
The reported activity of HMFO on furfuryl alcohols and aldehydes makes this enzyme a suitable biocatalyst for the production of FDCA from HMF, which takes place through the hydrated 2,5-diformylfuran (DFF) and 2,5-formylfurancarboxylic acid (FFCA) gem-diol intermediates (Fig. 1) (14). Other oxidases, such as galactose oxidase (GAO) from Dactylium dendroides and glyoxal oxidase (GLOX) from Pycnoporus cinnabarinus, have also been reported to act on furfural derivatives, but their activity is restricted to the alcohol or the aldehyde groups, respectively (15). A recent study has demonstrated the ability of aryl-alcohol oxidase (AAO) from Pleurotus eryngii to catalyze the separate oxidation of HMF, DFF, and FFCA, although the complete oxidation of HMF by AAO stops at the FFCA level, due to inhibition of the last oxidation step by the peroxide generated in the first two reactions (16). More recently, a new copper radical oxidase from Colletotrichum graminicola has been described as an AAO-type enzyme and reported to oxidize HMF into DFF and 5-hydroxymethylfurancarboxylic acid (HMFCA) into FFCA without detected activity on the aldehyde groups of these compounds (17). Due to the above-named characteristics, efficient production of FDCA by the latter oxidases requires the combination of several enzymes, such as the AAO/chloroperoxidase (18), AAO/peroxygenase (15, 19), GAO/peroxygenase (15, 20), GLOX/AAO (21), or AAO/catalase (16) couples already reported. Therefore, MetspHMFO is the only enzyme described to date able to perform by itself (i.e., without the concourse of a second enzyme) the three oxidation steps for FDCA production from HMF. In recent years, several strategies have been followed to increase the performance of MetspHMFO for FDCA bioproduction, such as its coexpression with the HmfH enzyme, which oxidizes HMF by the HMFCA route (3), in whole-cell systems (22), directed mutagenesis of MetspHMFO (23), or its combination with lipase in an enzymatic cascade (24).
FIG 1.
Reactions for conversion of HMF into FDCA by MetspHMFO. First, the alcohol group of HMF is oxidized, resulting in DFF. Then, oxidation of the hydrated form (gem-diol) of DFF gives FFCA. Finally, hydrated FFCA is converted into FDCA.
Taking into account the good performance and promising use of HMFO for FDCA production, in the present study, we focused first on the search for new HMFOs in databases. Then, as a result of Escherichia coli expression trials with four HMFO genes, two new enzymes, from Pseudomonas nitroreducens and an unidentified Pseudomonas strain (25), were purified and characterized, and their oxidative activity on HMF was analyzed in short- and long-term reactions.
RESULTS
Identification of new HMF-oxidizing enzymes.
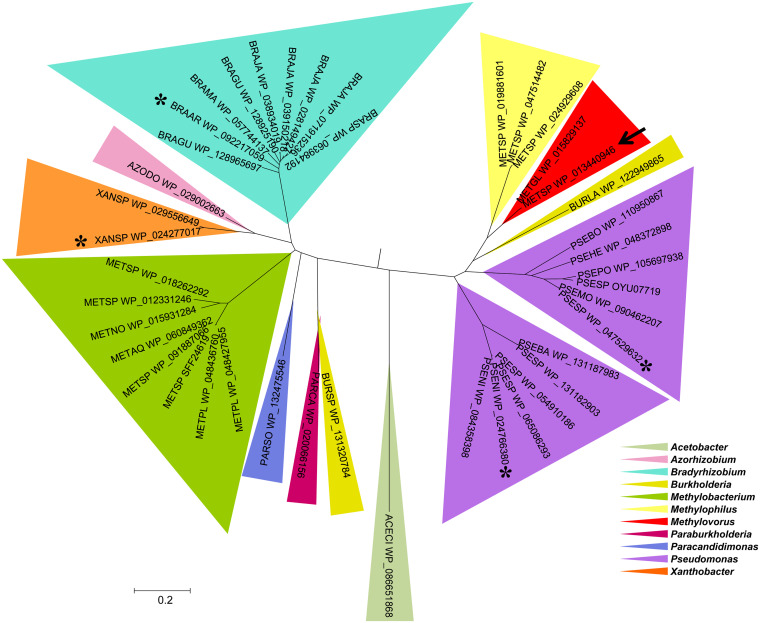
BLAST of the amino acid sequence of MetspHMFO (UniProt E4QP00) against the nonredundant protein sequences in GenBank revealed 41 entries with identities of >45%, all of them from the phylum Proteobacteria (Fig. 2). The largest numbers of HMFOs were found in the Pseudomonas (12 sequences), Bradyrhizobium (9 sequences), and Methylobacterium (8 sequences) genera, and those with the highest homology with MetspHMFO were HMFOs from Methylophylus, Pseudomonas, and Burkholderia species, with more than 60% identity with the query (Table S1 in the supplemental material).
FIG 2.
Maximum-likelihood phylogenetic tree of 41 HMFO-like sequences in 11 bacterial genera, from searching GenBank. The sequence used as the BLAST query is indicated with an arrow, and the four sequences optimized for E. coli expression are indicated with asterisks.
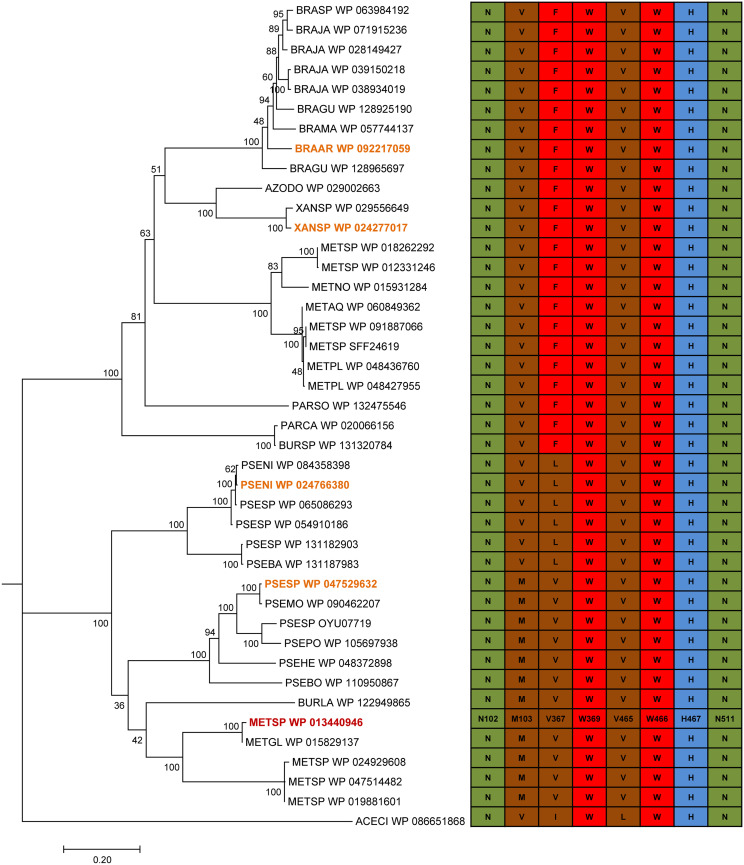
All of the sequences described above conserve the ADP-binding domain and the consensus sequences PS00623 and PS00624, typical of the GMC oxidoreductase superfamily (Fig. S1) (1). A more detailed analysis of conserved active-site amino acids (Fig. 3) reveals that all of them include the same histidine and asparagine residues, MetspHMFO His467 and Asn511, at the catalytic positions described for the latter enzyme (12). Asn102, Trp369, and Trp466 are also fully conserved, while substitutions are often observed at Met103, involved in the correct orientation of the substrate with respect to the active site, and at Val367, described as a site for improving the activity on carboxyl-containing substrates (12). From this repertoire of enzymes, four HMFO sequences from the two main HMFO clusters (Fig. 2)—one from P. nitroreducens (PseniHMFO), one from Pseudomonas sp. strain 11/12A (PsespHMFO), one from Xanthobacter sp. strain 126 (XanspHMFO), and one from Bradyrhizobium arachidis (BraarHMFO)—were selected and optimized for E. coli expression to produce them as recombinant proteins, together with MetspHMFO.
FIG 3.
Conservation of residues around the HMFO active site after alignment of sequences included in the phylogenetic tree. Nonpolar residues are colored in brown, aromatic residues are colored in red, basic residues are colored in blue, and acidic residues are colored in green. The four new HMFO sequences optimized for gene expression in E. coli and that of MetspHMFO are labeled in orange and red, respectively.
Expression and purification of two new HMFOs.
The four optimized genes (Table S2) were cloned into the pET23b and/or the His-tagged pET28a plasmid and transformed into E. coli C41(DE3). XanspHMFO and BraarHMFO could not be expressed using either the pET23b or the pET28a vector, while PseniHMFO and PsespHMFO were purified and characterized after pET28a and pET23b expression, respectively. To evaluate whether the His tag affects the properties of HMFOs, the optimized gene encoding MetspHMFO (Table S2) was also cloned into both plasmids, and the tagged/untagged forms were characterized in parallel. The four proteins were expressed as soluble, active enzymes by slow induction at 16°C during 72 to 96 h.
Untagged MetspHMFO and PsespHMFO were purified to electrophoretic homogeneity (Fig. S2a and c, respectively) by two anion exchange chromatographic steps (see conditions in Table S3), yielding 34 and 10 mg of protein per liter of culture, respectively (Table 1). For the His-tagged proteins, anionic chromatography after the affinity chromatographic step provided pure enzyme (Fig. S2b and d), with yields of 17 and 47 mg of protein per liter of culture of MetspHMFOHis and PseniHMFOHis, respectively (Table 1). The specific activities of the purified enzymes were 3.3 to 3.6 U · mg−1 for MetspHMFO (similar for the untagged and the His-tagged forms), 2.0 U · mg−1 for PsespHMFO, and 0.2 U · mg−1 for PseniHMFOHis. Vanillyl alcohol was used for the activity estimations described above, since it is oxidized with the highest catalytic efficiency, as described below.
TABLE 1.
Summary of purification processes of recombinant HMFOsa
| HMFO | Purification step | Total protein (mg) | Total activity (U)b | Sp act (U/mg) | Degree of purification (fold) | Yield (%) |
|---|---|---|---|---|---|---|
| MetspHMFO | Crude extract | 1,660 | 284 | 0.17 | 1.0 | 100 |
| ResourceQ | 71 | 187 | 2.62 | 15.4 | 66 | |
| MonoQ | 34 | 120 | 3.57 | 21.0 | 42 | |
| MetspHMFOHisc | Crude extract | 1,910 | 100 | 0.05 | 1.0 | 100 |
| HiTrap | 31 | 81 | 2.65 | 50.4 | 81 | |
| MonoQ | 17 | 56 | 3.30 | 62.9 | 56 | |
| PsespHMFO | Crude extract | 1,720 | 138 | 0.08 | 1.0 | 100 |
| ResourceQ | 50 | 71 | 1.42 | 17.8 | 52 | |
| MonoQ | 10 | 20 | 2.02 | 25.2 | 15 | |
| PseniHMFOHisc | Crude extract | 1,650 | 12 | 0.01 | 1.0 | 100 |
| HiTrap | 103 | 10 | 0.09 | 12.6 | 79 | |
| MonoQ | 47 | 8 | 0.16 | 22.2 | 63 |
Data are for protein from 1 liter of E. coli culture.
Measured with vanillyl alcohol in 50 mM Tris-HCl, pH 7.0, at 25°C.
His-tagged protein.
The molecular weights (MW) of the different HMFOs, measured by matrix-assisted laser desorption and ionization-time of flight (MALDI-TOF) mass spectrometry, were ∼57 kDa (Table 2), in good agreement with the theoretical masses calculated from their amino acid sequences (Table S1). This indicates that HMFOs are not purified as covalently bound dimeric enzymes. Also, the experimental isoelectric point (pI) values of 6.0 to 6.5 were similar to the theoretical ones (as shown in the tables mentioned above).
TABLE 2.
Spectroscopic and physicochemical properties of recombinant HMFOsa
| HMFO | Value for indicated property |
||||||
|---|---|---|---|---|---|---|---|
| Spectroscopicb |
Physicochemicalc |
||||||
| λband II (nm) | λband I (nm) | εband I (M−1 cm−1) | A278/Aband I | MW (Da) | pI | Mean Tm ± SD (°C) | |
| MetspHMFO | 387 | 457 | 11,271 | 10.7 | 57,547.0 | 6.5 | 47.93 ± 0.02 |
| MetspHMFOHis | 387 | 457 | 11,667 | 10.9 | 57,854.3 | 6.6 | 47.77 ± 0.06 |
| PsespHMFO | 387 | 457 | 11,432 | 10.4 | 56,945.2 | 6.1 | 40.05 ± 0.18 |
| PseniHMFOHis | 382 | 456 | 12,060 | 10.1 | 58,863.5 | 6.5 | 43.80 ± 0.21 |
Measured in 50 mM Tris-HCl, pH 7.0, at 25°C.
λband, wavelength of the corresponding flavin band; εband, extinction coefficient at the corresponding flavin band. See Fig. S3b.
MW, pI, and Tm values were estimated by MALDI-TOF, isoelectric focusing, and ThermoFAD assay, respectively.
HMFO spectral properties.
The secondary structures of the enzymes were evaluated by circular dichroism (CD) (Fig. S3a). The far-UV spectra showed minimums at ∼222 nm, typical of α-helix structures, being more intense in the MetspHMFO and PsespHMFO spectra. The contents in α-helixes (27 to 47%), β-sheets (11 to 20%), and turns (20 to 24%) were obtained by deconvolution of the CD spectra (Table S4). As shown for MetspHMFOHis, the molar ellipticity per residue (MER) intensity revealed an α-helix decrease (from 37% to 27%) and an increase of unordered structure (from 26% to 37%) in the His-tagged enzymes.
The enzymes showed UV-visible spectra characteristic of FAD-containing proteins (Table 2 and Fig. S3b). Two peaks at 382 to 387 nm (band II) and 456 to 457 nm (band I) with a shoulder at 483 nm and A279/A457 ratios of ∼10 were observed, indicating the proper incorporation of the cofactor. The calculated extinction coefficients at the band I maximums (εband I) of MetspHMFO, MetspHMFOHis, PsespHMFO, and PseniHMFOHis were 11,271, 11,667, 11,432, and 12,060 M−1 cm−1, respectively. These values were used to estimate the corresponding enzyme concentrations.
Temperature and pH stability.
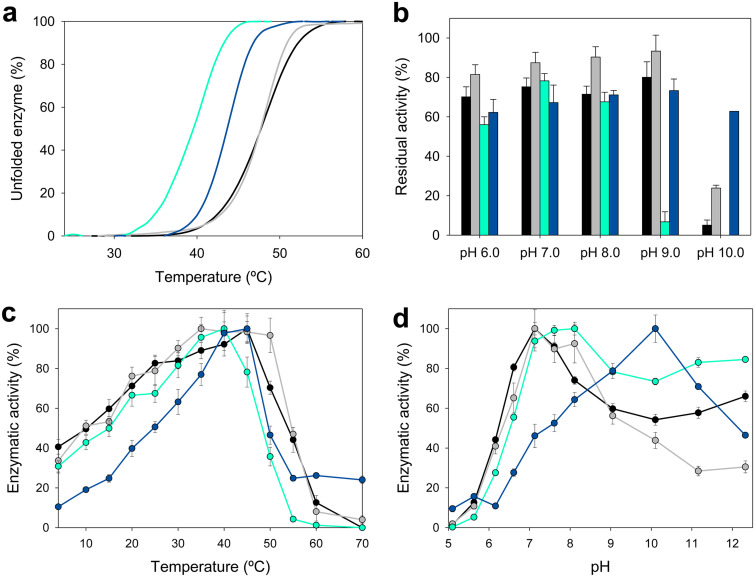
As an estimation of thermostability, the melting temperature (Tm) for each enzyme was calculated from the changes of FAD fluorescence using the ThermoFAD assay (26). The denaturation curves indicated a Tm of 48°C for the enzyme from Methylovorus (both MetspHMFO and MetspHMFOHis), while PsespHMFO and PseniHMFO appeared to be 8°C and 4°C less stable, respectively (Fig. 4a and Table 2).
FIG 4.
Thermal stability measured as melting temperature (a), pH stability after 72-h incubation at 25°C in B&R buffer (pH 6 to 9) (b), and optimal temperature (c) and pH (d) (referred to the optimal value) for MetspHMFO (black), MetspHMFOHis (gray), PsespHMFO (cyan), and PseniHMFOHis (blue). All activities (b to d) were measured with vanillyl alcohol as the substrate. Mean values and standard deviations are shown.
To determine the range of pHs in which the enzymes are active, we measured the residual activity on vanillyl alcohol after 72 h of incubation under different pH conditions. The enzyme from Methylovorus (both MetspHMFO and MetspHMFOHis) was quite stable at pH 6 to 9, conserving 70 to 85% activity (Fig. 4b). At pH 10, the activities of MetspHMFO and MetspHMFOHis decreased to only 5% and 24% activity, respectively, while PseniHMFOHis kept ∼65 to 70% of its initial activity between pH 6 and 10. The range in which PsespHMFO was stable was slightly narrower, pH 6 to 8, with only ∼10% activity and total activity loss at pH 9 and pH 10, respectively. Outside these pH ranges, the activity of all the enzymes decreased abruptly in the first 24 h (data not shown).
Optimal temperature and pH.
The pH and temperature optima were also calculated with vanillyl alcohol as the substrate. PsespHMFO and PseniHMFOHis attained their maximal activities at 35 to 40°C and 40 to 45°C, respectively, and decreased to 50% below 15°C and 25°C, respectively, and above 50°C for both (Fig. 4c). Similar profiles were observed for MetspHMFO, which attained its maximal activity at 45°C and kept more than 80% activity between 25 and 45°C (20 to 50°C for MetspHMFOHis).
With regard to pH, none of the enzymes were active below pH 5 (Fig. 4d). The optimum for MetspHMFO (both for the untagged and His-tagged enzyme) was at pH 7.0, with the enzyme retaining ∼70% of its activity at pH 6.5 to 8.0. Similarly, PsespHMFO showed its maximal activity at pH 7.5 to 8.0 and still showed 80% of its activity at pH 7 to 12. Although at basic pHs, the initial rates for MetspHMFO and PsespHMFO were high, the velocities decayed abruptly after a few seconds due to enzyme inactivation. However, PseniHMFOHis exhibited a quite different pH behavior, since it was more active at basic pH, with the maximal value at pH 10.0. Below pH 9.0 and above pH 11.0, its activity decreased to less than 80%.
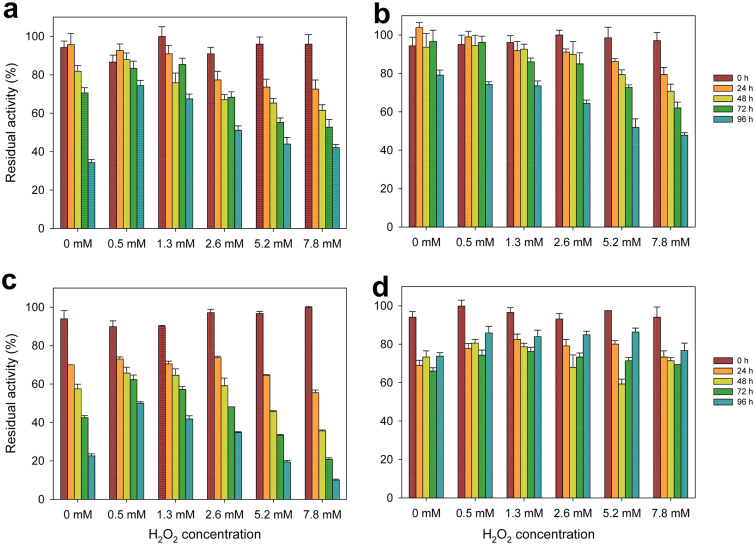
Effect of H2O2 on HMFOs’ stability.
Since during HMFO activity, hydrogen peroxide is produced from O2 reduction, we evaluated the effect that different amounts of H2O2 (0.5 to 7.8 mM) had on the enzymes’ stability. The results for MetspHMFO and MetspHMFOHis suggest that the His tag has a positive effect on enzyme stability, even in the absence of H2O2 (Fig. 5a and b). Also, PseniHMFOHis was more stable to peroxide exposure than MetspHMFO, keeping around 70% of its activity after 96 h in all the concentrations tested (Fig. 5d). In contrast, PsespHMFO was the least stable enzyme (Fig. 5c). In fact, low peroxide amounts (0.5 to 2.6 mM) had a slight stabilizing effect on PsespHMFO, while at the highest concentration, its activity decreased to only 10%.
FIG 5.
Effect of H2O2 on the stability of MetspHMFO (a), MetspHMFOHis (b), PsespHMFO (c), and PseniHMFOHis (d). Residual activities were measured with vanillyl alcohol after different incubation times with increasing amounts of H2O2 in 50 mM Tris-HCl, pH 7.0, at 25°C. Mean values and standard deviations are shown.
Kinetic parameters for oxidation of HMF and its DFF and FFCA derivatives.
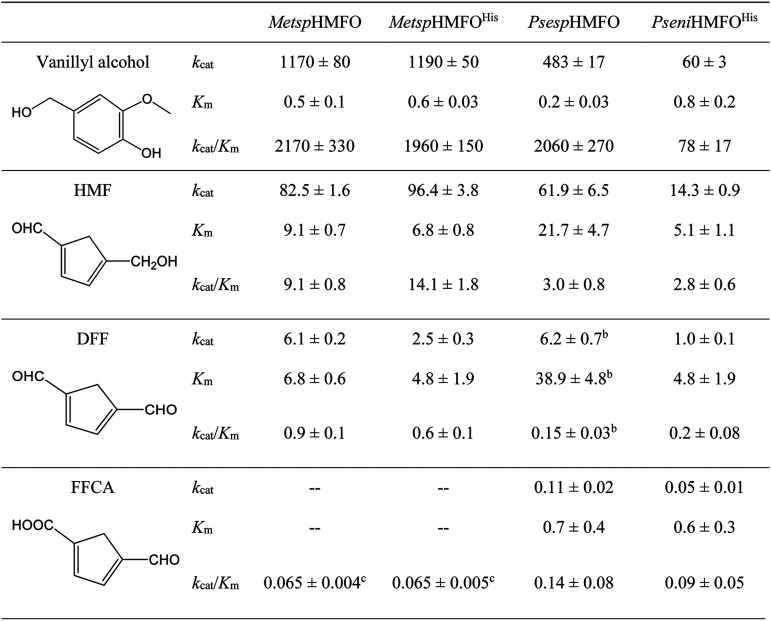
The kinetic parameters for oxidation of vanillyl alcohol and furfuryl substrates HMF, DFF, and FFCA were determined for the four enzyme preparations (Table 3). The untagged and His-tagged MetspHMFO showed similar kcat and Km values for vanillyl alcohol (measured by direct spectrophotometric estimation of the aldehyde product). PsespHMFO oxidized vanillyl alcohol with a catalytic efficiency (kcat/Km) similar to that of the enzyme from Methylovorus sp., while PseniHMFOHis was less efficient in oxidizing this substrate (with ∼28-fold-lower values than the other enzymes).
TABLE 3.
Steady-state kinetic constants for the oxidation of vanillyl alcohol, HMF, DFF, and FDCA by HMFOsa
Measured at 25°C in 50 mM Tris-HCl, pH 7.0, except for vanillyl alcohol oxidation by PsespHMFO and PseniHMFOHis, which were measured at optimal pHs of 8 and 10, respectively. Units of measure are as follows: kcat, min−1; Km, mM; kcat/Km, mM−1 min−1.
Data fitted to Hill equation (n = 2.4 ± 0.5).
Due to substrate inhibition, MetspHMFO and MetspHMFOHis were not saturated and only a kobs/[FFCA] value was obtained.
Oxidation of HMF and DFF was followed by measuring H2O2 release, using a coupled assay with horseradish peroxidase (HRP) at pH 7.0 (which is close to the HRP optimum). HMF was the best furfuryl substrate for all the enzymes. MetspHMFO and PsespHMFO showed similar kcat values, with a significant increase in the Km for PsespHMFO, while for PseniHMFOHis, the Km was lower. The efficiencies for HMF oxidation by PsespHMFO and PseniHMFO were ∼4-fold lower than for MetspHMFO. Concerning DFF oxidation, it was in all cases 10- to 14-fold slower than that of HMF, showing that the PsespHMFO enzyme had a positive cooperative effect (Hill coefficient, nH = 2.4) and, again, a Km value significantly higher than for the rest of the enzymes.
Due to the very slow FFCA oxidation, the kinetic parameters for this compound could not be calculated in a continuous assay. Therefore, the production of FDCA was followed by high-performance liquid chromatography (HPLC) during 48 h of incubation with increasing concentrations of FFCA at 25°C and pH 7.0. The enzyme from Methylovorus showed strong substrate inhibition at FFCA concentrations higher than 3 mM. This made it impossible to assess the kinetic parameters, as no saturation by FFCA was attained and only kobs (observed rate constant)/concentration values were estimated for comparison of efficiencies. The FFCA inhibitory effect was much milder for the enzymes from Pseudomonas, and saturation was attained due to the lower Km values. These results showed that the FFCA oxidation efficiencies of PseniHMFOHis and PsespHMFO are 1.4- and 2.2-fold higher, respectively, than those of MetspHMFO. However, the differences in catalytic efficiencies for FFCA oxidation with regard to those for HMF and DFF were significantly lower in PseniHMFOHis and PsespHMFO than in MetspHMFO. This suggests a more efficient overall oxidation of HMF to produce FDCA by the Pseudomonas enzymes.
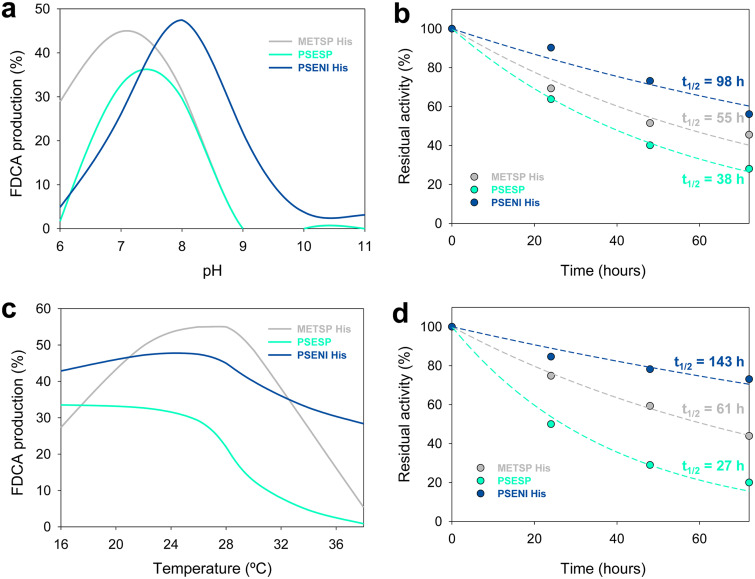
HMF conversion: effects of pH and temperature.
Taking into account the similar kinetic parameters of MetspHMFO and MetspHMFOHis and the higher stability under exposure to H2O2 of the latter form, the following experiments with this enzyme were performed with the His-tagged form. As for MetspHMFOHis, the enzymes from Pseudomonas were able to oxidize HMF to FDCA, catalyzing the three reaction steps. Taking into account their pH and temperature profiles (Fig. 4c and d), the conditions for HMF conversion by each enzyme were investigated in 72-h reactions (Fig. 6). Maximal FDCA production was achieved at pH 7.0, pH 7.5, and pH 8.0 for MetspHMFOHis, PsespHMFO, and PseniHMFOHis, respectively (Fig. 6a). Under optimal pH conditions, MetspHMFOHis and PsespHMFO showed half-lives (t1/2) of 55 and 38 h, respectively, while PseniHMFOHis was more stable, with a remarkable t1/2 of ∼100 h (Fig. 6b). Outside their optimal pH conditions, MetspHMFOHis and PsespHMFO had reduced half-lives, while PseniHMFOHis was quite stable in the range of pH 6 to 8 (Fig. S4, left).
FIG 6.
FDCA production after 72-h reaction of 1.5 mM HMF with 2.5 μM MetspHMFOHis (gray), PsespHMFO (cyan), and PseniHMFOHis (blue) under different pH (a) and temperature (c) conditions, and residual activities along the reactions at the optimal pH (b) and 28°C temperature (d).
Concerning temperature, PseniHMFOHis and PsespHMFO kept maximal FDCA production (around 45% and 30%, respectively) in the range of 16 to 28°C, while MetspHMFOHis showed maximal production in the 24 to 28°C range (Fig. 6c) (all reactions at optimal pH). At 28°C, the t1/2 values for the three HMFOs were 61, 27, and 143 h for MetspHMFOHis, PsespHMFO, and PseniHMFOHis, respectively (Fig. 6d). Above 28°C, MetspHMFOHis and PsespHMFO had decreased FDCA yields (Fig. 6c) due to enzyme inactivation, as shown by the t1/2 decreases (Fig. S4, right). PseniHMFOHis was the most thermostable enzyme for FDCA production (Fig. S4f), with t1/2 values of more than 100 h in the range of 16 to 28°C and around 90 h at 30 and 34°C. Even at 38°C, where the other two enzymes showed t1/2 values of ≤4 h, PseniHMFOHis had a t1/2 of 15 h.
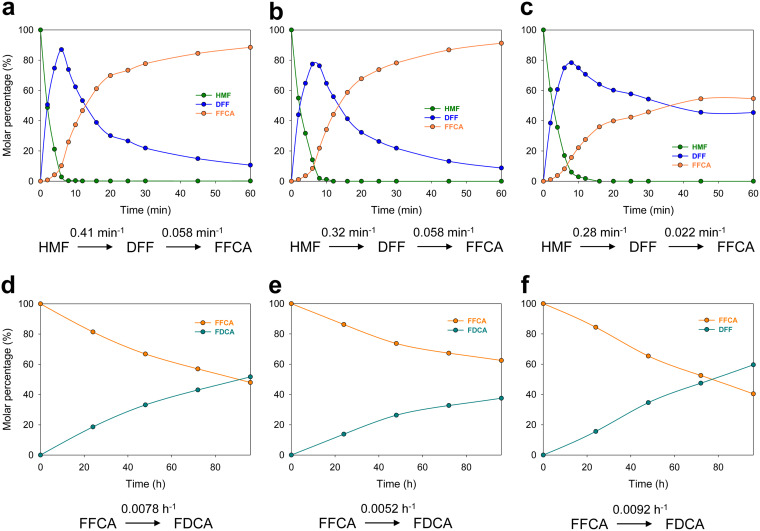
FDCA production: kinetics and long-term conversions.
Taking into account the results described above, oxidation of HMF by the different HMFOs was followed first during 1 h at 25°C and the optimal pH of each enzyme (Fig. 7). For all of them, the oxidation of HMF was faster than that of DFF [k(HMF→DFF)/k(DFF→FFCA) was ∼7-, 6-, and 13-fold faster for MetspHMFOHis, PsespHMFO, and PseniHMFOHis, respectively], with PseniHMFOHis being the least efficient in these conversions, as expected from the HMF and DFF steady-state parameters (Table 3). For FFCA oxidation, reactions were followed during 4 days under the same conditions. As expected, this reaction was seen to be the limiting step for FDCA production, with k(FFCA→FDCA) values of 0.0078, 0.0052, and 0.0092 h−1 for MetspHMFOHis, PsespHMFO, and PseniHMFOHis, respectively, indicating a more efficient FFCA oxidation by PseniHMFOHis, which would therefore be the best enzyme for application.
FIG 7.
Time course of reaction of 1.5 mM HMF (a to c) or FFCA (d to f) with 2.5 μM MetspHMFOHis (a, d), PsespHMFO (b, e), or PseniHMFOHis (c, f) in 50 mM Tris-HCl at the optimal pH for each enzyme and 25°C. Oxidation rates, estimated by fitting to equations , , and , are indicated.
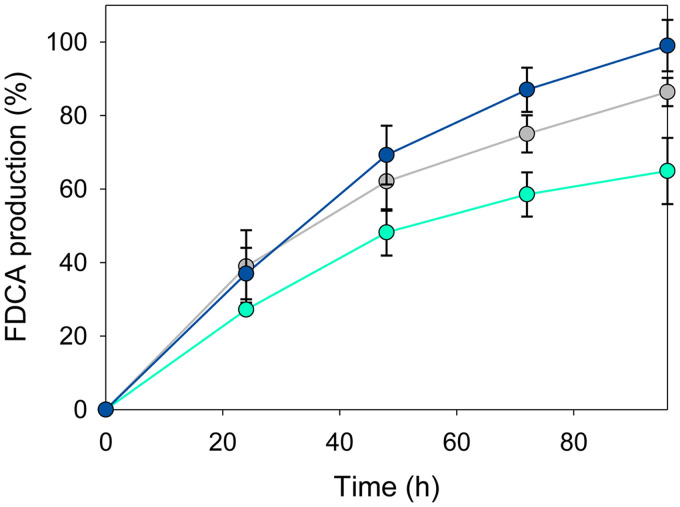
Finally, long-term conversion of HMF was followed during 4 days (Fig. 8) at 28°C and the optimal pH for each enzyme. With a substrate/enzyme molar ratio of 300, the highest FDCA production was achieved by PseniHMFOHis, with nearly complete conversion into FDCA after 96 h of reaction, while for MetspHMFOHis and PsespHMFO, the yields were 89% and 65%, respectively (Table 4). Interestingly, under these conditions, PseniHMFOHis showed a significantly longer half-life (150 h) than the other enzymes (∼40 to 60 h) and a higher total turnover number (TTN) for FDCA production.
FIG 8.
FDCA production from HMF (1.5 mM) by the MetspHMFO (gray), PsespHMFO (cyan), and PseniHMFO (blue) enzymes (5 μM) in 50 mM Tris-HCl at 28°C and the optimal pH for each of them. Mean values and standard deviations are shown.
TABLE 4.
Catalytic performance parameters for the production of FDCA from HMF by HMFOs in Tris-HCla
| HMFO | Optimal pH | Half-life (h) | TTNb | % FDCA production (mean ± SD) |
|---|---|---|---|---|
| MetspHMFOHis | 7.0 | 61 | 802 | 89 ± 4 |
| PsespHMFO | 7.5 | 40 | 584 | 65 ± 9 |
| PseniHMFOHis | 8.0 | 150 | 891 | 99 + 7 |
Production of FDCA from HMF at 1.5 mM by HMFOs at 5 μM in 50 mM Tris-HCl at 28°C after 96 h of reaction.
TTN, total turnover number.
DISCUSSION
HMFO screening, heterologous expression, and purification.
The HMFO from Methylovorus sp. (MetspHMFO) is the only enzyme described to date that is directly related to HMF oxidation. This connection was established due to its homology with the related HmfH protein that forms part of a gene cluster involved in HMF metabolism in C. basilensis (3, 6). In the present study, new HMFOs from P. nitroreducens and a Pseudomonas sp. strain were described, following their production as recombinant proteins. These two HMFOs, together with others from a Xanthobacter sp. strain and B. arachidis that could not be produced in E. coli, were selected from a total of 41 HMFO-like sequences analyzed after a database search for homology with MetspHMFO. Although an earlier attempt to express the gene responsible for HMF oxidation by C. basilensis and a homologous gene from Paraburkholderia phytofirmans was made, only MetspHMFO had been successfully produced as a functional protein (6) to date. Here, the two sequences from Pseudomonas strains, with 63 to 64% identity with the MetspHMFO sequence, were heterologously expressed in E. coli as active, soluble enzymes, together with MetspHMFO. The three recombinant HMFOs appeared as monomeric proteins, in agreement with a previous report on the latter enzyme (12), and were purified with good yields, increasing to three the number of members of the HMFO family currently available.
New information on MetspHMFO.
Comparison of the results obtained here for the Methylovorus enzyme purified without and with a His tag (MetspHMFO and MetspHMFOHis) indicated that the tag does not perturb the incorporation of the cofactor, as shown by the UV-visible spectra. Moreover, deconvolution of the CD spectra indicated an increase in the unordered structure, consistent with 90% of the His tag being disordered (27), and a slight decrease in the α-helix content. Neither activity nor the main physicochemical parameters were affected by the presence of the His tag, but a positive effect on stability under exposure to H2O2 was found for the tagged MetspHMFO enzyme. This would represent an advantage for enzymes generating H2O2, which, as reported for other oxidases of the same superfamily, can inhibit the enzyme reactions or the enzymes themselves (28, 29).
The kinetic parameters for vanillyl alcohol oxidation by MetspHMFO were similar to those previously reported for the same enzyme (14). However, the reported kcat values for HMF and DFF were significantly higher (594 min−1 and 96 min−1, respectively) than those obtained in the present study. Considering the steady-state kinetic parameters obtained here, the estimated times for HMF and DFF consumption would be 7 and 98 min, respectively, which is consistent with the results obtained in 1-h reactions (1.5 mM HMF and 2.5 μM HMFO). In contrast, Dijkman et al. (14) still detected DFF after 1 h of reaction between 2 mM HMF or DFF and 1 μM HMFO, while according to their kcat value, all DFF should be consumed in ∼20 min. In any case, the kcat and Km values for oxidation of these furfural derivatives by the MetspHMFO produced here are of the same order as those previously reported for other oxidases acting on these substrates, such as AAO (19) and GLOX (21). Regarding FFCA, the catalytic efficiency, estimated as kobs/[FFCA], was of the same order as that previously reported for MetspHMFO (12), although the strong inhibition by FFCA (above 3 mM) has not been described before.
Properties of two new HMFOs.
The new enzymes from Pseudomonas strains were able to directly produce FDCA from HMF, with slight differences in their optimal reaction conditions, such as the higher activity and stability of PseniHMFO under slightly alkaline conditions. Although the new HMFOs do not show more activity on HMF and DFF than MetspHMFO, it is important to mention that they show higher efficiencies for FFCA oxidation, the rate-limiting step in FDCA production by HMFO (14) and other oxidases (16).
Strikingly, in terms of efficiency, PsespHMFO showed values for FFCA oxidation similar to those observed for DFF oxidation. The cooperative effect for DFF oxidation by this enzyme can be attributed to slow conformational changes that accompany substrate binding or product release. Similar cooperative effects have been described for other monomeric enzymes with single ligand-binding sites (30, 31).
In the case of PseniHMFO, the efficiency for FFCA oxidation was only half of that of DFF oxidation. The results described above contrast with the much lower efficiency observed for MetspHMFO (FFCA oxidation 10-fold lower than DFF oxidation). The comparatively higher speed in rate-limiting FFCA oxidation by PseniHMFO results in higher final yields of FDCA in HMF bioconversions under optimized conditions. Moreover, its higher tolerance of the H2O2 produced during the three oxidation steps results in a higher enzymatic stability during turnover, increasing its half-life. This stability provides an important advantage, since oxidases are often inhibited by large amounts of H2O2. Thus, our data reveal PseniHMFO as a robust and suitable biocatalyst for prolonged incubations, due to its longer half-life in wider ranges of pHs, temperatures, and H2O2 concentrations.
Analysis of HMFO molecular models.
Molecular modeling of PsespHMFO and PseniHMFO was undertaken at the Swiss-Model server (32), using the MetspHMFO crystal structure (PDB code 4UDP) (12) as the template. The predicted secondary structures were similar, as expected from their far-UV CD spectra.
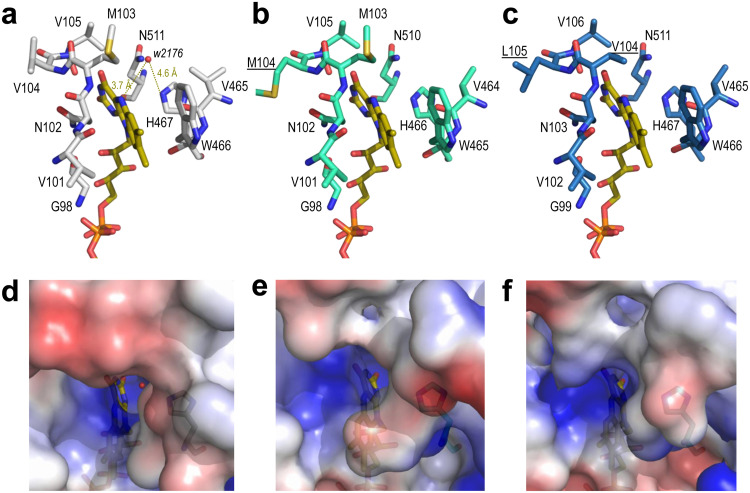
However, inspection of their active sites revealed subtle differences in the environment (less than 6 Å) of the reactive N5 of FAD. Interestingly, in the MetspHMFO crystal structure, a water molecule is located at 3.7 Å from the above-named N atom and 4.6 Å from the catalytic His467 (Fig. 9a), at a position that is most probably occupied by the substrates during catalysis. Concerning neighbor residues, the pair Met103-Val104 in MetspHMFO (Fig. 9a) is replaced by Met103-Met104 in PsespHMFO (Fig. 9b) and by Val104-Leu105 in PseniHMFO (Fig. 9c). These two positions follow an active-site asparagine that is totally conserved in the GMC superfamily (1) as part of the conserved PS00623 sequence. Directed mutagenesis of MetspHMFO revealed how an M103A substitution (a changed of Met to Ala at position 103) significantly modified the enzyme turnover (for vanillyl alcohol) due to substrate pocket modification (12). A free space, potentially affecting the position of HMF and its partially oxidized derivatives (DFF and FFCA) with respect to the flavin cofactor, could also exist in PseniHMFO with a valine at this position.
FIG 9.
Details of the active sites of MetspHMFO (a, d), PsespHMFO (b, e), and PseniHMFO (c, f). (a to c) Residues at less than 6 Å from the reactive N5 atom of the FAD cofactor (CPK-colored sticks) are indicated, with those differing from MetspHMFO underlined. Distances from His467 and flavin ring to water2176 (red sphere) are shown in panel a. (d to f) Semitransparent solvent-accessible surfaces colored by electrostatic potential (red, negative; blue, positive) around the flavin access channel. Water2176 inside the channel is shown in panel d. From the crystal structure with PDB code 4UDP (12) (a and d) and molecular homology models (32) (b, c, e, and f).
The above-described differences do not include the presence of residues that could promote carbonyl activation in FFCA (33), as found in classical aldehyde-oxidizing oxidoreductases. However, changes in the flavin environment of PsespHMFO and PseniHMFO could modify the interaction of the flavin with the protein. Such interactions can modulate the redox potential of the oxidized isoalloxazine ring, promoting the enzyme’s activity on less reactive substrates (34).
Pointing to the active-site cavity, Val367 is found in MetspHMFO. This residue is conserved in PsespHMFO, while Leu367 replaces the valine in PseniHMFO (Fig. 3). Interestingly, it has been reported that mutating this valine to arginine in MetspHMFO results in higher activity on FFCA (12), as found for PseniHMFO. However, the effect in the MetspHMFO variant is due to the introduction of a basic (arginine) residue, contributing to adequate positioning of the above-mentioned acidic substrate, while the natural change in PseniHMFO would only slightly reduce the pocket size.
On the other hand, the electrostatic surface potentials of the three HMFOs showed differences in charge distribution around the entrance of the flavin-access channel (Fig. 9d to f), which could also modulate the access of substrates. Namely, the electronegative region partially covering the channel entrance in MetspHMFO could impede the access of acidic FFCA to the active site. However, the less electronegative entrances in PsespHMFO and especially in PseniHMFO would facilitate the entrance of this substrate, resulting in the observed higher activity of the new enzymes in catalyzing the rate-limiting step in HMF conversion to FDCA. The solvent access surfaces also showed that, although the O2 substrate would more easily diffuse to attain the flavin ring of the buried cofactor in HMFOs, the entrance of the furfuryl-reducing substrates through the narrow access channel would most probably involve some side chain rearrangement at the residues forming the channel, as described in a related AAO (35, 36).
Concluding remarks.
Two enzymes from two Pseudomonas species have been added to the repertoire of HMFOs available, where only one member was purified and characterized to date. The enzyme from P. nitroreducens is a promising candidate for HMF oxidation due to its higher catalytic efficiency for FFCA oxidation, the bottleneck for FDCA production, compared to that of the previously described MetspHMFO. More importantly, in the context of industrial production of FDCA, additional advantages of the P. nitroreducens enzyme are its stability under exposure to H2O2 and its robustness for long-term incubations, as shown by its high t1/2 values under wider ranges of pH and temperature conditions. This feature would facilitate the enzyme’s reuse and applicability in continuous operation for the production of FDCA as a building block for the production of bioplastics.
MATERIALS AND METHODS
Chemicals.
HMF was kindly provided by AVA Biochem. DFF, FDCA, vanillyl alcohol, and vanillin were purchased from Sigma-Aldrich (St. Louis, MO, USA). FFCA was purchased from TCI America (Portland, OR, USA). AmplexRed and horseradish peroxidase were obtained from Invitrogen (Waltham, MA, USA).
Sequence search, phylogenetic analysis, and structure modeling.
The screening of HMFO-like sequences was performed by BLAST of the amino acid sequence of MetspHMFO against the nonredundant protein sequences in GenBank. Multiple alignments of the enzymes found with Clustal Omega (37) were used to identify the conserved motifs (ADP binding domain and Prosite PS00623 and PS00624 sequences) in GMC proteins. A maximum-likelihood phylogenetic tree was constructed by MEGA X with 100-iteration bootstrapping (38) using the Whelan and Goldman (39) model of evolution using gamma-distributed rate variation with empirical amino acid frequencies and invariant sites (WAG+F+I+G). Finally, structural homology models of the protein sequences were generated using the Swiss-Model server (https://swissmodel.expasy.org) (32), and PyMOL version 2.033 (40) was used to examine the molecular structures obtained and generate solvent-accessible electrostatic surfaces.
Heterologous expression of HMFOs in E. coli.
The codifying DNA sequences of the HMFOs from Methylovorus sp. M668, P. nitroreducens, Pseudomonas sp. 11/12A, Xanthobacter sp. 126, and Bradyrhizobium arachidis (NCBI accession numbers WP_013440946, WP_024766380, WP_047529632, WP_024277017, and WP_092217059, respectively) were manually optimized for E. coli expression and synthesized by ATG:biosynthetics. The sequences were subcloned from the pGH vector into the pET23b(+) or pET28a(+) expression vector. C41(DE3) E. coli cells were used for expression. E. coli cells containing the expression vectors were grown overnight in LB broth supplemented with the corresponding antibiotic at 37°C under continuous shaking. These precultures were used to inoculate 1-liter amounts of LB (supplemented with the antibiotic) and the cultures grown for 3 to 4 h at 37°C under continuous shaking until reaching an optical density at 500 nm of 0.9. Cultures were induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside, grown for 72 to 96 h at 16°C, and then harvested by centrifugation. Protein expression was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Purification of recombinant HMFOs.
The bacterial pellets were resuspended in the corresponding lysis buffer (Table S3 in the supplemental material), and the mixtures were incubated with 0.1 mg/ml of DNase I and 2 mg/ml lysozyme for 45 min. The solutions were sonicated and centrifuged to eliminate insoluble debris.
MetspHMFO and PsespHMFO (untagged proteins) were purified in a ResourceQ 6-ml column (GE Healthcare) with a linear NaCl gradient (60 to 160 mM in 4 column volumes [CV]) in 50 mM Tris-HCl, pH 8.0, followed by a MonoQ 5/50 GL column in which the protein fractions were eluted with a NaCl linear gradient (0 to 70 mM in 10 CV) in 50 mM Tris-HCl, pH 8.0 (Table S3).
His-tagged fusion proteins (MetspHMFOHis and PseniHMFOHis) were purified by affinity chromatography in a HiTrap IMAC FF column (GE Healthcare) with a linear imidazole gradient (20 m to 300 mM in 6 CV) in 500 mM NaCl, 20 mM Tris-HCl, pH 7.5, followed by anionic exchange chromatography with a ResourceQ column (GE Healthcare) with an NaCl linear gradient (0 to 500 mM in 5 CV) in 50 mM Tris-HCl, pH 7.5 (Table S3).
The purified HMFOs were dialyzed against 50 mM Tris-HCl, pH 7.0, and stored at −80°C, being stable for several months.
Spectroscopic measurements.
The UV-visible spectra of purified HMFOs were recorded between 250 and 700 nm in a Cary4000 spectrophotometer. Their extinction coefficients were calculated by heat denaturation (41) and estimation of the free FAD released (εband I = 11,300 mM−1 cm−1) (42).
Far-UV CD spectra of HMFOs (5 μM) were recorded in a Jasco J-815 spectropolarimeter at 25°C in 5 mM Tris-HCl, pH 7.0, with a 0.1-cm path length. The spectra were analyzed with the CDPro programs SELCON3 (43), CDSSTR (44), and CONTINLL (45) to determine the secondary structure.
MW and pI determination.
MW was analyzed by MALDI-TOF mass spectrometry at the Proteomics and Genomics Facility of CIB (CSIC), a member of the ProteoRed-ISCIII network. The experiments were performed in an Autoflex III MALDI–tandem-TOF instrument (Bruker Daltonics, Bremen, Germany) with a smart beam laser. Samples dissolved in 50 mM Tris-HCl, pH 7.0, were mixed with the matrix solution (3.5-dimethoxy-4-hydroxycinnamic acid dissolved in a mixture of acetonitrile–01% trifluoroacetic acid [1:2, vol/vol]). External calibration was performed using bovine albumin (Sigma), covering the range from 20,000 to 70,000 Da.
The pI was determined by 2-dimensional (2-D) electrophoresis. The first dimension was run on immobilized pH gradient strips (pH 3 to 10, linear, 7 cm) (Bio-Rad), and the second dimension was run on 12% SDS–PAGE gels. Protein bands were stained with a colloidal blue staining kit (Invitrogen).
pH and H2O2 stability.
The pH and H2O2 stabilities were estimated by incubating the enzymes (5 to 40 μM) in 100 mM Britton and Robinson (B&R) buffer at different pHs (pH 4.0 to 9.0) or in the presence of different H2O2 concentrations (0.5 to 7.8 mM) in 50 mM Tris-HCl, pH 7.0. Residual activities were estimated by following the oxidation of saturating concentrations of vanillyl alcohol (3 mM) in 50 mM Tris-HCl, pH 7.0, at 25°C, immediately after mixing and after 24 h, 48 h, and 72 h of incubation at 25°C. For each enzyme, the highest activity after mixing was taken as 100% activity, and the percentages of residual activity at the different times and conditions were calculated according to this maximal value.
Thermal stability.
The melting temperatures of the enzymes were analyzed by the ThermoFAD method (26). The increase in fluorescence due to FAD released as a consequence of enzyme unfolding was monitored with a real-time PCR thermocycler iQ5 (Bio-Rad). Denaturation curves were fitted to equation 1, describing a two-step thermal denaturation equilibrium:
| (1) |
where , FN and FD are the fluorescence signals of the native and denatured states, mN and mD are the slopes that describe their dependences with temperature, T and Tm are the temperature and the melting temperature, respectively, R is the universal gas constant, and ΔG, ΔH, and ΔCp are the free energy, enthalpy, and specific heat of denaturation, respectively.
Optimal pH and temperature.
The optimal temperatures and pHs for vanillyl alcohol oxidation by HMFOs were determined by measuring the oxidation of saturating concentrations of the substrate (3 mM) in 50 mM Tris-HCl buffer, pH 7.0, at different temperatures between 4°C and 70°C or in 100 mM B&R buffer in the range of pHs from 2.0 to 12.0. It was observed that the UV-visible spectra of vanillyl alcohol and vanillin changed with pH (Fig. S5a and b) (46). Thus, in the case of the optimal pH, the activity was measured using the extinction coefficient for vanillyl alcohol oxidation to vanillin calculated for each pH (Fig. S5c).
Steady-state kinetics.
Enzymatic activity was estimated by following the production of vanillin upon oxidation of vanillyl alcohol at 25°C in 50 mM Tris-HCl at the indicated pH in each case, using the corresponding extinction coefficient for vanillyl alcohol oxidation to vanillin (Fig. S5c).
Steady-state kinetic parameters for HMF and DFF oxidation by HMFOs were calculated by monitoring the production of H2O2 during oxidation of the different substrates, using an HRP-coupled assay with AmplexRed as the final substrate in 50 mM Tris-HCl, pH 7.0, as previously described (16).
The oxidation of FFCA was measured in end-time mode by incubating different concentrations of FFCA (0.4 to 6 mM) with the enzymes (1 μM) at 25°C in 50 mM Tris-HCl buffer, pH 7.0. After 48 h, reactions were stopped by the addition of 1 M HCl up to pH 2 to 3, and the products were quantified by HPLC as described below.
In all cases, kinetic parameters were determined by fitting the initial reaction rates at different alcohol or aldehyde concentrations to the Michaelis-Menten equation (equation 2) or to the Hill equation (equation 3) using SigmaPlot software.
| (2) |
| (3) |
where E and S are the concentrations of enzyme and substrate, respectively, kcat is the catalytic constant, Km is the Michaelis-Menten constant, and n is the Hill coefficient.
HMF transformations.
Long-term HMF oxidation by HMFOs was assayed using 2.5 to 5 μM enzyme and 1.5 mM HMF in 50 mM Tris-HCl, at the pHs and temperatures indicated in Results for each experiment, under continuous shaking (300 rpm). Samples were taken at different times, and reactions were stopped by HCl addition. The products of the reactions were analyzed by HPLC, using an ion exchange Supelcogel C-610H column (300 by 7.8 mm, 9-μm particle size; Sigma). Compounds were eluted with 5 mM H2SO4 at a flow rate of 0.75 ml/min and 30°C. Under these conditions, the retention times of FDCA, FFCA, HMF, and DFF were 15.5, 21.3, 30.2, and 37.3 min, respectively. Detection of the different compounds was done at 264 nm, and their quantification was performed using the corresponding calibration curves (16). Controls consisting of the substrates in the absence of enzyme were treated in the same way to monitor eventual spontaneous oxidation.
Residual activity of HMFOs along the reactions was measured just after taking aliquots, before the addition of HCl, by monitoring their activities against vanillyl alcohol. The activity decay as a function of time was calculated from equation 4, allowing the estimation of the half-life (equation 5):
| (4) |
| (5) |
where λ is the activity decay constant and t1/2 is the half-life.
Data availability.
All data are included here and in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
This work has been funded by the H2020 BBI-JU (http://www.bbi-europe.eu) project EnzOx2 (H2020-BBI-PPP-2015-2-720297). M.E. acknowledges grant number PEJD-2016/BIO-2329 from the Programa de Garantía Juvenil (Consejería de Educación, Juventud y Deporte, Comunidad de Madrid).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Cavener DR. 1992. GMC oxidoreductases. A newly defined family of homologous proteins with diverse catalytic activities. J Mol Biol 223:811–814. doi: 10.1016/0022-2836(92)90992-s. [DOI] [PubMed] [Google Scholar]
- 2.Papageorgiou GZ, Tsanaktsis V, Bikiaris DN. 2014. Synthesis of poly(ethylene furandicarboxylate) polyester using monomers derived from renewable resources: thermal behavior comparison with PET and PEN. Phys Chem Chem Phys 16:7946–7958. doi: 10.1039/c4cp00518j. [DOI] [PubMed] [Google Scholar]
- 3.Koopman F, Wierckx N, de Winde JH, Ruijssenaars HJ. 2010. Identification and characterization of the furfural and 5-(hydroxymethyl)furfural degradation pathways of Cupriavidus basilensis HMF14. Proc Natl Acad Sci U S A 107:4919–4924. doi: 10.1073/pnas.0913039107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wierckx N, Koopman F, Bandounas L, de Winde JH, Ruijssenaars HJ. 2010. Isolation and characterization of Cupriavidus basilensis HMF14 for biological removal of inhibitors from lignocellulosic hydrolysate. Microb Biotechnol 3:336–343. doi: 10.1111/j.1751-7915.2009.00158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crigler J, Eiteman MA, Altman E. 2020. Characterization of the furfural and 5-hydroxymethylfurfural (HMF) metabolic pathway in the novel isolate Pseudomonas putida ALS1267. Appl Biochem Biotechnol 190:918–930. doi: 10.1007/s12010-019-03130-x. [DOI] [PubMed] [Google Scholar]
- 6.Dijkman WP, Fraaije MW. 2014. Discovery and characterization of a 5-hydroxymethylfurfural oxidase from Methylovorus sp. strain MP688. Appl Environ Microbiol 80:1082–1090. doi: 10.1128/AEM.03740-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ewing TA, Dijkman WP, Vervoort JM, Fraaije MW, van Berkel W. 2014. The oxidation of thiols by flavoprotein oxidases: a biocatalytic route to reactive thiocarbonyls. Angew Chem Int Ed Engl 53:13206–13209. doi: 10.1002/anie.201407520. [DOI] [PubMed] [Google Scholar]
- 8.Fan F, Gadda G. 2005. On the catalytic mechanism of choline oxidase. J Am Chem Soc 127:2067–2074. doi: 10.1021/ja044541q. [DOI] [PubMed] [Google Scholar]
- 9.Hernández-Ortega A, Lucas F, Ferreira P, Medina M, Guallar V, Martínez AT. 2012. Role of active site histidines in the two half-reactions of the aryl-alcohol oxidase catalytic cycle. Biochemistry 51:6595–6608. doi: 10.1021/bi300505z. [DOI] [PubMed] [Google Scholar]
- 10.Wongnate T, Chaiyen P. 2013. The substrate oxidation mechanism of pyranose 2-oxidase and other related enzymes in the glucose-methanol-choline superfamily. FEBS J 280:3009–3027. doi: 10.1111/febs.12280. [DOI] [PubMed] [Google Scholar]
- 11.Wongnate T, Surawatanawong P, Chuaboon L, Lawan N, Chaiyen P. 2019. The mechanism of sugar C-H bond oxidation by a flavoprotein oxidase occurs by a hydride transfer before proton abstraction. Chemistry 25:4460–4471. doi: 10.1002/chem.201806078. [DOI] [PubMed] [Google Scholar]
- 12.Dijkman WP, Binda C, Fraaije MW, Mattevi A. 2015. Structure-based enzyme tailoring of 5-hydroxymethylfurfural oxidase. ACS Catal 5:1833–1839. doi: 10.1021/acscatal.5b00031. [DOI] [Google Scholar]
- 13.Dijkman WP. 2015. HMF oxidase. Characterization, application and engineering of 5-(hydroxymethyl)furfural oxidase. PhD thesis Rijksuniversiteit, Groningen, the Netherlands. [Google Scholar]
- 14.Dijkman WP, Groothuis DE, Fraaije MW. 2014. Enzyme-catalyzed oxidation of 5-hydroxymethylfurfural to furan-2,5-dicarboxylic acid. Angew Chem Int Ed Engl 53:6515–6518. doi: 10.1002/anie.201402904. [DOI] [PubMed] [Google Scholar]
- 15.Karich A, Kleeberg BS, Ullrich R, Hofrichter M. 2018. Enzymatic preparation of 2,5-furandicarboxylic acid (FDCA)—a substitute of terephthalic acid—by the joined action of three fungal enzymes. Microorganisms 6:5. doi: 10.3390/microorganisms6010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serrano A, Calviño E, Carro J, Sánchez-Ruiz MI, Cañada FJ, Martínez AT. 2019. Complete oxidation of hydroxymethylfurfural to furandicarboxylic acid by aryl-alcohol oxidase. Biotechnol Biofuels 12:217. doi: 10.1186/s13068-019-1555-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathieu Y, Offen WA, Forget SM, Ciano L, Viborg AH, Blagova E, Henrissat B, Walton PH, Davies GJ, Brumer H. 2020. Discovery of a fungal copper-radical oxidase with high catalytic efficiency towards 5-hydroxymethylfurfural and benzyl alcohols for green bioprocessing. ACS Catal 10:3042–3058. doi: 10.1021/acscatal.9b04727. [DOI] [Google Scholar]
- 18.Hanke PD. May 2012. Enzymatic oxidation of hydroxymethylfurfural. US patent 8,183,020 B2.
- 19.Carro J, Ferreira P, Rodríguez L, Prieto A, Serrano A, Balcells B, Ardá A, Jiménez-Barbero J, Gutiérrez A, Ullrich R, Hofrichter M, Martínez AT. 2015. 5-Hydroxymethylfurfural conversion by fungal aryl-alcohol oxidase and unspecific peroxygenase. FEBS J 282:3218–3229. doi: 10.1111/febs.13177. [DOI] [PubMed] [Google Scholar]
- 20.Kalum L, Morant MD, Lund H, Jensen J, Lapainaite I, Soerensen NH, Pedersen S, Østergaard L, Xu F. January 2014. Enzymatic oxidation of 5-hydroxymethylfurfural and derivatives thereof. International patent WO2014-015256A2.
- 21.Daou M, Yassine B, Wikee S, Record E, Duprat F, Bertrand E, Faulds CB. 2019. Pycnoporus cinnabarinus glyoxal oxidases display differential catalytic efficiencies on 5-hydroxymethylfurfural and its oxidized derivatives. Fungal Biol Biotechnol 6:4. doi: 10.1186/s40694-019-0067-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan H, Li J, Shin H-D, Du G, Chen J, Shi Z, Liu L. 2018. Improved production of 2,5-furandicarboxylic acid by overexpression of 5-hydroxymethylfurfural oxidase and 5-hydroxymethylfurfural/furfural oxidoreductase in Raoultella ornithinolytica BF60. Bioresour Technol 247:1184–1188. doi: 10.1016/j.biortech.2017.08.166. [DOI] [PubMed] [Google Scholar]
- 23.Martin C, Ovalle Maqueo A, Wijma HJ, Fraaije MW. 2018. Creating a more robust HMF oxidase by combining computational predictions with a novel effective library design. Biotechnol Biofuels 11:56. doi: 10.1186/s13068-018-1051-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu S, Liu Q, Tan H, Zhang F, Yin H. 3 March 2020. A novel 2,5-furandicarboxylic acid biosynthesis route from biomass-derived 5-hydroxymethylfurfural based on the consecutive enzyme reactions. Appl Biochem Biotechnol doi: 10.1007/s12010-020-03290-1. [DOI] [PubMed] [Google Scholar]
- 25.McTaggart TL, Shapiro N, Woyke T, Chistoserdova L. 2015. Draft genome of Pseudomonas sp. strain 11/12A, isolated from Lake Washington sediment. Genome Announc 3:e01587-14. doi: 10.1128/genomeA.01587-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forneris F, Orru R, Bonivento D, Chiarelli LR, Mattevi A. 2009. ThermoFAD, a thermofluor-adapted flavin ad hoc detection system for protein folding and ligand binding. FEBS J 276:2833–2840. doi: 10.1111/j.1742-4658.2009.07006.x. [DOI] [PubMed] [Google Scholar]
- 27.Carson M, Johnson DH, McDonald H, Brouillette C, Delucas LJ. 2007. His-tag impact on structure. Acta Crystallogr D Biol Crystallogr 63:295–301. doi: 10.1107/S0907444906052024. [DOI] [PubMed] [Google Scholar]
- 28.Bao J, Furumoto K, Yoshimoto M, Fukunaga K, Nakao K. 2003. Competitive inhibition by hydrogen peroxide produced in glucose oxidation catalyzed by glucose oxidase. Biochem Eng J 13:69–72. doi: 10.1016/S1369-703X(02)00120-1. [DOI] [Google Scholar]
- 29.Halada P, Brugger D, Volc J, Peterbauer CK, Leitner C, Haltrich D. 2016. Oxidation of Phe454 in the gating segment inactivates Trametes multicolor pyranose oxidase during substrate turnover. PLoS One 11:e0148108. doi: 10.1371/journal.pone.0148108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porter CM, Miller BG. 2012. Cooperativity in monomeric enzymes with single ligand-binding sites. Bioorg Chem 43:44–50. doi: 10.1016/j.bioorg.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Acosta H, Cáceres A, González-Marcano E, Quiñones W, Avilán L, Dubourdieu M, Concepción JL. 2014. Hysteresis and positive cooperativity as possible regulatory mechanisms of Trypanosoma cruzi hexokinase activity. Mol Biochem Parasitol 198:82–91. doi: 10.1016/j.molbiopara.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 32.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T. 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teng H, Segura E, Grubmeyer C. 1993. Conserved cysteine residues of histidinol dehydrogenase are not involved in catalysis—novel chemistry required for enzymatic aldehyde oxidation. J Biol Chem 268:14182–14188. [PubMed] [Google Scholar]
- 34.Mayhew SG. 1999. Potentiometric measurement of oxidation-reduction potentials, p 49–59. In Chapman SK, Reid GA (ed), Flavoprotein protocols. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 35.Hernández-Ortega A, Lucas F, Ferreira P, Medina M, Guallar V, Martínez AT. 2011. Modulating O2 reactivity in a fungal flavoenzyme: involvement of aryl-alcohol oxidase Phe-501 contiguous to catalytic histidine. J Biol Chem 286:41105–41114. doi: 10.1074/jbc.M111.282467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hernández-Ortega A, Borrelli K, Ferreira P, Medina M, Martínez AT, Guallar V. 2011. Substrate diffusion and oxidation in GMC oxidoreductases: an experimental and computational study on fungal aryl-alcohol oxidase. Biochem J 436:341–350. doi: 10.1042/BJ20102090. [DOI] [PubMed] [Google Scholar]
- 37.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whelan S, Goldman N. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 40.Schrödinger. 2017. The PyMOL molecular graphics system, version 2.0. Schrödinger, LLC, New York, NY. [Google Scholar]
- 41.Aliverti A, Curti B, Vanoni MA. 1999. Identifying and quantitating FAD and FMN in simple and in iron-sulfur-containing flavoproteins. Methods Mol Biol 131:9–23. doi: 10.1385/1-59259-266-X:9. [DOI] [PubMed] [Google Scholar]
- 42.Dawson RMC, Elliot DC, Elliot WH, Jones KM. 1986. Data for biochemical research. Oxford Science Publications, Oxford, UK. [Google Scholar]
- 43.Sreerama N, Woody RW. 1993. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal Biochem 209:32–44. doi: 10.1006/abio.1993.1079. [DOI] [PubMed] [Google Scholar]
- 44.Johnson WC. 1999. Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins 35:307–312. doi:. [DOI] [PubMed] [Google Scholar]
- 45.van Stokkum IH, Spoelder HJ, Bloemendal M, van Grondelle R, Groen FC. 1990. Estimation of protein secondary structure and error analysis from circular dichroism spectra. Anal Biochem 191:110–118. doi: 10.1016/0003-2697(90)90396-q. [DOI] [PubMed] [Google Scholar]
- 46.Englis DT, Wollermann LA. 1957. Significance of pH in determination of vanillin by ultraviolet absorption. Anal Chem 29:1151–1153. doi: 10.1021/ac60128a010. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included here and in the supplemental material.