The efficiently disseminating conjugative or mobilizable BHR plasmids play key roles in the horizontal spread of genetic information between closely related and phylogenetically distant species, which can be harmful from the medical, veterinary, or industrial point of view. Understanding the mechanisms determining the plasmid’s ability to function in diverse hosts is essential to help limit the spread of undesirable plasmid-encoded traits, e.g., antibiotic resistance. The range of a plasmid’s promiscuity depends on the adaptations of its transfer, replication, and stability functions to the various hosts. IncU plasmids, with the archetype plasmid RA3, are considered to constitute a reservoir of antibiotic resistance genes in aquatic environments; however, the molecular mechanisms determining their adaptability to a broad range of hosts are rather poorly characterized. Here, we present the transcriptional organization of the stability module and show that the gene transcript dosage effect is an important determinant of the stable maintenance of RA3 in different hosts.
KEYWORDS: RNAP read-through, broad-host-range plasmid, gene expression, stability functions, transcript dosage
ABSTRACT
The broad-host-range (BHR) conjugative plasmids have developed diverse adaptive mechanisms defining the range of their promiscuity. The BHR conjugative RA3 plasmid, the archetype of the IncU group, can transfer between, replicate in, and be maintained in representatives of Alpha-, Beta-, and Gammaproteobacteria. Its stability module encompasses ten open reading frames (ORFs) apparently organized into five operons, all transcribed in the same direction from several strong promoters that are tightly regulated either by autorepressors or by global plasmid-encoded regulators. In this paper, we demonstrate that owing to an efficient RNA polymerase (RNAP) read-through, the transcription from the first promoter, orf02p, may continue through the whole module. Moreover, an analysis of mRNA produced from the wild-type (WT) stability module and its deletion variants deprived of particular internal transcription initiation sites reveals that in fact each operon may be transcribed from any upstream promoter, giving rise to multicistronic transcripts of variable length and creating an additional level of gene expression control by transcript dosage adjustment. The gene expression patterns differ among various hosts, indicating that promoter recognition, regulation, and the RNAP read-through mechanisms are modulated in a species-specific manner.
IMPORTANCE The efficiently disseminating conjugative or mobilizable BHR plasmids play key roles in the horizontal spread of genetic information between closely related and phylogenetically distant species, which can be harmful from the medical, veterinary, or industrial point of view. Understanding the mechanisms determining the plasmid’s ability to function in diverse hosts is essential to help limit the spread of undesirable plasmid-encoded traits, e.g., antibiotic resistance. The range of a plasmid’s promiscuity depends on the adaptations of its transfer, replication, and stability functions to the various hosts. IncU plasmids, with the archetype plasmid RA3, are considered to constitute a reservoir of antibiotic resistance genes in aquatic environments; however, the molecular mechanisms determining their adaptability to a broad range of hosts are rather poorly characterized. Here, we present the transcriptional organization of the stability module and show that the gene transcript dosage effect is an important determinant of the stable maintenance of RA3 in different hosts.
INTRODUCTION
Horizontal gene transfer (HGT) is considered the most critical factor in bacterial adaptation and evolution (1–3). Conjugative plasmids play key roles as vehicles in HGT between both closely related and phylogenetically distant species. The broad-host-range (BHR) conjugative plasmids have developed numerous adaptive mechanisms allowing them not only to spread but also to replicate and be stably maintained in a diverse set of hosts (4). The three functional categories of the adaptive mechanisms are responsible, respectively, for conjugation host range, replication host range, and long-term host range, often quite variable in their scope (5). Thus, the conjugation host range refers to the ability to form mating pairs and to disseminate the plasmid genome among various species and is usually the widest host range (6). The replication host range specifies the hosts in which the plasmid can successfully replicate and function as an independent replicon. The long-term host range is generally the narrowest and defines the species in which the plasmid molecules are stably maintained extrachromosomally even in the absence of selective pressure (7).
Studies on the host adaptation mechanisms of BHR plasmids have been focused mainly on their replication systems, the structure of oriVs, Rep proteins, and their accessory proteins as well as the interactions between the plasmid and host replication machinery (8–17). Some research has also been done on the factors responsible for the promiscuous conjugative transfer, the role of the ubiquitous type IV secretion systems (T4SS), and the specificity of coupling proteins and primases (18–21). In contrast, relatively little attention has been paid to the adaptation of the stability mechanisms to various hosts in the long-term plasmid persistence (7, 22, 23).
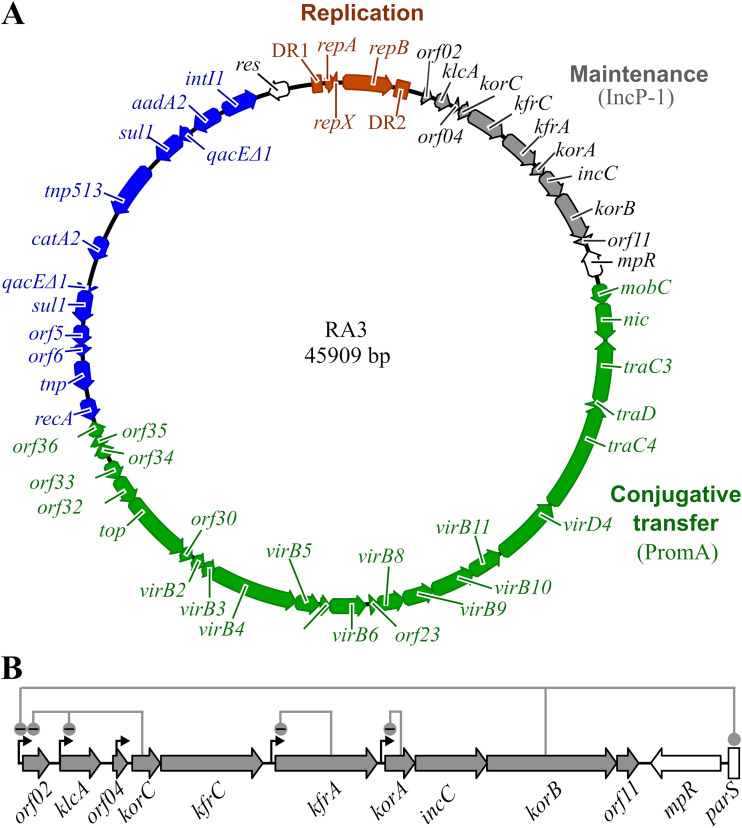
Our research model is the BHR conjugative plasmid RA3, the archetype of the IncU group, which is capable of replication and stable maintenance in, and transfer between, various representatives of Alpha-, Beta-, and Gammaproteobacteria (24). The RA3 genome, of 49.1 kb (GenBank accession no. DQ401103), has a mosaic modular structure where genes encoding proteins involved in the same process cluster into functional modules, and these modules demonstrate similarity to blocks of genes from various other BHR plasmids (Fig. 1A). The RA3 conjugative module resembles the one from PromA plasmids (25), whereas its stability module encodes proteins homologous to those of IncP-1 plasmids (26, 27).
FIG 1.
Genomic map of plasmid RA3. (A) Blocks of genes forming distinct functional modules are colored differently. The replication module (brown) is comprised of the repA-repB operon surrounded by long direct repeats DR1 and DR2. Genes of the stability module (gray) show homology to IncP-1 plasmids. The conjugative transfer module (green) resembles one found in PromA plasmids. Most accessory genes (blue), so called plasmid “genetic” load, belong to the class I integron (24). Genes res (invertase/recombinase) and mpR (putative zinc metallopeptidase) of unassigned plasmid functions are labeled in white (24). Arrows indicate the direction of transcription. (B) Close-up of the stability module. Thin black arrows indicate previously identified promoters (24, 28, 29). Regulatory circuits are shown as lines connecting the regulatory genes with their target sequences. KorB binds also to the parS centromere-like sequence, a cis-acting site of the partition complex.
The stability module of RA3 encompasses the region from nucleotide (nt) 2070 to nt 9830 (Fig. 1B), with 10 open reading frames (ORFs) transcribed in the same direction, i.e., orf02-klcA-orf04-korC-kfrC-kfrA-korA-incC-korB-orf11, followed by the centromere-like sequence parS. In silico DNA inspection suggested the presence of five transcription initiation signals in this module (24). Cloning of the predicted promoters in the promoter-probe vector and their analysis conducted in Escherichia coli confirmed the functionality of five promoter sequences located upstream of the orf02, klcA, korC, kfrA, and korA genes, respectively (Fig. 1B). Regulatory studies on individual promoters have revealed that four strong promoters are either controlled by global regulators (orf02p and klcAp) or efficiently autoregulated (kfrAp and korAp) (24, 28, 29). Only the weak korCp located within putative orf04 is transcribed constitutively (29).
The best-studied part of the module, the korA-incC-korB-orf11 operon (28), encodes an active partitioning system, where IncC and KorB belong to the ParA and ParB families of partitioning proteins, respectively, and KorA is a DNA binding protein, an autorepressor of korAp. Orf11 fulfills an accessory function in the partition (28). Besides its role in segrosome formation at parS, KorB also acts as a global transcriptional regulator controlling stability functions and conjugation by binding to three operator sites (OB), i.e., within orf02p, the first promoter of the stability module, and two promoters, mobCp and orf023p, from the conjugative transfer module (29–31). Upstream of the active partition operon are encoded two alpha-helical proteins, KfrA and KfrC, homologs of which were postulated to be involved in the segregation of IncP-1 plasmids (32, 33). KfrA is a DNA binding protein and autoregulates the monocistronic kfrA operon (24) whereas KfrC is encoded in the bicistronic korC-kfrC operon (Fig. 1B). KorC serves as the main global regulator of plasmid RA3, coordinating the expression of the replication (orf02prev), stability (orf02p and klcAp), and conjugative transfer (orf033p and orf034p) operons (29). Two monocistronic operons at the beginning of the stability module encode Orf02, a small protein exhibiting similarity to the N-terminal part of DnaA from Aeromonas spp., and KlcA, a putative antirestriction protein (34).
The roles of several products of the RA3 stability module have not been fully understood even in E. coli, where the majority of the experiments have been conducted. It also seemed important to decipher how the organization of this unidirectionally transcribed, highly compacted module might influence the expression of the genes in various hosts.
Here, we compared the gene expression patterns of the intact RA3 stability module and its deletion derivatives. Using this approach for the whole RA3 plasmid and also its stability module cloned into a heterologous replicon, we found that this module is organized as a long multicistronic operon with numerous internal promoters and a few terminators/attenuators modulating the expression of downstream genes. The RNA polymerase (RNAP) read-through and an impact of the multiple upstream promoters on downstream gene expression were also detected in hosts other than E. coli, although they varied in different species. The transcriptional studies on RA3 stability module deletion derivatives combined with stability assays revealed the important role of encoded proteins in plasmid maintenance in various hosts.
RESULTS
Transcriptional read-through from orf02p to orf11.
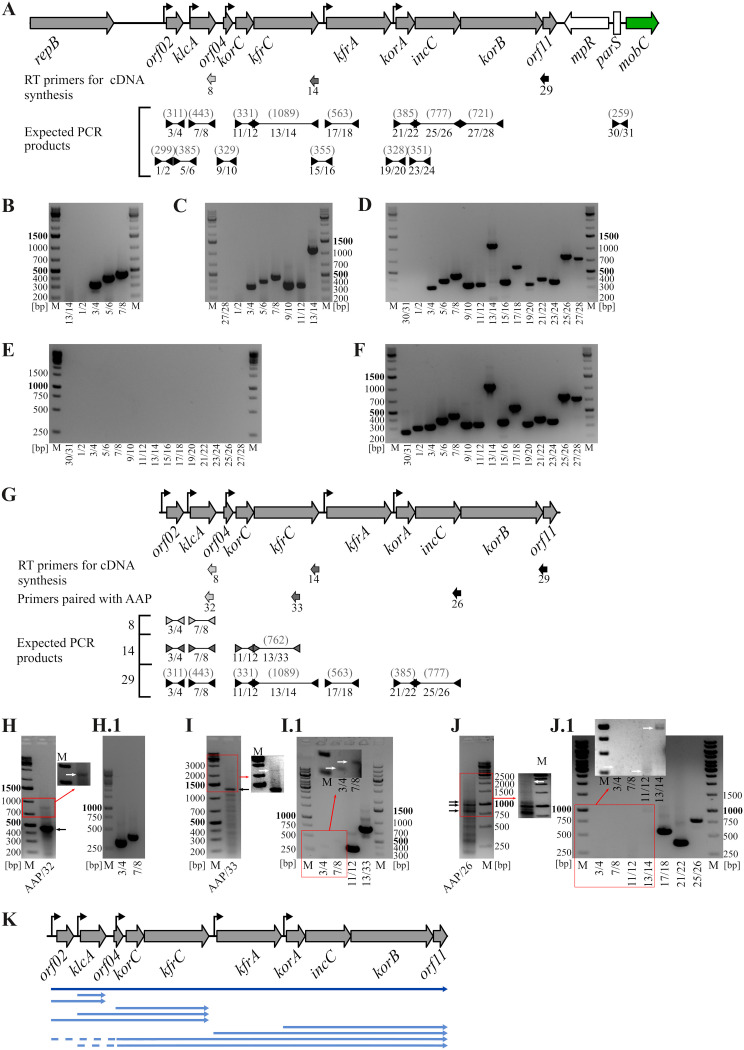
The unidirectional transcription and highly compacted arrangement of the stability module suggested the possibility of reading through from the upstream transcriptional signals into the downstream units. To verify this assumption, total RNA was isolated from the E. coli DH5α(RA3) strain and subjected to reverse transcription followed by PCR (RT-PCR). Three different primers were used in the RT reaction; these were complementary to the 3′ end of the klcA gene (primer 8), the 3′ end of the kfrC gene (primer 14), and the orf11 gene (primer 29). The cDNAs obtained in each reaction were then amplified with pairs of primers corresponding to the coding and intergenic parts of the analyzed region (Fig. 2A). PCR products synthesized on a cDNA template started from primer 8 encompassed orf02 and klcA (Fig. 2B), suggesting possibility of RNAP initiating transcription at orf02p and reading through the intergenic region orf02-klcA. PCR products obtained on cDNAs started from primer 14 indicated the reading through from orf02p to kfrC (Fig. 2C), whereas the results of PCRs on the cDNA template obtained from primer 29 confirmed that mRNA initiated at orf02p might extend through the whole stability region up to orf11 (Fig. 2D). Notably, no PCR products were obtained in any set of reactions with primers encompassing orf02p (primers 1/2), which excludes the transcription from the replication module (Fig. 1 and 2A) into the stability region. For each set of PCRs, an adequate pair of primers downstream of the start site for the given cDNA was used: 13/14 to amplify kfrC for the shortest cDNA, 27/28 to amplify korB, and 30/31 to amplify the parS region for two longer cDNAs (Fig. 2A). For all three cDNAs, the products of such control PCRs were not detected (Fig. 2B, C, and D). Two additional control sets of PCRs were conducted, with RNA as a template (negative control) to verify lack of DNA contamination in RNA samples (Fig. 2E) and with RA3 DNA as a template to demonstrate the efficiency of reaction with all the primers pairs (Fig. 2F).
FIG 2.
Transcripts of the RA3 stability module. (A to D) Analysis of mRNA by RT-PCR. Total RNA was isolated from E. coli DH5α(RA3) and used for cDNA synthesis. Three primers, 8, 14, and 29, annealing to different parts of the stability module were used in separate reverse transcription (RT) reactions for cDNAs synthesis. The cDNAs were used as templates for PCR with different pairs of primers. PCR-amplified fragments were separated on agarose gels, stained with ethidium bromide, and photographed. (A) Map of the stability module with flanking regions. The positions of RT primers and PCR primers as well as the expected PCR products are shown below. Sizes of PCR products are given in parentheses. (B to D) PCR products obtained with the pairs of primers indicated beneath the lanes and various cDNAs as templates, i.e., synthesized from primer 8 complementary to klcA mRNA (B), synthesized from primer 14 complementary to kfrC mRNA (C), and synthesized from primer 29 complementary to orf11 mRNA (D). Next to 1-kb DNA ladder (M) the appropriate control reaction mixture was loaded, i.e., products of PCR with a pair of primers annealing beyond the expected boundaries of the analyzed cDNAs. (E) RNA purity control. PCRs with total RNA as a template instead of cDNAs were run with the indicated pairs of primers. (F) Primer quality control. Products of PCRs with RA3 DNA as a template and the indicated pairs of primers are shown. (G to J) Analysis of mRNAs using 5′RACE. (G) Map of the stability module. The positions of the primers used for cDNA synthesis, the nested PCR primers used in combination with 5′RACE primer AAP (abridged anchor primer), and primers used in the second round of PCRs for three sets of cDNAs are indicated. The sizes of expected PCR products are shown in parentheses. (H to J) Analysis of 5′RACE products obtained in the first set of PCRs with AAP and appropriate nested primers on three cDNAs as templates, i.e., synthesized from primer 8 (H), from primer 14 (I), and from primer 29 (J). Marked sectors of the photographs were manipulated to intensify weak bands (white arrows). The reaction mixtures from panels H to J were used as templates for the second round of PCRs with pairs of specific primers (H.1 to J.1). (K) Schematic summary of the mRNA analysis presented above. Blue arrows depict identified variants of transcripts identified in the above-described experiments. Dashed lines represent deduced parts of the transcripts.
The RT-PCR results indicated the presence of long transcripts initiated at orf02p and continuing toward orf11, hence indicating cotranscription of the whole maintenance module. The fact that orf02, klcA, korC, kfrA, and korA are preceded by functional promoters (24, 28, 29) suggested the possibility of a polarized transcript dosage, with progressively more multiple transcript variants for the genes further downstream in the module. To check if indeed transcripts of various lengths exist for a given ORF, the 5′ rapid amplification of cDNA ends (5′RACE) procedure (35) was applied. Total RNA isolated from E. coli DH5α(RA3) was reverse transcribed with primers 8, 14, and 29 as described above. The cDNAs were 3′ tailed with a stretch of dCTPs and used as templates for PCRs with an abridged anchor primer (AAP) complementary to the dC tail paired with appropriate nested primers 32, 33, and 26, as shown in Fig. 2G. Parts of these RT-PCR mixtures were separated on agarose gels to visualize products corresponding to the cDNAs synthesized on the transcripts that were presumably of various lengths (Fig. 2H, I, and J). The remaining parts were diluted and used as templates for PCRs with pairs of primers specific to the ORFs located upstream of the sequences complementary to primers 32, 33, and 26 (Fig. 2H.1, I.1, and J.1). The best results were obtained for the potential cDNA mixture synthesized with primer 8 complementary to the 3′ end of klcA. Two products were visualized in the first set of PCRs with primers AAP and 32, confirming the presence of two transcripts for klcA starting at orf02p and klcAp (Fig. 2H), which was further verified by the subsequent round of PCRs with primers specific to orf02 and klcA, respectively (Fig. 2H.1). When the cDNAs obtained with the use of primer 14, complementary to the 3′ end of the kfrC gene, were used as a template, two products were detected after the first round of PCR with AAP and the nested primer 33 (Fig. 2I). The intense band corresponded to the product synthesized on cDNA with the 3′ end determined by the transcription start site (TSS) of korC. Another, much weaker band (inset in Fig. 2I) presumably corresponded to cDNA with the 3′ end determined by the TSS of klcA. Despite the fact that no product was visible for the cDNA extended up to orf02p, the next round of PCRs led to amplification of all four ORFs from kfrC up to orf02, although with various efficiencies (inset in Fig. 2I.1). The most ambiguous were the results for cDNA(s) obtained with primer 29, which annealed in the region of orf11. The PCRs with AAP and the “nested” primer 26 unexpectedly showed the main three products with sizes between 800 and 1,100 bp (Fig. 2J). Whereas a fragment of ca. 1,100 bp might correspond to cDNA ending at the TSS of korA, the two smaller products suggested an additional TSS(s) upstream of incC. The presence of these putative promoter sequences has been further analyzed (see below). A product of ca. 2,200 bp, corresponding to cDNA ending at the TSS of kfrA, was also visible after intensity enhancement (inset in Fig. 2J). The next round of PCRs using the specific pairs of primers for ORFs upstream of korB led to the amplification of fragments corresponding to ORFs from incC to orf02 (except for klcA), although with the efficiency of the reactions clearly inverse to the distance from the 3′ end of the stability module (Fig. 2J.1). Although the 5′RACE experiments did not directly and clearly show the whole spectrum of transcripts for particular ORFs, they supported the hypothesis of a gradient transcript dosage along the stability module (Fig. 2K). The numerous small PCR products obtained with AAP and “nested primers” likely resulted from nonspecific annealing of the AAP to the C stretches in the GC-rich sequences of the kfrC, kfrA, and incC genes (24).
Analysis of transcription termination signals.
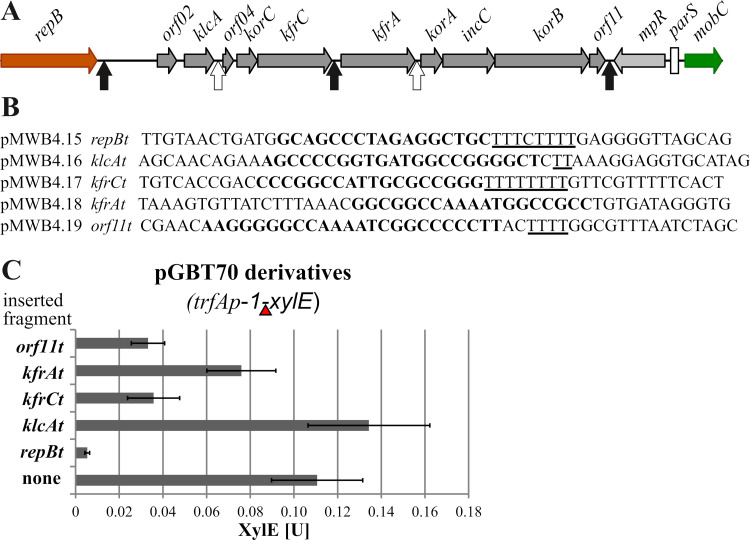
The expression of multigenic modules is regulated not only at their promoters but also by transcription terminators. In silico inspection of the RA3 replication and stabilization regions uncovered two putative unidirectional Rho-independent transcription terminators (GC-rich palindromic sequence followed by a run of Ts) after the kfrC and orf11 genes in the stabilization module and a third one after the repB gene separating the replication and stabilization modules (Fig. 3A and B). Also, the intergenic regions klcA-orf04 and kfrA-korA contain GC-rich palindromic sequences (however, without a long run of Ts) which could pause/modulate the progress of RNAP (Fig. 3A and B). The five relevant intergenic regions were cloned individually into pGBT70 (36) between the strong trfAp-1RK2 promoter and the xylE reporter gene to verify their putative terminator/modulator action. This plasmid had been successfully used to characterize transcription terminator sequences before (37). The plasmid constructs were introduced into E. coli C600K, and the activity of catechol 2,3-dioxygenase, encoded by xylE, was assayed. The regions downstream of repB, kfrC, and orf11 noticeably hampered the transcription initiated at trfAp-1, while the sequence downstream of klcA did not (Fig. 3C). The result for the region downstream of the kfrA gene was ambiguous, showing a 30% decrease in the XylE activity compared to that for the unmodified pGBT70 plasmid. This decrease may suggest E. coli RNAP pausing at this sequence, but that needs an independent confirmation. Thus, we experimentally demonstrated the presence of two efficient Rho-independent transcription terminators in the RA3 stability module: one in the middle of the module downstream of the kfrC gene and the other at the very end of the module, following orf11.
FIG 3.
Analysis of putative RA3 transcription terminators in and close to the RA3 stability module. (A) Positions of putative transcription terminators/attenuators in the stability module and flanking regions, with Rho-independent terminators indicated by black arrows and hypothetical transcriptional Rho-dependent terminators/attenuators by white arrows. (B) Putative terminator sequences with GC-rich inverted repeats (bold) and stretches of Ts (underlined) indicated. For repBt, a fragment of 157 bp encompassing this sequence was amplified by PCR on RA3 DNA; other sequences were obtained as synthetic double-stranded oligonucleotides. They were cloned into pGBT70 between the trfAp-1 promoter and the xylE cassette (insertion marked by red triangle in panel C). (C) Effect of putative terminators on xylE transcription. E. coli DH5α was transformed with pGBT70 (control without additional insert) or its derivatives indicated in panel B. XylE activity was assayed in extracts from exponential-phase cultures of transformants. Experiments were repeated at least three times, and mean values with standard deviations are shown.
Transcriptional analysis of the synthetic RA3 stability module and its mutated variants: effect of mutations on plasmid stability in various hosts.
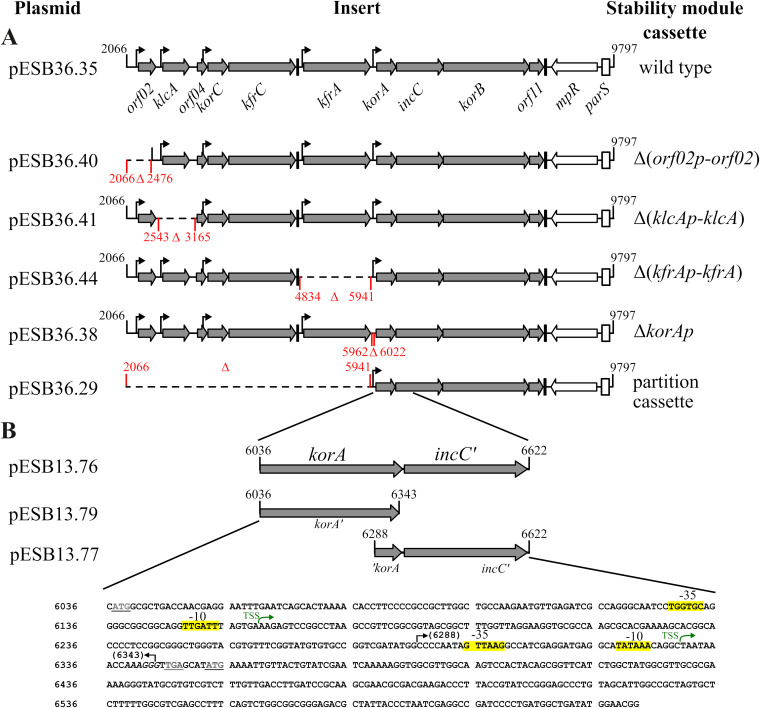
To assess the roles of the individual promoters in the expression of downstream genes/operons in the RA3 stability module, a set of deletion variants, deprived of one or a combination of several promoters and/or particular genes, was constructed (see the supplemental material). The obtained library of the module variants, cloned into an unstable BHR plasmid, allowed study of the roles of genes of interest in the maintenance of the plasmid in various hosts.
For these studies, the wild-type (WT) module and its Δ(orf02p-orf02), Δ(klcAp-klcA), Δ(kfrAp-kfrA), ΔkorAp, and Δ(orf02p-kfrA) (the partition operon on its own) variants were chosen (Fig. 4A). The rationale behind the deleting of the promoter regions with the adjacent ORFs in the case of the monocistronic and strongly regulated klcA and kfrA operons was to avoid altered expression of these presumably important ORFs from upstream promoters. As a precaution, no genetic manipulations were undertaken in the korC operon, since KorC is a potent transcriptional regulator of orf02p and klcAp (29). The constructed modules were cloned into a single-copy, extremely unstable pESB36 vector, a derivative of pABB32 (38). pESB36 is based on the RK2 minireplicon (pRK415), is mobilizable by the RK2 conjugative system owing to the presence of oriTRK2, and carries a convenient system for detection of the plasmid presence in colonies (repApRA3-lacZ transcriptional fusion). pESB36, pESB36.35 (WT stability module inserted), and its deletion derivatives (Fig. 4A) were introduced into two representative Gammaproteobacteria species (E. coli and Pseudomonas putida), two Alphaproteobacteria species (Agrobacterium tumefaciens and Paracoccus aminovorans), and one Betaproteobacteria species (Cupriavidus necator). Transformants/transconjugants were analyzed with respect to stability module gene expression and stable maintenance of the pESB36.35 derivatives during growth without selection. The plasmid segregation assay demonstrated that the pESB36 loss rate (LR) was variable in the hosts tested and varied from 2% per generation in P. putida to 18% per generation in E. coli and to more than 25% per generation in P. aminovorans (Table 1). Such differences in the vector stability could be due to the variations in the plasmid copy number (PCN) caused by differential gene expression of the miniRK2 replicon in the analyzed species. Estimation of the pESB36 copy number per chromosome in cells from stationary-phase cultures grown under selection confirmed this conjecture. The PCN for two hosts, P. aminovorans and E. coli, was only 0.12 to 0.15, whereas that for P. putida was 2.9 (Table 2).
FIG 4.
Deletion variants of the RA3 stability module used for studies on gene expression and plasmid maintenance in diverse hosts. (A) Schematic presentation of the stability module variants cloned into the low-copy-number pESB36. Thin black arrows indicate known promoter sequences, and black bars mark positions of Rho-independent transcriptional terminators. The extent of deletions is shown in red. (B) Identification of previously unanticipated promoters within the incC gene. The indicated regions were cloned into the promoter-probe vector pPT01 upstream of the xylE cassette and introduced into E. coli DH5α. Activity of the putative promoters was analyzed by XylE assay (see the text). The nucleotide sequence of the fragment cloned into pESB13.76 is presented. The black arrows delineate boundaries of the smaller fragments. The stop codon for korA and the start codon of incC are underlined. Green arrows depict two newly identified TSSs for incC in the korA coding sequence. Putative promoter motifs are highlighted in yellow.
TABLE 1.
Loss rate per generation and stability index of analyzed derivatives of pESB36 in various hostsa
| Plasmid | LR and SI in host: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
Escherichia coli |
Paracoccus aminovorans |
Agrobacterium tumefaciens |
Cupriavidus necator |
Pseudomonas putida |
||||||
| LR (%) | SI | LR (%) | SI | LR (%) | SI | LR (%) | SI | LR (%) | SI | |
| pESB36 | 18.0 | X | >25 | X | 2.7 | X | 4.9 | X | 2.0 | X |
| pESB36.35 (WT) | 3.5 | 5.1 | 0.4 | NC | 0.5 | 5.4 | 1.1 | 4.9 | 0.5 | 4.0 |
| pESB36.40 (Δorf02p-orf02) | 14.5 | 1.2 | 0.05 | NC | 1.8 | 1.5 | 4.6 | 1.1 | 2.2 | 0.9 |
| pESB36.41 (ΔklcAp-klcA) | 15.0 | 1.2 | 0.3 | NC | 1.0 | 2.7 | 5.8 | 0.8 | 2.0 | 1.0 |
| pESB36.44 (ΔkfrAp-kfrA) | 11.4 | 1.6 | 0.5 | NC | 1.2 | 2.3 | 5.0 | 1.0 | 2.4 | 0.8 |
| pESB36.38 (ΔkorAp) | 4.6 | 3.9 | ND | ND | ND | ND | ND | ND | 2.7 | 0.7 |
X, irrelevant; NC, not calculable; ND, not determined.
TABLE 2.
Estimated plasmid copy numbers in various hosts
| Plasmid | Mean (SD) copy no. in host: |
||||
|---|---|---|---|---|---|
| Escherichia coli | Paracoccus aminovorans | Agrobacterium tumefaciens | Cupriavidus necator | Pseudomonas putida | |
| pESB36 | 0.15 (0.02) | 0.12 (0.03) | 1.03 (0.12) | 2.62 (0.54) | 2.9 (0.68) |
| pESB36.35 | 1.31 (0.09) | 1.08 (0.17) | 3.08 (0.70) | 3.34 (0.67) | 3.3 (0.69) |
The presence of the RA3 stability module significantly increased the persistence of the plasmid (pESB36.35) in all the species tested, again to different extents. The plasmid was very stably maintained in P. aminovorans, P. putida, and A. tumefaciens, with a loss rate per generation of 0.4 to 0.5% (Table 1), and slightly less so in C. necator (LR, 1.1%). The highest plasmid loss rate, 3.5%, was found for E. coli. However, the ratio of the loss rates for the empty vector and pESB36.35 in a given strain, the so-called stability index (SI), was quite similar in all the hosts, with the exception of P. aminovorans, where it exceeded 60 (Table 1).
(i) Expression of the stability module and its deletion variants in E. coli. The E. coli EC1250 (Δlac) strain was transformed with pESB36, pESB36.35, and its derivatives, transformants were grown on rich medium with selection, and total RNA was isolated. cDNA was synthesized on the RNA with the use of a mix of random hexamer primers, and then quantitative real-time PCR (qPCR) was performed with pairs of primers specific for each ORF of the stability module, with the exception of the short orf04 overlapping korCp. The results were normalized to the amount of mRNA for the chromosomal marker cysG (39).
The expression of blocks of genes in the WT stability module varied significantly, confirming the functionality of the five previously identified promoters in the context of the whole module (Fig. 5A). The lowest levels of transcripts were found for orf02, the first ORF in the module, and for kfrA, both presumably encoded by the tightly regulated monocistronic operons (24). In turn, klcA expression was 10-fold higher than that of orf02, despite the expected strong repression of klcAp by KorC (29). The korC, kfrC, and korA genes showed an intermediate level of expression, and unexpectedly, the highest levels of transcripts were found for incC, korB, and orf11.The organization of the korA-incC-korB-orf11 region suggested a single operon structure (with closely packed or overlapped ORFs) whose expression was dependent on the strong but autorepressed korA promoter (24, 28). The observation here of decoupling of korA expression and three downstream genes raised the possibility of the existence of an internal promoter(s) in this operon. The presence of the promoters driving a korAp-independent expression of incC-korB-orf11 (at least in E. coli) was verified by cloning of a 586-bp DNA fragment (Fig. 4B) encompassing the korA coding sequence and the 5′ end of incC into the promoter-probe vector pPT01 (40) upstream of the xylE cassette (pESB13.76). Determination of the XylE activity in the extracts from exponentially growing E. coli C600K(pESB13.76) cells revealed a weak transcriptional activity of the cloned fragment [7.2 mU, versus 0.5 mU in control E. coli C600K(pPT01) cells]. A similar XylE activity (6.5 mU) was detected in extracts from E. coli C600K(pESB13.77) and C600K(pESB13.79) when the shortened fragments were cloned upstream of xylE, pointing out the localization of the incC promoter(s) in the 3′ end of korA (Fig. 4B). Importantly, the 5′RACE experiments also suggested the possibility of additional TSSs within korA (Fig. 2J). The products detected in the 5′RACE experiments were isolated and sequenced. The 5′ ends of these fragments localized two putative TSSs in korA, one corresponding to the 3′ end of korA and another 169 bp upstream, as marked in Fig. 4B together with corresponding promoter motifs. Importantly, these weak constitutive internal transcriptional signals seem to significantly increase the amounts of mRNAs for partitioning genes in comparison to korA mRNA (Fig. 5A). Such accumulation of shorter transcripts may result from the higher stability of these mRNAs than those initiated at korAp, as differential mRNA decay has been observed in other prokaryotic multicistronic operons (41, 42).
FIG 5.
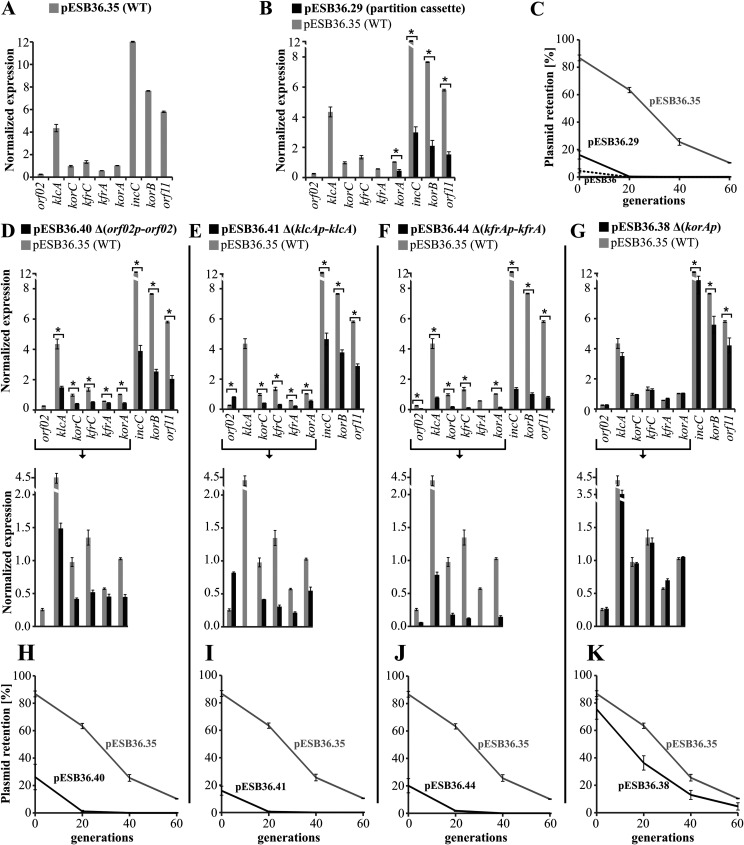
Transcription profiles of RA3 stability module deletion variants in E. coli. E. coli strain EC1250 was transformed with the pESB36 vector carrying RA3 stability module variants as indicated. Total RNA was isolated from three exponentially growing cultures of transformants in the presence of selection (biological replicates) and used as the template for cDNA synthesis with random hexameric primers. Three technical replicates of qPCRs with pairs of primers corresponding to individual ORFs were run for each cDNA lot. Target gene expression was normalized to the expression of a chromosomal reference gene, cysG. Mean values with standard deviations for at least three assays are shown. (A) Normalized expression of the ORFs in the WT stability module [E. coli EC1250(pESB36.35)]. (B and D to F) Normalized expression of ORFs in deletion variants (dark gray bars) was compared to that in the WT stability module (light gray bars). The bracketed parts of the diagrams were scaled up for clarity. E. coli EC1250 carrying the following plasmids was studied: pESB36.29 (Δorf02p-kfrA, partition operon), pESB36.40 (Δorf02p-orf02), pESB36.41 (ΔklcAp-klcA), pESB36.44 (ΔkfrAp-kfrA), and pESB36.38 (ΔkorAp). *, P < 0.05 in two-sided Student t test, assuming equal variance. (C and H to K) Plasmid segregation assays. Transformants of E. coli strain EC1250 were grown without selection for up to 60 generations. Every 20 generations, cultures were spread on L agar with X-Gal. Plasmid retention was expressed as the relative number of blue colonies.
The transcription profiling of the stability module variants lacking single or several promoters confirmed the participation of RNAP read-through in the establishing of this region’s expression pattern (Fig. 5B to F). It was most clearly seen when the expression of the korA-incC-korB-orf11 genes was compared between the intact WT stability module and the partition operon on its own (Δorf02p-kfrA variant, pESB36.29) (Fig. 5B). The transcripts for the partition operon genes were almost four times less abundant in the construct lacking all the DNA sequence upstream of korAp than in the WT stability module. Similarly, deletion of orf02p-orf02 at least halved the expression levels of the majority of downstream genes compared to the WT module (Fig. 5D). Deletion of the klcAp-klcA region decreased the expression of downstream genes 2- to 3-fold but, in parallel, increased the expression of orf02 3-fold (Fig. 5E). The apparent derepression of orf02p might be due to the lower level of expression of korC and korB, encoding orf02p repressors (29). Another plausible explanation of the observed “induction” might be a relief from a negative interference between the orf02p and klcAp transcriptional and regulatory signals, since these two promoter regions arose by duplication (24). The unexpected lack of transmission of the enhanced orf02p activity to the genes downstream of orf02 could be a result of the genetic manipulations leading to the deletion of the klcAp-klcA region.
In the absence of korAp, no change in expression of korA was detected and there was only a slight decrease in the expression of the partitioning genes (Fig. 5G), confirming significant participation of RNAP read-through from the upstream promoters into korA and a minor role of korAp in driving expression of downstream loci. The most unexpected result was the strong effect of the kfrAp-kfrA deletion on the expression of the whole stability module (Fig. 5F). Genes downstream of kfrA were even expressed 12-fold less abundantly despite the low level of transcriptional activity of kfrAp in the intact module. Intriguingly, the upstream genes also were strongly downregulated. Further studies should elucidate the consequences imposed on plasmid functions by the lack of kfrAp-kfrA.
E. coli EC1250 (Δlac) transformants carrying pESB36.35 variants were also tested for plasmid retention over 60 generations of growth in rich medium without selection. Analysis of plasmid stability showed that all deletion variants except pESB36.38 ΔkorAp were unstable in E. coli (Fig. 5C and H to K). Since only the ΔkorAp derivative demonstrated hardly any effect on expression of the partition operon, it may be concluded that any manipulation leading to the decreased transcription of incC-korB-orf11 led to an increase in the rate of loss of these plasmids (Fig. 5 and Table 1).
(ii) Expression of the stability module in hosts other than E. coli. The RA3 plasmid has a very wide range of hosts belonging to Alpha-, Beta-, and Gammaproteobacteria (24), and hence it was important to establish whether the transcriptional signals characterized in E. coli function in a similar way in the other hosts and whether the RNA polymerases of other bacteria were also capable of reading through the termination/attenuation signals in the stability module. The mRNA levels of the RA3 genes of interest were measured with respect to those of the recommended reference gene for each transconjugant, and the results are presented in Fig. 6, normalized to orf02 taken as 1. The analysis of the WT stability module expression (pESB36.35) by RT-qPCR demonstrated that in all transconjugants the levels of incC and korB transcription were the highest (or among the highest, as in C. necator), similarly to the case in E. coli, but the expression of the genes preceding the partition operon varied significantly depending on the host (Fig. 6A). The expression of orf02 and klcA seemed to be correlated and severalfold lower than that of korC and kfrC in P. aminovorans, A. tumefaciens, and P. putida, whereas in C. necator the first four genes were expressed at comparable levels. Expression of kfrA was like the expression of orf02 and klcA in A. tumefaciens and P. putida, lower in C. necator, and slightly higher in P. aminovorans. These results indicated substantial modulations of the abilities of the host transcriptional machinery to recognize different initiation signals (e.g., more efficient expression from korCp in all species other than E. coli) and presumably to respond to regulators such as KorC (orf02p and klcAp) or KfrA (kfrAp).
FIG 6.
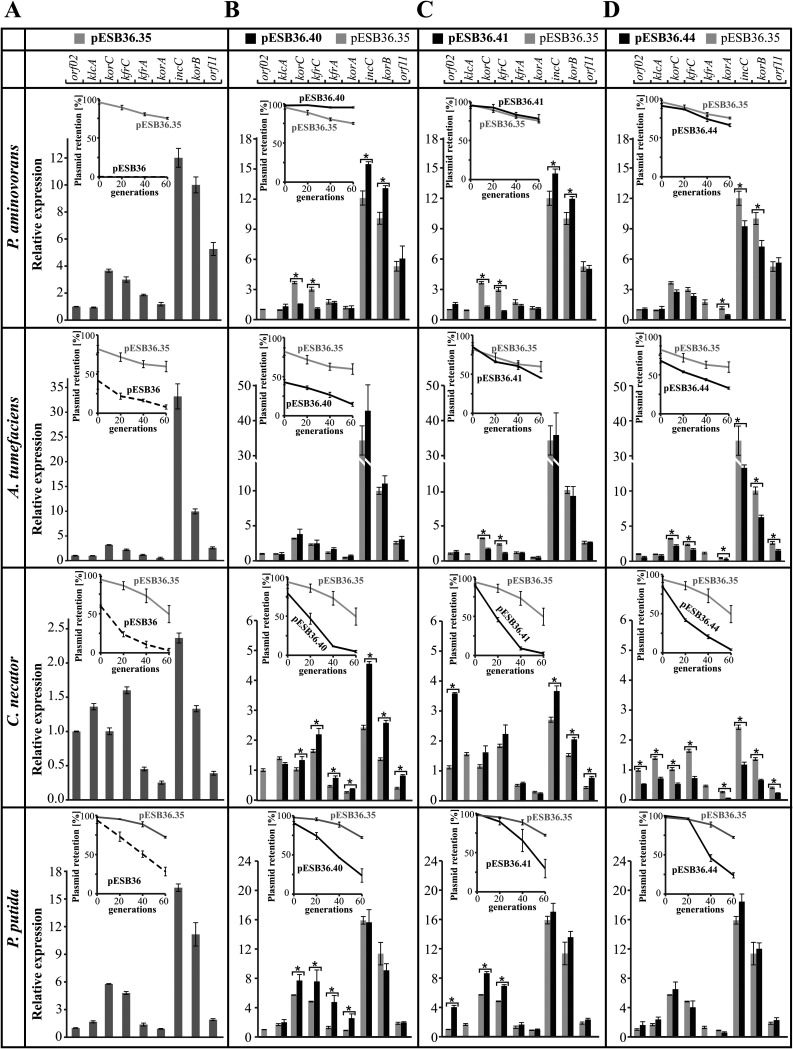
Transcriptional profiles of RA3 stability module variants in Paracoccus aminovorans, Agrobacterium tumefaciens, Cupriavidus necator, and Pseudomonas putida hosts. RT-qPCR analysis was conducted on RNA samples isolated from exponentially growing cultures of transconjugants of appropriate hosts. Expression of stability module ORFs was normalized to the rpoD reference gene in P. aminovorans, A. tumefaciens, and P. putida and to gyrB in C. necator. Mean values with standard deviations for at least three assays are shown. (A) Relative expression of the ORFs in the WT stability module (pESB36.35) in various hosts. For clarity of presentation, the normalized results (target gene versus reference gene for each strain) are shown relative to the expression of orf02 in each strain. Insets demonstrate pESB36.35 plasmid retention in comparison to vector pESB36 in the analyzed host. (B to D) Relative gene expressions in the WT stability module (pESB36.35, light gray bars) and tested deletion variants (dark gray bars) in each of the hosts. For clarity of presentation, the normalized results are shown relative to the expression of the orf02 from the WT stability module (pESB36.35) in each strain. Insets demonstrate retention of each plasmid variant in the analyzed host in comparison to vector pESB36.35 carrying the intact stability module (WT). *, P < 0.05 in two-sided Student t test, assuming equal variance.
To check whether read-through takes place in hosts other than E. coli, the expression of genes in the Δ(orf02p-orf02), Δ(klcAp-klcA), and Δ(kfrAp-kfrA) variants of the stability module was analyzed by RT-qPCR in the transconjugants of four hosts carrying pESB36.40 (Fig. 6B), pESB36.41 (Fig. 6C), and pESB36.44 (Fig. 6D), respectively. Plasmid retention in transconjugants was also monitored for 60 generations of growth without selection, and the results of the stability assays are shown in insets in Fig. 6. Additionally, the calculated plasmid loss rates and stability indexes (if feasible to calculate) are included in Table 1.
Expression of the truncated stability modules varied between the strains. Notably, the deletion of orf02p-orf02 did not have as strong a negative effect on the level of transcripts of the downstream genes as in E. coli (Fig. 5D and 6B). A decrease in transcription, but only of korC and kfrC, was observed in P. aminovorans (with a simultaneous increase in the transcription of the partition operon), and no change in expression of the stability module was observed in A. tumefaciens (Fig. 6B, two upper panels). In contrast, in C. necator the orf02p-orf02 deletion had a positive effect on the expression of almost all remaining genes (with the highest effect on the partition operon), whereas an increase in korC, kfrC, kfrA, and korA expression (but not that of incC-korB-orf11) was detected in P. putida (Fig. 6B, two bottom panels). This may suggest that RNAP read-through from orf02p encompasses only klcAp and korCp in P. aminovorans, plays no role in A. tumefaciens, and negatively interferes with the expression of downstream operons in C. necator and P. putida. Expression of the partition operon seems to be uncoupled to some extent from other parts of the module in these four species. Notably, deletion of the orf02p-orf02 fragment had a large impact on pESB36.40 stability in the analyzed hosts (Fig. 6B and Table 1). Similar to the case for E. coli (Fig. 5D), the construct was unstable in A. tumefaciens, C. necator, and P. putida (loss rate close to that of pESB36 vector). In E. coli the pESB36.40 loss might have resulted from the lower expression of the partition operon (or of other genes in the module) (Fig. 5D and H). In species in which the remaining genes in the stability module were expressed at the same level as in the wild-type fragment (A. tumefaciens) or even higher (C. necator and P. putida), the instability was likely caused by lack of the Orf02 product itself. The reverse effect of the orf02p-orf02 deletion on plasmid stability was observed in P. aminovorans, in which the pESB36.40 loss rate dropped 8-fold. This coincides with the increase of the partition operon expression but also with the decrease in the level of korC and kfrC transcripts. Further studies should define the role of Orf02 in regulation of gene expression and stability of the plasmid in these hosts.
The klcAp-klcA deletion led to a severalfold increase of the transcription from orf02p in E. coli (Fig. 5E). A similar effect was observed in C. necator and P. putida (Fig. 6C); however, in these hosts, in contrast to the case for E. coli, it was conveyed into higher expression of korC-kfrC operon, as expected for the RNAP reading through from orf02p into downstream genes. In two other species, P. aminovorans and A. tumefaciens, the lack of the klcAp-klcA DNA fragment lowered the transcription of the korC-kfrC operon, indicating significant participation of klcAp in the expression of this part of the stability module. Remarkably, the deletion of klcAp-klcA fragment either had no impact on the expression of the partition operon (A. tumefaciens and P. putida) or led to an increase of the expression of this operon (P. aminovorans and C. necator). In these four strains, the effect of klcAp-klcA deletion on the expression of the adjacent operon, korC-kfrC, varied from the effect on expression of the partition operon, confirming the uncoupling of two parts of the stability module (Fig. 6C).
The maintenance of the construct deprived of klcAp-klcA (pESB36.41) varied from being stable in P. aminovorans and A. tumefaciens to being unstable in C. necator and P. putida (insets in Fig. 6C and Table 1) and E. coli (Fig. 5I). Instability of the construct in E. coli might be explained by lowered expression of the partition operon; however, in C. necator the partition operon transcription was elevated, and in P. putida it was unchanged, in comparison with the WT module. Hence, either increased production of Orf02 or lack of KlcA may account for the plasmid instability in these hosts. Further studies are needed to discriminate between these options. It is worthwhile to notice that deletions of either orf02p-orf02 or klcAp-klcA had an identical impact on the stability module expression in P. aminovorans (Fig. 6B and C, upper panels); however, only the first deletion increased plasmid stability in this host. This favors the hypothesis of Orf02 playing the negative role in plasmid maintenance in this species.
Deletion of kfrAp-kfrA in E. coli had a significant influence on expression of the upstream and downstream genes in the module (Fig. 5F). A similar effect of Δ(kfrAp-kfrA) was detected in C. necator and A. tumefaciens (Fig. 6D). In P. aminovorans only expression of the partition operon was significantly lowered, but it did not affect the stability of pESB36.44 in this host. Modest instability of the construct was observed in A. tumefaciens, whereas the highest plasmid LR was observed in C. necator and P. putida (Fig. 6D and Table 1). The decrease in pESB36.44 stability in A. tumefaciens, C. necator, and E. coli (Fig. 5J) may result from the lower expression of the partition operon or other genes; however, in the case of P. putida, the lack of KfrA itself seems to be deleterious for the plasmid maintenance (Fig. 6D, insets, and Table 1).
DISCUSSION
Plasmids usually provide advantageous traits to their hosts but also impose a fitness cost on the cells. In the absence of selection for the plasmid-borne beneficial traits, the cells that have lost the plasmid easily outcompete the plasmid-bearing rivals (43), which should lead to plasmid extinction. However, laboratory and environmental studies have shown long-term plasmid persistence under nonselective conditions, a phenomenon dubbed the “plasmid paradox” (44). To ameliorate the fitness cost, the plasmid and host genomes undergo compensatory coevolution, which has been of great research interest in recent years (17, 43, 45–48). To increase the probability of the compensatory evolution events taking place, plasmids have to ensure lowering the rate of their loss (49, 50).
To this end, plasmids have evolved efficient copy number control mechanisms and tight regulation of gene expression to diminish the metabolic burden imposed on the host as well as evolved/acquired specific stability mechanisms to support their maintenance in the population, which is especially important for low-copy-number replicons. Besides the prevalent and well-known multimer resolution systems (51), active partition mechanisms (52), and postsegregational killing (toxin-antitoxin) systems (53, 54), additional factors may play significant roles during the establishment of broad-host-range plasmids in phylogenetically diverse hosts (7, 45, 55).
Above all, the promiscuous plasmids have to secure an adequate production of vital plasmid proteins regardless of the highly diversified efficiency of the transcriptional and translational machinery in these hosts (56). The question of how the broad-host-range plasmids cope with various host factors to ensure adequate levels of plasmid gene expression is far from being fully understood.
In general, gene expression depends on the RNAP recognition of promoters (57), the level of transcriptional regulators, transcriptional organization, global and local DNA topology controlled by host- or plasmid-encoded nucleoid-associated proteins (NAPs) (58, 59), and the position of the gene in the genome with respect to the origin of replication (60, 61). The initial concept of the bacterial operon defined it as a transcriptional unit (TU) that provided simultaneous expression of genes within the operon through production of a single polycistronic mRNA initiated at the promoter upstream of the first gene (62). Control of this unique promoter allowed a coordinated regulation of all genes. However, recent studies have revealed that bacterial transcriptomes are far more complex than previously thought (63–65). The architecture of bacterial operons may involve multiple internal differently regulated promoters and various terminators generating multiple TUs and leading to differential gene expression within these operons (66). It has been shown that various blocks of genes in an operon are often alternatively transcribed under diverse conditions; e.g., the E. coli polycistronic flagella operon fliF to fliR of thirteen genes may be split in up to seven various suboperons after heat shock or phosphorus starvation (67).
Genome-wide transcript analysis not only demonstrated variability of transcripts corresponding to the operon-clustered genes but also stressed the important regulatory roles that the stability of the mRNAs and their translation efficiency play in gene expression (42, 66, 68–71). First, it was assumed that the prokaryotic transcripts are processed only by 3′-to-5′ exonucleases, leading to an abundance of transcripts for the 5′ part of the multicistronic operons (polarity expression). Discovery of the endoribonucleases and their role in mRNA decay in combination with regulatory functions of RNA binding proteins and noncoding RNAs (ncRNAs) revealed a complex regulatory network responsible for differential transcript abundance even in the operons (72–76).
Similarly to the organization of their hosts’ genomes, the mosaic plasmids cluster the genes engaged in the same processes into functional modules, i.e., the replication, conjugation, and stability modules. This evolutionary trend has been usually explained by gene products forming complexes, e.g., replisome, relaxosome, or transferosome, and temporal/spatial requirements for the synthesis of such complexes (77). Even if various stability functions of the plasmids are encoded in several operons, they are often located next to each other, seemingly representing separate TUs integrated only by common regulators, such as in RK2 (GenBank accession no. BN000925) of IncP-1 (78), R388 (GenBank accession no. BR000038) of IncW (79), or R46 (GenBank accession no. AY046276) of IncN (80).
Our studies on plasmid RA3, the archetype of the IncU group, have revealed that this conjugative, low-copy-number, broad-host-range plasmid has a mosaic modular structure with functional modules intertwined by the global regulatory network (24, 28–31, 81). The present project focused on the expression of the RA3 stability module, comprising a unidirectionally transcribed set of ten ORFs split apparently into five differently regulated operons (24). In the course of these studies, additional, previously unanticipated TSSs as well as transcription terminator and attenuator signals were found in the RA3 stability module.
The active partition operon korA-incC-korB-orf11 has previously been identified as the main part of the stability module (24, 28), but the roles of the remaining six ORFs have not been known. Before attempting to analyze them, it seemed important to establish the transcriptional organization of the region and elucidate if and how the individual regions of the module influence the expression of other genes. The possibility of RNAP read-through and its putative effect on the relative abundance of individual transcripts along the module (gene transcript dosage) were investigated.
Thus, it was shown here that the whole RA3 stability module was built as a polycistronic operon, orf02 to orf11, but transcribed into alternative mRNAs due to at least five internal promoters and various terminators. Such a transcriptional organization may have evolved to ensure the relative expression levels of the individual genes that would meet the requirements of various hosts. The transcription analysis of the stability module in representative strains of Alpha-, Beta-, and Gammaproteobacteria in which RA3 replicated and seemed to be stably maintained (24) indicated differential expression patterns.
The transcriptional profiles of the RA3 stability module in E. coli and the four other hosts, P. putida, P. aminovorans, A. tumefaciens, and C. necator, showed similar abundances of transcripts for the partition genes (Fig. 5A and 6A) but a substantial variability for the upstream region of the module. The relatively high expression from korCp in P. putida, P. aminovorans, and A. tumefaciens suggests more efficient RNAP recognition of this transcription initiation signal, leading to a stronger repression of the first two promoters by KorC than in E. coli (Fig. 5D and 6B). In turn, in C. necator the expression of the first three operons was at a similar level, indicating yet another specificity of its transcriptional machinery.
Transcriptional analysis of pESB36.35 deletion derivatives in E. coli clearly demonstrated the impact of the upstream transcriptional signals on the expression of the downstream genes due to RNAP read-through. Some observations indicated that multiple transcripts originating in the stability module may be more prone to nucleolytic degradation than others, as was observed, e.g., for the active partition operon. The much higher level of incC mRNAs than of korA mRNAs not only seems to result from the presence of weak internal promoters within korA but indicates the distinct stability of transcripts starting at korAp versus incCp.
The transcriptional studies conducted in different hosts demonstrated the species-specific dependence of expression of particular cistrons on RNAP read-through, TSSs, and intrinsic and Rho-dependent terminators and also suggested differential mRNA decay. In various combinations of host and plasmid deletion variant, a high plasmid loss rate was observed (Table 1), but further studies are needed to define whether the plasmid instability is due to transcriptional dysfunction or lack or abundance of specific products.
Analysis of the stable maintenance of pESB36.35 deletion derivatives in various hosts combined with the transcriptional analysis by qRT-PCR allowed us to determine the important, though converse, roles of the Orf02 product in A. tumefaciens, C. necator, P. putida, and P. aminovorans. At this point, no conclusion about Orf02 function in E. coli may be drawn, since deletion of orf02p-orf02 diminished expression of all downstream genes and the effect on segregation might have been caused by lower expression of, e.g., the partition operon. As mentioned above, the destabilizing effect of KlcA deficiency in E. coli, C. necator, and P. putida is far more complicated to interpret due to the simultaneous increase in orf02 expression. Finally, KfrA is definitely important for plasmid establishment/maintenance in P. putida, E. coli, A. tumefaciens, and C. necator but not in P. aminovorans, and its role is under investigation.
Studies on functions of particular plasmid-encoded proteins in plasmid biology are usually carried out on deletion/substitution mutants. However, as shown here, such genetic modifications may introduce changes in the transcriptional organization (initiation and termination signals) and regulation of gene/operon expression locally but may also affect gene expression in other regions of the plasmid. To avoid erroneous conclusions, the analysis of plasmid functions should be complemented by a thorough transcriptional analysis. As demonstrated here, in the case of broad-host-range plasmids, such analysis should be conducted in diverse hosts.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The Escherichia coli strains used were C600K (thr-1 leu-6 thi-1 lacY1 supE44 ton21 galK) (82), DH5α [F− (ϕ80dlacZΔM15) recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169] (83), EC1250 [F− araD139 Δ(lacZYA-argF)U169 rpsL150 relA1 deoC1 ptsF25 rbsR flbB5301 trp-1] (84), and S17-1 (recA pro hsdR RP4-2-Tc::Mu-Km::Tn7) (85). Agrobacterium tumefaciens LBA1010R Rifr and Paracoccus aminovorans JCM7685 Rifr were kindly provided by D. Bartosik (University of Warsaw, Poland). Pseudomonas putida KT2442 Rifr was kindly provided by C. M. Thomas (University of Birmingham, United Kingdom). Cupriavidus necator (previously Ralstonia eutropha) 7MP228r Rifr was kindly provided by K. Smalla (Research Institute for Cultivated Plants, Germany).
Bacteria were grown in L broth (86) at 37°C (E. coli) or 28°C (A. tumefaciens, C. necator, P. aminovorans, and P. putida). L broth or L agar (L broth with 1.5% [wt/vol] agar) were supplemented with kanamycin (Km) (50 μg ml−1 for E. coli and 20 μg ml−1 for P. aminovorans and P. putida), chloramphenicol (Cm) (10 μg ml−1 for E. coli, 50 μg ml−1 for A. tumefaciens, and 150 μg ml−1 for C. necator), and rifampin (Rif) (100 μg ml−1). The L agar used for blue/white screening contained 40 μg ml−1 X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside).
Plasmid DNA isolation and analysis and DNA amplification and manipulation.
Plasmids used and constructed in this study are listed in Table 3 (those referred to in the main text) and in Table S1 in the supplemental material (intermediate constructs). Plasmid DNA manipulations were carried out by standard procedures (87) and in accordance with the manufacturers’ instructions. Standard PCRs (88) were performed with appropriate pairs of primers listed in Table 4 and the RA3 DNA template. Complementary oligonucleotides corresponding to the putative transcriptional terminator sequences or introducing restriction sites were annealed by heating to 95°C, slowly cooled, and cloned into appropriately digested plasmids. All new plasmid inserts were verified using dye terminator sequencing at the Laboratory of DNA Sequencing and Oligonucleotide Synthesis, Institute of Biochemistry and Biophysics, Polish Academy of Science.
TABLE 3.
Plasmids used and constructed in this study that are mentioned in the text
| Plasmid | Relevant features or description (reference)a |
|---|---|
| pABB32 | Mini-RK2 derivative, cat (Cmr) klcApRA3-xylE lacO-MCS-lacO (38) |
| pGBT70 | oriSC101 Kmr trfAp-1RK2-xylE, here used as a terminator-probe vector (36) |
| pPT01 | oriSC101 Kmr, promoterless xylE, promoter-probe vector (40) |
| pUC18 | oriMB1 bla (Apr), cloning vector (97) |
| RA3 | IncU Cmr Smr Sur (24) |
| pESB2.88 | pUC18 with ′korA-incC, PCR fragment amplified on RA3 template with primers 45 and 26 inserted between SphI and SalI restriction sites of pUC18 (RA3 coordinates 6288–7114) |
| pESB13.76 | pPT01 with incCp-1-xylE, PCR fragment amplified on RA3 template with primers 44 and 24 inserted between PaeI and BamHI restriction sites of pPT01 (RA3 coordinates 6036–6622) |
| pESB13.77 | pPT01 with incCp-2-xylE, PCR fragment amplified on RA3 template with primers 45 and 24 inserted between PaeI and BamHI restriction sites of pPT01 (RA3 coordinates 6288–6622) |
| pESB13.79 | pPT01 with incCp-4-xylE, PCR fragment amplified on RA3 template with primers 44 and 46 inserted between PaeI and BamHI restriction sites of pPT01 (RA3 coordinates 6036–6343) |
| pESB36 | pABB32 derivative with oriTRK2 repAp-lacZ lacIq-tacp-korBRK2, Cmr Kmr, unstable, RK2 mobilizable vectorb |
| pESB36.29 | pESB36 with the synthetic RA3 partition cassette (korAp-korA-incC-korB-orf11-mpR-parS)b |
| pESB36.35 | pESB36 with the synthetic RA3 WT stability module (orf02p-orf02-klcAp-klcA-orf04-korCp-korC-kfrC-kfrAp-kfrA-korAp-korA-incC-korB-orf11-mpR-parS)b |
| pESB36.38 | pESB36 with the synthetic RA3 ΔkorAp stability module (orf02p-orf02-klcAp-klcA-orf04-korCp-korC-kfrC-kfrAp-kfrA-korA-incC-korB-orf11-mpR-parS)b |
| pESB36.40 | pESB36 with the synthetic RA3 Δ(orf02p-orf02) stability module (′orf02-klcAp-klcA-orf04-korCp-korC-kfrC-kfrAp-kfrA-korAp-korA-incC-korB-orf11-mpR-parS)b |
| pESB36.41 | pESB36 with the synthetic RA3 Δ(klcAp-klcA) stability module (orf02p-orf02-orf04-korCp-korC-kfrC-kfrAp-kfrA-korAp-korA-incC-korB-orf11-mpR-parS)b |
| pESB36.44 | pESB36 with the synthetic RA3 Δ(kfrAp-kfrA) stability module (orf02p-orf02-klcAp-klcA-orf04-korCp-korC-kfrC-korAp-korA-incC-korB-orf11-mpR-parS)b |
| pMWB5.15 | pGBT70 with trfAp-1RK2-repBtc-xylE, PCR fragment amplified on RA3 template with primers 34 and 35 inserted between KpnI and NcoI sites of pGBT70 (RA3 coordinates 1353–1509) |
| pMWB5.16 | pGBT70 with trfAp-1RK2-klcAt-xylE, annealed oligonucleotides 36 and 37 inserted between KpnI and NcoI sites of pGBT70 (RA3 coordinates 3061–3110) |
| pMWB5.17 | pGBT70 with trfAp-1RK2-kfrCt-xylE, annealed oligonucleotides 38 and 39 inserted between KpnI and NcoI sites of pGBT70 (RA3 coordinates 4771–4820) |
| pMWB5.18 | pGBT70 with trfAp-1RK2-kfrAt-xylE, annealed oligonucleotides 40 and 41 inserted between KpnI and NcoI sites of pGBT70 (RA3 coordinates 5981–6030) |
| pMWB5.19 | pGBT70 with trfAp-1RK2-orf11t-xylE, annealed oligonucleotides 42 and 43 inserted between KpnI and NcoI sites of pGBT70 (RA3 coordinates 8701–8750) |
MCS, multiple-cloning site. ′, truncated ORF (position of the symbol indicates 5′ or 3′ end deletion). A list of primers is presented in Table 4.
A list of the intermediate constructs and a detailed description of plasmid construction are presented in the supplemental material.
repBt, putative Rho-independent transcriptional terminator downstream of the indicated gene.
TABLE 4.
Oligonucleotides used in this study
| Category and no. | Name | Sequence (5′→3′)a |
|---|---|---|
| Primers used in RT-PCR and 5′RACE | ||
| 1 | 10G | cggaattcGGTGGCCCATTTCGTACGTA |
| 2 | orf02pR | cgggatccCGATCACGCTCCCAGGTCAA |
| 3 | Orf02F | cagaattcATGATCCACACAGCTAACCG |
| 4 | SalOrf02 | cgcgtcgacATAGGCCAAATCGGCCTACT |
| 5 | klcApL | gcgcatgcGGGAGCGTGATCGTTACGGT |
| 6 | klcApR | gcggatccATTGCAGCCATACGGCGAGG |
| 7 | EcoklcAL | gcgaattcATGATGCACACAGAACTTAATC |
| 8 | SalklcAR | cgcgtcgacCTAGTCTATTGCGGCCAAGA |
| 9 | KorCpF | cgcagatctGAAATGGTGCCCCTGGTATG |
| 10 | PkorCp | gcggatccCAATCTTCAGCAAACGGCCT |
| 11 | korCRA3L | gcgaattcATGATTAGACCTGAAACGCT |
| 12 | korCRA3R | cggtcgacTTATGTTCGGTCATGGTTTC |
| 13 | KFRCFL | gcaagcttgGAAttCATGACCGAACATAAGGCCGA |
| 14 | KFRCFP | aacccggggCCGCTCTAGATCGTCTTCAT |
| 15 | prkfrA1 | gcggatccgcatgcCTCGCTGATAACCTGGCCCT |
| 16 | prkfrA2 | gcggatccCTCGCGCACCTGCTCATTTG |
| 17 | Ia | cgccaattgagatctgaatccATGACCATGATTAAGCCTG |
| 18 | IIb | gcggtcgactacgtaAACGACTCGATAGACTGG |
| 19 | korApL | gcgaatccAGTTGACTAAGCCAACAGCG |
| 20 | korApR | cgggatCCTTCCTAACCAAAGCCGCTAC |
| 21 | korAL | cggaattcATGGCGCTGACCAACGAGGA |
| 22 | korAR | cggtcgacGTAGTGGACTTGCCAACGCC |
| 23 | IncBF1 | cggcagcatgCCCAATAGTTAAGGCCATCG |
| 24 | IncCprP | gcggatccGTTCCATATCAGCCATC |
| 25 | incCL | cggaatccATGAAAATTGTTACTGTATC |
| 26 | incCRA3R | cggtcgacTTTACCACTCATTCAGCCAC |
| 27 | korBL | cggaattcATGAGTGGTAAAGGGGCAGA |
| 28 | korBGSP2 | TCTGCCGTTGTCAGTTCGTC |
| 29 | korBR | cggtcgacCGAACAACAACAGCACTCCA |
| 30 | OriRA3NF | cggaatccacatgtAGTTAGGGGAAGCCGACGAG |
| 31 | OriRA3D | cgcgtcgacacatgtCGATAGCTCTTTGCCATTAAC |
| 32 | klcAGSP2 | AATCCCGCAAGCTGTGATAC |
| 33 | kfrCGSP2 | AGCTCCGCTTTTGCCCATTC |
| AAP | GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG | |
| AUAP | GGCCACGCGTCGACTAGTAC | |
| Primers used for cloning | ||
| 34 | TERREPBF | ccgaattcggtaccACAGGCGGCTAGGTGTAAAG |
| 35 | TERREPBR | ccgtcgacacatgtAGGTGGAAGATTAAGCGGGT |
| 36 | TERKLCAF | cAGCAACAGAAAGCCCCGGTGATGGCCGGGGCTCTTAAAGGAGGTGCATAGa |
| 37 | TERKLCAR | catgtCTATGCACCTCCTTTAAGAGCCCCGGCCATCACCGGGGCTTTCTGTTGCTggtac |
| 38 | TERKFRCF | cTGTCACCGACCCCGGCCATTGCGCCGGGTTTTTTTTGTTCGTTTTTCACTa |
| 39 | TERKFRCR | catgtAGTGAAAAACGAACAAAAAAAACCCGGCGCAATGGCCGGGGTCGGTGACAggtac |
| 40 | TERKFRAF | cTAAAGTGTTATCTTTAAACGGCGGCCAAAATGGCCGCCTGTGATAGGGTGa |
| 41 | TERKFRAR | catgtCACCCTATCACAGGCGGCCATTTTGGCCGCCGTTTAAAGATAACACTTTAggtac |
| 42 | TERORF11F | cCGAACAAGGGGGCCAAAATCGGCCCCCTTACTTTTGGCGTTTAATCTAGCa |
| 43 | TERORF11R | catgtGCTAGATTAAACGCCAAAAGTAAGGGGGCCGTTTTGGCCCCCTTGTTCGggtac |
| 44 | IncCprL | cgcgcgcatgcATGGCGCTGACCAACGAG |
| 45 | IncCBF1 | cggcagcatgcCCAATAGTTAAGGCCATCG |
| 46 | IncCAR | gcggattcTTTGGTTTATTAGCCTGT |
| 47 | OriTG | acggtcgacacatgtCTGGTTGGCTTGGTTTCATC |
| 48 | OriTD | cgGAATTCACATGTTTGCCAAAGGGTTCGTGTAG |
| 49 | ODGSN | accatggtCATG |
| 50 | Ant5 | cgggatccTAGCTGCTGCCAGGATAAAC |
| 51 | repAprF | gcgaattcagatcttGCGGGCCTGATCTATTGTTG |
| 52 | Oligo1G | aACTAAGCCAACAGCGAAACGCCAACAGAAAAAAGACGCAGCCGACACCAAAGAGTAAgatctCTGCA |
| 53 | Oligo1D | GagatcTTACTCTTTGGTGTCGGCTGCGTCTTTTTTCTGTTGGCGTTTCGCTGTTGGCTTAGTt |
| 54 | 3G | cggaattcCGCGACTCGCTGATAACCTG |
| 55 | 3D | cgctgcagcaattgggaTCCCTGTATTGTATGTA |
| 56 | 4D | cgctgcagcaattgggatccctTACAATACAACGGAGTGA |
| 57 | 7G | cgagaattcGTGCCCCTGGTAT |
| 58 | 7D | cggGATCcGCATTTGAGTTTGTACGACC |
| 59 | 8G | cggaattcTCAATtCCGGAGAACTCCGA |
| 60 | 8DNOWY2 | cggtcgacGGGGCACCAaTTgTTTTTCTA |
| 61 | 9GNOWY | cggaattcctcgagGCAGCCTGGAGCTcAATAAA |
| 62 | 9D | cgctgcagTCGGAGTTCaCCGGTATTGA |
| 63 | 10D | cgTTTATTgAGCTCCAGGCTGC |
| 64 | ORF02R | gcgtcgactccggAATAGGCCAAATCGGCCTACT |
| 65 | 1A | gcgaattCGTTaACCATCGACGGTGCCCCGATTG |
| 66 | 2B | gcgtcgACAGtACTCCAACGGCAACAGCAGC |
| 67 | 3A | gcgaattcAGcGCTGTTGTTGTTCGCGGCCTATC |
| 68 | 3B | gcgtcgaccccgGGTGCAATTTTAGCACA |
| 69 | 6A | gcgaattcagatctATAGGGTGAAGGTCATGGCG |
| 70 | 6B | gcgtcgaccaattgACCCCCATCATTCAGCCACCCCCATTT |
| 71 | 7A | gcgaattcagatctGACACCAAAGAGTAACCCC |
| Primers used in qPCR analysisb | ||
| 72 | orf02F | ATGATCCACACAGCTAACCG |
| 73 | orf02R | TAAAACAGACCGACGAGACG |
| 74 | klcAF | TGCAATGCCTCGCCATGTAG |
| 75 | klcAR | CGTTCGACAGCTCCCACATAAG |
| 76 | korCF | GGTGTTGGAGCTGATTAGGC |
| 77 | korCR | GAAGGTTCGGGTTCCCTTTC |
| 78 | kfrCF | CCTGTCCTGGCTGTTGAGTC |
| 89 | kfrCR | TGTACGACCGACCTTTTCCG |
| 80 | kfrAF | ATCAGGAGCGTGATGAAGCC |
| 81 | kfrAR | TCAACCTCTGAAGCCAAGCC |
| 82 | korAF | AACGAGGAATTTGAATCAGCAC |
| 83 | korAR | CTTAGGCCGGACTCTTTCAC |
| 84 | incCF | ACGAAGACCCTTACCGTATCC |
| 85 | incCR | CCCGTTCCATATCAGCCATC |
| 86 | korBF1 | TCCGTTCAAGCCTTGGCTATC |
| 87 | korBR1 | GTGCTCGGGTTCTTCAGGTC |
| 88 | orf11F | TTGTTGTTCGCGGCCTATCC |
| 89 | orf11R | AACGTCGCCTGGTAAAAGCTG |
| 90 | CysGF | TTGTCGGCGGTGGTGATGTC |
| 91 | CysGR | ATGCGGTGAACTGTGGAATAAACG |
| 92 | PArefF7 | CCAAGCTGGCTGTCCTCTTC |
| 93 | PArefR7 | CCGAAGAACTGGCCGAAAAG |
| 94 | AgrF | TAGGTGTCGGCAATGGTGTC |
| 95 | AgrR | ACGAGGATGTGACTGACGTG |
| 96 | RalstFP | GCCTGCACCACCTTGTCTTC |
| 97 | RalstRP | TGTGGATGGTGACCTGGATCT |
| 98 | PutRefF | TCCGGAGCACTCTCGAATAC |
| 99 | PutRefR | CGCAACAGCAGTCTCGTATC |
| 100 | EcCysGzF | GCATTAGCGTTTATTCCACAG |
| 101 | EcCysGzR | GAGAAGGCTTTCATCAAATGG |
| 102 | pEStfAzF | CGATCACCTTCACGTTCTAC |
| 103 | pEStfAzR | CGGCCTTCGTGTAATACC |
Restriction enzyme recognition sites or overhangs are underlined, other sequences noncomplementary to the template are shown in lowercase, and an additional Shine-Dalgarno sequence is shown in italic.
Primers used in qPCR for the reference genes (90 to 99) are as follows: primers 90/91, cysG of E. coli; primers 92/93, rpoD of P. aminovorans; primers 94/95, rpoD of A. tumefaciens; primers 96/97, gyrB of C. necator; primers 98/99, rpoD of P. putida. Primers used in qPCR for plasmid copy determination (100 to 103) are as follows: primers 100/101, reference chromosomal gene cysG of E. coli; primers 102/103, plasmid gene trfA.
Plasmid construction.
The medium-copy-number promoter-probe vector pPT01, based on the pSC101 replicon (40), was used to monitor the promoter activity of the cloned DNA fragments. PCR products were cut with BamHI and PaeI and ligated into pPT01 upstream of the promoterless xylE cassette. To test the activity of putative transcriptional terminators, pGBT70 trfAp-1-xylE (36), a pPT01 derivative, was used. The point mutation in the −10 box of a very strong trfAp of RK2 (trfAp-1) lowers its transcriptional activity more than 10-fold, making it suitable for assays of weak transcription termination signals (37). PCR-amplified fragments or double-stranded oligonucleotides corresponding to the putative transcriptional terminators/attenuators were inserted between KpnI and NcoI restriction sites in the test vector to separate trfAp-1 from the xylE cassette. Catechol-2,3-dioxygenase (XylE) activity assays were indicative of functionality of the inserts in modulation of xylE transcription.
Previously constructed pABB32 (38), an unstable broad-host-range (BHR) test vector based on the RK2 minireplicon from the IncP-1α incompatibility group, was further modified to extend its use in various bacterial hosts. First, oriTRK2 was PCR amplified on the RK2 template and cloned as a PscI restriction fragment into the unique NcoI cleavage site of pABB32 to facilitate conjugative mobilization of the obtained pESB29 to chosen species, with the use of the RK2 conjugative system integrated into the E. coli S17-1 genome. Next, the lacIq tacp-korBRK2 cassette was inserted between the BamHI and NruI restriction sites of pESB29 to further downregulate the trfA gene expression and to decrease the copy number of the resulting pESB33 construct. Subsequently, the BamHI-PscI restriction fragment of pESB33, carrying the klcApRA3-xylE cassette, was replaced by the repApRA3-lacZ transcriptional fusion for easy monitoring of the resulting pESB35 plasmid segregation in an even broader range of hosts (white/blue colonies on L agar with X-Gal). Finally, the kanamycin resistance cassette (aphA1) was cloned into the MunI restriction site of pESB35 next to cat (Cmr) to give pESB36.
The synthetic RA3 stability module was constructed in two pieces (part 1, orf02p-orf02-klcAp-klcA-orf04-korCp-korC-kfrC-kfrAp-kfrA; part 2, korAp-korA-incC-korB-orf11-mpR-parS) by a multistep procedure of joining restriction and PCR-amplified fragments in the high-copy-number vectors. This strategy, described in detail in the supplemental material, allowed for the construction of various deletion derivatives deprived of particular promoter regions and ORFs. After thorough verification of the DNA sequence, especially at junction points, variants of the RA3 stability module were recloned into the BHR unstable vector pESB36 and used in RT-qPCR assays of transcriptional activities as well as in the plasmid stable maintenance experiments.
Bacterial transformation.
Competent cells of E. coli were prepared by the standard CaCl2 method (87).
Conjugation procedure.
E. coli strain S17-1 was transformed with pESB36, pESB36.35, and its deletion variants, and such transformants were used as donors in conjugation with the following recipient strains: A. tumefaciens LBA1010R Rifr, P. aminovorans JCM7685 Rifr, P. putida KT2442 Rifr, or C. necator 7MP228r Rifr. Cells from the stationary-phase cultures of recipients and donors (grown under antibiotic selection) were washed twice with L broth and resuspended in the initial volume of the medium. Aliquots (100 μl) of donor and recipient were mixed, spotted on an L agar plate, and incubated for approximately 24 h at 28°C. Cells were suspended in 2 ml of L broth, and 100 μl of serial dilutions was plated on L agar with rifampin, X-Gal, and the appropriate selective antibiotic and incubated at 28°C.
RNA isolation and analysis.
(i) RT reaction followed by PCR. An overnight E. coli DH5α(RA3) culture was diluted 1:100 into fresh L broth supplemented with chloramphenicol and propagated with shaking until the optical density at 600 nm (OD600) reached 0.4 to 0.6. Two-milliliter samples were mixed with 4 ml of RNAprotect Bacteria reagent (Qiagen). Total RNA was isolated using the RNeasy minikit (Qiagen) according to the manufacturer’s instruction and treated with the Turbo DNase kit (Ambion) to remove DNA contamination. The control PCRs were conducted on purified RNA to ensure lack of a DNA template. The RNA concentration was estimated using the NanoDrop ND-1000 spectrophotometer (Thermo Scientific). The integrity and overall quality of RNA preparation were assessed by native agarose gel electrophoresis (87) (see Fig. S1 in the supplemental material). cDNA synthesis and purification were performed with the 5′RACE System for Rapid Amplification of cDNA Ends, version 2.0 (Invitrogen). Briefly, 4 μg of total RNA was used per reaction with SuperScript II reverse transcriptase (RT) and primer 8, 14, or 29 (Table 4). The bulk of the RNA was removed with a mixture of RNases T1 and H. cDNA was then used as a template for PCRs with appropriate pairs of primers. PCR products were analyzed by agarose gel electrophoresis, and gels were stained with ethidium bromide and photographed.
(ii) RT-qPCR analysis. Three independent cultures of each analyzed strain, i.e., E. coli EC1250, A. tumefaciens LBA1010R Rifr, P. aminovorans JCM7685 Rifr, P. putida KT2442 Rifr, or C. necator 7MP228r Rifr carrying pESB36.35 (WT RA3 stability module) or its derivatives (pESB36.29, pESB36.38, pESB36.40, pESB36.41, or pESB36.44), were grown until the stationary phase of growth at 37°C (E. coli) or 28°C (other strains) in L broth supplemented with appropriate antibiotics. Cultures were diluted 1:100 into fresh L broth with antibiotic selection and propagated to an OD600 of 0.4 to 0.6. Total RNA was isolated from logarithmic-phase cultures and prepared as described above. The first-strand cDNA was synthesized with the TranScriba kit (A&A Biotechnology) using 1 to 3 μg of total RNA per reaction with random hexamer primers. RT-qPCRs were performed in the LightCycler 480 system (Roche Life Sciences, Penzberg, Germany) using Hot FIREPol EvaGreen qPCR Mix Plus (Solis Biodyne) according to the manufacturer’s instructions. The reactions were carried out with 1 μl of 6-times-diluted cDNA in a total volume of 19 μl, and three technical replicates were done for each gene/primer combination. Primers used to amplify the reference and target genes were checked for specificity and efficiency (only primers with amplification factor between 1.95 and 2 were used). Target gene expression was normalized to the reference gene, i.e., cysG (E. coli) (39), rpoD (P. putida, P. aminovorans, and A. tumefaciens) (89), or gyrB (C. necator) (89, 90), and calculated using the Pfaffl method (91). The mean values with standard deviations from three independent biological samples for each strain/plasmid combination were reported.
(iii) 5′RACE method. The 5′ rapid amplification of cDNA ends (5′RACE) technique (35) was done using the 5′RACE System for Rapid Amplification of cDNA Ends, version 2.0 (Invitrogen), as outlined in the manufacturer’s protocol.
(a) Identification of multiple transcripts for ORFs in the RA3 stability module. Total RNA from E. coli DH5α(RA3) and cDNAs (with a gene-specific [GSP1] primer [8, 14, or 29]) were prepared as described above for the RT-PCR technique. Next, homopolymeric tails were added to the 3′ ends of the cDNAs using terminal deoxynucleotidyl transferase (TdT) and dCTP. The tailed cDNA was PCR amplified with the abridged anchor primer (AAP), containing a run of Gs at the 3′ end, and an appropriate nested (GSP2) primer (32, 33, or 26). To increase the sensitivity of detection, the obtained PCR products were then used as templates in subsequent reactions with appropriate pairs of nested primers homologous to the studied sequence (3/4, 7/8, 11/12, 13/14, 13/33, 17/18, 21/22, and 25/26). Following PCRs, products were analyzed by agarose gel electrophoresis, and gels were stained with ethidium bromide and photographed.
(b) Identification of transcription start sites of incC. Total RNA isolated from E. coli DH5α(RA3) was primed with primer 29 and used for cDNA synthesis. cDNAs were 3′ dC tailed as described above and used as templates in PCRs with primers AAP and 26 (Fig. 2J). The primary PCR products then were reamplified using the abridged universal amplification primer (AUAP), which is identical to the 5′ part of AAP, paired with primer 24 (Fig. 2A). Following amplification, 5′RACE products were separated by agarose gel electrophoresis, extracted from the gel using the Gel-out kit (A&A Biotechnology), and sequenced.
Determination of catechol 2,3-dioxygenase (XylE) activity.
Overnight cultures of E. coli C600K transformants of the promoter-probe (pPT01) or terminator-probe (pGBT70) vectors and their derivatives were diluted 1:50 into fresh L broth supplemented with an appropriate antibiotic and grown to an OD600 of 0.5 to 0.8. Catechol 2,3-dioxygenase activity was assayed spectrophotometrically in cleared supernatants of sonicated cells as described previously (92). The reaction was initiated by addition of catechol (final concentration, 15 μM). One unit of catechol 2,3-oxygenase activity is defined as the amount of enzyme needed to convert 1 μmol of catechol to 2-hydroxymuconic semialdehyde per mg of protein in 1 min. Protein concentration was determined using the Bradford method (93). Experiments were performed in triplicate, and the mean values with standard deviations were reported.
Determination of plasmid stability.
Three independent cultures of each analyzed strain, i.e., E. coli EC1250, A. tumefaciens LBA1010R Rifr, P. aminovorans JCM7685 Rifr, P. putida KT2442 Rifr, or C. necator 7MP228r Rifr carrying pESB36, pESB36.35, pESB36.29, pESB36.38, pESB36.40 pESB36.41, or pESB36.44, were cultivated until the stationary phase of growth at 37°C (E. coli) or 28°C (other strains) in L broth supplemented with an appropriate antibiotic. Subsequently, the cultures were diluted 105-fold into fresh L broth (without antibiotic selection) and grown for up to 20 generations as described above. The procedure was then repeated twice (after every 20 generations) until at least 60 generations of growth without selection. In parallel, with every passage step, 100-μl aliquots of the 10−6 dilution of cultures were plated on L agar supplemented with 40 μg ml−1 X-Gal and incubated at 28°C or 37°C to obtain approximately 100 to 350 colonies. Plasmid retention was calculated as a percentage of blue colonies. The plasmid loss rate (LR) per generation (%) was calculated using the formula (, where n is the number of generations, Fi is the fraction of cells containing plasmid at the initial time point, and Ff is the fraction of cells containing plasmid at the final time point. The stability index (SI) for each construct was calculated as the ratio of the rate of loss of pESB36 to the rate of loss of the vector with the RA3 stability module variant in each host (94).
Plasmid copy number.
The copy number of pESB36 and pESB36.35 in various hosts was estimated by real-time qPCR. E. coli EC1250, A. tumefaciens LBA1010R Rifr, P. aminovorans JCM7685 Rifr, P. putida KT2442 Rifr, and C. necator 7MP228r Rifr carrying pESB36 or pESB36.35 were grown on L broth with a selective antibiotic to the stationary phase. Total bacterial DNA was extracted using a modification of the method of Chen and Kuo (95, 96). Briefly, cells were collected from 5 ml of the cultures by centrifugation and suspended in 400 μl of TESO lysis buffer (40 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0], 1% [wt/vol] SDS, 20 mM sodium acetate) and 132 μl of 5 M NaCl. The samples were vortexed for 1 min and then centrifuged at 4°C for 10 min at 16,000 × g. One volume of phenol-chloroform (1:1 [vol/vol]) was added to the supernatants, and samples were vortexed for approximately 1 min and centrifuged as before. The extraction procedure was repeated for the upper phase, with an equal volume of chloroform. Subsequently, the aqueous fraction was collected, and DNA was precipitated with 2 volumes of 98% ethanol (87). Fifty and 100 nanograms of each DNA template were used in qPCRs with 5× Hot FIREPol EvaGreen qPCR Mix Plus (Solis Biodyne), and reactions were carried out according to the manufacturer’s instructions in the LightCycler 480 Instrument II (Roche). Single-copy genes on the chromosomes were chosen as the reference genes: cysG for E. coli, rpoD for P. putida, P. aminovorans, and A. tumefaciens, and gyrB for C. necator. trfA was used as a plasmid target for pESB36 and pESB36.35 (pairs of primers are listed in Table 4). The primers were checked for specificity and efficiency (only primers with an amplification factor between 1.95 and 2 were used). All qPCRs were done in triplicates. The PCN, defined as the number of plasmid amplicons relatively to the number of chromosome amplicons, was calculated considering the amplification efficiencies of the primers used (95, 96). The average results of at least four biological replicates with standard deviation were reported.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by Polish National Science Centre grants 2011/03/B/NZ1/06540 and 2015/17/B/NZ2/01160 to G.J.-B. (in the latter case as a member of consortium).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Gogarten JP, Doolittle WF, Lawrence JG. 2002. Prokaryotic evolution in light of gene transfer. Mol Biol Evol 19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- 2.Jain R, Rivera MC, Moore JE, Lake JA. 2003. Horizontal gene transfer accelerates genome innovation and evolution. Mol Biol Evol 20:1598–1602. doi: 10.1093/molbev/msg154. [DOI] [PubMed] [Google Scholar]
- 3.Koonin EV, Wolf YI. 2008. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res 36:6688–6719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jain A, Srivastava P. 2013. Broad host range plasmids. FEMS Microbiol Lett 348:87–96. doi: 10.1111/1574-6968.12241. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki H, Yano H, Brown CJ, Top EM. 2010. Predicting plasmid promiscuity based on genomic signature. J Bacteriol 192:6045–6055. doi: 10.1128/JB.00277-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas CM, Smith CA. 1987. Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Annu Rev Microbiol 41:77–101. doi: 10.1146/annurev.mi.41.100187.000453. [DOI] [PubMed] [Google Scholar]
- 7.De Gelder L, Ponciano JM, Joyce P, Top EM. 2007. Stability of a promiscuous plasmid in different hosts: no guarantee for a long-term relationship. Microbiology 153:452–463. doi: 10.1099/mic.0.2006/001784-0. [DOI] [PubMed] [Google Scholar]
- 8.Caspi R, Pacek M, Consiglieri G, Helinski DR, Toukdarian A, Konieczny I. 2001. A broad host range replicon with different requirements for replication initiation in three bacterial species. EMBO J 20:3262–3271. doi: 10.1093/emboj/20.12.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doran KS, Helinski DR, Konieczny I. 1999. Host-dependent requirement for specific DnaA boxes for plasmid RK2 replication. Mol Microbiol 33:490–498. doi: 10.1046/j.1365-2958.1999.01491.x. [DOI] [PubMed] [Google Scholar]
- 10.Fernández-Tresguerres ME, Martín M, García de Viedma D, Giraldo R, Díaz-Orejas R. 1995. Host growth temperature and a conservative amino acid substitution in the replication protein of pPS10 influence plasmid host range. J Bacteriol 177:4377–4384. doi: 10.1128/jb.177.15.4377-4384.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maestro B, Sanz JM, Faelen M, Couturier M, Díaz-Orejas R, Fernández-Tresguerres E. 2002. Modulation of pPS10 host range by DnaA. Mol Microbiol 46:223–234. doi: 10.1046/j.1365-2958.2002.03155.x. [DOI] [PubMed] [Google Scholar]
- 12.Maestro B, Sanz JM, Díaz-Orejas R, Fernández-Tresguerres E. 2003. Modulation of pPS10 host range by plasmid-encoded RepA initiator protein. J Bacteriol 185:1367–1375. doi: 10.1128/jb.185.4.1367-1375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pagotto F, Dillon J-A. 2001. Multiple origins and replication proteins influence biological properties of β-lactamase-producing plasmids from Neisseria gonorrhoeae. J Bacteriol 183:5472–5481. doi: 10.1128/JB.183.19.5472-5481.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakai H, Komano T. 1996. DNA replication of IncQ broad-host-range plasmids in gram-negative bacteria. Biosci Biotechnol Biochem 60:377–382. doi: 10.1271/bbb.60.377. [DOI] [PubMed] [Google Scholar]
- 15.Shah DS, Cross MA, Porter D, Thomas CM. 1995. Dissection of the core and auxiliary sequences in the vegetative replication origin of promiscuous plasmid RK2. J Mol Biol 254:608–622. doi: 10.1006/jmbi.1995.0642. [DOI] [PubMed] [Google Scholar]
- 16.Scherzinger E, Ziegelin G, Bárcena M, Carazo JM, Lurz R, Lanka E. 1997. The RepA protein of plasmid RSF1010 is a replicative DNA helicase. J Biol Chem 272:30228–30236. doi: 10.1074/jbc.272.48.30228. [DOI] [PubMed] [Google Scholar]
- 17.Yano H, Deckert GE, Rogers LM, Top EM. 2012. Roles of long and short replication initiation proteins in the fate of IncP-1 plasmids. J Bacteriol 194:1533–1543. doi: 10.1128/JB.06395-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chatfield LK, Orr E, Boulnois GJ, Wilkins BM. 1982. DNA primase of plasmid ColIb is involved in conjugal DnA synthesis in donor and recipient bacteria. J Bacteriol 152:1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomis-Rüth FX, de la Cruz F, Coll M. 2002. Structure and role of coupling proteins in conjugal DNA transfer. Res Microbiol 153:199–204. doi: 10.1016/s0923-2508(02)01313-x. [DOI] [PubMed] [Google Scholar]
- 20.Meyer R. 2009. Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid 62:57–70. doi: 10.1016/j.plasmid.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas J, Hecht DW. 2007. Interaction of Bacteroides fragilis pLV22a relaxase and transfer DNA with Escherichia coli RP4-TraG coupling protein. Mol Microbiol 66:948–960. doi: 10.1111/j.1365-2958.2007.05967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts RC, Burioni R, Helinski DR. 1990. Genetic characterization of the stabilizing functions of a region of broad-host-range plasmid RK2. J Bacteriol 172:6204–6216. doi: 10.1128/jb.172.11.6204-6216.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sia EA, Roberts RC, Easter C, Helinski DR, Figurski DH. 1995. Different relative importances of the par operons and the effect of conjugal transfer on the maintenance of intact promiscuous plasmid RK2. J Bacteriol 177:2789–2797. doi: 10.1128/jb.177.10.2789-2797.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulinska A, Czeredys M, Hayes F, Jagura-Burdzy G. 2008. Genomic and functional characterization of the modular broad-host-range RA3 plasmid, the archetype of the IncU group. Appl Environ Microbiol 74:4119–4132. doi: 10.1128/AEM.00229-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van der Auwera GA, Król JE, Suzuki H, Foster B, Van Houdt R, Brown CJ, Mergeay M, Top EM. 2009. Plasmids captured in C. metallidurans CH34: defining the PromA family of broad-host-range plasmids. Antonie Van Leeuwenhoek 96:193–204. doi: 10.1007/s10482-009-9316-9. [DOI] [PubMed] [Google Scholar]
- 26.Pansegrau W, Lanka E, Barth PT, Figurski DH, Guiney DG, Haas D, Helinski DR, Schwab H, Stanisich VA, Thomas CM. 1994. Complete nucleotide sequence of Birmingham IncPα plasmids. Compilation and comparative analysis. J Mol Biol 239:623–663. doi: 10.1006/jmbi.1994.1404. [DOI] [PubMed] [Google Scholar]
- 27.Thorsted PB, Macartney DP, Akhtar P, Haines AS, Ali N, Davidson P, Stafford T, Pocklington MJ, Pansegrau W, Wilkins BM, Lanka E, Thomas CM. 1998. Complete sequence of the IncPβ plasmid R751: implications for evolution and organisation of the IncP backbone. J Mol Biol 282:969–990. doi: 10.1006/jmbi.1998.2060. [DOI] [PubMed] [Google Scholar]
- 28.Kulinska A, Cao Y, Macioszek M, Hayes F, Jagura-Burdzy G. 2011. The centromere site of the segregation cassette of broad-host-range plasmid RA3 is located at the border of the maintenance and conjugative transfer modules. Appl Environ Microbiol 77:2414–2427. doi: 10.1128/AEM.02338-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ludwiczak M, Dolowy P, Markowska A, Szarlak J, Kulinska A, Jagura-Burdzy G. 2013. Global transcriptional regulator KorC coordinates expression of three backbone modules of the broad-host-range RA3 plasmid from IncU incompatibility group. Plasmid 70:131–145. doi: 10.1016/j.plasmid.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 30.Godziszewska J, Kulińska A, Jagura-Burdzy G. 2014. MobC of conjugative RA3 plasmid from IncU group autoregulates the expression of bicistronic mobC-nic operon and stimulates conjugative transfer. BMC Microbiol 14:235. doi: 10.1186/s12866-014-0235-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kulinska A, Godziszewska J, Wojciechowska A, Ludwiczak M, Jagura-Burdzy G. 2016. Global transcriptional regulation of backbone genes in broad-host-range plasmid RA3 from the IncU group involves segregation protein KorB (ParB family). Appl Environ Microbiol 82:2320–2335. doi: 10.1128/AEM.03541-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jagura-Burdzy G, Thomas CM. 1992. kfrA gene of broad host range plasmid RK2 encodes a novel DNA-binding protein. J Mol Biol 225:651–660. doi: 10.1016/0022-2836(92)90392-w. [DOI] [PubMed] [Google Scholar]
- 33.Adamczyk M, Dolowy P, Jonczyk M, Thomas CM, Jagura-Burdzy G. 2006. The kfrA gene is the first in a tricistronic operon required for survival of IncP-1 plasmid R751. Microbiology 152:1621–1637. doi: 10.1099/mic.0.28495-0. [DOI] [PubMed] [Google Scholar]
- 34.Liang W, Xie Y, Xiong W, Tang Y, Li G, Jiang X, Lu Y. 2017. Anti-restriction protein, KlcAHS, promotes dissemination of carbapenem resistance. Front Cell Infect Microbiol 7:150. doi: 10.3389/fcimb.2017.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frohman MA, Dush MK, Martin GR. 1988. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A 85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jagura-Burdzy G, Khanim F, Smith CA, Thomas CM. 1992. Crosstalk between plasmid vegetative replication and conjugative transfer: repression of the trfA operon by trbA of broad host range plasmid RK2. Nucleic Acids Res 20:3939–3944. doi: 10.1093/nar/20.15.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bartosik AA, Markowska A, Szarlak J, Kulińska A, Jagura-Burdzy G. 2012. Novel broad-host-range vehicles for cloning and shuffling of gene cassettes. J Microbiol Methods 88:53–62. doi: 10.1016/j.mimet.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 38.Bartosik AA, Glabski K, Kulinska A, Lewicka E, Godziszewska J, Markowska A, Jagura-Burdzy G. 2016. Convenient broad-host-range unstable vectors for studying stabilization cassettes in diverse bacteria. BMC Microbiol 16:59. doi: 10.1186/s12866-016-0674-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou K, Zhou L, Lim Q‘E, Zou R, Stephanopoulos G, Too H-P. 2011. Novel reference genes for quantifying transcriptional responses of Escherichia coli to protein overexpression by quantitative PCR. BMC Mol Biol 12:18. doi: 10.1186/1471-2199-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thorsted PB, Shah DS, Macartney D, Kostelidou K, Thomas CM. 1996. Conservation of the genetic switch between replication and transfer genes of IncP plasmids but divergence of the replication functions which are major host-range determinants. Plasmid 36:95–111. doi: 10.1006/plas.1996.0037. [DOI] [PubMed] [Google Scholar]
- 41.Båga M, Göransson M, Normark S, Uhlin BE. 1988. Processed mRNA with differential stability in the regulation of E. coli pilin gene expression. Cell 52:197–206. doi: 10.1016/0092-8674(88)90508-9. [DOI] [PubMed] [Google Scholar]
- 42.Dar D, Sorek R. 2018. Extensive reshaping of bacterial operons by programmed mRNA decay. PLoS Genet 14:e1007354. doi: 10.1371/journal.pgen.1007354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.San Millan A, MacLean RC. 2017. Fitness costs of plasmids: a limit to plasmid transmission. Microbiol Spectr 5:MTBP-0016-2017. doi: 10.1128/microbiolspec.MTBP-0016-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harrison E, Brockhurst MA. 2012. Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol 20:262–267. doi: 10.1016/j.tim.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 45.De Gelder L, Williams JJ, Ponciano JM, Sota M, Top EM. 2008. Adaptive plasmid evolution results in host-range expansion of a broad-host-range plasmid. Genetics 178:2179–2190. doi: 10.1534/genetics.107.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harrison E, Guymer D, Spiers AJ, Paterson S, Brockhurst MA. 2015. Parallel compensatory evolution stabilizes plasmids across the parasitism-mutualism continuum. Curr Biol 25:2034–2039. doi: 10.1016/j.cub.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 47.Porse A, Schønning K, Munck C, Sommer M. 2016. Survival and evolution of a large multidrug resistance plasmid in new clinical bacterial hosts. Mol Biol Evol 33:2860–2873. doi: 10.1093/molbev/msw163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wong A. 2017. Epistasis and the evolution of antimicrobial resistance. Front Microbiol 8:246. doi: 10.3389/fmicb.2017.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harrison E, Dytham C, Hall JPJ, Guymer D, Spiers AJ, Paterson S, Brockhurst MA. 2016. Rapid compensatory evolution promotes the survival of conjugative plasmids. Mob Genet Elements 6:e1179074. doi: 10.1080/2159256X.2016.1179074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heuer H, Fox RE, Top EM. 2007. Frequent conjugative transfer accelerates adaptation of a broad-host-range plasmid to an unfavorable Pseudomonas putida host. FEMS Microbiol Ecol 59:738–748. doi: 10.1111/j.1574-6941.2006.00223.x. [DOI] [PubMed] [Google Scholar]
- 51.Austin S, Ziese M, Sternberg N. 1981. A novel role for site-specific recombination in maintenance of bacterial replicons. Cell 25:729–736. doi: 10.1016/0092-8674(81)90180-x. [DOI] [PubMed] [Google Scholar]
- 52.Ebersbach G, Gerdes K. 2005. Plasmid segregation mechanisms. Annu Rev Genet 39:453–479. doi: 10.1146/annurev.genet.38.072902.091252. [DOI] [PubMed] [Google Scholar]
- 53.Hayes F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301:1496–1499. doi: 10.1126/science.1088157. [DOI] [PubMed] [Google Scholar]
- 54.Díaz-Orejas R, Espinosa M, Yeo CC. 2017. The importance of the expendable: toxin–antitoxin genes in plasmids and chromosomes. Front Microbiol 8:1479. doi: 10.3389/fmicb.2017.01479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bahl MI, Hansen LH, Sørensen SJ. 2009. Persistence mechanisms of conjugative plasmids. Methods Mol Biol 532:73–102. doi: 10.1007/978-1-60327-853-9_5. [DOI] [PubMed] [Google Scholar]
- 56.Dorman MJ, Dorman CJ. 2018. Regulatory hierarchies controlling virulence gene expression in Shigella flexneri and Vibrio cholerae. Front Microbiol 9:2686. doi: 10.3389/fmicb.2018.02686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feklístov A, Sharon BD, Darst SA, Gross CA. 2014. Bacterial sigma factors: a historical, structural, and genomic perspective. Annu Rev Microbiol 68:357–376. doi: 10.1146/annurev-micro-092412-155737. [DOI] [PubMed] [Google Scholar]
- 58.Suzuki-Minakuchi C, Hirotani R, Shintani M, Takeda T, Takahashi Y, Matsui K, Vasileva D, Yun C-S, Okada K, Yamane H, Nojiri H. 2015. Effects of three different nucleoid-associated proteins encoded on IncP-7 plasmid pCAR1 on host Pseudomonas putida KT2440. Appl Environ Microbiol 81:2869–2880. doi: 10.1128/AEM.00023-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dillon SC, Dorman CJ. 2010. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol 8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 60.Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E. 2011. Linking RNA polymerase backtracking to genome instability in E. coli. Cell 146:533–543. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Merrikh H, Zhang Y, Grossman AD, Wang JD. 2012. Replication-transcription conflicts in bacteria. Nat Rev Microbiol 10:449–458. doi: 10.1038/nrmicro2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jacob F, Monod J. 1961. On the regulation of gene activity. Cold Spring Harbor Symp Quant Biol 26:193–211. doi: 10.1101/SQB.1961.026.01.024. [DOI] [PubMed] [Google Scholar]
- 63.Cho B-K, Zengler K, Qiu Y, Park YS, Knight EM, Barrett CL, Gao Y, Palsson BØ. 2009. The transcription unit architecture of the Escherichia coli genome. Nat Biotechnol 27:1043–1049. doi: 10.1038/nbt.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chou W-C, Ma Q, Yang S, Cao S, Klingeman DM, Brown SD, Xu Y. 2015. Analysis of strand-specific RNA-seq data using machine learning reveals the structures of transcription units in Clostridium thermocellum. Nucleic Acids Res 43:e67. doi: 10.1093/nar/gkv177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mao X, Ma Q, Liu B, Chen X, Zhang H, Xu Y. 2015. Revisiting operons: an analysis of the landscape of transcriptional units in E. coli. BMC Bioinformatics 16:356. doi: 10.1186/s12859-015-0805-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Conway T, Creecy JP, Maddox SM, Grissom JE, Conkle TL, Shadid TM, Teramoto J, San Miguel P, Shimada T, Ishihama A, Mori H, Wanner BL. 2014. Unprecedented high-resolution view of bacterial operon architecture revealed by RNA sequencing. mBio 5:e01442-14. doi: 10.1128/mBio.01442-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li S, Dong X, Su Z. 2013. Directional RNA-seq reveals highly complex condition-dependent transcriptomes in E. coli K12 through accurate full-length transcripts assembling. BMC Genomics 14:520. doi: 10.1186/1471-2164-14-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dar D, Sorek R. 2018. High-resolution RNA 3′-ends mapping of bacterial Rho-dependent transcripts. Nucleic Acids Res 46:6797–6805. doi: 10.1093/nar/gky274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gardner PP, Barquist L, Bateman A, Nawrocki EP, Weinberg Z. 2011. RNIE: genome-wide prediction of bacterial intrinsic terminators. Nucleic Acids Res 39:5845–5852. doi: 10.1093/nar/gkr168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hui MP, Foley PL, Belasco JG. 2014. Messenger RNA degradation in bacterial cells. Annu Rev Genet 48:537–559. doi: 10.1146/annurev-genet-120213-092340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peters JM, Mooney RA, Kuan PF, Rowland JL, Keles S, Landick R. 2009. Rho directs widespread termination of intragenic and stable RNA transcription. Proc Natl Acad Sci U S A 106:15406–15411. doi: 10.1073/pnas.0903846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Higgins CF, Smith NH. 1986. Messenger RNA processing, degradation and the control of gene expression, p 179–198. In Booth IR, Higgins CF (ed), Regulation of gene expression—25 years on. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 73.Coburn GA, Mackie GA. 1999. Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog Nucleic Acids Res Mol Biol 62:55–108. doi: 10.1016/s0079-6603(08)60505-x. [DOI] [PubMed] [Google Scholar]
- 74.Misra TK, Apirion D. 1979. RNase E, an RNA processing enzyme from Escherichia coli. J Biol Chem 254:11154–11159. [PubMed] [Google Scholar]
- 75.Mohanty BK, Kushner SR. 2018. Enzymes involved in posttranscriptional RNA metabolism in Gram-negative bacteria. Microbiol Spectr 6:RWR-0011-2017. doi: 10.1128/microbiolspec.RWR-0011-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dar D, Sorek R. 2018. Bacterial noncoding RNAs excised from within protein-coding transcripts. mBio 9:e01730-18. doi: 10.1128/mBio.01730-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thomas CM. 2000. Paradigms of plasmid organization. Mol Microbiol 37:485–491. doi: 10.1046/j.1365-2958.2000.02006.x. [DOI] [PubMed] [Google Scholar]
- 78.Adamczyk M, Jagura-Burdzy G. 2003. Spread and survival of promiscuous IncP-1 plasmids. Acta Biochim Pol 50:425–453. doi: 10.18388/abp.2003_3696. [DOI] [PubMed] [Google Scholar]
- 79.Fernandez-Lopez R, del Campo I, Revilla C, Cuevas A, de la Cruz F. 2014. Negative feedback and transcriptional overshooting in a regulatory network for horizontal gene transfer. PLoS Genet 10:e1004171. doi: 10.1371/journal.pgen.1004171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Delver EP, Belogurov AA. 1997. Organization of the leading region of IncN plasmid pKM101 (R46): a regulation controlled by CUP sequence elements. J Mol Biol 271:13–30. doi: 10.1006/jmbi.1997.1124. [DOI] [PubMed] [Google Scholar]
- 81.Godziszewska J, Moncalián G, Cabezas M, Bartosik AA, de la Cruz F, Jagura‐Burdzy G. 2016. Concerted action of NIC relaxase and auxiliary protein MobC in RA3 plasmid conjugation. Mol Microbiol 101:439–456. doi: 10.1111/mmi.13401. [DOI] [PubMed] [Google Scholar]
- 82.McKenney K, Shimatake H, Court D, Schmeissner U, Brady C, Rosenberg M. 1981. A system to study promoter and terminator signals recognized by Escherichia coli RNA polymerase. Gene Amplif Anal 2:383–415. [PubMed] [Google Scholar]
- 83.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 84.Jagura-Burdzy G, Hulanicka D. 1981. Use of gene fusions to study expression of cysB, the regulatory gene of the cysteine regulon. J Bacteriol 147:744–751. doi: 10.1128/JB.147.3.744-751.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Simon R, O'Connell M, Labes M, Pühler A. 1986. Plasmid vectors for the genetic analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol 118:640–659. doi: 10.1016/0076-6879(86)18106-7. [DOI] [PubMed] [Google Scholar]
- 86.Kahn M, Kolter R, Thomas C, Figurski D, Meyer R, Remaut E, Helinski DR. 1979. Plasmid cloning vehicles derived from plasmids ColE1, F, R6K, and RK2. Methods Enzymol 68:268–280. doi: 10.1016/0076-6879(79)68019-9. [DOI] [PubMed] [Google Scholar]
- 87.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 88.Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. 1986. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harbor Symp Quant Biol 51:263–273. doi: 10.1101/sqb.1986.051.01.032. [DOI] [PubMed] [Google Scholar]
- 89.Rocha DJP, Santos CS, Pacheco L. 2015. Bacterial reference genes for gene expression studies by RT-qPCR: survey and analysis. Antonie Van Leeuwenhoek 108:685–693. doi: 10.1007/s10482-015-0524-1. [DOI] [PubMed] [Google Scholar]
- 90.Jugder B-E, Chen Z, Ping DTT, Lebhar H, Welch J, Marquis CP. 2015. An analysis of the changes in soluble hydrogenase and global gene expression in Cupriavidus necator (Ralstonia eutropha) H16 grown in heterotrophic diauxic batch culture. Microb Cell Fact 14:42. doi: 10.1186/s12934-015-0226-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zukowski MM, Gaffney DF, Speck D, Kauffmann M, Findeli A, Wisecup A, Lecocq JP. 1983. Chromogenic identification of genetic regulatory signals in Bacillus subtilis based on expression of a cloned Pseudomonas gene. Proc Natl Acad Sci U S A 80:1101–1105. doi: 10.1073/pnas.80.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 94.Roberts RC, Helinski DR. 1992. Definition of a minimal plasmid stabilization system from the broad-host-range plasmid RK2. J Bacteriol 174:8119–8132. doi: 10.1128/jb.174.24.8119-8132.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Škulj M, Okršlar V, Jalen Š, Jevševar S, Slanc P, Štrukelj B, Menart V. 2008. Improved determination of plasmid copy number using quantitative real-time PCR for monitoring fermentation processes. Microb Cell Fact 7:6. doi: 10.1186/1475-2859-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wein T, Hülter NF, Mizrahi I, Dagan T. 2019. Emergence of plasmid stability under non-selective conditions maintains antibiotic resistance. Nat Commun 10:2595. doi: 10.1038/s41467-019-10600-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yanisch-Perron C, Vieira J, Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.