Antibiotics have been reported to induce biofilm formation in many bacteria at subinhibitory concentrations. Accordingly, it is conceivable that the MIC against drug-sensitive bacteria may promote biofilm formation of resistant bacteria. Since drug-resistant bacteria have spread, it is important to understand the behavior of resistant bacteria. Streptococcus mutans is bacitracin resistant, and the 1/8× MIC of bacitracin, which is a cell wall-targeted antibiotic, induced eDNA-dependent biofilm formation. The ΔmbrC and ΔmbrD strains, which are not resistant to bacitracin, also formed biofilms in the presence of bacitracin at 1/2× MIC, and biofilms of both the wild type and mutants promoted horizontal gene transfer. Another cell wall-targeted antibiotic, vancomycin, showed effects on biofilms and gene transfer similar to those of bacitracin. Thus, treatment with cell wall-targeted antibiotics may promote the spread of drug-resistant genes in biofilms. Therefore, the behavior of resistant bacteria in the presence of antibiotics at sub-MICs should be investigated when using antibiotics.
KEYWORDS: bacitracin, biofilms, extracellular DNA, horizontal gene transfer, mbrCD, resistant genes, rgp
ABSTRACT
Antibiotics are used to treat or prevent some types of bacterial infection. The inappropriate use of antibiotics unnecessarily promotes antibiotic resistance and increases resistant bacteria, and controlling these bacteria is difficult. While the emergence of drug-resistant bacteria is a serious problem, the behavior of drug-resistant bacteria is not fully understood. In this study, we investigated the behavior of Streptococcus mutans, a major etiological agent of dental caries that is resistant to bacitracin, which is a cell wall-targeting antibiotic, and focused on biofilm formation in the presence of bacitracin. S. mutans UA159 most strongly induced extracellular DNA (eDNA)-dependent biofilm formation in the presence of bacitracin at 1/8× MIC. The ΔmbrC and ΔmbrD mutant strains, which lack bacitracin resistance, also formed biofilms in the presence of bacitracin at 1/2× MIC. This difference between the wild type and the mutants was caused by the induction of atlA expression in the mid-log phase. We also revealed that certain rgp genes involved in the synthesis of rhamnose-glucose polysaccharide related to cell wall synthesis were downregulated by bacitracin. In addition, glucosyltransferase-I was also involved in eDNA-dependent biofilm formation. The biofilm led to increased transformation efficiencies and promoted horizontal gene transfer. Biofilms were also induced by ampicillin and vancomycin, antibiotics targeting cell wall synthesis, suggesting that cell envelope stress triggers biofilm formation. Therefore, the expression of the atlA and rgp genes is regulated by S. mutans, which forms eDNA-dependent biofilms, promoting horizontal gene transfer in response to cell envelope stress induced by sub-MICs of antibiotics.
IMPORTANCE Antibiotics have been reported to induce biofilm formation in many bacteria at subinhibitory concentrations. Accordingly, it is conceivable that the MIC against drug-sensitive bacteria may promote biofilm formation of resistant bacteria. Since drug-resistant bacteria have spread, it is important to understand the behavior of resistant bacteria. Streptococcus mutans is bacitracin resistant, and the 1/8× MIC of bacitracin, which is a cell wall-targeted antibiotic, induced eDNA-dependent biofilm formation. The ΔmbrC and ΔmbrD strains, which are not resistant to bacitracin, also formed biofilms in the presence of bacitracin at 1/2× MIC, and biofilms of both the wild type and mutants promoted horizontal gene transfer. Another cell wall-targeted antibiotic, vancomycin, showed effects on biofilms and gene transfer similar to those of bacitracin. Thus, treatment with cell wall-targeted antibiotics may promote the spread of drug-resistant genes in biofilms. Therefore, the behavior of resistant bacteria in the presence of antibiotics at sub-MICs should be investigated when using antibiotics.
INTRODUCTION
Antibiotics have contributed to human health as an effective treatment for bacterial infections. However, due to inappropriate use, bacteria have evolved resistance mechanisms to survive in the presence of antibiotics, and now, there are resistant bacteria for all antibiotics (1). Therefore, it is possible that resistant bacteria and susceptible bacteria coexist in the same bacterial flora. In this situation, if an antibiotic regulated a particular susceptible pathogenic bacterium, it would decrease the target bacterium, and resistant bacteria would become dominant; eventually, the native flora would be destroyed. In general, the destroyed flora is recovered by complex mechanisms such as bacterial interactions and bacterium-host interactions. By understanding the behavior of resistant bacteria that survive after antibiotic use, it will be possible to effectively control bacterial infections.
Streptococcus mutans, a major etiological agent of dental caries and infective endocarditis, exerts pathogenicity by forming biofilms. Biofilms are formed by polysaccharide-dependent mechanisms and polysaccharide-independent mechanisms. S. mutans has three glucosyltransferases (GtfB, GtfC, and GtfD, encoded by gtfB, gtfC, and gtfD, respectively) and a fructosyltransferase (Ftf, encoded by sacB) as polysaccharide syntheses, and these enzymes synthesize polysaccharides (glucan and fructan) from sucrose (2–5). In addition, we reported that Ftf synthesizes fructan from raffinose as a substrate (6). Among these enzymes, GtfB and GtfC are involved in insoluble glucan synthesis and closely related to the pathogenicity of S. mutans. Extracellular DNA (eDNA) is involved in polysaccharide-independent biofilm formation (7, 8). eDNA is considered to be genomic DNA (gDNA) released from dead cells and is involved in the attachment of bacteria to surfaces and adhesion between bacteria (9, 10). Accordingly, cell death is important for the production of eDNA by S. mutans. atlA encodes a major autolysin that causes autolysis (11, 12). In addition, cell death is also induced by quorum sensing via competence-stimulating peptide (CSP).
S. mutans contains bacitracin resistance genes (13). Bacitracin is an antimicrobial peptide secreted by several Bacillus species and has bactericidal activity mainly against Gram-positive bacteria (14, 15). Bacitracin is used in clinical settings, but systemic administration can cause anaphylactic reactions (16). In contrast, oral administration of bacitracin is effective for the control of vancomycin-resistant enterococcal infections, since bacitracin is poorly absorbed in the gastrointestinal tract (17). However, oral bacitracin has been designated an orphan drug for the treatment of pseudomembranous colitis caused by Clostridium difficile. Bacitracin inhibits peptidoglycan synthesis by binding to the lipid carrier C55-undecaprenyl pyrophosphate (C55-PP), which is involved in bacterial peptidoglycan synthesis (18, 19). S. mutans contains mbrABCD resistance genes (20). These genes are conserved in S. mutans and used to isolate S. mutans from other oral streptococci (13). mbrAB encodes an ABC transporter, and mbrCD encodes a two-component regulatory system. The deletion of these genes resulted in an approximately 100-fold increase in bacitracin sensitivity (20, 21). The bacitracin resistance genes mbrABCD in S. mutans are homologous to bceABRS in Bacillus subtilis, and the resistance mechanism has been investigated in B. subtilis in detail (22). bceAB, encoding an ABC transporter, is responsible for the removal of bacitracin from the bacitracin attack site, and bceRS, encoding a two-component regulatory system, induces the expression of bceAB in response to bacitracin (22, 23).
Rhamnose-glucose polysaccharide (RGP) is a component of the cell wall in S. mutans and is linked to peptidoglycan. RGP is important for fitness under stress conditions and is also involved in bacitracin resistance (20, 24). S. mutans contains rgpABCDEFGHI, genes involved in RGP synthesis. RgpG transfers N-acetylglucosamine to membrane-linked lipid carriers. RgpA, RgpB, and RgpF are required for the assembly of l-rhamnose on lipid carriers. Glucose side chains are formed by glucosyltransferase, RgpE, RgpH, and RgpI. Mature RGP is transferred by the ABC transporter RgpCD (24). In addition, the mutant strains lacking rgpA, rgpB, rgpC, and rgpD exhibited an approximately 5-fold increase in bacitracin sensitivity (20).
In recent years, it has been reported that many bacteria induce eDNA-dependent biofilm formation in the presence of antimicrobial substances at sub-MICs (25–27). In S. mutans, sub-MICs of chlorhexidine, tea polyphenols, and sodium fluoride, which are known anticaries agents, upregulated the expression of biofilm-related genes (28). We found that S. mutans induced biofilm formation in the presence of sub-MICs of bacitracin. The ΔmbrC and ΔmbrD strains, which were not resistant to bacitracin, could also form biofilms in the presence of sub-MICs of bacitracin. However, the biofilm formation mechanism of the mutant strains was different from that of the wild-type (WT) strain. In this study, we show that bacitracin at sub-MICs triggers the cell envelope stress response and induces eDNA-dependent biofilm formation via regulation of the expression of the atlA and rgp genes in S. mutans. Furthermore, we also show that the biofilm can be a potential risk for horizontal resistance gene transfer.
RESULTS
Biofilm formation in the presence of sub-MICs of bacitracin.
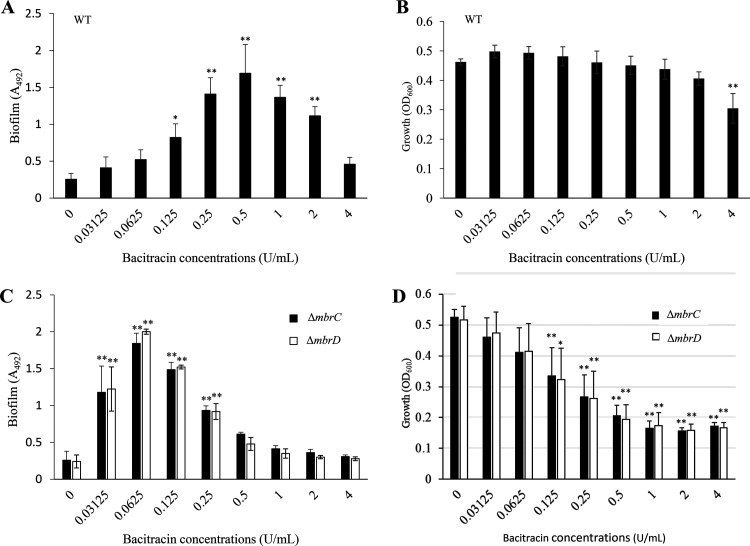
To conduct the biofilm formation assay in the presence of bacitracin at sub-MICs, we identified the MIC of bacitracin against the S. mutans UA159 WT strain by preparing a 2-fold dilution series of bacitracin in brain heart infusion (BHI) broth. In accordance with other reports (20), the MIC of bacitracin was determined to be 4 U/ml against the WT strain (Table 1). Thus, we conducted a biofilm formation assay in the presence of bacitracin at concentrations less than 4 U/ml. Since the biofilm mechanisms in S. mutans is generally divided into polysaccharide-dependent and polysaccharide-independent biofilm formation, we first examined the effect of sub-MICs of bacitracin on polysaccharide-dependent biofilm formation. In tryptic soy broth without dextrose but with sucrose (TSBs), which are conditions capable of causing polysaccharide-dependent biofilm formation, the biofilm formation levels in the presence of bacitracin concentrations from 1/32× to 1/2× MIC and in the absence of bacitracin did not differ (see Fig. S1 in the supplemental material). Since 1× MIC of bacitracin inhibited cell growth, biofilm formation was also decreased at this concentration (Fig. S1). This result showed that biofilm formation depended on whether growth was possible and that sub-MIC levels of bacitracin did not remarkably affect the biofilm. In turn, we examined the effect of bacitracin at sub-MICs on polysaccharide-independent biofilm formation in TSB with glucose (TSBg), which does not allow enough polysaccharide to be synthesized for biofilm formation, although TSB contains small amounts of substrates (sucrose and oligosaccharide) for polysaccharide synthesis (6). S. mutans could not induce biofilm formation in TSBg without bacitracin, but biofilm formation was significantly increased at 1/32× MIC to 1/2× MIC of bacitracin, and the biomass reached a maximum at 1/8× MIC of bacitracin (Fig. 1A). Additionally, no significant growth inhibition occurred in the presence of the concentrations that induced biofilm formation (Fig. 1B). It was suggested that S. mutans actively induced biofilm formation in the presence of bacitracin at concentrations that did not affect growth. Thus, in agreement with many reports on drug-sensitive bacteria (25–27), this result indicated that biofilm formation could be induced even in drug-resistant bacteria by the presence of sub-MICs of antibiotics.
TABLE 1.
MICs of bacitracin against the WT strain and the mbr mutants
| Strain | MIC (U/ml) | MBC (U/ml) |
|---|---|---|
| UA159 WT | 4 | 64 |
| ΔmbrC mutant | 0.125 | 8 |
| ΔmbrD mutant | 0.125 | 8 |
FIG 1.
Biofilm formation assay in TSBg with various concentrations of bacitracin. (A and C) Effect of bacitracin on 5-h biofilms formed by the WT (A) and mbr mutant strains (C). To evaluate the effect of bacitracin, we performed a biofilm formation assay using TSBg medium, which is a medium that does not induce biofilm formation. (B and D) Effect of bacitracin on 5-h growth in the WT (B) and mbr mutant strains (D). To investigate the relationship between biofilm induction and growth, the optical density (OD600) was measured when samples were cultured under the same conditions as biofilm formation. The data are the mean ± standard deviation (SD) of three independent experiments. The data were analyzed by one-way analysis of variance (ANOVA) and a Bonferroni’s posttest. The asterisks indicate a significant difference (**, P < 0.01; *, P < 0.05).
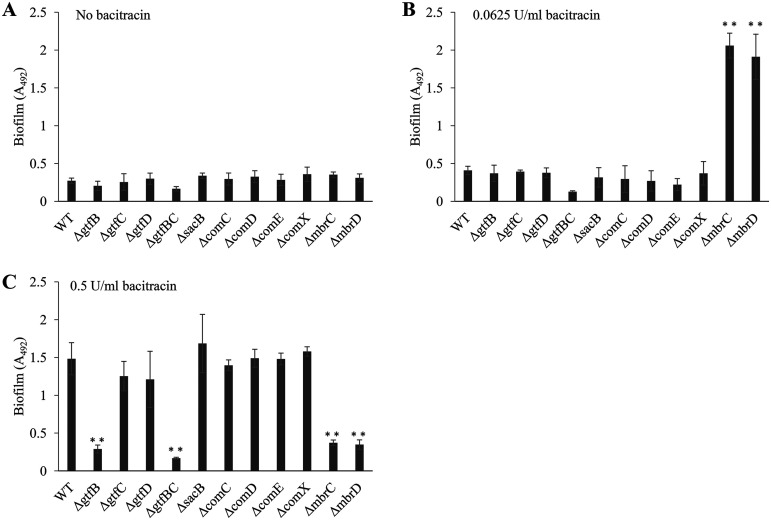
A previous report indicated that a subinhibitory concentration of β-defensin-3, which is a small defense peptide secreted by salivary glands and gingival and mucosal epithelia, induced biofilm formation in the WT strain but not in any of the bceABRS (mbrABCD) mutants that are not resistant to bacitracin (29). We considered that biofilm induction in the presence of bacitracin may be triggered by a bacitracin-specific response. We then constructed and analyzed bacitracin-sensitive mutant strains that were no longer responsive to bacitracin. mbrCD encodes a two-component regulatory system for bacitracin-specific responses. These genes are essential for bacitracin resistance, and the MIC of bacitracin decreased in strains lacking either of these genes (20, 30). To clarify whether mbrCD is involved in inducing biofilm, we conducted a biofilm formation assay using mutant strains with deletions of mbrC (ΔmbrC) and mbrD (ΔmbrD). First, we determined the MIC of bacitracin against the ΔmbrC and ΔmbrD mutants. The MIC of bacitracin against these mutants was decreased to 0.125 U/ml compared to the MIC against the WT strain (Table 1). Biofilm formation in the ΔmbrC and ΔmbrD mutant strains was induced in the presence of 1/4× MIC to 2× MIC of bacitracin, and the level of biofilm formation reached a maximum at 1/2× MIC of bacitracin (Fig. 1C). There was a difference in the biofilm formation of the strains: the WT strain most strongly induced biofilm formation at 1/8× MIC of bacitracin, whereas the ΔmbrC and ΔmbrD mutants most strongly induced biofilm formation at 1/2× MIC of bacitracin. Additionally, biofilm formation in the mbr mutant strains was induced even with bacitracin at concentrations over the MIC, although growth was significantly inhibited (Fig. 1D). This result suggested that there were some differences in the biofilm induction mechanism in the WT strain and the mbr mutant strains, not merely a shift in the biofilm-inducing concentrations due to a change in the MIC.
Analysis of the biofilm-inducing mechanism.
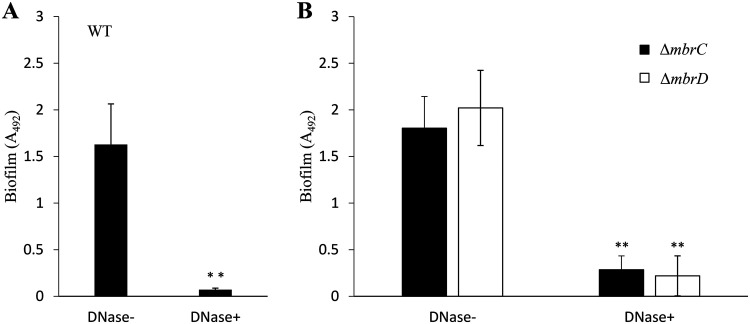
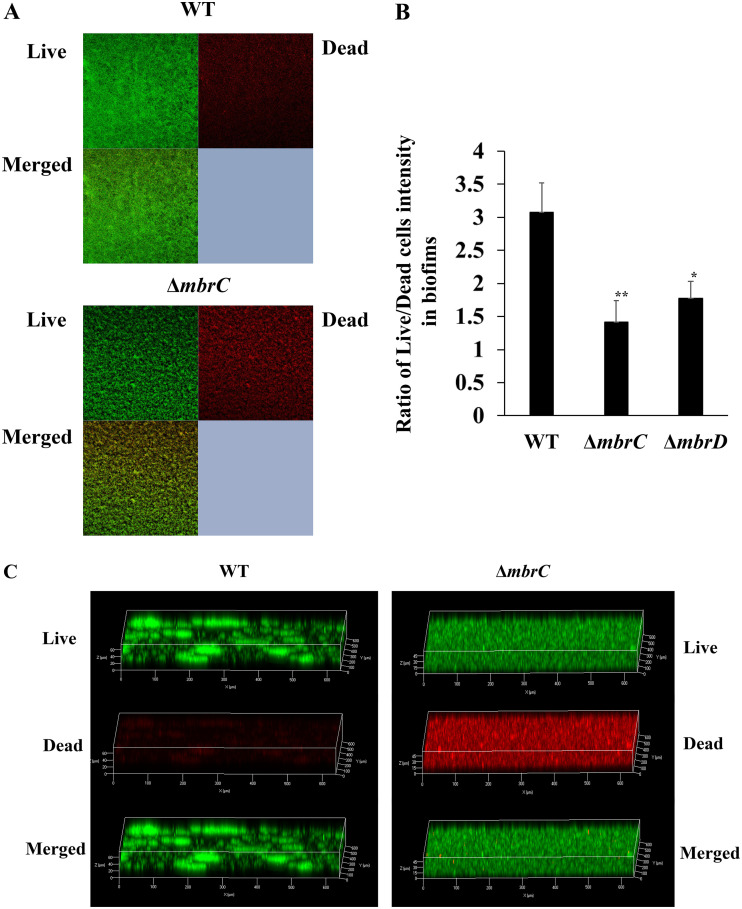
To understand the mechanism of biofilm induction in TSBg, we elucidated the biofilm formation mechanism of the WT strain in the presence of 1/8× MIC of bacitracin and the mbr mutant strains in the presence of 1/2× MIC of bacitracin, because these concentrations strongly induced biofilm formation. Since the main carbon source in TSBg is glucose, which is not available for polysaccharide synthesis, we predicted that substances other than polysaccharides are important in biofilm formation and considered the possibility that eDNA is a major component of the biofilm. We then indirectly assessed the contribution of eDNA to biofilm formation by examining the inhibition of biofilm formation when DNase I was added to the medium. When DNase I was added to the medium at 50 U/ml, the WT strain did not form biofilms (Fig. 2A). Additionally, the amount of eDNA was significantly higher in the presence of bacitracin than in the absence of bacitracin (Fig. S2). Biofilms formed by the ΔmbrC and ΔmbrD strains were also markedly reduced by the addition of DNase I (Fig. 2B). These results indicated that biofilm formation in both the WT and mbr mutant strains largely depended on eDNA. eDNA is generally considered genomic DNA released from dead cells (6, 10). Thus, it was predicted that cell lysis and release of DNA frequently occur under biofilm-inducing conditions. Confocal laser scanning microscopy (CLSM) analysis of the biofilm cells stained by the Live/Dead BacLight bacterial viability kit revealed that the WT and ΔmbrC biofilms both contained dead cells (Fig. 3A). The WT strain formed a biofilm in which the ratio of live cells to dead cells was high (Fig. 3B), and many aggregates and biofilm pieces were widely observed (Fig. 3C). On the other hand, the biofilm formed by the mutant strains had a lower ratio of live cells to dead cells than the WT strain, the cells were scattered in the biofilm, and no aggregates of cells were observed. These results showed that the WT strain actively forms a three-dimensional (3D) biofilm that is rich in live bacteria, while the mbr mutant strains form sediment-like biofilms that do not have the distinguishable 3D structure characteristics that the wild type has and are rich in dead cells.
FIG 2.
Inhibitory effect of DNase I on the biofilms. DNase I inhibition of biofilm formation by the WT strain (A) and mbrC and mbrD strains (B) is shown. To evaluate the contribution of eDNA to biofilm formation, we determined the inhibitory effect by adding 50 U/ml DNase I before incubation (DNase+) on the biofilm formations of the WT strain and mbrC and mbrD mutants induced by 0.5 U/ml (1/8× MIC for the WT) and 0.0625 U/ml (1/2× MIC for the mbr mutants) of bacitracin, respectively, and these biofilms were compared with biofilms formed without DNase I (DNase−). These data are the mean ± SD of three independent experiments. The asterisks indicate a significant difference between two groups (Student's t test; **, P < 0.01, versus DNase−).
FIG 3.
Observation of live and dead cells in the biofilm. (A and C) Two-dimensional (A) and three-dimensional (C) images of biofilm formed by the WT strain in TSBg with 0.5 U/ml of bacitracin (1/8× MIC for the WT) and by the ΔmbrC mutant strain in TSBg with 0.0625 U/ml of bacitracin (1/2× MIC for the mbr mutants) are shown. The biofilm cells were stained with the Live/Dead BacLight bacterial viability kit for 30 min and observed with CLSM. A representative image from three independent experiments is shown. (B) Ratios of Live/Dead staining intensities were also observed in biofilms from WT, ΔmbrC, and ΔmbrD strains. These data are the mean ± SD of three independent experiments. The asterisks indicate a significant difference between WT and ΔmbrC or ΔmbrD strains (Student's t test; **, P < 0.01; *, P < 0.05; versus WT).
To clarify the cause of lysis, we assessed whether bacitracin had direct bactericidal activity against the WT, ΔmbrC, and ΔmbrD strains at biofilm-inducing concentrations. The number of live cells was calculated by comparing the number of CFU at 5 h of incubation with bacitracin to that without bacitracin (Table 2). There was no difference in number of live cells, even with bacitracin at the concentration at which biofilm formation was most strongly induced. The minimum bactericidal concentrations (MBCs) of bacitracin against the WT and mbr mutant strains were determined to be 64 U/ml and 8 U/ml, respectively (Table 1). These concentrations were much higher than the concentration with the greatest biofilm induction (1/8× MIC for the WT strain and 1/2× MIC for the mbr mutant strains). Thus, we reasoned that biofilm formation at these concentrations was not associated with the direct bactericidal activity of bacitracin.
TABLE 2.
Changes in the number of the live cells by bacitracin treatmenta
| Strain | Change in no. of live cells |
|
|---|---|---|
| bac− | bac+ | |
| UA159 WT | (5.53 ± 2.79) × 108 | (5.58 ± 3.58) × 108 |
| ΔmbrC mutant | (5.91 ± 2.39) × 108 | (6.21 ± 0.7) × 108 |
| ΔmbrD mutant | (5.38 ± 0.87) × 108 | (5.27 ± 2.04) × 108 |
bac−, without bacitracin; bac+, with bacitracin.
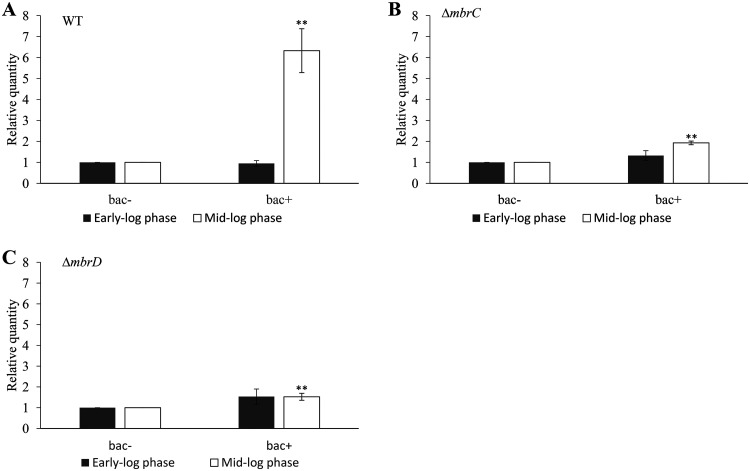
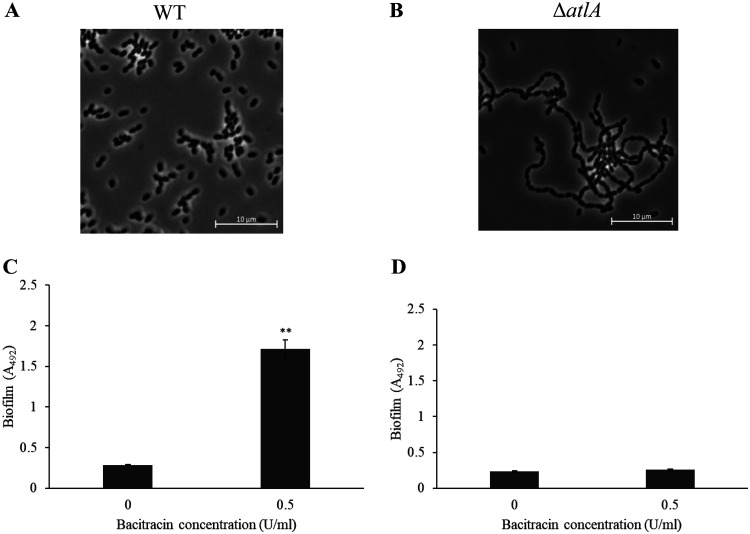
In turn, we focused on autolysis as a mechanism that led to the production of eDNA in biofilms. AtlA is the major autolysin in S. mutans (11). We predicted that this autolysin contributed to biofilm formation. We quantified the expression of the atlA gene in the early and mid-log phases by reverse transcription- quantitative PCR (RT-qPCR). In the early log phase, the expression levels of atlA in the WT strain were the same in the presence and absence of bacitracin (Fig. 4A). Meanwhile, the expression of atlA in the mid-log phase was higher in the presence of bacitracin than in the absence of bacitracin (Fig. 4A). These results suggest that autolysis caused by AtlA might be induced in response to bacitracin at the mid-log phase. On the other hand, the expression pattern of the atlA gene in the ΔmbrC and ΔmbrD strains was different from that in the WT strain. An increase in atlA expression was also observed in the presence of bacitracin even in the ΔmbrC and ΔmbrD mutant strains, but the increase in expression in these strains was less than that in the WT strain (Fig. 4B and C). These results showed that the induction of atlA expression was affected by the loss of responsiveness to bacitracin. In addition, this suggests that the difference in atlA expression may be responsible for the difference in the inducing concentrations and the morphology of the biofilm between the WT and mbr mutant strains. We then examined the role of atlA in biofilm formation using the atlA deletion mutant (ΔatlA). We first observed the cell morphology by phase-contrast microscopy and confirmed that in comparison with the WT strain, the ΔatlA strain formed a long chain (Fig. 5A and B). This result is in agreement with other reports (11). Biofilm formation induced by sub-MICs of bacitracin was not observed in the ΔatlA mutant (Fig. 5C and D). Therefore, the biofilm formation induced by sub-MICs of bacitracin was considered to be dependent on autolysis and cell separation after cell division by atlA, which is induced in the mid-log phase.
FIG 4.
Gene expression of atlA in the presence of sub-MIC of bacitracin. We determined the gene expression of atlA at an OD600 of 0.3 (early log phase) and an OD600 of 0.5 (mid-log phase). (A) The WT strain was cultivated in TSBg with 0.5 U/ml of bacitracin (1/8× MIC for the WT) (bac+) and without bacitracin (bac−). (B and C) The ΔmbrC (B) and ΔmbrD (C) mutant strains were cultivated in TSBg with 0.0625 U/ml of bacitracin (bac+) and without bacitracin (bac−). These figures indicate the relative expression levels of atlA compared with the expression in TSBg cultivated without bacitracin. These data were normalized to the expression of an endogenous control (lactate dehydrogenase, ldh). These data are the mean ± SD of three independent experiments. The asterisks indicate a significant difference between two groups (Student's t test; **, P < 0.01, versus bac−).
FIG 5.
Cell morphology and biofilm formation of the ΔatlA strain. The WT (A) and ΔatlA (B) strains cultivated in TSBg were observed by phase-contrast microscopy. A biofilm formation assay was performed in TSBg with 0.5 U/ml of bacitracin (1/8× MIC for the WT) using the WT (C) and the ΔatlA (D) strains. The data are the mean ± SD of three independent experiments. The asterisks indicate a significant difference between two groups (Student's t test; **, P < 0.01, versus bac−).
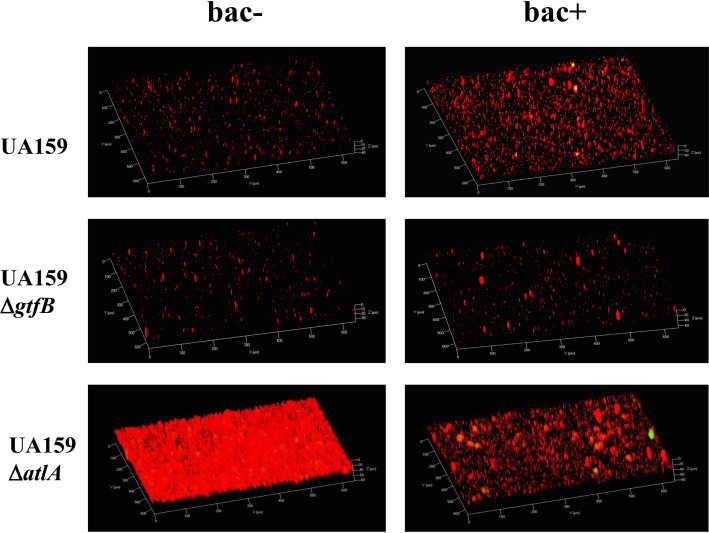
To confirm the involvement of polysaccharides, we performed a biofilm formation assay using deletion mutants of gtfB (ΔgtfB) (which encodes GtfB, an enzyme involved in the synthesis of an insoluble glucan), gtfC (ΔgtfC) (which encodes GtfC, an enzyme involved in the synthesis of an insoluble and soluble glucan), gtfBC (ΔgtfBC), gtfD (ΔgtfD) (which encodes GtfD, an enzyme involved in the synthesis of a soluble glucan), and sacB (ΔsacB) (which encodes Ftf, an enzyme involved in the synthesis of a soluble fructan). These strains deficient in polysaccharide synthesis did not form biofilms in the absence of bacitracin or in the presence of 1/2× MIC of bacitracin (Fig. 6A and B). The biofilm formation of the ΔgtfC, ΔgtfD, and ΔsacB strains was equivalent to that of the WT strain in the presence of 1/8× MIC of bacitracin, but biofilm formation was not induced in the ΔgtfB and ΔgtfBC strains (Fig. 6C). From this result, it was predicted that insoluble glucan synthesized by GtfB from sucrose was involved in inducing biofilm formation in the presence of sub-MICs of bacitracin, and TSB medium contained a small amount of sucrose. We then changed the biofilm formation medium from TSBg to semidefined medium plus 0.25% glucose (SDMg), a sucrose-free medium, and conducted the biofilm formation assay. Biofilm formation was not induced in the WT strain at any bacitracin concentration (Fig. S3). Therefore, it was suggested that polysaccharide synthesis by GtfB from sucrose, the abundance of which was low in TSB, is necessary for biofilm formation. To examine whether GtfB contributes to biofilm formation, we performed Live/Dead staining of the WT and ΔgtfB strains cultured in sucrose-free medium and observed cells by CLSM. The WT strain showed more aggregates formed by complexes of live and dead cells in the presence of bacitracin than in the absence of bacitracin (Fig. 7). However, the numbers of aggregates in the ΔgtfB mutant did not change with or without bacitracin (Fig. 7). ΔgtfC and ΔgtfD mutants showed results similar to those of the WT (data not shown). These results suggested that the physical presence of GtfB promoted the aggregation of cells, and the synthesis of insoluble glucan from the small amount of sucrose in TSB supported biofilm formation induced by sub-MICs of bacitracin.
FIG 6.
Effects of polysaccharides and Com regulation system on biofilm formation. (A to C) A biofilm formation assay was performed in TSBg without bacitracin (A), with 0.0625 U/ml of bacitracin (1/2× MIC for the mbr mutants) (B), and with 0.5 U/ml of bacitracin (C). To assess the involvement of polysaccharides and Com-dependent quorum sensing in biofilm formation, we examined the 5-h biofilm formed by the polysaccharide synthase and com gene mutants. The data are the mean ± SD of three independent experiments. The data were analyzed with one-way ANOVA and a Bonferroni’s posttest. The asterisks indicate a significant difference (**, P < 0.01).
FIG 7.
Observation of dead and live cells in the aggregates. CLSM analysis of aggregates was performed for the WT, ΔgtfB, and ΔatlA strains cultivated in BHI broth (bac−) or in BHI broth with 0.5 U/ml of bacitracin (1/8× MIC for the WT) (bac+). The aggregates were stained using the Live/Dead BacLight bacterial viability kit for 30 min and observed with CLSM. Live cells and dead cells are shown in green and red, respectively. A representative image from three independent experiments is shown.
Subsequently, we examined whether the Com regulation system, known as a cell death-inducing mechanism, was involved in biofilm formation and performed a biofilm formation assay using com gene mutant strains (ΔcomC, ΔcomD, ΔcomE, and ΔcomX) (31). comC, comD, comE, and comX encode a CSP precursor, a histidine kinase for CSP recognition, a response regulator for CSP response, and an alternative sigma factor for induction of a ComX regulon, respectively. Similar to the WT strain, the com mutant strains did not form biofilms in the absence of bacitracin or in the presence of 0.0625 U/ml of bacitracin (1/2× MIC for mbr mutants), but these mutants could form biofilms in the presence of 0.5 U/ml of bacitracin (1/8× MIC for the WT) (Fig. 6). Therefore, cell death controlled by the Com regulation system was not involved in the eDNA-dependent biofilm formation induced by sub-MICs of bacitracin.
Cell envelope stress involvement in biofilm induction.
If the biofilm formation induced by sub-MICs of bacitracin is associated with bacitracin-specific recognition by mbrCD, biofilm formation may be affected by the lack of responsiveness to bacitracin in the ΔmbrC and ΔmbrD strains. Nonetheless, the ΔmbrC and ΔmbrD strains retained the ability to form biofilms. This result suggested that biofilm formation was induced by a mechanism other than a bacitracin-specific response via the mbr system. We then hypothesized that S. mutans induced biofilm formation in response to cell envelope stress by bacitracin. S. mutans contains a gene, brpA, that participates in the regulation of the cell envelope stress response (32). We constructed a deletion mutant of brpA (ΔbrpA) and conducted a biofilm formation assay using the mutant. When the ΔbrpA strain was cultured in TSBg medium supplemented with various concentrations of bacitracin, the mutant did not form biofilms at any bacitracin concentration (Fig. S4). In contrast, biofilm formation in the ΔbrpA strain was induced at levels comparable to those in the WT strain when the mutant strain was cultivated in TSB medium, supporting that polysaccharide was synthesized (Fig. S5). We confirmed that the ΔbrpA strain displayed a long-chain phenotype, which was observed in another report (33) (Fig. S6). Therefore, the direct stimulation of biofilm formation induced by sub-MICs of bacitracin is dependent on cell envelope stress, and it has been suggested that the ΔbrpA strain failed to respond to the stress caused by sub-MICs of bacitracin, exhibiting a long-chain phenotype that adversely affects biofilm formation (34), and could not induce eDNA-dependent biofilm formation.
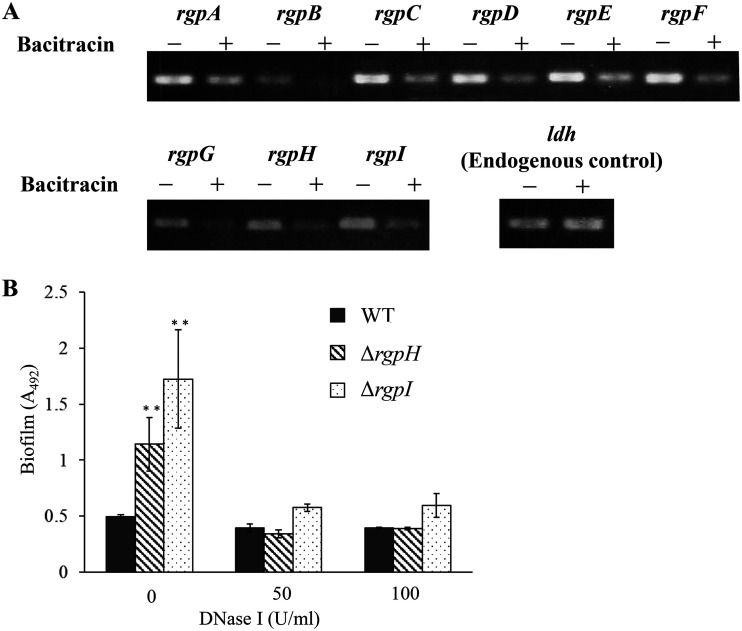
S. mutans produces RGPs surrounding the surface of the cells. The loss of RGPs resulted in an increased susceptibility to certain cell wall-targeting antibiotics, while a recent study suggested that RGP may influence biofilm formation (35). We performed reverse transcription-PCR (RT-PCR) to examine whether the presence of bacitracin affected the expression of the RGP genes. The expression of all rgp genes decreased in the presence of sub-MICs of bacitracin (Fig. 8A). We confirmed that RT-PCR worked properly by checking the expression of a housekeeping gene, ldh. Additionally, we constructed mutant strains with deletions in rgpH (ΔrgpH) and rgpI (ΔrgpI) and cultured these strains in the absence of bacitracin. The ΔrgpH and ΔrgpI strains formed biofilms, but the WT strain did not form biofilms (Fig. 8B). Biofilm formation by the ΔrgpH and ΔrgpI strains was inhibited by the addition of DNase I (Fig. 8B). Therefore, the reduced expression of rgp genes in the WT strain was thought to result in an increase in eDNA-dependent biofilm formation in the presence of sub-MICs of bacitracin.
FIG 8.
Expressions of rgp genes in biofilm stimulated with bacitracin. (A) The WT was cultured in TSBg or TSBg with 0.5 U/ml of bacitracin (1/8× MIC for the WT) until the early log phase (OD600 = 0.3), and RNA was extracted from the cells. RT-PCR was performed using the RNA, and aliquots of PCR products were electrophoresed on a 2% agarose gel. The housekeeping gene ldh was used as an endogenous control. (B) To evaluate the effect of RGP deficiency on eDNA-dependent biofilm formation, a biofilm formation assay was performed in TSBg with or without DNase I using the ΔrgpH and ΔrgpI strains. The data are the mean ± SD of three independent experiments. The data were analyzed with one-way ANOVA and a Bonferroni’s posttest. The asterisks indicate a significant difference (**, P < 0.01).
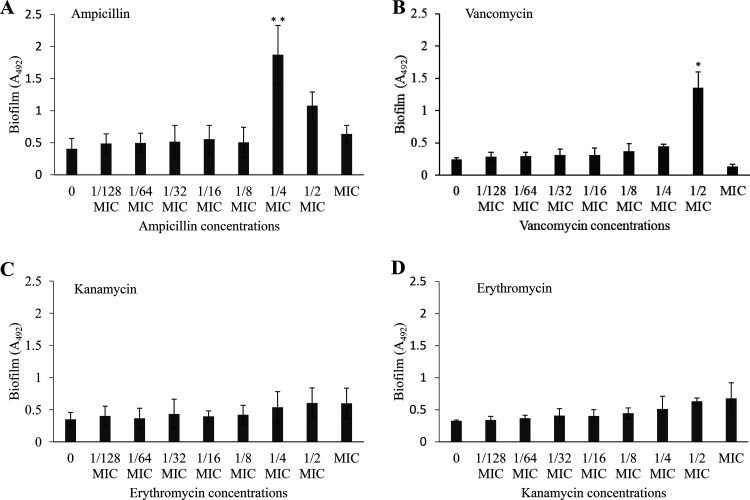
If cell envelope stress by bacitracin induces eDNA-dependent biofilm formation, biofilm formation should also be induced by antibiotics other than bacitracin, which stresses the bacterial cell envelope. Biofilm formation assays were then conducted in the presence of antibiotics with different mechanisms of action. Biofilm formation was also induced in the presence of sub-MICs of ampicillin and vancomycin, antibiotics that inhibit peptidoglycan synthesis (Fig. 9A and B). However, biofilm formation was not induced in the presence of sub-MICs of erythromycin and kanamycin, which inhibit protein synthesis (Fig. 9C and D). From these results, it seemed that antibiotics that inhibit peptidoglycan synthesis caused cell envelope stress in bacterial cells and induced eDNA-dependent biofilm formation. In contrast, sub-MICs of other types of antibiotics did not induce signals for biofilm formation.
FIG 9.
Biofilm formation assay in the presence of other antibiotics. To investigate whether antibiotics other than bacitracin induced biofilm formation, biofilm formation assays were conducted in the presence of antibiotics with different mechanisms of action: inhibition of peptidoglycan synthesis (ampicillin [MIC, 0.05 μg/ml] [A], vancomycin [1 μg/ml] [B]) and inhibition of protein synthesis (kanamycin [240 μg/ml] [C] and erythromycin [0.1 μg/ml] [D]). The data are the mean ± standard deviation (SD) of three independent experiments. The data were analyzed with one-way ANOVA and a Bonferroni’s posttest. The asterisks indicate a significant difference (**, P < 0.01; *, P < 0.05).
Increased risk of drug resistance by biofilm induction.
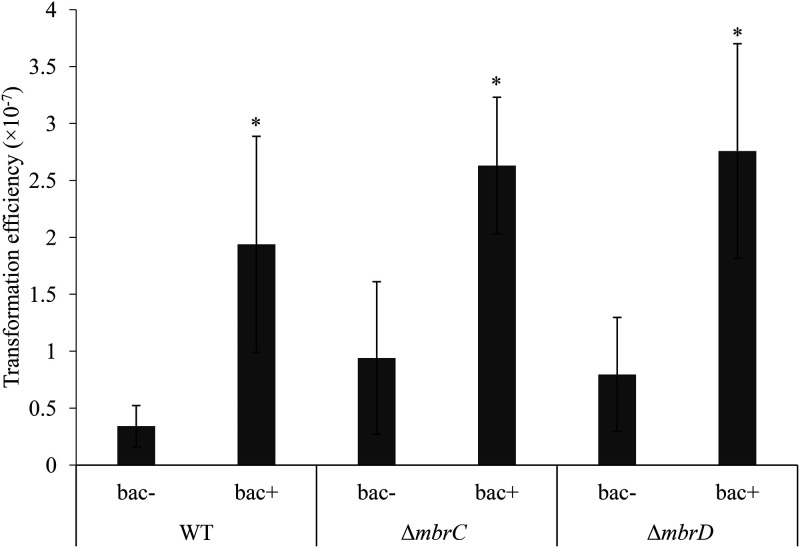
It has been pointed out that biofilms can contribute to horizontal gene transfer (36). We examined the possibility that S. mutans may facilitate the acquisition of new drug resistance genes by inducing biofilm formation. Transformation assays using the pDL278 plasmid carrying a spectinomycin resistance gene showed that transformation efficiencies were increased in both the WT and mbr mutant strains in the presence of sub-MICs of bacitracin (Fig. 10). The transformation efficiency was also increased by sub-MICs of vancomycin (Fig. S7). Therefore, it was shown that the induction of biofilm formation in response to cell envelope stress by sub-MICs of bacitracin and vancomycin promotes horizontal gene transfer.
FIG 10.
Transformation assay in the absence and presence of bacitracin. S. mutans was cultured in TSBg containing the pDL278 plasmid carrying a spectinomycin resistance gene. Bacitracin was added at the concentration at which each strain most strongly induced biofilm formation (bac+, 0.5 U/ml [1/8× MIC for the WT strain] and 0.0625 U/ml [1/2× MIC for the mbr mutant strains]). After culture for 5 h, the cells were cultured on a mitis salivarius agar plate with or without spectinomycin for 36 h. We measured the number of CFU and determined the transformation efficiency. The data are the mean ± SD of three independent experiments. The asterisks indicate a significant difference between two groups (Student's t test; *, P < 0.05, versus bac−).
DISCUSSION
In this study, we revealed that eDNA-dependent biofilm formation was induced at bacitracin concentrations below the MIC regardless of whether bacitracin resistance was present. Since the concentration that most strongly induced biofilm formation in the WT strain, 0.5 U/ml, was above the MIC of other bacitracin-sensitive and non-biofilm-forming bacteria in commensal oral flora, such as Streptococcus salivarius (37), the bacitracin concentration not only induces biofilm formation of S. mutans but may also change the balance of resistant and sensitive bacteria. Because the addition of antibiotics changed the prevalence and composition of the antibiotic-resistant microflora in dental plaque containing lactobacilli, streptococci, and Actinomyces spp. (38), it is important to understand the behavior of drug-resistant bacteria, especially those resistant to cell wall synthesis inhibitors, and take countermeasures against these bacteria. Data on drug-resistant bacteria will help to develop better bacterial control strategies.
The expression of atlA was increased in the mbr mutant strain at mid-log phase but not to a level as high as that of the WT strain. This suggests that WT strains more actively induce biofilm formation by changes in gene expression, whereas the induction mechanism of mbr mutant strains may be incomplete. In fact, based on CLSM, there were relatively many live cells in the biofilm formed by the WT strain, and the biofilm exhibited a microcolony-like morphology, which is one of the characteristics of the biofilm of S. mutans (6); however, the biofilm formed by the mbr mutants had relatively few live cells, and no prominent structures were observed. Since it was reported that the balance between lysed cells and living cells was important for drug-induced biofilm formation in Enterococcus faecalis (26), we examined cell death induction via the Com regulation system and the direct bactericidal effect of bacitracin, but these appeared to have little effect. Accordingly, it was predicted that the balance between live and dead cells due to the difference in the induction of atlA expression at mid-log phase resulted in the difference in the concentration that induced biofilm formation.
Biofilm formation in the presence of bacitracin was dramatically reduced with DNase I treatment, but biofilm formation was not completely polysaccharide independent because it was not induced in the ΔgtfB and ΔgtfBC strains. TSB contains a small amount of sucrose, which is a substrate for the synthesis of glucan (6). When cultured in sucrose-free medium, the WT strain aggregated, while the ΔgtfB mutant did not aggregate. These results suggested that Gtf-I encoded by gtfB is involved in the aggregation of cells at the protein level, not in the enzymatic activity, because the substrate (sucrose) is absent. We previously reported that Gtf-I is essential for eDNA-dependent biofilm formation (6). Therefore, it was suggested that Gtf-I physically and chemically contributes to eDNA-dependent biofilm formation in TSBg with bacitracin medium.
We also revealed that the stress response of the cell envelope and RGP, which is part of the cell wall, were necessary for biofilm induction. The deletion of brpA, which plays pivotal roles in the cell envelope stress response, leads to various phenotypes (32, 33). The ΔbrpA strain showed a long-chain phenotype consistent with other reports (33). It is known that abnormal chaining in streptococci results in a biofilm-defective phenotype (30). Our results also indicated that the ΔbrpA strain was unable to form biofilms in the presence of sub-MICs of bacitracin. In addition, it was considered that the decreased expression of the RGP synthetic genes in the presence of sub-MICs of bacitracin contributed to biofilm induction. In streptococci, cells not encapsulated by polysaccharides showed increased attachment to surfaces and formed aggregates and biofilms (39–43). In addition, deletion of rgpI decreased glucan and increased eDNA levels (44). We confirmed that the ΔrgpH and ΔrgpI strains can form biofilms without bacitracin. Therefore, it was shown that S. mutans induces biofilm formation by regulating the structure of the cell surface and responding to cell envelope stress. In support of the notion that stresses on the cell envelope trigger biofilm formation, we showed that the cell wall-inhibiting antibiotics ampicillin and vancomycin also induce biofilm formation at sub-MICs. In Gram-positive bacteria, including S. mutans, it has recently been reported that biofilm formation was induced by antimicrobial agents targeting the cell envelope, including β-lactams (25–28). Therefore, it is necessary to elucidate the common mechanisms that may underlie biofilm formation.
The accumulation of microorganisms as mono- or polymicrobial aggregates is commonly referred to as a biofilm, which can consist of a diverse community of bacteria (45). The formation of these sessile communities and their inherent resistance to antibiotics are at the root of persistent and chronic bacterial infections. If resistant bacteria or highly tolerant bacteria form a multispecies biofilm with susceptible bacteria, there is a potential risk that various genes, including drug resistance genes, will spread within the community due to horizontal gene transfer. Therefore, understanding the behavior of preexisting drug-resistant and biofilm-forming bacteria and the role of cell wall inhibitory antibiotics in horizontal gene transfer is important for treating infectious diseases.
In conclusion, we analyzed the response of bacitracin-resistant bacteria such as S. mutans to bacitracin and focused mainly on biofilm formation. We found that regulation of the expression of the atlA and rgp genes, which are involved in cell surface properties, contributes to biofilm formation induced by sub-MICs of bacitracin. GtfB, which is highly bound to the cell surface, was also essential for biofilm formation. Our results suggested that S. mutans induced eDNA-dependent biofilm formation in response to the cell envelope stress caused by sub-MICs of bacitracin and other cell wall inhibitory antibiotics. In addition, drug resistance genes may spread in biofilms, since horizontal gene transfer occurs more frequently in biofilm cells than in planktonic cells and biofilms contain a large amount of eDNA. Therefore, when using cell wall-targeted antibiotics at sub-MICs, attention may need to be paid to the behavior of resistant bacteria in bacterial infections.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 3. S. mutans UA159 and isogenic mutants were grown in brain heart infusion (BHI) broth (Difco Laboratories, Detroit, MI) at 37°C in an anaerobic atmosphere containing 5% CO2 (AnaeroPack-CO2; Mitsubishi Gas Chemical Company, Tokyo, Japan). These prepared cells were used in this study.
TABLE 3.
Bacterial strains and plasmids
| Strain or plasmid | Relevant propertiesa | Source or reference |
|---|---|---|
| S. mutans strains | ||
| UA159 | Wild type; Erms Kans Spcs | 53 |
| ΔmbrC mutant | UA159 mbrC Ermr | This study |
| ΔmbrD mutant | UA159 mbrD Ermr | This study |
| ΔgtfB mutant | UA159 gtfB Ermr | 6, 54 |
| ΔgtfC mutant | UA159 gtfC Kanr | 6, 54 |
| ΔgtfD mutant | UA159 gtfD Kanr | 6 |
| ΔsacB mutant | UA159 sacB Ermr | 6 |
| ΔgtfBC mutant | UA159 gtfB Ermr gtfC Kanr | 6, 54 |
| ΔbrpA mutant | UA159 brpA Ermr | This study |
| ΔatlA mutant | UA159 atlA Ermr | N. Obana, University of Tsukuba, unpublished |
| ΔrgpH mutant | UA159 rgpH Ermr | This study |
| ΔrgpI mutant | UA159 rgpI Ermr | This study |
| Plasmids | ||
| pUC19+Em | pUC19 containing Ermr | 6 |
| pResEmMCS10 | Streptococcal integration plasmid; Ermr | 30 |
| pDL278 | E. coli-Streptococcus shuttle vector; Spcr | 35 |
Erms, erythromycin susceptibility; Kans, kanamycin susceptibility; Spcs, spectinomycin susceptibility; Ermr, erythromycin resistance; Kanr, kanamycin resistance; Spcr, spectinomycin resistance.
Construction of mutants.
Primers used to construct mutants are listed in Table 4. Sequences of the target genes were obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/). Deletion mutants were generated by inserting an antibacterial resistance gene as described previously (6, 46). DNA fragments for generating ΔmbrC, ΔmbrD, ΔrgpH, and ΔrgpI deletion mutants were created with PCR and ligation (41). The upstream and downstream sequences of the target genes and the erythromycin resistance gene were amplified from S. mutans UA159 gDNA and pUC19+Em, which is a plasmid with the erythromycin resistance gene from pResEmMCS10 inserted in the multicloning site, respectively (47). The amplicons were digested with BamHI or BamHI and HindIII, and the resulting fragments were ligated. These fragments were introduced into S. mutans UA159 competent cells. The DNA fragment for generating the ΔbrpA mutant was constructed by overlap extension PCR. The upstream and downstream sequences of brpA were amplified from gDNA of S. mutans UA159. The erythromycin resistance gene was amplified from pUC19+Em. The amplicons were ligated with overlap extension PCR, and the resultant fragment was introduced into S. mutans UA159. The transformants were selected on mitis salivarius agar (Difco Laboratories, Detroit, MI) plates supplemented with erythromycin (10 μg/ml). Mutations in the target genes of the genomic DNA were confirmed by PCR.
TABLE 4.
Primers for mutant construction
| Primer | Sequencea |
|---|---|
| mbrC uF | cccccgaattcCTTCCTGCCTTTACTTATGGC |
| mbrC uR | cccccggtaccTGGGCATGGTCATGCTCTTTC |
| mbrC dF | ccccctctagaAAGAGACTATCCAATTGACTCC |
| mbrC dR | cccccaagcttTGTCAGACTGTTCCTGCAGC |
| mbrD uF | cccccgaattcGTGCTAACGAGTTCACTAAGC |
| mbrD uR | cccccggtaccCCGCCAGTATAAAATGAGAC |
| mbrD dF | ccccctctagaACTGCTGCAGGAACAGTCTGAC |
| mbrD dR | cccccaagcttCTTAATGGTGATCTGCCCAC |
| brpA uF | GTTGGTACTCTCATTGGTACAGGAG |
| brpA uR | cccatgcgtttgggcccCTCCTAAAGCTACCAAGC |
| brpA dF | cccaaggagatggccggCAGATTACAGTAGCAGCG |
| brpA dR | CCATGGCTATCATCTCCAAGTGCTC |
| ermR F | gggcccaaacgcatgggAATACGCAAACCGCCTCT |
| ermR R | ccggccatctccttgggGTGCCACCTGACGTCTAA |
| 832 uF | GGCAAGGGCTATACAAAA |
| 832 uR-Bam | ccggatccGGCGGTAATGATTCTTAATAAAA |
| 832 dF-Bam | ccggatccCGTTGATTATTTATACGG |
| 832 dR | AGCGTCCATAACCGTAAT |
| 833 uF | AGAATTATTTTTGTAACCGCAGA |
| 833 uR-Bam | ccggatccTGGTTTCTTCCTCATTATAACAA |
| 833 dF-Bam | ccggatccGGAAACCAAGAAAAGACCA |
| 833 dR | GTCGTACTAAATTATTGCCGAA |
Additional sequences which do not correspond to the sequences of the template DNA are indicated by lowercase letters.
Antimicrobial susceptibility assays.
A broth microdilution method was used with a 96-well microtiter plate to identify the MICs of the antibiotics bacitracin (Wako Pure Chemical Industries, Tokyo, Japan), ampicillin (Wako Pure Chemical Industries, Tokyo, Japan), vancomycin (Wako Pure Chemical Industries, Tokyo, Japan), kanamycin (Sigma Chemical, St. Louis, MO), and erythromycin (Sigma Chemical, St. Louis, MO) for the S. mutans UA159 strain. Two microliters of overnight culture was inoculated into 198 μl of BHI broth containing a 2-fold dilution series of antibiotics. The culture plates were incubated at 37°C for 24 h in an aerobic atmosphere containing 5% CO2. After incubation, the optical density at 620 nm (OD620) was measured on a plate reader (Thermo Bioanalysis Japan, Tokyo, Japan).
Minimal bactericidal concentrations (MBCs) of bacitracin were identified using MIC test plates. A total of 20 μl of culture was plated on a BHI agar plate without antibiotics, and the MBC was determined as the lowest concentration at which <5 CFU were observed after 24 h of incubation at 37°C in an aerobic atmosphere containing 5% CO2.
Human saliva collection.
Whole saliva samples from volunteers with good oral health were collected in ice-chilled sterile bottles. To remove debris, the whole saliva was centrifuged (4°C, 14,000 × g, 15 min). The supernatant was filtered with a 0.22-μm syringe filter (Millex-GP; Merck Millipore, Darmstadt, Germany) to sterilize the sample. The sterilized saliva was used immediately for biofilm formation assays.
Biofilm formation assays.
We used TSB (Difco Laboratories, Detroit, MI) supplemented with 0.25% (wt/vol) TSBg or TSBs or semidefined medium (SDM) with 0.25% glucose for biofilm formation (6). Overnight cultures were inoculated into these media at a ratio of 1:5 in a human saliva-coated 96-well microtiter plate (Sumitomo Bakelite, Tokyo, Japan). The biofilm formation assay was performed at 37°C for 5 h in an aerobic atmosphere containing 5% CO2 under static conditions. After incubation, the medium was removed, and the wells were washed two times with distilled water (DW). The plate was air dried, and the biofilm was stained with safranin (0.25% safranin and 0.5% ethanol in H2O; Muto Pure Chemicals, Tokyo Japan) for 15 min. The wells were then washed two times with DW to remove the excess dye and air dried. Safranin was extracted from the biofilm with 100 μl of 70% (vol/vol) ethanol, and the absorbance at 492 nm (A492) was measured.
To evaluate the contribution of eDNA to biofilm formation, we conducted a biofilm inhibition experiment by adding DNase I. Before the biofilm formation assay, 50 U/ml of DNase I (Roche Applied Science, Mannheim, Germany) was added to the medium to evaluate the inhibitory effect of eDNA degradation on biofilm formation. The biofilm formation assay and quantification were performed by the above-mentioned procedure.
Quantitative analysis of eDNA.
Quantification of eDNA was conducted as described elsewhere (48). Briefly, 1 ml of overnight culture was mixed with 5 ml of TSBg with or without bacitracin in a saliva-coated 6-well plate (Iwaki, Tokyo, Japan). The plate was incubated at 37°C for 5 h in an aerobic atmosphere containing 5% CO2 under static conditions. After incubation, the biofilm formed on the bottom of the culture plate was removed, and the biofilm and supernatant were treated with 5 U/ml dextranase (Sigma Chemical, St. Louis, MO) at 40°C for 30 min and proteinase K solution (Qiagen, Venlo, The Netherlands) at 56°C for 30 min to disperse the biofilm cells. The sample was filtered through a 0.22-μm syringe filter to remove the cells. The eDNA contained in the sample was isolated with the cetyltrimethylammonium bromide (CTAB) method as described elsewhere (49). The yield of eDNA was quantified by absorptiometry (NanoDrop Lite spectrophotometer; Thermo Fisher Scientific, Waltham, MA).
CLSM.
Biofilm cells were stained using the Live/Dead BacLight bacterial viability kit (Molecular Probes, Leiden, The Netherlands) after biofilms were formed. SYTO 9 and propidium iodide (PI) were used to stain bacteria with intact cell membranes (live cells, green fluorescence) and bacteria with damaged cell membranes (dead cells, red fluorescence), respectively. After 30 min of staining, the biofilm was washed two times with DW to remove excess dye. The biofilms were observed by confocal laser scanning microscopy (CLSM; LSM 700, Carl Zeiss, Jena, Germany), and three-dimensional (3D) images of biofilms were acquired by taking two-dimensional (2D) images with a Plan-Apochromat 10×/0.45 M27 lens objective. Zen software (Zeiss) was used to analyze the biofilms.
RNA extraction and RT-qPCR.
Total RNA was isolated from cells according to the method established by Cury et al. (50). To collect the cells at early and mid-log phase, 2 ml of overnight culture and 28 ml of TSBg with or without bacitracin were mixed in a 50-ml polyethylene tube and incubated at 37°C in an aerobic atmosphere containing 5% CO2 under static conditions until the early log (OD600 = 0.3) and mid-log (OD600 = 0.5) phases were reached. The cells were collected by centrifugation (5,500 × g, 10 min) and resuspended in cold phosphate-buffered saline (PBS). To separate the cells and extracellular substances, the cells were sonicated at 2 W for 30 s. These manipulations, including resuspension and sonication, were repeated two times. The bacterial cell pellet was suspended in NAES buffer (50 mM sodium acetate buffer, 10 mM EDTA, and 1% SDS [pH 5.0]). A total of 625 μl of acidified phenol and 125 μl of chloroform was added, and the sample was transferred to a 2-ml tube containing 0.5 g of 0.1-mm-diameter glass beads; the samples were shaken on a TissueLyser (Qiagen, Venlo, The Netherlands) for 3 min. After centrifugation (4°C, 14,000 × g, 5 min), the aqueous phase of the samples was transferred into a microtube, and 625 μl of acidified phenol and 125 μl of chloroform were added. These manipulations were repeated one more time. After centrifugation (4°C, 14,000 × g, 5 min), the aqueous phase of the samples was transferred to a microtube, and 750 μl of phenol-chloroform-isoamyl alcohol (25:24:1) was added to the solution. The aqueous phase of the samples was transferred to a microtube after centrifugation (4°C, 14,000 × g, 5 min). The total RNA in solution was precipitated with 2-propanol and washed with 70% ethyl alcohol (EtOH) three times. cDNA was synthesized with ReverTra Ace qPCR RT master mix with gDNA remover (Toyobo, Osaka, Japan) using 500 ng of total RNA as the template for reverse transcription. RT-PCR using the primers in Table 5 was performed by Tks Gflex DNA polymerase (TaKaRa, Shiga, Japan). We used the lactate dehydrogenase gene ldh, which is a housekeeping gene in S. mutans, as an endogenous control (51).
TABLE 5.
Primers for RT-qPCR and RT-PCR
| Primer | Sequence |
|---|---|
| ldh-Fw | TTGGCGACGCTCTTGATCTTAG |
| ldh-Rv | GTCAGCATCCGCACAGTCTTC |
| atlA-Fw | CAGCAAACACAGGAACAGTCTG |
| atlA-Rv | TCAGGAGTCTGCACAGAACTAG |
| rgpA-Fw | GCAGTTAACTTATCATGTGGCCTG |
| rgpA-Rv | CATATCATAAGCAATGACTCTGGCAG |
| rgpB-Fw | GCAGACTTTGCTAAGTCTGATGC |
| rgpB-Rv | CTGCCAAACATCATCTTGATCGC |
| rgpC-Fw | CGTTATCAAGGAAGCGCCATTG |
| rgpC-Rv | CCATACCCATCATGGTTGTTTCTTG |
| rgpD-Fw | GTTTGAGAACTGCCTTAGTCAATCG |
| rgpD-Rv | CAGAACCATTTCGGCCTACAATAC |
| rgpE-Fw | GAAGCTGGATTGGTGAAGCTATTG |
| rgpE-Rv | CTAGCTTTCTTACAAATATCGGACCAAG |
| rgpF-Fw | CGTTTATCAGTTGACTCAAATGAGATCC |
| rgpF-Rv | CCAGAATTCTGCCGTTGAATGAAG |
| rgpG-Fw | GTTGATAATCCAAATGCGAGACGG |
| rgpG-Rv | CAAAATAGGATTTTCCTCCGATTTGC |
| rgpH-Fw | GTTTTATTAAGAATCATTACCGCCTATTG |
| rgpH-Rv | GATCGTTTGATCTCCTGCATC |
| rgpI-Fw | GTTATAATGAGGAAGAAACCATTTACCC |
| rgpI-Rv | CTGTTAAAGTCTTATCACTTGAACCATC |
Transformation assay.
Cultures of the WT, ΔmbrC, and ΔmbrD strains were prepared by the same procedure described for the biofilm formation assay in TSBg. The concentrations of bacitracin were 0.5 U/ml (1/8× MIC) for the WT and 0.0625 U/ml (1/2× MIC) for the mbr mutant strains. The pDL278 plasmid was added to each medium at a final concentration of 2.5 μg/ml (52). After culture for 5 h under the same conditions as for biofilm formation, the cell suspensions were poured and cultured on mitis salivarius agar plates with or without spectinomycin (500 μg/ml) for 36 h. We counted the number of CFU and determined the transformation efficiency.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ryoma Nakao and Satoru Hirayama for their advice and valuable discussions, Mizuho Motegi for support, and Nozomu Obana for the gift of a deletion mutant strain.
This work was supported in part by Grants-in-Aid for the Development of Scientific Research (no. 21390506, 24659821, and 16K11537) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, by a Grant-in-Aid for Scientific Research (no. 18J21373) from the Japan Society for the Promotion of Science Research Fellowship for Young Scientists, by the Research Program on Emerging and Re-emerging Infectious Diseases from the Japan Agency for Medical Research and Development (AMED grant no. 40105502), and by the Japan Science and Technology Agency (ERATO grant no. JPMJER1502).
The manuscript was corrected by American Journal Experts.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Gajdács M. 2019. The concept of an ideal antibiotic: implications for drug design. Molecules 24:892. doi: 10.3390/molecules24050892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki H, Shiroza T, Hayakawa M, Sato S, Kuramitsu HK. 1986. Cloning of a Streptococcus mutans glucosyltransferase gene coding for insoluble glucan synthesis. Infect Immun 53:587–594. doi: 10.1128/IAI.53.3.587-594.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanada N, Kuramitsu HK. 1988. Isolation and characterization of the Streptococcus mutans gtfC gene, coding for synthesis of both soluble and insoluble glucans. Infect Immun 56:1999–2005. doi: 10.1128/IAI.56.8.1999-2005.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Critchley P, Wood JM, Saxton CA, Leach SA. 1967. The polymerisation of dietary sugars by dental plaque. Caries Res 1:112–129. doi: 10.1159/000259506. [DOI] [PubMed] [Google Scholar]
- 5.Velázquez-Hernández ML, Baizabal-Aguirre VM, Bravo-Patiño A, Cajero-Juárez M, Chávez-Moctezuma MP, Valdez-Alarcón JJ. 2009. Microbial fructosyltransferases and the role of fructans. J Appl Microbiol 106:1763–1778. doi: 10.1111/j.1365-2672.2008.04120.x. [DOI] [PubMed] [Google Scholar]
- 6.Nagasawa R, Sato T, Senpuku H. 2017. Raffinose induces biofilm formation by Streptococcus mutans in low concentrations of sucrose by increasing production of extracellular DNA and fructan. Appl Environ Microbiol 83:e00869-17. doi: 10.1128/AEM.00869-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das T, Sehar S, Manefield M. 2013. The roles of extracellular DNA in the structural integrity of extracellular polymeric substance and bacterial biofilm development. Environ Microbiol Rep 5:778–786. doi: 10.1111/1758-2229.12085. [DOI] [PubMed] [Google Scholar]
- 8.Liao S, Klein MI, Heim KP, Fan Y, Bitoun JP, Ahn S-J, Burne RA, Koo H, Brady LJ, Wen ZT. 2014. Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery. J Bacteriol 196:2355–2366. doi: 10.1128/JB.01493-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das T, Sharma PK, Krom BP, Van Der Mei HC, Busscher HJ. 2011. Role of eDNA on the adhesion forces between Streptococcus mutans and substratum surfaces: influence of ionic strength and substratum hydrophobicity. Langmuir 27:10113–10118. doi: 10.1021/la202013m. [DOI] [PubMed] [Google Scholar]
- 10.Rose SJ, Babrak LM, Bermudez LE. 2015. Mycobacterium avium possesses extracellular DNA that contributes to biofilm formation, structural integrity, and tolerance to antibiotics. PLoS One 10:e0128772. doi: 10.1371/journal.pone.0128772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shibata Y, Kawada M, Nakano Y, Toyoshima K, Yamashita Y. 2005. Identification and characterization of an autolysin-encoding gene of Streptococcus mutans. Infect Immun 73:3512–3520. doi: 10.1128/IAI.73.6.3512-3520.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung CJ, Hsu RB, Shun CT, Hsu CC, Chia JS. 2017. AtlA mediates extracellular DNA release, which contributes to Streptococcus mutans biofilm formation in an experimental rat model of infective endocarditis. Infect Immun 85:e00252-17. doi: 10.1128/IAI.00252-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gold OG, Jordan HV, van Houte J. 1973. A selective medium for Streptococcus mutans. Arch Oral Biol 18:1357–1364. doi: 10.1016/0003-9969(73)90109-x. [DOI] [PubMed] [Google Scholar]
- 14.Johnson BA, Anker H, Meleney FL. 1945. Bacitracin: a new antibiotic produced by a membrane of the B. subtilis group. Science 102:376–377. doi: 10.1126/science.102.2650.376. [DOI] [PubMed] [Google Scholar]
- 15.Storm DR. 1974. Mechanism of bacitracin action: a specific lipid-peptide interaction. Ann N Y Acad Sci 235:387–398. doi: 10.1111/j.1749-6632.1974.tb43278.x. [DOI] [PubMed] [Google Scholar]
- 16.Blas M, Briesacher KS, Lobato EB. 2000. Bacitracin irrigation: a cause of anaphylaxis in the operating room. Anesth Analg 91:1027–1028. doi: 10.1097/00000539-200010000-00049. [DOI] [PubMed] [Google Scholar]
- 17.O'Donovan CA, Fan-Havard P, Tecson-Tumang FT, Smith SM, Eng RHK. 1994. Enteric eradication of vancomycin-resistant Enterococcus faecium with oral bacitracin. Diagn Microbiol Infect Dis 18:105–109. doi: 10.1016/0732-8893(94)90074-4. [DOI] [PubMed] [Google Scholar]
- 18.Rao S, Venkateswerlu G. 1989. Metal ion influence on the growth inhibition of Neurospora crassa by bacitracin. Curr Microbiol 19:253–258. doi: 10.1007/BF01570171. [DOI] [Google Scholar]
- 19.Ming LJ, Epperson JD. 2002. Metal binding and structure-activity relationship of the metalloantibiotic peptide bacitracin. J Inorg Biochem 91:46–58. doi: 10.1016/s0162-0134(02)00464-6. [DOI] [PubMed] [Google Scholar]
- 20.Tsuda H, Yamashita Y, Shibata Y, Nakano Y, Koga T. 2002. Genes involved in bacitracin resistance in Streptococcus mutans. Antimicrob Agents Chemother 46:3756–3764. doi: 10.1128/aac.46.12.3756-3764.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitagawa N, Shiota S, Shibata Y, Takeshita T, Yamashita Y. 2011. Characterization of MbrC involved in bacitracin resistance in Streptococcus mutans. FEMS Microbiol Lett 318:61–67. doi: 10.1111/j.1574-6968.2011.02238.x. [DOI] [PubMed] [Google Scholar]
- 22.Radeck J, Fritz G, Mascher T. 2017. The cell envelope stress response of Bacillus subtilis: from static signaling devices to dynamic regulatory network. Curr Genet 63:79–90. doi: 10.1007/s00294-016-0624-0. [DOI] [PubMed] [Google Scholar]
- 23.Mascher T, Margulis NG, Wang T, Ye RW, Helmann JD. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol Microbiol 50:1591–1604. doi: 10.1046/j.1365-2958.2003.03786.x. [DOI] [PubMed] [Google Scholar]
- 24.Kovacs CJ, Faustoferri RC, Quivey RG. 2017. RgpF is required for maintenance of stress tolerance and virulence in Streptococcus mutans. J Bacteriol 199:e00497-17. doi: 10.1128/JB.00497-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mlynek KD, Callahan MT, Shimkevitch AV, Farmer JT, Endres JL, Marchand M, Bayles KW, Horswill AR, Kaplan JB. 2016. Effects of low-dose amoxicillin on Staphylococcus aureus USA300 biofilms. Antimicrob Agents Chemother 60:2639–2651. doi: 10.1128/AAC.02070-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu W, Hallinen KM, Wood KB. 2017. Interplay between antibiotic efficacy and drug-induced lysis underlies enhanced biofilm formation at subinhibitory drug concentrations. Antimicrob Agents Chemother 62:1–22. doi: 10.1128/AAC.01603-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doroshenko N, Tseng BS, Howlin RP, Deacon J, Wharton JA, Thurner PJ, Gilmore BF, Parsek MR, Stoodley P. 2014. Extracellular DNA impedes the transport of vancomycin in Staphylococcus epidermidis biofilms preexposed to subinhibitory concentrations of vancomycin. Antimicrob Agents Chemother 58:7273–7282. doi: 10.1128/AAC.03132-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong L, Tong Z, Linghu D, Lin Y, Tao R, Liu J, Tian Y, Ni L. 2012. Effects of sub-minimum inhibitory concentrations of antimicrobial agents on Streptococcus mutans biofilm formation. Int J Antimicrob Agents 39:390–395. doi: 10.1016/j.ijantimicag.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 29.Tian XL, Salim H, Dong G, Parcells M, Li YH. 2018. The BceABRS four-component system that is essential for cell envelope stress response is involved in sensing and response to host defence peptides and is required for the biofilm formation and fitness of Streptococcus mutans. J Med Microbiol 67:874–883. doi: 10.1099/jmm.0.000733. [DOI] [PubMed] [Google Scholar]
- 30.Ouyang J, Tian XL, Versey J, Wishart A, Li YH. 2010. The BceABRS four-component system regulates the bacitracin-induced cell envelope stress response in Streptococcus mutans. Antimicrob Agents Chemother 54:3895–3906. doi: 10.1128/AAC.01802-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dufour D, Lévesque CM. 2013. Cell death of Streptococcus mutans induced by a quorum-sensing peptide occurs via a conserved streptococcal autolysin. J Bacteriol 195:105–114. doi: 10.1128/JB.00926-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bitoun JP, Liao S, Yao X, Ahn S-J, Isoda R, Nguyen AH, Brady LJ, Burne RA, Abranches J, Wen ZT. 2012. BrpA is involved in regulation of cell envelope stress responses in Streptococcus mutans. Appl Environ Microbiol 78:2914–2922. doi: 10.1128/AEM.07823-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wen ZT, Burne RA. 2002. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl Environ Microbiol 68:1196–1203. doi: 10.1128/aem.68.3.1196-1203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arrigucci R, Pozzi G. 2017. Identification of the chain-dispersing peptidoglycan hydrolase LytB of Streptococcus gordonii. PLoS One 12:e0176117. doi: 10.1371/journal.pone.0176117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De A, Liao S, Bitoun JP, Roth R, Beatty WL, Wu H, Wen ZT. 2017. Deficiency of RgpG causes major defects in cell division and biofilm formation, and deficiency of LytR-CpsA-Psr family proteins leads to accumulation of cell wall antigens in culture medium by Streptococcus mutans. Appl Environ Microbiol 83:e00928-17. doi: 10.1128/AEM.00928-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madsen JS, Burmølle M, Hansen LH, Sørensen SJ. 2012. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol Med Microbiol 65:183–195. doi: 10.1111/j.1574-695X.2012.00960.x. [DOI] [PubMed] [Google Scholar]
- 37.Saputo S, Faustoferri RC, Quivey RG. 2017. Vitamin D compounds are bactericidal against Streptococcus mutans and target the bacitracin-associated efflux system. Antimicrob Agents Chemother 62:e01675-17. doi: 10.1128/AAC.01675-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ready D, Roberts AP, Pratten J, Spratt DA, Wilson M, Mullany P. 2002. Composition and antibiotic resistance profile of microcosm dental plaques before and after exposure to tetracycline. J Antimicrob Chemother 49:769–775. doi: 10.1093/jac/dkf005. [DOI] [PubMed] [Google Scholar]
- 39.Domenech M, García E, Moscoso M. 2009. Versatility of the capsular genes during biofilm formation by Streptococcus pneumoniae. Environ Microbiol 11:2542–2555. doi: 10.1111/j.1462-2920.2009.01979.x. [DOI] [PubMed] [Google Scholar]
- 40.Moscoso M, García E, López R. 2006. Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J Bacteriol 188:7785–7795. doi: 10.1128/JB.00673-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rukke HV, Hegna IK, Petersen FC. 2012. Identification of a functional capsule locus in Streptococcus mitis. Mol Oral Microbiol 27:95–108. doi: 10.1111/j.2041-1014.2011.00635.x. [DOI] [PubMed] [Google Scholar]
- 42.Tanabe S-I, Bonifait L, Fittipaldi N, Grignon L, Gottschalk M, Grenier D. 2010. Pleiotropic effects of polysaccharide capsule loss on selected biological properties of Streptococcus suis. Can J Vet Res 74:65–70. [PMC free article] [PubMed] [Google Scholar]
- 43.Di Xia F, Mallet A, Caliot E, Gao C, Trieu-Cuot P, Dramsi S. 2015. Capsular polysaccharide of group B Streptococcus mediates biofilm formation in the presence of human plasma. Microbes Infect 17:71–76. doi: 10.1016/j.micinf.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 44.Rainey K, Michalek SM, Wen ZT, Wu H. 2019. Glycosyltransferase-mediated biofilm matrix dynamics and virulence of Streptococcus mutans. Appl Environ Microbiol 85:e02247-18. doi: 10.1128/AEM.02247-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 46.Narisawa N, Kawarai T, Suzuki N, Sato Y, Ochiai K, Ohnishi M, Watanabe H, Senpuku H. 2011. Competence-dependent endogenous DNA rearrangement and uptake of extracellular DNA give a natural variant of Streptococcus mutans without biofilm formation. J Bacteriol 193:5147–5154. doi: 10.1128/JB.05240-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shiroza T, Kuramitsu HK. 1993. Construction of a model secretion system for oral streptococci. Infect Immun 61:3745–3755. doi: 10.1128/IAI.61.9.3745-3755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kawarai T, Narisawa N, Suzuki Y, Nagasawa R, Senpuku H. 2016. Streptococcus mutans biofilm formation is dependent on extracellular DNA in primary low pH conditions. J Oral Biosci 58:55–61. doi: 10.1016/j.job.2015.12.004. [DOI] [Google Scholar]
- 49.Corinaldesi C, Danovaro R, Dell'Anno A. 2005. Simultaneous recovery of extracellular and intracellular DNA suitable for molecular studies from marine sediments. Appl Environ Microbiol 71:46–50. doi: 10.1128/AEM.71.1.46-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cury JA, Seils J, Koo H. 2008. Isolation and purification of total RNA from Streptococcus mutans in suspension cultures and biofilms. Braz Oral Res 22:216–222. doi: 10.1590/s1806-83242008000300005. [DOI] [PubMed] [Google Scholar]
- 51.Merritt J, Kreth J, Qi F, Sullivan R, Shi W. 2005. Non-disruptive, real-time analyses of the metabolic status and viability of Streptococcus mutans cells in response to antimicrobial treatments. J Microbiol Methods 61:161–170. doi: 10.1016/j.mimet.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 52.LeBlanc DJ, Lee LN, Abu-Al-Jaibat A. 1992. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid 28:130–145. doi: 10.1016/0147-619X(92)90044-B. [DOI] [PubMed] [Google Scholar]
- 53.Ajdić D, McShan WM, McLaughlin RE, Savić G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, Lin S, Qian Y, Li S, Zhu H, Najar F, Lai H, White J, Roe BA, Ferretti JJ. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A 99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki Y, Nagasawa R, Senpuku H. 2017. Inhibiting effects of fructanase on competence-stimulating peptide-dependent quorum sensing system in Streptococcus mutans. J Infect Chemother 23:634–641. doi: 10.1016/j.jiac.2017.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.