TCC’s widespread use as an antimicrobial agent has made it a ubiquitous environmental pollutant despite its withdrawal due to ecological and toxicological concerns. With its antibacterial mechanism of action still being unknown, we undertook a comparative target analysis between TCC, PK150 (a recently discovered antibacterial compound with structural resemblance to TCC), and TCS (another widely employed chlorinated biphenyl antimicrobial) in the bacterium Staphylococcus aureus. We show that there are distinct differences in each compound’s mode of action, but also identify a shared target between TCC and PK150, the interference with menaquinone metabolism by inhibition of MenG. The prevailing differences, however, which also manifest in a remarkably better broad-spectrum activity of PK150, suggest that even high levels of TCC or TCS resistance observed by continuous environmental exposure may not affect the potential of PK150 or related N,N′-diaryl urea compounds as new antibiotic drug candidates against multidrug-resistant infections.
KEYWORDS: Staphylococcus aureus, triclocarban, triclosan, antimicrobial agents, mechanisms of action, proteomics
ABSTRACT
Triclocarban (TCC), a formerly used disinfectant, kills bacteria via an unknown mechanism of action. A structural hallmark is its N,N′-diaryl urea motif, which is also present in other antibiotics, including the recently reported small molecule PK150. We show here that, like PK150, TCC exhibits an inhibitory effect on Staphylococcus aureus menaquinone metabolism via inhibition of the biosynthesis protein demethylmenaquinone methyltransferase (MenG). However, the activity spectrum (MIC90) of TCC across a broad range of multidrug-resistant staphylococcus and enterococcus strains was much narrower than that of PK150. Accordingly, TCC did not cause an overactivation of signal peptidase SpsB, a hallmark of the PK150 mode of action. Furthermore, we were able to rule out inhibition of FabI, a confirmed target of the diaryl ether antibiotic triclosan (TCS). Differences in the target profiles of TCC and TCS were further investigated by proteomic analysis, showing complex but rather distinct changes in the protein expression profile of S. aureus. Downregulation of the arginine deiminase pathway provided additional evidence for an effect on bacterial energy metabolism by TCC.
IMPORTANCE TCC’s widespread use as an antimicrobial agent has made it a ubiquitous environmental pollutant despite its withdrawal due to ecological and toxicological concerns. With its antibacterial mechanism of action still being unknown, we undertook a comparative target analysis between TCC, PK150 (a recently discovered antibacterial compound with structural resemblance to TCC), and TCS (another widely employed chlorinated biphenyl antimicrobial) in the bacterium Staphylococcus aureus. We show that there are distinct differences in each compound’s mode of action, but also identify a shared target between TCC and PK150, the interference with menaquinone metabolism by inhibition of MenG. The prevailing differences, however, which also manifest in a remarkably better broad-spectrum activity of PK150, suggest that even high levels of TCC or TCS resistance observed by continuous environmental exposure may not affect the potential of PK150 or related N,N′-diaryl urea compounds as new antibiotic drug candidates against multidrug-resistant infections.
INTRODUCTION
Triclocarban (TCC) is a broad-spectrum anti-infective agent that has been used in personal care products such as soaps and lotions since the 1960s (1). In 2017, however, the Food and Drug Administration (FDA) banned the use of TCC together with that of triclosan (TCS), another halogenated biphenyl antimicrobial agent (Fig. 1), because of safety concerns. TCC and TCS can be absorbed by the body (2, 3) and were found to exhibit several mammalian off-target effects; both act as weak endocrine disruptors (4), and TCC was additionally found to exhibit anti-inflammatory effects by inhibiting soluble epoxide hydrolase and to alter cardiac function by interference with fatty acid metabolism in mouse models (3, 5, 6). TCS, in contrast, was demonstrated to depolarize the inner mitochondrial membrane and uncouple oxidative phosphorylation (7–9). Adding to these health-related concerns are environmental problems; due to their multiple chlorination sites, these compounds are persistent to biodegradation and were thus shown to accumulate in the environment and cause potentially toxic effects in soil and water organisms (10–15).
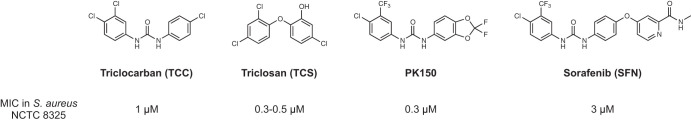
FIG 1.
Structures of the antibacterial agents triclocarban (TCC) and triclosan (TCS), structurally related PK150, and sorafenib (SFN). Their activities (MICs) in S. aureus NCTC 8325 are displayed below the respective structures.
In contrast to the very extensive studies on its impact on health and environment, surprisingly little is known about the antibacterial mode of action of TCC. While TCS is known to inhibit enoyl-acyl-carrier-protein reductase FabI (16–18) and has been proposed to further disturb the lipid bilayer nonspecifically at higher concentrations (19), the corresponding mechanisms for TCC are rather unclear, and studies into their elucidation are scarce; one study investigated the production of membrane leakage by several compounds but found no such effect for TCC (20). Comparison of TCC and TCS by molecular dynamics simulations, though, suggested that at high concentrations both compounds might destabilize the lipid bilayer membrane in a similar nonspecific manner (21). Finally, a study which aimed at generating improved derivatives of TCC investigated the mode of action by fluorescence staining and microscopy and found TCC to induce cell lysis and membrane deformation (22). However, a specific protein target as in the case of TCS has not been identified so far.
Recently, we published the development of a novel antibacterial compound named PK150, which was derived from the anticancer drug sorafenib (SFN) (Fig. 1) (23). Extensive mode of action analysis validated two independent protein targets in Staphylococcus aureus, which involved the overactivation of signal peptidase Ib (SpsB), a key enzyme in the bacterial secretory pathway, and inhibition of demethylmenaquinone methyltransferase (MenG), the last step in the biosynthesis of the essential electron carrier menaquinone. Given the structural similarity between PK150 and TCC, including the characteristic N,N′-diaryl urea motif, it is conceivable that some of these mechanistic features identified for PK150 might also apply to TCC. This is corroborated by some similarities that both compounds exert on target cells, including membrane deformation and cell lysis (23). Additionally, both compounds are only active in Gram-positive bacteria, and development of resistance has not yet been observed under laboratory conditions (23, 24).
In this work, we show that the target profile of TCC exhibits similar but also distinct features compared to PK150. While MenG is inhibited by both compounds, we rule out a contribution of SpsB to TCC’s mode of action. Furthermore, TCC does not inhibit FabI and also shows distinct differences to TCS in proteomic profiling. As such, this work underlines the versatility of N,N′-diaryl urea and diaryl ether antibiotics and proposes the contribution of a specific target to the overall mode of action of TCC.
RESULTS
PK150 and TCC differ in their spectrum of activity against multidrug-resistant staphylococci and enterococci.
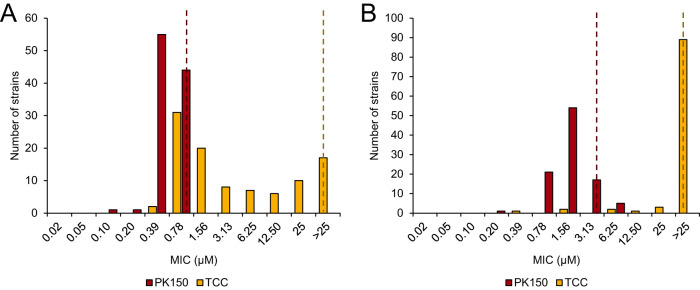
We first analyzed whether the two N,N′-diaryl urea compounds PK150 and TCC exert comparable activity profiles against a variety of multidrug-resistant clinical strains, among them bacteria resistant to last-resort antibiotics such as linezolid, vancomycin, or daptomycin. We determined the MIC values of both compounds in 100 different staphylococcus strains, mainly S. aureus, and 100 enterococcus strains, mainly Enterococcus faecium and Enterococcus faecalis, to define the MIC90 (Fig. 2; see also Data File S1 in the supplemental material). We found the activity profile of both compounds to be very different; PK150 exhibited activity against all tested strains, with an MIC90 of 0.78 μM against staphylococci and an MIC90 of 3.13 μM against enterococci. In contrast, TCC showed a remarkably broad MIC distribution and was inactive against 17% of staphylococci and 89% of enterococci (MIC90 of >25 μM in both cases). This not only demonstrates that the antibacterial properties of PK150 are superior to those of TCC but also suggests fundamental differences in their mode of action. We further looked into 40 staphylococcus strains exhibiting intermediate or high-level resistance against TCC (MIC ≥ 6.25 μM), but we did not observe any differences regarding their general resistance pattern compared to the full strain selection (Data File S1). However, we noted that none of those 40 strains belonged to the clonal complex subtype 5 ST225 (spa type t003, Rhine-Hesse epidemic strain), which is one of the predominant hospital-associated methicillin-resistant S. aureus (MRSA) strains in Germany, although 9% of our collection comprised this subtype (25, 26).
FIG 2.
TCC exhibits a different activity spectrum than that of PK150. MIC determination of PK150 and TCC against a panel of 100 strains of staphylococci (A) and enterococci (B) is shown. MICs were tested up to a concentration of 25 μM, and the distribution was plotted for each compound. Dotted lines indicate the respective MIC90 values. Experiments were performed in two biologically independent replicates, and for cases in which the MIC differed between the replicates the higher value was taken as the resulting MIC. See Data File S1 in the supplemental material for further details.
TCC and TCS show little to no effect on bacterial membrane integrity.
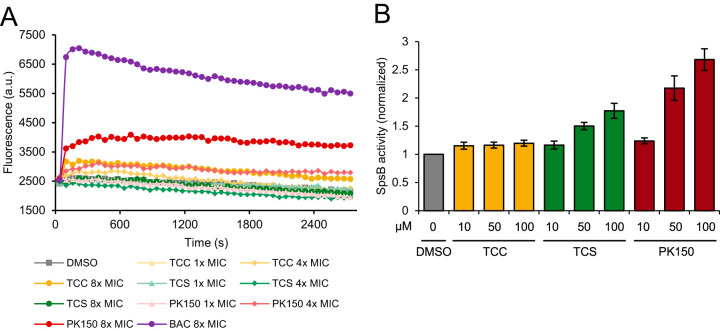
We next investigated the mechanistic reasons for these observed differences. First, we looked into the effect of TCC on the general integrity of the bacterial cell membrane. As mentioned above, evidence for induced membrane leakage was not found in previous experiments, but TCC is predicted to perturb the membrane at higher concentrations (20, 21). For our assay, we analyzed the interaction of the dye propidium iodide with bacterial DNA, which is only accessible if the cell membrane is permeabilized. As a positive control, the detergent benzalkonium chloride (27) was used and evoked an immediate and drastic increase in fluorescence (Fig. 3A). PK150 was previously shown to induce an increase in dye fluorescence, likely a consequence of autolysin-induced cell lysis (23), and a moderate, concentration-dependent effect was also observed here (Fig. 3A). Of note, the strain tested here differs from the strain used in the previous study, which could account for differences in total fluorescence increase. In contrast, TCC induced a much weaker effect than PK150, and TCS showed no change at all compared to the dimethyl sulfoxide (DMSO) control (Fig. 3A). This confirms that both TCC and TCS do not act via membrane perturbation at concentrations close to their MIC and further suggests the existence of specific targets for TCC and TCS (where FabI is a known target).
FIG 3.
Effects on membrane integrity and SpsB activation potential by TCC, TCS, and PK150. (A) Membrane integrity assay analyzing the permeability of S. aureus NCTC 8325 cells upon treatment with TCC, TCS, PK150, or benzalkonium chloride (BAC) as a positive control. Interaction of propidium iodide with DNA was measured over time. Data are representative for three biological replicates. (B) Förster resonance energy transfer (FRET)-based SpsB activity assay with membrane-bound endogenous SpsB from S. aureus NCTC 8325 (200 μg/ml total protein concentration) and different concentrations of TCC, TCS, and PK150 (positive control). Substrate cleavage rates are normalized to dimethyl sulfoxide (DMSO)-treated samples. Data represent mean values ± standard deviation (SD) of averaged triplicates of biologically independent experiments (n = 4).
TCC does not overactivate PK150’s target SpsB.
Given their structural similarity, we next examined whether TCC interacts with the known targets of PK150. One unprecedented property of PK150 is its capability to overactivate the signal peptidase SpsB. This can be measured in a Förster resonance energy transfer (FRET)-based peptidase assay using an artificial fluorescent substrate and endogenous SpsB embedded into native membranes of S. aureus (23). We performed this assay at three different concentrations of both TCC and TCS and also added PK150 as a positive control for an SpsB activator (Fig. 3B). TCC did not exhibit a stimulatory effect on SpsB activity, indicating an important mechanistic difference from PK150. Interestingly, TCS induced a moderate activation of the enzyme, despite its structure being less similar to PK150 than PK150 is to TCC.
TCC inhibits PK150’s other target, MenG.
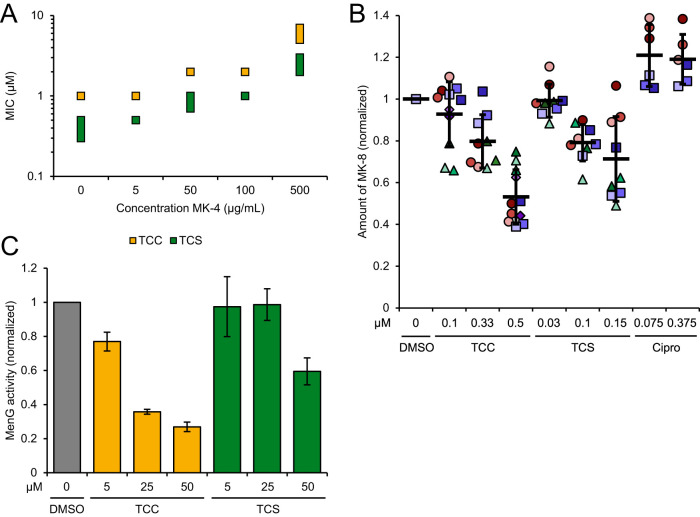
Regarding the second confirmed cellular mechanism of PK150, inhibition of MenG, we examined whether addition of menaquinone-4 (MK-4) rescued bacteria from TCC and TCS treatment. Interestingly, a moderate shift in MIC up to about 6-fold, dependent on MK-4 concentration, was observed (Fig. 4A) for both compounds. To exclude unspecific effects, such as aggregation of the compounds by high concentrations of MK-4 or a general fitness increase of the cells (due to the additional vitamin supply), which could interfere with the assay and contribute to these results, we performed further validation experiments: First, we quantified endogenous levels of menaquinone-8 (MK-8) in live S. aureus cells treated with TCC, TCS, or ciprofloxacin (Cipro) as the control. MK-8 levels were reduced in a concentration-dependent manner by up to 47% by TCC and up to 29% by TCS, on average, which was not observed with ciprofloxacin treatment (Fig. 4B). Second, we directly measured MenG inhibition by methylation of the artificial substrate demethylmenaquinone-2 (DMK-2) in lysates of S. aureus cells overexpressing MenG, extraction, and subsequent liquid chromatography-mass spectrometry (LC-MS) analysis. Again, TCC showed stronger effects and reduced MenG activity by 23% at 5 μM compound concentration and by up to 73% at 50 μM, while TCS inhibited MenG only at a concentration of 50 μM and to an extent of 41% (Fig. 4C). PK150, in comparison, had been shown to diminish endogenous MK-8 by up to 33% and to reduce MenG activity by 70% at the same respective concentrations (23).
FIG 4.
Interference with menaquinone biosynthesis and inhibition of MenG by TCC and TCS. (A) MIC shift by addition of menaquinone-4 (MK-4) to the bacterial growth medium. MICs of TCC and TCS were determined in the presence of various concentrations of MK-4 as indicated. The data represent results from three biologically independent experiments performed in triplicates. Where the MIC varied between replicates, the MIC is given as a range as displayed by the extension of the data point. (B) Metabolic profiling of endogenous menaquinone levels in S. aureus NCTC 8325 cells on compound treatment. Bacteria were treated with subinhibitory concentrations (0.1-fold to 0.5-fold of the respective MIC) of TCC, TCS, or ciprofloxacin (Cipro) and menaquinone-8 (MK-8) was extracted and quantified by LC–MS. MK-8 levels are normalized to DMSO-treated samples. Each color represents an individual, independent extraction experiment, where differently shaped symbols (circles, rectangles, triangles and diamonds) indicate independent biological samples. Error bars denote mean values ± SD. (C) Enzymatic assay monitoring the methylation of DMK-2 by cellular lysate of MenG-overexpressing S. aureus pRMC2-MenG (20 mg/ml total protein concentration). Production of MK-2 was quantified by liquid chromatography-mass spectrometry (LC-MS) and normalized to the respective DMSO-treated samples. Data represent the averaged values ± SD from three independent experiments.
Neither TCC nor PK150 inhibits TCS’s target FabI.
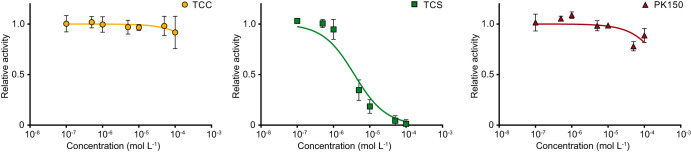
Given the similarity between TCS and TCC with regard to mammalian off-target effects and environmental behavior, and the fact that FabI is a confirmed target of TCS, we next investigated whether TCC might interact with FabI in a similar manner. We cloned and purified S. aureus FabI and assayed its activity on reducing trans-2-octenoyl N-acetylcysteamine thioester (t-o-NAC thioester), a common artificial substrate for this enzyme (28). We were able to reproduce the inhibition caused by TCS, whereas neither TCC nor PK150 exhibited a significant inhibitory effect (Fig. 5).
FIG 5.
Inhibition of FabI by TCC, TCS, and PK150. Recombinant FabI was preincubated with NADPH and compounds at different concentrations, and then the substrate t-o-NAC thioester was added and the decrease in absorbance at 340 nm was recorded. Enzyme activity was determined by the slope at t = 0, and relative activity was calculated by normalization to the DMSO-treated control (100% activity) and the heat-inactivated negative control (0% activity). The data were subsequently fitted to a sigmoidal equation. For TCS, the 50% inhibitory concentration (IC50) was calculated to be 3.76 ± 0.11 μM. Data represent mean values ± SD of averaged triplicates from three independent experiments.
Full proteome analysis reveals different patterns for TCC and TCS treatment.
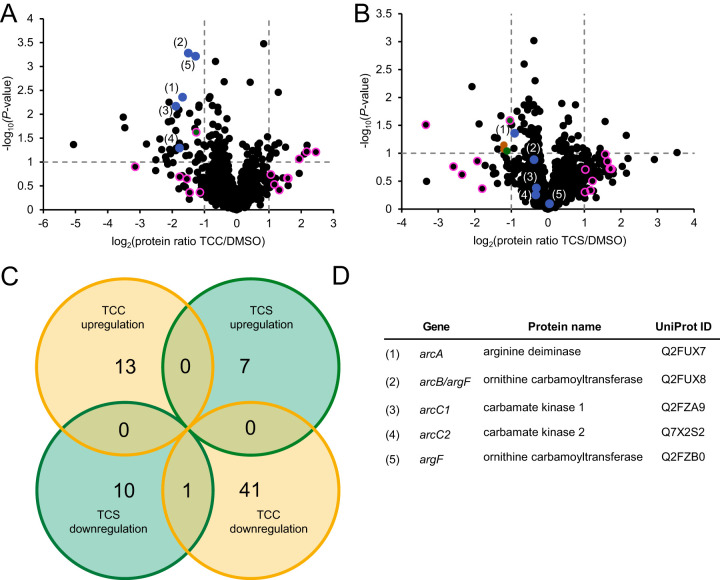
In order to analyze the effects evoked by the compounds on a global level, we performed whole-proteome analysis of S. aureus cells treated with subinhibitory concentrations (0.5× MIC) of TCC or TCS (Fig. 6A and B; see Data File S2 in the supplemental material). Bacterial proteins were digested with trypsin and subjected to analysis by liquid chromatography-tandem mass spectrometry following label free quantification of identified peptides (29). Analysis of proteins whose expression levels were significantly altered between DMSO- and compound-treated condition [|log(protein ratio)| > 1 and −log(P value) > 1] revealed only one overlapping protein between TCC and TCS treatment (phosphotransferase system [PTS] EIIBC component; GenPept accession number Q2G1G5), supporting the view that the underlying mechanisms of action are rather distinct from each other (Fig. 6C). We further tried to identify particular pathways that might be up- or downregulated by network analysis with STRING (Fig. 6A, B, and D; Data File S2) (30). Most strikingly, a significant downregulation of the entire arginine deiminase pathway (arginine deiminase [GenPept accession number Q2FUX7, encoded by arcA], carbamate kinase 1 [accession number Q2FZA9, encoded by arcC1], carbamate kinase 2 [accession number Q7X2S2, encoded by arcC2], and two ornithine carbamoyltransferases [accession number Q2FUX8, encoded by arcB {despite being annotated as argF in NCTC 8325}, and accession number Q2FZB0, encoded by argF]) was observed upon TCC treatment (Fig. 6D) (31). Regarding upregulated proteins, two proteins were found to be generally associated with dicarboxylic acid metabolism by GO (32, 33) annotation (Data File S2), but no significant interaction network was detected. For TCS treatment, there was no significant pathway enrichment among upregulated proteins, and only a few downregulated proteins were found to be involved in functional enrichment; these are associated with carbohydrate phosphotransferase activity or transmembrane transport by gene ontology (GO) (32, 33) and Pfam (34) annotation, but do not give rise to a comprehensive pathway (for details, see Data File S2).
FIG 6.
Full proteome analysis of S. aureus NCTC 8325 upon TCC and TCS treatment. (A and B) Volcano plots showing the log2 fold change of protein levels in the full proteome of S. aureus treated with subinhibitory concentrations (0.5× MIC) of TCC (A) or TCS (B). Colors denote pathways that were significantly enriched among downregulated proteins [log2(protein ratio) < −1 and −log(P value) > 1] by STRING (30) analysis. Blue dots represent the proteins associated with the arginine deiminase (ADI) pathway (see below) which were affected by TCC treatment. Green and brown dots represent proteins associated with the EIIB/EIIC phosphotransferase system and ABC transporter transmembrane domains, respectively. These terms were found to be enriched in the STRING analysis of TCS treatment. Pink circles represent all proteins—regardless of P value—that were up- or downregulated [|log2(protein ratio)| > 1] by both TCC and TCS treatment. The only protein also meeting the significance cutoff [−log(P value) > 1] is shown in green (PTS EIIBC component; GenPept accession number Q2G1G5). The data represent average values, and the P values were calculated using a two-sided two-sample t test; n = 4 independent experiments per group. See Data File S2 in the supplemental material for further details. (C) Venn diagram showing overlap in protein up- or downregulation between TCC and TCS treatment. The respective circles indicate the total number of proteins for each stated case [|log2(protein ratio)| > 1 and −log(P value) > 1]. (D) Table identifying proteins associated with the ADI pathway (31) in the volcano plots in panels A and B. All proteins are annotated within the KEGG pathway “arginine biosynthesis.” Proteins 1, 3, and 4 are directly annotated as elements of the ADI pathway in S. aureus NCTC 8325 by GO (33). The gene encoding protein 2 is located in the same gene cluster as protein 1, and its homologs in related S. aureus strains (such as COL, MW2 or USA300) are annotated as part of the ADI pathway in UniProt. Protein 5 catalyzes the same reaction as protein 2, but according to its annotation seems to be involved in arginine biosynthesis rather than catabolism.
DISCUSSION
Overactivation of SpsB has been identified as a key feature of the PK150 scaffold that leads to increased secretion of autolytic enzymes and subsequent cell lysis, but despite its structural similarity, we could not confirm the same activity for TCC. In addition, TCC was inactive against most tested enterococci and against a good proportion of staphylococci. Although the reasons for this observation might be complex and further work is required for their exact elucidation, it seems plausible that the differences in SpsB activation could contribute to these diverging effects. Furthermore, it indicates that the mechanisms leading to TCC resistance do not affect susceptibility toward PK150, suggesting that environmental exposure of bacteria to TCC is unlikely to evoke cross-resistance against PK150 or related N,N′-diaryl urea compounds.
However, the shift of MIC in the presence of MK-4, the reduction of endogenous menaquinone levels in live cells, and the direct inhibition of MenG activity in cellular lysate are comparable traits of both compounds (23). Based on these findings, it is conceivable that MenG inhibition is involved in TCC’s mode of action, which points toward it being the first ever identified molecular target of TCC. Of course, this does not exclude the existence of additional target mechanisms, which can include other proteins and, in principle, nonspecific effects such as membrane disruption. However, our finding that TCC barely affects membrane permeability, which is in accordance with its failure to induce potassium leakage in an earlier study (20), is contradictory to unspecific membrane damage and supports the view of a more specific mechanism.
Nevertheless, the proteomic data deliver a complex picture of the processes underlying TCC treatment. Of note, no effect on menaquinone metabolism was observed. Instead, the most significant finding was a downregulation of the arginine deiminase pathway. Interestingly, this pathway was found to be upregulated in the proteomes of S. aureus resistant to the PK150 parent compound SFN (23). The arginine deiminase pathway catabolizes arginine to carbamoyl phosphate, which is further converted to ammonia, carbon dioxide and ATP (31). It has been associated with enhanced fitness and virulence, as the MRSA strain USA300 carries a mobile genetic element containing an additional arginine deiminase operon, which serves as an epidemiological marker (35). Importantly, it represents a way to generate energy that is independent of menaquinone (31). Thus, while its upregulation makes sense for cells resistant to a disruptor of menaquinone biosynthesis, it is rather surprising that the exact opposite is found in the case of TCC and nonresistant cells. Such a response would actually aggravate the detrimental consequences of MenG inhibition. Hence, although the exact mechanisms inducing these changes remain unclear, they appear to strengthen the antibiotic effect of TCC and also provide an independent link for alterations in bacterial energy metabolism.
The proteomic data further revealed a completely different picture in the case of TCS treatment. Unfortunately, the pattern of up- and downregulated proteins does not give rise to a clearly targeted pathway. Nevertheless, together with the finding that TCC does not inhibit FabI, these data suggest that both compounds act by different mechanisms.
An interesting finding is that TCS exhibits a stimulation of SpsB activity as well as an inhibitory effect on menaquinone biosynthesis. Both effects are only weak to moderate and are comparable to what was observed with SFN, which is about ten times less potent than TCS (23). Of note, mutations in FabI, which are commonly referred to as conferring TCS resistance, do not produce fully resistant strains but merely decrease the activity of TCS by about an order of magnitude (18, 36). In addition, similarly TCS-resistant bacteria have also been isolated that show no alterations in the fabI gene (36). It is thus not surprising that the involvement of other, unknown targets for the mode of action of TCS has been proposed before (37). Although further experiments are needed for validation, our experiments point to the possibility that SpsB and MenG could represent such additional targets, particularly at higher TCS concentrations.
Taken together, we identified MenG as a putative molecular target of TCC, reminiscent of structurally related PK150, but also established distinct differences between the two compounds, as well as between TCC and TCS. Hence, our findings shed some light on the obscure mechanism of TCC and underline menaquinone biosynthesis inhibition as an important antibiotic target pathway. In addition, our work highlights the novelty of the PK150 structure as a promising entity with potential against multidrug-resistant pathogens, which is unlikely to be affected by environmentally acquired TCC resistance.
MATERIALS AND METHODS
Materials and strains.
TCC and TCS were purchased from Sigma-Aldrich. MK-4 and MK-9 were purchased from Santa Cruz Biotechnology and Carbosynth, respectively. PK150 and DMK-2 were synthesized as reported previously (23). Unless otherwise stated, S. aureus NCTC 8325 (Institute Pasteur, France) was used for laboratory experiments and was cultured in B medium (10 g/liter casein peptone, 5 g/liter NaCl, 5 g/liter yeast extract, and 1 g/liter K2HPO4 [pH 7.5]). The collection of staphylococci and enterococci for MIC90 determination was provided by the National Reference Centre for Staphylococci and Enterococci (Robert Koch Institute, Wernigerode, Germany). Further details on the specific strains can be found in Data File S1 in the supplemental material.
MIC determination, MIC90, and MK-4 shift assay.
Staphylococcus aureus cultures were grown at 37°C with gentle shaking (200 rpm) in B medium from overnight cultures until an optical density at 600 nm (OD600) of ∼0.4 to 0.6, and then diluted into B medium to give a final concentration of 105 CFU/ml. Compounds at various concentrations were added to diluted bacterial cultures (100 μl/well final volume; final concentration of DMSO from compound stocks was 1%) in triplicates. A growth control containing DMSO only and a sterile control containing medium were included. After incubation (37°C, 200 rpm, 24 h), the dilution series was analyzed for microbial growth as indicated by turbidity. The lowest concentration in the dilution series at which no growth of bacteria could be observed by eye was defined as the MIC of the compound. MIC values were determined by three independent experiments. Note that for the case of TCS, the MIC against S. aureus NCTC 8325 was found to vary between 0.3 μM and 0.5 μM. For experiments relating to this MIC, a value was determined beforehand and this same value taken for the subsequent respective experiment. These values were as follows: 0.3 μM for metabolic menaquinone profiling (Fig. 4B) and full proteome analysis (Fig. 6), 0.39 μM for membrane integrity assay (Fig. 3A), and 0.5 μM for MIC shift assay (Fig. 4A).
For MIC90 determination, clinical isolates were grown on Mueller-Hinton Broth (MHB) culture plates, and 3 to 4 single colonies were picked and used to inoculate 5 ml MHB. Cultures were grown until they reached an optical density corresponding to 0.5 McFarland units. These cultures were diluted 1:50 in 0.85% (wt/vol) NaCl solution to prepare the final inoculum. A 1-μl aliquot of a 3 mM stock solution in DMSO of the respective test compounds was placed in the first column of a round-bottomed 96-well plate. The wells of this column were filled up with 99 μl of MHB. Columns 2 to 12 were filled with 50 μl MHB, and 2-fold serial dilutions of the test compounds were made by transferring 50 μl from column 1 to column 2 and so forth until column 10. Column 11 served as a growth control and column 12 as a sterile control. The plate was inoculated by adding 10 μl of the freshly prepared inoculum to the wells of columns 1 through 11. Bacteria were grown at 37°C for 20 h without agitation. The MIC was determined as the lowest compound concentration with no observable bacterial growth after visual inspection. All MIC raw data and information on the characteristics of the tested strains have been uploaded to the supplemental material as an Excel file (Data File S1).
For MIC shift assays in the presence of exogenous menaquinone-4 (MK-4), the growth medium was additionally supplemented with different concentrations of MK-4 before addition of the bacteria, giving a total DMSO concentration in the final assay of 1.5%.
Membrane integrity assay.
To assess cell wall permeability upon treatment with compounds, cells (S. aureus NCTC 8325) were grown (37°C, 200 rpm) from respective overnight cultures until they reached an OD600 value of 0.4 to 0.8. The cells were then harvested (6,000 × g, 4°C, 5 min), washed with 5 mM HEPES-NaOH buffer (pH 7.2; supplemented with 5 mM glucose), and then resuspended in the same buffer to an OD600 of 0.4. The assay was conducted in 96-well plates (Nunc flat-bottomed transparent 96-well plates; Thermo Fisher Scientific) with 99 μl cell suspension per well. The assay was started by adding 1 μl of 1 mM propidium iodide (Sigma-Aldrich) in dimethylformamide (DMF) to each well (10 μM final concentration) and incubating at 37°C in the Tecan Infinite M200 Pro microplate reader to allow the propidium iodide to be integrated into the membranes. During incubation, fluorescence was measured (535-nm excitation and 617-nm emission). After 15 min, 1 μl of the respective compound or control was added to each well from DMSO stocks with the appropriate concentrations. The fluorescence was measured for 60 min at 37°C. As a positive control, 16 μg/ml (8-fold MIC) of the detergent benzalkonium chloride (BAC) was added to the cell suspension, while 1 μl DMSO was added for a baseline fluorescence control.
FRET-based SpsB activity assay.
SpsB activities were measured using membranes of S. aureus and a Förster resonance energy transfer (FRET) assay as described previously (23, 38). Briefly, S. aureus NCTC 8325 membranes containing native SpsB (0.2 mg/ml total protein concentration) were pretreated with different concentrations of compound (TCC, TCS, and positive-control PK150) or DMSO for 5 min at 37°C. A synthetic peptide substrate based on SceD modified by 4-(4-dimethylaminophenylazo)benzoic acid (DABCYL) and 5-((2-aminoethyl)amino)-1-naphthalenesulfonic acid (EDANS) was added (DABCYL-AGHDAHASET-EDANS; AnaSpec) to give 10 μM final substrate concentration (1:100 from DMF stock), and fluorescence was monitored by a Tecan Infinite 200Pro plate reader for 1 h at 37°C (345-nm excitation and 510-nm emission wavelength). SpsB activity was determined by the slope of the fluorescence increase and normalized to the DMSO control.
Metabolic profiling of endogenous MK-8.
The experiments were performed as described previously (23). Mid-exponential-phase cultures of S. aureus NCTC 8325 were diluted into B medium to give a final concentration of 105 CFU/ml. TCC, TCS, and control ciprofloxacin (Cipro) were added to various final concentrations (TCC and TCS: 0.1×, 0.33×, and 0.5× MIC; Cipro: 0.1× and 0.5× MIC), and bacteria were incubated for 24 h (or longer if the OD600 was ≪ 6) at 37°C, 200 rpm. Equivalent amounts of cells (equal to 4 ml at an OD600 of 6) were harvested (6,000 × g, 4°C, 10 min), washed with phosphate-buffered saline (PBS), and either stored at −80°C for later use or directly resuspended in 10 ml 2-propanol 70% (vol/vol), supplemented with 500 ng/ml menaquinone-9 (MK-9) as an internal standard, and sonicated (3 × 30 s, 80% intensity; Sonopuls HD 2070 ultrasonic rod; Bandelin electronic GmbH) for extraction. Cellular debris were removed by centrifugation (12,000 × g, 30 min, 4°C), and the supernatant was loaded onto a Chromabond C18 solid-phase extraction column (3 ml, 500 mg; Macherey-Nagel) preconditioned with 6 ml methanol and 6 ml water. Columns were washed with 3 ml water, then 3 ml 2-propanol 70%, then eluted with 10 ml methanol/2-propanol 3:1 (vol/vol). The eluate was concentrated to dryness and resolved in 200 μl acetonitrile. LC-MS analysis was performed on an LTQ-FT Ultra Fourier-transform ion cyclotron resonance mass spectrometer coupled with an UltiMate 3000 high-performance liquid chromatography (HPLC) system (Thermo Fisher Scientific) using atmospheric pressure chemical ionization (APCI). A 10-μl aliquot of sample was injected into an XBridge BEH300 C4 column (3.5 μm, 2.1 × 150 mm; Waters) and eluted with a gradient of 54% acetonitrile/water and 0.1% formic acid (FA) to 90% acetonitrile/water and 0.1% FA over 1 h at a flow rate of 0.2 ml/min. Single-ion monitoring (SIM) of MK-8 ([M+H], 717.56) and MK-9 ([M+H], 785.62) was performed in parallel, and the respective areas under the curve (AUC) were determined for quantification using XCalibur software (Thermo Fisher Scientific). AUC for MK-8 were first normalized to the internal standard MK-9 and then normalized to the DMSO-treated controls.
MenG activity assay.
Cellular lysates containing overexpressed MenG were prepared from S. aureus pRMC2-MenG as described previously (23). Briefly, cells were grown in B medium with chloramphenicol (10 μg/ml) until an OD600 of ∼0.5 to 0.6, induced with anhydrotetracycline (400 ng/ml), and incubated overnight. Cells were harvested, washed with PBS, resuspended in 20 mM Tris-HCl (pH 7.5), 1 mM EDTA, and 10 mM β-mercaptoethanol, and then lysed and adjusted to a final protein concentration of 50 mg/ml.
For the assay, 500 μl of solution containing 100 mM Tris-HCl (pH 8), 10 mM MgCl2, 10 mM dithiothreitol (DTT), 100 μM S-adenosyl methionine, 25 μM DMK-2, 200 μl MenG lysate, and compounds (TCC and TCS) at different concentrations (5, 25, and 50 μM) were incubated at 37°C and 200 rpm for 80 min. After that, 7 ml 2-propanol 70%, supplemented with 500 ng/ml MK-4 as an internal standard, was added and extraction and quantification of menaquinone was performed as for metabolic profiling of MK-8, with the following alterations: 3 ml H2O was added to the centrifuged supernatant prior to loading onto the solid-phase extraction columns, and washing of the columns was conducted with 3 ml H2O only. The gradient of the LC-MS analysis ranged from 18% acetonitrile/water and 0.1% FA to 90% acetonitrile/water and 0.1% FA over 1 h, and single-ion monitoring was performed for DMK-2 ([M+H], 295.17), MK-2 ([M+H], 309.18) and MK-4 ([M+H], 445.31) in parallel. Resulting data were acquired by three independent experiments.
FabI cloning and activity assay.
FabI was amplified from genomic S. aureus DNA using forward (ggggacaagtttgtacaaaaaagcaggctttATGTTAAATCTTGAAAACAAAACA; capital letters indicate overlap with the fabI gene) and reverse (ggggaccactttgtacaagaaagctgggtgTTATTTAATTGCGTGGAATCC) primers that introduced AttB sites for Gateway cloning. The PCR product was cloned into donor vector pDONR201 (Invitrogen) by BP reaction, transformed in Escherichia coli XL1 blue, and reisolated, and a subsequent LR reaction was performed to clone the sequence into pET300/NT-Dest destination vector (Invitrogen), giving a FabI construct with an N-terminal His tag. This construct was first transformed into E. coli XL1 blue, then into E. coli BL21(DE3) for overexpression. Integrity of the construct was confirmed by Sanger sequencing (Genewiz). For expression and purification, mid-exponential-phase cultures were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and incubated for 4 h at 37°C and 200 rpm. Cells were harvested, washed with PBS, resuspended in lysis buffer (50 mM Tris [pH 8], 300 mM NaCl, and 5 mM imidazole), and lysed by sonication (4 × 5 min, 80% intensity; Sonopuls HD 2070 ultrasonic rod; Bandelin electronic GmbH). Debris was removed by centrifugation (22,000 × g, 4°C, 30 min), and the lysate was purified by affinity chromatography on a Ni-nitrilotriacetic acid (NTA) agarose column (5-ml column volume; GE Healthcare) on an Äkta protein purification system (GE Healthcare), using the buffer described above with 20 mM imidazole for washing and 500 mM for elution. After buffer exchange to 50 mM Tris (pH 8.5), 200 mM NaCl, and 2 mM DTT by dialysis, a size exclusion chromatography with a Superdex 200 10/300 GL (120-ml column volume; GE Healthcare) was performed for final purification. Protein purity was checked by SDS-PAGE.
The substrate t-o-NAC thioester was synthesized as described in the literature (28). Conditions for the FabI assay were adapted and modified from the procedure described there. Briefly, 1.4 μM FabI, 0.8 mM NADPH and compounds at various concentrations (0.1 μM to 100 μM) or DMSO were incubated in 50 mM sodium acetate (pH 6.5) for 10 min at 30°C, then t-o-NAC thioester (1.6 mM final concentration) was added, and the decrease in absorbance at 340 nm was monitored for 30 min at 30°C. Inhibition was determined by the slope of the absorbance curve at t = 0, relative to the DMSO-treated control (100% activity) and the heat control (0% activity, after incubation at 95°C for 10 min). The 50% inhibitory concentration (IC50) values were determined by fitting to a sigmoidal equation.
Full-proteome analysis.
S. aureus NCTC 8325 overnight cultures were diluted into B medium to give a final OD600 of 0.1 (40 ml per biological replicate) and then grown (37°C, 200 rpm) for 5 h. Cells were harvested (6,000 × g, 4°C, 10 min), washed with PBS, and resuspended in B medium to give a final cell density of 1.5 × 109 CFU/ml. Ten-milliliter portions were aliquoted into polypropylene plastic tubes and incubated with TCC (0.5 μM, 0.5× MIC), TCS (0.15 μM, 0.5× MIC), or DMSO at 37°C and 200 rpm for 1.5 h. Cells were pelleted by centrifugation (6,000 × g, 4°C, 10 min) and washed with PBS. Cells were stored at −80°C for at least 16 h and then lysed. For cell lysis, bacterial cell pellets were resuspended in 200 μl 100 mM Tris (pH 7.4) and lysed by sonication (5 × 20 s at 65% intensity with 30-s rest on ice in between; Sonopuls HD 2070 ultrasonic rod; Bandelin electronic GmbH). Eighty microliters of 10% (wt/vol) SDS and 1.25% (wt/vol) sodium deoxycholate was added, and samples were heated for 10 min at 90°C. Lysates were sonicated for 10 s at 10% intensity to shear nucleic acids and centrifuged (13,000 × g, room temperature [RT], 10 min) to pellet debris. Protein concentrations were measured using the Pierce BCA protein assay kit (Thermo Fisher Scientific, Pierce Biotechnology) and full-proteome samples were adjusted to equal protein amounts. Subsequently, proteins were precipitated with ice-cold acetone (4 volumes) at −20°C overnight and centrifuged (16,900 × g, 4°C, 15 min), and the pellets washed twice with ice-cold methanol (1 ml). Protein pellets were air dried and dissolved in denaturation buffer (7 M urea, 2 M thiourea in 20 mM [pH 7.5] HEPES buffer), then reduced with Tris(2-carboxyethyl)phosphine hydrochloride (TCEP; 10 mM; 37°C, 1,200 rpm, 1 h) and subsequently alkylated with iodoacetamide (12.5 mM; 25°C, 1,200 rpm, 30 min). Alkylation was quenched by the addition of dithiothreitol (12.5 mM) and subsequent incubation (RT, 1,200 rpm, 30 min). For digestion, LysC (1:200 enzyme:protein ratio; 0.5 μg/μl) was added to each sample, and the samples were incubated at RT with shaking at 700 rpm for 2 h. Following dilution with 50 mM triethylammonium bicarbonate (TEAB) buffer (1:5), samples were further digested with trypsin (1:100 enzyme:protein ratio; 0.5 μg/μl) at 37°C overnight. The reaction was stopped by adding FA to a final pH of 2 to 3. Peptides were desalted on-column using 50 mg SepPak C18 Vac cartridges (Waters); SepPak C18 cartridges were equilibrated once with acetonitrile (ACN) (1 ml), elution buffer (1 ml; 80% ACN and 0.5% FA) and three times with aqueous 0.5% FA solution (1 ml). Subsequently, the samples were loaded by gravity flow, washed three times with aqueous 0.5% FA solution (1 ml), eluted with elution buffer (0.5 ml, 80% ACN, and 0.5% FA) and lyophilized using a vacuum centrifuge.
Prior to mass spectrometry, peptides were reconstituted in 0.5% (vol/vol) FA and filtered through 0.22-μm polyvinylidene difluoride (PVDF) filters (Millipore). Samples were analyzed via HPLC-tandem mass spectrometry (MS/MS) using an UltiMate 3000 nano HPLC system (Dionex) equipped with an Acclaim C18 PepMap100 trap (75-μm internal diameter × 2 cm) and Acclaim C18 PepMap RSLC separation column (75-μm internal diameter × 50 cm) coupled to a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific). Peptides were loaded on the trap column at a flow rate of 5 μl/min with aqueous 0.1% trifluoroacetic acid (TFA) and then transferred onto the separation column at 0.4 μl/min. Buffers for the nanochromatography pump were aqueous 0.1% FA (buffer A) and 0.1% FA in ACN (buffer B). Samples were separated using a gradient that raised buffer B from 5% to 22% in 112 min, followed by a buffer B increase to 32% within 10 min. Buffer B content was further raised to 90% within the next 10 min and held for another 10 min at 90%. Subsequently, buffer B was decreased to 5% and held until the end of the run (total, 152 min). The MS instrument was operated in a TOP12 data-dependent mode. MS full scans were performed at a resolution of 140,000 in the orbitrap and the scan range was set from 300 to 1,500 m/z. The AGC target was set to 3.0 × 106, the maximum ion injection time was 50 ms, and internal calibration was performed using the lock mass option. Monoisotopic precursor selection as well as dynamic exclusion (exclusion duration, 60 s) was enabled. Precursors with charge states of >1 and intensities greater than 1.6 × 104 were selected for fragmentation. Isolation was performed in the quadrupole using a window of 1.6 m/z and an AGC target of 1.0 × 105 with a maximum injection time of 50 ms. Fragments were generated using higher-energy collisional dissociation (HCD) (normalized collision energy, 27%) and detected in the orbitrap.
Peptide and protein identifications were performed using MaxQuant (version 1.6.0.1) with Andromeda (39) as the search engine, using the following parameters: carbamidomethylation of cysteines as fixed and oxidation of methionine as dynamic modifications, trypsin/P as the proteolytic enzyme, 4.5 ppm for precursor mass tolerance (main search ppm), and 0.5 Da for fragment mass tolerance (ion trap mass spectrometry [ITMS] MS/MS tolerance). Searches were performed against the UniProt database for S. aureus NCTC 8325 (taxon identifier 93061, downloaded 19 August 2019). Quantification was performed using MaxQuant’s LFQ algorithm (29). The “I = L,” “requantify,” and “match between runs” (default settings) options were used. Identification was done with at least 2 unique peptides and quantification with unique peptides only.
For statistics with Perseus (version 1.6.0.0) (40), four biological replicates were analyzed. Putative contaminants, reverse hits, and proteins identified by site only (i.e., only identified by a peptide carrying a modified residue) were removed. LFQ intensities were log2(x) transformed and filtered to contain minimum three valid values in at least one condition. Missing values were imputed on the basis of a normal distribution (width = 0.3, downshift = 1.8). P values were obtained by a two-sided two-sample t test over the four biological replicates.
For pathway enrichment analysis using STRING (30), all proteins that showed a log2(protein enrichment) ratio of compound versus DMSO-treated control of more than 1 or less than −1 and a log10(P value) of >1 were entered into the database (https://string-db.org/; multiple proteins analysis), and the resulting enriched pathways were inspected further.
The processed tables for protein group analysis in Perseus and the results of the STRING analysis are included in the supplemental material as an Excel file (see Data File S2 in the supplemental material).
Data availability.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (41) partner repository under data set identifier PXD018347.
Supplementary Material
ACKNOWLEDGMENTS
R.M. was supported by a doctoral fellowship of the Boehringer Ingelheim Fonds. S.A.S. was funded by the Center for Integrated Protein Science Munich (CIPSM) and by Deutsche Forschungsgemeinschaft (DFG). M.W.H. and C.F. were funded by the Federal Ministry for Education and Research (BMBF) under the framework program VIP+ (project aBACTER).
We thank Philipp Le for critical discussions, Sabrina Schönberger for excellent experimental support, and Mona Wolff, Katja Bäuml, Katja Gliesche, and Franziska Erdmann for excellent technical support.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.MacKenzie AR. 1970. Effectiveness of antibacterial soaps in a healthy population. JAMA 211:973–976. doi: 10.1001/jama.1970.03170060037007. [DOI] [PubMed] [Google Scholar]
- 2.Calafat AM, Ye X, Wong LY, Reidy JA, Needham LL. 2008. Urinary concentrations of triclosan in the U.S. population: 2003–2004. Environ Health Perspect 116:303–307. doi: 10.1289/ehp.10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schebb NH, Inceoglu B, Ahn KC, Morisseau C, Gee SJ, Hammock BD. 2011. Investigation of human exposure to triclocarban after showering and preliminary evaluation of its biological effects. Environ Sci Technol 45:3109–3115. doi: 10.1021/es103650m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christen V, Crettaz P, Oberli-Schrämmli A, Fent K. 2010. Some flame retardants and the antimicrobials triclosan and triclocarban enhance the androgenic activity in vitro. Chemosphere 81:1245–1252. doi: 10.1016/j.chemosphere.2010.09.031. [DOI] [PubMed] [Google Scholar]
- 5.Liu JY, Qiu H, Morisseau C, Hwang SH, Tsai HJ, Ulu A, Chiamvimonvat N, Hammock BD. 2011. Inhibition of soluble epoxide hydrolase contributes to the anti-inflammatory effect of antimicrobial triclocarban in a murine model. Toxicol Appl Pharmacol 255:200–206. doi: 10.1016/j.taap.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie W, Zhang W, Ren J, Li W, Zhou L, Cui Y, Chen H, Yu W, Zhuang X, Zhang Z, Shen G, Li H. 2018. Metabonomics indicates inhibition of fatty acid synthesis, beta-oxidation, and tricarboxylic acid cycle in triclocarban-induced cardiac metabolic alterations in male mice. J Agric Food Chem 66:1533–1542. doi: 10.1021/acs.jafc.7b05220. [DOI] [PubMed] [Google Scholar]
- 7.Ajao C, Andersson MA, Teplova VV, Nagy S, Gahmberg CG, Andersson LC, Hautaniemi M, Kakasi B, Roivainen M, Salkinoja-Salonen M. 2015. Mitochondrial toxicity of triclosan on mammalian cells. Toxicol Rep 2:624–637. doi: 10.1016/j.toxrep.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teplova VV, Belosludtsev KN, Kruglov AG. 2017. Mechanism of triclosan toxicity: mitochondrial dysfunction including complex II inhibition, superoxide release and uncoupling of oxidative phosphorylation. Toxicol Lett 275:108–117. doi: 10.1016/j.toxlet.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Popova LB, Nosikova ES, Kotova EA, Tarasova EO, Nazarov PA, Khailova LS, Balezina OP, Antonenko YN. 2018. Protonophoric action of triclosan causes calcium efflux from mitochondria, plasma membrane depolarization and bursts of miniature end-plate potentials. Biochim Biophys Acta Biomembr 1860:1000–1007. doi: 10.1016/j.bbamem.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 10.Ying GG, Yu XY, Kookana RS. 2007. Biological degradation of triclocarban and triclosan in a soil under aerobic and anaerobic conditions and comparison with environmental fate modelling. Environ Pollut 150:300–305. doi: 10.1016/j.envpol.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Stasinakis AS, Mamais D, Thomaidis NS, Danika E, Gatidou G, Lekkas TD. 2008. Inhibitory effect of triclosan and nonylphenol on respiration rates and ammonia removal in activated sludge systems. Ecotoxicol Environ Saf 70:199–206. doi: 10.1016/j.ecoenv.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 12.Chalew TEA, Halden RU. 2009. Environmental exposure of aquatic and terrestrial biota to triclosan and triclocarban. J Am Water Works Assoc 45:4–13. doi: 10.1111/j.1752-1688.2008.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dann AB, Hontela A. 2011. Triclosan: environmental exposure, toxicity and mechanisms of action. J Appl Toxicol 31:285–311. doi: 10.1002/jat.1660. [DOI] [PubMed] [Google Scholar]
- 14.Gao L, Yuan T, Cheng P, Bai Q, Zhou C, Ao J, Wang W, Zhang H. 2015. Effects of triclosan and triclocarban on the growth inhibition, cell viability, genotoxicity and multixenobiotic resistance responses of Tetrahymena thermophila. Chemosphere 139:434–440. doi: 10.1016/j.chemosphere.2015.07.059. [DOI] [PubMed] [Google Scholar]
- 15.Hedrick-Hopper TL, Koster LP, Diamond SL. 2015. Accumulation of triclosan from diet and its neuroendocrine effects in Atlantic croaker (Micropogonias undulatus) under two temperature regimes. Mar Environ Res 112:52–60. doi: 10.1016/j.marenvres.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 16.McMurry LM, Oethinger M, Levy SB. 1998. Triclosan targets lipid synthesis. Nature 394:531–532. doi: 10.1038/28970. [DOI] [PubMed] [Google Scholar]
- 17.Heath RJ, Yu YT, Shapiro MA, Olson E, Rock CO. 1998. Broad spectrum antimicrobial biocides target the FabI component of fatty acid synthesis. J Biol Chem 273:30316–30320. doi: 10.1074/jbc.273.46.30316. [DOI] [PubMed] [Google Scholar]
- 18.Heath RJ, Li J, Roland GE, Rock CO. 2000. Inhibition of the Staphylococcus aureus NADPH-dependent enoyl-acyl carrier protein reductase by triclosan and hexachlorophene. J Biol Chem 275:4654–4659. doi: 10.1074/jbc.275.7.4654. [DOI] [PubMed] [Google Scholar]
- 19.Villalain J, Mateo CR, Aranda FJ, Shapiro S, Micol V. 2001. Membranotropic effects of the antibacterial agent triclosan. Arch Biochem Biophys 390:128–136. doi: 10.1006/abbi.2001.2356. [DOI] [PubMed] [Google Scholar]
- 20.Walsh SE, Maillard JY, Russell AD, Catrenich CE, Charbonneau DL, Bartolo RG. 2003. Activity and mechanisms of action of selected biocidal agents on Gram-positive and -negative bacteria. J Appl Microbiol 94:240–247. doi: 10.1046/j.1365-2672.2003.01825.x. [DOI] [PubMed] [Google Scholar]
- 21.Orsi M, Noro MG, Essex JW. 2011. Dual-resolution molecular dynamics simulation of antimicrobials in biomembranes. J R Soc Interface 8:826–841. doi: 10.1098/rsif.2010.0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pujol E, Blanco-Cabra N, Julián E, Leiva R, Torrents E, Vázquez S. 2018. Pentafluorosulfanyl-containing triclocarban analogs with potent antimicrobial activity. Molecules 23:2853. doi: 10.3390/molecules23112853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le P, Kunold E, Macsics R, Rox K, Jennings MC, Ugur I, Reinecke M, Chaves-Moreno D, Hackl MW, Fetzer C, Mandl FAM, Lehmann J, Korotkov VS, Hacker SM, Kuster B, Antes I, Pieper DH, Rohde M, Wuest WM, Medina E, Sieber SA. 2020. Repurposing human kinase inhibitors to create an antibiotic active against drug-resistant Staphylococcus aureus, persisters and biofilms. Nat Chem 12:145–158. doi: 10.1038/s41557-019-0378-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walsh S, Maillard JY, Russell A, Catrenich C, Charbonneau D, Bartolo R. 2003. Development of bacterial resistance to several biocides and effects on antibiotic susceptibility. J Hosp Infect 55:98–107. doi: 10.1016/s0195-6701(03)00240-8. [DOI] [PubMed] [Google Scholar]
- 25.Schulte B, Bierbaum G, Pohl K, Goerke C, Wolz C. 2013. Diversification of clonal complex 5 methicillin-resistant Staphylococcus aureus strains (Rhine-Hesse clone) within Germany. J Clin Microbiol 51:212–216. doi: 10.1128/JCM.01967-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engelthaler DM, Kelley E, Driebe EM, Bowers J, Eberhard CF, Trujillo J, Decruyenaere F, Schupp JM, Mossong J, Keim P, Even J. 2013. Rapid and robust phylotyping of spa t003, a dominant MRSA clone in Luxembourg and other European countries. BMC Infect Dis 13:339. doi: 10.1186/1471-2334-13-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gravel J, Paradis-Bleau C, Schmitzer AR. 2017. Adaptation of a bacterial membrane permeabilization assay for quantitative evaluation of benzalkonium chloride as a membrane-disrupting agent. Medchemcomm 8:1408–1413. doi: 10.1039/c7md00113d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng CJ, Yoo JS, Lee TG, Cho HY, Kim YH, Kim WG. 2005. Fatty acid synthesis is a target for antibacterial activity of unsaturated fatty acids. FEBS Lett 579:5157–5162. doi: 10.1016/j.febslet.2005.08.028. [DOI] [PubMed] [Google Scholar]
- 29.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. 2014. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering C. 2019. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makhlin J, Kofman T, Borovok I, Kohler C, Engelmann S, Cohen G, Aharonowitz Y. 2007. Staphylococcus aureus ArcR controls expression of the arginine deiminase operon. J Bacteriol 189:5976–5986. doi: 10.1128/JB.00592-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. 2000. Gene ontology: tool for the unification of biology. Nat Genet 25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The Gene Ontology Consortium. 2018. The Gene Ontology Resource: 20 years and still going strong. Nucleic Acids Res 47:D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE, Finn RD. 2019. The Pfam protein families database in 2019. Nucleic Acids Res 47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 36.Brenwald NP, Fraise AP. 2003. Triclosan resistance in methicillin-resistant Staphylococcus aureus (MRSA). J Hosp Infect 55:141–144. doi: 10.1016/s0195-6701(03)00222-6. [DOI] [PubMed] [Google Scholar]
- 37.McDonnell G, Pretzer D. 1998. Action and targets of triclosan. ASM News 64:670–674. [Google Scholar]
- 38.Rao S, Bockstael K, Nath S, Engelborghs Y, Anné J, Geukens N. 2009. Enzymatic investigation of the Staphylococcus aureus type I signal peptidase SpsB—implications for the search for novel antibiotics. FEBS J 276:3222–3234. doi: 10.1111/j.1742-4658.2009.07037.x. [DOI] [PubMed] [Google Scholar]
- 39.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 40.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. 2016. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 41.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Pérez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, Vizcaíno JA. 2019. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (41) partner repository under data set identifier PXD018347.