Abstract
Background
Circular RNAs (circRNAs) are critical regulators of many diseases, including esophageal squamous cell carcinoma (ESCC). A recent study has shown that circLPAR3 is highly expressed in ESCC, but its role and mechanism in ESCC are still unclear.
Methods
The expression levels of circLPAR3, microRNA-375 (miR-375), miR-433, and high-mobility group box 1 (HMGB1) were measured by quantitative real-time polymerase chain reaction (qRT-PCR). The circular characteristic and localization of circLPAR3 were identified by Ribonuclease R (RNase R) and nuclear-cytoplasmic separation assay. Also, cell proliferation was detected by cell counting kit-8 (CCK-8) and colony formation assays. Cell migration and invasion were tested by transwell assay. Moreover, Western blot (WB) analysis was used to test the levels of proliferation and metastasis-related protein, as well as the HMGB1 protein. Besides, mice xenograft models were constructed to assess the effect of circLPAR3 on ESCC tumor growth in vivo. In addition, dual-luciferase reporter and RNA pull-down assays were used to identify the mechanism of circLPAR3.
Results
CircLPAR3 was upregulated in ESCC tissues and cells, and its high expression was related to the poor prognosis of ESCC patients. CircLPAR3 was a stable cyclic transcript, mainly located in the cytoplasm, and its knockdown hindered the proliferation, migration and invasion of ESCC cells and inhibited ESCC tumor growth in vivo. MiR-375/miR-433 could be sponged by circLPAR3, and their inhibitors could reverse the suppression effect of silenced circLPAR3 on ESCC progression. HMGB1 could be targeted by miR-375/miR-433, and its overexpression also could invert the inhibition effect of circLPAR3 knockdown on ESCC progression.
Conclusion
CircLPAR3 might play an oncogenic role in ESCC through sponging miR-375/miR-433 to promote HMGB1 expression, which might provide a theoretical basis for circLPAR3 to become a biomarker for ESCC therapy.
Keywords: ESCC, circLPAR3, miR-375, miR-433, HMGB1
Introduction
Esophageal squamous cell carcinoma (ESCC) is a malignant lesion caused by abnormal hyperplasia of esophageal squamous epithelium, which is a common histological type of esophageal cancer (EC) and one of the most common malignant tumors in the world.1,2 Currently, surgical treatment is the preferred treatment for ESCC, which can be combined with radiotherapy, chemotherapy and comprehensive treatment. Nevertheless, the 5-year survival rate for ESCC patients was less than 20%.3,4 Therefore, it is necessary to explore new biomarkers of ESCC to develop the effective treatment of ESCC.
Circular RNAs (circRNAs) are new types of small molecule RNAs, which are characterized by covalently closed loop structure and have high stability.5,6 Abnormal expression of circRNAs has been found to be involved in many diseases, including cancer.7 Many circRNAs have been found to be abnormally expressed in ESCC, and are associated with poor prognosis and progression of ESCC, such as hsa_circ_0000337, circ-PRKCI and circ-SMAD7.8–10 In a recent study, microarray analysis showed that circLPAR3 (hsa_circ_0004390) was significantly upregulated in ESCC tissues.11 However, its role in ESCC has not been studied. At present, circRNAs have been proved to play a regulatory role through a variety of mechanisms, among which its function as microRNAs (miRNAs) sponges has been proved most.12 For example, circIFT80 can sponge miR-1236-3p to promote the progression of colorectal cancer,13 and circ-SMARCA5 restrain multiple myeloma progression through sponging miR-767-5p.14 Therefore, the circRNA/miRNA/mRNA axis is an important pathway to elucidate the molecular mechanism of circRNA.
The purpose of this study is to explore the role of circLPAR3 in ESCC and its potential mechanism, so as to provide new effective targets for the treatment and diagnosis of ESCC. In our study, we found that circLPAR3 was highly expressed in ESCC, so we speculated that it might play an important role in the development of ESCC. Therefore, we conducted loss-functional experiments to verify this. In addition, through bioinformatics analysis, we found that circLPAR3 regulated the expression of high-mobility group box 1 (HMGB1) by targeting miR-375 and miR-433 in ESCC. Our results enrich the research content of circLPAR3, and provide theoretical basis for the clinical treatment of ESCC.
Materials and Methods
Samples and Peripheral Blood Collection
We recruited 50 ESCC patients from The First People’s Hospital of Yunnan Province and collected their clinical and pathological features data and peripheral blood. None of the patients had received any treatment before the operation, and all of them had completed the written informed consent. A total of 50 pairs of ESCC and adjacent normal tissues were collected and stored at −80°C until use. In addition, the peripheral blood of 31 healthy peoples who came to the hospital for routine physical examination was collected for this study. All procedures of this study were approved by the Ethical Committee of The First People’s Hospital of Yunnan Province and were in accordance with the Declaration of Helsinki.
Cell Culture
Human ESCC cell lines (EC109, EC9706, KYSE30 and KYSE150) and human esophageal epithelial cell line (HET-1A) were bought from Jining Shiye (Shanghai, China) and cultured in RPMI-1640 medium (Hyclone, Logan, UT, USA) containing 10% fetal bovine serum (FBS, Hyclone), 100 U/mL penicillin and 100 μg/mL streptomycin (Procell, Wuhan, China) at 37°C in an incubator with 5% CO2. All cell lines purchased in this study have been authenticated by STR.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNAs were extracted from tissues and cells using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and reverse-transcribed complementary DNA (cDNA) using the cDNA Synthesize Kit (Beyotime, Shanghai, China). QRT-PCR was performed using SYBR Green (Takara, Dalian, China). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and U6 were used as internal controls. The primers were listed as follows: circLPAR3, F 5ʹ-GGTGTACCTGCGGATCTACG-3ʹ, R 5ʹ-AGCTTTGTTCCTGTCCAGTCA-3ʹ; HMGB1, F 5ʹ-GAACAACACTGCTGCGGATG-3ʹ, R 5ʹ-CCCTTTTTCGCTGCATCAGG-3ʹ; GAPDH, F 5ʹ-ACCACAGTCCATGCCATCAC-3ʹ, R 5ʹ-TCCACCACCCTGTTGCTGTA-3ʹ; miR-375, F 5ʹ-AGCCGTTTGTTCGTTCGGCT-3ʹ, R 5ʹ-GTGCAGGGTCCGAGGT-3ʹ; miR-433, F 5ʹ-TCGGCAATCATGATGGGCTCCTC-3ʹ, R 5ʹ-CTCAACTGGTGTCGTGGAGTC-3ʹ; U6, F 5ʹ-GCAGGAGGTCTTCACAGAGT-3ʹ, R 5ʹ-TCTAGAGGAGAAGCTGGGGT-3ʹ. The 2−ΔΔCt method was used to calculate the fold changes.
CircRNA Identification and Localization
Ribonuclease R (RNase R; Geneseed, Guangzhou, China) was used to identify the circular characteristic of circLPAR3. In briefly, the extracted RNA was treated with RNase R, followed by qRT-PCR to determine the expression of circLPAR3 and GAPDH. GAPDH was served as the representative of linear RNA. Cytoplasmic & Nuclear RNA Purification Kit (Amyjet, Wuhan, China) was performed to extract the cytoplasmic and nuclear RNA of KYSE30 and KYSE150 cells. Then, qRT-PCR was used to assess the expression levels of circLPAR3, U6 and GAPDH in cytoplasmic and nuclear. GAPDH and U6 were served as the cytoplasm control and nuclear control, respectively.
Cell Transfection
Lentiviral short hairpin RNA (shRNA) targeting circLPAR3 (sh-circLPAR3#1/2) and its negative control (sh-NC), miR-375/miR-433 mimic and inhibitor (miR-375/miR-433 and anti-miR-375/miR-433) or their negative controls (miR-NC and anti-NC), HMGB1 overexpression plasmid (HMGB1) and its negative control (vector) were all synthesized by Ribobio (Guangzhou, China). Lipofectamine 3000 (Invitrogen) was used to transfect all plasmid vectors into KYSE30 and KYSE150 cells.
Cell Counting Kit-8 (CCK-8) Assay
KYSE30 and KYSE150 cells were seeded into 96-well plates. At a specific time (0 h, 24 h, 48 h and 72 h) after transfection, the cells were incubated with 10 μL CCK-8 (Amyjet) for 4 h. Then, the absorbance at 450 nm was determined to evaluate cell viability using a microplate reader (Mindray, Shenzhen, China).
Colony Formation Assay
KYSE30 and KYSE150 cells were seeded into 6-well plates. After transfection, cells were incubated for 2 weeks. Then, KYSE30 and KYSE150 cells were fixed with methanol and stained with crystal violet to count the number of colonies (> 50 cells) under a microscope (Leica, Wetzlar, Germany).
Cell Migration and Invasion Assays
Transwell chambers (Corning Inc., Corning, NY, USA) were used to perform this assay. KYSE30 and KYSE150 cells were seeded into the upper chambers with or without Matrigel (BD Biosciences, San Jose, CA, USA) to detect cell invasion and migration, respectively. The upper chambers were added with serum-free medium, while the lower chambers were added with serum medium. After 24 h, cells were fixed and stained, and the number of migrated and invaded cells were imaged and counted under a microscope (Leica).
Western Blot (WB) Analysis
Total proteins of KYSE30 and KYSE150 cells were extracted using RIPA buffer (Beyotime). The same amount of protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). After blocked with nonfat milk, the membranes were incubated with primary antibodies against proliferating cell nuclear antigen (PCNA; 1:1000, Bioss, Beijing, China), Ki-67 (1:200, Bioss), matrix metalloproteinase (MMP2; 1:1000, Bioss), MMP9 (1:500, Bioss), HMGB1 (1:750, Bioss) or GAPDH (1:500, Bioss) at 4°C overnight. Then, the membranes were incubated with secondary antibody (1:1000, Bioss) for 1 h, and the protein signals were visualized using an enhanced chemiluminescence solution (Beyotime).
Mice Xenograft Models
BALB/c male nude mice were bought from Shanghai Laboratory Animal Center (Shanghai, China). KYSE150 cells transfected sh-circLPAR3 or sh-NC were subcutaneously injected into nude mice. The length and width of tumors in nude mice were measured weekly to calculate tumor volume using the formula: volume (mm3) = width2 × length/2. After 5 weeks, mice were sacrificed and the tumors were taken for further study. Our research was approved by the Ethical Committee of The First People’s Hospital of Yunnan Province. Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. All animals received humane care according to the National Institutes of Health (USA) guidelines.
Dual-Luciferase Reporter Assay
CircLPAR3-WT/MUT or HMGB1 3ʹUTR-WT/MUT was inserted into the pmirGLO reporter vectors (Enzyme Research, Shanghai, China). KYSE30 and KYSE150 cells were plated in 24-well plates for 24 h. The cells were co-transfected with the above reporter plasmid and miR-375/miR-433 mimic or miR-NC using Lipofectamine 3000 (Invitrogen). After 48 h, Dual-Luciferase Reporter Assay Kit (Beyotime) was used to assess the relative luciferase activity.
RNA Pull-Down Assay
KYSE30 and KYSE150 cells were lysed with RIPA buffer (Beyotime) and lysates were collected. Biotin-labeled circLPAR3 (Bio-circLPAR3) or negative control (Bio-NC) probe (synthesized from Sangon Inc., Shanghai, China) was incubated with streptavidin magnetic beads (Invitrogen) at 4°C overnight. After that, cell lysates were incubated with the magnetic beads mixture at room temperature for 1 h. Total RNA was purified by RNA simple Kit (Tiangen Biotech, Beijing, China) and qRT-PCR was performed to detect the enrichment of miR-375 or miR-433.
Statistical Analysis
Data were analyzed by GraphPad Prism 5.0 software (GraphPad Software, San Diego, CA, USA) and displayed as mean ± standard deviation (SD). Student’s t-test or one-way analysis of variance was used for statistical analysis. P < 0.05 was considered to be statistically significant.
Results
circLPAR3 Was Highly Expressed in ESCC and Linked to Poor Prognosis
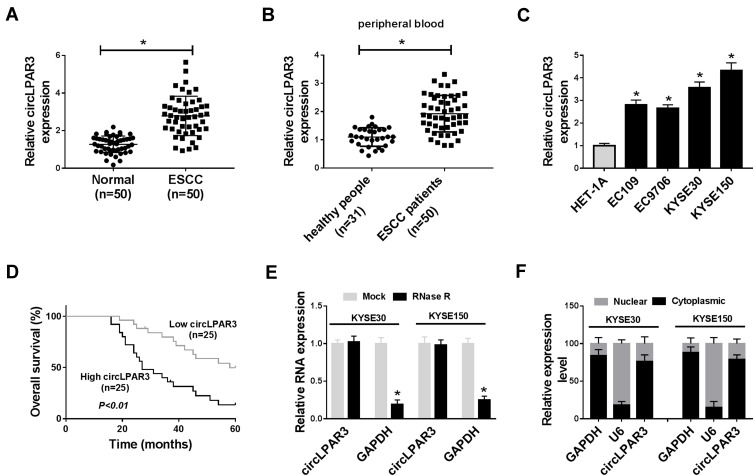
Firstly, we tested the expression of circLPAR3 in ESCC tissues and cells. QRT-PCR results showed that circLPAR3 was remarkably overexpressed in ESCC tissues when compared with adjacent normal tissues (Figure 1A). Further, we also found that the expression of circLPAR3 in the peripheral blood of ESCC patients was higher than that in healthy peoples (Figure 1B). Also, circLPAR3 was upregulated in ESCC cell lines (EC109, EC9706, KYSE30 and KYSE150) compared to HET-1A cells (Figure 1C). According to the median value of circLPAR3 expression in ESCC patients’ tissues, circLPAR3 expression was divided into high expression group and low expression group. The relationship between circLPAR3 and clinical and pathological features of ESCC patients showed that high circLPAR3 expression was positively correlated with N classification and tumor nodes metastasis (TNM) stage in ESCC patients (P < 0.05, Table 1). Moreover, shorter overall survival times were observed in the high circLPAR3 expression group as compared with the low circLPAR3 expression group, as demonstrated by Kaplan-Meier analysis (Figure 1D). Besides, we used RNase R to confirm the circular characteristics of circLPAR3. As shown in Figure 1E, circLPAR3 was resistant to RNase R, while linear RNA GAPDH could be digested by RNase R, indicating that circLPAR3 was circular. Also, we detected the localization of circLPAR3 and found that circLPAR3 was mainly distributed in the cytoplasm of ESCC cells (Figure 1F). Therefore, these results suggested that circLPAR3 might play a vital role in ESCC.
Figure 1.
The expression of circLPAR3 was increased in esophageal squamous cell carcinoma (ESCC) tissues and cells. (A) The expression of circLPAR3 in ESCC tissues (ESCC) and adjacent normal tissues (Normal) was detected by qRT-PCR. (B) The expression of circLPAR3 in the peripheral blood of ESCC patients and healthy peoples was measured by qRT-PCR. (C) QRT-PCR was used to determine the circLPAR3 expression in ESCC cell lines (EC109, EC9706, KYSE30 and KYSE150) and HET-1A cells. (D) Kaplan-Meier analysis was performed to examine the correlation between circLPAR3 expression and overall survival of ESCC patients. (E) The relative expression levels of circLPAR3 and Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in KYSE30 and KYSE150 cells were assessed by qRT-PCR after treatment with RNase R. (F) The expression levels of circLPAR3, U6 and GAPDH in the nuclear and cytoplasmic of KYSE30 and KYSE150 cells were detected by qRT-PCR after nuclear-cytoplasmic separation. *P < 0.05.
Table 1.
The Relationship Between circLPAR3 and Clinical and Pathological Features of ESCC Patients
| Characteristic | circLPAR3 Expression | P-value | |
|---|---|---|---|
| High (n = 25) | Low (n = 25) | ||
| Gender | 0.333 | ||
| Male | 17 | 20 | |
| Female | 8 | 5 | |
| Age (yr) | 0.552 | ||
| <60 | 2 | 1 | |
| ≥60 | 23 | 24 | |
| Differentiation | 0.306 | ||
| Good/Moderate | 18 | 21 | |
| Poor | 7 | 4 | |
| T classification | 0.047 | ||
| T1+T2 | 8 | 15 | |
| T3+T4 | 17 | 10 | |
| N classification | 0.005* | ||
| Positive | 7 | 17 | |
| Negative | 18 | 8 | |
| TNM stages | 0.001* | ||
| I/II | 18 | 5 | |
| III/IV | 7 | 20 | |
Note: *P < 0.05.
Knockdown of circLPAR3 Inhibited ESCC Cell Proliferation, Migration and Invasion
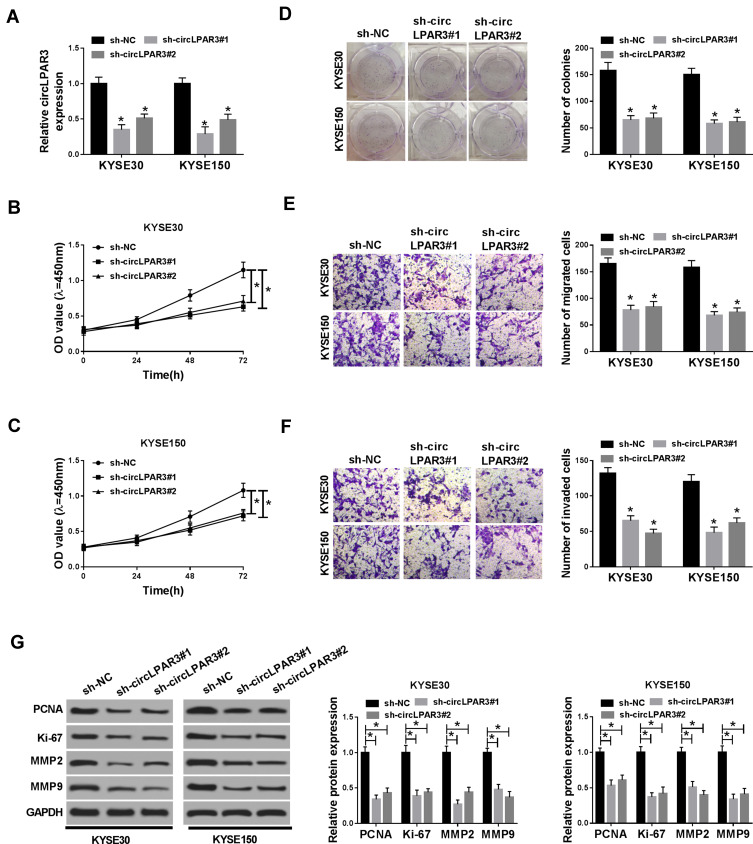
To explore the biological effect of circLPAR3 on ESCC cells, we transfected sh-circLPAR3#1/2 into KYSE30 and KYSE150 cells, and the qRT-PCR results revealed that both of them could effectively reduce the expression level of circLPAR3 in KYSE30 and KYSE150 cells (Figure 2A). Subsequently, we assessed the proliferation, migration, and invasion abilities of ESCC cells after circLPAR3 silencing. CCK-8 results showed that circLPAR3 knockdown suppressed the viability of KYSE30 and KYSE150 cells (Figure 2B and C). Also, colony formation assay results indicated that the number of colonies KYSE30 and KYSE150 cells was reduced in the sh-circLPAR3#1/2 group (Figure 2D). These results suggested that silenced-circLPAR3 restrained the proliferation of ESCC cells. Furthermore, the number of migrated and invaded KYSE30 and KYSE150 cells was also decreased after circLPAR3 knockdown (Figure 2E and F), indicating that circLPAR3 silencing inhibited the migration and invasion of KYSE30 and KYSE150 cells. Meanwhile, the levels of proliferation-related proteins (PCNA and Ki-67) and metastasis-related proteins (MMP2 and MMP9) were detected, and the results determined that silencing of circLPAR3 could markedly reduce their protein levels in KYSE30 and KYSE150 cells (Figure 2G), which once again confirmed the conclusion that circLPAR3 inhibited proliferation, migration and invasion in ESCC cells. All data revealed that circLPAR3 expression was crucial to ESCC progression.
Figure 2.
Effect of circLPAR3 knockdown on esophageal squamous cell carcinoma (ESCC) progression. KYSE30 and KYSE150 cells were transfected with (short hairpin) sh-circLPAR3#1/2 or short hairpin-negative control (sh-NC). (A) The expression of circLPAR3 was detected by qRT-PCR to evaluate transfection efficiency. (B, C) CCK-8 assay was used to assess the viability of KYSE30 and KYSE150 cells. (D) Colony formation assay was performed to measure the number of colonies in KYSE30 and KYSE150 cells. (E, F) The number of migrated and invaded KYSE30 and KYSE150 cells was determined by transwell assay. (G) Western blot (WB) analysis was performed to assess the protein levels of proliferating cell nuclear antigen (PCNA), Ki-67, matrix metalloproteinase2 (MMP2) and MMP9 in KYSE30 and KYSE150 cells. *P < 0.05.
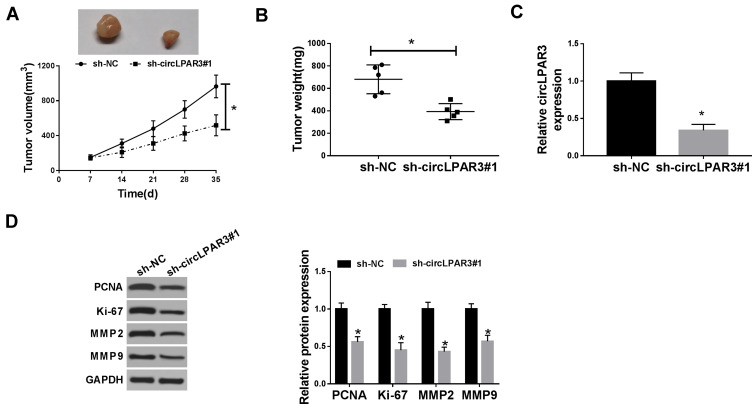
Silencing of circLPAR3 Reduced ESCC Tumor Growth in vivo
To further investigate the role of circLPAR3 in ESCC, we transfected sh-circLPAR3#1 into mice to perform in vivo experiments. After 35 d of monitoring, we found that the growth rate of tumor volume was remarkably restrained in the circLPAR3 knockdown group compared with that in the control group (Figure 3A). Besides, the tumor weight of the circLPAR3 knockdown group was also markedly decreased (Figure 3B). To determine the effectiveness of circLPAR3 knockdown, we performed qRT-PCR to test circLPAR3 expression. The results showed that circLPAR3 expression was significantly decreased in the sh-circLPAR3 group (Figure 3C). In addition, we also detected the protein levels of PCNA, Ki-67, MMP2 and MMP9 and found that silenced-circLPAR3 obviously suppressed their protein levels in tumors (Figure 3D), indicating that circLPAR3 might restrain proliferation and metastasis thereby inhibiting the tumor growth of ESCC. Hence, these data confirmed that circLPAR3 might play an oncogenic role in ESCC.
Figure 3.
Effect of circLPAR3 silencing on esophageal squamous cell carcinoma (ESCC) tumor growth in vivo. (A) Tumor volume was calculated with length × width2/2 method at the indicated time point. (B) Tumor weight was measured in mice. (C) The expression of circLPAR3 was detected by qRT-PCR to evaluate transfection efficiency. (D) The protein levels of proliferating cell nuclear antigen (PCNA), Ki-67, matrix metalloproteinase2 (MMP2) and MMP9 were tested by Western blot (WB) analysis. *P < 0.05.
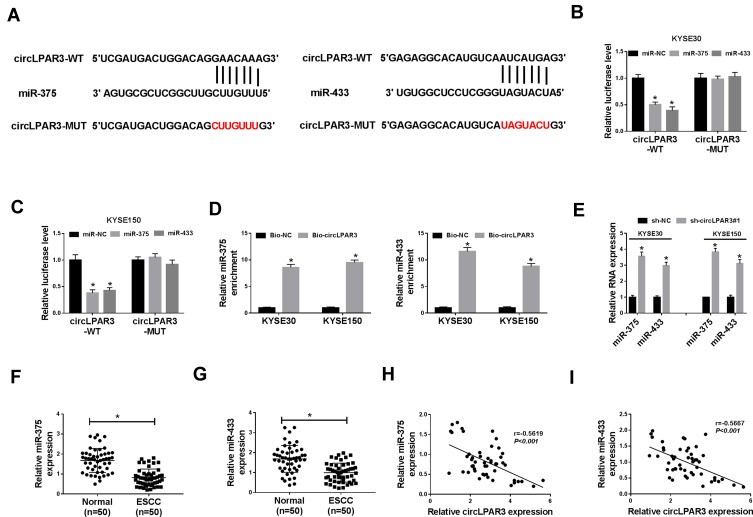
circLPAR3 Could Sponge miR-375/miR-433
To explore the mechanism of circLPAR3 function as a miRNA sponge, we predicted the potential targeted miRNAs of circLPAR3 using the Circinteractome tool and found that circLPAR3 could target multiple miRNAs (miR-1238, miR-198, miR-370, miR-375, miR-411, miR-433, miR-561 and miR-570). However, we detected the predicted miRNA level after knocking down circLPAR3 in KYSE30 cells, and finally found the most significant increase in miR-375 and miR-433 levels (Supplement Figure 1A). Therefore, miR-375 and miR-433 were selected for this study. The complementary binding sites between miR-375/miR-433 and circLPAR3 were shown in Figure 4A. To confirm their targeting relationship, we performed the dual-luciferase reporter assay. The results showed that miR-375 or miR-433 overexpression could significantly hinder the luciferase activity of circLPAR3-WT in KYSE30 and KYSE150 cells, but had no effect on circLPAR3-MUT (Figure 4B and C). Besides, RNA pull-down assay results also revealed that miR-375 and miR-433 were markedly enriched in Bio-circLPAR3 compared with that in Bio-NC (Figure 4D), confirming the interaction between circLPAR3 and miR-375 or miR-433 in ESCC. Meanwhile, we also found that silenced circLPAR3 promoted miR-375 and miR-433 expression in KYSE30 and KYSE150 cells (Figure 4E). Through detecting the expression of miR-375 and miR-433 in ESCC tissues, we discovered that miR-375 and miR-433 were lower expressed in ESCC tissues compared with adjacent normal tissues (Figure 4F and G). In addition, we also analyzed the correlation between circLPAR3 and miR-375 or miR-433, and the results showed that the expression of miR-375 or miR-433 was negatively correlated with circLPAR3 in ESCC tissues (Figure 4H and I). These results confirmed that miR-375 and miR-433 could be absorbed by circLPAR3 in ESCC.
Figure 4.
CircLPAR3 could sponge miR-375/miR-433. (A) The sequences of circLPAR3 containing the miR-375/miR-433 binding sites or mutant binding sites were shown. (B, C) Dual-luciferase reporter assay was used to detect the interaction between miR-375/miR-433 and circLPAR3 in KYSE30 and KYSE150 cells. (D) RNA pull-down assay was performed to determine the enrichment of miR-375 and miR-433 in Bio-circLPAR3 or Bio-(negative control) NC (E) The effects of sh-circLPAR3#1 on the expression levels of miR-375 and miR-433 in KYSE30 and KYSE150 cells were detected by qRT-PCR. (F, G) The expression levels of miR-375 and miR-433 in ESCC tissues (ESCC) and adjacent normal tissues (Normal) were determined by qRT-PCR. (H, I) The correlation between circLPAR3 and miR-375 or miR-433 was measured by Pearson correlation analysis. *P < 0.05.
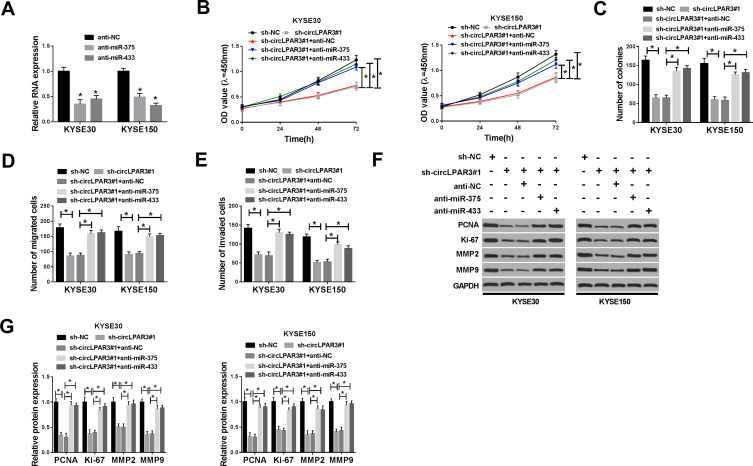
miR-375 or miR-433 Inhibitor Could Reverse the Inhibition Effect of circLPAR3 Knockdown on ESCC Progression
To further explore the function of miR-375 and miR-433, we synthesized anti-miR-375/miR-433 and tested their transfection efficiency. QRT-PCR results showed that anti-miR-375 and anti-miR-433 had a good inhibitory effect on miR-375 and miR-433 expression in ESCC cells, and follow-up experiments could be conducted (Figure 5A). Subsequently, we co-transfected sh-circLPAR3#1 and anti-miR-375 or anti-miR-433 into KYSE30 and KYSE150 cells to verify whether circLPAR3 played a role in ESCC through miR-375 and miR-433. CCK-8 and colony formation assays results revealed that the inhibition effect of silenced-circLPAR3 on the viability and colonies of KYSE30 and KYSE150 cells could be inverted by miR-375 or miR-433 inhibitor (Figure 5B and C), suggesting that miR-375 or miR-433 inhibitor reversed the suppression effect of circLPAR3 knockdown on proliferation in KYSE30 and KYSE150 cells. Besides, transwell assay results determined that miR-375 or miR-433 inhibitor also recovered the suppression effect of circLPAR3 knockdown on the number of migrated and invaded KYSE30 and KYSE150 cells to promote the migration and invasion in ESCC cells (Figure 5D and E). Furthermore, the promotion effect of anti-miR-375 or anti-miR-433 on the protein levels of PCNA, Ki-67, MMP2 and MMP9 in KYSE30 and KYSE150 cells also confirmed that miR-375 or miR-433 inhibitor could reverse the inhibition function of circLPAR3 silencing on proliferation and metastasis in ESCC cells (Figure 5F and G). Therefore, all data illustrated that circLPAR3 regulated ESCC progression through miR-375 and miR-433.
Figure 5.
Effects of miR-375 and miR-433 inhibitors on esophageal squamous cell carcinoma (ESCC) progression. (A) The expression levels of miR-375 and miR-433 were detected by qRT-PCR to evaluate the transfection efficiency of anti-miR-375 and anti-miR-433. KYSE30 and KYSE150 cells were co-transfected with sh-circLPAR3#1 and anti-miR-375 or anti-miR-433. (B) The viability of KYSE30 and KYSE150 cells was measured by CCK-8 assay. (C) Colony formation assay was used to assess the number of colonies in KYSE30 and KYSE150 cells. (D, E) Transwell assay was performed to determine the number of migrated and invaded KYSE30 and KYSE150 cells. (F, G) The protein levels of proliferating cell nuclear antigen (PCNA), Ki-67, matrix metalloproteinase2 (MMP2) and MMP9 in KYSE30 and KYSE150 cells were tested by Western blot (WB) analysis. *P < 0.05.
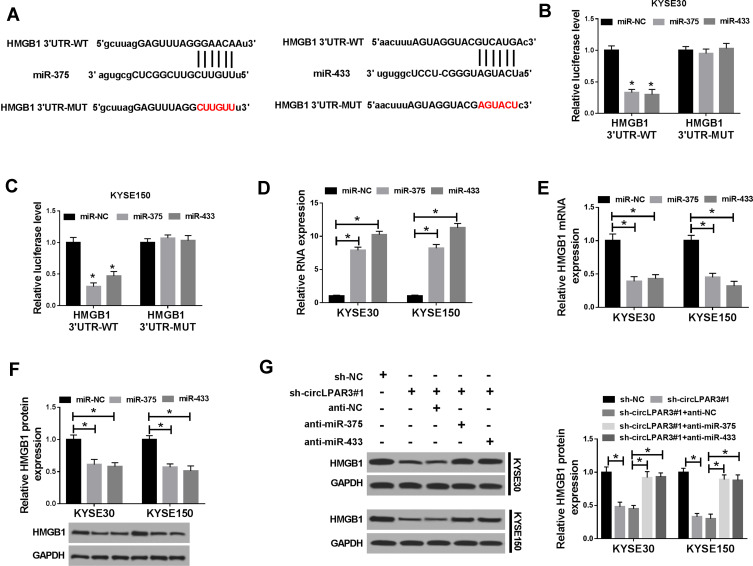
HMGB1 Was a Target of miR-375/miR-433
To identify the downstream target genes of miR-375 and miR-433, we used the StarBase tool to predict the common target genes of miR-375 and miR-433 and found that both miR-375 and miR-433 could target 6 genes (E2F3, E2F7, EZH1, HMGB1, HMGB3 and RUNX2). After overexpression of miR-375 and miR-433, the mRNA expression of HMGB1 in the 6 target genes was significantly inhibited (Supplement Figure 1B). Therefore, HMGB1 was chose for this study. The binding sites of miR-375/miR-433 to HMGB1 3ʹUTR were exhibited in Figure 6A. Dual-luciferase reporter assay results revealed that the overexpression of both miR-375 and miR-433 remarkably suppressed the luciferase activity of HMGB1 3ʹUTR-WT in KYSE30 and KYSE150 cells, while had no effect on the luciferase activity of HMGB1 3ʹUTR-MUT (Figure 6B and C). Besides, we explored the effect of miR-375 and miR-433 on HMGB1 expression in KYSE30 and KYSE150 cells using miR-375 and miR-433 mimics. Through detecting the expression of miR-375 and miR-433, we discovered that the transfection efficiency of miR-375 and miR-433 mimics was excellent (Figure 6D). The decreased mRNA and protein expression of HMGB1 in KYSE30 and KYSE150 cells indicated that miR-375 and miR-433 overexpression obviously suppressed the HMGB1 expression (Figure 6E and F). Moreover, knockdown of circLPAR3 could markedly inhibit HMGB1 expression, and miR-375 or miR-433 inhibitor could reverse the inhibition effect of circLPAR3 knockdown on HMGB1 expression (Figure 6G). Thus, these results suggested that HMGB1 was a downstream target gene of the circLPAR3/miR-375 or circLPAR3/miR-433 axis in ESCC.
Figure 6.
HMGB1 was a target of miR-375/miR-433. (A) The binding sites and mutant binding sites between high-mobility group box 1 (HMGB1) and miR-375/miR-433 were shown. (B, C) Dual-luciferase reporter assay was used to detect the interaction between HMGB1 and miR-375/miR-433 in KYSE30 and KYSE150 cells. (D) The expression levels of miR-375 and miR-433 were determined by qRT-PCR to evaluate the transfection efficiency of miR-375 and miR-433 mimics. (E, F) The mRNA and protein levels of HMGB1 were measured by qRT-PCR and WB analysis to evaluate the effect of miR-375/miR-433 overexpression on HMGB1 expression. KYSE30 and KYSE150 cells were co-transfected with (short hairpin) sh-circLPAR3#1 and anti-miR-375 or anti-miR-433. (G) The protein level of HMGB1 was tested by Western blot (WB) analysis in KYSE30 and KYSE150 cells. *P < 0.05.
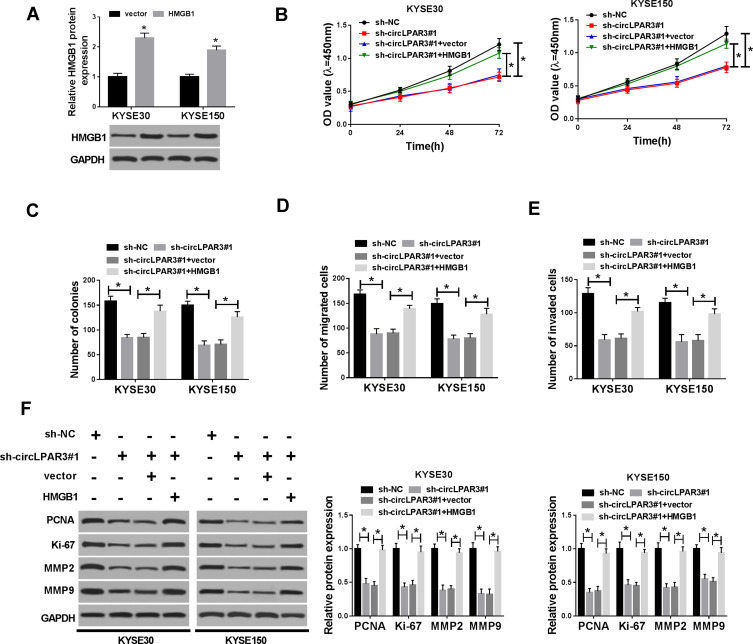
Overexpression of HMGB1 Reversed the Suppression Effect of circLPAR3 Knockdown on ESCC Progression
To confirm the role of HMGB1 on ESCC, we first detected the transfection efficiency of HMGB1 overexpression plasmid in KYSE30 and KYSE150 cells. WB results showed that HMGB1 overexpression plasmid could effectively promote HMGB1 expression, so that follow-up experiments could be conducted (Figure 7A). We co-transfected sh-circLPAR3 and HMGB1 overexpression plasmid into KYSE30 and KYSE150 cells to assess the role of HMGB1 in ESCC. The results revealed that HMGB1 overexpression counteracted the inhibition effect of silenced-circLPAR3 on the progression of KYSE30 and KYSE150 cells, as demonstrated by CCK-8 and colony formation assays (Figure 7B and C). Similarly, transwell assay results indicated that overexpression of HMGB1 could also significantly eliminate the inhibition of circLPAR3 knockdown on the migration and invasion of KYSE30 and KYSE150 cells (Figure 7D and E). The promoting effect of HMGB1 overexpression on the protein levels of PCNA, Ki-67, MMP2 and MMP9 also confirmed that HMGB1 could reverse the inhibition effect of circLPAR3 on ESCC progression (Figure 7F). Altogether, these data suggested that circLPAR3 exerted the oncogenic role in ESCC through HMGB1.
Figure 7.
Effect of HMGB1 overexpression on esophageal squamous cell carcinoma (ESCC) progression. (A) The protein level of high-mobility group box 1 (HMGB1) was detected by Western blot (WB) analysis to evaluate the transfection efficiency of HMGB1 overexpression plasmid. KYSE30 and KYSE150 cells were co-transfected with (short hairpin) sh-circLPAR3#1 and HMGB1 overexpression plasmid. (B) CCK-8 assay was performed to measure the viability of KYSE30 and KYSE150 cells. (C) Colony formation assay was used to test the number of colonies in KYSE30 and KYSE150 cells. (D, E) The number of migrated and invaded KYSE30 and KYSE150 cells was assessed by transwell assay. (F) The protein levels of proliferating cell nuclear antigen (PCNA), Ki-67, matrix metalloproteinase2 (MMP2) and MMP9 in KYSE30 and KYSE150 cells were determined by WB analysis. *P < 0.05.
Discussion
At present, the etiology of ESCC is not clear, which may be related to genetics, diet and other factors. CircRNAs are novel endogenous noncoding RNAs (ncRNAs) that are widely involved in the development of tumors and have been shown to be a biomarker for the diagnosis and prognosis of many cancers.15,16 CircLPAR3 is reported to be markedly highly expressed in ESCC, but its underlying mechanism has not been clarified. Here, we confirmed the upregulation of circLPAR3 expression in ESCC tissues and cells. Furthermore, high circLPAR3 expression was closely associated with clinical and pathological features and poor prognosis of ESCC patients. Besides, we demonstrated that circLPAR3 was a stable cyclic transcript and mainly located in the cytoplasm. Functional experiments showed that circLPAR3 knockdown inhibited the proliferation, migration and invasion of ESCC cells in vitro and reduced ESCC tumor growth in vivo. Further mechanism studies indicated that circLPAR3 promoted HMGB1 expression mainly by competitive binding with miR-375 and miR-433. Therefore, our findings highlighted the importance of circRNA expression in ESCC and revealed the role of a new circRNA, circLPAR3 in ESCC. The carcinogenicity of circLPAR3 was expected to be a biomarker of molecular therapy for ESCC patients.
As the research progressed, researchers discovered that circRNAs are abundant in eukaryotes and have an aberrant expression in many diseases, including cancer.17 Many circRNAs features had been proven, for example, circ_0000218 could promote proliferation and metastasis in colorectal cancer through regulating the miR-139-3p/RAB1A axis, thereby playing a carcinogenic role in colorectal cancer.18 Hsa_circ_0028502 and hsa_circ_0076251 expression levels were related to the clinical and pathological characteristics of hepatocellular carcinoma (HCC) patients and could be used as novel biomarkers for HCC.19 Although some circRNAs have been investigated functionally, there are still many circRNAs that have not been studied. In our study, we explored a new circRNA associated with ESCC, circLPAR3, which might better emphasize the important role of circRNAs in cancer.
Given that one of the circRNA mechanisms act as a miRNA sponge, we investigated miRNAs that can be adsorbed by circLPAR3. We found that circLPAR3 could directly bind to two miRNAs, miR-375 and miR-433. Previous studies have shown that miR-375 and miR-433 have been shown to be differentially expressed in many cancers and involved in the regulation of cancer progression.20,21 Moreover, the results of Xu et al and Shi et al suggested that miR-375 and miR-433 expression were decreased in ESCC.22,23 Therefore, miR-375 and miR-433 might function as the tumor suppressor in ESCC. Moreover, we also discovered that HMGB1 was the common target of miR-375 and miR-433. HMGB1, a member of damage-associated molecular patterns (DAMPs) family, can bind to DNA and participate in transcriptional regulation.24 Many studies have confirmed that HMGB1 is highly expressed in various cancers and promotes the development of cancers.25,26 Di et al reported that HMGB1 is upregulated in ESCC and modulates the proliferation and radioresistance of ESCC.27 In our study, we observed that circLPAR3 silencing could hinder HMEB1 expression, whereas this repression could be reversed by inhibition of miR-375 or miR-433. Besides, miR-375 and miR-433 inhibitors or HMGB1 overexpression also inverted the suppression of silenced circLPAR3 on the progression of ESCC, implying that the circLPAR3-miR-375/miR-433-HMGB1 axis does exist in ESCC. At present, most studies focused on the interaction between circRNAs and single miRNA, which markedly weakened the biological effect of circRNAs.28–30 Our study found that circLPAR3 could target miR-375 and miR-433, which remarkably increased the biological capacity of circLPAR3 and provided more references for future studies.
Of note, miR-375 and miR-433 inhibitors or HMGB1 overexpression could not completely counteract the suppressed proliferation, migration and invasion in ESCC cells caused by circLPAR3 knockdown, indicating that in addition to miR-375/miR-433 and HMGB1, there might be other miRNAs and target genes involved in the regulation of circLPAR3 on ESCC progression. Therefore, the molecular mechanism of circLPAR3 needed further exploration and improvement.
In conclusion, our study demonstrated that circLPAR3 enhanced HMGB1 expression to promote the proliferation, migration and invasion of ESCC by adsorbing miR-375/miR-433, which provided strong evidence for circLPAR3 as a promising prognostic biomarker of ESCC.
Disclosure
The authors report no funding and no conflicts of interest for this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 3.Tang W-R, Fang J-Y, Wu K-S, et al. Epidemiological characteristics and prediction of esophageal cancer mortality in China from 1991 to 2012. Asian Pac J Cancer Prev. 2014;15(16):6929–6934. doi: 10.7314/APJCP.2014.15.16.6929 [DOI] [PubMed] [Google Scholar]
- 4.Pennathur A, Gibson MK, Jobe BA, et al. Oesophageal carcinoma. Lancet. 2013;381(9864):400–412. doi: 10.1016/S0140-6736(12)60643-6 [DOI] [PubMed] [Google Scholar]
- 5.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi: 10.1080/15476286.2015.1020271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. doi: 10.1016/j.molcel.2018.06.034 [DOI] [PubMed] [Google Scholar]
- 7.Ng WL, Mohd Mohidin TB, Shukla K. Functional role of circular RNAs in cancer development and progression. RNA Biol. 2018;15(8):995–1005. doi: 10.1080/15476286.2018.1486659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song H, Xu D, Shi P, et al. Upregulated circ RNA hsa_circ_0000337 promotes cell proliferation, migration, and invasion of esophageal squamous cell carcinoma. Cancer Manag Res. 2019;11:1997–2006. doi: 10.2147/CMAR.S195546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi N, Shan B, Gu B, et al. Circular RNA circ-PRKCI functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-3680-3p in esophageal squamous cell carcinoma. J Cell Biochem. 2019;120(6):10021–10030. doi: 10.1002/jcb.28285 [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Wang Q, Zhu D, et al. Up-regulation of circ-SMAD7 inhibits tumor proliferation and migration in esophageal squamous cell carcinoma. Biomed Pharmacother. 2019;111:596–601. doi: 10.1016/j.biopha.2018.12.116 [DOI] [PubMed] [Google Scholar]
- 11.Shi Y, Guo Z, Fang N, et al. hsa_circ_0006168 sponges miR-100 and regulates mTOR to promote the proliferation, migration and invasion of esophageal squamous cell carcinoma. Biomed Pharmacother. 2019;117:109151. doi: 10.1016/j.biopha.2019.109151 [DOI] [PubMed] [Google Scholar]
- 12.Shang Q, Yang Z, Jia R, et al. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18(1):6. doi: 10.1186/s12943-018-0934-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng W, Gong H, Wang Y, et al. circIFT80 functions as a ceRNA of miR-1236-3p to promote colorectal cancer progression. Mol Ther Nucleic Acids. 2019;18:375–387. doi: 10.1016/j.omtn.2019.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Liu H, Wu Y, Wang S, et al. Circ-SMARCA5 suppresses progression of multiple myeloma by targeting miR-767-5p. BMC Cancer. 2019;19(1):937. doi: 10.1186/s12885-019-6088-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patop IL, Kadener S. circRNAs in cancer. Curr Opin Genet Dev. 2018;48:121–127. doi: 10.1016/j.gde.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang M, Xin Y. Circular RNAs: a new frontier for cancer diagnosis and therapy. J Hematol Oncol. 2018;11(1):21. doi: 10.1186/s13045-018-0569-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pei FL, Cao MZ, Li YF. Circ_0000218 plays a carcinogenic role in colorectal cancer progression by regulating miR-139-3p/RAB1A axis. J Biochem. 2020;167(1):55–65. [DOI] [PubMed] [Google Scholar]
- 19.Jiang Z, Shen L, Wang S, et al. Hsa_circ_0028502 and hsa_circ_0076251 are potential novel biomarkers for hepatocellular carcinoma. Cancer Med. 2019. doi: 10.1002/cam4.2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang F-Z, Zhang M-Q, Zhang L, et al. Long non-coding RNA ROR1-AS1 enhances colorectal cancer metastasis by targeting miR-375. Eur Rev Med Pharmacol Sci. 2019;23(16):6899–6905. doi: 10.26355/eurrev_201908_18729 [DOI] [PubMed] [Google Scholar]
- 21.Li J, Chen M, Yu B. miR-433 suppresses tumor progression via Smad2 in non-small cell lung cancer. Pathol Res Pract. 2019;215(10):152591. doi: 10.1016/j.prp.2019.152591 [DOI] [PubMed] [Google Scholar]
- 22.Xu H, Jiang J, Zhang J, et al. MicroRNA-375 inhibits esophageal squamous cell carcinoma proliferation through direct targeting of SP1. Exp Ther Med. 2019;17(3):1509–1516. doi: 10.3892/etm.2018.7106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi Q, Wang Y, Mu Y, et al. MiR-433-3p inhibits proliferation and invasion of esophageal squamous cell carcinoma by targeting GRB2. Cell Physiol Biochem. 2018;46(5):2187–2196. doi: 10.1159/000489548 [DOI] [PubMed] [Google Scholar]
- 24.Fu L, Nishibori M. The role of high mobility group box-1 in epileptogenesis. Acta Med Okayama. 2019;73(5):383–386. doi: 10.18926/AMO/57367 [DOI] [PubMed] [Google Scholar]
- 25.Fang J, Ge X, Xu W, et al. Bioinformatics analysis of the prognosis and biological significance of HMGB1, HMGB2, and HMGB3 in gastric cancer. J Cell Physiol. 2019;235(4):3438–3446. [DOI] [PubMed] [Google Scholar]
- 26.Lv DJ, Song XL, Huang B, et al. HMGB1 promotes prostate cancer development and metastasis by interacting with brahma-related gene 1 and activating the Akt signaling pathway. Theranostics. 2019;9(18):5166–5182. doi: 10.7150/thno.33972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di X, He G, Chen H, et al. High-mobility group box 1 protein modulated proliferation and radioresistance in esophageal squamous cell carcinoma. J Gastroenterol Hepatol. 2019;34(4):728–735. doi: 10.1111/jgh.14371 [DOI] [PubMed] [Google Scholar]
- 28.Dong Y, Xu T, Zhong S, et al. Circ_0076305 regulates cisplatin resistance of non-small cell lung cancer via positively modulating STAT3 by sponging miR-296-5p. Life Sci. 2019;239:116984. [DOI] [PubMed] [Google Scholar]
- 29.Song H, Bian ZX, Li HY, et al. Characterization of hsa_circ_0000594 as a new biomarker and therapeutic target for hepatoblastoma. Eur Rev Med Pharmacol Sci. 2019;23(19):8274–8286. [DOI] [PubMed] [Google Scholar]
- 30.Li T, Sun X, Chen L. Exosome circ_0044516 promotes prostate cancer cell proliferation and metastasis as a potential biomarker. J Cell Biochem. 2020;121(3):2118–2126. [DOI] [PubMed] [Google Scholar]