Abstract
V. minor contains monomeric eburnamine-type of indole alkaloids having utilization as a neuro-medicinal plant. The biosynthetic pathway studies using miRNAs has been the focal point for plant genomic research in recent years and this technique is utilized to get an insight into a possible pathway level study in V. minor as understanding of genes in this prized medicinal plant is meagrely understood. The de novo transcriptomic analysis using Illumina Next gen sequencing has been performed in glasshouse shifted plant and transformed roots to elucidate the possible non confirmed steps of terpenoid indole alkaloids (TIAs) pathway in V. minor. A putative TIA pathway is elucidated in the study including twelve possible TIAs biosynthetic genes. The specific miRNA associated with TIAs pathway were identified and their roles were discussed for the first time in V. minor. The comparative analysis of transcriptomic data of glasshouse shifted plant and transformed roots showed that the raw reads of transformed roots were higher (83,740,316) compared to glasshouse shifted plant (67,733,538). The EST-SSR prediction showed the maximum common repeats among glasshouse shifted plant and transformed roots, although small variation was found in trinucleotide repeats restricted to glasshouse shifted plant. The study reveals overall 37 miRNAs which were observed to be true and can have a role in pathway as they can regulate the growth and alkaloid production. The identification of putative pathway genes plays an important role in establishing linkage between Aspidosperma and Eburnamine alkaloids.
Electronic supplementary material
The online version of this article (10.1007/s12298-020-00842-x) contains supplementary material, which is available to authorized users.
Keywords: de novo transcriptomic analysis, miRNA, EST-SSR, Terpenoid indole alkaloids, Vinca minor
Introduction
Vinca minor (family Apocyanaceae) is a known medicinal plant from ancient times that is used for its astringent, wound-healing, antidermatosic and antigalactic properties in various health care systems (Verma et al. 2012a). The major secondary metabolite considered important in V. minor is vincamine. It is a monomeric eburnamine- type of indole alkaloid (Farahanikia et al. 2011). The vincamine is used in treatment of neuronal homeostasis and in brain circulation as it has modulatory effect on these cells. The attributes of neuro-protective potencies and antihypoxic activities of vincamine were found to be responsible for such modulatory effects (Vas and Gulyás 2005). Due to such properties, vincamine has shown positive results against certain disorders of brain in elderly patients like transient ischemic deficits, vertigo and memory disturbances as shown by clinical trials. Increased cerebral blood flow, oxygen consumption and glucose utilization are major actions which are enhanced by vincamine positively (Belal et al. 2009; Molchan et al. 2012; Verma et al. 2014a, b). Inspite of its wide utilization as a neuro medicine, V. minor is not a thoroughly studied herb as the biogenetic pathway leading to the synthesis of vincamine and other bioactive alkaloids is not adequately elucidated at the level of enzymes and genes. The early steps of the pathway which are similar to that of other TIAs producing members of family Apocyanaceae like Catharanthus and Rauwolfia species has been deciphered so far. The initial steps starts with formation of a common precursor molecule strictocidine that is formed by interaction between indole ring donor tryptamine and a terpenoid moiety donor secologanin by the action of strictosidine synthase (Fig. 1). More importantly, the formation of strictosidine is the first indicative step of carbon flux movement to secondary metabolite production from primary metabolite production. It is further reacted upon by the enzyme strictosidine β- glucosidase (SGD) to yield a highly reactive unstable aglycon that can be channelized to yield more than 50 types of TIAs in V. minor (Szabó 2008). It is important to mention here that pathway level understandings in this plant system could pave the way to produce vincamine from an alternative source such as V. minor transformed roots. Previous studies in our lab have shown that Agrobacterium rhizogenes-mediated transformed hairy root cultures of V. minor provide better in vitro platforms for vincamine production (Verma et al. 2014a, b) and expression of pathway genes in them is amenable to up-regulation by biotic and abiotic elicitations. Possibility of their up-scaling in bioreactor is also quite prominent and accessible. Thus, the hairy root cultures in V. minor plant system could be considered as a potential tool not only as an alternative system of vincamine production but also for providing insight into important studies such as transcriptomic level analysis which may lead to better understanding of functions of particular genes as well as pathway level understanding.
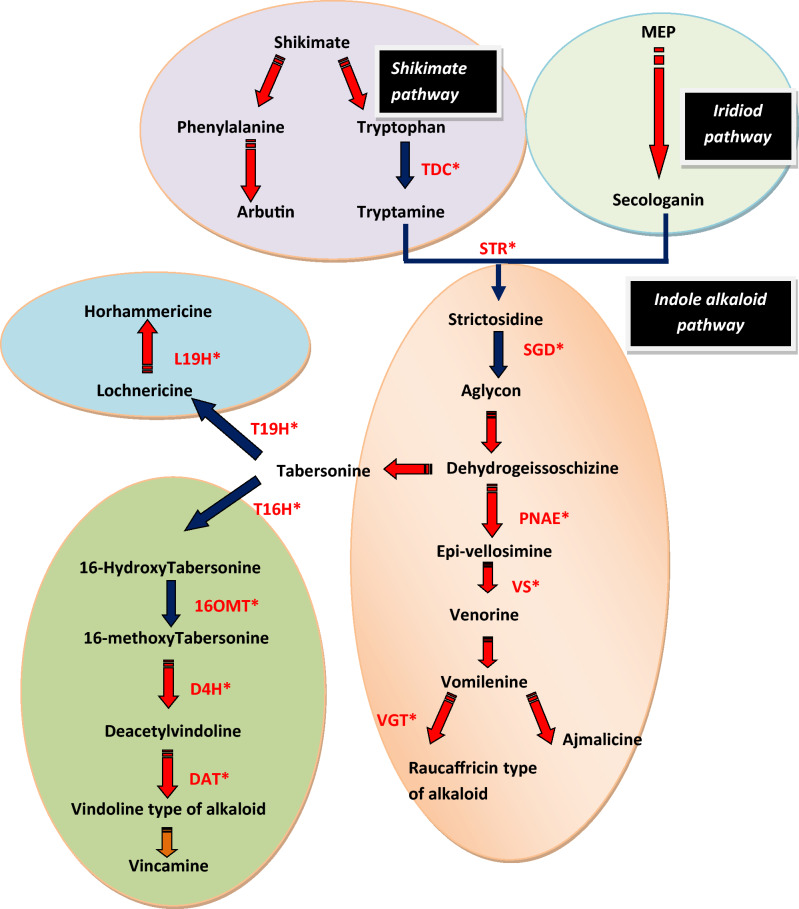
Fig. 1.
Putative TIAs pathway operating in Vinca minor. TDC: tryptophan decarboxylase; STR: Strictosidine synthase; SGD: Strictosidine-o-beta-D glusosidase; T16H: Tabersonine 16 hydroxylase6; T19H: Tabersonin 19 hydroxylase; PNAE: Polyeuridine aldehyde esterase; DAT: Deacetylvindoline-o-acetyltransferase; VGT: Vomilenine glucosyltransferase
Biotechnological interventions in medicinal plant research in recent years have shown that entire biosynthetic pathway can be manipulated by potential use of microRNAs (miRNAs). These miRNAs are small RNA molecules (21–24-nucleotide-long) which are involved in various developmental processes associated with hormonal signaling (Mallory et al. 2005; Wang et al. 2005), plant’s root/shoot growth (Bazin et al. 2012; Khaldun et al. 2015; Yu et al. 2015), floral development (Wang et al. 2009; Luo et al. 2013) and reproduction and stress response (Guleria et al. 2011; Budak et al. 2014). Recent developments in this area of research has suggested their involvement in production and regulation of secondary metabolites (Li et al. 2015; Singh et al. 2016a). The present study takes an effort to identify the conserved miRNAs from two of the V. minor plant materials (i) glasshouse shifted plant that were established under in vitro conditions, rooted and were shifted to glasshouse and (ii) the transformed roots that were alternative source of vincamine production.
The main objective behind studying these two different morphogenic level of plant system is to understand the common and exclusive miRNA regulating the secondary metabolite production in V. minor plant system as both the tissue were able to produce vincamine. Apart from it, the transcriptomic studies of transformed roots could provide an insight into identification of new genes and glasshouse shifted plant can provide a proper comparative study as the native rooted plants. In addition to the above mentioned objectives, this study also focuses on the use of microsatellites in V. minor plant system to identify certain simple sequence repeats (EST-SSRs) which may help in identifying putative genes with new functions.
The microsatellites (SSRs) are found distributed in eukaryotes such as animals, plants and microorganism, which are short DNA motif repeats (1–6 nucleotide). These microsatellites are used as a marker to assess genetic variation with significantly better advantages as compared to restriction fragment length polymorphism (RFLPs), random amplification of polymorphic DNA (RAPD) and amplified fragment length polymorphism (AFLP). They can be genomic SSRs or EST-SSRs. The use of EST-SSRs as markers have added advantages as they are easily reproducible, cheap, highly polymorphic in nature and more importantly located on expressed sequenced tags. Slowly, EST-SSRs based marker usage is catching momentum as these are nowadays used in breeding application due to such attributes and are proving to be an ideal molecular marker for studies (Qu and Liu 2013). With improvements in molecular and bioinformatics fields, characterization of more EST-SSRs are being completed which may have possible function (Wei et al. 2011).
Therefore in the present study, de novo transcriptomic analysis using Illumina Next gen sequencing has been performed in glasshouse shifted plant as well as in the transformed roots to explore the chances of elucidating unknown steps of terpenoid indole alkaloids (TIAs) pathway in V. minor which till recent times have not been achieved. This study discusses for the first time miRNA identification associated with TIAs pathway in V. minor plant system which is a novel study in itself. The pathway linked important EST-SSRs in V. minor has also been discussed herewith which may help in understanding the pathway level gene functions in a improved way in this prized medicinal plant with promising future potential.
Materials and methods
Material and library preparation
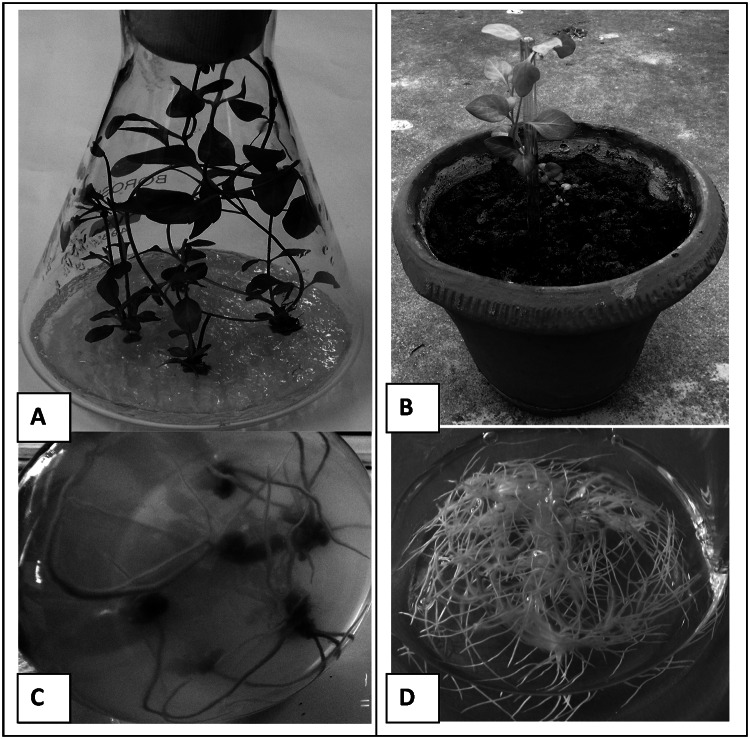
The in vitro established plants of V. minor were maintained and rooted on MS basal medium (Murashige and Skoog 1962) with 1.0 mg/l 6-benzyladenine (BA) and 0.1 mg/l naphthalene acetic acid (NAA). The thoroughly washed rooted plants were further acclimatized in glasshouse conditions where new buds were emerged within 15 days. At that stage, plants were taken for transcriptomic studies. Agrobacterium rhizogenes mediated transformed roots PVG (Verma et al. 2014a) have been maintained on one-fourth strength B5 liquid medium (Gamborg et al. 1968) (Fig. 2). RNA was isolated from both the samples following TRIZOL based method. For RNA isolation, small rooted twig of glasshouse established plants were taken. The preparation of library was done at Genotypic Technology’s Genomics facility following Illumina Tru Seq RNA library protocol outlined in “TruSeq RNA Sample Preparation Guide” (Part # 15008136; Rev. A; Nov 2010). The fragmentation of the purified m-RNA was carried out till 8 min at high temperature (94˚C) with divalent cation presence followed by reverse transcription with the help of Superscript III Reverse transcriptase enzyme through random hexamers priming. In the presence of DNA polymerase I and RNaseH, the c-DNA second strand was synthesized. The High Prep PCR (MAGBIO, Cat# AC-60050) was used to clean the c-DNA followed by ligation with illumina adapter to the c-DNA molecules after the completion of end repair and A base addition. After the completion of the ligation process, SPRI cleanup was carried out. A total of 8 cycles of PCR were used for library amplification and quantification of the prepared library was done by using Qubit followed by validation for its quality by subjecting aliquot to high sensitivity Bio analyzer chip (Agilent).
Fig. 2.
Tissue used for generating transcriptomic data. aIn vitro rooted plants growing on MS basal medium fortified with 1.0 mg/l 6-benzyladenine (BA) and 0.1 mg/l naphthalene acetic acid (NAA); b bottom view of in vitro grown plant ready to shift to glasshouse conditions showing roots; c Glasshouse shifted plant used for isolating RNA; dAgrobacterium rhizogenes mediated transformed root PVG
Analysis tools used for de-novo assembly and transcript annotation
For the quality check of raw reads, FASTQC (http://www.bioinformatiSample1.babraham.ac.uk/projects/fastqc/) was used. Perl codes were used to process raw data including adapter, B-block and low quality base filtering. For de-novo assembly of the glass house shifted plant and transformed root, Trinity (Grabherr et al. 2011) software was used. Transcript annotation was done by ncbi-BLAST-2.2.29 + tool using homology search (Altschul et al. 1990). Differential gene expression analysis was performed using Deseq tool (Anders and Huber 2010). The transcripts which were greater than or equal to 500 bp were annotated against all Viridiplantae kingdom protein sequences (from uniprot protein database) and the transcripts below 500 bp were annotated with pfam database.
EST-SSR Identification
EST-SSR prediction was done by using MIcroSAtellite (MISA) software (http://pgrc.ipk-gatersleben.de/misa). MISA algorithm obligatory identify mononucleotide repeats as 10 or more units, dinucleotide as 6 or more units and trinucleotide, tetranucleotide, pentanucleotide or hexanucleotide repeats as 5 or more units. Compound EST-SSRs are referred to combining independent EST-SSRs that are separated by less than 100 nucleotides.
Identification of microRNAs
Pre-miRNAs were predicted by considering two features (sequence conservation and structural stability). For sequence conservation, only four mismatches were allowed for homolog prediction against reported miRNAs of Arabidopsis thaliana using BLAST search (E = 10). To filter coding sequence from the assemble data of both samples, BLASTX search (e-value ≤ 1e−8) was executed against UniProtKB/Swiss-Prot and UniProtKB/TrEMBL database. Stability of secondary structure was also evaluated in the terms of number and size of bulges (≤ 1), MFE (Minimum Free Energy) and MFEI (Minimum Free Energy Index ≤ 0.9 kcal/mol). An embedded module UNAFold was used for primary and precursor miRNA folding. UNAFold parameters were applied, maximum base pair distance was 3000, maximum bulge/interior loop size was 30, and single thread run at 37 °C temperature. Other criteria used for study were same as described previously (Singh et al. 2016a) using C-mii (Numnark et al. 2012).
In addition, KAAS server was used for analyzing the identified miRNA regulation process pathway of secondary metabolites. Network approach was applied to reveal the co-regulation of predicted miRNAs. A phylogenetic analysis of identified miRNAs was performed. ClustalW was used to align the sequences. Phylogenetic tree was constructed by MEGA 6.0 (Tamura et al. 2013). Maximum Likelihood method based on the Kimura 2-parameter model was used.
Results and discussion
De novo assembly and annotation
The library was found suitable for Illumina (NextSeq 500) based on the population observed in the profile as the library showed a peak at the range of 250–600 bp. The adaptor flanked inserts were of size 120 bp and effective sequencing insert size was 130–480 bp. A total of 67,733,538 and 83,740,316 raw reads were generated for glass house shifted plant and transformed roots, respectively. The FASTQC based processed reads were found to be 53220084 and 64979786 of glass house shifted plant and transformed root, respectively. Trinity assembly gene generated a total of 83286 (glass house shifted plant) and 89620 (transformed root) unigenes (Table 1). The minimum transcript length was found to be similar for both samples (301). Interestingly, maximum transcript length of glass house shifted plant was higher (15690) than transformed roots (12125) with an average length of 1060 and 1038.9, respectively. A total number of 33778 and 58613 transcripts of glass house shifted plant were over 1000 bp and 500 bp, respectively. Meanwhile, the assembly of transcript of transformed roots revealed 34719 and 62097 transcripts were over 1000 bp and 500 bp, respectively (NCBI SRA no. SRR5581424 and SRR5581425).
Table 1.
De-novo assembly statistics of sequenced transcriptome data of glasshouse shifted plant and transformed roots of V.minor
| Organism Name | Vinca minor | |
|---|---|---|
| Sample Name | Glasshouse shifted Plant | Transformed roots |
| Tool used | Trinity | |
| Hash length | 25 | |
| Transcripts Generated | 83286 | 89620 |
| Maximum Transcript Length | 15690 | 12125 |
| Minimum Transcript Length | 301 | 301 |
| Average Transcript Length | 1060 | 1038.9 |
| Median Transcript Length | 1033.5 | 1405.5 |
| Total Transcripts Length | 88282740 | 93105316 |
| Total Number of Non-ATGC Characters | 0 | 0 |
| Transcripts ≥ 500 b | 58613 | 62097 |
| Transcripts > 1 Kb | 33778 | 34719 |
| Transcripts > 10 Kb | 8 | 3 |
| N50 value | 1457 | 1430 |
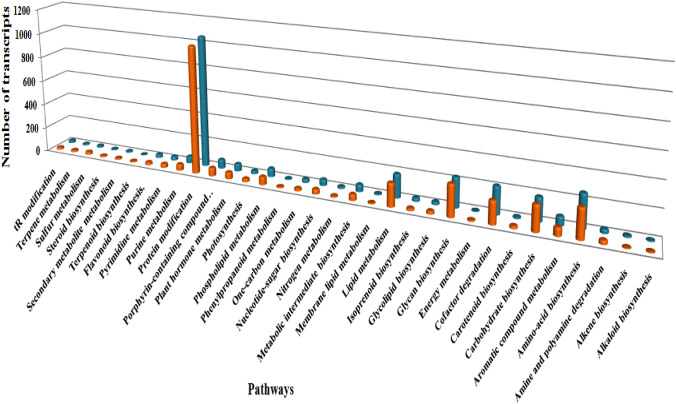
In case of glass house shifted plant, a total of 40537 transcripts were annotated from uniprot database, while 7587 transcript were noted as un-annotated. Similarly, in the case of transformed roots, a total number of 41419 transcripts were considered as annotated, whereas 9936 transcripts were un-annotated. With concern to the annotation results obtained from pfam database, a total number of 22842 annotated and 3064 un-annotated transcripts were found in glass house shifted plant. Likewise, in the case of transformed roots, 25526 and 3154 number of transcripts were annotated and un-annotated respectively. It is important to note here that annotated transcripts of both glass house shifted plant and transformed roots were depicting their involvement in different metabolic pathway viz;, biological process, molecular function and cellular component. A comparative analysis of the annotated transcripts from uniprot database involved in different metabolic pathways were also performed (Fig. 3) in which protein modification, lipid metabolism, amino-acid biosynthesis, glycan and carbohydrate biosynthesis were the major pathways observed in both the samples. It is pertinent to mention here a small variation was observed in the case of number of transcripts involved. The protein modification metabolic pathway was found most abundant in both the samples, in which 1025 transcripts were from glass house shifted plant and 1059 transcripts were from transformed roots. Similarly, in the case of secondary metabolite metabolism variation of one transcript was observed. Terpene metabolism has shown involvement of 17 and 13 transcripts from glass house shifted plant and transformed roots and steroid biosynthesis revealed 12 transcripts from glass house shifted plant and 10 from transformed roots. Adding to it, the flavonoid biosynthesis showed the involvement of 28 transcripts from transformed root and 23 from glass house shifted plant.
Fig. 3.
Comparative study of the annotated transcripts involved in different metabolic pathways from uniprot database. Orange and blue colors shows the transcripts of glasshouse shifted plant and transformed root, respectively
The immense potential of vincamine pharmaceutically, less occurrence in the plant, non-availability of chemical alternatives, high market price and costing demands an in depth prospection of V. minor at biogenetic pathway level (Verma et al. 2014a). Such information is required not only to devise appropriate breeding strategies to up-regulate their in planta synthesis, but also for engineering alternate production platforms for desired metabolites in homologous to heterologous in vitro culture systems. Plant secondary metabolism as such, is a highly dynamic process. At any point of time, thousands of such compounds are formed, stored, and often catabolised under several developmental and environmental influences (Facchini and De Luca 2008). In terms of cellular resource economy and energy budgeting, these metabolites are expensive to be produced and stored. Moreover, the metabolic pathways are intensively crossed-linked and hence, their synthesis is often rigidly regulated not only at gene level but also at the levels of precursor availability, enzyme activity, inter- and intra-cellular compartmentation and metabolite transport mechanisms across organs and organelles (Verma et al. 2012b). In recent times, the NGS platforms are explored variously for particular gene identification or locating genomic intervals for the purpose of targeted sequencing in plant systems. This emerging technique has the possibility to compare the transcriptomic analysis of several plants. Also, this has made successful achievement in the cataloging and characterizing a particular cell/tissue’s transcriptome which in turn helps in identifying particular gene or the pattern of splicing of mRNA. Added benefits with NGS based techniques are the quantification of transcripts differentially in different pathological and physiological conditions (Wei et al. 2013).
EST-SSR identification
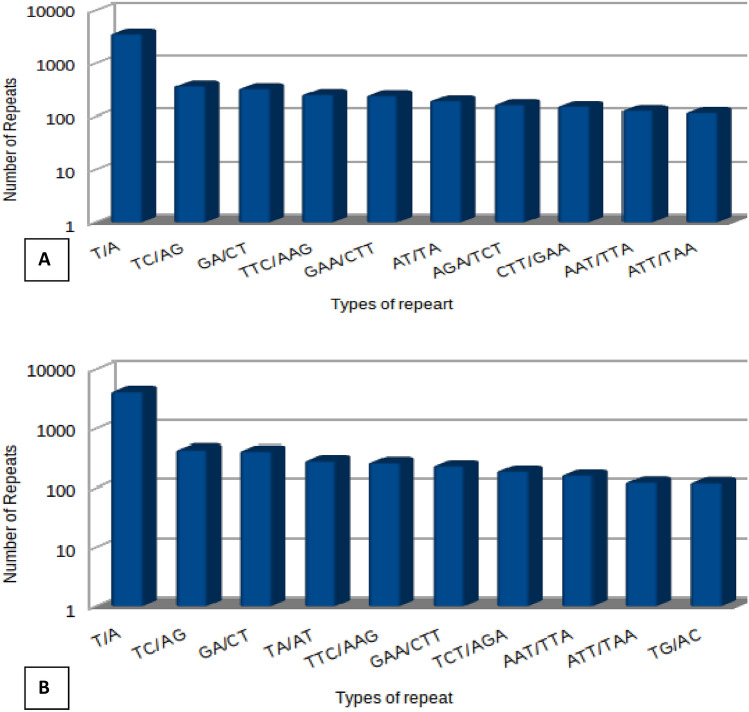
EST-SSRs are considered to be one of the most powerful tools in genetic studies and could be proven a beneficial tool in unexplored plant such as V. minor. The attempt for identification of EST-SSRs in V. minor transformed roots and glass house shifted plant was carried out with an objective that this plant system has not been fully explored at molecular marker level studies and a comparative analysis between these two samples may lead to possibility of identification of possible pathway identification in this plant system. Notably, a total number of EST-SSRs found in glass house shifted plant was less when compared to the complete number of EST-SSRs which were found in the transformed roots of the plant. Out of 77248 sequences recognized in transformed roots, 14082 EST-SSRs were found whereas in case of glass house shifted plant 13950 EST-SSRs were found in 71231 sequences. Number of sequences having more than 1 EST-SSR in glass house shifted plant was 12381 and in transformed roots it was observed to be 1612. More importantly, mono-nucleotide repeats were found to be the most abundant EST-SSR motif in both sample followed by di-nucleotide, tri-nucleotide, tetra-nucleotide, penta-nucleotide and hexa-nucleotide motifs. The number of compound EST-SSRs present in glass house shifted plant and transformed root were 546 and 690, respectively. In both the sample maximum repeats were commonly observed (T/A, TC/AG, GA/CT, TA/AT, TTC/AAG, GAA/CTT, AGA/TCT, ATT/TTA and ATT/TAA). A small variation was observed in the case of tri nucleotide repeat (CTT/GAA) which was specific to glass house shifted plant (Fig. 4), which may also be responsible for its functional variation in biological condition.
Fig. 4.
The statistics for 10 random repeats is represented EST-SSRs identified in a glasshouse shifted plants and b transformed roots
The microsatellite markers are known for their key role in functional genomics. Microsatellites can be genomic SSRs or EST-SSRs based on the source of generation i.e. genome or expressed sequence tags based SSRs, respectively. EST-SSRs are positioned in the coding region and are documented from transcribed RNA sequences which are generated by transcriptomic studies. A RNA-sequencing data is more beneficial than whole genome due to high reproducibility and less systematic differences among technical replicates. EST-SSRs are highly recommended for biodiversity studies due to their low-cost development, enhanced hosting sequence conservation, elevated related taxa transferability and higher genetic diversity (Taheri et al. 2018).
In this particular study, the EST-SSRs are considered over genomic SSRs as they have more potential to assess changes occurring in gene during domestication. Moreover, only limited number of EST-SSR markers are enough for resolving phylogenetic relationships instead of using large number of genomic SSR markers (Tabbasam et al. 2014). In a study on P. violascen in which EST-SSRs were generated, that helped in identifying sixteen different varieties and ten species of Phyllostachys (Cai et al. 2019).
EST-SSRs based marker is an emerging technique used in different field of studies such as construction of fingerprint, diversity analysis among species and more importantly in breeding which are assisted by molecular markers. The EST-SSRs identification through advanced techniques like next generation sequencing in important medicinal plants such as V. minor may play an important role in generating large amount of data which in turn could be a considerable resource in assessing the genetic diversity and population structures among strictly related TIAs producing plants and also help in resolving multifarious associations among them. The novel EST-SSRs data generation studies conducted in this study can provide ample chances to corelate many related plants which belong to Vinca family such as Rauwolfia, Catharanthus, as similar kind of studies have been conducted previously in Mentha in which new EST-SSRs were determined for 13 accessions of M. piperita and they were also utilized to evaluate cross-species transferability in four Mentha species (Kumar et al. 2015).
Putative TIAs biosynthetic route in V. minor
TIAs biosynthesis in V. minor family as per the targets determined in the transcriptomic data of both the samples (glass house shifted plant and transformed roots) was found very similar to other two indole alkaloid containing Apocyanaceae members Rauwolfia and Catharanthus. A total of twelve enzymes from the TIAs and associated pathways have been observed. Their transcript profile in both the samples (glass house shifted plant as well as transformed roots) has been shown in Table 2. Based on these observations, a putative model of TIA pathway in V. minor has been proposed (Fig. 1). It is proposed from the conducted studies that like other members of Apocyanaceae, strictosidine—the condensation product of tryptamine and secologanin, catalyzed by enzyme strictosidine β-glucosidase (SGD) which leads to exceedingly reactive unstable aglycon. This aglycon opens up a new branch point towards the synthesis of dehydrogeissoschizine that can be channelized in two directions. It can either be fluxed towards the synthesis of tabersonine that can be further converted into horhammericine in the plant roots by the action of T19H (tabersonine 19 hydroxylase) or acted upon by T16H (tabersonine 16 hydroxylase) and form 16-hydroxy tabersonine that is pushed towards vindoline synthesis (via involvement of D4H [Deacetoxyvindoline 4-hydroxylase] and DAT [Deacetylvindoline-o-acetyltransferase]) in the leaves. Further rearrangements of vindoline type of alkaloid lead to formation of vincamine. Alternately, dehydrogeissoschizine can be diverted towards the synthesis of epi-vellosimine by the enzyme polyeuridine aldehyde esterase (PNAE) which further metabolizes into vomilenine. From this point, either ajmalicine is formed in the roots or by the action of vomilenine glucosyltransferase (VGT), raucaffricine type of alkaloids are formed.
Table 2.
Expression profile of genes associated with putative TIAs pathway in Vinca minor
| Gene name | Transcripts | |||||
|---|---|---|---|---|---|---|
| Up-regulated | Down_regulated | Neutral | Expressed in Glasshouse shifted plants only | Expressed in transformed roots only | Total | |
| Deacetoxyvindoline 4-hydroxylase (EC 1.14.11.20) | 6 | 17 | 28 | 2 | 0 | 53 |
| Tabersonine 16-hydroxylase (EC 1.14.13.73) (Cytochrome P45071D12) (Fragment) | 4 | 3 | 6 | 0 | 0 | 13 |
| Vinorine synthase (EC 2.3.1.160) | 4 | 4 | 7 | 1 | 0 | 16 |
| Vomilenine glucosyltransferase (EC 2.4.1.219) | 8 | 14 | 19 | 1 | 1 | 43 |
| Tabersonine 16-O-methyltransferase (EC 2.1.1.94) | 0 | 1 | 0 | 0 | 0 | 1 |
| Deacetylvindoline O-acetyltransferase (EC 2.3.1.107) | 2 | 3 | 4 | 0 | 0 | 9 |
| Hydroquinone glucosyltransferase (EC 2.4.1.218) | 4 | 5 | 2 | 0 | 0 | 11 |
| Polyneuridine-aldehyde esterase (EC 3.1.1.78) | 6 | 5 | 21 | 0 | 2 | 34 |
| Strictosidine-O-beta-D-glucosidase (EC 3.2.1.105) | 3 | 13 | 11 | 4 | 0 | 31 |
| Tryptophan decarboxylase (EC 4.1.1.28) | 0 | 4 | 4 | 0 | 0 | 8 |
| Tabersonine/lochnericine 19-hydroxylase (EC 1.14.14.) | 0 | 8 | 0 | 0 | 0 | 8 |
C. roseus and R. serpentina are closely related to V. minor. Interpretation done by combining the present data and previous studies performed in the C. roseus, a putative step involved in TIAs pathway in V. minor has been elucidated. The putative pathway showed two branch points from the dehydrogeissoschizine intermediate. One was found to be similar with Rauwolfia, where raucaffricin type of alkaloids are formed through the involment of polyeuridine aldehyde esterase (PNAE) and vomilenine glucotransferase (VGT). Another route was found to be similar with vindoline pathway operated in C. roseus. The involvement of cytochrome P 450 is a must for this rearrangement process which is needed for vindoline biosynthesis. This enzyme has the capability of catalyzing the epoxide formation which can further lead to vincamine- eburnamine backbone formation after rearrangements. Here, by this formation, it provides indicative evidence to a long lasting believed hypothesis that eburnamine and aspidosperma- type alkaloids have a relationship biosynthetically (Kellner et al. 2015).
Characteristic of the conserved miRNA genes of glass house shifted plant and transformed root
Initially, 176 conserved precursor miRNAs were predicted using both sample data, out of which 86 were from glass house shifted plant and 90 were from transformed root. To overcome the rate of false prediction, all predicted pre-miRNAs were evaluated at sequential and structural level. In total, 37 miRNAs were selected as true candidates (Table 3).
Sequential level evaluation
Table 3.
Predicted miRNAs from the glasshouse shifted plants and transformed roots of V.minor
| miRNA families | miRNAs | NB | BZ | MFE* | MFEI* | PL | GC | All Content | miRNA sequence | Tissue |
|---|---|---|---|---|---|---|---|---|---|---|
| miR156 | miR156a | 1 | 1 | − 41.5 | − 1.185 | 78 | 44.87 | A(20), U(23), G(18), C(17), N(0) | 5′:UGACAGAAGAUAGAGAGCAC:3′ | GSP |
| miR156h | 1 | 1 | − 37.9 | − 1.184 | 77 | 41.55 | A(17), U(28), G(17), C(15), N(0) | 5′:UGACAGAAGAUAGAGGGCAC:3′ | TR | |
| miR157 | miR157a | 1 | 1 | − 41.7 | − 1.191 | 79 | 44.3 | A(20), U(24), G(18), C(17), N(0) | 5′:UUGACAGAAGAUAGAGAGCAC:3′ | GSP |
| miR157d | 1 | 1 | − 37.9 | − 1.184 | 77 | 41.55 | A(17), U(28), G(17), C(15), N(0) | 5′:UGACAGAAGAUAGAGGGCAC:3′ | TR | |
| miR159 | miR159a | 0 | 0 | − 78.4 | − 0.956 | 174 | 47.12 | A(44), U(48), G(44), C(38), N(0) | 5′:CUUGGACUGAAGGGAGCUCCC:3′ | GSP |
| miR167 | miR167c | 1 | 1 | − 31.9 | − 1.029 | 83 | 37.34 | A(20), U(32), G(16), C(15), N(0) | 5′:GAAGCUGCCAGCAUGAUCUAA:3′ | GSP |
| miR168 | miR168a | 0 | 0 | − 55.8 | − 0.914 | 147 | 41.49 | A(34), U(52), G(34), C(27), N(0) | 5′:UCGCUUGAUGCAGGUCGGGAA:3′ | GSP |
| miR170 | miR170 | 0 | 0 | − 31.1 | − 0.971 | 75 | 42.66 | A(17), U(26), G(17), C(15), N(0) | 5′:UGAUUGAGCCGUGCCAAUAUC:3′ | TR |
| miR171 | miR171b | 0 | 0 | − 27.9 | − 0.9 | 81 | 38.27 | A(19), U(31), G(18), C(13), N(0) | 5′:UUGAGCCGUGCCAAUAUCACG:3′ | GSP |
| miR171b | 0 | 0 | − 35.4 | − 0.983 | 81 | 44.44 | A(19), U(26), G(20), C(16), N(0) | 5′:UUGAGCCGUGCCAAUAUCACG:3′ | TR | |
| miR1888 | miR1888 | 0 | 0 | − 28.3 | − 1.01 | 104 | 26.92 | A(39), U(37), G(19), C(9), N(0) | 5′:UAAGUUAAGAUUUCCCAAGAA:3′ | TR |
| miR319 | miR319a | 0 | 0 | − 73.3 | − 0.916 | 192 | 41.66 | A(47), U(65), G(45), C(35), N(0) | 5′:UUGGAUUGAAGGGAGCUCUAC:3′ | GSP |
| miR391 | miR391 | 1 | 1 | − 38.7 | − 0.9 | 99 | 43.43 | A(27), U(29), G(21), C(22), N(0) | 5′:UACGCAGGAGAGAUGGCGCCC:3′ | GSP |
| miR391 | 1 | 1 | − 43.2 | − 1.028 | 85 | 49.41 | A(18), U(25), G(22), C(20), N(0) | 5′:UACGCAGGAGAGAUGGCGCCG:3′ | TR | |
| miR393 | miR393a | 1 | 1 | − 36.4 | − 0.933 | 99 | 39.39 | A(25), U(35), G(19), C(20), N(0) | 5′:UCCAAAGGGAUCGCAUUGAUCU:3′ | GSP |
| miR394 | miR394a | 0 | 0 | − 39.4 | − 0.96 | 82 | 50 | A(14), U(27), G(20), C(21), N(0) | 5′:UUGGCAUUCUGUCCACCUCC:3′ | GSP |
| miR396 | miR396a | 0 | 0 | − 37.2 | − 0.907 | 92 | 44.56 | A(19), U(32), G(24), C(17), N(0) | 5′:UUCCACGGCUUUCUUGAACUG:3′ | GSP |
| miR396a | 0 | 0 | − 21.9 | − 0.912 | 59 | 40.67 | A(16), U(19), G(15), C(9), N(0) | 5′:UUCCACAGCUUUAGUUGACUG:3′ | TR | |
| miR398 | miR398a | 0 | 0 | − 34.5 | − 0.958 | 81 | 44.44 | A(23), U(22), G(20), C(16), N(0) | 5′:UGUGUUCUCAGGUCGCCCCUG:3′ | GSP |
| miR398a | 0 | 0 | − 38.7 | − 1.045 | 93 | 39.78 | A(22), U(34), G(19), C(18), N(0) | 5′:UGUGUUCUCAGGUCGCCCCUG:3′ | TR | |
| miR408 | miR408 | 0 | 0 | − 38.6 | − 0.941 | 83 | 49.39 | A(16), U(26), G(20), C(21), N(0) | 5′:AUGCACUGCCUCUUCCCUGGC:3′ | TR |
| miR414 | miR414 | 0 | 0 | − 28.1 | − 1.17 | 55 | 43.63 | A(14), U(17), G(15), C(9), N(0) | 5′:UCUUCUUCGUCCUCAUCUUCA:3′ | TR |
| miR5015 | miR5015b | 0 | 0 | − 22.9 | − 1.145 | 66 | 30.3 | A(21), U(25), G(16), C(4), N(0) | 5′:UCUGUUGUUGUUGUUCUUCUU:3′ | GSP |
| miR5015b | 0 | 0 | − 22.9 | − 1.145 | 66 | 30.3 | A(21), U(25), G(16), C(4), N(0) | 5′:UCUGUUGUUGUUGUUCUUCUU:3′ | TR | |
| miR5021 | miR5021 | 0 | 0 | − 897 | − 2.379 | 1110 | 33.96 | A(367), U(366), G(189), C(188), N(0) | 5′:GAAGAAGAAGACGAAGAAGA:3′ | GSP |
| miR5021 | 0 | 0 | − 15.9 | − 1.223 | 36 | 36.11 | A(10), U(13), G(6), C(7), N(0) | 5′:UCUGAAGAAGAAGUAGAAGA:3′ | TR | |
| miR5651 | miR5651 | 0 | 0 | − 28.4 | − 0.916 | 69 | 44.92 | A(15), U(23), G(17), C(14), N(0) | 5′:UGGUGCGGUUCAAUUAGAAAA:3′ | TR |
| miR5658 | miR5658 | 0 | 0 | − 17.9 | − 0.994 | 55 | 32.72 | A(17), U(20), G(6), C(12), N(0) | 5′:CUGAUGAUGAAGAUGAAGAAA:3′ | GSP |
| miR5658 | 0 | 0 | − 28.6 | − 0.953 | 105 | 28.57 | A(26), U(49), G(21), C(9), N(0) | 5′:ACCAUGAUGAUGAUGAUGGAA:3′ | TR | |
| miR828 | miR828 | 0 | 0 | − 36.1 | − 0.975 | 94 | 39.36 | A(25), U(32), G(21), C(16), N(0) | 5′:UCUUGCUCAAAUGAGUAUUCCA:3′ | TR |
| miR829.2 | miR829.2 | 0 | 0 | − 20.4 | − 1.073 | 119 | 15.96 | A(54), U(46), G(10), C(9), N(0) | 5′:AAAAAUAAAGCUUAAAGGUAA:3′ | TR |
| miR837-5p | miR837-5p | 0 | 0 | − 381.4 | − 1.417 | 715 | 37.62 | A(219), U(227), G(143), C(126), N(0) | 5′:AACGUUUUCUUCUUCGUUUCA:3′ | GSP |
| miR838 | miR838 | 0 | 0 | − 20.2 | − 1.188 | 46 | 36.95 | A(17), U(12), G(9), C(8), N(0) | 5′:UCUUCUUCUACUUCUUCUUCA:3′ | TR |
| miR854a | miR854a | 0 | 0 | − 38.7 | − 1.105 | 73 | 47.94 | A(18), U(20), G(21), C(14), N(0) | 5′:GAUGAGGAUAGGAACGGGGAU:3′ | TR |
| miR865-3p | miR865-3p | 1 | 1 | − 8.3 | − 0.922 | 43 | 20.93 | A(16), U(18), G(3), C(6), N(0) | 5′:UUUUGCUACAAAUUUAUCCAA:3′ | GSP |
| miR866-3p | miR866-3p | 0 | 0 | − 41.9 | − 1.102 | 98 | 38.77 | A(28), U(32), G(20), C(18), N(0) | 5′:ACGGAAUCCGUCUUUCAAUA:3′ | GSP |
| miR866-3p | 0 | 0 | − 39.2 | − 0.98 | 98 | 40.81 | A(27), U(31), G(21), C(19), N(0) | 5′:ACGGAAUCCGUCUUUCAAUA:3′ | TR |
NB, Number of bulges, BZ, Bulge size,*kcal/mol (MFE and MFEI values), PL, length of precursor miRNAs, GSP, glasshouse shifted plants, TR, transformed roots
The alignment of homolog and predicted miRNAs were not allowed to have more than 4 mismatches. The length of precursor miRNAs was observed to have significant variation. Maximum miRNA candidates (Total 25) fell in the range of 35–95 nt. Similarly, mature miRNAs were also observed with the range of 20–22 nt. A total of 28 miRNAs had 21nt length, while for 20 and 22nt, only 8 and 1 miRNA (miR393a) candidates were observed, respectively. In order to predict pre-miRNAs, GC content was also evaluated and it was found that the average of the all predicted GC content was 39.24, which also proposed that predicted miRNAs may have higher AU content as compared to GC content.
Structural level evaluation
Stability of the secondary structure of pre-miRNA is the vital attribute of miRNA prediction study which has been assessed in terms of Minimum Free Enegy (MFE) and Minimum Free Energy Index (MFEI). The MFEI is also another parameter to signify structural stability.
In present study, 29 out of 37 miRNAs were observed to have 20 to 50 (-kcal/mol) rang of MFE. Maximum miRNA candidates (19 out of 37) demonstrated to have 0.9 (-kcal/mol). Only one miRNA was shown to contain 2.0 (-kcal/mol), while rest of the miRNAs depicted 1.0 (-kcal/mol) of MFEI values. It is important to mention here that previous case studies have also taken only MFE (Minimum Free Energy) and MFEI (Minimum Free Energy Index) parameter to assess the stability of secondary structure (Singh and Sharma 2014). In presented study, number of bulges (NB) and bulge size (BZ) were also customized to minimum value (≥ 1).
Target gene prediction of V. minor miRNAs and their annotation
A target search was performed individually for glass house shifted plant as well as for transformed roots. Only predicted miRNA families were used for the analysis. A total of 350 and 423 targets were annotated for glass house shifted plant and transformed roots, respectively. All these targets were observed to be involved in several biological process viz, signaling, reproduction, localization, response to stimulus, growth and developmental process. The miRNA–target folding module of the tool suggests that, there were several un-annotated target genes which were identified. The secondary metabolic pathways of the identified targets, regulated by predicted miRNAs were the focal point of the present study.
Comparative study of the conserved miRNA genes of glass house shifted plant and transformed roots
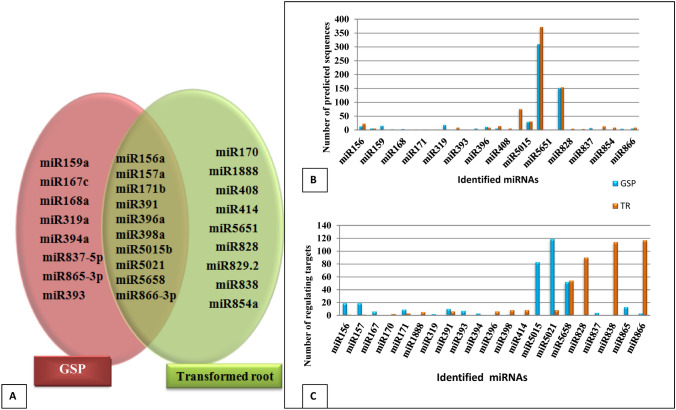
From predicted and evaluated 37 miRNA candidates (Table 3), 18 were identified from glass house shifted plant, while 19 were from transformed roots amongst which 10 miRNA families (miR156a, miR157a, miR171b, miR391, miR396a, miR398a, miR5015b, miR5021, miR5658, miR866-3p) were common between both the tissues (Fig. 5a). Findings also suggest that there are 8 and 9 unique miRNAs for glass house shifted plant and transformed root, respectively.
Fig. 5.
Comparative analysis of both samples of Vinca minor;a unique and common miRNAs between both tissues b showed the number of predicted miRNAs and c showed the number of the targets regulated by Vinca-miRNAs. Blue and orange colors were represent the glasshouse shifted plant (GSP) and transformed root (TR), respectively
In the case of miRNA prediction, miR5021 and miR5658 have high rate of prediction in both tissue, but relatively higher in transformed root as compared to glass house shifted plant (Fig. 5b,c). On the other hand, in the case of target prediction, miR5021 was observed to regulate maximum targets in the glass house shifted plant as compared to the transformed root. A very small variation was observed for the number of targets for miR5658 in both the tissues.
In the case of secondary metabolic pathway regulation, several pathways assumptions were identified. The miR5021 and miR5658 were observed as pre-dominant miRNAs in both the tissues (Table 4 and 5). These candidates were investigated to play a role in the diterpenoid, flavonoid, isoflavonoid and metabolism of xenobiotics by cytochrome P450. Tryptophan metabolism was regulated by different miRNA family in both tissues modulating the expression of same gene NAD-aldehyde dehydrogenase (EC: 1.2.1.3).
Table 4.
miRNAs involved in different metabolic pathways of Vinca minor
| Pathway | Ezyme ID | Pathway ID | Tissue | miRNAs |
|---|---|---|---|---|
| Diterpenoid biosynthesis | EC:1.14.11.15 | map00904 | TR | miR5658 |
| EC:1.14.11.13 | map00904 | TR | miR5658 | |
| Flavonoid biosynthesis | EC:1.14.11.22 | map00941 | TR | miR5021 |
| EC:1.14.11.22 | map00941 | GSP | miR5021 | |
| Isoflavonoid biosynthesis | EC:1.14.11.22 | map00943 | TR | miR5021 |
| EC:1.14.11.22 | map00943 | GSP | miR5021 | |
| Limonene and pinene degradation | EC:1.2.1.3 | map00903 | GSP | miR5021 |
| EC:1.2.1.3 | map00903 | TR | miR838 | |
| Metabolism of xenobiotics by cytochrome P450 | EC:1.2.1.5 | map00980 | GSP | miR5021 |
| EC:2.4.1.17 | map00980 | GSP | miR5021, miR865-3p | |
| EC:2.5.1.18 | map00980 | TR | miR838 | |
| EC:1.1.1.1 | map00980 | TR | miR838 | |
| EC:1.2.1.5 | map00980 | TR | miR838 | |
| EC:2.4.1.17 | map00980 | TR | miR865-3p | |
| EC:1.1.1.1 | map00980 | GSP | miR865-3p | |
| Nicotinate and nicotinamide metabolism | EC:1.2.1.24 | map00760 | GSP | miR5021 |
| EC:3.6.1.9 | map00760 | GSP | miR5658 | |
| EC:1.2.1.24 | map00760 | TR | miR838 | |
| Phenylpropanoid biosynthesis | EC:1.11.1.7 | map00940 | GSP | miR5021 |
| EC:1.2.1.68 | map00940 | GSP | miR5021 | |
| EC:1.2.1.68 | map00940 | TR | miR838 | |
| Steroid biosynthesis | EC:5.5.1.9 | map00100 | TR | miR865-3p |
| EC:5.5.1.9 | map00100 | GSP | miR865-3p | |
| Steroid hormone biosynthesis | EC:2.4.1.17 | map00140 | GSP | miR5021, miR865-3p |
| EC:2.4.1.17 | map00140 | TR | miR865-3p | |
| Tryptophan metabolism | EC:1.2.1.3 | map00380 | GSP | miR5021 |
| EC:1.2.1.3 | map00380 | TR | miR838 |
GSP, glasshouse shifted plants; TR, transformed roots
Table 5.
miRNA associated with alkaloid production in Vinca minor
| miRNA | Tissue | Transcription factor/proteins/enzymes associated with alkaloid production |
|---|---|---|
| miR5658 | Transformed roots | Ethylene-responsive transcription factor ABR1 |
| Transcription factor AS1 | ||
| Zinc finger CCCH domain containing protein 63 | ||
| Transcription factor MYB 48 | ||
| Glasshouse Shifted Plants | Ethylene-responsive transcription factor 13, 1,1A,2 | |
| miR838 | Transformed roots | bHLH 104, 147 |
| Zinc finger CCCH domain containing protein 32, 58 | ||
| Ethylene- responsive TF 1B | ||
| Transcription factor 1CE1 | ||
| Transcription factor MYB 3 | ||
| Transcription factor TCP 20,7 | ||
| miR414 | Transformed roots | BIM1 |
| Zinc finger CCCH domain containing protein 1 | ||
| Heat stress transcription factor A-6b | ||
| TT8 | ||
| miR5021 | Transformed roots | Zinc finger CCCH domain containing protein 30, 58 |
| Ethylene- responsive TF 5, 1B, RAP2-4 | ||
| Dof zinc finger protein DOF3.7 | ||
| Transcription factor 1CE1 | ||
| Transcription factor MYB 39, 44 | ||
| WRKY transcription factor 6 | ||
| Glasshouse Shifted Plants | bHLH 147, 148, 149, 150 | |
| Zinc finger CCCH domain containing protein 30, 24, 50, 56, 66 | ||
| Chorismate mutase | ||
| Ethylene- responsive TF 5, ERF 104, 105 | ||
| Transcription factor MYB 48, 59, 305; MYB related proteins AS1, AS2 | ||
| Transcription factor TCP8, 22, 23, 14, PCF3 | ||
| Probable WRKY transcription factor 23, 48, 8, 71, 28 | ||
| miR828 | Transformed roots | Transcription factor MYB 3 |
| miR5015b | Glasshouse Shifted Plants | Ethylene-responsive transcription factor ERF104, ERF105, ERF-115, ERF 114, ERF 113, ERF 110, ERF11, ERF 010, ERF 008, ABR1, RAP2-10, RAP2-1 |
| Transcription factor 1CE1 SCREAM2, bHLH 61, 93 | ||
| miR319a | Glasshouse Shifted Plants | Transcription factor GAMYB |
| miR156a | Glasshouse Shifted Plants | Squamosa promoter-binding protein 1 |
The high throughput sequencing and in silico identification are used in non-model plants for identification of mi-RNA and their target regions. The next lead is conforming the design and interaction of bioengineering strategies experimentally which will be a positive step in improving the secondary metabolites with the help of mi-RNA (Bulgakov and Avramenko 2015). This is the first report of determination of miRNAs associated with various pathways related to alkaloid production in V. minor. A total of 37 miRNAs were reported which are found to be true. These miRNAs can regulate growth and alkaloid production. The mi-RNA mediated manipulations will help in increase of desired secondary metabolite concentration by permitting re-direction of the flow of metabolites in plant cells within a particular biosynthetic pathway.
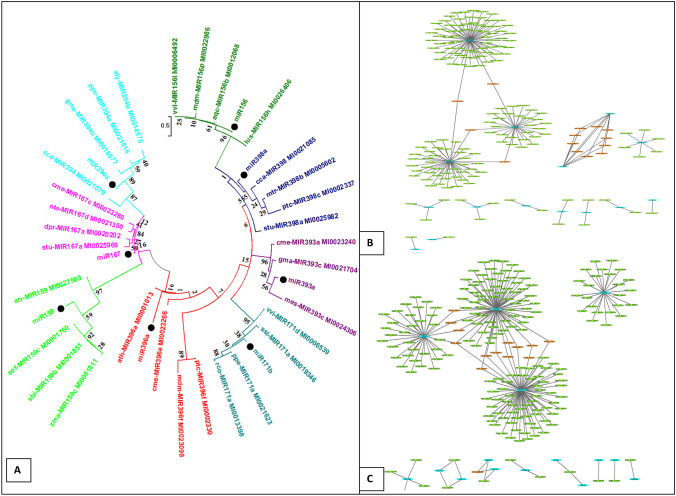
Phylogenetic and network analysis
Phylogenetic analysis was performed for randomly selected miRNAs. The precursor sequences were downloaded from miRBase for each individual miRNA family. In this particular study, identification of 37 miRNAs belonging to 20 distinct miRNA families successfully achieved. More importantly much of the proportion of sequence similarity was observed among the plant families with respect to the predicted miRNAs.
To analyze the co-regulated targets by more than one miRNA family, a biological network was constructed (Fig. 6). In the case of glass house shifted plant, Trihelix Transcription Factor (TGT-2, GT-2) and DEAD-box ATP-dependent RNA helicase ISE2 were the targets, co-regulated by miR5015b and miR5658. The TGT-2 was earlier reported for cold and salt stresses (Xi et al. 2012). Thus results also suggest that miR5015b and miR5658 showed response to stimuli by modulating the expression of TGT-2. The ERF5 was shared by miR5021 and miR5015b, while IRX14 was shared by miR5021 and miR5658.
Fig. 6.
a Phylogenetic tree of identified miRNAs along with their closely related miRNA families. Every cluster of miRNA family was represented by different color. Black dot represents the Vinca-miRNAs. vvi (Vitis vinifera), aqc (Aquilegia caerulea), lus (Linum usitatissimum), mdm (Malus domestica), ath (Arabidopsis thaliana), nta (Nicotiana tabacum), bra (Brassica rapa), aly (Arabidopsis lyrata), zma (Zea mays), sof (Saccharum officinarum), sbi (Sorghum bicolor), atr (Amborella trichopoda), stu (Solanum tuberosum), cme (Cucumis melo), dpr (Digitalis purpurea), sly (Solanum lycopersicum), ppe (Prunus persica), rco (Ricinus communis), ssl (Salvia sclarea), pta (Pinus taeda), sly (Solanum lycopersicum), ata (Aegilops tauschii), hbr (Hevea brasiliensis), gma (Glycine max), mes (Manihot esculenta), ppe (Prunus persica), cca (Cynara cardunculus). miRNA mediated gene regulatory network of b glasshouse shifted plant and c transformed root
In the case of transformed roots, a total of 14 targets were commonly observed among the miRNA families. A total of three targets were found to be co regulated by miR414 and miR838 and nine targets were shared by miR838 and miR5021. KC1D was demonstrated as a single target, regulated by three miRNA families viz. miR414, miR838 and miR5021.
By the knowledge generated over more than 30 years of research work, the indications have now been made that environmental and developmental factors play role in TIA biosynthesis and is highly regulated by these two factors. It is also noteworthy to mention that TIA biosynthesis works through several step enzymatic network complex. A definite cross talk spatially and temporarily between various plant organs has been indicated and these cross talk occur at intra as well as at intercellular level (Verma et al. 2012b). Table 5 depicts miRNAs regulating transcription factors/proteins/enzymes associated with alkaloid production in V. minor. It was observed that miR5658 and miR5021 regulates the alkaloid production in glass house shifted plant as well as transformed roots while miR828, miR414, miR838, miR5015b, miR319a & miR156a were found to be unique for transformed roots and glass house shifted plant, respectively.
Phylogenetic analysis of predicted miRNAs reveals the differential evolutionary rate for each miRNA family (Fig. 6) as reported earlier (Singh and Sharma 2014; Singh et al. 2016a, b). There are few report of miRNA analysis for plant family Apocynaceae in miRBase (Griffiths-Jones 2010). Only, few conserved as well as novel miRNAs were reported for the plant Catharanthus roseus of the same family (Pani and Mahapatra 2013; Prakash et al. 2014). These findings may enhance the understanding of miRNA and their regulation in Apocynaceae. In the case of network analysis, miR156 and miR157 were observed to share all (10) the targets. Squamosa-promoter binding protein-like (SPL) was demonstrated as most prominent target for mi156 and miR157 in both tissues. The regulation of SPL by miR156 was reported earlier for defining an endogenous flowering pathway in Arabidopsis thaliana (Wang et al. 2009). The role of miR156 by modulating the expression level of SPL can also control the lateral root development process of plant (Yu et al. 2015).
Conclusions
Present study showed gene catalog for TIAs biosynthesis in Vinca minor glass house shifted plant and transformed roots via de novo transcriptome assembly, which will help in discovery of new genes in TIA pathway by working as a useful resource. Comparison of transcriptomic data of glass house shifted plant and transformed root tissues of V. minor showed that raw reads of transformed root was relatively higher in comparison to glass house shifted plant. After quality check and assembly process, processed reads were obtained, which lead to EST-SSR prediction and pathway analysis. The EST-SSRs prediction results showed, maximum repeats were common in both sample with small variation in tri nucleotide repeats (CTT/GAA) specific to glass house shifted plant. Annotation results and pathway analysis of both the samples suggests putative TIAs pathway operating in Vinca minor. Present studies have testified this potential by reconstituting for the first time a putative TIAs synthesis pathway operating in V. minor. This study gives a sound molecular basis to the hypotheis about biosynthetic relationship between the aspidosperma- and eburnamine type alkaloids and also the study provides information regarding the formation of epoxide that can undergo rearrangement to yield the vincamine–eburnamine backbone in vindoline synthesis. Provided that metabolite profiling is carried out with transcriptome analysis, further knowledge about complex metabolic pathways could be obtained. This has been anticipated that this study will take TIAs biosynthesis research one step ahead in right direction. The miRNAs reported in present study may be highly focused targets for modulating secondary metabolism and associated growth factors in V. minor.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The work presented here has been supported by DST-FAST TRACK SERC/LS-261/2012 and YSS/2015/001417. Grateful appreciation also goes to the Council of Scientific & Industrial Research (CSIR), New Delhi, India, for the financial support in the form of CSIR-SRA (Pool Scientist) at CSIR-NCL to the senior author. We highly acknowledge the help rendered by Dr. Quessen Akhtar in interpreting transcriptomic data. The financial support of the Department of Science and Technology (DST), New Delhi in the form of DST-INSPIRE-SRF (INSPIRE Fellow Code: IF120740) to the author Noopur Singh is gratefully acknowledged.
Author contribution
PV & SAK conceptualized the problem and carried out experiments. NS performed miRNA analysis; PV performed the pathway analysis. AKM & AS assisted in interpretation part. Manuscript has been written by PV, SAK, FJ & NS and reviewed by AKM & AS.
Compliance with ethical standards
Conflict of interests
Author declares that they have no conflict of interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Priyanka Verma and Noopur Singh have contributed equally to this work.
Contributor Information
Priyanka Verma, Email: priyankaurbest@gmail.com.
Noopur Singh, Email: singh.rajpoot.noopur@gmail.com.
Shamshad Ahmad Khan, Email: shamshadkhanptc@gmail.com.
Ajay Kumar Mathur, Email: akmcath@gmail.com.
Ashok Sharma, Email: ashoksharma@cimap.res.in.
Farrukh Jamal, Email: farrukhrmlau@gmail.com.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin J, Bustos-Sanmamed P, Hartmann C, Lelandais-Brière C, Crespi M. Complexity of miRNA-dependent regulation in root symbiosis. Philos Trans R Soc Lond B Biol Sci. 2012;367:1570–1579. doi: 10.1098/rstb.2011.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belal TS, Barary MH, Ibrahim MEAL, Sabry SM. Kinetic spectrophotometric analysis of naftidrofuryl oxalate and vincamine in pharmaceutical preparations using alkaline potassium permanganate. J Food Drug Anal. 2009;17:415–423. [Google Scholar]
- Budak H, Khan Z, Kantar M. History and current status of wheat miRNAs using next-generation sequencing and their roles in development and stress. Brief Funct Genomics. 2014 doi: 10.1093/bfgp/elu021. [DOI] [PubMed] [Google Scholar]
- Bulgakov VP, Avramenko TV. New opportunities for the regulation of secondary metabolism in plants: focus on microRNAs. Biotechnol Lett. 2015;37:1719–1727. doi: 10.1007/s10529-015-1863-8. [DOI] [PubMed] [Google Scholar]
- Cai K, Zhu L, Zhang K, Li L, Zhao Z, Zeng W, Lin X. Development and characterization of EST-SSR markers from RNA-seq data in Phyllostachys violascens. Front Plant Sci. 2019;10:50. doi: 10.3389/fpls.2019.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchini PJ, De Luca V. Opium poppy and Madagascar periwinkle: model non-model systems to investigate alkaloid biosynthesis in plants. Plant J. 2008;54:763–784. doi: 10.1111/j.1365-313X.2008.03438.x. [DOI] [PubMed] [Google Scholar]
- Farahanikia B, Akbarzadeh T, Jahangirzadeh A, Yassa N, Shams Ardekani MR, Mirnezami T, Hadjiakhoondi A, Khanavi M. Phytochemical Investigation of Vinca minor Cultivated in Iran. Iran J Pharm Res. 2011;10:777–785. [PMC free article] [PubMed] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K. Nutrient requirements of suspension cultures of soyabean root cells. Exp Cell Res. 1968;50:151–158. doi: 10.1016/0014-4827(68)90403-5. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S (2010) miRBase: microRNA sequences and annotation. Curr Protoc Bioinforma Ed. Board Andreas Baxevanis Al Chapter 12, Unit 12.9.1-10 [DOI] [PubMed]
- Guleria P, Mahajan M, Bhardwaj J, Yadav SK. Plant small RNAs: biogenesis, mode of action and their roles in abiotic stresses. Genom Proteo Bioinform. 2011;9:183–199. doi: 10.1016/S1672-0229(11)60022-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner F, Geu-Flores F, Sherden H, Brown S, et al. Discovery of a P450-catalyzed step in vindoline biosynthesis: a link between the aspidosperma and eburnamine alkaloids. Chem Commun. 2015;51:7626. doi: 10.1039/C5CC01309G. [DOI] [PubMed] [Google Scholar]
- Khaldun ABM, Huang W, Liao S, Lv H, Wang Y. Identification of MicroRNAs and target genes in the fruit and shoot tip of Lycium chinense: a traditional chinese medicinal plant. PLoS ONE. 2015;10:e011633. doi: 10.1371/journal.pone.0116334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar B, Kumar U, Yadav HK. Identification of EST–SSRs and molecular diversity analysis in Mentha piperita. Crop J. 2015;3:335–342. doi: 10.1016/j.cj.2015.02.002. [DOI] [Google Scholar]
- Li F, Wang W, Zhao N, Xiao B, Cao P, et al. Regulation of nicotine biosynthesis by an endogenous target mimicry of microRNA in Tobacco. Plant Physiol. 2015;169:1062–1071. doi: 10.1104/pp.15.00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Guo Z, Li L. Evolutionary conservation of microRNA regulatory programs in plant flower development. Dev Biol. 2013;380:133–144. doi: 10.1016/j.ydbio.2013.05.009. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Bartel DP, Bartel B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005;17:1360–1375. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molchan O, Romashko S, Yurin V. L-tryptophan decarboxylase activity and tryptamine accumulation in callus cultures of Vinca minor L. PCTOC. 2012;108:535–539. doi: 10.1007/s11240-011-0060-2. [DOI] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Numnark S, Mhuantong W, Ingsriswang S, Wichadakul D. C-mii: a tool for plant miRNA and target identification. BMC Genom. 2012;13:S16. doi: 10.1186/1471-2164-13-S7-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pani A, Mahapatra RK. Computational identification of microRNAs and their targets in Catharanthus roseus expressed sequence tags. Genomics Data. 2013;1:2–6. doi: 10.1016/j.gdata.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash P, Ghosliya D, Gupta V. Identification of conserved and novel microRNAs in Catharanthus roseus by deep sequencing and computational prediction of their potential targets. Gene. 2014;554:181–195. doi: 10.1016/j.gene.2014.10.046. [DOI] [PubMed] [Google Scholar]
- Qu J, Liu J. A genome-wide analysis of simple sequence repeats in maize and the development of polymorphism markers from next-generation sequence data. BMC Res Notes. 2013;6:403. doi: 10.1186/1756-0500-6-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Sharma A. In-silico identification of miRNAs and their regulating target functions in Ocimum basilicum. Gene. 2014;552:277–282. doi: 10.1016/j.gene.2014.09.040. [DOI] [PubMed] [Google Scholar]
- Singh N, Srivastava S, Sharma A. Identification and analysis of miRNAs and their targets in ginger using bioinformatics approach. Gene. 2016;575:570–576. doi: 10.1016/j.gene.2015.09.036. [DOI] [PubMed] [Google Scholar]
- Singh N, Srivastava S, Shasany AK, Sharma A. Identification of miRNAs and their targets involved in the secondary metabolic pathways of Mentha spp. Comput Biol Chem. 2016;64:154–162. doi: 10.1016/j.compbiolchem.2016.06.004. [DOI] [PubMed] [Google Scholar]
- Szabó LF. Rigorous biogenetic network for a group of indole alkaloids derived from strictosidine. Molecules. 2008;13:1875–1896. doi: 10.3390/molecules13081875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabbasam N, Zafar Y, Rahman M. Pros and cons of using genomic SSRs and EST-SSRs for resolving phylogeny of the genus Gossypium. Plant Syst Evol. 2014;300:559–575. doi: 10.1007/s00606-013-0891-x. [DOI] [Google Scholar]
- Taheri S, Abdullah TL, Yusop MR, Hanafi MM, Sahebi M, Azizi P, Shamshiri RR. Mining and development of novel SSR markers using next generation sequencing (NGS) data in plants. Molecules. 2018;23:399. doi: 10.3390/molecules23020399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Bio Evolut. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vas A, Gulyás B. Eburnamine derivatives and the brain. Med Res Rev. 2005;25:737–757. doi: 10.1002/med.20043. [DOI] [PubMed] [Google Scholar]
- Verma P, Mathur AK, Shankar K. Enhanced vincamine production in selected tryptophan-overproducing shoots of Vinca minor. PCTOC. 2012;111:239–245. doi: 10.1007/s11240-012-0185-y. [DOI] [Google Scholar]
- Verma P, Mathur AK, Srivastava A, Mathur A. Emerging trends in research on spatial and temporal organization of terpenoid indole alkaloids pathway in Catharanthus roseus: a literature up-date. Protoplasma. 2012;249:255–268. doi: 10.1007/s00709-011-0291-4. [DOI] [PubMed] [Google Scholar]
- Verma P, Khan SA, Mathur AK, Shanker K, Kalra A. Fungal endophytes enhanced the growth and production kinetics of Vinca minor hairy roots and cell suspensions grown in bioreactor. PCTOC. 2014;118:257–268. doi: 10.1007/s11240-014-0478-4. [DOI] [Google Scholar]
- Verma P, Khan SA, Mathur AK, Shanker K, Lal RK. Regulation of vincamine biosynthesis and associated growth promoting effects through elicitation, cyclooxygenase inhibition and precursor feeding of bioreactor grown Vinca minor hairy roots. Appl Biochem Biotech. 2014;173:663–672. doi: 10.1007/s12010-014-0883-5. [DOI] [PubMed] [Google Scholar]
- Wang J-W, Wang L-J, Mao Y-B, Cai W-J, Xue H-W, et al. Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. Plant Cell. 2005;17:2204–2216. doi: 10.1105/tpc.105.033076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J-W, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138:738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- Wei W, Qi X, Wang L, Zhang Y, Hua W, Li D, Lv H, Zhang X. Characterization of the sesame (Sesamum indicum L) global transcriptome using Illumina paired-end sequencing and development of EST-SSR markers. BMC Genomics. 2011;12:451. doi: 10.1186/1471-2164-12-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Xiao M, Hayward A, et al. Applications and challenges of next-generation sequencing in Brassica species. Planta. 2013;238:1005–1024. doi: 10.1007/s00425-013-1961-6. [DOI] [PubMed] [Google Scholar]
- Xi J, Qiu Y, Du L, Poovaiah BW. Plant-specific trihelix transcription factor AtGT2L interacts with calcium/calmodulin and responds to cold and salt stresses. Plant Sci Int J Exp Plant Biol. 2012;185–186:274–280. doi: 10.1016/j.plantsci.2011.11.013. [DOI] [PubMed] [Google Scholar]
- Yu N, Niu Q-W, Ng K-H, Chua N-H. The role of miR156/SPLs modules in Arabidopsis lateral root development. Plant J Cell Mol Biol. 2015 doi: 10.1111/tpj.12919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.