Highlights
-
•
Testing supplies for COVID-19 are limited worldwide.
-
•
Validity of alternative testing supplies is needed.
-
•
This article supports the use of non-flocked swabs of the oropharynx/nares for COVID-19 testing.
-
•
These data support the use of PBS as transport media for COVID-19 testing.
-
•
Performance of non-flocked swabs in PBS is similar to that of previously-validated swabs.
Keywords: COVID-19, SARS-CoV-2, Swab, Oropharyngeal, Nares, PCR
Abstract
The COVID-19 pandemic has led to a worldwide shortage of nasopharyngeal swabs and universal transport media. This study evaluated a combined oropharynx/nares (OP/Na) sample collection using two readily-available non-flocked swabs, transported in phosphate-buffered saline, and demonstrates equivalent performance in SARS-CoV-2 detection compared to a previously-validated OP/Na collection kit.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the etiological agent of 2019 novel coronavirus disease (COVID-19) that quickly spread as a pandemic (Zhu et al., 2020; Zhou et al., 2020; Gorbalenya et al., 2020). Like other respiratory viruses, molecular testing is the mainstay of COVID-19 diagnosis (Tang et al., 2020; LeBlanc et al., 2020a; Corman et al., 2020), the preferred specimens being from the upper respiratory tract, collected with a flocked nasopharyngeal (NP) swab placed into universal transport medium (UTM) (Marty et al., 2020). During the pandemic, global supply chain disruptions of NPs and UTM limited testing capacity worldwide, and alternatives were sought. A commercial collection kit commonly used for sexually transmitted infection (STI) testing, the Aptima Multitest Kit in specimen transport media (STM) (Hologic Inc), has been validated for SARS-CoV-2 detection (LeBlanc et al., 2020b; Avaniss-Aghajani et al., 2020). In a recent study, a combined sampling of the posterior oropharynx and bilateral anterior nares (OP/Na) using the Aptima swabs in STM was shown to be equivalent to NP swabs transported in UTM (LeBlanc et al., 2020b). However, with increased demand for SARS-CoV-2 testing in clinical laboratories, availability of the Aptima swab collection kits has also become limited. Other recent studies demonstrated that phosphate buffered saline (PBS) was a suitable media for specimen transport as it supported the recovery of SARS-CoV-2 RNA for molecular detection (Perchetti et al., 2020; Radbel et al., 2020; Rodino et al., 2020). This study evaluated the feasibility of using PBS as transport medium following an OP/Na collection with one of two non-flocked swabs: 1) the M40 Transystem (Copan Italia, Brescia, Italy), a swab commonly used for bacterial culture (Morosini et al., 2006; Tano and Melhus, 2011); and 2) the BD ProbeTec Qx Collection Kit for Endocervical and Lesion Specimens (Becton, Dickinson and Company, Sparks MD, USA), a swab used for molecular detection of STIs on the BD Viper instrument (Tunsjø et al., 2014; Lang et al., 2014).
2. Methods
Real-time reverse transcription polymerase chain reaction (rRT-PCR) was performed using the SARS-CoV-2 assay on the Cobas 6800 system (Roche Diagnostics), and a positive result was defined as amplification of at least one of two genetic targets (Orf1a or E gene). To ensure compatibility of PBS on the instrument, analytical sensitivity was estimated using 10-fold serial dilutions of a SARS-CoV-2 derived from a positive clinical specimen, which were spiked 1:10 (v/v) into each transport medium (i.e. UTM, STM, and PBS). Experiments are the results of triplicate values obtained in three independent experiments using the same clinical specimen. For clinical specimens or viral dilutions prepared in UTM, 600 μL was processed directly on the Cobas 6800 instrument, as recommended by the manufacturer. Specimens or viral dilutions in PBS mirrored the processing of those in UTM. For specimens or viral dilutions in STM, a pre-processing dilution step (1:6) was required to overcome the deleterious effects its high concentrations of detergent (LeBlanc et al., 2020b). Briefly, 200 μL of STM was diluted into 1 mL of Cobas Omni Specimen Diluent (Roche Diagnostics), mixed with gentle pipetting to avoid the formation of bubbles, and specimens were processed within 2 h of preparation. Viral dilutions were quantified relative to a standard curve generated from quantified in vitro transcribed RNA which was provided in-kind by the National Microbiology Laboratory (Winnipeg, MB) (Table S1).
To assess the clinical performance of the two alternative non-flocked swabs (i.e. M40 Transystem and the BD ProbeTec Qx Collection Kit), 15 patients previously identified as COVID-positive with mild to moderate disease (living in long-term care or admitted to acute care) were enrolled into the study, after obtaining informed consent. OP/Na collections were performed sequentially on the same patient, using the M40 Transystem featuring a plastic shaft (Copan Italia, Brescia, Italy), the BD ProbeTec Qx Collection Kit for Endocervical and Lesion Specimens with a polyurethane tip (Becton, Dickinson and Company, Sparks MD, USA), and the Aptima Multitest Kit (Hologic Inc) was used as the reference method (LeBlanc et al., 2020b). Following collection, both the M40 Transystem and the BD ProbeTec Qx endocervical swabs were placed in a 15 mL conical tube (Falcon, Corning Incorporated, Corning, NY, USA) containing 3 mL of sterile 1 × PBS, pH 7.4 (Gibco, ThermoFisher Scientific), whereas the Aptima Multitest swab was placed into 2.9 mL of STM provided in the collection kit. Samples were transported to the laboratory and were processed in parallel within 4 h, or alternatively, held at 4 °C until processed within 12 h of collection.
3. Results
The analytical sensitivity for SARS-CoV-2 was not impacted by the different transport media, and the limit of detection at 95% for each was below 1 copy/mL (Table S1). While not significant, STM showed a lower proportion of viral detection in dilution near the limit of detection, which might be attributed to the 1:6 dilution required for preprocessing of specimens in STM (LeBlanc et al., 2020b).
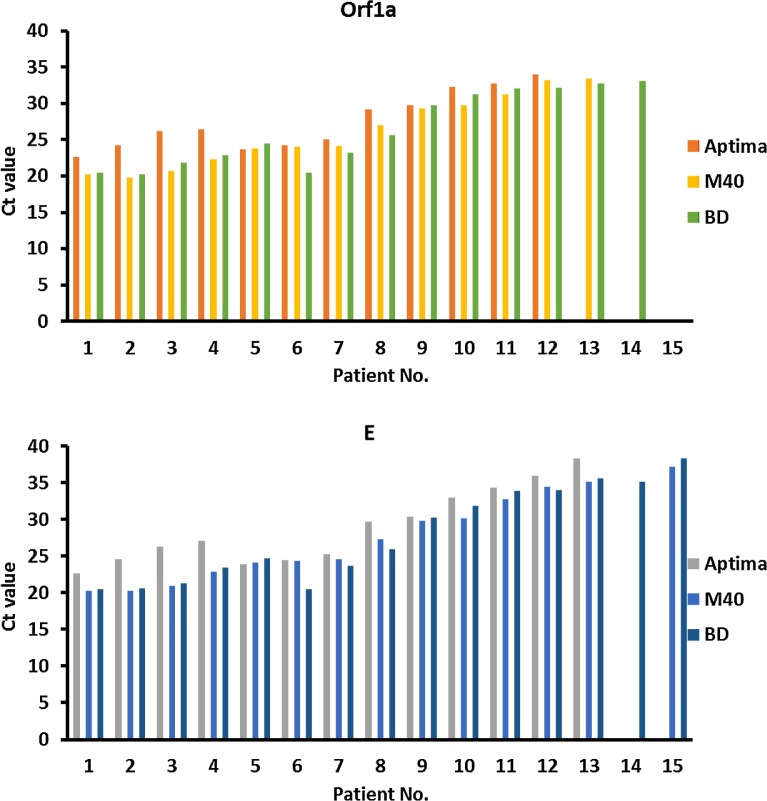
For clinical validation of OP/Na collection using the M40 and BD swabs compared to the Aptima swab, fifteen COVID-19 positive patients participated, with an average age of 74.2 years (range 48–92 years). The mean number of days since their initial positive result was 3.9 (median 2 days; range 2–13 days), 93.3% (14/15) of whom were originally diagnosed using the Aptima Multitest swab and one having been diagnosed with a NP swab in UTM. Of the participants, 86.7% (13/15) had consistent results across all three swabs. Fourteen patients (93.3%) were positive by two swab types. Interestingly, the reference method (i.e. Aptima swab) failed to detect SARS-CoV-2 in two patients (Fig. 1 and Table S2). In the first patient, both the M40 and BD swabs detected the E gene target in a specimen that was negative with the Aptima swab collection. In the second patient, SARS-CoV-2 was solely detected using a M40 swab collection, but both genetic targets were positive (Fig. 1 and Table S2).
Fig. 1.
Ct values for SARS-CoV-2 RT-PCR from 15 known positive patients. Two gene targets’ (Orf1a and E gene) Ct values obtained on the Cobas 6800 RT-PCR assay. Triplicate combined oropharyngeal/nares swabs obtained using Aptima Multitest kits, M40 bacterial swabs in phosphate-buffered saline (PBS), and BD ProbeTec Qx swabs in PBS.
4. Discussion
This study assessed the feasibility of an OP/Na collection with either M40 or BD swabs placed in PBS. While this study could not perform a head-to-head comparison of M40 and BD swabs in PBS against the preferred collection device (i.e. NP in UTM) due to global supply chain shortages, the swabs collected in PBS were compared to the Aptima Multitest swab in STM - a reference collection shown not to be significantly different than NP swabs in UTM when using the SARS-CoV-2 assay on the Roche 6800 instrument (LeBlanc et al., 2020b).
The study initially assessed the ability to recover SARS-CoV-2 RNA from PBS, compared to both the study reference (i.e. STM) and traditional reference media (i.e. UTM). Overall, PBS had comparable analytical sensitivity to UTM. During testing using a 10-fold serial dilution of SARS-CoV-2 virus, the proportion of viral dilutions in STM detected were lower than PBS or UTM, but differences did not achieve significance. A possible explanation for this observation might be the 1:6 dilution required for pre-processing of specimens in STM, given the high concentration of detergents (LeBlanc et al., 2020b). More importantly, the analytical sensitivity for SARS-CoV-2 detection in PBS was equivalent to UTM. The ability to use PBS as a transport medium for the detection of SARS-CoV-2 is congruent with previous reports (Perchetti et al., 2020; Radbel et al., 2020; Rodino et al., 2020). Rodino et al. (2020) showed that PBS was a reasonable substitute to viral transport media, as SARS-CoV-2 RNA could be recovered for up to 7 days. Similarly, a study of swabs from endotracheal secretions of COVID-19 patients demonstrated stability and recovery of SARS-CoV-2 RNA from PBS when compared to UTM, even after 18 h at room temperature (Radbel et al., 2020). A more recent study compared the performance of PBS transport for SARS-CoV-2 at various temperatures and extended periods of time (Perchetti et al., 2020). While temperature and stability analyses were not performed in this study, under conditions for PBS transport (4 °C), minimal degradation of SARS-CoV-2 RNA occurred up to 28 days (Perchetti et al., 2020). This far exceeds the maximum transport time required for transport to hospital or public health laboratories performing SARS-CoV-2 rRT-PCR in Canada (LeBlanc et al., 2020a).
For the clinical validation, OP/Na collection using M40 and BD swabs with PBS transport media were compared to a similar collection with Aptima swabs in STM (LeBlanc et al., 2020b). This study targeted OP/Na collections in known positive patients with mild to moderate disease, as patients progressing to more severe COVID-19 disease might only be detected in lower respiratory tract specimens (Hanson et al., 2020). In the 15 patients enrolled in the study, Ct values for all swabs spanned a large range, with values spanning the mid-twenties to values near the assay limit of detection in the high thirties. Ct values of the two alternative swabs were strikingly similar, and were consistently lower than those of the accompanying reference swab. This is likely owing to the requirement of a 1:6 dilution during pre-processing of specimens collected in STM, which could lower the analytical sensitivity and possibly impact detection of patients with low viral loads (i.e. early or late disease). In fact, the Aptima swab collection missed the identification of 2 cases. The Ct values in discrepant results were near the limit of detection, ranging from 33.1–38.3, suggesting low viral loads in the upper respiratory tract. Interestingly, the collection using ProbeTec Qx swabs in PBS identified all previously known cases of COVID-19. The M40 swabs in PBS identified 93.3% (14/15), missing only a single case with a low viral load. At low viral loads, the molecular detection of SARS-CoV-2 molecular lacks reproducibility (LeBlanc et al., 2020a). Low viral loads have been demonstrated early in infection, and late in disease, possibly leading to false-negative results (Kucirka et al., 2020), highlighting the importance of repeat testing in those with initial negative results but high clinical suspicion (Watson et al., 2020).
Though limited to known-positive patients with mild-moderate symptoms, and by a low number of study participants, this study provided clinical evidence that an OP/Na collection using non-flocked swabs designed for bacterial culture or cervical investigations can perform as well for the diagnosis of COVID-19 as the previously validated Aptima Multitest Kit. This report supports the use of PBS as a transport medium. Our results are supportive of those recently published, demonstrating the diagnostic reliability of cotton-tipped plastic swabs for NP sampling as compared to rayon-tipped swabs, transported in TRIS-EDTA (Freire-Paspuel et al., 2020) Repurposed commonly used non-flocked swabs, paired with a readily available buffer provides a solution for COVID-19 testing during times of UTM and flocked NP swab shortages.
Funding
No funding was received for this work. While commercial kits were used in the study, no industry sponsors were involved in the study concept, design, data analysis, or writing of the manuscript.
Ethics
This project was a quality assurance initiative and did not require research ethics board review.
CRediT authorship contribution statement
Glenn Patriquin: Conceptualization, Methodology, Writing - review & editing. Ian Davis: Conceptualization, Methodology, Writing - review & editing. Charles Heinstein: Investigation, Resources, Writing - review & editing. Jimmy MacDonald: Investigation, Resources, Writing - review & editing. Todd F. Hatchette: Conceptualization, Methodology, Investigation, Resources, Writing - review & editing. Jason J. LeBlanc: Conceptualization, Methodology, Investigation, Resources, Writing - review & editing.
Declaration of Competing Interest
The authors have no conflicts to declare.
Acknowledgements
The authors are indebted to the members of the Division of Microbiology, Department of Pathology and Laboratory Medicine, Nova Scotia Health Authority (NSHA), who were instrumental for sample processing and laboratory testing.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jviromet.2020.113948.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Avaniss-Aghajani E., Sarkissian A., Fernando F., Avaniss-Aghajani A. Validation of the hologic’s aptima unisex and multitest specimen collection kits used for endocervical and male urethral swab specimen (Aptima swab) for sample collection of SARS-CoV-2. J. Clin. Microbiol. 2020;2020 doi: 10.1128/JCM.00753-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire-Paspuel B., Vega-Mariño P., Velez A., Castillo P., Gomez-Santos E.E., Cruz M., Garcia-Bereguiain M.A. Cotton-tipped plastic swabs for SARS-CoV-2 RT-qPCR diagnosis to prevent supply shortages. Front. Cell. Infect. Microbiol. 2020 doi: 10.3389/fcimb.2020.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J. The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson K.E., Caliendo A.M., Arias C.A., Englund J.A., Lee M.J., Loeb M., Patel R., El Alayli A., Kalot M.A., Falck-Ytter Y., Lavergne V., Morgan R.L., Murad M.H., Sultan S., Bhimraj A., Mustafa R.A. 2020. Infectious Diseases Society of America Guidelines on the Diagnosis of COVID-19.www.idsociety.org/COVID19guidelines/dx [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucirka L.M., Lauer S.A., Laeyendecker O., Boon D., Lessler J. Variation in false-negative rate of reverse transcriptase polymerase chain reaction–based SARS-CoV-2 tests by time since exposure. Ann. Intern. Med. 2020 doi: 10.7326/M20-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang A.L.S., Roberts C., Mazzulli T., Hatchette T.F., LeBlanc J.J. Detection and differentiation of herpes simplex viruses by use of the Viper platform: advantages, limitations, and concerns. J. Clin. Microbiol. 2014;52(6):2186–2188. doi: 10.1128/JCM.03636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc J.J., Gubbay J.B., Li Y., Needle R., Arneson S.R., Marcino D., Charest H., Desnoyers G., Dust K., Fattouh R., Garceau R., German G., Hatchette T.F., Kozak R.A., Krajden M., Kuschak T., Lang A.L.S., Levett P., Mazzulli T., McDonald R., Mubareka S., Prystajecky N., Rutherford C., Smieja M., Yu Y., Zahariadis G., Zelyas N., Bastien N. On behalf of the COVID-19 pandemic diagnostics investigation team of the canadian public health laboratory network (CPHLN) respiratory virus working group. Real-time PCR-based SARS-CoV-2 detection in canadian laboratories. J. Clin. Virol. 2020;104433 doi: 10.1016/j.jcv.2020.104433. Published online 2020 May 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc J.J., Heinstein C., MacDonald J., Pettipas J., Hatchette T.F., Patriquin G. A combined oropharyngeal/nares swab is a suitable alternative to nasopharyngeal swabs for the detection of SARS-CoV-2. J. Clin. Virol. 2020;128(July):104442. doi: 10.1016/j.jcv.2020.104442. Published online 2020 May 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty F.M., Chen K., Verrill K.A. How to obtain a nasopharyngeal swab specimen. N. Engl. J. Med. 2020;382:e76. doi: 10.1056/NEJMvcm2010260. https://www.nejm.org/doi/full/10.1056/NEJMvcm2010260#full [DOI] [PubMed] [Google Scholar]
- Morosini M.-I., Loza E., Gutiérrez O., Almaraz F., Baquero F., Cantón R. Evaluation of 4 swab transport systems for the recovery of ATCC and clinical strains with characterized resistance mechanisms. Diagn. Microbiol. Infect. Dis. 2006;56(1):19–24. doi: 10.1016/j.diagmicrobio.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Perchetti G.A., Huang M.L., Peddu V., Jerome K.R., Greninger A.L. Stability of SARS-CoV-2 in PBS for molecular detection. J. Clin. Microbiol. 2020;(May 15) doi: 10.1128/JCM.01094-20. JCM.01094-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radbel J., Jagpal S., Roy J., Brooks A., Tischfield J., Sheldon M., Bixby C., Witt D., Gennaro M.L., Horton D.B., Barrett E.S., Carson J.L., Panettieri Jr R.A., Blaser M.J. Detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is comparable in clinical samples preserved in saline or viral transport medium. J. Mol. Diagn. 2020 doi: 10.1016/j.jmoldx.2020.04.209. S1525-1578(20)30323-30328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodino K.G., Espy M.J., Buckwalter S.P., Walchak R.C., Germer J.J., Fernholz E., Boerger A., Schuetz A.N., Yao J.D., Binnicker M.J. Evaluation of saline, phosphate buffered saline and minimum essential medium as potential alternatives to viral transport media for SARS-CoV-2 testing. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00590-20. JCM.00590-20, jcm;JCM.00590-20v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.W., Schmitz J.E., Persing D.H., Stratton C.W. The laboratory diagnosis of COVID-19 infection: current issues and challenges. J. Clin. Microbiol. 2020;(April 3) doi: 10.1128/JCM.00512-20. [Epub ahead of print] pii: JCM.00512-00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tano E., Melhus A. Evaluation of three swab transport systems for the maintenance of clinically important bacteria in simulated mono- and polymicrobial samples. APMIS. 2011;119(3):198–203. doi: 10.1111/j.1600-0463.2010.02710.x. [DOI] [PubMed] [Google Scholar]
- Tunsjø H.S., Smedsrud E., Holm P.C., Rokvam K., Schie C., Rognlien V., Augustin I., Fostervold A. Purification of DNA eluates from the ProbeTec GC Q(x) Assay on BD Viper XTR allows for further analysis and confirmation of gonorrhea. Eur. J. Clin. Microbiol. Infect. Dis. 2014;33(1):49–54. doi: 10.1007/s10096-013-1927-4. [DOI] [PubMed] [Google Scholar]
- Watson J., Whiting P.F., Brush J.E. Interpreting a COVID-19 test result. BMJ. 2020;369:m1808. doi: 10.1136/bmj.m1808. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. China Novel Coronavirus Investigating and Research Team. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.