Abstract
Accumulation of macrophage “foam” cells, laden with cholesterol and cholesteryl ester, within the intima of large arteries, is a hallmark of early “fatty streak” lesions which can progress to complex, multicellular atheromatous plaques, involving lipoproteins from the bloodstream and cells of the innate and adaptive immune response. Sterol accumulation triggers induction of genes encoding proteins mediating the atheroprotective cholesterol efflux pathway. Within the arterial intima, however, this mechanism is overwhelmed, leading to distinct changes in macrophage phenotype and inflammatory status. Over the last decade marked gains have been made in understanding of the epigenetic landscape which influence macrophage function, and in particular the importance of small non-coding micro-RNA (miRNA) sequences in this context. This review identifies some of the miRNA sequences which play a key role in regulating “foam” cell formation and atherogenesis, highlighting sequences involved in cholesterol accumulation, those influencing inflammation in sterol-loaded cells, and novel sequences and pathways which may offer new strategies to influence macrophage function within atherosclerotic lesions.
Keywords: Coronary heart disease, Atherosclerosis, Macrophage “foam” cell, Cholesterol, Inflammation, MicroRNA
Core tip: Micro-RNA (miRNA) sequences are short non-coding RNAs which play a key role in epigenetic regulation of gene transcription and translation. Significant changes in miRNA expression occur in macrophage “foam” cells, laden with cholesterol and cholesteryl ester, which contribute not only to macrophage phenotype and inflammatory status, but also to novel pathways which may influence the development of atherosclerotic lesions, the principal underlying cause of coronary heart disease. The rapid expansion of this field of research is leading to new therapeutic targets and strategies for treatment of this progressive and highly complex global disease.
INTRODUCTION
The purpose of this review is to identify and contextualise the emerging roles of micro-RNA (miRNA) sequences involved in epigenetic regulation of cholesterol deposition within macrophage “foam” cells, a rapidly developing area of key interest to researchers and clinicians developing new therapeutic strategies to combat coronary heart disease (CHD). CHD, a major cause of global morbidity and mortality, is principally caused by atherosclerosis, a complex, progressive chronic inflammatory disease. Genetic factors contribute to atherosclerosis, in combination with environmental, metabolic and behavioural triggers including elevated serum lipid levels, diabetes, obesity, hypertension and smoking[1]. Atherosclerotic lesions originate at non-random locations of the vasculature[2,3], where alterations in haemodynamic blood flow, such as decreased shear stress and turbulent flow, are sensed by endothelial cells, disrupting homeostatic cellular organisation, increasing permeability of the arteries and enabling the accumulation of circulating cholesterol-rich low-density lipoprotein (LDL) in the intima[2-4]. Local inflammation in endothelial cells is mediated by activation of the pro-inflammatory transcription factor, nuclear factor-κB (NF-κB), in part due to shear stress-mediated inhibition of the anti-inflammatory transcription factor Kruppel-like factor 2[5,6]. This leads to increased expression of adhesion molecules, E-selectin, intercellular adhesion molecule-1 (ICAM-1) and vascular cell adhesion molecule-1, and of pro-inflammatory cytokines and chemoattractants[6]. Further, endothelial expression of lipoxygenase enzymes and production of reactive oxygen species (ROS) oxidatively modify proteoglycan-bound LDL (oxLDL)[7]. This amplifies the local inflammatory response mediated by receptors such as lectin-like oxidised LDL receptor 1 (LOX-1) and toll-like receptor 4 (TLR4) present on endothelial and smooth muscle cells[8-10].
Endothelial expression of adhesion molecules, and chemokine-chemokine receptor interactions, recruit circulating monocytes to the intima, where they differentiate into macrophages in response to macrophage colony-stimulating factor[11]. Intimal macrophages recognise modified components of oxLDL and internalise oxLDL and LDL, becoming “foam” cells due to accumulation of droplets of cholesteryl ester. This occurs via interaction with scavenger receptors (SRs), such as SR-A1, SR-B1, cluster of differentiation 36 (CD36, SR-B2), CD68 (SR-D1), LOX-1 (SR-E1), TLR4, and the LDL receptor (LDLR)[7,12-14]. Lipoprotein lipase (LPL) is also implicated in foam cell formation in distinct ways: Inhibition of LPL activity by angiopoietin-like protein 4 decreases lipid uptake in macrophages, whereas genetic deletion of this protein increases lipid uptake, expression of lipid-induced genes and respiration[15]. Additional receptor independent mechanisms such as macro- and micropinocytosis can also lead to the uptake of these lipoproteins[16,17]. The influx of cholesterol-rich lipoproteins through these various mechanisms, as well as the rate limited process of cholesterol efflux (below), leads to the generation of lipid-laden “foam” cells with reduced capacity to migrate from the intima[18-20].
Accumulation of macrophages and lipid-laden foam cells is accompanied by plaque enrichment with additional immune cells[21]. T-helper cells, activated by oxLDL-induced maturation of dendritic cells (DCs), recognise epitopes on apolipoprotein B100 (ApoB100) in native LDL and oxLDL[22,23]. Phenotypically, these TH cells are primarily of the TH1 subset, producing pro-inflammatory cytokines such as interferon gamma (IFN-γ) and tumour necrosis factor alpha (TNF-α), but atheroprotective anti-inflammatory regulatory T cells (Tregs) are also present[24,25]. The combination of hyperlipidaemia, endothelial expression of adhesion molecules and chemokines, and deposition of chemokines on endothelial cells by activated platelets, recruits and activates additional immune subsets, including neutrophils[26-30]. Neutrophils express myeloperoxidase that produces hypochlorous acid, that promotes LDL oxidation and foam cell formation and increases retention of LDL in the intima via binding to LPL[7,26-31].
As the complexity of the arterial microenvironment increases, atherosclerotic plaques develop a number of key features. Responding to signals such as growth factors, cytokines and oxidised phospholipids (oxPL), vascular smooth muscle cells (VSMCs) undergo phenotypic switching from contractile quiescent VSMCs to synthetic, migratory and proliferative VSMC[32,33]. This leads to dramatic vascular remodelling and arterial thickening via production of matrix degrading metalloproteinases and a shift in production from type I and III collagen to type VIII collagen[34-36]. Further, intimal VSMCs accumulate lipids and can take on a foam cell phenotype which, under endoplasmic reticulum (ER) stress or in the presence of increased intracellular free cholesterol, leads to apoptosis and necrosis of both macrophage-derived and VSMC-derived foam cells, forming a hypoxic, necrotic core and extracellular lipid pools[37-41]. Hypoxia inducible factor 1 enhances neo-vascularisation via induction of vascular endothelial growth factor A expression in macrophages and VSMC[42,43], and by increased expression of macrophage SRs and pro-inflammatory mediators and decreased expression of ATP binding cassette (ABC) transporters responsible for cellular cholesterol efflux[44-46].
Macrophage sub-populations within atherosclerotic lesions
Within the arterial intima, macrophages exhibit notable phenotypic plasticity in response to multiple signals from this complex microenvironment and can exhibit pro- or anti-atherosclerotic responses (Figure 1). Pro-inflammatory (M1) macrophages can be generated in vitro in response to a variety of stimuli associated with a TH1 response, such as lipopolysaccharide (LPS) and IFN-γ[47,48], resulting in increased expression of pro-inflammatory mediators such as interleukin (IL)-1β, IL-6, TNF-α, IL-12 and IL-23, and ROS[47,49,50]. Oxidized LDL induces activation of the NF-κB pathway which enhances the pro-inflammatory response in M1-like macrophages and expression of pro-inflammatory mediators in macrophages polarised to the anti-inflammatory (M2) phenotype[51-53]. Further, oxLDL and individual components derived from oxLDL, such as free cholesterol and cholesterol crystals, cholesteryl ester hydroperoxides, and 7-ketocholesteryl-9-carboxynonanoate (Figure 1), have been shown to activate pro-inflammatory pathways including the nod-like receptor protein 3 (NLRP3) inflammasome[54], mitogen activated protein kinase pathway[55,56] and NF-κB signalling[57,58].
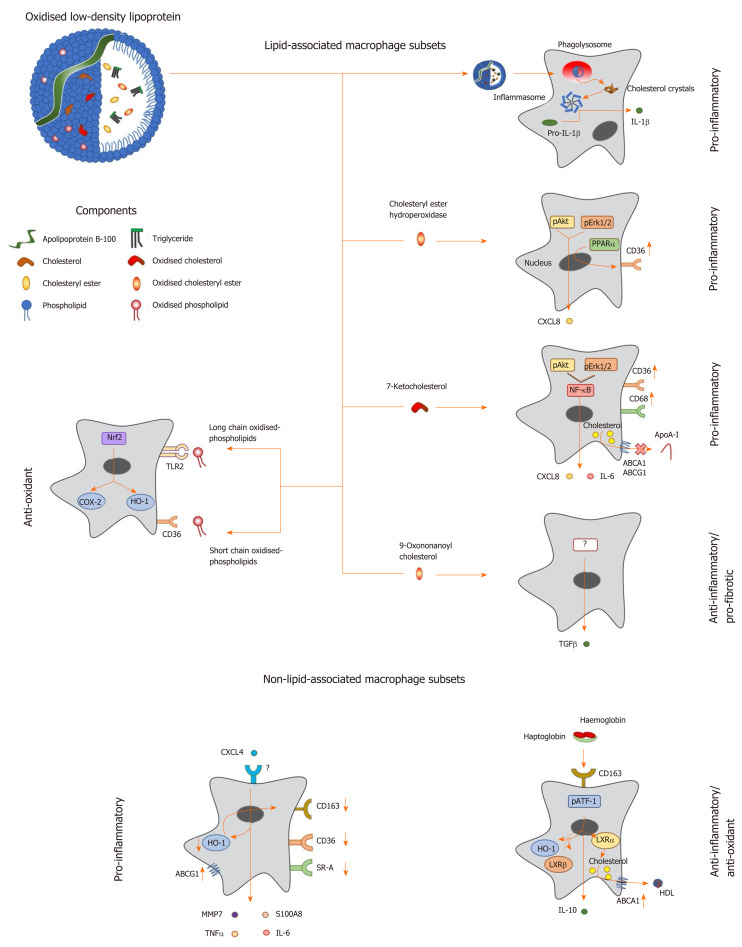
Figure 1.
Macrophage phenotype in response to lipids and other factors within atherosclerotic lesions. Oxidized low density lipoprotein particles are composed of a number of components including (modified) apolipoprotein B, (oxidized) free cholesterol, (oxidized) cholesteryl esters and (oxidized) phospholipids, which induce a range of unique transcriptional responses in macrophages (indicated by arrows), resulting in altered expression of secreted cytokines and cell surface receptors, and differing macrophage phenotypes (highlighted in bold). Macrophages can be polarised to a pro-inflammatory phenotype by components of oxidized LDL in a number of differing ways. Cholesterol crystals activate the inflammasome to increase expression of interleukin-1 (IL-1), while cholesteryl ester hydroperoxides activate phospho-extracellular signal-regulated protein kinases 1 and 2 (pERK1/2) and phospho-protein kinase B (pAkt) pathways, resulting in induction of peroxisome proliferator activated receptor alpha and increased expression of C-X-C motif ligand 8 (CXCL8) and the scavenger receptor, cluster of differentiation 36 (CD36). The modified cholesterol metabolite, 7-ketocholesterol, also triggers pAkt and pERK1/2 signalling, resulting in activation of nuclear factor kappa B (NF-κB), and enhanced output of pro-inflammatory cytokines such as IL-6 and CXCL8; this oxysterol also increases expression of CD36 and CD68, potentiating uptake of modified lipoproteins, and represses expression of ATP binding cassette transporter A1 (ABCA1) and ABCG1, limiting cholesterol removal via apolipoprotein A-I (apoA-I) and high density lipoprotein. By contrast, other components of oxLDL can induce an anti-inflammatory phenotype: For example, 9-oxononanoyl cholesterol induces expression of transforming growth factor β and stimulates fibrosis. Short chain oxidized phospholipids interact with CD36 to activate cyclooxygenase-2 (COX-2) and heme oxygenase-1 (HO-1), while long chain oxidized phospholipids bind toll-like receptor-2, resulting in activation of nuclear factor erythroid-2-related factor 2 which enhances expression of antioxidant genes, including HO-1, to protect cells against further oxidative damage. Other factors found within atherosclerotic lesions can also influence macrophage phenotype: For example, the chemokine CXCL4 induces the expression of pro-inflammatory factors such IL-6, tumor necrosis factor α, matrix metalloproteinase 7 and S100 calcium binding protein A8, while repressing the expression of ABCG1, HO-1, CD36, scavenger receptor-A and CD163. Alternatively an anti-inflammatory and antioxidant phenotype can be induced by the interaction of hemoglobin:haptoglobin complexes with CD163, which activates phospho-activating transcription factor-1, resulting in changes in LXR expression, induction of HO-1 and increased expression of IL-10 and ABCA1. IL: Interleukin; TGF: Transforming growth factor; TNF: Tumor necrosis factor; HO-1: Heme oxygenase-1; MMP: Matrix metalloproteinase; LXR: Liver X receptor; ABCA1: ATP binding cassette transporter A1; HDL: High density lipoprotein; pAkt: Phospho-protein kinase B; NF-κB: Nuclear factor kappa B; pATF-1: Phospho-activating transcription factor-1; PPAR-α: Peroxisome proliferator activated receptor alpha; CXCL: C-X-C motif ligand; ABCG1: ATP binding cassette transporter G1.
Macrophages, however, can adopt a variety of additional immunoregulatory subtypes within the phenotypic spectrum. A subset of anti-inflammatory macrophages (M2a), generated in response to cytokines such as IL-4 and IL-13, are produced as part of the TH2 response[59,60]. These cells are associated with wound healing via production of factors such as fibronectin and transforming growth factor β (TGF-β)[59,60]. Exposure to immune complexes, and to TLR ligands, generates another subset, termed M2b, which have both protective and detrimental roles and express high levels of the anti-inflammatory cytokine IL-10, low levels of pro-inflammatory IL-12, but also other pro-inflammatory mediators including chemoattractant C-C motif chemokine ligand 1 (CCL1)[61-63]. Stimulation of macrophages with glucocorticoids and IL-10 induce macrophage phenotype (M2c), which play a predominant role in clearance of apoptotic cells[64,65]. Finally, a pro-angiogenic population, termed M2d, is induced in murine macrophages in response to adenosine agonists in conjunction with TLR-signalling[66,67].
Interestingly, while some components of oxLDL activate inflammatory pathways, oxysterols and the product of cholesteryl ester oxidation, 9-oxonanoyl cholesterol, induce expression of the anti-inflammatory and pro-fibrotic cytokine TGF-β[68,69]. Consequently, treatment of macrophages with oxLDL can also induce an anti-inflammatory phenotype[70]. This apparent discrepancy between inflammatory versus anti-inflammatory signalling may be due to the degree of oxidation of the LDL particle[71], or the extent of lipid accumulation within cells[72]. Oxidized phospholipids (oxPL) (Figure 1) also induce a distinct macrophage phenotype (Mox) in murine models[73-75]; Mox macrophages exhibit reduced expression of M1, as well as M2-like, macrophage markers and enhanced expression of nuclear factor erythroid 2-related factor 2 (Nrf2) dependent anti-oxidant genes[73]. Further, macrophage subsets found within the plaque microenvironment include M4 macrophages, which are induced by chemokine (C-X-C) motif ligand 4 (CXCL4)[76], MHb macrophages, induced by hemoglobin: Haptoglobin complexes[77], and Mhem macrophages, induced by heme[78,79]; both MHb and Mhem macrophages exhibit resistance to cholesterol loading[77-79].
Cholesterol accumulation in macrophage “foam” cells: Responses to excess (oxy)sterol
Macrophages play a vital role in handling excess LDL-cholesterol and/or toxic oxLDL metabolites, via esterification to cytosolic lipid droplets by Acyl CoA: Cholesterol Acyltransferase (ACAT-1-/2) or lysosomal sequestration[80]. Autophagy also contributes to lipid droplet formation, with Beclin-1 inhibiting the formation of droplets in response to modified LDL in naïve cells but not in inflammatory activated macrophages[81]. Accumulation of sterol is central to activation of nuclear liver X receptors (LXRs), which lie under the control of oxysterol metabolites of cholesterol; the extent of activation of these transcription factors may also contribute to the heterogeneity of macrophage responses to sterol accumulation[72,82]. LXRs control the expression of ABC transporters (ABCA1, ABCG1/G4) that actively efflux cholesterol from cells to acceptors, such as apolipoprotein (apo) A-I, apoE and high-density lipoprotein (HDL). Binding of apoA-I to ABCA1 triggers an array of cell signalling pathways[83], and mobilisation of stored cholesteryl esters via cholesteryl ester hydrolases, releases cholesterol which trafficks to the plasma membrane for efflux as nascent HDL[80,84]. In addition, ABCG1 and ABCG4 aid the formation of more mature forms of HDL, so they work in concert with ABCA1 to initiate the process of reverse cholesterol transport which can return cholesterol to the liver for excretion via the classical and alternative bile acid pathways[80,84].
Liver X nuclear receptors form obligate heterodimers with retinoid X receptors which bind directly to the LXR response element, a direct repeat 4 (DR4) motif of the six base pair sequence AGTTCA separated by four base pairs[85-88]. Ligand binding triggers a conformational change in the heterodimer, dissociating nuclear receptor co-repressors [NCOR1/NCOR2 (SMRT)] proteins which undergo ubiquitination and proteasomal degradation, and engaging coactivator proteins (steroid receptor coactivators, PPAR-γ coactivator 1 and nuclear receptor coactivator 6)[87]. Genetic deletion of LXRs in bone marrow derived macrophages, however, reveal a more complex picture: Depending on the target gene, LXR deletion can up- or down-regulate, or effect no change in, gene expression[87].
LXRs contribute to the inactivation of the counter-regulatory system operated by sterol regulatory binding proteins (SREBPs) which belong to the basic helix-loop-helix leucine zipper (bHLH-Zip) family of transcription factors[89]. Three SREBP isoforms exist, encoded by two genes: SREBF1 (SREBP-1a and SREBP-1c) and SREBF2 (SREBP-2) which target sterol response elements (SRE). Unlike SREBP-1a which is constitutively expressed and targets all SRE with low specificity, SREBP-1c and SREBP-2 are inducible and regulate the expression of genes encoding proteins involved in fatty acid and cholesterol metabolism, respectively[89,90]; SREBP-2 also transcriptionally regulates the LDL receptor which mediates endocytosis of LDL from the circulation. In sterol-replete cells, SREBP transcription factors remain inactive, sequestered at the ER by binding to a chaperone, SREBP cleavage activating protein (SCAP), which contains a five transmembrane sterol sensing domain and interacts with the ER anchor, insulin-induced gene (INSIG-1/-2). As cholesterol levels fall, the interaction of SCAP with INSIG is lost, allowing SCAP-SREBP to traffick to the Golgi apparatus via inclusion in COPII-vesicles[89,90]. Golgi site-1 and site-2 proteases (SP-1, SP-2) cleave the amino terminal of SREBP-2, releasing a transcriptionally active fragment which is imported into the nucleus to target sterol-responsive genes, including SREBF2 itself; rapid degradation of nuclear (nSREBP) serves to terminate this signalling pathway[89,90].
LXRs operate in functional opposition to SREBP-2, repressing cholesterol biosynthesis via novel negative LXR DNA-response elements in the promoter region of genes encoding squalene synthase and lanosterol 14-demethylase[91-93], and promoting the degradation of LDL receptors by increasing the expression of proprotein convertase subtilisin/kexin type 9 (PCSK9)[94,95]. The E3 ubiquitin ligase, inducible degrader of the LDL receptor (IDOL) is an LXR target gene: IDOL dimers interact with members of the ubiquitin-conjugating enzyme (UBE) 2D family of E2 ubiquitin ligases to transfer ubiquitin to the cytoplasmic tail of members of the LDL receptor family, promoting receptor degradation[94,95]. Oxysterols also bind to Insig-1/2, sequestering SREBPs at the ER, further ensuring repression of cholesterol biosynthesis and uptake[88]. By contrast, LXR agonists induce gene expression of SREBP-1c; fatty acid synthase, and a number of desaturase and elongase enzymes in the fatty acid biosynthetic pathway, are also directly regulated by LXR[87]. Thus, efficient delivery of cholesterol to the ER is needed to inhibit proteolytic processing of SREBP-1c to an active transcription factor[89,90], and to limit increased biosynthesis of fatty acids[87].
LXRs also exert anti-inflammatory effects, some of which are indirect and due to the increased expression of ABCA1[96], and production of anti-inflammatory HDL[97,98]. HDL inhibit TLR signalling in macrophages and cytokine signalling in bone marrow progenitors by removal of cholesterol from lipid rafts[99,100] and induce activating transcription factor 3, suppressing the expression of pro-inflammatory genes[97,98]. Multiple mechanisms exist by which LXRs modulate inflammatory responses, some of which involve transactivation and others transrepression[86-88]. Pathway-specific responses occur: LXR activation inhibits NF-κB dependent induction of pro-inflammatory genes in response to LPS and responses triggered by TLR4 and TNF-α but exerts minimal impact on the pathway mediated by TLR3[86-88]. LXRs also regulate apoptosis and enhance survival of macrophages within lesions, while IFN-γ promotes neointimal hyperplasia and macrophage apoptosis by promoting ubiquitin-dependent LXR degradation[101,102].
However, it should be recognised that the pharmacology of oxysterols is highly complex: A large number of nuclear, and G-protein coupled (GPR), receptors bind these bioactive lipids [e.g., retinoid-related orphan receptors, ER, Epstein-Barr virus induced GPR (EBI2/GPR183) and IL-8 receptor (CXCR2)][93]. This, combined with the complexity of oxysterol metabolism, enzymatic conversion to other species such as esters, bile acids and 3-sulphate derivatives, and tissue- and species-specific effects, makes deciphering the (patho)physiological impact of these molecules particularly challenging[93].
EPIGENETIC MECHANISMS CONTRIBUTING TO “foam cell” FORMATION: THE EMERGING ROLE OF MICRORNA
It is increasingly clear that epigenetic mechanisms such as DNA methylation, histone post-translational modification and changes in expression of non-coding RNA, such as long non-coding RNA (lncRNA) and miRNA, are important contributors to macrophage phenotype and the pathogenesis of “foam cell” formation. Alterations in chromatin structure and gene expression exert both acute and chronic effects on a wide array of biological processes which influence macrophage lipid accumulation and inflammatory responses. For example, DNA methyltransferases catalyse methylation of the 5’-position of cytosine residues, using S-adenosyl-methionine as the methyl donor, resulting in hypermethylation of CpG islands and stable repression of transcription[103,104]. Chromatin histone post-translational modifiers, such as histone acetyltransferases and deacetylases and histone methyltransferases, target lysine and/or arginine residues to induce or repress gene expression, dynamically fine-tuning gene expression by controlling the access of transcription factors to promoter and enhancer regions[105,106].
An additional layer of epigenetic regulation is provided by non-coding RNA sequences, including lncRNA sequences, longer than 200 nucleotides, and miRNA sequences (20-25 nucleotides in size), the focus of this review article, which fine-tune expression of multiple (networks of) genes in response to environmental factors, including oxLDL[107-109]. Sequences encoding miRNA can be found singly or in clusters throughout the genome, located in intron-exon portions of protein-encoding genes or intergenic regions[110,111]. Transcription is dependent on the activity of RNA polymerase II/III and expression, in relation to intergenic miRNA, can be dependent or independent of host gene expression[111-114]. MicroRNA are frequently found in clusters and can be co-transcribed and separated by splicing, or expressed independently[115]. Transcription and generation of miRNA occurs through both canonical and non-canonical pathways, with less information available on the latter[116]. The canonical pathway involves generation of a hairpin-containing primary miRNA (pri-miRNA) transcript containing a 5’ methylated cap and a 3’ polyadenylated tail required for pri-miRNA processing and transport[112,117]. Processing occurs via a microprocessor complex consisting of the double-stranded RNA-binding protein DiGeorge syndrome critical region gene 8 which recognises methyl motifs present in the pri-miRNA[118,119]. This interaction serves as an anchor for a ribonuclease II (RNase III), known as Drosha, which cleaves the hairpin structure from the pri-miRNA transcript generating precursor miRNA (pre-miRNA)[120-122].
Export of pre-miRNA (around 70 nucleotides in length) from the nucleus involves the nucleocytoplasmic transporter factor exportin-5 and Ras-related nuclear protein (Ran)GTP[123]. Recognition and binding to exportin-5 occurs primarily through interaction with the 3’ overhanging sequence of pre-miRNA. Blunt ended pre-miRNA remain capable of interaction while RanGTP is bound to the hairpin structure, following release into the cytoplasm[124,125]; hydrolysis of GTP to GDP results in release of the pre-miRNA[118]. Once localised in the cytoplasm, pre-miRNA is processed by a second RNase III enzyme, Dicer, to a mature miRNA duplex (19-25 nucleotides) through removal of the stem-loop structure[126,127]. The guide strand, which has lower base pairing stability, is loaded onto the RNA-induced silencing complex (RISC) composed of Dicer, transactivation response (TAR) RNA binding protein (TRBP) and Argonaute proteins (1 to 4). After integration into the active RISC complex, miRNAs base pair with their complementary mRNA molecules, guided by their miRNA recognising element[128,129].
Degradation of target mRNA occurs only when the miRNA and the target mRNA match exactly (perfect match) or are nearly exactly complementary to each other; this process is the same as the RNA interference induced by artificial small interfering RNA (siRNA)[130,131]. By contrast, if the complementarity between miRNA and target mRNA is only partial (imperfect match), then more moderate reductions in mRNA levels accompanied by translational repression will occur. Targeting occurs through binding of the seed sequence of RISC-incorporated miRNA to conserved complementary regions found in the 3’UTR of target mRNA[130,131]. Factors such as AU-rich regions near seed region binding sites, and auxiliary binding of the miRNA to transcript can also play a role in determining target specificity, reducing translational efficiency or inducing mRNA destabilisation via deadenylation[132-134]. miRNA also exert regulatory functions on gene expression in the nucleus, paradoxically promoting gene expression in certain conditions[135,136]. In eukaryotic cells, miRNA molecules can bind several target sequences, mainly within the 3’-UTR of mRNA with varying degrees of complementarity, so that each single miRNA is able to interact with and regulate a large number of genes. Computational prediction suggests that more than 60% of all mammalian protein-coding genes are conserved targets of miRNA, while each miRNA has target sites in hundreds of different genes[137,138]; miRNAs also display tissue-specific expression[139] and concentration-dependent effects in pathologically affected organs and tissues[140-142].
miRNA not only regulate the transcriptional landscape of the cell, but some sequences exist in the extracellular environment in a variety of different forms; degradation of miRNA is avoided through association with Argonaute RISC catalytic component 2 (Ago2), and to a lesser extent nucleophosmin 1 (NMP1)[143-145]. miRNA are found enriched in extracellular vesicles such as exosomes, microvesicles, and lipoproteins such as HDL[146-148], and represent novel biomarkers of atherosclerosis[149]. Secreted miRNA may elicit pro- and anti-atherosclerotic functions: EC when placed under conditions of atherogenic shear stress release Ago2-bound miR-126-3p which in turn downregulate contractile VSMC markers[150], while delivery of miR-223 by HDL to EC leads to downregulation of ICAM-1[151,152].
miRNA sequences implicated in macrophage “foam” cell formation
Over the last five years, there has been an explosion of interest in the role of miRNA involved in macrophage biology, and in “foam” cell formation in particular. Table 1[153,155-204] indicates some of the miRNA sequences identified by interrogation of the NCBI PubMed database, as either altered by uptake of modified LDL by macrophages, or implicated in the pathogenesis of foam cell formation. Many of the genes targeted by these sequences play established roles in either lipid metabolism or inflammation, but a significant number have no prior links to either process, highlighting the importance of miRNA research in driving the discovery of novel cellular processes contributing to disease.
Table 1.
MicroRNA sequences associated with macrophage ”foam cell” formation and atheroma
| MicroRNA (↑↓) | Macrophage | Stimulus | Target | Outcomes in vitro | Outcomes in vivo | Ref. |
| Lethal: (let)-7g-5p (↓) | Human: THP-1 macrophages | Oxidized low density lipoprotein (oxLDL) | Nuclear factor kappa beta (NF-κB) (canonical and non-canonical pathways) | Inhibits phosphorylation of inhibitor kappa B kinase (IKK-κB and inhibitor (I) κB, downregulates sterol regulatory element binding transcription factor (SREBF2) and upregulates ATP binding cassette (ABC) transporter A1 (ABCA1); reduces expression of mitogen-activated protein kinase kinase kinase 1 (MEKK1), IKK-κ and inhibits IKK-κ phosphorylation | Overexpression of let-7 g in apolipoprotein (apo)E-/- mice fed a high fat diet (HFD) reduces macrophage accumulation and aortic plaque area; let-7 g sponge accelerates aortic macrophage accumulation in the same model | Wang et al[155], 2017 |
| miR-7-5p | Human: THP-1 macrophages | oxLDL ± puerarin | Serine/threonine kinase 11 (STK11) | Mimic significantly decreases cholesterol efflux and promotes cholesterol deposition | - | Li et al[156], 2017 |
| miR-9-5p | Human: THP-1 macrophages | oxLDL | Acyl CoA: Cholesterol acyltransferase (ACAT-1; SOAT1) | Mimic decreases levels of ACAT-1 protein, reduces cholesterol esterification and blocks foam cell formation | - | Xu et al[157], 2013 |
| miR-10a-5p | Murine: ApoE-/- macrophages | Deletion of dicer | - | Mimic rescues defective oxidation of fatty acids in alternatively activated Dicer-deficient macrophages, limiting foam cell formation and inflammation | Levels of hsa-miR-10a are negatively linked to atheroma progression; blockade of miR-10a exacerbates atheroma in apoE-/- mice (HFD) | Wei et al[158], 2018 |
| miR-16-5p (↓) | Murine: RAW 264.7 macrophages | OxLDL | Programmed cell death 4 (PDCD4) | Mimic decreases expression and secretion of pro-inflammatory cytokines (interleukin (IL)-6, Tumour necrosis factor (TNF-α), promotes that of anti-inflammatory IL-10, and suppresses NF-κB expression; inhibitor achieves the reverse | Levels decreased in atheroma in apoE-/- (HFD) | Liang et al[159], 2016 |
| miR-17-5p | Human: THP-1 macrophages | oxLDL | Beclin-1 | Mimic inhibits the enhancement of autophagy and cholesterol efflux induced by interferon-stimulated gene 15 (ISG15) | - | Huang et al[160], 2018 |
| miR-19a-3p | - | - | HMG-Box transcription factor-1 (HBP-1) | - | Serum levels are elevated in atherosclerotic patients, and in aortae susceptible to atherosclerosis. Inhibition decreases atheroma and aortic lipid accumulation in apoE-/- (HFD) mice | Chen et al[161], 2017 |
| miR-19b-3p (↑) | Human: THP-1 macrophages; Murine: Peritoneal macrophages | Acetylated LDL (AcLDL) | ABCA1 | Mimic inhibits cholesterol efflux to ApoA-I, increasing cholesterol mass | Overexpression of miR-19b decreases reverse cholesterol transport (RCT) in vivo; mimic reduces high density lipoprotein (HDL) levels, increases lesion area and lipid content in apoE-/- mice fed a Western diet (WD); inhibitor achieves the reverse | Lv et al[162], 2014 |
| miR-19b-3p (↓) | Human: THP-1 macrophages; Murine: Peritoneal macrophages | AcLDL + diosgenin | ABCA1 | Inhibitor enhances ABCA1 cholesterol efflux | Inhibitor promotes RCT in vivo, elevates HDL levels, reduces aortic lipid deposition and plaque area in apoE-/- mice (WD) | Lv et al[163], 2015 |
| miR-20a/b (-5p) | Human: THP-1 macrophages; Murine: RAW264.7 macrophages | oxLDL | ABCA1 | Mimic decreases cholesterol efflux to apoA-I, and increases macrophage cholesterol content | Mimic reduces hepatic expression of ABCA1 and high-density lipoprotein (HDL) levels, impairs reverse cholesterol transport and promotes atherogenesis in apoE-/- mice | Liang et al[164], 2017 |
| miR-21-5p | Murine: RAW 264.7 macrophages | OxLDL ± LPS (lipopoly-saccharide) | Toll like receptor 4 (TLR4); NF-κB | LPS stimulation of miR-21 inhibits foam cell formation and reduces secretion of IL-6, IL-12, TNF-α | - | Feng et al[165], 2014 |
| miR-21-5p | Murine: Bone-marrow derived macrophages | AcLDL | Mitogen-activated protein kinase kinase 3 (MKK3) | MiR21-/- macrophages exhibit increased ABCG1 degradation and decreased cholesterol efflux, enhancing foam cell formation. | Most abundant miR in murine macrophages; levels elevated in aortic plaque macrophages isolated from LDL receptor knockout (Ldlr-/-) (WD) mice; knockout of miR-21 enhances arterial macrophage accumulation, production of inflammatory cytokines | Canfrán-Duque et al[166], 2017 |
| miR-23a-5p (↑) miR-23a-3p | Murine: RAW 264.7 macrophages | oxLDL | ABCA1/G1 | Inhibitor enhances cholesterol efflux and decreases foam cell formation via upregulation of ABCA1/G1 expression | Plasma levels correlate with plaque progression and vulnerability in patient with acute ischemic stroke. Long-term systemic delivery of antagomir reduces atheroma and promotes plaque stability (apoE-/- mice) | Yang et al[167], 2018 |
| miR-24-3p | Primary: Human monocyte-derived macrophages | Colony stimulating factor (CSF) | Matrix metallo-proteinase (MMP)-14 | Inhibitor increases macrophage invasive capacity | Hsa-miR-24 levels inversely correlate with MMP-14 protein, and lesion instability Inhibitor increases lesion size and MMP-14 levels in apoE-/- mice (HFD) | Di Gregoli et al[168], 2014 |
| miR-27a/b (-3p) | Human: THP-1 macrophages; Murine: RAW264.7 macrophages | AcLDL | ABCA1 | Mimic decreases cholesterol efflux, and increases free cholesterol content in macrophages, but blocks uptake of oxLDL [Lipoprotein lipase (LPL), cluster of differentiation (CD36)] and inhibits cholesterol esterification | - | Zhang et al[169], 2014 |
| miR-28a-5p (↑) | Murine: RAW264.7 macrophages | oxLDL | LDL receptor class A domain containing 3 (LRAD3) | - | - | Li et al[170], 2018 |
| miR-30c-5p | Human: THP-1 macrophages | oxLDL | Caspase 3 | Cluster differentiation (CD) 36-dependent uptake of oxLDL reduces miR-30c-5p, enhancing IL secretion | Low serum levels of hsa-miR-30c-5p predict carotid atherosclerosis. Antagomir impairs endothelial healing following carotid injury (C57BL/6J mice) | Ceolotto et al[171], 2017 |
| miR-30c-1-3p (↑) | Murine: RAW264.7 macrophages | oxLDL | Oxidized LDL (lectin-like) receptor 1 (LOX-1) | - | - | Li et al[170], 2018 |
| miR-33a/b (-5p) (↑) | Human: THP-1 macrophages | oxLDL ± C. pneumoniae | ABCA1 | Inhibitor promotes cholesterol efflux compared with C. pneumoniae control | - | Zhao et al[172], 2014 |
| miR-33a-5p | Human: THP-1 macrophages | Vaspin ± LPS | ABCA1 | Vaspin decreases expression of miR-33a via inhibition of NF-κB, enhancing cholesterol efflux | - | Gao et al[173], 2018 |
| miR-33 | Human: THP-1 macrophages; Murine: Peritoneal macrophages | - | Peroxisome proliferator activator receptor coactivator 1 (PGC1A), pyruvate dehydrogenase kinase 4 (PDK4), solute carrier family 25 member 25 (SLC25A25), nuclear respiratory factor 1 (NRF1), transcription factor A, mitochondrial (TFAM) | Inhibitor enhances mitochondrial respiration, and cholesterol efflux to apoA-I | Antagomir reduces atheroma in apoE-/- mice (WD) | Karunakaran et al[154], 2015 |
| miR-33 | Murine: Peritoneal macrophages | AcLDL | Autophagy protein 5 (Atg5); autophagy-related 12 (Atg12), microtubule-associated protein light chain 3 (Map11c3b), AMP-activated protein kinase κ1 (Prkaa1), lysosomal associated membrane protein 1 (Lamp1), transcription factor EB (TFEB), Forkhead box O-3 (FOXO3) | Mimic inhibits the breakdown of lipid droplets by repressing effectors of macrophage autophagy. Silencing promotes lipid droplet catabolism, aiding ABCA1-dependent cholesterol efflux | Inhibition restores defective autophagy in aorta and macrophages of Ldlr-/- mice | Ouimet et al[153], 2017 |
| miR-33a-5p (↑) | Human: THP-1 macrophages | oxLDL | ABCA1 | Mimic decreases the expression of ABCA1 and promotes lipid accumulation in macrophages, while the inhibitor achieves the reverse | Levels are elevated in individuals at risk of atherosclerosis | Kim et al[174], 2017 |
| miR-34a-5p (↓) | Human: THP-1 macrophages | oxLDL + Hcy | Histone deacetylase 1 (HDAC1) | Overexpression of miR-34a reduces HDAC1 levels in foam cells, while knockdown achieves the reverse; HDAC1 induces homocysteine (Hcy) dependent foam cell formation | Levels of miR-34a decrease, and expression of HDAC1 increases, in the aorta of apoE-/- mice fed a high methionine diet | Zhao et al[175], 2017 |
| miR-98-5p (↓) | Murine: Peritoneal macrophages | oxLDL | Oxidized LDL (lectin-like) receptor 1 (LOX-1) | Mimic reduces expression of LOX-1 and inhibits foam cell formation; inhibitor achieves the reverse | Mimic decreases expression of LOX-1 and lipid accumulation in the aortic root in apoE-/- mice (HFD); inhibitor achieves the reverse | Dai et al[176], 2018 |
| miR-125b-5p (↑) | Murine: RAW 264.7 macrophages | LPS | NF-κB | Silencing of CD40 downregulates levels of miR-125; LPS stimulates miR-125b expression | miR-125b levels are increased in atherosclerosis; siRNA-CD40 apoE-/- mice exhibit reductions in lesion area | Hueso et al[177], 2016 |
| miR-128- (↓) | Murine: RAW 264.7 macrophages | oxLDL | - | Mimic reverses the pro-atherogenic impact of long non-coding (lnc)RNA NEAT1 on foam cell formation | - | Chen et al[178], 2018 |
| miR-133a | Murine: RAW 264.7 macrophages | oxLDL | Testicular orphan nuclear receptor 4 (TR4) | Mimic prevents TR-4 mediated enhancement of lipid uptake via CD36 | - | Peng et al[179], 2016 |
| miR-134-5p | Human: THP-1 macrophages | oxLDL | Angiopoietin (ANGTPL)/lipoprotein lipase (LPL) | LPL activity and protein, inflammatory cytokines and cholesterol mass enhanced by miR-134 mimic; inverse achieved using an inhibitor | - | Lan et al[180], 2016 |
| miR-134-5p | - | - | ANGTPL4/LPL | - | Mimic increases atherosclerotic lesions, release of proinflammatory cytokines and peritoneal macrophage lipid accumulation in apoE-/-(HFD) mice; inhibitor achieves the reverse | Ye et al[181], 2018 |
| miR-144-3p | Human: THP-1 macrophages;Murine: Peritoneal and J774.1 macrophages | Liver X receptor ligand T0901317 | ABCA1 | Mimic reduces cholesterol efflux to apoA-I in macrophages | Mimic reduces HDL levels in vivo (C57BL/6); inhibitor achieves the reverse | Ramirez et al[182], 2013 |
| miR-144-3p | Human: THP-1 macrophages | oxLDL | ABCA1 | Mimic reduces cholesterol efflux and enhances expression of cytokines (IL-1, TNF-α, IL-6) | Agomir inhibits RCT in vivo, and accelerates atherosclerosis in apoE-/- mice (HFD). Circulating levels of miR-144-3p correlate with acute myocardial infarction | Hu et al[183], 2014 |
| miR-146a-5p | Murine: Peritoneal macrophages (wild type and apoE-/-) | NF-κB | Increases in miR-146a inhibit pro-inflammatory responses in macrophages (TNF-α) | miR-146a mimic inhibits inflammation and plaque development in apoE-/- x Ldlr-/- and Ldlr-/- mice (HFD) | Li et al[184], 2015 | |
| miR-146b-5p (↑) | Human: THP-1 macrophages | oxLDL | TNF receptor (TNFR) associated factor 6 (TRAF6) | Inhibition promotes inflammation and lipid uptake during formation of foam cells | Levels are elevated in foam cells, and clinical specimens from patients with atherosclerosis | Lin et al[185], 2017 |
| miR-148a-5p miR-152-3p | - | - | DNA methyl-transferase 1 (DNMT1) | Viral overexpression reduces expression of DNMT1, increases levels of adipocyte differentiation related protein (ADRP) and enhances cholesterol accumulation in foam cells; down-regulation achieves the reverse | Aortic levels increased in hyperhomo-cysteinaemic apoE-/- mice | Yang et al[186], 2017 |
| miR-150-5p (↑) | Human: THP-1 macrophages | oxLDL | Adiponectin receptor 2 (ADIPOR2) | Mimic inhibits lipid accumulation, increasing cholesterol efflux to apoA-I and HDL; an inhibitor achieves the reverse. Down-regulation of ADIPOR2 replicates the impact of the miR-150 mimic | - | Li et al[187], 2016 |
| miR-155-5p (↑) | Human: Peripheral blood monocytes | oxLDL | - | - | - | Chen et al[178], 2009 |
| miR-155-5p (↑) | Murine: RAW 264.7 macrophages | oxLDL | HMG-box transcription factor 1 (HBP1) | Enhances lipid uptake and reactive oxygen species production by macrophages | Antagomir decreases lipid accumulation in macrophages and lesion formation in apoE-/- mice (HFD). Level is up-regulated in CD14+ monocytes from coronary heart disease (CHD) patients | Tian et al[188], 2014 |
| miR-155-5p (↑) | Human: THP-1 macrophages | oxLDL | Calcium-regulated heat stable protein 1 (CARHPS1)-TNF-α | Mimic blocks lipid uptake and suppresses expression of TNF-α; inhibitor achieves the reverse | Elevated in clinical samples (plaque and plasma) from patients with atherosclerosis | Li et al[189], 2016 |
| miR-155-5p | Human: THP-1 macrophages | - | T-cell immunoglobulin and mucin domain-3 (Tim-3) | Mimic enhances expression of cholesteryl ester (CE) hydrolase (CEH). Overexpression inhibits foam cell formation, intracellular CE accumulation and enhances efflux of cholesterol | - | Zhang et al[190], 2018 |
| miR-181a-5p (↓) | Human: THP-1 macrophages | oxLDL | Mitogen-activated protein kinase kinase 1 (MEK1) | Activates MEK/ERK/NF-κB, upregulates NLR family leucine-rich repeat protein 3 (NRLP3) inflammasome-related proteins (NRLP, caspase-1, IL-18, IL-1) | - | Song et al[191], 2019 |
| miR-181a-5p (↑) | Human: THP-1 macrophages | oxLDL | Toll-like receptor 4 (TLR4) | Decreases expression of CD36 protein, and lipid [Total cholesterol (TC), Triglyceride (TG)] accumulation. Inhibits THP-1 apoptosis, and increases Il-6, IL-1, TNF-α protein expression | - | Du et al[192], 2018 |
| miR-188-3p (↑) | - | - | - | - | Overexpression induces intravascular lipid accumulation, suppresses oxidation and macrophage inflammation in apoE-/- mice, and reduces serum levels of Regulated upon activation normal T-cell expressed and secreted (RANTES), LOX1 and inducible Nitric Oxide Synthase (iNOS) | Zhang et al[193], 2018 |
| miR-212-3p | Human: THP-1 macrophages | oxLDL | Sirtuin 1 (SIRT1) | Overexpression promotes lipid accumulation during foam cell formation, and reduces ABCA1 expression and cholesterol efflux; depletion achieves the reverse | Levels are decreased in atheroma and macrophages in apoE-/- mice (HFD) | Miao et al[194], 2018 |
| miR-216a-5p | Human: THP-1 macrophages | oxLDL | Cystathionine-γ-lyase (CSE) | Mimic inhibits expression of ABCA1, decreases phosphorylation of phosphatidyl-inositol 3-kinase (PI3K) and protein kinase B (AKT), and reduces ABCA1 expression and cholesterol efflux, promoting lipid accumulation; inverse occurs with the inhibitor | - | Gong et al[195], 2016 |
| miR-217 | - | - | - | - | Serum levels are negatively correlated with plaque development in apoE-/- mice (HFD). Mimic reduces intimal media thickness, reduces levels of pro-atherogenic lipoproteins and inhibits inflammation in the same model | Liu et al[196], 2018 |
| miR-221-3p (↓) | Murine: RAW 264.7 macrophages | oxLDL | A disintegrin and metalloprotease-22 (ADAM22) | Mimic reduces foam cell formation and apoptosis; inhibitor achieves the reverse | - | Zhuang et al[197], 2019 |
| miR-223 (↓) | Murine: Bone marrow derived macrophages; Murine: RAW264.7 macrophages | LPS, oxLDL | TLR4 | Overexpression reduces foam cell formation, and production of pro-inflammatory cytokines via repression of NK-κB signaling; inhibitor achieves the reverse | Elevated levels in aortic lesions in apoE-/- mice (HFD) | Wang et al[198], 2015 |
| mir-302a-3p (↓) | Primary: Mouse macrophages | oxLDL, AcLDL | ABCA1 | Mimic decreases cholesterol efflux to apoA-I | Inhibitor enhances ABCA1 in hepatic and aortic tissues of Ldlr-/- mice (HFD), increases HDL and reduces plaque size and inflammation | Meiler et al[199], 2015 |
| miR-361-5p | Human: THP-1 macrophages | oxLDL | LPL | Mir-361-5p is upregulated by apelin, resulting in suppression of LPL translation, inhibition of lipid accumulation and proinflammatory cytokine secretion | - | Zhang et al[200], 2017 |
| miR-378-3p (↑) | Human: THP-1 macrophages; Murine: J774.1 macrophages | oxLDL ± coenzyme Q10 | ABCG1 | Coenzyme Q10 (Q10) protects cholesterol efflux by reducing expression of miR-378 | Q10 promotes RCT and reduces atheroma in apoE-/- mice (HFD) | Wang et al[201], 2014 |
| miR-382-5p (↑) | Human: THP-1 macrophages | oxLDL, AcLDL | Nuclear factor 1A (NFIA) | Mimic increases cholesterol content and reduces cholesterol efflux; enhances LPS-stimulated production of pro-inflammatory cytokines (IL-6, TNF-α, IL-1) | - | Hu et al[108], 2015 |
| miR-486-5p | Human: THP-1 macrophages | oxLDL | Histone acetyl-transferase 1 (HAT1) | Mimic downregulates the expression of ABCA1, limiting cholesterol efflux and promoting foam cell formation; inhibitor achieves the reverse | - | Liu et al[202], 2016 |
| miR-497-5p (↑) | Human: THP-1 macrophages | oxLDL | Apelin | Overexpression of miR-497 promotes cholesterol efflux and decreases cholesterol efflux; inhibitor achieves the reverse | - | Cui et al[203], 2017 |
| miR-758-5p | Human: THP-1 macrophages | oxLDL | CD36 | Mimic decreases uptake of DiI-labelled OxLDL via modulation of CD36; inhibitor achieves the reverse | - | Li et al[204], 2017 |
TargetScan and miRDB were used to confirm mouse miRNA target prediction in humans; where (↑↓) is not indicated, the level of miRNA was not confirmed altered by macrophage lipid accumulation. ABCA1: ATP binding cassette transporter A1; ABCG1: ATP binding cassette transporter G1; oxLDL: Oxidized low density lipoprotein; AcLDL: Acetylated low density lipoprotein; ADAM22: A disintegrin and metalloprotease-22; ADIPOR2: Adiponectin receptor 2); ADRP: Adipocyte differentiation related protein; AKT: Protein kinase B; ANGTPTL4: Angiopoetin-like 4; ApoE: Apolipoprotein E; ATG5: Autophagy protein 5; ATG12: Autophagy-related 12; CARHPS1: Calcium-regulated heat stable protein 1; CD: Cluster of differentiation; CEH: Cholesteryl ester hydrolase 1; CSE: Cystathionine-γ-lyase; CSF: Colony stimulating factors; DNMT1: DNA methyltransferase 1; ERK: extracellular signal-regulated kinase; FOXO3: Forkhead protein O3; HAT1: Histone acetyltransferase 1; HBP-1: HMG-Box transcription factor 1; Hcy: Homocysteine; HDAC1: Histone deacetylase 1; HFD: High fat diet; IL: Interleukin; ISG15: Interferon-stimulated gene 15; IKKα/β: Inhibitor kappa B kinase alpha/beta; iNOS: Inducible nitric oxide synthase; Lamp1: Lysosomal associated membrane protein 1; lncRNA: Long noncoding RNA; LPL: Lipoprotein lipase; LPS: Lipopolysaccharide; LRAD3: Low density lipoprotein receptor class A domain containing 3; Map11c3b: LC3 microtubule-associated protein light chain 3; MEK1: Mitogen activated protein kinase kinase 1; MEKK1: Mitogen-activated protein kinase kinase kinase 1; MKK3: Mitogen activated protein kinase kinase 3; MMP-14: Matrix metalloproteinase-14; NF-κB: Nuclear factor kappa B; NFAM: Transcription factor A, mitochondrial; NFIA: Nuclear factor 1A; NLRP3: NLR-family, leucine-rich repeat protein 3; NRF1: Nuclear respiratory factor 1; OLR1/LOX1: Oxidized low density lipoprotein (lectin-like) receptor 1; PDCD4: Programmed cell death 4; PDK4: Pyruvate dehydrogenase kinase 4; PGC1-γ: Peroxisome proliferator activated receptor-γ coactivator 1; PI3K: Phosphatidylinositol 3-kinase; Prkaa1: AMP-activated protein kinase-α1; RANTES: Regulated upon activation normal T cell expressed and secreted; SIRT1: Sirtuin 1, NAD-dependent protein deacetylase sirtuin 1; SLC25A25: Solute carrier family 25, member 25; SREBF: Sterol regulatory element-binding transcription factor; STK11: Serine/threonine kinase 11; TC: Total cholesterol; TFEB: Transcription factor EB; TG: Triglyceride; TLR4: Toll-like receptor 4; Tim-3: T-cell immunoglobulin and mucin domain-3; TNF-α: Tumour necrosis factor alpha; TR4: Testicular orphan nuclear receptor 4; TRAF6: TNF receptor (TNFR) associated factor 6; Vaspin: Visceral adipose tissue-derived serine protease inhibitor; WD: Western diet.
Multiple miRNA sequences target genes involved in macrophage cholesterol homeostasis
It is well established that miR-33, encoded by an intronic sequence within SREBF2, plays a role in modulating cholesterol metabolism, in part by repressing expression of ABCA1[152]. However, this sequence also represses effectors of macrophage autophagy[153] thereby inhibiting the breakdown of lipid droplets, and targets genes central to mitochondrial respiration, which are needed for effective cholesterol efflux to apoA-I[154]. The expressions of ABCA1 and/or ABCG1 within the cholesterol efflux pathway are also targeted by miR-19b[162], miR-20a/b[164], miR-23a-5p[167], miR-27a/b[169], miR-144[182] and miR-378[201] (Table 1), highlighting the complexity of the epigenetic regulation mediated by microRNA sequences. Equally, proteins involved in uptake of modified LDL are also modulated by miRNA sequences: TLR-4 by miR-21[165], miR-181a[192] and miR-223[198], LOX-1 by miR-30[170] and miR-98[176], CD36 by miR-181a[192] and miR-758[204], while LPL is targeted by miR-134[180,181] and miR-361[200] (Figure 2). Storage of cholesterol as droplets of cholesteryl ester is modified by miR-9 targeting of SOAT1[157], while cholesterol removal by autophagy is reduced by miR-17-5p dependent repression of Beclin-1[160].
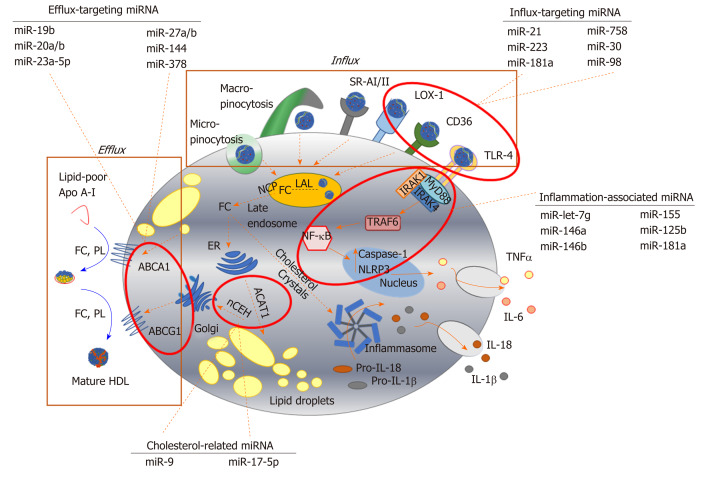
Figure 2.
Key pathways involved in foam cell formation regulated by microRNA. Native low-density lipoprotein (LDL) and oxidized LDL is processed in the late endosome/lysosomes by lysosomal acid lipase, generating free cholesterol which is stored in the form of lipid droplets after esterification via acyl-CoA cholesteryl acyl transferase or sterol O-acyltransferase 1 at the endoplasmic reticulum. Cholesterol can be removed from the cell by trafficking from the late endosomes, or by hydrolysis of lipid droplets and transport to the plasma membrane to be transferred to lipid-poor apolipoprotein A-I or nascent high-density lipoprotein via ABC transporters (ATP binding cassette transporter A1, ATP binding cassette transporter G1). Inflammatory signalling pathways can be activated in foam cells via recognition of oxLDL by toll-like receptor 4 (TLR4), or by intracellular free cholesterol forming cholesterol crystals, leading to the expression and release of inflammatory cytokines. Since their initial discovery, numerous miRNA sequences have been shown to play a role in targeting each of these key steps in the formation of foam cells. For example, influx of modified lipoproteins into macrophages via oxidized low-density lipoprotein (lectin-like) receptor 1, cluster of differentiation 36 and TLR4, is reported to be regulated by miR-21, miR-30, miR-98, miR-181a, miR-223 and miR-758. The cholesterol efflux pathway is modulated by distinct sequences, including miR-19b, miR-20a/b, miR-23, miR-27a/b, mir-155 and miR-378, while miR-9 and miR-17 regulate the esterification and hydrolysis of cholesterol droplets. MicroRNA-181a regulates both lipid metabolism and inflammatory response, while let-7g, miR-146a/b, miR125b and miR-155 influence macrophage inflammatory phenotype. Dotted arrows indicate the paths for cholesterol derived from the extracellular environment within the macrophage; solid arrows indicate inflammatory signalling pathways. HDL: High density lipoprotein; ABCA1: ATP binding cassette transporter A1; ABCG1: ATP binding cassette transporter G1; ACAT-1: Acyl-CoA cholesteryl acyl transferase or sterol O-acyltransferase 1; ApoA-I: Apolipoprotein A-I; CD: Cluster of differentiation; ER: Endoplasmic reticulum; FC: Free cholesterol; IL: Interleukin; IRAK: Interleukin 1 receptor associated kinase 4; LAL: Lysosomal acid lipase; LOX-1: Oxidized low-density lipoprotein (lectin-like) receptor 1; MyD88: Myeloid differentiation primary response 88; nCEH: Neutral cholesteryl ester hydrolase; NPC: Niemann-Pick disease type C; NF-κB: Nuclear factor kappa beta; PL: Phospholipid; SR: Scavenger receptor; TLR: Toll-like receptor; TNF-α: Tumour necrosis factor alpha.
Notably, a mimic of miR-134, which enhances LPL activity and protein expression and increases macrophage cholesterol mass, also promotes the production of inflammatory cytokines[180] and increases atheroma formation in the apoE-/- murine model of atheroma[181]. Sequences repressing ABCA1 (miR-144[182], miR-302[199]) also enhance cytokine expression; a mimic of miR-144[183] accelerates lesion development in vivo, and circulating levels of this sequence correlate with acute myocardial infarction[183], while an inhibitor of miR-302 increases aortic and hepatic expression of ABCA1 and reduces plaque size and inflammation in Ldlr-/- mice fed a high fat diet[199].
miRNA sequences linking inflammation with cholesterol accumulation in macrophages
miRNA sequences which target the expression of proteins within cell signalling pathways mediating inflammatory responses have also been shown to reduce cholesterol accumulation in macrophages (Table 1)[153,155-204]. For instance, let-7g inhibits both canonical (RelA/p50) and non-canonical (RelB/p52) NF-κB signalling pathways, limiting inflammatory (IL-1, IL-6, MCP-1) and apoptotic responses, and decreasing macrophage foam cell formation[155] in vitro and in vivo. Further, let-7g inhibition of nuclear translocation of RelA/p50 in macrophages treated with OxLDL prevents NF-κB dependent upregulation of SREBF2 and miR-33a, and results in up-regulation of ABCA1[155]. Indeed, aberrant expression of members of the lethal-7 (let-7) miRNA family have been linked with a number of diseases, including atherosclerosis and cancer[205-207]: Reductions in expression of let-7, which can be mediated by RNA binding protein Lin-28 homolog A (Lin-28), is observed in human carotid plaques from diabetic individuals, and diabetic apoE-/- mice[207].
miR-146a, which also targets the NF-κB pathway, inhibits the production of TNF-α by macrophages in vitro, and limits inflammation and plaque development in murine models of atheroma[184]. Plaque development and inflammation are also inhibited by miR-146a which targets the tumour necrosis factor receptor-associated factor (TRAF6)-NF-κB signalling axis[185] thought to underlie many cardiovascular pathologies[208]. Equally, the loss of miR-21, which targets mitogen-activated protein (MAP) kinase kinase 3 (MKK3) within the p38 MAP kinase pathway[166], promotes the degradation of ABCG1, reducing cholesterol removal and promoting the formation of foam cells in vitro. In vivo, deletion of miR-21 increases the number of macrophages within arterial lesions, and enhances the production of inflammatory cytokines[166]. Reductions in expression of miR-181a, which targets mitogen-activated protein kinase kinase 1 (MEK1) in the extracellular signal-regulated kinase (ERK)-1/2 pathway, have been linked to upregulation of NRLP3 inflammasome-related proteins[191], while increased expression of this sequence is associated with decreases in macrophage lipid accumulation[192].
Unsurprisingly, given their roles in regulating inflammatory responses, a number of miRNA sequences have been linked with regulating macrophage polarisation to differing phenotypes, recently reviewed by Essandoh et al[209]. Notably, miR-9, miR-125b and miR-155 are sequences linked with polarization towards the M1 phenotype[209]; miR-125b and miR-155 are induced by exposure to oxLDL in human macrophages, but mimics of miR-9 and miR-155 are linked with inhibition of foam cell formation by repression of SOAT1[157], enhanced expression of cholesteryl ester hydrolase[190], blockade of lipid uptake[189] and increased cholesterol efflux[190], suggesting divergence from the inflammation-lipid accumulation axis. Macrophages are induced to the M2 phenotype by several sequences, including miR-146a and miR-223[209]. miRNA-146a inhibits inflammatory responses in murine macrophages, and also reduces inflammation and plaque formation in murine models of atheroma[184]. Levels of miR-223 are reduced by OxLDL, and LPS, but elevated in murine atherosclerotic lesions, and overexpression of this sequence prevents both foam cell formation and production of inflammatory cytokines[198]. However, much less is known about the impact of miRNA mimics or inhibitors involved in phenotypic modulation after induction of lipid accumulation in macrophages, and whether these molecules can induce phenotypic plasticity or aid lesion regression remains a key question.
Novel and emerging pathways associated with foam cell formation
Importantly, research into microRNA sequences modulated during foam cell formation has highlighted a number of previously unrecognised pathways contributing to this process, which may also prove useful therapeutic targets (Figure 3). For example, the study of miR-155 revealed a previously unsuspected role for calcium-regulated heat stable protein 1 (CARHSP1/CRHSP-24) in foam cell formation[189]. This cytoplasmic protein, a cold shock domain (CSD) protein family member, is found within processing bodies or exosome granules, and was first identified as the physiological substrate for calcineurin (PP2B)[189,210,211]. The conserved CSD domain binds to the AU-rich element (ARE) in the 3-UTR of TNF-α, increasing mRNA stability and enhancing inflammation[210,211]. NF-κB induction of miR-155 by oxLDL in human macrophages is mirrored by increased levels in plasma and atherosclerotic lesions of patients with atherosclerosis[189]. MicroRNA-155 binds directly to the 3’-UTR of CARHSP1 to reduce expression of this protein and TNF-α in macrophage foam cells; knockdown of CARHSP1 inhibits lipid accumulation and TNF-α production, while overexpression of CARHSP1 reverses the protective effects of miR-155[189].
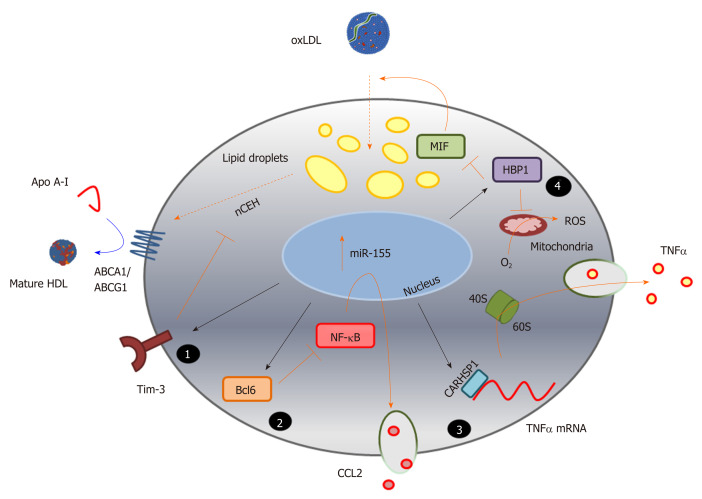
Figure 3.
Identification of novel pathways associated with foam cell formation. MicroRNA sequences altered during macrophage foam cell formation reveal novel pathways involved in this process, and highlight the complexity of miRNA function in targeting multiple gene pathways, exemplified here by miR-155 (Table 1). Inhibition of expression of the cell surface immunoregulatory glycoprotein, T-cell immunoglobulin and mucin-domain containing-3 (Tim-3), by miR-155 (1) increases hydrolysis of cholesteryl esters by neutral cholesteryl ester hydrolase, and promotes efflux of this lipid via ATP binding cassette transporter A1 and ATP binding cassette transporter G1, to apolipoprotein A-I and high density lipoprotein respectively. MicroRNA-155 also represses expression of the transcriptional repressor B-cell lymphoma 6 protein[236], which increases nuclear factor kappa beta activity and enhances production of the chemokine C-C motif ligand 2 (2); Repression of the cytoplasmic protein, calcium-regulated heat stable protein (CARHSP1) by miR-155, results in reduced binding of this protein to the 3’UTR of the TNFα gene transcript, and to reduced mRNA stability and decreased output of this cytokine (3); MicroRNA-155 also directly targets (represses) HMG-Box transcription factor 1, thereby increasing production of reactive oxygen species and loss of inhibition of macrophage migration inhibitory factor, leading to increased oxidatively modify proteoglycan-bound low density lipoprotein uptake and lipid accumulation (4). The black arrows represent direct targeting by miR-155; orange bars and arrows represent the functions of miR-155 targets in the absence of this miRNA sequence. HDL: High density lipoprotein; ABCA1: ATP binding cassette transporter A1; ABCG1: ATP binding cassette transporter G1; ACAT-1: Acyl-CoA cholesteryl acyl transferase or sterol O-acyltransferase 1; ApoA-I: Apolipoprotein A-I; Bcl6: B-cell lymphoma 6 protein; CARHSP1: Calcium-regulated heat stable protein; CCL2: C-C motif chemokine ligand 2; HBP1: HMG-Box transcription factor 1; MIF: Macrophage migration inhibitory factor; NF-κB: Nuclear factor kappa beta; ROS: Reactive oxygen species; TNF-α: Tumour necrosis factor alpha.
Equally, insight into the hitherto uncharacterised role of programmed cell death 4 (PDC4) in foam cell formation and atherosclerosis was revealed by investigation of the function of miR-16[159]. Expression of PDCD4, which can act as a tumour suppressor, is induced by apoptosis and is known to regulate both inflammatory and apoptotic responses[212-214]. MicroRNA-16 suppresses the activation of inflammatory macrophages by directly targeting the 3’-UTR of PDCD4[159]. Levels of miR-16 decline in macrophages treated with oxLDL and in aortic lesions of apoE-/- mice fed a high fat diet, which also exhibit greater levels of PDCD4 protein. Either knockdown of PDCD4, or transfection with a miR-16 mimic, inhibits the expression and secretion of pro-inflammatory cytokines, and enhances expression and release of the anti-inflammatory factor IL-10, while an inhibitor of miR-16 achieves the reverse; these outcomes are also associated with modulation of ERK, p38 MAP kinase and NF-κB[212].
In other studies, the mechanism of action of molecules such as puerarin, the major bioactive ingredient isolated from Pueraria lobata and known as Gegen in traditional Chinese medicine, have been revealed by studies using miRNA[156]. Targeting the 3-UTR region of serine/threonine kinase 11 (STK11) using a miR-7 mimic, revealed that this drug enhances ABCA1-dependent cholesterol efflux via a mechanism which involves STK11 activation of AMP kinase and enhanced expression of PPAR-γ-LXR-ABCA1. Finally, it is clear that the contribution of some miRNA targets in foam cells remain to be established. For example, miR-28-5p, which is upregulated in murine macrophages treated with oxLDL, targets LDL receptor class A domain containing 3 (LRAD3)[170], but the contribution of this novel lipoprotein receptor to foam cell formation has not been investigated: At present, this protein has been linked with amyloid precursor protein trafficking in neurons[215] and with activation of E3 ubiquitin-protein ligase Itchy homolog (Itch) and E3 ubiquitin-protein ligase NEDD4 that promote proteasomal degradation[216].
Pathways targeted by miRNA sequences altered in human macrophage “foam” cells: DIANA/KEGG predictive analysis
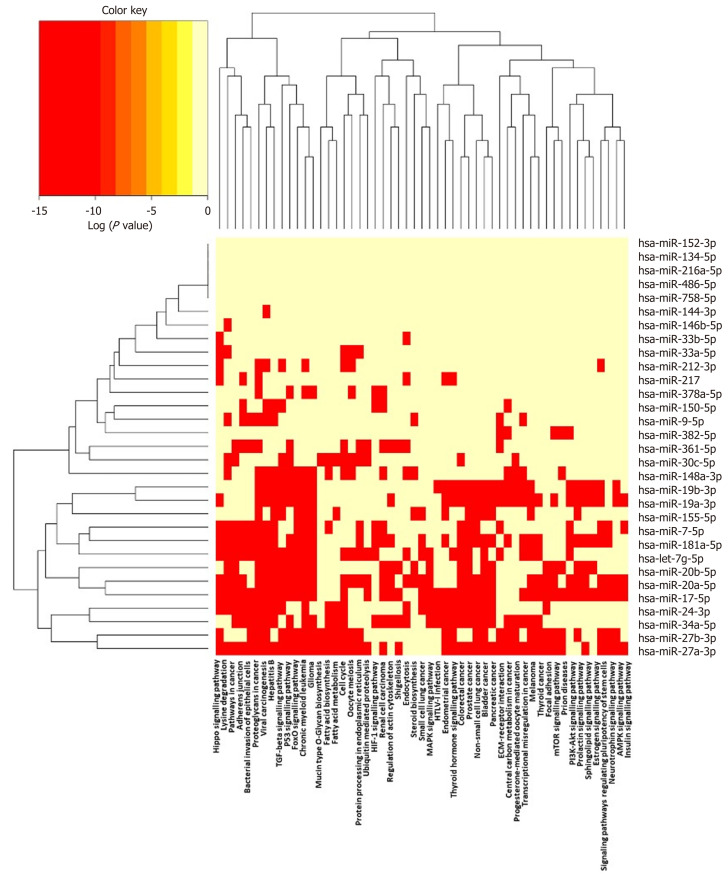
It is increasingly recognised that networks of miRNA sequences, and their combined effects on multiple pathways, are important epigenetic determinants of complex phenotypes, just as genome-wide association studies have revealed shared genes and pathways in human disease[217,218]. The (human) sequences described in Table 1 were analysed using DIANA-miRPATH v3.0, and the miRNA versus GO/GOSlim/KEGG entries heat map is shown in Figure 4. This functionality enables identification of miRNA belonging to similar functional categories, and identification of pathways lying under the regulation of similar miRNAs[219].
Figure 4.
Pathways targeted by miRNA sequences altered in human macrophage “foam” cells: DIANA/KEGG predictive analysis. The human miRNA sequences described in Table 1 were analysed using DIANA-miRPATH v3.0, yielding a heat map of miRNA vs Gene Ontology/GOSlim/Kyoto Encyclopedia for Genes and Genomes entries.
Several well-established pathways, targeted by multiple and distinct miRNA sequences/clusters, and known to regulate vascular function and atherogenesis, emerge from this predictive analysis. These include adherens junctions, which are a key part of the common signalling network linking age-related disease proteins (ARDPs) and longevity-associated proteins (LAPs) in the human interactome[220]. The endothelial adherens junction complex, formed of vascular endothelial (VE)-cadherin and associated catenins, is a key determinant of arterial permeability, dysregulation contributing to vascular inflammation and atherosclerosis[221], while attenuating intraplaque vascular leakage reduces macrophage accumulation, necrotic core size and intraplaque haemorrhage[222]. Identification of the TGF-β signalling pathway as a target of miRNA sequences altered in macrophage foam cells is equally unsurprising, given the widely recognised, and extensively reviewed, role of this cytokine in controlling macrophage phenotype[223], atherosclerosis[224] and cardiovascular function[225].
More intriguingly, the Hippo signalling pathway emerges as highly targeted by multiple miRNA sequences implicated in foam cell formation (Figure 4); originally discovered in Drosophila and highly conserved in mammalian cells, this pathway regulates cell survival, proliferation and apoptosis[226]. Li et al[170] first showed that target genes of differentially expressed miRNA sequences are enriched in this pathway, in murine RAW264.7 macrophages treated with OxLDL; the Hippo/Yes-associated protein (YAP) signalling pathway is linked with vascular remodelling, pulmonary hypertension, aortic aneurysm, restenosis and angiogenesis, and atherosclerosis[226,227]. Notably, the atheroprotective effect of steady laminar flow in major arteries is linked with inhibition of Hippo/YAP effector function[228] while activation of this pathway is linked with vascular remodelling, and switching of arterial smooth muscle cells to the “synthetic” proliferative phenotype in response to biochemical stretch[229]. The effector function of YAP is linked with accelerated atherosclerosis in apoE-/- mice[230], while the herbal extract Scutellarin can protect against atherosclerosis in rats by modulating the Hippo-YAP-Forkhead box (FOXO)3A transduction pathway[231].
Another pathway enriched in targets of multiple miRNA sequences is that involved in bacterial invasion of epithelial cells (Figure 4). Infection and systemic inflammation are linked with atherogenesis in a number of epidemiological studies[232] and vascular cells and macrophages are subject to invasion by bacteria via a number of mechanisms, including evasion of autophagy and internalisation via lipid rafts. In turn, this has led to the notion of vascular tissue providing a “privileged niche” in which bacteria can persist in dormancy for extended periods of time before becoming activated in phagocytic cells, contributing to the chronic and unresolved inflammation which characterises atherosclerosis[232]. The epigenetic miRNA profile found in macrophage “foam” cells which may modulate susceptibility to bacterial invasion may also suggest key proteins (and pathogens) implicated in this process, and/or highlight possible therapeutic strategies designed to limit the impact of vascular “infectology”[232].
THERAPEUTIC OPTIONS: CLINICAL APPLICATIONS OF MIRNA (TARGETS)
miRNA pathways are excellent candidates for pharmacological manipulation, and have been invoked as biomarkers, diagnostics or therapeutics for a number of disease conditions[140,233,234]. For example, Caruso et al[233] monitored dynamic changes in microRNA profiles in lung tissue during the development of pulmonary arterial hypertension (PAH) in hypoxic rodents. The same authors discovered that miR-145 is a useful indicator of hypoxia in mice, and of heritable and idiopathic pulmonary arterial hypertension in patients; down-regulation of miR-145 also had utility in protecting against development of PAH in mice[234]. Further, the “ThyraMIR” testing platform, which examines the expression of a panel of ten miRNA in conjunction with selected disease-associated genes, has been approved for diagnostic use in thyroid cancer when malignancy risk cannot be determined by conventional cytology[235].
Treatments involving miRNAs focus on the concept of specifically influencing levels of miRNAs in certain diseases – including suppression of miRNAs, as well as raising miRNA levels or substituting artificially generated copies[140]. Mimics can be used for gene silencing, by generating artificial, double-stranded miRNA-like RNA fragments, which bind specifically to target mRNA, activating the RISC complex; this results in down-regulation of specific mRNAs and gene suppression (above). Equally, chemically engineered oligonucleotides are capable of silencing single endogenous miRNAs, binding to the target mature miRNA, leading to reduced activation of RISC and up-regulation of specific mRNAs and gene expression. Other approaches involve “target mimicry” using miRNA sponges, masking or erasers[140].
Delivery of disease-specific miRNA mimics or inhibitors remains in the developmental stage with multiple miRNA therapeutics currently in clinical trials. The most advanced trial, currently in Phase II, employs a chemically engineered inhibitor for miR-122 (Miravirsen) which, under normal conditions, binds to the 5’-UTR region of the hepatitis C virus and enhances its transcription[236-243]. By hybridizing to mature miR-122, Miravirsen has been shown to effectively inhibit viral replication with minimal “off target” effects[236-239]. MicroRNA-based clinical trials are also underway for the development of novel treatments for various cancers. Currently in Phase I and Phase II trials, the efficacy of an inhibitor (MRG-106) targeting miR-155 is being investigated for treatment of a variety of lymphomas, reflecting the recognised role of this sequence in driving malignant lymphocyte proliferation[240,241].
Despite the encouraging progression of miRNA therapies, significant challenges have also been highlighted in some clinical trials. One such promising miRNA therapeutic, the miR-34 mimic “MRX34”, was employed in a Phase I trial for patients with advanced liver cancer[242-244]; despite dose-dependent modulation of miR-34 target oncogenes, the study was halted due to serious adverse effects in a small cohort of subjects[242-244]. This study also highlighted a challenging area for the development of miRNA-based therapies: The preclinical studies demonstrated that the liposomal delivery system resulted in elevated miR-34 in multiple tissues in non-human primates[244]. While this may be beneficial for miRNA therapeutics used to treat diseases that can arise in several anatomical locations, in the case of tissue specific diseases, such as atherosclerosis, site-specific homing could dramatically reduce potential off-target effects.
To overcome this issue, liposomes enriched in specific amino acid sequences have been developed which result in increased tissue-specific accumulation. The efficacy of this system, for the delivery of short, siRNA, has been demonstrated in vivo in osteoporotic mice[245]. Use of a lipid nanoparticle containing C-C chemokine receptor type 2 (CCR2)-targeting siRNA resulted in high levels of localisation in bone marrow and spleen, significant reductions in monocyte CCR2 expression, decreased myeloid cell infiltration in the plaque and an overall reduction in lesion size in ApoE-/- mice[246]. While this system targets atherosclerotic plaque indirectly, additional delivery mechanisms have been employed in animal studies that may facilitate plaque-directed delivery of miRNA-based therapeutics. Notably, reconstituted HDL (rHDL) can act as a carrier particle for delivery of drugs and microRNA: In ApoE-/- mice, rHDL was used to delivery simvastatin to plaque regions, resulting in reduced local inflammation[247], while miR-223 incorporated into rHDL in vitro was able to selectively target cells expressing SR-BI[148].
Thus, many factors need careful consideration in developing miRNA therapeutics for atherosclerosis, including effective vectors and delivery options, and the nature of “off-target” side-effects and/or toxicities which may occur due to disruption of multiple target genes and/or cell signalling networks[248,249]. However, since differing microRNA sequences impact on distinct stages of the atherogenic process[248,249], delivery of a pool of mimics and/or inhibitors may be an attractive therapeutic strategy for treatment of this complex, multicellular disease[248,249]. Defined stages of the disease process could be targeted by distinct miRNA mimics/inhibitors, predicated by serum levels of secreted miRNA sequences. Such approaches, if fully validated, might be used to provide personalised treatment, or be beneficial in targeting asymptomatic patients, or those in whom statin use is contraindicated or ineffective[250-252].
CONCLUSION
Huge advances have been made in understanding the epigenetic factors, and particularly the role of small non-coding miRNA sequences, in regulating macrophage “foam” cell formation and function over the last decade. Networks of genes regulated by multiple miRNA sequences have been revealed, and new pathways discovered which contribute to the atherogenic process, which may ultimately lead to RNA-based therapeutics capable of preventing or regressing the formation of complex atherosclerotic lesions by targeting macrophage function.
Footnotes
Manuscript source: Unsolicited manuscript
Specialty type: Cardiac and cardiovascular systems
Country/Territory of origin: United Kingdom
Peer-review report’s scientific quality classification
Grade A (Excellent): A, A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
Conflict-of-interest statement: Prof. Graham A and Dr. Dempsie Y have received research funding from Heart Research UK (RG2651) in support of PhD research student, Mr Lightbody RJ; Prof. Graham A, Dr. Dempsie Y and Dr. Taylor JMW are employees of Glasgow Caledonian University.
Peer-review started: January 30, 2020
First decision: April 18, 2020
Article in press: June 10, 2020
P-Reviewer: Barik R, S Berezin AE, Kharlamov AN S-Editor: Yan JP L-Editor: A E-Editor: Zhang YL
Contributor Information
Richard James Lightbody, Department of Biological and Biomedical Sciences, School of Health and Life Sciences, Glasgow Caledonian University, Glasgow G4 0BA, United Kingdom.
Janice Marie Walsh Taylor, Department of Biological and Biomedical Sciences, School of Health and Life Sciences, Glasgow Caledonian University, Glasgow G4 0BA, United Kingdom.
Yvonne Dempsie, Department of Biological and Biomedical Sciences, School of Health and Life Sciences, Glasgow Caledonian University, Glasgow G4 0BA, United Kingdom.
Annette Graham, Department of Biological and Biomedical Sciences, School of Health and Life Sciences, Glasgow Caledonian University, Glasgow G4 0BA, United Kingdom. ann.graham@gcu.ac.uk.
References
- 1.Batty GD, Kivimäki M, Bell S. Comparison of risk factors for coronary heart disease morbidity versus mortality. Eur J Prev Cardiol. 2019:2047487319882512. doi: 10.1177/2047487319882512. [DOI] [PubMed] [Google Scholar]
- 2.Asakura T, Karino T. Flow patterns and spatial distribution of atherosclerotic lesions in human coronary arteries. Circ Res. 1990;66:1045–1066. doi: 10.1161/01.res.66.4.1045. [DOI] [PubMed] [Google Scholar]
- 3.Dai G, Kaazempur-Mofrad MR, Natarajan S, Zhang Y, Vaughn S, Blackman BR, Kamm RD, García-Cardeña G, Gimbrone MA., Jr Distinct endothelial phenotypes evoked by arterial waveforms derived from atherosclerosis-susceptible and -resistant regions of human vasculature. Proc Natl Acad Sci USA. 2004;101:14871–14876. doi: 10.1073/pnas.0406073101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skålén K, Gustafsson M, Rydberg EK, Hultén LM, Wiklund O, Innerarity TL, Borén J. Subendothelial retention of atherogenic lipoproteins in early atherosclerosis. Nature. 2002;417:750–754. doi: 10.1038/nature00804. [DOI] [PubMed] [Google Scholar]
- 5.Parmar KM, Larman HB, Dai G, Zhang Y, Wang ET, Moorthy SN, Kratz JR, Lin Z, Jain MK, Gimbrone MA, Jr, García-Cardeña G. Integration of flow-dependent endothelial phenotypes by Kruppel-like factor 2. J Clin Invest. 2006;116:49–58. doi: 10.1172/JCI24787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang C, Baker BM, Chen CS, Schwartz MA. Endothelial cell sensing of flow direction. Arterioscler Thromb Vasc Biol. 2013;33:2130–2136. doi: 10.1161/ATVBAHA.113.301826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshida H, Kisugi R. Mechanisms of LDL oxidation. Clin Chim Acta. 2010;411:1875–1882. doi: 10.1016/j.cca.2010.08.038. [DOI] [PubMed] [Google Scholar]
- 8.Yang K, Zhang XJ, Cao LJ, Liu XH, Liu ZH, Wang XQ, Chen QJ, Lu L, Shen WF, Liu Y. Toll-like receptor 4 mediates inflammatory cytokine secretion in smooth muscle cells induced by oxidized low-density lipoprotein. PLoS One. 2014;9:e95935. doi: 10.1371/journal.pone.0095935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sawamura T, Kume N, Aoyama T, Moriwaki H, Hoshikawa H, Aiba Y, Tanaka T, Miwa S, Katsura Y, Kita T, Masaki T. An endothelial receptor for oxidized low-density lipoprotein. Nature. 1997;386:73–77. doi: 10.1038/386073a0. [DOI] [PubMed] [Google Scholar]
- 10.Sambandam T, Baker JR, Christner JE, Ekborg SL. Specificity of the low density lipoprotein-glycosaminoglycan interaction. Arterioscler Thromb. 1991;11:561–568. doi: 10.1161/01.atv.11.3.561. [DOI] [PubMed] [Google Scholar]
- 11.Moore KJ, Koplev S, Fisher EA, Tabas I, Björkegren JLM, Doran AC, Kovacic JC. Macrophage Trafficking, Inflammatory Resolution, and Genomics in Atherosclerosis: JACC Macrophage in CVD Series (Part 2) J Am Coll Cardiol. 2018;72:2181–2197. doi: 10.1016/j.jacc.2018.08.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.PrabhuDas MR, Baldwin CL, Bollyky PL, Bowdish DME, Drickamer K, Febbraio M, Herz J, Kobzik L, Krieger M, Loike J, McVicker B, Means TK, Moestrup SK, Post SR, Sawamura T, Silverstein S, Speth RC, Telfer JC, Thiele GM, Wang XY, Wright SD, El Khoury J. A Consensus Definitive Classification of Scavenger Receptors and Their Roles in Health and Disease. J Immunol. 2017;198:3775–3789. doi: 10.4049/jimmunol.1700373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Traber MG, Kayden HJ. Low density lipoprotein receptor activity in human monocyte-derived macrophages and its relation to atheromatous lesions. Proc Natl Acad Sci USA. 1980;77:5466–5470. doi: 10.1073/pnas.77.9.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi SH, Harkewicz R, Lee JH, Boullier A, Almazan F, Li AC, Witztum JL, Bae YS, Miller YI. Lipoprotein accumulation in macrophages via toll-like receptor-4-dependent fluid phase uptake. Circ Res. 2009;104:1355–1363. doi: 10.1161/CIRCRESAHA.108.192880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oteng AB, Ruppert PMM, Boutens L, Dijk W, van Dierendonck XAMH, Olivecrona G, Stienstra R, Kersten S. Characterization of ANGPTL4 function in macrophages and adipocytes using Angptl4-knockout and Angptl4-hypomorphic mice. J Lipid Res. 2019;60:1741–1754. doi: 10.1194/jlr.M094128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kruth HS, Jones NL, Huang W, Zhao B, Ishii I, Chang J, Combs CA, Malide D, Zhang WY. Macropinocytosis is the endocytic pathway that mediates macrophage foam cell formation with native low density lipoprotein. J Biol Chem. 2005;280:2352–2360. doi: 10.1074/jbc.M407167200. [DOI] [PubMed] [Google Scholar]
- 17.Anzinger JJ, Chang J, Xu Q, Buono C, Li Y, Leyva FJ, Park BC, Greene LE, Kruth HS. Native low-density lipoprotein uptake by macrophage colony-stimulating factor-differentiated human macrophages is mediated by macropinocytosis and micropinocytosis. Arterioscler Thromb Vasc Biol. 2010;30:2022–2031. doi: 10.1161/ATVBAHA.110.210849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo Y, Duan H, Qian Y, Feng L, Wu Z, Wang F, Feng J, Yang D, Qin Z, Yan X. Macrophagic CD146 promotes foam cell formation and retention during atherosclerosis. Cell Res. 2017;27:352–372. doi: 10.1038/cr.2017.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park YM, Drazba JA, Vasanji A, Egelhoff T, Febbraio M, Silverstein RL. Oxidized LDL/CD36 interaction induces loss of cell polarity and inhibits macrophage locomotion. Mol Biol Cell. 2012;23:3057–3068. doi: 10.1091/mbc.E11-12-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Llodrá J, Angeli V, Liu J, Trogan E, Fisher EA, Randolph GJ. Emigration of monocyte-derived cells from atherosclerotic lesions characterizes regressive, but not progressive, plaques. Proc Natl Acad Sci USA. 2004;101:11779–11784. doi: 10.1073/pnas.0403259101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galkina E, Kadl A, Sanders J, Varughese D, Sarembock IJ, Ley K. Lymphocyte recruitment into the aortic wall before and during development of atherosclerosis is partially L-selectin dependent. J Exp Med. 2006;203:1273–1282. doi: 10.1084/jem.20052205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nickel T, Schmauss D, Hanssen H, Sicic Z, Krebs B, Jankl S, Summo C, Fraunberger P, Walli AK, Pfeiler S, Weis M. oxLDL uptake by dendritic cells induces upregulation of scavenger-receptors, maturation and differentiation. Atherosclerosis. 2009;205:442–450. doi: 10.1016/j.atherosclerosis.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 23.Stemme S, Faber B, Holm J, Wiklund O, Witztum JL, Hansson GK. T lymphocytes from human atherosclerotic plaques recognize oxidized low density lipoprotein. Proc Natl Acad Sci USA. 1995;92:3893–3897. doi: 10.1073/pnas.92.9.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frostegård J, Ulfgren AK, Nyberg P, Hedin U, Swedenborg J, Andersson U, Hansson GK. Cytokine expression in advanced human atherosclerotic plaques: dominance of pro-inflammatory (Th1) and macrophage-stimulating cytokines. Atherosclerosis. 1999;145:33–43. doi: 10.1016/s0021-9150(99)00011-8. [DOI] [PubMed] [Google Scholar]
- 25.Kimura T, Kobiyama K, Winkels H, Tse K, Miller J, Vassallo M, Wolf D, Ryden C, Orecchioni M, Dileepan T, Jenkins MK, James EA, Kwok WW, Hanna DB, Kaplan RC, Strickler HD, Durkin HG, Kassaye SG, Karim R, Tien PC, Landay AL, Gange SJ, Sidney J, Sette A, Ley K. Regulatory CD4+ T Cells Recognize Major Histocompatibility Complex Class II Molecule-Restricted Peptide Epitopes of Apolipoprotein B. Circulation. 2018;138:1130–1143. doi: 10.1161/CIRCULATIONAHA.117.031420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drechsler M, Megens RT, van Zandvoort M, Weber C, Soehnlein O. Hyperlipidemia-triggered neutrophilia promotes early atherosclerosis. Circulation. 2010;122:1837–1845. doi: 10.1161/CIRCULATIONAHA.110.961714. [DOI] [PubMed] [Google Scholar]
- 27.Huo Y, Schober A, Forlow SB, Smith DF, Hyman MC, Jung S, Littman DR, Weber C, Ley K. Circulating activated platelets exacerbate atherosclerosis in mice deficient in apolipoprotein E. Nat Med. 2003;9:61–67. doi: 10.1038/nm810. [DOI] [PubMed] [Google Scholar]
- 28.Daugherty A, Dunn JL, Rateri DL, Heinecke JW. Myeloperoxidase, a catalyst for lipoprotein oxidation, is expressed in human atherosclerotic lesions. J Clin Invest. 1994;94:437–444. doi: 10.1172/JCI117342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hazell LJ, Stocker R. Oxidation of low-density lipoprotein with hypochlorite causes transformation of the lipoprotein into a high-uptake form for macrophages. Biochem J. 1993;290(Pt 1):165–172. doi: 10.1042/bj2900165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Leeuwen M, Gijbels MJ, Duijvestijn A, Smook M, van de Gaar MJ, Heeringa P, de Winther MP, Tervaert JW. Accumulation of myeloperoxidase-positive neutrophils in atherosclerotic lesions in LDLR-/- mice. Arterioscler Thromb Vasc Biol. 2008;28:84–89. doi: 10.1161/ATVBAHA.107.154807. [DOI] [PubMed] [Google Scholar]
- 31.Auerbach BJ, Bisgaier CL, Wölle J, Saxena U. Oxidation of low density lipoproteins greatly enhances their association with lipoprotein lipase anchored to endothelial cell matrix. J Biol Chem. 1996;271:1329–1335. doi: 10.1074/jbc.271.3.1329. [DOI] [PubMed] [Google Scholar]
- 32.Alexander MR, Murgai M, Moehle CW, Owens GK. Interleukin-1β modulates smooth muscle cell phenotype to a distinct inflammatory state relative to PDGF-DD via NF-κB-dependent mechanisms. Physiol Genomics. 2012;44:417–429. doi: 10.1152/physiolgenomics.00160.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cherepanova OA, Pidkovka NA, Sarmento OF, Yoshida T, Gan Q, Adiguzel E, Bendeck MP, Berliner J, Leitinger N, Owens GK. Oxidized phospholipids induce type VIII collagen expression and vascular smooth muscle cell migration. Circ Res. 2009;104:609–618. doi: 10.1161/CIRCRESAHA.108.186064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galis ZS, Muszynski M, Sukhova GK, Simon-Morrissey E, Unemori EN, Lark MW, Amento E, Libby P. Cytokine-stimulated human vascular smooth muscle cells synthesize a complement of enzymes required for extracellular matrix digestion. Circ Res. 1994;75:181–189. doi: 10.1161/01.res.75.1.181. [DOI] [PubMed] [Google Scholar]
- 35.Lopes J, Adiguzel E, Gu S, Liu SL, Hou G, Heximer S, Assoian RK, Bendeck MP. Type VIII collagen mediates vessel wall remodeling after arterial injury and fibrous cap formation in atherosclerosis. Am J Pathol. 2013;182:2241–2253. doi: 10.1016/j.ajpath.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacobsen K, Lund MB, Shim J, Gunnersen S, Füchtbauer EM, Kjolby M, Carramolino L, Bentzon JF. Diverse cellular architecture of atherosclerotic plaque derives from clonal expansion of a few medial SMCs. JCI Insight. 2017;2:e95890. doi: 10.1172/jci.insight.95890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allahverdian S, Chehroudi AC, McManus BM, Abraham T, Francis GA. Contribution of intimal smooth muscle cells to cholesterol accumulation and macrophage-like cells in human atherosclerosis. Circulation. 2014;129:1551–1559. doi: 10.1161/CIRCULATIONAHA.113.005015. [DOI] [PubMed] [Google Scholar]
- 38.Ball RY, Stowers EC, Burton JH, Cary NR, Skepper JN, Mitchinson MJ. Evidence that the death of macrophage foam cells contributes to the lipid core of atheroma. Atherosclerosis. 1995;114:45–54. doi: 10.1016/0021-9150(94)05463-s. [DOI] [PubMed] [Google Scholar]
- 39.Hegyi L, Skepper JN, Cary NR, Mitchinson MJ. Foam cell apoptosis and the development of the lipid core of human atherosclerosis. J Pathol. 1996;180:423–429. doi: 10.1002/(SICI)1096-9896(199612)180:4<423::AID-PATH677>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 40.Tabas I. Free cholesterol-induced cytotoxicity a possible contributing factor to macrophage foam cell necrosis in advanced atherosclerotic lesions. Trends Cardiovasc Med. 1997;7:256–263. doi: 10.1016/S1050-1738(97)00086-8. [DOI] [PubMed] [Google Scholar]
- 41.Seimon TA, Nadolski MJ, Liao X, Magallon J, Nguyen M, Feric NT, Koschinsky ML, Harkewicz R, Witztum JL, Tsimikas S, Golenbock D, Moore KJ, Tabas I. Atherogenic lipids and lipoproteins trigger CD36-TLR2-dependent apoptosis in macrophages undergoing endoplasmic reticulum stress. Cell Metab. 2010;12:467–482. doi: 10.1016/j.cmet.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, Semenza GL. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16:4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sluimer JC, Gasc JM, van Wanroij JL, Kisters N, Groeneweg M, Sollewijn Gelpke MD, Cleutjens JP, van den Akker LH, Corvol P, Wouters BG, Daemen MJ, Bijnens AP. Hypoxia, hypoxia-inducible transcription factor, and macrophages in human atherosclerotic plaques are correlated with intraplaque angiogenesis. J Am Coll Cardiol. 2008;51:1258–1265. doi: 10.1016/j.jacc.2007.12.025. [DOI] [PubMed] [Google Scholar]
- 44.Parathath S, Mick SL, Feig JE, Joaquin V, Grauer L, Habiel DM, Gassmann M, Gardner LB, Fisher EA. Hypoxia is present in murine atherosclerotic plaques and has multiple adverse effects on macrophage lipid metabolism. Circ Res. 2011;109:1141–1152. doi: 10.1161/CIRCRESAHA.111.246363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crucet M, Wüst SJ, Spielmann P, Lüscher TF, Wenger RH, Matter CM. Hypoxia enhances lipid uptake in macrophages: role of the scavenger receptors Lox1, SRA, and CD36. Atherosclerosis. 2013;229:110–117. doi: 10.1016/j.atherosclerosis.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 46.Folco EJ, Sukhova GK, Quillard T, Libby P. Moderate hypoxia potentiates interleukin-1β production in activated human macrophages. Circ Res. 2014;115:875–883. doi: 10.1161/CIRCRESAHA.115.304437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dalton DK, Pitts-Meek S, Keshav S, Figari IS, Bradley A, Stewart TA. Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science. 1993;259:1739–1742. doi: 10.1126/science.8456300. [DOI] [PubMed] [Google Scholar]
- 48.Boldrick JC, Alizadeh AA, Diehn M, Dudoit S, Liu CL, Belcher CE, Botstein D, Staudt LM, Brown PO, Relman DA. Stereotyped and specific gene expression programs in human innate immune responses to bacteria. Proc Natl Acad Sci USA. 2002;99:972–977. doi: 10.1073/pnas.231625398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verreck FA, de Boer T, Langenberg DM, Hoeve MA, Kramer M, Vaisberg E, Kastelein R, Kolk A, de Waal-Malefyt R, Ottenhoff TH. Human IL-23-producing type 1 macrophages promote but IL-10-producing type 2 macrophages subvert immunity to (myco)bacteria. Proc Natl Acad Sci USA. 2004;101:4560–4565. doi: 10.1073/pnas.0400983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng Z, Shi F, Zhou Z, Sun F, Sun MH, Sun Q, Chen L, Li D, Jiang CY, Zhao RZ, Cui D, Wang XJ, Jing YF, Xia SJ, Han BM. M1 macrophage mediated increased reactive oxygen species (ROS) influence wound healing via the MAPK signaling in vitro and in vivo. Toxicol Appl Pharmacol. 2019;366:83–95. doi: 10.1016/j.taap.2019.01.022. [DOI] [PubMed] [Google Scholar]
- 51.Hirose K, Iwabuchi K, Shimada K, Kiyanagi T, Iwahara C, Nakayama H, Daida H. Different responses to oxidized low-density lipoproteins in human polarized macrophages. Lipids Health Dis. 2011;10:1. doi: 10.1186/1476-511X-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Tits LJ, Stienstra R, van Lent PL, Netea MG, Joosten LA, Stalenhoef AF. Oxidized LDL enhances pro-inflammatory responses of alternatively activated M2 macrophages: a crucial role for Krüppel-like factor 2. Atherosclerosis. 2011;214:345–349. doi: 10.1016/j.atherosclerosis.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 53.Bae YS, Lee JH, Choi SH, Kim S, Almazan F, Witztum JL, Miller YI. Macrophages generate reactive oxygen species in response to minimally oxidized low-density lipoprotein: toll-like receptor 4- and spleen tyrosine kinase-dependent activation of NADPH oxidase 2. Circ Res. 2009;104:210–218, 21p following 218. doi: 10.1161/CIRCRESAHA.108.181040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duewell P, Kono H, Rayner KJ, Sirois CM, Vladimer G, Bauernfeind FG, Abela GS, Franchi L, Nuñez G, Schnurr M, Espevik T, Lien E, Fitzgerald KA, Rock KL, Moore KJ, Wright SD, Hornung V, Latz E. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. 2010;464:1357–1361. doi: 10.1038/nature08938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kang JH, Kim JK, Park WH, Park KK, Lee TS, Magae J, Nakajima H, Kim CH, Chang YC. Ascochlorin suppresses oxLDL-induced MMP-9 expression by inhibiting the MEK/ERK signaling pathway in human THP-1 macrophages. J Cell Biochem. 2007;102:506–514. doi: 10.1002/jcb.21312. [DOI] [PubMed] [Google Scholar]
- 56.Shao Q, Shen LH, Hu LH, Pu J, Jing Q, He B. Atorvastatin suppresses inflammatory response induced by oxLDL through inhibition of ERK phosphorylation, IκBα degradation, and COX-2 expression in murine macrophages. J Cell Biochem. 2012;113:611–618. doi: 10.1002/jcb.23388. [DOI] [PubMed] [Google Scholar]
- 57.Harkewicz R, Hartvigsen K, Almazan F, Dennis EA, Witztum JL, Miller YI. Cholesteryl ester hydroperoxides are biologically active components of minimally oxidized low density lipoprotein. J Biol Chem. 2008;283:10241–10251. doi: 10.1074/jbc.M709006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang Z, Li W, Wang R, Zhang F, Chi Y, Wang D, Liu Z, Zhang Y, Matsuura E, Liu Q. 7-ketocholesteryl-9-carboxynonanoate induced nuclear factor-kappa B activation in J774A.1 macrophages. Life Sci. 2010;87:651–657. doi: 10.1016/j.lfs.2010.09.028. [DOI] [PubMed] [Google Scholar]
- 59.Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med. 1992;176:287–292. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee CG, Homer RJ, Zhu Z, Lanone S, Wang X, Koteliansky V, Shipley JM, Gotwals P, Noble P, Chen Q, Senior RM, Elias JA. Interleukin-13 induces tissue fibrosis by selectively stimulating and activating transforming growth factor beta(1) J Exp Med. 2001;194:809–821. doi: 10.1084/jem.194.6.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sironi M, Martinez FO, D'Ambrosio D, Gattorno M, Polentarutti N, Locati M, Gregorio A, Iellem A, Cassatella MA, Van Damme J, Sozzani S, Martini A, Sinigaglia F, Vecchi A, Mantovani A. Differential regulation of chemokine production by Fcgamma receptor engagement in human monocytes: association of CCL1 with a distinct form of M2 monocyte activation (M2b, Type 2) J Leukoc Biol. 2006;80:342–349. doi: 10.1189/jlb.1005586. [DOI] [PubMed] [Google Scholar]
- 62.Gerber JS, Mosser DM. Reversing lipopolysaccharide toxicity by ligating the macrophage Fc gamma receptors. J Immunol. 2001;166:6861–6868. doi: 10.4049/jimmunol.166.11.6861. [DOI] [PubMed] [Google Scholar]
- 63.Edwards JP, Zhang X, Frauwirth KA, Mosser DM. Biochemical and functional characterization of three activated macrophage populations. J Leukoc Biol. 2006;80:1298–1307. doi: 10.1189/jlb.0406249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zizzo G, Hilliard BA, Monestier M, Cohen PL. Efficient clearance of early apoptotic cells by human macrophages requires M2c polarization and MerTK induction. J Immunol. 2012;189:3508–3520. doi: 10.4049/jimmunol.1200662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zizzo G, Cohen PL. IL-17 stimulates differentiation of human anti-inflammatory macrophages and phagocytosis of apoptotic neutrophils in response to IL-10 and glucocorticoids. J Immunol. 2013;190:5237–5246. doi: 10.4049/jimmunol.1203017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ferrante CJ, Pinhal-Enfield G, Elson G, Cronstein BN, Hasko G, Outram S, Leibovich SJ. The adenosine-dependent angiogenic switch of macrophages to an M2-like phenotype is independent of interleukin-4 receptor alpha (IL-4Rα) signaling. Inflammation. 2013;36:921–931. doi: 10.1007/s10753-013-9621-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pinhal-Enfield G, Ramanathan M, Hasko G, Vogel SN, Salzman AL, Boons GJ, Leibovich SJ. An angiogenic switch in macrophages involving synergy between Toll-like receptors 2, 4, 7, and 9 and adenosine A(2A) receptors. Am J Pathol. 2003;163:711–721. doi: 10.1016/S0002-9440(10)63698-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leonarduzzi G, Sevanian A, Sottero B, Arkan MC, Biasi F, Chiarpotto E, Basaga H, Poli G. Up-regulation of the fibrogenic cytokine TGF-beta1 by oxysterols: a mechanistic link between cholesterol and atherosclerosis. FASEB J. 2001;15:1619–1621. doi: 10.1096/fj.00-0668fje. [DOI] [PubMed] [Google Scholar]
- 69.Sottero B, Gamba P, Longhi M, Robbesyn F, Abuja PM, Schaur RJ, Poli G, Leonarduzzi G. Expression and synthesis of TGFbeta1 is induced in macrophages by 9-oxononanoyl cholesterol, a major cholesteryl ester oxidation product. Biofactors. 2005;24:209–216. doi: 10.1002/biof.5520240125. [DOI] [PubMed] [Google Scholar]
- 70.Rios FJ, Koga MM, Pecenin M, Ferracini M, Gidlund M, Jancar S. Oxidized LDL induces alternative macrophage phenotype through activation of CD36 and PAFR. Mediators Inflamm. 2013;2013:198193. doi: 10.1155/2013/198193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seo JW, Yang EJ, Yoo KH, Choi IH. Macrophage Differentiation from Monocytes Is Influenced by the Lipid Oxidation Degree of Low Density Lipoprotein. Mediators Inflamm. 2015;2015:235797. doi: 10.1155/2015/235797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim K, Shim D, Lee JS, Zaitsev K, Williams JW, Kim KW, Jang MY, Seok Jang H, Yun TJ, Lee SH, Yoon WK, Prat A, Seidah NG, Choi J, Lee SP, Yoon SH, Nam JW, Seong JK, Oh GT, Randolph GJ, Artyomov MN, Cheong C, Choi JH. Transcriptome Analysis Reveals Nonfoamy Rather Than Foamy Plaque Macrophages Are Proinflammatory in Atherosclerotic Murine Models. Circ Res. 2018;123:1127–1142. doi: 10.1161/CIRCRESAHA.118.312804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kadl A, Meher AK, Sharma PR, Lee MY, Doran AC, Johnstone SR, Elliott MR, Gruber F, Han J, Chen W, Kensler T, Ravichandran KS, Isakson BE, Wamhoff BR, Leitinger N. Identification of a novel macrophage phenotype that develops in response to atherogenic phospholipids via Nrf2. Circ Res. 2010;107:737–746. doi: 10.1161/CIRCRESAHA.109.215715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kadl A, Galkina E, Leitinger N. Induction of CCR2-dependent macrophage accumulation by oxidized phospholipids in the air-pouch model of inflammation. Arthritis Rheum. 2009;60:1362–1371. doi: 10.1002/art.24448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kadl A, Sharma PR, Chen W, Agrawal R, Meher AK, Rudraiah S, Grubbs N, Sharma R, Leitinger N. Oxidized phospholipid-induced inflammation is mediated by Toll-like receptor 2. Free Radic Biol Med. 2011;51:1903–1909. doi: 10.1016/j.freeradbiomed.2011.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Scheuerer B, Ernst M, Dürrbaum-Landmann I, Fleischer J, Grage-Griebenow E, Brandt E, Flad HD, Petersen F. The CXC-chemokine platelet factor 4 promotes monocyte survival and induces monocyte differentiation into macrophages. Blood. 2000;95:1158–1166. [PubMed] [Google Scholar]
- 77.Finn AV, Nakano M, Polavarapu R, Karmali V, Saeed O, Zhao X, Yazdani S, Otsuka F, Davis T, Habib A, Narula J, Kolodgie FD, Virmani R. Hemoglobin directs macrophage differentiation and prevents foam cell formation in human atherosclerotic plaques. J Am Coll Cardiol. 2012;59:166–177. doi: 10.1016/j.jacc.2011.10.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boyle JJ, Harrington HA, Piper E, Elderfield K, Stark J, Landis RC, Haskard DO. Coronary intraplaque hemorrhage evokes a novel atheroprotective macrophage phenotype. Am J Pathol. 2009;174:1097–1108. doi: 10.2353/ajpath.2009.080431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boyle JJ, Johns M, Kampfer T, Nguyen AT, Game L, Schaer DJ, Mason JC, Haskard DO. Activating transcription factor 1 directs Mhem atheroprotective macrophages through coordinated iron handling and foam cell protection. Circ Res. 2012;110:20–33. doi: 10.1161/CIRCRESAHA.111.247577. [DOI] [PubMed] [Google Scholar]
- 80.Howles PN. Cholesterol Absorption and Metabolism. Methods Mol Biol. 2016;1438:177–197. doi: 10.1007/978-1-4939-3661-8_11. [DOI] [PubMed] [Google Scholar]
- 81.Hadadi-Bechor S, Haim Y, Pecht T, Gat R, Tarnovscki T, Gericke M, Rudich A. Autophagy differentially regulates macrophage lipid handling depending on the lipid substrate (oleic acid vs. acetylated-LDL) and inflammatory activation state. Biochim Biophys Acta Mol Cell Biol Lipids. 2019;1864:158527. doi: 10.1016/j.bbalip.2019.158527. [DOI] [PubMed] [Google Scholar]
- 82.Chen W, Chen G, Head DL, Mangelsdorf DJ, Russell DW. Enzymatic reduction of oxysterols impairs LXR signaling in cultured cells and the livers of mice. Cell Metab. 2007;5:73–79. doi: 10.1016/j.cmet.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao GJ, Yin K, Fu YC, Tang CK. The interaction of ApoA-I and ABCA1 triggers signal transduction pathways to mediate efflux of cellular lipids. Mol Med. 2012;18:149–158. doi: 10.2119/molmed.2011.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yvan-Charvet L, Wang N, Tall AR. Role of HDL, ABCA1, and ABCG1 transporters in cholesterol efflux and immune responses. Arterioscler Thromb Vasc Biol. 2010;30:139–143. doi: 10.1161/ATVBAHA.108.179283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pehkonen P, Welter-Stahl L, Diwo J, Ryynänen J, Wienecke-Baldacchino A, Heikkinen S, Treuter E, Steffensen KR, Carlberg C. Genome-wide landscape of liver X receptor chromatin binding and gene regulation in human macrophages. BMC Genomics. 2012;13:50. doi: 10.1186/1471-2164-13-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee SD, Tontonoz P. Liver X receptors at the intersection of lipid metabolism and atherogenesis. Atherosclerosis. 2015;242:29–36. doi: 10.1016/j.atherosclerosis.2015.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schulman IG. Liver X receptors link lipid metabolism and inflammation. FEBS Lett. 2017;591:2978–2991. doi: 10.1002/1873-3468.12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fessler MB. The challenges and promise of targeting the Liver X Receptors for treatment of inflammatory disease. Pharmacol Ther. 2018;181:1–12. doi: 10.1016/j.pharmthera.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Eberlé D, Hegarty B, Bossard P, Ferré P, Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004;86:839–848. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 90.Sato R. Sterol metabolism and SREBP activation. Arch Biochem Biophys. 2010;501:177–181. doi: 10.1016/j.abb.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 91.Wang Y, Rogers PM, Su C, Varga G, Stayrook KR, Burris TP. Regulation of cholesterologenesis by the oxysterol receptor, LXRalpha. J Biol Chem. 2008;283:26332–26339. doi: 10.1074/jbc.M804808200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Traversari C, Russo V. Control of the immune system by oxysterols and cancer development. Curr Opin Pharmacol. 2012;12:729–735. doi: 10.1016/j.coph.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 93.Guillemot-Legris O, Mutemberezi V, Muccioli GG. Oxysterols in Metabolic Syndrome: From Bystander Molecules to Bioactive Lipids. Trends Mol Med. 2016;22:594–614. doi: 10.1016/j.molmed.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 94.Zhang L, Reue K, Fong LG, Young SG, Tontonoz P. Feedback regulation of cholesterol uptake by the LXR-IDOL-LDLR axis. Arterioscler Thromb Vasc Biol. 2012;32:2541–2546. doi: 10.1161/ATVBAHA.112.250571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sorrentino V, Zelcer N. Post-transcriptional regulation of lipoprotein receptors by the E3-ubiquitin ligase inducible degrader of the low-density lipoprotein receptor. Curr Opin Lipidol. 2012;23:213–219. doi: 10.1097/MOL.0b013e3283532947. [DOI] [PubMed] [Google Scholar]
- 96.Tang C, Houston BA, Storey C, LeBoeuf RC. Both STAT3 activation and cholesterol efflux contribute to the anti-inflammatory effect of apoA-I/ABCA1 interaction in macrophages. J Lipid Res. 2016;57:848–857. doi: 10.1194/jlr.M065797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.De Nardo D, Labzin LI, Kono H, Seki R, Schmidt SV, Beyer M, Xu D, Zimmer S, Lahrmann C, Schildberg FA, Vogelhuber J, Kraut M, Ulas T, Kerksiek A, Krebs W, Bode N, Grebe A, Fitzgerald ML, Hernandez NJ, Williams BR, Knolle P, Kneilling M, Röcken M, Lütjohann D, Wright SD, Schultze JL, Latz E. High-density lipoprotein mediates anti-inflammatory reprogramming of macrophages via the transcriptional regulator ATF3. Nat Immunol. 2014;15:152–160. doi: 10.1038/ni.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fotakis P, Kothari V, Thomas DG, Westerterp M, Molusky MM, Altin E, Abramowicz S, Wang N, He Y, Heinecke JW, Bornfeldt KE, Tall AR. Anti-Inflammatory Effects of HDL (High-Density Lipoprotein) in Macrophages Predominate Over Proinflammatory Effects in Atherosclerotic Plaques. Arterioscler Thromb Vasc Biol. 2019;39:e253–e272. doi: 10.1161/ATVBAHA.119.313253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Azzam KM, Fessler MB. Crosstalk between reverse cholesterol transport and innate immunity. Trends Endocrinol Metab. 2012;23:169–178. doi: 10.1016/j.tem.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fessler MB, Parks JS. Intracellular lipid flux and membrane microdomains as organizing principles in inflammatory cell signaling. J Immunol. 2011;187:1529–1535. doi: 10.4049/jimmunol.1100253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Röszer T. Transcriptional control of apoptotic cell clearance by macrophage nuclear receptors. Apoptosis. 2017;22:284–294. doi: 10.1007/s10495-016-1310-x. [DOI] [PubMed] [Google Scholar]
- 102.Zhao Q, Zhou D, You H, Lou B, Zhang Y, Tian Y, Guo N, Chen X, Liu Y, Wu Y, Yuan Z, Zhou J. IFN-γ aggravates neointimal hyperplasia by inducing endoplasmic reticulum stress and apoptosis in macrophages by promoting ubiquitin-dependent liver X receptor-α degradation. FASEB J. 2017;31:5321–5331. doi: 10.1096/fj.201700327R. [DOI] [PubMed] [Google Scholar]
- 103.Du HP, Li J, You SJ, Wang YL, Wang F, Cao YJ, Hu LF, Liu CF. DNA methylation in cystathionine-γ-lyase (CSE) gene promoter induced by ox-LDL in macrophages and in apoE knockout mice. Biochem Biophys Res Commun. 2016;469:776–782. doi: 10.1016/j.bbrc.2015.11.132. [DOI] [PubMed] [Google Scholar]
- 104.Chistiakov DA, Orekhov AN, Bobryshev YV. Treatment of cardiovascular pathology with epigenetically active agents: Focus on natural and synthetic inhibitors of DNA methylation and histone deacetylation. Int J Cardiol. 2017;227:66–82. doi: 10.1016/j.ijcard.2016.11.204. [DOI] [PubMed] [Google Scholar]
- 105.Wang X, Wang S, Yao G, Yu D, Chen K, Tong Q, Ye L, Wu C, Sun Y, Li H, Hermann DM, Doeppner TR, Jin F, Dai Y, Wu J. Identification of the histone lysine demethylase KDM4A/JMJD2A as a novel epigenetic target in M1 macrophage polarization induced by oxidized LDL. Oncotarget. 2017;8:114442–114456. doi: 10.18632/oncotarget.17748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bekkering S, Joosten LA, van der Meer JW, Netea MG, Riksen NP. The epigenetic memory of monocytes and macrophages as a novel drug target in atherosclerosis. Clin Ther. 2015;37:914–923. doi: 10.1016/j.clinthera.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 107.Shan K, Jiang Q, Wang XQ, Wang YN, Yang H, Yao MD, Liu C, Li XM, Yao J, Liu B, Zhang YY, J Y, Yan B. Role of long non-coding RNA-RNCR3 in atherosclerosis-related vascular dysfunction. Cell Death Dis. 2016;7:e2248. doi: 10.1038/cddis.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hu YW, Zhao JY, Li SF, Huang JL, Qiu YR, Ma X, Wu SG, Chen ZP, Hu YR, Yang JY, Wang YC, Gao JJ, Sha YH, Zheng L, Wang Q. RP5-833A20.1/miR-382-5p/NFIA-dependent signal transduction pathway contributes to the regulation of cholesterol homeostasis and inflammatory reaction. Arterioscler Thromb Vasc Biol. 2015;35:87–101. doi: 10.1161/ATVBAHA.114.304296. [DOI] [PubMed] [Google Scholar]
- 109.Giral H, Kratzer A, Landmesser U. MicroRNAs in lipid metabolism and atherosclerosis. Best Pract Res Clin Endocrinol Metab. 2016;30:665–676. doi: 10.1016/j.beem.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 110.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 114.Monteys AM, Spengler RM, Wan J, Tecedor L, Lennox KA, Xing Y, Davidson BL. Structure and activity of putative intronic miRNA promoters. RNA. 2010;16:495–505. doi: 10.1261/rna.1731910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ramalingam P, Palanichamy JK, Singh A, Das P, Bhagat M, Kassab MA, Sinha S, Chattopadhyay P. Biogenesis of intronic miRNAs located in clusters by independent transcription and alternative splicing. RNA. 2014;20:76–87. doi: 10.1261/rna.041814.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Abdelfattah AM, Park C, Choi MY. Update on non-canonical microRNAs. Biomol Concepts. 2014;5:275–287. doi: 10.1515/bmc-2014-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yeom KH, Lee Y, Han J, Suh MR, Kim VN. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 2006;34:4622–4629. doi: 10.1093/nar/gkl458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 122.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 123.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zeng Y, Cullen BR. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32:4776–4785. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang X, Xu X, Ma Z, Huo Y, Xiao Z, Li Y, Wang Y. Dynamic mechanisms for pre-miRNA binding and export by Exportin-5. RNA. 2011;17:1511–1528. doi: 10.1261/rna.2732611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 127.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 128.Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S, Liu Q, Tomari Y. ATP-dependent human RISC assembly pathways. Nat Struct Mol Biol. 2010;17:17–23. doi: 10.1038/nsmb.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 130.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 131.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 132.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Moore MJ, Scheel TK, Luna JM, Park CY, Fak JJ, Nishiuchi E, Rice CM, Darnell RB. miRNA-target chimeras reveal miRNA 3'-end pairing as a major determinant of Argonaute target specificity. Nat Commun. 2015;6:8864. doi: 10.1038/ncomms9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Benhamed M, Herbig U, Ye T, Dejean A, Bischof O. Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nat Cell Biol. 2012;14:266–275. doi: 10.1038/ncb2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 139.Deng Q, Hu H, Yu X, Liu S, Wang L, Chen W, Zhang C, Zeng Z, Cao Y, Xu-Monette ZY, Li L, Zhang M, Rosenfeld S, Bao S, Hsi E, Young KH, Lu Z, Li Y. Tissue-specific microRNA expression alters cancer susceptibility conferred by a TP53 noncoding variant. Nat Commun. 2019;10:5061. doi: 10.1038/s41467-019-13002-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Schulte C, Zeller T. microRNA-based diagnostics and therapy in cardiovascular disease-Summing up the facts. Cardiovasc Diagn Ther. 2015;5:17–36. doi: 10.3978/j.issn.2223-3652.2014.12.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Genemaras AA, Ennis H, Kaplan L, Huang CY. Inflammatory cytokines induce specific time- and concentration-dependent MicroRNA release by chondrocytes, synoviocytes, and meniscus cells. J Orthop Res. 2016;34:779–790. doi: 10.1002/jor.23086. [DOI] [PubMed] [Google Scholar]
- 142.De Rosa R, De Rosa S, Leistner D, Boeckel JN, Keller T, Fichtlscherer S, Dimmeler S, Zeiher AM. Transcoronary Concentration Gradient of microRNA-133a and Outcome in Patients With Coronary Artery Disease. Am J Cardiol. 2017;120:15–24. doi: 10.1016/j.amjcard.2017.03.264. [DOI] [PubMed] [Google Scholar]
- 143.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kapustin AN, Chatrou ML, Drozdov I, Zheng Y, Davidson SM, Soong D, Furmanik M, Sanchis P, De Rosales RT, Alvarez-Hernandez D, Shroff R, Yin X, Muller K, Skepper JN, Mayr M, Reutelingsperger CP, Chester A, Bertazzo S, Schurgers LJ, Shanahan CM. Vascular smooth muscle cell calcification is mediated by regulated exosome secretion. Circ Res. 2015;116:1312–1323. doi: 10.1161/CIRCRESAHA.116.305012. [DOI] [PubMed] [Google Scholar]
- 147.Li L, Zhu D, Huang L, Zhang J, Bian Z, Chen X, Liu Y, Zhang CY, Zen K. Argonaute 2 complexes selectively protect the circulating microRNAs in cell-secreted microvesicles. PLoS One. 2012;7:e46957. doi: 10.1371/journal.pone.0046957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Parahuleva MS, Lipps C, Parviz B, Hölschermann H, Schieffer B, Schulz R, Euler G. MicroRNA expression profile of human advanced coronary atherosclerotic plaques. Sci Rep. 2018;8:7823. doi: 10.1038/s41598-018-25690-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhu JJ, Liu YF, Zhang YP, Zhao CR, Yao WJ, Li YS, Wang KC, Huang TS, Pang W, Wang XF, Wang X, Chien S, Zhou J. VAMP3 and SNAP23 mediate the disturbed flow-induced endothelial microRNA secretion and smooth muscle hyperplasia. Proc Natl Acad Sci USA. 2017;114:8271–8276. doi: 10.1073/pnas.1700561114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tabet F, Vickers KC, Cuesta Torres LF, Wiese CB, Shoucri BM, Lambert G, Catherinet C, Prado-Lourenco L, Levin MG, Thacker S, Sethupathy P, Barter PJ, Remaley AT, Rye KA. HDL-transferred microRNA-223 regulates ICAM-1 expression in endothelial cells. Nat Commun. 2014;5:3292. doi: 10.1038/ncomms4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Näär AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ouimet M, Ediriweera H, Afonso MS, Ramkhelawon B, Singaravelu R, Liao X, Bandler RC, Rahman K, Fisher EA, Rayner KJ, Pezacki JP, Tabas I, Moore KJ. microRNA-33 Regulates Macrophage Autophagy in Atherosclerosis. Arterioscler Thromb Vasc Biol. 2017;37:1058–1067. doi: 10.1161/ATVBAHA.116.308916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Karunakaran D, Thrush AB, Nguyen MA, Richards L, Geoffrion M, Singaravelu R, Ramphos E, Shangari P, Ouimet M, Pezacki JP, Moore KJ, Perisic L, Maegdefessel L, Hedin U, Harper ME, Rayner KJ. Macrophage Mitochondrial Energy Status Regulates Cholesterol Efflux and Is Enhanced by Anti-miR33 in Atherosclerosis. Circ Res. 2015;117:266–278. doi: 10.1161/CIRCRESAHA.117.305624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang YS, Hsi E, Cheng HY, Hsu SH, Liao YC, Juo SH. Let-7g suppresses both canonical and non-canonical NF-κB pathways in macrophages leading to anti-atherosclerosis. Oncotarget. 2017;8:101026–101041. doi: 10.18632/oncotarget.18197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Li CH, Gong D, Chen LY, Zhang M, Xia XD, Cheng HP, Huang C, Zhao ZW, Zheng XL, Tang XE, Tang CK. Puerarin promotes ABCA1-mediated cholesterol efflux and decreases cellular lipid accumulation in THP-1 macrophages. Eur J Pharmacol. 2017;811:74–86. doi: 10.1016/j.ejphar.2017.05.055. [DOI] [PubMed] [Google Scholar]
- 157.Xu J, Hu G, Lu M, Xiong Y, Li Q, Chang CC, Song B, Chang T, Li B. MiR-9 reduces human acyl-coenzyme A:cholesterol acyltransferase-1 to decrease THP-1 macrophage-derived foam cell formation. Acta Biochim Biophys Sin (Shanghai) 2013;45:953–962. doi: 10.1093/abbs/gmt096. [DOI] [PubMed] [Google Scholar]
- 158.Wei Y, Corbalán-Campos J, Gurung R, Natarelli L, Zhu M, Exner N, Erhard F, Greulich F, Geißler C, Uhlenhaut NH, Zimmer R, Schober A. Dicer in Macrophages Prevents Atherosclerosis by Promoting Mitochondrial Oxidative Metabolism. Circulation. 2018;138:2007–2020. doi: 10.1161/CIRCULATIONAHA.117.031589. [DOI] [PubMed] [Google Scholar]
- 159.Liang X, Xu Z, Yuan M, Zhang Y, Zhao B, Wang J, Zhang A, Li G. MicroRNA-16 suppresses the activation of inflammatory macrophages in atherosclerosis by targeting PDCD4. Int J Mol Med. 2016;37:967–975. doi: 10.3892/ijmm.2016.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Huang C, Yu XH, Zheng XL, Ou X, Tang CK. Interferon-stimulated gene 15 promotes cholesterol efflux by activating autophagy via the miR-17-5p/Beclin-1 pathway in THP-1 macrophage-derived foam cells. Eur J Pharmacol. 2018;827:13–21. doi: 10.1016/j.ejphar.2018.02.042. [DOI] [PubMed] [Google Scholar]
- 161.Chen H, Li X, Liu S, Gu L, Zhou X. MircroRNA-19a promotes vascular inflammation and foam cell formation by targeting HBP-1 in atherogenesis. Sci Rep. 2017;7:12089. doi: 10.1038/s41598-017-12167-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lv YC, Tang YY, Peng J, Zhao GJ, Yang J, Yao F, Ouyang XP, He PP, Xie W, Tan YL, Zhang M, Liu D, Tang DP, Cayabyab FS, Zheng XL, Zhang DW, Tian GP, Tang CK. MicroRNA-19b promotes macrophage cholesterol accumulation and aortic atherosclerosis by targeting ATP-binding cassette transporter A1. Atherosclerosis. 2014;236:215–226. doi: 10.1016/j.atherosclerosis.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 163.Lv YC, Yang J, Yao F, Xie W, Tang YY, Ouyang XP, He PP, Tan YL, Li L, Zhang M, Liu D, Cayabyab FS, Zheng XL, Tang CK. Diosgenin inhibits atherosclerosis via suppressing the MiR-19b-induced downregulation of ATP-binding cassette transporter A1. Atherosclerosis. 2015;240:80–89. doi: 10.1016/j.atherosclerosis.2015.02.044. [DOI] [PubMed] [Google Scholar]
- 164.Liang B, Wang X, Song X, Bai R, Yang H, Yang Z, Xiao C, Bian Y. MicroRNA-20a/b regulates cholesterol efflux through post-transcriptional repression of ATP-binding cassette transporter A1. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862:929–938. doi: 10.1016/j.bbalip.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 165.Feng J, Li A, Deng J, Yang Y, Dang L, Ye Y, Li Y, Zhang W. miR-21 attenuates lipopolysaccharide-induced lipid accumulation and inflammatory response: potential role in cerebrovascular disease. Lipids Health Dis. 2014;13:27. doi: 10.1186/1476-511X-13-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Canfrán-Duque A, Rotllan N, Zhang X, Fernández-Fuertes M, Ramírez-Hidalgo C, Araldi E, Daimiel L, Busto R, Fernández-Hernando C, Suárez Y. Macrophage deficiency of miR-21 promotes apoptosis, plaque necrosis, and vascular inflammation during atherogenesis. EMBO Mol Med. 2017;9:1244–1262. doi: 10.15252/emmm.201607492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yang S, Ye ZM, Chen S, Luo XY, Chen SL, Mao L, Li Y, Jin H, Yu C, Xiang FX, Xie MX, Chang J, Xia YP, Hu B. MicroRNA-23a-5p promotes atherosclerotic plaque progression and vulnerability by repressing ATP-binding cassette transporter A1/G1 in macrophages. J Mol Cell Cardiol. 2018;123:139–149. doi: 10.1016/j.yjmcc.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 168.Di Gregoli K, Jenkins N, Salter R, White S, Newby AC, Johnson JL. MicroRNA-24 regulates macrophage behavior and retards atherosclerosis. Arterioscler Thromb Vasc Biol. 2014;34:1990–2000. doi: 10.1161/ATVBAHA.114.304088. [DOI] [PubMed] [Google Scholar]
- 169.Zhang M, Wu JF, Chen WJ, Tang SL, Mo ZC, Tang YY, Li Y, Wang JL, Liu XY, Peng J, Chen K, He PP, Lv YC, Ouyang XP, Yao F, Tang DP, Cayabyab FS, Zhang DW, Zheng XL, Tian GP, Tang CK. MicroRNA-27a/b regulates cellular cholesterol efflux, influx and esterification/hydrolysis in THP-1 macrophages. Atherosclerosis. 2014;234:54–64. doi: 10.1016/j.atherosclerosis.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 170.Li X, Feng S, Luo Y, Long K, Lin Z, Ma J, Jiang A, Jin L, Tang Q, Li M, Wang X. Expression profiles of microRNAs in oxidized low-density lipoprotein-stimulated RAW 264.7 cells. In Vitro Cell Dev Biol Anim. 2018;54:99–110. doi: 10.1007/s11626-017-0225-3. [DOI] [PubMed] [Google Scholar]
- 171.Ceolotto G, Giannella A, Albiero M, Kuppusamy M, Radu C, Simioni P, Garlaschelli K, Baragetti A, Catapano AL, Iori E, Fadini GP, Avogaro A, Vigili de Kreutzenberg S. miR-30c-5p regulates macrophage-mediated inflammation and pro-atherosclerosis pathways. Cardiovasc Res. 2017;113:1627–1638. doi: 10.1093/cvr/cvx157. [DOI] [PubMed] [Google Scholar]
- 172.Zhao GJ, Mo ZC, Tang SL, Ouyang XP, He PP, Lv YC, Yao F, Tan YL, Xie W, Shi JF, Wang Y, Zhang M, Liu D, Tang DP, Zheng XL, Tian GP, Tang CK. Chlamydia pneumoniae negatively regulates ABCA1 expression via TLR2-Nuclear factor-kappa B and miR-33 pathways in THP-1 macrophage-derived foam cells. Atherosclerosis. 2014;235:519–525. doi: 10.1016/j.atherosclerosis.2014.05.943. [DOI] [PubMed] [Google Scholar]
- 173.Gao JH, Zeng MY, Yu XH, Zeng GF, He LH, Zheng XL, Zhang DW, Ouyang XP, Tang CK. Visceral adipose tissue-derived serine protease inhibitor accelerates cholesterol efflux by up-regulating ABCA1 expression via the NF-κB/miR-33a pathway in THP-1 macropahge-derived foam cells. Biochem Biophys Res Commun. 2018;500:318–324. doi: 10.1016/j.bbrc.2018.04.066. [DOI] [PubMed] [Google Scholar]
- 174.Kim SH, Kim GJ, Umemura T, Lee SG, Cho KJ. Aberrant expression of plasma microRNA-33a in an atherosclerosis-risk group. Mol Biol Rep. 2017;44:79–88. doi: 10.1007/s11033-016-4082-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhao Q, Li S, Li N, Yang X, Ma S, Yang A, Zhang H, Yang S, Mao C, Xu L, Gao T, Yang X, Zhang H, Jiang Y. miR-34a Targets HDAC1-Regulated H3K9 Acetylation on Lipid Accumulation Induced by Homocysteine in Foam Cells. J Cell Biochem. 2017;118:4617–4627. doi: 10.1002/jcb.26126. [DOI] [PubMed] [Google Scholar]
- 176.Dai Y, Wu X, Dai D, Li J, Mehta JL. MicroRNA-98 regulates foam cell formation and lipid accumulation through repression of LOX-1. Redox Biol. 2018;16:255–262. doi: 10.1016/j.redox.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hueso M, De Ramon L, Navarro E, Ripoll E, Cruzado JM, Grinyo JM, Torras J. Silencing of CD40 in vivo reduces progression of experimental atherogenesis through an NF-κB/miR-125b axis and reveals new potential mediators in the pathogenesis of atherosclerosis. Atherosclerosis. 2016;255:80–89. doi: 10.1016/j.atherosclerosis.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 178.Chen T, Huang Z, Wang L, Wang Y, Wu F, Meng S, Wang C. MicroRNA-125a-5p partly regulates the inflammatory response, lipid uptake, and ORP9 expression in oxLDL-stimulated monocyte/macrophages. Cardiovasc Res. 2009;83:131–139. doi: 10.1093/cvr/cvp121. [DOI] [PubMed] [Google Scholar]
- 179.Peng XP, Huang L, Liu ZH. miRNA-133a attenuates lipid accumulation via TR4-CD36 pathway in macrophages. Biochimie. 2016;127:79–85. doi: 10.1016/j.biochi.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 180.Lan G, Xie W, Li L, Zhang M, Liu D, Tan YL, Cheng HP, Gong D, Huang C, Zheng XL, Yin WD, Tang CK. MicroRNA-134 actives lipoprotein lipase-mediated lipid accumulation and inflammatory response by targeting angiopoietin-like 4 in THP-1 macrophages. Biochem Biophys Res Commun. 2016;472:410–417. doi: 10.1016/j.bbrc.2015.10.158. [DOI] [PubMed] [Google Scholar]
- 181.Ye Q, Tian GP, Cheng HP, Zhang X, Ou X, Yu XH, Tan RQ, Yang FY, Gong D, Huang C, Pan YJ, Zhang J, Chen LY, Zhao ZW, Xie W, Li L, Zhang M, Xia XD, Zheng XL, Tang CK. MicroRNA-134 Promotes the Development of Atherosclerosis Via the ANGPTL4/LPL Pathway in Apolipoprotein E Knockout Mice. J Atheroscler Thromb. 2018;25:244–253. doi: 10.5551/jat.40212. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 182.Ramírez CM, Rotllan N, Vlassov AV, Dávalos A, Li M, Goedeke L, Aranda JF, Cirera-Salinas D, Araldi E, Salerno A, Wanschel A, Zavadil J, Castrillo A, Kim J, Suárez Y, Fernández-Hernando C. Control of cholesterol metabolism and plasma high-density lipoprotein levels by microRNA-144. Circ Res. 2013;112:1592–1601. doi: 10.1161/CIRCRESAHA.112.300626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hu YW, Hu YR, Zhao JY, Li SF, Ma X, Wu SG, Lu JB, Qiu YR, Sha YH, Wang YC, Gao JJ, Zheng L, Wang Q. An agomir of miR-144-3p accelerates plaque formation through impairing reverse cholesterol transport and promoting pro-inflammatory cytokine production. PLoS One. 2014;9:e94997. doi: 10.1371/journal.pone.0094997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Li K, Ching D, Luk FS, Raffai RL. Apolipoprotein E enhances microRNA-146a in monocytes and macrophages to suppress nuclear factor-κB-driven inflammation and atherosclerosis. Circ Res. 2015;117:e1–e11. doi: 10.1161/CIRCRESAHA.117.305844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lin N, An Y. Blockade of 146b-5p promotes inflammation in atherosclerosis-associated foam cell formation by targeting TRAF6. Exp Ther Med. 2017;14:5087–5092. doi: 10.3892/etm.2017.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yang A, Sun Y, Gao Y, Yang S, Mao C, Ding N, Deng M, Wang Y, Yang X, Jia Y, Zhang H, Jiang Y. Reciprocal Regulation Between miR-148a/152 and DNA Methyltransferase 1 Is Associated with Hyperhomocysteinemia-Accelerated Atherosclerosis. DNA Cell Biol. 2017;36:462–474. doi: 10.1089/dna.2017.3651. [DOI] [PubMed] [Google Scholar]
- 187.Li J, Zhang S. microRNA-150 inhibits the formation of macrophage foam cells through targeting adiponectin receptor 2. Biochem Biophys Res Commun. 2016;476:218–224. doi: 10.1016/j.bbrc.2016.05.096. [DOI] [PubMed] [Google Scholar]
- 188.Tian FJ, An LN, Wang GK, Zhu JQ, Li Q, Zhang YY, Zeng A, Zou J, Zhu RF, Han XS, Shen N, Yang HT, Zhao XX, Huang S, Qin YW, Jing Q. Elevated microRNA-155 promotes foam cell formation by targeting HBP1 in atherogenesis. Cardiovasc Res. 2014;103:100–110. doi: 10.1093/cvr/cvu070. [DOI] [PubMed] [Google Scholar]
- 189.Li X, Kong D, Chen H, Liu S, Hu H, Wu T, Wang J, Chen W, Ning Y, Li Y, Lu Z. miR-155 acts as an anti-inflammatory factor in atherosclerosis-associated foam cell formation by repressing calcium-regulated heat stable protein 1. Sci Rep. 2016;6:21789. doi: 10.1038/srep21789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zhang F, Zhao J, Sun D, Wei N. MiR-155 inhibits transformation of macrophages into foam cells via regulating CEH expression. Biomed Pharmacother. 2018;104:645–651. doi: 10.1016/j.biopha.2018.05.068. [DOI] [PubMed] [Google Scholar]
- 191.Song J, Yang S, Yin R, Xiao Q, Ma A, Pan X. MicroRNA-181a regulates the activation of the NLRP3 inflammatory pathway by targeting MEK1 in THP-1 macrophages stimulated by ox-LDL. J Cell Biochem. 2019;120:13640–13650. doi: 10.1002/jcb.28637. [DOI] [PubMed] [Google Scholar]
- 192.Du XJ, Lu JM, Sha Y. MiR-181a inhibits vascular inflammation induced by ox-LDL via targeting TLR4 in human macrophages. J Cell Physiol. 2018;233:6996–7003. doi: 10.1002/jcp.26622. [DOI] [PubMed] [Google Scholar]
- 193.Zhang XF, Yang Y, Yang XY, Tong Q. MiR-188-3p upregulation results in the inhibition of macrophage proinflammatory activities and atherosclerosis in ApoE-deficient mice. Thromb Res. 2018;171:55–61. doi: 10.1016/j.thromres.2018.09.043. [DOI] [PubMed] [Google Scholar]
- 194.Miao H, Zeng H, Gong H. microRNA-212 promotes lipid accumulation and attenuates cholesterol efflux in THP-1 human macrophages by targeting SIRT1. Gene. 2018;643:55–60. doi: 10.1016/j.gene.2017.11.058. [DOI] [PubMed] [Google Scholar]
- 195.Gong D, Cheng HP, Xie W, Zhang M, Liu D, Lan G, Huang C, Zhao ZW, Chen LY, Yao F, Tan YL, Li L, Xia XD, Zheng XL, Wang ZB, Tang CK. Cystathionine γ-lyase(CSE)/hydrogen sulfide system is regulated by miR-216a and influences cholesterol efflux in macrophages via the PI3K/AKT/ABCA1 pathway. Biochem Biophys Res Commun. 2016;470:107–116. doi: 10.1016/j.bbrc.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 196.Liu H, Li X, Song Y, Wang Z. MicroRNA-217 attenuates intima-media complex thickness of ascending aorta measured by ultrasound bio-microscopy and inhibits inflammation and lipid metabolism in atherosclerotic models of ApoE-/- mice. Lipids Health Dis. 2018;17:170. doi: 10.1186/s12944-018-0825-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhuang X, Li R, Maimaitijiang A, Liu R, Yan F, Hu H, Gao X, Shi H. miR-221-3p inhibits oxidized low-density lipoprotein induced oxidative stress and apoptosis via targeting a disintegrin and metalloprotease-22. J Cell Biochem. 2019;120:6304–6314. doi: 10.1002/jcb.27917. [DOI] [PubMed] [Google Scholar]
- 198.Wang J, Bai X, Song Q, Fan F, Hu Z, Cheng G, Zhang Y. miR-223 Inhibits Lipid Deposition and Inflammation by Suppressing Toll-Like Receptor 4 Signaling in Macrophages. Int J Mol Sci. 2015;16:24965–24982. doi: 10.3390/ijms161024965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Meiler S, Baumer Y, Toulmin E, Seng K, Boisvert WA. MicroRNA 302a is a novel modulator of cholesterol homeostasis and atherosclerosis. Arterioscler Thromb Vasc Biol. 2015;35:323–331. doi: 10.1161/ATVBAHA.114.304878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zhang X, Ye Q, Gong D, Lv Y, Cheng H, Huang C, Chen L, Zhao Z, Li L, Wei X, Zhang M, Xia X, Yu X, Zheng X, Wang S, Wang Z, Tang C. Apelin-13 inhibits lipoprotein lipase expression via the APJ/PKCα/miR-361-5p signaling pathway in THP-1 macrophage-derived foam cells. Acta Biochim Biophys Sin (Shanghai) 2017;49:530–540. doi: 10.1093/abbs/gmx038. [DOI] [PubMed] [Google Scholar]
- 201.Wang D, Yan X, Xia M, Yang Y, Li D, Li X, Song F, Ling W. Coenzyme Q10 promotes macrophage cholesterol efflux by regulation of the activator protein-1/miR-378/ATP-binding cassette transporter G1-signaling pathway. Arterioscler Thromb Vasc Biol. 2014;34:1860–1870. doi: 10.1161/ATVBAHA.113.302879. [DOI] [PubMed] [Google Scholar]
- 202.Liu D, Zhang M, Xie W, Lan G, Cheng HP, Gong D, Huang C, Lv YC, Yao F, Tan YL, Li L, Zheng XL, Tang CK. MiR-486 regulates cholesterol efflux by targeting HAT1. Biochem Biophys Res Commun. 2016;472:418–424. doi: 10.1016/j.bbrc.2015.11.128. [DOI] [PubMed] [Google Scholar]
- 203.Cui J, Ren Z, Zou W, Jiang Y. miR-497 accelerates oxidized low-density lipoprotein-induced lipid accumulation in macrophages by repressing the expression of apelin. Cell Biol Int. 2017;41:1012–1019. doi: 10.1002/cbin.10808. [DOI] [PubMed] [Google Scholar]
- 204.Li BR, Xia LQ, Liu J, Liao LL, Zhang Y, Deng M, Zhong HJ, Feng TT, He PP, Ouyang XP. miR-758-5p regulates cholesterol uptake via targeting the CD36 3'UTR. Biochem Biophys Res Commun. 2017;494:384–389. doi: 10.1016/j.bbrc.2017.09.150. [DOI] [PubMed] [Google Scholar]
- 205.Stahlhut C, Slack FJ. Combinatorial Action of MicroRNAs let-7 and miR-34 Effectively Synergizes with Erlotinib to Suppress Non-small Cell Lung Cancer Cell Proliferation. Cell Cycle. 2015;14:2171–2180. doi: 10.1080/15384101.2014.1003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Wu G, Huang P, Ju X, Li Z, Wang Y. Lin28B over-expression mediates the repression of let-7 by hepatitis B virus X protein in hepatoma cells. Int J Clin Exp Med. 2015;8:15108–15116. [PMC free article] [PubMed] [Google Scholar]
- 207.Brennan E, Wang B, McClelland A, Mohan M, Marai M, Beuscart O, Derouiche S, Gray S, Pickering R, Tikellis C, de Gaetano M, Barry M, Belton O, Ali-Shah ST, Guiry P, Jandeleit-Dahm KAM, Cooper ME, Godson C, Kantharidis P. Protective Effect of let-7 miRNA Family in Regulating Inflammation in Diabetes-Associated Atherosclerosis. Diabetes. 2017;66:2266–2277. doi: 10.2337/db16-1405. [DOI] [PubMed] [Google Scholar]
- 208.Abdullah M, Berthiaume JM, Willis MS. Tumor necrosis factor receptor-associated factor 6 as a nuclear factor kappa B-modulating therapeutic target in cardiovascular diseases: at the heart of it all. Transl Res. 2018;195:48–61. doi: 10.1016/j.trsl.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Essandoh K, Li Y, Huo J, Fan GC. MiRNA-Mediated Macrophage Polarization and its Potential Role in the Regulation of Inflammatory Response. Shock. 2016;46:122–131. doi: 10.1097/SHK.0000000000000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Pfeiffer JR, McAvoy BL, Fecteau RE, Deleault KM, Brooks SA. CARHSP1 is required for effective tumor necrosis factor alpha mRNA stabilization and localizes to processing bodies and exosomes. Mol Cell Biol. 2011;31:277–286. doi: 10.1128/MCB.00775-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Lindquist JA, Brandt S, Bernhardt A, Zhu C, Mertens PR. The role of cold shock domain proteins in inflammatory diseases. J Mol Med (Berl) 2014;92:207–216. doi: 10.1007/s00109-014-1136-3. [DOI] [PubMed] [Google Scholar]
- 212.Lankat-Buttgereit B, Göke R. The tumour suppressor Pdcd4: recent advances in the elucidation of function and regulation. Biol Cell. 2009;101:309–317. doi: 10.1042/BC20080191. [DOI] [PubMed] [Google Scholar]
- 213.Liu X, Cheng Y, Yang J, Krall TJ, Huo Y, Zhang C. An essential role of PDCD4 in vascular smooth muscle cell apoptosis and proliferation: implications for vascular disease. Am J Physiol Cell Physiol. 2010;298:C1481–C1488. doi: 10.1152/ajpcell.00413.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Vikhreva PN, Kalinichenko SV, Korobko IV. Programmed cell death 4 mechanism of action: The model to be updated? Cell Cycle. 2017;16:1761–1764. doi: 10.1080/15384101.2017.1371881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Ranganathan S, Noyes NC, Migliorini M, Winkles JA, Battey FD, Hyman BT, Smith E, Yepes M, Mikhailenko I, Strickland DK. LRAD3, a novel low-density lipoprotein receptor family member that modulates amyloid precursor protein trafficking. J Neurosci. 2011;31:10836–10846. doi: 10.1523/JNEUROSCI.5065-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Noyes NC, Hampton B, Migliorini M, Strickland DK. Regulation of Itch and Nedd4 E3 Ligase Activity and Degradation by LRAD3. Biochemistry. 2016;55:1204–1213. doi: 10.1021/acs.biochem.5b01218. [DOI] [PubMed] [Google Scholar]
- 217.Cora' D, Re A, Caselle M, Bussolino F. MicroRNA-mediated regulatory circuits: outlook and perspectives. Phys Biol. 2017;14:045001. doi: 10.1088/1478-3975/aa6f21. [DOI] [PubMed] [Google Scholar]
- 218.Gui H, Kwan JS, Sham PC, Cherny SS, Li M. Sharing of Genes and Pathways Across Complex Phenotypes: A Multilevel Genome-Wide Analysis. Genetics. 2017;206:1601–1609. doi: 10.1534/genetics.116.198150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Wolfson M, Budovsky A, Tacutu R, Fraifeld V. The signaling hubs at the crossroad of longevity and age-related disease networks. Int J Biochem Cell Biol. 2009;41:516–520. doi: 10.1016/j.biocel.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 221.Vestweber D. VE-cadherin: the major endothelial adhesion molecule controlling cellular junctions and blood vessel formation. Arterioscler Thromb Vasc Biol. 2008;28:223–232. doi: 10.1161/ATVBAHA.107.158014. [DOI] [PubMed] [Google Scholar]
- 222.Haasdijk RA, Den Dekker WK, Cheng C, Tempel D, Szulcek R, Bos FL, Hermkens DM, Chrifi I, Brandt MM, Van Dijk C, Xu YJ, Van De Kamp EH, Blonden LA, Van Bezu J, Sluimer JC, Biessen EA, Van Nieuw Amerongen GP, Duckers HJ. THSD1 preserves vascular integrity and protects against intraplaque haemorrhaging in ApoE-/- mice. Cardiovasc Res. 2016;110:129–139. doi: 10.1093/cvr/cvw015. [DOI] [PubMed] [Google Scholar]
- 223.Shapouri-Moghaddam A, Mohammadian S, Vazini H, Taghadosi M, Esmaeili SA, Mardani F, Seifi B, Mohammadi A, Afshari JT, Sahebkar A. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol. 2018;233:6425–6440. doi: 10.1002/jcp.26429. [DOI] [PubMed] [Google Scholar]
- 224.Toma I, McCaffrey TA. Transforming growth factor-β and atherosclerosis: interwoven atherogenic and atheroprotective aspects. Cell Tissue Res. 2012;347:155–175. doi: 10.1007/s00441-011-1189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Goumans MJ, Ten Dijke P. TGF-β Signaling in Control of Cardiovascular Function. Cold Spring Harb Perspect Biol. 2018;10:a022210. doi: 10.1101/cshperspect.a022210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13:877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Virbasius JV, Czech MP. Map4k4 Signaling Nodes in Metabolic and Cardiovascular Diseases. Trends Endocrinol Metab. 2016;27:484–492. doi: 10.1016/j.tem.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Xu S, Koroleva M, Yin M, Jin ZG. Atheroprotective laminar flow inhibits Hippo pathway effector YAP in endothelial cells. Transl Res. 2016;176:18–28.e2. doi: 10.1016/j.trsl.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Wang Y, Cao W, Cui J, Yu Y, Zhao Y, Shi J, Wu J, Xia Z, Yu B, Liu J. Arterial Wall Stress Induces Phenotypic Switching of Arterial Smooth Muscle Cells in Vascular Remodeling by Activating the YAP/TAZ Signaling Pathway. Cell Physiol Biochem. 2018;51:842–853. doi: 10.1159/000495376. [DOI] [PubMed] [Google Scholar]
- 230.Xiao J, Jin K, Wang J, Ma J, Zhang J, Jiang N, Wang H, Luo X, Fei J, Wang Z, Yang X, Ma D. Conditional knockout of TFPI-1 in VSMCs of mice accelerates atherosclerosis by enhancing AMOT/YAP pathway. Int J Cardiol. 2017;228:605–614. doi: 10.1016/j.ijcard.2016.11.195. [DOI] [PubMed] [Google Scholar]
- 231.Fu Y, Sun S, Sun H, Peng J, Ma X, Bao L, Ji R, Luo C, Gao C, Zhang X, Jin Y. Scutellarin exerts protective effects against atherosclerosis in rats by regulating the Hippo-FOXO3A and PI3K/AKT signaling pathways. J Cell Physiol. 2019;234:18131–18145. doi: 10.1002/jcp.28446. [DOI] [PubMed] [Google Scholar]
- 232.Kozarov E. Bacterial invasion of vascular cell types: vascular infectology and atherogenesis. Future Cardiol. 2012;8:123–138. doi: 10.2217/fca.11.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Caruso P, MacLean MR, Khanin R, McClure J, Soon E, Southgate M, MacDonald RA, Greig JA, Robertson KE, Masson R, Denby L, Dempsie Y, Long L, Morrell NW, Baker AH. Dynamic changes in lung microRNA profiles during the development of pulmonary hypertension due to chronic hypoxia and monocrotaline. Arterioscler Thromb Vasc Biol. 2010;30:716–723. doi: 10.1161/ATVBAHA.109.202028. [DOI] [PubMed] [Google Scholar]
- 234.Caruso P, Dempsie Y, Stevens HC, McDonald RA, Long L, Lu R, White K, Mair KM, McClure JD, Southwood M, Upton P, Xin M, van Rooij E, Olson EN, Morrell NW, MacLean MR, Baker AH. A role for miR-145 in pulmonary arterial hypertension: evidence from mouse models and patient samples. Circ Res. 2012;111:290–300. doi: 10.1161/CIRCRESAHA.112.267591. [DOI] [PubMed] [Google Scholar]
- 235.Sistrunk JW, Shifrin A, Frager M, Bardales RH, Thomas J, Fishman N, Goldberg P, Guttler R, Grant E. Clinical performance of multiplatform mutation panel and microRNA risk classifier in indeterminate thyroid nodules. J Am Soc Cytopathol. 2020 doi: 10.1016/j.jasc.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 236.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 237.Chang J, Guo JT, Jiang D, Guo H, Taylor JM, Block TM. Liver-specific microRNA miR-122 enhances the replication of hepatitis C virus in nonhepatic cells. J Virol. 2008;82:8215–8223. doi: 10.1128/JVI.02575-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Gebert LF, Rebhan MA, Crivelli SE, Denzler R, Stoffel M, Hall J. Miravirsen (SPC3649) can inhibit the biogenesis of miR-122. Nucleic Acids Res. 2014;42:609–621. doi: 10.1093/nar/gkt852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, King BD, Kauppinen S, Levin AA, Hodges MR. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 240.Seto AG, Beatty X, Lynch JM, Hermreck M, Tetzlaff M, Duvic M, Jackson AL. Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br J Haematol. 2018;183:428–444. doi: 10.1111/bjh.15547. [DOI] [PubMed] [Google Scholar]
- 241.Kopp KL, Ralfkiaer U, Gjerdrum LM, Helvad R, Pedersen IH, Litman T, Jønson L, Hagedorn PH, Krejsgaard T, Gniadecki R, Bonefeld CM, Skov L, Geisler C, Wasik MA, Ralfkiaer E, Ødum N, Woetmann A. STAT5-mediated expression of oncogenic miR-155 in cutaneous T-cell lymphoma. Cell Cycle. 2013;12:1939–1947. doi: 10.4161/cc.24987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Beg MS, Brenner AJ, Sachdev J, Borad M, Kang YK, Stoudemire J, Smith S, Bader AG, Kim S, Hong DS. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs. 2017;35:180–188. doi: 10.1007/s10637-016-0407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Hong DS, Kang YK, Borad M, Sachdev J, Ejadi S, Lim HY, Brenner AJ, Park K, Lee JL, Kim TY, Shin S, Becerra CR, Falchook G, Stoudemire J, Martin D, Kelnar K, Peltier H, Bonato V, Bader AG, Smith S, Kim S, O'Neill V, Beg MS. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br J Cancer. 2020;122:1630–1637. doi: 10.1038/s41416-020-0802-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Kelnar K, Bader AG. A qRT-PCR Method for Determining the Biodistribution Profile of a miR-34a Mimic. Methods Mol Biol. 2015;1317:125–133. doi: 10.1007/978-1-4939-2727-2_8. [DOI] [PubMed] [Google Scholar]
- 245.Zhang G, Guo B, Wu H, Tang T, Zhang BT, Zheng L, He Y, Yang Z, Pan X, Chow H, To K, Li Y, Li D, Wang X, Wang Y, Lee K, Hou Z, Dong N, Li G, Leung K, Hung L, He F, Zhang L, Qin L. A delivery system targeting bone formation surfaces to facilitate RNAi-based anabolic therapy. Nat Med. 2012;18:307–314. doi: 10.1038/nm.2617. [DOI] [PubMed] [Google Scholar]
- 246.Leuschner F, Dutta P, Gorbatov R, Novobrantseva TI, Donahoe JS, Courties G, Lee KM, Kim JI, Markmann JF, Marinelli B, Panizzi P, Lee WW, Iwamoto Y, Milstein S, Epstein-Barash H, Cantley W, Wong J, Cortez-Retamozo V, Newton A, Love K, Libby P, Pittet MJ, Swirski FK, Koteliansky V, Langer R, Weissleder R, Anderson DG, Nahrendorf M. Therapeutic siRNA silencing in inflammatory monocytes in mice. Nat Biotechnol. 2011;29:1005–1010. doi: 10.1038/nbt.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Duivenvoorden R, Tang J, Cormode DP, Mieszawska AJ, Izquierdo-Garcia D, Ozcan C, Otten MJ, Zaidi N, Lobatto ME, van Rijs SM, Priem B, Kuan EL, Martel C, Hewing B, Sager H, Nahrendorf M, Randolph GJ, Stroes ES, Fuster V, Fisher EA, Fayad ZA, Mulder WJ. A statin-loaded reconstituted high-density lipoprotein nanoparticle inhibits atherosclerotic plaque inflammation. Nat Commun. 2014;5:3065. doi: 10.1038/ncomms4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Laffont B, Rayner KJ. MicroRNAs in the Pathobiology and Therapy of Atherosclerosis. Can J Cardiol. 2017;33:313–324. doi: 10.1016/j.cjca.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Nazari-Jahantigh M, Wei Y, Noels H, Akhtar S, Zhou Z, Koenen RR, Heyll K, Gremse F, Kiessling F, Grommes J, Weber C, Schober A. MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in macrophages. J Clin Invest. 2012;122:4190–4202. doi: 10.1172/JCI61716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Esper RJ, Nordaby RA. Cardiovascular events, diabetes and guidelines: the virtue of simplicity. Cardiovasc Diabetol. 2019;18:42. doi: 10.1186/s12933-019-0844-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Kataoka Y, St John J, Wolski K, Uno K, Puri R, Tuzcu EM, Nissen SE, Nicholls SJ. Atheroma progression in hyporesponders to statin therapy. Arterioscler Thromb Vasc Biol. 2015;35:990–995. doi: 10.1161/ATVBAHA.114.304477. [DOI] [PubMed] [Google Scholar]
- 252.Nozue T, Yamamoto S, Tohyama S, Fukui K, Umezawa S, Onishi Y, Kunishima T, Sato A, Nozato T, Miyake S, Takeyama Y, Morino Y, Yamauchi T, Muramatsu T, Hibi K, Terashima M, Michishita I TRUTH Investigators. Comparison of arterial remodeling and changes in plaque composition between patients with progression versus regression of coronary atherosclerosis during statin therapy (from the TRUTH study) Am J Cardiol. 2012;109:1247–1253. doi: 10.1016/j.amjcard.2011.12.016. [DOI] [PubMed] [Google Scholar]