Graphical abstract
Keywords: Distant supervision, Medical concept normalization, Deep learning, Text embedding, Graph embedding
Highlights
-
•
Medical Concept Normalization maps informal phrases to formal medical concepts.
-
•
Training MCN models requires creating labeled datasets, which is effort intensive.
-
•
We overcome this problem by creating automatically labeled dataset from social media.
-
•
We augment our distant dataset with examples extracted from the SNOMED CT lexicon.
-
•
Our approach significantly outperforms the previous state-of-the-art on two datasets.
Abstract
We consider the task of Medical Concept Normalization (MCN) which aims to map informal medical phrases such as “loosing weight” to formal medical concepts, such as “Weight loss”. Deep learning models have shown high performance across various MCN datasets containing small number of target concepts along with adequate number of training examples per concept. However, scaling these models to millions of medical concepts entails the creation of much larger datasets which is cost and effort intensive. Recent works have shown that training MCN models using automatically labeled examples extracted from medical knowledge bases partially alleviates this problem. We extend this idea by computationally creating a distant dataset from patient discussion forums. We extract informal medical phrases and medical concepts from these forums using a synthetically trained classifier and an off-the-shelf medical entity linker respectively. We use pretrained sentence encoding models to find the k-nearest phrases corresponding to each medical concept. These mappings are used in combination with the examples obtained from medical knowledge bases to train an MCN model. Our approach outperforms the previous state-of-the-art by 15.9% and 17.1% classification accuracy across two datasets while avoiding manual labeling.
1. Background
Medical social media is a subset of social media focusing on medical topics. It primarily includes medical forums, blogs, and tweets. It draws participation from a variety of cohorts such as patients, caretakers, consultants, doctors, pharmacists, researchers, and journalists [1]. Recent studies have shown that medical social media can be leveraged to find the side effects of a particular drug [2], detect the spread of infectious diseases, monitor public health [3] and understand a patient’s experience in healthcare [4] [5]. However, automatically identifying such insights is challenging due to the lexical and grammatical variability of the language used in social media which contains informal language, non-standard grammar, and typographic errors. Due to the varied backgrounds and expertise levels of its users, medical social media also contains non-standard medical terminology, jargon and abbreviations [6]. The task of Medical Concept Normalization (MCN) aims to map a variable length phrase to a medical concept in some external coding system. Table 1 provides a few examples of social media phrases mapped to medical concepts.1 MCN has several applications in improving patient care, such as the understanding and answering of patients’ questions, early detection of patients requiring immediate attention, and digital disease surveillance [7].
Table 1.
Examples of mappings between social media phrases and medical concepts. SNOMED CT is a medical knowledge base.
| Social Media Phrase | Normalized Medical Concept |
|---|---|
| feel like i was hit by a train | Pain (SNOMED ID: 22253000) |
| no way i ’m gettin any sleep 2nite | Insomnia (SNOMED ID: 193462001) |
| imence pain in legs | Severe pain (SNOMED ID: 76948002) |
| the same gassy feeling | Bloating (SNOMED ID: 60728008) |
Medical entity linking, which is closely related to MCN, links medical entity mentions in the text with their corresponding entities in a medical Knowledge Base (KB) such as SNOMED CT [8]. While both tasks share the primary goal of disambiguation of a sequence of words, there exist a few substantial differences. Medical entity linking operates on medical entity mentions which have an entity type such as Disease, Drug, Symptom, Treatment, and Test whereas MCN operates on phrases which may or may not have an entity type. For instance, the phrase cant shut up for the whole day is not recognized as a medical entity by any medical entity recognizer and yet is mapped to Hyperactive Behavior, SNOMED ID: 44548000 by MCN models [9]. Furthermore, MCN, unlike medical entity linking, allows mapping between the source text and target concept to be loosely defined. For instance, no way i ’m gettin any sleep 2nite could be mapped to Insomnia, SNOMED ID: 193462001. Medical entity linking uses the context of an entity mention along with the information from the medical KB, to decide which entity is being referred to in the text [10]. Lack of such information makes MCN a relatively more challenging task than medical entity linking.
Most of the current deep learning models (Section 2) formulate MCN as a supervised text classification problem. This formulation has several major shortcomings. First, these have shown high performance across various MCN datasets containing small number of target concepts along with adequate number of training examples per concept. However, scaling these models to millions of medical concepts entails the creation of much larger datasets which is cost and effort intensive. Experts have to manually identify medical concept mentions and then map them to their corresponding target medical concepts. Second, these models fail to map a medical phrase to any concept if it is not present in the training set. Third, number of medical concepts increase with advancements in medical science. These models need to be retrained from scratch whenever new concepts are added to the target lexicon. Retraining these models is a computationally expensive process. In our recent work [6] we show that training MCN models using automatically labeled examples extracted from medical knowledge bases (such as SNOMED CT) partially alleviates these problems. In this work, we extend this idea further by computationally creating a distantly supervised dataset from patient discussion forum posts (Section 3). First, we develop synthetic examples to train a binary classifier which identifies if a given input phrase is medical or non-medical and use it to extract all medical phrases from the discussion posts (Section 3.2.1). We also discover the medical concepts within each post using an off-the-shelf medical entity linker (Section 3.2.2). We use pretrained sentence encoding models to find k-nearest phrases corresponding to each medical concept. These mappings are used in combination with examples obtained from the medical knowledge bases to train an MCN model. Experiments (Section 5) on two benchmark datasets (Section 4) reveal that our approach shows 15.9% and 17.1% improvement in classification accuracy over the previous state-of-the-art (Section 6) while eliminating the need for manual labeling. We discuss the performance improvements and the errors introduced by our approach (Section 7) and conclude this work by discussing future research directions (Section 8).
2. Related work
Existing approaches for MCN can be divided into three major categories. The first category formulates MCN as a monolingual translation problem, where the task is to map informal phrases () to formal medical concepts (). Limsopatham et al. [11] adapt phrase-based machine translation [12] to translate medical phrases from Twitter language to formal medical language. During inference, output of the machine translation model is mapped to one of the concepts in medical lexicons based on the ranked similarity of their word vector representations. Similar to [11], statistical machine translation based techniques have also been used to normalize non-medical phrases [13] [14] [15].
The second category poses MCN as a supervised text categorization problem, in which an informal medical phrase is categorized into one of the predefined categories wherein each category represents a unique concept in a medical lexicon. Limsopatham et al. [9] use Convolutional Neural Networks (CNNs) and Recurrent Neural Networks (RNNs) along with the pretrained word embeddings for normalizing medical concepts. Their approach outperforms lexical matching and machine translation based approaches by up to 44%. Lee et al. [7] also employ CNNs and RNNs for normalizing medical concepts. However, instead of using pretrained word embeddings trained on generic text they experiment with word embeddings trained on a variety of clinical sources which improves the performance of their model by up to 21.28%. Belousov et al. [16] use an ensemble of multinomial logistic regression and Bidirectional RNNs for this task and discover that the ensemble model yields the highest accuracy. Tutubalina et al. [17] use bidirectional RNNs with attention to convert a given input phrase into a semantic representative vector, which is then appended with a set of features extracted using cosine similarity between input phrase and medical concept descriptions obtained from the UMLS Metathesaurus. The resultant representation is fed to a softmax classifier which maps it to one of the predefined medical concepts. They discover that appending semantic features (extracted from UMLS) to the deep learning model improves its performance of the model by 5.2%. Almost all models discussed earlier use word embeddings and therefore fail to learn character structure features inside words and ignore the Out-of-Vocabulary (OOV) words. In order to overcome this problem, Niu et al. [18] present a multi-task character-level attentional network model for MCN. Miftahutdinov et al. [19] propose the use of multilayered bidirectional transformer encoder BERT [20] to extract the vector representation of input phrases which are subsequently used to train a softmax classifier.
The third type of MCN methods project the informal phrases and formal medical concepts into a common embedding space. During inference, cosine similarity based ranking is used to retrieve the most similar concept to a given input phrase. Metke-Jimenez et. al. [21] use TF-IDF representation to retrieve relevant medical concepts corresponding to paraphrased concept mentions. In our recent work [6] we propose a RoBERTa [22] based neural model which maps the input phrase and medical concepts into a common embedding space. We leverage existing medical knowledge bases (such as SNOMED CT) to first obtain embeddings for each medical concept using various text and graph based embeddings. Subsequently, we train a neural model to map informal phrases into the target embedding space. We have shown that this approach outperforms all the existing approaches and achieves an improvement of up to 6.3% compared to the previous state-of-the-art [19]. For the extent of this work, we regard [6] as the state-of-the-art for this task.
Most of the recent works [6], [9], [11], [17], [18], [19] use CADEC dataset for experimentation, while some of these [6], [19] also use PsyTAR dataset. Both CADEC and PsyTAR datasets were created from the patient discussion forum askapatient.com. Few works [9], [17], [18], [19], use TwADR-L dataset for experiments which consisted of medical phrases extracted from Twitter mapped to the medical concepts in the SIDER 4 medical lexicon.2 Almost all recent works [6], [9], [11], [12], [13], [14], [15], [16], [17], [18], [19], [21] use classification accuracy as the primary metric for measuring the performance of MCN models.
3. Approach
Main drawback of previous approaches such as [7], [9], [19] is that they rely on the availability of pairs of medical phrases and concepts for training MCN models. Creating such a training dataset entails the painstaking task of manually identifying social media phrases and mapping them to one of the concepts in a medical lexicon. In our prior work [6] we used two techniques to overcome this challenge. First, we encoded all target medical concepts into a common embedding space using a variety of text and graph embeddings methods and thereafter transformed an input phrase into a vector in the target embedding space. This allowed our model to map phrases to even those medical concepts which were not present in the training set. Second, we generated labeled examples for MCN using SNOMED CT synonyms by treating each synonym as a medical phrase. We have shown that our model trained exclusively on SNOMED CT synonyms demonstrates reasonable performance when compared to other approaches. Section 3.1 gives a brief overview of our prior work.
In this work, we propose a novel distant supervision based approach to generate training samples for MCN without any manual labeling or human intervention of any kind. We then augment these with the labeled examples obtained from SNOMED CT and use the resultant dataset for training MCN model. We use the same MCN architecture as proposed in our prior work (Section 3.1). Our main contribution in this work is the automatic creation of distantly supervised MCN dataset which is discussed in Section 3.2. For the extent of this work, we use our prior work [6] as a baseline.
3.1. Medical concept normalization by encoding target knowledge
In this Section, we describe our previous approach for MCN. We used a two staged approach for normalizing medical concept mentions. In the first stage, all medical concepts from a target lexicon (such as SNOMED CT) are encoded into fixed sized embeddings using a variety of text and graph based embedding methods such that similar medical concepts are closer in the target embedding space. In the second stage, each input phrase is transformed into a vector using pretrained RoBERTa model [22] which is then transformed into a vector in the target embedding space using the two layered feed forward neural network shown in Eqs. (1), (2), where and the weight matrices of the RoBERTa model are trainable parameters. All parameters are trained using AdamW [23] stochastic optimizer which aims to maximize the cosine similarity between the transformed vector and the corresponding target embedding. During inference, a new input phrase is mapped to a vector in the target concept embedding space which is then classified to a concept using the 1-NN (nearest neighbour) method.
| (1) |
| (2) |
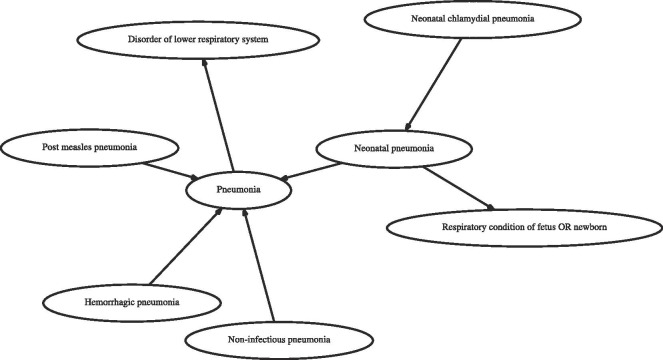
We now describe the process of obtaining target embeddings for medical concepts using text and graph based embedding methods. For text embedding methods we extract the concept description of each target concept ID by doing a lookup in SNOMED CT knowledge base. Table 2 shows few target concept IDs and their corresponding descriptions from SNOMED CT. The descriptions are given as inputs to pretrained sentence encoding models such as Universal Sentence Encoders [24] (denoted, USE) to obtain an embedding corresponding to target concept ID. Other sentence encoding models include Embeddings from Language models [25] (denoted, ELMo), Bidirectional Encoder Representations from Transformers [20] (denoted, BERT) and averaged word embeddings [26] (denoted, AvgEmb). The graph based embeddings are obtained by first constructing a graph using SNOMED CT wherein each vertex is a unique medical concept and related concepts are connected through unlabeled edges as depicted in Fig. 1 . This graph is given as input to graph embedding algorithms such as Deepwalk [27], Node2Vec [28], HARP [29] and LINE [30], each of which return an embedding for every vertex in the graph.
Table 2.
Examples of medical concept IDs and their descriptions obtained from SNOMED CT.
| Concept ID | Concept Description |
|---|---|
| 22298006 | Myocardial Infarction |
| 363518003 | Malignant tumor of kidney |
| 66071002 | Viral hepatitis type B |
| 35031000119100 | Acute aspiration pneumonia |
| 247761005 | Reduced Concentration |
| 79890006 | Loss of appetite |
Fig. 1.
A snapshot of SNOMED CT graph. Related concepts are depicted using unlabeled edges.
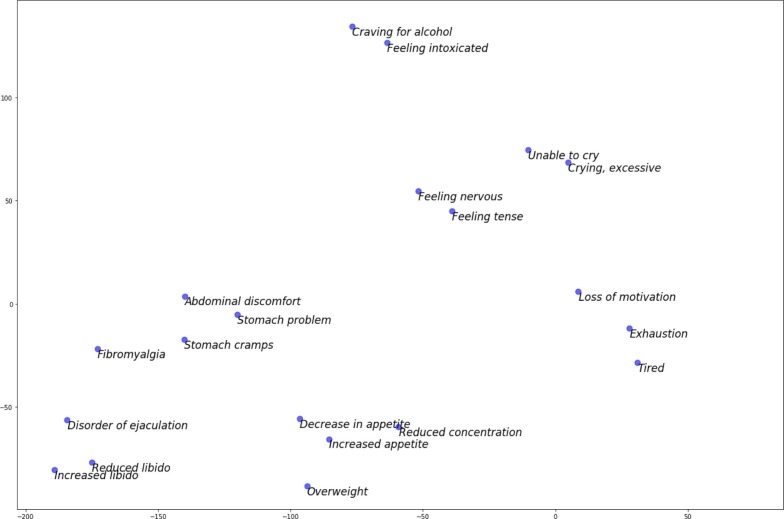
Both text and graph label embedding methods aim to map medical concepts into vectors, such that similar concepts are closer in the target embedding space. These embeddings are made publicly accessible at https://zenodo.org/record/3842143. Fig. 2 uses the T-distributed Stochastic Neighbor Embedding (t-SNE) technique [31] to depict the medical concept embeddings in a two dimensional space, wherein the embeddings were obtained using the pretrained Universal Sentence Encoder (USE) model. Observe that similar concepts such as abdominal discomfort, stomach problems and Stomach cramps are closer in the embedding space, whereas dissimilar concepts such as Craving for alcohol, Loss of motivation, decrease in appetite and Reduced libido are farther in the embedding space.
Fig. 2.
The t-SNE visualization of SNOMED CT concept embeddings obtained from the pretrained Universal Sentence Encoder model. Each point on the plot represents a unique concept in SNOMED CT.
3.2. Automatically creating distantly supervised dataset for MCN
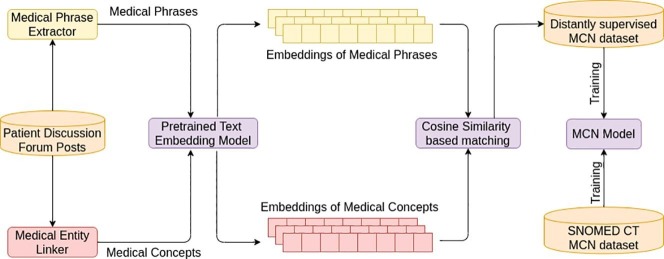
For training the model described in Section 3.1 we create a distantly supervised dataset consisting of automatically extracted pairs of informal medical phrases and concepts, thereby circumventing the need for manual labeling or human intervention of any kind. Fig. 3 shows the architecture of our proposed approach. We first crawl social media posts from patient discussion forums which contains informal medical expressions such as not able 2 sleep 2nite and ma head is bursting as well as formal medical concept descriptions such as Insomnia and Headache. The formal and informal medical phrases are extracted using a binary classifier described in Section 3.2.1 and the medical concepts associated with a post are discovered using an off-the-shelf medical entity linker described in Section 3.2.2. Table 3 shows an example of a social media post, the phrases extracted by medical phrase extractor and the concepts discovered by medical entity linker. The informal medical phrases extracted from a post are matched against the medical concepts described in the same post which results in several (phrase, concept) pairs. Further, k-nearest phrases corresponding to each medical concept are selected using the cosine similarity between their text embeddings, which are obtained by passing them through the pretrained universal sentence encoder model [24], while the remaining pairs are discarded. Consider the medical phrases belly aching, tummy upset, stomach ulcer, stomach cancer which are paired with the medical concept Stomach ache, 271681002 and are ranked according to their cosine similarity with Stomach ache, 271681002. Using , includes the pairs (belly aching, 271681002) and (tummy upset, 271681002) in the distantly supervised set and discards the remaining pairs. The value of k is empirically determined so as to maximize MCN accuracy on a hold-out validation set.
Fig. 3.
Architecture of the proposed Distant Supervision Approach.
Table 3.
A post from patient discussion forum along with the medical phrases and concepts extracted from it.
| Sample Post | Medical Phrases | SNOMED CT Concepts |
|---|---|---|
| I noticed some swollen lymph nodes on the right side of my neck, including one under my chin which is really odd shape. I saw an ENT last November who chalked them up to sinus issues and told me to flush my sinuses twice a day |
|
|
An alternative method for selecting automatically labeled examples could be to select pairs with cosine similarity greater than a pre-designated threshold and discard the remaining pairs. Although this approach seems reasonable, we do not use it as it might introduce label imbalance in our dataset. For instance, common concepts such as Pain might have a high representation in our dataset when compared to rare concepts such as Aarskog syndrome, which is undesirable.
3.2.1. Medical phrase extractor
We train a binary classifier which classifies a textual phrase into medical or non-medical category. We extract all the noun phrases from patient discussion forum posts using Stanford CoreNLP library [32] and use this classifier to filter out non-medical phrases while retaining medical phrases. This results in a collection of formal and informal medical phrases corresponding to each post. We avoid human labeling by using synthetically generated examples to train this classifier. For the medical category, synthetic examples are obtained by extracting all medical concept descriptions from SNOMED CT. For non-medical category, synthetic examples are obtained by extracting noun phrases from a news corpora [33] (non-medical corpora) using Stanford CoreNLP library [32]. We use binary Support Vector Machine (SVM) with radial basis function kernel to categorize the social media phrases, where each phrase is represented by the averaged word vector of the words present in it. The embeddings for each word were obtained from the pretrained Word2Vec model [26].
3.2.2. Medical entity linking
We use MetaMap [34] as an off-the-shelf medical entity linker. It identifies concepts from text and links them to entries of KBs such as SNOMED CT and MeSH [35]. MetaMap assigns one or more semantic types such as, sign or symptom and disease or syndrome to each identified concept. Existing study shows that MetaMap exhibits high precision and poor recall when used to extract concepts from medical social media posts [36]. In this work, precision is of paramount importance for generating high quality distant data. Therefore, we further improve the precision using the following two heuristics. First, medical entities which correspond to one of the major 18 semantic types described by [36] are retained while others are discarded. Second, the most frequent incorrect mappings are pruned using a manually created list. For instance, the generic word Hi, which is highly frequent, is wrongly identified as a disease by MetaMap and is mapped to the medical concept Haemophilus Influenzae, SNOMED ID: 44470000.
4. Datasets
CADEC: CSIRO Adverse Drug Event Corpus (CADEC) [37] is an MCN dataset created from publicly available medical forum posts. All the posts discussing adverse drug reactions resulting from the usage of Diclofenac and Lipitor drugs were obtained from askapatient.com. Human experts were asked to identify medical phrases such as feeling that i was hit by a train. Each phrase was then manually mapped to a medical concept in SNOMED CT lexicon resulting in 6,754 distinct phrases mapped to 1029 unique concepts. Tutubalina et al. [17] created a five fold dataset from these mappings which were made publicly accessible.3
PsyTAR: Psychiatric Treatment Adverse Reactions (PsyTAR) corpus [38] is an MCN dataset created from askapatient.com. In this dataset, all the posts which mention the medications Cymbalta, Effexor, Lexapro and Zoloft were obtained. Similar to the CADEC dataset, human experts were asked to manually discover medical phrases and map to SNOMED CT concepts which resulted in 6,556 medical phrases mapped to 618 concepts. Miftahutdinov et al. [19] created a five fold dataset from these mappings which were made publicly accessible.4 Pattisapu et al. [6] used the publicly accessible folds of the CADEC and PsyTAR datasets to evaluate the performance of their MCN model. For a fair comparison, we use the same folds to evaluate the performance of our approach.
SNOMED CT Synonyms: The SNOMED CT lexicon consists of a collection of medical concepts wherein each medical concept has a unique SNOMED ID. Every concept in this lexicon is associated with its fully specified name and its synonyms. Currently, SNOMED CT contains over 350,000 medical concepts. Table 4 shows few examples of medical concepts obtained from SNOMED CT. Pattisapu et al. [6] were the first to leverage this resource for training their MCN model. They extract the synonyms of medical concepts present in PsyTAR and CADEC datasets and create a automatically labeled dataset by treating each synonym as a medical phrase.
Table 4.
SNOMED-CT Medical Concepts and their Synonyms.
| SNOMED ID | Fully Specified Name | Synonyms |
|---|---|---|
| 271681002 | Stomach ache | belly ache, tummy ache, stomach discomfort, sore tummy, stomach upset |
| 61462000 | Malaria | paludism, plasmodiosis |
| 363518003 | Malignant tumor of kidney | CA - cancer of kidney, renal malignant tumour, CA - renal cancer, renal cancer |
| 424206003 | Genus Ebolavirus | Ebola-like viruses, Ebolavirus, Ebola virus |
| 840539006 | Disease caused by 2019 novel coronavirus | Disease caused by Wuhan coronavirus, Disease caused by 2019-nCoV |
Distantly supervised dataset: For creating the distantly supervised dataset, we first downloaded 6,07,107 posts from the patient discussion forum https://patient.info/forums. These posts primarily consisted of patient conversations about various diseases, drugs and symptoms. The posts that contained non-standard encodings or less than hundred characters were discarded, which resulted in a total of 4,23,782 posts. We extracted 11,381 unique medical concepts using the medical entity linker described in Section 3.2.2. We extracted 1.97 million medical phrases using the medical phrase extractor described in Section 3.2.1. We used the approach discussed in Section 3.2 to create a distantly supervised dataset from the phrase, concept pairs. Table 5 shows a few labeled examples obtained using distant supervision method. Note that it also contains some noisy labels, for instance, the phrase heart burn is wrongly associated with the medical concept Abdominal discomfort with a cosine similarity of 0.5194. We observed that compared to the SNOMED CT synonyms dataset, this dataset has a better coverage of phrases containing slang words, non-standard terminology, acronyms and spelling errors.
Table 5.
Sample labeled examples obtained from distant data.
| Medical Phrase | Medical Concept | Cosine similarity |
|---|---|---|
| Manic episode | manic mood | 0.7119 |
| digestive system and/or stomach cramps | Stomach cramps | 0.7201 |
| arm twitches | Muscle twitch | 0.6102 |
| n panicky | Panic | 0.5171 |
| heart burn | Upset stomach | 0.5194 |
5. Experiments
Our main objective in this work is to build an MCN model which does not use manually labeled examples. We therefore use the SNOMED CT synonyms dataset and the distantly supervised dataset described in Section 4 to train our model. We do not use the training folds of CADEC and PsyTAR datasets. Our baseline is the MCN model proposed by Pattisapu et al. [6] which is trained using SNOMED CT synonyms dataset. To train our MCN model, we initialize its parameters (Section 3.1) with random values. We first train the model on SNOMED CT synonyms dataset and then fine tune it by training it on the distant dataset. We find that setting , i.e. selecting nine distant examples per target medical concept gives the optimal results on a hold-out validation set. At this stage, our model has seen all the labeled examples of SNOMED CT synonyms and distantly supervised datasets. However, as discussed in Section 4, the distantly supervised dataset contains several noisy examples which might adversely affect the performance of our model during inference. To overcome this, we fine-tune our model again on SNOMED CT synonyms dataset. During inference, we use the trained model to map a given input phrase to a vector in the target embedding space and further map it to its nearest medical concept. In this work, we separately experiment with individual target embedding types. It would be interesting to experiment with heterogeneous target embeddings by combining various text and graph based target embeddings, we leave it as a future work. For evaluating the baseline and our approach, we use only those samples of CADEC and PsyTAR datasets whose target concept IDs are also present in the distantly supervised dataset. This results in a dataset which contains nearly 25% of the concepts present in CADEC and PsyTAR datasets. This dataset size gap can be reduced by increasing the coverage of our distant supervised set such that it spans higher number of medical concepts, thereby causing a higher overlap. For evaluating the performance of both the models we compute the average classification accuracy across the test folds.
6. Results
Table 6 shows the comparison of our approach with the baseline [6] across multiple target embedding methods. Our model outperforms the baseline by a significant margin. Our model achieves the best classification accuracy of 74.39% on PsyTAR and 76.72% on CADEC datasets, whereas the best accuracy using the baseline is 63.81% and 65.48% respectively. The best performance across both datasets and approaches was achieved using graph embedding methods (Deepwalk and HARP). Across all text embedding methods, we find that Universal Sentence Encoder (USE) consistently gives better performance.
Table 6.
Performance comparison of our approach with the baseline (MCN model trained on SNOMED CT synonyms).
|
MCN trained on SNOMED CT |
MCN trained on SNOMED CT and Distant Data |
|||
|---|---|---|---|---|
| PsyTAR | CADEC | PsyTAR | CADEC | |
| AvgEmb | 59.32 | 55.78 | 70.28 | 68.28 |
| BERT | 42.90 | 50.48 | 62.53 | 62.91 |
| ELMo | 48.80 | 47.46 | 67.22 | 68.40 |
| USE | 64.18 | 52.81 | 73.43 | 70.97 |
| Deepwalk | 58.09 | 65.48 | 72.27 | 76.72 |
| HARP | 63.81 | 61.04 | 74.39 | 73.38 |
| LINE | 55.59 | 61.35 | 68.50 | 71.45 |
| Node2Vec | 51.74 | 59.38 | 71.34 | 70.98 |
| Std. deviation | 6.92 | 5.75 | 3.60 | 3.79 |
7. Analysis and discussion
7.1. Performance improvement across datasets and target embedding methods
We find that the performance improvement across target embedding methods is inconsistent. We attribute this to the suitability of the target embedding for this task. We observe that the improvement is higher for low performing embeddings (such as ELMo) as compared to high performing embeddings (such as HARP). Additionally, we observe that the proposed approach improves the performance while reducing the standard deviation across multiple target embedding methods. In a way, distant supervision reduces the impact of the choice of target embedding method. We also find that the performance improvement is not consistent across datasets. The main reason for this is that both datasets contain different medical concepts. In fact, there are only twelve common target medical concepts in CADEC and PsyTAR datasets and the medical phrases corresponding to these concepts are different in CADEC and PsyTAR datasets. Moreover, in the proposed distant supervision approach we add a fixed (k) number of labeled examples corresponding to each target concept. This further increases the difference between the training datasets which in turn results in an inconsistent performance improvement (of a particular target embedding method) across datasets.
7.2. Impact of the size of distantly supervised dataset
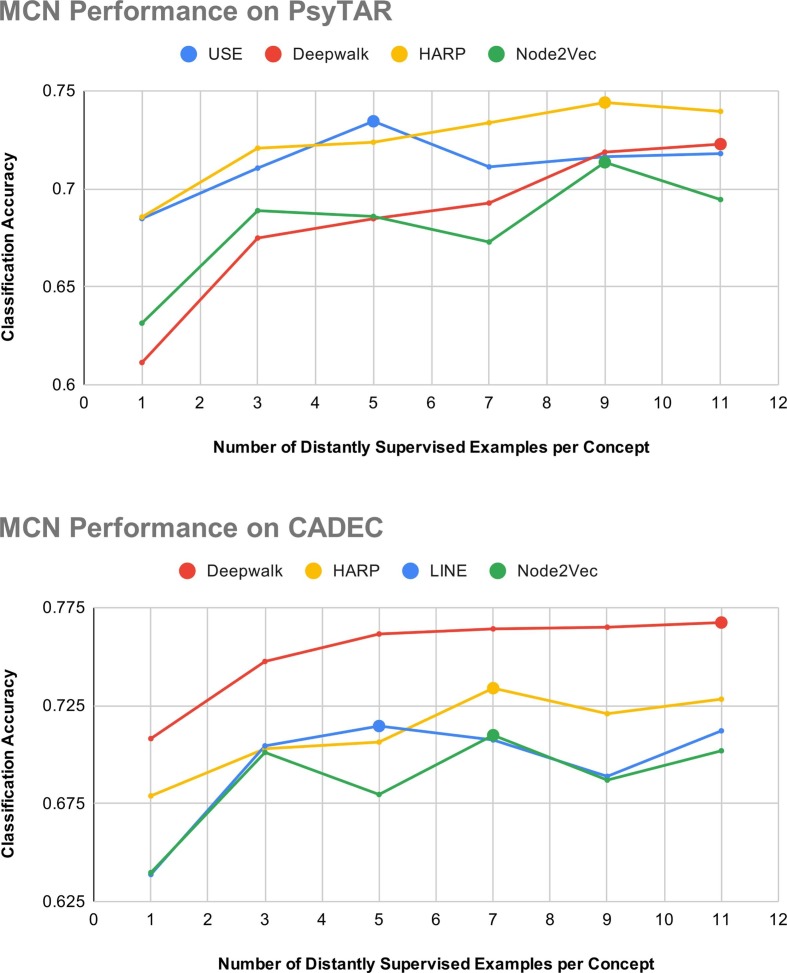
As discussed in Section 3, distant examples are selected based on the similarity between the source medical phrases and target medical concepts. In order to avoid label imbalance, we select equal number of examples corresponding to every target medical concept in the evaluation dataset. In this Section, we analyze the performance of our approach by varying the number of distantly supervised examples shown to the MCN model. Fig. 4 depicts our model’s performance across the four best performing target embedding types for PsyTAR and CADEC datasets. Across both datasets, the classification accuracy improves initially, but degrades after a particular point. We observe that beyond a point increasing the number of distant examples leads to the selection of incorrectly labeled pairs which adversely affects the model’s performance. We also observe that the performance improvement is not consistent across all target embedding methods. We attribute this to the quality of target embeddings and the nature of evaluation dataset.
Fig. 4.
Performance comparison of best performing label embedding methods across varying number of distantly supervised examples. Color viewing advised. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
7.3. Performance and error analysis
In this Section, we analyze the performance gain or loss caused by using distant supervision. To study this, we manually examine the medical concepts for which we observed a significant performance improvement or deterioration after being trained on distantly supervised examples. Table 7 shows the percentage improvement obtained by using the distant supervision corresponding to few selected medical concepts. We observe that for many medical concepts such as Abdominal Discomfort, there are very few labeled examples in SNOMED CT synonyms dataset. Our proposed method increases the labeled examples corresponding to such concepts by extracting related medical phrases, such as mild abdomen pain, random abdominal pains, abdominal cavity tiredness, fatigue, extream fatigue from social media posts. Therefore, improvement in performance can be attributed to the increase in the size and diversity of the training set. On the other hand, we also observed cases wherein distant supervision adds noisy examples to the training set thereby adversely affecting the performance. For instance, the phrases ear spray, nasal spray, gaucoma eye and laser treatment are wrongly associated with the concept Buzzing in ear.
Table 7.
Sample medical phrases in SNOMED CT and Distant Datasets. %Imp represents percentage improvement in performance.
| Concept | %Imp | Sample Medical phrases in SNOMED CT | Sample Medical phrases in Distant Data |
|---|---|---|---|
| Reduced libido | 147 | reduced libido, decreased libido, low libido | low libido, low sex drive, sex libido, non-existent sex drive, |
| Abdominal discomfort | 100 | abdominal discomfort | mild abdomen pain, random abdominal pains, abdominal cavity |
| Tired | 200 | tired, feeling tired | tiredness, fatigue, extream fatigue |
| Buzzing in ear | −33.3 | buzzing in ear | ear spray, outer ear infection, nasal spray, gaucoma eye, laser treatment, ibs |
| Foot pain | −40.9 | foot pain, podalgia | foot pain sounds, ankle pain, large toe pain, foot, leg pain, pain, foot tends |
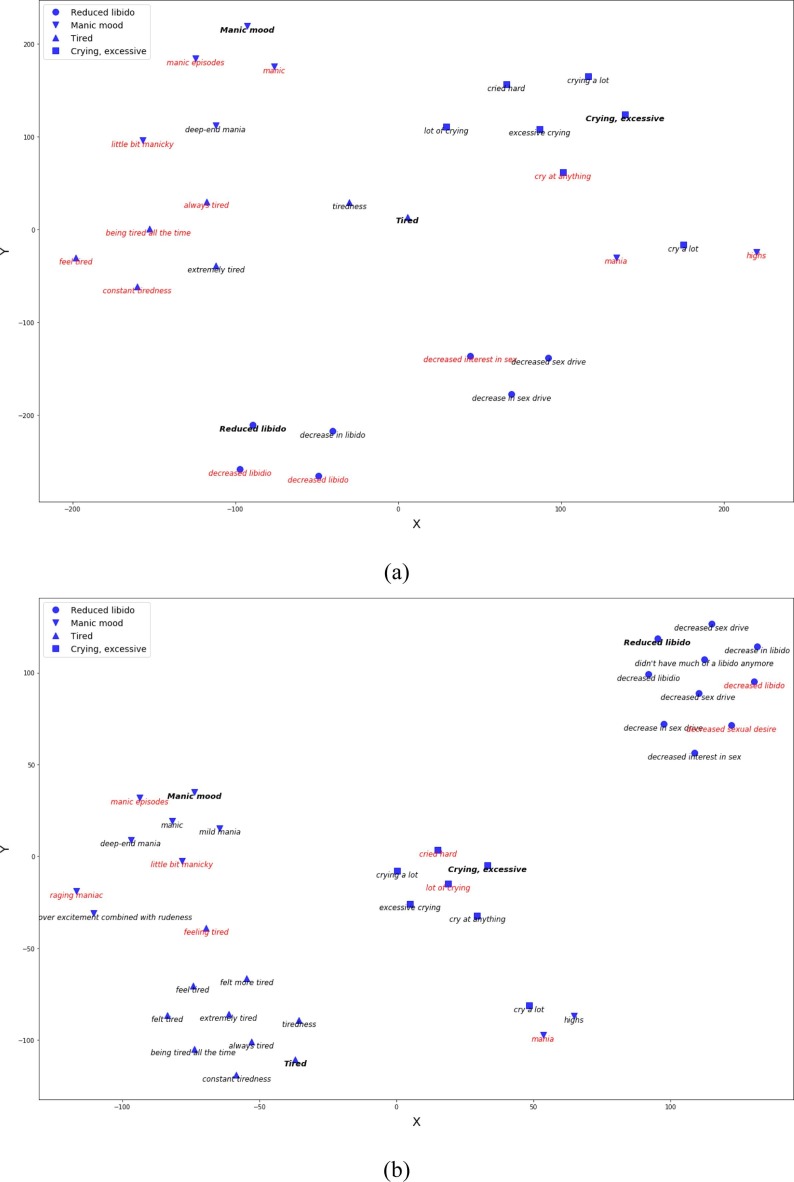
We observed that, despite using distant supervision, about 25% of the test examples were wrongly classified. We notice three different types of errors which are briefly discussed below. First, there were several phrases in the test set which could be mapped to multiple concepts, whereas the human labeled examples associate each medical phrase with exactly one medical concept. Therefore, our model is penalized if it maps an input phrase to a correct target concept which does not match with the corresponding human labeled concept. We find this to be the most common error type, Table 8 lists several examples of this error type. Second, some phrases contain abbreviations of medical concepts. Our model fails to correctly map phrases that contain abbreviations of medical concepts. For instance, our model wrongly categorizes the phrases rls, fms, felt like on an lsd. Lastly, we find that our model, as well as the baseline MCN model, do not efficiently categorize long medical phrases such as felt as if i needed to be doing something all of the time or difficult time being interested on an intimate level. Although our model shows relatively better performance across lengthy phrases, there is still a large scope for improvement. In general, we observed that distant supervision boosts the performance of our model in most cases. Fig. 5 shows the t-SNE representation of medical concepts and phrase embeddings obtained using the baseline and the proposed approach. We observe that the MCN model trained using distant supervision maps the medical phrases closer to their corresponding concepts.
Table 8.
MCN Examples which were wrongly classified by our model.
| Input Phrase | Target Concept | Predicted Concept |
|---|---|---|
| balance my mood | Moody | Manic mood |
| feel like a junkie | Drugged state | Feeling intoxicated |
| feels like to be on crack | Drugged state | Feeling intoxicated |
| difficulty to concentrate | Reduced concentration | Unable to concentrate |
| concentration is poor | Poor concentration | Unable to concentrate |
| difficulty concentratiing | Poor concentration | Unable to concentrate |
| always feeling tired | Exhaustion | Tired |
| pain really bad | Severe pain | Excruciating pain |
| intense, horrid pain | Severe pain | Excruciating pain |
| at first, trembling belly | Upset stomach | Stomach cramps |
| stomach distress | Upset stomach | Stomach ache |
Fig. 5.
t-SNE representation of medical phrases and concepts embeddings obtained using (a) baseline (b) distant supervision. The concepts depicted in this plot were encoded using LINE target embedding method. Red color indicates wrongly categorized phrase. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
8. Conclusions
In this work, we address the problem of Medical Concept Normalization (MCN) in social media which aims to map a variable length social media phrase to a medical concept in some external coding system. Most of the current approaches pose MCN as a supervised text classification task which requires a large number of medical phrase, concept pairs for training. Creating such a training dataset entails the painstaking task of manually identifying social media phrases and mapping them to one of the concepts in a medical lexicon. Pattisapu et al. [6] have leveraged medical knowledge bases (such as SNOMED CT) to extract synonyms of medical concepts and use the synonym, concept pairs as training data. Their model trained on SNOMED CT synonyms dataset alone shows a reasonable performance accuracy across multiple datasets. In this work, we extend this idea further by augmenting the SNOMED CT dataset with an automatically constructed distantly supervised dataset created from patient discussion forum posts. Experimental results across multiple datasets show that the MCN model trained using the proposed approach outperforms the baseline by a significant margin.
In future, we would like to increase the coverage of medical concepts in our distant data by obtaining multiple posts from a variety of medical social media forums. In our current work, we use generic pretrained models such as Universal Sentence Encoders to measure the similarity between medical phrases and concepts. In future, we would like to use the model trained on SNOMED CT synonym dataset to select medical phrase, concept pairs. Our current work explores the use of several text and graph based embeddings. It would be interesting to experiment with heterogeneous label embeddings. We would also want to experiment with graph convolutional networks [39] to obtain target embeddings for medical concepts. In our current work, we use AvgEmb based approach to extract medical phrases (Section 3.2.1) from patient discussion forums. It would be interesting to see if we can further improve the performance of our medical phrase extractor (and thereby the performance of our MCN model) by using other sentence encoders discussed in Section 3.1.
CRediT authorship contribution statement
Nikhil Pattisapu: Conceptualization, Writing - original draft, Methodology, Software. Vivek Anand: Data curation, Resources, Visualization. Sangameshwar Patil: Validation, Formal analysis, Writing - review & editing. Girish Palshikar: Investigation, Funding acquisition. Vasudeva Varma: Supervision, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
We use italics to denote medical phrases and typewriter to denote medical concepts.
References
- 1.Pattisapu N., Gupta M., Kumaraguru P., Varma V. Proceedings of the 2017 IEEE/ACM International Conference on Advances in Social Networks Analysis and Mining, 2017. 2017. Medical persona classification in social media, in; pp. 377–384. [Google Scholar]
- 2.Nikfarjam A., Sarker A., O’connor K., Ginn R., Gonzalez G. Pharmacovigilance from social media: mining adverse drug reaction mentions using sequence labeling with word embedding cluster features. J. Am. Med. Inform. Assoc. 2015;22(3):671–681. doi: 10.1093/jamia/ocu041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.M.J. Paul, A. Sarker, J.S. Brownstein, A. Nikfarjam, M. Scotch, K.L. Smith, G. Gonzalez, Social media mining for public health monitoring and surveillance, in: Biocomputing 2016: Proceedings of the Pacific symposium, World Scientific, 2016, pp. 468–479.
- 4.Attai D.J., Cowher M.S., Al-Hamadani M., Schoger J.M., Staley A.C., Landercasper J. Twitter social media is an effective tool for breast cancer patient education and support: patient-reported outcomes by survey. J. Med. Internet Res. 2015;17(7) doi: 10.2196/jmir.4721. e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hawn C. Take two aspirin and tweet me in the morning: how twitter, facebook, and other social media are reshaping health care. Health Affairs. 2009;28(2):361–368. doi: 10.1377/hlthaff.28.2.361. [DOI] [PubMed] [Google Scholar]
- 6.N. Pattisapu, S. Patil, G. Palshikar, V. Varma, Medical concept normalization by encoding target knowledge, in: Machine Learning for Health Workshop, NeurIPS 2019, 2020, pp. 246–259.
- 7.Lee K., Hasan S.A., Farri O., Choudhary A., Agrawal A. Healthcare Informatics (ICHI), 2017 IEEE International Conference on, IEEE. 2017. Medical concept normalization for online user-generated texts, in; pp. 462–469. [Google Scholar]
- 8.Donnelly K. Snomed-ct: The advanced terminology and coding system for ehealth. Stud. Health Technol. Informat. 2006;121:279. [PubMed] [Google Scholar]
- 9.N. Limsopatham, N. Collier, Normalising medical concepts in social media texts by learning semantic representation, in: Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), vol. 1, 2016, pp. 1014–1023.
- 10.Shen W., Wang J., Han J. Entity linking with a knowledge base: Issues, techniques, and solutions. IEEE Trans. Knowl. Data Eng. 2014;27(2):443–460. [Google Scholar]
- 11.N. Limsopatham, N. Collier, Adapting phrase-based machine translation to normalise medical terms in social media messages, 2015, arXiv preprint arXiv:1508.02285.
- 12.P. Koehn, F.J. Och, D. Marcu, Statistical phrase-based translation, in: Proceedings of the 2003 Conference of the North American Chapter of the Association for Computational Linguistics on Human Language Technology, vol. 1, Association for Computational Linguistics, 2003, pp. 48–54.
- 13.Pennell D., Liu Y. Proceedings of 5th International Joint Conference on Natural Language Processing. 2011. A character-level machine translation approach for normalization of sms abbreviations, in; pp. 974–982. [Google Scholar]
- 14.T. Schlippe, C. Zhu, J. Gebhardt, T. Schultz, Text normalization based on statistical machine translation and internet user support, in: Eleventh Annual Conference of the International Speech Communication Association, 2010.
- 15.Schlippe T., Zhu C., Lemcke D., Schultz T. in: 2013 IEEE International Conference on Acoustics, Speech and Signal Processing, IEEE. 2013. Statistical machine translation based text normalization with crowdsourcing; pp. 8406–8410. [Google Scholar]
- 16.M. Belousov, W. Dixon, G. Nenadic, Using an ensemble of generalised linear and deep learning models in the smm4h 2017 medical concept normalisation task, in: Proceedings of the Second Workshop on Social Media Mining for Health Applications (SMM4H). Health Language Processing Laboratory, 2017.
- 17.Tutubalina E., Miftahutdinov Z., Nikolenko S., Malykh V. Medical concept normalization in social media posts with recurrent neural networks. J. Biomed. Informat. 2018;84:93–102. doi: 10.1016/j.jbi.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 18.Niu J., Yang Y., Zhang S., Sun Z., Zhang W. Multi-task character-level attentional networks for medical concept normalization. Neural Process. Lett. 2019;49(3):1239–1256. [Google Scholar]
- 19.Z. Miftahutdinov, E. Tutubalina, Deep neural models for medical concept normalization in user-generated texts, 2019, arXiv preprint arXiv:1907.07972.
- 20.J. Devlin, M.-W. Chang, K. Lee, K. Toutanova, Bert: Pre-training of deep bidirectional transformers for language understanding, 2018, arXiv preprint arXiv:1810.04805.
- 21.A. Metke-Jimenez, S. Karimi, Concept extraction to identify adverse drug reactions in medical forums: A comparison of algorithms, 2015, arXiv preprint arXiv:1504.06936.
- 22.Y. Liu, M. Ott, N. Goyal, J. Du, M. Joshi, D. Chen, O. Levy, M. Lewis, L. Zettlemoyer, V. Stoyanov, Roberta: A robustly optimized bert pretraining approach, 2019, arXiv preprint arXiv:1907.11692.
- 23.I. Loshchilov, F. Hutter, Decoupled weight decay regularization, 2018.
- 24.Cer D., Yang Y., Kong S.-Y., Hua N., Limtiaco N., John R.S., Constant N., Guajardo-Cespedes M., Yuan S., Tar C. Universal sentence encoder. 2018 arXiv preprint arXiv:1803.1117. [Google Scholar]
- 25.Peters M.E., Neumann M., Iyyer M., Gardner M., Clark C., Lee K., Zettlemoyer L. Deep contextualized word representations. 2018 arXiv preprint arXiv:1802.0536. [Google Scholar]
- 26.Mikolov T., Chen K., Corrado G., Dean J. Efficient estimation of word representations in vector space. 2013 arXiv preprint arXiv:1301.378. [Google Scholar]
- 27.Perozzi B., Al-Rfou R., Skiena S. Proceedings of the 20th ACM SIGKDD international conference on Knowledge discovery and data mining, ACM. 2014. Deepwalk: Online learning of social representations; pp. 701–710. [Google Scholar]
- 28.Grover A., Leskovec J. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, ACM. 2016. node2vec: Scalable feature learning for networks; pp. 855–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.H. Chen, B. Perozzi, Y. Hu, S. Skiena, Harp: Hierarchical representation learning for networks, in: Thirty-Second AAAI Conference on Artificial Intelligence, 2018.
- 30.J. Tang, M. Qu, M. Wang, M. Zhang, J. Yan, Q. Mei, Line: Large-scale information network embedding, in: Proceedings of the 24th international conference on world wide web, International World Wide Web Conferences Steering Committee, 2015, pp. 1067–1077.
- 31.Maaten L.v.d., Hinton G. Visualizing data using t-sne. J. Mach. Learn. Res. 2008;9:2579–2605. [Google Scholar]
- 32.Manning C.D., Surdeanu M., Bauer J., Finkel J.R., Bethard S., McClosky D. Proceedings of 52nd Annual Meeting of the Association for Computational Linguistics: System Demonstrations. 2014. The stanford corenlp natural language processing toolkit, in; pp. 55–60. [Google Scholar]
- 33.T. Rose, M. Stevenson, M. Whitehead, The reuters corpus volume 1-from yesterday’s news to tomorrow’s language resources., in: Lrec, vol. 2, Las Palmas, 2002, pp. 827–832.
- 34.A.R. Aronson, Effective mapping of biomedical text to the umls metathesaurus: the metamap program., in: Proceedings of the AMIA Symposium, American Medical Informatics Association, 2001, p. 17. [PMC free article] [PubMed]
- 35.Lipscomb C.E. Medical subject headings (mesh) Bull. Med. Libr. Assoc. 2000;88(3):265. [PMC free article] [PubMed] [Google Scholar]
- 36.Denecke K. Proceedings of the Fourth Workshop on Building and Evaluation Resources for Health and Biomedical Text Processing. 2014. Extracting medical concepts from medical social media with clinical nlp tools: a qualitative study, in. [Google Scholar]
- 37.Karimi S., Metke-Jimenez A., Kemp M., Wang C. Cadec: A corpus of adverse drug event annotations. J. Biomed. Informat. 2015;55:73–81. doi: 10.1016/j.jbi.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 38.Zolnoori M., Fung K.W., Patrick T.B., Fontelo P., Kharrazi H., Faiola A., Wu Y.S.S., Eldredge C.E., Luo J., Conway M. A systematic approach for developing a corpus of patient reported adverse drug events: a case study for ssri and snri medications. J. Biomed. Informat. 2019;90 doi: 10.1016/j.jbi.2018.12.005. 103091. [DOI] [PubMed] [Google Scholar]
- 39.Kipf T.N., Welling M. Semi-supervised classification with graph convolutional networks. 2016 arXiv preprint arXiv:1609.0290. [Google Scholar]