Summary
Microbial sulfate reduction and sulfur oxidation are vital processes to enhance organic matter degradation in sediments. However, the diversity and composition of sulfate‐reducing bacteria (SRB) and sulfur‐oxidizing bacteria (SOB) and their environmental driving factors are still poorly understood in aquaculture ponds, which received mounting of organic matter. In this study, bacterial communities, SRB and SOB from sediments of aquaculture ponds with different sizes of grass carp (Ctenopharyngodon idellus) were analysed using high‐throughput sequencing and quantitative real‐time PCR (qPCR). The results indicated that microbial communities in aquaculture pond sediments of large juvenile fish showed the highest richness and abundance of SRB and SOB, potentially further enhancing microbial sulfur cycling. Specifically, SRB were dominated by Desulfobulbus and Desulfovibrio, whereas SOB were dominated by Dechloromonas and Leptothrix. Although large juvenile fish ponds had relatively lower concentrations of sulfur compounds (i.e. total sulfur, acid‐volatile sulfide and elemental sulfur) than those of larval fish ponds, more abundant SRB and SOB were found in the large juvenile fish ponds. Further redundancy analysis (RDA) and linear regression indicated that sulfur compounds and sediment suspension are the major environmental factors shaping the abundance and community structure of SRB and SOB in aquaculture pond sediments. Findings of this study expand our current understanding of microbial driving sulfur cycling in aquaculture ecosystems and also provide novel insights for ecological and green aquaculture managements.
In aquaculture pond sediments, sulfate‐reducing bacteria (SRB) produced AVS through sulfate reduction, and sulfur‐oxidizing bacteria was especially important to reduce AVS. High concentrations of sulfur compounds (TS, AVS, and ES) could inhibit SRB and sulfate reduction. Disturbance to sediments by fish activities could enhance the sulfate reduction and sulfur oxidation in sediments.
Introduction
As a key part of global biogeochemical cycles, sulfur cycling mediated by sulfate reduction and sulfur oxidation plays a critical role in nutrient metabolism processes (Li et al., 2018). Sulfur compounds are important oxidants or reductants for microbial respiration in sediments, which are mainly catalysed by SRB and SOB (Santander‐De Leon et al., 2013). On the one hand, dissimilatory sulfate reduction mediated by SRB is the dominant anaerobic mineralization pathway of organic matter degradation in sediments (Watanabe et al., 2013), which consequently release hydrogen sulfide simultaneously into ecosystems (Tian et al., 2017). On the other hand, sulfur oxidation mediated by SOB can convert hydrogen sulfide to elemental sulfur or sulfate, and remove toxic and foul‐smelling compounds such as H2S (Cheng et al., 2018). Generally, high sulfate concentrations and organic matter deposition in marine sediments enhance sulfate reduction, releasing sulfide into pore water (Kondo et al., 2004; Aller, 2014). However, high oxidation efficiency of sulfide could restrict translocation of dissolved sulfide into water–sediment interfaces (Dyksma et al., 2016), and limited oxidants in sediments may lead to a formation of various intermediate sulfur compounds (e.g. thiosulfate and elemental sulfur) (Van Den Ende and Gemerden, 2010). The importance of sulfur cycling in sediments has been increasingly recognized (Choi et al., 2006), but studies of microbial sulfur cycling mainly focused on marine (Böttcher, 2001) and freshwater environments (Bryukhanov et al., 2018). However, little is known about high‐density aquaculture ecosystems in coastal areas, which has been quickly developed in the past decade (FAO, 2018).
Coastal aquaculture has high‐nutrient loads and low dissolved oxygen situation (Sachidanandamurthy and Yajurvedi, 2006). A low efficiency of feed utilization in aquaculture ponds leads to a great accumulation of organic matters such as uneaten formulated feeds and fish faeces in sediments (Holmer and Kristensen, 1996; Asami et al., 2005). Ultimately, these accumulative organic matters will be degraded by SRB, as the sulfate reduction is a major driver in transforming organic carbon to CO2 (Anantharaman et al., 2018). Moreover, sulfate usually is the most abundant water‐soluble electron acceptor for SRB (Huycke and Gaskins, 2004; Knossow et al., 2015). This is especially true in ponds located at estuarine areas along coastline of South China. SRB in aquaculture pond sediments also can use other electron donors such as short chain fatty acids (SCFA), which are abundant in formulated feed (Hansen and Blackburn, 1995). The degradation and mineralization of these organic matters can lead to serious deterioration of water quality, resulting in eutrophication of aquaculture ponds (Krishnani et al., 2010b). Furthermore, sulfur compounds produced by SRB can be oxidized to sulfate by SOB (Barton et al., 2014; Ihara et al., 2017) and therefore maintain sulfide concentrations at a safe level (Fernandes et al., 2010). Although sulfur cycling plays such important roles in nutrient cycling and environmental safety in aquaculture ecosystems, and current research mainly focused on the composition of SRB and SOB (Rubio‐Portillo et al., 2019). However, the regulatory mechanisms of SRB and SOB remain poorly understood in aquaculture ponds.
Sulfur cycling could be affected by various factors in aquaculture pond sediments (Duc et al., 2018). Generally, amounts of feed supply and feed utilization efficiency are the major factors that influence fish growth (Biswas et al., 2005; Kondo et al., 2012). Therefore, excessive nutrient input is the most common strategy in the aquaculture industry to increase production, which also will significantly affect compositions and bio‐functions of bacterial communities in sediments (Leflaive et al., 2008). Besides, suspended sediments due to activities of aquatic animals not only increase turbidity in water column (Croel and Kneitel, 2011), but also disturb microbial communities in sediments (Mendoza‐Lera and Mutz, 2013). In aquaculture ponds, larger fish usually have strong disturbances to water–sediment interfaces, which will further influence the bacterial communities in sediments (Jochum et al., 2017). Combined with field observations, a previous study found that fish size could impact the accumulation and consumption of sulfur‐containing substances (Barnes and Jennings, 2007).
In this study, we aimed to understand how feeding practices and fish sizes will affect sulfur‐cycling microbial communities in the sediment of aquaculture ponds. We hypothesized that sulfur‐cycling microbial communities would differ among aquaculture ponds with different sizes of fish, and nutrient accumulation and sediment suspension due to fish activities would enhance the sulfur‐cycling microbial communities in the sediment. To test these hypotheses, we examined sulfur‐cycling microbial communities in aquaculture ponds cultured with three sizes of grass carp by high‐throughput sequencing and qPCR of 16S rRNA (Griffiths et al., 2000), dsrB and soxB (Zhang et al., 2017) genes. We found that aquaculture ponds with large grass carp showed more abundant sulfur‐cycling microorganisms than those with smaller sizes of fish, and sulfur compounds and sediment suspension were the major driving factors. This study provides novel knowledge that strong fish activities (bioturbation) decreasing toxic sulfur compounds, changing sulfur‐cycling microbial communities in aquaculture pond sediments.
Results
Environmental parameters in grass carp aquaculture ponds
Our results showed clear differences about physicochemical parameters of sediment and water in aquaculture ponds with different sizes of fish (Table 1). With the increase in fish sizes, total sulfur (TS), elemental sulfur (ES) and acid‐volatile sulfur (AVS) in the sediment all significantly (P < 0.05) decreased. However, total organic carbon (TOC) and total nitrogen (TN) in the sediment were not significantly influenced by fish size. Total suspended solids (TSS) in the water were significantly (P < 0.05) higher in large juvenile fish ponds than larval and small juvenile fish ponds. By contrast, the transparency was significantly (P < 0.05) lower in large juvenile fish ponds. TN and nitrate in the water were also significantly (P < 0.05) increased with the increase in fish sizes.
Table 1.
Summary of sediment and water physicochemical properties in aquaculture ponds with different sizes of grass carp.
| Habitat | Parameter | Ponds | ||
|---|---|---|---|---|
| L | SJ | LJ | ||
| Sediment | pH | 7.66 ± 0.52 | 7.49 ± 0.43 | 7.41 ± 0.33 |
| AVS (mg g−1) | 0.44 ± 0.19a | 0.40 ± 0.19a | 0.11 ± 0.05b | |
| TS (mg g−1) | 4.10 ± 1.34a | 2.45 ± 1.58b | 1.16 ± 0.25c | |
| ES (mg g−1) | 0.63 ± 0.24 a | 0.43 ± 0.11ab | 0.25 ± 0.16b | |
| Sulfate (mM l−1) | 2.28 ± 0.96 | 2.77 ± 1.04 | 2.27 ± 0.34 | |
| TOC (mg g−1) | 20.85 ± 3.48 | 18.06 ± 7.06 | 19.75 ± 3.38 | |
| TN (mg g−1) | 1.89 ± 0.44 | 1.63 ± 0.79 | 1.58 ± 0.29 | |
| Water | TSS (mg l−1) | 2.47 ± 0.75b | 2.72 ± 1.01b | 4.05 ± 0.17a |
| Transparency (m) | 0.22 ± 0.08a | 0.18 ± 0.05a | 0.12 ± 0.02b | |
| TN (mg l−1) | 5.48 ± 1.15c | 7.01 ± 1.84b | 8.72 ± 0.62a | |
| Nitrate (mg l−1) | 0.19 ± 0.12c | 1.64 ± 1.60b | 3.55 ± 0.86a | |
Data showed as Mean ± SD, n = 12. AVS, acid‐volatile sulfur; ES, elemental sulfur; L, larval; LJ, large juvenile; SJ, small juvenile; TN, total nitrogen; TOC, total organic carbon; TS, total sulfur; TSS, total suspended solids. Values with different superscript letters mean significantly different (P < 0.05) among ponds, and same letters or without letters indicate no significant difference.
Fish growth increased bacterial diversity in aquaculture pond sediments
Our high‐throughput sequencing and qPCR analysis were used to examine changes in bacterial diversity and abundance among different fish ponds. In particular, the total number of OTUs obtained from 16S rRNA, drsB and soxB genes was 14 926, 518 and 483 respectively. The large juvenile fish ponds showed significantly (P < 0.05) higher number of OTUs and Shannon indices for bacterial community and SOB compared to larval and small juvenile fish ponds. However, for the SRB, Shannon indices had no significantly difference among larval, small juvenile and large juvenile fish ponds (Fig. S1). The bacterial community structure, as well as SRB and SOB composition in large juvenile fish ponds, was quite different from those in larval and small juvenile fish ponds visualized by the PCoA (Fig. 1). The qPCR analysis showed that the abundance of 16S rRNA, dsrB and soxB genes in large juvenile fish ponds was the highest (Fig. 2). Specifically, the abundance of dsrB gene in the large juvenile fish ponds accounted for 0.25% of the 16S rRNA gene abundance, while the soxB gene accounted for 3.5% of total bacterial abundance in large juvenile fish ponds.
Fig. 1.
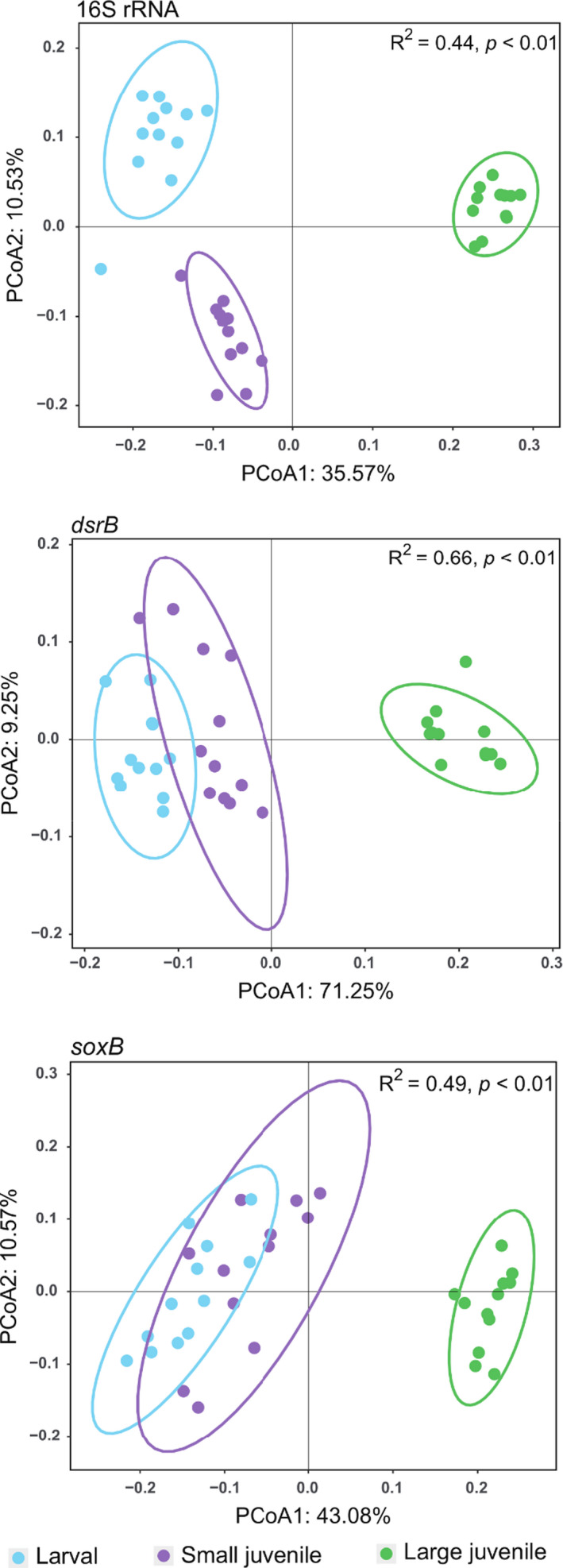
Principal coordination analysis (PCoA) showing the dissimilarity of microbial communities according to sequencing results of 16S rRNA, dsrB and soxB genes. Anosim test was employed to indicate the significance of dissimilarity.
Fig. 2.
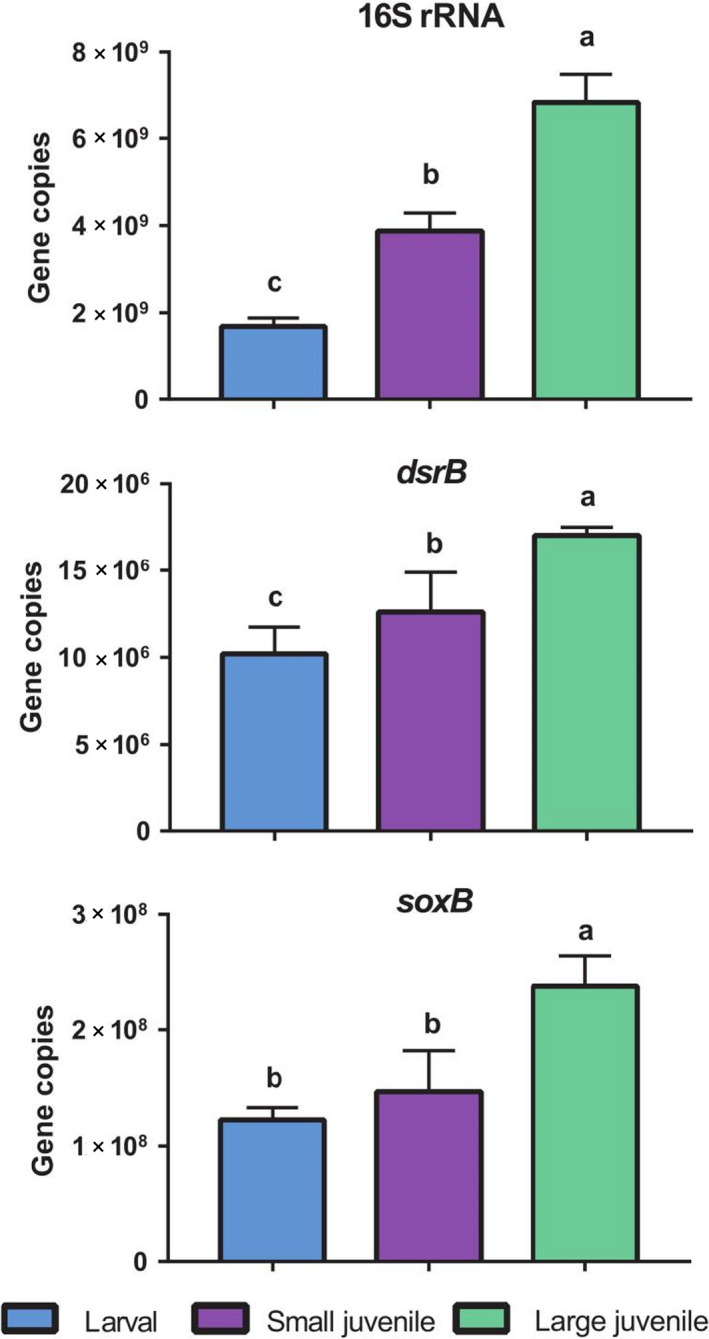
Abundances of 16S rRNA, dsrB and soxB genes (copies per g wet sediment) in ponds cultured with larval, small juvenile and large juvenile grass carp. Data presented as mean ± SD (n = 12)
The composition of bacterial communities, SRB and SOB in aquaculture pond sediments
To understand dominant sulfur‐cycling microorganisms and compare their relative abundance among aquaculture ponds with different sizes of fish, we filtered unidentified OTUs at the genus level and focused on the classified top 10 genera (Fig. S2). In the 16S rRNA gene data set, Geobacter and Anaerolinea were dominant genera in all samples. Anaerolinea, Desulfatiglans and Geobacter were dominant genera in large juvenile fish ponds, while Candidatus Anammoximicrobium, Desulfobacca, Pirellula and Coxiella were more abundant in larval and small juvenile fish ponds than those in large juvenile fish ponds (P < 0.05). For SRB, Desulfobulbus and Desulfovibrio were dominant genera in all samples, and Desulfovibrio, Desulfomoniel, Desolfococcus and Desulfobulbus were more abundant in small juvenile and large juvenile fish ponds than those in larval fish ponds (P < 0.05). For SOB, Dechloromonas and Leptothrix were dominant genera in all samples, and Dechloromonas was more abundant in larval and small juvenile fish ponds, while Thiobacillus showed higher abundance in large juvenile fish ponds (P < 0.05).
Although we identified some specific species according to the Silva database of bacteria and FunGene database (http://fungene.cme.msu.edu/) of SRB and SOB, about 58% OTUs of bacteria, 80% OTUs of SRB and 20% OTUs of SOB were unclassified. We further used the neighbour‐joining method to identify the phylogenetic position of those unidentified OTUs according to the sequence similarity. In particular, we picked up the OTUs with their abundance over 0.5% of 16S rRNA, dsrB and soxB gene sequences, then constructed the phylogenetic tree with 21, 40 and 34 OTUs remained for 16S rRNA, dsrB and soxB genes respectively (Fig. 3). For bacteria, most OTUs failed to annotate to the species level, and most of them belonged to different orders. In addition, these dominant bacteria were more abundant in large juvenile fish pond sediments. For SRB, only OTU_52 (Desulfobulbus propionicus), OTU_62 (Desulfobulbus propionicus) and OTU_449 (bacterium enrichment culture clone S2S‐F7) were well matched to the SRB database. OTU_445 and OTU_175 were abundant in all types of sediments, and they appeared to be closely related to Desulfovibrionales. Also, we found that a novel dsrB cluster (including OTU_1136, OTU_1137, OTU_1140, OTU_1141, OTU_1143, OTU_1149 and OTU_1175) was more abundant in larval and small juvenile fish ponds compared to large juvenile fish ponds. For SOB, 27 OTUs were annotated. Dechloromonas aromatica, OTU_163 (closely related to Acidithiobacillales), Thiobacillus denitrificans, Halochromatium glycolicum and Leptothrix cholodnii were abundant in all types of sediments.
Fig. 3.
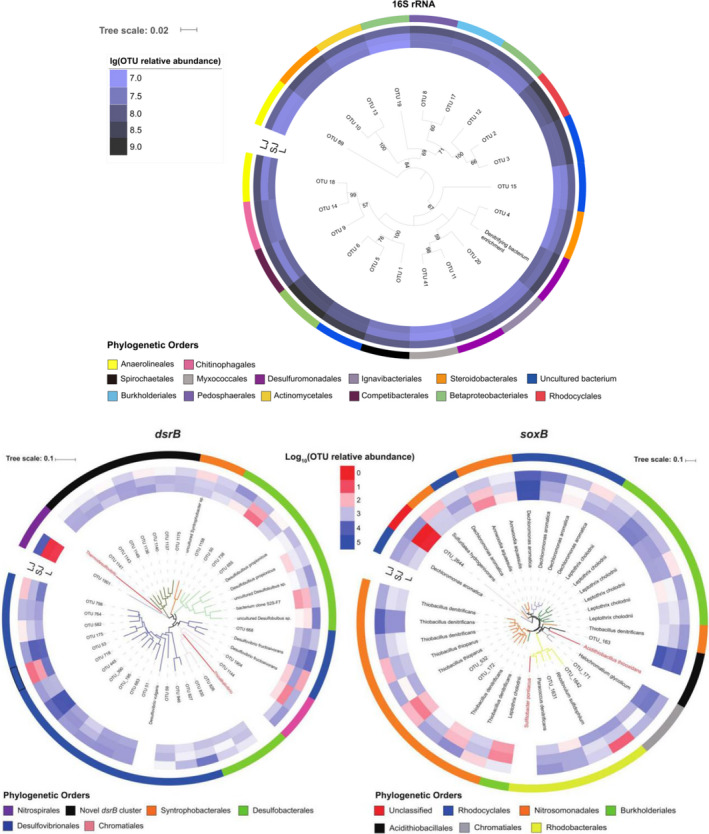
Neighbour‐joining phylogenetic trees generated according to the 16S rRNA, dsrB and soxB genes. The trees were constructed based on the inferred amino acids of representative nucleotide sequences of OTUs. Only OTUs containing ≥ 0.5% of the total abundances were presented, and bootstrap values over 50% based on 1000 replicates were shown. L, larval; LJ, large juvenile; SJ, small juvenile.
Sulfur compound accumulation and sediment suspension shaped the microbial communities in aquaculture pond sediments
RDA analysis was used to find key factors driving microbial community variations across the three different types of aquaculture pond sediments. Several environmental variables were removed by forward‐selection algorithm in order to develop a robust model (Fig. 4). Final RDA showed that TOC and TN explained few variations in community structure of bacteria, SRB and SOB (P > 0.1). While TSS significantly affected the distribution of bacterial communities (F = 3.00, P = 0.01), SRB (F = 5.50, P = 0.003), and SOB (F = 2.30, P = 0.03). The AVS was another most important environmental factor that affected the distribution of bacterial communities (F = 8.96, P = 0.001), SRB (F = 16.30, P = 0.001) and SOB (F = 9.19, P = 0.001). More specifically, TSS was strongly associated with microbial communities in large juvenile fish ponds, whereas ES and AVS were closely correlated with microbial communities of larval and small juvenile fish ponds. Mantel tests further verified that TS, AVS, ES and TSS were significantly correlated with the community structure of bacteria SRB and SOB (P < 0.05), and TOC and TN were only correlated with the community structure of SOB (Fig. S3). To better quantify the relative contributions of subgroups of environmental parameters to the variations in community structure of bacteria, SRB and SOB (at OTU level), we conducted a variance partitioning analysis (VPA) on three subgroups of factors (i.e. nutrient loading, sulfur compounds, sediment suspension). Overall, sulfur compounds and sediment suspension explained more variations than nutrient loading for variations in community structure of bacteria (Fig. 5A), SRB (Fig. 5B) and SOB (Fig. 5C).
Fig. 4.
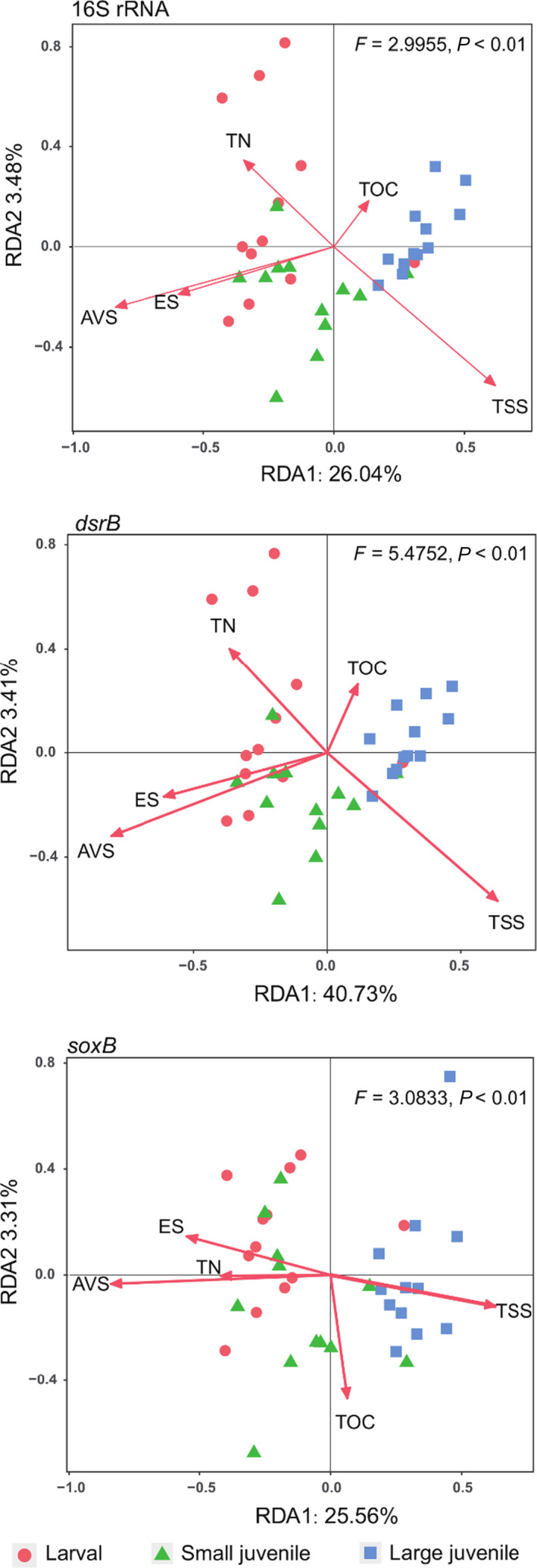
Redundancy analysis (RDA) ordination showing the relationships between bacterial communities (16S rRNA)/sulfate‐reducing bacteria (dsrB)/sulfur‐oxidizing bacteria (soxB) and environmental factors. AVS, acid‐volatile sulfur; ES, elemental sulfur; TN, total nitrogen; TOC, total organic carbon; TSS, total suspended solids.
Fig. 5.
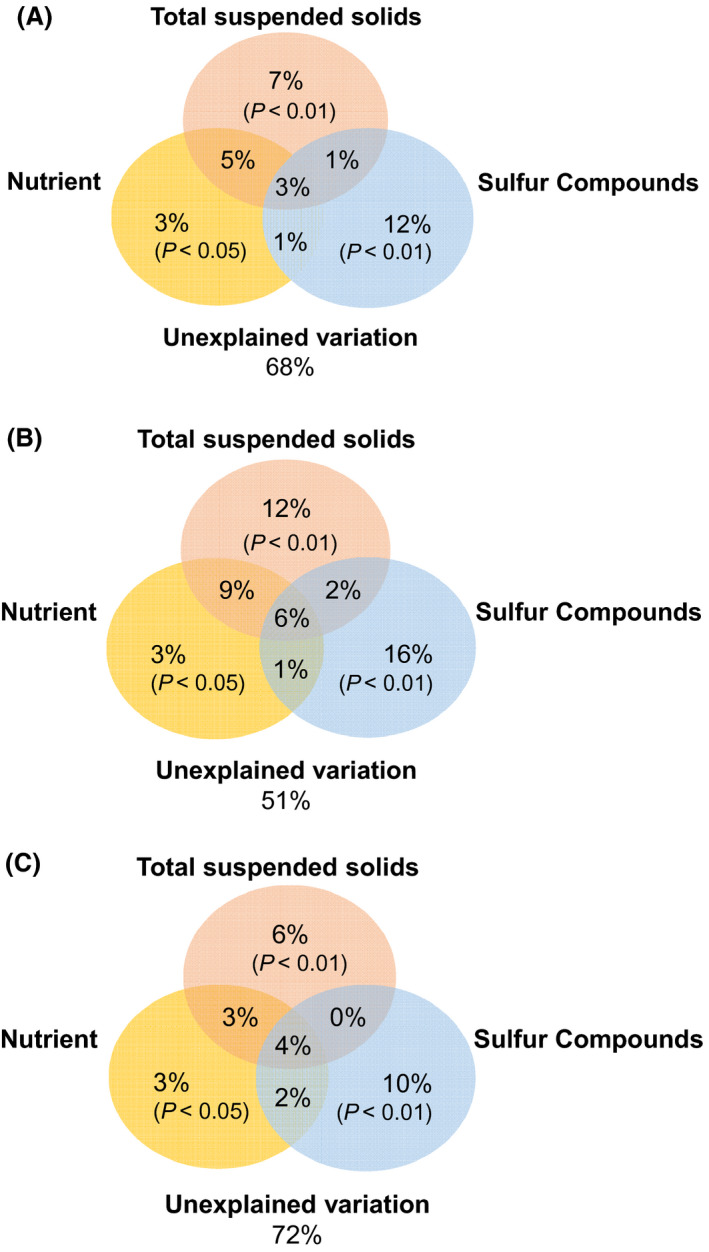
Variation partition analysis (VPA) showing the effects of environmental factors on the bacterial communities (A), sulfate‐reducing bacteria (B) and sulfur‐oxidizing bacteria (C) in sediments. Nutrient loading including total nitrogen (TN) and total organic carbon (TOC); sulfur compounds including total sulfur (TS), acid‐volatile sulfur (AVS) and elemental sulfur (ES); sediment suspension reflected by total suspended solids (TSS).
Furthermore, linear regression was used to examine how sediment suspension and sulfur compounds influenced abundances of SRB and SOB. Results indicated that abundances of SRB and SOB increased with increasing of sediment suspension, whereas the abundance of SRB decreased with increasing of sulfur compounds (Fig. S4).
Discussion
Sulfur cycling mediated by SRB and SOB plays an important role in sediments of aquaculture ecosystems. We found that aquaculture ponds cultured with large juvenile fish (grass carp) showed lower content of sulfur compounds but higher sediment suspension, which resulted in higher abundances of SRB and SOB and different microbial community structures in sediments compared to ponds cultured with larval fish.
The abundances and structures of sulfur‐cycling microbial communities differed among ponds cultured with different sized fish, which could be due to environmental discrepancies between pond sediments. AVS pool in sediments is highly dynamic (Morse and Rickard, 2004) and varies with sediment‐type, especially in high sulfidation environments (Rickard and Morse, 2005). However, concentration of AVS in sediments depends on the sulfate reduction rate of SRB (Liu et al., 2010). In this study, alpha diversity indexes and qPCR results all showed the ponds with large juvenile fish harboured more abundant SRB compared to those for larval fish ponds. According to the environmental standards of AVS for evaluating aquatic systems (Japan Fisheries Resource Conservation Association, 1995), ponds with larval and small juvenile fish were classified as ‘moderately polluted’, whereas ponds with large juvenile fish could be ‘unpolluted’. Theoretically, SRB should be more abundant in larval and small juvenile fish ponds due to high concentrations of AVS. Meanwhile, sulfate concentration showed no significant (P > 0.05) differences among ponds, and that was also not significantly correlated with microbial communities. Therefore, sulfate may not be the primary substrate to produce AVS in these aquaculture ponds. For instance, some volatile sulfur compounds can be produced from sulfur‐containing amino acids by methanogens and SRB, which is rich in formulated feeds (Holmer and Storkholm, 2001; Lu et al., 2013; Sun et al., 2015). Beyond that, AVS may be derived from soluble sulfide (Morse et al., 1987). Thus, the sulfur‐containing organic matters may be an important source of AVS except for sulfate in aquaculture pond sediments. However, this conclusion still needs further experiments to confirm.
The microbial community in farming sediments could be affected by organic enrichment (White, 2003). However, a previous study demonstrated that marine sediments with different levels of organic enrichments showed no prominent differences in SRB (Yoshida et al. , 1982). Our Mantel tests and RDA ordination showed that TOC and TN only weakly correlated with community structures of SRB and SOB, which indicated that accumulation of uneaten feeding food had little correlation with sulfur‐cycling microorganisms in aquaculture sediments. There may have two reasons. First, fish disturbance to sediment can inhibit organic matter accumulation (Brummett, 2000). Second, large fish ponds have more feed input but high utilization efficiency (Einen and Roem, 1997). All of these can reduce the difference of organic matter depositions in sediments among different aquaculture ponds. Instead, sulfide generated from microbial metabolism would influence the microbial activity in sediments (Visscher et al., 1995; Lyimo et al., 2009). We found that sulfur compounds showed strong correlations with microbial communities via RDA plot and VPA analysis, especially SRB and SOB. Moreover, linear regression indicated that high levels of sulfur compounds led to low abundance of SRB, as some SRB species could be restrained by sulfide at high concentrations (Reis et al., 1992). Sulfide also can inhibit SRB even at a low concentration (Takahashi et al., 2008). Thus, sulfur compounds like TS and AVS were more important than TOC and TN in influencing the community structure of SRB. High concentrations of sulfur compounds in aquaculture pond sediments would inhibit the abundance of SRB.
Disturbance to sediments by grass carp reshaped microbial communities in sediments. Since aquaculture ponds are closed systems and usually have no obvious disturbances to sediments expect fish activities. Animal disturbance to water–sediment interfaces frequently happened in ponds and lakes (Kristensen et al., 2012), and fish activities could increase the water turbidity (Adamek and Marsalek, 2013). Furthermore, bioturbation would accelerate the release of nutrients (like total nitrogen) into overlying water (Hansen et al., 1998), the suspended particles could also promote nitrification in the water (Xia et al., 2004). In this study, TSS, TN and nitrate contents in the water were significantly higher in large juvenile fish ponds, indicating strong disturbances caused by larger grass carps. The RDA and VPA results also showed the strong disturbance caused by fish would affect the distribution of SRB and SOB in aquaculture ponds. Meysman et al. (2005) found that the rate of sulfate reduction enhanced with increasing perturbation. Our linear regression results also showed that sediment suspension caused by fish activities increased the abundance of SRB and SOB. Fish disturbance disturbed the anoxic microenvironment and oxidized the chemical compounds through sulfide oxidation (Bertics and Ziebis, 2009), thus extended micro‐niches into an oxidized zone (Zorn et al., 2006). Although SRB are generally described as obligate anaerobes, some groups like Desulfovibrio possess a complex aggregation of enzymes to defence against oxidative stress (Dolla et al., 2006), and some SRB even could use O2 as a terminal electron acceptor for their aerobic growth (Lefèvre et al., 2016). A previous study found that disturbance could increase SRB abundance even in an aerobic environment (Bertics and Ziebis, 2009). In addition, SOB can use oxygen as an oxidizing agent during the oxidation of sulfide (Pokorna and Zabranska, 2015), which could be facilitated by fish disturbance. Furthermore, SOB can transform hazardous sulfur compounds and serve as bioremediation strategies (Krishnani et al., 2010a). Our results indicated that fish disturbance increased the abundance of SRB and SOB in upper sediment layers, further enhanced the sulfate reduction and sulfur oxidation processes.
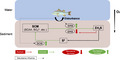
Based on findings of this study and our current knowledge, we proposed a putative model of sulfur cycling dominated by SRB and SOB in aquaculture ponds (Fig. 6). SRB transformed sulfur‐containing materials (SCM) into sulfide through sulfate reduction process, and most sulfides were oxidized to sulfate through sulfur oxidation by SOB. The sulfate reduction process was affected by sulfur compounds in sediments, and a high concentration of sulfur compounds would inhibit SRB activity. Meanwhile, sulfur cycling mediated by SRB and SOB would be facilitated by the fish disturbance to sediments, thus activated the sulfur cycling.
Fig. 6.
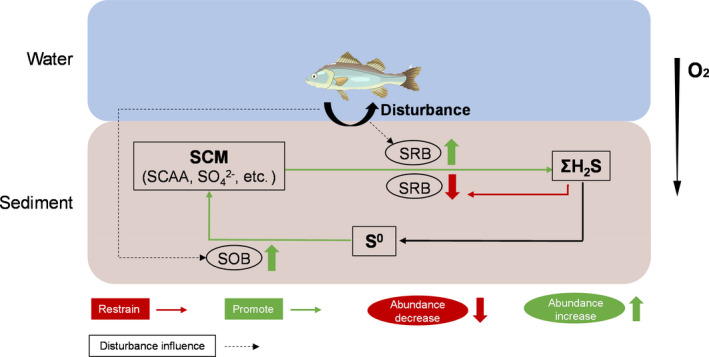
Putative model showing the sulfur cycling dominated by sulfate‐reducing bacteria (SRB) and sulfur‐oxidizing bacteria (SOB), and their major influence factors in aquaculture ponds. SCM, sulfur‐containing materials; SCAA, sulfur‐containing amino acids.
Conclusions
We found that the abundance of SRB and SOB and microbial community structure were significantly different in aquaculture ponds cultured with different sizes of grass carp. In aquaculture pond sediments, SRB produced AVS through sulfate reduction, and SOB was especially important to reduce AVS. High concentrations of sulfur compounds (TS, AVS and ES) could inhibit SRB and sulfate reduction. Disturbance to sediments by fish activities could enhance the sulfate reduction and sulfur oxidation in sediments. These findings help us to better understand microbial sulfur cycling in aquaculture ponds and provide new insights into microbial manipulation of healthy aquaculture environment.
Experimental procedures
Study area and sampling procedures
Samples were collected from grass carp aquaculture ponds at Nansha (22°60′97.88″N, 113°62′18.05″E), Guangdong province of China on 18 April and 31 May 2018. Three kinds of ponds cultured with different sizes of grass carps, i.e., larval fish (L), small juvenile fish (SJ) and large juvenile fish (LJ) were sampled. Each pond is 1.5 km2 in area, and about 2.0 m in‐depth without water exchanging during our sampling period. In all ponds, the clays were removed and the sediments were further treated with quicklime before culturing grass carp. These ponds used for culture already lasted for nearly one year before sampling. During the one year of culturing, each pond only used for culturing one sized fish, and the fish size grew to big enough would be transferred to another corresponding type of ponds. On the first sampling time, the culture of larval fish had just begun three days ago, and the culture of small and large juvenile fish had lasted for half a month. The benthic organisms were not common in the pond sediment. During 18 April and 31 May, in larval fish ponds, the fish initially weighted ~ 0.2 g and increased by 0.8 g, the density was 750 fish per m2; in small juvenile fish ponds, the fish initially weighted ~ 50 g and increased by 150 g, the density was 30 fish per m2; in large juvenile fish ponds, the fish initially weighted ~ 400 g and increased by 180 g, the density was 5 fish per m2. The feed supply is commercial crumbled (larval fish) or pelleted (juvenile fish) formulated feed with a ratio of 5% body weight per day. For each size of grass carp, three ponds were sampled as biological templates, and three surface water (1 l each) and three sediment (500 g each) samples were collected from the left, centre and right of each pond. Then, mixed three sediment samples and three water samples in each pond for the sampling occasion of 18 April; however, the sampling occasion of 31 May was not mixed, and thus, totally there were 36 water samples and 36 sediment samples for further analysis. The water (about 50 cm below water level) and upper sediments (0–8 cm) were obtained by using 5‐litre hydrophore sampler and Van Veen Grab Sampler respectively. All samples were kept in ice box and brought back to the laboratory. A subset of ~ 50 g of each sediment sample was separated and frozen at −80°C for further DNA extraction, and the rest of sediments and water samples were used for physicochemical parameter tests.
Physicochemical analyses
A subset of 10 g of each sediment sample was separated for measuring total organic carbon (TOC), total nitrogen (TN) and total sulfur (TS). Specifically, sediment was oven‐dried at 65 °C till a constant weight and sieved through a 150‐μm sieve. Then, 200 mg dry sediment of each sample was mixed thoroughly with 1 ml of 0.5 mol l−1 hydrochloric acid and then dried at 80°C for 4–5 h. TOC was examined by using a PRIMACSMCS TOC Analyzer (Skalar, Netherlands). For measuring TN and TS, about 6 mg dry sediment of each sample was loaded into a CHNS/O Elemental analyser (Vario EL cube, Germany). Total suspended solids (TSS) were measured using a modified method to avoid plankton influence in the water, and the TSS were used to represent the degree of sediment suspension. Specifically, a glass fibre filter with pore size of 0.45 μm was dried at 400°C for 4 h and weighed as a blank control, 200 ml water sample was filtered through such filter was dried at 400°C for 4 h and weighed, and TSS was calculated from their subtraction. Transparency was measured by using a Secchi disk. Total nitrogen of water was analysed according to the method as described by Parsons and Takahashi (1973), and nitrate was analysed by using the method described previously (Ebina et al., 1983).
Fresh sediments were used to measure pH, sulfate (SO4 2−), acid‐volatile sulfide (AVS) and elemental sulfur (ES). Briefly, pH was determined by using a pH electrode (SevenCompact™ pH Meter S210; Mettler Toledo, Swiss) (Ihara et al., 2017). For analysing the SO4 2−, about 20 g wet sediments were centrifuged in a tube at 4000 × g for 10 min to obtain pore water. The pore water was filtered through H‐pretreatment columns and C18‐pretreatment columns (ANPEL, Shanghai) to remove metal ion and organic matters respectively. Then, SO4 2− was measured by ion chromatography (Thermo Fisher, ICS‐600). AVS measured here included all sulfide that can release H2S if treated with acid (e.g. H2S, HS−, FeS) (Rickard and Morse, 2005). For measuring the AVS, 6 g fresh sediments were transferred into 250‐ml polyethylene bottle with a small vial containing 10 ml 3% alkaline zinc solution. After purging with nitrogen gas for 5 min to remove oxygen, 10 ml 9 M HCl was injected, then closed syringe stopcock of the polyethylene bottle immediately (Brouwer and Murphy, 2010). An additional 1 ml 1 M ascorbic acid was injected into the bottle to avoid interference of ferric ions. After diffusion and reaction at room temperature for 4 h, sulfide precipitates in the alkaline zinc solution were collected and measured by iodometric titration method (Hsieh and Yang, 1989; Hsieh and Shieh, 1997). ES was defined as the sulfur extracted with methanol from sediment samples and measured by Reversed‐Phase HPLC. About 1 g fresh sediment was transferred into 50‐mL polyethylene centrifuge tube with 25 ml pure methanol, and shook for 24 h in a rotary shaker (25°C, 160 r min−1). After centrifuged (3000 rpm min−1) for 5 min, the supernatant was filtered through 0.22‐μm membrane and analysed by HPLC using a C18 reversed‐phase column (Thermo, Eclipse XDB‐C18 5 μm, 150 × 4.6 mm I.D). The eluent was composed of ratio at methanol/water = 95:5 (v/v) with a flow rate of 0.8 ml min−1, and a UV detection performed at 254 nm wavelengths to quantify elemental sulfur (Zopfi et al., 2004; Knossow et al., 2015).
DNA extraction and PCR amplification
Only sediment samples were involved in microbial analysis. Total microbial DNA was extracted by using a freeze‐grinding method with sodium dodecyl sulfate (SDS) (Zhou et al., 1996), followed by PowerSoil DNA Kit (Mo Bio Laboratories Inc., Carlsbad, CA, USA) according to the manufacturer’s instructions. The concentration and quality of extracted DNA were determined by using NanoDrop One spectrophotometer (Thermo Fisher Scientific, MA, USA), diluted to the same concentration (10 ng μl−1) and stored at −80°C until subsequent analyses.
The V4 region of the 16S rRNA gene, dsrB and soxB genes was amplified by primer sets used previously (Table S1). Amplicons for Illumina sequencing were prepared following the barcoded PCR procedures: 50 µl reaction system including 10 µl buffer (5×), 0.2 µl Q5 High‐Fidelity DNA Polymerase, 10 µl High GC Enhancer, 1 µl dNTP, 10 µM of each primer and 60 ng genomic DNA. The amplification of 16S rRNA gene was conducted with an initial denaturation at 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, 53°C for 45 s and 72°C for 1 min, and a final extension at 72°C for 5 min. PCR cycling procedures for the dsrB gene are 30 cycles of 95°C for 30 s, 50°C for 30 s and 72°C for 30 s; for the soxB gene are 30 cycles of 95°C for 30 s, 55°C for 30 s and 72°C for 40 s. The positive PCR products were purified through VAHTSTM DNA Clean Beads.
A second round Solexa PCR was performed in a 40 µl reaction which contained 20 µl (2×) Phµsion HF MM, 8 µl ddH2O, 10 µM of each primer and 10 µl PCR products from the first step. Thermal cycling conditions were as follows: an initial denaturation at 98°C for 30 s, followed by 10 cycles at 98°C for 10 s, 65°C for 30 s and 72°C for 30 s, with a final extension at 72°C for 5 min. PCR products were visualized using 1.8% agarose gels to obtain target band, purified as the previous step, then quantified by Quant‐iT™dsDNA HS Reagent and mixed fully. All mixtures were added to a 1.8% agarose gel, the gel electrophoresis was performed with 120 V for 40 min, and the specific band was excised and purified.
High‐throughput sequencing and data processing
High‐throughput sequencing was performed on Illumina platforms in the Biomarker Technologies Corporation, Beijing. Specifically, fragments of the 16S rRNA gene (291 bp, V4 region) and dsrB gene (350 bp) were sequenced by Illumina HiSeq PE250 platform, and the soxB gene (470 bp) was sequenced by Illumina MiSeq PE300 platform.
The obtained raw paired‐end 16S rRNA, dsrB and soxB gene sequences were filtered ambiguous base(s) (“N”). After trimming and identifying sequencing regions, the obtained high‐quality fragments were aligned in Quantitative Insights Into Microbial Ecology (QIIME) v1.9.1 (http://qiime.org/). The aligned 16S rRNA gene sequences without chimeras were compared based on the Ribosomal Database Project (RDP) database by using USEARCH v11 (Edgar, 2013). The operational taxonomic units’ (OTUs) table was generated by using UNOISE method with 97% cut‐off (Edgar, 2016). OTUs were grouped into different taxonomy by using Silva database release 132. The frameshifts of the dsrB and soxB genes were corrected used the RDP FrameBot tool in Galaxy pipeline (http://mem.rcees.ac.cn:8080/), and only reads whose translated proteins mapped to reference protein sequences were retained (Wang et al., 2013). After removing chimeras, the high‐quality sequences of functional genes were clustered into OTUs at a 90% identity threshold by QIIME and VSEARCH (Pelikan et al., 2015). The reference database for annotation was downloaded from FunGene Pipeline (http://fungene.cme.msu.edu/), and the comparison identity for taxonomy assignments was 0.9 and 0.7 for the dsrB and soxB respectively. Raw OTUs obtained from the dsrB and soxB gene sequences were filtered with an average relative abundance of lower than 1/10,000. Afterwards, microbial alpha‐diversity (observed OTUs and Shannon) was calculated by QIIME (Caporaso et al., 2010). A phylogenetic tree was constructed in MegaX and iTOL (https://itol.embl.de/) using a neighbour‐joining algorithm with 1000 bootstrap (Kumar et al., 2018).
Quantitative real‐time PCR analysis (qPCR)
The abundance of 16S rRNA, dsrB and soxB genes was profiled by qPCR. All PCR products were purified by using DiaSpin DNA Gel Kit (Sangon, Shanghai, China), and cleaned targeted gene fragments were extracted from Trans1‐T1 using pEASY®‐T1 Simple Cloning Kit (TransGen, Beijing, China). Plasmids were sequenced and verified using the basic local alignment search tool (BLAST) (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The concentrations of plasmids containing target sequences were measured by Qubit™ Assays fluorometer (Thermo Fisher Scientific). The initial stand curves were diluted by gradient starting from 10‐fold dilutions of plasmids carrying the targeted genes.
The qPCR was performed using the primers used previously (Table S1) according to the methods as described by Jiang et al. (2015) and He et al. (2015). All qPCR experiments were conducted on a BIO‐RAD CFX96™ Real‐Time System with SYBR Green method with triplicate replicates. The 12‐µl volume of qPCR mixture contained 6 µl iTaq™ Universal SYBR Green Supermix (BIO‐RAD), 0.3 µM each primer and 1.6 µl template DNA. The amplification procedure was as follows: initial denaturation for 5 min at 95°C, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 52°C for 30 s (16S rRNA), or at 60°C for 40 s (dsrB), or 56.3°C for 45 s at (soxB), and extension at 72°C for 45 s. The data were analysed using the CFX Maestro software, and all correlation coefficients (R 2) of standard curves were > 0.99. The amplification efficiencies (E) of 16S rRNA, dsrB and soxB genes were 110%, 85.7% and 93.4% respectively.
Statistical analysis
Microbial community dissimilarities were visualized by principal coordinate analysis (PCoA) based on Bray–Curtis distance using vegan package (Yao et al., 2014). Pearson’s correlation coefficient was used to determine the relationship between environmental factors. The relationships between microbial communities and environmental factors were determined by the Mantel tests and redundancy analysis (RDA) using vegan package. The statistical significance of the RDA model was tested by Monte Carlo permutation tests (with 999 permutations). Variation partitioning analysis (VPA) was performed to quantify the relative contributions of environmental variables using the varpart procedure in the vegan package. All the analyses as described above were performed using the R statistical programs v3.4.4. Linear correlation of bacterial abundance and environmental variances were conducted using software graphpad prism 7. Differences of environmental factors and taxonomy between different ponds were performed by one‐Way ANOVA and Tukey’s multiple comparison test in graphpad prism 7. The significant level of all data was set at P < 0.05.
Conflict of interest
All authors declare no conflict of interest. None of the materials presented in this manuscript has been previously published, nor are they under consideration for publication elsewhere.
Finding information
This work was supported by the National Natural Science Foundation of China (31672262, 31802350, 31800417), the Fundamental Research Funds for the Central Universities (19lgzd28, 19lgpy164), the Hundred Talents Program through Sun Yat‐sen University (38000‐18821107) and the Foundation of the State Key Laboratory of Applied Microbiology Southern China (SKLAM005‐2018).
Supporting information
Table S1. Primers used for amplification of 16S rRNA, dsrB and soxB genes.
Fig. S1. Microbial richness and diversity in sediments of aquaculture ponds with different sizes of grass carp. Mean values were plotted with the standard deviations (n = 12). Significance (P < 0.05) was tested according to one‐way ANOVA, followed by Tukey’ s multiple comparison test. The presence of different letters denoted significant differences, and the same letter indicated no significant differences.
Fig. S2. Relative abundances of top 10 genera of bacterial communities (16S rRNA), sulfate‐reducing bacteria (dsrB) and sulfur‐oxidizing bacteria (soxB). L: larval; SJ: small juvenile; LJ: large juvenile.
Fig. S3. Mantel tests showing the relationships between microbial communities and environmental factors. The thickness of connecting lines represent correlation level, and wider lines indicate stronger correlation. The Pearson’s test showing the relationship between key environmental factors, only values matched |r| > 0.5 and P < 0.05 were retained, otherwise r value transformed to 0. TSS: total suspended solids; TS: total sulfur; AVS: acid‐volatile sulfur; ES: elemental sulfur; TOC: total organic carbon; TN: total nitrogen.
Fig. S4. Linear regression analysis showing the relationships between abundances of sulfate‐reducing bacteria (reflected by dsrB gene) or sulfur‐oxidizing bacteria (reflected by soxB gene) and key environmental factors. TSS: total suspended solids; TS: total sulfur; AVS: acid‐volatile sulfide.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31672262, 31802350, 31800417), the Fundamental Research Funds for the Central Universities (19lgzd28, 19lgpy164), the Hundred Talents Program through Sun Yat‐sen University (38000‐18821107) and the Foundation of the State Key Laboratory of Applied Microbiology Southern China (SKLAM005‐2018).
Microbial Biotechnology (2020) 13(5), 1597–1610
Funding information
This work was supported by the National Natural Science Foundation of China (31672262, 31802350, 31800417), the Fundamental Research Funds for the Central Universities (19lgzd28, 19lgpy164), the Hundred Talents Program through Sun Yat‐sen University (38000‐18821107) and the Foundation of the State Key Laboratory of Applied Microbiology Southern China (SKLAM005‐2018).
Contributor Information
Xiafei Zheng, Email: zhengxiafei@hotmail.com.
Qingyun Yan, Email: yanqy6@mail.sysu.edu.cn.
Data availability statement
The raw sequencing data can be found at the National Centre for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under accession numbers of SUB6464626 (16S rRNA), SUB6467726 (dsrB) and SUB6467851 (soxB).
References
- Adamek, Z. , and Marsalek, B. (2013) Bioturbation of sediments by benthic macroinvertebrates and fish and its implication for pond ecosystems: a review. Aquacult Int 21: 1–17. [Google Scholar]
- Aller, R.C. (2014) Sedimentary diagenesis, depositional environments, and benthic fluxes. Treatise on Geo 8: 293–334. [Google Scholar]
- Anantharaman, K. , Hausmann, B. , Jungbluth, S.P. , Kantor, R.S. , Lavy, A. , Warren, L.A. , et al (2018) Expanded diversity of microbial groups that shape the dissimilatory sulfur cycle. ISME J 12: 1715–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asami, H. , Aida, M. , and Watanabe, K. (2005) Accelerated sulfur cycle in coastal marine sediment beneath areas of intensive shellfish aquaculture. Appl Environ Microbiol 71: 2925–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, C. , and Jennings, S. (2007) Effect of temperature, ration, body size and age on sulphur isotope fractionation in fish. Rapid Commun Mass Spectrom 21: 1461–1467. [DOI] [PubMed] [Google Scholar]
- Barton, L. , Fardeau, M.L. , and Fauque, G. (2014) The metal‐driven biogeochemistry of gaseous compounds in the environment. Dordrecht, Netherlands: Springer. [Google Scholar]
- Bertics, V. , and Ziebis, W. (2009) Biodiversity of benthic microbial communities in bioturbated coastal sediments is controlled by geochemical microniches. ISME J 3: 1269–1285. [DOI] [PubMed] [Google Scholar]
- Biswas, A.K. , Seoka, M. , Inoue, Y. , Takii, K. , and Kumai, H. (2005) Photoperiod influences the growth, food intake, feed efficiency and digestibility of red sea bream (Pagrus major). Aquaculture 250: 666–673. [Google Scholar]
- Böttcher, M.E. (2001) Sulfur isotope fractionation in the biogeochemical sulfur cycle of marine sediments. Isotopes Environ Health Stud 37: 97–99. [DOI] [PubMed] [Google Scholar]
- Brouwer, H. , and Murphy, T.P. (2010) Diffusion method for the determination of acid‐volatile sulfides (AVS) in sediment. Environ Toxicol Chem 13: 1273–1275. [Google Scholar]
- Brummett, R.E. (2000) Food organism availability and resource partitioning in organically or inorganically fertilized Tilapia rendalli ponds. Aquaculture 183: 57–71. [Google Scholar]
- Bryukhanov, A.L. , Vlasova, M.A. , Malakhova, T.V. , Perevalova, A.A. , and Pimenov, N.V. (2018) Phylogenetic diversity of the sulfur cycle bacteria in the bottom sediments of the Chersonesus Bay. Microbiology 87: 372–381. [Google Scholar]
- Caporaso, J.G. , Kuczynski, J. , Stombaugh, J. , Bittinger, K. , Bushman, F.D. , Costello, E.K. , et al (2010) QIIME allows analysis of high‐throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y. , Yuan, T. , Deng, Y. , Lin, C. , Zhou, J. , Lei, Z. , et al (2018) Use of sulfur‐oxidizing bacteria enriched from sewage sludge to biologically remove H2S from biogas at an industrial‐scale biogas plant. Bioresource Technol Rep 3: 43–50. [Google Scholar]
- Choi, J.H. , Park, S.S. , and Jaffé, P.R. (2006) Simulating the dynamics of sulfur species and zinc in wetland sediments. Ecol Modell 199: 315–323. [Google Scholar]
- Croel, R.C. , and Kneitel, J.M. (2011) Ecosystem‐level effects of bioturbation by the tadpole shrimp Lepidurus packardi in temporary pond mesocosms. Hydrobiologia 665: 169–181. [Google Scholar]
- Dolla, A. , Fournier, M. , and Dermoun, Z. (2006) Oxygen defense in sulfate‐reducing bacteria. J Biotechnol 126: 87–100. [DOI] [PubMed] [Google Scholar]
- Duc, V.L. , Song, B. , Ito, H. , Hama, T. , Otani, M. , and Kawagoshi, Y. (2018) High growth potential and nitrogen removal performance of marine anammox bacteria in shrimp‐aquaculture sediment. Chemosphere 196: 69–77. [DOI] [PubMed] [Google Scholar]
- Dyksma, S. , Bischof, K. , Fuchs, B.M. , Hoffmann, K. , Meier, D. , Meyerdierks, A. , et al (2016) Ubiquitous Gammaproteobacteria dominate dark carbon fixation in coastal sediments. ISME J 10: 1939–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina, J. , Tsutsui, T. , and Shirai, T. (1983) Simultaneous determination of total nitrogen and total phosphorus in water using peroxodisulfate oxidation. Water Res 17: 1721–1726. [Google Scholar]
- Edgar, R.C. (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10: 996. [DOI] [PubMed] [Google Scholar]
- Edgar, R.C. (2016) UNOISE2: improved error‐correction for Illumina 16S and ITS amplicon sequencing. bioRxiv: 081257. [Google Scholar]
- Einen, O. , and Roem, A. (1997) Dietary protein/energy ratios for Atlantic salmon in relation to fish size: growth, feed utilization and slaughter quality. Aquacult Nutr 3: 115–126. [Google Scholar]
- FAO . (2018) The State of World Fisheries and Aquaculture 2018. Rome, Italy: State of World Fisheries and Aquaculture. [Google Scholar]
- Fernandes, S.O. , Kulkarni, S. , Shirodkar, R.R. , Karekar, S.V. , Kumar, R.P. , Rayadurga, S. , et al (2010) Water quality and bacteriology in an aquaculture facility equipped with a new aeration system. Environ Monit Assess 164: 81–92. [DOI] [PubMed] [Google Scholar]
- Griffiths, R.I. , Whiteley, A. , O'Donnell, A. , and Bailey, M.J. (2000) Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA‐ and rRNA‐based microbial community composition. Appl Environ Microbiol 66: 5488–5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, L.S. , and Blackburn, T.H. (1995) Amino acid degradation by sulfate‐reducing bacteria: evaluation of four methods. Limnol Oceanogr 40: 502–510. [Google Scholar]
- Hansen, K.S. , Mouridsen, S. , and Kristensen, E. (1998) The impact of Chironomus plumosus larvae on organic matter decay and nutrient (N, P) exchange in a shallow eutrophic lake sediment following a phytoplankton sedimentation. Hydrobiologia 364: 65–74. [Google Scholar]
- He, H. , Zhen, Y. , Mi, T. , Xu, B. , Wang, G. , Zhang, Y. , et al (2015) Community composition and distribution of sulfate‐ and sulfite‐reducing prokaryotes in sediments from the Changjiang estuary and adjacent East China Sea. Estuar Coast Shelf Sci 165: 75–85. [Google Scholar]
- Holmer, M. , and Kristensen, E. (1996) Seasonality of sulfate reduction and pore water solutes in a marine fish farm sediment: the importance of temperature and sedimentary organic matter. Biogeochemistry 32: 15–39. [Google Scholar]
- Holmer, M. , and Storkholm, P. (2001) Sulphate reduction and sulphur cycling in lake sediments: a review. Freshw Biol 46: 431–451. [Google Scholar]
- Hsieh, Y.P. , and Shieh, Y.N. (1997) Analysis of reduced inorganic sulfur by diffusion methods: improved apparatus and evaluation for sulfur isotopic studies. Chem Geol 137: 255–261. [Google Scholar]
- Hsieh, Y.P. , and Yang, C.H. (1989) Diffusion methods for the determination of reduced inorganic sulfur species in sediments. Limnol Oceanogr 34: 1126–1130. [Google Scholar]
- Huycke, M. , and Gaskins, R. (2004) Commensal bacteria, redox stress, and colorectal cancer: mechanisms and models. Exp Biol Med 229: 586–597. [DOI] [PubMed] [Google Scholar]
- Ihara, H. , Hori, T. , Aoyagi, T. , Takasaki, M. , and Katayama, Y. (2017) Sulfur‐oxidizing bacteria mediate microbial community succession and element cycling in launched marine sediment. Front Microbiol 8: 152–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Japan Fisheries Resource Conservation Association. (1995) Environmental standards for aquaculture, 1995 (in Japanese). Tokyo, Japan: Japan Fisheries Resource Conservation Association. [Google Scholar]
- Jiang, W. , Liang, P. , Wang, B. , Fang, J. , Lang, J. , Tian, G. , et al (2015) Optimized DNA extraction and metagenomic sequencing of airborne microbial communities. Nat Protoc 10: 768–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochum, L.M. , Chen, X. , Lever, M.A. , Loy, A. , Jørgensen, B.B. , Schramm, A. , et al (2017) Depth distribution and assembly of sulfate‐reducing microbial communities in marine sediments of Aarhus Bay. Appl Environ Microbiol 83: e0154717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knossow, N. , Blonder, B. , Eckert, W. , Turchyn, A.V. , Antler, G. , and Kamyshny, A. Jr (2015) Annual sulfur cycle in a warm monomictic lake with sub‐millimolar sulfate concentrations. Geochem Trans 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo, R. , Nedwell, D.B. , Purdy, K.J. , and Silva, S.Q. (2004) Detection and enumeration of sulphate‐reducing bacteria in estuarine sediments by competitive PCR. Geomicrobiol J 21: 145–157. [Google Scholar]
- Kondo, R. , Mori, Y. , and Sakami, T. (2012) Comparison of sulphate‐reducing bacterial communities in Japanese fish farm sediments with different levels of organic enrichment. Bull Jap Soc Microb Ecol 27: 193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnani, K. , Gopikrishna, G. , Pillai, S.M. , and Gupta, B.P. (2010a) Abundance of sulphur‐oxidizing bacteria in coastal aquaculture using soxB gene analyses. Aquacult Res 41: 1290–1301. [Google Scholar]
- Krishnani, K.K. , Kathiravan, V. , Natarajan, M. , Kailasam, M. , and Pillai, S.M. (2010b) Diversity of sulfur‐oxidizing bacteria in greenwater system of coastal aquaculture. Appl Biochem Biotechnol 162: 1225–1237. [DOI] [PubMed] [Google Scholar]
- Kristensen, E. , Penha‐Lopes, G. , Delefosse, M. , Valdemarsen, T. , Quintana, C.O. , and Banta, G. (2012) What is bioturbation? The need for a precise definition for fauna in aquatic science. Mar Ecol Prog Ser 446: 285–302. [Google Scholar]
- Kumar, S. , Stecher, G. , Li, M. , Knyaz, C. , and Tamura, K. (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefèvre, C.T. , Howse, P.A. , Schmidt, M.L. , Sabaty, M. , Menguy, N. , Luther, G.W. , et al (2016) Growth of magnetotactic sulfate‐reducing bacteria in oxygen concentration gradient medium. Environ Microbiol Rep 8: 1003–1015. [DOI] [PubMed] [Google Scholar]
- Leflaive, J. , Danger, M. , Lacroix, G. , Lyautey, E. , Oumarou, C. , and Ten‐Hage, L. (2008) Nutrient effects on the genetic and functional diversity of aquatic bacterial communities. FEMS Microbiol Ecol 66: 379–390. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Jing, H. , Xia, X. , Cheung, S. , Suzuki, K. , and Liu, H. (2018) Metagenomic insights into the microbial community and nutrient cycling in the western subarctic pacific ocean. Front Microbiol 9: 623–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Yan, C. , Kate, L.S. , Zhang, R. , and Lu, H. (2010) The distribution of acid‐volatile sulfide and simultaneously extracted metals in sediments from a mangrove forest and adjacent mudflat in Zhangjiang Estuary, China. Mar Pollut Bull 60: 1209–1216. [DOI] [PubMed] [Google Scholar]
- Lu, X. , Fan, C. , He, W. , Deng, J. , and Yin, H. (2013) Sulfur‐containing amino acid methionine as the precursor of volatile organic sulfur compounds in algea‐induced black bloom. J Environ Sci 25: 33–43. [DOI] [PubMed] [Google Scholar]
- Lyimo, T.J. , Pol, A. , Harhangi, H.R. , Jetten, M.S.M. , and Op den Camp, H.J.M. (2009) Anaerobic oxidation of dimethylsulfide and methanethiol in mangrove sediments is dominated by sulfate‐reducing bacteria. FEMS Microbiol Ecology 70: 483–492. [DOI] [PubMed] [Google Scholar]
- Mendoza‐Lera, C. , and Mutz, M. (2013) Microbial activity and sediment disturbance modulate the vertical water flux in sandy sediments. Freshw Sci 32: 26–38. [Google Scholar]
- Meysman, F.J.R. , Boudreau, B.P. , and Middelburg, J.J. (2005) Modeling reactive transport in sediments subject to bioturbation and compaction. Geochim Cosmochim Acta 69: 3601–3617. [Google Scholar]
- Morse, J.W. , and Rickard, D. (2004) Peer reviewed: chemical dynamics of sedimentary acid volatile sulfide. Environ Sci Technol 38: 131–136. [DOI] [PubMed] [Google Scholar]
- Morse, J.W. , Millero, F.J. , Cornwell, J.C. , and Rickard, D. (1987) The chemistry of the hydrogen sulfide and iron sulfide systems in natural waters. Earth Sci Rev 24: 1–42. [Google Scholar]
- Parsons, T.R. , and Takahashi, M. (1973) Biological oceanographic processes. Oxford, UK: Pergamon. [Google Scholar]
- Pelikan, C. , Herbold, C. , Hausmann, B. , Müller, A. , Pester, M. , and Loy, A. (2015) Diversity analysis of sulfite‐ and sulfate‐reducing microorganisms by multiplex dsrA and dsrB amplicon sequencing using new primers and mock community‐optimized bioinformatics. Environ Microbiol 18: 2994–3009. [DOI] [PubMed] [Google Scholar]
- Pokorna, D. , and Zabranska, J. (2015) Sulfur‐oxidizing bacteria in environmental technology. Biotechnol Adv 33: 1246–1259. [DOI] [PubMed] [Google Scholar]
- Reis, M.A.M. , Almeida, J.S. , Lemos, P.C. , and Carrondo, M.J.T. (1992) Effect of hydrogen sulfide on growth of sulfate reducing bacteria. Biotechnol Bioeng 40: 593–600. [DOI] [PubMed] [Google Scholar]
- Rickard, D. , and Morse, J.W. (2005) Acid volatile sulfide (AVS). Mar Chem 97: 141–197. [Google Scholar]
- Rubio‐Portillo, E. , Villamor, A. , Fernandez‐Gonzalez, V. , Antón, J. , and Sanchez‐Jerez, P. (2019) Exploring changes in bacterial communities to assess the influence of fish farming on marine sediments. Aquaculture 506: 459–464. [Google Scholar]
- Sachidanandamurthy, K.L. , and Yajurvedi, H.N. (2006) A study on physicochemical parameters of an aquaculture body in Mysore city, Karnataka, India. J Environ Biol 27: 615–618. [PubMed] [Google Scholar]
- Santander‐De Leon, S.M.S. , Okunishi, S. , Kihira, M. , Nakano, M. , Nunal, S.N. , Hidaka, M. , et al (2013) Characterization of the bacterial community in the sediment of a brackish lake with oyster aquaculture. Biocontrol Sci 18: 29–40. [DOI] [PubMed] [Google Scholar]
- Sun, J. , Hu, S. , Sharma, K.R. , Ni, B.J. , and Yuan, Z. (2015) Degradation of methanethiol in anaerobic sewers and its correlation with methanogenic activities. Water Res 69: 80–89. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y. , Suto, K. , Inoue, C. , and Chida, T. (2008) Polysulfide reduction using sulfate‐reducing bacteria in a photocatalytic hydrogen generation system. J Biosci Bioeng 106: 219–225. [DOI] [PubMed] [Google Scholar]
- Tian, H. , Gao, P. , Chen, Z. , Li, Y. , Li, Y. , Wang, Y. , et al (2017) Compositions and abundances of sulfate‐reducing and sulfur‐oxidizing microorganisms in water‐flooded petroleum reservoirs with different temperatures in China. Front Microbiol 8: 143–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Ende, F.P. , and Gemerden, H.V. (2010) Sulfide oxidation under oxygen limitation by a Thiobacillus thioparus isolated from a marine microbial mat. FEMS Microbiol Ecol 13: 69–77. [Google Scholar]
- Visscher, P.T. , Taylor, B.F. , and Kiene, R.P. (1995) Microbial consumption of dimethyl sulfide and methanethiol in coastal marine sediments. FEMS Microbiol Ecol 18: 145–154. [Google Scholar]
- Wang, Q. , Quensen, J.F. , Fish, J.A. , Kwon Lee, T. , Sun, Y. , Tiedje, J.M. , et al (2013) Ecological patterns of nifH genes in four terrestrial climatic zones explored with targeted metagenomics using framebot, a new informatics tool MBio 4: e00592‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, T. , Kojima, H. , Takano, Y. , and Fukui, M. (2013) Diversity of sulfur‐cycle prokaryotes in freshwater lake sediments investigated using aprA as the functional marker gene. Syst Appl Microbiol 36: 436–443. [DOI] [PubMed] [Google Scholar]
- White, P. (2003) Organic enrichment of sediments from salmon farming in Norway: environmental factors, management practices, and monitoring techniques. Aquaculture 226: 165–180. [Google Scholar]
- Xia, X.H. , Yang, Z.F. , Huang, G.H. , Zhang, X.Q. , Yu, H. , and Rong, X. (2004) Nitrification in natural waters with high suspended‐solid content‐a study for the Yellow River. Chemosphere 57: 1017–1029. [DOI] [PubMed] [Google Scholar]
- Yao, M. , Rui, J. , Li, J. , Dai, Y. , Bai, Y. , Heděnec, P. , et al (2014) Rate‐specific responses of prokaryotic diversity and structure to nitrogen deposition in the Leymus chinensis steppe. Soil Biol Biochem 79: 81–90. [Google Scholar]
- Yoshida, Y. , Murakami, A. , and Nozawa, K. (1982) Eutrophication and biological indicators in coastal waters (in Japanese). Tokyo, Japan: Koseisha‐Koseikaku. [Google Scholar]
- Zhang, Y. , Wang, G.X. , Zhen, Y. , Mi, Z.T. , He, H. , and Yu, G.Z. (2017) Microbial diversity and community structure of sulfate‐reducing and sulfur‐oxidizing bacteria in sediment cores from the East China Sea. Front Microbiol 8: 2133–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Bruns, M.V. , and Tiedje, J.M. (1996) DNA recovery from soils of diverse composition. Appl Environ Microbiol 62: 316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zopfi, J. , Ferdelman, T.G. , and Fossing, H. (2004) Distribution and fate of sulfur intermediates–sulfite, thiosulfate, and elemental sulfur–in marine sediments. Geol Soc Am 379: 97–116. [Google Scholar]
- Zorn, M.E. , Lalonde, S.V. , Gingras, M.K. , Pemberton, S.G. , and Konhauser, K.O. (2006) Microscale oxygen distribution in various invertebrate burrow walls. Geobiology 4: 137–145. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primers used for amplification of 16S rRNA, dsrB and soxB genes.
Fig. S1. Microbial richness and diversity in sediments of aquaculture ponds with different sizes of grass carp. Mean values were plotted with the standard deviations (n = 12). Significance (P < 0.05) was tested according to one‐way ANOVA, followed by Tukey’ s multiple comparison test. The presence of different letters denoted significant differences, and the same letter indicated no significant differences.
Fig. S2. Relative abundances of top 10 genera of bacterial communities (16S rRNA), sulfate‐reducing bacteria (dsrB) and sulfur‐oxidizing bacteria (soxB). L: larval; SJ: small juvenile; LJ: large juvenile.
Fig. S3. Mantel tests showing the relationships between microbial communities and environmental factors. The thickness of connecting lines represent correlation level, and wider lines indicate stronger correlation. The Pearson’s test showing the relationship between key environmental factors, only values matched |r| > 0.5 and P < 0.05 were retained, otherwise r value transformed to 0. TSS: total suspended solids; TS: total sulfur; AVS: acid‐volatile sulfur; ES: elemental sulfur; TOC: total organic carbon; TN: total nitrogen.
Fig. S4. Linear regression analysis showing the relationships between abundances of sulfate‐reducing bacteria (reflected by dsrB gene) or sulfur‐oxidizing bacteria (reflected by soxB gene) and key environmental factors. TSS: total suspended solids; TS: total sulfur; AVS: acid‐volatile sulfide.
Data Availability Statement
The raw sequencing data can be found at the National Centre for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under accession numbers of SUB6464626 (16S rRNA), SUB6467726 (dsrB) and SUB6467851 (soxB).


