This is the first study to quantify the dependence on wind velocity of airborne bacterial emission fluxes from soil. It demonstrates that manure bacteria get aerosolized from fertilized soil more easily than soil bacteria, and it applies bacterial genomic sequencing for the first time to trace environmental fecal contamination back to its source in the chicken barn.
Summary
This is the first study to quantify the dependence on wind velocity of airborne bacterial emission fluxes from soil. It demonstrates that manure bacteria get aerosolized from fertilized soil more easily than soil bacteria, and it applies bacterial genomic sequencing for the first time to trace environmental faecal contamination back to its source in the chicken barn. We report quantitative, airborne emission fluxes of bacteria during and following the fertilization of agricultural soil with manure from broiler chickens. During the fertilization process, the concentration of airborne bacteria culturable on blood agar medium increased more than 600 000‐fold, and 1 m3 of air carried 2.9 × 105 viable enterococci, i.e. indicators of faecal contamination which had been undetectable in background air samples. Trajectory modelling suggested that atmospheric residence times and dispersion pathways were dependent on the time of day at which fertilization was performed. Measurements in a wind tunnel indicated that airborne bacterial emission fluxes from freshly fertilized soil under local climatic conditions on average were 100‐fold higher than a previous estimate of average emissions from land. Faecal bacteria collected from soil and dust up to seven weeks after fertilization could be traced to their origins in the poultry barn by genomic sequencing. Comparative analyses of 16S rRNA gene sequences from manure, soil and dust showed that manure bacteria got aerosolized preferably, likely due to their attachment to low‐density manure particles. Our data show that fertilization with manure may cause substantial increases of bacterial emissions from agricultural land. After mechanical incorporation of manure into soil, however, the associated risk of airborne infection is low.
Introduction
Ambient air carries diverse bacteria at average concentrations of ~ 104 m−3 (Despres et al., 2012). Airborne bacteria have been studied for their potential adverse health effects (Fröhlich‐Nowoisky et al., 2016) and for their impact on precipitation, as they may function as effective nuclei for cloud condensation and ice formation (Möhler et al., 2007). Most airborne bacteria are attached to other particles, e.g. soil fragments or agglomerates of other bacterial cells, with a median aerodynamic diameter of 4 µm (Despres et al., 2012). Due to their small size, bacteria remain suspended in the atmosphere for an average period of several days, and they can get transported over long distances (up to thousands of kilometres) during this time (Despres et al., 2012). For example, the long‐distance transport of large quantities of dust derived from arid soils in Africa and Asia and its high microbial load upon deposition has been documented (Kellog and Griffin, 2006). Molecular studies detected considerable microbial diversity associated with such desert dust (Yamaguchi et al., 2012; Barberan et al., 2014), and viable bacteria have been cultivated from it, including opportunistic pathogens (Kellog et al., 2004; Prospero et al., 2005; Hervas et al., 2009). However, bacterial emission fluxes from biologically more active regions, including grasslands and plant‐covered agricultural fields, are many times stronger than those from desert soils, as indicated by direct measurements (Lindemann et al., 1982; Lighthart and Shaffer, 1994) and by flux modelling (Burrows et al., 2009b).
Estimates for global bacterial emissions range from 0.7 to 28 million tons annually (Despres et al., 2012). While concentrations and specific identities of airborne bacteria vary greatly over space and time (Burrows et al., 2009a; Bowers et al., 2011; Caliz et al., 2018), the spatial and temporal variation of emission rates has been studied very little, limiting the precision of current numerical models of microbial transport (Burrows et al., 2009b; Despres et al., 2012). Moreover, aerosolized microorganisms have rarely been linked to their specific sources (Bowers et al., 2011), hindering investigations into the mechanisms and dynamics of microbial emission, transport and deposition. As a consequence, microbiology currently lacks a quantitative and mechanistic understanding of the aerial dispersal of environmental bacteria.
In the European Union, 14.5 million tons of chicken meat are produced annually and there are an additional 400 million laying hens (https://ec.europa.eu/agriculture), resulting in the production of approximately 22 million tons of chicken litter per year (Chastain et al., 2002). The majority of this manure is disposed of as fertilizer onto agricultural land without any prior treatment. Solid manure from poultry production is prone to dust emissions due to its high dry matter content and small particle sizes (Kabelitz et al., 2020). It is currently not clear, however, which quantities of faecal bacteria from chicken manure get aerosolized during and after soil fertilization, over which distances these microorganisms may get transported atmospherically, or if they pose a risk of infection.
We here present quantitative measurements of bacterial emissions from soil fertilized with broiler manure, based on field trials and wind tunnel experiments. We assess the contribution of agricultural fertilization to bacterial emissions by modelling emission fluxes and transport routes of airborne bacteria and demonstrate how genomic sequencing can be applied to trace emitted faecal bacteria back to their sources. Finally, we provide an assessment of infection risk based on this data.
Results
Pathogenic bacteria in chicken faeces and manure
We sampled chicken faeces from two intensive broiler fattening farms and tested them for the presence of a variety of pathogenic bacteria by enrichment and by cultivation on selective agar media following dilution to extinction. All faeces samples carried Ent. faecium and ESBL‐producing E. coli. No MRSA was detected, even by using a two‐step enrichment procedure. As expected, manure delivered to the field site 13 days later contained abundant enterococci (Table 1). Interestingly, however, we were unable to recover any ESBL‐producing E. coli from this manure, even though they had been abundant in chicken faeces just a few days before. Results from cultivation‐independent analyses are described below.
Table 1.
Bacterial concentrations in soil, manure and dust.
| Total bacteria (microscopy) | Blood agar counts (CFU) | Enterococci (CFU) | |
|---|---|---|---|
| Manure, per g | 2.4 × 1010 ± 1.4 × 1010 | 5.2 × 108 ± 1.4 × 108 | 8.6 × 106 ± 3.6 × 106 |
| Unfertilized soil, per g | 1.0 × 109 ± 3.9 × 108 | 1.9 × 106 ± 4.0 × 105 | 1.5 × 103 ± 5.7 × 102 |
| Fertilized soil, per g | 1.3 × 109 ± 3.1 × 108 | 5.3 × 106 ± 1.3 × 106 | 1.1 × 105 ± 6.4 × 104 |
| Background air, per m3 | 1.2 × 106 b | 113 b | < Detection limit |
| Dust, manure application, per m3 | 4.9 × 108 ± 1.5 × 108 | 7.3 × 107 ± 1.8 × 107 | 2.9 × 105 ± 6.2 × 104 |
| Dust, manure incorporation, per m3 | 2.9 × 107 ± 3.0 × 106 | 8.4 × 105 ± 1.6 × 105 | 9.4 × 103 a |
| Dust from manure application, per g | 3.2 × 1013 ± 7.5 × 1012 | 5.1 × 1012 ± 1.1 × 1012 | 2.1 × 1010 ± 4.8 × 109 |
| Dust from manure incorporation, per g | 1.7 × 1011 ± 2.6 × 1010 | 4.5 × 109 ± 6.0 × 108 | 8.7 × 107 a |
Means and standard errors from three replicate samples are indicated.
For enterococcal concentrations in dust during manure incorporation, triplicate measurements were not available due to contamination of agar plates.
Background air was sampled only once.
Emissions during manure application and incorporation
We measured emissions of dust and associated microorganisms during the application and incorporation of poultry manure in a field experiment. Details on the characteristics of particulate emissions from this experiment will be reported elsewhere (Münch et al., in press). Briefly, the total measured PM10 concentrations consisted of the regional background and additional dust from agricultural activities. The background was measured and used to adjust PM10 concentrations for the share released by manure application and incorporation. Each time the dust plume emitting from the tractor hit the measuring instruments, PM10 concentrations increased for a few seconds, to maximally 160 µg m−3 during manure application and to maximally 1300 µg m−3 during manure incorporation respectively (Fig. S2). Concentrations in dust plumes were used to calculate source strengths (Maffia et al., 2020) (Equations 2 and 3), and total PM10 emissions were inferred considering processing times, assuming constant dust release (Table 2).
Table 2.
PM10 emissions and bacterial emission fluxes. a
| PM10 | Bacterial cell counts (epifluor. microscopy) | Blood agar CFU | KAA agar CFU (Enterococci) | ||
|---|---|---|---|---|---|
| Fertilizer application | per ha | 0.18 ± 0.12 kg | 5.5 × 1016 ± 3.6 × 1016 | 8.5 × 1015 ± 5.7 × 1015 | 3.5 × 1013 ± 2.4 × 1013 |
| Tillage | per ha | 1.15 ± 0.59 kg | 1.8 × 1014 ± 9.3 × 1013 | 5.0 × 1012 ± 2.6 × 1012 | 2.9 × 1010 ± 1.5 × 1010 |
| Wind‐driven emission, immediately following fertilization b | per ha per month per m2 per second | 6.1 kg (41 kg) |
9.6 × 1014 (6.4 × 1015) 3.6 × 104 (2.5 × 105) |
2.7 × 1013 (1.8 × 1014) 1 × 103 (7.1 × 103) |
1.5 × 1011 (1.0 × 1012) 5.9 (38) |
| Wind‐driven emission, four weeks after fertilization c |
per ha per month per m2 per second |
6.1 kg (41 kg) |
4.7 × 1014 (3.2 × 1015) 1.8 × 104 (1.2 × 105) |
3.9 × 1011 (2.6 × 1012) 15 (100) |
5.3 × 109 (3.6 × 1010) 0.2 (1.3) |
Emissions during fertilizer application and tillage were measured in the field. Wind‐driven emissions were measured in a wind tunnel and projected to the field site considering long‐term weather data.
Long‐term mean (maximum) for March (Fig. 6_PM10release), based on bacterial load measured during tillage.
Long‐term mean (maximum) for March (Fig. 6_PM10release), based on bacterial load measured 4 weeks after fertilization.
In parallel to recording particulate emissions, air samples were collected for microbiological analyses by impingement. The concentration of bacteria culturable on agar plates (blood agar counts) increased 600 000‐fold to 7.3 × 107 per m3 of air during application activity and 7000‐fold to 8.4 × 105 per m3 during incorporation of fertilizer into soil respectively (Table 1). Enterococci had not been detected at all in background air samples, but were found at concentrations of up to 2.9 × 105 per m3 of air during manure application and 9.4 × 103 per m3 during tillage (Table 1). Bacterial concentrations per gram of dust were consistently higher than per gram of soil or manure, respectively (Table 1), indicating the preferred aerosolization of bacteria.
Microscopic cell counts and agar counts decreased with increasing distance from the dust sources (Fig. 1). Of note, enterococcal and blood agar counts were almost 10‐fold higher during manure application than during subsequent incorporation into the soil, even though PM10 emissions were lower (Fig. 1). Based on dust concentrations and a Gaussian dispersion model, 5.5 × 1016 ± 3.6 × 1016 total bacteria (as counted microscopically), including 3.5 × 1013 ± 2.4 × 1013 culturable enterococci, were emitted per hectare during the fertilizer application process, and 1.8 × 1014 ± 9.3 × 1013 bacteria and 2.9 × 1010 ± 1.5 × 1010 enterococci were emitted per hectare during subsequent tillage activities (Table 2).
Fig. 1.
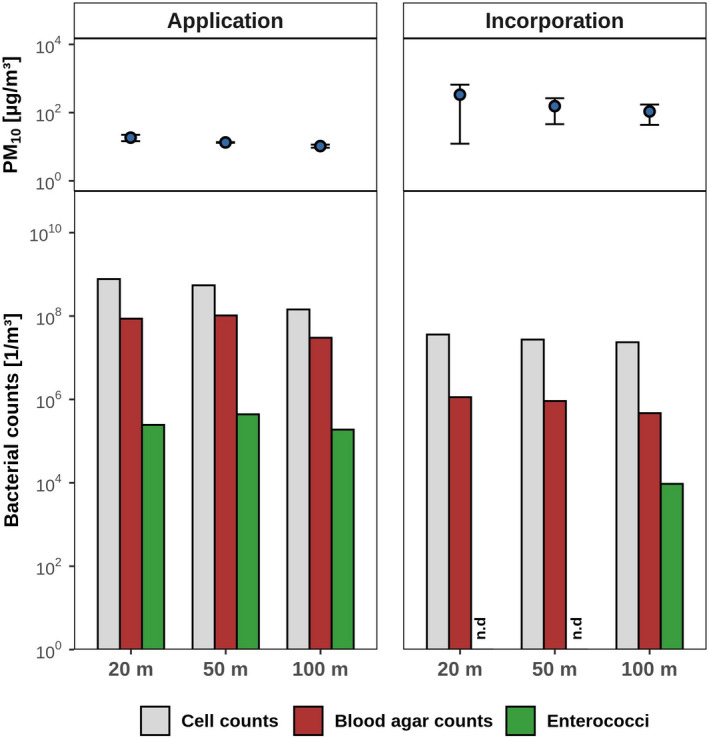
PM10 (top) and microbial emissions (bottom) captured by the measuring instruments during manure application and incorporation processes at increasing distances to the tractor engine. Air samples were collected over 10 min for each distance, and particulate concentrations were monitored in parallel at a height of 1.5 m. Total bacteria were determined as microscopic cell counts, culturable bacteria as blood agar counts and enterococci as counts on KAA medium.
Modelling of dust transport
The transport of dust emitted during manure application and incorporation was simulated by using a dispersion model (Fig. 2). The turbulent nature of the atmospheric boundary layer resulted in an upward mixing of particles to up to 2500 m above ground level, and particles were removed from the atmosphere due to their gravitational settling and turbulent processes depending on their size. The results showed that the maximum number of dust particles released during manure application got deposited within a distance of 20 km, whereas this maximum was at 200 km during subsequent manure incorporation due to a more mature atmospheric boundary layer in the afternoon (Fig. 2). However, during both manure application and incorporation, approximately 10% of dust got transported over > 300 km into a Southeastern direction, and 0.4% or 0.5%, respectively, got transported over > 1000 km (Fig. 2).
Fig. 2.
Simulation of dust transport during field experiment. Probability densities for the height over ground and the distance from the emission source are shown.
A. Simulation for emissions during manure application in the morning (10:49–12:15) of 31 May 2017.
B. Simulation for emissions during incorporation of manure into soil (13:33–14:45).
Estimation of the potential for wind erosion induced PM10 emissions from the test field
We measured PM10 emissions and the associated release of bacteria from fertilized soil in wind tunnel experiments, assuming wind erosion occurs shortly after application and incorporation of manure (Fig. 3). Above the field‐specific threshold wind velocity of 7.0 m s−1, particles were released that initiated the so‐called avalanching effect, resulting in an exponential increase of transport intensity at increasing wind velocity (Figs 3 and 4A). Based on this correlation, we used historic weather data to calculate potential PM10 emissions from this field at an hourly basis for the period 1992 to 2018. Large differences between years were evident, with 0–354 kg of PM10 emitted from this field annually (Fig. 4B). Calculated PM10 emissions correlated with the yearly course of the wind speed, with the highest values inferred for winter and spring (October to April; Fig. 4C). Fertilization is usually performed in spring (March to April) in this area. During this time period, we estimated that 6 ± 15 kg (April; mean, standard deviation) to 13 ± 23 kg (March) of PM10 got emitted from the test field per month in previous years (Fig. 4C). Due to weather variation between years, these estimates ranged from 0 kg to 268 kg per month and therefore were associated with large standard errors when averaged over 27 years. If the field had been fertilized with poultry manure as investigated here, these average dust emissions had been associated with emission fluxes of 3.6 × 104 m−2s−1 total bacteria (based on microscopic counts; range, 0–2.5 × 105 m−2s−1), 103 m−2s−1 bacteria culturable on blood agar (range, 0–7.1 × 103 m−2s−1) and 5.9 m−2s−1 enterococci (range, 0–38) respectively (Table 2).
Fig. 3.
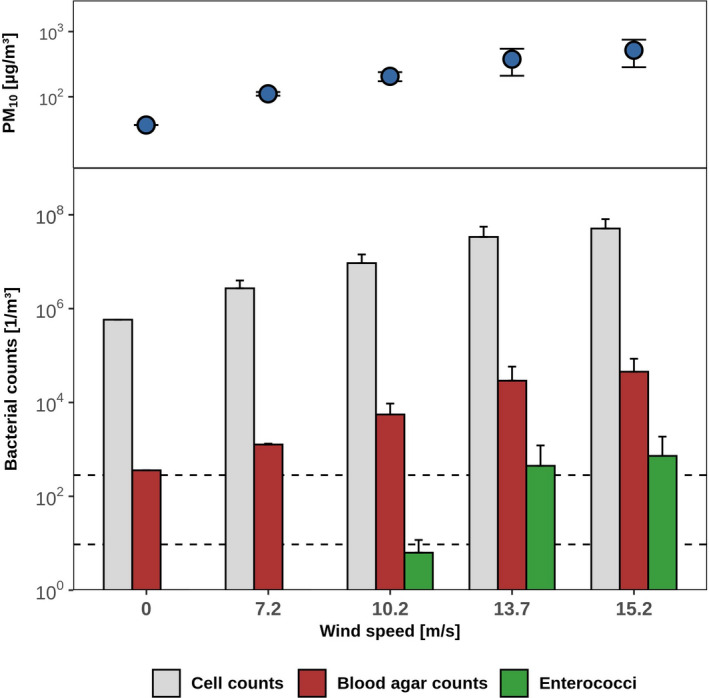
Wind tunnel simulation of wind‐driven release of bacteria from soil four weeks after fertilization. PM10 concentrations, microscopic cell counts and CFU for bacteria on blood agar and for enterococci at four different wind velocities and for background measurements (at 0 m s−1) are depicted; means and standard deviations from three replicate experiments.
Fig. 4.
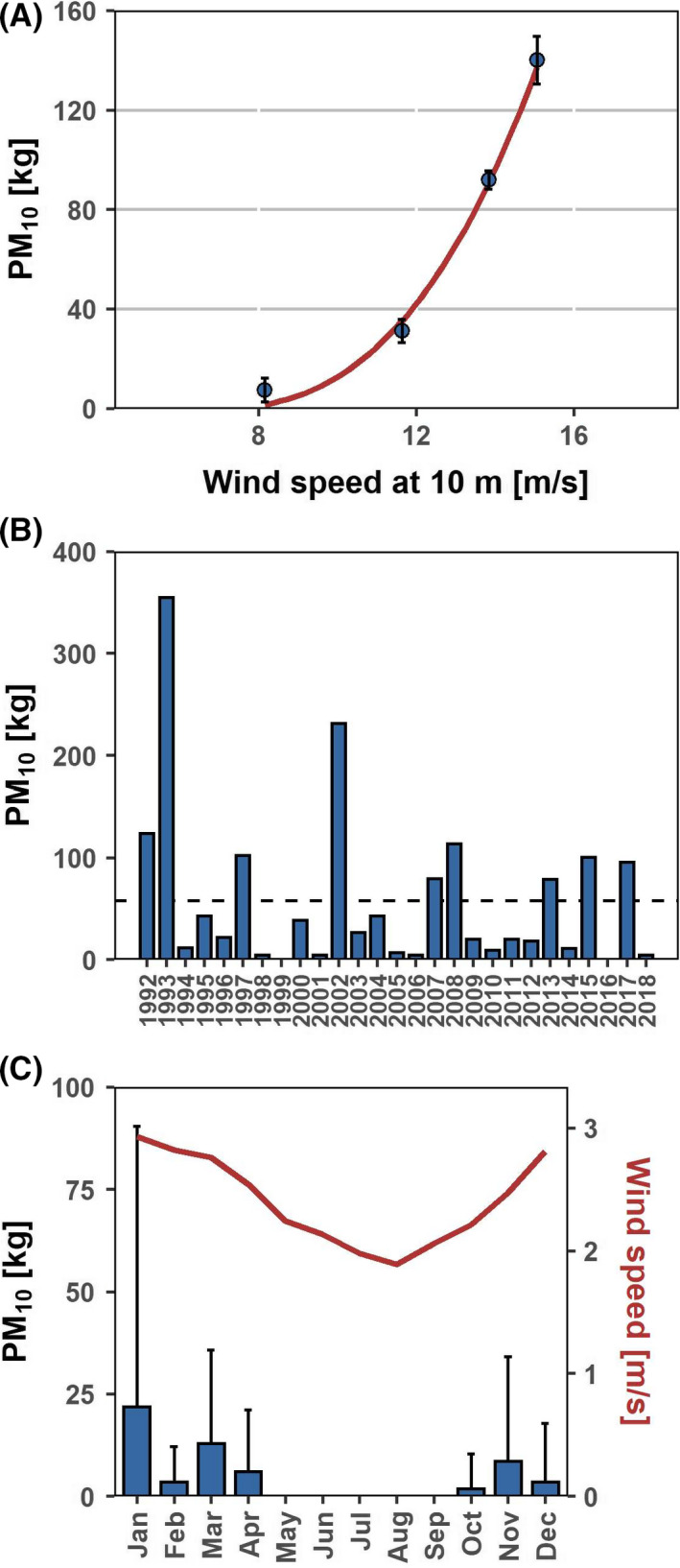
Emissions of PM10 from the test field. (A) Calculated PM10 emission from wind tunnel experiments was plotted against wind velocity. (B,C) Estimated PM10 emissions from the test field (2.1 ha) in the last 26 years. (B) Annual sum, the dashed line indicates the mean annual emission, (C) monthly mean.
Survival of manure‐derived enterococci in soil
We monitored numbers of bacteria (microscopy, blood agar counts) and enterococci (KAA agar) in soil prior to fertilization and thereafter in 2–5 weeks intervals until 19 weeks after fertilization (Fig. 5). In manure, microscopic cell counts were 50‐fold larger than blood agar counts, indicating around 2% culturability of manure bacteria (Table 1). In contrast, culturability of soil bacteria was approximately 0.2% (Table 1). The number of enterococci was 8.6 × 106 ± 3.6 × 106 CFU g−1 in manure and 1.5 × 103 ± 5.7 × 102 CFU g−1 in non‐fertilized soil (Table 1). Fertilization increased the number of enterococci in soil by more than 70‐fold to 1.1 × 105 ± 6.4 × 104 CFU g−1 (Table 1). Seven weeks later, enterococci counts had decreased to background level (Figure 5).
Fig. 5.
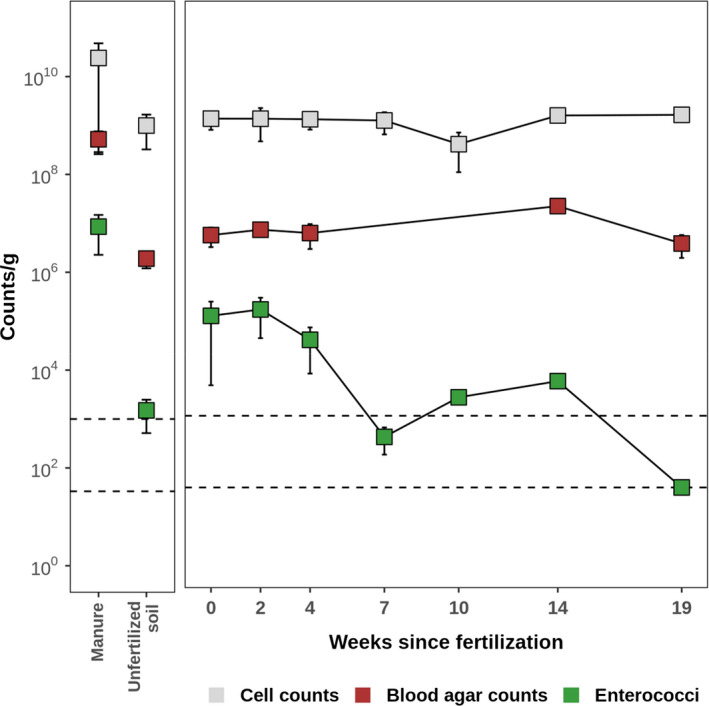
Survival of enterococci in soil after fertilization. Mean bacterial counts from samples collected from three distinct spots on the field site; error bars: standard deviation. LoD, limit of detection; LoQ, limit of quantification.
The enteric pathogen Ent. faecium had not been found in soil samples prior to fertilization, but was consistently identified by species‐specific PCR among enterococcal isolates from fertilized soil until 50 days after fertilization. Other predominant enterococci species were Ent. faecalis and Ent. casseliflavus (identified by mass spectroscopy). Genomes from 92 Ent. faecium isolates, which had been collected from chicken stables (faeces), manure, fertilized soil and dust, got sequenced by using Illumina technology. While genome analyses indicated considerable phylogenetic diversity, encompassing multiple multilocus sequence types (STs), the Ent. faecium isolates were not closely related to strains which frequently cause healthcare‐associated human infections (i.e. STs 17, 18, 117, 80, 192; Fig. 6). Importantly, we found multiple identical or near‐identical Ent. faecium genomes (maximum difference, one core‐genome allele; clades I to VI in Fig. 6) across all samples, including faeces, manure, fertilized soil and dust. Therefore, viable Ent. faecium detected in fertilized soil and dust indeed originated from applied manure, rather than from any environmental sources.
Fig. 6.
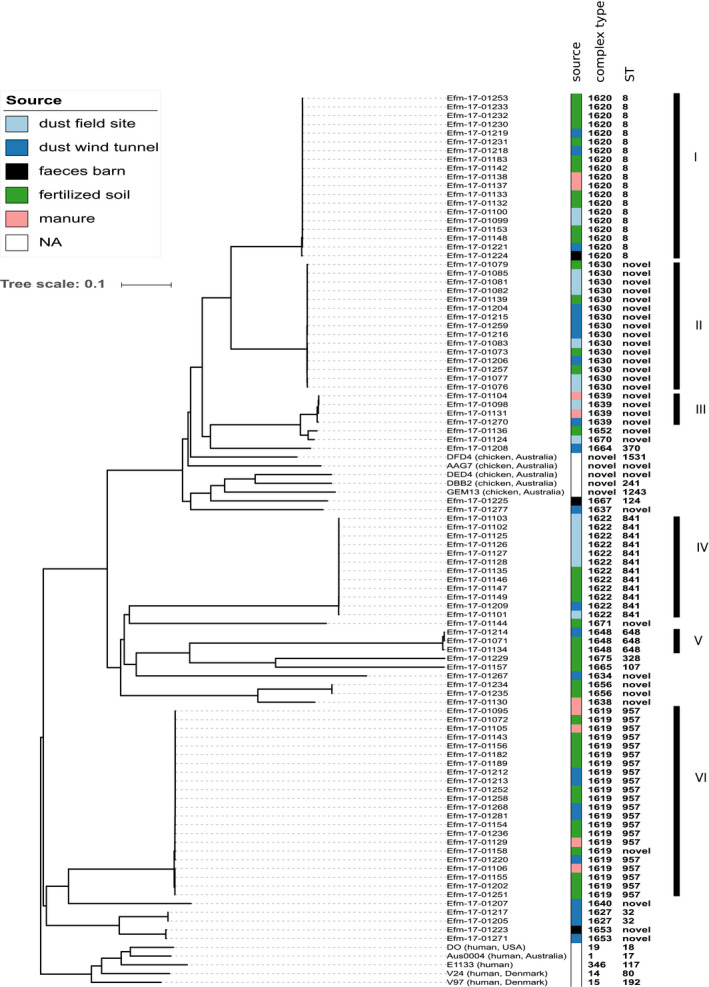
Genome‐based phylogenetic relationships among Ent. faecium isolates. A neighbour‐joining tree was reconstructed from a matrix of core‐genome allelic distances. Colours indicate the sampling sources as indicated. Six clusters with multiple near‐identical genomes each (distance ≤ 1 core‐genome allele) are labelled with roman numerals.
Bacterial diversity
To investigate the composition of bacterial communities in the different samples, we extracted their total DNA and PCR‐amplified and sequenced the V4 region of 16S rDNA (Caporaso et al., 2011). Background air did not provide sufficient DNA for PCR amplification (not shown). The resulting 4.68 × 106 quality‐filtered sequencing reads from 19 samples were assigned to 2.59 × 104 unique amplicon sequence variants (SV). Rarefaction analysis demonstrated that the sequencing depth for manure and almost all dust samples was sufficient to achieve saturation of the number of SVs (Fig. S3). Soil and dust (from wind tunnel experiments) carried greater bacterial diversity than chicken manure (Fig. 7). While only 775 ± 89 SVs were found in manure, numbers of SVs were consistently greater than 3000 in soil and soil‐derived dust (Fig. 7, Fig. S3). Large bacterial diversity in soil and dust samples was also reflected by Shannon and Simpson indices (Fig. 7) and by the number of bacterial phyla and genera detected (Figs S4 and S5).
Fig. 7.
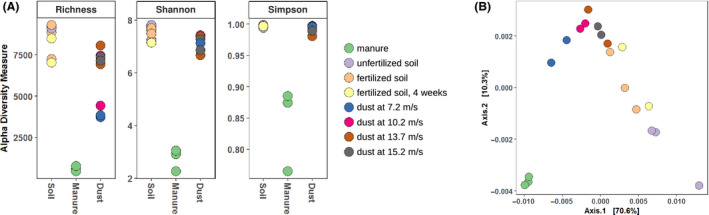
Diversity analyses based on 16S rRNA gene sequencing. Colours indicate types of samples.
A. Alpha diversity measures.
B. Principal coordinate analysis of weighted UniFrac distances between samples.
At the level of individual sequence variants (SVs), 47 predominant SVs, affiliated to nine bacterial families, together accounted for 96% of 16S rDNA sequences from manure (Fig. 8). These SVs accounted for < 0.2% of 16S rDNA from soil prior to fertilization and for 2% of 16S rDNA from freshly fertilized soil (Fig. 8). Four weeks after fertilization, these 16S rDNA sequences had decreased to 0.7% in soil, yet they got enriched to up to 12% in dust emitted from this soil (Fig. 8).
Fig. 8.
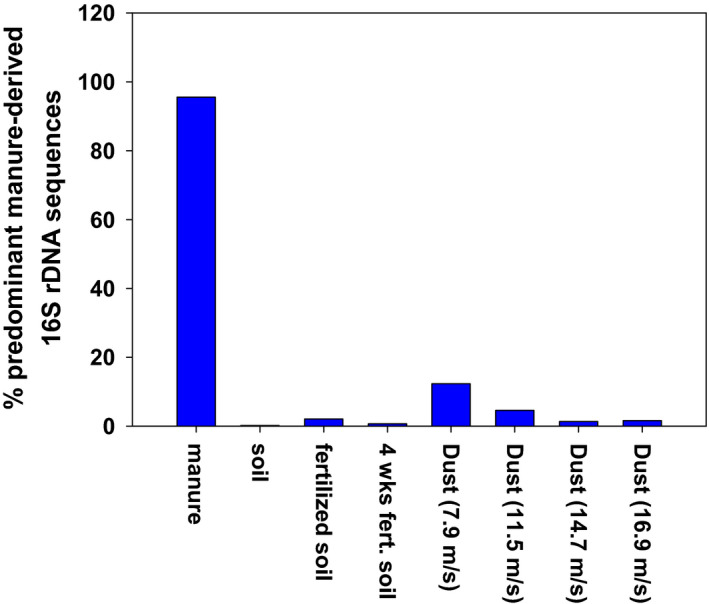
Aggregated abundances of 47 sequence variants that each accounted for > 0.1% of 16S rRNA sequencing reads in chicken manure. These sequence variants were affiliated to Staphylococcaceae, Lactobacillaceae, Leuconostocaceae, Brevibacteriaceae, Corynebacteriaceae, Dermabacteraceae, Nocardiopsaceae, Bacillaceae, Enterobacteriaceae, Ruminococcaceae and Streptococcaceae (in the order of their proportional abundance in manure).
Discussion
Dust‐associated bacterial emissions during fertilizer application and tillage operations
Emissions of PM10 during the process of manure application accounted for total losses of only 0.003% of the 12 tons of manure applied. Considerable numbers of viable bacteria were associated with these particulate emissions, however. Twenty metres downwind from the application site, the concentration in air of bacteria culturable on blood agar medium increased more than 105‐fold compared to background levels. One m3 of air carried 2.9 x 105 enterococci, which had been undetectable in background air.
Comparable data in the literature are scarce, and previously reported concentrations of aerosolized bacteria during the process of spreading liquid manure (cattle slurry) varied by five orders of magnitude, from 2 × 103 culturable bacteria per m3 (Boutin et al., 1988) to 108 enterobacteria per m3 (Hobbs et al., 2004). Our measurements were at the high range compared to this literature data, which were to be expected because poultry litter is dryer (29% water content) and therefore more prone to aerosolization than liquid slurry (Kabelitz et al., 2020).
Measurements of airborne bacterial emissions during tillage operations have not previously been reported. In our field experiment, the quantities of particulate emissions during tillage activities incorporating fertilizer into the soil were in the range to be expected for a sandy clay loam soil with a water content of 9%. Tillage aerosolized 6.4 times larger amounts of PM10 than the preceding fertilizer spreading procedure. This larger amount of dust, however, carried less than 1% of the microscopically counted bacterial cells and only small fractions (< 0.1%) of blood agar plate counts and enterococcal (i.e. KAA agar) plate counts, compared to dust from fertilizer spread (Table 2). Obviously, tillage resulted in a dilution of manure in the soil. As a consequence, the majority of dust released during tillage was derived from the soil and hence contained a large proportion of inanimate mineral particles.
Not surprisingly, our sequence analyses of bacterial 16S rRNA genes revealed drastically different taxonomic compositions of the bacterial communities in soil and manure (Figs 7 and 8). In the past, low culturability from soil dust had been ascribed to high proportions of dead bacterial cells (Lighthart, 1997). More recently, however, it has been well documented that the majority of soil bacteria cannot be readily cultivated on nutrient‐rich agar media (Hugenholtz et al., 1998; Janssen et al., 2002). While 2% of bacteria from manure were successfully cultivated on blood agar in our hands, culturability of bacteria from soil was 10 times lower, at about 0.2% (Table 1). Hence, while large amounts of dust got emitted during tillage operations, it carried comparatively low concentrations of bacteria and particularly low numbers of those microorganisms that could be readily cultivated under standard conditions, including faecal bacteria. We conclude that the atmospheric release of manure‐derived faecal bacteria during fertilizer spread was more quantitatively relevant than during subsequent tillage operations (more than 1000‐fold in our experiments).
Considering real weather conditions on the day of the field experiment, modelling of dust transport indicated that the residence time of aerosolized microorganisms depended strongly on the boundary layer conditions during the emission and during transport. Afternoon conditions favoured the elevation of aerosolized bacteria to greater altitudes. However, large proportions of dust particles released from both manure application and incorporation were predicted to have prevailed in the atmosphere for several days and got transported over several hundred kilometres (Fig. 2), and fractions of 0.01% reached the Black Sea (1,600 km from the source) within 48 h. Assuming a survival rate that was previously measured for airborne Pseudomonas fluorescens, 0.3‰ of aerosolized bacteria would survive a two days atmospheric journey (Amato et al., 2015). This model suggests that of the blood agar detectable bacteria that got aerosolized from our test field during manure application activities (Table 2), 3 × 107 reached the Black Sea in a viable state, including 1 × 105 enterococci. Likewise, 2 × 108 of the total bacteria (microscopic counts) would have had a chance to survive this aerial transport, even though it is not known what proportion of these organisms had been viable when they got lifted from the soil.
Strong wind‐driven bacterial emission fluxes from fertilized soil
Very few quantitative measurements of aerial microbial emission fluxes from soil have been reported throughout the literature. Available studies had presented point measurements, whereas the dependence on wind velocity and on weather conditions had not been investigated in the past. Our wind tunnel experiments provided soil‐specific parameters for the functional relationship between wind velocity and bacterial emissions. Considering long‐term precipitation and wind conditions, our data showed that bacterial emission fluxes from freshly fertilized soil, based on epifluorescence microscopy and averaged over 26 years, were 100‐fold higher than previous mean estimates of fluxes from land (Lindemann et al., 1982; Burrows et al., 2009b). Peak bacterial fluxes during high‐wind months were again almost seven‐fold higher than the long‐term average. Immediately following fertilization, even blood agar counts were eight‐fold higher than previous cultivation‐based estimates (Lindemann et al., 1982), although our cultivation conditions (blood agar, incubated aerobically at 37°C) were not ideal for soil bacteria.
Previous investigations that had relied on bacterial cultivation on agar media alone (Lindemann et al., 1982; Lighthart and Shaffer, 1994) likely underestimated bacterial emission fluxes from soil severely. Our microscopic bacterial cell counts were up to three orders of magnitude higher than cultivation‐based numbers (i.e. agar counts; Table 2), corroborating previous findings on the low culturability of soil bacteria by standard methods (Janssen et al., 2002). Our microscopic counts matched previously reported data from quantitative PCR, targeting bacterial 16S rRNA genes from aerosolized bacteria, which had been collected above agricultural soil following the spread of liquid dairy manure (Jahne et al., 2015).
Our data showed that wind‐driven bacterial emissions from manure‐fertilized soil were lower than those released during manure application, but were quantitatively about equally relevant as emissions caused by tillage (Table 2). Long‐term weather data suggested that conditions for particulate and bacterial emissions from soil were most favourable during winter and spring, i.e. at times, when protective vegetation cover is scarce and fertilization gets performed. Temporal variations of climatic conditions and vegetation may alter the emission strength of soil and thereby contribute to the global, seasonal fluctuations of bacterial air concentrations that had been observed previously (Burrows et al., 2009b). This seasonal variability has as yet not been incorporated in atmospheric models of bacterial transport. Additional investigations are warranted to elucidate the dependence of bacterial emissions on soil type and soil moisture, which are known to influence particulate emissions (Funk et al., 2008).
We conclude that current models (Burrows et al., 2009b) underestimate bacterial emission fluxes from manure‐fertilized agricultural soil about 100‐fold to 670‐fold, depending on actual weather conditions. Very likely, fertilization increased bacterial emission fluxes due to alterations of the soil composition. We observed that manure‐derived bacteria got aerosolized preferably, which was probably due to their attachment to organic manure particles. Wind‐driven erosion of soil causes organic matter to get suspended into the air more easily than mineral particles, leading to its relative enrichment at increasing height above ground (Iturri et al., 2017; Nerger et al., 2017).
Risk of infection
Enterococci are indicators of faecal contamination (Boehm and Sassoubre, 2014). Our comparative analysis of genome sequences demonstrated that Ent. faecium in fertilized soil up to seven weeks after manure application and in emitted dust originated from chicken faeces. While similar persistence of enterococci in manure‐fertilized soil had been reported previously (Stocker et al., 2015; Hodgson et al., 2016), bacterial genome sequencing had not been applied in the past to trace any faecal contamination back to their source. Genome sequencing provides significantly better discriminatory power and specificity than analyses of 16S rRNA genes, which are more commonly applied for this purpose (Unno et al., 2018).
Surprisingly, ESBL‐producing E. coli was below the detection limit in manure and dust, even though it had been abundant in litter collected from chicken barns. However, recent experiments confirmed a rapid decline of ESBL‐producing E. coli under aerobic composting conditions in manure heaps, suggesting that storage of poultry manure for 72 h was sufficient to effectively inactivate the majority of drug‐resistant E. coli (Siller et al., 2020).
Most previous investigations of infection risks associated with the application of manure as agricultural fertilizer considered environmental contamination due to microbial runoff and impairment of stream and ground water quality (Bradford et al., 2013; Blaustein et al., 2015). Only limited research has measured emissions of bioaerosols from cattle and pig slurry during its land application (Boutin et al., 1988; Hobbs et al., 2004). One recent study had recommended a minimum distance of 160 m between manure application sites and any fields growing food crops, to avoid contamination with pathogens from manure (Jahne et al., 2016). Our atmospheric modelling results suggested, however, that transport trajectories of emissions depend on prevailing atmospheric conditions, which could change during the day and result in the transport of manure particles over hundreds of kilometres. In another study, the same authors had assessed the risk of airborne infection from a field fertilized with cattle manure, considering the exposure to manure‐associated gastrointestinal pathogens through swallowing of inhaled bacteria after their deposition in the upper respiratory tract (Jahne et al., 2015). While that study reported a combined probability of infection of 1:500 at a distance of 100 m from the manure application site (Jahne et al., 2015), our measurements indicated 200‐fold higher concentrations of enterococci within the dust plume during fertilizer application, suggesting an increased risk for those directly exposed, e.g. agricultural workers. In contrast, emissions of faecal bacterial were much lower during tillage and afterwards, indicating that the risk of airborne infection from properly prepared fields was low. Incorporation of manure into soil within few hours after application is required by law in Germany, to minimize runoff and gaseous emissions. Obviously, this policy also reduces the risk of airborne infection effectively.
Experimental procedures
Field experiment
A field experiment was performed on 31 May 2017 on 2.1 hectares of fallow land near Müncheberg‐Friedrichshof (Brandenburg, Germany). The field consisted of sandy loam and had not been fertilized with animal waste for 15 years. Two days before the experiment, the test field was pretreated with a chisel plow to subvert the plant cover. Manure for the field experiment was obtained from an intensive poultry‐fattening farm in Brandenburg, housing about 19 000 animals per stable on wood pellets. Chicken housings had been sampled by collecting 30 chicken droppings from each stable. At the end of a fattening period, the stables were mucked out and the accumulated manure (water content, 30%) was transported to the field site, where it was stored in a heap overnight. From 10:49 h to 12:15 h (Central European Summer Time), 12 tons of manure got applied using a manure spreader ‘Strautmann BE4’ (spreading width 3 m). From 13:33 h to 14:45 h, the manure was incorporated into the soil by applying a field cultivator ‘Lemken Smaragd 9’ (working depth 8 cm, working width 3 m). Aerial samples were collected at the leeward side of the field when the tractor reached distances of 20, 50 and 100 m to the measuring point (Fig. S1). The experimental set‐up consisted of two dust monitors (Environmental Dust Monitor EDM #164; Grimm Aerosol‐Technik, Germany) at 1.50 m and 3.80 m height, each measuring particle concentrations continuously every 6 s at a flow rate of 1.2 l min−1, and an XMX‐CV aerosol collection system (Dycor, Canada) at 1.50 m, capturing particles by impingement into 5 ml phosphate‐buffered saline (PBS) at a flow rate of 530 l min−1 for 10 min. After 10 min, the collection buffer was exchanged. Air temperature and relative humidity, wind velocity and wind direction were recorded by using a mobile weather station. Prior to and following fertilization, soil samples were collected at three representative, equally separated spots on the field site. Each of these samples was a mixture of five shovels of soil taken in a star‐shaped pattern around the sampling spot. The same spots were sampled afterwards several times until October 2017 to determine the persistence of bacteria in the soil. Samples were stored at 4°C for up to 24 h until they were analysed for microbial content.
Gaussian dispersion model
To calculate the amount of dust produced during manure application and incorporation, a ‘Gaussian dispersion model’ was used, which describes the ambient air concentration of a pollutant from a continuous emitting point source including a deposition term (Weber, 1976; Anonymous, 1986; Seinfeld and Pandis, 2006):
| (1) |
where c [µg m−3] is the dust concentration, x [m], y [m] and z [m] are the distances from the source in downwind, crosswind and vertical directions, Q [µg s−1] is the source strength (manure spreader or cultivator, respectively), u h [m s−1] is the wind speed at height h [m], σy [m] and σz [m] are the dispersion of dust in horizontal and vertical directions, V di [m s−1] is the deposition velocity of particles with different size classes (V di(PM10) = 0.07 m s−1), and ξ is a non‐dimensional stability parameter. Further, a = σz 2 is for tillage operations and a = 2σz 2 for manure application. For tillage, the vertical dispersion parameter a is reduced to σz 2, because dust from tillage is emitted just above the ground and the dust plume develops only vertically upwards. Rearranging Equation (1) enables the determination of the source strength from the concentration measurements:
| (2) |
The released dust during each operation at the field is finally calculated by
| (3) |
where EF [kg ha−1] is the emission factor per unit area, t [s] is the duration of the operation, and A [ha] is the field size.
Modelling atmospheric transport of dust particles
Trajectory calculations are a commonly applied method to investigate the pathway of a particle through the atmosphere. In order to account for the atmosphere’s strong turbulent fluctuations such as changes in wind speed and direction over time and space, a set of trajectories is calculated. Thereby, a single trajectory represents one statistically possible path of a group of particles. Consequently, a large number of trajectories together represent the most likely transport pathway of all particles emitted from one individual source through the atmosphere. In other words, this results in a distribution of likely dispersion pathways. In the framework of this study, we used the Lagrangian particle simulation module ‘itpas’ (M. Faust and K. Schepanski, unpublished data) based on the COSMO‐Model trajectory module (Miltenberger et al., 2013) to calculate the dispersion of dust‐like particles (diameter, 1–10 µm) within the atmospheric boundary layer (lowest kilometres of the atmosphere). In order to describe the particle dust emission flux and particle size distribution, the model was initialized by particle size distributions and emission fluxes measured during the field experiment (Münch et al., in press). In order to represent the different meteorological conditions during manure application and incorporation, respectively, two 48 h simulations were performed: 31 May 2017 at 10:50 and 13:30 (local time). Meteorological boundary data were taken from the European Center for Medium‐range Weather Forecast ERA5 dataset.
Wind tunnel experiments
As measurements of wind erosion at the field site are quite uncertain due to unsteady weather conditions, we decided to investigate the test field soil's susceptibility to wind erosion in a wind tunnel as described previously (Funk et al., 2019). The boundary layer wind tunnel was operated as an open‐circuit system, receiving ambient air from outside of the building. Twenty‐eight days after manure application and incorporation, soil (500 l) from the upper 8 cm of the test field was collected and filled into the tunnel's working section of 7 m length × 0.7 m width, resulting in a soil layer of 0.1 m thickness. A EDM164 and a Dycor aerosol collector were placed in the tunnel's suspension chamber (Funk et al., 2019). For each experiment, dust (PM10) and bacterial emissions were measured at wind velocities of 7.2, 10.2, 13.7 and 15.2 m s−1. The wind velocity was increased within 30 s to the appropriate value and kept constant for 15 min. Because of the limited availability of erodible fractions of the soil, the surface was exhausted after a certain time, which always happened within the 10 min measuring interval of the Dycor. We estimated the wind velocity‐dependent emissions as sum of the PM10 emitted during the 15 min runs in relation to the total tunnel floor (4.9 m2) as mass per area. After each experiment, the soil surface was refreshed by mixing it with a rake.
Estimation of the potential for wind erosion induced PM10 emissions from the test field
Based on the wind tunnel tests, the following equation was derived, which follows the typical form of sand transport equations to describe the exponential increase of transport intensity q with increasing wind velocity u or friction velocity u* (Funk and Engel, 2015; Jarrah et al., 2020), with
| (4) |
where q, soil flux [kg m−2 s−1]; u*,u*t, friction velocity and threshold friction velocity [ms−1]; A, summarized coefficient of several empirical and physical parameter of influence.
As dust emissions arise from sand transport, there is a direct relationship between both transport modes (Marticorena and Bergametti, 1995). In the wind tunnel experiments, most parameters were kept constant, including soil composition and moisture, aggregation and roughness, so a simplified correlation between wind velocity (u) and PM10 emissions (F PM10), which represents a specific part of the total soil flux q, was obtained by non‐linear regression:
| (5) |
where F PM10 was calculated from concentration [µg m−3], air flow rate [m3 s−1], wind tunnel floor area (4.9 m2) and time [s]. A = 1, will change if initial parameters are changed.
The wind tunnel results were transferred to natural open land conditions by converting wind velocities measured in the wind tunnel to the standard wind velocity at a measuring height of 10 m. The threshold wind velocity ut was 7.0 m s−1, valid for that field with a dry surface and roughness as after seedbed preparation (Funk and Engel, 2015). The total PM10 emissions of the test field were estimated considering its area of 21 000 m2. On the basis of historic weather data [i.e. hourly values of wind velocity, precipitation, temperature, solar irradiation, air humidity from 1992 to 2018 (DWD, 2019)], we estimated the potential extent of PM10 emissions from the test field during the last 27 years. Wind erosion and associated PM10 emissions were assumed when wind velocity exceeded the threshold of 7.0 m s−1, and there had been no precipitation during the preceding 24 h (or when evapotranspiration was higher than precipitation). The vegetation cover was not considered for these estimations.
Cultivation‐based enumeration of bacteria from manure, soil and dust samples
For enumeration of bacteria in manure and soil samples, 10 g of sample material was mixed with 90 g LB medium (Carl Roth). Samples were homogenized for 30 s with a bag mixer (Interscience, St. Nom la Bretèche, France) and left for sedimentation of coarse particles (30 min, room temperature). Dilutions of the supernatants and of impingement suspensions from the aerosol collector were diluted to extinction and streaked on Columbian blood agar (Oxoid) for quantification of total aerobe colony forming units (CFU) and on kanamycin‐aesculin agar (KAA; Oxoid) for selection and quantification of Enterococci spp. Subsequent species identification of Enterococcus faecium was achieved by using a species‐specific PCR (Cheng et al., 1997) and verified by MALDI‐TOF mass spectroscopy. Samples for mass spectroscopy were prepared by ethanol‐formic acid extraction as described previously (Schumann and Maier, 2014) (Protocol 3), and spectra were recorded, evaluated and identified by using a MALDI Biotyper Smart System GP (Bruker Daltonik, Bremen, Germany). In addition, supernatants were streaked on ‘Chromagar VRE’ (Chromagar, France) for vancomycin‐resistant enterococci and on ‘Chromagar MRSA’ (Chromagar, France) for methicillin‐resistant Staphylococcus aureus (MRSA). Extended‐spectrum beta‐lactamase producing E. coli were isolated on McConkey agar No. 3 (Oxoid) supplemented with 1 mg l−1 cefotaxime, and species identification of E. coli was achieved by MALDI‐TOF mass spectroscopy.
The limit of detection (LoD, 1 count per agar plate) was 102 CFU g−1 for soil and manure samples and 9.43 CFU m−3 for dust impingement samples. Accordingly, the limit of quantification (LoQ, 30 counts per agar plate) was 3 × 103 CFU g−1 for soil and 283 CFU m−3 for dust.
Microscopy
For determination of total bacterial counts, an aliquot of each sample was fixed in 1% glutardialdehyde and stored at 4°C until analysis. Samples were stained with SYBR Green and filtered onto black 0.2 µm isopore membrane filters (Merck Millipore, Darmstadt, Germany) (Vieira et al., 2019). Filters were placed between object slide and coverslip and sealed with nail polish, and images were monitored employing a Nikon Eclipse Ti inverted microscope with GFP (485/20‐525/30) filters. Fluorescence z‐stacks were taken using a Nikon N Plan Apochromat λ × 100/1.45 oil objective and an ORCA FLASH 4.0 HAMAMATSU camera. At least 10 fields of view (17 404.19 µm2) per sample were recorded. For quantification, ten fields of view were analysed using the NIS‐Elements software 4.30 (Nikon). Bacterial cells were identified automatically by using the object count tools (smooth: 4×, clean: 4×, fill holes: on, separate: 4×) and manually examined to account for background variation.
DNA extraction and 16S rRNA gene sequencing
Extraction of DNA from manure, soil and dust samples and subsequent PCR amplification of 16S rRNA gene segments were performed according to the 16S rDNA amplicon protocol of the Earth Microbiome Project (http://www.earthmicrobiome.org/protocols‐and‐standards/16s/). In brief, DNA was extracted using the DNeasy PowerSoil Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Primers used for amplification of the V4 region of the 16S rRNA gene were published previously (Caporaso et al., 2011; Parada et al., 2016). Forward primers contained sample‐specific barcodes to enable multiplexing. PCR reactions were performed in triplicates, containing 0.8 x Platinum Hot Start PCR Master Mix (Thermo Fisher Scientific), 0.2 µM each of forward and reverse primer, and 1 ng of template DNA under the following conditions: 3 min at 94°C, 35 cycles at 94°C for 45 s, 50°C for 60 s, 72°C for 90 s and a final extension step at 72°C for 10 min. Equimolar amounts of merged triplicates were pooled, purified by using QIAquick PCR purification columns (Qiagen) and sequenced on the MiSeq platform (MiSeq Reagent Kit v3, 600 cycle; Illumina).
Processing of 16S rRNA gene sequences
Amplicon sequences were analysed via the plugin‐based qiime2 pipeline (version 2018.4; Bolyen, 2018). Demultiplexed forward reads were trimmed to 260 bases according to quality parameters using fastx_trimmer (FASTX Toolkit Version 0.0.14). Truncated reads were loaded into the Qiime2 environment and processed by using implemented software tools. By using Deblur (Amir et al., 2017), sequences were error‐filtered and unique sequence variants (SV) identified. Sequences were aligned with mafft (Katoh and Standley, 2013) and used to build a phylogenetic tree with fasttree (Price et al., 2009). Taxonomy was assigned to the respective SVs by alignment with the SILVA reference database, release 128 (Quast et al., 2013). Resulting BIOMtable, taxonomy and rooted tree were exported and further processed with R (version 3.4.3, available at https://www.R‐project.org/) using the R package ‘phyloseq’ (McMurdie and Holmes, 2013). Rarefaction analysis was performed by plotting the number of SVs versus sequencing depth with the R package ‘ranacapa’ (version 0.1.0). For normalization, sequence counts were rarefied to the read count of the sample with the lowest total number of reads after error filtering (1.23 × 105 reads). As alpha diversity estimates, richness (observed number of SVs per sample) and Shannon and Simpson indices were computed. For estimation of beta diversity, unweighted and weighted UniFrac distance matrices were determined and subjected to principal coordinate analysis (PCoA) using the ‘phyloseq’ package.
Genome sequencing and phylogenetic analysis
DNA was purified from plate‐grown Ent. faecium using the DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer's instructions, except that bacterial cell lysis was performed with lysozyme (90 000 U) in 1% Triton X‐100 for 45 min at 37°C. Sequencing libraries were prepared using a modified Nextera XT protocol (Baym et al., 2015; Steglich et al., 2018). The libraries were sequenced on a NextSeq machine with a NextSeq 500/550 midoutput v2 kit (Illumina). Illumina sequencing reads were assembled by using spades v3.13.0, and resulting contigs were processed in seqsphere 6.0.2 for cgMLST allele calling and for determination of cgMLST‐based complex types and classical MLST sequence types (STs; de Been et al., 2015). Distances between cgMLST allelic profiles were used to construct a neighbour‐joining phylogenetic tree, which was subsequently annotated with iTOL 4.0.3 (Letunic and Bork, 2016). Genome sequencing read data from 92 Enterococcus faecium isolates were submitted to the European Nucleotide Archive under accession number PRJEB36824.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
Fig. S1. Experimental set‐up. (A) Satellite image of test field in Brandenburg near Müncheberg. The blue arrow indicates the predominant wind direction on the day of the test field, the asterisk indicates the position of the measuring equipment, and the dotted lines mark the tractor tracks during air sample collection. (B) Tractor equipped with manure spreader during manure application. (C) Tractor equipped with field cultivator during manure incorporation. (D) Measuring equipment consisted of two dust monitors at different heights and an aerosol collection device. (E) Wind tunnel.
Fig. S2. PM10 concentrations measured during manure application and incorporation at heights of 3.80 m and 1.50 m, as indicated. Light blue shading indicates time periods of aerosol collection by impingement.
Fig. S3. Rarefaction curves indicating the number of 16S rDNA sequence variants (SV) detected as a function of sequencing depth. The dotted line highlights the number of reads used for rarefying (normalization, N = 120 000).
Fig. S4. Taxonomic composition of 16S rDNA sequencing data at phylum level.
Fig. S5. Taxonomic composition of 16S rDNA sequencing data at genus level.
Acknowledgements
We thank the farm owners for supporting this study, allowing sample collection at their facilities and donation of manure for experiments. We are grateful to U. Steiner for technical assistance, to J. Sikorski for help with Qiime2 and R and for helpful comments on the manuscript, D. Jacob for language editing, and to the Leibniz research alliance Infections '21 for support. This study was funded by the Leibniz Association (grant number: SAW‐2017‐DSMZ‐2). Open access funding enabled and organized by Projekt DEAL.
Microbial Biotechnology (2020) 13(5), 1631–1647
Funding information
This study was funded by the Leibniz Association (grant number: SAW‐2017‐DSMZ‐2).
References
- Amato, P. , Joly, M. , Schaupp, C. , Attard, E. , Möhler, O. , Morris, C.E. , et al (2015) Survival and ice nucleation activity of bacteria as aerosols in a cloud simulation chamber. Atmos Chem Phys 15: 6455–6465. [Google Scholar]
- Amir, A. , McDonald, D. , Navas‐Molina, J.A. , Kopylova, E. , Morton, J.T. , Zech Xu, Z. , et al (2017) Deblur rapidly resolves single‐nucleotide community sequence patterns. mSystems 2 e00191–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous (1986) Erste allgemeine Verwaltungsvorschrift zum Bundes‐Immissionsschutzgesetz – Technische Anleitung zur Reinhaltung der Luft (TA Luft) vom 7. Februar 1986. Gemeinsames Ministerialblatt (GMBl) G 3191: 95.
- Barberan, A. , Henley, J. , Fierer, N. , and Casamayor, E.O. (2014) Structure, inter‐annual recurrence, and global‐scale connectivity of airborne microbial communities. Sci Total Environ 487: 187–195. [DOI] [PubMed] [Google Scholar]
- Baym, M. , Kryazhimskiy, S. , Lieberman, T.D. , Chung, H. , Desai, M.M. , and Kishony, R. (2015) Inexpensive multiplexed library preparation for megabase‐sized genomes. PLoS One 10: e0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Been, M. , Pinholt, M. , Top, J. , Bletz, S. , Mellmann, A. , van Schaik, W. , et al (2015) Core genome multilocus sequence typing scheme for high‐resolution typing of Enterococcus faecium . J Clin Microbiol 53: 3788–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaustein, R.A. , Pachepsky, Y.A. , Shelton, D.R. , and Hill, R.L. (2015) Release and removal of microorganisms from land‐deposited animal waste and animal manures: a review of data and models. J Environ Qual 44: 1338–1354. [DOI] [PubMed] [Google Scholar]
- Boehm, A.B. , and Sassoubre, L.M. (2014) Enterococci as indicators of environmental fecal contamination In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Gilmore M.S., Clewell D.B., Ike Y., and Shankar N. (eds). Boston, MA: Massachusetts Eye and Ear Infirmary; https://www.ncbi.nlm.nih.gov/books/NBK190421/. [PubMed] [Google Scholar]
- Bolyen, E. (2018) Reproducible, interactive, scalable, and extensible microbiome data science. Peer J Preprints 6: e27295v27292. [Google Scholar]
- Boutin, P. , Torre, M. , Serceau, R. , and Rideau, P.‐J. (1988) Atmospheric bacterial contamination from landspreading of animal wastes: evaluation of the respiratory risk for people nearby. J Agric Eng Res 39: 149–160. [Google Scholar]
- Bowers, R.M. , McLetchie, S. , Knight, R. , and Fierer, N. (2011) Spatial variability in airborne bacterial communities across land‐use types and their relationship to the bacterial communities of potential source environments. ISME J 5: 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, S.A. , Morales, V.L. , Zhang, W. , Harvey, R.W. , Packman, A.I. , Mohanram, A. , and Welty, C. (2013) Transport and fate of microbial pathogens in agricultural settings. Crit Rev Environ Sci Technol 43: 775–893. [Google Scholar]
- Burrows, S.M. , Elbert, W. , Lawrence, M.G. , and Pöschl, U. (2009a) Bacteria in the global atmosphere ‐ part 1: review and synthesis of literature data for different ecosystems. Atmos Chem Phys 9: 9263–9280. [Google Scholar]
- Burrows, S.M. , Butler, T. , Jöckel, P. , Tost, H. , Kerkweg, A. , Pöschl, U. , and Lawrence, M.G. (2009b) Bacteria in the global atmosphere ‐ part 2: modeling of emissions and transport between different ecosystems. Atmos Chem Phys 9: 9281–9297. [Google Scholar]
- Caliz, J. , Triado‐Margarit, X. , Camarero, L. , and Casamayor, E.O. (2018) A long‐term survey unveils strong seasonal patterns in the airborne microbiome coupled to general and regional atmospheric circulations. Proc Natl Acad Sci USA 115: 12229–12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, J.G. , Lauber, C.L. , Walters, W.A. , Berg‐Lyons, D. , Lozupone, C.A. , Turnbaugh, P.J. , et al (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108: 4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastain, J.P. , Camberato, J.J. , and Skewes, P. (2002) Poultry manure production and nutrient content In Poultry Training Manual. Clemson, SC: Clemson University, College of Agriculture, Forestry and Life Sciences; https://www.clemson.edu/extension/camm/manuals/. [Google Scholar]
- Cheng, S. , McCleskey, F.K. , Gress, M.J. , Petroziello, J.M. , Liu, R. , Namdari, H. , et al (1997) A PCR assay for identification of Enterococcus faecium . J Clin Microbiol 35: 1248–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres, V.R. , Huffman, J.A. , Burrows, S.M. , Hoose, C. , Safatov, A.S. , Buryak, G. , et al (2012) Primary biological aerosol particles in the atmosphere: a review. Tellus Series B: Chem Phys Meteorol 64: 15598. [Google Scholar]
- DWD (2019) German Weather Service. Download from the Climate Data Center. URL https://www.dwd.de/EN/ourservices/cdcftp/cdcftp.html.
- Fröhlich‐Nowoisky, J. , Kampf, C.J. , Weber, B. , Huffman, J.A. , Pöhlker, C. , Andreae, M.O. , et al (2016) Bioaerosols in the Earth system: climate, health, and ecosystem interactions. Atmos Res 182: 346–376. [Google Scholar]
- Funk, R. , and Engel, W. (2015) Investigations with a field wind tunnel to estimate the wind erosion risk of row crops. Soil Till Res 145: 224–232. [Google Scholar]
- Funk, R. , Reuter, H.I. , Hoffmann, C. , Engel, W. , and Oettl, D. (2008) Effect of moisture on fine dust emission from tillage operations on agricultural soils. Earth Surf Proc Land 33: 1851–1863. [Google Scholar]
- Funk, R. , Papke, N. , and Hör, B. (2019) Wind tunnel tests to estimate PM10 and PM2.5 emissions from complex substrates of open‐cast strip mines in Germany. Aeol Res 39: 23–32. [Google Scholar]
- Hervas, A. , Camarero, L. , Reche, I. , and Casamayor, E.O. (2009) Viability and potential for immigration of airborne bacteria from Africa that reach high mountain lakes in Europe. Environ Microbiol 11: 1612–1623. [DOI] [PubMed] [Google Scholar]
- Hobbs, P. , Davies, D. , Williams, J. , Bakewell, H. , and Smith, K. (2004) Measurement of pathogen transfer in aerosols following land application of manure In Sustainable Organic Waste Management for Environmental Protection and Food Safety, vol II: Proceedings of the 11th International Conference of the FAO ESCORENA Network on the Recycling of Agricultural, Municipal and Industrial Residues in Agriculture (RAMIRAN), Murcia, Spain. [Google Scholar]
- Hodgson, C.J. , Oliver, D.M. , Fish, R.D. , Bulmer, N.M. , Heathwaite, A.L. , Winter, M. , and Chadwick, D.R. (2016) Seasonal persistence of faecal indicator organisms in soil following dairy slurry application to land by surface broadcasting and shallow injection. J Environ Manage 183: 325–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz, P. , Goebel, B.M. , and Pace, N.R. (1998) Impact of culture‐independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol 180: 4765–4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturri, L.A. , Funk, R. , Leue, M. , Sommer, M. , and Buschiazzo, D.E. (2017) Wind sorting affects differently the organo‐mineral composition of saltating and particulate materials in contrasting texture agricultural soils. Aeol Res 28: 39–49. [Google Scholar]
- Jahne, M.A. , Rogers, S.W. , Holsen, T.M. , and Grimberg, S.J. (2015) Quantitative microbial risk assessment of bioaerosols from a manure application site. Aerobiologia 31: 73–87. [Google Scholar]
- Jahne, M.A. , Rogers, S.W. , Holsen, T.M. , Grimberg, S.J. , Ramler, I.P. , and Kim, S. (2016) Bioaerosol deposition to food crops near manure application: quantitative microbial risk assessment. J Environ Qual 45: 666–674. [DOI] [PubMed] [Google Scholar]
- Janssen, P.H. , Yates, P.S. , Grinton, B.E. , Taylor, P.M. , and Sait, M. (2002) Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia . Appl Environ Microbiol 68: 2391–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrah, M. , Mayel, S. , Tatarko, J. , Funk, R. , and Kuka, K. (2020) A review of wind erosion models: data requirements, processes, and validity. Catena 187: 104388. [Google Scholar]
- Kabelitz, T. , Ammon, C. , Funk, R. , Münch, S. , Biniasch, O. , Nübel, U. , et al (2020) Functional relationship of particulate matter (PM) emissions, animal species, and moisture content during manure application. Environ Int 143: 105577 [DOI] [PubMed] [Google Scholar]
- Katoh, K. , and Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellog, C.A. , and Griffin, D.W. (2006) Aerobiology and the global transport of desert dust. Trends Ecol Evol 21: 638–644. [DOI] [PubMed] [Google Scholar]
- Kellog, C.A. , Griffin, D.W. , Garrison, V.H. , Peak, K.K. , Royall, N. , Smith, R.R. , and Shinn, E.A. (2004) Characterization of aerosolized bacteria and fungi from desert dust events in Mali, West Africa. Aerobiologia 20: 99–110. [Google Scholar]
- Letunic, I. , and Bork, P. (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44: W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lighthart, B. (1997) The ecology of bacteria in the alfresco atmosphere. FEMS Microbiol Ecol 23: 263–274. [Google Scholar]
- Lighthart, B. , and Shaffer, B.T. (1994) Bacterial flux from Chaparral into the atmosphere in mid‐summer at a high desert location. Atmos Environ 28: 1267–1274. [Google Scholar]
- Lindemann, J. , Constantinidou, H.A. , Barchet, W.R. , and Upper, C.D. (1982) Plants as sources of airborne bacteria, including ice nucleation‐active bacteria. Appl Environ Microbiol 44: 1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffia, J. , Dinuccio, E. , Amon, B. , and Balsari, P. (2020) PM emissions from open field crop management: emission factors, assessment methods and mitigation measures – a review. Atmos Environ 226: 117381. [Google Scholar]
- Marticorena, B. , and Bergametti, G. (1995) Modeling the atmospheric dust cycle: 1. Design of a soil‐derived dust emission scheme. J Geophys Res 100: D8: 16,415–416,430. [Google Scholar]
- McMurdie, P.J. , and Holmes, S. (2013) Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8: e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miltenberger, A.K. , Pfahl, S. , and Wernli, H. (2013) An online trajectory module (version 1.0) for the nonhydrostatic numerical weather prediction model COSMO. Geosci Model Dev 6: 1989–2004. [Google Scholar]
- Möhler, O. , DeMott, P.J. , Vali, G. , and Levin, Z. (2007) Microbiology and atmospheric processes: the role of biological particles in cloud physics. Biogeosciences 4: 1059–1071. [Google Scholar]
- Münch, S. , Papke, N. , Thiel, N. , Nübel, U. , Siller, P. , Roesler, U. , et al. The effect of farmyard manure application on dust emissions from arable soils. Atmospheric Pollution Research (in press). 10.1016/j.apr.2020.06.007 [DOI] [Google Scholar]
- Nerger, R. , Funk, R. , Cordsen, E. , and Fohrer, N. (2017) Application of a modeling approach to designate soil and soil organic carbon loss to wind erosion on long‐term monitoring sites (BDF) in Northern Germany. Aeol Res 25: 135–147. [Google Scholar]
- Parada, A.E. , Needham, D.M. , and Fuhrman, J.A. (2016) Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18: 1403–1414. [DOI] [PubMed] [Google Scholar]
- Price, M.N. , Dehal, P.S. , and Arkin, A.P. (2009) Fasttree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26: 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prospero, J.M. , Blades, E. , Mathison, G. , and Naidu, R. (2005) Interhemispheric transport of viable fungi and bacteria from Africa to the Caribbean with soil dust. Aerobiologia 21: 1–19. [Google Scholar]
- Quast, C. , Pruesse, E. , Yilmaz, P. , Gerken, J. , Schweer, T. , Yarza, P. , et al (2013) The SILVA ribosomal RNA gene database project: improved data processing and web‐based tools. Nucleic Acids Res 41: 590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann, P. , and Maier, T. (2014) Chapter 13 ‐ MALDI‐TOF mass spectrometry applied to classification and identification of bacteria In Methods in Microbiology. Goodfellow M.(ed). Cambridge, MA: Academic Press, pp. 275–306. [Google Scholar]
- Seinfeld, J.H. , and Pandis, S.N. (2006) Atmospheric Chemistry and Physics: From Air Pollution to Climate Change. Hoboken, NJ: John Wiley & Sons. [Google Scholar]
- Siller, P. , Daehre, K. , Thiel, N. , Nübel, U. , and Roesler, U. (2020) Impact of short‐term storage on the quantity of extended‐spectrum beta‐lactamase‐producing Escherichia coli in broiler litter under practical conditions. Poultry Sci 99: 2125–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steglich, M. , Hoffmann, J.D. , Helmecke, J. , Sikorski, J. , Spröer, C. , Riedel, T. , et al (2018) Convergent loss of ABC transporter genes from Clostridioides difficile genomes is associated with impaired tyrosine uptake and p‐cresol production. Front Microbiol 9: 901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker, M.D. , Pachepsky, Y.A. , Hill, R.L. , and Shelton, D.R. (2015) Depth‐dependent survival of Escherichia coli and enterococci in soil after manure application and simulated rainfall. Appl Environ Microbiol 81: 4801–4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unno, T. , Staley, C. , Brown, C.M. , Han, D. , Sadowsky, M.J. , and Hur, H.G. (2018) Fecal pollution: new trends and challenges in microbial source tracking using next‐generation sequencing. Environ Microbiol 20: 3132–3140. [DOI] [PubMed] [Google Scholar]
- Vieira, S. , Sikorski, J. , Gebala, A. , Boeddinghaus, R.S. , Marhan, S. , Rennert, T. , et al (2019) Bacterial colonization of minerals in grassland soils is selective and highly dynamic. Environ Microbiol 22: 917–933. [DOI] [PubMed] [Google Scholar]
- Weber, A.H. (1976) Atmospheric Dispersion Parameters in Gaussian Plume Modeling. Part 1: Review of Current Systems and Possible Future Developments. Raleigh, NC: North Carolina State University. [Google Scholar]
- Yamaguchi, N. , Ichijo, T. , Sakotani, A. , Baba, T. , and Nasu, M. (2012) Global dispersion of bacterial cells on Asian dust. Sci Rep 2: 525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Experimental set‐up. (A) Satellite image of test field in Brandenburg near Müncheberg. The blue arrow indicates the predominant wind direction on the day of the test field, the asterisk indicates the position of the measuring equipment, and the dotted lines mark the tractor tracks during air sample collection. (B) Tractor equipped with manure spreader during manure application. (C) Tractor equipped with field cultivator during manure incorporation. (D) Measuring equipment consisted of two dust monitors at different heights and an aerosol collection device. (E) Wind tunnel.
Fig. S2. PM10 concentrations measured during manure application and incorporation at heights of 3.80 m and 1.50 m, as indicated. Light blue shading indicates time periods of aerosol collection by impingement.
Fig. S3. Rarefaction curves indicating the number of 16S rDNA sequence variants (SV) detected as a function of sequencing depth. The dotted line highlights the number of reads used for rarefying (normalization, N = 120 000).
Fig. S4. Taxonomic composition of 16S rDNA sequencing data at phylum level.
Fig. S5. Taxonomic composition of 16S rDNA sequencing data at genus level.


