Abstract
Purpose
Lung cancer is a leading cause of cancer-related death, with lung adenocarcinoma (LUAD) representing the most common subtype. Recently, exosome-based biomarkers have provided new diagnostic approaches for malignancies. We aimed to identify specific exosomal microRNAs (miRNAs) as noninvasive biomarkers for LUAD.
Patients and Methods
A total of 110 participants were enrolled and randomly divided into two sets: the discovery set (n=20) and the validation set (n=90). Exosomes were isolated from serum, and miRNAs were subsequently extracted. Candidate miRNAs (miR-21, miR-221-3p, miR-222-3p, miR-223, miR-638 and miR-1290) were detected by quantitative real-time PCR (qRT-PCR) in the discovery set. The upregulated miR-1290 was then selected for further analysis in the validation set along with three tumor markers (CEA, CYFRA21-1 and NSE). The diagnostic and prognostic value of exosomal miR-1290 were estimated through receiver-operating characteristic (ROC) and survival analysis.
Results
Serum exosomal miR-1290 was significantly upregulated in LUAD patients compared to healthy controls (P<0.001) and decreased after resection (P=0.0029). Its expression level was associated with tumor stage, tumor size, lymph node and distant metastasis (all P <0.05). Exosomal miR-1290 had a higher diagnostic efficacy than CEA, CYFRA21-1 and NSE, with a sensitivity of 80.0% and specificity of 96.7% (AUC: 0.937, 95% CI: 0.890–0.985; P<0.001). Moreover, LUAD patients with a high level of exosomal miR-1290 had significantly poorer progression-free survival (PFS) than those with a low level of exosomal miR-1290 (mean PFS: 14 months vs 37 months, P<0.001). Cox proportional hazards model analysis demonstrated that exosomal miR-1290 could be an independent risk factor for the prognosis of LUAD (HR=7.80, P=0.017).
Conclusion
Serum exosomal miR-1290 could be a potential diagnostic and prognostic biomarker for LUAD.
Keywords: lung adenocarcinoma, circulating miRNA, exosome, biomarker
Introduction
As the most common fatal malignancy, lung cancer has become the leading cause of cancer-related death, accounting for twenty-five percent of cancer-related mortalities.1,2 Lung adenocarcinoma (LUAD) is the most common histological subtype of lung cancer, and its incidence and mortality have recently increased.3 Although the development of targeted therapy and immunotherapy has improved the prognosis of LUAD patients, the 5-year survival rate remains below 20%, which is partly due to delayed diagnosis. Low-dose computed tomography (LDCT), as the currently recommended screening method, has been found to reduce lung cancer mortality in individuals who smoke. However, its high false-positive rate may result in unnecessary radiation harm and high costs.1 Moreover, traditional tumor markers for lung cancer, such as carcinoembryonic antigen (CEA), cytokeratin 19 fragment (CYFRA21-1), and neuron-specific enolase (NSE), has shown insufficient specificity or sensitivity for a reliable evaluation.4 Hence, effective biomarkers of LUAD remain required.
Exosomes are membranous extracellular vesicles with a diameter of 30 to 100 nm and are secreted by various cell types.5 They are demonstrated to be critical mediators of intercellular communications by transferring biomolecules such as proteins, lipids and nucleic acids.6 Tumor-derived exosomes can be delivered into the local microenvironment or spread away through the circulation, thus facilitating tumor progression and metastasis.7 MicroRNAs (miRNAs) have been identified in exosomes.8 They are non-coding RNAs of approximately 18–25 nucleotides in length and act as posttranscriptional regulators of gene expression.9 Specific oncogenic and tumor suppressive miRNAs in exosomes may provide diagnostic or prognostic potential in cancer due to their differential expression between cancer cells and normal cells.8–11 Furthermore, the stable presence of exosomal miRNAs (exo-miRs) in multifarious body fluids also enhances their usefulness as tumor markers.12–14
Recent studies have indicated the diagnostic and prognostic potential of circulating exosomal miRNAs in several cancers, including lung cancer.15 Cazzoli et al identified two exosomal miRNA panels for screening and discriminating LUAD patients from healthy smokers and granuloma patients.16 Jin et al performed next-generation sequencing and found that lung cancer histotypes could be distinguished by a 4-miRNA panel with high sensitivity and specificity.17 Exosomal miRNAs have also been utilized as prognostic predictors in lung cancer. The levels of exosomal miR-21 and miR-4257 may predict non-small-cell lung cancer (NSCLC) recurrence.18 Wei et al reported that exosomal miR-222-3p may predict gemcitabine sensitivity in NSCLC patients.19 However, inconsistent results among different studies and the lack of reliable diagnostic cut-off values limit their clinical application. Therefore, our study aimed to identify the dysregulated serum exosomal miRNAs in LUAD patients and determine their absolute concentrations. Moreover, their diagnostic and prognostic value in LUAD was evaluated.
Patients and Methods
Patients and Clinical Specimens
A total of 70 LUAD patients and 40 healthy controls (HCs) were recruited from the First Affiliated Hospital of Nanjing Medical University (Nanjing, China) between January 2016 and January 2019. They were randomly assigned to the discovery set or the validation set. The discovery set consisted of 10 LUAD patients and 10 HCs. The validation set included 30 HCs, 30 early-stage LUAD patients and 30 advanced-stage LUAD patients (Table 1). LUAD patients were diagnosed through histological examination, and the tumor stage was estimated based on the IASLC eighth edition of TNM classification. No patients received chemotherapy, radiotherapy or other antitumor therapy before serum collection. In addition, 20 paired post-operative serum samples were collected one week after radical surgery.
Table 1.
Demographic and Clinical Features of the Study Populations
| Discovery Set (n=20) | Validation Set (n=90) | ||||
|---|---|---|---|---|---|
| LUAD | HC | Early-Stage LUAD | Advanced-Stage LUAD | HC | |
| N | 10 | 10 | 30 | 30 | 30 |
| Age | 56.7±11.2 | 61.4±10.2 | 58.5±9.6 | 60.8±10.7 | 52.8±12.7 |
| Gender | |||||
| Male | 6 (60%) | 5 (50%) | 11 (36.7%) | 17 (56.7%) | 18 (60%) |
| Female | 4 (40%) | 5 (50%) | 19 (63.3%) | 13 (43.3%) | 12 (40%) |
Abbreviations: LUAD, lung adenocarcinoma; HC, healthy control.
This study was approved by the Research and Ethical Committee of the hospital (No. 2017-SRFA-027) and conducted in accordance with the Declaration of Helsinki. Participants were informed about the study and signed informed consents prior to sample collection.
Serum was separated from whole blood by centrifugation at 1500 g for 10 min. Subsequently, the supernatant was centrifuged at 12,000 g for 10 min at 4°C. The cell-free serum samples were then stored at −80°C until exosome isolation.
Exosomes and miRNA Extraction
Exosomes were isolated from serum using Invitrogen™ Total Exosome Isolation Kits (Cat. No. 4478360, Invitrogen, NYC, USA). Briefly, 300 µL of serum was added with 60µL of exosome isolation reagent and mixed well by vortexing. The mixture was incubated at 4°C for 30 min followed by centrifugation at 12,000 g for 10 min at room temperature. Subsequently, the supernatant was discarded, and the remaining pellet was resuspended in 100 µL phosphate-buffered saline (PBS).
The miRcute miRNA Isolation Kit (Tiangen, Beijing, China) was used to extract miRNA from exosomes. We followed the manufacturer’s protocol. The obtained miRNA was stored at −80°C until further analysis.
Transmission Electron Microscopy (TEM)
Exosome samples were fixed with 1% glutaraldehyde and put into a carbon-coated copper grid. The uranyl-oxalate solution (pH 7.0) was then covered on the grid for 5 min. Finally, TEM (Tecnai G2, FEI, USA) was used to observe exosomes.
Nanoparticle Tracking Analysis (NTA)
The size and concentration of exosomes were defined using the ZetaView instrument (Particle Metrix, Germany). Samples were diluted appropriately in PBS and measured according to the protocol of manufacture.
Western Blotting
Specific exosome surface marker CD9 was detected by Western blotting. Exosomes were denatured by boiling in SDS loading buffer (Beyotime, Shanghai, China) and then separated by SDS-PAGE electrophoresis and transferred onto PVDF membranes (Millipore, USA). After blocking, the membrane was incubated with a rabbit primary antibody against CD9 (Cell Signaling, USA, 1:1000 dilution) overnight at 4°C. Following incubation with HRP-conjugated antibodies (ZSBIO, Beijing, China, 1:3000 dilution), the proteins were visualized using Tanon Chemiluminescence Imager (Tanon, Shanghai, China) with ECL blotting detection reagents (Millipore, USA).
Relative Quantification of miRNA
To explore the differentially expressed exosomal miRNAs in LUAD, the levels of six candidate miRNAs were examined using a relative quantification method. The extracted miRNA was reverse-transcribed with adding a poly (A) tail using the Mir-X miRNA First-Strand Synthesis Kit (TaKaRa Bio, Japan). The reaction mixture was incubated at 37°C for 60 min, and then 85°C for 5 min. qRT-PCR was performed using TB-Green Premix Ex Taq II Reagent (TaKaRa Bio, Japan). The mRQ 3′ Primer (TaKaRa Bio, Japan) was used as the reverse primer for each miRNA, and the sequences of miRNA-specific primers are provided in Table S1. The conditions of the amplification reaction were as follows: 95°C for 10 s, 40 cycles of 95°C for 5 s, and 60°C for 32 s. Each sample was analyzed in duplicate. Relative miRNA expression levels were calculated using the ΔCT method with U6 snRNA as a reference (CtmiRNA - CtU6).
Absolute Quantification of miRNA
MiR-1290 was selected for downstream validation, and its absolute concentration was determined using the Hairpin-itTM miRNAs RT-PCR Quantitation Kit (GenePharma, Shanghai, China) following the manufacturer’s protocol. Briefly, serially diluted synthetic miR-1290 oligonucleotides (5ʹ-UGGAUUUUUGGAUCAGGGA-3ʹ) ranging from 102 fmol/L to 105 fmol/L were applied to qRT-PCR. Subsequently, the Ct values were plotted against corresponding synthetic miRNA concentrations (in log10 form) to establish the calibration curves. The miRNA abundance of the serum samples was estimated based on comparing Ct values to the standard curve. The specific primers for miRNA were purchased from GenePharma Company (Shanghai, China). Reverse transcription was applied using the following temperature profile: 25°C for 30 min, 45°C for 30 min, and 85°C for 5 min. PCR was carried out on an ABI 7500 system (Applied Biosystems, USA) under the following conditions: 95°C for 3 min, 40 cycles of 95°C for 12 s and 62°C for 40 s. All samples were detected in duplicate.
Detection of Traditional Tumor Markers
The levels of CEA, CYFRA21-1, and NSE were determined by electrochemiluminescence immunoassay (ECLIA) using the Cobas® analyzer (Roche Diagnostics, Mannheim, Germany) with corresponding kits.
Statistical Analysis
Data are presented as medians with the interquartile range. The Mann–Whitney U-test was performed to compare the differences in exosomal miRNA levels between LUAD patients and healthy controls. Differences among multiple groups were assessed by the Kruskal–Wallis test. The paired t-test was used in the pre- and postoperative groups. Diagnostic efficacy was evaluated using receiver operating characteristic (ROC) curves and the area under the ROC curve (AUC). The survival analysis was performed using the Kaplan-Meier (KM) method. Univariate and multivariate Cox proportional hazards model analysis was used to identify prognostic factors of LUAD. All statistical tests above were performed using SPSS software (Version 24.0, IBM Corp, USA) and GraphPad Prism Software (San Diego, USA). Two-sided P < 0.05 was considered statistically significant.
Results
Characterization of Serum Exosomes
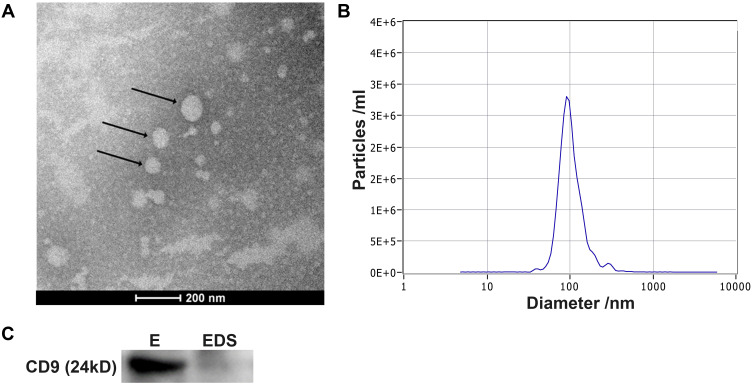
TEM, NTA and Western blotting were performed to identify the exosomes (Figure 1). TEM images revealed that the vesicles were round in shape with diameters ranging from 100 nm to 120 nm (Figure 1A). NTA showed that the average size of the vesicles was 100 nm with a concentration of approximately 2.5 × 107/mL (Figure 1B). Finally, Western blot analysis demonstrated that CD9, a typical exosomal surface marker, was present on the isolated vesicles but not in the remaining exosome-depleted serum (Figure 1C). These results suggest that the vesicles isolated from the serum were exosomes and were consistent with previous studies.
Figure 1.
Characterization of isolated exosomes. (A) Exosomes were visualized by TEM. Typical exosomes were indicated with the black arrows. (B) NTA analysis revealed that the size distribution of exosomes was 50–150 nm. (C) The exosomal protein marker CD9 was analyzed in exosomes (E) and exosome-depleted serum (EDS) by Western blotting.
Exosomal miR-1290 Was Upregulated in LUAD Patients
Based on our previous study and other reports,19–24 six exo-miRs (miR-21, miR-221-3p, miR-222-3p, miR-223, miR-638, miR-1290) were selected as candidate miRNAs. We detected their expression levels in the discovery set by qRT-PCR. As shown in Figure 2, exosomal miR-1290 was significantly upregulated in the serum of LUAD patients compared to that of healthy controls (P=0.0014).
Figure 2.
Expression levels of the miRNA candidates in the discovery set was assessed by qRT-PCR. Relative expression levels of miR-1290 (A), miR-21 (B), miR-638 (C), miR-221-3p (D), miR-222-3p (E) and miR-223 (F) in the healthy controls and LUAD patients are shown. U6 snRNA served as the internal control. **P<0.01.
Abbreviation: NS, non-significant.
Overexpression of Serum Exosomal miR-1290 in LUAD Patients Was Validated
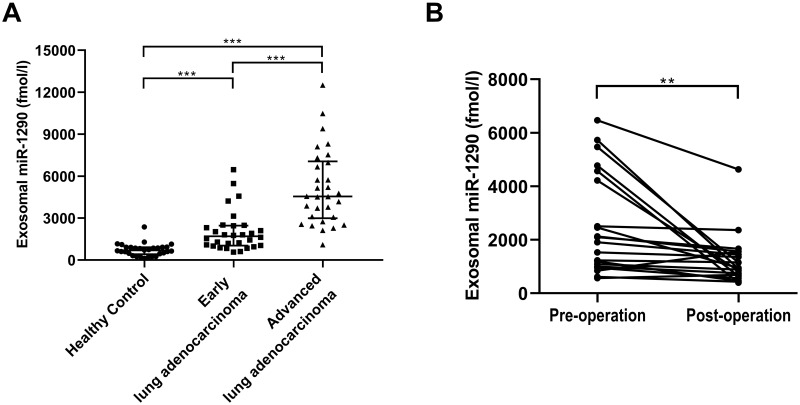
To confirm the expression of serum exosomal miR-1290 in LUAD, the absolute concentration of miR-1290 was subsequently determined in a validation set (Table 1). As shown in Figure 3A, elevated exosomal miR-1290 concentrations were observed in patients with early-stage LUAD [1697.0 (1032.3, 2462.0) fmol/l, P < 0.001] and advanced-stage LUAD [4547.7 (2987.4, 7060.3) fmol/l, p < 0.0001] compared to healthy controls [689.8 (420.0, 915.2) fmol/l].
Figure 3.
Expression of serum exosomal miR-1290 in the validation set. The absolute concentrations of serum miR-1290 were measured in healthy controls (HCs), early LUAD and advanced LUAD patients (A) (all P<0.01, ANOVA). The levels of serum exosomal miR-1290 in LUAD patients significantly decreased after surgery (B). **P<0.01, ***P<0.001.
Moreover, we examined 20 pairs of pre- and post-operative serum specimens collected from LUAD patients who underwent radical resection. The exosomal miR-1290 levels were significantly decreased after resection (p = 0.0029, Figure 3B). The exosomal miR-1290 concentrations in the pre- and post-operative groups were 2001.8 (1019.0, 4479.2) fmol/l and 928.8 (652.9, 1526.4) fmol/l, respectively.
Correlations Between Exosomal miR-1290 and Clinical Characteristics in LUAD
We further investigated the association between serum exosomal miR-1290 levels and clinical pathology characteristics in LUAD (Table 2). The LUAD patients were divided into high- (n=30) and low-expression (n=30) groups based on the median value of serum exosomal miR-1290. We noted that circulating exosomal miR-1290 was elevated in males (P=0.038). In addition, a higher exosomal miR-1290 level was significantly related to an advanced TNM stage (P<0.001). Moreover, we found that the exosomal miR-1290 level was positively correlated with tumor size (P<0.001), lymph node metastasis (P=0.003), distant metastasis (P<0.001), smoking (P=0.015) and drinking (P=0.005). However, no significant correlation was observed between serum exosomal miR-1290 level and age, differentiation degree, hypertension, diabetes mellitus, and tuberculosis.
Table 2.
Correlation Between Serum Exo-miR-1290 Levels and Clinical Characteristics
| Characteristics | miR-1290 Low (n=30) | miR-1290 High (n=30) | P value |
|---|---|---|---|
| Gender (male) | 10 | 18 | 0.038 |
| Age | 58.50±10.25 | 60.83±9.74 | 0.392 |
| Tumor stage | <0.001 | ||
| I–II | 25 | 5 | |
| III–IV | 5 | 25 | |
| Tumor size | <0.001 | ||
| >3.0 cm | 4 | 16 | |
| ≤3.0 cm | 24 | 9 | |
| NA | 2 | 5 | |
| Differentiation | 0.073 | ||
| Well | 10 | 4 | |
| Poor | 6 | 11 | |
| NA | 14 | 15 | |
| Lymph node metastasis | 0.003 | ||
| Yes | 11 | 21 | |
| No | 18 | 6 | |
| NA | 1 | 3 | |
| Distant metastasis | <0.001 | ||
| Yes | 5 | 18 | |
| No | 24 | 10 | |
| NA | 1 | 2 | |
| Hypertension | 0.590 | ||
| Yes | 6 | 8 | |
| No | 23 | 22 | |
| NA | 1 | 0 | |
| Diabetes mellitus | 0.670 | ||
| Yes | 3 | 2 | |
| No | 25 | 27 | |
| NA | 2 | 1 | |
| Smoking | 0.015 | ||
| Yes | 6 | 16 | |
| No | 20 | 13 | |
| NA | 4 | 1 | |
| Drinking | 0.005 | ||
| Yes | 1 | 11 | |
| No | 23 | 15 | |
| NA | 6 | 4 | |
| Tuberculosis | 1.000 | ||
| Yes | 1 | 2 | |
| No | 26 | 25 | |
| NA | 3 | 3 |
Note: All data with P-value <0.05 are bolded.
Abbreviations: Low, low expression (≤ median); High, high expression (> median); NA, not available.
Diagnostic Efficiency of Exosomal miR-1290 in LUAD
The diagnostic performance of exosomal miR-1290 for LUAD was evaluated by performing a ROC analysis. Exosomal miR-1290 showed better diagnostic efficiency than CEA, CYFRA21-1 and NSE in discriminating LUAD patients from HCs (Figure 4, Table 3). The AUC of exosomal miR-1290 was 0.937 (95% CI: 0.890–0.985), with a sensitivity of 80.0% and specificity of 96.7%, as well as a cut-off value of 1286.0 fmol/l. The AUCs of CEA, CYFRA21-1 and NSE were 0.725 (95% CI: 0.623–0.828), 0.495 (95% CI: 0.371–0.619), and 0.770 (95% CI: 0.671–0.869), respectively. In addition, we found that CEA and CYFRA21-1 exhibited relatively high specificity (93.3% and 96.7%, respectively) but poor sensitivity (54.2% and 24.1, respectively). NSE showed relatively good sensitivity (76.3%), while low specificity (26.7%).
Figure 4.
ROC analysis to discriminate LUAD patients from healthy controls according to serum exosomal miR-1290, CEA, CYFRA21-1 and NSE.
Table 3.
Diagnostic Performance of Serum Exosomal miR-1290 and Clinical Biomarkers
| Biomarkers | P value | AUC | 95% CI | Cut-off value | Sensitivity | Specificity |
|---|---|---|---|---|---|---|
| miR-1290 | <0.001 | 0.937 | 0.890–0.985 | 1286.0 fmol/l | 80.0% | 96.7% |
| CEA | 0.001 | 0.725 | 0.623–0.828 | 2.9 ng/mL | 54.2% | 93.3% |
| CYFRA21-1 | 0.940 | 0.495 | 0.371–0.619 | 3.2 ng/mL | 24.1% | 96.7% |
| NSE | <0.001 | 0.770 | 0.671–0.869 | 14.2 ng/mL | 76.3% | 26.7% |
Note: All data with P-value <0.05 are bolded.
Abbreviation: AUC, area under the ROC curve.
Exosomal miR-1290 is an Independent Prognostic Factor of LUAD
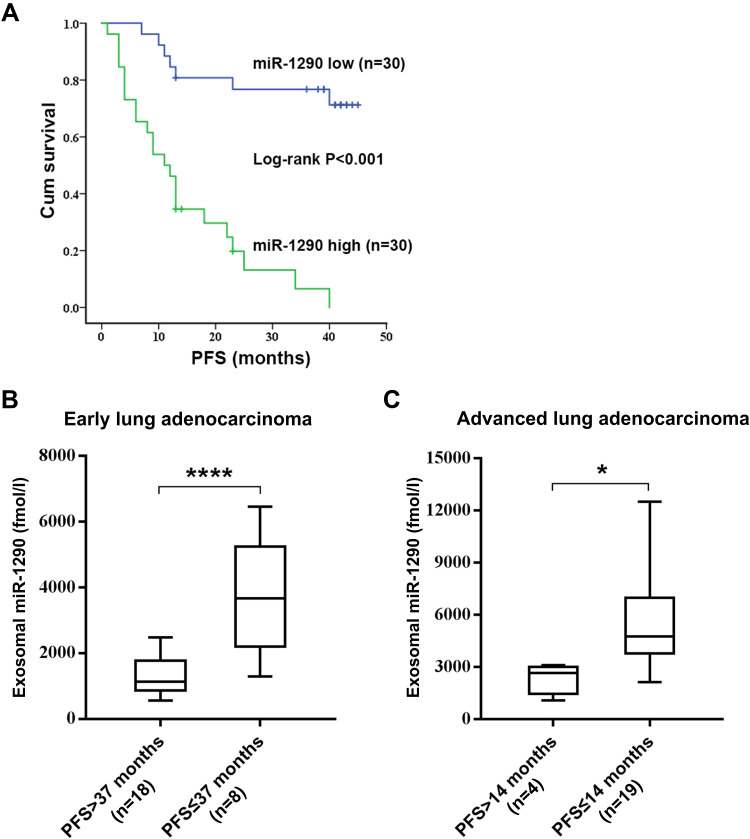
To investigate the prognostic value of serum exosomal miR-1290, LUAD patients in the validation set were followed-up, and progression-free survival (PFS) data were collected. According to the median value of exosomal miR-1290, LUAD patients were classified into high expression (n=30) or low expression (n=30) groups. Kaplan–Meier survival analysis indicated that patients with elevated exosomal miR-1290 had shorter PFS than those with lower levels (P<0.001; Log rank test; Figure 5A). The mean PFS was 14 months and 37 months in the high expression and low expression groups, respectively. Furthermore, for patients with stage I–II LUAD, exosomal miR-1290 levels were higher in patients with a PFS of no more than 37 months (P < 0.0001, Figure 5B). For advanced LUAD patients, higher serum exosomal miR-1290 levels were found in those with PFS shorter than 14 months (P = 0.0379, Figure 5C).
Figure 5.
Kaplan–Meier survival analysis of patients with LUAD based on serum exosomal miR-1290 levels. The median serum exosomal miR-1290 level (2530.71 fmol/l) was set as the cut-off value. LUAD patients with high serum exosomal miR-1290 levels showed significantly lower progression-free survival (PFS) than those with low serum exosomal miR-1290 levels (P <0.001; Log rank test) (A). Analysis of PFS according to the expression levels of exosomal miR-1290 in early-stage LUAD patients (B) and advanced-stage LUAD patients (C). *P<0.05, ****P<0.0001.
Univariate Cox proportional hazards regression was employed to identify the potential factors that affect the PFS of LUAD patients. The results showed that tumor stage, tumor size, lymph node and distant metastasis, gender, drinking status, CYFRA21-1, and exosomal miR-1290 levels may be associated with the PFS of LUAD patients (all P<0.05, Table 4). Multivariate analysis revealed that exosomal miR-1290 level was an independent prognostic factor in LUAD patients (P=0.017, HR=7.80, 95% CI:1.44–42.21, Table 4).
Table 4.
Univariate and Multivariate Cox Proportional Hazards Regression Model Analysis for Prediction of PFS in LUAD from the Validation Set
| Parameters | Univariate Analysis | Multivariate Analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Age (≤60) | 1.14 | 0.69–2.90 | 0.346 | |||
| Gender | 0.29 | 0.14–0.63 | 0.001 | 0.22 | 0.06–0.86 | 0.030 |
| Tumor stage | 7.70 | 3.21–18.45 | <0.001 | |||
| Tumor size | 5.50 | 2.42–12.53 | <0.001 | |||
| Differentiation | 5.70 | 0.74–43.67 | 0.094 | |||
| Lymph node metastasis | 5.52 | 2.33–13.04 | <0.001 | |||
| Distant metastasis | 4.68 | 1.98–11.05 | <0.001 | |||
| Hypertension | 0.85 | 0.32–2.25 | 0.749 | |||
| Diabetes mellitus | 0.98 | 0.30–3.25 | 0.975 | |||
| Smoking | 1.79 | 0.84–3.84 | 0.133 | |||
| Drinking | 3.01 | 1.31–6.92 | 0.009 | |||
| Tuberculosis | 1.18 | 0.28–4.97 | 0.827 | |||
| miR-1290 | 7.84 | 3.18–19.31 | <0.001 | 7.80 | 1.44–42.41 | 0.017 |
| CEA | 1.92 | 0.89–4.17 | 0.098 | |||
| CYFRA21-1 | 3.13 | 1.40–6.97 | 0.005 | |||
| NSE | 1.24 | 0.60–2.55 | 0.557 | |||
Note: All data with P-value <0.05 are bolded.
Abbreviation: HR, hazard ratio.
Discussion
Novel biomarkers with high sensitivity and specificity are required for the timely and effective treatment of LUAD. Emerging evidence suggests that exosomal miRNAs are promising tumor markers with the advantage of high sensitivity, specificity and stability.25 Many recent studies have reported that exosomal miRNAs might be used for cancer diagnosis and prognosis. However, only a few studies have been conducted on serum exosomal miRNAs in LUAD, and their results showed inconsistencies.25,26 In addition, in these studies, the miRNAs were measured by a relative quantitative method, hindering the potential for clinical application.
In this study, we initially explored the expression of six miRNA candidates in the discovery set. Consequently, exosomal miR-1290 was observed to be markedly elevated in LUAD patients, and its absolute concentrations were further detected in the validation set. As a result, we found that exosomal miR-1290 was elevated in both early- and advanced-stage LUAD patients. Notably, exosomal miR-1290 could significantly discriminate LUAD patients at early stages from those at advanced stages. Furthermore, we observed significant positive correlations between exosomal miR-1290 and tumor stage, tumor size, and lymph node and distant metastasis. These data suggested that overexpression of serum exosomal miR-1290 might indicate the malignant behavior of LUAD. In addition, elevated levels of serum exosomal miR-1290 in LUAD patients were indicated to be tumor-derived because of their downtrend after malignancy resection (P = 0.0029). These results demonstrated the close correlation between exosomal miR-1290 and the malignant degree of cancer and further suggested the putative diagnostic value of exosomal miR-1290 for LUAD.
MiR-1290 was first discovered as an oncogene in human embryonic stem cells.27 Subsequently, miR-1290 was found to be involved in the progression of various tumors, including lung cancer. Zhang et al indicated that miR-1290 was a crucial driver for tumor initiation and cancer progression in NSCLC.28 Kim et al found that the epithelial-mesenchymal transition (EMT) and invasiveness of NSCLC cells could be suppressed by inhibiting miR-1290.29 In addition, interferon regulatory factor 2 (IRF2) was reported to be a target of miR-1290, thereby promoting NSCLC proliferation and invasion.30 Our previous study also confirmed the overexpression of miR-1290 in LUAD cells. Moreover, in vitro and in vivo experiments revealed that miR-1290 could activate the PI3K/AKT and JAK/STAT3 pathways, thereby strengthening LUAD cell proliferation, invasion and metastasis.24 Coupled with the fact that the exosomal miRNAs reflect the state of their original cells to some degree, these reports further corroborated the enormous potential of exosomal miR-1290 as a biomarker for LUAD.
Routine tumor markers of lung cancer, such as CEA, CYFRA21-1, NSE and SCCA, are not sensitive or specific enough for a reliable evaluation.4 Although the combined detection of multiple biomarkers could improve the sensitivity, it leads to problems such as decreased specificity and increased cost, which make it difficult to meet clinical requirements. Therefore, many studies have focused on searching for more sensitive and specific biomarkers of lung cancer. In this study, we evaluated the diagnostic performance of exosomal miR-1290 along with CEA, NSE and CYFRA21-1 to discriminate LUAD patients from HCs. The results showed an AUC of 0.937 with a sensitivity of 80% and a specificity of 96.7% for exosomal miR-1290, which was markedly better than that for CEA, NSE and CYFRA21-1. Therefore, exosomal miR-1290 may serve as a diagnostic biomarker for LUAD. Moreover, the survival analysis demonstrated that LUAD patients with elevated exosomal miR-1290 had shorter PFS than those with lower levels (14 months vs 37 months). Multivariate Cox regression analysis showed that exosomal miR-1290 level was an independent prognostic factor in LUAD patients. Thus, exosomal miR-1290 could also be utilized as a potential prognostic predictor of LUAD.
Although this study and others all indicate that exosomal miRNAs are potential biomarkers for clinical application, there are still some weaknesses in current exosomal miRNA test approaches. First, the detection techniques for exosomal miRNAs are complicated and time-consuming. Second, a uniform and standardized method for the isolation and absolute quantification of exosomal miRNAs is still lacking. More simple and accurate detection methods for exosomal miRNAs need to be developed for future clinical applications.
There are some limitations of our study needed to be addressed. First, this is a single-center study with a small sample size. In addition, patients with benign lung disease were not included. The findings in this study need to be further validated with large-scale samples in a wider population.
Conclusion
Exosomal miR-1290 is significantly upregulated in LUAD serum, and it may serve as a potential biomarker for the diagnosis and prognosis of LUAD.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant number: 81772269) and the Key Laboratory for Laboratory Medicine of Jiangsu Province of China (No. ZDXKB2016005).
Disclosure
The authors report no conflicts of interest for this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 3.Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thoracic Oncol. 2011;6(2):244–285. doi: 10.1097/JTO.0b013e318206a221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duffy MJ, O’Byrne K. Tissue and blood biomarkers in lung cancer: a review. Adv Clin Chem. 2018;86. [DOI] [PubMed] [Google Scholar]
- 5.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–579. doi: 10.1038/nri855 [DOI] [PubMed] [Google Scholar]
- 6.Pant S, Hilton H, Burczynski ME. The multifaceted exosome: biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem Pharmacol. 2012;83(11):1484–1494. doi: 10.1016/j.bcp.2011.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126(4):1208–1215. doi: 10.1172/JCI81135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun Z, Shi K, Yang S, et al. Effect of exosomal miRNA on cancer biology and clinical applications. Mol Cancer. 2018;17(1):147. doi: 10.1186/s12943-018-0897-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 10.Iqbal MA, Arora S, Prakasam G, Calin GA, Syed MA. MicroRNA in lung cancer: role, mechanisms, pathways and therapeutic relevance. Mol Aspects Med. 2019;70:3–20. doi: 10.1016/j.mam.2018.07.003 [DOI] [PubMed] [Google Scholar]
- 11.Salehi M, Sharifi M. Exosomal miRNAs as novel cancer biomarkers: challenges and opportunities. J Cell Physiol. 2018;233(9):6370–6380. doi: 10.1002/jcp.26481 [DOI] [PubMed] [Google Scholar]
- 12.Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820(7):940–948. doi: 10.1016/j.bbagen.2012.03.017 [DOI] [PubMed] [Google Scholar]
- 13.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110(1):13–21. doi: 10.1016/j.ygyno.2008.04.033 [DOI] [PubMed] [Google Scholar]
- 14.Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10(1):42–46. doi: 10.3816/CLC.2009.n.006 [DOI] [PubMed] [Google Scholar]
- 15.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478):6478. doi: 10.1126/science.aau6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cazzoli R, Buttitta F, Di Nicola M, et al. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thoracic Oncol. 2013;8(9):1156–1162. doi: 10.1097/JTO.0b013e318299ac32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin X, Chen Y, Chen H, et al. Evaluation of Tumor-Derived Exosomal miRNA as Potential Diagnostic Biomarkers for Early-Stage Non-Small Cell Lung Cancer Using Next-Generation Sequencing. Clin Cancer Res. 2017;23(17):5311–5319. doi: 10.1158/1078-0432.CCR-17-0577 [DOI] [PubMed] [Google Scholar]
- 18.Dejima H, Iinuma H, Kanaoka R, Matsutani N, Kawamura M. Exosomal microRNA in plasma as a non-invasive biomarker for the recurrence of non-small cell lung cancer. Oncol Lett. 2017;13(3):1256–1263. doi: 10.3892/ol.2017.5569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei F, Ma C, Zhou T, et al. Exosomes derived from gemcitabine-resistant cells transfer malignant phenotypic traits via delivery of miRNA-222-3p. Mol Cancer. 2017;16(1):132. doi: 10.1186/s12943-017-0694-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Luo F, Wang B, et al. STAT3-regulated exosomal miR-21 promotes angiogenesis and is involved in neoplastic processes of transformed human bronchial epithelial cells. Cancer Lett. 2016;370(1):125–135. doi: 10.1016/j.canlet.2015.10.011 [DOI] [PubMed] [Google Scholar]
- 21.Chen X, Jin Y, Feng Y. Evaluation of plasma extracellular vesicle microRNA signatures for lung adenocarcinoma and granuloma with monte-carlo feature selection method. Front Genet. 2019;10:367. doi: 10.3389/fgene.2019.00367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sozzi G, Boeri M, Rossi M, et al. Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: a correlative MILD trial study. J clin oncol. 2014;32(8):768–773. doi: 10.1200/JCO.2013.50.4357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang F, J-f L, Cao Y, et al. miR-638 is a new biomarker for outcome prediction of non-small cell lung cancer patients receiving chemotherapy. Exp Mol Med. 2015;47(5):e162. doi: 10.1038/emm.2015.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao X, Yang D, Gong X, Mo D, Pan S, Xu J. miR-1290 promotes lung adenocarcinoma cell proliferation and invasion by targeting SOCS4. Oncotarget. 2018;9(15):11977–11988. doi: 10.18632/oncotarget.24046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fortunato O, Gasparini P, Boeri M, Sozzi G. Exo-miRNAs as a New Tool for Liquid Biopsy in Lung Cancer. Cancers. 2019;11(6):6. doi: 10.3390/cancers11060888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vanni I, Alama A, Grossi F, Dal Bello MG, Coco S. Exosomes: a new horizon in lung cancer. Drug Discov Today. 2017;22(6):927–936. doi: 10.1016/j.drudis.2017.03.004 [DOI] [PubMed] [Google Scholar]
- 27.Morin RD, O’Connor MD, Griffith M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18(4):610–621. doi: 10.1101/gr.7179508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang WC, Chin TM, Yang H, et al. Tumor-initiating cell-specific miR-1246 and miR-1290 expression converge to promote non-small cell lung cancer progression. Nat Commun. 2016;7(1):11702. doi: 10.1038/ncomms11702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim G, An H-J, Lee M-J, et al. Hsa-miR-1246 and hsa-miR-1290 are associated with stemness and invasiveness of non-small cell lung cancer. Lung Cancer. 2016;91:15–22. doi: 10.1016/j.lungcan.2015.11.013 [DOI] [PubMed] [Google Scholar]
- 30.Jin -J-J, Liu Y-H, Si J-M, Ni R, Wang J. Overexpression of miR-1290 contributes to cell proliferation and invasion of non small cell lung cancer by targeting interferon regulatory factor 2. Int J Biochem Cell Biol. 2018;95:113–120. doi: 10.1016/j.biocel.2017.12.017 [DOI] [PubMed] [Google Scholar]