Summary
Immunotherapies have emerged as highly promising approaches to treat cancer patients. Allogeneic haematopoietic cell transplantation (HCT) is the most validated tumour immunotherapy available to date but its clinical efficacy is limited by toxicities, such as graft-versus-host disease (GVHD) and treatment resistance leading to relapse. The problems with new cellular therapies and checkpoint inhibitors are similar. However, development of biomarkers post-HCT, particularly for toxicities, has taken off in the last decade and has expanded greatly. Thanks to the advances in genomics, transcriptomics, proteomics and cytomics technologies, blood biomarkers have been identified and validated in promising diagnostic tests, prognostic tests stratifying for future occurrence of GVHD, and predictive tests for responsiveness to GVHD therapy and non-relapse mortality. These biomarkers may facilitate timely and selective therapeutic intervention. This review outlines a path from biomarker discovery to first clinical correlation, focusing on soluble STimulation-2 (sST2) – the interleukin (IL)-33-decoy receptor – which is the most validated biomarker.
Keywords: haematopoietic stem cell transplantation, graft-versus-host disease, biomarkers, blood and marrow transplant immunology, interleukins
Immunotherapies have emerged as highly promising approaches to treat cancer patients. Allogeneic haematopoietic cell transplantation (HCT) is the most validated tumour immunotherapy available to date. While HCT can induce beneficial graft-versus-leukaemia (GVL) effects, the adverse effect of graft-versus-host disease (GVHD), which is closely linked to GVL, is a major source of morbidity and mortality following HCT. Clinically, significant acute GVHD (aGVHD) continues to affect up to 50% of HCT recipients (Zeiser & Blazar, 2017). Also, the incidence of chronic GVHD (cGVHD) has been as high as 70% in HCT recipients who survived 100 days (Cooke et al., 2016). Severe cGVHD results in high non-relapse mortality (NRM) reaching 12%, significant morbidity, organ dysfunction, impaired quality of life and increased incidence of secondary malignancies (Wingard et al., 2011). The conditioning regimen, underlying primary disease, alloreactivity induced by donor T cells, and prophylactic immunosuppressive drugs, lead to other less common but still potentially fatal complications post-HCT. These can be engraftment syndrome, hepatic sinusoidal obstruction syndrome (SOS) – previously known as veno-occlusive disease, thrombotic microangiopathy (TMA), and idiopathic pneumonia syndrome (IPS). Markers of the beneficial GVL will be extremely valuable. Until recently, available diagnostic and staging tools frequently failed to identify patients at elevated risk of disease progression or death, but the past decade has seen an explosive evolution of ‘-omics’ technologies. Building on these opportunities, blood biomarkers have been identified and validated in several cohorts for the main post-transplantation outcomes. This review article summarises definitions of the different types of biomarkers, current information on biomarkers for post-transplantation outcomes, and proposes future directions for biomarker-based clinical trials and ultimately biomarker use in standard practice. Furthermore, it will focus on soluble STimulation-2 (sST2), the interleukin (IL)-33-decoy receptor – the most validated biomarker, which is not only mechanistic but also druggable and thus represents an ideal biomarker.
What are the definitions of the different types of biomarkers?
The 2014 NIH (National Institutes of Health) Chronic Graft-Versus-Host Disease Consensus Biomarker Working Group (which focused on GVHD and haematopoietic stem cell transplant) and FDA (US Food and Drug Administration) experts defined the different types of biomarkers (Paczesny et al., 2015a). Briefly, there are four types of markers: (i) diagnostic biomarkers identify the presence of a disease as compared to similar presentation of other etiology, (ii) prognostic biomarkers identify likelihood of a clinical event, disease recurrence or progression in patients who have the disease or medical condition of interest, (iii) predictive biomarkers identify individuals who are more likely than similar individuals without the biomarker to experience a favourable or unfavourable effect from exposure to a medical treatment, and (iv) response to treatment biomarkers show that a biological response has occurred in an individual who has been exposed to a medical treatment. In 2015, the FDA-NIH Joint Leadership Council developed the BEST (Biomarkers, EndpointS and other Tools) resources which further explained these definitions (FDA-NIH Biomarker Working Group, 2016). The BEST glossary aims to capture distinctions between biomarkers and clinical assessments, and to describe their distinct roles in biomedical research, clinical practice and medical product development. Importantly, safety biomarkers were defined as biomarkers which are measured before or after an exposure to a medical product to indicate the likelihood, presence or extent of toxicity as an adverse effect.
What sample type and clinical information should be in one’s biobank?
Ideal clinical tests involve longitudinal non-invasive sample collection, and thus plasma, sera or urine are preferable. These sample types are also relatively easy to collect, process and store. They are a good source of information related to systemic diseases such as GVHD, since the levels of individual blood proteins represent a summation of multiple events which occur in every organ system. Another important question to ask is whether one’s sample collection should be calendar- versus event-driven, or both? The main advantage of an event-driven collection is the granularity of the clinical information that can be obtained at the time of diagnosis of GVHD. There is no interference with GVHD treatment as the sample is collected before the treatment is started, but this type of collection is limited by the need of a dedicated team that can collect and process the samples, as well as enter clinical data in real time every day of the week. A calendar-driven collection is particularly interesting for prognostic markers and demands less logistics as the samples are planned. Ideally, if resources allow, both calendar- and event-driven samples should be collected. It is of course obvious that linking this samples collection to a comprehensive clinical database maintained by skilled data managers is key to the success of a biorepository. It should allow for samples to be de-identified for multicentre studies. Several investigators have established such a biobank and more recently a large US multicentre cohort has been accrued by the Blood and Marrow Transplant Clinical Trials Network (BMTCTN), collecting prospectively biological samples on 1710 HCT recipients, including 200 African American and 200 paediatric participants. Serum, plasma on EDTA (ethylenediamine tetraacetic acid) and heparin, PAXgene lysates and buffy coat were collected at pre-HCT and at days 7, 14, 21, 28, 42, 56 and 90 post-HCT with all the pre-HCT characteristics as well as 12 major outcomes (https://web.emmes.com/study/bmt2/protocol/1202_protocol/1202_protocol.html) (BMTCTN 1202, NCT01879072).
Discovery tools: genomics, transcriptomics and proteomics
Genomics
Host genomics.
Strategies to improve outcomes after HCT can be split into those which reduce pre-transplantation risk and those which facilitate the diagnosis and prognosis of post-transplantation complications. Advances in pre-transplantation risk stratification have been made through detailed evaluations based on HLA (human leucocyte antigen) genetics (Petersdorf, 2017), as well as genome-wide association studies (GWAS) of polymorphisms, which either increase transplantation risk or protect against complications. A recent GWAS showed that the number of minor histocompatibility antigen mismatches doubles in unrelated versus sibling HLA-matched transplants, but has less impact on aGVHD than mismatching at HLA-DP (Martin et al., 2017). Another recent GWAS, including approximately 3000 donor–recipient pairs (Discovery-BMT study), showed that functional single nucleotide polymorphisms (SNPs) in the major histocompatibility complex (MHC) class II region are associated with overall survival (OS) after HLA-matched unrelated donor HCT (Sucheston-Campbell et al., 2016). However, studies of candidate-genetic polymorphisms in large cohorts have been unable to replicate findings from previous smaller studies for both aGVHD and cGVHD. This suggests that most published SNP associations have not held up or have not been reproducible, either because they were non-functional or in linkage with more important genetic elements (Martin et al., 2016; Karaesmen et al., 2017; Tang et al., 2019). As an exception, donor SNPs in IL1RL1 showed strong correlations with pre-transplantation serum/plasma concentrations of soluble Stimulation-2 (sST2), also called interleukin (IL)-33 receptor, as well as an association with the risk of aGVHD with potential implications for donor selection (Karaesmen et al., 2019).
Microbiome genomics.
Recently, intestinal tract bacterial floral diversity, as represented by the inverse Simpson index, was suggested as a risk-stratification biomarker. Faecal specimens were collected from 80 HCT recipients at stem cell engraftment, and the low-diversity group (inverse Simpson <2) had the highest rate of transplant-related death (Taur et al., 2014). The presence of specific species such as Blautia that correlated with reduced death from GVHD has also been proposed as a potential biomarker (Jenq et al., 2015). Microbiome-host interactions and their potential as biomarkers were recently extensively reviewed (Andermann et al., 2018).
Transcriptomics
As with genomic analysis, studies of gene expression signatures of GVHD can be categorised as candidate studies and omics-driven biomarker discovery studies, which may offer a less biased approach to identifying genes, pathways and gene expression networks active in this disease. In the past decade, large transcriptomic initiatives have enabled major discoveries in the fields of infectious disease, vaccinology and solid organ transplantation (Chaussabel & Pulendran, 2015; Nakaya et al., 2015; Bontha et al., 2017). Transcriptomic analysis is mainly performed on bulk peripheral blood mononuclear cells (PBMCs), avoiding contamination by granulocytes seen with whole blood approaches. In HCT, a 20-gene set classifier distinguishing tolerant and non-tolerant subjects was discovered, although not validated independently (Pidala et al., 2017). A whole PBMC approach can scan all circulating cells, but the resulting transcriptome is often dominated by the largest cell population, which does not always represent pathogenic cell types. Thus, some groups have purified cell populations prior to RNA (ribonucleic acid) isolation. Studies related to aGVHD have used sorted T cells, given their prominent role in disease pathogenesis. Using sorted CD3+ T cells in non-human primates (NHPs) in unsupervised gene expression analyses, ‘hypera-cute’ aGVHD was shown to be driven by Th/Tc1-mediated dysfunction and ‘breakthrough’ aGVHD, driven by inflammatory IL17-dominated pathways (Furlan et al., 2016). Aurora Kinase A, OX40:OX40L and CD28 pathways were also shown to mediate aGVHD, induced in both NHP and human allo-reactive T cells, which can be blocked in combination with mTOR inhibition with sirolimus, or CD28 blockade to induce long-term control of aGVHD (Furlan et al., 2015; Tkachev et al., 2017; Watkins et al., 2018). In cGVHD, gene expression in circulating monocytes identified two upregulated pathways: interferon (IFN)-inducible genes (MX1, CXCL9, CXCL10) and innate receptors for cellular damage (TLR7 and DDX58) (Hakim et al., 2016). The knowledge gained from the studies described above was almost exclusively derived from gene array experiments performed on bulk cell populations. In the coming years, new techniques, particularly single-cell RNASeq (scRNASeq), will provide insights for mechanistic questions, only answerable by single-cell analysis, allowing studies on low-frequency cells, particularly in low-cell input samples, such as gut biopsies (Zheng et al., 2017).
Proteomics
While genomics and transcriptomics techniques have become routine, proteomics is still performed in specialised laboratories. However, discovery of a protein disease marker is immensely valuable, as it represents the actual state of disease (Mann & Jensen, 2003). Here, we focus on the use of proteomics in large-scale studies that investigated qualitative and quantitative differences in complete protein profiles among samples from patients with and without GVHD or other complications post-HCT. Both non-MS (mass spectrometry) – such as antibody arrays–and MS-based proteomic approaches have been employed. Although antibody arrays are quantitative and highly sensitive for low abundance proteins such as cytokines, their main disadvantage is the restricted number of antibodies on the array, which thus limits the candidates to ‘usual suspects’. In contrast, next-generation MS is a powerful tool for qualitative and quantitative characterisation of proteins in complex protein mixtures (Altelaar et al., 2013). At present, these approaches are too time-consuming for use in validation, but they remain efficient methods for biomarker discovery as shown in Fig 1. During the validation process, bias in the prioritisation of candidate markers often exists because of the lack of highly specific sandwich enzyme-linked immunosorbent assay (ELISA). ELISAs are quantitative, high-throughput and highly reproducible, limiting both inter- and intra-assay variability.
Fig 1.
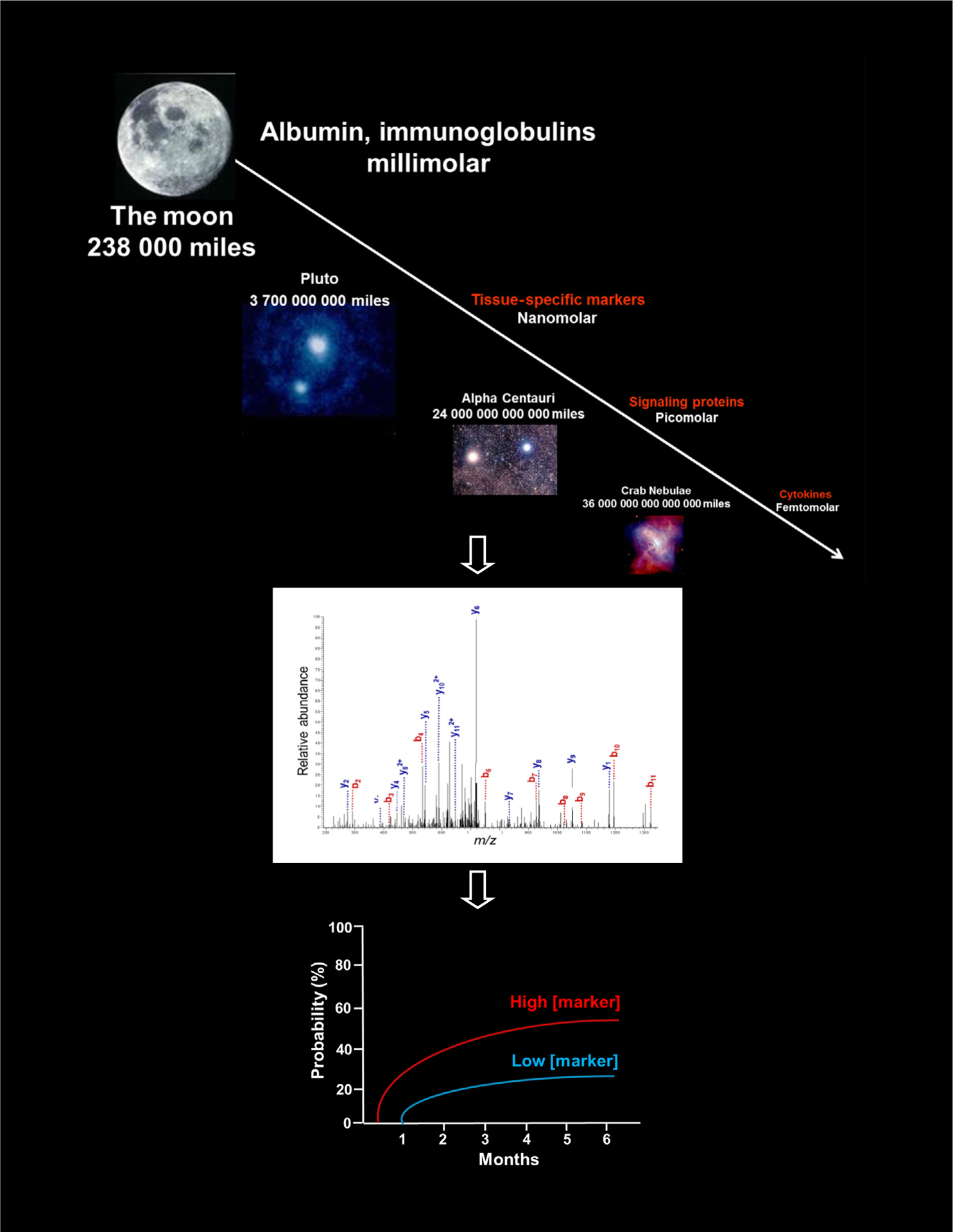
Proteomics workflow from the complexity of the plasma matrix to the discovery of clinically relevant biomarkers of post-HCT outcomes using tandem mass spectrometry. The plasma has a 1012 dynamic range. Tissue proteins, signaling proteins, and cytokines are potential biomarkers but of low abundance. To see them, scientists built the equivalent of a Hubble Space Telescope for the proteome with current mass spectrometry workflow. A good prognostic biomarker will identify the disease before it occurs clinically. If a patient has high biomarkers, its probability to develop the disease will be increased [Colour figure can be viewed at wileyonlinelibrary.com].
Cytomics
Profiles of immune cell populations are obtained by high-throughput flow cytometry or mass cytometry. CyTOF is a time-of-flight (TOF) MS approach for measuring many markers on cells, similar to flow cytometry, except the antibodies are labeled with heavy metal ion tags instead of fluorochromes. Its main advantage over flow cytometry is the combination of more antibody specificities in a single sample, without significant spillover between channels. This technology and its software tools permit discovery studies of new populations. Flow cytometry and mass cytometry have enabled identification of several important immune cells: regulatory T cells (Tregs) (Zorn et al., 2005; Magenau et al., 2010; Koreth et al., 2011), B cells (Sarantopoulos et al., 2009; Flynn et al., 2015), T follicular helper (TFH) cells (Forcade et al., 2016), T follicular regulatory (TFR) cells (Kamihara et al., 2017) and iNKT (invariant natural killer T) cells (Mavers et al., 2017). Proteomics with cytometry has been used to discover new cell populations in aGVHD, such as CD146+ CD4+ T cells or blood mucosal-associated T cells (CD161+ TCRVα7.2+ T cells) and CD38+ T cells in cGVHD (Li et al., 2016; Forcade et al., 2017; Stikvoort et al., 2017). Although the frequencies and absolute numbers of such immune cells provide insight into the pathophysiology of GVHD, and these cells may represent excellent therapeutic targets, the relatively low throughput of cytomics, lack of a standard curve for quantification and the need for a large volume of samples makes them less ideal biomarkers than soluble factors measurable by ELISA.
The robustness of the assay used for the test matter
Robustness is a measure of the assay capacity to remain unaffected by small changes in test conditions (i.e.: incubation time, temperature and sample). Robustness is established after several validation steps are followed.
Assay optimisation (pre-validation) determines how a range of matrix and sample elements, as well as assay conditions, affect assay parameters and assay performance. These data, along with scientific judgement, set the acceptance criteria for the assay validation. It is important to establish acceptance criteria before executing the validation protocol.
Assay qualification is an experimental protocol that demonstrates that an accepted method will provide meaningful data for the specific conditions, matrix and samples that the procedure is intended for. Assay qualification may not require validation of accuracy and reliability of the method (sensitivity), but may merely verify the suitability of the protocol under actual conditions (generally, specificity).
Assay validation – comprehensive experiments evaluate and document the quantitative performance of an assay, including sensitivity, specificity, accuracy, precision, detection limit, and range and limits of quantitation. Full assay validation will include inter-assay and inter-laboratory assessments of assay repeatability and robustness.
These assay analytical performance characteristics are summarised in Table I. After the nine ‘steps’ of assay/method validation listed in this table, the tests meet all of the requirements needed for CLIA-88, the NYSDOH CLEP Standards, and the quality standards of the College of American Pathologists. Only a few GVHD biomarkers have followed this rigorous validation, one of them being the ST2 ELISA assay.
Table I.
Assay analytical parameters definitions.
|
Respecting the major phases of biomarker development matter
Importantly, biomarker development entails multiple phases, from the identification of promising molecular targets to routine use in clinical practice. The 2014 NIH Chronic GVHD Consensus Biomarker Working Group summarised an ideal framework for biomarker development (Paczesny et al., 2015a; Barrett, 2017). However, only a few GVHD biomarker studies have followed this recommended framework, and included patients derived from multiple centres or independent cohorts of patients (Vander Lugt et al., 2013; Yu et al., 2016; Abu Zaid et al., 2017; Hartwell et al., 2017). It is therefore critical to restate the necessary phases of biomarker development. These phases parallel the assay validation steps.
Phase 1: identification
The initial phase aims at identifying candidate biomarkers in a small experiment of well-matched cases and controls selected from the populations in which the biomarker is intended for use. In this phase, it is important to define the clinical context of use and the reported outcomes that will be captured to assess a clinical endpoint, for example, non-relapse mortality, relapse mortality, aGVHD grades, or NIH cGVHD grades. The type of biomarker that is looked at should be defined at this point – the most appropriate time point post-HCT, for the type of biomarker, as well as the most appropriate controls for the cases. For example, if we want a diagnosis marker of cGVHD, we will need to look at samples at the time of onset of cGVHD. This is typically after day 180 post-HCT. Patients should be naïve of cGVHD treatment, and controls should be matched for demographics and sample time points, which might be difficult to obtain if there are none on the study. For a cGVHD prognostic biomarker, time points should be defined before the clinical signs, so estimated at day 90–100 post-HCT. Importantly, it should not be assumed that the same controls are appropriate for different clinical contexts. Although it is worth testing a diagnosis marker for its prognosis potential, it might not be seen, as was the case for CCL15 (Du et al., 2018). In contrast, ST2 is a predictive biomarker (Vander Lugt et al., 2013; Levine et al., 2015; Pidala et al., 2020), as well as a prognostic (Vander Lugt et al., 2013; Nelson et al., 2014; McDonald et al., 2015; Ponce et al., 2015; Abu Zaid et al., 2017; Hartwell et al., 2017; Kanakry et al., 2017) and a response biomarker (McDonald et al., 2017; Major-Monfried et al., 2018).
Phase 2: qualification also called verification
This phase confirms the analytical validity of an assay, as explained above. The assay practicality should also be considered: is the potential sample to be measured easy to obtain, is the sample stable until the test can be performed, and is it cost-effective? Once the type of biomarker, the clinical context, the type of sample, the time point, the control and the assay are validated and all parameters detailed in Table I are defined, these are locked down (finalised) and cannot be changed without reverification of the test under the revised conditions.
Phase 3: validation
This phase assesses the robustness of the test in all samples from the intended use population for the specific outcome defined earlier. For this phase, the cohort used should be independent from the cohort previously studied in phases 1 and 2, as long as they are consistent with the intended use population. If, however, the demographics are too different from the population for which the biomarker’s use is intended, qualification testing could fail inappropriately (Table II).
Table II.
Three major phases for biomarker development
|
Statistical considerations
During the advancement of the different biomarker phases, statistics will be used. A ‘statistical hypothesis test’ is a formal scientific method to examine the plausibility of a specific statement regarding the comparison of an outcome between two or more groups. The statement regarding the comparison is typically formulated as a ‘null hypothesis’, stating that there is no difference in outcome between groups. Statistical significance and clinical significance are not the same thing. The magnitude and direction of the effect must be considered. Thus, confidence interval (CI) is more helpful than statistical test to assess the presence or absence of the clinical significance. Even if there are many results that are statistically significant, but not likely due to the play of chance, they may be irrelevant due to the small clinical effect. Conversely, a lack of statistical significance should not be confused with a negative result; it may arise from a lack of statistical power due to a limited sample size. When a statistical test is performed, one of four outcomes will occur, depending on whether the null hypothesis is true or false and whether the statistical test rejects or does not reject the null hypothesis: (i) the procedure rejects a true null hypothesis (a false-positive type I error), (ii) the procedure does not reject a true null hypothesis (a true negative), (iii) the procedure rejects a false null hypothesis (a true positive), or (iv) the procedure does not reject a false null hypothesis (a false-negative type II error); represented as sensitivity and specificity (see Table III). As proteomic biomarkers are often quantified against a standard curve, they will render continuous variable values, with several potential cutoffs. In this case, receiver-operating characteristic (ROC) curve analysis is used, because the accuracy is not distorted by fluctuations caused by the use of arbitrarily chosen cutoffs (Gu & Pepe, 2011). The area under the curve (AUC) is the derived summary measure of accuracy, and determines the inherent ability of the test to discriminate between cases and controls. Using this analysis as a measure of performance, one can compare individual tests or judge whether various combinations of tests can improve accuracy (Paczesny et al., 2009). However, one point of caution with multiplicity is that it can inflate a type I error when more than one test is used. When 10 statistically independent tests are performed, each with a significance level of 0·05, the chance of at least one test being significant is no longer 0·05, but approximately 0·40 = 1 − (1 – 0·05)10 (Bauer, 1991). To accommodate the issue, it can be necessary to control the overall type I error to less than the threshold of 0·05 in a confirmatory clinical study. In practical settings, multiplicity would arise in the following situations: testing for multiple endpoints; exploration of multiple biomarkers; subgroup analyses.
Table III.
Statistical analyses for evaluating test(s)
|
The use of sensitivity versus positive predictive value (PPV) for complication post-HCT is debatable. However, sensitivity is usually more useful for the physician to make a call on how to use the biomarker in a clinical trial. Indeed, PPV is dependent on the prevalence of the complication and as the prevalence increases, the PPV also increases while the NPV decreases. Prevalence measures the proportion of subjects who are in a specific health-related state at a point in time. In other words, it is the incidence of the complication multiplied by the duration of the disease, which depends on death and cure rates. Not surprisingly, it has been extremely difficult to estimate accurately the prevalence of most complications in the HCT population. With this limitation in mind, estimates of PPV have been performed for acute GVHD where incidence rate is roughly estimated at 50% (McDonald et al., 2015). This can be performed for rare complications such as IPS but with statistical manipulation (Seo et al., 2018). Table III summarises key statistical analyses for evaluating a new test(s).
Most validated biomarkers
Table IV summarises the proteins that have moved from candidate to biomarker for different post-transplantation outcomes.
Table IV.
Plasma biomarkers for post-HCT outcomes.*
| Protein | Study | No. of patients in the study | Association direction | Diagnosis time point (median day post-HCT) | Prognostic time point (median day post-HCT) | Refs |
|---|---|---|---|---|---|---|
| Acute GVHD | ||||||
| Four-protein panel (sIL-2Rα, TNFR1, HGF, IL-8) | Paczesny (2009) | 42 + 282† + 142‡ | Increased | 28 | ND | Paczesny et al (2009) |
| ST2 | Vander Lugt (2013) | 20 + 38I† + 673‡ + 75‡ | Increased | 28 | 14 | Vander Lugt et al (2013) |
| Levine (2015) | 328 + 164† + 300f | Increased | 28 | ND | Levine et al (2015) | |
| McDonald (2015) | 74† + 76† | Increased | 28 | Not significant | McDonald et al (2015) | |
| Abu Zaid (2016) | 211 (independent cohort following validation) | Increased | 28 | ND | Abu Zaid et al (2017) | |
| Hartwell (2017) | 620 + 309‡ + 358‡ | Increased | ND | 7 | Hartwell et al (2017) | |
| TIM3 | Hansen (2013) | 20 + 127† + 22f | Increased | 28 | ND | Hansen et al (2013) |
| McDonald (2015) | 74† + 76† + 167‡ | Increased | 28 | 14 | McDonald et al (2015) | |
| Abu Zaid (2016) | 211 (independent cohort following validation) | Increased | 28 | ND | Abu Zaid et al (2017) | |
| IL-6 | Kennedy (2014) | 53 (one cohort but subsequently validated) | Increased (3–14) then decreased | 30 | 7–14 | Kennedy et al (2014) |
| McDonald (2015) | 74† + 76† | Increased | 28 | Not significant | McDonald et al (2015) | |
| Gl-specific | ||||||
| Reg3α | Ferrara (2011) | 20 + 871† + 143† | Increased | 28 | ND | Ferrara et al (2011) |
| TIM3 | Hansen (2013) | 20 + 127† + 22† | Increased | 28 | ND | Hansen et al (2013) |
| Liver-specific | ||||||
| Reg3α >HGF and KRT18 | Luft (2007) Harris (2011) | 55 + 826† + 128† | Increased | 28 | ND | Harris et al (2012, Luft et al (2007) |
| Skin-specific | ||||||
| (Elafin) | Paczesny (2010) | 20 + 492† | Increased | 28 | ND | Paczesny et al (2010) |
| Bruggen (2015) | 59 | Increased in skin | 28 | ND | Bruggen et al (2015) | |
| Late acute GVHD | ||||||
| AREG/EGF ratio | Holtan (2016) | 105 + 50† | Increased | 160 | ND | Holtan et al (2016) |
| Chronic GVHD | ||||||
| (sBAFF) | Sarantopoulos (2007) | 104 | Increased | 480 | NA | Sarantopoulos et til. (2007) |
| Fujii (2008) | 80 (paediatric) | Increased | 171 (early), 429 (late) | NA | Fujii et al. (2008) | |
| Kitko (2014) | 35 + 109† + 21I† | Increased, and not validated in independent cohort | 154, 256 (early), 619 (late) | NA | Kitko et al. (2014) | |
| Kariminia (2016) | 23 + 198† + 83† | Increased | 203, 174 | NA | Kariminia et al. (2016) | |
| CXCL9 | Kitko (2014) | 35 + 109† + 211† | Increased | 154, 256 (early), 619 (late) | NA | Kitko et al. (2014) |
| Yu (2016) | 53 + 211† + 180 | Increased | 210, 203 | 100 | Yu et al. (2016) | |
| Kariminia (2016) | 23 + 198† + 83† | Increased, and not validated in independent cohort | 203, 174 | NA | Kariminia et al. (2016) | |
| Hakim (2016) | 26 + 83† | Increased | 132 | NA | Hakim et al. (2016) | |
| Abu Zaid (2016) | 211‡ | Increased | NA | 100, 180, 365 (time dependent analysis) | Abu Zaid et al. (2017) | |
| CXCL10 | Kariminia (2016) | 23 + 198† + 83† | Increased | 203, 174 | NA | Kariminia et al. (2016) |
| Hakim (2016) | 26 + 83† | Increased | 132 | NA | Hakim et al. (2016) | |
| Four-protein panel | Yu (2016) | 53 + 211† + 180‡ | Increased | 210, 203 | 100 | Yu et al. (2016) |
| (CXCL9, ST2, OPN, MMP3) | ||||||
| (MMP3) | Liu (2016) | 76 (BOS) | Increased | 531 | NA | Liu et al. (2016) |
| (CCL15) | Du (2018) | 211† + 792‡ | Increased at onset but not prognostic | 203 | 100 | Du et al. (2018) |
| CRS | ||||||
| ST2, sIL-2Rα, TNFR1 | Chang (2014) | 927 | Increased | 14 | NA | Chang et al. (2014) |
| SOS | ||||||
| ST2, ANG2, HA, VCAM1 L-ficolin | Akil (2015) | 40 + 45† + 35† | All increased except for L- ficolin which was decreased | 14 | NA | Akil et al. (2015) |
| HA, VCAM1 L-ficolin | Akil (2015) | 26‡ + 24‡ | All increased except for L- ficolin which was decreased | NA | 0 | Akil et al. (2015) |
| L-ficolin | Abu Zaid (2017) | 211 | Decreased | 28 | NA | Abu Zaid et al. (2017) |
| TMA | ||||||
| ST2 | Rotz (2017) | 95‡ + 110‡ + 107‡ | Increased | NA | 14 | Rotz et al. (2017) |
| EASIX | Luft (2017) | 239 + 141† + 173† + 89† | Increased (significant only in reduced intensity | conditioning) | 30–44 | |
| ND | Luft et al. (2017) | |||||
| IPS | ||||||
| ST2, IL-6, TNFR1 | Seo (2017) | 240 | Increased ST2 and IL-6 vs. controls, TNFR1 vs. viral pneumonia | 23 | 7 | Seo et al. (2018) |
| PTDM | ||||||
| ST2 | Johnpulle (2017) | 36 + 26 + 12 (adults) | Increased | 30 | 14 (engraftment) | Johnpulle et al. (2017) |
| ST2 | Rowan (2019) | 55 children | Increased | NA | 14 | Rowan et al. (2019) |
ND, not done; NA, not applicable.
This table includes only proteins that have been discovered with a large-scale proteomics platform, are identifiable, and which have reached the point of validation as biomarkers with the same reproducible assay in at least two independent cohorts of sufficient sample sizes from different institutions according to the 2014 NIH consensus on biomarkers criteria. Candidate biomarkers of interest that have not met these criteria are indicated in parenthesis.
Patient number in validation cohort 1 and cohort 2.
Prognostic cohort.
Acute GVHD biomarkers
Before the development of biomarkers, aGVHD diagnosis relied entirely on clinical signs in one of three major target organs: skin, liver and/or gastrointestinal (GI) tract, as confirmed by biopsy (Mowat & Socie, 2004).
The first biomarker panel identified and validated for aGVHD diagnosis is a four-protein biomarker panel [IL-2 receptor α chain (sIL-2Rα/sCD25), tumour necrosis factor receptor-1 (TNFR1), IL-8 and hepatocyte growth factor (HGF)], discovered by screening aGVHD patient plasma samples by competitive hybridisation to arrays of antibodies for 130 proteins (Paczesny et al., 2009).
- ST2 is the most validated biomarker for aGVHD and has been tested for several clinical outcomes.
- ST2 as a predictive marker. ST2 was identified and validated as a biomarker for the risk of therapy-resistant aGVHD and death in two independent cohorts from two centres. Using state-of-the art tandem mass spectrometry proteomics, a comparison was made between plasma obtained a median of 16 days after therapy initiation from patients with a complete response by day 28, and patients with progressive aGVHD during therapy. Of the 12 lead candidate markers, ST2 was as significant as the other 12 markers in a panel for predicting resistance to aGVHD therapy and subsequent death without relapse. As compared with patients with low ST2 values at therapy initiation, patients with high ST2 values were 2·3 times as likely to have treatment-resistant GVHD (95%, 1·5–3·6) and 3·7 times as likely to die within six months after therapy (95% CI, 2·3–5·9). Furthermore, for the first time for aGVHD markers, ST2 was predictive regardless of the clinical onset aGVHD grade (Vander Lugt et al., 2013). Since then, ST2 has been validated in large cohorts totaling >1000 HCT patients (Levine et al., 2015). It has now been implemented in several clinical trials, including the Blood and Marrow Transplant Clinical Trials Network (BMTCTN) study 1501 (NCT02806947). This was a randomised phase II multicentre open-label study, evaluating sirolimus and prednisone in patients with Minnesota standard-risk and low-risk biomarker-confirmed aGVHD, which shows that sirolimus (a steroid-free treatment) provides similar day 28 complete/partial response rates as prednisone in initial therapy of standard-risk acute GVHD (Pidala et al., 2020). This means that biomarkers already help the clinician decide on a less toxic aGVHD treatment. What more? Trials with high-risk biomarkers using intensified treatment are also under development for patients with newly diagnosed aGVHD.
- ST2 as a prognostic marker. Plasma ST2 values measured early post-transplantation before the clinical signs of aGVHD were associated with six-month mortality without relapse, and improved risk stratification for death without relapse after transplantation (Vander Lugt et al., 2013). This has been further validated in several cohorts totaling ~3000 HCT patients (McDonald et al., 2015; Abu Zaid et al., 2017; Hartwell et al., 2017). Furthermore, ST2 as a prognostic marker has been validated in different HCT settings: i) a cohort of patients receiving cyclophosphamide-fludarabine non-myeloablative HCT (Nelson et al., 2014), ii) a cohort of patients receiving cord blood allo-HCT (Ponce et al., 2015), iii) three cohorts of patients treated with PTCy following HLA-haploidentical or HLA-matched-related or -unrelated allo-HCT (Kanakry et al., 2017), and d) a cohort of patients in a multicentre phase 3 trial with uniform clinical and transplant characteristics post-allo-HCT (Abu Zaid et al., 2017). However, although ST2 was validated on large retrospective sets, it has not yet been implemented prospectively into pre-emptive treatment, as commented on in Clinical Chemistry (Paczesny, 2017).
- ST2 as a response to treatment marker. ST2 in combination with TIM3 tested 14 days post-prednisone, and ST2 in combination with Reg3α tested seven days post-steroids, showed convincing statistical values comparing responders versus non-responders (McDonald et al., 2017; Major-Monfried et al., 2018).
Interleukin (IL)-6 levels were measured from day 3 to 60 post-transplant HCT in 53 patients, and found that they elevated early during the transplant course (Kennedy et al., 2014).
- Target-specific biomarkers that can differentiate skin GVHD from other rashes and GI GVHD from other forms of enteritis were discovered and could replace invasive biopsies in this fragile population.
- Regenerating islet-derived 3-alpha (Reg3α) and T-cell immunoglobulin mucin-3 (TIM3) were discovered as gastrointestinal (GI) GVHD biomarkers, and were also validated as prognostic biomarkers of aGVHD when measured at day 7 and 14 respectively (McDonald et al., 2015; Hartwell et al., 2017).
- Elafin was discovered as a diagnostic marker utilising next-generation proteomics and plasma samples from skin aGVHD, and was validated in skin biopsies (Paczesny et al., 2010; Bruggen et al., 2015).
- HGF and cytokeratin-18 fragments (KRT18) were correlated with liver GVHD, although Reg3α had a much better AUC for diagnosis of liver GVHD than these two biomarkers, probably due to the frequent association of gut and liver aGVHD and the better sensitivity of Reg3α (Luft et al., 2007; Harris et al., 2012).
Urine proteomics identified patterns of peptides that correlated with aGVHD and cGVHD in European cohorts (Weissinger et al., 2007; Weissinger et al., 2017), but were not validated in current US cohorts. In addition, a multicentre, double-blinded, placebo-controlled trial of preemptive treatment for aGVHD, using the aGVHD pattern, showed no differences between groups (Weissinger et al., 2017). One possible explanation is that classifiers using a machine learning-based algorithm can be over-fitted. Thus, these classifiers have not yet met criteria for FDA approval as biomarkers. Low urinary levels of indoxyl-sulfate, a metabolite of indole that reflects GI microbiome diversity, have been correlated with poor outcome in a single-centre cohort of 131 patients (Weber et al., 2015).
Faecal proteins such as calprotectin and alpha-1-antitrypsin have been suggested as candidate biomarkers, as they correlated with response to corticosteroids in GIGVHD, in a single-centre cohort of 72 patients (Rodriguez-Otero et al., 2012). Although increases in faecal proteins were reported by multiple studies, these were small sample size, single-centre cohort studies employing different tests. Thus, these proteins have yet to be confirmed as biomarkers.
Circulating angiogenic factors were correlated with late aGVHD with some inconsistencies in findings between cohorts, GI biopsies and experimental models (Holtan et al., 2016; Ramadan et al., 2017; Amin et al., 2018). Importantly, the authors compared AREG/EGF ratio in classic aGVHD and cGVHD, and found that AREG/EGF ratio was also elevated in classic aGVHD, but not in cGVHD (Holtan et al., 2016).
CRS post-HCT typically debuts at the time of engraftment but sometimes earlier. It has been associated with hyperacute GVHD and subsequent severe aGVHD. ST2 measured at day 14 post-HCT has been shown to be the most significant marker associated with CRS in a cohort of 927 patients and predicted poor outcomes (Chang et al., 2014).
Chronic GVHD biomarkers.The clinical manifestations of cGVHD often resemble those of autoimmune diseases. Its diagnosis is based on clinical symptoms (i.e. inflammatory and fibrotic components) involving almost any target organ (e.g. skin, nails, mouth, eyes, genitalia, skeletal muscle, GI tract, liver and lung). Blood biomarkers (cellular and protein) have been evaluated. Some noteworthy and novel biomarkers reported since the 2014 NIH consensus biomarker and biology papers (Paczesny, 2015; Cooke et al., 2016) are listed below.
High levels of soluble B cell activating factor and the balance of B cell subsets during B cell reconstitution were the first biomarkers correlated with cGVHD (Sarantopoulos & Ritz, 2015).
Prolonged imbalance of CD4+ CD25+ FOXP3+ Tregs versus conventional CD4+ T cells post-HCT was associated with a loss of tolerance and significant cGVHD manifestations (Alho et al., 2016).
Using a quantitative proteomics approach, a biomarker panel of four proteins [ST2, CXCL9, matrix metalloproteinase 3 (MMP-3) and osteopontin] showed significant correlation with cGVHD diagnosis. Moreover, at day + 100 post-HCT, this panel allowed patient stratification according to cGVHD risk (Yu et al., 2016). MMP-3 was also correlated with bronchiolitis obliterans diagnosis (Liu et al., 2016). Recently, both CXCL9 and CXCL10 were significantly correlated with cGVHD diagnosis in the first replication cohort, but only CXCL10 was in the second (Kariminia et al., 2016). In another study, gene expression profiling of circulating monocytes from cGVHD patients revealed significant up-regulation of IFN-inducible (including CXCL9 and CXCL10) and damage-response genes in cGVHD patients, compared to controls. These pathways were confirmed in plasma ELISAs, showing elevated CXCL9 and CXCL10 levels (Hakim et al., 2016). Together, the IFN-inducible chemokines CXCL9 and CXCL10, which are responsible for CXCR3-expressing Th1/NK lymphocyte recruitment (Paczesny & Abu Zaid, 2016), are upregulated at diagnosis and warrant further testing in prospective studies.
An activated Th17-prone T cell subset expressing both CD146 and CCR5 was found to be involved in cGVHD, and sensitive to pharmacological inhibition (Forcade et al., 2017).
Circulating TFH cells were shown to correlate with cGVHD and exhibit a Th17 profile (Forcade et al., 2016).
Plasma CD163 concentration was associated with de novo-onset cGVHD (Inamoto et al., 2017).
Among 42 patients who received ibrutinib after failure of prior therapy, responders had decreased levels of sIL-2Ra, CX3CL1, CXCL9, CXCL10, CCL22 and CCL4 (Miklos et al., 2017).
CCL15 was recently discovered as a novel biomarker in patients via murine cGVHD proteome profiling (Du et al., 2018).
Hepatic SOS is a major complication during the early post-HCT period, caused by both toxic injury of conditioning therapy to sinusoidal endothelial cells and inflammation, with clinical symptoms of hyperbilirubinemia, tender hepatomegaly, ascites and weight gain. The incidence and severity of SOS have decreased significantly in recent years, but SOS-related deaths are still observed in clinical practice. Biomarkers for SOS diagnosis [ST2, angiopoietin-2 (ANG2), L-ficolin, hyaluronic acid (HA) and vascular cell adhesion molecule-1 (VCAM1)] and prognosis [L-ficolin, HA and VCAM1] were identified by a proteomics study and validated in several cohorts (Akil et al., 2015; Abu Zaid et al., 2017).
TMA is associated with endothelial injury in vivo and was recently linked to complement activation in vitro. ST2 was shown to be a reliable early biomarker of TMA, independent of aGVHD in several cohorts (Rotz et al., 2017). Routine laboratory measurements (lactate dehydrogenase, creatinine and thrombocytes) can be used to create a formula called the Endothelial Activation and Stress Index (EASIX), which was found to be a predictor of survival in patients with reduced intensity conditioning (Luft et al., 2017).
IPS is a non-infectious pulmonary post-HCT complication that is difficult to diagnose. A recent study showed that ST2 and IL-6 are diagnostic and prognostic biomarkers of IPS, and TNFR1 is a marker for differential diagnosis from viral pneumonia. ST2 at onset and at day 7 post-HCT had the highest positive predictive value for IPS occurrence (Seo et al., 2018).
New-onset PTDM occurs commonly post-HCT and is associated with reduced survival (Engelhardt et al., 2012). High ST2 at engraftment predicts increased PTDM and correlates with poor outcomes in adults and children (Johnpulle et al., 2017; Rowan et al., 2019).
GVHD-free GVL biomarkers. A recent study has shown that it was possible to identify a GVHD-free antitumoural signature by proteomics and systems biology after allogeneic Donor Lymphocyte Injection (DLI), comparing plasma proteome post-DLI of patients who experienced GVL and GVHD, with the proteome of patients who experienced GVL without GVHD. The approach provided a 61-protein signature that largely validated a single-cell profiling experiment of activated T cells. Novel markers, such as RPL23, ILF2, CD58 and CRTAM were identified and could be extended to other antitumoural responses after validation in independent cohorts (Kansagra et al., 2019).
Mechanistic biomarkers
If a biomarker is routed in the pathogenesis of the disease, it will provide insight into disease pathogenesis, making this marker even more relevant. For example, during experimental aGVHD, intestinal stromal cells and intestinal T cells producing interferon (IFN)γ and IL17 are major sources of sST2 which sequesters IL33, limiting its availability to cytoprotective T cells expressing the transmembrane molecule form of ST2 [mostly T helper 2 (Th2) cells and ST2+ FoxP3+ regulatory T cells (Tregs)] (Zhang et al., 2015). Another example is Reg3α which has been shown to prevent crypt apoptosis and to control aGVHD (Zhao et al., 2018).
Druggable biomarkers
The next level of attractiveness for a biomarker is if it can be targeted with drugs. In the field of rheumatologic diseases, cytokines have long been involved as markers, and have been targeted directly or via intracellular signaling with Janus kinase (JAK) inhibitors (jakinibs) (Gadina et al., 2019). In the field of cancer, the signal transducer and activator of transcription 3 (STAT3) has been proposed as an attractive focus for cancer therapy, and has recently been targeted with a small-molecule degrader to achieve complete tumour regression in vivo (Bai et al., 2019). For GVHD, therapeutic approaches have been largely limited to non-specific targeting of effector cells with corticosteroids that remain the first-line treatment for patients presenting GVHD symptoms. Biomarkers can represent promising targets for new therapeutics which will be aGVHD-specific drugs, because they will target the appropriate effector T cells to increase efficacy and lower toxicity. For this reason, ST2 again represents a good example of a druggable biomarker. Indeed, blockade of sST2 in the peritransplantation period with a neutralising monoclonal antibody or small-molecule inhibitors reduces aGVHD severity and mortality by increasing plasma IL33, and decreasing the numbers of gut-infiltrating and IFNγ-producing T cells, and increasing the numbers of cytoprotective T cells (Zhang et al., 2015; Ramadan et al., 2018). Adoptive transfer of cells expressing membrane-bound ST2 (Tregs, IL-9-expressing T cells, innate lymphoid cells type 2) leads to similar results in murine models and is currently in trials (Bruce et al., 2017; Ramadan et al., 2017).
Conclusions and future perspective of biomarkers in transplant and cellular therapy research
The reasons why ST2 became a golden nugget biomarker are that it: (i) was discovered through a relatively unbiased proteomics approach, (ii) used a non-invasive cost-efficient sample (plasma), (iii) has been validated as a predictive, prognostic and response biomarker for aGVHD but also for other complications of alloreactivity post-HCT, (iv) is measured using a robust ELISA assay, (v) has followed the major phases of biomarker development including validation in large independent cohorts, (vi) is, in addition, a mechanistic biomarker and, finally, (vii) is druggable. Future directions include trials to assess the effectiveness of aGVHD biomarker-based pre-emption. Efforts to discover better cGVHD biomarkers and target-specific cGVHD biomarkers are underway through American and European initiatives (Paczesny et al., 2015a; Wolff et al., 2018). In view of the increased number of approved cellular therapies that have been associated with short- and long-term toxicities – most commonly CRS and immune effector cell-associated neurotoxicity syndrome (ICANS) (Kansagra et al., 2019), there are initiatives to develop biomarkers for cellular therapy toxicity and efficacy that would benefit from the work already done in the HCT field.
Acknowledgements
The authors are supported by grants from the National Institutes of Health: National Cancer Institute (R01CA168814), the National Heart, Lung, and Blood Institute (R21HL139934), and the Eunice Kennedy Shriver National Institute of Child Health and Human Development of Health (R01HD074587).
Footnotes
Conflict of interest
S.P. is an inventor on a patent on ‘Methods of detection of graft-versus-host disease’ (13/573766).
References
- Abu Zaid M, Wu J, Wu C, Logan BR, Yu J, Cutler C, Antin JH, Paczesny S & Choi SW (2017) Plasma biomarkers of risk for death in a multicenter phase 3 trial with uniform transplant characteristics post-allogeneic HCT. Blood, 129, 162–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akil A, Zhang Q, Mumaw CL, Raiker N, Yu J, Velez de Mendizabal N, Haneline LS, Robertson KA, Skiles J, Diaz-Ricart M, Carreras E, Renbarger J, Hanash S, Bies RR & Paczesny S (2015) Biomarkers for diagnosis and prognosis of sinusoidal obstruction syndrome after hematopoietic cell transplantation. Biology of Blood and Marrow Transplantation, 21, 1739–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alho AC, Kim HT, Chammas MJ, Reynolds CG, Matos TR, Forcade E, Whangbo J, Nikiforow S, Cutler CS, Koreth J, Ho VT, Armand P, Antin JH, Alyea EP, Lacerda JF, Soiffer RJ & Ritz J (2016) Unbalanced recovery of regulatory and effector T cells after allogeneic stem cell transplantation contributes to chronic GVHD. Blood, 127, 646–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altelaar AF, Munoz J & Heck AJ (2013) Next-generation proteomics: towards an integrative view of proteome dynamics. Nature Reviews Genetics, 14, 35–48. [DOI] [PubMed] [Google Scholar]
- Amin K, Usman Y, Schultz B, Vaughn B, Howard J, Khoruts A, Defor TE, Forster C, Rashidi A, Weisdorf DJ, MacMillan ML, Blazar BR, Mortari A & Holtan SG(2018) Low amphiregulin expression in intestinal biopsies of patients with acute graft-versus-host disease. In: BMT Tandem Meetings. Biology of Blood and Marrow Transplantation, Salt Lake City, UT. [Google Scholar]
- Andermann T, Peled J, Ho C, Reddy P, Riches M, Storb R, Teshima T, van den Brink M, Alousi A, Balderman S, Chiusolo P, Clark W, Holler E, Howard A, Kean L, Koh A, McCarthy P, McCarty J, Mohty M, Nakamura R, Rezvani K, Segal B, Shaw B, Shpall E, Sung A, Weber D, Whangbo J, Wingard J, Wood W, Perales MA, Jenq R & Bhatt A (2018) Microbiome-host interactions in hematopoietic stem-cell transplant recipients. Biology of Blood and Marrow Transplantation, 24, 1322–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Zhou H, Xu R, Zhao Y, Chinnaswamy K, McEachern D, Chen J, Yang CY, Liu Z, Wang M, Liu L, Jiang H, Wen B, Kumar P, Meagher JL, Sun D, Stuckey JA & Wang S (2019) A potent and selective small-molecule degrader of STAT3 achieves complete tumor regression in vivo. Cancer Cell, 36, e417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett AJ (2017) Transplant biomarkers ready for the clinic? Blood, 129, 137–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer P (1991) Multiple testing in clinical trials. Statistics in Medicine, 10, 871–890, discussion 889–890. [DOI] [PubMed] [Google Scholar]
- Bontha SV, Maluf DG, Mueller TF & Mas VR (2017) systems biology in kidney transplantation: the application of multi-omics to a complex model. American Journal of Transplantation, 17, 11–21. [DOI] [PubMed] [Google Scholar]
- Bruce DW, Stefanski HE, Vincent BG, Dant TA, Reisdorf S, Bommiasamy H, Serody DA, Wilson JE, McKinnon KP, Shlomchik WD, Armistead PM, Ting JPY, Woosley JT, Blazar BR, Zaiss DMW, McKenzie ANJ, Coghill JM & Serody JS (2017) Type 2 innate lymphoid cells treat and prevent acute gastrointestinal graft-versus-host disease. Journal of Clinical Investigation, 127, 1813–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggen MC, Petzelbauer P, Greinix H, Contassot E, Jankovic D, French L, Socie G, Rabitsch W, Kuzmina Z, Kalhs P, Knobler R, Stingl G & Stary G (2015) Epidermal elafin expression is an indicator of poor prognosis in cutaneous graft-versus-host disease. The Journal of Investigative Dermatology, 135, 999–1006. [DOI] [PubMed] [Google Scholar]
- Chang L, Frame D, Braun T, Gatza E, Hanauer DA, Zhao S, Magenau JM, Schultz K, Tokala H, Ferrara JL, Levine JE, Reddy P, Paczesny S & Choi SW (2014) Engraftment syndrome after allogeneic hematopoietic cell transplantation predicts poor outcomes. Biology of Blood and Marrow Transplantation, 20, 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaussabel D & Pulendran B (2015) A vision and a prescription for big data-enabled medicine. Nature Immunology, 16, 435–439. [DOI] [PubMed] [Google Scholar]
- Cooke KR, Luznik L, Sarantopoulos S, Hakim FT, Jagasia M, Fowler DH, van den Brink MR, Hansen JA, Parkman R, Miklos DB, Martin PJ, Paczesny S, Vogelsang G, Pavletic S, Ritz J, Schultz KR & Blazar BR (2016) The biology of chronic graft-versus-host disease: a task force report from the national institutes of health consensus development project on criteria for clinical trials in chronic graft-versus-host disease. Biology of Blood and Marrow Transplantation, 23, 211–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Flynn R, Paz K, Ren HG, Ogata Y, Zhang Q, Gafken PR, Storer BE, Roy NH, Burkhardt JK, Mathews W, Tolar J, Lee SJ, Blazar BR & Paczesny S (2018) Murine chronic graft-versus-host disease proteome profiling discovers CCL15 as a novel biomarker in patients. Blood, 131, 1743–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt BG, Jagasia SM, Crowe JE Jr, Griffith ML, Savani BN, Kassim AA, Lu P, Weitkamp JH, Moore DJ, Yoder SM, Rock MT & Jagasia M (2012) Predicting posttransplantation diabetes mellitus by regulatory T-cell phenotype: implications for metabolic intervention to modulate alloreactivity. Blood, 119, 2417–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA-NIH Biomarker Working Group (2016) BEST (Biomarkers, EndpointS, and other Tools) Resource. Food and Drug Administration (US), Silver Spring, MD. [PubMed] [Google Scholar]
- Ferrara JL, Harris AC, Greenson JK, Braun TM, Holler E, Teshima T, Levine JE, Choi SW, Huber E, Landfried K, Akashi K, Vander Lugt M, Reddy P, Chin A, Zhang Q, Hanash S & Paczesny S (2011) Regenerating islet-derived 3-alpha is a biomarker of gastrointestinal graft-versus-host disease. Blood, 118, 6702–6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn R, Allen JL, Luznik L, MacDonald KP, Paz K, Alexander KA, Vulic A, Du J, Panoskaltsis-Mortari A, Taylor PA, Poe JC, Serody JS, Murphy WJ, Hill GR, Maillard I, Koreth J, Cutler CS, Soiffer RJ, Antin JH, Ritz J, Chao NJ, Clynes RA, Sarantopoulos S & Blazar BR (2015) Targeting Syk-activated B cells in murine and human chronic graft-versus-host disease. Blood, 125, 4085–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forcade E, Kim HT, Cutler C, Wang K, Alho AC, Nikiforow S, Ho VT, Koreth J, Armand P, Alyea EP, Blazar BR, Soiffer RJ, Antin JH & Ritz J (2016) Circulating T follicular helper cells with increased function during chronic graft-versus-host disease. Blood, 127, 2489–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forcade E, Paz K, Flynn R, Griesenauer B, Amet T, Li W, Liu L, Bakoyannis G, Jiang D, Chu HW, Lobera M, Yang J, Wilkes DS, Du J, Gartlan K, Hill GR, MacDonald KP, Espada EL, Blanco P, Serody JS, Koreth J, Cutler CS, Antin JH, Soiffer RJ, Ritz J, Paczesny S & Blazar BR (2017) An activated Th17-prone T cell subset involved in chronic graft-versus-host disease sensitive to pharmacological inhibition. JCI Insight, 2, 92111. 10.1172/jci.insight.92111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Cuvelier G, She K, Aslanian S, Shimizu H, Kariminia A, Krailo M, Chen Z, McMaster R, Bergman A, Goldman F, Grupp SA, Wall DA, Gilman AL & Schultz KR (2008) Biomarkers in newly diagnosed pediatric-extensive chronic graft-versus-host disease: a report from the Children’s Oncology Group. Blood, 111, 3276–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlan SN, Watkins B, Tkachev V, Flynn R, Cooley S, Ramakrishnan S, Singh K, Giver C, Hamby K, Stempora L, Garrett A, Chen J, Betz KM, Ziegler CG, Tharp GK, Bosinger SE, Promislow DE, Miller JS, Waller EK, Blazar BR & Kean LS (2015) Transcriptome analysis of GVHD reveals aurora kinase A as a targetable pathway for disease prevention. Science Translational Medicine, 7, 315ra191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlan SN, Watkins B, Tkachev V, Cooley S, Panoskaltsis-Mortari A, Betz K, Brown M, Hunt DJ, Schell JB, Zeleski K, Yu A, Giver CR, Waller EK, Miller JS, Blazar BR & Kean LS (2016) Systems analysis uncovers inflammatory Th/Tc17-driven modules during acute GVHD in monkey and human T cells. Blood, 128, 2568–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadina M, Le MT, Schwartz DM, Silvennoinen O, Nakayamada S, Yamaoka K & O’Shea JJ (2019) Janus kinases to jakinibs: from basic insights to clinical practice. Rheumatology (Oxford), 58, i4–i16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W & Pepe MS (2011) Estimating the diagnostic likelihood ratio of a continuous marker. Biostatistics, 12, 87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim FT, Memon S, Jin P, Imanguli MM, Wang H, Rehman N, Yan XY, Rose J, Mays JW, Dhamala S, Kapoor V, Telford W, Dickinson J, Davis S, Halverson D, Naik HB, Baird K, Fowler D, Stroncek D, Cowen EW, Pavletic SZ & Gress RE (2016) Upregulation of IFN-inducible and damage-response pathways in chronic graft-versus-host disease. The Journal of Immunology, 197, 3490–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JA, Hanash SM, Tabellini L, Baik C, Lawler RL, Grogan BM, Storer B, Chin A, Johnson M, Wong CH, Zhang Q, Martin PJ & McDonald GB (2013) A novel soluble form of Tim-3 associated with severe graft-versus-host disease. Biology of Blood and Marrow Transplantation, 19, 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AC, Ferrara JL, Braun TM, Holler E, Teshima T, Levine JE, Choi SW, Landfried K, Akashi K, Vander Lugt M, Couriel DR, Reddy P & Paczesny S (2012) Plasma biomarkers of lower gastrointestinal and liver acute graft-versus-host disease. Blood, 12, 2960–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell MJ, Ozbek U, Holler E, Renteria AS, Major-Monfried H, Reddy P, Aziz M, Hogan WJ, Ayuk F, Efebera YA, Hexner EO, Bunworasate U, Qayed M, Ordemann R, Wolfl M, Mielke S, Pawarode A, Chen YB, Devine S, Harris AC, Jagasia M, Kitko CL, Litzow MR, Kroger N, Locatelli F, Morales G, Nakamura R, Reshef R, Rosler W, Weber D, Wudhikarn K, Yanik GA, Levine JE & Ferrara JL (2017) An early-biomarker algorithm predicts lethal graft-versus-host disease and survival. JCI Insight, 2, e89798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtan SG, Khera N, Levine JE, Chai X, Storer B, Liu HD, Inamoto Y, Chen GL, Mayer S, Arora M, Palmer J, Flowers MED, Cutler CS, Lukez A, Arai S, Lazaryan A, Newell LF, Krupski C, Jagasia MH, Pusic I, Wood W, Renteria AS, Yanik G, Hogan WJ, Hexner E, Ayuk F, Holler E, Watanaboonyongcharoen P, Efebera YA, Ferrara JLM, Panoskaltsis-Mortari A, Weisdorf D, Lee SJ & Pidala J (2016) Late acute graft-versus-host disease: a prospective analysis of clinical outcomes and circulating angiogenic factors. Blood, 128, 2350–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamoto Y, Martin PJ, Paczesny S, Tabellini L, Momin AA, Mumaw CL, Flowers MED, Lee SJ, Carpenter PA, Storer BE, Hanash S & Hansen JA (2017) Association of plasma CD163 concentration with de novo-onset chronic graft-versus-host disease. Biology of Blood and Marrow Transplantation, 23, 1250–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenq RR, Taur Y, Devlin SM, Ponce DM, Goldberg JD, Ahr KF, Littmann ER, Ling L, Gobourne AC, Miller LC, Docampo MD, Peled JU, Arpaia N, Cross JR, Peets TK, Lumish MA, Shono Y, Dudakov JA, Poeck H, Hanash AM, Barker JN, Perales MA, Giralt SA, Pamer EG & van den Brink MR (2015) Intestinal blautia is associated with reduced death from graft-versus-host disease. Biology of Blood and Marrow Transplantation, 21, 1373–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnpulle RA, Paczesny S, Jung DK, Daguindau E, Jagasia MH, Savani BN, Chin-ratanalab W, Cornell RF, Goodman S, Greer JP, Kassim AA, Sengsayadeth S, Byrne MT & Engelhardt BG (2017) Metabolic complications precede alloreactivity and are characterized by changes in suppression of tumorigenicity 2 signaling. Biology of Blood and Marrow Transplantation, 23, 529–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamihara Y, Forcade E, Koreth J, Liu H, Kubo T, Whangbo J, Hirakawa M, Nikiforow S, Ho VT, Armand P, Cutler CS, Edwin Alyea III E, Blazar B, Antin JH, Soiffer RJ & Ritz J (2017) Low-dose Interleukin-2 Therapy Activates Circulating T Follicular Regulatory Cells And Suppresses Circulating T Follicular Helper Cells in Patients with Chronic Gvhd. American Society of Hematology Blood, Atlanta, GA. [Google Scholar]
- Kanakry CG, Bakoyannis G, Perkins SM, McCurdy SR, Vulic A, Warren EH, Daguindau E, Olmsted T, Mumaw C, Towlerton AM, Cooke KR, O’Donnell PV, Symons HJ, Paczesny S & Luznik L (2017) Plasma-derived proteomic biomarkers in HLA-haploidentical or HLA-matched bone marrow transplantation using post-transplantation cyclophosphamide. Haematologica, 102, 932–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kansagra AJ, Frey NV, Bar M, Laetsch TW, Carpenter PA, Savani BN, Heslop HE, Bol-lard CM, Komanduri KV, Gastineau DA, Chabannon C, Perales MA, Hudecek M, Aljurf M, Andritsos L, Barrett JA, Bachanova V, Bonini C, Ghobadi A, Gill SI, Hill J, Kenderian S, Kebriaei P, Nagler A, Maloney D, Liu HD, Shah NN, Kharfan-Dabaja MA, Shpall EJ, Mufti GJ, Johnston L, Jacoby E, Bazarbachi A, DiPersio JF, Pavletic SZ, Porter DL, Grupp SA, Sadelain M, Litzow MR, Mohty M & Hashmi SK (2019) Clinical utilization of chimeric antigen receptor T cells in B cell acute lymphoblastic leukemia: an expert opinion from the European society for blood and marrow transplantation and the American society for blood and marrow transplantation. Biology of Blood and Marrow Transplantation, 25, e76–e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaesmen E, Rizvi AA, Preus L, McCarthy PL, Pasquini MC, Onel K, Zhu X, Spellman S, Haiman CA, Stram DO, Pooler L, Sheng X, Zhu Q, Yan L, Liu Q, Hu Q, Webb A, Brock G, Clay-Gilmour AI, Battaglia S, Tritchler D, Liu S, Hahn T & Sucheston-Campbell LE (2017) Replication and validation of genetic polymorphisms associated with survival after allogeneic blood or marrow transplant. Blood, 130, 1585–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaesmen E, Hahn T, Dile AJ, Rizvi AA, Wang J, Wang T, Haagenson MD, Preus L, Zhu Q, Liu Q, Yan L, Liu S, Haiman CA, Stram D, Pooler L, Sheng X, Van Den Berg D, Brock G, Webb A, McCarthy PL, Pasquini MC, Spellman SR, Lee SJ, Paczesny S & Sucheston-Campbell LE (2019) Multiple functional variants in the IL1RL1 region are pre-transplant markers for risk of GVHD and infection deaths. Blood Advances, 3, 2512–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariminia A, Holtan SG, Ivison S, Rozmus J, Hebert MJ, Martin PJ, Lee SJ, Wolff D, Subrt P, Abdossamadi S, Sung S, Storek J, Levings M, Aljurf M, Arora M, Cutler C, Gallagher G, Kuruvilla J, Lipton J, Nevill TJ, Newell LF, Panzarella T, Pidala J, Popradi G, Szwajcer D, Tay J, Toze CL, Walker I, Couban S, Storer BE & Schultz KR (2016) Heterogeneity of chronic graft-versus-host disease biomarkers: association with CXCL10 and CXCR3+ NK cells. Blood, 127, 3082–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy GA, Varelias A, Vuckovic S, Le Texier L, Gartlan KH, Zhang P, Thomas G, Anderson L, Boyle G, Cloonan N, Leach J, Sturgeon E, Avery J, Olver SD, Lor M, Misra AK, Hutchins C, Morton AJ, Dur-rant ST, Subramoniapillai E, Butler JP, Curley CI, MacDonald KP, Tey SK & Hill GR (2014) Addition of interleukin-6 inhibition with tocilizumab to standard graft-versus-host disease prophylaxis after allogeneic stem-cell transplantation: a phase 1/2 trial. The Lancet Oncology, 15, 1451–1459. [DOI] [PubMed] [Google Scholar]
- Kitko CL, Levine JE, Storer BE, Chai X, Fox DA, Braun TM, Couriel DR, Martin PJ, Flowers ME, Hansen JA, Chang L, Conlon M, Fiema BJ, Morgan R, Pongtornpipat P, Lamiman K, Ferrara JL, Lee SJ & Paczesny S (2014) Plasma CXCL9 elevations correlate with chronic GVHD diagnosis. Blood, 123, 786–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koreth J, Matsuoka K, Kim HT, McDonough SM, Bindra B, Alyea EP 3rd, Armand P, Cutler C, Ho VT, Treister NS, Bienfang DC, Prasad S, Tzachanis D, Joyce RM, Avigan DE, Antin JH, Ritz J & Soiffer RJ (2011) Interleukin-2 and regulatory T cells in graft-versus-host disease. New England Journal of Medicine, 365, 2055–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JE, Braun TM, Harris AC, Holler E, Taylor A, Miller H, Magenau J, Weisdorf DJ, Ho VT, Bolanos-Meade J, Alousi AM, Ferrara JL & Blood & Marrow Transplant Clinical Trials Network (2015) A prognostic score for acute graft-versus-host disease based on biomarkers: a multicentre study. The Lancet Haematology, 2, e21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Liu L, Gomez A, Zhang J, Ramadan A, Zhang Q, Choi SW, Zhang P, Greenson JK, Liu C, Jiang D, Virts E, Kelich SL, Chu HW, Flynn R, Blazar BR, Hanenberg H, Hanash S & Paczesny S (2016) Proteomics analysis reveals a Th17-prone cell population in presymptomatic graft-versus-host disease. JCI Insight, 1, 86660. 10.1172/jci.insight.86660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yue Z, Yu J, Daguindau E, Kushekhar K, Zhang Q, Ogata Y, Gafken PR, Inamoto Y, Gracon A, Wilkes DS, Hansen JA, Lee SJ, Chen JY & Paczesny S (2016) Proteomic characterization reveals that MMP-3 correlates with bronchiolitis obliterans syndrome following allogeneic hematopoietic cell and lung transplantation. American Journal of Transplantation, 16, 2342–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luft T, Conzelmann M, Benner A, Rieger M, Hess M, Strohhaecker U, Görner M, Hegen-bart U, Ho A & Dreger P (2007) Serum cytokeratin-18 fragments as quantitative markers of epithelial apoptosis in liver and intestinal graft-versus-host disease. Blood, 110, 4535–4542. [DOI] [PubMed] [Google Scholar]
- Luft T, Benner A, Jodele S, Dandoy CE, Storb R, Gooley T, Sandmaier BM, Becker N, Radujkovic A, Dreger P & Penack O (2017) EASIX in patients with acute graft-versus-host disease: a retrospective cohort analysis. The Lancet Haematology, 4, e414–e423. [DOI] [PubMed] [Google Scholar]
- Magenau JM, Qin X, Tawara I, Rogers CE, Kitko C, Schlough M, Bickley D, Braun TM, Jang PS, Lowler KP, Jones DM, Choi SW, Reddy P, Mineishi S, Levine JE, Ferrara JL & Paczesny S (2010) Frequency of CD4(+)CD25(hi)FOXP3(+) regulatory T cells has diagnostic and prognostic value as a biomarker for acute graft-versus-host-disease. Biology of Blood and Marrow Transplantation, 16, 907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major-Monfried H, Renteria AS, Pawarode A, Reddy P, Ayuk F, Holler E, Efebera YA, Hogan WJ, Wolfl M, Qayed M, Hexner EO, Wudhikarn K, Ordemann R, Young R, Shah J, Hartwell MJ, Chaudhry MS, Aziz M, Etra A, Yanik GA, Kroger N, Weber D, Chen YB, Nakamura R, Rosler W, Kitko CL, Harris AC, Pulsipher M, Reshef R, Kowalyk S, Morales G, Torres I, Ozbek U, Ferrara JLM & Levine JE (2018) MAGIC biomarkers predict long-term outcomes for steroid-resistant acute GVHD. Blood, 131, 2846–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M & Jensen ON (2003) Proteomic analysis of post-translational modifications. Nature Biotechnology, 21, 255–261. [DOI] [PubMed] [Google Scholar]
- Martin PJ, Fan W, Storer BE, Levine DM, Zhao LP, Warren EH, Flowers ME, Lee SJ, Carpenter PA, Boeckh M, Hingorani S, Yan L, Hu Q, Preus L, Liu S, Spellman S, Zhu X, Pasquini M, McCarthy P, Stram D, Sheng X, Pooler L, Haiman CA, Sucheston-Campbell L, Hahn T & Hansen JA (2016) Replication of associations between genetic polymorphisms and chronic graft-versus-host disease. Blood, 128, 2450–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin PJ, Levine DM, Storer BE, Warren EH, Zheng X, Nelson SC, Smith AG, Mortensen BK & Hansen JA (2017) Genome-wide minor histocompatibility matching as related to the risk of graft-versus-host disease. Blood, 129, 791–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavers M, Maas-Bauer K & Negrin RS (2017) Invariant natural killer T cells as suppressors of graft-versus-host disease in allogeneic hematopoietic stem cell transplantation. Frontiers in Immunology, 8, 900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald GB, Tabellini L, Storer BE, Lawler RL, Martin PJ & Hansen JA (2015) Plasma biomarkers of acute GVHD and nonrelapse mortality: predictive value of measurements before GVHD onset and treatment. Blood, 126, 113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald GB, Tabellini L, Storer BE, Martin PJ, Lawler RL, Rosinski SL, Schoch HG & Hansen JA (2017) Predictive value of clinical findings and plasma biomarkers after fourteen days of prednisone treatment for acute graft-versus-host disease. Biology of Blood and Marrow Transplantation, 23, 1257–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklos D, Cutler CS, Arora M, Waller EK, Jagasia M, Pusic I, Flowers ME, Logan AC, Nakamura R, Blazar BR, Li Y, Chang S, Lal I, Dubovsky J, James DF, Styles L & Jaglowski S (2017) Ibrutinib for chronic graft-versus-host disease after failure of prior therapy. Blood, 130, 2243–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat A & Socie G (2004) Intestinal graft-vs.-host disease. In: Graft-vs.-Host Disease (eds. by Ferrara JLM, Cooke KR & Deeg HJ), pp. 279–327. Marcel Dekker, New York. [Google Scholar]
- Nakaya HI, Hagan T, Duraisingham SS, Lee EK, Kwissa M, Rouphael N, Frasca D, Gersten M, Mehta AK, Gaujoux R, Li GM, Gupta S, Ahmed R, Mulligan MJ, Shen-Orr S, Blomberg BB, Subramaniam S & Pulendran B (2015) Systems analysis of immunity to influenza vaccination across multiple years and in diverse populations reveals shared molecular signatures. Immunity, 43, 1186–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RP Jr, Khawaja MR, Perkins SM, Elmore L, Mumaw CL, Orschell C & Paczesny S (2014) Prognostic biomarkers for acute graft-versus-host disease risk after cyclophosphamide-fludarabine nonmyeloablative allotransplantation. Biology of Blood and Marrow Transplantation, 20, 1861–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paczesny S (2015) Graft-versus-host disease in children after hematopoietic cell transplantation: potential clinical utility of biomarkers. International Journal of Hematologic Oncology, 4, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paczesny S (2017) Acute graft-versus-host disease prognosis: are biomarkers ready for preemptive clinical trials? Clinical Chemistry, 63, 1561–1563. [DOI] [PubMed] [Google Scholar]
- Paczesny S & Abu Zaid M (2016) CXCL10: most consistent cGVHD biomarker? Blood, 127, 2950–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paczesny S, Krijanovski OI, Braun TM, Choi SW, Clouthier SG, Kuick R, Misek DE, Cooke KR, Kitko CL, Weyand A, Bickley D, Jones D, Whitfield J, Reddy P, Levine JE, Hanash SM & Ferrara JL (2009) A biomarker panel for acute graft-versus-host disease. Blood, 113, 273–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paczesny S, Braun TM, Levine JE, Hogan J, Crawford J, Coffing B, Olsen S, Choi SW, Wang H, Faca V, Pitteri S, Zhang Q, Chin A, Kitko C, Mineishi S, Yanik G, Peres E, Hanauer D, Wang Y, Reddy P, Hanash S & Ferrara JL (2010) Elafin is a biomarker of graft-versus-host disease of the skin. Science Translational Medicine, 2, 13ra12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paczesny S, Hakim FT, Pidala J, Cooke K, Lathrop J, Griffith LM, Hansen J, Jagasia M, Miklos D, Pavletic S, Parkman R, Russek-Cohen E, Flowers ME, Lee S, Martin P, Vogelsang G, Walton M & Schultz KR (2015a) National Institutes of Health consensus development project on criteria for clinical trials in chronic graft-versus-host disease: III. the 2014 biomarker working group report. Biology of Blood and Marrow Transplantation, 21, 780–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersdorf EW (2017) Role of major histocompatibility complex variation in graft-versus-host disease after hematopoietic cell transplantation. F1000Research, 6, 617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidala J, Sigdel TK, Wang A, Hsieh S, Inamoto Y, Martin PJ, Flowers ME, Hansen JA, Lee SJ & Sarwal MM (2017) A combined biomarker and clinical panel for chronic graft versus host disease diagnosis. The Journal of Pathology: Clinical Research, 3, 3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidala J, Hamadani M, Dawson P, Martens M, Alousi AM, Jagasia M, Efebera YA, Chhabra S, Pusic I, Holtan SG, Ferrara JL, Levine JE, Mielcarek M, Anasetti C, Antin JH, Bolanos-Meade J, Howard A, Logan BR, Leifer E, Pritchard TS, Horowitz MM & MacMillan ML (2020) Randomized multicenter trial of sirolimus vs. prednisone as initial therapy for standard risk acute GVHD: BMT CTN 1501. Blood, 135, 97–107. 10.1182/blood.2019003125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce DM, Hilden P, Mumaw C, Devlin SM, Lubin M, Giralt S, Goldberg JD, Hanash A, Hsu K, Jenq R, Perales MA, Sauter C, van den Brink MR, Young JW, Brentjens R, Kernan NA, Prockop SE, O’Reilly RJ, Scaradavou A, Paczesny S & Barker JN (2015) High day 28 ST2 levels predict for acute graft-versus-host disease and transplant-related mortality after cord blood transplantation. Blood, 125, 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadan A, Griesenauer B, Adom D, Kapur R, Hanenberg H, Liu C, Kaplan MH & Paczesny S (2017) Specifically differentiated T cell subset promotes tumor immunity over fatal immunity. Journal of Experimental Medicine, 214, 3577–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadan AM, Daguindau E, Rech JC, Chinnaswamy K, Zhang J, Hura GL, Griesenauer B, Bolten Z, Robida A, Larsen M, Stuckey JA, Yang CY & Paczesny S (2018) From proteomics to discovery of first-in-class ST2 inhibitors active in vivo. JCI Insight, 3, 99208. 10.1172/jci.insight.99208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Otero P, Porcher R, Peffault de Latour R, Contreras M, Bouhnik Y, Xhaard A, Andreoli A, Ribaud P, Kapel N, Janin A, Socie G & Robin M (2012) Fecal calprotectin and alpha-1 antitrypsin predict severity and response to corticosteroids in gastrointestinal graft-versus-host disease. Blood, 119, 5909–5917. [DOI] [PubMed] [Google Scholar]
- Rotz SJ, Dandoy CE & Davies SM (2017) ST2 and endothelial injury as a link between GVHD and microangiopathy. New England Journal of Medicine, 376, 1189–1190. [DOI] [PubMed] [Google Scholar]
- Rowan CM, Teagarden AM, Cater DT, Moser EAS, Bakoyannis G & Paczesny S (2019) Early high plasma ST2, the decoy IL-33 receptor, in children undergoing hematopoietic cell transplantation is associated with the development of post-transplant diabetes mellitus. Haematologica, haematol.2019.222992. [Epub ahead of print]. 10.3324/haematol.2019.222992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarantopoulos S & Ritz J (2015) Aberrant B-cell homeostasis in chronic GVHD. Blood, 125, 1703–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarantopoulos S, Stevenson KE, Kim HT, Bhuiya NS, Cutler CS, Soiffer RJ, Antin JH & Ritz J (2007) High levels of B-cell activating factor in patients with active chronic graft-versus-host disease. Clinical Cancer Research, 13, 6107–6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarantopoulos S, Stevenson KE, Kim HT, Cutler CS, Bhuiya NS, Schowalter M, Ho VT, Alyea EP, Koreth J, Blazar BR, Soiffer RJ, Antin JH & Ritz J (2009) Altered B-cell homeostasis and excess BAFF in human chronic graft-versus-host disease. Blood, 113, 3865–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Yu J, Jenkins IC, Leisenring WM, Steven-Ayers T, Kuypers JM, Huang ML, Jerome KR, Boeckh M & Paczesny S (2018) Diagnostic and prognostic plasma biomarkers for idiopathic pneumonia syndrome after hematopoietic cell transplantation. Biology of Blood and Marrow Transplantation, 24, 678–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stikvoort A, Chen Y, Radestad E, Torlen J, Lakshmikanth T, Bjorklund A, Mikes J, Achour A, Gertow J, Sundberg B, Remberger M, Sundin M, Mattsson J, Brodin P & Uhlin M (2017) combining flow and mass cytometry in the search for biomarkers in chronic graft-versus-host disease. Frontiers in Immunology, 8, 717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucheston-Campbell L, Preus L, Spellman S, Pasquini MC, McCarthy PL, Onel K, Zhu X, Haiman C, Stram DO, Pooler L, Sheng X, Zhu Q, Yan L, Liu Q, Hu Q, Liu S, Clay A, Battaglia S & Hahn T (2016) Functional single nucleotide polymorphisms (SNPs) in the major histocompatibility complex (MHC) class II region are associated with overall survival (OS) after HLA matched unrelated donor BMT: results from the discovery-BMT study. Biology of Blood and Marrow Transplantation, 22, S72–S73. [Google Scholar]
- Tang H, Hahn T, Karaesmen E, Rizvi AA, Wang J, Paczesny S, Wang T, Preus L, Zhu Q, Wang Y, Haiman CA, Stram D, Pooler L, Sheng X, Van Den Berg D, Brock G, Webb A, Pasquini MC, McCarthy PL, Spellman SR & Sucheston-Campbell LE (2019) Validation of genetic associations with acute GVHD and nonrelapse mortality in DISCOV-eRY-BMT. Blood Advances, 3, 2337–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taur Y, Jenq RR, Perales MA, Littmann ER, Morjaria S, Ling L, No D, Gobourne A, Viale A, Dahi PB, Ponce DM, Barker JN, Giralt S, van den Brink M & Pamer EG (2014) The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood, 124, 1174–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkachev V, Furlan SN, Watkins B, Hunt DJ, Zheng HB, Panoskaltsis-Mortari A, Betz K, Brown M, Schell JB, Zeleski K, Yu A, Kirby I, Cooley S, Miller JS, Blazar BR, Casson D, Bland-Ward P & Kean LS (2017) Successfully achieving immune balance after transplant: Combined OX40L and mTOR blockade controls effector T cell activation while preserving Treg reconstitution. Science Translational Medicine, 9, eaan3085. 10.1126/scitranslmed.aan3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Lugt MT, Braun TM, Hanash S, Ritz J, Ho VT, Antin JH, Zhang Q, Wong CH, Wang H, Chin A, Gomez A, Harris AC, Levine JE, Choi SW, Couriel D, Reddy P, Ferrara JL & Paczesny S (2013) ST2 as a marker for risk of therapy-resistant graft-versus-host disease and death. New England Journal of Medicine, 369, 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins BK, Tkachev V, Furlan SN, Hunt DJ, Betz K, Yu A, Brown M, Poirier N, Zheng HB, Taraseviciute A, Colonna L, Mary C, Blancho G, Soulillou JP, Panoskaltsis-Mortari A, Sharma P, Garcia A, Strobert E, Hamby K, Garrett A, Deane T, Blazar BR, Vanhove B & Kean LS (2018) CD28 blockade controls T cell activation to prevent graft-versus-host disease in primates. J Clin Invest, 128, 3991–4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber D, Oefner PJ, Hiergeist A, Koestler J, Gessner A, Weber M, Hahn J, Wolff D, Stammler F, Spang R, Herr W, Dettmer K & Holler E (2015) Low urinary indoxyl sulfate levels early after transplantation reflect a disrupted microbiome and are associated with poor outcome. Blood, 126, 1723–1728. [DOI] [PubMed] [Google Scholar]
- Weissinger EM, Schiffer E, Hertenstein B, Ferrara JL, Holler E, Stadler M, Kolb HJ, Zander A, Zurbig P, Kellmann M & Ganser A (2007) Proteomic patterns predict acute graft-versus-host disease after allogeneic hematopoietic stem cell transplantation. Blood, 109, 5511–5519. [DOI] [PubMed] [Google Scholar]
- Weissinger EM, Jochen M, Schleuning M, Schmid C, Messinger D, Beutel G, Wagner EM, Schetelig J, Baurmann H, Rank A, Stolzl F, Schäfer-Eckhard K, Westphal K, Bethge W, Bunjes D, Heidenreich D, Klein S, Wolff D, Holler E, Kreipe H, Jonigk D, Türüchanow I, Rothmann C, Raad J, Durban A, Hamwi I, Ehrlich S, Schweier P, Krauter J, Stadler M & Ganser A (2017) The Multicentre, Double-Blinded, Placebo-Controlled Clinical-Trial (Pre-GvHD) for Prediction and Pre-Emptive Treatment of Acute GvHD. American Society of Hematology, Blood, Atlanta, GA. [Google Scholar]
- Weissinger EM, Human C, Metzger J, Ham-bach L, Wolf D, Greinix HT, Dickinson AM, Mullen W, Jonigk D, Kuzmina Z, Kreipe H, Schweier P, Bohm O, Turuchanow I, Ihlenburg-Schwarz D, Raad J, Durban A, Schiemann M, Konecke C, Diedrich H, Holler E, Beutel G, Krauter J, Ganser A & Stadler M (2017) The proteome pattern cGvHD_MS14 allows early and accurate prediction of chronic GvHD after allogeneic stem cell transplantation. Leukemia, 31, 654–662. [DOI] [PubMed] [Google Scholar]
- Wingard JR, Majhail NS, Brazauskas R, Wang Z, Sobocinski KA, Jacobsohn D, Sorror ML, Horowitz MM, Bolwell B, Rizzo JD & Socie G (2011) Long-term survival and late deaths after allogeneic hematopoietic cell transplantation. Journal of Clinical Oncology, 29, 2230–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff D, Greinix H, Lee SJ, Gooley T, Paczesny S, Pavletic S, Hakim F, Malard F, Jagasia M, Lawitschka A, Hansen JA, Pulanic D, Holler E, Dickinson A, Weissinger E, Edinger M, Sarantopoulos S & Schultz KR (2018) Biomarkers in chronic graft-versus-host disease: quo vadis? Bone Marrow Transplantation, 53, 832–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Storer BE, Kushekhar K, Abu Zaid M, Zhang Q, Gafken PR, Ogata Y, Martin PJ, Flowers ME, Hansen JA, Arora M, Cutler C, Jagasia M, Pidala J, Hamilton BK, Chen GL, Pusic I, Lee SJ & Paczesny S (2016) Biomarker panel for chronic graft-versus-host disease. Journal of Clinical Oncology, 34, 2583–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiser R & Blazar BR (2017) Acute graft-versus-host disease – biologic process, prevention, and therapy. New England Journal of Medicine, 377, 2167–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ramadan AM, Griesenauer B, Li W, Turner MJ, Liu C, Kapur R, Hanenberg H, Blazar BR, Tawara I & Paczesny S (2015) ST2 blockade reduces sST2-producing T cells while maintaining protective mST2-expressing T cells during graft-versus-host disease. Science Translational Medicine, 7, 308ra160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Kim YH, Jeong S, Greenson JK, Chaudhry MS, Hoepting M, Anderson ER, van den Brink MR, Peled JU, Gomes AL, Slingerland AE, Donovan MJ, Harris AC, Levine JE, Ozbek U, Hooper LV, Stappen-beck TS, Ver Heul A, Liu TC, Reddy P & Ferrara JL (2018) Survival signal REG3alpha prevents crypt apoptosis to control acute gastrointestinal graft-versus-host disease. Journal of Clinical Investigations, 128, 4970–4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ & Bielas JH (2017) Massively parallel digital transcriptional profiling of single cells. Nature Communications, 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn E, Kim HT, Lee SJ, Floyd BH, Litsa D, Arumugarajah S, Bellucci R, Alyea EP, Antin JH, Soiffer RJ & Ritz J (2005) Reduced frequency of FOXP3+ CD4+CD25+ regulatory T cells in patients with chronic graft-versus-host disease. Blood, 106, 2903–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]


