Abstract
Clonally expanded blood cells with somatic mutations (clonal hematopoiesis, CH) are commonly acquired with age and increase risk of blood cancer1–9. The blood clones identified to date contain diverse large-scale mosaic chromosomal alterations (mCAs: deletions, duplications, and copy-neutral loss of heterozygosity [CN-LOH]) on all chromosomes1,2,5,6,9, but the sources of selective advantage that drive expansion of most clones remain unknown. To identify genes, mutations and biological processes that give selective advantage to mutant clones, we analyzed genotyping data from the blood-derived DNA of 482,789 UK Biobank participants10, identifying 19,632 autosomal mCAs which we analyzed for relationships to inherited genetic variation. Fifty-two inherited, rare, large-effect coding or splice variants in seven genes associated with greatly increased (odds ratios 11 to 758) vulnerability to CH with specific acquired CN-LOH mutations. Acquired mutations systematically replaced the inherited risk alleles (at MPL) or duplicated them to the homologous chromosome (at FH, NBN, MRE11, ATM, SH2B3, and TM2D3). Three of the seven genes (MRE11, NBN, and ATM) encode components of the MRN-ATM pathway, which limits cell division after DNA damage and telomere attrition11−13; another two (MPL, SH2B3) encode proteins that regulate stem cell self-renewal14–16. In addition to these monogenic inherited forms of CH, we found a common and surprisingly polygenic form: CN-LOH mutations across the genome tended to cause chromosomal segments with alleles that promote hematopoietic cell expansions to replace their homologous (allelic) counterparts, increasing polygenic drive for blood-cell proliferation traits. Readily-acquired mutations that replace chromosomal segments with their homologous counterparts appear to interact with pervasive inherited variation to create a challenge for lifelong cytopoiesis.
Mosaic chromosomal alterations in UK Biobank
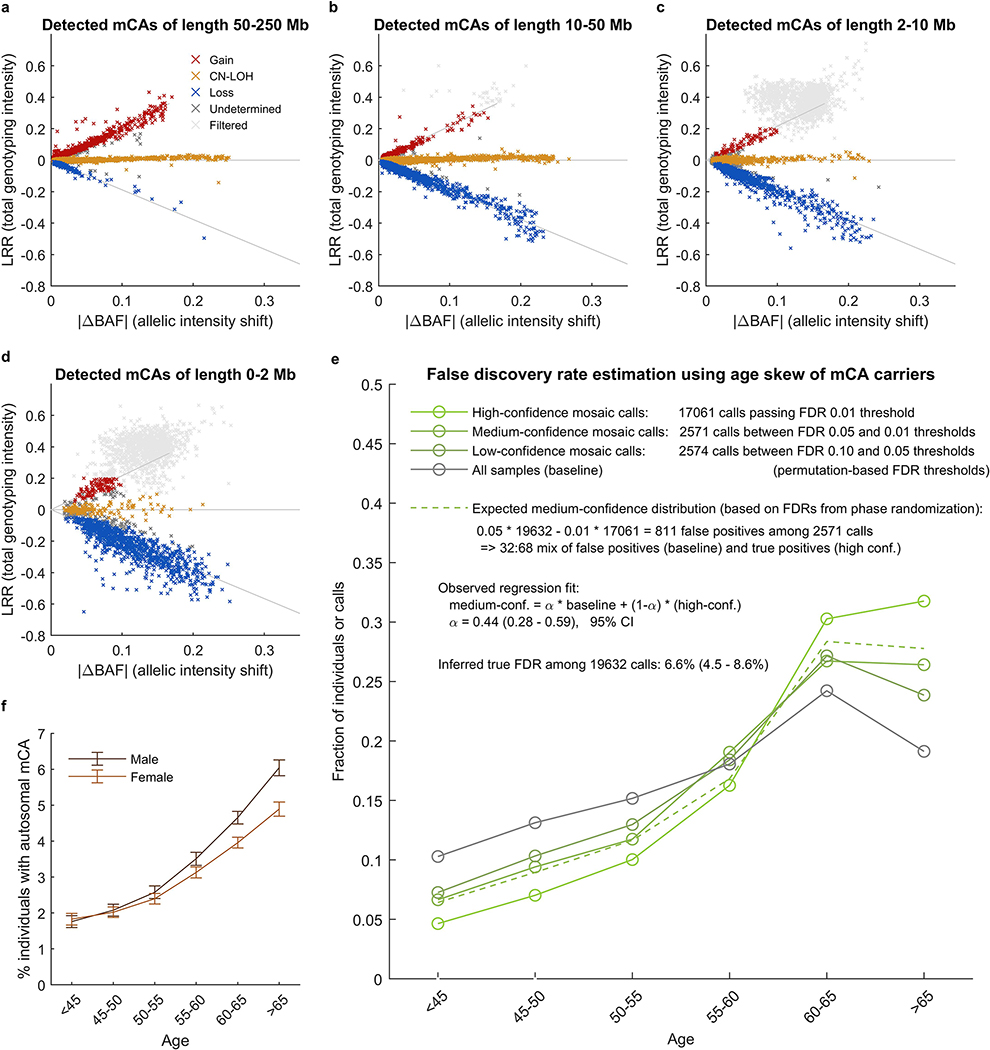
We identified 17,111 CH cases involving 19,632 autosomal mCAs (Extended Data Figures 1–2 and Supplementary Tables 1–3) by analyzing SNP-array intensity data from 482,789 UK Biobank (UKB) participants 40–70 years of age10. To identify these cases, we applied a method we recently described and applied to the UKB interim release (∼31% of the current cohort)9; our approach finds imbalances in the abundances of homologous chromosomal segments by combining allele-specific intensity data with long-range chromosomal phase information17,18 (Methods and Supplementary Note 1). We classified 73% of the detected mCAs as either loss (3,718 events), gain (2,389 events), or CN-LOH (8,185 events), the replacement of one chromosomal segment by its homologous (allelic) counterpart (Supplementary Table 1). (Another 5,340 mCAs could not be confidently classified, as power to detect imbalances exceeded power to distinguish copy-neutral from copy-number-altering mCAs9; Extended Data Fig. 2a–d.) Of the 19,632 detected mCAs, 12,683 were present at cell fractions from 0.7% to 5%, and 6,949 were present at cell fractions >5%. Consistent with previous work1,2,5,6,9, mCAs on different chromosomes exhibited different recurrence rates and size distributions (Extended Data Fig. 1a) and a range of tendencies to be more common in one sex (usually males, though with clear exceptions) and the elderly (Extended Data Fig. 1b and Supplementary Table 4). Clones also tended to be found in individuals with anomalous counts for one or more blood-cell types (Extended Data Fig. 1c and Supplementary Table 5).
Monogenic inherited forms of clonal hematopoiesis
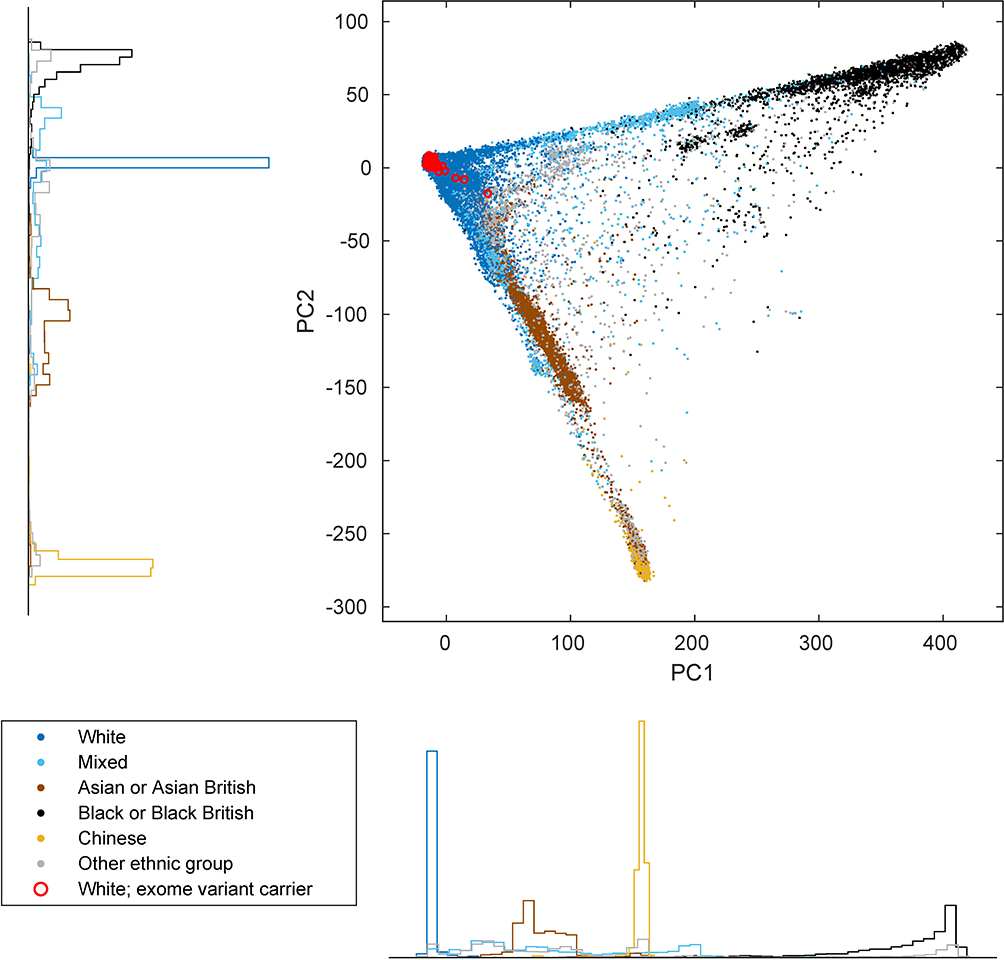
We next sought to identify specific genes and variants that might propel clonal selection. We recently identified three loci (MPL, ATM, and TM2D3–TARSL2) at which inherited rare variants increase the risk of developing clones with acquired CN-LOH mutations that affect the rare inherited risk allele in a predictable way9. To detect loci targeted by CN-LOH mutations in this manner, and to identify likely-causal inherited variants at these loci, we searched the genome for associations between inherited variants and CN-LOH mutations acquired in cis. (To avoid potential confounding from population stratification, we restricted these analyses to 455,009 individuals who reported European ancestry; Extended Data Fig. 3 and Supplementary Note 2.)
Inherited rare variants at seven loci (MPL, ATM, TM2D3, FH, NBN, MRE11, and SH2B3) associated at genome-wide significance with the development of blood clones in which an acquired CN-LOH mutation had affected the inherited risk allele in a consistent way (Fig. 1, Extended Data Table 1, Extended Data Fig. 4, and Supplementary Table 6). At six loci (all loci other than MPL), the inherited rare alleles were overwhelmingly made homozygous by somatic CN-LOH mutations (149 of 153 cases; binomial P=3.9×10–39). Associations at all seven loci appeared to be driven by rare coding variants with large effect sizes (ORs 11–555; 95% CIs 5.8–724): the lead associated variants at six of the seven loci were coding mutations, and the lead variant at the remaining locus, MRE11 (rs762019591; Fisher’s exact P=3.0×10–11), tagged a nonsense SNP in MRE11 (rs587781384; Extended Data Table 1).
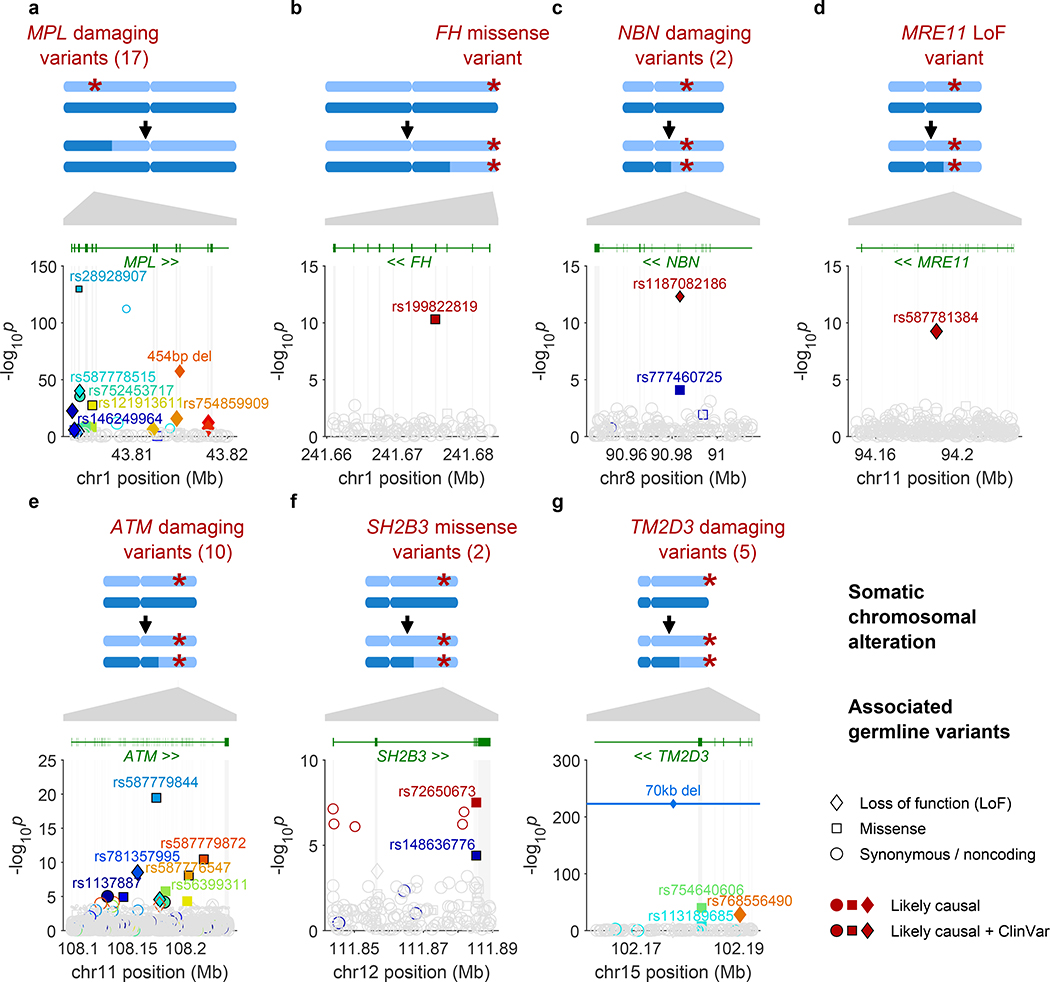
Figure 1: Fine-mapped inherited sequence alleles associated with the acquisition/selection of CN-LOH mutations in cis.
(a) MPL, (b) FH, (c) NBN, (d) MRE11, (e) ATM, (f) SH2B3, (g) TM2D3. At each locus, the CN-LOH mutations acquired by expanded clones tend to have deleted (a) or duplicated (b–g) the inherited alleles in a predictable manner as shown. Each panel is organized in the following way: top, genomic modifications observed in clones; bottom, association P-values (two-sided Fisher’s exact test on n≥378,307 individuals; Methods) vs. chromosomal position. All variants with filled symbols are likely causal coding or splice variants (Extended Data Table 1); black marker edges indicate evidence of pathogenicity in ClinVar21. Distinct colors are used to indicate the statistical independence of variants; any variants in linkage disequilibrium with likely causal variants (R2>0.2 in cases) are indicated with open symbols with a border color matching that of the likely causal variant. Symbol shapes indicate the effects of the indicated variant on encoded protein (LoF, missense, etc.); symbol sizes scale inversely with minor allele frequency.
The functions of five of the seven implicated genes converged upon two likely mechanisms of clonal advantage. Three implicated genes (MRE11, NBN, ATM) encode proteins that act together to limit cell growth after DNA damage and telomere attrition13. Specifically, MRE11 and NBN encode two of the three proteins in the MRN complex, which recognizes double-strand breaks and activates the checkpoint kinase encoded by ATM11,12. Thus, strong-effect inherited variants (including protein-truncating variants) at MRE11, NBN, and ATM—made homozygous by CN-LOH (Fig. 1c,d,e)—appear to disrupt a key pathway that limits proliferation in cells that have experienced DNA damage or telomere shortening.
Two other implicated genes encode proteins that regulate self-renewal of hematopoietic stem cells: MPL, which encodes the myeloproliferative leukemia protein, which positively regulates stem cell self-renewal14,15 in addition to its roles in thrombocytopoiesis; and SH2B3, which encodes a signaling protein (LNK) that negatively regulates hematopoietic signaling through MPL16. Clonally selected CN-LOH mutations appeared to have opposite effects upon rare inherited (putative function-reducing) variants in MPL and SH2B3: the acquired 1p CN-LOH mutations eliminated rare inherited variants (including protein-truncating variants) in MPL (Fig. 1a), while the acquired 12q CN-LOH mutations duplicated SH2B3 variants to the other homolog (Fig. 1f). The primary SH2B3 risk allele (rs72650673:A) has been previously found to increase platelet counts in carriers19, suggesting roles at multiple levels of hematopoietic differentiation.
Inherited mutations in FH, which encodes the fumarate hydratase protein (which functions in the Krebs cycle), are an established cause of benign and malignant neoplasms in multiple tissues20. The molecular function(s) of TM2D3 are unknown.
To identify additional variants at all seven loci for which CN-LOH mutations led to subsequent clonal proliferation, we performed fine-mapping analyses comprehensively examining coding and splice variants in these genes by integrating information from SNP arrays, imputation, and exome sequencing (Methods, Extended Data Fig. 5, and Supplementary Notes 3 and 4). Among 616 missense, predicted loss-of-function (LoF), or likely pathogenic21 variants tested (Methods), 52 variants associated independently with CN-LOH mosaicism in cis (at FDR<0.05 significance per locus; ORs 11–758, 95% CIs 4–2,618), including multiple variants in MPL (28 variants), ATM (13), TM2D3 (5), NBN (2), and SH2B3 (2); 38 of the 52 individual variants reached Bonferroni significance (Fisher’s exact P<8.1×10–5 for 616 variants tested; Fig. 1, Extended Data Table 1, and Supplementary Tables 7 and 8). All 52 variants were rare (population allele frequency <0.2%). Intriguingly, 23 of the 52 variants had been reported as clinically significant21 in hereditary blood disorders (eight variants in MPL and one in SH2B3) or cancer (11 variants in ATM and one each in MRE11, NBN, and FH). All 28 MPL variants were removed from the genomes of expanded clones by CN-LOH mutations (244 of 244 cases, binomial P=7.1×10–74), consistent with a model in which the inherited alleles (with reduced MPL function) have a hypo-proliferative effect that is rescued by CN-LOH9. The 24 inherited variants at the other six loci were systematically made homozygous by CN-LOH (233 of 239 cases, binomial P=5.6×10–61), consistent with pro-proliferative effects of reduced ATM, MRE11, NBN, SH2B3, TM2D3, and FH function (Extended Data Table 1, Fig. 1 and Supplementary Table 7). Sharing of long haplotypes among individuals with 1p CN-LOH mutations spanning MPL and among individuals with 11q CN-LOH mutations spanning ATM indicated that while the risk variants we identified (Extended Data Table 1 and Supplementary Table 7) are likely to be the primary drivers of heritable CH risk at these loci, the full allelic series likely include many more risk variants (Extended Data Figures 6–7).
To detect additional potential risk variants and to estimate the fraction of CN-LOH clones attributable to inherited protein-altering variants (including still-rarer variants) at each locus, we examined exome sequence data available for 49,960 of the UK Biobank participants22. Among 271 exome-sequenced individuals of European ancestry with unexplained mosaic CN-LOH events spanning the seven loci (i.e., not carrying any of the 52 variants already identified), 22 individuals carried 21 distinct ultra-rare coding or splice variants that altered the encoded proteins (vs. 1.28 individuals expected by chance, P=2.8×10–20; “ultra-rare” refers to population allele frequency less than 0.0001; Methods and Supplementary Tables 9–11). Collectively, MPL variants identified by these association and burden analyses were present in 39 of 71 exome-sequenced individuals with 1p CN-LOH events spanning MPL (vs. 0.5 expected), suggesting that ∼54% of acquired 1p CN-LOH events are driven by inherited coding or splice variants at MPL (Supplementary Table 11). Similarly, inherited variants at ATM, NBN, SH2B3, and TM2D3 appeared to drive ∼17–33% of CN-LOH events spanning these loci (Supplementary Table 11). Altogether we estimate that about 5% of clones with CN-LOH arose from one of these seven monogenic inherited vulnerabilities.
Common inherited variants at five loci conferred more modest mCA risk (ORs of 1.07–1.24). Common variants at TCL1A and DLK1 on 14q associated with acquired 14q CN-LOH mutations (Supplementary Table 12 and Supplementary Note 5), whereas common variants at TERC, SP140, and the previously-implicated TERT locus7 broadly increased the risk of CH involving any autosomal mCA (Supplementary Table 13 and Supplementary Note 6). Notably, TERC and TERT both encode proteins with key roles in the maintenance and elongation of telomeres (Supplementary Table 14).
Some CN-LOH events provided “second hits” to acquired point mutations. At the frequently-mutated DNMT3A, TET2, and JAK2 loci3,4, ∼24–60% of CN-LOH mutations appeared to provide second hits to somatic point mutations detectable from exome sequencing reads (Extended Data Fig. 8 and Supplementary Table 11; additional CN-LOH events spanning these loci might be explained by point mutations present at lower cell fractions we could not detect among ∼10–40 sequencing reads per haplotype; Methods). Among 33 exome-sequenced individuals with 9p CN-LOH events, 20 individuals had at least one read suggesting JAK2 V617F mutation; conversely, 18 of 46 individuals with JAK2 V617F calls had a detectable mCA on 9p (15 CN-LOH events and three chromosome 9 duplications). Together, the putative “second-hit” clones at these loci accounted for about 0.3% of all detected CN-LOH clones.
Clonal CN-LOH mutations increase polygenic drive
The great majority of the 17,111 hematopoietic clones we observed in UKB still had unknown causes; most clones had CN-LOH mutations, which were numerous on every chromosome arm (Extended Fig. 1a). This posed the intriguing question of what genetic effect propels detected clones in a manner that is so distributed across the genome. Recent work in human and agricultural genetics has revealed that many phenotypes are shaped by polygenic effects from alleles of modest effect at hundreds of loci spread across all chromosomes23–25.
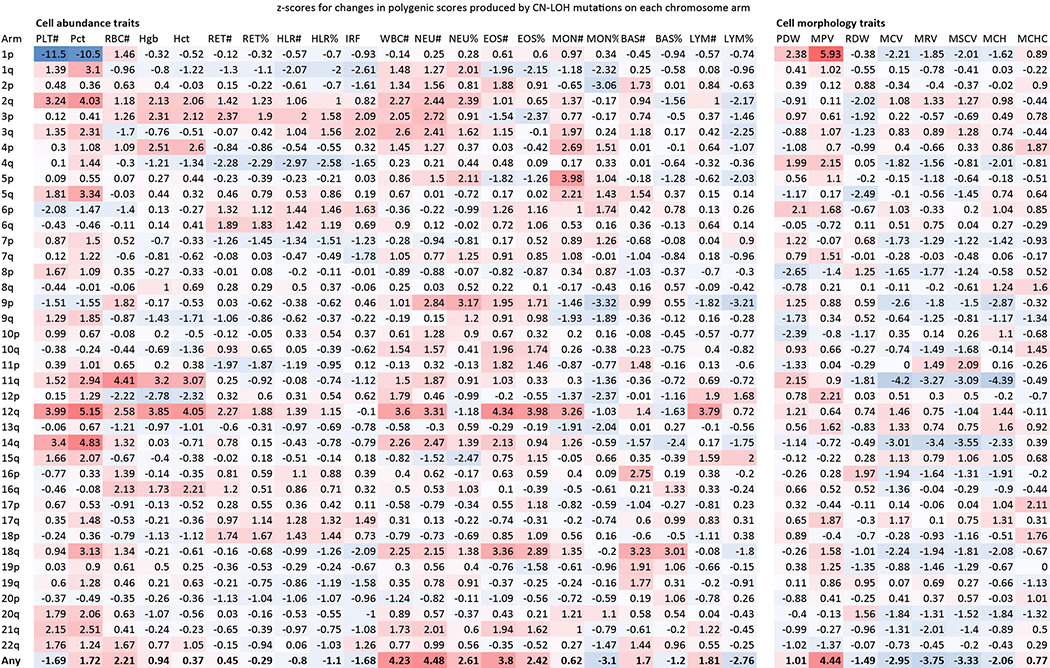
We hypothesized that inherited haplotypes along a chromosome arm can themselves be instruments for clonal selection (Fig. 2a). To evaluate this possibility, we tested whether the haplotypes duplicated and deleted by likely-CN-LOH mutations (Methods) tended to differ systematically in polygenic drive for blood-cell abundance phenotypes, as estimated from combinations of many inherited alleles and these alleles’ relationships to blood-cell abundances in the general population. We evaluated this by building polygenic statistical models26 for blood-cell abundance traits (using data on blood-cell counts from UKB participants) and for clonal Y chromosome loss, a frequent marker of hematopoietic clones27. Based on these models, we estimated “hematopoietic polygenic risk scores” (HPRS) for the combinations of common alleles along the haplotypes gained and lost by CN-LOH mutations in expanded clones (Methods).
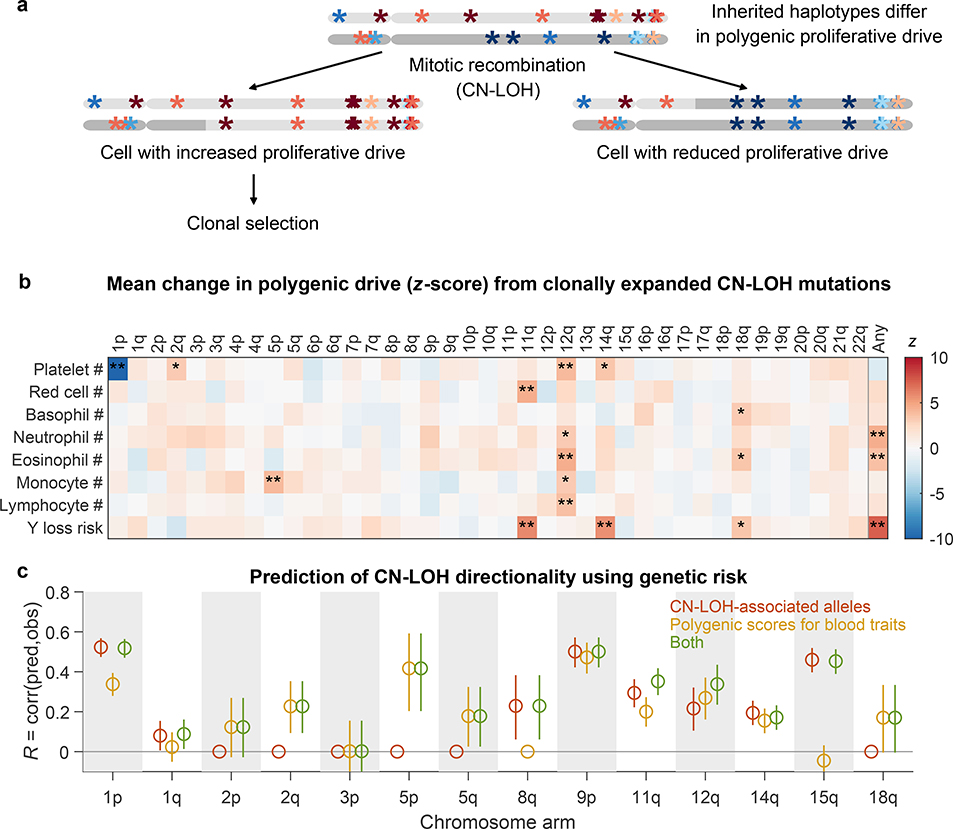
Figure 2: Polygenic and monogenic influences on clonal proliferation of cells with CN-LOH mutations.
(a) Two cellular outcomes of a CN-LOH mutation (mitotic recombination) involving homologous chromosome arms that bear inherited alleles with differing proliferative potentials. In one cell, the CN-LOH mutation has duplicated the chromosomal arm that has alleles that more strongly promote proliferation; proliferative polygenic drive increases, potentially resulting in clonal selection of the mutant cell. By contrast, the cell with the complementary CN-LOH mutation may have reduced tendency to proliferate. (b) CN-LOH mutations in expanded clones broadly increase polygenic risk scores for increased blood-cell counts and risk of mosaic Y chromosome loss (a marker for clonal hematopoiesis27). The heatmap displays changes in polygenic scores for each trait, averaged across all ascertained (expanded) CN-LOH mutations observed on each chromosome arm (color bar, z-score; ∗, significant at FDR<0.05; ∗∗, Bonferroni-corrected P<0.05). (c) Prediction of the direction of CN-LOH mutations (in expanded clones) from inherited alleles on the affected chromosome arms. Prediction accuracy (the correlation between predicted and observed CN-LOH direction) is plotted for predictions made using: only CN-LOH-associated alleles (Extended Data Table 1 and Supplementary Table 7) (red); polygenic score differentials on affected chromosomal segments (orange); or both sources of information (green). Error bars, 95% CIs. Results are plotted for 14 chromosome arms for which at least one predictor was available. Numeric data and sample sizes are provided in Supplementary Tables 15 and 18. Analyses of polygenic scores for control traits such as height and BMI are provided in Supplementary Table 16.
CN-LOH mutations in expanded clones tended to have caused chromosomal segments with higher HPRS to replace homologous (allelic) counterparts with lower HPRS (Fig. 2b). Averaging across all autosomal CN-LOH events, the allelic substitutions produced by CN-LOH mutations significantly increased polygenic scores for clonality with Y chromosome loss (P=1.2×10–13; P=4.3×10–8 and P=5.2×10–7 for CN-LOH in men and women separately) and also tended to increase polygenic scores for the individual blood-cell abundance traits (most significant: neutrophil counts, P=7.5×10–6; eosinophil counts, P=1.4×10–4). This effect was observed throughout the genome: 14 distinct combinations of chromosome arms and cell-abundance traits exhibited significant upward shifts in HPRS (at an FDR of 0.05), and 209 of all 312 combinations exhibited a positive mean increase (P=2.0×10–9, sign test; Fig. 2b and Supplementary Table 15). These effects were specific to polygenic scores for blood cell abundance traits: CN-LOH mutations did not tend to affect polygenic scores for control traits such as height and BMI (Supplementary Table 16), and results were mixed for blood cell morphology traits (Extended Data Fig. 9). CN-LOH mutations did appear to act on risk alleles for myeloproliferative neoplasms (Supplementary Table 17 and Supplementary Note 7).
These results raised the intriguing possibility that the direction of mosaic CN-LOH mutations—i.e., which haplotype has been made homozygous in a clone that rises to detectable frequency—can be predicted from inherited variation. To test this idea, we performed cross-validated prediction using logistic regression on either (i) the CN-LOH-associated alleles we had found (Extended Data Table 1); (ii) polygenic score differentials on chromosomal segments affected by CN-LOH; or (iii) both CN-LOH-associated alleles and polygenic score differentials (Methods). Polygenic scores and specific inherited CN-LOH-associated alleles each helped predict CN-LOH directions; combining both sources of information yielded the most predictive information, reaching significance (FDR<0.05) for 12 of 14 chromosome arms tested (Fig. 2c and Supplementary Table 18; we tested the 14 arms for which the prediction algorithm nominated at least one predictor for testing in a non-overlapping data set; Methods and Supplementary Table 19). The directions of CN-LOH mutations were correctly predicted for 59% (P=2.3×10–44) of 5,582 CN-LOH events on these 14 arms (range 50-–70%). Stronger inherited imbalances correlated with greater predictability: upon restricting to events involving larger imbalances in polygenic scores (top quintile), prediction accuracy increased to 72% (P=1.1×10–82).
Cancer and cardiovascular risk associated with mCAs
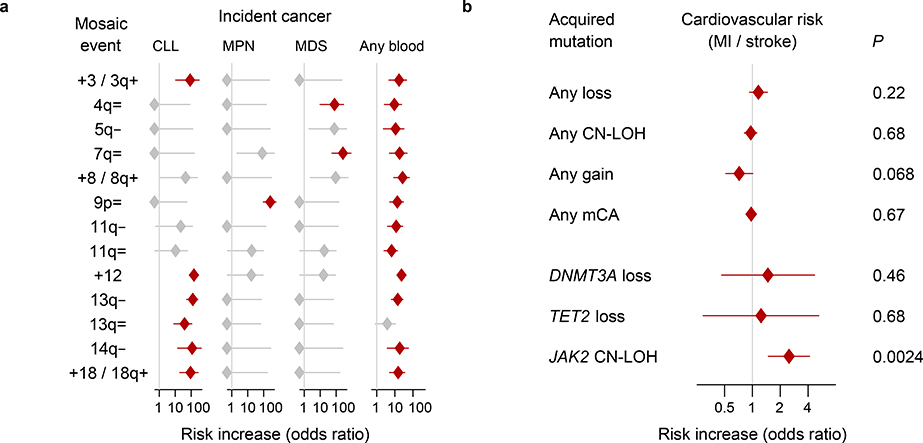
Clonal hematopoiesis increases risk of adverse health outcomes, including blood cancers, cardiovascular disease, and mortality1–4,8,28. The size of the full UK Biobank data set allowed us to further appreciate the extent to which different mCAs associate with distinct health outcomes (Methods). Thirteen specific mCAs significantly associated (FDR<0.05) with subsequent hematological cancer diagnoses during 4–9 years of follow-up. The +12, 13q–, and 14q– events conferred >100-fold higher risk of chronic lymphocytic leukemia (CLL), and JAK2-related 9p CN-LOH events conferred 260-fold (89–631-fold) higher risk of myeloproliferative neoplasms, replicating previous results9; 4q and 7q CN-LOH events conferred >70-fold higher risk of myelodysplastic syndromes (Fig. 3a and Supplementary Table 20). The +12 and 13q LOH events exhitibited shared genetic risk with CLL (Supplementary Table 21 and Supplementary Note 8). The far-more-common CN-LOH events on other chromosome arms also significantly increased blood cancer risk (aggregate hazard ratio=2.84 (2.14–3.78), even after excluding the very-strong-effect 9p/JAK2 events). (We corrected these analyses for age and sex and restricted to individuals with normal blood counts at assessment, no previous cancer diagnoses, and no cancer diagnoses within one year of assessment.) We did not find a significant increase in cardiovascular risk among individuals with most categories of clones—with the notable exception of JAK2-related 9p CN-LOH events (Fig. 3b and Supplementary Table 22)—suggesting that the relationship between clonal hematopoiesis and cardiovascular disease4,28 arises from clones that harbor specific mutations.
Figure 3: Associations of mCAs with incident cancers and cardiovascular disease.
(a) Clones with specific mCAs confer increased risk of incident blood cancers diagnosed >1 year after DNA collection in individuals with normal blood counts at assessment (Cochran-Mantel-Haenszel test adjusting for age and sex; error bars, 95% CIs). Seven of nine associations we previously reported9 (all but 16p= and 20q–) replicate here; “=” is shorthand for CN-LOH. (b) Loss, CN-LOH, and gain events (on any autosome) do not broadly increase risk for incident myocardial infarction or stroke, but CN-LOH events on 9p (containing JAK2) do increase cardiovascular risk28 (two-sided Fisher’s exact test on cases and controls matched for assessment year, age, sex, smoking, hypertension, BMI, and type 2 diabetes status; error bars, 95% CIs). Statistical tests are detailed in Methods. Numeric data and sample sizes are provided in Supplementary Tables 20 and 22.
Discussion
These results illuminate the clonal advantages conferred by CN-LOH, the common substitution of one chromosome arm for its homologous counterpart, which was present in most of the clones ascertained by mCAs (Extended Data Fig. 1a). Although the first-order gene-dosage effects of deletions and duplications are clear1,2,5,6,29, clonal expansions of copy-neutral mutations are more common (Extended Data Fig. 1a) and have been more mysterious: the substitution of one chromosome arm for its inherited homolog does not modify gene dosage, so why would a cell that has undergone such a mutation gain a proliferative advantage? Our results, obtained from many genomic loci, point to a core principle: clonally expanded CN-LOH events routinely replace inherited chromosomal segments with homologous segments that more strongly promote proliferation. Examples of potent CN-LOH events have previously been observed in disease studies at a few loci where CN-LOH events provide second hits to acquired mutations30, disrupt imprinting31, or revert pathogenic mutations in rare monogenic disorders of the skin and blood32,33. We recently observed that CN-LOH mutations can also lead to clonal selection in healthy blood by modifying allelic dosage of inherited rare variants at three loci9. The analyses described here suggest that this proliferative mechanism is in fact at work throughout the genome: we identified six more loci (FH, NBN, MRE11, SH2B3, TCL1A, and DLK1) at which CN-LOH mutations gain advantage from at least 50 inherited alleles (some with sufficiently large effects to produce multiple clonal expansions in the same individual; Supplementary Table 23 and Supplementary Note 9), and we observed a pervasive polygenic effect attributable to combinations of inherited alleles along chromosome arms. The finding here that the direction of 5,582 CN-LOH mutations (across 14 chromosome arms) could be predicted with 59% accuracy—based only on the alleles inherited on each arm—suggests that a substantial fraction of clonal expansions with CN-LOH (at least 59 – 41 = 18%) are influenced by inherited alleles that cause maternal and paternal haplotypes to differ in their tendency to promote proliferation. Furthermore, this underestimates the strength and prevalence of polygenic selective pressure; as polygenic risk scores are informed by larger samples and lower-frequency alleles, their predictive accuracy tends to greatly increase24,25,34.
We were initially surprised that even a modest fraction of an individual’s polygenic risk—arising from a single chromosome arm—could apparently create substantial clonal advantage. We believe that this results from an important aspect of clonal evolution: mutated cells compete with nearly isogenic cells in a common, shared environment. Estimates of the effects of common alleles and polygenic risk—which are usually made in the context of diverse genetic backgrounds and abundant environmental variation—are likely to underestimate the potential of such alleles to become instruments for clonal selection.
Because human populations harbor abundant heterozygosity, and mitotic recombination events occur frequently over an individual’s lifetime9,32, imbalances in the proliferative potential of the homologous chromosome arms inherited from one’s two parents provide a context in which clonal selection is almost inevitable. Managing this dynamic may present challenges for cytopoiesis throughout the lifespan in any genetically diverse species.
Methods
UK Biobank cohort and genotyping
The UK Biobank is a very large prospective study of individuals aged 40–70 years at assessment35. Participants attended assessment centers between 2006 and 2010, where they contributed blood samples for genotyping and blood analysis and answered questionnaires about medical history and environmental exposures. In the years since assessment, health outcome data for these individuals (e.g., diagnoses of cancer and cardiovascular disease) have been accruing via UK national registries and hospital records managed by the NHS.
We analyzed genetic data from the full UK Biobank cohort, which consists of 488,377 individuals genotyped on the Affymetrix UK BiLEVE and UK Biobank Axiom arrays. The BiLEVE and Biobank arrays have >95% overlap and contain a total of 784,256 unique autosomal variants; 49,950 individuals were genotyped on the BiLEVE array36 and the remaining individuals on the Biobank array. We restricted our analyses to 487,409 individuals passing previous genotyping QC and previously imputed to ∼93 million autosomal variants10; we re-phased these individuals using Eagle218 to improve phasing accuracy and imputed them to the union of the BiLEVE and Biobank arrays using Minimac337 (Supplementary Note 3). We further removed 427 individuals with low genotyping quality (B-allele frequency s.d.>0.11 at heterozygous sites), 4,111 individuals with evidence of possible sample contamination (Supplementary Note 1), and 82 individuals who had withdrawn consent, leaving 482,789 individuals for analysis. We performed data processing using plink38.
We additionally analyzed exome sequencing data available for 49,960 individuals22. To extend our rare variant association analyses to include variants identified in exome-sequenced individuals, we phased these individuals using Eagle2 and imputed into the full cohort using Minimac4 (Supplementary Note 3).
Detection of mCAs using genotyping intensities and long-range haplotype phase
As in our previous work9, we detected mCAs in genotyping intensity data from blood DNA samples using an approach that leverages the chromosome-scale accuracy of statistical phasing in the UK Biobank cohort17,18 (Supplementary Note 3). In brief, our approach harnesses long-range phase information to search for local imbalances between maternal and paternal allelic fractions in a cell population, enabling considerable gains in sensitivity for detection of large events at low cell fractions9. A full description of the method and a detailed exploration of its statistical properties compared to previous approaches is presented in the Supplementary Notes of ref.9. As before, we applied our approach to genotyping intensities that we transformed to log2 R ratio (LRR) and B-allele frequency (BAF) values39 (which measure total and relative allelic intensities) after affine-normalization and GC wave-correction1,9,40. We estimated cell fractions of mCAs using the formulas relating BAF to cell fraction presented in Table 1 of the supplement of ref.1.
In analyzing the full cohort, we made two minor modifications to our original approach. First, we halved the switch error rate parameter of our hidden Markov model (HMM) for BAF deviations, reflecting improved phasing accuracy in the full cohort. Second, we perfomed a few additional QC steps on the event calls to filter potential technical artifacts that we identified in the full data set; these filters affected <1% of the call set (Supplementary Note 1) and only affected four event calls from our previous analysis9.
Our detection procedure produced a final call set of 19,632 autosomal mCAs at a nominal FDR of 0.05 (based on our phase randomization approach to estimate statistic significance)9. We verified that our FDR was well-controlled using an independent FDR estimation procedure based on the age distribution of event carriers9; this approach produced a concordant FDR estimate of 6.6% (4.5–8.6%) (Extended Data Fig. 2e and Supplementary Note 1.3). We also verified that rates of mosaic events on each chromosome were very consistent with our previous call set on the interim UKB data9. We note that for our current study, we re-analyzed the interim samples for mosaicism using improved haplotype phasing in the full UK Biobank cohort; the increased phasing accuracy led to slightly higher detection sensitivity, such that the overall autosomal mCA detection rate increased by ∼10%. As before, we observed that lower-confidence events tended to have uncertain copy number (because our power to detect allelic imbalances exceeds our power to distinguish CN-LOH from copy-number alterations) and less-precise event boundaries9; we provide information on the uncertainty of each event call in Supplementary Data. We note that our replication here of results we previously reported from the interim UK Biobank release (e.g., genomic distribution of mCAs, age and sex distribution of mCAs, relationships to blood cell indices, mCA risk loci, and associations with hematological cancers) lends support to the validity of our methods.
Identifying variants associated with CN-LOH mutations in cis
We performed two types of association tests to identify inherited variants that influence mosaic CN-LOH mutations in cis. First, for each variant, we performed a Fisher test for association with a case-control phenotype specific to that variant: we considered samples to be cases if they carried a likely CN-LOH event containing the variant or within 4Mb (to allow for uncertainty in event boundaries). We considered an event to be a likely CN-LOH event if it either (i) was called as a CN-LOH event or (ii) had undetermined copy number, extended to a telomere, and had |LRR|<0.02. We performed this test on all typed and imputed variants and applied a genome-wide significance threshold of 5×10–8 for coding variants and 1×10–9 for all other variants.
Second, we searched for variants for which CN-LOH mutations in individuals heterozygous for the variant tended to preferentially duplicate one allele and remove the other allele from the genome. For each variant, we examined heterozygous individuals with a likely CN-LOH event overlapping the variant, and then performed a binomial test to check whether the CN-LOH direction tended to favor one allele versus the other. We restricted the binomial test to individuals in which the variant was confidently phased relative to the mosaic event, i.e., there was no disagreement in five random resamples from the HMM used to call the event).
Given that the two association tests described above are independent, the second test provided a means of validating associations identified by the first test, as any spurious associations from the first test would have no correlation with CN-LOH direction, whereas variants truly associated with CN-LOH mutations in cis typically have strong associations with CN-LOH direction (Extended Data Table 1). We also performed a combined test to identify common variants that did not reach genome-wide significance in the first test alone (which was underpowered for common variants due to small case counts) but reached significance using both tests together (Fisher’s combined P<1×10–8).
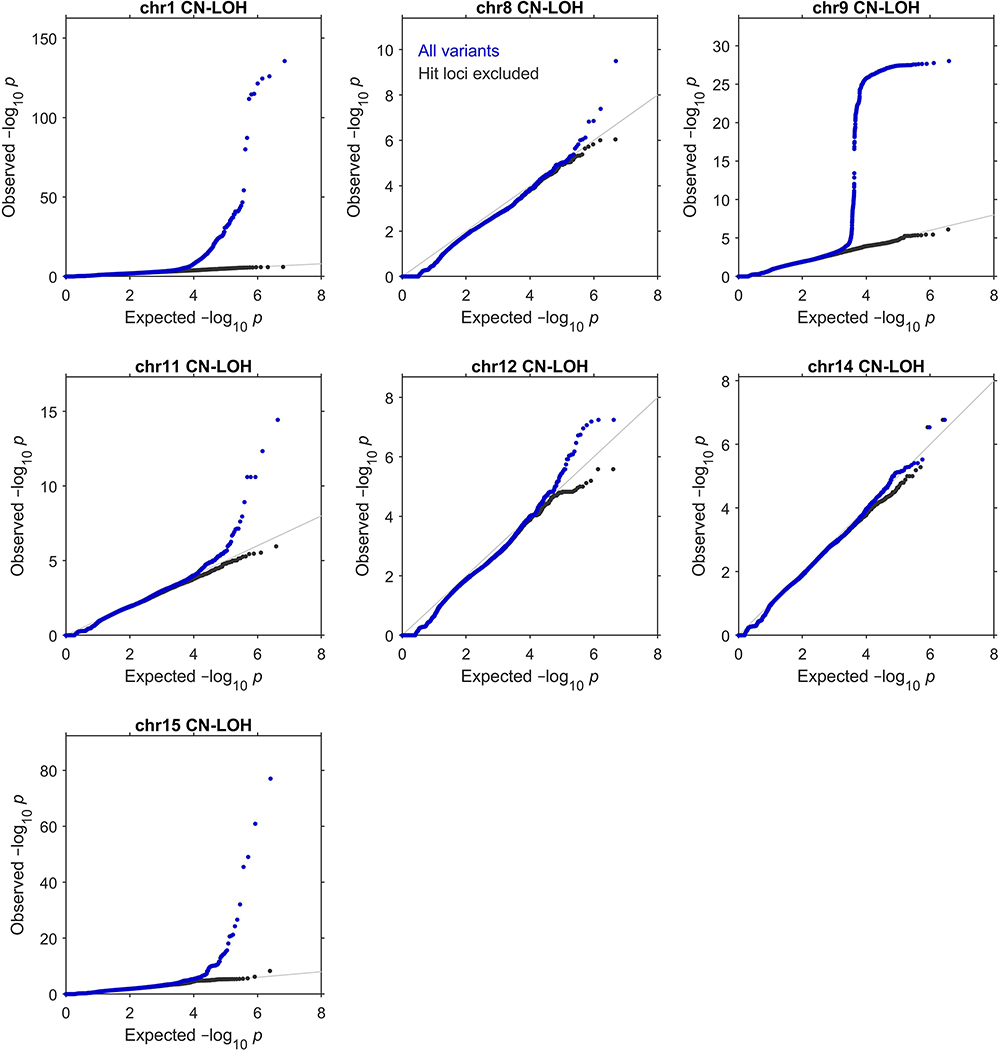
We restricted our association analyses to 455,009 individuals who reported European ancestry. Among these individuals, 96,590 pairs had previously been identified to be third-degree or closer relatives10,41. For each chromosome, we pruned the samples to an unrelated subset by removing one individual from each related pair, preferentially keeping (i) individuals with a likely CN-LOH on the chromosome and (ii) older controls. This pruning decreased total sample sizes to slightly less than 380,000 individuals (Supplementary Table 6). We verified that filtering on ancestry and relatedness in this way produced well-calibrated association test statistics (Extended Data Fig. 4 and Supplementary Note 2).
Fine-mapping loci associated with CN-LOH mutations in cis
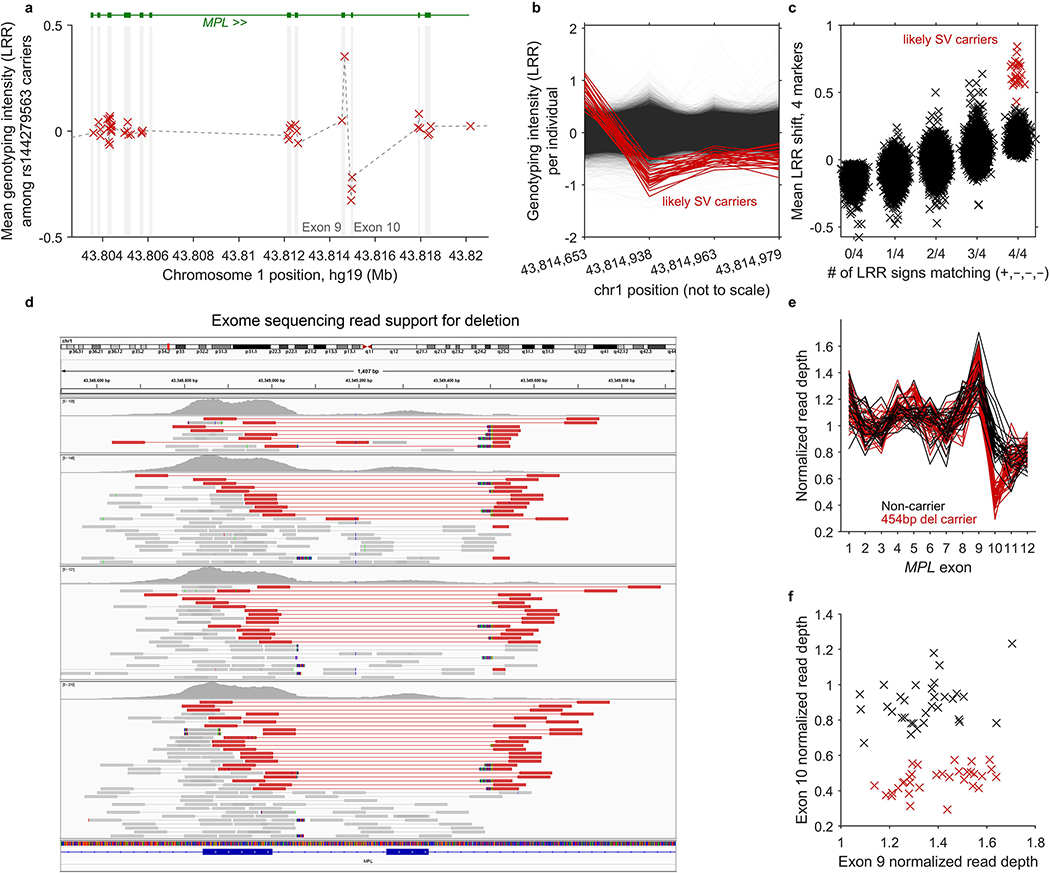
Given that our association analyses identified rare, large-effect coding variants in seven genes (FH, NBN, MRE11, SH2B3, MPL, ATM, and TM2D3), we undertook fine-mapping analyses at these loci to uncover additional coding or splice variants in these genes likely to be objects of clonal selection (upon modification of allelic load via CN-LOH mutation). We tested variants in these genes in three categories: (i) missense variants with CADD v1.3 score >20 (ref.42); (ii) predicted LoF variants (i.e., stop gained, frameshift, splice acceptor, or splice donor sites in any transcript annotated by VEP43); and (iii) likely pathogenic variants (according to ClinVar21, downloaded Mar 25, 2019). We restricted these analyses to rare variants with MAF between 5×10–6 and 0.01. For directly genotyped variants, we required missingness <0.01; for imputed variants, we required INFO>0.2 (for variants imputed by UK Biobank using IMPUTE410) or Minimac R2>0.4 (for variants we imputed; Supplementary Note 3). In addition to variants available from genotyping and imputation, we also tested two structural variants: a 454bp deletion that we discovered in MPL by analyzing exome sequencing reads using IGV44 and mosdepth45 (Extended Data Fig. 5 and Supplementary Note 4) and a ∼70kb deletion of TM2D3 that we previously identified9. In total, 616 variants across the seven loci satisfied these criteria.
Of these 616 variants, 38 variants reached Bonferroni significance (P<8.1×10–5; Extended Data Table 1) and 52 variants reached FDR<0.05 significance (assessed per gene; Supplementary Table 7). We determined that all 52 FDR-significant variants were likely to causally drive independent associations with CN-LOH events in cis, based on the following lines of evidence. First, CN-LOH events acted on all 52 variants in the expected direction (consistently removing rare variants in MPL and duplicating rare variants in the other six genes; Supplementary Table 7); in contrast, variants associated by chance would have random phase relative to CN-LOH events. Second, none of the 52 variants tagged other nearby variants with stronger associations (Fig. 1). On the contrary, nearby variants in linkage disequilibrium (computed in-sample) with the 52 variants had weaker associations explained by tagging of the 52 variants (Fig. 1), and we verified that the variants in the MPL and ATM loci that we previously reported9 each tagged one of the 52 variants (Supplementary Table 8). Third, none of the 52 variants tagged each other. The association signals at the 52 variants were driven by almost entirely non-overlapping sets of carriers who also had CN-LOH events in cis; the only overlap occurred between 11q CN-LOH individuals carrying the rs587779872 ATM missense variant (6 carriers with 11q CN-LOH) and the rs786204751 ATM stop gain variant (2 carriers with 11q CN-LOH, both also carrying rs587779872; Extended Data Fig. 7). The rs587779872 association remained significant in non-carriers of rs786204751, while the rs786204751 stop gain mutation nullified the effect of the rs587779872 missense mutation (occurring later in ATM), leading us to conclude that these associations were likely to be independent.
Burden analyses to detect ultra-rare variants targeted by CN-LOH events
To identify CN-LOH events potentially explained by variants too rare to reach significance in single-variant association analyses, we analyzed variant calls from exome sequencing of 49,960 UK Biobank participants22 for a burden of ultra-rare coding and splice variants in individuals with CN-LOH events. As in our other association analyses, we restricted to individuals who reported European ancestry. Because these variant calls potentially contained a small fraction of somatic variants that had risen to cell fractions higher than ∼20%, we included DNMT3A, TET2, and JAK2 in these analyses in addition to the seven genes at which we found inherited variants influencing CH. Beyond being frequently mutated in CH3,4, DNMT3A, TET2, and JAK2 are also frequently overlapped by CN-LOH events (Extended Data Fig. 1a), suggesting that some CN-LOH events act on previously-acquired point mutations in these genes.
As in our fine-mapping analyses, we considered variants annotated as (i) missense with CADD score >20, (ii) predicted LoF, or (iii) likely pathogenic in ClinVar. We restricted to ultra-rare variants (MAF<1×10–4), with the exception of JAK2 V617F, which was called in 46 exome-sequenced individuals (MAF=4.6×10–4). (For JAK2 and ATM, we used exome variant calls generated by UK Biobank using the “functionally equivalent” (FE) pipeline46, which we found provided slightly better power at these loci; for all other analyses, we used variant calls from Regeneron’s Seal Point Balinese (SPB) pipeline22.) For each gene, we examined individuals with CN-LOH events spanning the gene (not already explained by any of the 52 variants identified in our association analyses) and tabulated the number of such individuals who carried any of the rare variants under consideration (Supplementary Table 10). We then computed a burden P-value using a one-sided binomial test comparing the observed count to expectation (based on variant frequencies among 46,633 exome-sequenced individuals who reported European ancestry).
For each variant call potentially targeted by a CN-LOH event, we further examined allelic read depths from the exome sequencing data to assess whether the variant was likely to be of inherited or acquired origin. While read depths were generally insufficient to make a confident assessment on a per-variant level (and making this determination is complicated by mapping bias toward the reference allele3), the allelic depths broadly indicated that all or most variants implicated at our seven inherited risk loci were indeed inherited, while all or most variants implicated at DNMT3A, TET2, and JAK2 had been acquired somatically (Extended Data Fig. 8).
GWAS for trans associations with any autosomal mCA
We tested common variants for trans associations with the presence of any detectable autosomal mCA. We computed association test statistics using BOLT-LMM26,47 on 452,469 individuals (of which 16,366 were cases) who reported European ancestry and had imputation data available on autosomes and the X chromosome10. We included 20 principal components, age, age squared, sex, smoking status, genotyping array, and assessment center as covariates in the linear mixed model to guard against confounding and to improve power by removing phenotypic variance explained by covariates.
Polygenic scores for blood cell traits
We analyzed 29 blood count traits: counts and percentages of basophils, eosinophils, lymphocytes, monocytes, neutrophils, platelets, red cells, reticulocytes, and high light scatter reticulocytes; white cell count, platelet and red cell distribution widths, immature reticulocyte fraction, hemoglobin concentration, mean corpuscular hemoglobin (MCH), MCH concentration, mean corpuscular volume, mean platelet volume, mean reticulocyte volume, and mean sphered cell volume. (These traits constituted all available blood count traits except nucleated red blood cell indices, which were mostly zero.) We performed basic QC and normalization on these traits using the following steps: (i) remove outliers (>7 times farther from median than nearest quartile); (ii) stratify into males, pre-menopausal females, and post-menopausal females; (iii) within each stratum: (a) inverse-normal transform; (b) regress out age, age2, height, height2, BMI, BMI2, ethnic group, alcohol use, and smoking status; (c) inverse-normal transform again.
We computed polygenic score coefficients (i.e., “betas” in a linear predictor) for the traits listed above using the --predBetasFile option of BOLT-LMM26,47, which estimates polygenic score coefficients using a Bayesian linear mixed model that accounts for linkage disequilbrium among variants. We computed coefficients for 709,999 autosomal and X chromosome variants in the intersection of the Biobank and BiLEVE arrays that passed QC filters (allele frequency deviation <0.02 between the arrays, missingness <0.05, failed QC in at most one genotyping batch10). For each blood count phenotype, we restricted the sample set to individuals of self-reported European ancestry with non-missing phenotype (437,009–445,438 individuals depending on the phenotype). We ran BOLT-LMM using the same set of covariates we used in our trans GWAS. We computed polygenic risk coefficients for Y loss in blood cells using an analogous analysis restricted to males27.
We note that among the 29 blood count parameters we considered, some of the parameters corresponding to abundances of blood cell types might be surrogates for enhanced cellular fitness (in many cases of mitotic progenitors rather than the cell types themselves). However, we also considered other parameters that reflect cell size or morphology (some of which had polygenic scores that tended to be decreased in expanded CN-LOH clones; Extended Data Fig. 9). These relationships may reflect the production of abnormal cells by biologically altered stem cells, rather than cellular fitness itself (which may be a property of the unobserved hematopoietic stem cells); for example, mean platelet volume (MPV) has been reported to be a marker of myeloproliferative disorders. In our analyses predicting the direction of CN-LOH events, we allowed the logistic model to consider polygenic scores for all 29 parameters, the idea being that it would treat the polygenic scores as proxies for a variety of proliferative or cell-production tendencies and learn from the data how to weight them appropriately.
Polygenic score differentials for CN-LOH events
The polygenic score coefficients we computed for blood cell traits allowed us to estimate the extent to which CN-LOH mutations modified the genetic components of these traits. For each CN-LOH mutation, we computed the difference in polygenic score carried by the haplotype that was duplicated versus the haplotype that was removed. (This quantity is equal to the difference between the polygenic load of the mutant CN-LOH genome versus the original genome.) We determined which haplotype was duplicated and which was deleted using our hidden Markov model of phased BAF deviations9, averaging across five posterior samples from the HMM. To identify chromosome arms in which CN-LOH events tended to increase polygenic load for specific blood cell traits, we averaged polygenic score differentials across all CN-LOH events on each arm and computed means and z-scores (independently for each blood cell trait; Fig. 2b and Supplementary Table 15). To maximize power, we included all “likely-CN-LOH” events in these analyses (i.e., events called as CN-LOH as well as events with undetermined copy number that extended to a telomere and had |LRR|<0.02, as in our cis association analyses), comprising a total of 11,638 likely-CN-LOH events on 39 chromosome arms containing at least 20 such events.
Prediction of CN-LOH directions using CN-LOH-associated alleles and polygenic scores
To assess the extent to which the direction of a CN-LOH event (i.e., which affected haplotype is duplicated and which one is deleted) can be predicted based on the alleles inherited on each haplotype, we fit logistic models on the CN-LOH events on each chromosome arm using 10-fold cross-validation. For each fold, we performed logistic regression using stepwise forward selection on three possible sets of predictors: (i) a single variable containing the difference in the number of CN-LOH-associated alleles (Extended Data Table 1 and Supplementary Tables 7 and 12) carried by the two affected haplotypes; (ii) 31 variables containing polygenic score differentials (for the 29 blood count indices, the Y loss trait, and myeloproliferative neoplasms; Supplementary Note 7) between the two affected haplotypes; and (iii) all 32 variables together. We started forward selection using the “number of CN-LOH-associated alleles” variable in analyses (i) and (iii) and an empty set of variables in analysis (ii). We stopped forward selection when model improvement was no longer significant at a 0.01 level. We restricted our prediction analyses to chromosome arms for which at least one variable was selected (on average across folds).
For each chromosome arm, we merged prediction results across the 10 held-out folds and then assessed accuracy in two ways. First, we computed the Pearson correlation (R) between observed and predicted CN-LOH directions (using continuous-valued prediction probabilities from logistic regression). Second, we computed raw prediction accuracy (using binary, hard-called predictions). As in our analyses of polygenic score differentials, we included all likely-CN-LOH events (as defined above) to maximize power in these analyses.
We note that evaluating the ability of polygenic scores to predict CN-LOH directions in the same samples in which polygenic scores were computed does not result in overfitting. The reason is that we are evaluating a different kind of prediction accuracy: ability to predict which of an individual’s two haplotypes is more likely to be made homozygous by a clonal CN-LOH event. This “directionality” information is independent of the unphased genotype and phenotype information used to build the polygenic scores.
Enrichment of mCA types in specific blood lineages
To identify classes of mCAs linked to different blood cell types9, we first classified mCAs based on chromosomal location and copy number. For each autosome, we defined five disjoint categories of mCAs that comprised the majority of detected events: loss on p-arm, loss on q-arm, CN-LOH on p-arm, CN-LOH on q-arm, and gain. We subdivided loss and CN-LOH events by arm but did not subdivide gain events because most gain events are whole-chromosome trisomies (Extended Data Fig. 1a). (We excluded the chr17 gain category because nearly all of these events arise from i(17q) isochromosomes already counted as 17p– events9.)
For each mCA type, we computed enrichment among individuals with anomalous (top 1%) values of each of 14 normalized blood indices (counts and percentages of lymphocytes, basophils, monocytes, neutrophils, red cells, and platelets, as well as distribution widths of red cells and platelets) using Fisher’s exact test (two-sided; P-values reported throughout this manuscript are from two-sided statistical tests unless explicitly stated otherwise). We restricted these analyses to individuals who reported European ancestry, and reported significant enrichments passing an FDR threshold of 0.05 (Extended Data Fig. 1c and Supplementary Table 5).
UK Biobank cancer phenotypes
We analyzed UK cancer registry data provided by UK Biobank for 81,401 individuals in our sample set who had one or more prevalent or incident cancer diagnoses. Cancer registry data included date of diagnosis and ICD-O-3 histology and behavior codes, which we used to identify individuals with diagnoses of CLL, MPN, MDS, or any blood cancer48,49. Because our focus was on the prognostic power of mCAs to predict diagnoses of incident cancers >1 year after DNA collection, we excluded all individuals with cancers reported prior to this time (either from cancer registry data or self-report of prevalent cancers). We also restricted our attention to the first diagnosis of cancer in each individual, and censored diagnoses after September 30, 2014, as suggested by UK Biobank (resulting in a median follow-up time of 5.7 years, s.d. 0.8 years, range 4–9 years). Finally, we restricted analyses to individuals who reported European ancestry. These exclusions reduced the total counts of incident cases to 199 (CLL), 138 (MPN), 70 (MDS), and 1,383 (any blood cancer). In our primary analyses, we further eliminated individuals with any evidence of potential undiagnosed blood cancer based on anomalous relevant blood indices (lymphocyte count outside the normal range of 1–3.5×109/L, red cell count >6.1×1012/L for males or >5.4×1012/L for females, platelet count >450×109/L, red cell distribution width >15%), leaving incident case counts of 107 (CLL), 67 (MPN), 56 (MDS), and 1,055 (any blood cancer).
Estimation of cancer risk conferred by mCAs
To identify classes of mCAs associated with incident cancer diagnoses, we classified mCAs based on chromosomal location and copy number as described above. We then restricted our attention to the 78 classes with at least 30 carriers (to reduce our multiple hypothesis burden, given that we would be underpowered to detect associations with the rarer events). For each mCA class, we considered a sample to be a case if it contained only the mCA or if the mCA had highest cell fraction among all mCAs detected in the sample (i.e., we did not count carriers of subclonal events as cases). We computed odds ratios and P-values for association between mCA classes and incident cancers using Cochran-Mantel-Haenszel (CMH) tests to stratify by sex and by age in six 5-year bins. We used the CMH test to compute odds ratios (for incident cancer any time during follow-up) rather than using a Cox proportional hazards model to compute hazard ratios because both the mCA phenotypes and the incident cancer phenotypes were rare, violating assumptions of normality underlying regression. We reported significant associations passing an FDR threshold of 0.05 (Fig. 3a and Supplementary Table 20).
UK Biobank cardiovascular disease phenotypes
We analyzed algorithmically-defined cardiovascular events (myocardial infarction and stroke) identified by UK Biobank for 26,873 individuals in our sample set. Events had been identified based on information from baseline questionnaires and/or nurse-led interviews and from linked hospital admission and death registry data sets. We restricted our analyses to individuals with no missing cardiovascular covariates, self-reported European ancestry, and no prevalent cardiovascular disease, leaving 433,339 individuals, of which 8,094 had incident cardiovascular events during 5–10 years of follow-up.
Estimation of cardiovascular risk conferred by mCAs
To increase statistical power and limit the multiple hypothesis testing burden, we grouped all incident cardiovascular events into a single case-control phenotype and tested this phenotype for association with detectable mCAs. We considered mosaicism phenotypes defined by grouping all autosomal mCAs into one phenotype or by grouping mCAs by copy number (loss, CN-LOH, or gain), and we also examined specific mCAs related to common mosaic point mutations3,4,28: focal deletions at DNMT3A, focal deletions at TET2, and CN-LOH mutations on 9p (which often duplicate a JAK2 V617F mutation50–53) (Extended Data Fig. 1a). For each category of mCAs, we created a subsample of mCA carriers and noncarriers matched on assessment year, age (in 1-year bins), sex, smoking status (current/ever/never), hypertension status, BMI (<25, 25–30, >30), and type 2 diabetes status, selecting carrier-noncarrier ratios to maximize power. We estimated cardiovascular risk conferred by each category of mCAs by performing Fisher’s exact test on the matched sample sets.
Code availability
A standalone software implementation (MoChA) of the algorithm used to call mCAs is available at https://github.com/freeseek/mocha. Code used to perform the specific analyses in this study is available from the authors upon request (but unlike MoChA, this code is not immediately portable to other computing environments).
Data availability
Mosaic event calls are available in Supplementary Data in anonymized form. The mCA call set has also been returned to UK Biobank (as Return 2062) to enable individual-level linkage to approved UK Biobank applications. Access to the UK Biobank Resource is available by application (http://www.ukbiobank.ac.uk/).
Extended Data
Extended Data Figure 1: Mosaic chromosomal alterations detected among 482,789 UK Biobank participants.
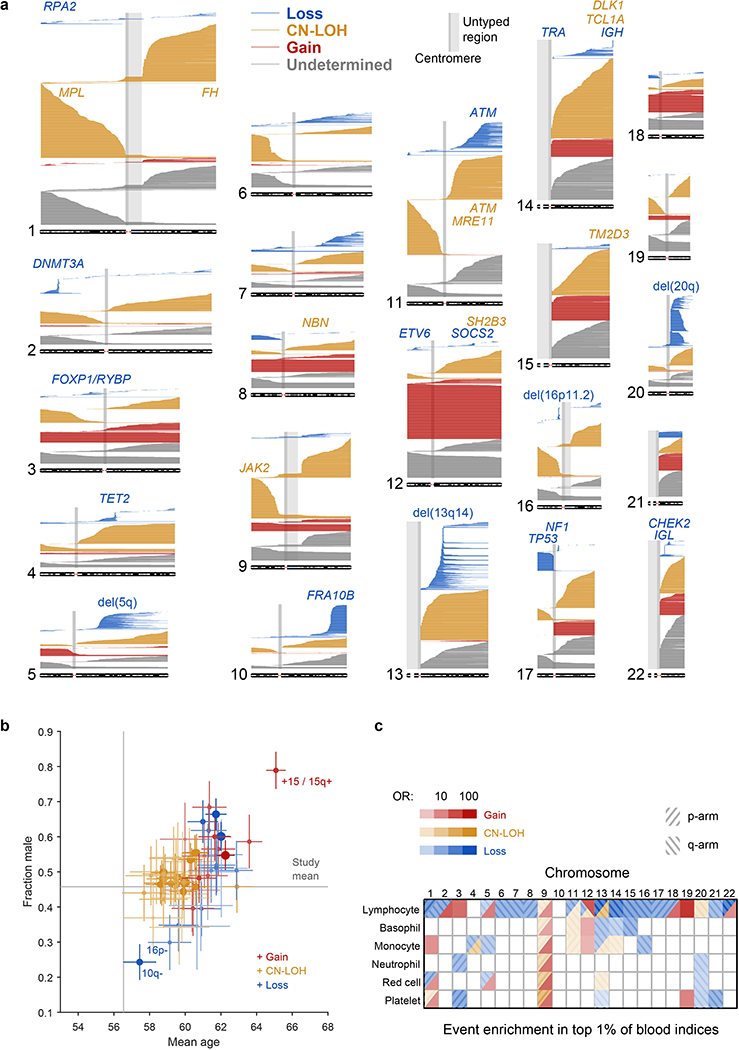
(a) Each horizontal line corresponds to an mCA; a total of 19,632 autosomal events in 17,111 unique individuals are displayed. Detected events are color-coded by copy number of the affected chromosome or segment (orange, LOH; blue, loss/deletion; red, gain/duplication). Focal deletions are labeled in blue with the names of putative target genes. Loci containing inherited variants influencing somatic events in cis are labeled in the same color as the corresponding mCA (orange for CN-LOH-associated loci, blue for losses). (b) Sex and age distributions of individuals with detected mosaic events. Marker size and color intensity increase with event frequency. Error bars, 95% CIs. Sample sizes are provided in Supplementary Table 1 and numeric data are provided in Supplementary Table 4. We previously reported three events with unusual sex biases (gains on chromosome 15, 16p11.2 deletions, and 10q terminal deletions)9, all of which replicated here. We have not identified a mechanism that could explain the sex biases. The overall tendency of male enrichment for most mCAs raises the possibility that environmental exposures could result in genomic insults that lead to mCAs; however, the heterogeneity of the level of male enrichment across different mCAs suggests that the mechanisms producing sex biases may be event-specific. (c) Enrichment of mosaic chromosomal alterations in individuals with anomalously high blood indices. Different mCAs are significantly enriched (FDR 0.05; one-sided Fisher’s exact test) among N=455,009 individuals with anomalous blood counts in different blood lineages (adjusted for age, sex, and smoking status). Events were grouped by chromosome and copy number, with loss and CN-LOH events subdivided by p-arm vs. q-arm. (We did not subdivide gain events by arm because most gain events are whole-chromosome trisomies.) Numeric data are provided in Supplementary Table 5.
Extended Data Figure 2: Copy number determination and QC of mosaic chromosomal alteration calls.
(a–d) Total vs. relative allelic intensities of mCAs detected on each chromosome. Mean log2 R ratio (LRR) of each detected mCA is plotted against estimated change in B allele frequency at heterozygous sites (|ΔBAF|). The data exhibit the characteristic “arrowhead” pattern in which LRR/|ΔBAF| approximately equals a positive constant for gain events, zero for CN-LOH events, and a negative constant for loss events. Possible constitutional duplications were filtered according to thresholds on LRR and |ΔBAF| defined in Supplementary Note 1. Constitutional duplications have expected |ΔBAF|=1/6 and have LRR≈0.36 in this data set. We chose exclusion thresholds to conservatively discard all calls that might belong to this cluster, applying more stringent filtering to shorter events because (i) most constitutional duplications are short and (ii) shorter events have noisier LRR and |ΔBAF| estimates. (e) Estimation of false discovery rate using age distributions of individuals with mCA calls. We generated age distributions for (i) “high-confidence” detected events passing a permutation-based FDR threshold of 0.01 (bright green), (ii) “medium-confidence” events below the FDR threshold of 0.01 but passing an FDR threshold of 0.05 (darker green), and (iii) “low-confidence” events below the FDR threshold of 0.05 but passing an FDR threshold of 0.10 (darkest green; excluded from our call set but plotted for context). We compared these distributions to the overall age distribution of UK Biobank participants (grey). Based on the numbers of events in each category, ≈32% of medium-confidence detected events are expected to be false positives. To estimate our true FDR, we regressed the medium-confidence age distribution on the high-confidence and overall age distributions, reasoning that the medium-confidence age distribution should be a mixture of correctly-called events (with age distribution similar to that of the high-confidence events) and spurious calls (with age distribution similar to the overall cohort). We observed a regression weight of 0.44 for the component corresponding to spurious calls, in good agreement with expectation, and implying a true FDR of 6.6% (4.5–8.6%, 95% CI based on regression fit on n=6 age bins). (f) Fractions of individuals with at least one detected autosomal mCA stratified by age and sex. Error bars, 95% CI. Numeric data are provided in Supplementary Table 3.
Extended Data Figure 3: Principal component plot of UK Biobank participants.
Individuals are plotted by their first two genetic principal component coordinates as computed by UK Biobank10 and colored according to self-reported ethnic background. Red circles indicate individuals identified in our exome analyses (of self-reported White individuals with mosaic CN-LOH events) as carriers of rare coding or splice variants in frequently-targeted genes. Marginal density histograms stratified by self-reported ethnic background are provided next to the PC1 and PC2 axes.
Extended Data Figure 4: Quantile-quantile plots of P-values produced by association analyses.
These plots verify the calibration of the statistical tests we used to identify the genome-wide significant associations reported in Extended Data Table 1 (see legend for details of statistical tests and sample sizes). In each plot, the blue dots correspond to an analysis of all variants tested, while the black dots correspond to an analysis in which regions surrounding significant associations were excluded. Specifically, the plots respectively exclude 1:35–55Mb (MPL), 1:239–244Mb (FH), 8:88–93Mb (NBN), 9:2.5–7.5Mb (JAK2), 11:92–97Mb (MRE11), 11:103–113Mb (ATM), 12:109–114Mb (SH2B3), 14:92.5–102.5Mb (TCL1A and DLK1), and 15:100Mb–qter (TM2D3). In all cases, exclusion of the hit regions (which account for a small fraction of the variants tested) resulted in a distribution close to the expected null.
Extended Data Figure 5: Identification and validation of an inherited MPL structural variant.
We suspected that an association between rs144279563 and acquired 1p CN-LOH mutations might tag a causal structural variant in MPL. (While rs144279563 is ∼1.5Mb downstream of MPL, it is sufficiently rare to be in linkage disequilibrium with variants several megabases away.) We therefore examined genotyping intensities at MPL from 49,950 individuals typed on the BiLEVE chip (which contains more probes within MPL than the Biobank chip, on which the remaining individuals were typed.) (a) Mean genotyping intensities over 42 carriers of the rs144279563 rare allele exhibit a sharp increase at the end of MPL exon 9 (1 genotyping probe) followed by a sharp decrease in exon 10 (3 genotyping probes). (b,c) Closer inspection of genotyping intensities at the 4 probes across all BiLEVE individuals enabled identification of 27 individuals likely to carry an inherited structural variant (20 of which carry the rs144279563 rare allele). We called this variant in the BiLEVE cohort using two criteria: (i) correct sign of LRR at the 4 probes (+,–,–,–); and (ii) mean signed LRR shift >0.4 over the 4 probes. (d) Read support for a 454bp deletion spanning MPL exon 10 in exome-sequenced individuals. We used IGV44 to plot paired-end reads aligning in or near MPL exons 9 and 10 in four exome-sequenced individuals imputed to carry the MPL structural variant (and also mosaic for 1p CN-LOH events). Read pairs highlighted in red have unusually long insert sizes, consistent with a deletion of genomic sequence between the aligned reads. Multicolored read segments indicate clipped reads in which one end of a read stops aligning to the reference genome. On the left side of the deletion, clipped reads align through hg19 base pair 43,814,728 (…AGGGACTGGG), with mismatches consistently occurring starting from 43,814,729 rightward (hg19: CGCCG…). On the right side of the deletion, clipped reads align starting from 43,815,178 (CTGGGACTCG…), with mismatches starting from 43,815,177 leftward (hg19: …CACCT). Examination of individual clipped reads revealed sequence matching …AGGGACTGGGACTCG…, indicating deletion of 5bp (CTGGG) in addition to the 449bp between aligning read segments. (Note that in this caption we have used hg19 coordinates for consistency with the rest of this manuscript; the IGV plot uses hg38 coordinates because reads had been aligned to hg38 (amounting to an offset of –465,671bp relative to hg19 at MPL.) (e,f) Decreased read depth at exon 10 in all 32 imputed carriers of the MPL exon 10 deletion who had been exome-sequenced. We used mosdepth45 to compute mean read depth across all 12 MPL exons in the 32 exome-sequenced imputed deletion carriers along with 32 controls. We normalized read depth in each individual by dividing by mean read depth across exons 1–8 and 11–12. All 32 imputed carriers of the exon 10 deletion had lower exon 10 normalized read depths than all 32 controls. We did not observe any evidence of increased read depth in exon 9 in carriers vs. controls.
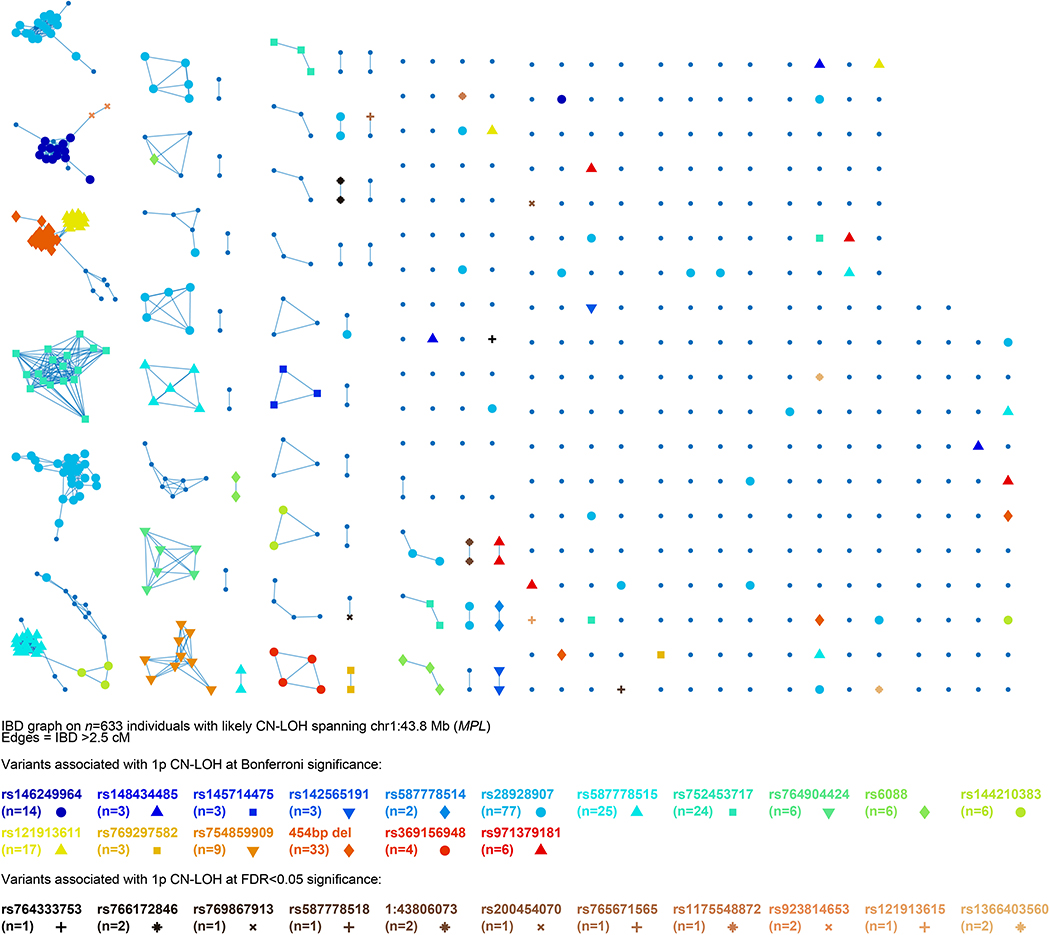
Extended Data Figure 6: Identity-by-descent (IBD) graph at MPL among individuals with likely 1p CN-LOH events spanning MPL.
We called IBD tracts using GERMLINE with haplotype extension54. Colored nodes indicate carriers of the 28 rare coding or splice variants we observed to be independently (and probably causally) associated with 1p CN-LOH mutations (always replacing the rare allele with the reference allele; Extended Data Table 1 and Supplementary Table 7). (Note that the numbers of carriers listed for each variant here are slightly higher than in the “Allelic shift” columns of Extended Data Table 1 and Supplementary Table 7 because allelic shifts could only be confidently ascertained for a subset of carriers.) The presence of additional IBD clusters not carrying any of the 28 highlighted variants suggests that even more causal variants in MPL remain to be discovered.
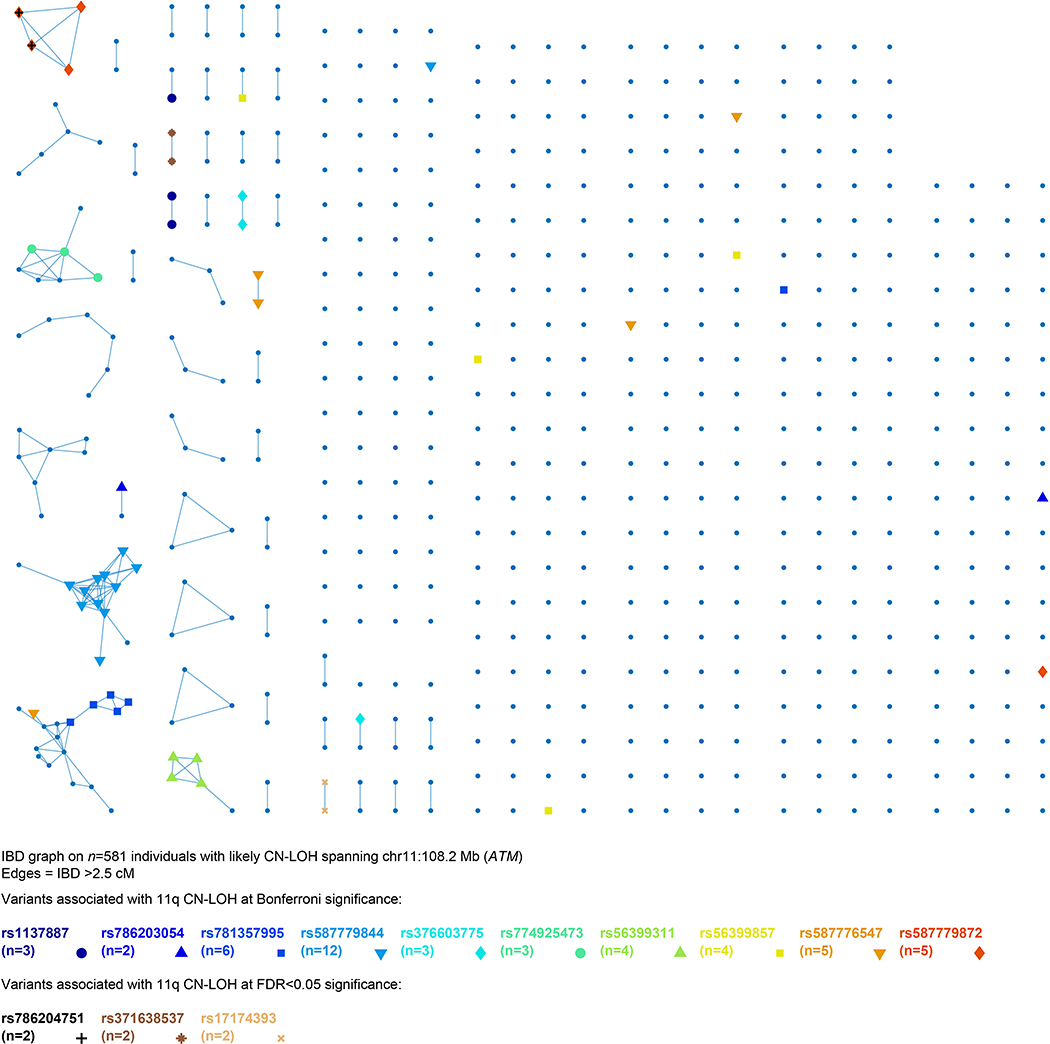
Extended Data Figure 7: Identity-by-descent (IBD) graph at ATM among individuals with likely 11q CN-LOH events spanning ATM.
We called IBD tracts using GERMLINE with haplotype extension54. Colored nodes indicate carriers of the eight rare coding or splice variants we observed to be independently (and probably causally) associated with 11q CN-LOH mutations (always making the rare allele homozygous; Extended Data Table 1 and Supplementary Table 7). The presence of additional IBD clusters not carrying any of the highlighted variants suggests that even more causal variants in ATM remain to be discovered. The two carriers of rs786204751 are also carriers of rs587779872, as discussed in Methods.
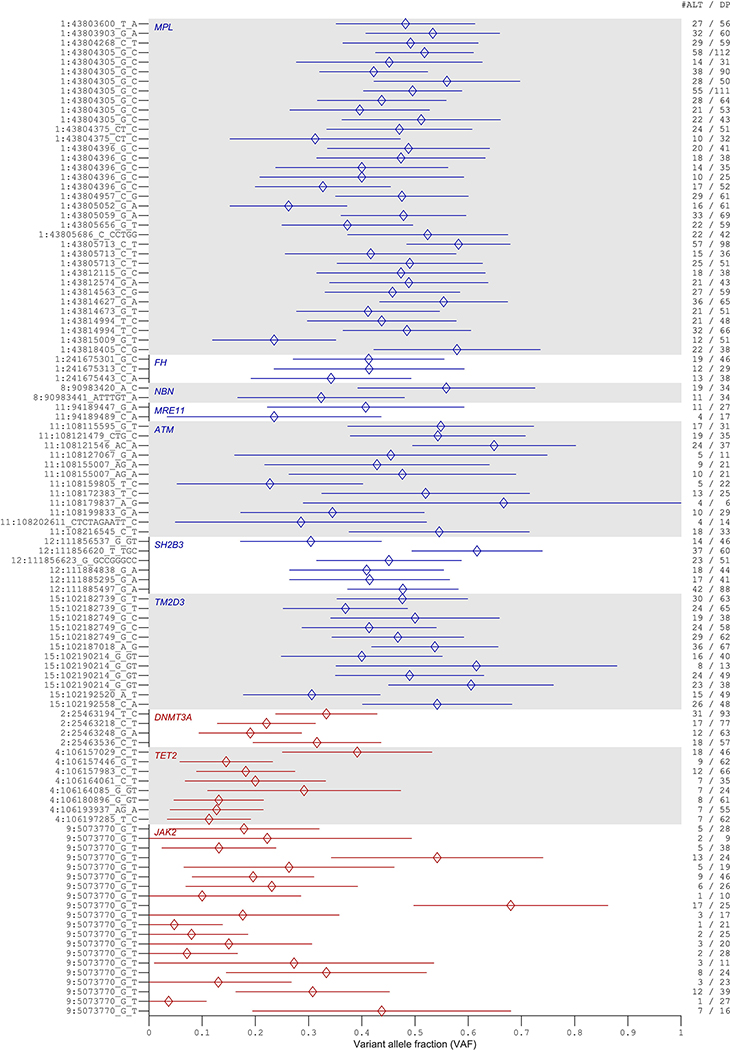
Extended Data Figure 8: Variant allele fractions of rare coding or splice variants likely to be targets of CN-LOH mutations in exome-sequenced individuals.
Variant allele fractions (VAF = number of reads matching the alternate allele divided by the total number of reads matching either the reference or the alternate allele) are plotted for each variant call identified as the potential target of a CN-LOH event (either from association analyses or burden analyses). Error bars, 95% CIs approximated using binomial standard errors multiplied by 1.96. Allelic read depths for variants identified at DNMT3A, TET2, and JAK2 are broadly indicative of somatic origin (VAF<0.5), while read depths for variants at the seven inherited risk loci are broadly consistent with inherited variation (VAF≈0.5). Read depths were generally insufficient to make a confident assessment of somatic vs. inherited origin on a per-variant level, as evidenced by wide VAF error bars; additionally, making this determination is further complicated by mapping bias toward the reference allele, which can produce VAF lower than 0.5 even for inherited variants3.
Extended Data Figure 9: Tendencies of CN-LOH mutations to modify polygenic scores for 29 blood cell parameters.
For each blood count parameter and each chromosome arm, the heatmap reports the z-score for the mean change in polygenic score across all CN-LOH mutations detected on the arm. Among the 29 blood count parameters we considered, some of the parameters corresponding to abundances of blood cell types might be surrogates for enhanced cellular fitness (in many cases of mitotic progenitors rather than the cell types themselves). Other parameters reflect cell size or morphology. Effects of CN-LOH mutations on polygenic scores for these parameters may reflect the production of abnormal cells by biologically altered stem cells, rather than cellular fitness itself (which may be a property of the unobserved hematopoietic stem cells). Columns: platelet count and crit (PLT#, Pct); red blood cell count (RBC#), hemoglobin (Hgb), and hematocrit (Hct) (both strongly correlated with RBC#); reticulocyte count and percent (RET#, RET%); high light scatter reticulocyte count and percent (HLR#, HLR%); immature reticulocyte fraction (IRF); white blood cell count (WBC#); neutrophil count and percent (NEU#, NEU%); eosinophil count and percent (EOS#, EOS%); monocyte count and percent (MON#, MON%); basophil count and percent (BAS#, BAS%); lymphocyte count and percent (LYM#, LYM%); platelet distribution width (PDW), mean platelet volume (MPV), RBC distribution width (RDW), mean corpuscular volume (MCV), mean reticulocyte volume (MRV), mean sphered cell volume (MSCV), mean corpuscular hemoglobin (MCH), and mean corpuscular hemoglobin concentration (MCHC).
Extended Data Table 1: Associations of mosaic CN-LOH mutations with inherited rare coding or splice variants in cis.
P-values from two independent statistical tests are reported: (i) a two-sided Fisher’s exact test treating individuals with a mosaic CN-LOH mutation in cis as cases (n≥378,307 individuals varying slightly among tests; Supplementary Table 6); and (ii) a binomial test for biased allelic imbalance in heterozygous cases. Loci reaching genome-wide significance in the first test are reported. At these loci, additional independently associated coding or splice variants reaching Bonferroni significance are also reported.
| GWAS | Allelic shift in hets | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Arm | Gene | Positiona | Variant | Effectb | Allelesc | AFd | P | OR (95% CI) | NREFe | NALT | P |
| Novel loci at which rare variants associate with CN-LOH events in cis | |||||||||||
| 1q | FH | 241675301 | rs199822819 | missense | G/C | 0.0003 | 4.9×10−11 | 28 (14–55) | 1 | 8 | 0.039 |
| 8q | NBN | 90983441 | rs1187082186 | frameshift | ATTTGT/A | 0.0002 | 4.8×10−13 | 210 (92–484) | 0 | 6 | 0.031 |
| 11q | MRE11 | 94189489 | rs587781384 | stop gained | C/A | 4×10−5 | 5.6×10−10 | 130 (50–338) | 0 | 5 | 0.062 |
| 12q | SH2B3 | 111885310 | rs72650673 | missense | G/A | 0.002 | 3.1×10−8 | 11 (5.8–20) | 1 | 8 | 0.039 |
| Previously reported loci at which rare variants associate with CN-LOH events in cis | |||||||||||
| 1p | MPL | 43804305 | rs28928907 | missense | G/C | 0.0006 | 1.9×10−130 | 142 (111–184) | 70 | 0 | 1.7×10−21 |
| 11q | ATM | 108172425 | rs587779844 | missense | C/T | 0.0001 | 3.5×10−20 | 96 (52–177) | 0 | 12 | 0.00049 |
| 15q | TM2D3 | 102151467 | 70kb delf | gene deletion | ref/del | 0.0003 | 9.8×10−224 | 555 (425–724) | 2 | 110 | 2.4×10−30 |
| Additional independently associated likely causal coding or splice variants | |||||||||||
| 1p | MPL | 43803600 | rs146249964 | splice donor | T/A | 0.0001 | 2.8×10−23 | 97 (55–171) | 12 | 0 | 0.00049 |
| 43803817 | rs148434485 | stop gained | C/T | 2×10−5 | 1.6×10−6 | 128 (37–446) | 2 | 0 | 0.5 | ||
| 43803824 | rs145714475 | missense | T/C | 2×10−5 | 1.9×10−6 | 120 (35–414) | 3 | 0 | 0.25 | ||
| 43803903 | rs142565191 | splice donor | G/A | 4×10−5 | 7.5×10−6 | 72 (22–238) | 3 | 0 | 0.25 | ||
| 43804234 | rs587778514 | frameshift | CCT/C | 1×10−5 | 3.9×10−5 | 199 (40–987) | 2 | 0 | 0.5 | ||
| 43804375 | rs587778515 | frameshift | CT/C | 0.0002 | 7.0×10−41 | 105 (68–161) | 24 | 0 | 1.2×10−7 | ||
| 43804396 | rs752453717 | splice modifier | G/C | 0.0003 | 5.8×10−36 | 74 (48–113) | 24 | 0 | 1.2×10−7 | ||
| 43804957 | rs764904424 | missense | C/G | 0.0001 | 2.1×10−8 | 35 (15–79) | 6 | 0 | 0.031 | ||
| 43805052 | rs6088 | missense | G/A | 9×10−5 | 8.3×10−10 | 61 (26–141) | 6 | 0 | 0.031 | ||
| 43805656 | rs144210383 | missense | G/T | 0.0001 | 5.3×10−9 | 44 (19–101) | 6 | 0 | 0.031 | ||
| 43805713 | rs121913611 | missense | C/T | 0.0002 | 3.3×10−28 | 102 (61–171) | 17 | 0 | 1.5×10−5 | ||
| 43812115 | rs769297582 | splice acceptor | G/C | 2×10−5 | 5.1×10−7 | 199 (54–737) | 3 | 0 | 0.25 | ||
| 43814627 | rs754859909 | stop gained | G/A | 7×10−5 | 1.7×10−16 | 126 (61–258) | 9 | 0 | 0.0039 | ||
| 43814729 | 454bp delg | exon 10 deletion | ref/del | 0.0002 | 3.6×10−58 | 153 (104–225) | 31 | 0 | 9.3×10−10 | ||
| 43817942 | rs369156948 | stop gained | C/T | 3×10−5 | 4.8×10−8 | 114 (39–333) | 4 | 0 | 0.12 | ||
| 43817973 | rs971379181 | frameshift | CG/C | 3×10−5 | 5.8×10−13 | 240 (93–618) | 6 | 0 | 0.031 | ||
| 8q | NBN | 90983420 | rs777460725 | missense | A/C | 0.0001 | 8.1×10−5 | 114 (28–465) | 0 | 2 | 0.5 |
| 11q | ATM | 108127067 | rs1137887 | splice modifier | G/A | 4×10−5 | 9.6×10−6 | 65 (20–214) | 0 | 2 | 0.5 |
| 108141801 | rs786203054 | missense | T/G | 7×10−6 | 1.2×10−5 | 437 (73–2618) | 0 | 2 | 0.5 | ||
| 108155007 | rs781357995 | frameshift | AG/A | 0.0001 | 3.0×10−9 | 48 (21–111) | 0 | 6 | 0.031 | ||
| 108175528 | rs376603775 | stop gained | C/T | 6×10−5 | 2.8×10−5 | 44 (14–143) | 0 | 4 | 0.12 | ||
| 108179837 | rs774925473 | splice modifier | A/G | 8×10−5 | 6.8×10−5 | 33 (10–104) | 0 | 3 | 0.25 | ||
| 108181006 | rs56399311 | missense | A/G | 8×10−5 | 1.7×10−6 | 44 (16–120) | 0 | 4 | 0.12 | ||
| 108201108 | rs56399857 | missense | T/G | 0.0002 | 4.9×10−5 | 18 (6.6–48) | 0 | 4 | 0.12 | ||
| 108202611 | rs587776547h | inframe deletion | C...T/C | 7×10−5 | 8.5×10−9 | 73 (29–183) | 0 | 5 | 0.062 | ||
| 108216545 | rs587779872 | missense | C/T | 2×10−5 | 3.6×10−11 | 251 (89–706) | 0 | 5 | 0.062 | ||
| 12q | SH2B3 | 111885295 | rs148636776 | missense | G/A | 0.0004 | 4.0×10−5 | 19 (7–50) | 0 | 5 | 0.062 |
| 15q | TM2D3 | 102182739 | rs113189685 | missense | G/T | 3×10−5 | 2.8×10−8 | 132 (45–389) | 1 | 3 | 0.62 |
| 102182749 | rs754640606 | missense | G/C | 5×10−5 | 1.2×10−40 | 544 (289–1025) | 0 | 19 | 3.8×10−6 | ||
| 102182761 | rs976377433 | missense | A/G | 3×10−5 | 2.3×10−8 | 140 (47–413) | 0 | 4 | 0.12 | ||
| 102190214 | rs768556490 | frameshift | G/GT | 3×10−5 | 8.2×10−29 | 758 (327–1759) | 1 | 11 | 0.0063 | ||
Base pair position in hg19 coordinates.
Variant effects (using evidence reported in ClinVar for splice variants).
Reference/alternate allele.
Alternate allele frequency (in UK Biobank individuals of European ancestry).
Number of mosaic individuals heterozygous for the variant in which the somatic event shifted the allelic balance in favor of the reference allele (by duplication of its chromosomal segment and loss of the homologous segment).
This ∼70kb deletion spans 15:102.15–102.22Mb, deleting TM2D3 and part of TARSL2.
This 454bp deletion spans 1:43,814,729–43,815,182, deleting MPL exon 10 (Extended Data Fig. 5).
This 9bp inframe deletion in ATM has alleles CTCTAGAATT/C.
Supplementary Material
Acknowledgments
We thank S. Bakhoum, S. Raychaudhuri, M. Sherman, S. Elledge, and C. Terao for helpful discussions. This research was conducted using the UK Biobank Resource under Application #19808. P.-R.L. was supported by US NIH grant DP2 ES030554, a Burroughs Wellcome Fund Career Award at the Scientific Interfaces, the Next Generation Fund at the Broad Institute of MIT and Harvard, a Glenn Foundation for Medical Research and AFAR Grants for Junior Faculty award, and a Sloan Research Fellowship. G.G. and S.A.M. were supported by US NIH grant R01 HG006855. G.G. was supported by US Department of Defense Breast Cancer Research Breakthrough Award W81XWH-16–1-0316. Computational analyses were performed on the O2 High Performance Compute Cluster, supported by the Research Computing Group, at Harvard Medical School (http://rc.hms.harvard.edu), and on the Genetic Cluster Computer (http://www.geneticcluster.org) hosted by SURFsara and financially supported by the Netherlands Scientific Organization (NWO 480–05-003 PI: Posthuma) along with a supplement from the Dutch Brain Foundation and the VU University Amsterdam.
Footnotes
The authors declare competing interests: patent application PCT/WO2019/ 079493 has been filed on the mCA detection method used in this work.
References
- [1].Jacobs KB et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nature Genetics 44, 651–658 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Laurie CC et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nature Genetics 44, 642–650 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Genovese G et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. New England Journal of Medicine 371, 2477–2487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jaiswal S et al. Age-related clonal hematopoiesis associated with adverse outcomes. New England Journal of Medicine 371, 2488–2498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Machiela MJ et al. Characterization of large structural genetic mosaicism in human autosomes. American Journal of Human Genetics 96, 487–497 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Vattathil S & Scheet P Extensive hidden genomic mosaicism revealed in normal tissue. American Journal of Human Genetics 98, 571–578 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zink F et al. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood 130, 742–752 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Abelson S et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 559, 400–404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Loh P-R et al. Insights into clonal haematopoiesis from 8,342 mosaic chromosomal alterations. Nature 559, 350–355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bycroft C et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Uziel T et al. Requirement of the MRN complex for ATM activation by DNA damage. The EMBO Journal 22, 5612–5621 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lee J-H & Paull TT ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308, 551–554 (2005). [DOI] [PubMed] [Google Scholar]
- [13].Deng Y, Guo X, Ferguson DO & Chang S Multiple roles for MRE11 at uncapped telomeres. Nature 460, 914 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kimura S, Roberts AW, Metcalf D & Alexander WS Hematopoietic stem cell deficiencies in mice lacking c-Mpl, the receptor for thrombopoietin. Proceedings of the National Academy of Sciences 95, 1195–1200 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Solar GP et al. Role of c-mpl in early hematopoiesis. Blood 92, 4–10 (1998). [PubMed] [Google Scholar]
- [16].Seita J et al. Lnk negatively regulates self-renewal of hematopoietic stem cells by modifying thrombopoietin-mediated signal transduction. Proceedings of the National Academy of Sciences 104, 2349–2354 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Loh P-R, Palamara PF & Price AL Fast and accurate long-range phasing in a UK Biobank cohort. Nature Genetics 48, 811–816 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Loh P-R et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nature Genetics 48, 1443–1448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Auer PL et al. Rare and low-frequency coding variants in CXCR2 and other genes are associated with hematological traits. Nature Genetics 46, 629 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Schultz KAP et al. PTEN, DICER1, FH, and their associated tumor susceptibility syndromes: clinical features, genetics, and surveillance recommendations in childhood. Clinical Cancer Research 23, e76–e82 (2017). [DOI] [PubMed] [Google Scholar]
- [21].Landrum MJ et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Research 46, D1062–D1067 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Van Hout CV et al. Whole exome sequencing and characterization of coding variation in 49,960 individuals in the UK Biobank. bioRxiv (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Meuwissen T, Hayes B & Goddard M Prediction of total genetic value using genome-wide dense marker maps. Genetics 157, 1819–1829 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Purcell SM et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Khera AV et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nature Genetics 50, 1219 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Loh P-R et al. Efficient Bayesian mixed model analysis increases association power in large cohorts. Nature Genetics 47, 284–290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Thompson DJ et al. Genetic predisposition to mosaic Y chromosome loss in blood. Nature 575, 652–657 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jaiswal S et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. New England Journal of Medicine 377, 111–121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Davoli T et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155, 948–962 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].O’Keefe C, McDevitt MA & Maciejewski JP Copy neutral loss of heterozygosity: a novel chromosomal lesion in myeloid malignancies. Blood 115, 2731–2739 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chase A et al. Profound parental bias associated with chromosome 14 acquired uniparental disomy indicates targeting of an imprinted locus. Leukemia 29, 2069–2074 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Choate KA et al. Mitotic recombination in patients with ichthyosis causes reversion of dominant mutations in KRT10. Science 330, 94–97 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tesi B et al. Gain-of-function SAMD9L mutations cause a syndrome of cytopenia, immunodeficiency, MDS and neurological symptoms. Blood 129, 2266–2279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ripke S et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sudlow C et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLOS Medicine 12, 1–10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wain LV et al. Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK Biobank. Lancet Respiratory Medicine 3, 769–781 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Das S et al. Next-generation genotype imputation service and methods. Nature Genetics 48, 1284–1287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chang CC et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4, 1–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Peiffer DA et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Research 16, 1136–1148 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Diskin SJ et al. Adjustment of genomic waves in signal intensities from whole-genome SNP genotyping platforms. Nucleic Acids Research 36, e126 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Manichaikul A et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Rentzsch P, Witten D, Cooper GM, Shendure J & Kircher M CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Research 47, D886–D894 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].McLaren W et al. The Ensembl Variant Effect Predictor. Genome Biology 17, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Thorvaldsdóttir H, Robinson JT & Mesirov JP Integrative genomics viewer (igv): high-performance genomics data visualization and exploration. Briefings in Bioinformatics 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Pedersen BS & Quinlan AR Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics 34, 867–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Regier AA et al. Functional equivalence of genome sequencing analysis pipelines enables harmonized variant calling across human genetics projects. Nature Communications 9, 4038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Loh P-R, Kichaev G, Gazal S, Schoech AP & Price AL Mixed-model association for biobank-scale datasets. Nature Genetics 50, 906–908 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Turner JJ et al. InterLymph hierarchical classification of lymphoid neoplasms for epidemiologic research based on the WHO classification (2008): update and future directions. Blood 116, e90–e98 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Arber DA et al. The 2016 revision to the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia. Blood 127, 2391–2405 (2016). [DOI] [PubMed] [Google Scholar]
- [50].Jones AV et al. JAK2 haplotype is a major risk factor for the development of myeloproliferative neoplasms. Nature Genetics 41, 446–449 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kilpivaara O et al. A germline JAK2 SNP is associated with predisposition to the development of JAK2V617F-positive myeloproliferative neoplasms. Nature Genetics 41, 455–459 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Olcaydu D et al. A common JAK2 haplotype confers susceptibility to myeloproliferative neoplasms. Nature Genetics 41, 450–454 (2009). [DOI] [PubMed] [Google Scholar]
- [53].Koren A et al. Genetic variation in human DNA replication timing. Cell 159, 1015–1026 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Gusev A et al. Whole population, genome-wide mapping of hidden relatedness. Genome Research 19, 318–326 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mosaic event calls are available in Supplementary Data in anonymized form. The mCA call set has also been returned to UK Biobank (as Return 2062) to enable individual-level linkage to approved UK Biobank applications. Access to the UK Biobank Resource is available by application (http://www.ukbiobank.ac.uk/).