Abstract
Inversions play an important role in disease and evolution but are difficult to characterize because their breakpoints map to large repeats. We increased by six-fold the number (n = 1,069) of previously reported great ape inversions using Strand-seq and long-read sequencing. We find that the X chromosome is most enriched (2.5-fold) for inversions based on its size and duplication content. There is an excess of differentially expressed primate genes near the breakpoints of large (>100 kb) inversions but not smaller events. We show that when great ape lineage-specific duplications emerge they preferentially (~75%) occur in an inverted orientation compared to their ancestral locus. We construct megabase-pair-scale haplotypes for individual chromosomes and identify 23 genomic regions that have recurrently toggled between a direct and inverted state over 15 million years. The direct orientation is most frequently the derived state for human polymorphisms that predispose to recurrent copy number variants associated with neurodevelopmental disease.
Inversions play an important role in disease and genome evolution as they suppress recombination1 and predispose to non-allelic homologous recombination (NAHR) associated with cancer and neurodevelopmental disease2. They are notoriously difficult to detect using both long- and short-read sequencing technologies3,4 because inversion breakpoints are typically embedded within highly identical segmental duplications (SDs)5–7 exceeding 50–100 kb in size8. It is estimated that more than 50% of inversions within human genomes are flanked by such inaccessible SDs4,6,9,10. True inversions are also difficult to distinguish from repeat sequences that have mobilized and inserted in an inverted orientation11. As a result, inversions are now recognized as one of the most under-ascertained forms of structural variation in human4 and nonhuman primate genomes, limiting our understanding of their evolution12.
Among apes, the largest cytogenetically visible inversions were first documented by Yunis and Prakash13, and most subsequent studies have inferred a subset of events using indirect genomic approaches12,14–18. For example, smaller inversions embedded in a unique sequence were readily detected using paired-end sequencing19, linked reads12,20, and assembly-based approaches12,21. These approaches especially fail to detect events that are flanked by SDs exceeding the length of the library inserts or sequence read length17.
Here, we apply Strand-seq22,23 to discover a comprehensive set of inversions in the great ape lineage and leverage long-read sequencing data to validate novel events that could not be confirmed by other approaches. Strand-seq is a single-cell sequencing technique that preserves directionality of single-stranded DNA at chromosome-length scale, allowing inversions to be readily detected and genotyped4,7. We apply this approach to provide a comprehensive framework for understanding the evolution and recurrence of inversions in the ape lineage.
RESULTS
Great ape inversion discovery
To systematically detect inversions in nonhuman primates (NHPs), we generated strand-specific sequence (Strand-seq) data from a representative of each great ape species22,23. We selected NHP individuals that differed from those where whole-genome assemblies were recently generated12, although this complicates the validation of heterozygous events not fixed in each species. We generated 62 high-quality single-cell libraries for chimpanzee (Dorien), 51 for bonobo (Ulindi), 81 for gorilla (GGO9), and 60 for orangutan (PPY10) (Table 1, Supplementary Fig. 1a, and Methods). Because genome coverage for each single-cell Strand-seq library is low (~0.02x) (Supplementary Fig. 1b), we increased the resolution for smaller inversions (1–50 kb) by concatenating all directional reads across all selected Strand-seq libraries into NHP-specific composite files4,7 (Fig. 1a, Supplementary Fig. 2, and Methods). Using composite files aligned to the human reference assembly (GRCh38), we detected inverted NHP loci as short as 1 kb in length by tracking changes in read directionality along each chromosome24 (Methods).
Table 1 |.
Summary of Strand-seq inversion callset
| # of Strand-seq libraries | Depth of coverage | # of simple inversions | # of inverted duplications | |
|---|---|---|---|---|
| Chimpanzee | 62 | 1.28 | 159 | 71 |
| Bonobo | 51 | 0.97 | 153 | 63 |
| Gorilla | 81 | 1.68 | 160 | 122 |
| Orangutan | 60 | 1.68 | 210 | 131 |
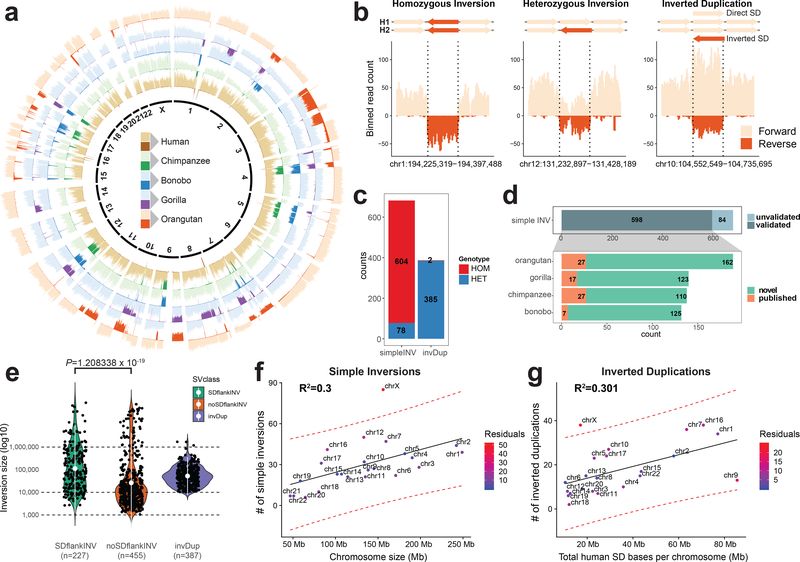
Figure 1 |. Inversion call summary.
a, Circular representation of composite files for each member of a great ape family. Genome of each individual is divided into 500-kb bins, and the number of reads mapped in forward (light color) and reverse (dark color) orientation in each bin is depicted as a bar along each chromosome. b, Example of inversion classes mapped in orangutan. Directional reads are binned into 10-kb bins (step 5 kb), and the number of reads mapped in forward (light color) and reverse (dark color) orientation is depicted as a vertical bar along a given genomic region. Inverted loci are highlighted by dashed lines. c, Summary of all inversions and inverted duplications mapped in this study. Inner circle summarizes the number of events found for each SV class (simple inversion, INV; inverted duplication, invDup). d, (i) Summary of validated simple inversions by other orthogonal technologies. (ii) Summary of validated simple inversions that appear to be novel in comparison to previously published data (green, novel; orange, published). e, Size distribution of simple inversions flanked by segmental duplications (SDs) (SDflankINV n = 227), simple inversions not flanked by SDs (noSDflankINV n = 455), and inverted duplications (invDup n = 387). White dot shows the mean of each distribution along with IQR range (Wilcoxon rank sum test). f, Scatterplot of the number of simple inversions (n = 682) given the chromosome length. g, Scatterplot showing the number of inverted duplications (n = 387) given the total length of known human SDs per chromosome. For f and g, regression line is added as a solid black line and 95% confidence intervals are highlighted as red dashed lines. Deviation from an expected number of inversions is expressed in the number of residuals.
We distinguish three classes of inversions. A homozygous inversion present on both homologs appears as a complete switch in reads mapping in reference orientation to all reads mapping in an inverted orientation (Fig. 1b). A heterozygous inversion resides on a single homolog and results in a 1:1 ratio of reads mapping in reference and inverted orientation. If there is no associated change in the underlying copy number, we group heterozygous and homozygous inversions as “simple” inversions (Fig. 1b). These are distinct from inverted duplications, where a change in copy number accompanies the localized change in read directionality. This class is often associated with lineage-specific SDs where at least one copy of a given locus resides in the genome in inverted orientation (Fig. 1b).
Among the NHPs, we detected 682 simple inversions and 387 inverted duplications (Fig. 1c, Supplementary Fig. 3, and Methods) with the number of events increasing with phylogenetic distance (Table 1). The vast majority of simple inversions (n = 604) are homozygous and likely represent fixed differences between humans and NHPs. The remainder (n = 78) are heterozygous, indicative of inversion polymorphisms within a great ape lineage (Supplementary Fig. 4b). As expected, nearly all (385 out of 387) inverted duplications appeared “heterozygous”, suggesting the duplicated locus occurs in an inverted orientation compared to the human locus and is on the same chromosome. These were easily distinguishable from simple heterozygous inversions by the increased sequence read depth over ancestral loci.
We performed extensive validations of both simple inversions and inverted duplications using a variety of orthogonal sequencing and mapping technologies (Supplementary Table 1 and Methods). Using fluorescence in situ hybridization (FISH), for example, we tested five large inversions (between 500 kb and 2.7 Mb in size) and confirmed that all were inverted in the predicted great ape (Supplementary Fig. 5, Supplementary Table 2, and Methods). We considered an inversion validated if it overlapped (50% reciprocal overlap) with an inversion call made by an orthogonal technology or an inversion was already published12,13,18,21,25 (Supplementary Fig. 6). Additionally, we attempted to assemble the breakpoints of 119 inversions using a recent phased long-read assembly approach4,26. This approach confirmed 27 inversions and provided sequence resolution of the inversion breakpoints (Supplementary Fig. 7, Supplementary Table 3, and Supplementary Note). Altogether, we validated 88% of our simple inversions (Fig. 1d), including most fixed events. Of the inversions that lack validation, 80% are either heterozygous (and therefore likely polymorphic in the lineage) or flanked by SDs and thus difficult to ascertain by other technologies. We estimate we have increased the number of validated simple inversions more than six-fold (78 vs. 521) when compared to previous studies (Fig. 1d).
Size and chromosomal distribution
Simple inversions ranged from 1,055 bp to 9.1 Mb in length (Supplementary Fig. 4a). Those flanked by SDs (n = 227; median 71,873 bp) were significantly larger (Wilcoxon rank sum test, two-sided, P = 1.21 × 10−19) (Fig. 1e) when compared to inversions not flanked by SDs (n = 455; median 12,476 bp). We note that this difference is unlikely due to ascertainment biases associated with previous studies12,27. Additionally, we found inversion size correlated positively with the size of SDs flanking the inversion28 (Supplementary Fig. 8). Strand-seq detection is much more sensitive than short-read-pair mapping approaches because inversion detection does not depend on the mapping of discordant reads to the reference genome17,29. Instead, the directionality with respect to the reference is embedded in every sequencing read, allowing for the unambiguous detection of inversions even when flanked by complex SDs7. Similarly, inverted duplications, which likely arise by duplicative transposition, show a wide size distribution (range 10,171–1,708,343 bp; median 48,421 bp) but rarely exceed 1 Mb in length, suggesting an upper bound for SD formation (Fig. 1e). Of note, we set a lower limit for inverted duplication calls at 10 kb.
While the number of simple inversions generally correlates with chromosome length (R2 = 0.3) (Fig. 1f,g), the X chromosome is an exception with ~2.5-fold more inversions when compared to the autosomal length (z-score = 3.57, P = 0.000177, one-sided) (Fig. 1f). This difference is even more pronounced for heterozygous inversions (4.6-fold), consistent with elevated rates of inversion polymorphism on the X chromosome (Supplementary Fig. 9). We also note that X chromosome inversions show a tighter size distribution (up to ~100 kb) compared to autosomes (Supplementary Fig. 10), possibly due to differences in the underlying architecture of SDs.
Unlike simple inversions, the number of inverted duplications correlates less strongly with chromosome size (R2 = 0.1) (Supplementary Fig. 11) but instead with SD content in the human genome (R2 = 0.301) (Fig. 1g). This is expected since lineage-specific duplications are 10-fold more likely to arise adjacent to ancestral duplicated sequences shared between two ape species30,31. If such a duplication arises in an inverted orientation, it will appear as an inverted duplication. For example, human chromosomes 5, 7, 10, 16 and 17 are among the most SD-rich chromosomes and similarly showed the greatest density of ape inverted duplications, often in close proximity to known human SDs (Supplementary Fig. 12). Once again, the clear exception is the X chromosome, which shows an excess of inverted duplications (Fig. 1g) with respect to autosomes given the chromosomal SD content.
Phylogenetic reconstruction
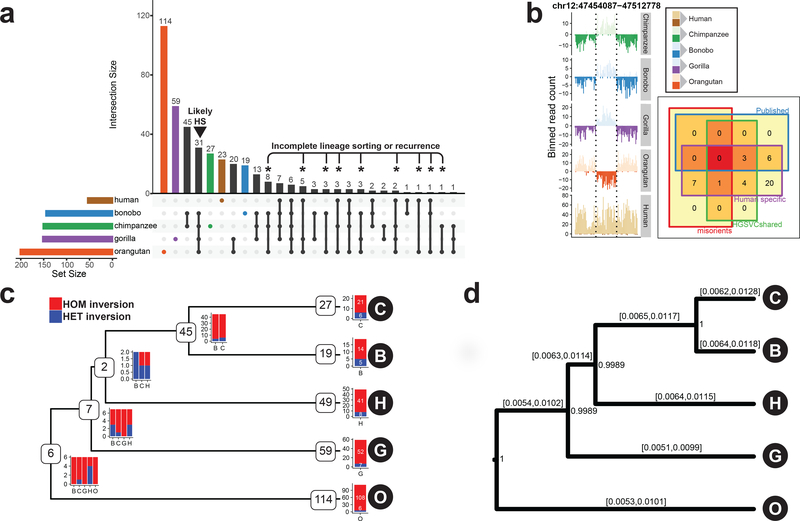
We compared the distribution of inversions among all great apes, including an African human sample4 (NA19240) (Fig. 2a, colored bars). Human-specific inversions are identifiable as loci that were inverted in all NHPs compared to the “direct” orientation in the human genome (Fig. 2b, left). We identified 26 total human-specific inversions, of which only 6 were previously reported (Fig. 2b, right, Methods)12,18. Excluding human reference genome misassemblies4, we classified all human-specific inversions as ancestral or lineage-specific parsimoniously assigning them to an ape phylogenetic tree (Methods). We placed 60 inversions on ancestral branches of the great ape phylogeny with the majority (n = 45), occurring on the ancestral Pan lineage (Fig. 2c). This was expected due to the recent divergence of chimpanzee and bonobo. Approximately 27% (16/60) of all ancestral inversions are heterozygous in one or more ape species and are likely polymorphic.
Figure 2 |. Lineage-specific simple inversions and their evolutionary rates.
a, An upsetR73 plot showing the number of shared inversions between members of the great ape family (≥50% reciprocal overlap). Black arrowhead points to putative human-specific inversions. Asterisks highlight inversions with recurrent or incomplete-lineage-sorting signatures. b, Example of a human-specific inversion predicted based on Strand-seq data. Inverted region is highlighted by dashed lines. Human-specific inversion is deemed as a region inverted in all NHPs in respect to the flanking region, but in direct orientation in humans. Inset: Venn diagram showing predicted human-specific inversions with respect to known genome minor alleles/misorients, human inversion polymorphisms4, and already published human-specific inverted loci. c, A tree constructed based on shared simple inversions (≥50% reciprocal overlap) using hierarchical clustering. Each branching node contains a number of shared inversions in a given subtree together with a barplot showing inversion genotypes per individual (B, bonobo; C, chimpanzee; G, gorilla; H, human; O, orangutan). Tips of the tree contain the number of inversions without a significant overlap (<50%) with any other inversion and are likely species specific. Barplot showing inversion genotypes for such species-specific inversions is plotted at each tip of the tree (Methods). d, A rooted MCMC evolutionary tree constructed based on a nonredundant set of 358 autosomal simple inversions among great apes. Inversion rates are reported for each branch as 95% highest posterior density confidence intervals. Numbers at each branching node provide posterior support for this tree topology based on 10,000 MCMC trees sampled from an MCMC chain of 10,000,000 samples constructed from these data.
Using a nonredundant dataset of simple autosomal inversions (n = 358), we also constructed a Bayesian evolutionary tree (Fig. 2d and Methods), and we estimate the rate of fixation of simple inversions as ~7 autosomal inversions per million years of evolution. No ape lineage showed evidence of inversion acceleration, with branch rates ranging from 0.0075–0.0093 inversions per locus (Supplementary Table 4); however, we observe variable inversion rates after accounting for the number of inverted base pairs per single-base-pair substitution (range: 0.05–17.13) (Supplementary Table 4). Interestingly, we identified 27 inverted loci that show evidence of homoplasy (Fig. 2a, black asterisks) either due to recurrent mutation or incomplete lineage sorting. For instance, 5 inversions shared between human, gorilla, and orangutan were absent in the Pan lineage, 11 out of the 27 likely recurrent loci reside on chromosome X, and 85% (23/27) are flanked by known human SDs.
Human polymorphism and inversion hotspots
We compared the 388 nonredundant simple ape inversions (including X chromosome) to 150 simple human inversions recently described for six humans of diverse ancestry4 (Supplementary Fig. 13 and Methods). Strikingly, we found one-third (49/150) of the human polymorphic inversions overlapped with an inversion detected in an NHP (Fig. 3a). Of these, 43% (21/49) mapped to the X chromosome (Fig. 3b, top track) and 31% (15/49) corresponded to the aforementioned recurrent ape inversion sites (n = 27). Notably, more than half (27/49) of these loci were heterozygous in an NHP lineage (Supplementary Fig. 14), evidence of polymorphism across multiple ape lineages. The majority (38/49) of these inversions were flanked by highly homologous SDs (Fig. 3b, bottom track), with 10 of these regions being polymorphic in a larger genotyping panel of the human population32 (Supplementary Fig. 15).
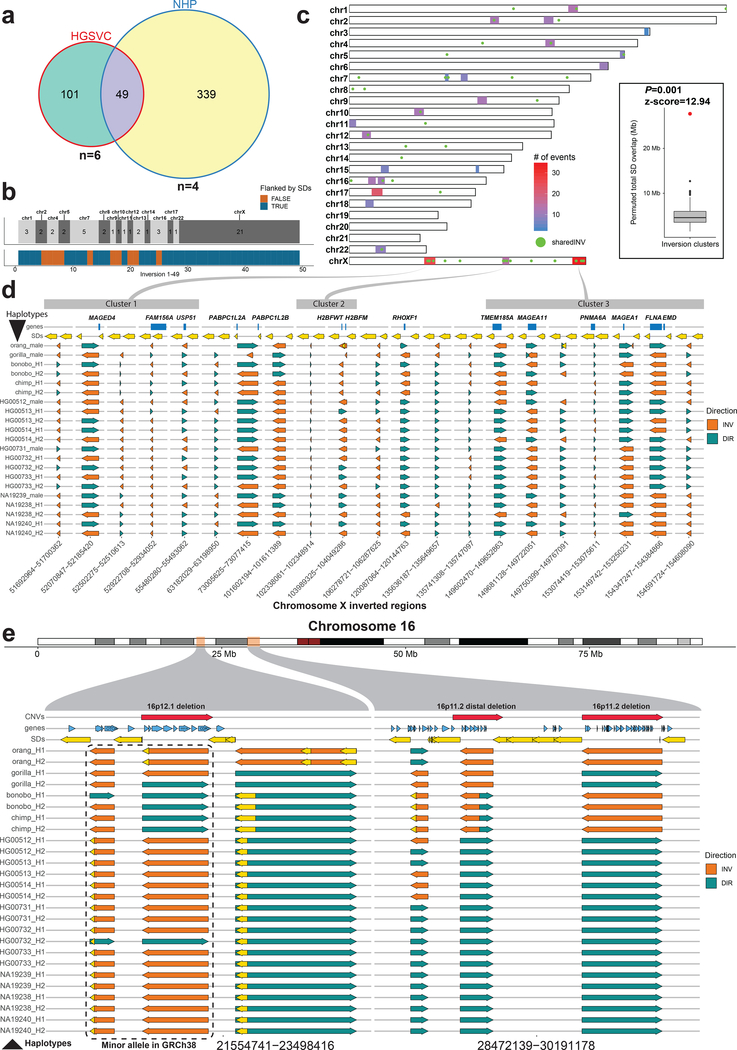
Figure 3 |. Shared inversions and inversion hotspots.
a, Venn diagram showing overlapping simple inversions (50% reciprocal overlap) between HGSVC nonredundant dataset and NHP redundant dataset. b, Top tracks: number of shared inversions between HGSVC and NHP datasets from a shown as counts per chromosome. Bottom track: inversions flanked by segmental duplications (SDs) are colored blue and those not flanked are orange. c, A genome-wide map of detected inversion breakpoint clusters based on simple inversions from HGSVC and NHPs. A set of inversions (n = 49) from a is plotted over this genome-wide map as green dots. Inset: compares the total number of SD base pairs mapping to the 23 breakpoint clusters (red dot, observed = 29,138,268) compared to a random genome-wide simulation (n = 1,000 permutations, RegioneR70 ‘permTEST’, min: 1,653,112, 1stQ: 3,757,630, median: 5,301,424, 3rdQ: 7,084,019, max: 11,940,452). d, Each row represents a haplotype with all tested inversions phased along the whole X chromosome. Inverted direction is shown by an orange arrow and direct orientation by a teal arrow. Top track plots protein-coding genes (blue rectangles) that overlap with either the inversion itself or with flanking SDs, shown as yellow arrows. Previously defined inversion breakpoint clusters are shown as gray rectangles at the top of the figure and are linked to their location on chromosome X in c. e, Each row represents a haplotype with all tested inversions phased along the whole chromosome 16. Inverted direction is shown by an orange arrow and direct orientation by a teal arrow. Top track plots protein-coding genes (blue arrows) that overlap either the inversion itself or with a flanking SD, shown as yellow arrows. Previously published35 pathogenic CNVs are shown as red arrows in the top track.
Inversion breakpoints were not randomly distributed but clustered into 23 discrete genomic regions (median size 5.5 Mb) (Fig. 3c and Supplementary Fig. 16) enriched for human female meiotic recombination hotspots (P = 0.021, z-score = 2.438) (Supplementary Fig. 17a). Twelve of these clusters harbor half (25/49) of the inversions shared between humans and NHPs. As expected, breakpoint clusters are enriched ~5.6-fold for SDs (Fig. 3c inset) with chromosomes 16, 17 and X harboring the greatest number. For example, we observe three distinct inversion clusters on chromosome X that encompass 21 inversions shared between humans and NHPs (Supplementary Fig. 18). Using the phase information embedded in Strand-seq data, we ordered and phased all 21 inversions, along the entire length of the chromosome X (Fig. 3d, Supplementary Fig. 19, and Methods), which revealed a remarkable degree of evolutionary toggling between humans and NHPs with SDs bracketing recurrently inverting regions and frequently containing protein-coding genes (Fig. 3d, top track, and Supplementary Fig. 20). Each human haplotype in these regions shows a unique combination of inverted and directly orientated loci (n = 21) (Supplementary Fig. 21) and is not significantly different from a random inversion state at these loci (Mantel statistic, P = 0.162; low bootstrap support; Supplementary Note). A similar pattern of inversion toggling was observed in two regions on chromosome 16 (Fig. 3e). Interestingly, both X chromosome and the reported regions on chromosome 16 are biased towards female meiotic recombination (Supplementary Fig. 17b).
Because inversion polymorphic regions have been associated with particular recurrent rearrangements8,33,34, we investigated 36 recurrent large-scale copy number variants (CNVs) associated with neurodevelopmental disorders in humans35 and found that 47% (17/36) of these overlap (50% reciprocal overlap) with our map of NHP inversions. This represents a ~14-fold enrichment when compared to a random simulation of pathogenic CNVs (z-score = 17.2, P = 2.09 × 10−66, two-sided, 100 iterations) (Supplementary Fig. 22a, Supplementary Table 5, and Methods). Two of these inversions are classified as occurring specifically in the human lineage, two inversions are known to be polymorphic in humans, while the remaining 13 are observed as simple NHP inversions (Supplementary Fig. 22b). At the species level, orangutan shows the greatest correspondence with nearly 42% (15/36) of recurrent CNVs overlapping an inversion and the highest frequency (0.88) of inverted loci at these regions (Supplementary Fig. 22c). In about half of these cases (8/15), orangutan represents the ancestral configuration based on synteny analysis with macaque and mouse (Supplementary Table 6). Interestingly, most of these CNV hotspot regions are in an inverted orientation in at least one NHP while most human haplotypes are in a direct orientation with respect to the human reference (Supplementary Figs. 23 and 24).
Inverted orientation bias for lineage-specific duplications
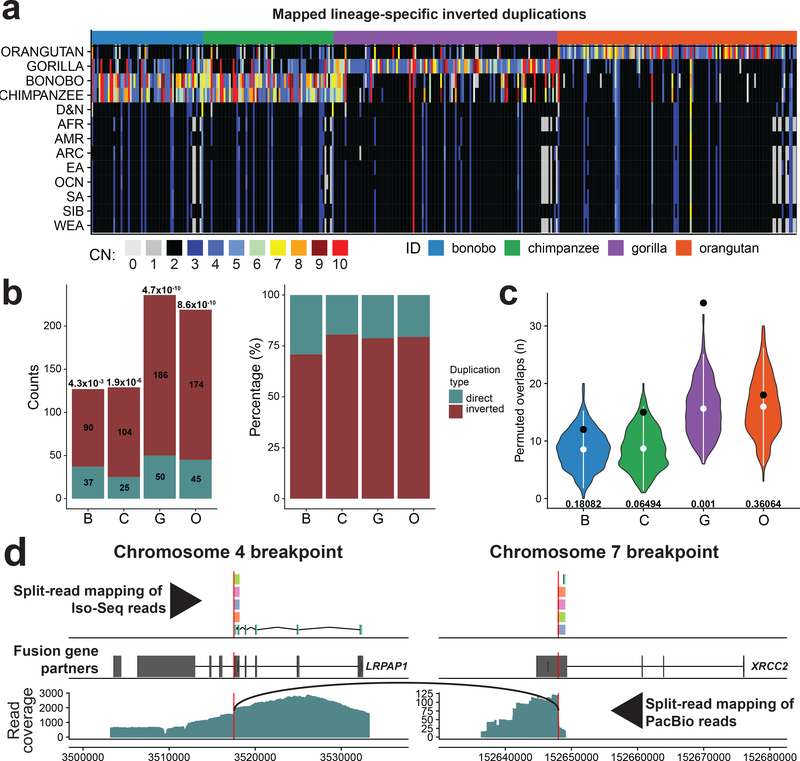
A relatively unique feature of the Strand-seq assay is the ability to distinguish simple inversions from inverted duplications associated with a copy number change4 (Fig. 1b). Unlike simple inversions that accumulate relatively uniformly between ape lineages, we observe a slight excess of inverted duplications in gorilla and orangutan when compared to bonobo and chimpanzee (Supplementary Fig. 25a), although the number of lineage-specific duplications generally recapitulates the ape phylogeny (Supplementary Fig. 25b). Taking advantage of short-read sequencing data from 286 human, ape, and archaic hominin genomes, we genotyped copy number and assayed lineage specificity for 387 inverted duplications (Methods). The majority of orangutan (93%) and gorilla (79%) copy number increases are lineage specific in comparison to the chimpanzee and bonobo, where >50% of the inverted duplications are shared (50% reciprocal overlap) due to their more recent divergence (Fig. 4a and Supplementary Fig. 25d). We highlight a human-specific duplication of GPRIN2 that was recently shown to be missing from the human reference (GRCh38)36 (Supplementary Fig. 26). Using an independent map of great ape–specific duplicatons31,37, we investigated if SDs show a preferential bias in their orientation (Methods). Excluding interchromosomal events (Supplementary Fig. 27 and Methods), we find that ~78% of lineage-specific duplications map in an inverted orientation (P < 0.005, Bonferroni corrected) (Fig. 4b). If we limit the analysis to only those lineage-specific duplications with no more than one or two additional copies (n = 3 or 4 copy number estimate in a diploid genome), this bias remains significant with ~75% of lineage-specific duplications occurring in an inverted orientation. In addition to this orientation bias, it should be noted that we predict an enrichment of inverted duplications mapping near the ends of chromosomes (last 5% of a chromosomal arm) with this difference being the most pronounced in gorilla (P = 0.001) (Fig. 4c)38.
Figure 4 |. Evolutionary impact of inverted duplications.
a, Heatmap of estimated copy number (mean CN) per inverted duplication (columns) in multiple human populations and NHPs (rows). b, Left: number of mapped duplicated regions in inverted versus direct orientation. Significance of observed differences between inverted and direct duplications is reported above each bar as P-value (chi-squared with Bonferroni correction). Right: each bar shows the proportions of inverted and direct duplications per NHP (colored as denoted in a). c, Enrichment analysis of inverted duplication in 0.05 fraction of each chromosome end (1–22 and X). Observed counts are shown by a black dot and the distributions of permuted counts (n = 1,000 permutations, RegioneR70 ‘permTEST’,) are depicted by violin plots. White dots show the mean of each distribution (B-8.54, C-8.69, G-15.6, O-16). At the bottom of each distribution there is a P-value showing the significance of difference between observed (B-12, C-15 G-34 O-18) and permuted counts. d, Predicted gene fusion between XRCC2 on chromosome 7 and LRPAP1 on chromosome 4. Upper track: split-read mappings of Iso-Seq reads over the predicted breakpoint (red vertical line). Iso-Seq reads that belong to the same transcript share the same color. Middle track: gene models of above mentioned genes (Exons, wide boxes; Introns, lines in between). Bottom track: split-read mapping of PacBio reads over the fusion breakpoint on chromosome 4. Black arc line connects ends of PacBio reads with split-read mappings.
Rearrangement and NHP gene expression differences
The association of inversions and SDs creates the potential for the formation of novel fusion transcripts and genes during evolution. We examined all NHP inverted regions searching for the presence of novel fusion genes based on a comparison of long-read genome sequence data and full-length non-chimeric (FLNC) transcripts generated for the different NHP species (Supplementary Table 8). We detected 15 putative fusion transcripts, of which three were further supported by long-read Pacific Biosciences (PacBio) data (Supplementary Table 9 and Methods). We identified a fusion gene specific to the gorilla lineage that was created by inverted duplication and reintegration of a segment of DNA between chromosomes 4 and 7 (Fig. 4d). This fusion is supported by both split-read mapping of FLNC transcripts and long PacBio reads.
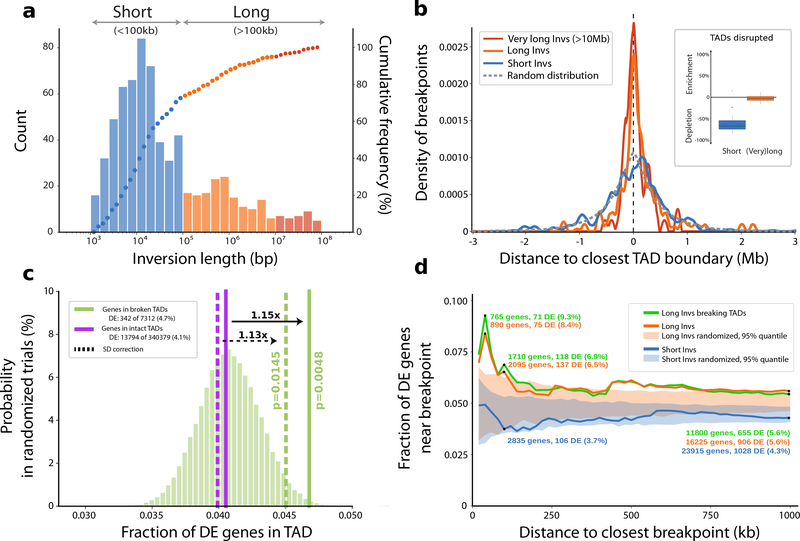
Inversions also carry the potential to rearrange gene regulatory regions and thus perturb gene-enhancer interactions, for example, by disrupting the structure of topologically associating domains (TADs), as previously reported in the context of human diseases39,40 (Supplementary Fig. 28). Notably, we find that breakpoints of larger inversions (>100 kb) tend to co-localize with human-defined TAD boundaries41 (Fig. 5a,b), whereas shorter (<100 kb) inversions do not show such tendency and instead their breakpoints appear to be strongly depleted from TAD boundaries (Fig. 5b, inset). Next, we investigated the effect of these large-scale balanced rearrangements on primate gene expression by analyzing bulk RNA-seq data from 21 human and 47 NHP samples spanning six tissues42. Per tissue, we observed a median of 1,499 differentially expressed (DE) genes in each NHP (compared to the corresponding human tissue). We found DE genes were located more frequently (~1.15-fold increase, P = 0.0048, one-sided permutation test; Methods) in TADs disrupted by an inversion compared to intact TADs that did not contain an inversion breakpoint (Fig. 5c). When testing differential expression with respect to inversion breakpoints, we observe more DE genes near the breakpoints of large inversions (>100 kb), when compared to small inversions (<100 kb) (Fig. 5d). We further investigated this effect using other recently published datasets42–45 with a specific emphasis on brain genes. We preselected protein-coding genes with disrupted gene-enhancer interaction at breakpoints of 388 nonredundant simple inversions. In total, we found 249 candidate genes, of which 102 are DE genes in at least one from the above-mentioned datasets, with 30 genes confirmed by two datasets (Supplementary Fig. 30a, Supplementary Table 10, and Supplementary Note), including neurodevelopmental disease genes (e.g., SETD7 or CTNNA3). In line with the previous analysis46, we continue to observe the trend of more DE genes located near the breakpoints of larger inversions (>100 kb) (Supplementary Fig. 30b, see circle sizes).
Figure 5 |. Impact of copy-neutral inversions on genome topology and differential gene expression.
a, Length distribution of all 387 nonredundant simple inversions, classified as ‘Short’ (<100 kb; blue) or ‘Long’ (>100 kb, orange). The histogram illustrates absolute counts of binned inversion lengths and the overlaid dots represent the cumulative frequency of inversions corresponding to each bin. (bp, base pair; kb, kilobase). b, Distance of each inversion breakpoint (centered at 0) to the closest topologically associating domain (TAD) boundary, stratified by inversion length (color coding according to a). The expected distance distribution for randomly placed breakpoints is indicated by the gray dotted line (Mb, Megabase). The inlay displays the proportion of inversions (stratified by length) that disrupt TADs (median short: −67.1%, median long: −2.4%). Percent ‘enrichment’ or ‘depletion’ is shown as the ratio of observed over expected disruptions calculated after randomizing inversion locations (Methods). c, Proportion of differentially expressed (DE) genes in TADs classified as either ‘broken’ (solid green horizontal line) or ‘intact’ (solid purple horizontal line). The underlying histogram depicts the expected DE frequency after randomizing TAD labels. Dotted lines represent the DE proportion after excluding genes in segmental duplications (SDs). One-sided permutation testing was used to derive P-values (Methods). d, Proportion of DE genes relative to inversion breakpoints and stratified by inversion length or whether the inversion disrupts a TAD. The shaded areas show the expected DE proportion measured in matched randomized breakpoints.
DISCUSSION
Inversions have been long thought to be a driving force in human evolution with the potential to reduce recombination, create fusion genes, and alter patterns of gene expression47,48. We assessed the latter by comparing regions of ape inversion with corresponding NHP bulk RNA-seq data42 with previously defined TADs41. Notably, we observe evidence that large (>100 kb) inversions may mediate gene regulatory changes in NHP evolution, unlike smaller inversions, which rarely associate with NHP gene expression changes. Irrespective of this, inversions are responsible only for a relatively small number of DE changes (~1.15-fold enrichment of DE genes), suggesting more complex gene regulatory relationships46. We further find that 15 out of 26 human-specific inversions have known enhancer regions49 within 5 kb distance (Supplementary Fig. 31a). For example, a human-specific inversion on chromosome 12 repositioned an enhancer in the vicinity of SLC48A1 that was previously shown to be upregulated in human neuronal cells (excitatory and inhibitory neurons) and radial glia44. SLC48A1 shows the highest expression in the spinal cord and enhances tumorigenic functions of non-small-cell lung cancer cells and tumor growth50 (Supplementary Fig. 31b).
Our analysis shows that inversions are among the most biased forms of genetic variation showing a highly nonrandom distribution. The X chromosome is the greatest outlier with approximately 2.5-fold more inversions based on its size and duplication content when compared to ape autosomes (Fig. 1f). This difference is most pronounced for heterozygous inversions suggesting elevated rates of inversion polymorphism for the X chromosome (Supplementary Fig. 9). It has been hypothesized that X chromosome hemizygosity and the absence of male recombination (outside the pseudoautosomal region) may be responsible for the abundance of X chromosome inversions by promoting NAHR for unpaired X chromosomes during meiosis51. It is possible that regions of sex-biased recombination may be particularly prone to inversions if such regions are more likely to fail to pair homologously during meiosis, allowing preferential intrachromosomal or interchromatidal exchange of genomic regions between duplicated sequences. Importantly, regions of inversion toggling, such as chromosome 16, are also known to be hotspots for NAHR associated with recurrent rearrangement, commonly seen in neurodevelopmental delay35 (Fig. 3e, red arrows). It is also interesting that the size distribution of simple inversions on the X chromosome is more tightly distributed than autosomes with an upper limit of ~100 kb (Supplementary Fig. 10). This size constraint may be the consequence of the relatively unique SD organization on the X chromosome, where closely distributed pairwise SDs provide the substrates for NAHR as opposed to autosomes where recent duplications are more interspersed52. Alternatively, selective effects on sex chromosomes may be playing a role53,54 eliminating such events in males.
Within a chromosome, there is also clear regional clustering and we identify 23 discrete regions where we observe an excess of ape inversions. These inversion breakpoint clusters are enriched ~6-fold for the presence of SDs (Fig. 3b, inset) with regions on chromosomes 16, 17 and X showing some of the largest intervals (Supplementary Fig. 22). Interestingly, chromosomes 16 and X are particularly biased for female recombination where genetic estimates suggest a 10-fold reduction in male recombination55 (Supplementary Fig. 17b). Targeted sequencing of large-insert BAC clones from orangutan, chimpanzee, and human confirm an excess of fixed and inverted polymorphisms with breakpoints mapping to these SDs56. Related to this feature, we also observe 27 shared inversions among the different ape species suggesting either recurrent inversions or incomplete lineage sorting during evolution (Fig. 2a)57. Several lines favor recurrent hotspots of mutation—85% (23/27) of these hotspots, for example, are flanked by SDs that would promote recurrent mutation by NAHR. We find that inversions flanked by SDs are much more likely to be polymorphic when compared to ape inversions not flanked by SDs. When we separately analyzed 150 validated human inversion polymorphisms4, we find 33% (49/150) overlap those detected in NHPs with 77% flanked by SDs and many mapping to the predicted 23 inversion breakpoint clusters. Once again, the X chromosome is disproportionately enriched carrying more than a third of these likely recurrent sites (11/27).
Phasing of individual human and NHP haplotypes reveals a remarkable pattern of inversion toggling extending previous observations of individual loci8,51,58 to entire chromosomal regions (Fig. 3d). One of these inversion hotspots on the X chromosome, for example, corresponds to the previously described FLNA-EMD inversion, which has been estimated to have undergone at least 10 independent inversion events based on a comparative sequencing study of 27 eutherian mammals51. The dynamics of recombination, linkage disequilibrium, and allele frequency of such ancient evolutionary polymorphisms can now be more systematically evaluated and is especially interesting in light of the fact that these inversion hotspots frequently contain protein-coding genes (e.g., MAGEA11, H2BFWT, HWBFRM, etc.) (Fig. 3d).
The association between SDs, inversion polymorphisms, and microdeletion and microduplication syndromes is long standing2,4,8,33,34. Recurrent inversions on the X chromosome have also been associated with Factor VIII deficiency observed both in humans and dogs, which appears to be mediated by inverted repeats arising independently or homogenized by conversion at the same regions58. In a few cases where the underlying mechanism has been investigated59–62, individuals carrying inverted haplotypes appear predisposed to higher rates of NAHR either because of SDs evolved in the flanking regions in direction orientation or because SDs become configured to predispose to interchromosomal rearrangement in the heterozygous state34,61. Related to this, one of the important findings of this study is the ability to distinguish simple inversions from inverted duplications by comparing SD and Strand-seq datasets31. In so doing, we determined that the preferred (75%) orientation for emergence of lineage-specific duplications is in the inverted orientation as opposed to the direct. While this is selectively advantageous in the short-term for reducing NAHR-mediated copy number changes, inverted duplications do set the stage for cascading and recurrent inversion toggling leading to simple inversions and ultimately more complex SDs predisposing to recurrent rearrangement and neurodevelopmental disease.
METHODS
Strand-seq library preparation and sequencing
Strand-seq libraries were prepared from B-cell lymphoblastic cell lines previously generated for a female Western chimpanzee (Pan troglodytes; Dorien), female bonobo (Pan paniscus; Ulindi), male Western gorilla (Gorilla gorilla; GGO9), and male orangutan (Pongo abelii; PPY10). All lines were maintained in RPMI-1640 with 10% FBS, 1% Glutamax and 1% penicillin/streptomycin. BrdU (Bromodeoxyuridine; Sigma, B5002) was added to log-phase cell cultures at 40 μM or 100 μM concentrations for a period of 18 or 24 hours. Single nuclei were prepared and sorted using the BD FACSMelody cell sorter into 96-well plates for Strand-seq library production, as previously described22,23. The Strand-seq protocol was implemented on a Biomek FXP liquid handling robotic system, and pooled single-cell libraries were sequenced on the NextSeq5000 platform (MID-mode, 75 bp paired-end protocol). After demultiplexing, Strand-seq reads were aligned to the human reference assembly GRCh38 (GCA_000001405.15_GRCh38_no_alt_analysis_set.fna) using the default parameters of BWA-MEM (version 0.7.15-r1140). Aligned BAM files were sorted by genomic position using SAMtools (version 1.7) and duplicated reads marked using sambamba (version 0.6.6). After alignment, each single library was evaluated to select only high-quality Strand-seq data for downstream analyses. Specifically, libraries with visible background reads (i.e., reads mapped to opposite direction on chromosomes that inherited template strands with the same directionality) and libraries with low (<50,000 reads) or uneven coverage were excluded, as detailed previously23,63.
Inversion detection from Strand-seq data
To increase the sensitivity of inversion calling and inversion breakpoint resolution, we constructed composite files for each individual great ape genome. As previously described4,7, composite files were generated by merging Strand-seq data for each chromosome based on shared strand inheritance patterns in order to produce a high-coverage directional file for the genome. Briefly, we concatenated reads from multiple Strand-seq libraries from each chromosomal region genotyped as either Watson-Watson (WW, inverted orientation) or Crick-Crick (CC, reference orientation) state. To match the reference orientation, we have reverse-complemented regions genotyped as WW prior to merging subsequent Strand-seq libraries. From a composite file, read directionality can then be assigned as either ‘reference’ and in the same (forward) orientation as the reference assembly or ‘inverted’ and in the opposite (reverse) orientation of the assembly. The ape-specific composite files produced in this study are available as UCSC formatted BED files (Data availability).
In each composite file, we then called inversions using the Bioconductor package breakpointR24 (Code availability). We used the ‘runBreakpointr’ function with the following parameters: [windowsize = 10000, binMethod = “multi”, background = 0.1, peakTh = 0.25, trim = 10, zlim = 3.291, minReads = 20, min.mapq = 10]. To run the same version of breakpointR as in this paper, please refer to github (Code availability). Inversion breakpoint resolution highly depends on the underlying genome architecture on each side of the inversion. Because short Strand-seq reads have difficulty mapping within SDs flanking the inversion, the breakpoint is typically placed within the SD range. In such regions, breakpoint prediction is less accurate and thus might not represent the exact breakpoint position. Details of breakpoint resolution achieved by breakpointR have been discussed previously24.
We curated every inversion breakpoint detected by breakpointR manually in the UCSC Genome Browser64 and classified events as simple inversion calls without an evidence for increased copy number (‘INV’) and more complex inversions with increase in copy (WSSD)65 as inverted duplication (‘invDup’). We assigned each inversion a genotype as either ‘HOM’ where vast majority of reads map in inverted orientation (Watson, minus strand) or ‘HET’ where there is approximately 1:1 ratio between reads in inverted (Watson, minus strand) and reference (Crick, plus strand) orientation.
Smaller changes in directionality not detected by breakpointR were included into the final callset only if they were supported either by the PacBio or Illumina callset. Because of limited coverage of Strand-seq data, we excluded simple inversions with less than 1 kb of unique sequence and inverted duplications smaller than 10 kb to ensure the high quality of our callset.
Long-read alignment parameters
Raw PacBio and Iso-Seq reads were obtained from previous studies12 (Supplementary Table 11). PacBio reads were aligned to GRCh38 using minimap2 (version 2.14-r883) using recommended minimap2 parameters by PBSV pipeline (--MD -t 8 -x map-pb -a --eqx -L -O 5,56 -E 4,1 -B 5 --secondary=no -z 400,50 -r 2k -Y). Iso-Seq reads were mapped to GRCh38 using minimap2 (version 2.14-r883) using the following parameters: -ax splice -uf -C5 --secondary=no –eqx.
Inversion validations
Strand-seq inversion callsets were validated using multiple orthogonal datasets, such as PacBio, Illumina, and Bionano optical maps (Supplementary Table 1). We called inversions in PacBio data using PBSV (version 2.0.2) and SNIFFLES (version 1.0.10) with default parameters. We used DELLY (version 0.7.9) to call inversions in short Illumina reads. Bionano inversion calls were obtained using an analysis pipeline provided by the vendor (Supplementary Note). Furthermore, we used previously published and validated NHP inversions12,18,25. We used the primatR package function ‘getReciprocalOverlaps’ in order to find for each Strand-seq inversion the best matching inversion call from any of the orthogonal dataset. Strand-seq inversions having ≥50% reciprocal overlap with any orthogonal dataset were deemed as validated. We attempted to validate the remaining unvalidated inversions by manual inspection of Bionano alignments and by projecting NHP de novo assemblies12 (Supplementary Table 12) against GRCh38 using dotplot analysis. In the case of bonobo, we used long-read data generated from the Mhudiblu cell line examining local assemblies of the inversion breakpoints. Lastly, we attempted to validate a selected number of inversions using FISH (Supplementary Note).
Phylogenetic analyses
In order to identify human-specific inversions and eliminate reference artefacts, we repeated the Strand-seq analysis with data generated for the Yoruban individual NA192404 using the same parameters. Human-specific inversions were defined as regions that are homozygously inverted in all NHPs with respect to a human reference (homozygous reference orientation) and confirmed with NA19240. We also removed all previously reported misassemblies in the human reference (Supplementary Table 13)4. We used 50% reciprocal overlap to delineate shared and lineage-specific inversions among great apes. We constructed a simple matrix where individuals (rows) that share any given loci (columns) based on 50% reciprocal overlap are assigned a value of one; otherwise they are assigned zero. Next, we compute the Hamming distance between all great apes, which is then used by hierarchical clustering to reconstruct the phylogeny purely based on the presence or absence of shared loci. In this analysis we do not take into account heterozygosity.
Estimating inversion rates in the great ape lineage
We computed three different rate estimates: the mean fixation rate of simple inversions per million years, branch rate estimates, and the rate per inverted base per single-nucleotide substitution.
The mean fixation rate of simple inversions per million years assumes a clockwise inversion rate across the great ape phylogeny and thus is defined as the total number of simple species-specific inversions divided by the sum of divergence times (in million years) among species. To infer the branch-specific rates and the phylogenetic relationships among primates of interest using the 358 autosomal inversion calls, we performed the Lewis Markov k model66 implemented in Bayesian phylogenetic-based (BEAST v2.5.0) analyses. We modeled the evolution of individual inversions as changes in separate discrete traits, where each trait has three states: homozygous human reference, heterozygous inverted, and homozygous inverted orientations. To run BEAST, we used Lewis Mk, with GAMMA Category Count=3 for the site model and a random local clock for clock model parameter to explicitly test mutation rate on individual branches in the tree. For tree priors, we used the birth-death model with default parameters but added a prior for the calibration of human–gorilla divergence using a log-normal distribution (M = 2.1, S = 0.085). We performed five independent runs to infer the phylogeny using a chain length of 10,000,000 samples and recorded every 1,000 samples. We used the accompanying program Tracer (v.1.7.1) to determine the quality of each run and used the first 10% as burn-in. All phylogenetic trees were plotted using Figtree (v1.4.3) and DensiTree (v2.2.6).
Finally, to estimate the rate of simple inversions relative to that of single-nucleotide variants (SNVs) on each branch of the inferred phylogeny as proposed by Sudmant et al.31, we computed the rate of inverted bases per substitution for each branch with the following formula: # inverted bases per substitution of a branch = (# total inverted bases on the branch / 2.87 × 109) / the substitution rate of inversion, where 2.87 × 109 is the genome size after excluding simple repeats and the rate of inversion is estimated by BEAST as listed in Supplementary Table 4.
Inverted duplication analysis
Besides inverted duplications, we list the number of direct duplications in the NHP genomes. We did this by scanning Strand-seq composite files in the UCSC Genome Browser and reporting regions of increased read depth (based on WSSD track) and reads mapped preferentially in the reference orientation (Supplementary Table 7). We further genotyped all lineage-specific duplications detected previously31. Of all 11,260 lineage-specific duplications, we retained only regions ≥10 kb that did not appear in humans. Note that a strength of Strand-seq is that it distinguishes directionality of intrachromosomal duplications in the majority of cases, including clustered duplications. In such cases seeing a mixture of direct and inverted reads mapping over the duplicated loci is evidence that at least one copy of this loci is in inverted orientation (Fig. 1b). However, the directionality of interchromosomal duplications is more difficult to reliably assess using Strand-seq. Because strand-state of chromosomes where a corresponding duplication resides might differ within a single Strand-seq library based on assortment, read directionality of these duplicated copies will reflect the strand-state of the chromosome they reside in. To avoid evaluating low-confidence inverted duplication sites, we removed regions that overlap with interchromosomal links predicted by PacBio split-read mappings (see below). This left us with 504 nonredundant regions. Of these, one region failed to lift from GRCh36 to GRCh38 coordinates. Next, we genotyped these regions as HET – heterozygous, HOM – homozygous inverted, or REF – homozygous reference (using primatR ‘genotypeRegions’ function with min.reads=5, alpha=0.05). Last, we calculated proportions between inverted and direct duplications for each NHP and established the significance of this difference using a chi-square test. Resultant P-values were corrected for multiple testing using Bonferroni correction. The same significance tests were repeated with 387 inverted and 88 direct duplications reported in this study (Supplementary Fig. 25c).
We attempted to validate inverted duplications using discordantly mapped BAC-end sequences with signatures of an inversion, deletion, or insertion. Inverted duplications that overlapped with at least 5% of discordantly mapped BAC ends and at least two discordantly mapped BAC ends in total were marked as supported by BAC-end mappings. In addition, we attempted to validate inverted duplications using de novo assembly and SDA36 for each NHP. We aligned assembled contigs against the human reference using minimap2 (version 2.17) to obtain genomic locations where given contigs map. Next, we used nucmer (version 3.1) to align all contigs against specific loci in the human reference with the following parameters: --mumreference -c 100 -g 1000 -l 5. Such alignments were visualized as dotplots and regions showing clear inversion patterns were marked as supported by de novo assembly (Supplementary Table 7).
Mapping inverted duplication loci
To identify the putative integration sites of lineage-specific duplications, we constructed a pseudo mate-pair read using PacBio reads that extend over inverted duplication breakpoints. Specifically, we split individual long PacBio reads using a k-mer size of 2 kb and a step size of 1 kb. For instance, a PacBio read of 12 kb in length is cut such that we initially create a 2-kb portion on the left and leave the rest of the PacBio read (10 kb) on the right. Then we move the cut site by 1 kb to the right, creating the left portion of the PacBio read of 4 kb and leaving the remaining 8-kb portion on the right. We iterated this procedure until the left mate read equals 2 kb (Supplementary Fig. 27a). The resulting pseudo mate-pairs were mapped to the human reference genome (GRCh38) in a paired-end fashion using BWA-MEM (version 0.7.15-r1140) with ‘-x pacbio’ parameter. Discordant read pairs that map to different chromosomal locations point to the sites where duplicated sequences integrate in the genome. We required a minimum of 10 unique PacBio reads to support such interchromosomal connections.
Human inversion callset and overlap
We compared NHP inversion data to a set of 150 human polymorphic inversions identified and phased from three 1000 Genomes Project trios of Han Chinese, Puerto Rican, and Yoruban Ibadan origin4. To detect inverted loci shared between NHPs and humans, we constructed a nonredundant dataset of NHP simple inversions. The set of human polymorphic inversions4 was filtered for events with ≥1 kb of unique sequence. We detected shared inversions between the HGSVC and NHP callsets based on 50% reciprocal overlap. Next, we re-genotyped shared NHP inversions based on Strand-seq composite files and reported the inverted loci frequency for both HGSVC and NHP individuals based on the number of inverted loci (HOM = 2 inverted loci, HET = 1 inverted loci, and REF = 0 inverted loci). To see how many of these regions are flanked by known human SDs, we downloaded a UCSC Genome Browser track of known human SDs and calculated the distance of each inversion breakpoint to the closest SD. We set inversions where both breakpoints are no further than 5 kb away from the closest SD as being flanked by SDs.
Overlap between simple inversions and pathogenic CNVs
The list of human pathogenic CNVs was obtained from a previous study that identified regions showing an excess of large deletions and duplications in cases of pediatric developmental delay when compared to normal population controls35. We searched for 50% reciprocal overlap between pathogenic CNVs (n = 36) and simple inversions (n = 682) using the primatR ‘getReciprocalOverlaps’ function. Those pathogenic CNVs that overlapped with simple inversions have been re-genotyped (primatR ‘genotypeRegions’ function) in all NHPs in order to compute the frequency of inverted loci in these regions (HOM = 2 inverted loci, HET = 1 inverted loci, and REF = 0 inverted loci). To estimate the level of enrichment of pathogenic CNVs in NHP simple inversions, we randomly shuffled these pathogenic CNVs 100 times and each time we evaluated 50% reciprocal overlap. This randomization was performed using the primatR ‘randomizeRanges’ function. Each pathogenic CNV was shuffled within its chromosome of origin and we excluded assembly gaps and centromeres from the randomization process.
Assigning human and NHP inversion to haplotypes
In order to assign all inversions (homozygous and heterozygous) to their corresponding haplotypes, we used phasing information embedded in Strand-seq data63. We used RTG tool67 (RTG Core Non-Commercial version 3.9.1) to call SNVs in Strand-seq data merged in a single BAM file. We used following RTG parameters: --min-mapq 10 --min-base-quality 10 --snps-only --no-calibration --machine-errors illumina --max-coverage 30. After obtaining the set of heterozygous SNVs, we used StrandPhaseR to phase single-cell haplotypes and to split all Strand-seq reads into their respective haplotypes68. Next we used the read-depth profile of haplotype-specific reads in order to assign inverted and reference alleles, in heterozygous conformation, into their respective haplotypes (Supplementary Fig. 19). We visualized the order and orientation of inverted regions using CRAN package ‘gggenes’ (version 0.4.0, https://cran.r-project.org/web/packages/gggenes/).
Fusion gene detection
To detect putative fusion genes, we used a tool called ‘cDNA_Cupcake’69 (https://github.com/Magdoll/cDNA_Cupcake/) and its function called ‘fusion_finder.py’ to perform fusion gene prediction based on recommended settings at https://github.com/Magdoll/cDNA_Cupcake/wiki/. In order to remove excess false positive calls we narrowed down initially predicted gene fusions to only those that lie in the vicinity (+/-1 kb) of predicted simple inversion and inverted duplication breakpoints. We further investigated split-read mapping signatures of Iso-Seq (FLNC) reads that map to different chromosomes of the human reference genome. To reduce the level of false positive calls, we further removed fusion predictions that do not overlap with known genes from the GENCODE database (v29) at both donor and acceptor sites as well as sites that overlap with known SD regions on either donor or acceptor sites. Lastly, we attempted to validate these fusion gene predictions based on PacBio split-read mappings as described in ‘Detection of inverted duplication transposition.’
Defining inverted breakpoint clusters
To detect regions of clustered inversion breakpoints, we merged together nonredundant HGSVC and NHP inversion callsets. We extracted inversion breakpoints for each inversion and submitted a sorted list of inversion breakpoints to the primatR ‘hotspotter’ function24 (parameters: bw=2000000, pval=5e-10). This function searches for regions of increased density of inversion breakpoints around the genome by using the density function to perform a KDE (Kernel Density Estimation). A P-value was calculated by comparing the density profile of the genomic events with the density profile of a randomly subsampled set of genomic events (bootstrapping).
Analysis of TAD disrupting inversions
A set of human-specific TAD boundaries was obtained from the study of Dixon et al.41. Coordinates of these boundaries were translated into the GRCh38 reference assembly using the liftOver tool available from the UCSC Genome Browser. All but one TAD were successfully mapped to the new reference genome (GRCh38). We measured the distance of TAD boundaries to breakpoints of simple inversions (nonredundant set n=388) separately for various inversion sizes (<100 kb, >100 kb and <10 Mb, and >10 Mb) (we excluded inverted duplications as we did not want copy number changes to affect the differential expression analyses). The distribution of distances to the closest TAD boundaries for each inversion size category was drawn as a KDE fitted curve. TADs were further marked as ‘disrupted’ in a scenario when only one breakpoint of a given inversion was positioned within the TAD (Supplementary Fig. 28), otherwise the TAD was classified as ‘intact’. Rates of disrupted TADs for different inversion size categories were examined as follows: the number of disrupted TADs per inversion category was counted and compared to values after inversion positions were randomized within each chromosome (excluding gaps and centromeric regions, and preserving inversion lengths and their relative distances) 100 times using regioneR70 (version 1.16.2) ‘circularRandomizeRegions’ function. This resulted in an estimate for the fold enrichment of broken TADs compared to randomly expected levels.
We further report genes whose differential expression is likely caused by an inversion that disrupts predicted gene-enhancer interaction. A gene-enhancer interaction was considered disturbed if one but not both inversion breakpoints fell between a gene and its associated enhancer. For this analysis, we used gene-enhancer interactions obtained from the geneHancer (v4.8)49 track from the UCSC Genome Browser. Only so-called ‘double elite’ gene-enhancer interactions derived from more than one experimental or computational method have been considered.
Differential gene expression analysis
Our differential expression considered 16,524 1:1:1:1:1 orthologs provided by ENSEMBL v91. We excluded genes that were non-expressed consistently across all samples (fpkm < 1 across all samples and tissues). We also excluded a list of 91 genes escaping X inactivation (obtained from71 due to expected gender-specific expression bias), which left us with 15,117 genes. The level of differential expression per gene was calculated using DEseq272 (version 1.24.0), with gender information included as a cofactor. Gene-wise read counts derived from RNA-seq data were obtained from Brawand et al.42. All NHPs were tested separately against human, resulting in a list of DE genes for each species. We consistently performed between-species DE analyses for matched tissues (e.g., human brain vs. chimpanzee brain, human brain vs. bonobo brain, …., human kidney vs. orangutan kidney). There was no data available for orangutan testis, and accordingly we performed 23 DE comparisons overall (4 species × 6 tissues, minus orangutan testis). Genes with an absolute shrunken fold-change larger than 2 and an adjusted Shannon information value (aka ‘surprisal (s) value’, a standard feature of DEseq2) below 0.005 were considered as ‘differentially expressed.’ Supplementary Figure 29 depicts differential expression in brain as an example. Overall DE levels per ape genome are consistent with the NHP phylogeny and species divergence (Supplementary Fig. 29b).
Differential expression in broken versus intact TADs
Genes were assigned to two groups based on whether or not they fell into a broken TAD (mediated by a balanced inversion), and the ratio of DE genes over total genes was calculated for each group separately. All genes were counted once for each tissue and species, resulting in 15,117 × 23 = 347,691 tests. A permutation test was used to test for statistical significance of the enrichment of DE genes in broken TADs. In 50,000 repetitions, genes were randomly assigned to the two groups (preserving the number of genes in both), and DE ratios were calculated after each permutation. The P-values were derived from the percentile of the observed versus randomized DE ratio. Distances of DE genes to the closest inversion breakpoint were obtained across all genes and for all 23 DE comparisons, and randomization was pursued by shuffling each inversion randomly on the chromosome that inversion had been observed in (shuffling was pursued 1,000 times).
Reporting Summary
Further information on research design is available in the Nature Research Life Sciences Reporting Summary linked to this article.
External datasets
Set of TADs in human41.
Bulk RNA-seq data for all NHPs42.
Set of X-inactivation escape genes71.
Brain organoids sequencing data was obtained from GEO under ID: GSE124299 and dbGaP phs000989.v344.
Raw Strand-seq data for NA19240 can be obtained at ENA (European Nucleotide Archive) under ID: PRJEB12849.
The raw 10X Genomics data are available on NCBI under BioProject PRJNA593056.
CODE AVAILABILITY
primatR package: https://github.com/daewoooo/primatR
breakpointR package: https://github.com/daewoooo/breakpointR, (devel branch)
Custom scripts: https://github.com/daewoooo/ApeInversion_paper/tree/master/Custom_scripts
Software releases at the publication date are available at zenodo, DOI: 10.5281/zenodo.3556774
DATA AVAILABILITY
Strand-seq data aligned to GRCh38 and ape-specific composite files are available at zenodo, DOI: 10.5281/zenodo.3818043
PacBio and Bionano datasets are reported in Supplementary Tables 11 and 14.
Supplementary data: https://github.com/daewoooo/ApeInversion_paper
PacBio and Bionano inversion callset: https://github.com/daewoooo/ApeInversion_paper/tree/master/Supplementary_datasets
Supplementary Material
ACKNOWLEDGEMENTS
We thank T. Brown for assistance in editing this manuscript. In addition, we thank S. Pääbo for generously providing the bonobo (Ulindi) and chimpanzee (Dorien) cell lines used in this study, along with H. Kaessmann and E. Leushkin for access to the ape RNA-seq data. We appreciate technical assistance from A. Pang and A. Hastie who provided Bionano inversion calls for NHPs. We also thank the EMBL Genomics Core facility, particularly V. Benes and J. Zimmermann, for assistance with automating Strand-seq library generation. This work was supported, in part, by grants from the U.S. National Institutes of Health (NIH HG002385 and HG010169 to E.E.E.). A.D.S. was supported by an Alexander von Humboldt Foundation Research Fellowship. P.H. was supported by the NIH Pathway to Independence Award (NHGRI, K99HG011041). A.S. was supported by the NIH Genome Training Grant (T32 HG000035-23). J.O.K. was supported by an ERC Consolidator grant (773026). S.C. was supported by a National Health and Medical Research Council (NHMRC) CJ Martin Biomedical Fellowship (#1073726). E.E.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
COMPETING INTERESTS
E.E.E. is on the scientific advisory board (SAB) of DNAnexus, Inc.
REFERENCES
- 1.Sturtevant AH Genetic factors affecting the strength of linkage in Drosophila. Proc. Natl. Acad. Sci. U. S. A 3, 555–558 (1917). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antonacci F et al. Characterization of six human disease-associated inversion polymorphisms. Hum. Mol. Genet 18, 2555–2566 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaisson MJP, Wilson RK & Eichler EE Genetic variation and the de novo assembly of human genomes. Nat. Rev. Genet 16, 627–640 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaisson MJP et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun 10, 1784 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kidd JM et al. Mapping and sequencing of structural variation from eight human genomes. Nature 453, 56–64 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kidd JM et al. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell 143, 837–847 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanders AD et al. Characterizing polymorphic inversions in human genomes by single-cell sequencing. Genome Res. 26, 1575–1587 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zody MC et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat. Genet 40, 1076–1083 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vicente-Salvador D et al. Detailed analysis of inversions predicted between two human genomes: errors, real polymorphisms, and their origin and population distribution. Hum. Mol. Genet 26, 567–581 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Giner-Delgado C et al. Evolutionary and functional impact of common polymorphic inversions in the human genome. Nat. Commun 10, 4222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tuzun E et al. Fine-scale structural variation of the human genome. Nat. Genet 37, 727–732 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Kronenberg ZN et al. High-resolution comparative analysis of great ape genomes. Science 360, eaar6343 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yunis J & Prakash O The origin of man: a chromosomal pictorial legacy. Science 215, 1525–1530 (1982). [DOI] [PubMed] [Google Scholar]
- 14.Kehrer-Sawatzki H, Sandig CA, Goidts V & Hameister H Breakpoint analysis of the pericentric inversion between chimpanzee chromosome 10 and the homologous chromosome 12 in humans. Cytogenet. Genome Res 108, 91–97 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Kehrer-Sawatzki H et al. Breakpoint analysis of the pericentric inversion distinguishing human chromosome 4 from the homologous chromosome in the chimpanzee (Pan troglodytes). Hum. Mutat 25, 45–55 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Ventura M et al. The evolution of African great ape subtelomeric heterochromatin and the fusion of human chromosome 2. Genome Res. 22, 1036–1049 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucas Lledó JI & Cáceres M On the power and the systematic biases of the detection of chromosomal inversions by paired-end genome sequencing. PLoS One 8, e61292 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Catacchio CR et al. Inversion variants in human and primate genomes. Genome Res. 28, 910–920 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alkan C, Coe BP & Eichler EE Genome structural variation discovery and genotyping. Nat. Rev. Genet 12, 363–376 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasekh ME et al. Discovery of large genomic inversions using long range information. BMC Genomics 18, 65 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feuk L et al. Discovery of human inversion polymorphisms by comparative analysis of human and chimpanzee DNA sequence assemblies. PLoS Genet. 1, e56 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falconer E et al. DNA template strand sequencing of single-cells maps genomic rearrangements at high resolution. Nat. Methods 9, 1107–1112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanders AD, Falconer E, Hills M, Spierings DCJ & Lansdorp PM Single-cell template strand sequencing by Strand-seq enables the characterization of individual homologs. Nat. Protoc 12, 1151–1176 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Porubsky D et al. breakpointR: an R/Bioconductor package to localize strand state changes in Strand-seq data. Bioinformatics 36, 1260–1261 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Szamalek JM et al. The chimpanzee-specific pericentric inversions that distinguish humans and chimpanzees have identical breakpoints in Pan troglodytes and Pan paniscus. Genomics 87, 39–45 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Sulovari A et al. Human-specific tandem repeat expansion and differential gene expression during primate evolution. Proc. Natl. Acad. Sci. U. S. A 116, 23243–23253 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newman TL et al. A genome-wide survey of structural variation between human and chimpanzee. Genome Res. 15, 1344–1356 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shao H et al. npInv: accurate detection and genotyping of inversions using long read sub-alignment. BMC Bioinformatics 19, 261 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rausch T et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28, i333–i339 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng Z et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature 437, 88–93 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Sudmant PH et al. Evolution and diversity of copy number variation in the great ape lineage. Genome Res. 23, 1373–1382 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giner-Delgado C et al. Evolution and functional impact of common polymorphic inversions in the human genome. Nat. Commun 10, 4222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osborne LR et al. A 1.5 million-base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat. Genet 29, 321–325 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giglio S et al. Olfactory receptor–gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am. J. Hum. Genet 68, 874–883 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coe BP et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat. Genet 46, 1063–1071 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vollger MR et al. Long-read sequence and assembly of segmental duplications. Nat. Methods 16, 88–94 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marques-Bonet T et al. A burst of segmental duplications in the genome of the African great ape ancestor. Nature 457, 877–881 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ventura M et al. Gorilla genome structural variation reveals evolutionary parallelisms with chimpanzee. Genome Res. 21, 1640–1649 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spielmann M, Lupiáñez DG & Mundlos S Structural variation in the 3D genome. Nat. Rev. Genet 19, 453–467 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Lupiáñez DG et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012–1025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dixon JR et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brawand D et al. The evolution of gene expression levels in mammalian organs. Nature 478, 343–348 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Sousa AMM et al. Molecular and cellular reorganization of neural circuits in the human lineage. Science 358, 1027–1032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pollen AA et al. Establishing cerebral organoids as models of human-specific brain evolution. Cell 176, 743–756.e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanton S et al. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature 574, 418–422 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Ghavi-Helm Y et al. Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat. Genet 51, 1272–1282 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hey J Speciation and inversions: chimps and humans. Bioessays 25, 825–828 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Navarro A & Barton NH Chromosomal speciation and molecular divergence--accelerated evolution in rearranged chromosomes. Science 300, 321–324 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Fishilevich S et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database 2017, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sohoni S et al. Elevated heme synthesis and uptake underpin intensified oxidative metabolism and tumorigenic functions in non-small cell lung cancer cells. Cancer Res. 79, 2511–2525 (2019).. [DOI] [PubMed] [Google Scholar]
- 51.Cáceres M, National Institutes of Health Intramural Sequencing Center Comparative Sequencing Program, Sullivan, R. T. & Thomas, J. W. A recurrent inversion on the eutherian X chromosome. Proc. Natl. Acad. Sci. U. S. A 104, 18571–18576 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bailey JA et al. Recent segmental duplications in the human genome. Science 297, 1003–1007 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Corbett-Detig RB & Hartl DL Population genomics of inversion polymorphisms in Drosophila melanogaster. PLoS Genet. 8, e1003056 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Natri HM, Merilä J & Shikano T The evolution of sex determination associated with a chromosomal inversion. Nat. Commun 10, 145 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kong A et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature 467, 1099–1103 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Nuttle X et al. Emergence of a Homo sapiens-specific gene family and chromosome 16p11.2 CNV susceptibility. Nature 536, 205–209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fuller ZL, Leonard CJ, Young RE, Schaeffer SW & Phadnis N Ancestral polymorphisms explain the role of chromosomal inversions in speciation. PLoS Genet. 14, e1007526 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lozier JN et al. The Chapel Hill hemophilia A dog colony exhibits a factor VIII gene inversion. Proc. Natl. Acad. Sci. U. S. A 99, 12991–12996 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Itsara A et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am. J. Hum. Genet 84, 148–161 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Antonacci F et al. Palindromic GOLGA8 core duplicons promote chromosome 15q13.3 microdeletion and evolutionary instability. Nat. Genet 46, 1293–1302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mohajeri K et al. Interchromosomal core duplicons drive both evolutionary instability and disease susceptibility of the Chromosome 8p23.1 region. Genome Res. 26, 1453–1467 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maggiolini FAM et al. Genomic inversions and GOLGA core duplicons underlie disease instability at the 15q25 locus. PLoS Genet. 15, e1008075 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Porubský D et al. Direct chromosome-length haplotyping by single-cell sequencing. Genome Res. 26, 1565–1574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kent WJ et al. The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sudmant PH et al. Diversity of human copy number variation and multicopy genes. Science 330, 641–646 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lewis PO A likelihood approach to estimating phylogeny from discrete morphological character data. Syst. Biol 50, 913–925 (2001). [DOI] [PubMed] [Google Scholar]
- 67.Cleary JG et al. Joint variant and de novo mutation identification on pedigrees from high-throughput sequencing data. J. Comput. Biol 21, 405–419 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Porubsky D et al. Dense and accurate whole-chromosome haplotyping of individual genomes. Nat. Commun 8, 1293 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weirather JL et al. Characterization of fusion genes and the significantly expressed fusion isoforms in breast cancer by hybrid sequencing. Nucleic Acids Res. 43, e116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gel B et al. regioneR: an R/Bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics 32, 289–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tukiainen T et al. Landscape of X chromosome inactivation across human tissues. Nature 550, 244–248 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Conway JR, Lex A & Gehlenborg N UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strand-seq data aligned to GRCh38 and ape-specific composite files are available at zenodo, DOI: 10.5281/zenodo.3818043
PacBio and Bionano datasets are reported in Supplementary Tables 11 and 14.
Supplementary data: https://github.com/daewoooo/ApeInversion_paper
PacBio and Bionano inversion callset: https://github.com/daewoooo/ApeInversion_paper/tree/master/Supplementary_datasets