Abstract
Silk spinning offers an evolution-based manufacturing strategy for industrial polymer manufacturing, yet remains largely inaccessible as the manufacturing mechanisms in biological and synthetic systems, especially at the molecular level, are fundamentally different. The appealing characteristics of silk spinning include the sustainable sourcing of the protein material, the all-aqueous processing into fibers, and the unique material properties of silks in various formats. Substantial progress has been made to mimic silk spinning in artificial manufacturing processes, despite the gap between natural and artificial systems. This report emphasizes the universal spinning conditions utilized by both spiders and silkworms to generate silk fibers in nature, as a scientific and technical framework for directing molecular assembly into high-performance structures. The preparation of regenerated silk feedstocks and mimicking native spinning conditions in artificial manufacturing are discussed, as is progress and challenges in fiber spinning and three-dimensional (3D) printing of silk-composites. Silk spinning is a biomimetic model for advanced and sustainable artificial polymer manufacturing, offering benefits in biomedical applications for tissue scaffolds and implantable devices.
Keywords: silk, spinning, biomimetics, self-assembly, sustainability, liquid crystal
Graphical Abstract

Silk spinning by both silkworms and spiders embodies a “living” and largely inaccessible nanotechnology for manufacturing strong materials with eco-friend and energy-minimum processing conditions. To fully unleash the potential of the silk spinning requires continuous biomimetic efforts, which, notably, is beyond the scope of the mere reconstruction of natural hierarchical structures of silks.
1. Introduction
Silk spinning has evolved for hundreds of millions of years and represents a striking engineering marvel.[1–3] Many arthropods spin fibers with silk, representing a large category of proteins with similarities in molecular design. Among these organisms, silkworms and spiders are best-known for using silk for manufacturing protective cocoons and a prey-capturing orb-webs, respectively (Figure 1). Mulberry silkworm (Bombyx mori) cocoons have long been harvested and utilized in weaving textiles with an elegant and glossy appearance, soft feel and durability; silk textiles date back to the second century BC in ancient China (Figure 1B)[4] and remain popular in fashion designs to date. Silk textiles have been important commodities for intercontinental trade, such as the Silk Road until the onset of low cost and versatile synthetic fibers (e.g., Nylon, polyester, others). Similar to silkworm silks, spider silks have been harvested for fishing lines and wound dressings, although they are available in significantly lower quantities due to the cannibalistic nature of spiders and the difficulties of domestication.
Figure 1.
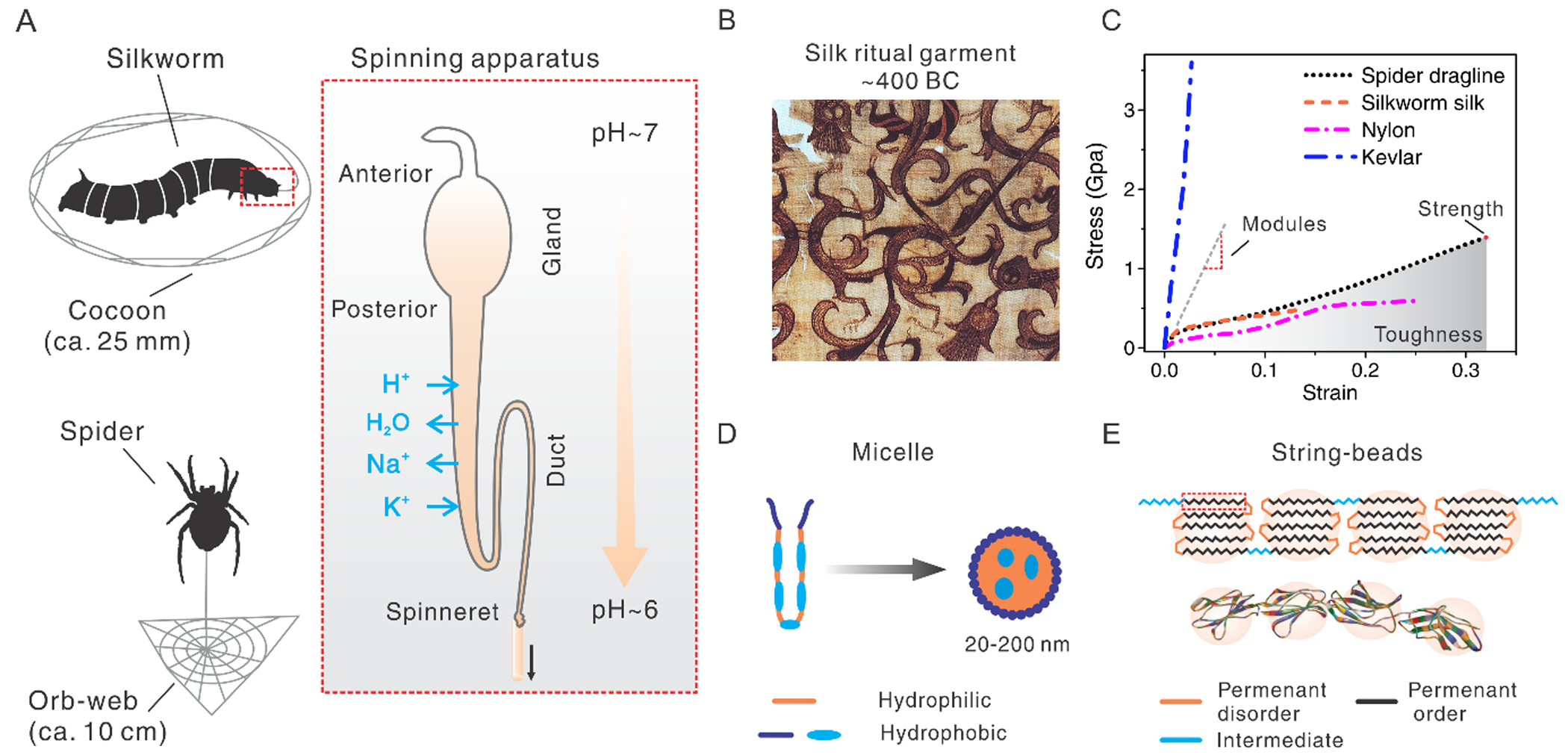
A. Schematic illustration of universal spinning conditions of both silkworms and spiders. Reproduced with permission.[119] Copyright 2019, John Wiley and Sons. B. An embroidered silk gauze ritual garment. Reproduced with permission.[4] Copyright 2019, Wikimedia Commons, the free media repository. C. A brief comparison in tensile performance between silks and other polymers. D. Schematic illustration of silk fibroin assembles into a micelle. Reproduced with permission.[2] Copyright 2003, Springer Nature. E. Schematic illustration of the “string of beads” model to show hairpin folding morphology. This model explains the fractions of disorder and order, as well as the mechanical strength of silk fibers. Reproduced with permission.[52] Copyright 2006, The Royal Society of Chemistry.
Beyond the historical implications of silks in luxury textiles, silks are increasingly recognized as a useful biomaterial with excellent mechanical performance, superior or comparable to a variety of natural and synthetic polymers (Figure 1C).[5, 6] Particularly, spider dragline silk (tensile toughness, ~160 J g−1) is tougher than most commercial high-performance polymers, including Kevlar 49 (~50 J g−1),[7] Nomex (~44 J g−1),[8] Nylon 66 (~80.5 J g−1),[8] polyester (~50 J g−1),[8] and ultra-high molecular weight polyethylene (47–70 J g−1),[9] as well as fibers in development, including multifibrillar polyacrylonitrile yarn (137±21 J g−1),[10] cellulose nanofibrils (~55 MJ m–3)[11] and graphene yarn (11.5–23.75 J g−1).[12] Only a handful of synthetic fibers have been reported to surpass the toughness of the spider dragline silk, including polyvinyl alcohol (PVA)/single-walled nanotubes (570 J g−1),[13] PVA/multiple-walled nanotubes (870 J g−1),[14] PVA/carbon nanotubes/reduced graphene oxide flakes (1000 J g−1),[15] and polyacrylonitrile nanofibers (~500 J g−1 in a diameter of 100 nm).[8] This high toughness of spider dragline silk results from the combination of high strength (~1.4 GPa) and extensibility (~30–150%). High extensibility is unfavorable in maintaining shape, but particularly useful to support loads normal to the long axis of the fiber,[16] for example, flying insects. Thus, silk-like materials are highly desired for a variety of applications, including athletic gear and reinforced polymer composites.[17] In addition, silk is a remarkable biomaterial with inherent biocompatibility and biodegradability, in contrast to most synthetic petroleum-derived materials. Silk consists of amino acid building blocks,[18] thus supporting proteolytic degradation [19] and in vivo compatibility in terms of eliciting low inflammation, low immune responses, and low blood clotting, all desirable features for biomaterial systems.[20] The degradation mechanisms of silk materials have been intensively studied[19, 21, 22] and summarized in a recent review paper.[23] Briefly, proteolytic degradation by the host immune system, especially macrophages and foreign-body giant cells, is key. It should be noted that the β-sheet crystals in the silk materials are different from the pathogenic β-amyloid structures; the silk β-sheet and its degraded product show no cellular toxicity.[21] The biocompatibility of silks provides benefits to a wide range of biomedical applications from tissue scaffolds[18, 24] to bioelectronic devices.[25–31] Furthermore, silks possess other useful material properties, such as optical transparency for applications of bio-optics [32–36] and controlled water uptake related to actuation.[37, 38] The biomedical applications of silk materials have rapidly emerged in recent years, which has been extensively summarized in several excellent reviews.[20, 39–42]
While silk is a remarkable and versatile material, silk spinning as a manufacturing processing, is even more remarkable and scientifically valuable. Silk spinning, in combination with the unique chemistry and sequence in the silk proteins, results in the superior properties of the silk in material formats. Silk spinning is also notably energy-efficient and environmentally benign (sustainability), which represents a fundamentally different mechanism from traditional industrial manufacturing. The all-aqueous and ambient conditions for the silk spinning are in contrast to the high temperature, high pressure, and organic solvent used for the manufacturing synthetic polymers. These non-sustainable conditions of industrial polymer manufacturing are often criticized because of the energy requirements, and adverse environmental impact.[43, 44] In particular, silk spinning uses minimum energy due to the evolutionary pressure of survival. An in vitro experiment of shear-induced fibrillation suggested that silk spinning can be ~90% more efficient in energy use in comparison to processing high-density polyethylene at 125°C.[45]
From the perspective of energy-information-conversion, silk spinning relies on the incredibly rich chemical information encoded into the primary amino acid sequence of the protein in order to optimize self-assembly (thus reduce energy requirements) and promote processing in water. Besides, silk spinning also relies on the information for solvent conditions in the spinning process that is tightly and dynamically regulated by biological systems (especially by the cells lining the gland) (Figure 1A). The silk spinning process is based on directed hierarchical molecular assembly. This evolutionary-based strategy is also found with other structural proteins, including collagenous tendons,[46] cytokeratin hair, and amyloid fibrils.[47] However, there is a notable difference between the manufacturing of silks and other protein structures: silk fibers are spun on-demand in the time scale of seconds or less, while other protein structures are grown most often in time scales of days, weeks or longer. The rapid spinning processing with silks assures the efficiency of manufacturing. Thus, silk spinning provides a particular biomimetic ideal and a source of inspiration for advanced and sustainable artificial polymer manufacturing.[48, 49]
Silk spinning has inspired diverse and substantial efforts towards artificial manufacturing with regenerated silk proteins and even other polymers.[50, 51] To translate silk spinning into artificial manufacturing, the underlying manufacturing mechanisms at the molecular level need to be understood. Several mutually inclusive models have been proposed (Figure 1), including string-beads [48, 52, 53] and micelle models (Figure 1D–E).[2, 54, 55] These mechanisms provided insight into the relationships between molecular conformation and mechanical performance, the formation of crystalline and amorphous regions, the orientation of the molecular chains, as well as the storage of highly concentrated protein solution in a metastable state; however, besides these key aspects, the molecular landscape of silk spinning remains incomplete and thus requires further insight.
In this report, we focus on recent and universal (e.g., silkworms and spiders) mechanistic discoveries regarding silk spinning. We also discuss the current developments towards biomimetic fiber spinning of recombinant and regenerated silks. The technical considerations for preparing silk feedstocks for artificial manufacturing from silkworm cocoons are emphasized. Finally, we discuss the recent development, limitations, and future trends in the 3D printing of silks. Silk spinning is an example of biofabrication[56] and a remarkable source of inspiration, especially for developing sustainable manufacturing techniques with tremendous versatility in material formats, properties, functionalization, chemistry, and opportunities.
2. Natural silk spinning
2.1. Silkworms and spiders
Mulberry B. mori silkworms and golden web spiders are the two predominately used arthropods for studying silk and silk spinning. Because silkworms and spiders have evolved in distinct pathways over millions of years, the comparative study of the two species provides key insights to the general principles that underlie silk spinning. A brief comparison of silk spinning between spiders and B. mori silkworms is summarized in Table 1. There are species-dependent differences in fiber performance as well as the process of silk spinning and. For example, the tensile strength of B. mori silkworm silk is about almost half that of silks from spiders, such as Araneus diadematus and Nephila clavipes. Of note, insect and spider silks have many variants; some of the insect silks (bagworm silks) show extraordinary strength and toughness that are comparable and even superior to most spider silks.[57] Several studies have revealed the critical relationship between the physical properties and the molecular design among silk variants.[16, 58–60] One particular insight is that the design of repetitive sequences determines the size, ratio, and distribution of β-sheet crystallites, thus leading to different mechanical performance.[58] In general, more homogeneously distributed crystallites leads to higher mechanical strength.[58] In addition, the silk spinning gland originates from a salivary gland in silkworms, while this is from the abdominal area in spiders. Despite these differences, however, there is considerable similarity in silk spinning between the two groups of organisms,[61] thus offering a general scientific framework for silk spinning, which is desired for developing biomimetic manufacturing techniques.
Table 1.
Comparison of silk spinning between spiders and B. mori silkworms
| Spider | Silkworm | ||
|---|---|---|---|
| Spinning conditions | Shear stress | Yes (Higher) | Yes (Lower) |
| Kosmotropic ion-Potassium | Yes | Yes | |
| Acidification (pH) | 7.6 to 5.7 | 8.2 to 6.2 | |
| Dehydration | Yes | Yes | |
| Spinning gland | Tapering Geometry (for Shear stress) | 100 μm to <10 μm | 400 μm to 50 μm |
| Carbonic anhydrase and ATPase driven proton pumps (for pH) | Yes | Yes | |
| The microvilli (for removing water) | Yes | Yes | |
| Spinning dope | Concentration | High (>30%) | High (>30%) |
| Liquid crystal spinning | Yes | Yes | |
| Molecular conformation and orientation | Yes | Yes | |
| Repeated motif | AAAAAA | GAGAGS | |
| Beta-sheet crystal size | Smaller | Larger | |
| Polymer melts-like Rheology | Yes | Yes | |
2.2. Spinning dope
The spinning dope is the highly concentrated aqueous solution (>30 wt%) of silk proteins within the spinning gland. Several properties of the spinning dope, including the molecular weight of the proteins utilized, amino acid content and patterns, liquid crystal phases, and rheological behavior, are compared by species and further discussed in the context of polymer manufacturing.
2.2.1. Large molecular weight
Silks are among the largest proteins in nature, with molecular weights larger than 300 kDa. The main structural component of B. mori silkworm silk is fibroin, which is composed of a heavy chain (~390 kDa), a light chain (~26 kDa), and a linking protein, P25 (~30 kDa).[62] In the silk fiber, the silk fibroin is coated with a layer of sericin proteins (~30–400 kDa). The dragline silk of spider N. clavipes is composed of two major ampullate spidroin proteins 1 and 2 (MaSp1 and MaSp2, ~300 kDa each). The large molecular weight (size) is associated with the high tensile strength of silk fibers due to increased interchain interactions and fewer chain-end defects, the same principle as in artificial polymer manufacturing.[63]
2.2.2. Amino acid sequence
Both silks contain short repetitive domains that are rich in glycine (G) and alanine (A), such as GAGAGX for silkworm silks and AAAAAA for spider silks (Figure 1E). Glycine and alanine have the smallest sidechains, a hydrogen and a methyl group, respectively. The small size of the sidechains is beneficial for tight packing or hairpin folding of the chains. The short repetitive domains are hydrophobic and interspersed between smaller hydrophilic non-repetitive domains. Thus, silk proteins resemble amphiphilic block co-polymers, which allows the use of polymer theory to explain the folding and assembly of silk proteins, for example, in the formation of micelles.[2]
The design of the amino acid sequence also determines the fraction of order and disorder[52, 64] as well as the size of the folded molecular structure (the beads),[52, 65] both of which tightly relate to the mechanical performance of the natural and the regenerated silks, as semi-crystalline materials generated from these processes.
2.2.3. Liquid crystal phase
Both native silk dopes of silkworms[66–69] and spiders[68, 70] are reported to include liquid crystal phases (Figure 2); the silk dope is flowable while containing well-aligned molecules in a crystal-like manner. The characteristic birefringence to show the alignment of molecules is observed in the native silk spinning gland[67, 70] and in the nematic schlieren textures of native silk secretions after water-evaporation.[68] These findings imply liquid crystal spinning of silks.[1] Liquid crystal phases are particularly important to manufacture high-performance fibers such as Kevlar and Vectran, providing a precursor to aligned molecular chains.[71]
Figure 2.
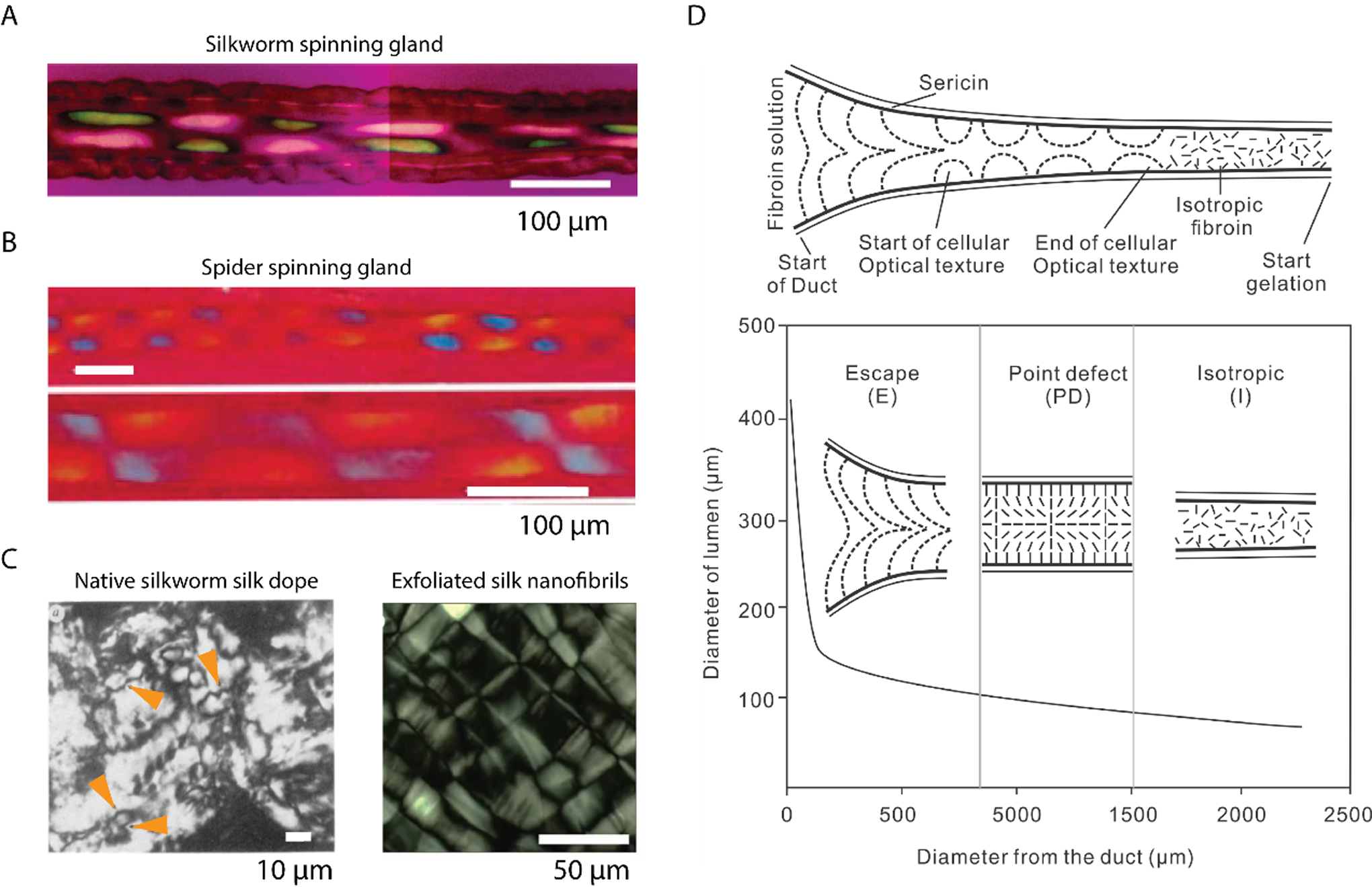
A. Polarizing micrographs show the cellular optical texture of the silk fibroin in situ within the duct. A first-order red compensator is used, and the slow axis is 45° to the longitudinal direction of the duct. Reproduced with permission.[66] Copyright 2007, American Chemical Society. B. Polarizing micrographs show the cellular optical texture of the spider silks in situ within the duct. The slow axis of the first-order red compensator is parallel to the longitudinal direction of the duct. Reproduced with permission.[70] Copyright 1999, The Royal Society. C. Left: a nematic schlieren texture of N. clavipes major ampullate gland secretion after partial drying between a glass microscope slide and a coverslip. Four disclinations are indicated by orange arrowheads. Reproduced with permission.[68] Copyright 1991, Nature. Right: a mosaic lamellar texture of exfoliated silk nanofibrils. Reproduced with permission.[127] Copyright 2017, Springer Nature. D. Texture transformation along the duct section of the silkworm spinning gland with flanked sericin and decreased diameter. There are three textures: escape (E), point defect (PD), and isotropic (I). The PD consists of alternating +1 and −1 point defects separated by a distance that scales with the duct diameter. Reproduced with permission.[72] Copyright 2010, The Royal Society of Chemistry.
Of note, industrial liquid crystal polymers are thermotropic, thus based on temperature and pressure to transit isotropic polymer solutions into liquid crystal phases that are then spun into high-performance fibers. While the native silk dope is a lyotropic liquid crystal, this material relies on solvent conditions to induce the phase transition. By using solvent conditions instead of temperature, lyotropic transitions require much less energy in comparison with thermotropic ones. The liquid crystal spinning of silks was modeled by using nematodynamics, nematostatics, and interfacial thermodynamics, and the resulting semi-quantitative prediction was consistent with the birefringence observed in the native spinning gland (Figure 2D).[72, 73]
2.2.4. Rheological behaviors
Native silk dope is a non-Newtonian liquid and demonstrates two interesting rheological behaviors: 1) shear-thinning[74] and 2) a cross-over point of modulus in frequency sweeps (orange arrows in Figure 3A).[75, 76] Shear-thinning refers to the decrease of viscosity as a result of increased static shear stress; this property is particularly beneficial to extruding viscous liquids from a small die, for example, the fiber spinning of silks, because the viscosity is reduced during extrusion, leading to decreased extruding pressure requirements and thus lower energy expenditures.
Figure 3.
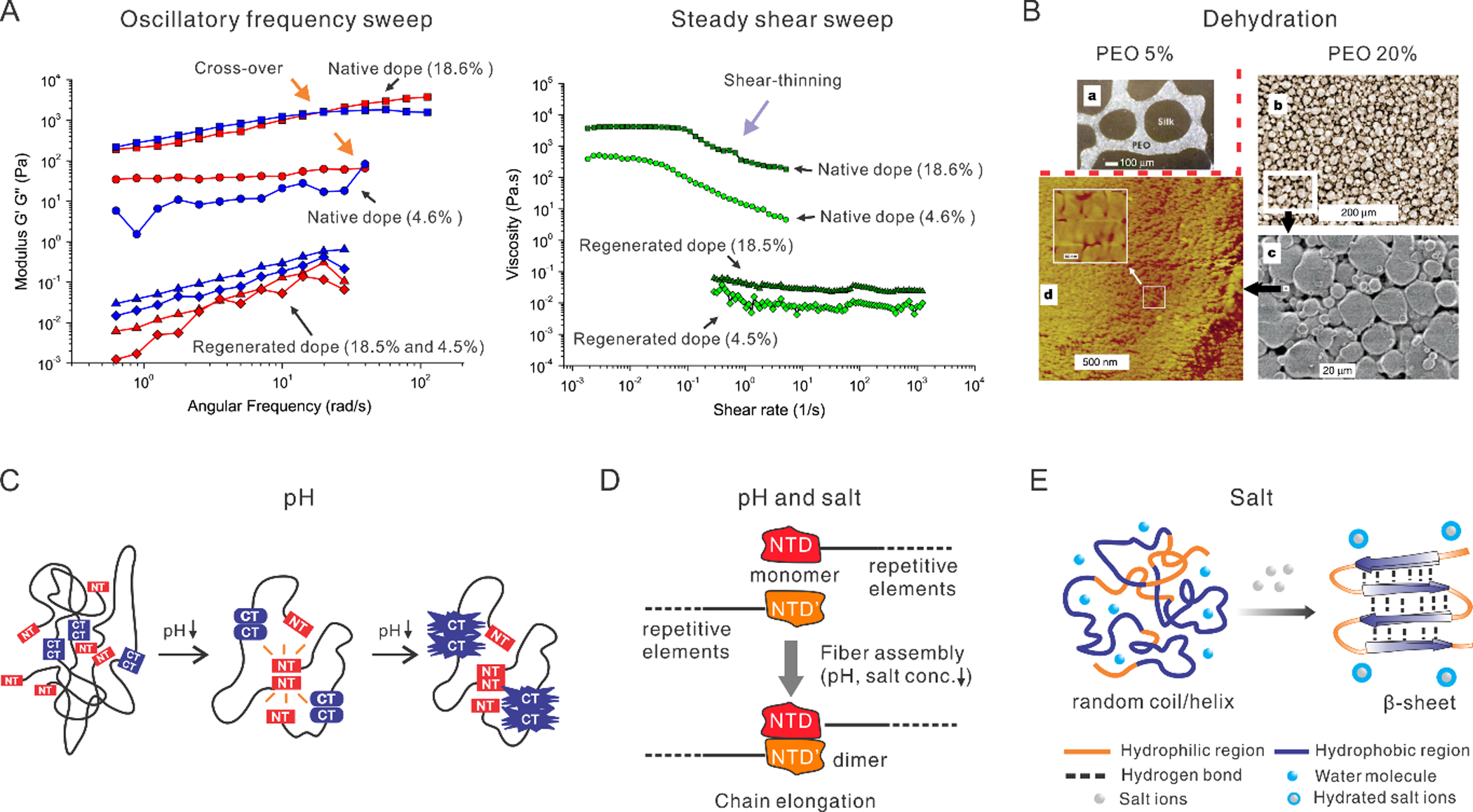
A. Rheological behavior of native and regenerated dopes of silk fibroin. Left: oscillatory frequency sweep. Cross-over points (indicated by orange arrows) exist in the native silk dopes but not in the regenerated silk dopes. Right: steady shear sweep. The shear-thinning behavior (purple arrow) exists in the native silk dopes but not in the regenerated silk dopes. The percentages in the brackets indicate silk protein dry weight concentration in weight per volume. Reproduced with permission.[165] Copyright 2007, John Wiley and Sons. B. An in vitro experiment of the dehydration of silk solution. A higher concentration of polyethylene oxide (PEO) leads to smaller globular structures that are similar to the native silk dope after treating with methanol. Reproduced with permission.[2] Copyright 2003, Springer Nature. C. Schematic illustration of a lock-and-trigger mechanism of spider silk for spinning, where N-terminal (NT) domain, as a lock, initially is dynamic but becomes increasingly stable as pH drops and eventually forms dimers; meanwhile, C-terminal (CT) domain gets destabilized, unfolds and forms amyloid-like fibrils that may trigger fiber formation. The black lines represent the repetitive regions. Reproduced with permission.[97] Copyright 2015, Springer Nature. D. The NT of spider dragline silk shows a pH and salt-dependent monomer-dimer equilibrium. The decrease of pH and sodium ions promote the formation of antiparallel dimers. Reproduced with permission.[102] Copyright 2011, John Wiley and Sons. E. A high concentration of salts captures the water molecules on the surface of proteins, facilitating the inter- and intra- molecularr interaction and the transition from random coil to β-sheet. Reproduced with permission.[104] Copyright 2005, Elsevier.
The cross-over point of silk proteins resembles that of undiluted melts of polymers with high molecular weights.[75, 76] In particular, at the low frequency of oscillation, the loss modulus of the silk dope is higher than the storage modulus, implying a liquid state; while at high frequency, the storage modulus is higher than the loss modulus, implying a solid. Thus, an increase of oscillatory frequency results in fibril formation and gelation of silk proteins from the soluble state.[75, 76] Fibril formation under mild shear forces presents an energy-efficient strategy, in sharp contrast to the fibril formation of synthetic polymers by temperature-driven phase transitions.[45]
2.3. Spinning conditions
Solvent conditions are tightly controlled by cells along the spinning gland to direct the hierarchical assembly of the silk proteins and the change of molecular conformations, which enables the phase-transition from silk dope to water-insoluble silk fibers. The widely accepted conformal change is from random coil/helix (disorder) to β-sheet crystalline (order), while intermediate conformation silk I (β-turn) is also proposed through an extensive investigation of the structure using nuclear magnetic resonance (NMR).[77]
The role of solvent conditions in the silk spinning is as important as the information encoded in the amino acid sequence of the proteins. Of note, several conditions appear to be universal during silk processing in the native systems when comparing among spiders and silkworms, including mechanical forces,[75] acidification,[78, 79] gradients of salt ions (like potassium and sodium)[54, 80, 81] and dehydration.[2]
2.3.1. Mechanical forces
There are two distinct mechanical forces involved in silk spinning. The first is shear forces that are generated by the flowing silk dope within the spinning duct. The spinning duct has a gradual decrease toward the spinneret in terms of diameter from ~100 μm to <10 μm in spiders,[82] and from ~400 μm to ~50 μm in silkworms.[66] Despite the different sizes, the change in diameter is similar in both silkworms and spiders, fit by the same second-order exponential decay.[66] The range of spinning speed is around 10–20 mm/s in both spiders and silkworms. Shear stress above a critical level of around 1–10 s−1 [83] is believed to extend and align the protein chains and induce fibril assembly[84, 85] and beta-sheet formation,[86] as the extended molecules tend to expose their hydrophobic domains to enable interchain interaction.
The other mechanical force is the pulling force imposed on nascent silk fiber immediately after exiting the spinneret. The pulling force is primarily generated by the bodyweight for the dragline of the spider and the head movement for B. mori silkworm silk. The pulling force is believed to foster the alignment of the backbone and side-chains of silk protein molecules along the longitudinal direction, crucial to mechanical performance.[87]
The exact roles of the shear forces and the pulling forces in silk spinning remain elusive. The pulling force has been suggested to be more dominant than the shear force; however, the shear force still seems necessary for the silk spinning. Recently, simulation and experimental evidence showed that the silk fiber is mainly pulled instead of pushed (sheared).[88] In one study, the head of the silkworm was fixed to remove the pulling force by head movement.[67] As a result, the silkworm extruded liquid instead of spun fibers. The extrusion of liquid may be achieved by two pairs of muscles around the middle part of the spinneret, which are the only muscles found around the spinning gland. In addition, the absence of muscles in the peristalsis of the spinning gland also suggests the dominant role of pulling forces in the silk spinning.[88]
2.3.2. pH
Along with the spinning gland in both silkworms and spiders, the pH gradually decreases, i.e., acidification. In the silkworm, the pH decreases from 8.2 to 6.2;[78] while in spiders, from 7.6 to 5.7.[89] The maintenance of this pH gradient is likely through epithelial cellular secretions of proton and carbonic anhydrase that converts water and carbon oxide to carbonic acid.
Several in vitro experiments indicated that pH influenced the stability and dimerization of the non-repetitive terminal domains (Figure 3C),[78, 90] molecular conformation [91], and rheology [92] of silk proteins. The pH-induced effects are usually relevant at low isoelectric points (PI) and the presence of amino acids with acidic side chains. For example, B. mori silk has a low PI of 4.2, and the acidic amino acids mainly exist in the N-terminal domain as well as in the multiple interspersed hydrophilic domains. At neutral pH, the acidic amino acids have negative charges, which prevents interactions between protein chains; while at lower pH, the electrostatic repulsion becomes less effective, permitting hydrophobic interactions and facilitating conformational changes and gelation.[92] The programmed acidification along the spinning gland has been suggested as a lock-and-trigger mechanism for the on-demand formation of silk fibrils (Figure 3C).[79] Here, the incorporation of the N-terminal domains from the spider Euprosthenops australis prevented the formation of fibrils at high pH, while allowing fibrillation at low pH. The fibrillation was triggered by the assembly of the N-terminal domains into dimers.
2.3.3. Salt ions
Salt (metallic) ions have significant effects on the folding and assembly of a variety of proteins, especially silk proteins, interacting with hydrated surfaces of the backbone and side chains of proteins.[81] According to the capability to capture water molecules from proteins, common salt ions are compared in the Hofmeister series,[93] and categorized into kosmotropic and chaotropic ions. For most proteins, kosmotropic ions facilitate aggregation (salting out), while chaotropic ions promote dissolution (salting in).
In the spinning glands of both spiders and silkworms, several metallic ions are involved: Na+, K+, Mg2+, Ca2+, Cu2+, and Zn2+. The two most abundant metallic ions are potassium and calcium in silkworms, as well as potassium and sodium in spiders. Along with the spinning gland, the content of kosmotropic ions such as potassium increases, while chaotropic ions such as sodium decrease (Figure 3D).[94, 95] From the silk dope within the spinning gland to the silk fibers of the spider N. ephila, K+ increases from 750×10−6 to 2900×10−6 mol/L and Na+ decreases from 3130×10−6 to 300×10−6 mol/L.[96] The opposite variation of kosmotropic and chaotropic ions along the spinning gland is desired for the silk spinning, in order to promote the gradual aggregation of the protein. It is noteworthy that restricted by current analytical techniques, the exact concentration of salt ions in the spinning duct remains largely unknown.[97] The exact mechanism of the salt effects in silk spinning are challenging to understand using conventional techniques, such as circular dichroism, NMR and Raman spectroscopy,[94, 98–100] because of the high concentration of the silk dope, as well as the large size and the repetition of short domains of the native silk proteins.
Nevertheless, some insights have been reported. A series of studies showed the effect of salt ion on the storage and fibril assembly of silk proteins.[54, 101–103] For example, high ionic strength of potassium (>1 M) generated silk particles while low ionic strength gave rise to bulk gelation. Sodium (up to 500 mM) helped to solubilize silk proteins against thermal and chemical denaturation and allowed long-term storage of the silk dope. By contrast, Na+ over the same range of concentrations (up to 400 mM) promoted the aggregation of silk fibroin and the generation of β-sheet crystallinity. The seemingly conflicting results are likely attributed to the modes of preparation of the silk samples. In the former work, the silk sample was prepared as a solution and then stored, while in the latter work, the silk sample was dried completely to form a solid film. Of note, the drying (evaporation of solvent) elevates the concentrations of the silk protein and the salt. Thus, in the silk film, the concentration of sodium can be much higher than in solution; at high concentration, sodium effectively changes the molecular conformation and aggregates the proteins, as demonstrated by salt-leaching to generate silk sponges (Figure 3E).[104]
2.3.4. Dehydration
The solid content (silk proteins) in the silk dope increases along with the spinning gland from ~12 wt% in the posterior division, to ~30 wt% in the anterior division,[105] and ~100% in the silk fibers (depending on environmental humidity).[106] The enrichment of silk proteins, accompanied by the removal of water, i.e., dehydration, is achieved by two steps, active reabsorption of water within the spinning gland and passive evaporation in the air after exiting the spinneret. The apical microvilli of cells lining the spinning gland increases surface area and favor dehydration. An in vitro experiment showed that dehydration by polyethylene oxide (PEO) influenced the size of the globular structures assembled by silk fibroin (Figure 3B).[2] Moreover, dehydration was not restricted within the spinning gland; as by infrared spectra, the water content of the immediately spun (<1 second) silk fiber was close to the unspun silk dope.[107] Because some silks are spun underwater,[108] dehydration may not be a prerequisite for silk spinning; however, the water content of the silk fibers relates to the mechanical performance of silks.[109, 110]
3. Artificial spinning of silks
Natural silk spinning has inspired many efforts towards developing fabrication techniques for artificially manufacturing silk proteins as well as other synthetic polymers.[50, 51, 111, 112] These techniques include fiber spinning,[113–116] casting/molding,[104] lithography with electron beam,[117, 118] and 3D printing/additive manufacturing.[119–122] Among these techniques, fiber spinning is the most widely explored due to the similarity to the natural spinning process. A brief comparison between the artificial and natural spinning of silk is illustrated in Figure 4. Despite these efforts, the artificially spun fibers remain mechanically inferior to spider dragline silks and suffer from heterogeneity.[123] In addition to fiber spinning, other techniques for manufacturing silks have demonstrated unique features, as reviewed in the context of microfabrication[124] and nanomaterials.[125]
Figure 4.
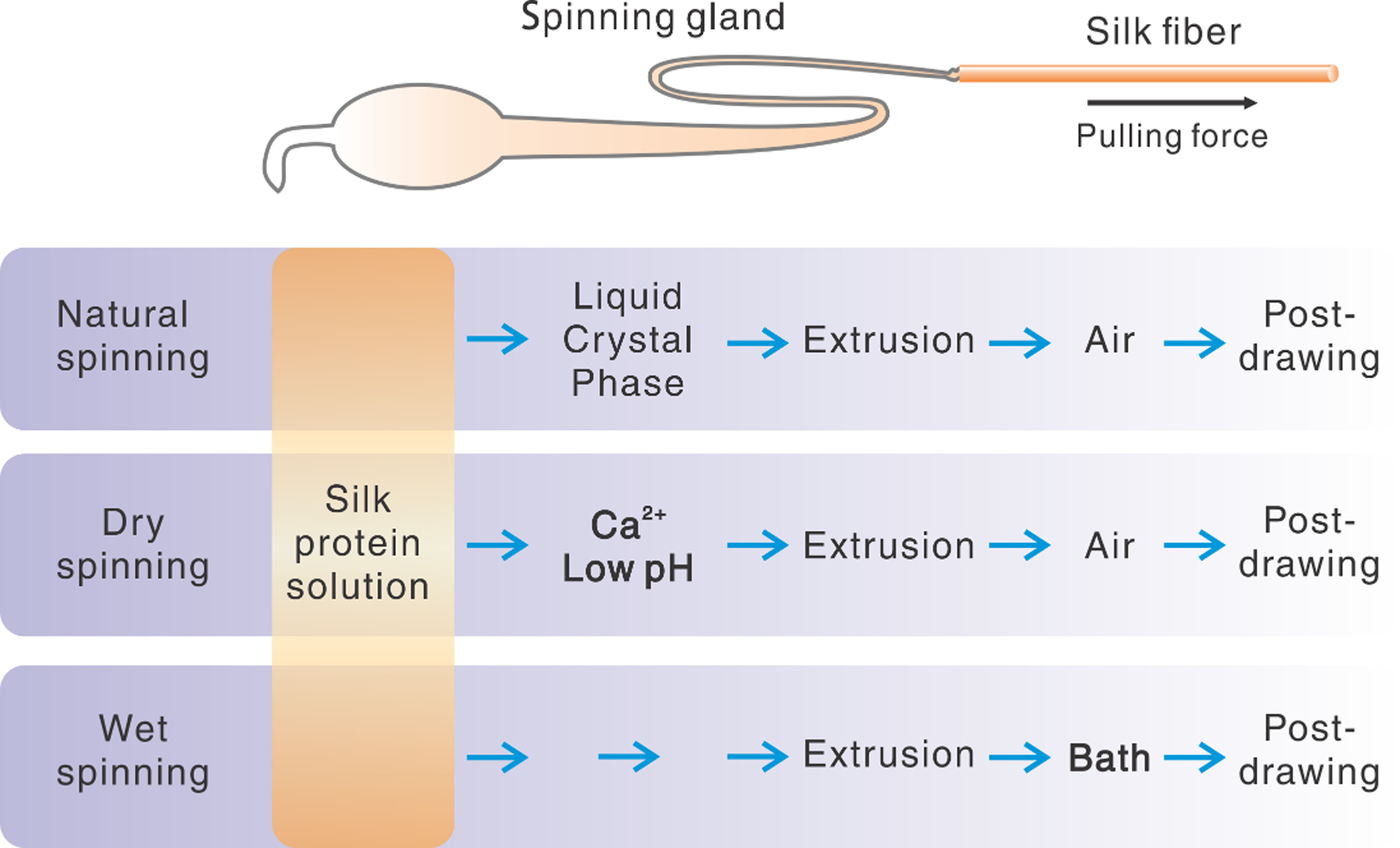
Comparison between natural and artificial silk spinning in stepwise procedures.
In this section, we will emphasize the technical considerations for preparing silk feedstocks and the reconstruction of natural spinning conditions.
3.1. Silk feedstocks
Silk feedstocks from different species of silkworms and spiders require different preparation techniques. Domesticated B. mori silkworms are the largest and probably the easiest source for preparing silk feedstocks because sericulture worldwide produces a massive number of cocoons. The silkworm cocoons are sequentially degummed and dissolved to regenerate silk solution feedstocks. Degumming refers to boiling cocoons in alkaline solutions to remove the coating layer, including waxes and sericins; the degummed silk is then dissolved in hydrogen-bond-destroying solvents, such as 9.3 M lithium bromide solution.[126] The chemicals used in the degumming and the dissolution are removed by dialysis against DI water to obtain a solution of silk proteins. An alternative to the solution of silk protein (molecule), solutions of silk nanofibrils can also be prepared and used for spinning fibers. These structures are prepared directly from silkworm cocoons by exfoliation in either hexafluoroisopropanol (HFIP)[127] or CaCl2-formic acid.[128] Of note, the spinning of silk nanofibrils is not covered here, as the spinning mechanism is different from native silk spinning but similar to the spinning of cellulose nanofibrils.[129, 130]
The feedstocks of spider silks are prepared by genetic engineering techniques due to the difficulties to domesticate spiders. The spider silk feedstocks are usually generated through a range of hosts, including Escherichia coli., plants, mammalian cells, yeasts, and transgenic animals.[131–135] Detailed protocols for producing recombinant spider silks for artificial spinning have been presented.[136] In addition, transgenic silkworms encoding chimeric silkworm/spider silk genes have been developed to spin artificial fibers that are either stronger[137] or tougher[138] than their natural counterparts.
3.1.1. Molecular weight and concentration
It has been a long-standing interest to generate silk feedstocks with high molecular weight (MW) and high concentrations in aqueous solutions, related to the mechanical strength of artificially spun fibers.
For recombinant spider silks, the large gene sizes are restricted due to the instability of long and repetitive DNA sequences in heterologous hosts, low efficiency of RNA translation due to complex secondary structures, as well as the high demand for specific amino acids in the silk sequences, such as glycine and alanine.[139] Metabolically engineered E. coli can address some of these limitations and have been used to produce native-sized recombinant spider silk proteins (250–320 kDa), leading to 20% w/v solutions in an organic solvent, HFIP for spinning fibers.[132] A synthetic biology approach, combining standardized DNA assembly and split intein-mediated ligation, was utilized to produce recombinant spider silks with an even higher MW, 556 kDa.[140] They also prepared silk feedstocks in HFIP (17% w/v), the artificial silk fibers spun from these proteins were mechanically comparable to natural spider silks.
Of note, HFIP was used for dissolving the recombinant silks, because of their low solubility in aqueous solution (0.4–2% w/v).[141, 142] The low aqueous solubility, partly resulting from the deviation from the conformations of native silk proteins, represents a challenge to the preparation of spider silk feedstocks. The use of organic solvents, such as HFIP, significantly alters the spinning conditions as well as compromises the sustainability of the process and potentially the biocompatibility of the materials. One solution to increase the solubility is to fuse the recombinant spider silk proteins with a highly hydrophilic domain. For example, a recombinant chimeric spider silk protein was generated containing the N-terminus (NT) from E. australis, major ampullate spidroins (MaSp1) and a C-terminus (CT) from Araneus ventricosus minor ampullate spidroins (MiSp), bracketing a short repetitive region from E. australis (NT2RepCT). The recombinant protein was soluble in aqueous solution at a high concentration up to 500 mg/ml (>50% w/v)[114] due to the solubility of both the NT and CT domains.
For silkworm silks (fibroin), MW is inherently smaller than the natural counterparts, because both degumming and dissolution cleave backbone peptide bonds and thus reduce the MW. Also, cleavage occurs at random sites, which broadens the polydispersity of the protein. Of note, dissolution seems to have a lower impact on the reduction of MW in comparison with degumming.[143] Nevertheless, by tuning the processing conditions, such as the processing time and the reagents, the MW can be controlled.[143, 144] For example, longer boiling time during degumming corresponds to lower MW, and vice versa (5 min, ~300 kDa; 30 min, ~100 kDa and 60 min, ~50 kDa).[144] Normally, regenerated silkworm silks have a much higher solubility than recombinant spider silks. After dialysis, the concertation of silk fibroin is around 8–10% w/v. Two techniques, reverse dialysis against polyethylene glycol (PEG) solution[145] and air drying in cool and humid environments,[146, 147] are often used to achieve higher concentrations, such as 30% w/v and above. Of note, the concentration of silk proteins impacts gelation rate and viscosity related to spinning, constituting challenges in reproducible results in processing and spinning. Na+ and Ca2+ ions delay gelation and are useful to achieve high concentrations of silk dopes for spinning.[54, 146] Moreover, the concentration of silk fibroin solution is inversely related to the MW, a trade-off as both concentration and MW are favorable for mechanical performance.
3.1.2. Processing biocompatibility
All-aqueous processing is a characteristic of silk spinning and desired for the artificial manufacturing of polymers. This water-based process also helps the integration of functional biomolecules into the spun silk structures.[148] Because biomolecules are often sensitive to denaturation or loss of activity in organic solvents, the use of toxic organic solvents, such as HFIP,[149] formic acid[150, 151] and methanol,[152, 153] should be minimized to achieve these goals. Lithium bromide, as an inorganic salt, dissociates completely in water/body fluids into lithium and bromide ions, implying the convenient and complete removal by water extraction during the preparation of silk materials. Second, lithium and bromide are essential micronutrients to humans. For example, bromide ion exists in seawater (65–67 mg/kg),[154] plants (8–43 mg/kg dry weight)[154] as well as human blood (5.3 ± 1.4 mg/L, whole blood).[155] Lithium is found naturally in the aquatic and terrestrial environment.[156] The recommended total daily intake of bromide and lithium is 24 mg/person per day[157] and 1 mg/ day for a 70 kg person,[158] respectively. Overall, lithium bromide shows very low acute toxicity upon oral administration[157] and low environmental toxicity and low bioaccumulation in the human body,[156] in sharp contrast to heavy metals of public health significance, including arsenic, cadmium, chromium, lead, and mercury.[159]
Alternatively, lithium bromide can be replaced with other solvents that destroy hydrogen-bonds,[143] including ternary solutions (Ca2+: water: methanol)[123, 152, 153, 160] and ionic liquids.[143, 161] All of these solvents, especially the non-organic solvents, are useful for wet-spinning[123, 149, 160] and electro-spinning of silks.[151, 162] Thus, there are multiple options for the dissolution of the degummed silks for meeting different levels of biocompatibility and environment protection.
3.1.3. Rheological characterization
Rheological characterization of regenerated silk feedstocks provides a quantitative manner to measure the interactions between the chains and to compare to the native silk dope.[119, 163–169] However, rheological results can be inconsistent. For example, low concentration silk feedstocks (3.8–5%) show similar viscosity at the shear rate of 1 (s−1): 0.03 Pa·s,[164] 0.01 Pa·s,[165] and 0.02 Pa·s.[120] However, only two of the results showed shear-thinning behavior;[120, 164] while the third did not.[165] For highly concentrated silk feedstocks (29–30 wt%, close to the concentration of native silk dope), similar inconsistencies exist. The shear-thinning effect of the silk solution was negligible in the two studies[120, 168], while in other studies, this effect was significant.[119, 169] Of note, native silk dope shows a considerable shear-thinning effect.[74] According to classical polymer rheology theory, shear-thinning refers to the decrease of viscosity under increase shear stress.[170] The strength of the molecular interactions determines whether the shear stress can disentangle and align molecules, which in turn allows molecules to slide past each other easily, lowering the overall viscosity.
We thus attribute the conflicting rheological behaviors reported in various studies to the different states of aggregation of silk protein molecules, which may be associated with different silk solution preparation and storage times and methods. The two studies reporting the negligible shear-thinning effects used reverse dialysis against concentrated PEG/PEO solutions;[120, 168] while the other studied used cooled airflow.[119, 169] A major difference between these two methods is the dynamics of water removal: the former is rapid while the latter is slow. The rapid removal of water may promote the gelation of the silk feedstocks.
Regenerated silk feedstocks are often dissimilar to native silk dopes in terms of rheological behavior. For example, the characteristic cross-over point between elastic and viscous modulus,[165] as well as the shear-induced nanofibrils.[171] These rheological features are relevant to fiber spinning; the cross-over point implies the transition from solution to gel, and shear-induced fibrils may be the precursors for the silk fibers. The differences in rheological behavior imply the importance of the reconstitution process of silk feedstocks in analogy to native silk dopes. As mentioned earlier, the native silk dope contains specific pH, salt ions and water content, which are known to influence rheological behavior [119] and fibril formation,[54] but are often missed in the silk feedstocks utilized in rheological studies as well as the artificial spinning of silks.
3.1.4. Other considerations
Both spider and silkworm silks have been widely adopted for making silk feedstocks for artificial manufacturing structures and devices. However, the inferior mechanical strength and toughness of natural silkworm silks in comparison to spider silks is a common question (Figure 1C). This difference, however, may be due to the distinct spinning processes, such as pulling (spinning) direction and speed. Silkworms pull the silk by the head movement in the eight-figure with multiple turnings,[52] leading to periodic weak points in the fibers (as seen chemically via alkaline digestion[172] by contrast, spiders pull dragline silk in a straight manner, thus avoiding fiber defects. In addition, silkworms pull the silk out at a relatively low speed, around 9.5 mm/s.[173] By increasing the pulling (reeling) speed to 27 mm/s, the strength of silkworm silks was comparable to that of spider silks.[174] Thus, silk feedstocks from either silkworm silks or spider silks should be readily useful for artificial manufacturing of strong and tough structures, as long as the spinning direction and speed are optimized.
3.2. Spinning conditions
3.2.1. solvent conditions
The spinning techniques of the silk feedstocks are often classified into two groups, wet and dry spinning, mainly based on whether a coagulation solvent bath is used (Figure 4).[123] In drying spinning, the silk feedstock, i.e., the solution of pure silk proteins, is first constituted by adding calcium chloride and lowering the pH, and then extrusion into the air;[175–177] the transition from the liquid solution to solid fiber is often achieved by the evaporation of the solvent. For wet spinning, the spinning dope of silk solution is first extruded into a “coagulation” bath to achieve the phase transition, followed by the combination of several post-treatments, including water rinsing, post-drawing, heating, and air drying.
For wet spinning, the coagulation bath usually consists of ammonium sulfate[115, 178, 179] and alcohol (methanol and isopropanol);[132, 134, 140, 180–183] The two reagents are used to precipitate proteins by salt and solvation effects, respectively.[184] The salt ions or miscible organic solvent added in the protein solution remove water from the surfaces of the silk proteins, i.e., hydration layer, which favors inter- and intra-chain interactions leading to aggregation. The role of potassium and sodium in silk spinning can be roughly explained by the Hofmeister series, as mentioned earlier.[185, 186] These salt and solvation effects are involved in native silk spinning; however, the ammonium sulfate and alcohol solutions are not present in native spinning, which highlights the distinct solvent conditions and, perhaps, different mechanisms between the artificial and native spinning. Particularly, alcohol is an organic and toxic solvent, thus challenging for cell-based spinning and also for large-scale industrial applications. In addition, methanol rapidly removes water from the surface of silk proteins and generates β-sheet crystals, in sharp contrast to the slow removal of water (such as water annealing) that leads to intermediate β-turn structures.[187, 188]
Besides the use of ammonium sulfate and alcohol for silk spinning, other aqueous solutions are less frequently used, including sodium bisulfate,[189] zinc and ferric ions,[183] and sodium acetate.[114] Sodium bisulfate was used to spin cellulose fibers and silks. Zinc and ferric ions were chosen on the basis of experimental optimization in terms of silk phase-transitions and solidification. Sodium acetate was recently employed to mimic the acidification along with the spinning gland. The resulting as-spun silk fibers were almost as tough as native silkworm silks. Of note, the solution of sodium acetate does not mimic other spinning conditions, as discussed earlier, such as ion effects and dehydration.
3.2.2. liquid crystal precursor
Despite the observation of liquid crystal phases in the native silk spinning glands of both silkworms and spiders, the experimental utility of this phase in regenerated and recombinant silk feedstocks remains limited. However, without showing liquid crystal phases during artificial spinning, strong silk fibers were artificially spun.[115, 134, 160, 182] The liquid crystal phase has been suggested as not required for fiber assembly.[186] Thus, the role of liquid crystalline phases in silk spinning remains controversial and requires further theoretical and experimental efforts.[190] The artificial spinning of silks with liquid crystal phases has been shown (Figure 2C).[127] However, there were significant differences in this artificial process to natural spinning: 1) silk nanofibrils, instead of soluble silk molecules, were employed, thus implying a different assembly mechanism into macroscopic fibers; 2) the observed mosaic-like birefringence texture implied a lamellar phase, which is distinct from the schlieren texture of nematic phases in native silk dopes (Figure 2C).[68]
4. 3D printing of silks
3D printing provides advantages in comparison to traditional manufacturing, such as manufacturing automation, in terms of the ability to generate sophisticated geometries and precise spatial-deposition of materials.[191] 3D printing has been applied to a wide range of polymers and materials, such as thermoplastics, photocurable resins,[192] aluminum alloy,[193] liquid crystal polymers,[194] optically transparent glass,[195, 196] hydrogels such as extracellular matrix (collagen) and synthetic polymers.[197, 198]
Notably, silk spinning is surprisingly similar to the extrusion-based 3D printing in many aspects. Silk fibers are spun naturally, similar to the extrusion during 3D printing; silk fibers as structural components constitute 3D structures such as orb-webs and cocoons; orb webs consist of different silk fibers from distinct glands, representing a natural version of multiple material 3D printing systems. Thus, it is both logical and promising to translate the mechanisms and techniques of silk spinning into 3D printing processes and to benefit a range of applications from tissue engineering scaffolds to smart devices.[199–201] Indeed, native silk dopes have inspired the development of concentrated polyelectrolyte inks for 3D printing.[112]
However, the 3D printing of silks, just like other protein materials including collagen/gelatin, is restricted due to low structural integrity and mechanical performance in comparison to their natural counterparts,[202–204] and thus often require structural enhancement by using supporting and sacrificial materials.[205, 206] Nevertheless, silk protein-based inks have been developed for 3D printing. The majority of silk inks are prepared by blending silk feedstocks with other structural components to enhance rheology and printability.[207] Pure silk proteins can be 3D printed by only a handful of techniques to date.
4.1. 3D printing of silk composites
The most common way to print silk in extrusion-based 3D processes is to blend silk with other solution/structure-enhancing materials/dopants, including the use of agar,[180] hydroxypropyl methylcellulose,[208] gelatin,[180, 209–211] PEG,[212] glycerol[210] and Konjac gum.[213] The dopants usually increase the viscosity and thus ameliorate the rheological behavior of the silk-composite inks, which helps maintain the shape of the prints. This approach may be physiologically relevant, as many living tissues present composite structures, including bone, cartilage, ligaments, tendons, or skin. Silk-composite prints offer a promising approach to generate different material platforms (Table 2). 3D printing with silk-based composites generates a versatile capability for the formation of a wide range of cells and tissues, including bone, skin, blood vessels, cartilage, cardiac, and brain tissues (Figure 5A).
Table 2.
Summary of silk-composite inks for tissue engineering
| Bioink Composition | Printing Method | Cell Types | Suggested Applications | Refs. |
|---|---|---|---|---|
| Alginate | Inkjet Bioprinting | NIH 3T3 fibroblasts | Vasculature | [234] |
| Bioactive glass | Extrusion printing | hBMSCs | Bone | [235] |
| Bioactive glass | Indirect additive manufacturing | hBMSCs | Bone | [236] |
| Bioactive glass + Gelatin | Extrusion printing | TVA-BMSCs | Bone | [237] |
| Carbon nanotubes | Extrusion printing - Coaxial Needle | N/A | Interactive biocompatible electronics | [238] |
| Cartilage acellular matrix | Extrusion printing | BMSCs | Cartilage | [239] |
| Chitosan | Extrusion printing | BMSCs | Cartilage | [240] |
| Collagen | Extrusion printing | BMSCs | Cartilage | [240] |
| Gelatin | Indirect additive manufacturing | BMSCs | Cartilage | [211] |
| Gelatin | Extrusion printing | Child foreskin fibroblasts | Skin tissue engineering | [209] |
| Glycerol/gelatin | Extrusion printing | N/A | Soft tissue reconstruction | [210] |
| Glycidyl methacrylate | Digital light processing | NIH 3T3 fibroblasts | Vasculature | [121] |
| Hydroxyapatite | Extrusion printing | hMSCs | Bone | [218] |
| Hydroxyapatite + Sodium Alginate | Extrusion printing | hBMSCs | Bone | [241] |
| Polyethylene glycol | Digital light processing | NIH 3T3 fibroblasts / keratinocytes | Bone | [242] |
| Polylactic acid + Hydroxyapatite | Extrusion printing | NIH 3T3 fibroblasts / MC3T3 osteoblasts | Bone fixation | [243] |
| Polyols | Extrusion printing | N/A | General tissue engineering | [180] |
| Polyvinyl alcohol | Indirect additive manufacturing | Chondrocytes | Cartilage | [244] |
Figure 5.
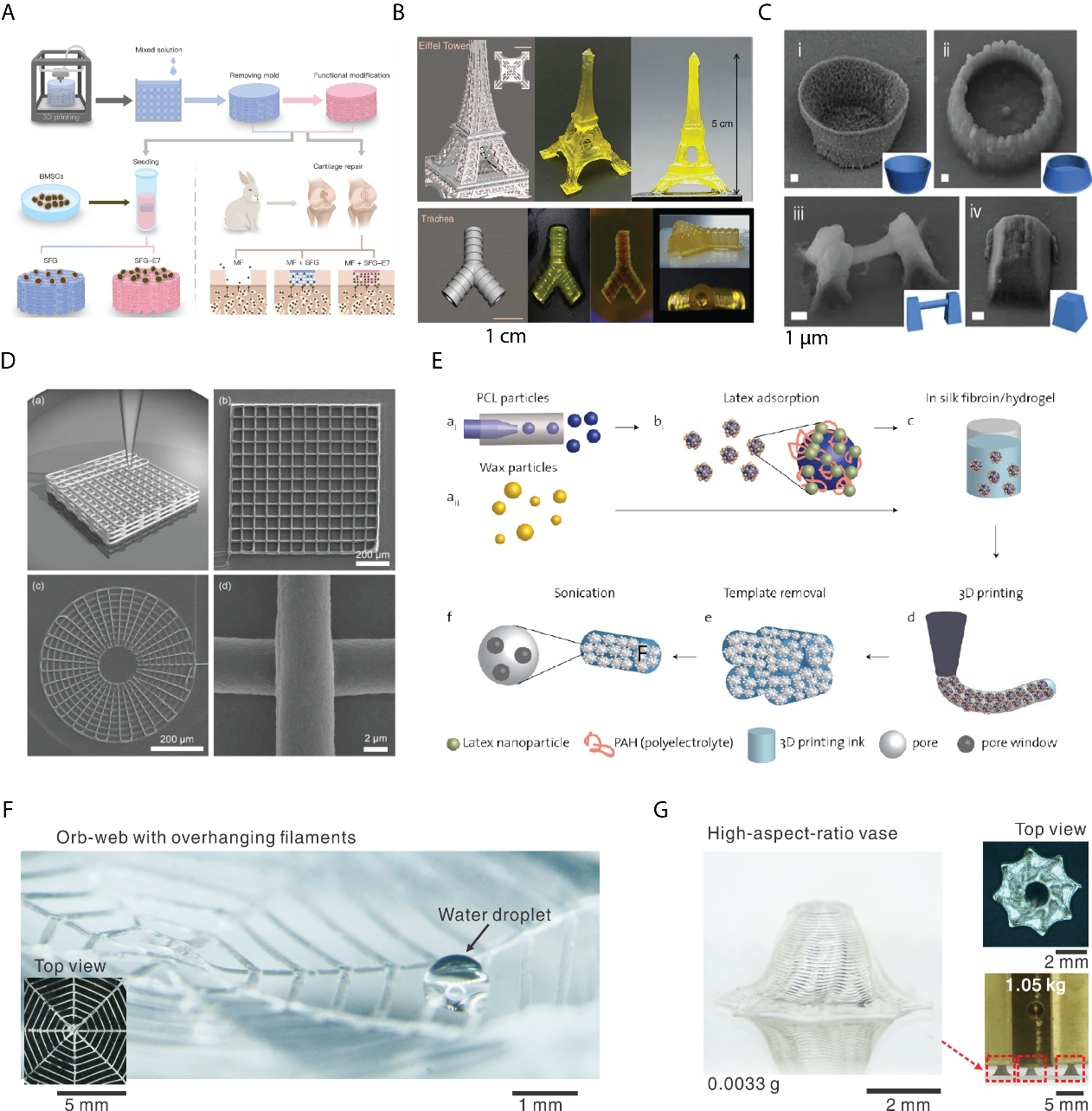
A. Schematic illustration of 3D printing of silk fibroin-gelatin composite inks for repairing cartilage injury in vivo and in vitro. Reproduced with permission.[211] Copyright 2017, John Wiley and Sons. B. The 3D prints in the shape of the Eiffel tower and Trachea by photocurable silk-MA. From left to right, CAD design, and the real prints. Reproduced with permission. [121]Copyright 2018, Springer Nature. C. SEM images of all-silk-based 3D prints by femtosecond laser-induced polymerization. (i) A microbowl; (ii) Another microbowl; (iii) A overhanging microwire; (iv) A truncated pyramid. Reproduced with permission.[122] Copyright 2015, Springer Nature. D. Schematic illustration and images (square lattice and circular web) of 3D direct ink writing of silk fibroin in a methanol bath. Reproduced with permission.[120] Copyright 2017, John Wiley and Sons. E. Schematic illustration of 3D printing with silk fibroin-Konjac gum composite ink with architectural control over multiple levels of hierarchy from macroscale to nanoscale. Latex nanoparticles, PCL, and wax particles are used as sacrificial templates, which can be removed by dissolution and ultrasonication and lead to open porous structures. Reproduced with permission.[213] Copyright 2007, American Chemical Society. F. 3D printing of monolithic silk fibroin using biomimetic and rationally designed aqueous salt bath. A printed two-layer overhanging orb-web composed of one arithmetic spiral and four radial straight lines in the width of ca. 100 μm. A water droplet sits across two filaments. Reproduced with permission.[119] Copyright 2019, John Wiley and Sons. G. A printed vase (≈0.0033 g) with high-aspect-ratio wall (Ca. 26) and inward inclination (63°). Three vases in a total of ca. 0.01 g can support a six-order heavier load (1050 g) without breaking or delamination, suggesting the desired mechanical stability. Reproduced with permission.[119] Copyright 2019, John Wiley and Sons.
Importantly, many silk-composite inks can contain cells for the direct construction of 3D cell-laden structures. In particular, silk/PEG bioinks were used to print a variety of tissue constructs with high resolution and homogeneity.[212] The cell-loaded constructs maintained their shape for at least 12 weeks in culture. Further, a specific concentration silk solution (10 wt%) facilitated cell growth, suggesting that these silk/PEG bioink gels may provide suitable scaffold environments for cell printing. In the efforts to mimic the natural hierarchical structure of silkworm and spider silk, micrometer-sized wax particles and nanoparticles were used as sacrificial materials to control the porosity at multiple scales in 3D printed silk structures (Figure 5C).[213]
Silk materials in the format of fibers and particles are useful as reinforcements for other materials used in 3D printing. The integration of silk particles in a 3D printed chitosan hydrogel resulted in a 5-fold increase in compressive modulus.[214] Likewise, integrating silk microfibers and nanofibers into chitosan hydrogels increased the mechanical properties of the 3D printed constructs without introducing cytotoxicity to human fibroblasts.[215] Micro- and nano-silk fibers can be used to reinforce silk hydrogels, with potential applications for the 3D printing of monolithic silk structures.[172, 216]
Besides blending structure-enhancing materials with silk for 3D printing, the silk protein can be modified to be photo-polymerized for light-based 3D printing.[117, 121, 122, 217] Digital light processing was developed to shape silk into complex organ structures, including Eiffel towers and tracheas (Figure 5B).[121] The silk ink was functionalized with glycidyl methacrylate and demonstrated mechanical robustness, structural stability, and cytocompatibility. In this case, the mechanical strength comes from the chemical cross-linking rather than the directed self-assembly of proteins. This process is quite different from native silk spinning, but proves useful, nonetheless.
4.2. Monolithic silk ink
The first 3D printing using inks of pure silk solution was based on the use of silk fibroin (28–30 wt%) (Figure 5D).[120, 218] This work resulted from the combination of the advanced technical infrastructure of extrusion-based 3D printing (direct-writing system) [112] and the widely employ methanol solvent (85%) for silk spinning. The technical platform of direct-writing enabled high resolution of the printed silk filaments with diameters of 5 μm. The 3D printed silk scaffolds inherited the well-recognized capability of silk materials to regenerate bone tissue, thus supporting the adhesion and growth of human bone marrow-derived mesenchymal stem cells.[219] The use of methanol (85%) as a coagulation bath significantly differentiated this 3D printing process from natural silk spinning, as described earlier. The methanol also prevents the integration of organic solvent-sensitive biomolecules into the 3D printed structures.
A supporting bath consisting of synthetic nanoclay (Laponite) and PEG was also used for printing pure silk solutions.[220] The laponite was mainly used as a granular gel media to support the 3D prints.[198] PEG was used to induce the physical cross-linking of silk inks.[221] Geometrical complexity in the supporting bath was demonstrated, while the mechanical performance of the prints was a limitation due to the assumed soft gels generated in this process.
To replace the methanol bath and to generate 3D prints with both mechanical and geometry robustness, the mechanism of the silk spinning was utilized as a guide, where the pH, salt ions and dehydration are harnessed together in a systematic way to control the phase-transition (solidification and gelation) of silk proteins.[119] Although exact in vivo solvent conditions remain unclear, we formulated an aqueous bath with a de novo chemical composition rationally tailored for 3D printing of pure silk proteins (Figure 5F, G).[119] The aqueous bath contained 0.5 m dipotassium phosphate and 4 m sodium chloride. These salt ions exist in the spinning gland and impose specific salt effects on silk proteins. The phosphate ions result in a slightly acidic environment (pH ~6) that mimics the anterior part of the spinning gland. The high concentration of salts provides high osmolarity (>8 M, as one sodium chloride molecule disassociates into two ions), as a general principle found in animals for dehydration, i.e., concentrating urine.[222] The aqueous bath is thus “biomimetic” and recapitulates the cell-regulated, complex, and dynamic solvent conditions in native spinning glands. We used this biomimetic bath to print silk proteins in 3D successfully. In particular, we printed overhanging filaments30 mm long and 85 μm in diameter and only mechanically supported at ends. Thus, the solidification dynamics of the ink must be rapid to prevent sagging, thus imposing stringent requirements on the printing process. The ratio between length and diameter of the filaments was around 375, significantly higher than previous results of 20 (polyelectrolyte ink)[223] and 33 (Carbomer-laden hydrogel).[224]
The 3D printing of silk fibroin is fundamentally different from the long-standing 3D printing of alginate,[225] albeit aqueous salt baths are used in both (Table 3). For 3D printing of alginate, the salt bath is normally composed of divalent cations, such as calcium ions, which crosslink alginate by binding to the guluronate blocks on alginate chains, termed “ionic crosslinking.”[226, 227] The salt ions thus constitute an essential structural component of the cross-linked alginate. The 3D prints of alginate will dissolve immediately after the loss of the cations, for example, by treatment with chelating agents (e.g., ethylenediaminetetraacetic acid).[228] Furthermore, complex geometries of the 3D printed alginate, such as an overhanging filament and a perfusable channel, require supporting materials like granular gels, which adds operational complexity.[225, 229]
Table 3.
Comparison of 3D printing of alginate and silk fibroin
| Alginate | 3D Silk printing[119] | ||
|---|---|---|---|
| Ink composition | Molecule | Polysaccharide | Protein |
| Concentration | ~4% w/v | ~30 wt% | |
| Aqueous bath | Salt ions | Calcium, magnesium… | Sodium, Potassium, Chloride and phosphate |
| Cation valent | Divalent | Monovalent | |
| Salt is a crosslinking agent and structural component | Yes | No | |
| 3D Printing | Printing technique | Extrusion-based 3D printing | Extrusion-based 3D printing |
| Phase-transition | Ionic bonding | Hydrogen bond | |
| Dissolving solvent | Chelating reagents | Strong-hydrogen-destroying solvents | |
| Printing capability | Supporting materials for hollow structures | Yes | No |
| Mechanic performance | Low | High | |
By contrast, the salt bath for 3D printing silk fibroin is composed of monovalent cations, such as sodium and potassium. Moreover, the salt is used to mimic the natural spinning conditions to induce the assembly of silk protein molecules, rather than forming ionic bonds as a structural component. Thirdly, the silk prints are solid structures, while the alginate print is in the form of hydrogels and contains a large amount of water. Finally, the ultimate tensile strength of silk prints is at least two orders of magnitude than alginate. The 3D printing of silk fibroin via directed molecular assembly thus implies a new paradigm for 3D bioprinting.
5. Outlook and Conclusions
As with the variety of biological structural materials, including bone, tendon, shells, and wood, silk has been widely explored for developing biomimetic materials. Notably, the biomimetic study of the silk spinning places more emphasis on processing rather than structure alone. Thus, the underlying scientific principles of the silk spinning can be drawn from the observed relationships between processing, structure, and function.[230, 231] The processing merits of silk spinning are particularly desirable and worth mimicking for industrial polymer manufacturing. In addition, silk spinning embodies sustainable manufacturing and can be recognized as a “living” nanotechnology.[232] The hierarchical assembly of molecules has been approximated in artificial systems,[233] but the mechanical superiority remains to be achieved with good process control and facile approaches amenable to scale up and industrialization.
Toward the silk spinning-inspired manufacturing of polymers, challenges lie in the incomplete understanding of silk spinning, especially the solvent conditions-directed assembly of the full-length silk protein. In addition, despite some work to mimic the aqueous conditions during silk spinning,[114, 119] simplified engineering systems to mimic the inherently complicated biological systems involved await creative solutions. Overall, future work will need to mesh mechanistic studies using a variety of biotechnological tools, computational modeling, microfluidics, and 3D printing with integral engineering platforms that exploit the advantages of the silk proteins and silk spinning. Thus, synergy in these approaches is essential, mimicking the co-evolutionary drivers for silk fibers in general, mecanical robustness, and aqueous and ambient processing. These are key goals to embrace in moving forward with sustainability and medicine as key and rewarding outcomes.
Acknowledgments
We thank the AFOSR (FA9550-17-1-0333), the NIH (P41EB002520, R01EB021264, R01NS092847, U01EB014976) for support of the work as foundations for this review.
Biographies

Xuan Mu is a Postdoctoral Research Associate working with Prof. David L. Kaplan at Tufts University. He received his Ph.D. from East China University of Science and Technology in Shanghai, China. He worked at National Center for Nanoscience and Technology and Peking Union Medical College (PUMC), both in Beijing, China, and Brigham Women’s Hospital in Boston, MA. His research focuses on microfluidics, Organs-on-chips, 3D printing, disease modeling, and clinical diagnosis.

Vincent Fitzpatrick obtained his Ph.D. in 2017 from Université Grenoble Alpes (France). His research focused on micropatterns of extracellular matrix proteins and morphogens on biomimetic films, for the single cell control of osteoblastic differentiation. Since 2018, he is a postdoctoral fellow with Professor David Kaplan at Tufts University (United States). His current research interests lie in the design of bone scaffolds and the 3D printing of silk-based materials for bone and cartilage tissue engineering.

David Kaplan holds an Endowed Chair, the Stern Family Professor of Engineering, at Tufts University. He is Professor and Chair of the Department of Biomedical Engineering and also holds faculty appointments in the School of Medicine, School of Dental Medicine, Department of Chemistry, and Department of Chemical and Biological Engineering. He directs the NIH P41 Tissue Engineering Resource Center and is Editor-in-Chief for ACS Biomaterials Science and Engineering.
References
- [1].Vollrath F, Knight DP, Nature 2001, 410, 541. [DOI] [PubMed] [Google Scholar]
- [2].Jin H-J, Kaplan DL, Nature 2003, 424, 1057. [DOI] [PubMed] [Google Scholar]
- [3].Gosline JM, Mechanical Design of Structural Materials in Animals, Princeton University Press, Princeton, NJ: 2018. [Google Scholar]
- [4].W. C. contributors, File:Chinese silk, 4th Century BC.JPG, https://commons.wikimedia.org/w/index.php?title=File:Chinese_silk,_4th_Century_BC.JPG&oldid=244498641, accessed: 26 October 2019 14:27 UTC.
- [5].Mittal N, Ansari F, Gowda K. Brouzet V,C, Chen P, Larsson PT, Roth SV, Lundell F, Wagberg L, Kotov NA, ACS Nano 2018, 12, 6378. [DOI] [PubMed] [Google Scholar]
- [6].Omenetto FG, Kaplan DL, Science 2010, 329, 528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cheng M, Chen W, Weerasooriya T, J. Eng. Mater. Technol 2005, 127, 197. [Google Scholar]
- [8].Papkov D, Zou Y, Andalib MN, Goponenko A, Cheng SZ, Dzenis YA, ACS Nano 2013, 7, 3324. [DOI] [PubMed] [Google Scholar]
- [9].Kromm F-X, Lorriot T, Coutand B, Harry R, Quenisset J-M, Polymer Testing 2003, 22, 463. [Google Scholar]
- [10].Liao X, Dulle M, de Souza e Silva JM, Wehrspohn RB, Agarwal S, Förster S, Hou H, Smith P, Greiner A, Science 2019, 366, 1376. [DOI] [PubMed] [Google Scholar]
- [11].Mittal N, Jansson R, Widhe M, Benselfelt T, Håkansson KMO, Lundell F, Hedhammar M, Söderberg LD, ACS Nano 2017, 11, 5148. [DOI] [PubMed] [Google Scholar]
- [12].Xu Z, Sun H, Zhao X, Gao C, Adv. Mater 2013, 25, 188. [DOI] [PubMed] [Google Scholar]
- [13].Dalton AB, Collins S, Munoz E, Razal JM, Ebron VH, Ferraris JP, Coleman JN, Kim BG, Baughman RH, Nature 2003, 423, 703. [DOI] [PubMed] [Google Scholar]
- [14].Miaudet P, Badaire S, Maugey M, Derre A, Pichot V, Launois P, Poulin P, Zakri C, Nano Lett 2005, 5, 2212. [DOI] [PubMed] [Google Scholar]
- [15].Shin MK, Lee B, Kim SH, Lee JA, Spinks GM, Gambhir S, Wallace GG, Kozlov ME, Baughman RH, Kim SJ, Nat. Commun 2012, 3, 650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gosline J, Guerette P, Ortlepp C, Savage K, J. Exp. Biol 1999, 202, 3295. [DOI] [PubMed] [Google Scholar]
- [17].Shah DU, Porter D, Vollrath F, Compos. Sci. Technol 2014, 101, 173. [Google Scholar]
- [18].Vepari C, Kaplan DL, Prog. Polym. Sci 2007, 32, 991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Horan RL, Antle K, Collette AL, Wang Y, Huang J, Moreau JE, Volloch V, Kaplan DL, Altman GH, Biomaterials 2005, 26, 3385. [DOI] [PubMed] [Google Scholar]
- [20].Asakura T, Tanaka T, Tanaka R, ACS Biomater. Sci. Eng 2019. [DOI] [PubMed] [Google Scholar]
- [21].Numata K, Cebe P, Kaplan DL, Biomaterials 2010, 31, 2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang Y, Rudym DD, Walsh A, Abrahamsen L, Kim H-J, Kim HS, Kirker-Head C, Kaplan DL, Biomaterials 2008, 29, 3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li C, Guo C, Fitzpatrick V, Ibrahim A, Zwierstra MJ, Hanna P, Lechtig A, Nazarian A, Lin SJ, Kaplan DL, Nature Reviews Materials 2019, 1. [Google Scholar]
- [24].Altman GH, Diaz F, Jakuba C, Calabro T, Horan RL, Chen J, Lu H, Richmond J, Kaplan DL, Biomaterials 2003, 24, 401. [DOI] [PubMed] [Google Scholar]
- [25].Hwang S-W, Tao H, Kim D-H, Cheng H, Song J-K, Rill E, Brenckle MA, Panilaitis B, Won SM, Kim Y-S, Science 2012, 337, 1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang X, Gu Y, Xiong Z, Cui Z, Zhang T, Adv. Mater 2014, 26, 1336. [DOI] [PubMed] [Google Scholar]
- [27].Tao H, Brenckle MA, Yang M, Zhang J, Liu M, Siebert SM, Averitt RD, Mannoor MS, McAlpine MC, Rogers JA, Adv. Mater 2012, 24, 1067. [DOI] [PubMed] [Google Scholar]
- [28].Ling S, Wang Q, Zhang D, Zhang Y, Mu X, Kaplan DL, Buehler MJ, Adv. Funct. Mater 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Shi J, Liu S, Zhang L, Yang B, Shu L, Yang Y, Ren M, Wang Y, Chen J, Chen W, Chai Y, Tao X, Adv. Mater 2019, 0, 1901958. [DOI] [PubMed] [Google Scholar]
- [30].Zhu B, Wang H, Leow WR, Cai Y, Loh XJ, Han MY, Chen X, Adv. Mater 2016, 28, 4250. [DOI] [PubMed] [Google Scholar]
- [31].Wang C, Xia K, Zhang Y, Kaplan DL, Acc. Chem. Res 2019, DOI: 10.1021/acs.accounts.9b00333. [DOI] [PubMed] [Google Scholar]
- [32].Kim S, Mitropoulos AN, Spitzberg JD, Tao H, Kaplan DL, Omenetto FG, Nat. Photonics 2012, 6, 818. [Google Scholar]
- [33].Omenetto FG, Kaplan DL, Nat. Photonics 2008, 2, 641. [Google Scholar]
- [34].Wang Y, Li W, Li M, Zhao S, De Ferrari F, Liscidini M, Omenetto FG, Adv. Mater 2019, 31, 1805312. [DOI] [PubMed] [Google Scholar]
- [35].Monks JN, Yan B, Hawkins N, Vollrath F, Wang Z, Nano Lett 2016, 16, 5842. [DOI] [PubMed] [Google Scholar]
- [36].Huby N, Vié V, Renault A, Beaufils S, Lefèvre T, Paquet-Mercier F, Pézolet M, Bêche B, Appl. Phys. Lett 2013, 102, 123702. [Google Scholar]
- [37].Liu D, Tarakanova A, Hsu CC, Yu M, Zheng S, Yu L, Liu J, He Y, Dunstan D, Buehler MJ, Sci. Adv 2019, 5, eaau9183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jia T, Wang Y, Dou Y, Li Y, Jung de Andrade M, Wang R, Fang S, Li J, Yu Z, Qiao R, Adv. Funct. Mater 2019, 29, 1808241. [Google Scholar]
- [39].Holland C, Numata K, Rnjak‐Kovacina J, Seib FP, Adv. Healthcare Mater 2019, 8, 1800465. [DOI] [PubMed] [Google Scholar]
- [40].Aigner TB, DeSimone E, Scheibel T, Adv. Mater 2018, 30, 1704636. [DOI] [PubMed] [Google Scholar]
- [41].Abascal NC, Regan L, Royal Society Open Biology 2018, 8, 180113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Janani G, Kumar M, Chouhan D, Moses JC, Gangrade A, Bhattacharjee S, Mandal BB, ACS Applied Bio Materials 2019. [DOI] [PubMed]
- [43].Robertson ID, Yourdkhani M, Centellas PJ, Aw JE, Ivanoff DG, Goli E, Lloyd EM, Dean LM, Sottos NR, Geubelle PH, Nature 2018, 557, 223. [DOI] [PubMed] [Google Scholar]
- [44].Witik RA, Gaille F, Teuscher R, Ringwald H, Michaud V, Månson J-AE, J. Cleaner Prod 2012, 29, 91. [Google Scholar]
- [45].Holland C, Vollrath F, Ryan AJ, Mykhaylyk OO, Adv. Mater 2012, 24, 105. [DOI] [PubMed] [Google Scholar]
- [46].Kadler KE, Hill A, Canty-Laird EG, Curr. Opin. Cell Biol 2008, 20, 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Knowles TP, Mezzenga R, Adv. Mater 2016, 28, 6546. [DOI] [PubMed] [Google Scholar]
- [48].Porter D, Vollrath F, Adv. Mater 2009, 21, 487. [Google Scholar]
- [49].Kaplan DL, Polym. Degrad. Stab 1998, 59, 25. [Google Scholar]
- [50].O’Brien JP, Fahnestock SR, Termonia Y, Gardner KH, Adv. Mater 1998, 10, 1185. [Google Scholar]
- [51].Wu Y, Shah DU, Liu C, Yu Z, Liu J, Ren X, Rowland MJ, Abell C, Ramage MH, Scherman OA, Proc. Natl. Acad. Sci 2017, 114, 8163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Vollrath F, Porter D, Soft Matter 2006, 2, 377. [DOI] [PubMed] [Google Scholar]
- [53].Vollrath F, Porter D, Polymer 2009, 50, 5623. [Google Scholar]
- [54].Hagn F, Eisoldt L, Hardy JG, Vendrely C, Coles M, Scheibel T, Kessler H, Nature 2010, 465, 239. [DOI] [PubMed] [Google Scholar]
- [55].Lu Q, Zhu H, Zhang C, Zhang F, Zhang B, Kaplan DL, Biomacromolecules 2012, 13, 826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Groll J, Boland T, Blunk T, Burdick JA, Cho D-W, Dalton PD, Derby B, Forgacs G, Li Q, Mironov VA, Biofabrication 2016, 8, 013001. [DOI] [PubMed] [Google Scholar]
- [57].Yoshioka T, Tsubota T, Tashiro K, Jouraku A, Kameda T, Nat. Commun 2019, 10, 1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Malay AD, Sato R, Yazawa K, Watanabe H, Ifuku N, Masunaga H, Hikima T, Guan J, Mandal BB, Damrongsakkul S, Sci. Rep 2016, 6, 27573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Guo C, Zhang J, Jordan JS, Wang X, Henning RW, Yarger JL, Biomacromolecules 2018, 19, 906. [DOI] [PubMed] [Google Scholar]
- [60].Numata K, Sato R, Yazawa K, Hikima T, Masunaga H, Polymer 2015, 77, 87. [Google Scholar]
- [61].Andersson M, Johansson J, Rising A, Int. J. Mol. Sci 2016, 17, 1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Koh L-D, Cheng Y, Teng C-P, Khin Y-W, Loh X-J, Tee S-Y, Low M, Ye E, Yu H-D, Zhang Y-W, Prog. Polym. Sci 2015, 46, 86. [Google Scholar]
- [63].Kumar A, Gupta RK, Fundamentals of polymer engineering, CRC Press, 2018. [Google Scholar]
- [64].Vollrath F, Porter D, Appl. Phys. A 2006, 82, 205. [Google Scholar]
- [65].Keten S, Xu Z, Ihle B, Buehler MJ, Nat. Mater 2010, 9, 359. [DOI] [PubMed] [Google Scholar]
- [66].Asakura T, Umemura K, Nakazawa Y, Hirose H, Higham J, Knight D, Biomacromolecules 2007, 8, 175. [DOI] [PubMed] [Google Scholar]
- [67].Magoshi J, Magoshi Y, Nakamura S, in Silk Polymers, ACS Publications, 1993, 292. [Google Scholar]
- [68].Kerkam K, Viney C, Kaplan D, Lombardi S, Nature 1991, 349, 596. [Google Scholar]
- [69].Willcox PJ, Gido SP, Muller W, Kaplan DL, Macromolecules 1996, 29, 5106. [Google Scholar]
- [70].Knight D, Vollrath F, Proc. R. Soc. B 1999, 266, 519. [Google Scholar]
- [71].Ciferri A, Polymer liquid crystals, Elsevier, 2012. [Google Scholar]
- [72].Rey AD, Soft Matter 2010, 6, 3402. [Google Scholar]
- [73].Rey AD, Herrera‐Valencia EE, Biopolymers 2012, 97, 374. [DOI] [PubMed] [Google Scholar]
- [74].Laity P, Gilks S, Holland C, Polymer 2015, 67, 28. [Google Scholar]
- [75].Holland C, Terry A, Porter D, Vollrath F, Nat. Mater 2006, 5, 870. [DOI] [PubMed] [Google Scholar]
- [76].Laity PR, Holland C, Biomacromolecules 2016, 17, 2662. [DOI] [PubMed] [Google Scholar]
- [77].Asakura T, Okushita K, Williamson MP, Macromolecules 2015, 48, 2345. [Google Scholar]
- [78].Domigan L, Andersson M, Alberti K, Chesler M, Xu Q, Johansson J, Rising A, Kaplan D, Insect Biochem. Mol. Biol 2015, 65, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Askarieh G, Nordling K, Saenz A, Casals C, Rising A, Johansson J, Knight SD, Nature 2010, 465, 236. [DOI] [PubMed] [Google Scholar]
- [80].Degtyar E, Harrington MJ, Politi Y, Fratzl P, Angew. Chem., Int. Ed 2014, 53, 12026. [DOI] [PubMed] [Google Scholar]
- [81].Okur HI, Hladílková J, Rembert KB, Cho Y, Heyda J, Dzubiella J, Cremer PS, Jungwirth P, J. Phys. Chem. B 2017, 121, 1997. [DOI] [PubMed] [Google Scholar]
- [82].Davies G, Knight D, Vollrath F, Tissue and Cell 2013, 45, 306. [DOI] [PubMed] [Google Scholar]
- [83].Breslauer DN, Lee LP, Muller SJ, Biomacromolecules 2008, 10, 49. [DOI] [PubMed] [Google Scholar]
- [84].Holland C, Urbach J, Blair D, Soft Matter 2012, 8, 2590. [Google Scholar]
- [85].Moriya M, Roschzttardtz F, Nakahara Y, Saito H, Masubuchi Y, Asakura T, Biomacromolecules 2009, 10, 929. [DOI] [PubMed] [Google Scholar]
- [86].Yucel T, Cebe P, Kaplan DL, Biophys. J 2009, 97, 2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Rousseau M-E, Lefevre T, Beaulieu L, Asakura T, Pézolet M, Biomacromolecules 2004, 5, 2247. [DOI] [PubMed] [Google Scholar]
- [88].Sparkes J, Holland C, Nat. Commun 2017, 8, 594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Andersson M, Chen G, Otikovs M, Landreh M, Nordling K, Kronqvist N, Westermark P, Jörnvall H, Knight S, Ridderstråle Y, PLoS biology 2014, 12, e1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Kronqvist N, Otikovs M, Chmyrov V, Chen G, Andersson M, Nordling K, Landreh M, Sarr M, Jörnvall H, Wennmalm S, Nat. Commun 2014, 5, 3254. [DOI] [PubMed] [Google Scholar]
- [91].Zhou P, Xie X, Knight DP, Zong X-H, Deng F, Yao W-H, Biochemistry 2004, 43, 11302. [DOI] [PubMed] [Google Scholar]
- [92].Terry AE, Knight DP, Porter D, Vollrath F, Biomacromolecules 2004, 5, 768. [DOI] [PubMed] [Google Scholar]
- [93].Jungwirth P, Cremer PS, Nature chemistry 2014, 6, 261. [DOI] [PubMed] [Google Scholar]
- [94].Zhou L, Chen X, Shao Z, Huang Y, Knight DP, J. Phys. Chem. B 2005, 109, 16937. [DOI] [PubMed] [Google Scholar]
- [95].Knight DP, Vollrath F, Naturwissenschaften 2001, 88, 179. [DOI] [PubMed] [Google Scholar]
- [96].Chen X, Huang Y-F, Shao Z-Z, Huang Y, Zhou P, Knight D, Vollrath F, Chem. Res. Chin. Univ 2004, 25, 1163. [Google Scholar]
- [97].Rising A, Johansson J, Nat. Chem. Biol 2015, 11, 309. [DOI] [PubMed] [Google Scholar]
- [98].Ruan Q-X, Zhou P, J. Mol. Struct 2008, 883, 85. [Google Scholar]
- [99].Ruan QX, Zhou P, Hu BW, Ji D, The FEBS journal 2008, 275, 219. [DOI] [PubMed] [Google Scholar]
- [100].Rembert KB, Paterová J, Heyda J, Hilty C, Jungwirth P, Cremer PS, J. Am. Chem. Soc 2012, 134, 10039. [DOI] [PubMed] [Google Scholar]
- [101].Eisoldt L, Hardy JG, Heim M, Scheibel TR, J. Struct. Biol 2010, 170, 413. [DOI] [PubMed] [Google Scholar]
- [102].Hagn F, Thamm C, Scheibel T, Kessler H, Angew. Chem., Int. Ed 2011, 50, 310. [DOI] [PubMed] [Google Scholar]
- [103].Lammel AS, Hu X, Park S-H, Kaplan DL, Scheibel TR, Biomaterials 2010, 31, 4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Kim U-J, Park J, Kim HJ, Wada M, Kaplan DL, Biomaterials 2005, 26, 2775. [DOI] [PubMed] [Google Scholar]
- [105].Percot A, Colomban P, Paris C, Dinh HM, Wojcieszak M, Mauchamp B, Vib. Spectrosc 2014, 73, 79. [Google Scholar]
- [106].Yazawa K, Ishida K, Masunaga H, Hikima T, Numata K, Biomacromolecules 2016, 17, 1057. [DOI] [PubMed] [Google Scholar]
- [107].Boulet-Audet M, Holland C, Gheysens T, Vollrath F, Biomacromolecules 2016, 17, 3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Case ST, Thornton JR, Int. J. Biol. Macromol 1999, 24, 89. [DOI] [PubMed] [Google Scholar]
- [109].Vollrath F, Edmonds DT, Nature 1989, 340, 305. [Google Scholar]
- [110].Plaza GR, Corsini P, Pérez‐Rigueiro J, Marsano E, Guinea GV, Elices M, J. Appl. Polym. Sci 2008, 109, 1793. [Google Scholar]
- [111].Gu L, Jiang Y, Hu J, Adv. Mater 2019, 0, 1904311. [DOI] [PubMed] [Google Scholar]
- [112].Lewis JA, Adv. Funct. Mater 2006, 16, 2193. [Google Scholar]
- [113].Koeppel A, Holland C, ACS Biomater. Sci. Eng 2017, 3, 226. [DOI] [PubMed] [Google Scholar]
- [114].Andersson M, Jia Q, Abella A, Lee X-Y, Landreh M, Purhonen P, Hebert H, Tenje M, Robinson CV, Meng Q, Plaza GR, Johansson J, Rising A, Nat. Chem. Biol 2017, 13, 262. [DOI] [PubMed] [Google Scholar]
- [115].Zhou G, Shao Z, Knight DP, Yan J, Chen X, Adv. Mater 2009, 21, 366. [Google Scholar]
- [116].Liu Y, Ren J, Ling S, Compos. Commun 2019, 13, 85. [Google Scholar]
- [117].Jiang J, Zhang S, Qian Z, Qin N, Song W, Sun L, Zhou Z, Shi Z, Chen L, Li X, Adv. Mater 2018, 30, 1705919. [DOI] [PubMed] [Google Scholar]
- [118].Kim S, Marelli B, Brenckle MA, Mitropoulos AN, Gil E-S, Tsioris K, Tao H, Kaplan DL, Omenetto FG, Nat. Nanotechnol 2014, 9, 306. [DOI] [PubMed] [Google Scholar]
- [119].Mu X, Wang Y, Guo C, Li Y, Ling S, Huang W, Cebe P, Hsu HH, De Ferrari F, Jiang X, Macromol. Biosci 2019, 1900191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Ghosh S, Parker ST, Wang X, Kaplan DL, Lewis JA, Adv. Funct. Mater 2008, 18, 1883. [Google Scholar]
- [121].Kim SH, Yeon YK, Lee JM, Chao JR, Lee YJ, Seo YB, Sultan MT, Lee OJ, Lee JS, Yoon S.-i., Nat. Commun 2018, 9, 1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Sun Y-L, Li Q, Sun S-M, Huang J-C, Zheng B-Y, Chen Q-D, Shao Z-Z, Sun H-B, Nat. Commun 2015, 6, 8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Ha S-W, Tonelli AE, Hudson SM, Biomacromolecules 2005, 6, 1722. [DOI] [PubMed] [Google Scholar]
- [124].Zhou Z, Zhang S, Cao Y, Marelli B, Xia X, Tao TH, Adv. Mater 2018, 30, 1706983. [DOI] [PubMed] [Google Scholar]
- [125].Wang Y, Guo J, Zhou L, Ye C, Omenetto FG, Kaplan DL, Ling S, Adv. Funct. Mater 2018, 28, 1805305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Rockwood DN, Preda RC, Yücel T, Wang X, Lovett ML, Kaplan DL, Nat. Protoc 2011, 6, 1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Ling S, Qin Z, Li C, Huang W, Kaplan DL, Buehler MJ, Nat. Commun 2017, 8, 1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Zhang F, Lu Q, Yue X, Zuo B, Qin M, Li F, Kaplan DL, Zhang X, Acta Biomater 2015, 12, 139. [DOI] [PubMed] [Google Scholar]
- [129].Lundahl MJ, Klar V, Wang L, Ago M, Rojas OJ, Ind. Eng. Chem. Res 2016, 56, 8. [Google Scholar]
- [130].Håkansson KM, Fall AB, Lundell F, Yu S, Krywka C, Roth SV, Santoro G, Kvick M, Wittberg LP, Wågberg L, Nat. Commun 2014, 5, 4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Prince JT, McGrath KP, DiGirolamo CM, Kaplan DL, Biochemistry 1995, 34, 10879. [DOI] [PubMed] [Google Scholar]
- [132].Xia X-X, Qian Z-G, Ki CS, Park YH, Kaplan DL, Lee SY, Proc. Natl. Acad. Sci 2010, 107, 14059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Edlund AM, Jones J, Lewis R, Quinn JC, New Biotechnol 2018, 42, 12. [DOI] [PubMed] [Google Scholar]
- [134].Lazaris A, Arcidiacono S, Huang Y, Zhou J-F, Duguay F, Chretien N, Welsh EA, Soares JW, Karatzas CN, Science 2002, 295, 472. [DOI] [PubMed] [Google Scholar]
- [135].Simmons JR, Xu L, Rainey JK, ACS Biomater. Sci. Eng 2019, 5, 4985. [DOI] [PubMed] [Google Scholar]
- [136].Teulé F, Cooper AR, Furin WA, Bittencourt D, Rech EL, Brooks A, Lewis RV, Nat. Protoc 2009, 4, 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Zhang X, Xia L, Day BA, Harris TI, Oliveira P, Knittel C, Licon AL, Gong C, Dion G, Lewis RV, Jones JA, Biomacromolecules 2019, 20, 2252. [DOI] [PubMed] [Google Scholar]
- [138].Teulé F, Miao Y-G, Sohn B-H, Kim Y-S, Hull JJ, Fraser MJ, Lewis RV, Jarvis DL, Proc. Natl. Acad. Sci 2012, 109, 923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Chung H, Kim TY, Lee SY, Curr. Opin. Biotechnol 2012, 23, 957. [DOI] [PubMed] [Google Scholar]
- [140].Bowen CH, Dai B, Sargent CJ, Bai W, Ladiwala P, Feng H, Huang W, Kaplan DL, Galazka JM, Zhang F, Biomacromolecules 2018, 19, 3853. [DOI] [PubMed] [Google Scholar]
- [141].Xu L, Rainey JK, Meng Q, Liu X-Q, PLoS One 2012, 7, e50227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Stark M, Grip S, Rising A, Hedhammar M, Engström W, Hjälm G, Johansson J, Biomacromolecules 2007, 8, 1695. [DOI] [PubMed] [Google Scholar]
- [143].Wang Q, Chen Q, Yang Y, Shao Z, Biomacromolecules 2012, 14, 285. [DOI] [PubMed] [Google Scholar]
- [144].Wray LS, Hu X, Gallego J, Georgakoudi I, Omenetto FG, Schmidt D, Kaplan DL, J. Biomed. Mater. Res., Part B 2011, 99, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Yan J, Zhou G, Knight DP, Shao Z, Chen X, Biomacromolecules 2009, 11, 1. [DOI] [PubMed] [Google Scholar]
- [146].Luo J, Zhang L, Peng Q, Sun M, Zhang Y, Shao H, Hu X, Int. J. Biol. Macromol 2014, 66, 319. [DOI] [PubMed] [Google Scholar]
- [147].Li C, Hotz B, Ling S, Guo J, Haas DS, Marelli B, Omenetto F, Lin SJ, Kaplan DL, Biomaterials 2016, 110, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Marelli B, Patel N, Duggan T, Perotto G, Shirman E, Li C, Kaplan DL, Omenetto FG, Proc. Natl. Acad. Sci 2017, 114, 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Yao J, Masuda H, Zhao C, Asakura T, Macromolecules 2002, 35, 6. [Google Scholar]
- [150].Um IC, Kweon H, Park YH, Hudson S, Int. J. Biol. Macromol 2001, 29, 91. [DOI] [PubMed] [Google Scholar]
- [151].Min B-M, Lee G, Kim SH, Nam YS, Lee TS, Park WH, Biomaterials 2004, 25, 1289. [DOI] [PubMed] [Google Scholar]
- [152].Ajisawa A, J. Seric.Sci. Jpn 1998, 67, 91. [Google Scholar]
- [153].Yamada H, Nakao H, Takasu Y, Tsubouchi K, Mater. Sci. Eng. C 2001, 14, 41. [Google Scholar]
- [154].Van Leeuwen FR, Sangster B, Hildebrandt AG, CRC crit. Rev. Toxicol 1987, 18, 189. [DOI] [PubMed] [Google Scholar]
- [155].Olszowy HA, Rossiter J, Hegarty J, Geoghegan P, Haswell-Elkins M, J. Anal. Toxicol 1998, 22, 225. [DOI] [PubMed] [Google Scholar]
- [156].Aral H, Vecchio-Sadus A, Ecotoxicol. Environ. Saf 2008, 70, 349. [DOI] [PubMed] [Google Scholar]
- [157].Bromide, https://www.who.int/water_sanitation_health/dwq/chemicals/bromide.pdf, accessed.
- [158].Schrauzer GN, J. Am. Coll. Nutr 2002, 21, 14. [DOI] [PubMed] [Google Scholar]
- [159].Tchounwou PB, Yedjou CG, Patlolla AK, Sutton DJ, in Molecular, clinical and environmental toxicology, Springer; 2012, p. 133. [Google Scholar]
- [160].Ha S-W, Park YH, Hudson SM, Biomacromolecules 2003, 4, 488. [DOI] [PubMed] [Google Scholar]
- [161].Phillips DM, Drummy LF, Conrady DG, Fox DM, Naik RR, Stone MO, Trulove PC, De Long HC, Mantz RA, J. Am. Chem. Soc 2004, 126, 14350. [DOI] [PubMed] [Google Scholar]
- [162].Cao H, Chen X, Huang L, Shao Z, Mater. Sci. Eng. C 2009, 29, 2270. [Google Scholar]
- [163].Matsumoto A, Lindsay A, Abedian B, Kaplan DL, Macromol. Biosci 2008, 8, 1006. [DOI] [PubMed] [Google Scholar]
- [164].Le TT, Park Y, Chirila TV, Halley PJ, Whittaker AK, Biomaterials 2008, 29, 4268. [DOI] [PubMed] [Google Scholar]
- [165].Holland C, Terry A, Porter D, Vollrath F, Polymer 2007, 48, 3388. [Google Scholar]
- [166].Zhu J, Zhang Y, Shao H, Hu X, Polymer 2008, 49, 2880. [Google Scholar]
- [167].Wang Q, Yang Y, Chen X, Shao Z, Biomacromolecules 2012, 13, 1875. [DOI] [PubMed] [Google Scholar]
- [168].Hodgkinson T, Chen Y, Bayat A, Yuan X-F, Biomacromolecules 2014, 15, 1288. [DOI] [PubMed] [Google Scholar]
- [169].Zhu J, Shao H, Hu X, Int. J. Biol. Macromol 2007, 41, 469. [DOI] [PubMed] [Google Scholar]
- [170].Osswald T, Rudolph N, Polymer Rheology - Fundamentals and Applications, Hanser Publishers, 2015. [Google Scholar]
- [171].Koebley SR, Thorpe D, Pang P, Chrisochoides P, Greving I, Vollrath F, Schniepp HC, Biomacromolecules 2015, 16, 2796. [DOI] [PubMed] [Google Scholar]
- [172].Mandal BB, Grinberg A, Gil ES, Panilaitis B, Kaplan DL, Proc. Natl. Acad. Sci 2012, 109, 7699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Kiyosawa M, Ito E, Shirai K, Kanekatsu R, Miura M, Kiguchi K, Zool. Sci 1999, 16, 215. [Google Scholar]
- [174].Shao Z, Vollrath F, Nature 2002, 418, 741. [DOI] [PubMed] [Google Scholar]
- [175].Jin Y, Zhang Y, Hang Y, Shao H, Hu X, J. Mater. Res 2013, 28, 2897. [Google Scholar]
- [176].Wei W, Zhang Y, Zhao Y, Shao H, Hu X, J. Funct. Polym 2009, 3. [Google Scholar]
- [177].Jin Y, Hang Y, Zhang Y, Shao H, Hu X, Mater. Res. Innovations 2014, 18, S2. [Google Scholar]
- [178].Fang G, Huang Y, Tang Y, Qi Z, Yao J, Shao Z, Chen X, ACS Biomater. Sci. Eng 2016, 2, 1992. [DOI] [PubMed] [Google Scholar]
- [179].Chen Z, Zhang H, Lin Z, Lin Y, van Esch JH, Liu XY, Adv. Funct. Mater 2016, 26, 8978. [Google Scholar]
- [180].Jose RR, Brown JE, Polido KE, Omenetto FG, Kaplan DL, ACS Biomater. Sci. Eng 2015, 1, 780. [DOI] [PubMed] [Google Scholar]
- [181].Nick Pace C, Trevino S, Prabhakaran E, Martin Scholtz J, Philos. Trans. R. Soc., B 2004, 359, 1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Heidebrecht A, Eisoldt L, Diehl J, Schmidt A, Geffers M, Lang G, Scheibel T, Adv. Mater 2015, 27, 2189. [DOI] [PubMed] [Google Scholar]
- [183].Lin Z, Deng Q, Liu XY, Yang D, Adv. Mater 2013, 25, 1216. [DOI] [PubMed] [Google Scholar]
- [184].Wingfield PT, Curr. Protoc. Protein Sci 2016, 84, A. 3F. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Slotta UK, Rammensee S, Gorb S, Scheibel T, Angew. Chem., Int. Ed 2008, 47, 4592. [DOI] [PubMed] [Google Scholar]
- [186].Heim M, Keerl D, Scheibel T, Angew. Chem., Int. Ed 2009, 48, 3584. [DOI] [PubMed] [Google Scholar]
- [187].Jin HJ, Park J, Karageorgiou V, Kim UJ, Valluzzi R, Cebe P, Kaplan DL, Adv. Funct. Mater 2005, 15, 1241. [Google Scholar]
- [188].Lu Q, Hu X, Wang X, Kluge JA, Lu S, Cebe P, Kaplan DL, Acta Biomater 2010, 6, 1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Esselen GJ, Google Patents, 1933.
- [190].Walker AA, Holland C, Sutherland TD, Proc. R. Soc. B 2015, 282, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Truby RL, Lewis JA, Nature 2016, 540, 371. [DOI] [PubMed] [Google Scholar]
- [192].Tumbleston JR, Shirvanyants D, Ermoshkin N, Janusziewicz R, Johnson AR, Kelly D, Chen K, Pinschmidt R, Rolland JP, Ermoshkin A, Science 2015, 347, 1349. [DOI] [PubMed] [Google Scholar]
- [193].Martin JH, Yahata BD, Hundley JM, Mayer JA, Schaedler TA, Pollock TM, Nature 2017, 549, 365. [DOI] [PubMed] [Google Scholar]
- [194].Gantenbein S, Masania K, Woigk W, Sesseg JP, Tervoort TA, Studart AR, Nature 2018, 561, 226. [DOI] [PubMed] [Google Scholar]
- [195].Kotz F, Arnold K, Bauer W, Schild D, Keller N, Sachsenheimer K, Nargang TM, Richter C, Helmer D, Rapp BE, Nature 2017, 544, 337. [DOI] [PubMed] [Google Scholar]
- [196].Klein J, Stern M, Franchin G, Kayser M, Inamura C, Dave S, Weaver JC, Houk P, Colombo P, Yang M, 3D Print. Addit. Manuf 2015, 2, 92. [Google Scholar]
- [197].Grigoryan B, Paulsen SJ, Corbett DC, Sazer DW, Fortin CL, Zaita AJ, Greenfield PT, Calafat NJ, Gounley JP, Ta AH, Science 2019, 364, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Lee A, Hudson A, Shiwarski D, Tashman J, Hinton T, Yerneni S, Bliley J, Campbell P, Feinberg A, Science 2019, 365, 482. [DOI] [PubMed] [Google Scholar]
- [199].Jose RR, Rodriguez MJ, Dixon TA, Omenetto F, Kaplan DL, ACS Biomater. Sci. Eng 2016, 2, 1662. [DOI] [PubMed] [Google Scholar]
- [200].DeBari MK, Keyser MN, Bai MA, Abbott RD, Connect. Tissue Res 2018, DOI: 10.1080/03008207.2018.1553959. [DOI] [PubMed]
- [201].Gregory DA, Zhang Y, Smith PJ, Zhao X, Ebbens SJ, Small 2016, 12, 4048. [DOI] [PubMed] [Google Scholar]
- [202].Włodarczyk-Biegun MK, del Campo A, Biomaterials 2017, 134, 180. [DOI] [PubMed] [Google Scholar]
- [203].Moroni L, Burdick JA, Highley C, Lee SJ, Morimoto Y, Takeuchi S, Yoo JJ, Nat. Rev. Mater 2018, 3, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Schacht K, Jüngst T, Schweinlin M, Ewald A, Groll J, Scheibel T, Angew. Chem., Int. Ed 2015, 54, 2816. [DOI] [PubMed] [Google Scholar]
- [205].Visser J, Melchels FP, Jeon JE, Van Bussel EM, Kimpton LS, Byrne HM, Dhert WJ, Dalton PD, Hutmacher DW, Malda J, Nat. Commun 2015, 6, 6933. [DOI] [PubMed] [Google Scholar]
- [206].Kang H-W, Lee SJ, Ko IK, Kengla C, Yoo JJ, Atala A, Nat. Biotechnol 2016, 34, 312. [DOI] [PubMed] [Google Scholar]
- [207].Chawla S, Midha S, Sharma A, Ghosh S, Adv. Healthcare Mater 2018, 7, 1701204. [DOI] [PubMed] [Google Scholar]
- [208].Dong T, Mi R, Wu M, Zhong N, Zhao X, Chen X, Shao Z, J. Mater. Chem. B 2019, 7, 4328. [Google Scholar]
- [209].Xiong S, Zhang X, Lu P, Wu Y, Wang Q, Sun H, Heng BC, Bunpetch V, Zhang S, Ouyang H, Sci. Rep 2017, 7, 4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Rodriguez MJ, Brown J, Giordano J, Lin SJ, Omenetto FG, Kaplan DL, Biomaterials 2017, 117, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Shi W, Sun M, Hu X, Ren B, Cheng J, Li C, Duan X, Fu X, Zhang J, Chen H, Adv. Mater 2017, 29, 1701089. [DOI] [PubMed] [Google Scholar]
- [212].Zheng Z, Wu J, Liu M, Wang H, Li C, Rodriguez MJ, Li G, Wang X, Kaplan DL, Adv. Healthcare Mater 2017, 7, 1701026. [DOI] [PubMed] [Google Scholar]
- [213].Sommer MR, Schaffner M, Carnelli D, Studart AR, ACS Appl. Mater. Interfaces 2016, 8, 34677. [DOI] [PubMed] [Google Scholar]
- [214].Zhang J, Allardyce BJ, Rajkhowa R, Zhao Y, Dilley RJ, Redmond SL, Wang X, Liu X, ACS Biomater. Sci. Eng 2018, 4, 3036. [DOI] [PubMed] [Google Scholar]
- [215].Zhang J, Allardyce BJ, Rajkhowa R, Kalita S, Dilley RJ, Wang X, Liu X, Mater. Sci. Eng. C 2019, 109784. [DOI] [PubMed] [Google Scholar]
- [216].Yodmuang S, McNamara SL, Nover AB, Mandal BB, Agarwal M, Kelly T-AN, Chao P.-h. G., Hung C, Kaplan DL, Vunjak-Novakovic G, Acta Biomater 2015, 11, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Dickerson MB, Dennis PB, Tondiglia VP, Nadeau LJ, Singh KM, Drummy LF, Partlow BP, Brown DP, Omenetto FG, Kaplan DL, ACS Biomater. Sci. Eng 2017, 3, 2064. [DOI] [PubMed] [Google Scholar]
- [218].Sun L, Parker ST, Syoji D, Wang X, Lewis JA, Kaplan DL, Adv. Healthcare Mater 2012, 1, 729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Sofia S, McCarthy MB, Gronowicz G, Kaplan DL, J. Biomed. Mater. Res 2001, 54, 139. [DOI] [PubMed] [Google Scholar]
- [220].Rodriguez MJ, Dixon TA, Cohen E, Huang W, Omenetto FG, Kaplan DL, Acta Biomater 2018, 71, 379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Wang X, Partlow B, Liu J, Zheng Z, Su B, Wang Y, Kaplan DL, Acta Biomater 2015, 12, 51. [DOI] [PubMed] [Google Scholar]
- [222].Berl T, Trans. Am. Clin. Climatol. Assoc 2009, 120, 389. [PMC free article] [PubMed] [Google Scholar]
- [223].Smay JE, Cesarano J, Lewis JA, Langmuir 2002, 18, 5429. [Google Scholar]
- [224].Chen Z, Zhao D, Liu B, Nian G, Li X, Yin J, Qu S, Yang W, Adv. Funct. Mater 2019, 29, 1900971. [Google Scholar]
- [225].Hinton TJ, Jallerat Q, Palchesko RN, Park JH, Grodzicki MS, Shue H-J, Ramadan MH, Hudson AR, Feinberg AW, Sci. Adv 2015, 1, e1500758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Lee KY, Mooney DJ, Prog. Polym. Sci 2012, 37, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Kuo CK, Ma PX, Biomaterials 2001, 22, 511. [DOI] [PubMed] [Google Scholar]
- [228].Andersen T, Auk-Emblem P, Dornish M, Microarrays 2015, 4, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Bhattacharjee T, Zehnder SM, Rowe KG, Jain S, Nixon RM, Sawyer WG, Angelini TE, Sci. Adv 2015, 1, e1500655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Aizenberg J, Fratzl P, Adv. Mater 2009, 21, 387. [Google Scholar]
- [231].Fratzl P, Journal of the Royal Society Interface 2007, 4, 637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Grzybowski BA, Huck WT, Nat. Nanotechnol 2016, 11, 585. [DOI] [PubMed] [Google Scholar]
- [233].Qiu H, Hudson ZM, Winnik MA, Manners I, Science 2015, 347, 1329. [DOI] [PubMed] [Google Scholar]
- [234].Compaan AM, Christensen K, Huang Y, ACS Biomater. Sci. Eng 2016, 3, 1519. [DOI] [PubMed] [Google Scholar]
- [235].Du X, Wei D, Huang L, Zhu M, Zhang Y, Zhu Y, Mater. Sci. Eng. C 2019, 103, 109731. [DOI] [PubMed] [Google Scholar]
- [236].Bidgoli MR, Alemzadeh I, Tamjid E, Khafaji M, Vossoughi M, Mater. Sci. Eng. C 2019, 103, 109688. [DOI] [PubMed] [Google Scholar]
- [237].Midha S, Kumar S, Sharma A, Kaur K, Shi X, Naruphontjirakul P, Jones JR, Ghosh S, Biomed. Mater 2018, 13, 055012. [DOI] [PubMed] [Google Scholar]
- [238].Zhang M, Zhao M, Jian M, Wang C, Yu A, Yin Z, Liang X, Wang H, Xia K, Liang X, Matter 2019, 1, 20. [Google Scholar]
- [239].Jung CS, Kim BK, Lee J, Min B-H, Park S-H, Tissue Eng. Regener. Med 2018, 15, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Sun K, Li R, Li H, Li D, Jiang W, Int. J. Polym. Mater. Polym. Biomater 2018, 67, 480. [Google Scholar]
- [241].Huang T, Fan C, Zhu M, Zhu Y, Zhang W, Li L, Appl. Surf. Sci 2019, 467, 345. [Google Scholar]
- [242].Kwak H, Shin S, Lee H, Hyun J, J. Ind. Eng. Chem 2019, 72, 232. [Google Scholar]
- [243].Yeon YK, Park HS, Lee JM, Lee JS, Lee YJ, Sultan MT, Seo YB, Lee OJ, Kim SH, Park CH, J. Biomater. Sci., Polym. Ed 2018, 29, 894. [DOI] [PubMed] [Google Scholar]
- [244].Lee J, Sultan M, Kim S, Kumar V, Yeon Y, Lee O, Park C, Int. J. Mol. Sci 2017, 18, 1707. [DOI] [PMC free article] [PubMed] [Google Scholar]


