Abstract
Histoplasma capsulatum is a dimorphic fungus that most frequently causes pneumonia, but can also disseminate and proliferate in diverse tissues. H. capsulatum has a complex secretion system that mediates the release of macromolecule-degrading enzymes and virulence factors. The formation and release of extracellular vesicles (EVs) are an important mechanism for non-conventional secretion in both ascomycetes and basidiomycetes. H. capsulatum EVs contain diverse proteins associated with virulence and are immunologically active. Despite the growing knowledge of EVs from H. capsulatum and other pathogenic fungi, the extent that changes in the environment impact the sorting of organic molecules in EVs has not been investigated. In this study, we cultivated H. capsulatum with distinct culture media to investigate the potential plasticity in EV loading in response to differences in nutrition. Our findings reveal that nutrition plays an important role in EV loading and formation, which may translate into differences in biological activities of these fungi in various fluids and tissues.
Keywords: Histoplasma capsulatum, histoplasmosis, dimorphic fungus, culture media, proteomics, lipidomics, metabolomics, BHI, Ham’s F12, RPMI, extracellular vesicles, USA
Introduction
Extracellular vesicles (EVs) have been identified in all biological kingdoms. Fungal EVs were first described in Cryptococcus neoformans, in 2007, as a trans-cell wall transport mechanism that effectively delivers macromolecules to the extracellular environment (1). The following year, this process was extended to several additional fungi, which demonstrates that this remarkable process is ancient, given that both Basidiomycota and Ascomycota generated EVs (2). Furthermore, research on fungal EVs reveals that their contents contain diverse components associated with fungal virulence, and are recognized by individuals previously infected with the specific fungus (1, 2). Since then, EVs have been shown to modify host effector responses, including altering macrophage activation, macrophage polarization and host susceptibility to fungal challenge (3). Histoplasmosis, caused by the dimorphic fungus Histoplasma capsulatum, is a significant cause of mortality and morbidity on the global stage, and it is the most common cause of fungal pneumonia in the U.S.A (4, 5). Thermally dimorphic fungi are remarkable among fungal pathogens because they cause disease in humans with normal or impaired immune defenses (6). H. capsulatum persists in soil as well as bird and bat excretes (7). Disruption of soil by natural events or by human activities, such as construction, leads to aerosolization of conidia and hyphal fragments. When inhaled into the lungs of a mammalian host (37°C), these infectious propagules convert into pathogenic yeast to cause pneumonia. Once infection is established in the lungs, the yeast can widely disseminate and produce EVs into the host environment (8). We have previously demonstrated that H. capsulatum produces EVs containing a vast array of proteins, lipids and carbohydrates (2). Remarkably, the packaging and release of H. capsulatum EVs can be dynamically regulated by the binding of antibody to the cell surface of this yeast (8, 9) which suggests that other stressors may dramatically impact EV biogenesis and, consequently, composition.
H. capsulatum is commonly cultivated in several different rich or nutritionally restrictive media. However, during infection, the fungus can reside and flourish in fluid and tissue environments throughout the body, ranging from the blood and cerebral spinal fluid to the lung and brain, and variables such as the pH, oxygen content and available nutrients are different in these diverse locations. In this study, we examined the impact of three standard growth media on EVs content loading, which we hypothesized would alter EVs characteristics and payloads. Given that there are significantly different nutritional conditions that H. capsulatum encounters during disseminated disease, exploring variations in EV production in different nutritional environments may lead to insights into EV plasticity and pathogenesis.
Experimental Procedures
Fungal culture
The H. capsulatum wild-type strain G217B (ATCC 26032) was used for all experiments. The strain has been maintained at −80°C and cultivated in liquid medium at 37°C prior to use. The media used for H. capsulatum cultivation were: brain heart infusion (BHI; BD Diagnostic System), Ham’s F-12 Nutrient Mixture (Thermofisher) supplemented with glucose (1.82g/L), glutamic acid (1 g/L), HEPES (6.5 g/L) and L-Cysteine (8.4 mg/L) referenced from a defined medium called Histoplasma macrophage medium (10), and Roswell Park Memorial Institute 1640 (RPMI; Corning Cellgro) supplemented with 1% Nutridoma (Roche Applied Science). BHI and Ham’s F12 are generally considered rich nutritional growth media whereas RPMI is a standard medium used for cell culture.
EV isolation
H. capsulatum G217B was inoculated into Ham’s F-12 medium and incubated at 37°C with rotary agitation at 150 rpm. Fifty mL of yeast cell culture in each media were adjusted to 0.05 Optical Density (OD) and further cultivated for 4 days at 37°C with rotary agitation. Culture supernatants were collected by centrifugation for 10 minutes at 4,000 x g for 20 minutes at 4°C. The supernatants were then filtered through a 0.45 micrometer filter, then concentrated (~15 times) in an Amicon filtration system with a 100-kDa cutoff membrane (Millipore). The collected concentrates were then subjected to 150.000 × gultracentrifugation (Beckman Coulter Optima TLX) with a TLA 100.3 rotor at 4°C, (Beckman Coulter) 2 times for 1 h each, with a suspension step after the first hour. EV pellets were suspended in 200 μL of sterile PBS.
EV Analysis
After isolation, EVs were quantified for protein and ergosterol by indirect methods. Protein quantification was determined by BCA Protein Assay Reagent kit (Pierce) and sterol quantification determined by Amplex Red Sterol assay kit (Invitrogen). EVs were analyzed by Dynamic Light Scattering (DLS) for vesicle size (2).
Multi-omics analysis
EVs were submitted to Metabolite, Protein and Lipid Extraction (MPLEx) and analyzed well-established a protocol as previously tested for different samples, including EVs and fungal cells (11–14). Briefly, EVs were resuspended in 25 μL of 50 mM NH4HCO3 and extracted with 5 volumes of chloroforrmmethanol solution (2:1, v:v). Samples were vortexed, incubated on ice for 5 min and centrifuged for 5 min at 12,000 xg at 4°C to separate the extracts into different solvent layers. Metabolites, proteins and lipids were collected into separate vials and submitted to specific mass spectrometry analysis pipelines. The hydrophilic metabolite layer was dried in a vacuum centrifuge prior to be derivatized with methoxyamine and N-methyl-N-(trimethylsilyl)trifluoroacetamide, and analyzed in a GC 7890A coupled with a single quadrupole MSD 5975C (Agilent Technologies, Inc) (15). Data files were processed with Metabolite Detector (16) for extracting peaks and deconvoluting the fragmentation spectra. Identifications were done by matching against the Fiehn Metabolomics Retention Time Locked (RTL) Library (17). Lipids were dried in a vacuum centrifuged and dissolved in pure methanol before being analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) on an Orbitrap mass spectrometer (Thermo Fisher Scientific) and identifications were done with the LIQUID software (18). Peak areas of manually validated lipid species were extracted with MZmine 2.0 (19). The protein layer is reduced with dithiothreitol, alkylated with iodoacetamide and digested with trypsin (9). Digested peptides were analyzed LC-MS/MS on a Q-Exactive mass spectrometer (Thermo Fisher Scientific). Collected data was processed with MaxQuant (v1.5.5.1) (20) by searching against the H. capsulatum G186AR sequences downloaded from the Uniprot Knowledge Base on August 15, 2016. Methionine oxidation and protein N-terminal acetylation were considered as variable modifications, while cysteine carbamidomethylation was set as fixed modifications. The label-free quantification (LFQ) values were used for quantitative analysis.
Statistical Analysis
Statistical analyses were done by using one-way ANOVA, followed by Tukey’s comparison test and analysis. Function-enrichment analysis was done by annotating H. capsulatum sequences by blasting against the KEGG database (21) testing for enrichment using Fisher’s exact test. Lipid ontology analysis was done using Lipid Mini-On (22). Asterisks presented in figures represent P-values, one asterisk for a value < 0.05, and two asterisks for a value < 0.01.
Results
Protein and ergosterol content and size of EV
EVs isolated from H. capsulatum cultivated in BHI, Ham’s F-12, and RPMI are referred hereafter as EV-BHI, EV-Ham’s, and EV-RPMI, respectively. EV-Ham’s presented the highest yield of both total protein and ergosterol. EV-BHI and EV-RPMI had comparable amounts of protein and ergosterol, which were less than that observed for EV-Ham’s (Fig. 1A and 1B).
Figure 1. Protein and ergosterol content of EVs from H. capsulatum grown in distinct media.
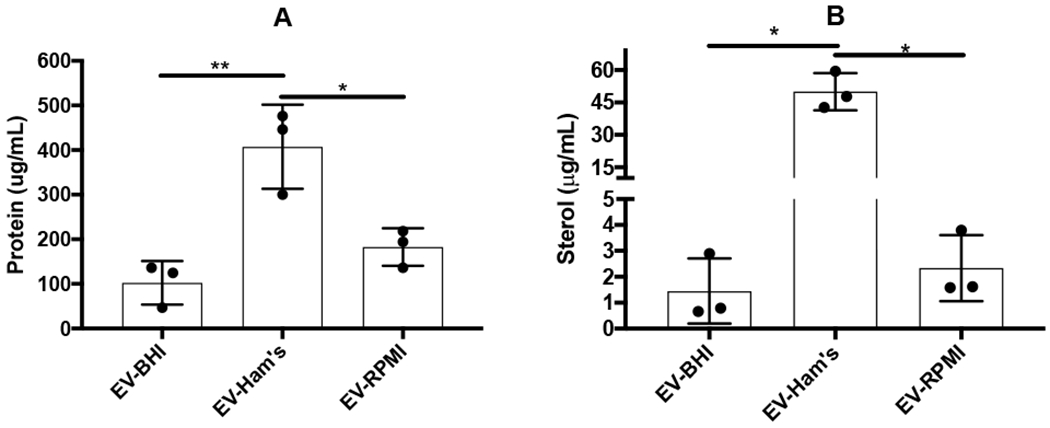
The graph shows the protein (A) and ergosterol (B) quantifications of EV isolated from each culture supernatant. Graphs represent average and standard deviations for at least 3 independent EV isolations. * = p < 0.05, ** = p < 0.01, by Tukey multiple comparison test.
Figure 2 presents the size of the EVs isolated from each medium. DLS analysis of fungal EVs reveals 2 population sizes of EVs in the three conditions, which is similar to prior reports for H. capsulatum and other fungi (23). The small EVs population range from 40 to 70 nm, and the larger from 200 to 500 nm. There was a difference in population sizes of the small and large EVs between EV-BHI and EV-Ham’s, otherwise all EVs are within range of previously described fungal EVs.
Figure 2. Size of EVs cultivated in distinct media by DLS.
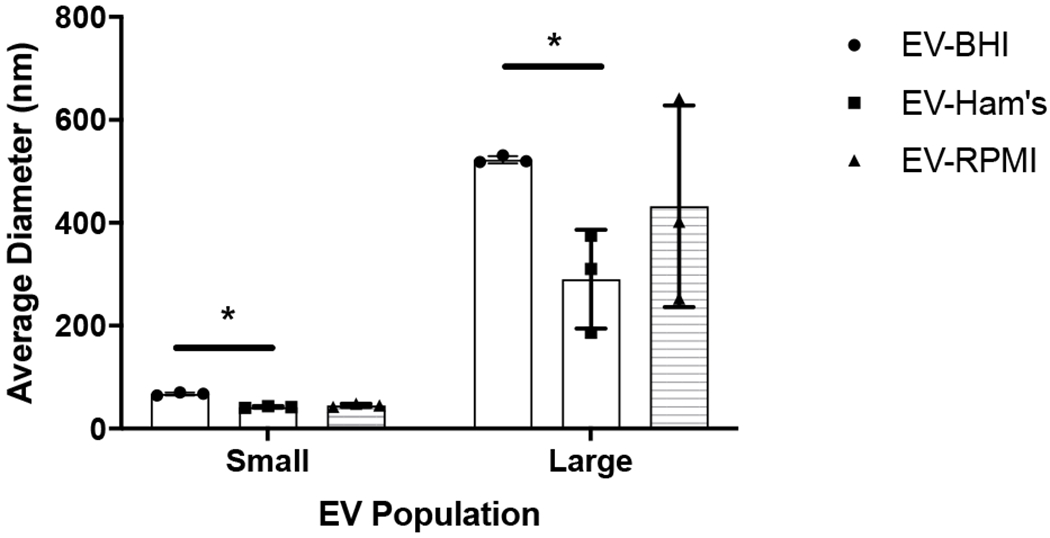
After their isolation from medium supernatant, EVs were analyzed by Dynamic Light Scattering. The graph shows average and standard deviation for samples from 3 independent experiments. * = p < 0.05 by ANOVA followed by Tukey’s multiple comparison test.
Proteomic analysis of EVs
Fungal EVs contain diverse proteins in their cargo, including molecules that act as virulence factors (24). Hence, we analyzed the protein content of EVs released from H. capsulatum cultured in distinct media conditions to determine if differences in nutrition altered the presence of potentially biologically active proteins. The proteomic analysis identified approximately 2,000 proteins, and 270 were significantly altered among the distinct growth conditions. Some of these proteins were found to be involved in key pathways, particularly cellular product synthesis and metabolism. Table 1 shows proteins included in important KEGG pathways, classified by fold enrichment, some of which we will refer to later in the text.
Table 1.
KEGG pathways affected by distinct culture conditions
| Map Description | Fold Enrichment | Fisher test |
|---|---|---|
| Other types of O-glycan biosynthesis | 25.59 | 9.7E-05 |
| Fatty acid biosynthesis | 10.24 | 2.4E-03 |
| Fatty acid degradation | 10.04 | 9.1E-05 |
| Pyruvate metabolism | 9.75 | 8.6E-07 |
| Arginine biosynthesis | 9.48 | 1.2E-04 |
| Citrate cycle (TCA cycle) | 7.58 | 9.6E-05 |
| Alanine, aspartate and glutamate metabolism | 7.06 | 1.5E-04 |
| Oxidative phosphorylation | 6.52 | 5.3E-10 |
| Amino sugar and nucleotide sugar metabolism | 6.46 | 7.4E-05 |
| Fatty acid metabolism | 6.32 | 8.8E-04 |
| 2-Oxocarboxylic acid metabolism | 5.17 | 2.2E-03 |
| Propanoate metabolism | 5.12 | 1.7E-02 |
| Glyoxylate and dicarboxylate metabolism | 5.06 | 6.4E-03 |
| Ribosome | 4.88 | 7.4E-07 |
| Peroxisome | 4.88 | 4.4E-04 |
| Starch and sucrose metabolism | 4.88 | 7.3E-03 |
| Valine, leucine and isoleucine degradation | 4.71 | 8.2E-03 |
| Glycolysis / Gluconeogenesis | 4.27 | 1.1E-02 |
| Phenylalanine metabolism | 4.27 | 2.7E-02 |
| Carbon metabolism | 4.22 | 2.0E-05 |
| Aminoacyl-tRNA biosynthesis | 4.16 | 5.4E-03 |
| Endocytosis | 4.14 | 5.3E-04 |
| Protein processing in endoplasmic reticulum | 4.10 | 2.6E-04 |
| Biosynthesis of antibiotics | 3.94 | 6.5E-09 |
| Biosynthesis of amino acids | 3.86 | 4.7E-05 |
| Biosynthesis of secondary metabolites | 3.73 | 1.5E-10 |
| Metabolic pathways | 3.49 | 3.9E-23 |
The proteins were grouped into four different clusters according to their abundance in BHI [group 4], RPMI [group 3], Ham’s [group 2] and BHI and Ham’s [group 1]. There were more abundant proteins in EV-Ham’s, and a greater variance in EV-BHI within the replicates.
EV-BHI and EV-Ham’s presented similar protein yields in group 4, which contains most of the protein yields. However, the majority of altered proteins were significantly abundant in EV-Ham’s compared to EV-BHI and EV-RPMI. The most variation in protein alteration between the EV-BHI, EV-Ham’s, and EV-RPMI occurred in group 1. Proteins from EV-RPMI were most abundant in group 2 (Figure 2).
The proteins abundant in both EV-Ham’s and EV-BHI were in the largest cluster in the heatmap (Fig. 2). In this cluster, important proteins involved with chitin metabolism were found, including chitin synthase (C0NZK3, C0NSP8 and C0NDJ0) and chitinase (C0NSB6). Also, enzymes involved with host response modulation were detected, such as peroxidase (C0NND7) and alkaline phosphatase (C0NBI7). Lipid metabolism enzymes were also abundant in these EVs, such as carnitine (C0NWZ6), long-chain-fatty-acid-CoA ligase (C0P0B7), acetyl-CoA carboxylase (C0NM24). EV-BHI presented an abundance of sugar metabolism-related enzymes, including alpha-glucosidase (C0NEI5) and glucosidase I (C0NHJ7). Interestingly, EV-RPMI didn’t present abundance of enzymes involved in cell wall remodeling and the only lipid-related abundant protein in EV-RPMI was the Inositol-3-phosphate synthase (C0NLK4). Among the proteins exported in EV-Ham’s, the high affinity copper transporter (C0NSW0), abundant only in EV-Ham’s. EV-Ham’s had the highest abundance of siderochrome-iron uptake transporter (C0NX70), as well as V-type proton ATPase subunit A (C0NJV7).
Lipidomic analysis of EVs
The lipidomic analysis detected 100 lipids, with significant alterations in 44 lipids among the different media conditions. Among the significantly altered lipids, the greatest variety was found in EV-Ham’s, in mostly species of phosphatidylethanolamine (PE) and phosphatidylcholine (PC). Figure 4A shows a heatmap with the significantly altered lipids. The heatmap shows 2 distinct clusters of altered lipids. The biggest cluster shows lipids that were abundant in EV-Ham’s, and the smaller cluster shows EV-RPMI abundant lipids. Lipids abundant in EV-BHI are present in both clusters, except for the sphingomyelins, which were abundant only in EV-BHI.
Figure 4.
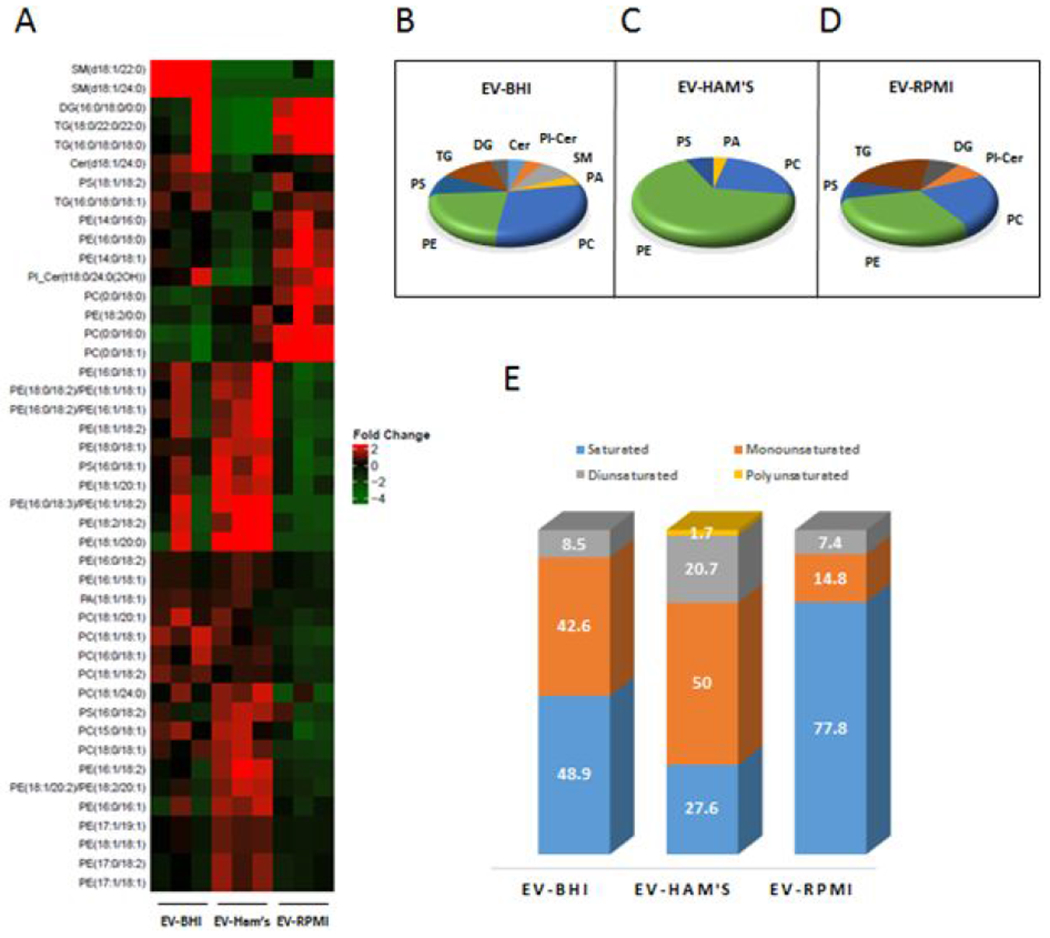
Heatmap with lipids significantly altered in EVs released by H. capsulatum cultured under different media.
EV-BHI presented the largest variety in class of lipids among the tested conditions, including several species of sphingomyelin, one ceramide, and a phosphoinositol ceramide. Phosphoinositol ceramide was also abundant in both EV-BHI and EV-RPMI. EV-Ham’s yielded significantly lower abundances of triacylglycerides (TG) and diacylglycerides (DG) compared to EV-BHI and EV-RPMI (Fig. 4B–4C). Most of the abundant lipids found in EV-RPMI (~78%) had saturated fatty acids (FA), while ~70% of the FA found in lipids abundant in EV-Ham’s were unsaturated. Regarding the presence of unsaturated lipids, EV-BHI’s lipids were in an intermediary level between the other conditions. (Fig. 3E).
Figure 3. EV proteins in different media conditions.
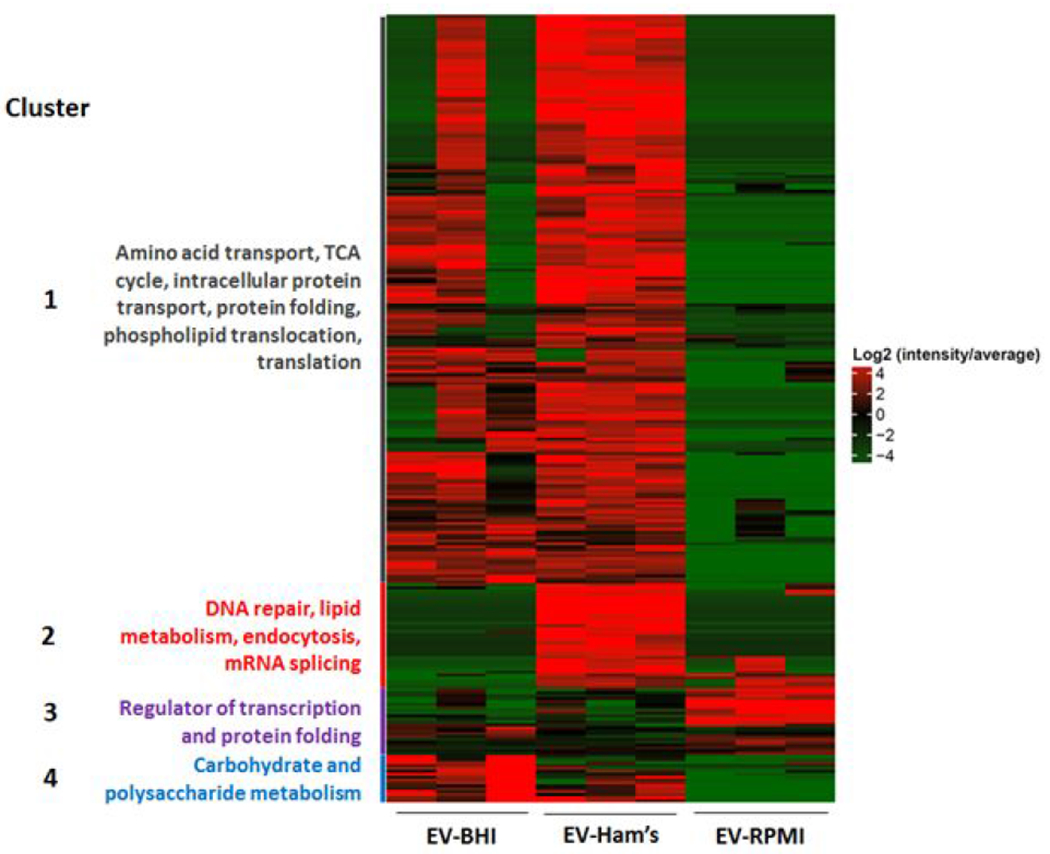
The heatmap shows differential abundance of proteins in EVs produced by H. capsulatum yeast cells cultivated in distinct media. Proteins were grouped based on function. Group 1: Amino acid transport, TCA cycle, intracellular protein transport, phospholipid translocation, and translation. Group 2: DNA repair, lipid metabolism, endocytosis, and mRNA splicing. Group 3: Transcription regulators and protein folding. Group 4: Carbohydrate and polysaccharide metabolism
Lysophospholipids were more abundant in EV-RPMI, especially lysophosphatidylcholine (LysoPC). LysoPCs were more abundant in EV-RPMI and EV-Ham’s compared to EV-BHI (Figure 5). One species of lysophosphatidylethanolamine (LysoPE) was present in EV-RPMI and EV-Ham’s. The few odd-chain FA found to be abundant in our analysis were present mostly in EV-Ham’s, and none of them were lysophospholipids.
Figure 5. Lysophospholipids in EVs from H. capsulatum grown in different media.

This figure shows the abundance of distinct lysophospholipids found in each condition. Significant differences were found between EV-RPMI and EV-BHI. * = p < 0.05 by one-way ANOVA followed by Dunn’s multiple comparisons test.
Metabolomic analysis of EVs
Carbohydrate metabolites vary in the EVs from each condition. EV-Ham’s presents the most abundant D-glucose content (Figure 6); however, the amount of D-glucose present in the media BHI and RPMI was not translated in the abundance of this carbohydrate in EV generated in their presence. Carbohydrates such as maltose, isomaltose, D-allose, and pyruvic acid, also associated with energy storage and production, were most abundantly found in EV-Hams. Although EV-Ham’s presents a majority of the carbohydrate metabolites, EV-BHI presents more abundance of trehalose and fructose. EV-BHI presents the most abundance of trehalose, and the least abundance of glucose and pyruvic acid compared to EV-Hams and EV-RPMI. EV-BHI had the highest abundance of L-ornithine even though the abundance of arginase was the same among all tested conditions. The high abundance of ethanolamine in EV-Ham’s reflects the lipidomic profile of these EV, where phospholipids containing ethanolamine represent more than half of the lipid classes found in this experimental condition.
Figure 6. Metabolic pathways affected by different media.
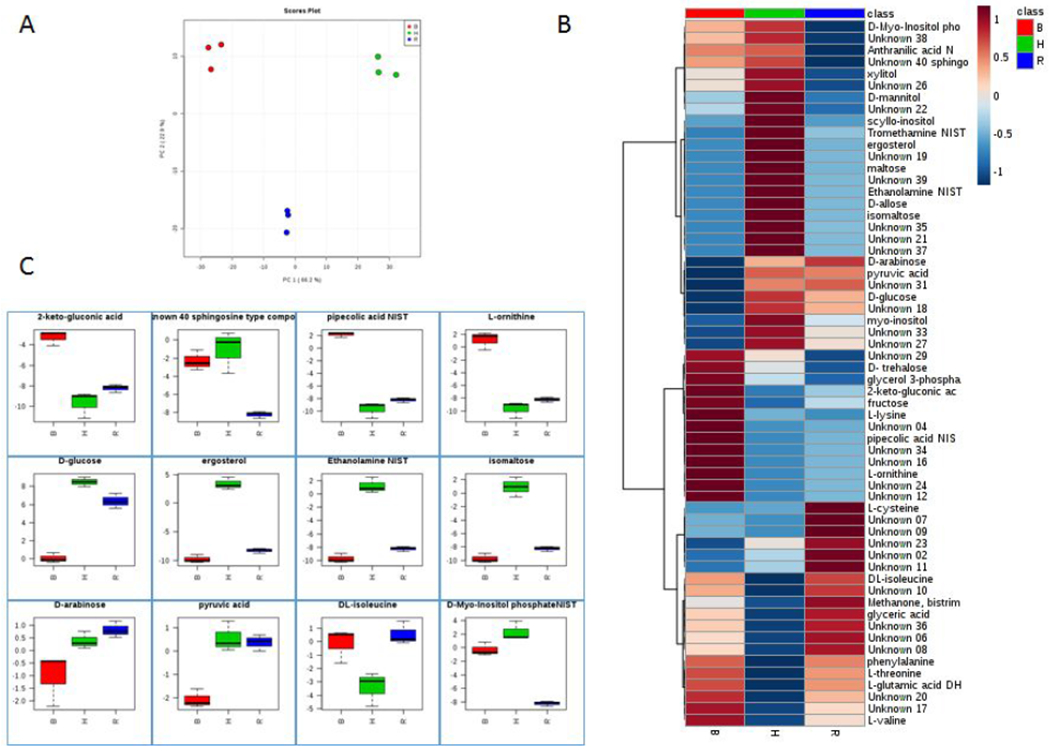
The PCA plot (A) illustrates the distinction between the metabolic profiles of EVs from different media. In the heatmap (B) the metabolites detected in EVs from H. capsulatum cultivated in different media. Some of the detected metabolites were depicted (C) to show their variation among EVs from different growth conditions.
Discussion
Compositional analyses of fungal EVs are essential to the understanding of how these extracellular compartments exert their functions. For instance, there is now concrete evidence that fungal EVs modulate virulence transfer between distinct fungal strains (25) activation of immune cells (26–29), prion transfer(30), and drug resistance in biofilms(31) among other functions. However, in the absolute majority of the studies showing fundamental biological functions of EVs, the bioactive vesicular components remained unknown. Therefore, knowledge on the compositional diversity of fungal EVs directly impact the understanding of their functions in nature, but also the design of EV-based biotechnological tools, including vaccines and therapeutic agents.
The initial description of EVs in C. neoformans has been followed by numerous interesting reports of EVs produced in other important human pathogenic fungi. Although EVs can also be released from non-pathogenic fungi, as well as in organisms from other kingdoms, their release by pathogenic fungi has called the attention of microbiologists mainly because these EVs can carry virulence factors (24). Although many aspects about fungal EV biology and their effects on hosts have been addressed to date, it is still poorly understood whether environmental conditions modulate EV cargo. Previously we partially explored this question, comparing EVs from H. capsulatum released in the presence or absence of monoclonal antibodies against a heat-shock protein exposed on the surface of H. capsulatum and found that antibody dynamically altered EV cargo in a dose-dependent manner (32). In the current work we analyzed the content of EV released from H. capsulatum cultivated in distinct media conditions, to evaluate EV plasticity regarding their cargo and constitution in response to nutrition.
Each of the medium tested in our study has a different chemical composition. BHI is a nutritious, buffered culture medium that contains infusions of brain and heart tissue and peptones to supply protein and other nutrients necessary to support the growth of fastidious and nonfastidious microorganisms (33). Ham’s F-12 was designed for serum-free single-cell plating of Chinese Hamster Ovary (CHO) cells, and has been used for a wide range of mammalian cells as well as bacteria and fungi. Ham’s F-12 Nutrient Mixture medium supplements were referenced from a defined medium called Histoplasma macrophage medium aforementioned. RPMI is unique from other medium because it contains the reducing agent glutathione and high concentrations of vitamins, inositol and choline. A full description of media components has been included in supplemental Table S1. Nutridoma, was added to RPMI as a supplement. It consists as a biochemically defined replacement for Fetal Bovine Serum (FBS). We observed the growth of H. capsulatum based on OD and were able to determine when the fungus reached stationary phase growth. This stage of growth was determined to be 96 hours after initial inoculation in each of the three media.
Proteins and lipids, among other organic molecules, are present in H. capsulatum EVs, some of which are closely linked to virulence during infection and disease. EV-Ham’s had the highest amount of both protein and ergosterol. However, these EVs were the smallest in size among the tested conditions, being smaller than EV-BHI and statistically similar to EV-RPMI, suggesting that EV-Ham’s are either produced in higher amounts than the other conditions, or that they are “denser” than EVs from the other media. Application of methods that could directly measure EV counts in each medium could answer this question in the future. Following the total protein content, the proteomic analysis showed that most of the altered proteins were abundantly found in EV-Ham’s, as EV-RPMI had only a few proteins significantly more abundant than the other conditions. Interestingly, EV-RPMI had lower abundance of proteins associated with carbohydrate and polysaccharide metabolism in Cluster 4 (Figure 3) even though RPMI medium contains higher levels of glucose (10%) compared to Ham’s F-12. EV-BHI showed some inconsistencies between its replicates, probably because of the nature of the medium, where different animals’ brain and heart material are present, so variations between distinct lots might occur. It may be possible that a few of the proteins analyzed were directly from the media itself. However, when we subjected the different media without fungal inoculation to our EV isolation protocol and tested residual fluid by DLS, there were no EVs detected (data not shown). In comparison, Ham’s F-12 medium was the most consistent condition where the yeast cells exported, through EVs, the largest variety of proteins. This feature might reflect how suitable this medium is on the growth of H. capsulatum for experiments that involve host-pathogen interactions. Copper and some other micronutrients in the host environment can become toxic to host pathogens. Among the proteins exported in EV-Ham’s, the high affinity copper transporter (C0NSW0), abundant only in EV-Ham’s, is an important immune subversion protein mechanism as one of the strategies used by host phagocytes to inhibit the viability of intracellular H. capsulatum is to reduce the access of copper into the phagosome (34). Therefore, the presence of this metal transporter can protect the host pathogen by inhibiting the use of copper in host defense cells. The presence of this metal transporter can be due to the fact that Ham’s F-12 medium contains the micronutrient metal cupric sulfate, while the other media do not. EV-Ham’s had the highest abundance of siderochrome-iron uptake transporter (C0NX70), a protein essential for the fungus to survive in environments with low iron availability. Iron acquisition in H. capsulatum also relies on the cooperative action of proton ATPases as the V-type proton ATPase subunit A (C0NJV7), which is also consistently abundant in EV-Ham’s.
The lipid profile of EV cultivated in distinct media was remarkably distinct. EV-BHI incorporated or synthesized different classes of lipids, as ceramides and sphingomyelins were present almost in an exclusive way in this condition. Ham’s F12 and RPMI are more defined media, while BHI has brain and heart material different animals, and it is the only one of these media that actually carries lipids, with the exception of linoleic acid present in Ham’s. Sphingomyelins are not synthesized by yeast cells due to the lack of sphingomyelin synthase in fungi (35), but they are probably up taken from media and were loaded into EVs. Nevertheless, polyunsaturated FA were consistently found only in EV-Ham’s, as well as the highest abundance of diunsaturated FA. The condition with fewer unsaturated FA was also found in EV-Ham’s. Almost 80% of the abundant FA in EV-RPMI were unsaturated, and accordingly the ergosterol presence in these EV was quite low. EV-Hams having fewer unsaturated FA is presumably balanced by the highest amount of ergosterol, limiting the EVs membrane fluidity. EV-RPMI also presented the highest abundance of lysophospholipids, despite the equal abundance of phospholipases in all the conditions. This data suggests that in this condition, the lysis of phospholipids would take place before the assembling of EV by intracellular phospholipases, or even in the extracellular milieu by secreted phospholipases non associated with EVs. In any case, differences in lipid composition induced by growth conditions might impact virulence and/or immunological potential. For instance, fungal EVs enriched with the immunogenic glycolipid ergosterylglucoside induced protection of the invertebrate host Galleria mellonella against a lethal challenge with C. neoformans (26).
Carbohydrate metabolites found within the EVs were also interesting. The glucose in each medium is described as follows: BHI medium provides 3 g/L of dextrose, Ham’s medium is supplemented with 1.82 g/L of glucose, and RPMI 1640 has 2 g/L of glucose. EV-Ham’s presented the highest amount of carbohydrates associated with energy production and storage. This could possibly be due to the high amounts received from the medium. Besides energy production, EV-RPMI also presented high amounts of carbohydrates associated with energy production. Trehalose, a stress tolerance related carbohydrate (36, 37), had increased abundance in EV-BHI. It has also been reported to be involved in mechanisms like substituting water molecules in cell membrane to protect from desiccation and freezing damage, conferring thermotolerance by stabilizing proteins during heat shock, and acting as a compatible osmolyte (38). Other functions of trehalose include being an alternative carbon source for filamentous fungi, especially in glycogen deficient environments (39). The high abundance of trehalose in EV-BHI can be related to the medium composition of mammalian ingredients, compounded with host temperate growth conditions. The presence of L-ornithine in EV-BHI does not reflect the abundance of arginase in these EVs, as this enzyme was equally abundant among all conditions. This pattern suggests that although it is possible to have L-ornithine synthesis inside EVs, the differential abundance found in EV-BHI was probably due to an increased synthesis of this metabolite inside the yeast cells or even a differential sorting during EV biogenesis.
Fungal EV cargo can be modulated by host factors, such as specific antibody binding to fungal cell surface proteins (9). However, the impact of media condition on EV biogenesis has not been examined. With this study, we demonstrate remarkable alterations of fungal EV cargo loading and EV release based on the available nutrients for fungal growth, and begin to reveal how these conditions can be related to cell-host interactions. These findings provide a strong proof of concept that that different host and environmental conditions, in this case media, can induce changes in the EV composition that may potentially lead to a more or less virulent state of a fungus.
Supplementary Material
Acknowledgements
J.D.N. was supported in part by NIH R01AI052733 and R21AI124797. E.S.N. was partially supported by NIH R21AI124797. Parts of this work were performed in the Environmental Molecular Science Laboratory, a U.S. Department of Energy (DOE) national scientific user facility at Pacific Northwest National Laboratory (PNNL) in Richland, WA. MLR and LN are supported by grants from the Brazilian agency Conselho Nacional de Desenvolvimento Cientifico e Tecnologico (CNPq, grants 405520/2018-2, 440015/2018-9, and 301304/2017-3 to M.L.R.; 311179/2017-7 and 408711/2017-7 to L.N.) and Fiocruz (grants VPPCB-007-FIO-18-2-57 and VPPIS-001-FIO-18-66 to M.L.R,). We also acknowledge support from the Instituto Nacional de Ciência e Tecnologia de Inovação em Doenças de Populações Negligenciadas (INCT-IDPN). M.L.R. is currently on leave from the position of Associate Professor at the Microbiology Institute of the Federal University of Rio de Janeiro, Brazil.
Footnotes
Conflict of Interest
We declare that there are no conflicts of interest.
References
- 1.Rodrigues ML, Nimrichter L, Oliveira DL, Frases S, Miranda K, Zaragoza O, et al. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot Cell. 2007;6(1):48–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albuquerque PC, Nakayasu ES, Rodrigues ML, Frases S, Casadevall A, Zancope-Oliveira RM, et al. Vesicular transport in Histoplasma capsulatum: an effective mechanism for trans-cell wall transfer of proteins and lipids in ascomycetes. Cellular microbiology. 2008;10(8):1695–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zamith-Miranda D, Nimrichter L, Rodrigues ML, Nosanchuk JD. Fungal extracellular vesicles: modulating host-pathogen interactions by both the fungus and the host. Microbes Infect. 2018;20(9–10):501–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cleare LG, Zamith-Miranda D, Nosanchuk JD. Heat Shock Proteins in Histoplasma and Paracoccidioides. Clinical and vaccine immunology : CVI. 2017;24(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu JH, Feudtner C, Heydon K, Walsh TJ, Zaoutis TE. Hospitalizations for endemic mycoses: a population-based national study. Clin Infect Dis. 2006;42(6):822–5. [DOI] [PubMed] [Google Scholar]
- 6.Gauthier GM. Fungal Dimorphism and Virulence: Molecular Mechanisms for Temperature Adaptation, Immune Evasion, and In Vivo Survival. Mediators of inflammation. 2017;2017:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prevention CfDCa. Fungal Disease 2014. [updated September 30, 2014 Available from: https://www.cdc.gov/fungal/global/index.html.
- 8.Baltazar LM, Zamith-Miranda D, Burnet MC, Choi H, Nimrichter L, Nakayasu ES, et al. Concentration-dependent protein loading of extracellular vesicles released by Histoplasma capsulatum after antibody treatment and its modulatory action upon macrophages. Scientific reports. 2018;8(1):8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matos Baltazar L, Nakayasu ES, Sobreira TJ, Choi H, Casadevall A, Nimrichter L, et al. Antibody Binding Alters the Characteristics and Contents of Extracellular Vesicles Released by Histoplasma capsulatum. mSphere. 2016;1(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Worsham PL, Goldman WE. Quantitative plating of Histoplasma capsulatum without addition of conditioned medium or siderophores. Journal of medical and veterinary mycology : bi-monthly publication of the International Society for Human and Animal Mycology. 1988;26(3):137–43. [PubMed] [Google Scholar]
- 11.Nakayasu ES, Nicora CD, Sims AC, Burnum-Johnson KE, Kim YM, Kyle JE, et al. MPLEx: a Robust and Universal Protocol for Single-Sample Integrative Proteomic, Metabolomic, and Lipidomic Analyses. mSystems. 2016; 1(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coelho C, Brown L, Maryam M, Vij R, Smith DFQ, Burnet MC, et al. Listeria monocytogenes virulence factors, including listeriolysin O, are secreted in biologically active extracellular vesicles. The Journal of biological chemistry. 2019;294(4):1202–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zamith-Miranda D, Heyman HM, Cleare LG, Couvillion SP, Clair GC, Bredeweg EL, et al. Multi-omics Signature of Candida auris, an Emerging and Multidrug-Resistant Pathogen. mSystems. 2019;4(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicora CD, Sims AC, Bloodsworth KJ, Kim YM, Moore RJ, Kyle JE, et al. Metabolite, Protein, and Lipid Extraction (MPLEx): A Method that Simultaneously Inactivates Middle East Respiratory Syndrome Coronavirus and Allows Analysis of Multiple Host Cell Components Following Infection. Methods Mol Biol. 2020;2099:173–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ansong C, Schrimpe-Rutledge AC, Mitchell HD, Chauhan S, Jones MB, Kim YM, et al. A multi-omic systems approach to elucidating Yersinia virulence mechanisms. Mol Biosyst. 2013;9(1):44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiller K, Hangebrauk J, Jager C, Spura J, Schreiber K, Schomburg D. MetaboliteDetector: comprehensive analysis tool for targeted and nontargeted GC/MS based metabolome analysis. Anal Chem. 2009;81(9):3429–39. [DOI] [PubMed] [Google Scholar]
- 17.Kind T, Wohlgemuth G, Lee DY, Lu Y, Palazoglu M, Shahbaz S, et al. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal Chem. 2009;81(24):10038–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kyle JE, Crowell KL, Casey CP, Fujimoto GM, Kim S, Dautel SE, et al. LIQUID: an-open source software for identifying lipids in LC-MS/MS-based lipidomics data. Bioinformatics. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pluskal T, Castillo S, Villar-Briones A, Oresic M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11(12):2301–19. [DOI] [PubMed] [Google Scholar]
- 21.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J Mol Biol. 2016;428(4):726–31. [DOI] [PubMed] [Google Scholar]
- 22.Clair G, Reehl S, Stratton KG, Monroe ME, Tfaily MM, Ansong C, et al. Lipid Mini-On: Mining and ontology tool for enrichment analysis of lipidomic data. Bioinformatics. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joffe LS, Nimrichter L, Rodrigues ML, Del Poeta M. Potential Roles of Fungal Extracellular Vesicles during Infection. mSphere. 2016;1(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodrigues ML, Godinho RM, Zamith-Miranda D, Nimrichter L. Traveling into Outer Space: Unanswered Questions about Fungal Extracellular Vesicles. PLoS pathogens. 2015;11(12):e1005240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bielska E, Sisquella MA, Aldeieg M, Birch C, O’Donoghue EJ, May RC. Pathogen-derived extracellular vesicles mediate virulence in the fatal human pathogen Cryptococcus gattii. Nature communications. 2018;9(1):1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colombo AC, Rella A, Normile T, Joffe LS, Tavares PM, de SAGR, et al. Cryptococcus neoformans Glucuronoxylomannan and Sterylglucoside Are Required for Host Protection in an Animal Vaccination Model. MBio. 2019;10(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vargas G, Rocha JD, Oliveira DL, Albuquerque PC, Frases S, Santos SS, et al. Compositional and immunobiological analyses of extracellular vesicles released by Candida albicans. Cellular microbiology. 2015;17(3):389–407. [DOI] [PubMed] [Google Scholar]
- 28.Bitencourt TA, Rezende CP, Quaresemin NR, Moreno P, Hatanaka O, Rossi A, et al. Extracellular Vesicles From the Dermatophyte Trichophyton interdigitale Modulate Macrophage and Keratinocyte Functions. Frontiers in immunology. 2018;9:2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peres da Silva R, Heiss C, Black I, Azadi P, Gerlach JQ, Travassos LR, et al. Extracellular vesicles from Paracoccidioides pathogenic species transport polysaccharide and expose ligands for DC-SIGN receptors. Scientific reports. 2015;5:14213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu S, Hossinger A, Hofmann JP, Denner P, Vorberg IM. Horizontal Transmission of Cytosolic Sup35 Prions by Extracellular Vesicles. MBio. 2016;7(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zarnowski R, Sanchez H, Covelli AS, Dominguez E, Jaromin A, Bernhardt J, et al. Candida albicans biofilm-induced vesicles confer drug resistance through matrix biogenesis. PLoS biology. 2018;16(10):e2006872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guimaraes AJ, Frases S, Gomez FJ, Zancope-Oliveira RM, Nosanchuk JD. Monoclonal antibodies to heat shock protein 60 alter the pathogenesis of Histoplasma capsulatum. Infection and immunity. 2009;77(4):1357–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenow EC. Focal infection with special reference to oral sepsis. Journal of Dental Research. 1919:205–49. [Google Scholar]
- 34.Shen Q, Beucler MJ, Ray SC, Rappleye CA. Macrophage activation by IFN-gamma triggers restriction of phagosomal copper from intracellular pathogens. PLoS pathogens. 2018;14(11):e1007444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christie WW. Sphingomyelin and Related Sphingophospholipids Dundee, Scotland: LIPID MAPS; 2019. [Available from: https://www.lipidmaps.org/resources/lipidweb/index.php?page=lipids/sphingo/sph/index.htm. [Google Scholar]
- 36.Estruch F Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiology Reviews. 2000;24(4):469–86. [DOI] [PubMed] [Google Scholar]
- 37.François J, Parrou JL. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiology Reviews. 2001;25(1):125–45. [DOI] [PubMed] [Google Scholar]
- 38.Hare PD, Cress WA, Van Staden J. Dissecting the roles of osmolyte accumulation during stress. Plant, Cell & Environment. 1998;21(6):535–53. [Google Scholar]
- 39.Thammahong A, Puttikamonkul S, Perfect JR, Brennan RG, Cramer RA. Central Role of the Trehalose Biosynthesis Pathway in the Pathogenesis of Human Fungal Infections: Opportunities and Challenges for Therapeutic Development. Microbiology and Molecular Biology Reviews. 2017;81(2):e00053–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


