Summary
Adult mammalian hematopoietic stem cells (HSCs) reside in the bone marrow (BM) but can be mobilized into blood for use in transplantation. HSCs interact with BM niche cells producing growth factor c-Kit ligand (Kitl/SCF) and chemokine CXCL12, and were thought to be static and sessile. We used two-photon laser scanning microscopy to visualize genetically labeled HSCs in the BM of live mice for several hours. The majority of HSCs showed dynamic non-spherical morphology and significant motility, undergoing slow processive motion interrupted by short stretches of confined motion. HSCs moved in the perivascular space and showed intermittent close contacts with SCF-expressing perivascular stromal cells. In contrast, mobilization-inducing blockade of CXCL12 receptor CXCR4 and integrins rapidly abrogated HSC motility and shape dynamics in real time. Our results reveal an unexpectedly dynamic nature of HSC residence in the BM and interaction with the SCF+ stromal niche, which is disrupted during HSC mobilization.
Graphical Abstract
eTOC Blurb
Hematopoietic stem cells (HSCs) produce all blood cell types throughout the adult life, but their behavior in their native bone marrow niche is poorly understood. Upadhaya and colleagues show that HSCs exhibit dynamic morphology and complex motility patterns that involve serial interactions with stromal cells in the bone marrow.
Introduction
Hematopoietic stem cells (HSCs) in the mammalian bone marrow (BM) are multipotent, self-renewing stem cells that can generate all blood cell lineages and maintain long-term hematopoiesis when transplanted into conditioned recipients (Eaves, 2015; Laurenti and Gottgens, 2018). In the steady state, HSCs also contribute to all hematopoietic lineages (Busch et al., 2015) and serve as a major source of unperturbed hematopoiesis in mice (Chapple et al., 2018; Sawai et al., 2016) and humans (Scala and Aiuti, 2019). The unique capacity of HSCs for hematopoietic reconstitution enables BM transplantation (BMT) to treat hematologic malignancies and other life-threatening diseases (Copelan, 2006). The rarity of HSCs in the BM puts a major limitation on the success of BMT, necessitating the strategies to mobilize HSCs from the BM into peripheral circulation (Hopman and DiPersio, 2014).
HSCs reside in a unique BM environment, termed the niche, which enables their long-term maintenance and multipotency (Crane et al., 2017; Wei and Frenette, 2018). HSCs are localized perivascularly, with both sinusoidal vessels and arterioles proposed as important sites of HSC localization (Acar et al., 2015; Chen et al., 2016; Itkin et al., 2016; Kunisaki et al., 2013). Critical local signals for the maintenance of HSCs and progenitors in the BM are provided by the growth factor c-Kit ligand (KITL, also known as stem cell factor or SCF) and the chemokine CXCL12 (also known as stroma-derived factor-1 or SDF-1). Both factors are highly expressed by perivascular stromal cells located primarily around the sinusoidal vessels in the BM (Ding and Morrison, 2013; Ding et al., 2012; Greenbaum et al., 2013; Omatsu et al., 2010). The interaction of HSCs with their BM niche is a critical target of HSC mobilization: in particular, pharmacological targeting of the CXCL12 receptor CXCR4 alone (Broxmeyer et al., 2005) or in combination with other receptors such as integrins (Cao et al., 2016; Rettig et al., 2012) causes rapid HSC mobilization that can be harnessed in clinical BMT.
In previous studies on the visualization of HSCs and progenitors, these cells were isolated from the donor BM, labeled with fluorescent dyes and transferred into recipient mice, followed by intravital microscopy shortly thereafter (Tjin et al., 2019). Purified murine or human HSCs were transplanted into irradiated recipient mice, whereupon they showed spheric morphology and no motility (Foster et al., 2015; Lo Celso et al., 2009; Rashidi et al., 2014). However, irradiation causes a major disruption of the BM microenvironment including the loss of vascular integrity (Lo Celso et al., 2009), and HSC transplantation involves non-physiological events such as intravasation. Recently, intravital microscopy of genetically labeled endogenous HSCs suggested their limited motility, which was increased by long-term treatment with cytotoxic drugs and/or cytokine G-CSF (Christodoulou et al., 2020). However, only few cells were examined in the steady state, warranting the analysis of a broader HSC population over longer periods of time. Furthermore, the interaction of HSCs with BM stromal cells and behavior during rapid mobilization have not been observed so far.
We have previously described a mouse model for genetic labeling of HSCs in vivo, based on inducible HSC-specific Cre recombination. The labeled HSCs reside at the top of hematopoietic hierarchy as judged by their undifferentiated phenotype and transcriptome, superior reconstitution capacity during transplantation and sustained multilineage contribution to long-term hematopoiesis in the steady state (Sawai et al., 2016; Upadhaya et al., 2018). Here we applied this model for specific labeling of HSCs and their visualization along with the SCF+ stromal cells in the BM of live adult mice. We report that HSCs show a dynamic morphology and a complex pattern of motile behavior while maintaining close contact with SCF+ cells. The motility of HSCs is disrupted during their rapid mobilization from the BM, suggesting that it represents a fundamental feature of HSC interaction with their functional niche.
Results
A system for intravital imaging of endogenous HSCs
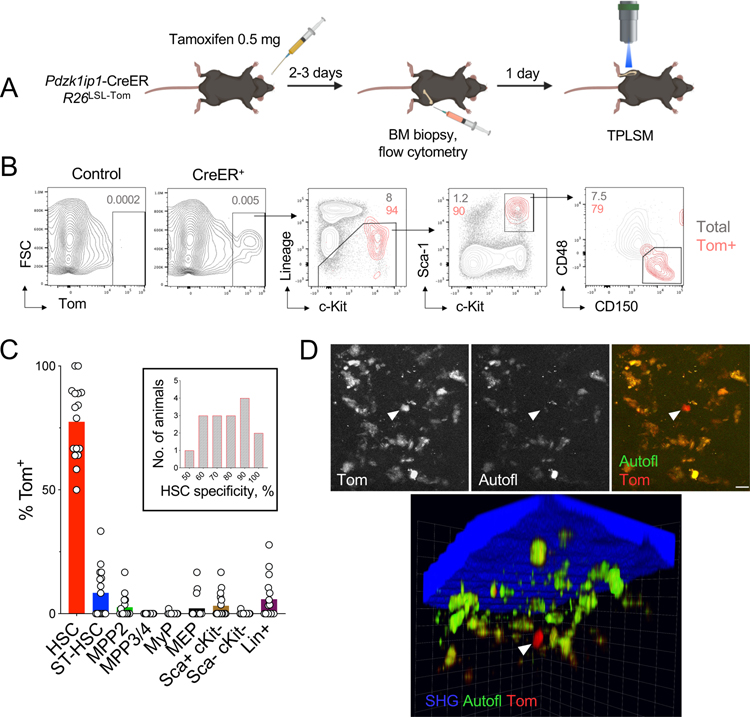
Our HSC labeling system is based on tamoxifen-inducible Cre recombinase (Cre-estrogen receptor fusion or CreER) expressed from the HSC-specific Pdzk1ip1 gene in the bacterial artificial chromosome (BAC) transgene. In Pdzk1ip1-CreER BAC transgenic mice crossed with the Rosa26-Lox-Stop-Lox(LSL)-tdTomato (R26LSL-Tom) reporter allele, the administration of tamoxifen induces the expression of red fluorescent protein tdTomato (Tom) in ~20–30% of phenotypic HSCs (Sawai et al., 2016). To visualize HSCs, we administered tamoxifen to Pdzk1ip1-CreER R26LSL-Tom mice, performed BM biopsy on one hind leg 2–3 days later, analyzed Tom expression by flow cytometry and selected only the animals with HSC-specific labeling (Fig. 1A). The single low dose of tamoxifen (0.5 mg) was 10-fold lower than used in HSC tracing (Busch et al., 2015; Chapple et al., 2018) and substantially below the cumulative amounts that can affect hematopoiesis (Nakada et al., 2014; Sanchez-Aguilera et al., 2014). As reported (Sawai et al., 2016; Upadhaya et al., 2018), Tom was expressed in ~23% of HSCs defined as Lineage-negative (Lin−) Sca-1+ c-Kit+ CD150+ CD48− (SLAM-LSK), with only minimal expression in other progenitors (Fig. S1A,B). Conversely, the majority of Tom+ cells comprised SLAM-LSK HSCs (average 77%), with six animals achieving ≥90% specificity (Fig. 1B,C). To further confirm the specificity of labeling, we crossed Pdzk1ip1-CreER R26LSL-Tom mice with Fgd5ZsGreen mice that express the green fluorescent protein (GFP) ZsGreen from the HSC-specific Fgd5 gene (Gazit et al., 2014). We found that Fgd5-driven GFP was expressed in the majority of LSK cells including CD48+ multipotent progenitors (MPP). In contrast, Tom was expressed exclusively in SLAM-LSK HSCs, and the majority of Tom+ cells were also GFP+ (Fig. S1C).
Fig. 1. A system for intravital imaging of endogenous HSCs.
(A) Experimental design of HSC observation by intravital microscopy.
(B) The expression of Tom in CreER reporter or CreER-negative (Control) animals 2–3 days after induction as determined by flow cytometry. Shown are representative staining profiles of total BM cells (gray) and gated Tom+ cells (red).
(C) The specificity of labeling shown as the fraction of each cell population among Tom+ cells. Symbols represent individual animals (n = 16); bars represent median. The inset shows the distribution of HSC labeling specificity across animals.
(D) Representative micrographs from TPLSM with signals from Tom (arrowhead), autofluorescent macrophages (autofl.) and bone (second harmonic generation, SHG). Scale bar, 8 μm.
The next day after the biopsy, the animals were anesthetized, the tibia of a contralateral hind leg was surgically exposed and thinned by microdrilling (Kohler et al., 2009), and the BM was examined by two-photon laser scanning microscopy (TPLSM). Compared to the commonly used calvarial bone (Mazo et al., 2011), tibia may be more representative of the BM depot in the body (Lassailly et al., 2013), contains more HSCs due to the deeper BM cavity, and facilitates stable TPLSM over long periods. Two-photon excitation at 920 nm allowed the detection of green and red fluorescence and the visualization of bone through second harmonic generation from collagen. We observed rare Tom+ cells along with resident BM macrophages identified by autofluorescence in red and green channels (Rakhymzhan et al., 2017) (Fig. 1D). Tom+ cells could be detected within 200 μm from the bone surface (Fig. 1D) and showed no specific enrichment near or away from the bone. Tom+ cells could be similarly detected by TPLSM when Lin− cells from induced reporter mice were injected i.v. into wild-type animals (Fig. S1D). Thus, our setup allows visualization of fluorescently labeled HSCs in the long bones of live animals.
Analysis of HSC behavior in the steady-state bone marrow by TPLSM
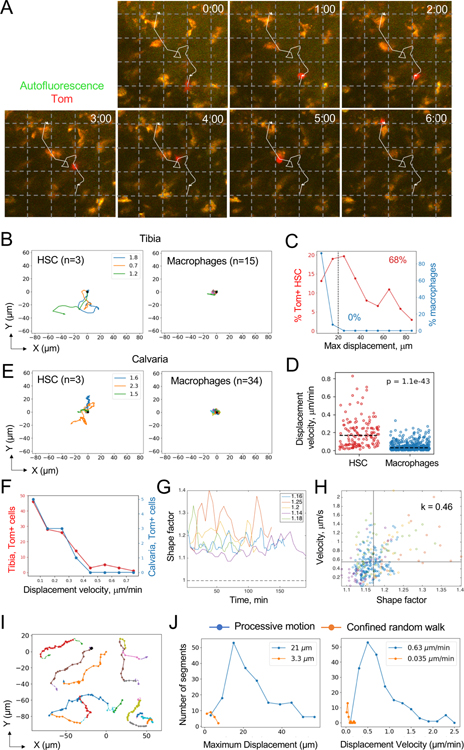
We observed Tom+ HSCs by TPLSM continuously for 1–6 hours (average observation length 3.6 hours). Multiple fields with Tom+ cells were identified in the same tibia and scanned sequentially every 2–10 min, capturing 139 cells in 16 independent animals. As illustrated by representative time-lapse still images (Fig. 2A) and videos (Video S1), HSCs exhibited dynamic non-spherical morphology with protrusions and prominent motility relative to the sessile macrophages. Track reconstruction in the XY plane showed that the majority of observed HSCs were motile, including HSCs in animals with 90–100% labeling specificity (Fig. 2B and S2A). Reflecting their sessile nature, >90% of observed macrophages showed maximum displacements (MD) of <15 μm in the XY plane, and none showed MD >20 μm. In contrast, only 13% of HSCs showed MD of <15 μm, whereas 68% of HSCs showed MD of >20 μm (25–85 μm, average 35 μm) (Fig. 2C). The fraction of HSCs with MD >20 μm in each animal correlated with the specificity of HSC labeling in that animal (Fig. S2B), suggesting that long-range movement is a representative behavior of HSCs. Accordingly, HSCs reached average displacement velocity of 0.15 μm/min, compared to 0.02 μm/min for macrophages (Fig. 2D).
Fig. 2. Analysis of HSC behavior in the steady-state BM by TPLSM.
(A) TPLSM snapshots at the indicated times (hours:minutes) demonstrating a representative Tom+ HSC (red) among autofluorescent macrophages (orange). The trajectory of the HSC during the observation period is indicated. Grid, 25 μm.
(B) Two-dimensional (XY) tracks of individual Tom+ HSCs imaged in the tibia of a representative animal. Times of HSC tracking in hours are indicated by the colored legends; right panel shows tracks of autofluorescent macrophages from the same animal.
(C) The distribution of maximum displacements of Tom+ HSCs (red) or autofluorescent macrophages (blue). Fractions of cells above the threshold 20 μm displacement (dashed line) are indicated.
(D) Displacement velocities of individual Tom+ HSCs (n = 139) and macrophages (n = 582). Dashed lines indicate medians.
(E) Two-dimensional (XY) tracks of individual Tom+ HSCs imaged in the calvaria of a representative animal. Data are presented as in panel B.
(F) The distribution of displacement velocities of Tom+ HSCs in the tibia (red) or calvaria (blue).
(G) Change in shape factor over time in representative HSCs from five individual animals; average values for each cell are indicated.
(H) Correlation between shape factor and cell velocity for cells from panel G; each symbol represents a separate time point.
(I) Representative examples of processive motion (PM) segments (solid colored lines) for six different HSCs from five individual animals.
(J) Distribution of movement parameters for PM (blue) and CRW segments (orange) across all Tom+ HSCs; numbers on each plot represent median values for the respective parameters.
To extend our results to other BM depots, we also recorded 12 Tom+ cells in the unmanipulated calvarial bone of 3 independent animals. Notably, these cells manifested the same motile behavior as HSCs in tibia (Fig. 2E and Video S2). Moreover, the distribution of displacement velocities of HSCs in the tibia and calvaria were comparable (Fig. 2F and S2C), confirming that our results were not grossly affected by the site of observation or mechanical thinning of the tibia.
We characterized HSC morphology by deriving cell perimeter (L) and area (A) from XY images and defining the shape factor (SF) as L2/(4πA), with SF=1 for a perfectly round cell. As shown in Fig. 2G, HSC shapes fluctuated within 10–20 min, with SF largely in 1.1–1.3 range (1.18 ±0.06) but never approaching 1. Importantly, SF was highly correlated to HSC velocity (Pearson’s correlation coefficient 0.5 ±0.1, Fig. 2H), highlighting the association between HSC motility and dynamic cell morphology.
The plot of mean squared displacement (MSD) versus time is commonly used in TPLSM analysis to define cellular behaviors such as a random walk (RW), a confined random walk (CRW) or a processive motion (PM) (Fooksman et al., 2010). However, MSD analysis of tracked HSCs revealed their widely different behaviors even within the same animal (Fig. S2D,E), and failed to establish a clear pattern across all analyzed HSCs (Fig. S2F). To better characterize HSC dynamics, we built upon the tools from single molecule imaging studies (Saxton, 1993) and their subsequent application to protein dynamics (Bouzigues and Dahan, 2007; Simson et al., 1995). The resulting algorithm performed unbiased segmentation of a cell trajectory and determined probabilities of each segment being a RW or CRW; segments with low probabilities of either behavior were identified as PM (Fig. S2G). When applied to HSCs, the algorithm segmented and parsed the tracks into stretches of PM separated by occasional short stretches of CRW (Fig. 2I). This approach revealed that 71% of HSCs contained at least one PM segment. On average, PM segments reached MD of 44 ±18 μm and displacement velocities of 0.77 ±0.43 μm/min (Fig. 2J). The remaining HSCs did not undergo PM during the observation period, displaying lower MD (11 ±4 μm) and velocities (0.03 ±0.02 μm/min). The stretches of PM covered distances of 30 ±18 μm and lasted for 44 ±28 min (versus 3.5 ±1.9 μm and 114 ±66 min, respectively, for stretches of CRW) (data not shown). These results suggest that the majority of HSCs exhibit a complex motile behavior with intermittent stretches of processive motion.
Exploring the phenotype and niche interactions of motile HSCs
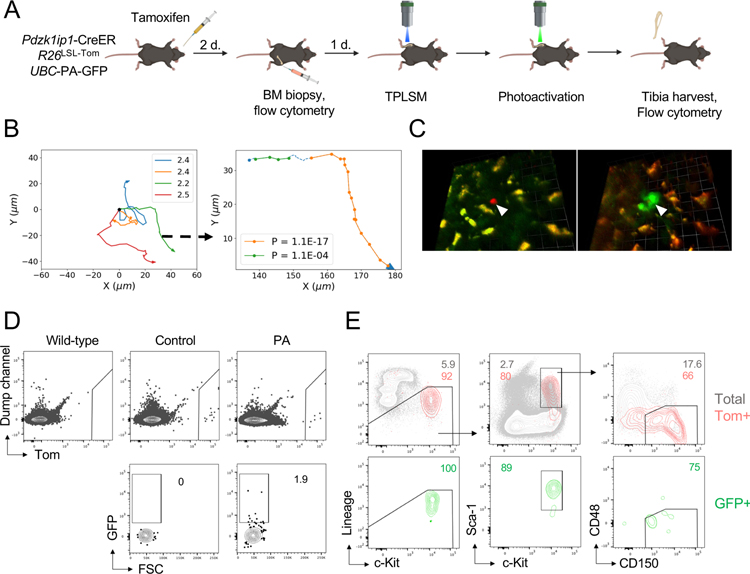
To directly confirm the identity of motile Tom+ cells, we used a mouse strain expressing the photoactivatable variant of GFP (PA-GFP) (Victora et al., 2010). Two-photon irradiation at 720–840 nm renders PA-GFP detectable by TPLSM with 940 nm excitation or by flow cytometry with 488 nm excitation. Pdzk1ip1-CreER R26LSL-Tom reporter mice carrying the ubiquitously expressed UBC-PA-GFP allele were induced with tamoxifen, confirmed for specific labeling by BM biopsy and examined by TPLSM (Fig. 3A). Tracking of Tom+ cells in the tibia for 2.5 hours confirmed their motility with long stretches of PM (Fig. 3B). Precise volumes (1200–1600 μm3) surrounding only these motile Tom+ cells were then illuminated with an 840 nm laser, converting PA-GFP molecules to GFP in Tom+ and their neighboring (Tom−) cells (Fig. 3C). The animals were sacrificed and BM cells from the entire photoactivated tibia were analyzed by flow cytometry. Tom+ cells in the photoactivated tibia, but not in control contralateral tibia, contained rare GFP+ cells (Fig. 3D). Importantly, the majority of GFP+ Tom+ cells had the SLAM-LSK phenotype (Fig. 3E), confirming that motility with PM represents an inherent behavior of HSCs in the steady state.
Fig. 3. The phenotype of motile HSCs as determined by photoactivation.
(A) Schematic illustrating the experimental design of photoactivation, including tamoxifen administration, BM biopsy, and intravital TPLSM before and after photoactivation. Only Tom+ that were motile were selected for photoactivation.
(B) XY tracks of individual HSCs before photoactivation (left) and a representative example of track segmentation of a single HSC (right). Numbers on the plot on the right represent P(Rnm) values for each PM segment.
(C) Representative fluorescent micrograph image from a photoactivation experiment. Shown is a compressed z-stack image of a Tom+ HSC (white arrowhead) immediately before (left) and following photoactivation (right). Autofluorescent macrophages are distinguished in orange. Scale bars 12.7 μm (left) and 14.2 μm (right).
(D-E) The detection and analysis of Tom and GFP signals cells in Pdzk1ip1-CreER R26LSL-Tom UBC-PA-GFP animals either photoactivated tibia (PA) or contralateral tibia (Control). CreER-negative (Wild-type) animal used as negative control. Shown are representative FACS plots showing expression of Tom and GFP within the total BM cells (D) and the phenotypes of Tom+ and GFP+ cells (E). Data concatenated from two individual experiments from two separate animals.
To visualize HSCs relative to blood vessels, we used Pdzk1ip1-CreER R26LSL-Tom mice carrying the Fgd5ZsGreen reporter (Fig. S1C) that is strongly expressed in the endothelium (Gazit et al., 2014). Double-reporter mice were induced with tamoxifen and examined by TPLSM as above to visualize labeled HSCs (red), endothelial cells (green) and macrophages (red and green autofluorescence). This analysis confirmed their expected perivascular localization (Fig. S3A) and movement in the perivascular space (data not shown). Next, we used the knock-in reporter strain expressing GFP from the SCF-encoding Kitl locus (Ding et al., 2012). In the resulting tamoxifen-induced Pdzk1ip1-CreER R26LSL-Tom KitlGFP/+ mice, we were able to detect HSCs among the sprawling network of stromal cells with thin processes; importantly, HSCs exhibited the same motile behavior as observed in single-color reporters. Moreover, HSCs appeared to form close transient contacts with SCF+ cells throughout their movement, including serial contacts with multiple SCF+ cells along the track (Fig. S3B and Video S3). Consistent with the high density of sinusoids and perivascular stromal cells in the BM (Gomariz et al., 2018), we did not observe a preferential enrichment of HSCs in proximity to SCF+ cells (data not shown). Collectively, these results reveal an unexpectedly dynamic HSC behavior and sequential interactions with multiple cells SCF+ cells in the BM niche.
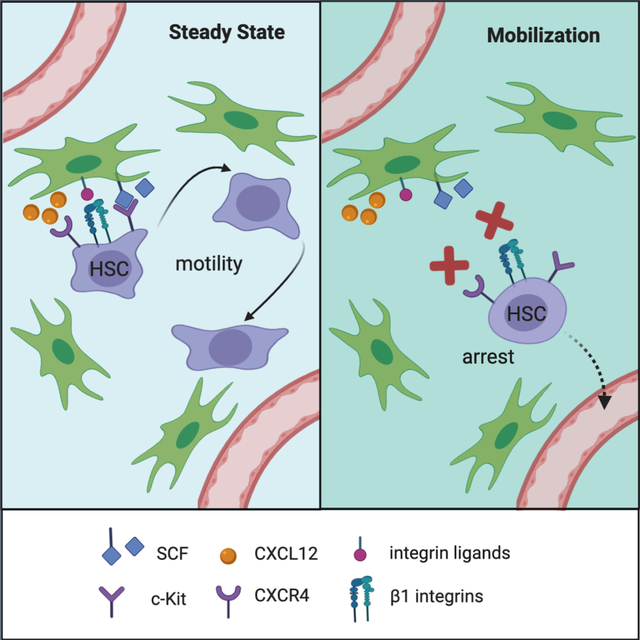
Analysis of HSC behavior during mobilization
To visualize HSC behavior following the disruption of HSC-niche interactions, we used AMD3100 (Plerixafor) and the N-benzenesulfonyl proline-based compound BOP, which block CXCR4 and a4b1/a9b1 integrins, respectively, and induce efficient HSC mobilization within ~30 minutes (Cao et al., 2016). We tracked Tom+ HSCs in tamoxifen-induced anesthetized reporter mice by TPLSM for 3 hours, administered AMD3100+BOP s.c. and continued cell tracking for additional 4 hours (Fig. 4A). Because only a minor fraction of BM HSCs is mobilized into the blood, actual mobilization events are rare and could not be captured. Nevertheless, AMD3100+BOP administration reduced HSC motility (Fig. 4B and Video S4), resulting in lower maximal displacement (Fig. 4C). Furthermore, displacement velocities of HSCs were significantly reduced by AMD3100+BOP (Fig. 4D), as was the fraction of PM stretches (Fig. 4E,F). Likewise, shape fluctuations decreased by about 30% as reflected by reduced standard deviation of SF (data not shown), and the correlation between SF and cell velocity diminished significantly after treatment (Fig. 4G, S4A,B). Thus, the blockade of CXCR4 and integrin signaling rapidly abrogated HSC motility, suggesting that HSC movement and BM retention are linked; disrupting one process disrupts both.
Fig. 4. Analysis of HSC behavior during mobilization.
(A) Experimental design of TPLSM before and after administration of AMD3100 and BOP (Rx).
(B) TPLSM snapshots at the indicated times demonstrating the trajectory of a Tom+ HSC before (cyan, Pre-Rx) and after (magenta, Post-Rx) treatment. Scale bar, 17μm.
(C) Representative XY trajectories of two Tom+ cells (labeled “a” and “b”) from three separate experiments before (cyan) and after (magenta) the treatment. Hours of tracking for each cell before and after the treatment are indicated.
(D) Displacement velocities before and after the treatment for 14 cells from 5 individual animals.
(E) Track segmentation of a representative Tom+ HSC before and after treatment. Associated P(Rnm) values for each PM segment before (Pre-Rx) and after treatment (Post-Rx) are indicated.
(F) Fraction of track segments comprising PM and CRW before (blue) and after treatment (green).
(G) 1Shape factor and velocity (V) of a single representative HSC before (blue) and after (red) the treatment. Dashed lines represent average values; correlation coefficients (k) are indicated.
Discussion
We found that the majority of HSCs in their native BM environment exhibit dynamic morphology and substantial motility. A fraction (13%, Fig. 2C) of static HSCs was also observed and may represent insufficient observation time or a distinct functional subset (Zhao et al., 2019). Notably, our system labels actively contributing HSCs as well as a small subset of HSCs that do not differentiate (Sawai et al., 2016), and therefore is likely to capture the entire spectrum of HSC behavior. The motility of HSCs may have gone undetected in previous transplantation-based intravital microscopy studies because of recipient irradiation, which severely perturbs the BM compartment. Indeed, hematopoietic progenitors transferred into non-irradiated mice showed increased motility (Foster et al., 2015) and transient protrusions (Bixel et al., 2017; Kohler et al., 2009), although HSCs were not specifically examined in these settings. A recent study genetically labeled ~12% of endogenous HSCs and reported their limited motility in the calvarial BM in the steady state (Christodoulou et al., 2020). The discrepancy between our respective studies is not due to the site of TPLSM, as we observed higher HSC motility even in the calvaria (Fig. S2C). It is possible that the genetic model of (Christodoulou et al., 2020) preferentially labels a minor distinct subset of HSCs, e.g. those that are less motile in our observations as mentioned above. Notably, the data of Christodoulou et al. represent 16 cells from 3 animals with six captured frames per cell; this limited dataset, along with the equally limited data from the calvarial BM reported herein, may be subject to batch effects and warrants additional observations. Indeed, representative videomicrocopy data presented in (Christodoulou et al., 2020) suggest HSC movement and shape dynamics that would be consistent with our data.
All mature cell types that originate from the HSCs show motility in the BM, including B cell progenitors (Beck et al., 2014; Fistonich et al., 2018), mature B cells (Cariappa et al., 2005), T cells (Milo et al., 2013), NK cells (Milo et al., 2018) and plasmacytoid dendritic cells (Spaulding et al., 2016). The observed motility of HSCs resembles these cell types but proceeds at much lower speeds, e.g. compared to pro/pre-B cells that move at ~2 mm/min (Fistonich et al., 2018). Accordingly, the pattern of HSC migration could not be adequately described by a single motion type, as also observed with other cell types over long periods of time (Banigan et al., 2015; Fricke et al., 2016). Instead, HSCs exhibited a complex pattern of intermittent processive motion, which may represent migration towards gradients of chemotactic factors such as CXCL12, and/or persistent search for limited growth factors such as the membrane-associated SCF. Notably, the perisinusoidal BM space features high sheer hydrodynamic forces, and immature B cells use active CXCR4-dependent migration to resist these forces and remain in the BM (Beck et al., 2014). Similarly, motility may actively retain HSCs in the BM, and its observed disruption during the mobilization via CXCR4 and integrin blockade may increase the likelihood of HSC exit into the blood.
In addition to its possible role in BM retention, the observed steady-state motility of HSCs has important implications for the nature of their BM niche. Instead of a static lodging with the nearest stromal cell, HSC motility allows dynamic interactions with multiple putative niche constituents (Crane et al., 2017; Wei and Frenette, 2018). This scenario is consistent with other specific anatomical niches such as the splenic marginal zone, whose resident cells are motile and retained dynamically (Arnon et al., 2013). It is also supported by our initial observations of HSCs undergoing dynamic interactions with SCF-expressing stromal cells. The network of BM stromal cells producing CXCL12 and/or SCF is dense, and it was proposed that HSC association with them may arise by pure chance (Gomariz et al., 2018). While our data do not imply a preferential association of HSCs with these stromal cells, they document prolonged interactions of HSCs with multiple SCF+ cells. Such interactions may be critical for the delivery of HSC-maintaining local signals such as membrane-bound SCF, integrin ligands and so on.
Limitations of the study
In addition to common technical caveats of TPLSM-based studies, such as the effects of anesthesia and of laser irradiation, our system comprises several features that may affect cell behavior. These include the potential effects of tamoxifen and of bone drilling, although these were mitigated by the low tamoxifen dose and by parallel observations in intact calvarial bones, respectively. In addition, the effects of high-level tdTomato expression on HSC behavior cannot be ruled out, although this expression is compatible with the normal expression profile and functionality of HSCs (Sawai et al., 2016; Upadhaya et al., 2018).
With respect to the analysis of HSC behavior during mobilization, the relative contributions of CXCR4 and specific integrins to HSC motility have not been addressed here and remain to be established. Given that the blockade of either pathway alone produces a relatively weak mobilization (Cao et al., 2016), this may require more sensitive detection methods. Finally, the preliminary visualization of HSCs and niche components presented herein needs to be expanded and subjected to robust quantification of cell-cell interactions. Future imaging at higher resolution should elucidate the relationship between the type and speed of HSC motion and their interaction with SCF+ cells and other components of the niche.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Boris Reizis (boris.reizis@nyulangone.org).
Materials Availability
The Pdzk1ip1-CreER transgenic strain is available from the Lead Contact’s lab upon request and following the completion of a Material Transfer Agreement. All other strains are available from mouse repositories as described in the Key Resources Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-mouse B220-eFluor 450 | ThermoFisher Scientific | Clone: RA3-6B2 RRID:AB_1548761 |
| anti-mouse CD11b-eFluor 450 | ThermoFisher Scientific | Clone: M1/70 RRID:AB_1582236 |
| anti-mouse CD150-PE/Cy7 | BioLegend | Clone: TC15-12F12.2 RRID:AB_439796 |
| anti-mouse CD41-APC or -APC/Cy7 | BioLegend | Clone: MWReg30 RRID:AB_11125581 |
| anti-mouse CD48-AF700 | BioLegend | Clone: HM48-1 RRID:AB_10612754 |
| anti-mouse cKit-APC or -APC/Cy7 | ThermoFisher Scientific | Clone: 2B8 RRID:AB_469429 |
| anti-mouse Gr-1-eFluor 450 | ThermoFisher Scientific | Clone: RB6-8C5 RRID:AB_1548788 |
| anti-mouse NK1.1-eFluor 450 | ThermoFisher Scientific | Clone: PK136 RRID: AB_2043877 |
| anti-mouse Sca-1-PerCP/Cy5.5 | ThermoFisher Scientific | Clone: D7 RRID: AB_914372 |
| anti-mouse TCRb-eFluor 450 | ThermoFisher Scientific | Clone: H57-597 RRID:AB_11062012 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| AMD3100 | Sigma-Aldrich | CAS# 155148-31-5 |
| N-(Benzenesulfonyl)-L-prolyl-L-O-(1-pyrrolidinylcarbonyl)tyrosine (BOP) | Tocris | CAS# 1947348-42-6 |
| Tamoxifen | Sigma-Aldrich | CAS# 10540-29-1 CHEBI:41774 |
| Ketamine | Covetrus | cat # 056344 NIFSTD:DB01221 |
| Xylasine | Covetrus | cat # 033198 |
| Acepromazine | Covetrus | cat # 003845 CHEBI:44932 |
| Experimental Models: Organisms/Strains | ||
| Pdzk1ip1-CreER | Reizis lab | N/A |
| B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | Jackson Laboratories | Stock No: 007914 RRID:IMSR_JAX:007914 |
| B6.Cg-Ptprca Tg(UBC-PA-GFP)1Mnz/J | Jackson Laboratories | Stock No: 022486 RRID:IMSR_JAX:022486 |
| Kitltm1.1Sjm/J | Jackson Laboratories | Stock No: 017860 RRID:IMSR_JAX:017860 |
| C57BL/6N-Fgd5tm2Djr/J | Jackson Laboratories | Stock No: 027788 RRID:IMSR_JAX:027788 |
| Software and Algorithms | ||
| Volocity 6.3 | PerkinElmer | RRID:SCR_002668 |
| FlowJo | FlowJo, LLC | RRID:SCR_008520 |
| Prism | GraphPad Software | RRID: SCR_002798 |
| Matlab | The MathWorks, Inc. | RRID:SCR_001622 |
| Python | Python.org | RRID:SCR_008394 |
| Matlab-based code and files | this paper | https://github.com/okrichev/Matlab-Files-HSC-project |
| Python-based code and files | this paper | https://github.com/okrichev/Python-Files-HSC-project |
Data and Code Availability
All computer codes used in the data analysis are available from the GitHub software repository.
Python files: https://github.com/okrichev/Python-Files-HSC-project
Matlab files: https://github.com/okrichev/Matlab-Files-HSC-project
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Pdzk1ip1-CreER R26LSL-Tom/LSL-Tom reporter animals have been described previously (Sawai et al., 2016; Upadhaya et al., 2018). Adult (8 to 16 weeks) mice of both sexes were used. Where indicated, the mice were crossed to a photoactivatable GFP (UBC-PA-GFP) transgenic mouse strain (B6.Cg-Ptprca Tg(UBC-PA-GFP)1Mnz/J), Kitl knock-in GFP reporter strain (Kitltm1.1Sjm/J) or Fgd5 knock-in GFP (ZsGreen) reporter strain (C57BL/6N-Fgd5tm2Djr/J) obtained from the Jackson Laboratories. All animal studies were performed according to the investigators’ protocols approved by the Institutional Animal Care and Use Committees of New York University School of Medicine and the Albert Einstein College of Medicine.
METHOD DETAILS
Animal Procedures
To induce recombination in the reporter mice, a single dose of 20–30 mg/kg tamoxifen (Sigma-Aldrich) was administered by gavage. BM biopsy was performed on day 2 or 3 after tamoxifen administration. Briefly, following isoflurane-induced anesthesia, a 28½ G insulin syringe needle was inserted into the joint surface of the femur through the patellar tendon and then into the bone cavity. Up to 20 ml of the bone marrow suspension was collected.
Cell Staining and Flow Cytometry
For stem and progenitor cell identification in the BM, cell suspensions were subjected to red blood cell lysis and stained with antibodies against CD150, CD48, c-Kit, Sca-1 and CD41 in addition to a cocktail of lineage markers (CD11b, NK1.1, B220, TCRb, Gr-1 and Ter119). Samples were acquired on an Attune NxT flow cytometer (Thermo Fisher Scientific). A detailed list of antibodies is provided in the Key Resource Table. Conventional flow cytometry analysis was performed using FlowJo software (FlowJo, LLC). Cell populations in the BM were defined as follows: HSC, Lin− Sca-1+ c-Kit+ (LSK) CD150+ CD48−; ST-HSC, LSK CD150− CD48−; MPP2, LSK CD150+ CD48+; MPP3/4, LSK CD150− CD48+; MyP, Lin− Sca-1− c-Kit+ CD150− CD41−; MEP, Lin− Sca-1−c-Kit+ CD150+ CD41−; MkP, Lin− Sca-1− c-Kit+ CD150+ CD41+.
Intravital TPLSM
One day following BM biopsy, mice were anesthetized using KXA (Ketamine, Xyalzine, Acepromazine) solution (4 μl/g) and secured on an externally heated imaging plate in a supine position. The medial region of tibia (contralateral to the femur used for biopsy) was surgically exposed and thinned to approximately 200 μm thickness using a mechanical microdrill. The leg was immobilized using a custom-built apparatus which contained an opening for the drilled bone to be exposed to the microscope objective. Images were collected on an Olympus FV1000-MPE upright laser scanning microscope fitted with a 25× 1.05NA water immersion objective and Mai Tai DeepSee Ti:Sapphire laser (Spectra-Physics) for two-photon excitation. For photoactivation experiments, selected cells were exposed to a brief pulse of laser tuned to 830 nm then subsequently imaged at 920 – 940 nm. Animal were kept under anesthesia using isoflurane inhalation during the entire intravital imaging session. 100–200 μl of lactated Ringer’s solution was periodically administered by subcutaneous injection to ensure proper hydration.
AMD3100 and BOP Treatment
For in-vivo HSC mobilization studies, a combination of AMD3100 (Sigma-Aldrich), 4 – 10 mg/kg and N-(Benzenesulfonyl)-L-prolyl-L-O-(1-pyrrolidinylcarbonyl)tyrosine (BOP, Tocris), 10–15 mg/kg was administered via subcutaneous injection.
QUANTIFICATION AND STATISTICAL ANALYSIS
Image Analysis
3D microscopy has much higher resolution in lateral dimensions than in the axial one, therefore, we limited our analysis to the cell motion in XY plane (i.e. the XY component of the overall motion). Thus, we avoided relatively large errors in the determined Z coordinates infringing on the accuracy of our method. Collected images were analyzed using Volocity 6.3 (PerkinElmer). Displacement velocity was defined as a maximal distance in XY plane between any two points on the track (i.e. largest displacement on the trajectory) divided by the whole track observation time. Another possible definition of the displacement velocity, as a distance between the initial and the final points on the track divided by the track observation time, produced similar results in all experiments. However, it tends to underestimate the dynamics of multiple HSCs that undergo a large circular motion that ends in the vicinity of the starting point.
General principles.
The basic premise of our analyses is that the simplest type of motion that a cell could perform is Random Walk (RW). In simple terms, RW represents uncorrelated and unbiased movements, i.e., every progressive step is dependent only the step preceding it and the direction of each step is completely random. The statistics of RW are well known, and any significant deviation of the actual trajectory statistics from that of RW necessitates the introduction of additional models. We used two such models for our analyses: 1) processive motion (PM) dynamics, where a cell largely preserves the general direction of its velocity, and 2) confined random walk (CRW) or confined motion (CM) dynamics, where cell exhibits RW that cannot (or has a low probability of) escaping from some space e.g. surrounded by obstacles.
Kinetics of RW is characterized by its diffusion coefficient D that determines how fast (on average) a random walker moves from its initial location. More specifically, for RW, D describes the rate of change of mean-square displacement 〈r2〉 (MSD) with time t. For motion in XY plane, MSD is represented as:
| (1) |
Distance exceeding by a significant factor is highly unlikely to be caused by RW and thus would indicate PM. In contrast, motion of a trajectory covering distance significantly smaller than , has a high likelihood of assuming CM. These likelihoods have been evaluated in (26). The probability P(R) of finding a random walker at any distance larger than R is given by:
| (2) |
For identification of confinements, the probability P(d) of RW to be fully confined within the distance d from the origin is (Saxton, 1993):
| (3) |
Tracks segmentation into processive and confined motions (PM and CRW).
The application of Eqs. 2&3 assumes knowledge of D. This can be measured only on RW and CRW segments which at this stage are not identified yet. Thus, we proceed iteratively. We first estimate D for each trajectory using its MSD and Eq. 1. Since there are PM stretches in the trajectory this approach overestimates D and therefore, is only used to exclude PM segments (excluded were segments for which P(R) as given by Eq.2 is smaller than P*=0.05) preliminarily so that D can be measured on the rest of the track. MSD (t) measured after PM is excluded were fitted with Eq. 1 for RW and Eq. 4 (below) for CRW dynamics respectively to obtain new estimates for D:
| (4) |
where a represents confinement radius. The value D given by the better of the two fits was used in further calculations. For the tracks that had too short or no RW and CRW segments, the median value of D over all tracks was used.
PM identification:
given D, for every two time points tm and tn on a track, we evaluated the probability P(Rnm) that the measured displacement Rnm between the two points is achieved by a simple RW dynamics within |tm – tn| time (Eq. 2). An example of P(Rnm) shown in the “heat map” in Fig. 3D (top inset). Low P(Rnm) values point to PM dynamics. Local minima with values below 0.005 were used: their n and m values identify the beginning and the end of a PM segment. Associated parameters such as distance covered (length), velocity, and time spent in each segment is also calculated.
CRW identification:
Track segments in between PM were tested for the CRW dynamics. For each pair tm and tn we determine confinement dnm as the largest distance between locations in the [tn, tm] interval and evaluate the probability P(dnm) of RW staying within the confinement (Eq. 3). An example of the corresponding “heat map” is shown in Fig. 3D (bottom inset). Low values of P(dnm) point to CRW dynamics. Local minima with values below 0.005 were used: their n and m values identify the beginning and the end of a CRW segment. Segments containing only a few points (typically below 10) are not considered.
All of the algorithms were tested on tracks generated by numerical simulations. Analysis was done in a program written in Python and all of the parameters discussed in this section can be varied. None of the conclusions reached in this work depends critically on the chosen parameters within a reasonably wide range.
Morphology analysis
Morphology analysis was performed in a custom-built Matlab code: 2D images of cells were used to avoid noisiness of the 3D images. Planes crossing HSCs were summed up, passed through a Gaussian filter (width of ~ 2 μm) and thresholded using Otsu’s algorithm applied to an area of ~30×30 μm2 around the cell. The perimeter L and area A of HSC binary images were measured using standard Matlab commands. The shape factor (SF) was calculated as L2/(4πA). Such defined SF is generally larger than one, being equal to one for a perfectly round cell.
Statistical analysis
Normal distribution of data was not assumed and statistical significance of differences between experimental groups was determined by non-parametric Mann-Whitney test using Prism software (GraphPad, La Jolla, CA) or by using the scipy.stats package in Python.
Supplementary Material
Video S1. Videomicroscopy of tdTomato+ HSCs in the tibia of live mice. Related to Figure 2. Representative examples of HSC movement in three individual animals are shown. Unprocessed image (left) shows Tom+ HSCs (red) and macrophages (orange). The processed image (right) shows cell contour along with its track (white). Scale bar is indicated on lower left. Scale in the upper right indicates time in hours:minutes:seconds:milliseconds.
Video S2. Videomicroscopy of tdTomato+ HSCs in the calvarium of live mice. Related to Figure 2. Representative example of a Tom+ HSC (red) in the calvarium of a live animal. Unprocessed image (left) shows Tom+ HSCs (red) and macrophages (orange). The processed image (right) shows cell contour along with its track (white). Scale bar is indicated on lower left. Scale in the upper right indicates time in hours:minutes:seconds:milliseconds.
Video S3. Videomicroscopy of tdTomato+ HSCs and GFP+ stromal cells in the tibia of live SCF reporter mice. Related to Figures 2–3. Unprocessed image (left) shows Tom+ HSCs (red), GFP+ stromal cells (blue) and macrophages (orange). The processed image (right) shows HSC contour along with its track (white) among GFP+ stromal cells (green). Scale in the upper right indicates time in hours:minutes:seconds:milliseconds.
Video S4. Videomicroscopy of tdTomato+ HSCs in the tibia of live mice before and after mobilization with AMD3100+BOP. Related to Figure 4. Representative examples of HSC movement in two individual animals before and after treatment with AMD3100 (AMD) and BOP. Unprocessed image (left) shows Tom+ HSCs (red) and macrophages (orange). The processed image (right) shows cell contour along with its track before (cyan) and after treatment (magenta). Time of acquisition before and after treatment is indicated on the right; scale in the upper right indicates total time in hours:minutes:seconds:milliseconds.
Highlights.
Endogenous HSCs in the bone marrow of live adult mice were observed for many hours
HSCs show dynamic morphology and complex motile behavior
Motile HSCs closely interact with perivascular SCF-expressing stromal cells
Mobilization-inducing blockade of CXCR4 and integrins rapidly arrests HSC motility
Acknowledgments
This work was supported by the NIH grant AG049074 (B.R.) and the Israel Science Foundation grant 1912/14 (O.K.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare no competing financial interest.
References
- Acar M, Kocherlakota KS, Murphy MM, Peyer JG, Oguro H, Inra CN, Jaiyeola C, Zhao Z, Luby-Phelps K, and Morrison SJ (2015). Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature 526, 126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon TI, Horton RM, Grigorova IL, and Cyster JG (2013). Visualization of splenic marginal zone B-cell shuttling and follicular B-cell egress. Nature 493, 684–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banigan EJ, Harris TH, Christian DA, Hunter CA, and Liu AJ (2015). Heterogeneous CD8+ T cell migration in the lymph node in the absence of inflammation revealed by quantitative migration analysis. PLoS Comput Biol 11, e1004058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck TC, Gomes AC, Cyster JG, and Pereira JP (2014). CXCR4 and a cell-extrinsic mechanism control immature B lymphocyte egress from bone marrow. J Exp Med 211, 2567–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bixel MG, Kusumbe AP, Ramasamy SK, Sivaraj KK, Butz S, Vestweber D, and Adams RH (2017). Flow Dynamics and HSPC Homing in Bone Marrow Microvessels. Cell Rep 18, 1804–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzigues C, and Dahan M (2007). Transient directed motions of GABA(A) receptors in growth cones detected by a speed correlation index. Biophys J 92, 654–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broxmeyer HE, Orschell CM, Clapp DW, Hangoc G, Cooper S, Plett PA, Liles WC, Li X, Graham-Evans B, Campbell TB, et al. (2005). Rapid mobilization of murine and human hematopoietic stem and progenitor cells with AMD3100, a CXCR4 antagonist. J Exp Med 201, 1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch K, Klapproth K, Barile M, Flossdorf M, Holland-Letz T, Schlenner SM, Reth M, Hofer T, and Rodewald HR (2015). Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature 518, 542–546. [DOI] [PubMed] [Google Scholar]
- Cao B, Zhang Z, Grassinger J, Williams B, Heazlewood CK, Churches QI, James SA, Li S, Papayannopoulou T, and Nilsson SK (2016). Therapeutic targeting and rapid mobilization of endosteal HSC using a small molecule integrin antagonist. Nature communications 7, 11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cariappa A, Mazo IB, Chase C, Shi HN, Liu H, Li Q, Rose H, Leung H, Cherayil BJ, Russell P, et al. (2005). Perisinusoidal B cells in the bone marrow participate in T-independent responses to blood-borne microbes. Immunity 23, 397–407. [DOI] [PubMed] [Google Scholar]
- Chapple RH, Tseng YJ, Hu T, Kitano A, Takeichi M, Hoegenauer KA, and Nakada D (2018). Lineage tracing of murine adult hematopoietic stem cells reveals active contribution to steady-state hematopoiesis. Blood advances 2, 1220–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JY, Miyanishi M, Wang SK, Yamazaki S, Sinha R, Kao KS, Seita J, Sahoo D, Nakauchi H, and Weissman IL (2016). Hoxb5 marks long-term haematopoietic stem cells and reveals a homogenous perivascular niche. Nature 530, 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulou C, Spencer JA, Yeh SA, Turcotte R, Kokkaliaris KD, Panero R, Ramos A, Guo G, Seyedhassantehrani N, Esipova TV, et al. (2020). Live-animal imaging of native haematopoietic stem and progenitor cells. Nature 578, 278–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copelan EA (2006). Hematopoietic stem-cell transplantation. N Engl J Med 354, 1813–1826. [DOI] [PubMed] [Google Scholar]
- Crane GM, Jeffery E, and Morrison SJ (2017). Adult haematopoietic stem cell niches. Nat Rev Immunol 17, 573–590. [DOI] [PubMed] [Google Scholar]
- Ding L, and Morrison SJ (2013). Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature 495, 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Saunders TL, Enikolopov G, and Morrison SJ (2012). Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 481, 457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaves CJ (2015). Hematopoietic stem cells: concepts, definitions, and the new reality. Blood 125, 2605–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fistonich C, Zehentmeier S, Bednarski JJ, Miao R, Schjerven H, Sleckman BP, and Pereira JP (2018). Cell circuits between B cell progenitors and IL-7(+) mesenchymal progenitor cells control B cell development. J Exp Med 215, 2586–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fooksman DR, Schwickert TA, Victora GD, Dustin ML, Nussenzweig MC, and Skokos D (2010). Development and migration of plasma cells in the mouse lymph node. Immunity 33, 118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster K, Lassailly F, Anjos-Afonso F, Currie E, Rouault-Pierre K, and Bonnet D (2015). Different Motile Behaviors of Human Hematopoietic Stem versus Progenitor Cells at the Osteoblastic Niche. Stem cell reports 5, 690–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke GM, Letendre KA, Moses ME, and Cannon JL (2016). Persistence and Adaptation in Immunity: T Cells Balance the Extent and Thoroughness of Search. PLoS Comput Biol 12, e1004818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazit R, Mandal PK, Ebina W, Ben-Zvi A, Nombela-Arrieta C, Silberstein LE, and Rossi DJ (2014). Fgd5 identifies hematopoietic stem cells in the murine bone marrow. J Exp Med 211, 1315–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomariz A, Helbling PM, Isringhausen S, Suessbier U, Becker A, Boss A, Nagasawa T, Paul G, Goksel O, Szekely G, et al. (2018). Quantitative spatial analysis of haematopoiesis-regulating stromal cells in the bone marrow microenvironment by 3D microscopy. Nature communications 9, 2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum A, Hsu YM, Day RB, Schuettpelz LG, Christopher MJ, Borgerding JN, Nagasawa T, and Link DC (2013). CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature 495, 227–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopman RK, and DiPersio JF (2014). Advances in stem cell mobilization. Blood Rev 28, 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin T, Gur-Cohen S, Spencer JA, Schajnovitz A, Ramasamy SK, Kusumbe AP, Ledergor G, Jung Y, Milo I, Poulos MG, et al. (2016). Distinct bone marrow blood vessels differentially regulate haematopoiesis. Nature 532, 323–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Schmithorst V, Filippi MD, Ryan MA, Daria D, Gunzer M, and Geiger H (2009). Altered cellular dynamics and endosteal location of aged early hematopoietic progenitor cells revealed by time-lapse intravital imaging in long bones. Blood 114, 290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisaki Y, Bruns I, Scheiermann C, Ahmed J, Pinho S, Zhang D, Mizoguchi T, Wei Q, Lucas D, Ito K, et al. (2013). Arteriolar niches maintain haematopoietic stem cell quiescence. Nature 502, 637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassailly F, Foster K, Lopez-Onieva L, Currie E, and Bonnet D (2013). Multimodal imaging reveals structural and functional heterogeneity in different bone marrow compartments: functional implications on hematopoietic stem cells. Blood 122, 1730–1740. [DOI] [PubMed] [Google Scholar]
- Laurenti E, and Gottgens B (2018). From haematopoietic stem cells to complex differentiation landscapes. Nature 553, 418–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Celso C, Fleming HE, Wu JW, Zhao CX, Miake-Lye S, Fujisaki J, Cote D, Rowe DW, Lin CP, and Scadden DT (2009). Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature 457, 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazo IB, Massberg S, and von Andrian UH (2011). Hematopoietic stem and progenitor cell trafficking. Trends Immunol 32, 493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo I, Blecher-Gonen R, Barnett-Itzhaki Z, Bar-Ziv R, Tal O, Gurevich I, Feferman T, Drexler I, Amit I, Bousso P, et al. (2018). The bone marrow is patrolled by NK cells that are primed and expand in response to systemic viral activation. Eur J Immunol 48, 1137–1152. [DOI] [PubMed] [Google Scholar]
- Milo I, Sapoznikov A, Kalchenko V, Tal O, Krauthgamer R, van Rooijen N, Dudziak D, Jung S, and Shakhar G (2013). Dynamic imaging reveals promiscuous crosspresentation of blood-borne antigens to naive CD8+ T cells in the bone marrow. Blood 122, 193–208. [DOI] [PubMed] [Google Scholar]
- Nakada D, Oguro H, Levi BP, Ryan N, Kitano A, Saitoh Y, Takeichi M, Wendt GR, and Morrison SJ (2014). Oestrogen increases haematopoietic stem-cell self-renewal in females and during pregnancy. Nature 505, 555–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omatsu Y, Sugiyama T, Kohara H, Kondoh G, Fujii N, Kohno K, and Nagasawa T (2010). The essential functions of adipo-osteogenic progenitors as the hematopoietic stem and progenitor cell niche. Immunity 33, 387–399. [DOI] [PubMed] [Google Scholar]
- Rakhymzhan A, Leben R, Zimmermann H, Gunther R, Mex P, Reismann D, Ulbricht C, Acs A, Brandt AU, Lindquist RL, et al. (2017). Synergistic Strategy for Multicolor Two-photon Microscopy: Application to the Analysis of Germinal Center Reactions In Vivo. Sci Rep 7, 7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashidi NM, Scott MK, Scherf N, Krinner A, Kalchschmidt JS, Gounaris K, Selkirk ME, Roeder I, and Lo Celso C (2014). In vivo time-lapse imaging shows diverse niche engagement by quiescent and naturally activated hematopoietic stem cells. Blood 124, 79–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rettig MP, Ansstas G, and DiPersio JF (2012). Mobilization of hematopoietic stem and progenitor cells using inhibitors of CXCR4 and VLA-4. Leukemia 26, 34–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Aguilera A, Arranz L, Martin-Perez D, Garcia-Garcia A, Stavropoulou V, Kubovcakova L, Isern J, Martin-Salamanca S, Langa X, Skoda RC, et al. (2014). Estrogen Signaling Selectively Induces Apoptosis of Hematopoietic Progenitors and Myeloid Neoplasms without Harming Steady-State Hematopoiesis. Cell Stem Cell 15, 791–804. [DOI] [PubMed] [Google Scholar]
- Sawai CM, Babovic S, Upadhaya S, Knapp DJ, Lavin Y, Lau CM, Goloborodko A, Feng J, Fujisaki J, Ding L, et al. (2016). Hematopoietic Stem Cells Are the Major Source of Multilineage Hematopoiesis in Adult Animals. Immunity 45, 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton MJ (1993). Lateral diffusion in an archipelago. Single-particle diffusion. Biophys J 64, 1766–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scala S, and Aiuti A (2019). In vivo dynamics of human hematopoietic stem cells: novel concepts and future directions. Blood advances 3, 1916–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simson R, Sheets ED, and Jacobson K (1995). Detection of temporary lateral confinement of membrane proteins using single-particle tracking analysis. Biophys J 69, 989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaulding E, Fooksman D, Moore JM, Saidi A, Feintuch CM, Reizis B, Chorro L, Daily J, and Lauvau G (2016). STING-Licensed Macrophages Prime Type I IFN Production by Plasmacytoid Dendritic Cells in the Bone Marrow during Severe Plasmodium yoelii Malaria. PLoS Pathog 12, e1005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjin G, Flores-Figueroa E, Duarte D, Straszkowski L, Scott M, Khorshed RA, Purton LE, and Lo Celso C (2019). Imaging methods used to study mouse and human HSC niches: Current and emerging technologies. Bone 119, 19–35. [DOI] [PubMed] [Google Scholar]
- Upadhaya S, Sawai CM, Papalexi E, Rashidfarrokhi A, Jang G, Chattopadhyay P, Satija R, and Reizis B (2018). Kinetics of adult hematopoietic stem cell differentiation in vivo. J Exp Med 215, 2815–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, and Nussenzweig MC (2010). Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell 143, 592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, and Frenette PS (2018). Niches for Hematopoietic Stem Cells and Their Progeny. Immunity 48, 632–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Tao F, Venkatraman A, Li Z, Smith SE, Unruh J, Chen S, Ward C, Qian P, Perry JM, et al. (2019). N-Cadherin-Expressing Bone and Marrow Stromal Progenitor Cells Maintain Reserve Hematopoietic Stem Cells. Cell Rep 26, 652–669 e656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Videomicroscopy of tdTomato+ HSCs in the tibia of live mice. Related to Figure 2. Representative examples of HSC movement in three individual animals are shown. Unprocessed image (left) shows Tom+ HSCs (red) and macrophages (orange). The processed image (right) shows cell contour along with its track (white). Scale bar is indicated on lower left. Scale in the upper right indicates time in hours:minutes:seconds:milliseconds.
Video S2. Videomicroscopy of tdTomato+ HSCs in the calvarium of live mice. Related to Figure 2. Representative example of a Tom+ HSC (red) in the calvarium of a live animal. Unprocessed image (left) shows Tom+ HSCs (red) and macrophages (orange). The processed image (right) shows cell contour along with its track (white). Scale bar is indicated on lower left. Scale in the upper right indicates time in hours:minutes:seconds:milliseconds.
Video S3. Videomicroscopy of tdTomato+ HSCs and GFP+ stromal cells in the tibia of live SCF reporter mice. Related to Figures 2–3. Unprocessed image (left) shows Tom+ HSCs (red), GFP+ stromal cells (blue) and macrophages (orange). The processed image (right) shows HSC contour along with its track (white) among GFP+ stromal cells (green). Scale in the upper right indicates time in hours:minutes:seconds:milliseconds.
Video S4. Videomicroscopy of tdTomato+ HSCs in the tibia of live mice before and after mobilization with AMD3100+BOP. Related to Figure 4. Representative examples of HSC movement in two individual animals before and after treatment with AMD3100 (AMD) and BOP. Unprocessed image (left) shows Tom+ HSCs (red) and macrophages (orange). The processed image (right) shows cell contour along with its track before (cyan) and after treatment (magenta). Time of acquisition before and after treatment is indicated on the right; scale in the upper right indicates total time in hours:minutes:seconds:milliseconds.
Data Availability Statement
All computer codes used in the data analysis are available from the GitHub software repository.
Python files: https://github.com/okrichev/Python-Files-HSC-project
Matlab files: https://github.com/okrichev/Matlab-Files-HSC-project