Abstract
Inflammation and altered glucose metabolism are two pathways implicated in the pathophysiology of major depressive disorder (MDD). We have previously shown that high inflammation as measured by C-reactive protein (CRP) in MDD patients is associated with symptoms of anhedonia, a core symptom of MDD, along with deficits in dopaminergic reward circuitry. Increased inflammation can shift metabolic demand and reprogram cellular energy sources, which may collectively impact the brain and reward processing to contribute to symptoms of anhedonia. To determine whether immunometabolic gene signatures were enriched in immune cells of depressed patients with increased inflammation and anhedonia, we examined whole-blood gene expression microarray (Illumina HumanHT-12) data from unmedicated, medically-stable patients with MDD (n=93). Patients were considered to have increased inflammation based on High (>3mg/L) versus Low (≤3mg/L) plasma CRP, and further classified as having a self-reported phenotype of High (n=30, 33rd percentile) versus Low (n=32, 67th percentile) Anhedonia. Functional enrichment of gene pathways was assessed by Gene Set Enrichment Analysis (GSEA) using the KEGG pathway database. Pathways related to glucose metabolism (insulin signaling, insulin resistance, HIF-1, PI3K/AKT signaling), cancer (e.g., genes related to insulin and PI3K/AKT signaling), and inflammation (B cell, T cell and Fc receptor signaling) were specifically enriched in patients with both High CRP and High Anhedonia (all FDR q<0.25). Within patients with High CRP in GSEA, the insulin signaling pathway was the top enriched pathway in patients with High versus Low Anhedonia (n=10 and 9 respectively), which was driven by genes expressed predominantly in monocytes (z=2.95, p<0.01). Conversely, within patients with High Anhedonia, in addition to enrichment of immunometabolic pathways, the tyrosine metabolism pathway was also reduced in patients with High versus Low CRP (n=20 and 10 respectively). Of note, enrichment of immunometabolic pathways was confirmed in complementary linear regression analyses examining pathways associated with a CRP-by-Anhedonia interaction term while controlling for clinical covariates in all patients (n=93). These results indicate that increased glucose and low tyrosine metabolism define a subset of depressed patients with high inflammation and anhedonia. Enrichment of cancer-related pathways driven by metabolic genes also suggests a shift in immune cell metabolism from oxidative phosphorylation to glycolysis. Together these data suggest that anhedonia in MDD with high CRP involves both immunometabolic shifts and reduced dopamine precursor availability.
Keywords: anhedonia, depression, inflammation, C-reactive protein, gene expression, glucose metabolism, insulin, dopamine, tyrosine
Introduction
Evidence indicates that inflammation and metabolic disturbances may interact to contribute to the pathophysiology of major depressive disorder (MDD) and particularly to symptoms of anhedonia (Treadway et al., 2019). A significant proportion of MDD patients exhibit evidence of peripheral and central nervous system inflammation, e.g., elevated inflammatory cytokines and the acute phase reactant C-reactive protein (CRP)(Felger et al., 2018), as well as metabolic disturbances such as insulin resistance (IR)(Li et al., 2016). Increased inflammation and metabolic disturbances are associated with reduced responsiveness to standard antidepressant medications (Furman et al., 2018; Haroon et al., 2018; Jha et al., 2018), likely through known consequences on neurotransmitter systems and related neurocircuits. Indeed, both endogenous and experimentally-induced inflammation has been reliably shown to impact mesolimbic dopamine and corticostriatal reward circuits in association with altered motivational states and symptoms of anhedonia (Felger and Treadway, 2017). Insulin has also been shown to affect mesolimbic dopamine, and IR may further contribute to depressive symptoms like anhedonia (Hamer et al., 2019), a multi-faceted symptom domain with distinct underlying pathophysiologies (Cooper et al., 2018). Thus, interactions between inflammation and metabolism and their impact on dopaminergic reward pathways may represent one pathophysiologic mechanism of anhedonia presentation (Pan et al., 2017; Swardfager et al., 2016).
We previously found that the tumor necrosis factor antagonist infliximab reduced depressive symptoms in patients with treatment-resistant MDD who exhibited high baseline plasma CRP, and anhedonia (engagement in work and activities) was the most improved symptom (Raison et al., 2013). Gene expression analyses revealed that, in addition to inflammatory pathways, baseline expression of genes related to glucose and lipid metabolism were also predictive of antidepressant response to infliximab (Mehta et al., 2013), findings which were validated by measurement of circulating glucose and lipid biomarkers (Bekhbat et al., 2018). Both metabolism-related genes and plasma markers were in turn affected by infliximab, consistent with bidirectional relationships between inflammation and metabolism. Whereas increased adiposity and high body mass index (BMI) contribute to inflammation (Capuron et al., 2017), inflammation has also been linked to cellular re-programing of glucose metabolism and development of IR. Functional and metabolic demands of immune cells activated by pathogens or danger signals in response to psychological or cellular stress cause rapid shifts in glucose metabolism from slow, energy-maximizing oxidative phosphorylation (OXPHOS) to rapid but less energy-yielding glycolysis (Ganeshan and Chawla, 2014). The resulting energy shortage is then amplified by inflammatory cytokines which promote IR, further limiting energy obtained through glycolysis (Shoelson et al., 2006).
Due to growing interest in the potential combined impact of inflammation and metabolic disturbance on the brain in the context of MDD (Lacourt et al., 2018; Treadway et al., 2019), and because whole-blood gene expression can uncover a broad range of immunometabolic pathways related to high inflammation and anhedonia, we conducted gene set enrichment analysis (GSEA) –a powerful approach to detect robust between-group transcriptomic changes (Subramanian et al., 2005)– in medication-free, medically-stable MDD patients. We hypothesized that shifts in gene expression linked to cellular re-programing of glucose metabolism would be observed specifically in patients with high plasma CRP and high anhedonia symptoms.
Methods
Ninety-three participants (age 21–65) with either a primary diagnosis of MDD (n=88) or bipolar disorder-current episode depressed (n=5) as determined by Structured Clinical Interview for Diagnostic and Statistical Manual-IV-TR (SCID-IV) were enrolled. Data from 62 of 93 patients were analyzed for GSEA, all patients (n=93) were used for linear regression (see below). Subjects were free of psychotropic and anti-inflammatory medications and excluded based on evidence of infection, autoimmune or inflammatory disease, uncontrolled medical illness or other psychiatric diagnoses including substance abuse (see Supplement).
Patient Subgrouping for GSEA Analysis
Because GSEA requires between-group contrasts (Subramanian et al., 2005), patients were classified based on levels of inflammation and anhedonia. Plasma CRP >3 versus ≤3mg/L defined “high” versus “low” inflammation (per American Heart Association). Within CRP categories, patients were subsequently classified as having Low (score range: 1–3, bottom 33rd percentile) versus High (score range: 6–9, top 67th percentile) Anhedonia based on an anhedonia subscale from the Inventory of Depressive Symptoms-Self Report (IDS-SR; see Supplement) previously associated with CSF inflammatory markers and inflammation-related deficits in reward circuity (Felger et al., 2018; Felger et al., 2016). Therefore, 62 patients with either Low or High Anhedonia were used for GSEA, 19 with High CRP (n=10 High and n=9 Low Anhedonia) and 43 with Low CRP (n=20 High and n=23 Low Anhedonia) (group contrasts, Fig S1).
Plasma hsCRP and Gene Expression
Plasma high sensitivity CRP was assessed via immunoturbidimetric assay (Sekisui Diagnostics, Exton, PA) and Beckman AU480 analyzer (Felger et al., 2018). Whole blood microarray was conducted as previously published (Mehta et al., 2013)(see Supplement).
Pathway Analysis
To identify immunometabolic pathways in patients with high inflammation and/or anhedonia, a series of between-group comparisons (Fig S1) were made using GSEA (Broad Institute). GSEA produces robust pathway enrichment results based on the cumulative effect of subtle expression changes across many genes within an entire pathway (Wang and Cairns, 2013). Functional enrichment of metabolic pathways was assessed using Kyoto Encyclopedia of Genes and Genomes (KEGG) gene sets included in Molecular Signatures Database (MSigDB)(Ogata et al., 1999) augmented with four additional metabolic pathways (Insulin resistance, HIF-1 signaling, AMPK signaling, and PI3K/AKT signaling) from KEGG Orthology, a routinely maintained database of molecular functional pathways (Kanehisa et al., 2016). Significance thresholding was nominal p-value <0.05 and FDR q-value <0.25, per recommendations by Broad Institute (Subramanian et al., 2005). To complement and confirm findings from GSEA, linear models including covariates were used to identify gene pathways associated with a CRP-by-Anhedonia interaction term using the entire sample (n=93; see Supplement).
Statistical Analysis
Descriptive statistics (mean, percent and standard deviation) summarized independent and dependent variables. Analyses were conducted using SPSS 25.0 (IBM Corp., Armonk, NY). To identify cellular source of genes driving enrichment of metabolic pathways, transcript origin analysis was conducted as previously published (Mehta et al., 2013)(see Supplement).
Results
Tables S1 and S2 show demographic and clinical variables for the study sample grouped by inflammation and anhedonia status.
Immunometabolic pathways were associated with high anhedonia in depressed patients with high CRP
Within depressed patients with High CRP, thirteen KEGG pathways were significantly enriched in patients with High (n=10) versus Low (n=9) Anhedonia (FDR<0.25)(Table 1, Contrast A). Four of the thirteen enriched pathways were related to glucose metabolism (insulin signaling, insulin resistance, hypoxia-inducible factor (HIF-1) signaling, and type 2 diabetes mellitus pathways) and three were cancer pathways (acute myeloid leukemia, renal cell carcinoma, and prostate cancer). Additionally, inflammatory pathways (B cell receptor, Fc gamma R mediated phagocytosis, and Fc epsilon RI signaling pathways) were enriched in patients with High Anhedonia. In contrast, within patients with Low CRP, only the extracellular matrix (ECM)-receptor interaction pathway was enriched in High (n=20) versus Low (n=23) Anhedonia (FDR<0.25)(Table 1, Contrast B).
Table 1.
Enrichment of KEGG pathways.
| Contrast | Enriched in | # | KEGG pathway | NES | p-value | FDR q-value |
|---|---|---|---|---|---|---|
| A. Within High CRP | High vs. Low Anhedonia | 1 | Insulin signaling pathway* | 2 | <0.001 | 0.011 |
| 2 | Insulin resistance* | 1.85 | <0.001 | 0.052 | ||
| 3 | Acute myeloid leukemia† | 1.8 | 0.001 | 0.068 | ||
| 4 | HIF-1 signaling pathway* | 1.66 | 0.003 | 0.134 | ||
| 5 | ERBB signaling pathway | 1.67 | 0.012 | 0.142 | ||
| 6 | Renal cell carcinoma† | 1.6 | 0.027 | 0.143 | ||
| 7 | Fc gamma R mediated phagocytosis | 1.6 | 0.008 | 0.151 | ||
| 8 | Fc epsilon RI signaling pathway | 1.68 | 0.002 | 0.152 | ||
| 9 | Neurotrophin signaling pathway | 1.6 | 0.019 | 0.164 | ||
| 10 | B cell receptor signaling pathway | 1.61 | 0.032 | 0.165 | ||
| 11 | VEGF signaling pathway | 1.62 | 0.001 | 0.166 | ||
| 12 | Type II diabetes mellitus* | 1.68 | 0.009 | 0.189 | ||
| 13 | Prostate cancer† | 1.53 | 0.03 | 0.241 | ||
| B. Within Low CRP | High vs. Low Anhedonia | 1 | ECM-receptor interaction | 1.89 | 0.005 | 0.082 |
| C. Within High Anhedonia | High vs. Low CRP | 1 | Tyrosine metabolism‡ | −1.87 | 0.001 | 0.085 |
| 2 | Glycosaminoglycan biosynthesis chondroitin sulfate | 1.49 | 0.042 | 0.192 | ||
| 3 | Pathways in cancer† | 1.5 | 0.016 | 0.196 | ||
| 4 | Melanoma† | 1.5 | 0.04 | 0.208 | ||
| 5 | T cell receptor signaling pathway | 1.45 | 0.049 | 0.219 | ||
| 6 | Long term depression | 1.44 | 0.04 | 0.226 | ||
| 7 | Regulation of autophagy | 1.52 | 0.03 | 0.243 | ||
| 8 | PI3K-AKT signaling pathway* | 1.41 | 0.034 | 0.243 |
Whole blood transcriptome of depressed patients was assessed for enrichment of metabolic pathways in GSEA using the KEGG curated database from MSigDB and four additional metabolic pathways (see methods). Group contrasts were: A) Within High CRP: High vs. Low Anhedonia (n=10 and n=9 respectively), B) Within Low CRP: High vs. Low Anhedonia (n=20 and n=23 respectively), and C) within High Anhedonia: High vs. Low CRP (n=10 and n=20 respectively). Pathways displaying significant enrichment (nominal p<0.05 and FDR q<0.25) are shown. Among depressed patients with High CRP, pathways related to glucose metabolism (*) and cancer (†) were significantly enriched in those with High vs. Low Anhedonia (A). Within patients with High Anhedonia, metabolism-related PI3K-AKT and cancer pathways were enriched in those with High vs. Low CRP (C). Among depressed patients with High Anhedonia, the tyrosine metabolism pathway (‡) was enriched in patients with Low vs. High CRP (C). KEGG: Kyoto Encyclopedia of Genes and Genomes; NES: Normalized Enrichment Score; FDR: False Discovery Rate; CRP: C-reactive Protein.
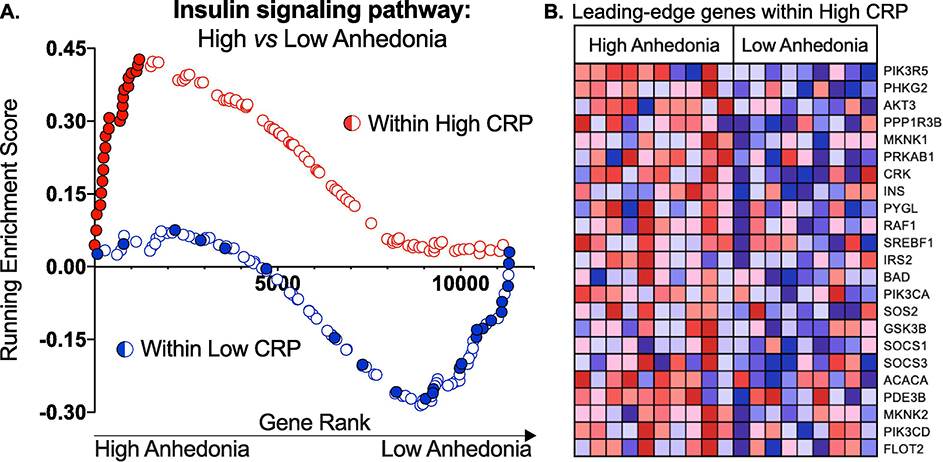
Insulin signaling was the top pathway associated with High Anhedonia among patients with High CRP (Normalized Enrichment Score=2.00, FDR q=0.01)(Fig 1A, red enrichment plot). This pathway was not differentially enriched in those with High versus Low Anhedonia within patients with Low CRP (Normalized Enrichment Score=−1.16, FDR q=1.00)(Fig 1A, blue enrichment plot), indicating specificity for patients with both elevated anhedonia severity and CRP. Furthermore, “leading-edge” genes driving enrichment of the insulin signaling pathway in patients with High CRP and High Anhedonia (genes shown in Fig 1B) were predominantly expressed by circulating monocytes (z=2.95, p<0.01), computed by transcript origin analysis.
Fig 1. High Anhedonia is associated with enrichment of the insulin signaling pathway among patients with high, but not low, inflammation.
A) Two GSEA enrichment plots of the insulin signaling pathway resulting from Contrast A: High vs. Low Anhedonia within High CRP (red enrichment plot) and Contrast B: High vs. Low Anhedonia within Low CRP (blue enrichment plot) are juxtaposed. Per Table 1, GSEA revealed that the insulin signaling pathway was enriched in High Anhedonia among depressed patients with High CRP (Normalized Enrichment Score=2.00, FDR q=0.01), but not in those with Low CRP (Normalized Enrichment Score=−1.16, FDR q=1.00). In each contrast, genes were ranked on the x-axis according to those most strongly expressed in High Anhedonia (low-number rank) to least (high-number rank). The running enrichment score (y axis) is plotted by each gene’s individual rank. Within patients with High CRP, genes in the leading-edge (contributing most to the enrichment score) are shown in solid red, and those same genes are shown in solid blue within the Low CRP group, indicating no enrichment of the insulin pathway in High vs. Low Anhedonia in these patients. B) Heatmap of leading-edge genes driving enrichment of the insulin signaling pathway in patients with High vs. Low Anhedonia among patients with High CRP. NES: Normalized Enrichment Score; FDR: False Discovery Rate; CRP: C-reactive Protein.
Increased PI3K/AKT and low tyrosine metabolism pathways were associated with high CRP in depressed patients with high anhedonia
To further test the specificity of immunometabolic gene signatures for the high inflammation and anhedonia phenotype, patients with High versus Low CRP were compared within the High and Low Anhedonia groups. Within patients with High Anhedonia, phosphoinositide 3-kinase/Akt (PI3K-AKT) signaling, cancer (pathways in cancer, melanoma) and T cell receptor signaling were among seven pathways enriched in patients with High (n=10) versus Low (n=20) CRP (Table 1, Contrast C). Moreover, the tyrosine metabolism pathway was significantly lower within patients with High Anhedonia who had High versus Low CRP (Table 1). No significant pathways were uncovered in High vs Low CRP within Low Anhedonia (Contrast D, see Fig S1).
Immunometabolic pathways associated with CRP-by-Anhedonia interaction were robust to covariate adjustment
Complementary linear analyses controlling for covariates (e.g., BMI) showed that gene probes positively associated with a CRP-by-Anhedonia interaction term in all patients (n=93) enriched both glucose metabolism-related (e.g., Pentose Phosphate Metabolism) and inflammatory (e.g., JAK-STAT, cancer) pathways. Adjustment for depression severity further uncovered enrichment of Insulin Resistance and Adipocytokine Signaling pathways (Table S3).
Discussion
This study describes a set of metabolic changes uniquely associated with anhedonic severity in the context of MDD patients with high inflammation. Here, we found that gene pathways related to glucose metabolism, IR and cancer were enriched specifically in the subset of depressed patients with both high inflammation and anhedonia. Furthermore, a metabolic pathway for the dopamine precursor tyrosine was reduced in high versus low inflammation within patients with a high anhedonia phenotype. These results demonstrate that anhedonia associated with high inflammation involves shifts in expression toward restructuring of cellular energy sources and away from dopamine synthetic pathways.
Specifically, we found that depressed patients with both high inflammation and anhedonia had significant enrichment of insulin, IR, PI3K/AKT, and Type 2 diabetes mellitus gene pathways compared to depressed patients with low inflammation and/or low anhedonia. A CRP-by-Anhedonia interaction in the full sample was associated with enrichment of insulin resistance, adipocytokine signaling, and pentose phosphate metabolism pathways in linear models controlling for covariates. Insulin regulates crucial energy homeostasis functions including glucose and lipid metabolism, and IR plays a key role in metabolic disorders such as Type 2 diabetes. Insulin’s regulation of glucose metabolism is primarily mediated through the PI3K/AKT cascade, which promotes absorption of glucose from blood into tissues and cells (Saltiel and Kahn, 2001). Importantly, inflammatory stimuli and byproducts of cellular stress induce IR by inhibiting insulin signal transduction (Shoelson et al., 2006).
Two studies (Nefs et al., 2012; Singh et al., 2019) previously found associations between impaired glucose metabolism and anhedonia symptoms in patients with metabolic disorders. Our results extend this literature by demonstrating that anhedonia in medically-stable MDD patients is characterized by insulin and cancer-related gene expression, likely reflecting shifts in the cellular metabolism of immune cells. Five pathways related to cancer were enriched in patients with high inflammation and anhedonia, primarily driven by genes controlling metabolism (e.g., in the PI3K/AKT pathway). Similar cancer pathways were associated with CRP-by-Anhedonia interaction in linear models with all subjects. Given that history of cancer was exclusionary, these pathways may reflect genes involved in the Warburg effect -metabolic shifting from OXPHOS to the faster, less energy-yielding glycolysis. First described in rapidly proliferating cancer cells, the Warburg effect is now well-characterized in activated immune cells. Findings that monocytic gene expression drove enrichment of the insulin signaling pathway is interesting considering that classically-activated M1 rely on glycolytic metabolism whereas alternatively-activated M2 utilize OXPHOS metabolism (Mills and O’Neill, 2016). Consistently, patients with high anhedonia and high inflammation also displayed enrichment of the HIF-1 pathway, involved in glycolytic shifts in myeloid cells (Palazon et al., 2014). Among MDD patients with high anhedonia, the tyrosine metabolism pathway was lower in patients with high versus low CRP. We have previously shown that inflammatory cytokines impair the conversion of phenylalanine to tyrosine (Felger et al., 2013), thus potentially limiting tyrosine availability for dopamine synthesis.
Strengths include use of medically-stable MDD patients off psychotropic and anti-inflammatory medications. Additionally, cholesterol-/diabetes-controlling medications were uncommon (n=3 total) and similar across groups. Change in appetite (per IDS-SR) was unrelated to anhedonia in all patients and after CRP stratification (data not shown), suggesting that links between metabolic pathways and anhedonia were not due to altered eating patterns. Limitations include small sample sizes after stratification by CRP and anhedonia in order to examine robust and cumulative expression of all genes in a given pathway in GSEA (Wang and Cairns, 2013). However, results were confirmed in linear models using all subjects. Anhedonia was measured using a single anhedonia subscale from IDS-SR. This scale previously correlated with inflammation and functional integrity of reward circuitry (Felger et al., 2018; Felger et al., 2016), and probes “motivational” and “consummatory” aspects of anhedonia. Future studies employing objective measures will explore this distinction.
In conclusion, our results support a framework by which the combined effects of inflammation and metabolic shifts toward IR and glycolysis hinder optimal energy generation and reduce dopamine availability to contribute to anhedonia in MDD. Understanding causal relationships among inflammation, glucose metabolism and anhedonia may reveal novel therapeutic approaches targeting immunometabolism for motivational deficits in psychiatric and medical illnesses.
Supplementary Material
Highlights.
Inflammation and metabolic disturbances are linked to anhedonia in MDD
Gene expression analyzed in MDD patients with High vs. Low CRP and/or Anhedonia
Immunometabolic pathways were enriched in the High CRP and High Anhedonia phenotype
Insulin resistance was the top pathway enriched in High CRP and High Anhedonia
High CRP and High Anhedonia also had reduced tyrosine metabolism pathway genes
Acknowledgments
Funding Sources
This study was supported by grants R01MH087604 (Miller), R01MH109637 (Felger), R01MH107033 (Haroon), R01MH112076 (Miller/Haroon), R01MH108605 (Treadway), and K23MH114037 (Goldsmith) from the National Institute of Mental Health; and grant CADF49143 from the Dana Foundation (Felger). In addition, the study was supported in part by PHS Grants UL1TR000454, UL1TR002378, KL2TR000455, and TL1TR002382 from the Clinical and Translational Science Award program, by the NIH/NCI under award number P30CA138292, and the Emory Integrated Genomics Core (EIGC), which is subsidized by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities.
Footnotes
Financial Disclosure
All authors declare no conflicts of interest. In the past year, Dr. Felger has consulted for Otsuka on a topic unrelated to this research. In the past three years Dr. Treadway has been a paid consultant to Avanir Pharmaceuticals and Blackthorn Therapeutics. None of these entities provided any funding for the current work and all views expressed are solely those of the authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bekhbat M, Chu K, Le NA, Woolwine BJ, Haroon E, Miller AH, Felger JC, 2018. Glucose and lipid-related biomarkers and the antidepressant response to infliximab in patients with treatment-resistant depression. Psychoneuroendocrinology 98, 222–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capuron L, Lasselin J, Castanon N, 2017. Role of Adiposity-Driven Inflammation in Depressive Morbidity. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology 42, 115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA, Arulpragasam AR, Treadway MT, 2018. Anhedonia in depression: biological mechanisms and computational models. Curr Opin Behav Sci 22, 128–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felger JC, Haroon E, Patel TA, Goldsmith DR, Wommack EC, Woolwine BJ, Le NA, Feinberg R, Tansey MG, Miller AH, 2018. What does plasma CRP tell us about peripheral and central inflammation in depression? Mol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felger JC, Li L, Marvar PJ, Woolwine BJ, Harrison DG, Raison CL, Miller AH, 2013. Tyrosine metabolism during interferon-alpha administration: association with fatigue and CSF dopamine concentrations. Brain Behav Immun 31, 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felger JC, Li Z, Haroon E, Woolwine BJ, Jung MY, Hu X, Miller AH, 2016. Inflammation is associated with decreased functional connectivity within corticostriatal reward circuitry in depression. Mol Psychiatry 21, 1358–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felger JC, Treadway MT, 2017. Inflammation Effects on Motivation and Motor Activity: Role of Dopamine. Neuropsychopharmacology 42, 216–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman JL, Soyombo A, Czysz AH, Jha MK, Carmody TJ, Mason BL, Scherer PE, Trivedi MH, 2018. Adiponectin Moderates Antidepressant Treatment Outcome in the Combining Medications to Enhance Depression Outcomes Randomized Clinical Trial. Pers Med Psychiatry 9–10, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganeshan K, Chawla A, 2014. Metabolic regulation of immune responses. Annu Rev Immunol 32, 609–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer JA, Testani D, Mansur RB, Lee Y, Subramaniapillai M, McIntyre RS, 2019. Brain insulin resistance: A treatment target for cognitive impairment and anhedonia in depression. Experimental neurology 315, 1–8. [DOI] [PubMed] [Google Scholar]
- Haroon E, Daguanno AW, Woolwine BJ, Goldsmith DR, Baer WM, Wommack EC, Felger JC, Miller AH, 2018. Antidepressant treatment resistance is associated with increased inflammatory markers in patients with major depressive disorder. Psychoneuroendocrinology 95, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha MK, Wakhlu S, Dronamraju N, Minhajuddin A, Greer TL, Trivedi MH, 2018. Validating pre-treatment body mass index as moderator of antidepressant treatment outcomes: Findings from CO-MED trial. Journal of affective disorders 234, 34–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M, 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44, D457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacourt TE, Vichaya EG, Chiu GS, Dantzer R, Heijnen CJ, 2018. The High Costs of Low-Grade Inflammation: Persistent Fatigue as a Consequence of Reduced Cellular-Energy Availability and Non-adaptive Energy Expenditure. Frontiers in behavioral neuroscience 12, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Shelton RC, Chassan RA, Hammond JC, Gower BA, Garvey TW, 2016. Impact of Major Depressive Disorder on Prediabetes by Impairing Insulin Sensitivity. J Diabetes Metab 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta D, Raison CL, Woolwine BJ, Haroon E, Binder EB, Miller AH, Felger JC, 2013. Transcriptional signatures related to glucose and lipid metabolism predict treatment response to the tumor necrosis factor antagonist infliximab in patients with treatment-resistant depression. Brain Behav Immun 31, 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EL, O’Neill LA, 2016. Reprogramming mitochondrial metabolism in macrophages as an anti-inflammatory signal. Eur J Immunol 46, 13–21. [DOI] [PubMed] [Google Scholar]
- Nefs G, Pouwer F, Denollet J, Kramer H, Wijnands-van Gent CJ, Pop VJ, 2012. Suboptimal glycemic control in type 2 diabetes: a key role for anhedonia? J Psychiatr Res 46, 549–554. [DOI] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M, 1999. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 27, 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazon A, Goldrath AW, Nizet V, Johnson RS, 2014. HIF transcription factors, inflammation, and immunity. Immunity 41, 518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z, Rosenblat JD, Swardfager W, McIntyre RS, 2017. Role of Proinflammatory Cytokines in Dopaminergic System Disturbances, Implications for Anhedonic Features of MDD. Curr Pharm Des 23, 2065–2072. [DOI] [PubMed] [Google Scholar]
- Raison CL, Rutherford RE, Woolwine BJ, Shuo C, Schettler P, Drake DF, Haroon E, Miller AH, 2013. A randomized controlled trial of the tumor necrosis factor antagonist infliximab for treatment-resistant depression: the role of baseline inflammatory biomarkers. JAMA Psychiatry 70, 31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltiel AR, Kahn CR, 2001. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414, 799–806. [DOI] [PubMed] [Google Scholar]
- Shoelson SE, Lee J, Goldfine AB, 2006. Inflammation and insulin resistance. J Clin Invest 116, 1793–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh MK, Leslie SM, Packer MM, Zaiko YV, Phillips OR, Weisman EF, Wall DM, Jo B, Rasgon N, 2019. Brain and behavioral correlates of insulin resistance in youth with depression and obesity. Horm Behav 108, 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swardfager W, Rosenblat JD, Benlamri M, McIntyre RS, 2016. Mapping inflammation onto mood: Inflammatory mediators of anhedonia. Neurosci Biobehav Rev 64, 148–166. [DOI] [PubMed] [Google Scholar]
- Treadway MT, Cooper JA, Miller AH, 2019. Can’t or Won’t? Immunometabolic Constraints on Dopaminergic Drive. Trends Cogn Sci 23, 435–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Cairns MJ, 2013. Gene set enrichment analysis of RNA-Seq data: integrating differential expression and splicing. BMC Bioinformatics 14 Suppl 5, S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.