Abstract
Genome editing opens up a new frontier in developing personalized therapeutic solutions. With the unprecedented advance in the discovery and engineering of gene editing nucleases, it has now become potentially feasible to therapeutically influence up to 90% of all human genetic mutations. Hearing loss is one of the most well studied fields from the genetics perspective, with more than one hundred identified deafness genes. Novel viral and non-viral vectors have been established as safe and efficient modalities to deliver transgenes into cells of the cochlea and to the vestibular system in animal models. Recent studies demonstrated proof-of-concept for therapeutic genome and base editing in the mammalian inner ear and preclinical development is ongoing. This review summarizes important advances and future challenges for this transformative therapeutic modality for genetic and non-genetic hearing loss.
1. Introduction on genome editing technologies
Gene therapy is revolutionizing medicine and previously untreated diseases become manageable. The recent approval of several gene therapy drugs by the Food and Drug Administration (FDA) for inherited blindness (voretigene neparvovec (Russell et al., 2017), Luxturna®), spinal muscular atrophy (onasemnogene abeparvovec (Mendell et al., 2017), Zolgensma®) and B-cell malignancies (axicabtagene ciloleucel (Neelapu et al., 2017), Yescarta® and tisagenlecleucel (Schuster et al., 2019), Kymriah®) sets the stage for this treatment modality to reach broader patient populations. Gene therapy was conceptualized in 1972 (Friedmann and Roblin, 1972) and it took five decades to finally be at a stage where safety and efficiency of this technology have been established. During these years, various viral vectors have been developed that enable the transport of foreign DNA into target cells (Miyoshi et al., 1998; Samulski et al., 1983, 1982; Zufferey et al., 1998). Non-viral vectors represent alternatives to viruses and are less immunogenic and are being considered safer.
Historically and currently, the majority of gene therapy applications rely on the delivery of the normal coding sequence of the gene of interest into the target cells. This approach is called ‘gene addition’ or ‘gene supplementation’. These gene addition therapies however, have several limitations. The coding sequence of some genes may be too large to fit into the applied viral vectors. Furthermore, viral-mediated gene delivery results in overexpression of the gene and lacks physiological regulation. In addition, gene addition usually cannot be applied in dominant diseases, where silencing or knocking-out of the mutant allele would be required.
With the advent of genome editing it now became a possibility to therapeutically intervene genetic diseases at the root cause of genomic DNA level. The recent introduction of the CRISPR (clustered regularly interspaced short palindromic repeats) system allows simple targeting and editing of virtually any sequence of interest in the genome (F Ann Ran et al., 2013). The simplicity of this system comes from the fact that it is based on targeting the genomic DNA using a complimentary RNA motif (Kleinstiver et al., 2016; F Ann Ran et al., 2013). This contrasts CRISPR with previous generations of gene editing systems, such as zinc-finger nucleases (Gaj et al., 2013), TALENSs (Gaj et al., 2013), or meganucleases (Silva et al., 2011), all of which are based on unique protein motifs that interact with a specific sequence in the genomic DNA. CRISPR is a two component system that uses a short RNA motif (guide RNA, gRNA) to deploy an active nuclease, the Cas9, to the target genomic site. The default action of all gene editing nucleases is an induction of a double stranded DNA break (DSBs), which will be – depending on the cellular state – repaired by different mechanisms. Next generation engineered genome editing systems however do not rely on DSBs, but use the CRISPR system to deploy certain effectors onto a specific locus. These effectors then trigger certain forms of DNA repair and lead to site-specific changes in the genomic sequence. These systems include epigenome editors (Holtzman and Gersbach, 2018), base editors (Gaudelli et al., 2017; Komor et al., 2016a) and the more recently developed prime editors (Anzalone et al., 2019), among others. In this review we summarize recent studies using gene editing systems in the inner ear, point out future avenues of research and discuss therapeutic applications.
2. RNA-guided nucleases (RGNs)
In this paragraph, we discuss the most commonly used gene editing systems, the RNA-guided nucleases, in more detail. As the name implies, these genome editing enzymes are guided by a short RNA motif to the target DNA site by classical Watson-Crick base pairing. RGNs are based on the bacterial CRISPR system and currently encompass three different types of enzymes, the Cas9, the Cas12a (Zetsche et al., 2015) (formerly known as Cpf1) and Cas12e (formerly known as CasX (Liu et al., 2019)). The ~100 bp guide RNA (gRNA) component has a ~20 nt complimentary region to the target DNA (Ann Ran et al., 2013). The first RGN was the CRISPR/Cas9 system from Staphylococcus pyogenes (SpCas9), and its mechanism of action has been studied in detail (Cong et al., 2013; Mali et al., 2013). The Cas9 enzyme binds to a so-called protospacer adjacent motif (PAM) site present on the genomic DNA adjacent to the gRNA hybridization region. In the case of the SpCas9 enzyme, this short PAM motif is an NGG sequence, where N can be any nucleotide. The PAM site is an important determinant of the functionality of the RGNs, however it also restricts its activity; as only genomic sites with an appropriate PAM site can be targeted. Recent efforts have been made to broaden the targeting range of Cas9. The first strategy has been to engineer the SpCas9 system to alter PAM specificity (Kleinstiver et al., 2015a). Kleinstiver et al. provided excellent examples for the versatility of this approach by engineering both Cas9 (Kleinstiver et al., 2015b) and also an enzyme from the Cas12a (Kleinstiver et al., 2019) family. The second strategy was to explore CRISPR/Cas9 enzymes in other bacterial species. This has led to the discovery of several novel gene editing systems, including the Staphylococcus aureus Cas9 (SaCas9) (Ran et al., 2015) or the Neisseria meningitidis Cas9 (Hou et al., 2013). The ever-expanding space of new editor variants essentially now permit the targeting of virtually any sequence of the genome.
3. Gene editing outcomes
Gene editing can be defined as inducing certain chemical changes at the genomic DNA. The final outcome of the genetic change is dependent on the type of the nuclease used and whether double-stranded break, single-stranded break (nick) or no break is induced. Furthermore, the outcome will also depend on the state of the cell. In general, double-stranded breaks induced by the CRISPR/Cas9 system are repaired by either non-homologous end joining (NHEJ) or homology-directed repair (HDR) (Ann Ran et al., 2013). In non-dividing cells, such as cochlear hair cells, NHEJ is the predominant mechanism, which usually leads to small insertions and/or deletions (indels) and therefore to heterogenous gene editing outcomes (Gyögy et al., 2019b). HDR in contrast allows for the induction of precise genetic changes, but this process is not active in non-dividing cells (Yeh et al., 2018), making HDR less useful for inner ear applications. NHEJ-based strategies applying CRISPR/Cas9 have already reached the clinic, however they rely on special circumstances, such as inactivating a splice mutation (Maeder et al., 2019) or integration of a therapeutic gene into the albumin locus in the liver (Sharma et al., 2015). NHEJ can also be utilized to knock out a dominant gene without disrupting the wild-type allele (see below). However, for the majority of genetic diseases due to the loss-of-function mutations, the NHEJ-mediated DNA-repair is not optimal. In order to translate gene editing to broad clinical applications, precise and highly efficient editing systems with more control on gene editing outcomes are required.
Next-generation gene editing systems take advantage of the Cas9 enzyme to anchor other effector domains onto target site, instead of relying on the endogenous activity of Cas9 to induce DSBs. For example, epigenome editors induce or repress gene expression (Holtzman and Gersbach, 2018). DNA base editors developed by the Liu lab induce deamination of either cytosines (Gaudelli et al., 2017) or adenines (Komor et al., 2016b) to allow for the repair of transitions mutations (C-T or A-G changes). More recently, a novel and highly promising approach, termed prime editing, can induce pre-determined precise changes, such as transitions, transversions, precise insertions, deletions and complex genetic changes (Anzalone et al., 2019). The outcomes of base and prime editing are more homogenous compared to gene editing outcomes following a DNA break. With the continuous development of the gene editing toolbox, it will be feasible to develop gene editing based therapies to target virtually any pathological mutation and provide precise correction in a not too distant future.
4. Types of human disease-causing mutations and their relevance to gene editing therapies
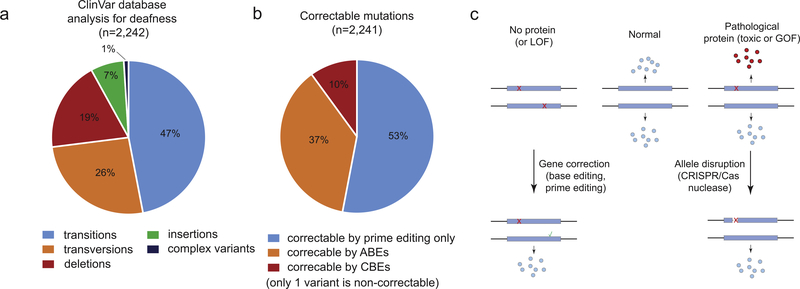
In order to develop therapeutic strategies required to overcome the effects of mutations, it is important to discuss the types of pathogenic genetic changes. The ClinVar database is an excellent source of human genetic variation and includes annotations on the pathogenicity of the mutations. Based on the ClinVar database, half of all pathogenic genetic changes arise from single nucleotide variations (Anzalone et al., 2019). We have analyzed variants listed in the ClinVar database that are associated with hearing loss and determined whether these variants are correctable using base- or prime editing (Fig. 1a, Supplementary Methods, Supplementary Table 1). We found that among variants associated with hearing loss, 47% are transitions (A-G or C-T changes), 26% are transversions (purin to pyrimidin changes) and 26% are small insertions or deletions. Based on this analysis we conclude that potentially 10% and 37% of mutations can be repaired by cytosine and adenine base editors, respectively (Fig. 1b). Interestingly, 53% of all mutations can be only be repaired by prime editors; these include transversions and small indels. Altogether, current genome editing technologies can address up to 99.9% of genetic defects associated with hearing loss. Non-correctable mutations include copy number variations, larger deletions and insertions; however, we have only found one single variant that is non-correctable with base- or prime editing.
Fig. 1.
(a) Types of mutations causing hearing loss. ClinVar database was accessed on the 10/21/2019. (b) Targetable mutations by different strategies. (c) The effect of mutations and therapeutic implications. LOF: loss-of-function, GOF: gain-of-function.
The other important factor to consider is whether a mutation has loss-of-function or gain-of-function (dominant negative) effect. Recessively inherited diseases arise when two loss-of-function mutations are present in each of two chromosomes (Fig. 1c). In order to provide a therapeutic solution with gene editing, one of the two mutations needs to be corrected/repaired in order to restore normal protein levels or function. Therefore, it is highly desirable to apply gene editing systems such as base editors or prime editors, which do not induce double-stranded breaks. In contrast, mutations that lead to a production of a pathological protein result in dominantly inherited diseases, where only one allele is mutated while the other allele is normal (Fig. 1c). In addition to the correction of the monoallelic mutation, the disruption of the mutant allele is also a viable therapeutic option; unless haploinsufficiency (one allele does not produce enough protein level) is involved in the pathogenesis. Allele-specific gene inactivation is an emerging strategy (B. György et al., 2018; György et al., 2019b; P. Li et al., 2018) to treat dominantly inherited diseases. The general challenge with a gene disruption strategies, however, is that most dominant mutations are single nucleotide changes (usually missense mutations) and selective inactivation of the mutant allele may be difficult due to limited specificity of the gRNA (György et al., 2019b). We will discuss potential options to overcome this hurdle in detail below.
5. Delivery modalities for gene and base editing
Efficient delivery of editing agents into diverse inner ear cell types is the prerequisite for the application of editing as therapy. The inner ear is ideal for local delivery, due to the fact that it has a confined space with a small volume and is separated from other organs. Furthermore, the number of inner ear cells to target is relatively small. Despite the ideal features of the inner ear for gene therapies, there are important concerns that have to be taken into account when selecting a delivery modality. Most importantly, unwanted editing in non-target cells is a major concern that needs to be minimized (Heidenreich and Zhang, 2016; Komor et al., 2017). Furthermore, genotoxicity and immunity associated with editing in the inner ear have to be understood and managed. Previously, CRISPR/Cas9 and base editor complexes have been delivered by multiple modalities including viral vectors, liposomes and nanoparticles (lipid, biopolymer and gold). The complexes can be delivered as DNA, as ribonucleoproteins (RNPs) or as proteins; all of which have different characteristics (Wang et al., 2017). We will discuss these different modalities in detail below.
5.1. Cationic liposomes
Cationic liposome-complexed anionic RNPs have been shown to be effective for delivering gene editing agents into inner ear auditory hair cells. Commercially available liposomes, such as Lipofectamine 2000 and RNAiMax, have been used to deliver SpCas9 and GFP-gRNA into the neonatal cochlea, resulting in editing in 20% of hair cells as assayed by abolished GFP signals due to NHEJ (Zuris et al., 2015). Using the same approach, Lipofectamine 2000 has been used to deliver SpCas9 and gRNA to target a single nucleotide mutation in the Tmc1 gene in the mouse model Beethoven (Bth) of dominant human genetic hearing loss DFNA36 (Gao et al., 2018). Disruption of the Bth mutation led to hearing preservation and to increased hair cell survival in the injected inner ears of mutant mice (Gao et al., 2018). Furthermore, liposome-mediated delivery significantly reduced genome-wide off-target effects compared to DNA-based CRISPR delivery. Lipofectamine 2000 has also been used for delivery of base editing agents that included a gRNA and cytosine base editor protein (BE3) in order to install a single nucleotide change in the Ctnnb1 gene in neonatal mouse cochlear supporting cells (Yeh et al., 2018).
The inner ear is a complex tissue with several different cell types. Furthermore, neonatal and mature cells have different characteristics from the targeting perspective. It is important for future work to expand liposome targets in the inner ear so they can be developed into effective delivery vehicles. While liposome-based delivery has been effective in neonatal hair cells and in some supporting cells, so far it has failed to target other inner ear cell types and cells in the fully mature inner ear (unpublished data). Continuous screening for novel different delivery vehicles of editing agents is important so that gene editing can be used as a therapy for deafness.
5.2. Nanoparticles
An emerging field of delivery is the application of nanoparticles. Nanoparticles can be formulated through compositions including lipids, biopolymers, cell-penetrating peptides (CPP) and gold, with each of these components offering different characteristics. Compared to other delivery systems, nanoparticles are capable of delivering editing complexes in diverse forms of DNA, RNP and mRNA. Nanoparticles have the capacity of delivering multiple components without size limitations, with reduced toxicity and minimal immune response. Nanoparticles can be efficiently manufactured for large scale production. Finally, nanoparticle-mediated delivery is transient, thus the editing agents (RNP and mRNA) will be degraded shortly after entering the cells, which leads to greatly reduced off-target effects (Glass et al., 2017; Lee et al., 2018, 2017; Mout et al., 2017; Sun et al., 2015). Given the transient nature of nanoparticle mediated delivery, recently there has been accelerated efforts to discover novel nanoparticles for the delivery of editing agents to treat diseases. Lipid based nanoparticles (LNP) have shown promising results in delivering editing agents into diverse mouse organs including heart, liver, lung and the CNS. Bioreducible LNPs have the advantage that in the intracellular environment, LNPs are rapidly degraded after releasing their content (Altinoglu et al., 2015). LNP based delivery has been shown to be able to target inner ear hair cells with higher efficiency, target diverse inner ear cell types including stria vascularis and neurons and have reduced toxicity (unpublished data). LNP are complex structures with different domains, including the head, linker, and tail domains. All of these domains can be modified by combinatory approach allowing for versatile engineering (Li et al., 2019b, 2019a; Y. Li et al., 2018; Wang et al., 2016, 2014) (Fig. 2). As a result of a combinatory library, large numbers of LNPs with different properties can be produced and screened for inner ear applications. This will likely result in identification of new LNPs capable of delivering a wide range of editing agents.
Fig. 2. Construction of synthetic biodegradable lipid library (LNPs) for delivery of editing agents.
(a) Typical structure of synthetic lipid consists of a head, a linker and a tail domain. (b) Each of a synthetic lipid domain can be easily tuned to produce a library with diverse structural variants, including variants with improved target specificity, cargo selectivity, reduced toxicity and packaging capacity. (c) Bioreducible property allows rapid disassembly of LNPs upon entry into the cells for release of the cargo. The TEM shows the size of LNP and RNP nanocomplex of 150–300 nm.
Other forms of nanoparticles have been also used successfully to deliver editing agents to target different tissues and cells to treat diseases, illustrating that some major barriers in delivery could be overcome by nanoparticle approaches. For example, gold nanoparticles have been used to deliver editing RNPs into brain to target diverse brain cell types including neurons, astrocytes and microglia without any apparent toxicity. ‘Such delivery resulted in efficient editing of metabotropic glutamate receptor 5 gene that rescued a mouse model of fragile-X syndrome (Lee et al., 2018). Recently, engineered amphiphilic peptide nanoparticles have been used to deliver SpCas9 and Cas12a in the form of RNP into cultured differentiated human airway epithelia cells and mouse lung (Krishnamurthy et al., 2019).
Similar to other nanoparticle delivery systems, CPP nanoparticle-mediated delivery did not show toxicity at the dose tested and was able to target a large number of cells along the respiratory track. Recently glutathione (GSH)-cleavable covalently crosslinked polymer coating, termed nanocapsule (NC) has been shown to deliver editing RNPs to target mouse tissues including retinal pigment epithelium and skeletal muscles in vivo (Chen et al., 2019).
In comparison to RNP delivery, the delivery of mRNA for editing has been challenging (Kowalski et al., 2019). This may be in part due to that after mRNA delivery, there is a time lag before the editor protein can be translated from the mRNA, and during this time some of the gRNAs may be degraded. In the case of RNP delivery, the editor protein and gRNA are preassembled and thus are able to exert their function immediately after the entry into the nucleolus. Modifications of gRNAs by changing the RNA 2′OH group to 2′OMe has shown to extend the stability and improve the editing efficiency when co-delivered with Cas9 mRNA (Mir et al., 2018; Yin et al., 2017). Intellia Therapeutics has recently reported that an ionizable lipid nanoparticle formulation mediated the delivery of Cas9 mRNA and single guide RNA that led to high-efficient editing of transthyretin gene in the mouse liver (Finn et al., 2018). This study demonstrated the feasibility of direct delivery of editor mRNA together with the gRNA for efficient editing in vivo.
5.3. Viral vectors
Viral vectors have also been used extensively to deliver gene editing agents with good outcomes in various tissues and organs. AAV vectors are widely used in clinical gene therapy applications, and they have been used frequently to deliver editing machinery to treat genetic disorders in various animal models for DMD, Hungtington’s chorea, cystic fibrosis (Vaidyanathan et al., 2019) and genetic blindness (Amoasii et al., 2018; Maeder et al., 2019; Monteys et al., 2017; Yang et al., 2017). Editors including SpCas9 and Cas12a together with their respective gRNAs have a combined size larger than 5 kb, thus they have to be packaged into different AAVs (Swiech et al., 2015). Dual AAVs have been used to deliver SpCas9 and gRNA separately to target tissues to treat disease in animal models (Bence György et al., 2018; Swiech et al., 2015). In contrast, SaCas9 is smaller in size (3.2 kb) (Friedland et al., 2015) and therefore can be packaged together with a gRNA into single AAV, resulting in efficient transduction and editing. The use of AAV for SaCas9 delivery and editing has been recently achieved by one of the authors to target the progressive hearing loss in Beethoven mice (György et al., 2019b). This treatment preserved hearing for an extended period of time, highlighting the potential for the AAVbased editing approach to treat genetic hearing loss (Gyögy et al., 2019b). For base editing that includes the dead Cas9 (dCas9), DNA deaminase and gRNA, the components have to be packaged into separate AAV vectors as the combined size is close to 6 kb, making it challenging to efficiently deliver the multiple components necessary for editing. A recent publication showed that intein-split base editors can be delivered using dual AAV vectors and base editing leads to efficient in vivo editing in various tissues (Levy et al., 2020). AAVs have been extensively evaluated in the inner ear of animal models including mouse and non-human primate (György et al., 2019a). However, in addition to size limit, much work is still needed to identify AAV vectors capable of efficiently targeting some major inner ear cell types including outer hair cells, supporting cells, stria vascularis and neurons, especially in the fully mature cochlea in larger animal models.
To conclude the delivery aspects, the inner ear is an ideal organ for local delivery. Future studies should test diverse delivery vehicles for different cargos for inner ear delivery, so that those with optimal delivery targets with reduced toxicity and off-targets can be identified for efficient inner ear editing. The delivery paradigms offered by different approaches will likely open new paths aiming at the delivery of diverse cargos to have effects on the inner ear.
6. Gene disruption for dominant hearing loss
The Cas9 enzyme is an active nuclease that upon deployment on the target DNA and creates a double-stranded DNA break (DSB). Depending on the state of the cell, this break can be repaired by different mechanisms, such as non-homologous end-joining (NHEJ), homology directed repair (HDR) or microhomology mediated end joining (MMEJ). In non-dividing cells, such as in cells of the mature inner ear, NHEJ is the predominant mechanism (Yeh et al., 2018). Therefore, application of the CRISPR system in the inner ear usually leads to insertions and deletions on the target gene (indels). These indels induce frameshift and lead to a knockout of the target gene by terminating the open reading frame. Although this is an imprecise outcome and different cells might have different genetic outcomes, this strategy still has high potential therapeutic value in the case of dominantly inherited mutations, where the inactivation of the pathological protein production is the desired outcome.
The most well studied animal model for dominant human hearing loss is the Beethoven model (Askew et al., 2015a; Beurg et al., 2015; Corns et al., 2016; Marcotti et al., 2006; Pan et al., 2013; Vreugde et al., 2002). The disease is caused by a T to A change (1235T→A) in the Tmc1 gene, a protein product of which is part of the mechanotransduction (MET) channel in inner ear hair cells (Pan et al., 2018, 2013). The mutation was discovered in the context of the large-scale N-ethyl-N-nitrosourea (ENU) mouse mutagenesis program in 2002 (Vreugde et al., 2002). The mutation leads to an amino acid change (M412K) that exhibits dominant inheritance as heterozygote animals show symptoms of progressive hearing loss. The Beethoven mutation causes a reduction in single MET-channel current amplitudes, reduces calcium permeability of the channel and increases the number of channels per hair cells (Pan et al., 2013). Based on these results it has been suggested that the pM412K mutation is a dominant gain-of-function mutation. Heterozygous animals demonstrate disorganization of hair bundles and hair cell loss ultimately leading to profound deafness. The deafness is progressive in nature and starts at high frequencies and ultimately affects all frequencies. The Beethoven model is an important model not only because it is very well characterized but also because it is equivalent to the human DNFA36 mutation (p.M418K), which has been first described in a large Chinese family (Zhao et al., 2014).
Several strategies have been attempted previously to overcome the toxic effects of the Beethoven mutation. These include overexpression of the wild-type Tmc2 (Askew et al., 2015b), artificial miRNA (Shibata et al., 2016) and genome editing (Gao et al., 2018; Gyögy et al., 2019b). AAV-mediated Tmc2 overexpression led to modest improvement of hearing thresholds in the Beethoven animals suggesting that wild-type Tmc2 can partly compensate for the Beethoven mutation and promotes survival of inner hair cells (Askew et al., 2015b). However, the hearing restoration was very limited as the best treated animals exhibited an ABR threshold of 95 dB at P25–30. An artificial miRNA was able to slow the progression of the disease, however the majority of the animals still progressed to profound deafness at all frequencies (Shibata et al., 2016). The same artificial miRNA also slowed degeneration when injected at later stages (P15–P16, 8 and 12 weeks), however animals ultimately progressed to deafness (Yoshimura et al., 2019). These results suggest that overexpression of a wild-type homologue or RNA silencing cannot overcome the severe effects of the Beethoven mutation.
Gene editing has the advantage that it works at the DNA and not at the RNA level with the aim to inactivate the single Beethoven allele. Two independent studies analyzed the efficacy of all elespecific gene disruption in the Beethoven model. Gao et al. showed disruption of the Tmc1Bth by a mutation targeting gRNA combined with the Streptococcus pyogenes Cas9 (SpCas9) (Gao et al., 2018). The SpCas9/gRNA complex was delivered to the inner ear as Cas9-gRNA-lipid complexes as described above. Injected animals demonstrated improved hair cell survival and improvement of auditory brainstem response thresholds at 4 weeks post-injection. However, the effect – although still statistically significant – diminished by 8 weeks suggesting that hair cell degeneration was not halted and animals ultimately progressed to profound deafness. These results suggest that only a fraction of the cells was targeted with the vector or allele-selectivity of this approach was not optimal.
A follow-up study has set out to find Cas9/gRNA combinations that discriminate between the WT and Beethoven allele (Gyögy et al., 2019b). In order to find a Cas9/gRNA combination that selectively disrupts the mutant allele the authors tested SpCas9 with 11 different gRNAs, high-fidelity Cas9 enzymes and a novel PAM variant from Staphylococcus aureus (SaCas9-KKH (Kleinstiver et al., 2015a)). SpCas9 and high-fidelity SpCas9 enzymes failed to specifically target the Beethoven allele as indel formation was apparent on the mutant and WT alleles as well. In contrast, the SaCas9-KKH differentiated between the Beethoven and WT alleles with complete selectivity, and indel formation was below background on the WT allele. This was due to the fact that the Beethoven allele, but not the WT allele, creates an NNNRRT PAM site for the SaCas9-KKH enzyme. These results suggest, that PAM targeting is more stringent then gRNA binding, and this approach can discriminate alleles that are different in only one single nucleotide. The SaCas9-KKH variant was packaged in a synthetic Anc80 AAV vector (Zinn et al., 2015) along with the gRNA cassette. This single AAV was then injected into the inner ears of newborn Tmc1Bth/−/ Tmc2−/− animals for physiological measurements and Tmc1Bth/+/Tmc2+/+ animals for sequencing, histology and hearing tests. Single-cell patch clamp recordings revealed that mechanotransduction is significantly abrogated in (close to undetectable) in Tmc1Bth/−/Tmc2−/− animals suggesting effective knockout of the single Beethoven allele.
In the Tmc1Bth/+/Tmc2+/+ animals various sequencing assays were performed. Indel rate was between 1 and 2%, however AAV-mediated SaCas9-KKH expression was specific for the Beethoven allele. Interestingly, indel rate was in a similar range in the case of the Tmc1 message, however the amount of Tmc1Bth message decreased significantly (the ratio of transcription level between Tmc1Bth and Tmc1WT control decreased by 30%). This suggests that indels on the messenger RNA lead to nonsense mediated decay, a mechanism that degrades RNA molecules with premature stop codons.
Functional testing of hearing revealed remarkable halting of hearing preservation in the heterozygous animals. A few treated animals, which were followed up to 1 year, exhibited near-normal hearing thresholds even at 1-year post-injection, the latest time point tested. Hearing thresholds were at WT levels (down to 20–30 dB) in the apex, however the hearing preservation was negligible at the base. This aligns well with previous observations on AAV transduction that shows a good transduction efficiency in the apex, which declines towards the base after RWM injection (Askew et al., 2015b; György et al., 2017; Landegger et al., 2017). Histology showed preservation of hair cells particularly in the apex and preservation of the organized staircase-like bundle morphology in treated animals.
These results altogether suggest that genome editing halts degeneration of hair cells and leads to near-complete normalization of hearing thresholds at lower frequencies. However, there are challenges and safety concerns with using genome editing nucleases. First it is possible that the long-term presence of the Cas9 will eventually lead to off-target effects or even disruption of the normal allele. Further studies are needed to evaluate whether gene editing can stop the degeneration once it has started or alternative delivery strategies (such as liposomes) should be assessed to overcome this limitation.
Although the Beethoven mutation is an important mutation and there is a human equivalent mutation in Tmc1, it is still a very rare form of hearing loss. The Gyorgy et al. study also performed a targetable space analysis using the ClinVar database and showed that ~21% of all human dominant mutations are potentially targetable with such a strategy, including 15 additional dominant deafness mutations.
7. Base editing for hearing loss
The major limitations to CRISPR/Cas9-mediated editing include the heterogenous gene editing outcomes after NHEJ, low efficiency by HDR in non-dividing cells, restricted PAM compatibility, sequence context preference and off-target editing. Indels caused by NHEJ are highly variable, with the deletions ranging from one base to many kilobases. Complexed rearrangements including crossover events could also occur (Kosicki et al., 2018). A highly efficient precise editing system would greatly improve some aspects of therapeutic applications.
Recently, David Liu and his co-workers have developed systems by which a modified Cas9 (dead Cas9 or Cas9 nickase), which does not cause double-strand breaks, is fused with a cytidine deaminase to form a cytosine base editor (CBE), or is fused with an adenine deaminase to form adenosine base editor (ABE). In the presence of gRNAs, CBE mediates a C•G to T•A conversion whereas ABE results in the conversion of A•T to G•C pairs in the genomic DNA (Gaudelli et al., 2017; Komor et al., 2016b). The action of nucleotide conversion enables precise editing that is required for the precise repair of genetic mutations. In contrast to HDR, which has low efficiency in postmitotic cells, base editing could achieve the efficiency of over 50% of base correction. Furthermore, off-target effects have been shown to be significantly reduced with this approach. Different version of BEs have been developed to include enzymes with alternative PAM requirements and relaxed PAM compatibilities. This increases the targeting space of base editing (Molla and Yang, 2019). Adenine base editing was shown to correct a mutation in a mouse model of Duchenne muscular dystrophy (Dmd) resulting in the expression of the dystrophin gene in 17% of myofibers, which was substantially higher than the 4% of dystrophin expression level required for improved muscle function (Ryu et al., 2018). CBE-mediated BE has been successfully applied to correct a genetic mutation in a mouse model of the human autosomal recessive liver disease phenylketonuria (PKU) (Villiger et al., 2018).
In addition to using BE to edit genetic mutations, BE can be applied to alter different biological processes. A recent study has demonstrated the utility of base editing in the inner ear. The β-catenin pathway has been shown to be important for hair cell regeneration through activation of Atoh1, an inner ear fate determining factor (Shi et al., 2010). Thus, activation of the β-catenin pathway is being explored for a hair cell regeneration approach. Using CBE base editing in a recent study, the codon TCT for the Ser33 was converted to the codon TTT for the Phe33 in the β-catenin gene, which increased the stability of β-catenin by blocking phosphorylation and ubiquitination. Such conversion in neonatal mice by lipofectamine 2000 mediated RNP delivery promoted supporting cell division and their subsequent transdifferentiation into hair cells (Yeh et al., 2018). It is likely that BE will lead to extensive inner ear applications to better understand inner ear biology and to develop treatment strategies for hearing loss.
8. Safety aspects and translational hurdles
Although gene editing is a highly promising strategy, there are several safety concerns that need to be adequately addressed before this technology can be widelyadopted in the clinic. Off-target effects represent one of the most important safety concerns. Previous studies have shown that SpCas9 can tolerate up to 7 mismatches between the gRNA (Tsai et al., 2015) and the target DNA sequence, and therefore CRISPR is prone to a high number of unwanted off-target cutting. In the past we have seen several strategies that decrease off-target effects. These include double Cas9 nickases (F. Ann Ran et al., 2013), dimeric Cas9 enzymes (Tsai et al., 2014) and high-fidelity Cas9 variants (Chen et al., 2017; Kleinstiver et al., 2016; Slaymaker et al., 2016). Another option is to limit Cas9 expression in time by using Cas9-RNA complexes, such as described in the cochlea (Gao et al., 2018; Zuris et al., 2015), or using promoters that have only temporary activity. Finally, an interesting strategy is to use a suicidal CRISPR construct that decreases its own activity via a self-targeting gRNA (Chen et al., 2016). In general, however, it is possible that CRISPR has very limited off-target effects in vivo in differentiated cells. Importantly, the majority of off-target assays work in cell lines or are biochemical assays, and in vivo off-target effects may be substantially different. A recent paper has found that CRISPR has no off-target effects in vivo with appropriate gRNA design (Akcakaya et al., 2018). Other issues include unwanted gene editing outcomes, such as AAV integration in the target site. A recent paper indicated high levels of AAV integration into CRISPR-induced double-stranded breaks upon AAV-mediated CRISPR delivery in the inner ear, brain and other tissues (Hanlon et al., 2019). Of note, this study also did not find in vivo off-target effects by assaying AAV sequences in the brain after intrahippocampal injection.
Other safety aspects of genome editing systems include immunogenicity of the vector and CRISPR components themselves. Cas9 is a bacterial protein, for which humans have been exposed to, thus it can be immunogenic. Studies have shown the presence of preexisting antibodies against SpCas9 could be detected in as many as 70% of healthy humans (Simhadri et al., 2018). This implies that immune response could reduce the editing efficacy or pose safety concerns. One way to circumvent the immunogenicity is to combine transient delivery of editing agents (e.g. LNP mediated RNP or mRNA delivery) with immunosuppression to mitigate safety concerns (Charlesworth et al., 2019; Wagner et al., 2019).
Gene editing-based inner ear therapy faces similar challenges as gene therapy. Editing efficiency is likely to be a key to sustained hearing recovery, which requires precise measurement of editing in the limited number of inner ear target cells. One approach is to isolate the target cells for indel analysis. We have demonstrated the utility by performing single-cell whole-genome amplification using isolated individual hair cells for indel detection (Gao et al., 2018). This method could be applied to other inner ear cell types provided they can be collected with little contamination from non-target cell types. The editing efficiency in inner ear could benefit from the application of modified gRNAs with improved stability and the inclusion of the elements such as NLS (nuclear localization signals) for enhanced nuclear transport of the Cas9 protein (Mir et al., 2018). There is an urgent need to identify new delivery vehicles to widen target cell types beyond hair cells such as supporting cells and stria fibrocytes important in GJB2 mutation caused hearing loss. One advantage of editing therapy is the ability to edit multiple mutations in different genes, which should provide a powerful tool to address hearing loss caused by bi-genic or multigenic mutations (Zheng et al., 2012, 2005).
Base editing, while is promising, has its own limitations. Most importantly, only transition mutations (C-T and A-G changes), but not transversions can be corrected. The base editing complex is substantially larger (6.1 kb) than an AAV vector can accommodate, therefore requires dual-AAV vector for delivery, which could reduce its efficiency. The application of non-viral delivery system such as LNPs could result in the delivery of large BE components in the form of proteins or mRNA, thus circumventing the size limitation bottleneck. While BE does not induce DSBs, CBE has high off-target effect on editing transcriptome-wide RNAs that are gRNAs independent (Grünewald et al., 2019; Zuo et al., 2019), a major concern that needs to be adequately addressed.
Finally, we have to discuss potential translational hurdles that limit the broad application of gene editing at the moment. Genetic forms of deafness are caused by mutations in more than 100 genes, and currently the ClinVar database lists 2242 pathogenic or potentially pathogenic mutations (as of 2019.10.21). Every mutation would minimally require a unique guide RNA and the production of a mutation-tailored AAV vector under clinical-grade good manufacturing practices. Also, theoretically a new clinical trial would be required for each mutation. However, it is possible that in the future, clinical trials will be organized in a way so that for each gene different gRNAs will not be considered as major changes provided one of the gRNAs has gone through a successful clinical development. It is highly important to perform preclinical experiments on humanized animal models to allow for the testing of the exact same gRNA targeting the human sequence.
Other translational hurdles including inter-species differences need to be considered. For example, promoters and AAV capsids might not translate between mice and humans. A recent work in the retina analyzing over 100 different promoters concluded that only 29% of promoters translate between mice and humans (Jüttner et al., 2019). This number was 65% in the case of non-human primates and human tissues, suggesting also some, although less extensive, translational hurdles when extrapolating results from primates to humans. Most of treatment paradigms is based on the studies in mouse models, whose inner ears are different from humans’ in development, anatomy and the route for delivery. The development of large animal models such as pig models of human hearing loss with characteristics mimicking human inner ears could offer the opportunities to develop better clinical treatment in humans (Hai et al., 2017).
9. Conclusions
Gene and base editing nucleases are innovative technologies that finally allow for genetic correction of several genetic diseases, including hereditary hearing loss. The ever-expanding space of RNA-guided nuclease-based applications allows targeting of virtually any mutation. Base editors and prime editors a next generation CRISPR based technologies that allow for precise gene editing outcomes even in non-dividing cells and differentiated tissues. Despite safety concerns and translational hurdles, this technology space advances rapidly into the clinic and hearing loss may also benefit from this unprecedented advance in genetic technology.
Supplementary Material
Acknowledgements
The authors acknowledge funding from NIH R01DC006908, R56DC006908 (Z-Y.C.), DOD W81XWH1810331 (Z-Y.C.), Fredrick and Ines Yeatts hair cell regeneration fellowship (Z-Y.C.) and David-Shulsky Foundation (Z-Y.C.).
Footnotes
Declaration of competing interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heares.2020.107958.
References
- Akcakaya P, Bobbin ML, Guo JA, Malagon-Lopez J, Clement K, Garcia SP, Fellows MD, Porritt MJ, Firth MA, Carreras A, Baccega T, Seeliger F, Bjursell M, Tsai SQ, Nguyen NT, Nitsch R, Mayr LM, Pinello L, BohloolyY M, Aryee MJ, Maresca M, Joung JK, 2018. In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature. 10.1038/s41586-018-0500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altinoglu S, Wang M, Xu Q, 2015. Combinatorial library strategies for synthesis of cationic lipid-like nanoparticles and their potential medical applications. Nanomedicine. 10.2217/nnm.14.192. [DOI] [PubMed] [Google Scholar]
- Amoasii L, Hildyard JCW, Li H, Sanchez-Ortiz E, Mireault A, Caballero D, Harron R, Stathopoulou TR, Massey C, Shelton JM, Bassel-Duby R, Piercy RJ, Olson EN, 2018. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science. 10.1126/science.aau1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ann Ran F, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F, 2013. Genome engineering using the CRIPR-Cas9 system. Nature. 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, Liu DR, 2019. Search-andreplace genome editing without double-strand breaks or donor DNA. Nature. 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askew C, Rochat C, Pan B, Asai Y, Ahmed H, Child E, Schneider BL, Aebischer P, Holt JR, 2015a. Tmc gene therapy restores auditory function in deaf mice. Sci. Transl. Med. 7 10.1126/scitranslmed.aab1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askew C, Rochat C, Pan B, Asai Y, Ahmed H, Child E, Schneider BL, Aebischer P, Holt JR, 2015b. Tmc gene therapy restores auditory function in deaf mice. Sci. Transl. Med. 7 10.1126/scitranslmed.aab1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beurg M, Goldring AC, Fettiplace R, 2015. The effects of Tmc1 Beethoven mutation on mechanotransducer channel function in cochlear hair cells. J. Gen. Physiol. 146, 233e243 10.1085/jgp.201511458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth CT, Deshpande PS, Dever DP, Camarena J, Lemgart VT, Cromer MK, Vakulskas CA, Collingwood MA, Zhang L, Bode NM, Behlke MA, Dejene B, Cieniewicz B, Romano R, Lesch BJ, GomezOspina N, Mantri S, Pavel-Dinu M, Weinberg KI, Porteus MH, 2019. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 10.1038/s41591-018-0326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Abdeen AA, Wang Y, Shahi PK, Robertson S, Xie R, Suzuki M, Pattnaik BR, Saha K, Gong S, 2019. A biodegradable nanocapsule delivers a Cas9 ribonucleoprotein complex for in vivo genome editing. Nat. Nanotechnol. 10.1038/s41565-019-0539-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, Sternberg SH, Joung JK, Yildiz A, Doudna JA, 2017. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550, 407e410 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Liu X, Zhang Y, Wang H, Ying H, Liu M, Li D, Lui KO, Ding Q, 2016. A self-restricted CRISPR system to reduce off-target effects. Mol. Ther. 10.1038/mt.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F, 2013. Multiplex genome engineering using CRISPR/ Cas systems. Science. 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corns LF, Johnson SL, Kros CJ, Marcotti W, 2016. Tmc1 point mutation affects Ca2+ sensitivity and block by dihydrostreptomycin of the mechanoelectrical transducer current of mouse outer hair cells. J. Neurosci. 36, 336e349 10.1523/JNEUROSCI.2439-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn JD, Smith AR, Patel MC, Shaw L, Youniss MR, van Heteren J, Dirstine T, Ciullo C, Lescarbeau R, Seitzer J, Shah RR, Shah A, Ling D, Growe J, Pink M, Rohde E, Wood KM, Salomon WE, Harrington WF, Dombrowski C, Strapps WR, Chang Y, Morrissey DV, 2018. A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 10.1016/j.celrep.2018.02.014. [DOI] [PubMed] [Google Scholar]
- Friedland AE, Baral R, Singhal P, Loveluck K, Shen S, Sanchez M, Marco E, Gotta GM, Maeder ML, Kennedy EM, Kornepati AVR, Sousa A, Collins MA, Jayaram H, Cullen BR, Bumcrot D, 2015. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 10.1186/s13059-015-0817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann T, Roblin R, 1972. Gene therapy for human genetic disease? Science 175, 949e955 10.1126/science.175.4025.949. [DOI] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF, 2013. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Tao Y, Lamas V, Huang M, Yeh WH, Pan B, Hu YJ, Hu JH, Thompson DB, Shu Y, Li Y, Wang H, Yang S, Xu Q, Polley DB, Liberman MC, Kong WJ, Holt JR, Chen ZY, Liu DR, 2018. Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature 553, 217e221 10.1038/nature25164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR, 2017. Programmable base editing of T to G C in genomic DNA without DNA cleavage. Nature. 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass Z, Li Y, Xu Q, 2017. Nanoparticles for CRISPR-Cas9 delivery. Nat. Biomed. Eng. 1, 854e855 10.1038/s41551-017-0158-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünewald J, Zhou R, Garcia SP, Iyer S, Lareau CA, Aryee MJ, Joung JK, 2019. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 10.1038/s41586-019-1161-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- György B, Lööv C, Zaborowski MP, Takeda S, Kleinstiver BP, Commins C,Kastanenka K, Mu D, Volak A, Giedraitis V, Lannfelt L, Maguire CA, Joung JK, Hyman BT, Breakefield XO, Ingelsson M, 2018. CRISPR/Cas9 mediated disruption of the Swedish APP allele as a therapeutic approach for early-onset alzheimer’s disease. Mol. Ther. Nucleic Acids 11 10.1016/j.omtn.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- György B, Meijer EJ, Ivanchenko MV, Tenneson K, Emond F, Hanlon KS, Indzhykulian AA, Volak A, Karavitaki KD, Tamvakologos PI, Vezina M, Berezovskii VK, Born RT, O’Brien M, Lafond JF, Arsenijevic Y, Kenna MA, Maguire CA, Corey DP, 2019a. Gene transfer with AAV9-PHP.B rescues hearing in a mouse model of usher syndrome 3A and transduces hair cells in a non-human primate. Mol. Ther. Clin. Dev. 10.1016/j.omtm.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- György B, Nist-Lund C, Pan B, Asai Y, Karavitaki KD, Kleinstiver BP, Garcia SP, Zaborowski MP, Solanes P, Spataro S, Schneider BL, Joung JK, Gel eoc GSG, Holt JR, Corey DP, 2019b. Allele-specific gene editing prevents deafness in a model of dominant progressive hearing loss. Nat. Med. 25, 1123e1130 10.1038/s41591-019-0500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- György B, Sage C, Indzhykulian AA, Scheffer DI, Brisson AR, Tan S, Wu X,Volak A, Mu D, Tamvakologos PI, Li Y, Fitzpatrick Z, Ericsson M, Breakefield XO, Corey DP, Maguire CA, 2017. Rescue of hearing by gene delivery to inner-ear hair cells using exosome-associated AAV. Mol. Ther. 10.1016/j.ymthe.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai T, Guo W, Yao J, Cao C, Luo A, Qi M, Wang Xianlong, Wang Xiao, Huang J, Zhang Y, Zhang H, Wang D, Shang H, Hong Q, Zhang R, Jia Q, Zheng Q, Qin G, Li Y, Zhang T, Jin W, Chen ZY, Wang H, Zhou Q, Meng A, Wei H, Yang S, Zhao J, 2017. Creation of miniature pig model of human Waardenburg syndrome type 2A by ENU mutagenesis. Hum. Genet. 10.1007/s00439-017-1851-2. [DOI] [PubMed] [Google Scholar]
- Hanlon KS, Kleinstiver BP, Garcia SP, Zaborowski MP, Volak A, Spirig SE, Muller A, Sousa AA, Tsai SQ, Bengtsson NE, Lööv C, Ingelsson M, Chamberlain JS, Corey DP, Aryee MJ, Joung JK, Breakefield XO, Maguire CA, György B, 2019. High levels of AAV vector integration into CRISPR-induced DNA breaks. Nat. Commun. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenreich M, Zhang F, 2016. Applications of CRISPR-Cas systems in neuroscience. Nat. Rev. Neurosci. 10.1038/nrn.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman L, Gersbach CA, 2018. Editing the epigenome: reshaping the genomic landscape. Annu. Rev. Genom. Hum. Genet. 10.1146/annurevgenom-083117-021632. [DOI] [PubMed] [Google Scholar]
- Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA, 2013. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. In: Proceedings of the National Academy of Sciences of the United States of America; 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jüttner J, Szabo A, Gross-Scherf B, Morikawa RK, Rompani SB, Hantz P, Szikra T, Esposti F, Cowan CS, Bharioke A, Patino-Alvarez CP, Keles Ö,Kusnyerik A, Azoulay T, Hartl D, Krebs AR, Schübeler D, Hajdu RI, Lukats A, Nemeth J, Nagy ZZ, Wu KC, Wu RH, Xiang L, Fang XL, Jin ZB, Goldblum D, Hasler PW, Scholl HPN, Krol J, Roska B, 2019. Targeting neuronal and glial cell types with synthetic promoter AAVs in mice, non-human primates and humans. Nat. Neurosci. 10.1038/s41593-019-0431-2. [DOI] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK, 2016. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490e495 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Nguyen NT, Topkar VV, Zheng Z, Joung JK, 2015a. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol. 33, 1293e1298 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales APW, Li Z, Peterson RT, Yeh JRJ, Aryee MJ, Joung JK, 2015b. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481e485 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Sousa AA, Walton RT, Tak YE, Hsu JY, Clement K, Welch MM, Horng JE, Malagon-Lopez J, Scarfo I, Maus MV, Pinello L, Aryee MJ, Joung JK, 2019. Engineered CRISPReCas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 10.1038/s41587-018-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Badran AH, Liu DR, 2017. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 10.1016/j.cell.2016.10.044. [DOI] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR, 2016a. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR, 2016b. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosicki M, Tomberg K, Bradley A, 2018. Repair of double-strand breaks induced by CRISPReCas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski PS, Rudra A, Miao L, Anderson DG, 2019. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol. Ther. 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy S, Wohlford-Lenane C, Kandimalla S, Sartre G, Meyerholz DK, Theberge V, Hall ee S, Duperr e AM, Del ‘Guidice T, Lepetit-Stoffaes JP, Barbeau X, Guay D, McCray PB, 2019. Engineered amphiphilic peptides enable delivery of proteins and CRISPR-associated nucleases to airway epithelia. Nat. Commun. 10.1038/s41467-019-12922-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landegger LD, Pan B, Askew C, Wassmer SJ, Gluck SD, Galvin A, Taylor R, Forge A, Stankovic KM, Holt JR, Vandenberghe LH, 2017. A synthetic AAV vector enables safe and efficient gene transfer to the mammalian inner ear. Nat. Biotechnol. 35, 280e284 10.1038/nbt.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Lee K, Panda S, Gonzales-Rojas R, Chong A, Bugay V, Park HM, Brenner R, Murthy N, Lee HY, 2018. Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nat. Biomed. Eng. 10.1038/s41551-018-0252-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Conboy M, Park HM, Jiang F, Kim HJ, Dewitt MA, Mackley VA, Chang K, Rao A, Skinner C, Shobha T, Mehdipour M, Liu H, Huang W-C, Lan F, Bray NL, Li S, Corn JE, Kataoka K, Doudna JA, Conboy I, Murthy N, 2017. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat. Biomed. Eng. 1, 889e901 10.1038/s41551-017-0137-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy JM, Yeh W-H, Pendse N, Davis JR, Hennessey E, Butcher R, Koblan LW, Comander J, Liu Q, Liu DR, 2020. Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat. Biomed. Eng. 4, 97e110 10.1038/s41551-019-0501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Kleinstiver BP, Leon MY, Prew MS, Navarro-Gomez D, Greenwald SH, Pierce EA, Joung JK, Liu Q, 2018. Allele-specific CRISPR-cas9 genome editing of the single-base P23H mutation for rhodopsin-associated dominant retinitis pigmentosa. CRISPR J. 10.1089/crispr.2017.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Bolinger J, Yu Y, Glass Z, Shi N, Yang L, Wang M, Xu Q, 2019a. Intracellular delivery and biodistribution study of CRISPR/Cas9 ribonucleoprotein loaded bioreducible lipidoid nanoparticles. Biomater. Sci. 10.1039/c8bm00637g. [DOI] [PubMed] [Google Scholar]
- Li Y, Chakraborty A, Chen J, Xu Q, 2019b. Combinatorial library of light-cleavable lipidoid nanoparticles for intracellular drug delivery. ACS Biomater. Sci. Eng. 10.1021/acsbiomaterials.9b00445. [DOI] [PubMed] [Google Scholar]
- Liu JJ, Orlova N, Oakes BL, Ma E, Spinner HB, Baney KLM, Chuck J, Tan D, Knott GJ, Harrington LB, Al-Shayeb B, Wagner A, Brötzmann J, Staahl BT,Taylor KL, Desmarais J, Nogales E, Doudna JA, 2019. CasX enzymes comprise a distinct family of RNA-guided genome. Nature. 10.1038/s41586-019-0908-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Stefanidakis M, Wilson CJ, Baral R, Barrera LA, Bounoutas GS, Bumcrot D, Chao H, Ciulla DM, DaSilva JA, Dass A, Dhanapal V, Fennell TJ, Friedland AE, Giannoukos G, Gloskowski SW, Glucksmann A, Gotta GM, Jayaram H, Haskett SJ, Hopkins B, Horng JE, Joshi S, Marco E, Mepani R, Reyon D, Ta T, Tabbaa DG, Samuelsson SJ, Shen S, Skor MN, Stetkiewicz P, Wang T, Yudkoff C, Myer VE, Albright CF, Jiang H, 2019. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat. Med. 10.1038/s41591-0180327-9. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM, 2013. RNA-guided human genome engineering via Cas9. Science. 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotti W, Erven A, Johnson SL, Steel KP, Kros CJ, 2006. Tmc1 is necessary for normal functional maturation and survival of inner and outer hair cells in the mouse cochlea. J. Physiol. (Lond.) 574, 677e698 10.1113/jphysiol.2005.095661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JR, Al-Zaidy S, Shell R, Arnold WD, Rodino-Klapac LR, Prior TW, Lowes L, Alfano L, Berry K, Church K, Kissel JT, Nagendran S, L’Italien J, Sproule DM, Wells C, Cardenas JA, Heitzer MD, Kaspar A, Corcoran S, Braun L, Likhite S, Miranda C, Meyer K, Foust KD, Burghes AHM, Kaspar BK, 2017. Single-dose gene-replacement therapy for spinal muscular atrophy. N. Engl. J. Med. 377, 1713e1722 10.1056/NEJMoa1706198. [DOI] [PubMed] [Google Scholar]
- Mir A, Alterman JF, Hassler MR, Debacker AJ, Hudgens E, Echeverria D, Brodsky MH, Khvorova A, Watts JK, Sontheimer EJ, 2018. Heavily and fully modified RNAs guide efficient SpyCas9-mediated genome editing. Nat. Commun. 10.1038/s41467-018-05073-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H, Blömer U, Takahashi M, Gage FH, Verma IM, 1998. Development of a self-inactivating lentivirus vector. J. Virol. 72, 8150e8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla KA, Yang Y, 2019. CRISPR/Cas-Mediated base editing: technical considerations and practical applications. Trends Biotechnol. 10.1016/j.tibtech.2019.03.008. [DOI] [PubMed] [Google Scholar]
- Monteys AM, Ebanks SA, Keiser MS, Davidson BL, 2017. CRISPR/Cas9 editing of the mutant huntingtin allele in vitro and in vivo. Mol. Ther. 10.1016/j.ymthe.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mout R, Ray M, Yesilbag Tonga G, Lee Y-W, Tay T, Sasaki K, Rotello VM, 2017. Direct cytosolic delivery of CRISPR/Cas9-Ribonucleoprotein for efficient gene editing. ACS Nano 11, 2452e2458 10.1021/acsnano.6b07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelapu SS, Locke FL, Bartlett NL, Lekakis LJ, Miklos DB, Jacobson CA, Braunschweig I, Oluwole OO, Siddiqi T, Lin Y, Timmerman JM, Stiff PJ, Friedberg JW, Flinn IW, Goy A, Hill BT, Smith MR, Deol A, Farooq U, McSweeney P, Munoz J, Avivi I, Castro JE, Westin JR, Chavez JC, Ghobadi A, Komanduri KV, Levy R, Jacobsen ED, Witzig TE, Reagan P, Bot A, Rossi J, Navale L, Jiang Y, Aycock J, Elias M, Chang D, Wiezorek J, Go WY, 2017. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N. Engl. J. Med. 377, 2531e2544 10.1056/NEJMoa1707447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B, Akyuz N, Liu XP, Asai Y, Nist-Lund C, Kurima K, Derfler BH, György B, Limapichat W, Walujkar S, Wimalasena LN, Sotomayor M, Corey DP, Holt JR, 2018. TMC1 forms the pore of mechanosensory transduction channels in vertebrate inner ear hair cells. Neuron. 10.1016/j.neuron.2018.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B, Gel eoc GS, Asai Y, Horwitz GC, Kurima K, Ishikawa K, Kawashima Y, Griffith AJ, Holt JR, 2013. TMC1 and TMC2 are components of the mechanotransduction channel in hair cells of the mammalian inner ear. Neuron 79, 504e515 10.1016/j.neuron.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F, 2015. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186e191 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell S, Bennett J, Wellman JA, Chung DC, Yu ZF, Tillman A, Wittes J, Pappas J, Elci O, McCague S, Cross D, Marshall KA, Walshire J, Kehoe TL, Reichert H, Davis M, Raffini L, George LA, Hudson FP, Dingfield L, Zhu X, Haller JA, Sohn EH, Mahajan VB, Pfeifer W, Weckmann M, Johnson C, Gewaily D, Drack A, Stone E, Wachtel K, Simonelli F, Leroy BP, Wright JF, High KA, Maguire AM, 2017. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet. 10.1016/S0140-6736(17)31868-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu SM, Koo T, Kim K, Lim K, Baek G, Kim ST, Kim HS, Kim DE, Lee H, Chung E, Kim JS, 2018. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat. Biotechnol. 10.1038/nbt.4148. [DOI] [PubMed] [Google Scholar]
- Samulski RJ, Berns KI, Tan M, Muzyczka N, 1982. Cloning of adeno-associated virus into pBR322: rescue of intact virus from the recombinant plasmid in human cells. Proc. Natl. Acad. Sci. U.S.A. 79, 2077e2081 10.1073/pnas.79.6.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samulski RJ, Srivastava A, Berns KI, Muzyczka N, 1983. Rescue of adeno-associated virus from recombinant plasmids: gene correction within the terminal repeats of AAV. Cell 33, 135e143 10.1016/0092-8674(83)90342-2. [DOI] [PubMed] [Google Scholar]
- Schuster SJ, Bishop MR, Tam CS, Waller EK, Borchmann P, McGuirk JP, Jäger U, Jaglowski S, Andreadis C, Westin JR, Fleury I, Bachanova V, Foley SR, Ho PJ, Mielke S, Magenau JM, Holte H, Pantano S, Pacaud LB, Awasthi R, Chu J, Anak Ö, Salles G, Maziarz RT, JULIET Investigators, 2019. Tisagenlecleucel in Adult Relapsed or Refractory Diffuse Large B-Cell Lymphoma. N. Engl. J. Med. 380, 45e56 10.1056/NEJMoa1804980. [DOI] [PubMed] [Google Scholar]
- Sharma R, Anguela XM, Doyon Y, Wechsler T, DeKelver RC, Sproul S, Paschon DE, Miller JC, Davidson RJ, Shivak D, Zhou S, Rieders J, Gregory PD, Holmes MC, Rebar EJ, High KA, 2015. In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood. 10.1182/blood-2014-12-615492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F, Cheng YF, Wang XL, Edge ASB, 2010. β-Catenin up-regulates Atoh1 expression in neural progenitor cells by interaction with an Atoh1 30 enhancer. J. Biol. Chem. 10.1074/jbc.M109.059055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata SB, Ranum PT, Moteki H, Pan B, Goodwin AT, Goodman SS, Abbas PJ, Holt JR, Smith RJH, 2016. RNA interference prevents autosomaldominant hearing loss. Am. J. Hum. Genet. 98, 1101e1113 10.1016/j.ajhg.2016.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva G, Poirot L, Galetto R, Smith J, Montoya G, Duchateau P, Paques F, 2011. Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr. Gene Ther. 10.2174/156652311794520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simhadri VL, McGill J, McMahon S, Wang J, Jiang H, Sauna ZE, 2018. Prevalence of pre-existing antibodies to CRISPR-associated nuclease Cas9 in the USA population. Mol. Ther. Clin. Dev. 10.1016/j.omtm.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F, 2016. Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84e88 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Ji W, Hall JM, Hu Q, Wang C, Beisel CL, Gu Z, 2015. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew. Chem., Int. Ed. Engl. 54, 12029e12033 10.1002/anie.201506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, Sur M, Zhang F, 2015. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 33, 102e106 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK, 2014. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 32, 569e576 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, Aryee MJ, Joung JK, 2015. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187e198 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan S, Salahudeen AA, Sellers ZM, Bravo DT, Choi SS, Batish A, Le W, De La O S, Kaushik MP, Galper N, Lee CM, Bao G, Chang EH, Wine JJ, Milla CE, Desai TJ, Nayak JV, Kuo CJ, Porteus MH, 2019. Highly efficient repair of the DF508 mutation in airway stem cells of cystic fibrosis patients with functional rescue of the differentiated epithelia. bioRxiv. 10.1101/561183. [DOI] [Google Scholar]
- Villiger L, Grisch-Chan HM, Lindsay H, Ringnalda F, Pogliano CB, Allegri G, Fingerhut R, Häberle J, Matos J, Robinson MD, Thöny B, Schwank G, 2018. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat. Med. 10.1038/s41591-018-0209-1. [DOI] [PubMed] [Google Scholar]
- Vreugde S, Erven A, Kros CJ, Marcotti W, Fuchs H, Kurima K, Wilcox ER, Friedman TB, Griffith AJ, Bailing R, Hrabe de Angelis M, Avraham KB, Steel KP, 2002. Beethoven, a mouse model for dominant, progressive hearing loss DFNA36. Nat. Genet. 30, 257e258 10.1038/ng848. [DOI] [PubMed] [Google Scholar]
- Wagner DL, Amini L, Wendering DJ, Burkhardt LM, Akyüz L, Reinke P, Volk HD, Schmueck-Henneresse M, 2019. High prevalence of Streptococcus pyogenes Cas9-reactive T cells within the adult human population. Nat. Med. 10.1038/s41591-018-0204-6. [DOI] [PubMed] [Google Scholar]
- Wang HX, Li M, Lee CM, Chakraborty S, Kim HW, Bao G, Leong KW, 2017. CRISPR/Cas9-Based genome editing for disease modeling and therapy: challenges and opportunities for nonviral delivery. Chem. Rev. 10.1021/acs.chemrev.6b00799. [DOI] [PubMed] [Google Scholar]
- Wang M, Alberti K, Varone A, Pouli D, Georgakoudi I, Xu Q, 2014. Enhanced intracellular siRNA delivery using bioreducible lipid-like nanoparticles. Adv. Healthc. Mat. 10.1002/adhm.201400039. [DOI] [PubMed] [Google Scholar]
- Wang M, Zuris JA, Meng F, Rees H, Sun S, Deng P, Han Y, Gao X, Pouli D, Wu Q, Georgakoudi I, Liu DR, Xu Q, 2016. Efficient delivery of genomeediting proteins using bioreducible lipid nanoparticles. In: Proceedings of the National Academy of Sciences of the United States of America; 10.1073/pnas.1520244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Chang R, Yang H, Zhao T, Hong Y, Kong HE, Sun X, Qin Z, Jin P, Li S, Li XJ, 2017. CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington’s disease. J. Clin. Invest. 10.1172/JCI92087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh WH, Chiang H, Rees HA, Edge ASB, Liu DR, 2018. In vivo base editing of post-mitotic sensory cells. Nat. Commun. 10.1038/s41467-01804580-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Song CQ, Suresh S, Wu Q, Walsh S, Rhym LH, Mintzer E, Bolukbasi MF, Zhu LJ, Kauffman K, Mou H, Oberholzer A, Ding J, Kwan SY, Bogorad RL, Zatsepin T, Koteliansky V, Wolfe SA, Xue W, Langer R, Anderson DG, 2017. structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol. 10.1038/nbt.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura H, Shibata SB, Ranum PT, Moteki H, Smith RJH, 2019. Targeted allele suppression prevents progressive hearing loss in the mature murine model of human TMC1 deafness. Mol. Ther. 10.1016/j.ymthe.2018.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, Van Der Oost J, Regev A, Koonin EV, Zhang F, 2015. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPRcas system. Cell. 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Wang D, Zong L, Zhao F, Guan L, Zhang P, Shi W, Lan L, Wang H, Li Q, Han B, Yang L, Jin X, Wang Jian, Wang Jun, Wang Q, 2014. A novel DFNA36 mutation in TMC1 orthologous to the Beethoven (Bth) mouse associated with autosomal dominant hearing loss in a Chinese family. PloS One 9 10.1371/journal.pone.0097064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng QY, Scarborough JD, Zheng Y, Yu H, Choi D, Gillespie PG, 2012. Digenic inheritance of deafness caused by 8J allele of myosin-VIIA and mutations in other Usher I genes. Hum. Mol. Genet. 10.1093/hmg/dds084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng QY, Yan D, Ouyang XM, Du LL, Yu H, Chang B, Johnson KR, Liu XZ, 2005. Digenic inheritance of deafness caused by mutations in genes encoding cadherin 23 and protocadherin 15 in mice and humans. Hum. Mol. Genet. 10.1093/hmg/ddi010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinn E, Pacouret S, Khaychuk V, Turunen HT, Carvalho LS, Andres-Mateos E, Shah S, Shelke R, Maurer AC, Maurer E, Xiao R, Vandenberghe LH, 2015. In silico reconstruction of the viral evolutionary lineage yields a potent gene therapy vector. Cell Rep. 12, 1056e1068 10.1016/j.celrep.2015.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L, Trono D, 1998. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol. 72, 9873e9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo E, Sun Y, Wei W, Yuan T, Ying W, Sun H, Yuan L, Steinmetz LM, Li Y, Yang H, 2019. Cytosine base editor generates substantial off-target singlenucleotide variants in mouse embryos. Science 10.1126/science.aav9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen ZY, Liu DR, 2015. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.