Abstract
Metabolism and inflammation are linked at many levels. Sickness behaviors are elicited by the immune system’s response to antigenic stimuli, and include changes in feeding and metabolism. The immune system is also regulated by the circadian (daily) clock, which generates endogenous rhythms, and synchronizes these rhythms to the light-dark cycle. Modern society has resulted in chronic misalignment or desynchronization of the circadian clock and the external environment. We have demonstrated that circadian desynchronization (CD) in mice alters metabolic function, and also affects both peripheral and central immune responses following a low-dose lipopolysaccharide (LPS) challenge. However, it is unclear how this altered immune response impacts sickness behaviors and metabolism following challenge. To test this, we housed male mice in circadian desynchronized (10-hours light:10-hours dark) or control (12-hours light:12-hours dark) conditions for 5–6 weeks. We then challenged mice with LPS (i.p., 400 μg/kg) or PBS and measured changes in body mass, feeding, drinking and locomotion using a comprehensive phenotyping system. Plasma, liver, and brain were collected 36h post-inoculation (hpi) and inflammatory messengers were measured via multiplex cytokine/chemokine array and qPCR. We find that recovery of locomotion and body mass is prolonged in CD mice following LPS challenge. Additionally, at 36 hpi the expression of several proinflammatory cytokines differ depending on pre-inoculation lighting conditions. Our findings add to the growing literature which documents how desynchronization of circadian rhythms can lead to disrupted immune responses and changes in metabolic function.
Keywords: biological rhythms, lipopolysaccharide, neuroimmune, metabolism
1. INTRODUCTION
Virtually all organisms on Earth exhibit oscillations in physiology and behavior over an approximate 24-hour day (Man et al., 2016). These 24-hour circadian rhythms in physiology and behavior enable organisms to anticipate and respond to the external environment driven by the rotation of the Earth about its axis (Man et al., 2016). Circadian rhythms are generated by molecular clocks in nearly every mammalian cell and tissue (Scheiermann et al., 2013). Under normal physiological conditions, these clocks are synchronized to each other by multiple pathways, including the autonomic nervous system and the hypothalamic-pituitary-adrenal (HPA) axis, which are directly regulated by the suprachiasmatic nucleus (SCN) of the hypothalamus (Kalsbeek et al., 2006).
Physiological and behavioral processes that are regulated by the circadian system include the sleep-wake cycle, locomotor activity, cardiovascular and digestive processes, hormone secretion, body temperature, metabolism, and immunity (Scheiermann et al., 2018). Considering the wide variety of processes that are regulated by the circadian system, it is perhaps not surprising that the desynchronization between the SCN and peripheral clocks is associated with various human pathologies including obesity, diabetes, cancer, and cardiovascular disease (Early and Curtis, 2016). With more than 15% of the United States’ working population regularly working night shifts, the development of these human pathologies in night shift workers is a considerable concern (Early and Curtis, 2016). Furthermore, circadian misalignment has become commonplace as more people travel across time zones and engage in activities at times when the circadian system promotes resting (Early and Curtis, 2016). Therefore, human pathologies associated with circadian desynchronization (CD) are not only a concern for night shift workers but also for the greater population. Our previous work has demonstrated that we can induce CD by housing mice in a 20h long light-dark cycle of 10h light, and 10h darkness (LD10:10). Relatively short-term housing in this environment (4–6wks) leads to metabolic dysregulation, including weight gain, as well as increased plasma leptin, insulin, and triglycerides (Karatsoreos et al., 2011). Remarkably, these effects are not associated with sleep deprivation, but instead changes in sleep timing and sleep quality (Phillips et al., 2015).
The immune system is regulated by the circadian clock. Some of the first experimental evidence that the immune system was regulated by the circadian clock was reported 60 years ago by Halberg et al. (1960), that revealed challenging the immune system of mice with bacterial lipopolysaccharide (LPS) at the end of the resting phase resulted in higher mortality rates than at other times of day. More recently, this phenomenon has been observed in non-pathogen induced inflammatory disease. For instance, there is a predictable time of symptom onset for immune conditions such as rheumatoid arthritis and asthma (Scheiermann et al., 2018). We now know that numerous immune cells contain intrinsic circadian clocks, including monocytes, macrophages, mast cells, neutrophils, eosinophils, natural killer cells, CD4+ T lymphocytes, and B lymphocytes (Scheiermann et al., 2018).
Emerging evidence supports the idea that the circadian clock prepares the mammalian immune system for defense against pathogens during the time that mammals are most likely to encounter pathogens, i.e., their active phase (Tognini et al., 2017). However, this increased resistance against pathogens comes with an increased susceptibility to these same pathogens at the beginning of the mammal’s resting phase (Tognini et al., 2017). The present studies demonstrate that CD prolongs recovery of locomotion and body mass, as measured by a comprehensive behavioral phenotyping system, in mice challenged with LPS. It also results in changes in both circulating cytokines and immune mediators, and expression of immune factors within the brain.
2. METHODS
2.1. Animals
Adult male C57/BL6Nhsd mice (5–6 weeks, n = 64; Envigo) were used in two independent experiments. Upon arrival, all mice were single housed in standard shoebox cages with food and water available ad libitum. The light cycles were maintained at 12-h light and 12-h dark (LD12:12) for at least 6 days to allow for acclimatization to the new environment. Following the acclimatization period, half of the mice (n = 16 from each experiment) were randomly assigned to CD (10-h light, 10-h dark, LD10:10), while the other half (n = 16 from each experiment) remained in Control (LD12:12) conditions. After five-to-six weeks of CD or Control housing, one half of the mice in each group were transferred in their home cages to a metabolic and behavioral phenotyping system (Promethion, Sable Systems, Las Vegas, NV) for real-time metabolic and behavioral analysis in their respective light conditions (Experiment 1: Behavior). The remaining mice stayed in their home cage, and were provided the identical cage enrichment tube as Promethion mice (Experiment 2: Immune). Experimental procedures were approved by Washington State University Animal Care and Use Committee.
2.2. LPS Immune Challenge
After at least 5 days of acclimatization (either Sable System or enrichment tube), mice were inoculated intraperitoneally with 0.4 mg/kg of LPS (Escherichia coli 026:B6, > 3,000,000 EU/mg, diluted in sterile PBS) (Sigma Aldrich, St. Louis, MO) or with sterile PBS. We used 0.4 mg/kg LPS since it is a mild, sub-septic dose capable of inducing an immune response and behavioral changes (Fonken et al., 2013; Phillips et al., 2015). The time-of-day of inoculation occurred toward the end of their inactive phase at ZT9. One LPS-treated CD mouse from the 36 hpi experiment died following inoculation. After LPS/PBS treatment, mice were monitored twice per day visually by the experimenter until experiment completion. Mice were euthanized either 7 dpi (Experiment 1: Behavior) or 36 hpi (Experiment 2: Immune) via cervical dislocation. Blood was collected in EDTA coated tubes and centrifuged at 1300 RCF for 15 minutes at 4°C to isolate plasma. Liver and brain were collected and flash frozen on dry ice. Plasma and tissues were stored at −80°C until further testing.
2.3. Promethion Behavioral and Metabolic System
The Promethion System was used to record body mass, locomotor, feeding and drinking activities of each mouse. Data was recorded continuously for 5 days before and 7 days following inoculation. In this cohort, one LPS-treated CD mouse was removed from all body mass and behavioral analyses post-inoculation because it did not respond to LPS treatment (likely due to a failed injection), and one PBS-treated control mouse was removed from all body mass and behavioral analyses where data was recorded by the Promethion System because it did not meet our behavioral recovery criteria (likely due to a technical issue with its recording cage).
2.3.1. Body mass
To determine how CD impacts changes in body mass over time following LPS challenge, we used body mass measurements recorded by hand (immediately before inoculation) and by the Promethion System. The body mass measurements recorded by the Promethion System occur when the mouse enters its enrichment tube. All mice had entered their weighing enrichment tube at least once by 48 hpi. Since 1/8 LPS-treated control and 3/7 LPS-treated CD mice did not return to their pre-inoculation body mass by study completion, we set our recovery criterion based on the mouse that was the greatest percentage away from returning to its pre-inoculation body mass. Thus, for survival analysis, the criterion was set as the time at which each mouse reached and sustained 93 percent of its pre-inoculation body mass for at least 2 consecutive body mass recordings.
2.3.2. Behavior
Baseline behaviors (Figure 1) were calculated as an average activity during the three (24-hour) days immediately prior to inoculation. Modified survival analysis was used to assess the effects of CD on recovery of activities related to sickness behavior such as locomotor, feeding, and drinking activities. First, for each activity analyzed, we determined the hourly rate of activity of each animal during a 24-hour period occurring exactly 1 day prior to inoculation and set this value as a within-animal baseline. We then set our recovery criteria as the point at which each animal sustained activity for at least two consecutive hourly bins equal to or greater than our baseline.
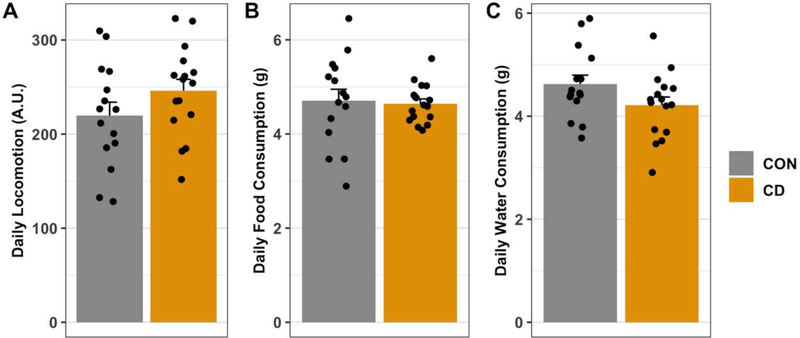
Fig. 1: Circadian desynchronization does not affect daily locomotor activity, food consumption, or water consumption.
After two days acclimatization to the behavioral phenotyping system locomotor activity, food consumption, and water consumption were measured. Average daily activity was calculated using data collected over the three (24-hour) days immediately preceding inoculation. No statistical differences between control (n = 15) and CD (n = 16) mice were detected for average locomotor activity (A), food consumption (B), and water consumption (C). Data are represented as mean ± SEM. Statistical analyses used Mann-Whitney U-tests.
2.4. Plasma Multiplex Cytokine Array
A multiplex cytokine assay was undertaken using a MagPix detection system (Luminex Corp., Austin, TX). The plasma multiplex cytokine/chemokine panel (ProcartaPlex Mo Th1/Th2 & Chemokine Panel 1 20plex, Lot #: 206290020) was completed following the manufacturer’s instructions (ThermoFisher, Waltham, MA). Briefly, this assay uses color-coded polystyrene beads that contain magnetite and cytokine/chemokine-specific detection antibodies. When the plasma sample is added, the detection antibodies on the magnetic beads bind to specific cytokines/chemokines present in the sample. The captured cytokines/chemokines are then sandwiched between the magnetic bead-conjugated detection antibodies and added biotinylated detection antibodies. The biotinylated detection antibodies are then bound by phycoerythrin (PE)-conjugated streptavidin. The amount of PE detected via the MagPix is directly proportional to the amount of bound cytokine/chemokine. Samples were run in duplicate and the intraassay coefficient of variation was 6.3%. Standards on the standard curve that did not meet a percent recovery between 80–120 percent were removed from the curve. The correlation coefficients (R2) of all standard curves were greater than 99%. Standard curves were calculated using Logistic 5P Weighted for all cytokines/chemokines measured except for GM-CSF and IL-18 whose standard curves were calculated using Logistic 4P Weighted.
2.5. Brain and Liver Gene Expression
Brain and liver tissues were collected from mice at 36 hpi and rapidly frozen in powdered dry ice and then stored at −80°C. One brain from a PBS-treated CD mouse was lost during collection. Hippocampal, hypothalamic, and medial prefrontal cortical punches were collected into Trizol reagent from each brain using a coring tool (inner diameter = 0.5 mm) and a freezing sliding microtome. Landmarks for these regions were identified using a mouse brain atlas. RNA was extracted from brain punches using Direct-zol™ RNA MicroPrep Kit (Zymo Research; cat. # R2060). For brain punches, aqueous phase obtained from Trizol/chloroform fractionation was used to load on the Zymo-Spin™ Column instead of Trizol reagent directly. Liver samples were homogenized in DNA/RNA Shield™ Reagent (Zymo Research; cat. # R1100–50). Liver RNA was isolated using Quick-RNATM MiniPrep Kit (Zymo Research; cat. # R1054). One RNA sample isolated from the liver of PBS-treated control mouse was not analyzed because of RNA degradation. RNA concentrations of both brain punches and liver were measured spectrophotometrically using NanoDrop (Thermo Scientific). Liver samples were diluted to equivalent concentrations for RT reaction. For brain, the whole RNA prep was used for reverse transcription. cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit with Rnase Inhibitor (Life Technologies, cat. # 4374966). Real-time qPCR was run using PerfeCTa® qPCR FastMix® II, Low ROXTM (QuantaBio; cat. # 97066–002) and the following off-the-shelf TaqMan Gene Expression Assays (Life Technologies): Rn18s (Mm04277571_s1), Casp1 (Mm00438023_m1), Il1b (Mm00434228_m1), Il6 (Mm00446190_m1), Nfkb1 (Mm00476361_m1), Pycard (Mm00445747_g1), and Tnf (Mm00443258_m1). Rn18s was used as the housekeeping gene to normalize between biological replicates. Samples were run in triplicate in 20 μl reaction mixture. 96-well plates were run on an Applied Biosystems ViiA7 RT-PCR machine. CT values were produced using ViiA 7 QuantStudioTM Real-Time PCR Software v1.3 (Applied Biosystems). Relative gene expression (Fold Change) was calculated using the comparative ΔΔCт method of relative quantification.
2.6. Statistical Analysis
Mann-Whitney U-tests were used to compare pre-inoculation locomotor activity, food consumption, and water consumption between control and CD mice. A one-way ANOVA (Type II) was used to detect group differences in pre-inoculation body mass. Survival curves for body mass and locomotor, feeding, and drinking activities were analyzed using a pairwise log-rank test with a BH correction for multiple comparisons. Two-way ANOVA analysis (Type II) was used to detect main effects of LPS treatment and environmental lighting condition and interactions on the hepatic, hypothalamic, hippocampal, and mPFC expression of proinflammatory genes (ΔCT), and plasma cytokines/chemokines. Post-Hoc Tukey HSD analysis followed the two-way ANOVA analyses for instances where interactions were detected. All analyses and figures were completed using R (R Core Team, 2019).
3. RESULTS
3.1. Baseline behavioral measures
We have previously shown that CD leads to increased body mass gain and this weight gain is associated with elevated plasma triglyceride, leptin and insulin levels (Karatsoreos et al., 2011). Using the Promethion metabolic and behavioral phenotyping system, we assessed whether there were changes in overall total locomotion, feeding and drinking activities. We did not observe a statistically significant difference in daily locomotion (Mann-Whitney U-test: p = 0.22), food consumption (Mann-Whitney U-test: p = 0.60) or water consumption (Mann-Whitney U-test: p = 0.09) averaged over a three-day period (Figure 1A–C).
3.2. Body mass
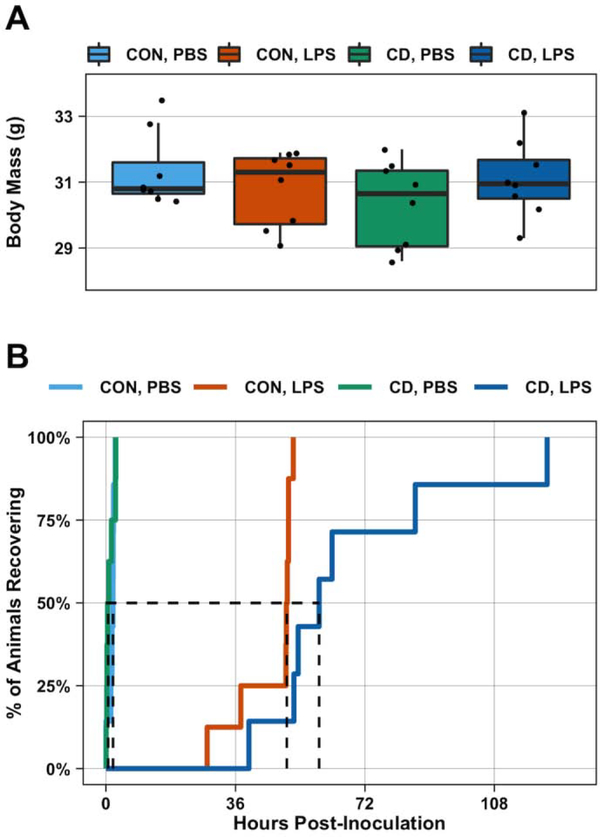
At time of inoculation there was no difference in average body mass between the groups (One-way ANOVA: p = 0.39, F1,28 = 1.03) (Figure 2A). To determine if differences existed in the rate of body mass recovery, we undertook survival analysis using the criterion described in the Methods. The recovery curves revealed no difference between PBS-treated control and CD mice (Log Rank, BH Correction for multiple comparisons: p = 0.95) (Figure 2B). However, recovery curves of LPS-treated mice were significantly different from the recovery curves of PBS-treated mice (Log Rank, BH Correction for multiple comparisons: p < 0.001) (Figure 2B). The recovery curves of LPS-treated control and CD mice were also significantly different from each other (Log Rank, BH Correction for multiple comparisons: p = 0.002). The median recovery time for LPS-treated CD mice (59 hours) was greater than the median recovery time of LPS-treated control mice (50 hours).
Fig. 2: Recovery of body mass following LPS challenge is affected by circadian desynchronization.
After 5 days acclimatization to the behavioral phenotyping system, control and CD mice were weighed (baseline body mass) and then challenged with PBS or LPS (0.4 mg/kg). Baseline body mass did not differ between groups (n = 8 per group) (A). Survival analysis was used to evaluate if lighting condition impacted recovery of body mass to baseline following LPS challenge. While the recovery curves of PBS-treated control (n = 7) and CD (n = 8) mice were not statistically different, there was a statistically significant difference between LPS-treated control (n = 8) and CD (n = 7) mice (B). Dashed line indicates median recovery times. Statistical analyses used one-way ANOVA (A) and pairwise log-rank test with a BH correction for multiple comparisons (B).
3.3. Sickness Behavior
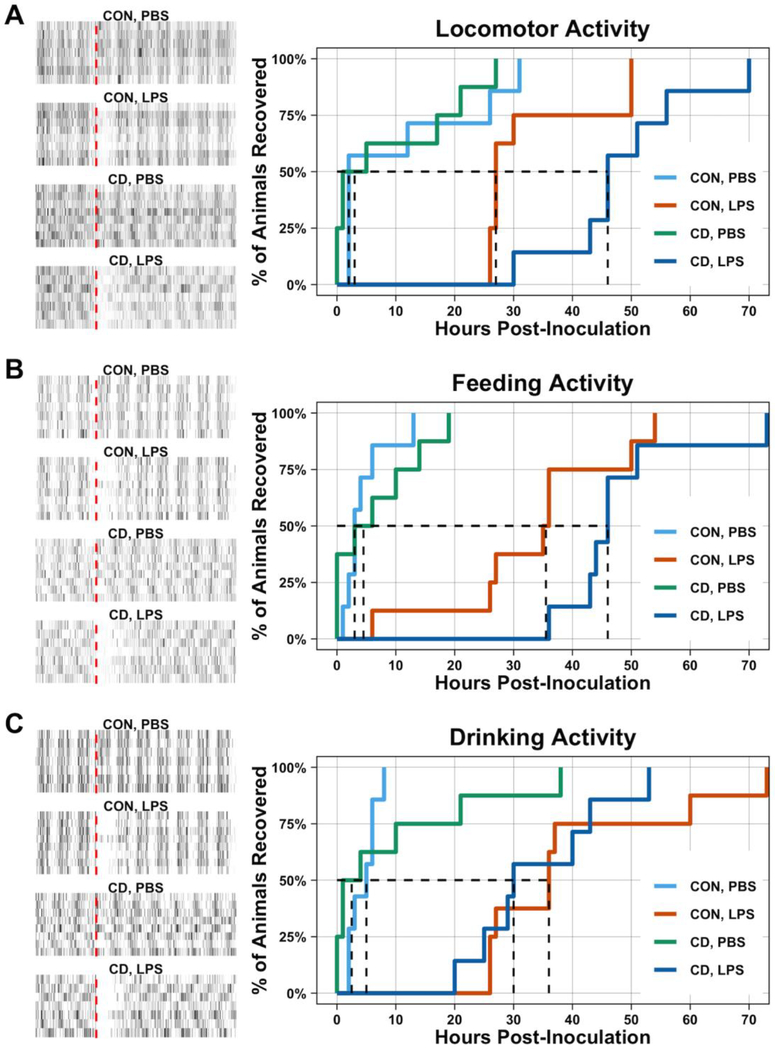
Next we aimed to determine how CD affected sickness behaviors. The time to reach our locomotor activity recovery criteria for CD mice was longer than the control mice. The recovery curve for the locomotor activity of LPS-treated CD mice was statistically different from LPS-treated control mice (Log Rank, BH Correction for multiple comparisons: p = 0.036) (Figure 3A), while the recovery for food consumption (Log Rank, BH Correction for multiple comparisons: p = 0.19) (Figure 3B) and water consumption (Log Rank, BH Correction for multiple comparisons: p = 0.57) (Figure 3C) was not statistically different between LPS-treated CD and control mice. The median recovery of locomotor and feeding activity occurred at 27 hours and 36 hours, respectively, for control mice challenged with LPS, while median recovery time was approximately 46 hours for both locomotor and feeding activities for LPS-treated CD mice. The median recovery time for water , F1,27 = 0.08) or a statistically significant interaction (Two-way ANOVA: TNFα, p = 0.93, F1,27 = 0.008; IL-1β, p = 0.093, F1,27 = 3.02) was observed for TNFα or IL-1β.consumption was 36 hours in LPS-treated control mice and 30 hours in LPS-treated CD mice.
Fig. 3: Circadian desynchronization prolongs recovery of locomotor activity in mice following LPS.
After 5 days acclimatization to the behavioral phenotyping system, control and CD mice were challenged with PBS or LPS (0.4 mg/kg). Behavior was recorded for a total of 7 dpi. Raw data of locomotor (A), feeding (B), and drinking (C) activity is depicted in the raster plots. The red-dashed line within the raster plots indicates time of inoculation. Survival plots are used to assess recovery following PBS or LPS challenge. No difference in recovery was observed for locomotor (A), feeding (B), or drinking (C) behavior following PBS treatment. However, following LPS challenge, CD mice required a longer recovery time to reach and sustain baseline locomotor activity levels compared to control mice (A). Although a similar trend was observed for feeding activity (B), the effect of LPS on feeding activity was not statistically significant. Furthermore, the effect of LPS on drinking activity was not dependent on lighting condition (C). Dashed line indicates median recovery times. Statistical analyses used pairwise log-rank test with a BH correction for multiple comparisons. Number of mice per group: CON, PBS = 7; CON, LPS = 8; CD, PBS = 8; CD, LPS = 7.
3.4. Immune Measures
Plasma and tissues (liver, brain) were collected from a separate cohort of mice 36 hpi. We chose 36 hpi because the locomotor and feeding activities of at least half of the LPS-treated control mice returned to their baseline levels of activity, while the LPS-treated CD mice still lagged behind.
3.4.1. Plasma Cytokine Measures
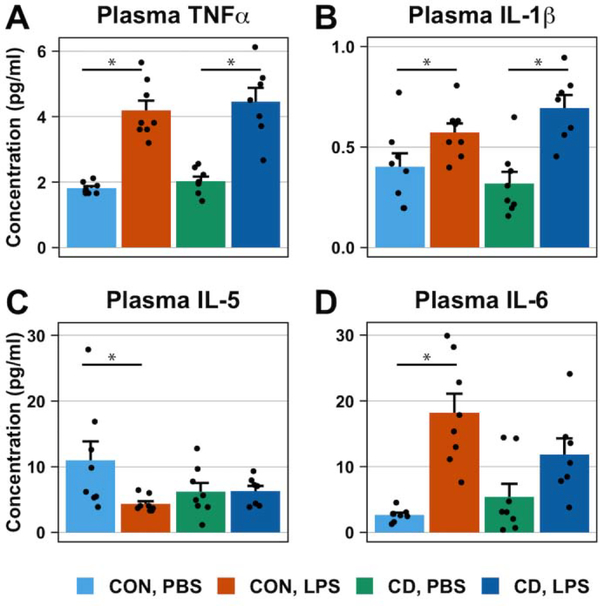
A comprehensive multiplex panel was used to measure the plasma concentrations of Th1-/Th2-associated cytokines and chemokines. We found a main effect of treatment on TNFα (Two-way ANOVA: p < 0.001, F1,27 = 85.79) and IL-1β (Two-way ANOVA: p < 0.001, F1,27 = 21.21) such that LPS treatment increased the plasma concentration of TNFα and IL-1β (Figure 4A, 4B). However, no main effect of lighting condition (Two-way ANOVA: TNFα p = 0.35, F1,27 = 0.91; IL-1β, p = 0.775, F1,27 = 0.08) or a statistically significant interaction (Two-way ANOVA: TNFα, p = 0.93, F1,27 = 0.008; IL-1β, p = 0.093, F1,27 = 3.02) was observed for TNFα or IL-1β.
Fig. 4. The inflammatory response of plasma cytokines to LPS treatment is affected by circadian desynchronization.
Bar plots represent the plasma cytokine concentrations 36 hours post-inoculation with PBS or LPS (0.4 mg/kg). LPS treatment increased the plasma concentration of TNFα (A) and IL-1β (B) in both control and CD mice. Conversely, LPS treatment decreased and increased plasma concentrations of IL-5 (C) and IL-6 (D), respectively, in control mice but not in CD mice. Data are represented as mean ± SEM. Statistical analyses on plasma concentration used two-way ANOVAs followed by post-hoc Tukey HSD tests for statistically significant interactions. Asterisks indicate that the difference between groups are statistically significant (p ≤ 0.05). Number of mice per group: CON, PBS = 8; CON, LPS = 8; CD, PBS = 8; CD, LPS = 7.
We also detected a main effect of treatment on plasma IL-5 and IL-6 (Two-way ANOVA: IL-5, p = 0.051, F1,27 = 4.16; IL-6, p < 0.001, F1,27 = 27.58), both of which, were accompanied by a statistically significant interaction (Two-way ANOVA: IL-5, p = 0.057, F1,27 = 3.96; IL-6, p = 0.042, F1,27 = 4.56). Post-hoc analysis indicates that LPS treatment decreases plasma concentrations of IL-5 in control mice (Post-Hoc Tukey HSD: p = 0.039) but not in CD mice (Post-Hoc Tukey HSD: p = 0.99) (Figure 4C). Conversely, LPS treatment increases plasma concentrations of IL-6 in control mice (Post-Hoc Tukey HSD: p < 0.001) but not in CD mice (Post-Hoc Tukey HSD: p = 0.17) (Figure 4D). No main effect of lighting condition on either plasma IL-5 or IL-6 was detected (Two-way ANOVA: IL-5, p = 0.37, F1,27 = 0.82; IL-6, p = 0.43, F = 0.63)
A main effect of treatment was also observed for several other plasma cytokines and chemokines including GM-CSF, Gro-α/KC, IL-4, IL-18, IP-10, MCP-1, MCP-3, MIP-1α, MIP-1β, MIP-2, and RANTES (see Table 1). No main effect of lighting condition or an interaction was observed for any of these cytokines or chemokines (see Table 1). We also did not find a main effect of treatment, a main effect of lighting condition, or an interaction for plasma Eotaxin, IFNγ, IL-12p70, or IL-13 (see Table 1).
Table 1.
Plasma cytokine and chemokine levels 36 hours following peripheral LPS inoculation in control and circadian desynchronized mice.
| Cytokine | Effect of LPS | Direction of effect | Effect of desynchronization | Direction of effect | Interaction |
|---|---|---|---|---|---|
| Eotaxin | p = 0.183, F = 1.87 | ns | p = 0.454, F = 0.58 | ns | p = 0.787, F = 0.07 |
| GM-CSF | p = 0.015, F = 6.79 | ↑ | p = 0.306, F = 1.09 | ns | p = 0.451, F = 0.58 |
| Gro-α/KC | p < 0.001, F = 30.91 | ↑ | p = 0.673, F = 0.18 | ns | p = 0.568, F = 0.33 |
| IFNγ | p = 0.452, F = 0.58 | ns | p = 0.939, F = 0.01 | ns | p = 0.252, F = 1.37 |
| IL-12p70 | p = 0.233, F = 1.49 | ns | p = 0.061, F = 3.82 | ns | p = 0.429, F = 0.65 |
| IL-13 | p = 0.528, F = 0.41 | ns | p = 0.073, F = 3.47 | ns | p = 0.596, F = 0.29 |
| IL-18 | p < 0.001, F = 139.21 | ↑ | p = 0.571, F = 0.33 | ns | p = 0.109, F = 2.75 |
| IL-1β | p < 0.001, F = 21.21 | ↑ | p = 0.775, F = 0.08 | ns | p = 0.094, F = 3.02 |
| IL-2 | p = 0.325, F = 1.01 | ns | p = 0.410, F = 0.70 | ns | p = 0.743, F = 0.11 |
| IL-4 | p = 0.008, F = 8.34 | ↑ | p = 0.704, F = 0.15 | ns | p = 0.728, F = 0.12 |
| IL-5 | p = 0.051, F = 4.16 | ↓ | p = 0.373, F = 0.82 | ns | p = 0.057, F = 3.96 |
| IL-6 | p < 0.001, F = 27.58 | ↑ | p = 0.435, F = 0.63 | ns | p = 0.042, F = 4.56 |
| IP-10 | p < 0.001, F = 166.85 | ↑ | p = 0.166, F = 2.02 | ns | p = 0.865, F = 0.03 |
| MCP-1 | p < 0.001, F = 61.93 | ↑ | p = 0.331, F = 0.98 | ns | p = 0.288, F = 1.18 |
| MCP-3 | p < 0.001, F = 69.35 | ↑ | p = 0.839, F = 0.04 | ns | p = 0.795, F = 0.07 |
| MIP-1α | p < 0.001, F = 51.66 | ↑ | p = 0.624, F = 0.25 | ns | p = 0.651, F = 0.21 |
| MIP-1β | p < 0.001, F = 103.78 | ↑ | p = 0.176, F = 1.93 | ns | p = 0.390, F = 0.76 |
| MIP-2 | p < 0.001, F = 26.01 | ↑ | p = 0.702, F = 0.15 | ns | p = 0.763, F = 0.09 |
| RANTES | p < 0.001, F = 117.39 | ↑ | p = 0.061, F = 3.82 | ns | p = 0.079, F = 3.34 |
| TNFα | p < 0.001, F = 85.79 | ↑ | p = 0.349, F = 0.91 | ns | p = 0.928, F = 0.01 |
Table presents results from plasma cytokine and chemokine multiplex array. Plasma was collected from control or circadian desynchronized mice 36 hours post-inoculation with PBS or LPS. Reported statistics (Two-way ANOVA, with p and F values) summarize main effects of LPS treatment and environmental lighting condition, and the interaction. Bolding indicates a statistically significant main effect or interaction.
3.4.2. Hepatic mRNA Expression
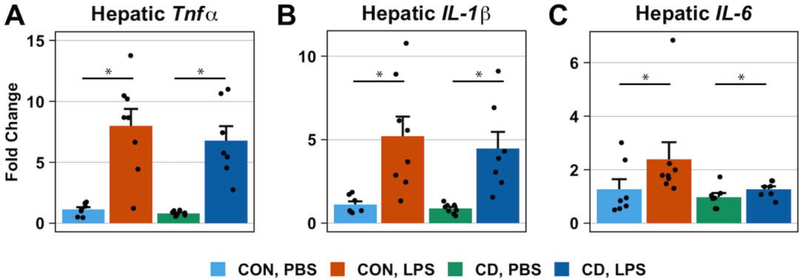
Since the liver is a key metabolic organ, shows a high level of circadian regulation, and has a prominent role in immune responses, we quantified expression of proinflammatory genes from hepatic tissue using qPCR. As expected, a main effect of treatment on the hepatic expression of proinflammatory cytokine genes TNFα, IL-1β, and IL-6 was detected (Two-way ANOVA: TNFα, p < 0.001, F1,26 = 99.38; IL-1β, p < 0.001, F1,26 = 55.06, IL-6, p = 0.01, F1,25 = 7.72) such that LPS treatment increased their expression in both control and CD mice (Figure 5A–C). However, we did not detect a main effect of lighting condition (Two-way ANOVA: TNFα, p = 0.42, F1,26 = 0.66; IL-1β, p = 0.45, F1,26 = 0.59; IL-6, p = 0.11, F1,25 = 2.75) or an interaction (Two-way ANOVA: TNFα, p = 0.69, F1,26 = 0.17; IL-1β, p = 0.83, F1,26 = 0.05; IL-6, p = 0.28, F1,25 = 1.20) on the expression of these proinflammatory cytokine genes.
Fig. 5. LPS treatment increases the hepatic expression of proinflammatory cytokine genes in both control and CD mice.
Bar plots represent the fold-change in proinflammatory cytokine mRNA levels 36 hours post-inoculation with PBS or LPS (0.4 mg/kg). mRNA levels were measured by quantitative PCR and fold change was calculated using the ΔΔCT method of relative quantification. No baseline difference of proinflammatory cytokine gene expression in the liver was observed. LPS treatment increased the hepatic expression of TNFα (A), IL-1β (B), and IL-6 (C). No statistically significant interactions were observed. Data are represented as mean ± SEM. Statistical analyses on ΔCT data used two-way ANOVAs. Asterisks indicate that the difference between groups are statistically significant (p ≤ 0.05). Number of mice per group: CON, PBS = 7; CON, LPS = 8; CD, PBS = 8 for TNFα and IL-1β, 7 for IL-6; CD, LPS = 7
We also found a main effect of LPS treatment on the hepatic expression of inflammasome-related genes Casp1 and Pycard (Two-way ANOVA: Casp1, p < 0.001, F1,26 = 22.03; Pycard, p < 0.001, F1,26 = 72.20) such that LPS treatment increased their expression in both control and CD mice (Supp. Figure 1A, 1B). However, there was no main effect of lighting condition (Two-way ANOVA: Casp1, p = 0.47, F1,26 = 0.53; Pycard, p = 0.63, F1,26 = 0.24) or an interaction (Two-way ANOVA: Casp1, p = 0.21, F1,26 = 1.63; Pycard, p = 0.74, F1,26 = 0.11) on these inflammasome-related genes. Additionally, we did not detect a main effect of LPS treatment (Two-way ANOVA: p = 0.25, F1,26 = 1.41), or a main effect of lighting condition (Two-way ANOVA: p = 0.92, F1,26 = 0.01), or an interaction (Two-way ANOVA: p = 0.26, F1,26 = 1.35) on the hepatic expression of Nf-κB (Supp. Figure 1C).
3.4.3. Hypothalamic, Hippocampal, and mPFC mRNA Expression
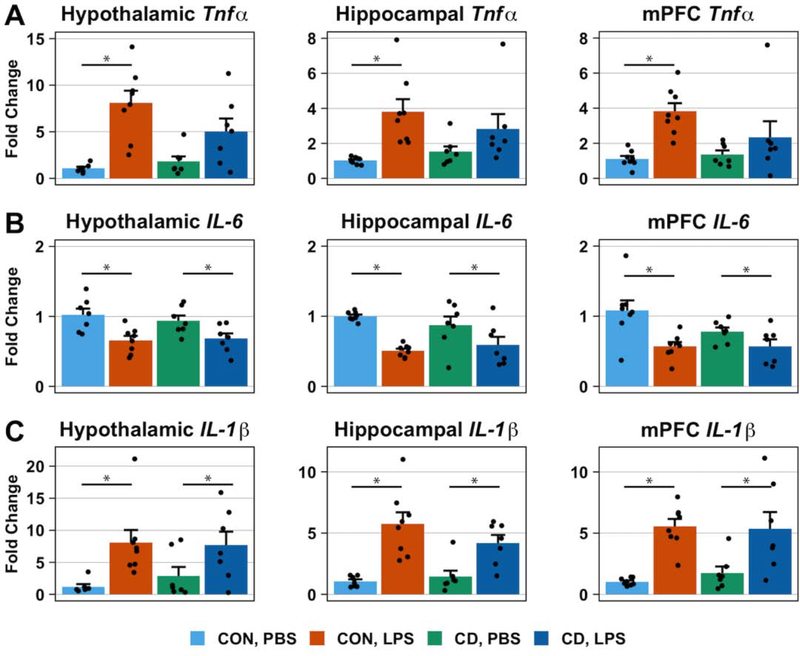
In the hypothalamus, hippocampus, and mPFC, a main effect of LPS treatment on TNFα expression (Two-way ANOVA: hypothalamus, p < 0.001, F1,25 = 31.63; hippocampus, p < 0.001, F1,26 = 30.53; mPFC, p = 0.005, F1,26 = 9.40) was accompanied by a statistically significant interaction (Two-way ANOVA: hypothalamus, Type II: p = 0.059, F1,25 = 3.91; hippocampus, p = 0.048, F1,26 = 4.29; mPFC, p = 0.038, F1,26 = 4.77). For all three brain regions, post-hoc analysis indicates that LPS treatment increases the expression of TNFα in control mice (Post-Hoc Tukey HSD: hypothalamus, p < 0.001; hippocampus, p < 0.001; mPFC, p = 0.005) but not in CD mice (Post-Hoc Tukey HSD: hypothalamus, p = 0.08; hippocampus, p = 0.13; mPFC, p = 0.96) (Figure 6A).
Fig. 6. Circadian desynchronization changes brain cytokine gene expression in response to LPS challenge.
Bar plots represent the fold-change in proinflammatory cytokine mRNA levels 36 hours post-inoculation with PBS or LPS (0.4 mg/kg). mRNA levels were measured by quantitative PCR and fold change was calculated using the ΔΔCT method of relative quantification. No baseline difference of proinflammatory cytokine mRNA levels in the hypothalamus, hippocampus, or mPFC was observed. The effect of LPS treatment on proinflammatory cytokine expression was consistent across brain regions. LPS treatment increased TNFα expression in control mice but not in CD mice (A). LPS treatment had opposite effects on the expression of IL-1β and IL-6. While LPS treatment reduced the expression of IL-6 in both control and CD mice (B), it also increased the expression of IL-1β in both control and CD mice (C). Data are represented as mean ± SEM. Statistical analyses on ΔCT data used two-way ANOVAs followed by post-hoc Tukey HSD tests for statistically significant interactions. Asterisks indicate that the difference between groups are statistically significant (p ≤ 0.05). Number of mice per group: CON, PBS = 8 for hippocampus and mPFC, 7 for hypothalamus; CON, LPS = 8; CD, PBS = 7; CD, LPS = 7.
A main effect of LPS treatment on the expression of IL-6 was detected in all three brain regions (Two-way ANOVA: hypothalamus, p < 0.001, F1,25 = 16.20; hippocampus, p < 0.001, F1,26 = 18.49; mPFC, p = 0.002, F1,26 = 12.32) such that LPS treatment reduced IL-6 expression (Figure 6B). We did not detect a main effect of lighting condition on IL-6 expression (Two-way ANOVA: hypothalamus, p = 0.82, F1,25 = 0.05; hippocampus, p = 0.54, F1,26 = 0.39; mPFC, p = 0.30, F1,26 = 1.12) or an interaction (Two-way ANOVA: hypothalamus, p = 0.57, F1,25 = 0.34; hippocampus, p = 0.28, F1,26 = 1.20; mPFC, p = 0.42, F1,26 = 0.67).
Similar to IL-6, we found a main effect of LPS treatment on IL-1β expression in all three brain regions (Two-way ANOVA: hypothalamus, p < 0.001, F1,25 = 17.61; hippocampus, p < 0.001, F1,26 = 52.61; mPFC, p < 0.001, F1,26 = 45.67). However, unlike IL-6, LPS treatment increased the expression of IL-1β (Figure 6C). We did not detect a statistically significant main effect of lighting condition on IL-1β expression (Two-way ANOVA: hypothalamus, p = 0.92, F1,25 = 0.008; hippocampus, p = 0.61, F1,26 = 0.26; mPFC, p = 0.78, F1,26 = 0.08) or an interaction (Two-way ANOVA: IL-1β, hypothalamus, p = 0.36, F1,25 = 0.86; hippocampus, p = 0.31, F1,26 = 1.06; mPFC, p = 0.21, F1,26 = 1.65).
We also probed for changes in the expression of inflammasome-related genes. We detected a main effect of LPS treatment on the expression of Casp1 in hypothalamus and hippocampus (Two-way ANOVA: hypothalamus, p = 0.004, F1,25 = 9.85; hippocampus p < 0.001, F1,26 = 19.64; mPFC, p = 0.38, F1,26 = 0.81) (Supp. Figure 2A). For Pycard there was a main effect of LPS treatment in all brain areas (Two-way ANOVA: hypothalamus, p < 0.001, F1,25 = 23.44; hippocampus, p < 0.001, F1,26 = 29.90; mPFC, p < 0.001, F1,26 = 30.85) (Supp. Figure 2B). We did not detect a statistically significant main effect of lighting condition on Casp1 (Two-way ANOVA: hypothalamus, p = 0.45, F1,25 = 0.60; hippocampus p = 0.72, F1,26 = 0.12; mPFC, p = 0.73, F1,26 = 0.11) or Pycard (Two-way ANOVA: hypothalamus, p = 0.996, F1,25 = 0.00; hippocampus, p = 0.93, F1,26 = 0.01; mPFC, p = 0.62, F1,26 = 0.25) expression or an interaction (Two-way ANOVA: Casp1, hypothalamus, p = 0.41, F1,25 = 0.71; hippocampus p = 0.72, F1,26 = 0.13; mPFC, p = 0.32, F1,26 = 1.02; Pycard, hypothalamus, p = 0.30, F1,25 = 1.12; hippocampus, p = 0.60, F1,26 = 0.29; mPFC, p = 0.22, F1,26 = 1.56).
For Nf-κB expression, we detected a main effect of LPS treatment for two out of three brain regions (Two-way ANOVA: hypothalamus, p = 0.015, F1,25 = 6.82; hippocampus, p = 0.16, F1,26 = 2.12; mPFC, p = 0.006, F1,26 = 9.01) such that LPS treatment increased hypothalamic and mPFC expression of Nf-κB in both control and CD mice (Supp. Figure 2C). We did not find a main effect of lighting condition on Nf-κB expression (Two-way ANOVA: hypothalamus, p = 0.93, F1,25 = 0.01; hippocampus, p = 0.08, F1,26 = 3.39; mPFC, p = 0.55, F1,26 = 0.37) or an interaction (Two-way ANOVA: Nf-κB, hypothalamus, p = 0.62, F1,25 = 0.25; hippocampus, p = 0.34, F1,26 = 0.94; mPFC, p = 0.26, F1,26 = 1.31).
4. DISCUSSION
Our previous results (Phillips et al., 2015) demonstrated that our CD paradigm alters the central and peripheral immune response to a low-dose LPS challenge 3 hpi. However, it was not known how this altered inflammatory response to LPS impacted physiological and behavioral responses. We hypothesized that this altered inflammatory response to LPS induced by CD would impact changes in body mass and sickness behavior following LPS challenge. Using survival analysis, we demonstrated that CD mice experience a prolonged recovery period of body mass and locomotor activity following LPS challenge compared to control mice. We also observed an increase in the median time required for LPS-treated CD mice to return to their pre-inoculation feeding activity compared to LPS-treated control mice.
It is well established that LPS inoculation leads to an initial loss in body mass followed by a recovery period (Dubos and Schaedler, 1961). We sought to determine if CD impacted the time required to reach recovery of body mass following LPS challenge. We detected a statistically significant difference in the overall rate by which each group of mice recovered to pre-inoculation body mass due to LPS treatment (Figure 2C), which was accompanied by a longer median time required for the body mass of half the CD mice to recover (59 hours) compared to the time required for half the control mice to recover (50 hours) following LPS challenge. Previous work by Fonken et al. (2013) has demonstrated that housing mice, which are nocturnal, in dim light during their active (night) phase, have increased anorectic responses following LPS (Fonken et al., 2013) but this work only focused on the first 24 hpi. In this study, food consumption was measured only daily, rather than the more high-resolution temporal analysis of the present findings. Moreover, our recovery data are based upon within subject changes, providing yet more new information about the process not only of the initial sickness responses, but also recovery over time. Thus, an important contribution of our work in this regard is the discovery that circadian desynchronization not only affects initial short-term responses to immune challenge, but that the consequences of this altered inflammatory response can be sustained. Understanding how circadian desynchronization changes the underlying physiological processes that regulate energy expenditure, food consumption, and body mass regulation will be important next steps to further untangle these relationships, and potentially mitigate such negative outcomes. Beyond the circadian range, interactions between environmental light-dark cycles and immune function are well known. In seasonally breeding vertebrates, there are clear links between photoperiod and immune function (Demas and Nelson, 1998; Nelson and Demas, 1996; Onishi et al., 2020; Stevenson and Prendergast, 2015). From wound healing (Cable et al., 2017), to immune stimulation (Demas and Nelson, 1998), to pathogen response (Schultz et al., 2017), environmental light conditions can clearly affect the immune system. It is noteworthy that many of these responses are also intricately linked with food availability and metabolic function (Drazen et al., 2001; Garcia et al., 2010).
We next wanted to probe if CD affected a subset of sickness behaviors, particularly those motivated behaviors that are most closely related to metabolic function. Broadly, sickness behaviors are a set of adaptive responses meant to rid the body of pathogens and preserve energy for mounting an immune and febrile response (Dantzer, 2017). In mammals, including rodents, sickness behavior is characterized by inactivity, sleepiness, decreased appetite, and social withdrawal (Dantzer, 2017). As such, sickness behaviors are thus strongly associated with metabolism and metabolic processes. In the present study, when referring to sickness behavior we are focusing on decreases in locomotor, feeding, and drinking activities that occur following LPS challenge. While recovery curves for locomotor activity were different, feeding and drinking behavior did not show statistical differences between LPS treated groups. However, in general, the rate by which mice recovered locomotor and feeding activity following LPS challenge was slower than mice that were treated with PBS (Figure 3A–3C). The median time required for the CD mice to recover to pre-inoculation locomotor (46 hours) and feeding (46 hours) activity levels was longer than the time required for half of the control mice to recover to their pre-inoculation activity levels (27 and 36 hours, respectively). Thus we posit that even a small non-statistically significant delay in the recovery of feeding, coupled with potential shifts in metabolic rate (outside the scope of the present studies) could lead to statistically significant shifts in body mass.
The recognition of LPS by immune cells and the subsequent immune response is adaptive, and its absence increases the susceptibility of organisms to bacterial infections. LPS is recognized by TLR-4 receptors of local immune cells, leading to the activation of Nf-κB, which promotes the expression and subsequent secretion of cytokines and chemokines. These act in an autocrine and a paracrine manner to prime the local area and signal circulating immune cells to migrate to the inflamed tissue. Some immune cells act as antigen-presenting cells (APCs), which following antigen recognition, will migrate to lymphoid organs and present the antigen to lymphocytes. The main classes of these cells are the T helper 1 (Th1) lymphocytes and T helper 2 (Th2) lymphocytes, which secrete different cytokines that impact how other immune cells function.
Using a Th1/Th2 multiplexed immunoassay, we found that LPS significantly altered nearly all measured parameters, while CD did not result in any baseline differences (Table 1). However, while LPS increased plasma IL-6 and decreased plasma IL-5 in control mice, these plasma cytokines did not statistically differ between PBS- and LPS-treated CD mice. Notably, we previously reported that CD mice had reduced concentrations of plasma IL-6 compared to CON mice at 3h following LPS challenge (Phillips et al., 2015). IL-6 plays an important role in the production of acute phase proteins by hepatocytes following LPS challenge (Rose-John et al., 2017). Additionally, IL-6 knockout mice have been demonstrated to be more susceptible to pathogens (Rose-John et al., 2017). Thus, it is plausible that consistently low circulating IL-6 observed in CD mice 3h and 36h following LPS challenge is implicated in a compromised host defense. IL-5 is generally considered a Th2 cytokine. Reduced circulating IL-5 in control mice at 36h following LPS challenge may be a result of normal counter-regulatory processes stemming from Th1-focused response. The fact that LPS treatment does not alter IL-5 in CD mice at 36 hpi may indicate a loss of this normal counter-regulation.
We then investigated how CD altered the response to LPS in different tissue compartments, namely the liver (a primary mediator of immune responses) and the brain (a key regulator of immune responses and sickness behaviors). In the liver, we found that at 36 hpi there were no differences between control and CD in any of the measured mRNA transcripts (Figure 5). In the brain, we examined three different regions, the prefrontal cortex, the hippocampus, and the hypothalamus, all regions known to show changes in local cytokine mRNA levels following immune challenge, and to express high levels of cytokine receptors (Dantzer, 2009; Skelly et al., 2013). We found that LPS-treated CD mice did not show the sustained elevated levels of TNFα mRNA in any of the regions explored, as compared to LPS-treated control mice. This finding is noteworthy since our previous work at 3 hpi showed that TNFα mRNA was not different in CD mice in the hippocampus, but was significantly elevated in CD mice in the hypothalamus (Phillips et al., 2015). While a caveat in directly comparing these two studies is the difference in time of inoculation, this general pattern of results suggests that there is a difference in the dynamic response of brain TNFα mRNA in response to LPS following CD. It has been well documented that TNFα can induce sickness behaviors in rodents (Bluthé et al., 1994, 1991; Palin et al., 2008, 2007). Within the hypothalamus, TNFα can reduce food intake (Moraes et al., 2006) potentially contributing to body mass loss. In addition, central administration of TNFα decreases social interaction behaviors and locomotion in mice (Palin et al., 2008). We posit that the early 3 hpi increase in TNFα transcripts in LPS-treated CD mice (Phillips et al., 2015) might contribute to the reduced recovery of CD mice. While the reduced TNFα transcript level in the brain of LPS treated CD mice at the 36 hpi time point seems counterintuitive, a potential explanation is a dysregulation of the normal regulatory, and counter regulatory, processes that occur following challenge with a pathogen. Specifically, given the well characterized interactions between TNFα and IL-1β 5/11/2020 6:21:00 PM, reduced TNFα transcripts in LPS-treated CD mice, while IL-1β mRNA remains elevated to the same degree as in LPS-treated control mice, might represent a potential shift in the coordinated activation of different components of the immune response. It is essential to reinforce the notion that the immune response does not occur in isolation, many pathways are activated together and in parallel, with significant interplay. Moreover, responses are dynamic, changing over time and space. With regards to our findings with Nf-κB, which might be interpreted as suggesting Nf-κB is not involved in the observed responses, it is important to note that pathogen driven Nf-κB pathway function relies only partially on de novo generation of Nf-κB transcripts. Instead, and perhaps more importantly, interacts with several other gene pathways (some of which are altered, see Supp.Fig.2) (Manning et al., 1995).
Our previous work demonstrated that IL-6 mRNA was elevated in the hypothalamus of CD mice but not control mice 3h after LPS challenge (Phillips et al., 2015). In the present study, at the 36h time point, we show that in all brain regions explored, IL-6 mRNA was reduced suggesting an anti-inflammatory targeting of IL-6. We suspect that this downregulation of IL-6 mRNA could be attributed to the inhibition of IL-6 trans-signaling via endogenous soluble gp130 (sgp130). IL-6 trans-signaling is a necessary communication pathway for IL-6 to signal various cells within the brain such as neurons and astrocytes (März et al., 1997; Van Wagoner et al., 1999). Moreover, following an intraperitoneal LPS challenge, the inhibition of IL-6 trans-signaling via sgp130 has been shown to reduce the expression of IL-6 mRNA and facilitate sickness behavior recovery (Burton et al., 2011). The primary source of IL-6 in the inflamed brain are astrocytes (Van Wagoner et al., 1999). We’ve previously shown that CD increases brain GFAP mRNA (Lananna et al., 2018), indicating astrocyte activation and presumably astrogliosis. Primed astrocytes in the hypothalamus could explain why we previously observed elevated hypothalamic IL-6 mRNA at 3h following peripheral LPS challenge in CD mice.
The results presented in this study leads to additional evidence that desynchronization of the circadian clock can lead to dysregulated innate immune responses. Our data go further in providing evidence that CD can also contribute to impaired recovery following exposure to a pathogen as measured by sickness behaviors including locomotor, feeding and drinking activity. These differences are associated with altered patterns of plasma cytokines, and hepatic and brain cytokine transcripts. Together, our findings present compelling evidence that desynchronization of the circadian clock can have significant effects on an individual’s behavioral and physiological recovery following exposure to a pathogen.
Supplementary Material
Supp. Fig. 1. LPS treatment increases the hepatic expression of inflammasome-related genes in control and CD mice.
Bar plots represent the fold-change in mRNA levels 36 hours post-inoculation with PBS or LPS (0.4 mg/kg). mRNA levels were measured by quantitative PCR and fold change was calculated using the ΔΔCT method of relative quantification. No baseline difference of Casp1 (A), Pycard (B), or Nf-κB (C) mRNA levels in the liver was observed. LPS treatment increased the hepatic expression of Casp1 (A) and Pycard (B) in both control and CD mice. There was no effect of LPS treatment on the hepatic expression of Nf-κB (C). No statistically significant interactions were observed. Data are represented as mean ± SEM. Statistical analyses on ΔCT data used two-way ANOVAs. Asterisks indicate that the difference between groups are statistically significant (p ≤ 0.05). Number of mice per group: CON, PBS = 7; CON, LPS = 8; CD, PBS = 8; CD, LPS = 7.
Supp. Fig. 2. LPS treatment increases the brain expression of proinflammatory genes essential for inflammasome and Nf-κB pathway function in control and CD mice.
Bar plots represent the fold-change in mRNA levels 36 hours post-inoculation with PBS or LPS (0.4 mg/kg). mRNA levels were measured by quantitative PCR and fold change was calculated using the ΔΔCT method of relative quantification. No baseline difference of Casp1 (A), Pycard (B) or Nf-κB (C) mRNA levels in the hypothalamus, hippocampus, or mPFC was observed. LPS treatment increased Casp1 expression in the hypothalamus and hippocampus, but not in the mPFC, in control and CD mice (A). LPS treatment increased the expression of Pycard in all three brain regions in both control and CD mice (B). LPS treatment increased the expression of Nf-κB in the hypothalamus and mPFC, but not in the hippocampus, in control and CD mice (C). No statistically significant interactions were detected. Data are represented as mean ± SEM. Statistical analyses on ΔCT data used two-way ANOVAs. Asterisks indicate that the difference between groups are statistically significant (p ≤ 0.05). Number of mice per group: CON, PBS = 8 for hippocampus and mPFC, 7 for hypothalamus; CON, LPS = 8; CD, PBS = 7; CD, LPS = 7.
Highlights.
Circadian desynchronization alters the metabolic and behavioral response to LPS.
Sickness behaviors are prolonged in circadian desynchronized mice.
Changes in brain and peripheral cytokines are associated with these effects.
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health (DK119811) and the National Science Foundation (CAREER 1553067) to INK. We would also like to acknowledge the skilled animal care staff at Washington State University, as well as Naomi Wallace who assisted with tissue collections, and Andy He for his general help in the laboratory.
Footnotes
Declarations of interest: none.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERNCES
- Bluthé R-M, Dantzer R, Kelley KW, 1991. Interleukin-1 mediates behavioural but not metabolic effects of tumor necrosis factor α in mice. European Journal of Pharmacology 209, 281–283. 10.1016/0014-2999(91)90184-R [DOI] [PubMed] [Google Scholar]
- Bluthé RM, Pawlowski M, Suarez S, Parnet P, Pittman Q, Kelley KW, Dantzer R, 1994. Synergy between tumor necrosis factor alpha and interleukin-1 in the induction of sickness behavior in mice. Psychoneuroendocrinology 19, 197–207. 10.1016/0306-4530(94)90009-4 [DOI] [PubMed] [Google Scholar]
- Burton MD, Sparkman NL, Johnson RW, 2011. Inhibition of interleukin-6 trans-signaling in the brain facilitates recovery from lipopolysaccharide-induced sickness behavior. Journal of Neuroinflammation 8, 54 10.1186/1742-2094-8-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cable EJ, Onishi KG, Prendergast BJ, 2017. Circadian rhythms accelerate wound healing in female Siberian hamsters. Physiol. Behav. 171, 165–174. 10.1016/j.physbeh.2016.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzer R, 2017. Neuroimmune Interactions: From the Brain to the Immune System and Vice Versa. Physiological Reviews 98, 477–504. 10.1152/physrev.00039.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzer R, 2009. Cytokine, Sickness Behavior, and Depression. Immunology and Allergy Clinics of North America, Psychoneuroimmunology 29, 247–264. 10.1016/j.iac.2009.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demas Gregory E., Nelson RJ, 1998. Photoperiod, Ambient Temperature, and Food Availability Interact to Affect Reproductive and Immune Function in Adult Male Deer Mice (Peromyscus maniculatus). J Biol Rhythms 13, 253–262. 10.1177/074873098129000093 [DOI] [PubMed] [Google Scholar]
- Demas GE, Nelson RJ, 1998. Short-day enhancement of immune function is independent of steroid hormones in deer mice (Peromyscus maniculatus). J. Comp. Physiol. B, Biochem. Syst. Environ. Physiol. 168, 419–426. 10.1007/s003600050161 [DOI] [PubMed] [Google Scholar]
- Drazen DL, Demas GE, Nelson RJ, 2001. Leptin effects on immune function and energy balance are photoperiod dependent in Siberian hamsters (Phodopus sungorus). Endocrinology 142, 2768–2775. 10.1210/endo.142.7.8271 [DOI] [PubMed] [Google Scholar]
- Dubos RJ, Schaedler RW, 1961. The effect of bacterial endotoxins on the water intake and body weight of mice. J Exp Med 113, 921–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early JO, Curtis AM, 2016. Immunometabolism: Is it under the eye of the clock? Seminars in Immunology, Immunometabolism 28, 478–490. 10.1016/j.smim.2016.10.006 [DOI] [PubMed] [Google Scholar]
- Fonken LK, Weil ZM, Nelson RJ, 2013. Mice exposed to dim light at night exaggerate inflammatory responses to lipopolysaccharide. Brain, Behavior, and Immunity 34, 159–163. 10.1016/j.bbi.2013.08.011 [DOI] [PubMed] [Google Scholar]
- Garcia NW, Greives TJ, Zysling DA, French SS, Chester EM, Demas GE, 2010. Exogenous insulin enhances humoural immune responses in short-day, but not long-day, Siberian hamsters (Phodopus sungorus). Proc. Biol. Sci. 277, 2211–2218. 10.1098/rspb.2009.2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halberg F, Johnson EA, Brown BW, Bittner JJ, 1960. Susceptibility Rhythm to E. coli Endotoxin and Bioassay. Proceedings of the Society for Experimental Biology and Medicine 103, 142–144. 10.3181/00379727-103-25439 [DOI] [PubMed] [Google Scholar]
- Kalsbeek A, Palm IF, La Fleur SE, Scheer FAJL, Perreau-Lenz S, Ruiter M, Kreier F, Cailotto C, Buijs RM, 2006. SCN Outputs and the Hypothalamic Balance of Life. J Biol Rhythms 21, 458–469. 10.1177/0748730406293854 [DOI] [PubMed] [Google Scholar]
- Karatsoreos IN, Bhagat S, Bloss EB, Morrison JH, McEwen BS, 2011. Disruption of circadian clocks has ramifications for metabolism, brain, and behavior. PNAS 108, 1657–1662. 10.1073/pnas.1018375108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lananna BV, Nadarajah CJ, Izumo M, Cedeño MR, Xiong DD, Dimitry J, Tso CF, McKee CA, Griffin P, Sheehan PW, Haspel JA, Barres BA, Liddelow SA, Takahashi JS, Karatsoreos IN, Musiek ES, 2018. Cell-Autonomous Regulation of Astrocyte Activation by the Circadian Clock Protein BMAL1. Cell Reports 25, 1–9.e5. 10.1016/j.celrep.2018.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man K, Loudon A, Chawla A, 2016. Immunity around the clock. Science 354, 999–1003. 10.1126/science.aah4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AM, Bell FP, Rosenbloom CL, Chosay JG, Simmons CA, Northrup JL, Shebuski RJ, Dunn CJ, Anderson DC, 1995. NF-kappa B is activated during acute inflammation in vivo in association with elevated endothelial cell adhesion molecule gene expression and leukocyte recruitment. J. Inflamm. 45, 283–296. [PubMed] [Google Scholar]
- März P, Herget T, Lang E, Otter U, Rose-John S, 1997. Activation of gp 130 by IL-6/soluble IL-6 receptor induces neuronal differentiation. European Journal of Neuroscience 9, 2765–2773. 10.1111/j.1460-9568.1997.tb01705.x [DOI] [PubMed] [Google Scholar]
- Moraes JC, Amaral ME, Picardi PK, Calegari VC, Romanatto T, Bermúdez-Echeverry M, Chiavegatto S, Saad MJ, Velloso LA, 2006. Inducible-NOS but not neuronal-NOS participate in the acute effect of TNF-α on hypothalamic insulin-dependent inhibition of food intake. FEBS Letters 580, 4625–4631. 10.1016/j.febslet.2006.07.042 [DOI] [PubMed] [Google Scholar]
- Nelson RJ, Demas GE, 1996. Seasonal changes in immune function. Q Rev Biol 71, 511–548. 10.1086/419555 [DOI] [PubMed] [Google Scholar]
- Onishi KG, Maneval AC, Cable EC, Tuohy MC, Scasny AJ, Sterina E, Love JA, Riggle JP, Malamut LK, Mukerji A, Novo JS, Appah-Sampong A, Gary JB, Prendergast BJ, 2020. Circadian and circannual timescales interact to generate seasonal changes in immune function. Brain, Behavior, and Immunity 83, 33–43. 10.1016/j.bbi.2019.07.024 [DOI] [PubMed] [Google Scholar]
- Palin K, Bluthé R-M, McCusker RH, Moos F, Dantzer R, Kelley KW, 2007. TNFα-induced sickness behavior in mice with functional 55 kD TNF receptors is blocked by central IGF-I. Journal of Neuroimmunology 187, 55–60. 10.1016/j.jneuroim.2007.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palin K, McCusker RH, Strle K, Moos F, Dantzer R, Kelley KW, 2008. Tumor necrosis factor-α-induced sickness behavior is impaired by central administration of an inhibitor of c-jun N-terminal kinase. Psychopharmacology 197, 629–635. 10.1007/s00213-008-1086-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips DJ, Savenkova MI, Karatsoreos IN, 2015. Environmental disruption of the circadian clock leads to altered sleep and immune responses in mouse. Brain, Behavior, and Immunity, Sleep, Brain, Behavior, and Immunity 47, 14–23. 10.1016/j.bbi.2014.12.008 [DOI] [PubMed] [Google Scholar]
- R Core Team, 2019. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Rose-John S, Winthrop K, Calabrese L, 2017. The role of IL-6 in host defence against infections: immunobiology and clinical implications. Nat Rev Rheumatol 13, 399–409. 10.1038/nrrheum.2017.83 [DOI] [PubMed] [Google Scholar]
- Scheiermann C, Gibbs J, Ince L, Loudon A, 2018. Clocking in to immunity. Nat Rev Immunol 18, 423–437. 10.1038/s41577-018-0008-4 [DOI] [PubMed] [Google Scholar]
- Scheiermann C, Kunisaki Y, Frenette PS, 2013. Circadian control of the immune system. Nat Rev Immunol 13, 190–198. 10.1038/nri3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz EM, Hahn TP, Klasing KC, 2017. Photoperiod but not food restriction modulates innate immunity in an opportunistic breeder, Loxia curvirostra. J. Exp. Biol. 220, 722–730. 10.1242/jeb.149898 [DOI] [PubMed] [Google Scholar]
- Skelly DT, Hennessy E, Dansereau M-A, Cunningham C, 2013. A Systematic Analysis of the Peripheral and CNS Effects of Systemic LPS, IL-1Β, TNF-α and IL-6 Challenges in C57BL/6 Mice. PLOS ONE 8, e69123 10.1371/journal.pone.0069123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson TJ, Prendergast BJ, 2015. Photoperiodic time measurement and seasonal immunological plasticity. Frontiers in Neuroendocrinology, Seasonal Changes in the Neuroendocrine System 37, 76–88. 10.1016/j.yfrne.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognini P, Thaiss CA, Elinav E, Sassone-Corsi P, 2017. Circadian Coordination of Antimicrobial Responses. Cell Host & Microbe 22, 185–192. 10.1016/j.chom.2017.07.007 [DOI] [PubMed] [Google Scholar]
- Van Wagoner NJ, Oh J-W, Repovic P, Benveniste EN, 1999. Interleukin-6 (IL-6) Production by Astrocytes: Autocrine Regulation by IL-6 and the Soluble IL-6 Receptor. J. Neurosci. 19, 5236–5244. 10.1523/JNEUROSCI.19-13-05236.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp. Fig. 1. LPS treatment increases the hepatic expression of inflammasome-related genes in control and CD mice.
Bar plots represent the fold-change in mRNA levels 36 hours post-inoculation with PBS or LPS (0.4 mg/kg). mRNA levels were measured by quantitative PCR and fold change was calculated using the ΔΔCT method of relative quantification. No baseline difference of Casp1 (A), Pycard (B), or Nf-κB (C) mRNA levels in the liver was observed. LPS treatment increased the hepatic expression of Casp1 (A) and Pycard (B) in both control and CD mice. There was no effect of LPS treatment on the hepatic expression of Nf-κB (C). No statistically significant interactions were observed. Data are represented as mean ± SEM. Statistical analyses on ΔCT data used two-way ANOVAs. Asterisks indicate that the difference between groups are statistically significant (p ≤ 0.05). Number of mice per group: CON, PBS = 7; CON, LPS = 8; CD, PBS = 8; CD, LPS = 7.
Supp. Fig. 2. LPS treatment increases the brain expression of proinflammatory genes essential for inflammasome and Nf-κB pathway function in control and CD mice.
Bar plots represent the fold-change in mRNA levels 36 hours post-inoculation with PBS or LPS (0.4 mg/kg). mRNA levels were measured by quantitative PCR and fold change was calculated using the ΔΔCT method of relative quantification. No baseline difference of Casp1 (A), Pycard (B) or Nf-κB (C) mRNA levels in the hypothalamus, hippocampus, or mPFC was observed. LPS treatment increased Casp1 expression in the hypothalamus and hippocampus, but not in the mPFC, in control and CD mice (A). LPS treatment increased the expression of Pycard in all three brain regions in both control and CD mice (B). LPS treatment increased the expression of Nf-κB in the hypothalamus and mPFC, but not in the hippocampus, in control and CD mice (C). No statistically significant interactions were detected. Data are represented as mean ± SEM. Statistical analyses on ΔCT data used two-way ANOVAs. Asterisks indicate that the difference between groups are statistically significant (p ≤ 0.05). Number of mice per group: CON, PBS = 8 for hippocampus and mPFC, 7 for hypothalamus; CON, LPS = 8; CD, PBS = 7; CD, LPS = 7.