Abstract
Background:
Pancreatic cancer is projected to become the second most common cause of cancer death over the next 5 years. Since inflammation is thought to be a common trajectory for disease initiation, we sought to prospectively characterize immune profiles using DNA methylation markers and examine DNA methylation levels previously linked to inflammation biomarkers to evaluate whether these immune markers play a key role in pancreatic cancer.
Methods:
In a nested case-control study pooling three U.S. prospective cohort studies, DNA methylation was measured in prediagnostic leukocytes of incident pancreatic cancer cases and matched controls using the Illumina MethylationEPIC array. Differentially methylated regions were used to predict immune cell types and CpGs previously associated with inflammatory biomarkers were selected for the analysis. DNA methylation data from a retrospective case-control study conducted in Spain (PanGenEU) was used for independent replication.
Results:
Immune cell proportions and ratio of cell proportions were not associated with pancreatic cancer risk in the nested case-control study. Methylation extent of CpGs residing in or near gene MNDA was significantly associated with pancreatic cancer risk in the nested case-control study and replicated in PanGenEU. Methylation level of a promoter CpG of gene PIM-1 was associated with survival in both studies.
Conclusions:
Using a targeted approach, we identified several CpGs that may play a role in pancreatic carcinogenesis in two large, independent studies with distinct study designs.
Impact:
These findings could provide insight into critical pathways that may help identify new markers of early disease and survival.
Keywords: Pancreatic cancer, DNA methylation, immune cell profiles, neutrophil-to-lymphocyte ratio (NLR), Illumina MethylationEPIC array
Introduction
In the absence of specific disease symptoms, pancreatic cancer is difficult to identify early in the course of the disease; only 10% of pancreatic tumors are localized at diagnosis (1). Overall mortality for pancreatic cancer is very high, with only 9% of patients surviving 5-years beyond diagnosis, primarily because over 50% of cases have metastasized by diagnosis (1), making tumors inoperable. Identifying pancreatic cancer at earlier stages could significantly improve survival with increased opportunities for surgery; however, due to poor diagnostic accuracy of existing detection methods, screening is currently not recommended for asymptomatic adults (2).
New high-dimensional arrays designed to measure DNA methylation levels at hundreds of thousands of CpG sites throughout the genome have opened opportunities to estimate immune cell proportions in frozen blood samples that were stored without the measurement of complete blood counts (CBC) or without assessing immune profiles (3). With this method, archived samples from prospective studies can be used to examine changes in immune cell proportions, and DNA methylation alterations associated with the immune response, in individuals who develop cancer months or years later, providing new opportunities to better understand biological mechanisms and, perhaps, identify biomarkers for early detection. This targeted approach can be used in parallel to agnostically testing associations with all 850K CpGs obtained from the DNA methylation arrays, known as epigenome wide association studies (EWAS).
Immune cell proportions, such as the ratio of neutrophil to lymphocyte (NLR), have been shown to accurately predict cancer survival (4,5), including pancreatic cancer (6), but no study has evaluated whether immune markers based on DNA methylation profiles are associated with risk of developing pancreatic cancer. To address this, we examined associations between known DNA methylation markers of immune response and pancreatic cancer risk using pre-diagnostic blood samples of cases and controls obtained from three large US cohort studies. The selected inflammation CpGs and immune cell proportions were also examined in relation to overall survival. CpGs identified in the pooled prospective study were then examined in a large Spanish case-control study; replication in a completely different study population using a different study design provides an opportunity to evaluate whether the immune markers were present, or amplified, at time of diagnosis.
Materials and Methods
The analysis described in this paper represents two different study designs: a nested case-control dataset sampled from 3 U.S prospective cohort studies, and a retrospective case-control study conducted in Spain (PanGenEU). The main analyses were conducted on pancreatic cancer cases and matched controls identified from the Nurses’ Health Study (NHS), the Physician’s Health Study (PHS), and the Health Professionals Follow-up Study (HPFS). Associations between the 50 CpGs of interest and pancreatic cancer risk were also examined in the Spanish component of the PanGenEU study, a multicenter pancreatic cancer case-control study based in Europe. No replication study could be conducted with other prospective data, given that no other prospective data exist with DNA methylation on pancreatic cancer cases and controls (to our knowledge); we conducted a replication using a retrospective case-control study, making the assumption that DNA methylation changes that would predispose to pancreatic cancer risk, or mark disease progression, would be detectable in blood at time of diagnosis. Replication in a retrospective study would also reduce reporting of chance findings.
In the cohort studies, 403 incident cases were confirmed to have pancreatic cancer among the participants who provided blood samples prior to cancer diagnosis. A control subject was matched to each case on cohort (which also matches on sex), age (+/− 1 year), date of blood draw (month 3+/− and year), smoking (never, past, current) and race (White/other). Incident density sampling was used for the selection of controls. A subset of participants had data on inflammatory markers (C-reactive protein, interleukin-6 and tumor necrosis factor-alpha) from a prior study in the same cohorts (7). The final dataset consisted of 393 cases and 431 controls. For the survival analysis, cases missing date of diagnosis (n=42) or date of death (n=9) were not included in the analysis.
The second dataset consisted of pancreatic cancer cases and controls obtained from the Spanish component of the European Study into Digestive Illnesses and Genetics (PanGenEU), a multicenter case-control study that was conducted between 2009–2014 in six European countries (Spain, Italy, Germany, United Kingdom, Sweden and Ireland) (8–11). For the methylation analyses, we selected a PanGenEU representative subset of 657 Spanish subjects, 357 cases and 300 controls. The final data set for this analysis included a total of 338 cases and 285 controls.
More details for each study are provided in the Supplementary Methods.
DNA methylation measurements
DNA extracted from buffy coats (nested case-control study) or granulocytes (PanGenEU) were bisulfite-treated and DNA methylation was measured with the Illumina Infinium MethylationEPIC BeadChip array (Illumina, Inc, CA, USA). Details on DNA methylation measurements and data processing are provided in the Supplementary Methods. Reproducibility of results from 850K Illumina array has been previously shown to be very high (r=0.997)(12). In addition, we previously conducted a pilot study to examine reproducibility of DNA methylation measured in peripheral blood over a 1-year period using this array and demonstrated that DNA methylation varies by site, but is stable across a large number of probes (13).
Estimation of immune cell composition
Leukocyte subtypes proportions (i.e., CD4T, CD8T, natural killer cells [NK], B cells, monocytes [Mono] and neutrophils) were estimated using the “estimateCellCounts2” function in the FlowSorted.Blood.EPIC Bioconductor package (14), which is based on previously published reference-based cell mixture deconvolution algorithm with reference library selection conducted using the IDOL methodology (15).
Inflammation-associated CpG sites
We selected 64 CpG sites that had been strongly associated with inflammation markers in previous studies to examine in this study (16,17). Eleven CpGs from Ahsan et al(16) were associated with multiple inflammatory blood markers among 698 individuals (listed in their Table 1), and 54 CpG sites reaching EWAS significance in a large study conducted to identify DNA methylation markers of C-reactive protein levels (an additional 4 CpG sites were not included in this study as they were not on the 850K array we used) (17). Of those, 1 CpG overlapped with the other publication. Finally, we removed 14 CpGs that had low intraclass correlations (ICCs< 0.4) in our pilot study (13). The remaining 50 CpGs we tested had ICCs ranging between 0.40 and 0.95 (calculated from the M values adjusted for age, cell composition and Combat adjusted). The CpGs with significant associations (in our results) had ICCs between 0.67 and 0.86.
Table 1.
Baseline characteristics for study population, by study and case-control status at end of follow-up
| Mean (SD) or N (%) | Total cohort studies (N=824) | NHS (N=370) | HPFS (N=297) | PHS (N=157) | PanGenEU (N=623) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | |
| N | 393 | 431 | 176 | 194 | 146 | 151 | 71 | 86 | 338 | 285 |
| Age at study entry | 60.6 (7.9) | 60.2 (7.7) | 59.2 (6.4) | 59.5 (6.2) | 63.8 (7.9) | 63.6 (7.8) | 57.5 (9.2) | 55.8 (8.3) | 66.3 (12.7) | 63.7 (13.2) |
| Time before diagnosis (year)a | 13.0 (6.2) | 14.3 (6.3) | 10.9 (5.6) | 13.8 (6.3) | N/A | |||||
| Female | 176 (44.8%) | 194 (45.0%) | all female | all male | all male | 142 (42.0) | 127 (44.6) | |||
| Raceb | ||||||||||
| White | 346 (94.0%) | 406 (94.9%) | 164 (93.2%) | 187 (96.4%) | 137 (93.8%) | 145 (96.0%) | 45 (97.8%) | 74 (89.2%) | 334 (98.8) | 276 (96.8) |
| Black | 4 (1.1%) | 1 (0.2%) | 1 (0.6%) | 1 (0.5%) | 2 (1.4%) | 0 | 1 (2.2%) | 0 | 1 (0.29) | 2 (0.70) |
| Other | 18 (4.6%) | 21 (4.9%) | 11 (6.3%) | 6 (3.1%) | 7 (4.8%) | 6 (4.0%) | 0 (0.0%) | 9 (10.5%) | ||
| Smokingc | ||||||||||
| Never | 159 (40.9%) | 173 (40.3%) | 73 (42.0%) | 81 (41.5%) | 55 (38.2%) | 58 (39.2%) | 31 (43.7%) | 34 (39.5%) | 137 (40.5) | 141 (49.5) |
| Past | 174 (44.7%) | 190 (44.3%) | 73 (42.0%) | 81 (41.5%) | 77 (53.5%) | 76 (51.4%) | 24 (33.8%) | 33 (38.4%) | 97 (28.7) | 80 (28.1) |
| Current | 57 (14.6%) | 65 (15.2%) | 29 (16.6%) | 32 (16.5%) | 12 (8.3%) | 14 (9.5%) | 16 (22.5%) | 19 (22.1%) | 100 (29.6) | 61 (21.4) |
| BMI (kg/m2)d | 26.0 (4.3) | 25.6 (3.9) | 26.2 (5.3) | 25.8 (4.7) | 25.9 (3.2) | 25.9 (3.3) | 25.9 (3.1) | 24.7 (2.6) | 27.1 (4.6) | 27.12 (5.6) |
| Diabetese | 19 (4.8%) | 11 (2.6%) | 11 (6.2%) | 7 (3.6%) | 4 (2.7%) | 2 (1.3%) | 4 (5.6%) | 2 (2.3%) | 108 (32.0) | 49 (17.2) |
| hsCRP (mg/L)f | 2.92 (5.53) | 2.83 (5.41) | 4.16 (7.48) | 3.48 (6.55) | 2.56 (4.30) | 2.46 (4.29) | 1.38 (1.24) | 2.25 (4.50) | ||
| TNF-αR2 (pg/mL)g | 2602 (677) | 2546 (645) | 2812 (691) | 2717 (640) | 2619 (665) | 2582 (698) | 2236 (505) | 2234 (455) | ||
| IL-6 (pg/mL)h | 2.2 (4.2) | 1.9 (3.3) | 2.7 (5.2) | 2.0 (4.8) | 1.8 (2.8) | 1.5 (1.0) | 1.9 (3.9) | 2.1 (2.5) | ||
| Adiponectin (ng/mL)i | 6674 (4759) | 6761 (4098) | 8716 (6080) | 8033 (4863) | 5158 (2464) | 6135 (3592) | 5380 (3248) | 5544 (2609) | ||
SD: standard deviation, BMI: body mass index, hsCRP: high sensitivity C-reactive protein, TNF-αR2: tumor necrosis factor-receptor, IL-6: interleukin-6.
11 missing values;
missing values for cohorts: 28, and PanGenEU: 10;
missing values for cohorts: 6, and PanGenEU: 7;
missing values for cohorts: 14, and PanGenEU: 44;
missing values for cohorts: 1, and PanGenEU: 4;
336 missing values;
349 missing values;
341 missing values; no data for PanGenEU for serum biomarkers.
Statistical analyses
All statistical analyses were performed in R (version 3.5.1). Immune cell ratios (e.g., CD4/CD8, neutrophil/lymphocyte, B cell/lymphocyte, T cell/lymphocyte) were calculated for each sample by taking the ratio of its predicted cell proportions described above. Quartiles were assigned according to distribution of immune cell ratios among controls. A series of unconditional multivariable logistic regression models were used to evaluate the association between immune cell ratio and pancreatic cancer case/control status (unconditional models were selected to maximize power by including controls without matched cases; results using conditional regression models were compared and no differences were observed for the ORs). Age at blood draw, cohort, smoking status (never, former, current), and date of blood draw (continuous) were adjusted for in each model. To minimize loss of cases/controls due to missing data, we did not include BMI as a covariate in the model; moreover, including BMI in sensitivity analyses did not alter associations (including associations with CpGs). Similar models were used to examine the association between inflammation-associated CpG sites (modeled as quartiles; study specific) and pancreatic cancer case/control status. In addition to adjusting for previously mentioned covariates, these models were additionally adjusted for cell composition (e.g., estimated proportions of CD4T, CD8T, NK, B cell and monocytes) given the potential for confounding by cell composition (18). Conditional and unconditional models were similar for the CpG analyses as well (only unconditional analyses are presented).
For the nested case-control study, Spearman’s rank correlation was used to calculate the correlation between methylation beta-values and C-reactive protein, IL-6 and TNF-alpha(7) (Supplemental Table 1), as the biomarker and methylation beta-values were not always normally distributed. Correlations between methylation beta-values of inflammation CpG probes were also estimated using Spearman’s rank correlation (Supplemental Figure 1).
We examined the association between survival time (calculated from date of cancer diagnosis to date of death or end of follow-up) and both immune cell ratios and the 50 inflammation CpGs among cases in the cohort studies using a series of multivariable Cox proportional hazard models. Age at blood draw, cohort, smoking status, date of blood draw, and time between blood draw and cancer diagnosis were adjusted for in the Cox proportional hazard models. Models testing for associations with inflammation-related CpG sites were additionally adjusted for estimated cell composition as described above. Associations with methylation levels were tested using tertiles and trends were tested using continuous variables. All deaths were included (overall survival analysis); however, the majority of deaths would most likely have been a result of pancreatic cancer.
Results
Characteristics of the participants included in this analysis are provided in Table 1; due to matching criteria in the cohorts, age and smoking status were similar in cases and controls. On average, participants in the nested case-control study were diagnosed with pancreatic cancer at 60.6 years old and provided blood samples an average of 13 years (range 6 months to 26 years) prior to diagnosis (Table 1 presents range for each study). Those who later developed pancreatic cancer had a slightly higher BMI than those who did not develop pancreatic cancer (BMI 26.0 vs 25.6 kg/m2, respectively), and 4.8% of cases had diabetes, compared to 2.6% of controls. Inflammatory markers at blood draw were not substantially different between cases and controls in each cohort, as previously reported (7). Pancreatic cancer cases from the PanGenEU study were older (mean 66.3 years old), and prevalence of current smoking and diabetes mellitus was also higher in that study (Table 1).
Immune cell proportions and pancreatic cancer risk
In the nested case-control study, immune cell proportions estimated from DNA methylation data did not vary by case-control status (Supplemental Figure 2). Furthermore, immune cell ratios for CD4/CD8, NLR, B-cell/lymphocyte, T-cell/lymphocyte, and monocyte/lymphocyte were not associated with risk of pancreatic cancer (Table 2). Associations were similar across cohorts, and among cases, the NLR remained stable as time from blood draw to diagnosis decreased (including blood draw ≤5 years prior to diagnosis). This analysis could not be conducted in the PanGenEU study as the DNA methylation was performed on granulocytes only (i.e, primarily neutrophils).
Table 2.
Odds ratios (OR) for immune cell ratio and pancreatic cancer risk in cohorts (nested case-control study).
| Age-adjusted Model |
Multivariate-adjusted Modela |
|||
|---|---|---|---|---|
| Cases/Controls | OR (95% CI) | Case/Controls | OR (95% CI) | |
| CD4/CD8 Ratio | ||||
| Q1 (< 1.25) | 97 / 108 | ref. | 96 / 108 | ref. |
| Q2 (1.26 – 1.88) | 101 / 107 | 1.06(0.72, 1.56) | 100 / 107 | 1.07(0.72, 1.58) |
| Q3 (1.89 – 2.73) | 91 / 108 | 0.95(0.64, 1.41) | 90 / 106 | 0.97(0.65, 1.45) |
| Q4 (≥ 2.74) | 104 / 108 | 1.08(0.73, 1.58) | 104 / 107 | 1.10(0.74, 1.62) |
| p for continuous = 0.84 | p for continuous = 0.76 | |||
| Neutrophil/Lymphocyte Ratio | ||||
| Q1 (< 1.29) | 97 / 108 | ref. | 97 / 108 | ref. |
| Q2 (1.30 – 1.69) | 82 / 107 | 0.85(0.57, 1.27) | 82 / 106 | 0.85(0.57, 1.27) |
| Q3 (1.70 – 2.26) | 108 / 108 | 1.11(0.76, 1.63) | 106 / 107 | 1.09(0.74, 1.61) |
| Q4 (≥ 2.27) | 106 / 108 | 1.09(0.75, 1.60) | 105 / 107 | 1.10(0.75, 1.61) |
| p for continuous = 0.40 | p for continuous = 0.41 | |||
| B cell /Lymphocyte Ratio | ||||
| Q1 (< 0.10) | 96 / 108 | ref. | 96 / 106 | ref. |
| Q2 (0.11 – 0.13) | 84 / 107 | 0.89(0.60, 1.32) | 83 / 106 | 0.87(0.59, 1.30) |
| Q3 (0.14 – 0.17) | 100 / 108 | 1.05(0.71, 1.55) | 100 / 108 | 1.04(0.71, 1.54) |
| Q4 (≥ 0.18) | 113 / 108 | 1.19(0.81, 1.75) | 111 / 108 | 1.15(0.78, 1.71) |
| p for continuous = 0.26 | p for continuous = 0.34 | |||
| T cell/Lymphocyte Ratio | ||||
| Q1 (< 0.58) | 103 / 108 | ref. | 102 / 108 | ref. |
| Q2 (0.59 – 0.64) | 95 / 107 | 0.94(0.64, 1.38) | 93 / 106 | 0.94(0.63, 1.39) |
| Q3 (0.65 – 0.69) | 101 / 108 | 0.99(0.68, 1.46) | 101 / 108 | 1.01(0.69, 1.50) |
| Q4 (≥ 0.70) | 94 / 108 | 0.93(0.63, 1.37) | 94 / 106 | 0.97(0.65, 1.44) |
| p for continuous = 0.78 | p for continuous = 0.97 | |||
Adjusted for age, cohort, date of blood draw, and smoking.
Inflammation-linked CpGs and pancreatic cancer risk
50 CpG sites whose methylation extents were previously associated with inflammatory markers were examined in relation to pancreatic cancer risk. Many of the CpGs examined were strongly correlated with CRP and IL-6 levels in our dataset (68% and 60%, respectively, of correlations were > |0.10| and statistically significant), but correlations were somewhat weaker for TNF-alphaR2 (Supplemental Table 1). In the nested case-control study, the methylation extents of 2 CpG sites (cg05304729 and cg06192883) were strongly associated with risk of pancreatic cancer (p<0.01 for continuous, without adjustment for multiple comparisons; Table 3). For cg05304729, associations with pancreatic cancer risk were much stronger when blood draw was closer to diagnosis (3-fold higher risk in top quartile vs bottom quartile for 0–5 and 5–10 years compared to 1.6-fold higher risk when blood was collected more than 10 years prior to diagnosis; Table 3).
Table 3.
Odds ratios for inflammatory-related CpGs and pancreatic cancer risk identified in the nested case-control study, stratified by study and time to diagnosis
| cg05304729 | cg06192883 | |||
|---|---|---|---|---|
| Cases / Controls | Multivariate OR (95% CI)a | Cases / Controls | Multivariate OR (95% CI)a | |
| Among All Cohorts | ||||
| Q1 | 72 / 108 | ref. | 75 / 107 | ref. |
| Q2 | 93 / 107 | 1.37(0.90, 2.07) | 91 / 107 | 1.21(0.79, 1.83) |
| Q3 | 100 / 105 | 1.52(1.00, 2.31) | 84 / 106 | 1.12(0.73, 1.73) |
| Q4 | 125 / 108 | 1.92(1.27, 2.91) | 140 / 108 | 1.82(1.18, 2.78) |
| p for continuous = 0.002 | p for continuous = 0.008 | |||
| Among Time to Diagnosis ≤ 5 Years | ||||
| Q1 | 6 / 108 | ref. | 6 / 107 | ref. |
| Q2 | 12 / 107 | 2.15(0.75, 6.16) | 11 / 107 | 1.83(0.63, 5.33) |
| Q3 | 12 / 105 | 2.14(0.74, 6.17) | 12 / 106 | 1.80(0.62, 5.24) |
| Q4 | 20 / 108 | 3.37(1.23, 9.18) | 21 / 108 | 2.57(0.92, 7.21) |
| p for continuous = 0.02 | p for continuous = 0.09 | |||
| Among Time to diagnosis 5 – 10 Years | ||||
| Q1 | 11 / 108 | ref. | 15 / 107 | ref. |
| Q2 | 26 / 107 | 2.75(1.25, 6.07) | 23 / 107 | 1.68(0.79, 3.60) |
| Q3 | 18 / 105 | 2.27(0.97, 5.29) | 15 / 106 | 1.06(0.46, 2.45) |
| Q4 | 28 / 108 | 3.34(1.48, 7.54) | 30 / 108 | 1.87(0.85, 4.10) |
| p for continuous = 0.01 | p for continuous = 0.25 | |||
| Among Time to diagnosis > 10 Years | ||||
| Q1 | 51 / 108 | ref. | 51 / 107 | ref. |
| Q2 | 53 / 107 | 1.11(0.68, 1.79) | 55 / 107 | 1.08(0.67, 1.74) |
| Q3 | 66 / 105 | 1.38(0.85, 2.21) | 56 / 106 | 1.13(0.69, 1.86) |
| Q4 | 76 / 108 | 1.64(1.02, 2.64) | 84 / 108 | 1.67(1.03, 2.72) |
| p for continuous = 0.03 | p for continuous = 0.03 | |||
| Among NHSb | ||||
| Q1 | 44 / 49 | ref. | 35 / 49 | ref. |
| Q2 | 39 / 48 | 0.88(0.48, 1.63) | 32 / 48 | 0.82(0.43, 1.57) |
| Q3 | 45 / 48 | 1.07(0.58, 1.98) | 42 / 48 | 1.09(0.57, 2.06) |
| Q4 | 47 / 49 | 1.04(0.56, 1.96) | 66 / 49 | 1.53(0.80, 2.95) |
| p for continuous = 0.75 | p for continuous = 0.11 | |||
| Among HPFSb | ||||
| Q1 | 20 / 38 | ref. | 31 / 37 | ref. |
| Q2 | 32 / 37 | 1.79(0.86, 3.73) | 44 / 36 | 1.49(0.74, 3.01) |
| Q3 | 35 / 36 | 1.95(0.92, 4.11) | 26 / 37 | 0.82(0.38, 1.76) |
| Q4 | 57 / 37 | 3.44(1.67, 7.12) | 43 / 38 | 1.33(0.64, 2.77) |
| p for continuous <0.001 | p for continuous = 0.85 | |||
| Among PHSb | ||||
| Q1 | 9 / 22 | ref. | 7 / 22 | ref. |
| Q2 | 22 / 21 | 2.58(0.94, 7.11) | 20 / 21 | 3.49(1.16, 10.55) |
| Q3 | 19 / 21 | 2.49(0.89, 7.01) | 24 / 21 | 5.07(1.66, 15.47) |
| Q4 | 21 / 22 | 2.62(0.93, 7.36) | 20 / 22 | 3.65(1.20, 11.10) |
| p for continuous = 0.12 | p for continuous = 0.03 | |||
|
Replication in PanGenEUb,c | ||||
| Q1 | 50 / 71 | ref. | 72 / 71 | ref |
| Q2 | 77 / 71 | 1.49(0.9, 2.49) | 60 / 71 | 0.82(0.49, 1.35) |
| Q3 | 86 / 71 | 1.48(0.88, 2.48) | 94 / 71 | 1.23(0.77, 1.98) |
| Q4 | 126 / 72 | 2.08(1.22, 3.57) | 112 / 72 | 1.33(0.80, 2.21) |
| p for continuous = 0.01 | p for continuous = 0.11 | |||
Adjusted for age, date of blood draw, smoking and cell proportions, and cohorts for combined analyses.
Used study-specified quartiles for methylation level;
PanGenEU model adjustments include age, sex, smoking and cell proportions.
The associations were consistently positive in at least 2 of the 3 cohorts, but weaker in the PHS and NHS cohorts, possibly because those two cohorts had more cases that were diagnosed more than 10 years after blood draw (Table 3). Similar associations were noted in overweight or normal weight participants for both CpG sites, indicating that not all the association was due to obesity.
The positive trend for methylation extent of cg05304729 was replicated in the PanGenEU study (where blood was collected after diagnosis) and a significant test for trend was observed (p=0.01), with a 2-fold increase in risk in the highest quartile of DNA methylation (Table 3). The associations were similar in men and women in the PanGenEU study (and statistically significant for each sex; females p=0.01 and males p=0.03). In contrast, methylation level at cg06192883 was not associated with pancreatic cancer risk in the PanGenEU study (Table 3). Statistically significant results for the inflammation CpGs in the PanGenEU study are provided in Supplemental Table 2.
Survival analysis
We also examined whether the immune cell ratios were associated with survival time among the cases in the nested case-control study (Table 4). Overall, the immune ratio measures were not associated with survival time, and associations were similar when stratifying on time between blood collection and date of diagnosis. Among the 50 CpGs tested, methylation level of 6 CpGs were statistically significant associated with overall survival at p≤0.05 (cg00159243, cg03957124, cg12785694, cg1818703, cg25325512, cg26804423; Table 4). Methylation level at two of these CpGs (cg00159243, cg25325512) was significantly associated with risk in PanGenEU (Supplemental Table 2), and methylation of cg25325512 was also associated with survival in PanGenEU (Q2 vs Q1: HR= 0.71, 95% CI 0.52–0.96; Q3 vs Q1: HR 0.72, 95% CI 0.51–1.00, p-continuous 0.057). Overall survival curves for methylation levels at this CpG in the cases from the cohort studies are presented in Figure 1. The ICC for cg25325512 was 0.86 in our pilot study (over a 1-year period), suggesting that methylation at this probe does not vary much over time, and thus provides a valid proxy for levels closer to diagnosis.
Table 4.
Association between immune cell counts ratio, inflammatory-related CpGs and overall survival time among cases from cohorts only (n = 342)
| Multivariate HR (95% CI) US cohorts |
|
|---|---|
| CD4/CD8 Ratioa | |
| Q1 [0.27, 1.45) | ref. |
| Q2 [1.48, 2.38) | 1.08 (0.82, 1.41) |
| Q3 [2.39, 32.33] | 0.96 (0.74, 1.26) |
| p for continuous = 0.79 | |
| Neutrophil/Lymphocyte Ratioa | |
| Q1 [0.54, 1.49) | ref. |
| Q2 [1.49, 2.09) | 0.88 (0.68, 1.16) |
| Q3 [2.09, 8.63] | 1.08 (0.83, 1.41) |
| p for continuous = 0.57 | |
| B cell /Lymphocyte Ratioa | |
| Q1 [0.01, 0.13) | ref. |
| Q2 [0.13, 0.17) | 1.13 (0.87, 1.48) |
| Q3 [0.17, 0.43] | 1.14 (0.86, 1.51) |
| p for continuous = 0.37 | |
| T cell /Lymphocyte Ratioa | |
| Q1 [0.38, 0.60) | ref. |
| Q2 [0.61, 0.68) | 0.85 (0.65, 1.10) |
| Q3 [0.68, 0.88] | 0.99 (0.76, 1.30) |
| p for continuous = 0.95 | |
| cg00159243a, b | |
| Q1 [0.24, 0.33) | ref. |
| Q2 [0.33, 0.37) | 1.29 (0.96, 1.74) |
| Q3 [0.37, 0.45] | 1.42 (1.00, 2.02) |
| p for continuous = 0.049 | |
| cg03957124a, b | |
| Q1 [0.40, 0.53) | ref. |
| Q2 [0.53, 0.58) | 0.81 (0.60, 1.08) |
| Q3 [0.58, 0.69] | 0.63 (0.42, 0.93) |
| p for continuous = 0.02 | |
| cg12785694a, b | |
| Q1 [0.07, 0.15) | ref. |
| Q2 [0.15, 0.20) | 1.00 (0.75, 1.34) |
| Q3 [0.20, 0.42] | 1.42 (1.02, 1.99) |
| p for continuous = 0.04 | |
| cg18181703a, b | |
| Q1 [0.34, 0.45) | ref. |
| Q2 [0.45, 0.50) | 0.92 (0.69, 1.22) |
| Q3 [0.50, 0.58] | 0.72 (0.54, 0.96) |
| p for continuous = 0.03 | |
| cg25325512a, b | |
| Q1 [0.25, 0.37) | ref. |
| Q2 [0.37, 0.42) | 0.90 (0.68, 1.18) |
| Q3 [0.42, 0.55] | 0.66 (0.49, 0.88) |
| p for continuous = 0.004 | |
| cg26804423a, b | |
| Q1 [0.61, 0.70) | ref. |
| Q2 [0.70, 0.74) | 1.13 (0.84, 1.52) |
| Q3 [0.74, 0.83] | 1.50 (1.04, 2.17) |
| p for continuous = 0.03 |
Adjusted for age, date of blood draw, time between blood draw and cancer diagnosis, smoking, and cohorts for combined analyses.
Further adjusted for cell proportions.
Figure 1.
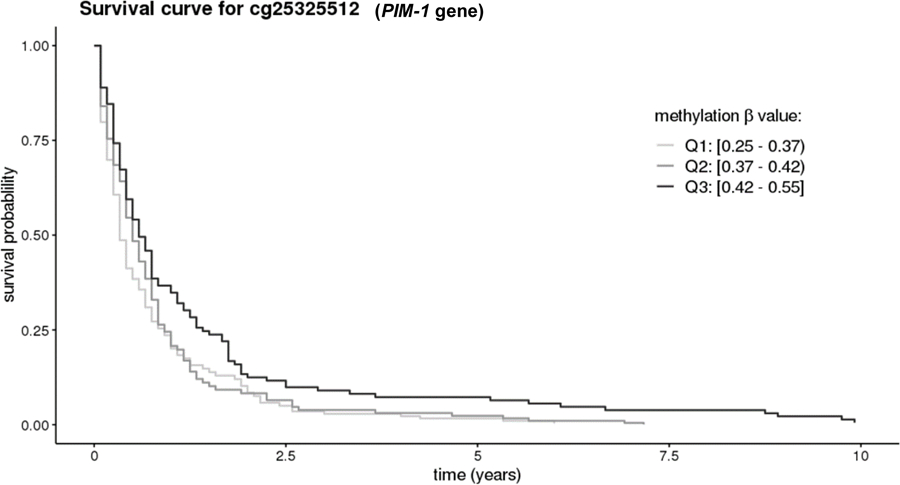
Overall survival curves among pancreatic cancer cases in the nested case-control study for the the CpG in PIM-1 promoter. Results for this CpG were consistent in the nested case-control study and PanGenEU. Curves are adjusted age, date of blood draw, time between blood draw and diagnosis, smoking, cohorts, and immune cell proportions.
Discussion
To our knowledge, this is the first study to examine associations between CpG methylation of inflammation markers, methylation derived immune cell composition, and risk of pancreatic cancer using pre-diagnostic blood samples. One of the goals of this study was to measure immune cell proportions in blood samples using established DNA methylation markers of immune cell types as flow cytometry could not be conducted on archived frozen blood. While we did not find any associations for ratios of immune cell proportions and risk of pancreatic cancer, we did identify and replicate an association with the DNA methylation level of a CpG previously associated with inflammation. We also identified an association with DNA methylation markers of inflammation and overall survival, but found no association for NLR and survival.
Our results do not provide support for an association between immune cell proportions and risk of pancreatic cancer or for overall survival. While no previous study had examined immune cell proportions and pancreatic cancer risk, numerous studies have reported a decrease in survival among pancreatic cancer cases with higher NLR (6). The difference between our findings and those from prior studies may be due to changes in cell proportions that occur closer to cancer diagnosis, rather than several years prior to diagnosis. The NLR analysis could not be performed in the cases from PanGenEU as DNA methylation was only measured in granulocytes.
Epigenetic-wide association studies (EWAS) using Illumina arrays to identify methylation sites associated with inflammatory blood markers have been carried out in two large studies (16,17). We selected 50 CpG sites that had met criteria for inclusion in this analysis (see Methods) and identified two (cg05304729 and cg06192883) that were statistically significantly associated with pancreatic cancer risk in the nested case-control study overall. For cg05304729, the associations were stronger as the collection of blood samples got closer to date of diagnosis, suggesting the inflammation increases closer to diagnosis, perhaps due to subclinical changes. The fact that the association was present more than 10 years prior to cancer diagnosis (Q4 vs Q1 OR =1.64, 95% CI = 1.02, 2.64; Table 3) suggests that the methylation level at that site is related to risk, rather than being sole consequence of the cancer. However, it is also noteworthy that the strength of the association increased as the time to diagnosis was shortened and that results were also observed in PanGenEU where blood collected was obtained at diagnosis. However, we did not observe an association for cg06192883 in PanGenEU.
Previous studies have reported strong associations between methylation at cg05304729 and levels of three different inflammation markers measured in blood (CXCL9(16), CXCL11(16) and TNFRSF6B(19)). In our study, methylation at cg05304729 was not correlated with CRP, TNFαR2, or IL-6 (Supplemental Table 1); the difference between the prior studies and our study might have been due to differences in inflammation markers measured. DNA methylation at both CpG sites have also been associated with BMI,(19) out of the 102 CpG sites tested in Myte et al., the two CpG sites identified in our current study were among the three most statistically significant associations with BMI in the prior study (p-values =0.0001). In addition, cg05304729 was identified as 1 of 20 probes associated with BMI in a separate EWAS study (FDR q = 0.015)(20) and cg06192883 was identified in another EWAS study on BMI (21). Given the known role of BMI in pancreatic cancer risk, the DNA methylation sites identified in this study may provide insight into the underlying biological pathways involved; importantly, the positive associations were also observed among subjects with normal BMI. Cg05304729 is located 200–1500 bases upstream of the transcriptional start site (Illumina annotation: TSS1500) for the myeloid nuclear differentiation antigen (MNDA) gene; expression of this gene has been previously associated with lymphoma, especially marginal zone derived lymphomas (22). This gene may also be involved in cell-specific response to interferons (23). More research will be necessary to understand the role of these pathways in pancreatic cancer.
Conducting a survival analysis, we identified methylation level for two CpG sites (cg00159243, cg25325512) that were significantly associated with overall survival in the nested case-control study (p≤0.05), and significantly associated with risk in PanGenEU (p<0.05). However, only the extent of methylation of cg25325512 was also associated with survival in PanGenEU (p=0.057). Cg25325512 is located on gene PIM1, an well-established oncogene (24) that has been widely targeted for anticancer drug discovery(25). Some studies have shown that high PIM-1 expression in pancreatic tumor tissue is associated with worse survival and, in a recent study, plasma PIM-1 level was associated with pancreatic cancer survival (HR = 1.87, 95% CI = 1.04–3.35) and risk (p<0.0001)(26). Given the implication of this finding, we went back to examine whether the association with risk existed in the nested case-control study (i.e., including controls); although the p-continuous was not significant, the highest quartile was borderline significant (HR = 0.68, 95% CI = 0.44 – 1.05, compared to the lowest) overall, and significant when blood was collected 10 years prior to diagnosis (HR = 0.55, 95% CI = 0.34 – 0.91, top to bottom quartile comparison). This finding is particularly interesting as it suggests DNA methylation at this site occurred many years prior to diagnosis and thus is not likely to be caused by the tumor development.
Our study strengths include use of pre-diagnostic blood and a large number of incident pancreatic cancer cases. Pre-diagnostic blood collection is critical to determine whether methylation states at different CpG sites were present prior to diagnosis, rather than identifying changes that might have occurred as a result of the cancer. By ruling out reverse causation, we could begin to identify pathways that play a role in the etiology of the disease but also identify early diagnosis markers. Being able to examine associations in a separate case-control study (PanGenEU) was an additional strength to this analysis as it provided an opportunity to evaluate the robustness of our findings in a completely different population, providing strong evidence of reproducibility. Other strengths of this study included adjustment for potential confounders, including age, race, smoking, BMI, and diabetes. Moreover, our data processing steps and random assignment of samples on plates removed potential technical biases.
Study limitations include our reliance on established DNA methylation markers for immune cell types, which are primarily limited to the main immune cell types. Subsets of immune cells that are more difficult to identify and may play a role in cancer, such as regulatory T-cells, could be associated with cancer risk, but were not available for this analysis. The EWAS results from this project are being published separately and represent an agnostic analysis versus this approach which was hypothesis driven.
This is the first prospective study examining the associations between immune cell proportions and risk of pancreatic cancer. While we did not observe associations with risk for several main known indicators of immune status previously associated with survival, such as NLR, we identified two CpGs that have been strongly associated with inflammation and BMI in prior studies. More research on MNDA and PIM-1 genes may reveal new area of research for pancreatic cancer risk, given that these genes have been previously implicated in other cancers, and PIM-1 expression has previously been associated with lower pancreatic cancer survival. Further research based on our findings may lead to identification of novel proteins that are differentially expressed prior to cancer diagnosis that could be tested in blood for early detection or for the identification of individuals at higher risk (without the need for DNA methylation measurements). Alternatively, our findings could lead to identification of pathways that may be targetable for treatment.
Supplementary Material
Acknowledgments
PanGenEU centers and investigators
Spanish National Cancer Research Centre (CNIO), Madrid, Spain: Núria Malats1, Francisco X Real1, Evangelina López de Maturana, Paulina Gómez-Rubio, Esther Molina-Montes, Lola Alonso, Mirari Márquez, Roger Milne, Ana Alfaro, Tania Lobato, Lidia Estudillo.
Verona University, Italy: Rita Lawlor1, Aldo Scarpa, Stefania Beghelli.
National Cancer Registry Ireland, Cork, Ireland: Linda Sharp1, Damian O’Driscoll.
Hospital Madrid-Norte-Sanchinarro, Madrid, Spain: Manuel Hidalgo1, Jesús Rodríguez Pascual.
Hospital Ramon y Cajal, Madrid, Spain: Alfredo Carrato1, Carmen Guillén-Ponce, Mercedes Rodríguez-Garrote, Federico Longo-Muñoz, Reyes Ferreiro, Vanessa Pachón, M Ángeles Vaz.
Hospital del Mar, Barcelona, Spain: Lucas Ilzarbe1, Cristina Álvarez-Urturi, Xavier Bessa, Felipe Bory, Lucía Márquez Mosquera, Ignasi Poves Prim, Fernando Burdío, Luis Grande, Mar Iglesias, Javier Gimeno.
Hospital Vall d´Hebron, Barcelona, Spain: Xavier Molero1, Luisa Guarner✝, Joaquin Balcells.
Technical University of Munich, Germany: Christoph Michalski1, Jörg Kleeff, Bo Kong.
Karolinska Institute, Stockholm, Sweden: Matthias Löhr1, Jiaqui Huang, Weimin Ye, Jingru Yu.
Hospital 12 de Octubre, Madrid, Spain: José Perea1, Pablo Peláez.
Hospital de la Santa Creu i Sant Pau, Barcelona, Spain: Antoni Farré1, Josefina Mora, Marta Martín, Vicenç Artigas, Carlos Guarner Argente, Francesc J Sancho, Mar Concepción, Teresa Ramón y Cajal.
The Royal Liverpool University Hospital, UK: William Greenhalf1, Eithne Costello.
Queen’s University Belfast, UK: Michael O’Rorke1, Liam Murray✝, Marie Cantwell.
Laboratorio de Genética Molecular, Hospital General Universitario de Elche, Spain: Víctor M Barberá1, Javier Gallego.
Instituto Universitario de Oncología del Principado de Asturias, Oviedo, Spain: Adonina Tardón1, Luis Barneo.
Hospital Clínico Universitario de Santiago de Compostela, Spain: Enrique Domínguez Muñoz1, Antonio Lozano, Maria Luaces.
Hospital Clínico Universitario de Salamanca, Spain: Luís Muñoz-Bellvís1, J.M. Sayagués Manzano, M.L. Gutíerrrez Troncoso, A. Orfao de Matos.
University of Marburg, Department of Gastroenterology, Phillips University of Marburg, Germany: Thomas Gress1, Malte Buchholz, Albrecht Neesse.
Queen Mary University of London, UK: Tatjana Crnogorac-Jurcevic1, Hemant M Kocher, Satyajit Bhattacharya, Ajit T Abraham, Darren Ennis, Thomas Dowe, Tomasz Radon
Scientific advisors of the PanGenEU Study: Debra T Silverman (NCI, USA) and Douglas Easton (U. of Cambridge, UK)
Financial Support:
The research reported in this publication was primarily supported by the NIH/National Cancer Institute grant R01 CA207110. In addition, other NIH funds contributed to the support of the investigators: P30 CA168525, and the Kansas IDeA Network of Biomedical Research Excellence Bioinformatics Core, supported in part by the National Institute of General Medical Science (NIGMS) Award P20GM103418.
The PanGenEU study was funded by: Fondo de Investigaciones Sanitarias (FIS), Instituto de Salud Carlos III, Spain (#PI11/01542, #PI0902102, #PI12/01635, #PI12/00815, #PI15/01573); European Cooperation in Science and Technology – COST Action #BM1204; EUPancreas EU-6FP Integrated Project (#018771-MOLDIAG-PACA), EU-FP7-HEALTH (#259737-CANCERALIA, #256974-EPC-TM-Net).
Footnotes
Principal Investigator in each center
Conflict of Interest
The authors have no conflicts of interests.
Data Availability Statement
All data from this study have been deposited in dbGAP and will be available on January 3, 2020 [“DNA Methylation Markers and Pancreatic Cancer Risk in 3 Cohort Studies (NHS, PHS, HPFS)” phs001917.v1.p1]. https://www.ncbi.nlm.nih.gov/projects/gapprev/gap/cgi-bin/study.cgi?study_id=phs001917.v1.p1
References
- 1.ACS. Cancer Facts & Figures 2020. Atlanta: American Cancer Society, Inc.; 2020. [Google Scholar]
- 2.Force USPST, Owens DK, Davidson KW, Krist AH, Barry MJ, Cabana M, et al. Screening for Pancreatic Cancer: US Preventive Services Task Force Reaffirmation Recommendation Statement. Jama 2019;322(5):438–44 doi 10.1001/jama.2019.10232. [DOI] [PubMed] [Google Scholar]
- 3.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC bioinformatics 2012;13:86 doi 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiencke JK, Koestler DC, Salas LA, Wiemels JL, Roy RP, Hansen HM, et al. Immunomethylomic approach to explore the blood neutrophil lymphocyte ratio (NLR) in glioma survival. Clinical epigenetics 2017;9:10 doi 10.1186/s13148-017-0316-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dolan RD, Laird BJA, Horgan PG, McMillan DC. The prognostic value of the systemic inflammatory response in randomised clinical trials in cancer: A systematic review. Critical reviews in oncology/hematology 2018;132:130–7 doi 10.1016/j.critrevonc.2018.09.016. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Y, Wei Q, Fan J, Cheng S, Ding W, Hua Z. Prognostic role of the neutrophil-to-lymphocyte ratio in pancreatic cancer: A meta-analysis containing 8252 patients. Clinica chimica acta; international journal of clinical chemistry 2018;479:181–9 doi 10.1016/j.cca.2018.01.024. [DOI] [PubMed] [Google Scholar]
- 7.Bao Y, Giovannucci EL, Kraft P, Qian ZR, Wu C, Ogino S, et al. Inflammatory plasma markers and pancreatic cancer risk: a prospective study of five U.S. cohorts. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2013;22(5):855–61 doi 10.1158/1055-9965.EPI-12-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomez-Rubio P, Pinero J, Molina-Montes E, Gutierrez-Sacristan A, Marquez M, Rava M, et al. Pancreatic cancer and autoimmune diseases: An association sustained by computational and epidemiological case-control approaches. International journal of cancer Journal international du cancer 2019;144(7):1540–9 doi 10.1002/ijc.31866. [DOI] [PubMed] [Google Scholar]
- 9.Gomez-Rubio P, Rosato V, Marquez M, Bosetti C, Molina-Montes E, Rava M, et al. A systems approach identifies time-dependent associations of multimorbidities with pancreatic cancer risk. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO 2017;28(7):1618–24 doi 10.1093/annonc/mdx167. [DOI] [PubMed] [Google Scholar]
- 10.Gomez-Rubio P, Zock JP, Rava M, Marquez M, Sharp L, Hidalgo M, et al. Reduced risk of pancreatic cancer associated with asthma and nasal allergies. Gut 2017;66(2):314–22 doi 10.1136/gutjnl-2015-310442. [DOI] [PubMed] [Google Scholar]
- 11.Molina-Montes E, Gomez-Rubio P, Marquez M, Rava M, Lohr M, Michalski CW, et al. Risk of pancreatic cancer associated with family history of cancer and other medical conditions by accounting for smoking among relatives. International journal of epidemiology 2018;47(2):473–83 doi 10.1093/ije/dyx269. [DOI] [PubMed] [Google Scholar]
- 12.Moran S, Arribas C, Esteller M. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics 2016;8(3):389–99 doi 10.2217/epi.15.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaimi I, Pei D, Koestler DC, Marsit CJ, De Vivo I, Tworoger SS, et al. Variation in DNA methylation of human blood over a 1-year period using the Illumina MethylationEPIC array. Epigenetics 2018;13(10–11):1056–71 doi 10.1080/15592294.2018.1530008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salas LA, Koestler DC, Butler RA, Hansen HM, Wiencke JK, Kelsey KT, et al. An optimized library for reference-based deconvolution of whole-blood biospecimens assayed using the Illumina HumanMethylationEPIC BeadArray. Genome biology 2018;19(64) doi 10.1186/s13059-018-1448-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koestler DC, Jones MJ, Usset J, Christensen BC, Butler RA, Kobor MS, et al. Improving cell mixture deconvolution by identifying optimal DNA methylation libraries (IDOL). BMC bioinformatics 2016;17:120 doi 10.1186/s12859-016-0943-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahsan M, Ek WE, Rask-Andersen M, Karlsson T, Lind-Thomsen A, Enroth S, et al. The relative contribution of DNA methylation and genetic variants on protein biomarkers for human diseases. PLoS genetics 2017;13(9):e1007005 doi 10.1371/journal.pgen.1007005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ligthart S, Marzi C, Aslibekyan S, Mendelson MM, Conneely KN, Tanaka T, et al. DNA methylation signatures of chronic low-grade inflammation are associated with complex diseases. Genome biology 2016;17(1):255 doi 10.1186/s13059-016-1119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adalsteinsson BT, Gudnason H, Aspelund T, Harris TB, Launer LJ, Eiriksdottir G, et al. Heterogeneity in white blood cells has potential to confound DNA methylation measurements. PloS one 2012;7(10):e46705 doi 10.1371/journal.pone.0046705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myte R, Sundkvist A, Van Guelpen B, Harlid S. Circulating levels of inflammatory markers and DNA methylation, an analysis of repeated samples from a population based cohort. Epigenetics 2019:1–11 doi 10.1080/15592294.2019.1603962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu K, Zhang X, Wang Z, Hu Y, Sinha R. Epigenome-wide association analysis revealed that SOCS3 methylation influences the effect of cumulative stress on obesity. Biol Psychol 2018;131:63–71 doi 10.1016/j.biopsycho.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wahl S, Drong A, Lehne B, Loh M, Scott WR, Kunze S, et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature 2016;541:81 doi 10.1038/nature2078410.1038/nature20784https://www.nature.com/articles/nature20784#supplementary-informationhttps://www.nature.com/articles/nature20784#supplementary-information . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanellis G, Roncador G, Arribas A, Mollejo M, Montes-Moreno S, Maestre L, et al. Identification of MNDA as a new marker for nodal marginal zone lymphoma. Leukemia 2009;23(10):1847–57 doi 10.1038/leu.2009.108. [DOI] [PubMed] [Google Scholar]
- 23.Mondini M, Vidali M, Airo P, De Andrea M, Riboldi P, Meroni PL, et al. Role of the interferon-inducible gene IFI16 in the etiopathogenesis of systemic autoimmune disorders. Ann N Y Acad Sci 2007;1110:47–56 doi 10.1196/annals.1423.006. [DOI] [PubMed] [Google Scholar]
- 24.Nawijn MC, Alendar A, Berns A. For better or for worse: the role of Pim oncogenes in tumorigenesis. Nature reviews Cancer 2011;11(1):23–34 doi 10.1038/nrc2986. [DOI] [PubMed] [Google Scholar]
- 25.Blanco-Aparicio C, Carnero A. Pim kinases in cancer: diagnostic, prognostic and treatment opportunities. Biochemical pharmacology 2013;85(5):629–43 doi 10.1016/j.bcp.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 26.Xu J, Xiong G, Cao Z, Huang H, Wang T, You L, et al. PIM-1 contributes to the malignancy of pancreatic cancer and displays diagnostic and prognostic value. J Exp Clin Cancer Res 2016;35(1):133 doi 10.1186/s13046-016-0406-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


