SUMMARY
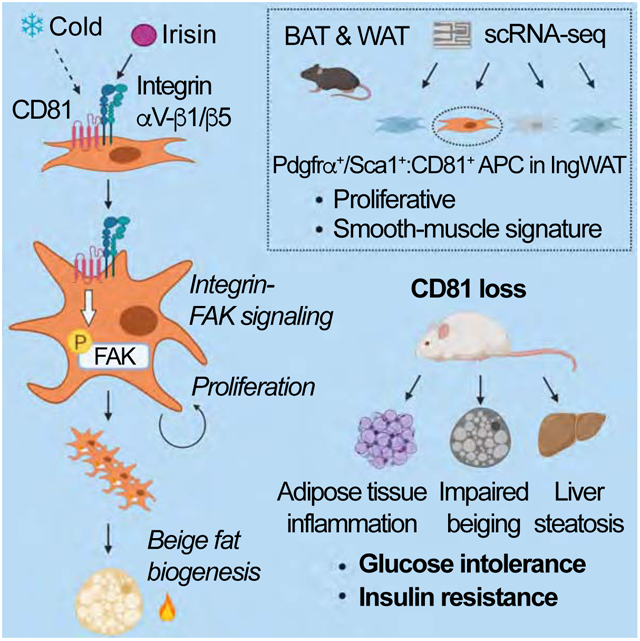
Adipose tissues dynamically remodel their cellular composition in response to external cues by stimulating beige adipocyte biogenesis; however, the developmental origin and pathways regulating this process remain insufficiently understood owing to adipose tissue heterogeneity. Here, we employed single-cell RNA-seq and identified a unique subset of adipocyte progenitor cells (APC) that possessed the cell-intrinsic plasticity to give rise to beige fat. This beige APC population is proliferative and marked by cell-surface proteins, including PDGFRα, Sca1, and CD81. Notably, CD81 is not only a beige APC marker, but also required for de novo beige fat biogenesis following cold exposure. CD81 forms a complex with αV/β1 and αV/β5 integrins, and mediates the activation of integrin-FAK signaling in response to irisin. Importantly, CD81 loss causes diet-induced obesity, insulin resistance, and adipose tissue inflammation. These results suggest that CD81 functions as a key sensor of external inputs and controls beige APC proliferation and whole-body energy homeostasis.
Keywords: Brown/beige fat, Adipocyte progenitors, Adipogenesis, Tissue remodeling, Metabolism, Metabolic disease
Graphical Abstract
INTRODUCTION
Adipose tissues comprise a dynamic metabolic organ that remodels the cellular size and composition in response to a variety of internal and external cues, including nutritional states and temperatures. Such metabolic adaptation that involves lipolysis, lipogenesis, adipogenesis, and thermogenesis, plays a central role in the regulation of energy homeostasis (Chouchani and Kajimura, 2019). A notable adaptive process within adipose tissues that has attracted particular attention is the “browning” or “beiging” of white adipose tissue (WAT), in which numerous mitochondria-enriched thermogenic adipocytes with multi-locular lipid droplets, a.k.a., beige adipocytes, emerge within WAT (Wu et al., 2012; Young et al., 1984). Importantly, adult humans possess beige fat: adult human BAT from the supraclavicular region contains mitochondria-enriched adipocytes that display a molecular signature resembling murine beige adipocytes (Lidell et al., 2013; Sharp et al., 2012; Shinoda et al., 2015a; Wu et al., 2012), and chronic cold acclimation or a β3-adrenergic receptor (β3-AR) agonist promotes the recruitment of new thermogenic fat (Finlin et al., 2018; van der Lans et al., 2013; Yoneshiro et al., 2013).
Emerging evidence suggests that increased beige fat biogenesis improves metabolic health in ways far beyond the induction of thermogenesis (Kajimura et al., 2015). For instance, selective activation of beige fat biogenesis improves systemic glucose tolerance and insulin sensitivity, reduces WAT inflammation and fibrosis, and protects against hepatic steatosis (Hasegawa et al., 2018; Ikeda et al., 2017; McDonald et al., 2015; Seale et al., 2011; Shinoda et al., 2015b). Conversely, impaired beige fat biogenesis results in insulin resistance, adipose tissue inflammation, fibrosis, and hepatic steatosis (Cohen et al., 2014; Wang et al., 2019). Thus, a better understanding of the regulatory circuits of beige fat biogenesis continues to be a significant area of research as it may lead to the development of therapeutic measures that improve metabolic health.
However, the developmental origins and regulation of beige fat are insufficiently understood. Lineage tracing analyses suggest that beige fat biogenesis involves both de novo differentiation from adipocyte progenitor cells (APC) and reinstatement of thermogenic activity in mature white adipocytes (Shao et al., 2019). As for de novo beige adipogenesis, previous studies report that progenitors expressing Acta2, Pax3, Pdgfra, or Pdgfrb in the stromal vascular fraction (SVF) of WAT give rise to beige adipocytes following cold exposure (Berry et al., 2016; Lee et al., 2012; Sanchez-Gurmaches and Guertin, 2014; Vishvanath et al., 2016). Of note, PDGFRα and Sca1 are expressed on the cell surface of adipose stromal cells that differentiate into both beige and white adipocytes (Berry and Rodeheffer, 2013; Lee et al., 2012; Schulz et al., 2011); however, both markers are also expressed in other stromal populations in adipose tissues and many organs. For example, a subset of PDGFRα+ cells in WAT expressing high levels of CD9 or ITGA5 are profibrogenic and drive adipose tissue fibrosis (Lin et al., 2018; Marcelin et al., 2017). Thus, it remains unclear what distinguishes beige APC from other stromal cell types. Accordingly, this study employed single-cell RNA-sequencing analysis (scRNA-seq) in order to identify beige APC.
RESULTS
CD81 is a cell surface molecule for a unique subset of APC.
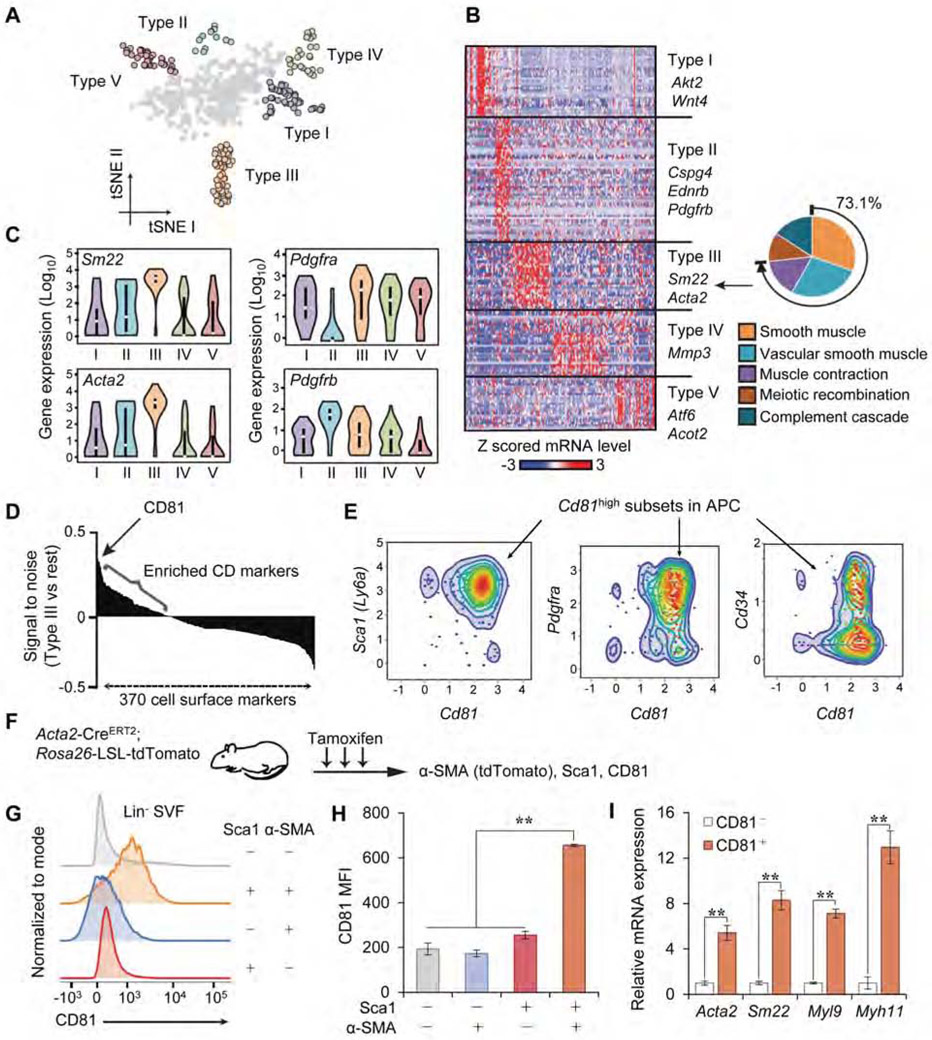
To characterize adipose stromal cells at single-cell resolution, we performed scRNA-seq analysis of lineage-negative (Lin−) stromal cells from murine interscapular BAT (iBAT), inguinal WAT (Ing WAT), and epididymal WAT (Epi WAT) (Figure S1A-D). We employed the Fluidigm C1 system that allowed for deeper sequencing (~1-2 million reads/cell) despite fewer number of cells, as compared to the 10X Genomics platform which has been applied to adipose tissues (Burl et al., 2018; Hepler et al., 2018; Merrick et al., 2019; Schwalie et al., 2018). This scRNA-seq analysis identified at least five distinct clusters (Types I-V) based on a tSNE map (Figure 1A). Among these groups, we found that stromal cells in the Type III cluster expressed a number of genes (73.1% of the total Type III-enriched genes) that were involved in vascular smooth muscle development and contraction (Figure 1B). For instance, violin plots showed that the Type III cells expressed high levels of Sm22 and Acta2 (encoding alpha-smooth muscle actin, α-SMA). Of note, most of these stromal cells, except Type II cells, expressed high levels of Pdgfra (Figure 1C). Instead, the majority of Type II cells expressed high levels of Pdgfrb and Ednrb, indicative of an endothelial signature (Hepler et al., 2018).
Figure 1. CD81 marks a subset of APC.
A) Classification of Lin stromal cells in mouse adipose tissues (Type I-V clusters) on a tSNE map.
B) Heat map of transcriptome in indicated cell clusters with representative genes. Right diagram:73.1% of the Type III cluster-enriched genes are involved in smooth muscle development and function.
C) Violin plots showing mRNA levels of indicated genes in Type I to V cell clusters.
D) Expression of cell surface genes, including CD81, that are enriched in Type III cluster.
E) Scatter plots showing the distribution of Cd81+ cells in Sca1+, Pdgfra+, or Cd34+ cells.
F) Illustration of the experiments in (G) and (H).
G) Distribution of CD81+ cells among Lin−: Sca1+: tdTomato+ cells from the Ing WAT of Acta2 reporter mice. n=3.
H) CD81 expression (mean fluorescence intensity, MFI) in indicated cells from mice in (G). **P < 0.01 by one-way ANOVA followed by Bonferroni’s post hoc test.
I) mRNA expression of smooth muscle-enriched genes in indicated cells from Ing WAT. CD81− cells, n=6; CD81+ cells, n=4. **P < 0.01 by unpaired Student’s t-test.
Data in (H, I) are represented as mean ± SEM.
This “vascular smooth muscle” gene signature in Type III cells caught our attention because stromal cells expressing Acta2 or Sm22 contribute to beige adipogenesis in Ing WAT (Berry et al., 2016; Long et al., 2014). To isolate live Type III cells by Fluorescence-Activated Cell Sorting (FACS), we first searched for cell surface markers that were enriched in this cluster (Figure 1D). Among 370 known cell surface molecules, several candidates, including Pdgfra, Ly6a (coding Sca1), and Cd81, were highly expressed in Type III cells relative to other groups (top 20 genes are listed in Table S1). Scatter plots from our scRNA-seq data showed that Cd81 was expressed in a subset of APCs that express Ly6a/Sca1 and Pdgfra (Figure 1E). A recent study reported that human CD34− APCs (Lin−: CD29+: CD34−) differentiate to beige adipocytes expressing high levels of Ucp1 and Cited1 (Raajendiran et al., 2019). Indeed, 57 % of the Cd81-expressing cells lacked Cd34 expression. Moreover, we analyzed a recently published scRNA-seq dataset of mouse APCs (Hepler et al., 2018) and validated the existence of a stromal population expressing high levels of Cd81, Sca1, Acta2, and Sm22 within this dataset (Figure S1E).
Accordingly, we aimed to isolate live Type III cells by using antibodies against these cell-surface markers, followed by FACS. To establish the method, we first isolated Lin− stromal cells expressing α-SMA in Ing WAT by using Acta2-CreERT2; Rosa26-LSL-tdTomato reporter mice that label all the α-SMA+ cells following tamoxifen (Figure 1F). We found that CD81 protein expression was significantly higher in Lin−: Sca1+: tdTomato+ (α-SMA+) cells than other populations (Figure 1G, H). Conversely, isolated CD81+ cells (Lin−: Sca1+: CD81+) from Ing WAT-derived SVFs expressed significantly higher levels of the smooth muscle-related genes, including Acta2, Sm22, Myl9, and Myh11, relative to CD81− cells (Lin−: Sca1+: CD81−) in Ing WAT (Figure 1I). Of note, the gene signature of isolated Lin− Sca1+ CD81+ cells from the Ing WAT resembled that of Type III cells (Figure S1F), and PDGFRα and Sca1 marked nearly the same CD81+ stromal population within the WAT of mice (Figure S1G). These data suggest that CD81 marks a subset of Sca1+ PDGFRα+ stromal cells possessing a smooth muscle-like signature, and serves as a useful cell surface marker to isolate the Type III cells from mouse WAT.
CD81+ stromal cells give rise to beige adipocytes in vivo.
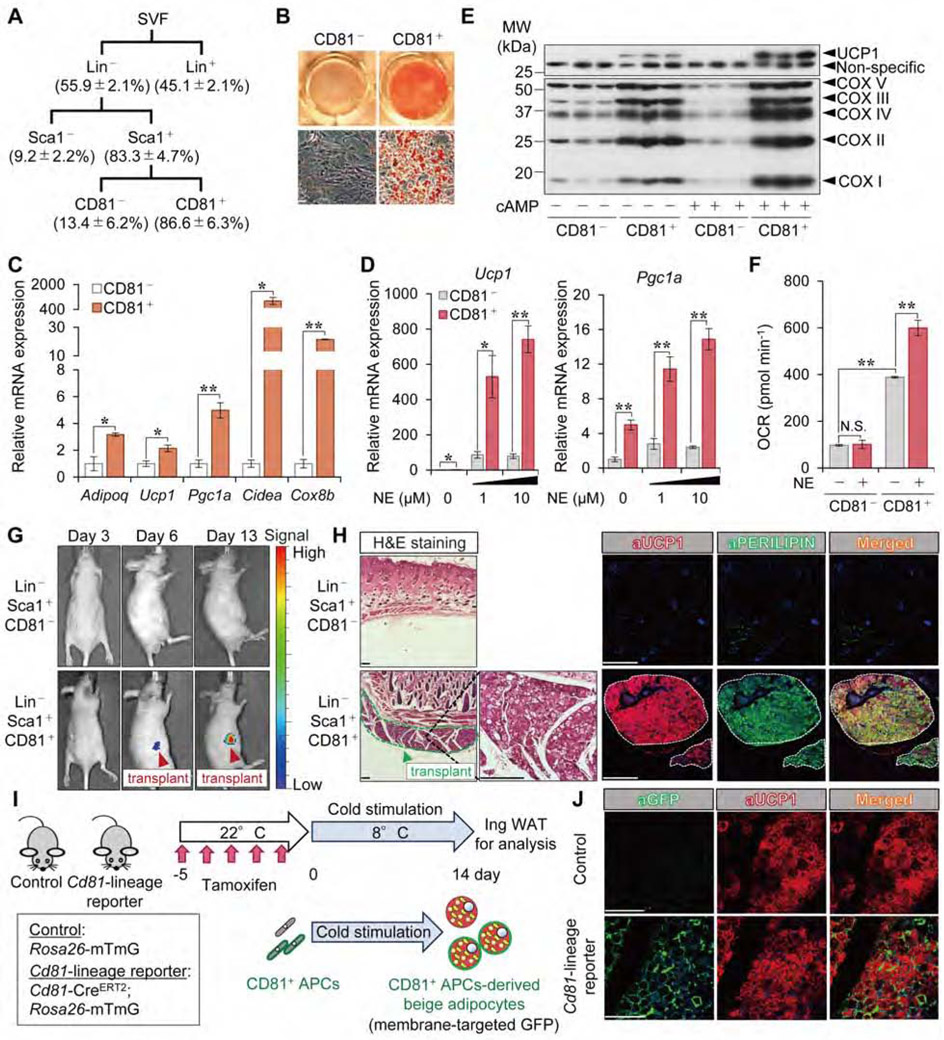
CD81, one of the tetraspanin family members, was originally identified as the target of an antiproliferative antibody for a human lymphoma cell line (Oren et al., 1990). To test if CD81 marks beige fat progenitors, we isolated CD81+ and CD81− cells from the SVF of mouse Ing WAT by FACS (Figure 2A). After reaching confluency in culture, the sorted cells were differentiated under adipogenic conditions (Figure S2A). Oil-Red-O staining showed that CD81+ cells differentiated into mature adipocytes, whereas CD81− cells showed limited adipogenic capacity even though they expressed Sca1 (Figure 2B). Differentiated CD81+ cells expressed many brown/beige fat-selective genes, such as Ucp1, Pgc1a, Cidea and Cox8b (Figure 2C). Moreover, differentiated CD81+ cells expressed significantly higher mRNA levels of Ucp1 and Pgc1a relative to CD81− cells, particularly after the cells were stimulated with norepinephrine (NE) (Figure 2D). It is worth noting that CD81+ cells possessed a cell-intrinsic capacity for beige adipogenesis without the browning cues (PPARγ agonists or NE): differentiated CD81+ cells expressed high levels of Ucp1, Pgc1a, Cidea, and Cox8b, even under the culture condition without rosiglitazone during the adipogenic induction phase (Figure S2B, S2C). Furthermore, differentiated CD81+ cells expressed higher protein levels of UCP1 and members of the mitochondrial OXPHOS complex than did CD81− cells, both in the presence and absence of forskolin (cAMP) (Figure 2E). Importantly, CD81+-derived differentiated adipocytes responded to NE by increasing both their total and uncoupled oxygen consumption rate (OCR), whereas CD81− cells did not have such responsiveness to NE (Figure 2F, Figure S2D).
Figure 2. CD81+ stromal cells give rise to beige adipocytes.
A) The scheme for the FACS analysis to isolate indicated cells from mouse inguinal WAT. The percentage yield of each population is based on the Ing WAT-derived SVFs after overnight culture. n=4.
B) Oil-Red-O staining of differentiated CD81− and CD81+ cells. Cells from the Ing WAT of BL6 mice were differentiated for 6 days under an adipogenic condition. Low (top) and high magnification (bottom) are shown. Scale bars = 50 μm.
C) mRNA expression of indicated genes in differentiated cells in (B). n=3.
D) mRNA expression of Ucp1 and Pgc1a in differentiated cells stimulated with or without NE for 4 hours. (C, D) *P < 0.05, **P < 0.01 by unpaired Student’s t-test.
E) Immunoblotting for UCP1 and indicated mitochondrial proteins in differentiated cells stimulated with or without forskolin (cAMP, 10 μM) for 6 hours.
F) OCR (pmol min−1) in differentiated cells treated with or without NE (10 μM). n=10. **P < 0.01 by one-way ANOVA followed by Bonferroni’s post hoc test.
G) Luciferase activity of transplants at indicated days after transplantation. Arrowhead indicates luciferase+ (UCP1+) transplants.
H) H&E staining and immunofluorescent staining for UCP1 and PERILIPIN in the transplants in (G). DAPI for counterstaining of immunofluorescent staining. Scale bars = 200 μm.
I) Illustration of the experiments in Cd81-lineage reporter mice.
J) Immunofluorescent staining for GFP and UCP1 in the Ing WAT from indicated mice. Scale bars = 100 μm.
Data in (A, C, D, F) are represented as mean ± SEM.
To determine if CD81+ cells give rise to beige fat in vivo, we isolated CD81+ cells and CD81− cells from the Ing WAT of UCP1-luciferase mice (ThermoMouse) in which luciferase activity reflects UCP1 expression (Galmozzi et al., 2014). The FACS-sorted cells were mixed in Hydromatrix gel and then transplanted into the subcutaneous region of immunodeficient Nude mice (Figures S2E, F). At six days after transplantation, we detected luciferase activity (i.e., UCP1 expression) in CD81+ cell transplants, but not in CD81− cell transplants (Figure 2G). The luciferase activity of CD81+ cell transplants was further increased by day 13 day after transplantation. Additionally, the CD81+ cell transplants derived from ThermoMouse contained multilocular adipocytes and expressed endogenous UCP1 and PERILIPIN (Figure 2H, Figure S2G). We also found that the CD81+ cell transplants expressed many thermogenic fat-selective genes at high levels similar to what is seen in cold-induced beige fat (Figure S2H). By contrast, CD81− transplants did not form visible fat tissues.
Next, we used lineage tracing of CD81+ cells within the adipose tissue to further determine the extent to which these cells differentiate into beige fat in vivo. To this end, we generated inducible Cd81-CreERT2 mice (knock-in mice that carry CreERT2 in the Cd81 gene locus at the N-terminus) and crossed them with Rosa26-mTmG reporter mice. Following pre-treatment with tamoxifen at ambient temperature for 5 consecutive days, the Cd81-lineage reporter mice (Cd81- CreERT2; Rosa26-mTmG) and control mice (Rosa26-mTmG mice) were acclimated to 8°C for 14 days to induce beige fat biogenesis (Figure 2I). Following cold exposure, a large number of cold-induced UCP1+ beige adipocytes in the Ing WAT were derived from CD81+ cells (Figure 2J, Figure S2I). Of note, some UCP1+ beige adipocytes in the Ing WAT of Cd81-lineage reporter mice did not express GFP (Figure S2J), consistent with works suggesting that some beige adipocytes are derived from mature adipocytes or other developmental lineages (Shao et al., 2019). On the other hand, the iBAT contained a very small fraction of GFP+ UCP1+ brown adipocytes after cold acclimation (Figure S2K), suggesting that CD81+ cells in iBAT are distinct from Myf5+ brown progenitors. In addition, CD81+-derived cells were distinct from PDGFRα+ fibroblasts in Ing WAT after cold acclimation (Figure S2L), reinforcing the results that CD81 marks a subset of PDGFRα+ stromal cells in the adipose tissue. Together, these data suggest that CD81 marks an APC population that possess the cell-intrinsic plasticity to give rise to beige adipocytes. Thus, we termed this population as beige APC.
CD81+ APC is a proliferative population in the inguinal WAT.
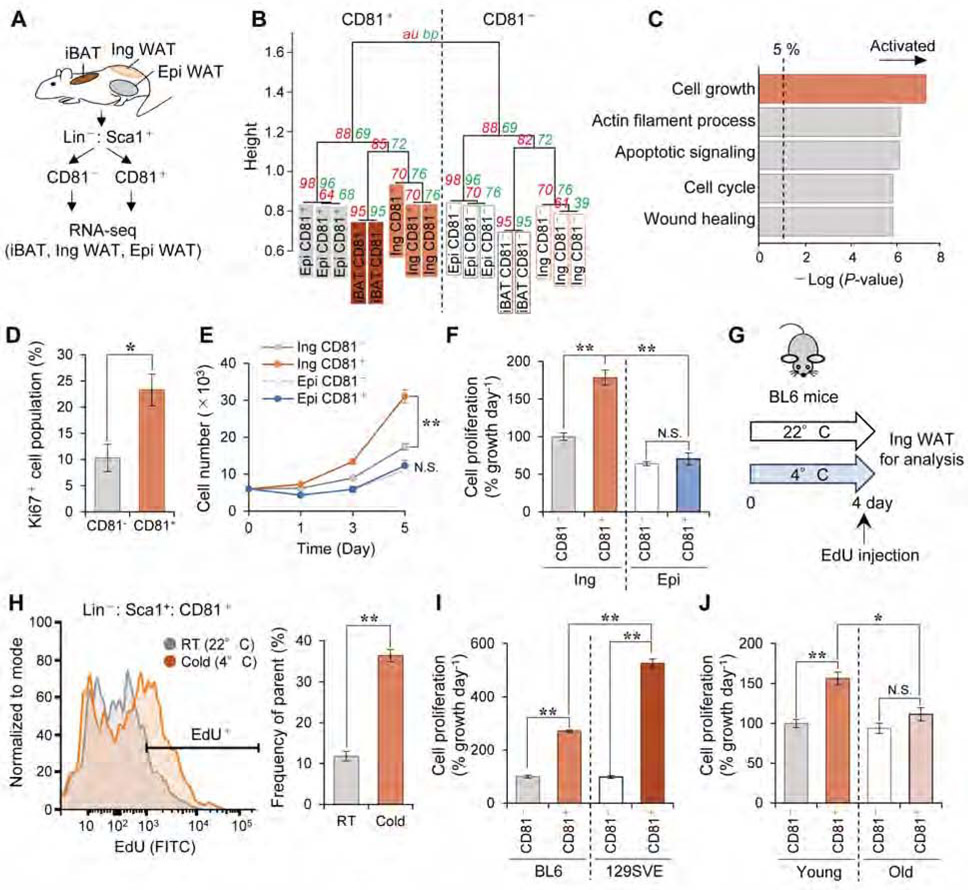
CD81 protein is expressed highly in adipose tissues and smooth muscle, and also expressed in other tissues at lower levels. Hence, we probed CD81+ APC in the Ing WAT for any functionally unique properties. To this end, we isolated CD81+ and CD81− cells from iBAT, Ing WAT, and Epi WAT of wild-type mice under the ambient temperature condition, and analyzed their transcriptome by RNA-seq (Figure 3A). Hierarchical clustering revealed that CD81+ APC exhibited a distinct molecular signature from CD81− cells in Ing WAT and other populations in the iBAT and Epi WAT (Figure 3B). Gene Ontology (GO) analysis of the transcriptome data suggested that biological pathways relating to cell growth, actin filament processes, apoptotic signaling, cell cycle, and wound healing, were significantly up-regulated in the Ing WAT-derived CD81+ cells (Figure 3C). These pathways were unique to Ing WAT-derived CD81+ cells because GO analysis of the transcriptome in CD81+ cells from the Epi WAT or iBAT did not highlight these biological pathways (Figure S3A). Consistent with the pathway analysis, we found a significantly more Ki67+ cells among Ing WAT-derived CD81+ cells than among Ing WAT-derived CD81− cells (Figure 3D). Furthermore, isolated Ing WAT-derived CD81+ cells grew significantly faster than did either their CD81− counterparts or CD81+ APCs from Epi WAT (Figure 3E, F).
Figure 3. CD81+ APC in the inguinal WAT is a proliferative stromal population.
A) Schematic illustration of the experiment. CD81+ cells (Lin−: Sca1+: CD81+) and CD81− cells (Lin−: Sca1+: CD81−) were isolated from indicated fat depots of 10-week-old BL6 mice.
B) Hierarchical clustering of transcriptomics. The horizontal distance represents similarities among each cluster as visualized by pvclust in R. Approximately Unbiased (au; red) P-value and Bootstrap Probability (bp; green) values as measures of certainty for clusters.
C) Upregulated biological pathways in CD81+ cells relative to CD81− cells from Ing WAT. n=3.
D) Ki67+ cell population (%) in CD81− and CD81+ cells from Ing WAT of BL6 mice. n=5. *P < 0.05 by unpaired Student’s t-test.
E) Cell number at indicated time points after seeding cells on non-coated culture plates. Cells were isolated from Ing WAT or Epi WAT of BL6 mice. n=4. **P < 0.01 by two-way repeated measures ANOVA.
F) Cell proliferation rate (% growth day−1) of indicated cells in (E). Data were presented as relative values to the growth rate of CD81− cell in Ing WAT.
G) Illustration of the experiments in (H).
H) EdU incorporation in CD81+ cells in (G). Right panel shows quantification of EdU+ cells. n=3. **P < 0.01 by unpaired Student’s t-test.
I) Cell proliferation rate of indicated cells from Ing WAT of BL6 and 129SVE male mice (12 weeks old). n=4.
J) Cell proliferation rate of indicated cells from Ing WAT of young (8-10 weeks) and old (60 weeks) male BL6 mice. Young, n=10; Old, n=5. (F, I, J) *P < 0.05, **P < 0.01 by one-way ANOVA followed by Bonferroni’s post hoc test.
Data in (D, E, F, H, I, J) are represented as mean ± SEM.
To ask how Ing WAT-derived CD81+ APC proliferation is regulated by cold exposure and genetic factors, we isolated CD81+ cells from the Ing WAT of wild-type C57BL/6J mice that were exposed to 4 °C for 4 days or 22°C before receiving an intraperitoneal injection of EdU (Figure 3G). Following the EdU pulse, we detected a robust EdU incorporation in Ing WAT-derived CD81+ APC, and this was further increased by cold exposure (Figure 3H). Next, we examined the impact of genetic background and age on the proliferation of CD81+ APC, because 129SVE mice possess higher amounts of beige adipocytes and are more resistant to diet-induced obesity than C57BL6/J mice (Guerra et al., 1998), while aging negatively impact beige fat biogenesis (Berry et al., 2017; Tajima et al., 2019). We found that the proliferative rate of CD81+ APC from the Ing WAT of 129SVE mice was significantly higher than corresponding APC from C57BL6/J mice, although CD81+ cells grew faster than did CD81− cells regardless of the genetic background (Figure 3I). Additionally, the cell growth rate of Ing WAT-derived CD81+ APC was significantly attenuated in 60-weeks-old mice, in which cold-induced beige fat biogenesis was also impaired (Figure 3J, Figure S3B). We also found that these 60-weeks-old mice had fewer CD81+ APC in the Ing WAT relative to young mice (8-10 weeks old) (Figure S3C). The data are aligned with the CD81+ APC transcriptomics data, as the cellular senescence pathway, often up-regulated by aging, was repressed in CD81+ APC from young mice (Figure S3D). These results suggest that CD81+ APC in Ing WAT is unique among other stromal cells in the high proliferative capacity, which is regulated by cold exposure, genetic background, and aging.
CD81 mediates irisin-induced FAK signaling by forming complexes with αV/β1 and αV/β5 integrins.
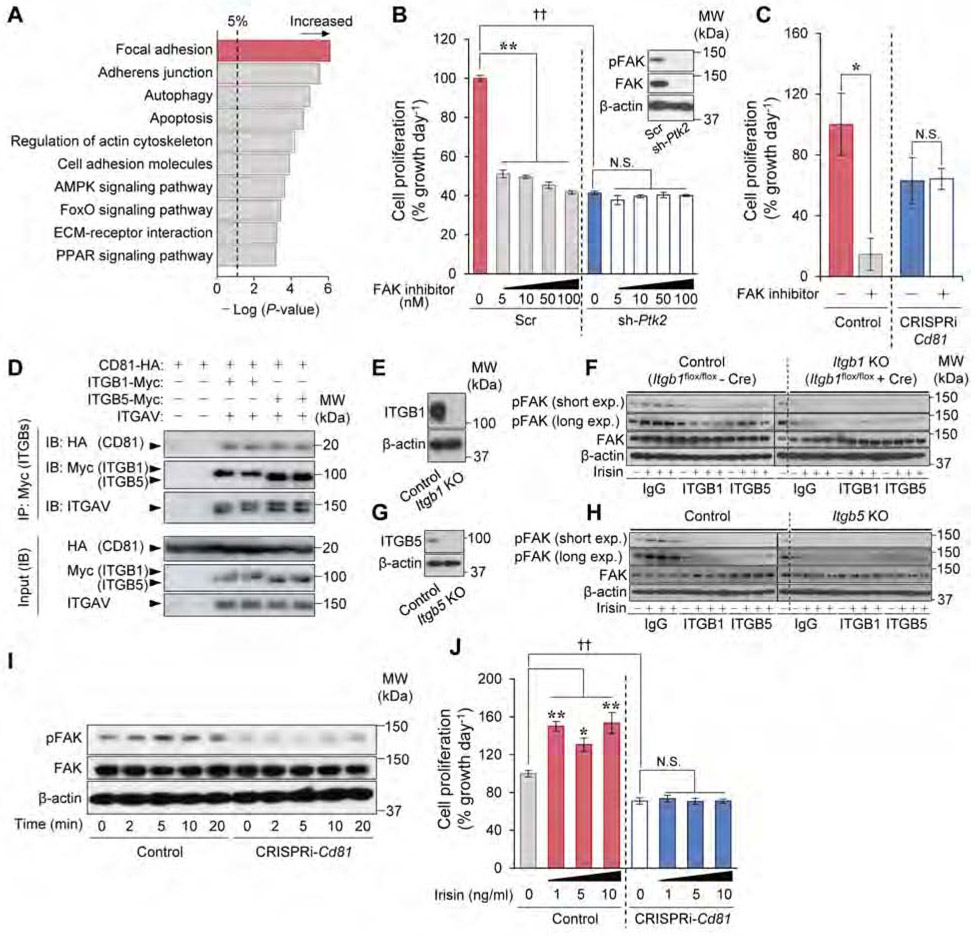
To determine the mechanisms underlying the highly proliferative nature of CD81+ APC in the Ing WAT, we next ran a functional enrichment analysis on the transcriptome data. The analysis showed that the focal adhesion pathway, including FAK (coded by the Ptk2 gene), was up-regulated in Ing WAT-derived CD81+ APC relative to CD81− cells (Figure 4A, Figure S4A). Thus, we examined the extent to which FAK controls Ing WAT-derived CD81+ APC proliferation by genetic and pharmacological approaches. We found that Ptk2 knockdown by lentiviral shRNA significantly inhibited cell proliferation in CD81+ cells. Furthermore, inhibition of FAK by PF-573228, a specific ATP-competitive inhibitor of FAK, potently reduced CD81+ APC proliferation over a wide concentration range, whereas this reduction was not seen when FAK was depleted in the cells (Figure 4B, Figure S4B). To test if CD81 is required for FAK signaling, we next depleted Cd81 by CRISPRi in which catalytically inactive Cas9 protein (dCas9) is fused to a Krüppel associated box (KRAB) domain. Transgenic mice expressing a gRNA targeting Cd81 were crossed with dCas9-KRAB mice to generate Cd81 deficient mice (CRISPRi-Cd81 mice) (Figure S4C-E). We found that the proliferation of Lin− Sca1+ stromal cells in the Ing WAT of CRISPRi-Cd81 mice was significantly lower than corresponding cells from control mice regardless of the culture conditions (plate-coating, FBS concentrations) (Figure S4F, G), while PF-573228 treatment did not diminish the proliferative rate of CD81-deficient cells (Figure 4C). These results indicate that FAK signaling is required for the proliferation of CD81+ APC in Ing WAT, and also suggest that rather than merely serving as a surface marker, CD81 is a functional regulator of beige APC proliferation.
Figure 4. CD81 forms complexes with αV/β1 and αV/β5 integrins and mediates irisin-induced FAK signaling.
A) The upregulated KEGG signaling pathways in CD81+ cells relative to CD81− cells from Ing WAT. n=3.
B) The effect of FAK inhibitor (PF-573228) on cell proliferation rate of CD81+ cells expressing shRNA targeting FAK (sh-Ptk2) or a scrambled control (Scr). Cells were treated with PF-573228 or vehicle (DMSO) for 4 days. n=4. Inset: immunoblotting for FAK and β-actin.
C) The effect of PF-573228 at 1 μM on cell proliferation rate of Lin−: Sca1+ stromal cells from CRISPRi-Cd81 mice and littermate controls. n=3. *P < 0.05 by unpaired Student’s t-test.
D) Immunoblotting for indicated proteins in HEK293T cells. Cell lysates were immunoprecipitated with Myc-tag beads (ITGB1 and ITGB5) and detected by antibodies against HA (CD81), Myc (ITGB1, ITGB5), and ITGAV. Inputs were shown in lower panels. The pellet and input were loaded at a ratio of 20:1.
E) Immunoblotting for ITGB1 and β-actin in control and Itgb1 KO CD81+ cells from the Ing WAT of Itgb1flox/flox mice.
F) Immunoblotting for FAK phosphorylation (pTyr397) (short and long exposure), total FAK, and β-actin in cells in (E) following irisin treatment. Cell lysates from control cells without irisin were included in the leftmost lane of the right panel (Itgb1 KO cells) as a reference.
G) Immunoblotting for ITGB5 and β-actin in CD81+ cells from the Ing WAT of Itgb5 KO or control mice.
H) Immunoblotting for indicated proteins in cells in (G) following irisin treatment.
I) Immunoblotting for indicated proteins in Lin−: Sca1+ stromal cells from the Ing WAT of CRISPRi-Cd81 mice and controls. Time point 0 is the cells prior to irisin treatment (control).
J) Cell proliferation rate of Lin−: Sca1+ cells in (I). Isolated cells were treated with irisin at indicated doses for 5 days. n=4. (B, J) *P < 0.05, **P < 0.01 by one-way ANOVA followed by Dunnett’s post hoc test. ††P < 0.01 by unpaired Student’s t-test with Bonferroni’s correction.
Data in (B, C, J) are represented as mean ± SEM.
CD81 is known to interact with integrins (Berditchevski et al., 1996; Termini and Gillette, 2017). Notably, Itgav (Integrin αV, a.k.a., CD51) was dominantly expressed in CD81+ APC from the Ing WAT at a significantly higher level than CD81− APC (Figure S5A). Among integrin β chains (ITGB), CD81+ cells expressed Itgb1 at the highest level, followed by Itgb5. This result caught our particular attention because a recent study showed that irisin, an exercise-induced myokine that promotes beige fat biogenesis, stimulates integrin-FAK signaling through αV/β5 and αV/β1 integrins, which are known to associate with CD81 (Chang and Finnemann, 2007; Kim et al., 2018). Accordingly, we hypothesized that CD81 forms a complex with αV/β1 and/or αV/β5 integrins in beige APC, and modulates integrin-FAK signaling in response to irisin. To test this hypothesis, we first examined the protein interactions between CD81 and αV/β1 or αV/β5 integrins by reconstituting these proteins in HEK293T cells. By immunoprecipitating ITGB1 or ITGB5 using a Myc-antibody in these cells, we found that both ITGB1 and ITGB5 interacted with integrin αV and CD81 (Figure 4D).
To test if αV/β1 or αV/β5 integrins in CD81+ APC are required for FAK signaling in response to irisin, we next isolated CD81+ APC from Ing WAT of Itgb1flox/flox mice in which Itgb1 was deleted by transducing the cells with a retroviral Cre (Figure 4E, Figure S5B). Subsequently, the Itgb1 KO and control cells (vector) were treated with antagonistic antibodies against ITGB1, ITGB5, or IgG control, prior to irisin treatment. We found that irisin stimulated FAK phosphorylation in control CD81+ APC, whereas genetic deletion of Itgb1 or the specific antagonistic antibody against ITGB1 completely blunted the effect of irisin to trigger FAK phosphorylation (Figure 4F). Antibody-based blockade of ITGB5 also potently, though not completely, blunted the irisin-induced FAK phosphorylation in CD81+ APC. To further probe the genetic requirement of ITGB5 for irisin-induced integrin-FAK signaling, we isolated CD81+ APCs from Ing WAT of Itgb5 null and wild-type control mice (Figure 4G, Figure S5C). Consistent with the above results, blockade of integrin ITGB1 by its antagonistic antibody completely blocked FAK phosphorylation following irisin treatment. We also found that genetic deletion of Itgb5 inhibited irisin-induced FAK phosphorylation in CD81+ APC (Figure 4H). Together, these data indicate that irisin-induced integrin-FAK signaling in CD81+ APC requires both β1 and β5 integrins.
Next, we examined the extent to which CD81 is required for irisin-induced FAK signaling in the Ing WAT of CRISPRi-Cd81 mice (Figure S5D). We found that irisin rapidly increased FAK phosphorylation in Lin− Sca1+ cells isolated from control mice within 5 min after the treatment, but this effect was completely lost in analogous cells from CRISPRi-Cd81 mice (Figure 4I). Conversely, we found that HEK293T cells ectopically expressing both αV/β5 integrins and CD81 responded to irisin at 1 pM to induce FAK phosphorylation, whereas cells expressing only αV/β5 integrins required irisin at 10 pM or higher, indicating that CD81 sensitized these cells to irisin (Figure S5E). Importantly, irisin at concentrations at or above 1 ng/ml stimulated the proliferation of Lin− Sca1+ cells from the Ing WAT of control mice, but failed to do so in analogous cells from CRISPRi-Cd81 mice (Figure 4J). These results indicate that CD81 is required for irisin-induced integrin-FAK signaling and APC proliferation.
CD81 is required for de novo beige fat biogenesis.
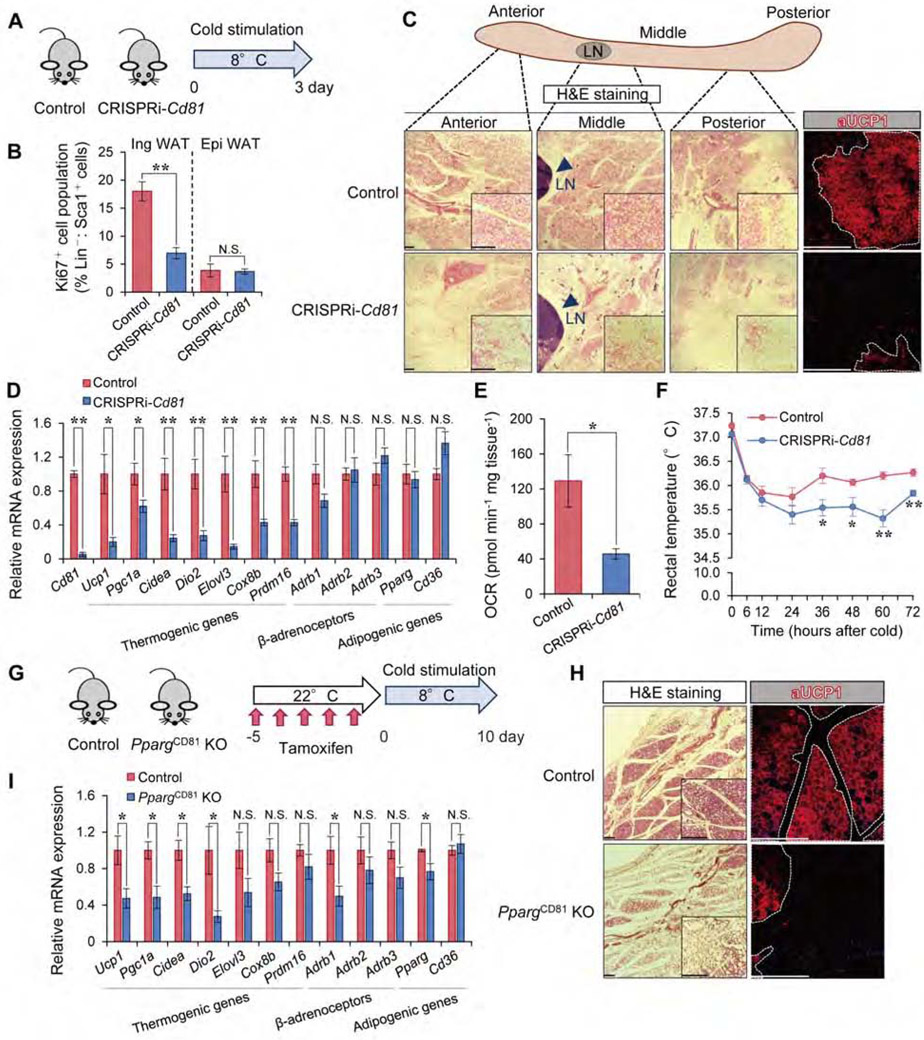
Our data highlighting CD81 as a regulator of beige APC proliferation prompted us to ask if CD81 is required for beige fat biogenesis. To this end, CRISPRi-Cd81 mice and littermate controls were exposed to 8°C temperature for 3 days (Figure 5A). Consistent with the cell proliferation data, the number of proliferative APC (Lin−: Sca1+: Ki67+) in the Ing WAT, but not in Epi WAT, of CRISPRi-Cd81 mice was significantly lower than that of control mice following cold exposure (Figure 5B). These data suggest that CD81 is required for cold-induced APC proliferation in the Ing WAT.
Figure 5. CD81 is required for de novo beige fat biogenesis.
A) Illustration of the experiments in (B-E).
B) Ki67+ cells (%) in Lin−: Sca1+ stromal cells from the Ing WAT and Epi WAT in (A). n=5.
C) H&E staining and UCP1 immunofluorescent staining in the Ing WAT in (A). Arrowhead indicates lymph node (LN). Scale bar = 200 μm.
D) mRNA expression of indicated genes in the Ing WAT in (A). n=5.
E) OCR in the Ing WAT in (A). Control, n=3; CRISPRi-Cd81, n=4.
F) Rectal core body temperature of indicated mice following cold exposure. Control, n=6; CRISPRi-Cd81, n=5. *P < 0.05, **P < 0.01 by two-way repeated measures ANOVA with post hoc test by unpaired Student’s t-test.
G) Illustration of the experiments in (H-I).
H) H&E staining and UCP1 immunofluorescent staining in the middle region of Ing WAT in (G). Scale bar = 200 μm.
I) mRNA expression of indicated genes in the Ing WAT in (G). Control, n=5; PpargCD81 KO mice, n=4. (B, D, E, I) *P < 0.05, **P < 0.01 by unpaired Student’s t-test.
Data in (B, D, E, F, I) are represented as mean ± SEM.
Next, we characterized the formation of cold-induced beige fat in these mice. Cold exposure stimulated the formation of beige adipocytes with multilocular lipid droplets and UCP1 expression in the Ing WAT of control mice. However, cold-induced beige fat biogenesis was strikingly impaired in the Ing WAT of CRISPRi-Cd81 mice (Figure 5C). Also, CD81 loss significantly reduced the expression of brown/beige fat-selective genes, but not general adipogenic genes and gene encoding β-adrenoceptors, in the Ing WAT following cold exposure (Figure 5D). Of note, we did not observe a notable difference in the expression of thermogenic genes in the iBAT between the genotypes (Figure S6A, B). The data are consistent with the results of our Cd81-lineage tracing result in which the majority of brown adipocytes in iBAT did not stem from CD81+ APC. Importantly, OCR in the Ing WAT, but not in the iBAT, of CRISPRi-Cd81 mice was significantly lower than that of control mice (Figure 5E, Figure S6C). Furthermore, the core body temperatures of CRISPRi-Cd81 mice were modestly but significantly lower than those of control mice following cold acclimation (Figure 5F), while cold-stimulated muscle shivering was not different between the genotypes (Figure S6D, E).
Since CRISPRi-Cd81 mice lack CD81 in all the cells, we next asked if the reduced beige fat biogenesis in CRISPRi-Cd81 mice was due to impaired de novo beige adipogenesis in CD81+ APC or reduced reinstallation of the thermogenic program in mature adipocytes. To this end, we generated inducible CD81+ cell-specific Pparg knockout mice (PpargCD81 KO, Cd81-CreERT2; Ppargflox/flox), in which Peroxisome proliferator-activated receptor-γ (PPARγ) was deleted in CD81+ APC (Figure 5G). In this mouse model, de novo beige adipogenesis in CD81+ APC would be selectively impaired following tamoxifen treatment, such that we could determine the contribution of CD81+ APC to de novo beige fat biogenesis. We thus pre-treated PpargCD81 KO mice and littermate control mice (Ppargflox/flox) with tamoxifen at ambient temperature for 5 consecutive days, and subsequently transferred these mice to 8°C for 10 days. Histological analysis of the Ing WAT showed that PpargCD81 KO mice had far fewer beige adipocytes and had lower UCP1 expression than did control mice, although some clusters of multilocular UCP1+ beige adipocytes were observed in PpargCD81 KO mice in the middle region of the Ing WAT (Figure 5H, Figure S6F). Furthermore, mRNA expression of brown/beige fat-selective genes in the Ing WAT of PpargCD81 KO mice was significantly lower than that of control mice (Figure 5I). Together, these results indicate that CD81 is required for cold-stimulated de novo beige fat biogenesis and thermogenesis.
CD81 loss leads to diet-induced obesity, glucose intolerance, and adipose tissue dysfunction.
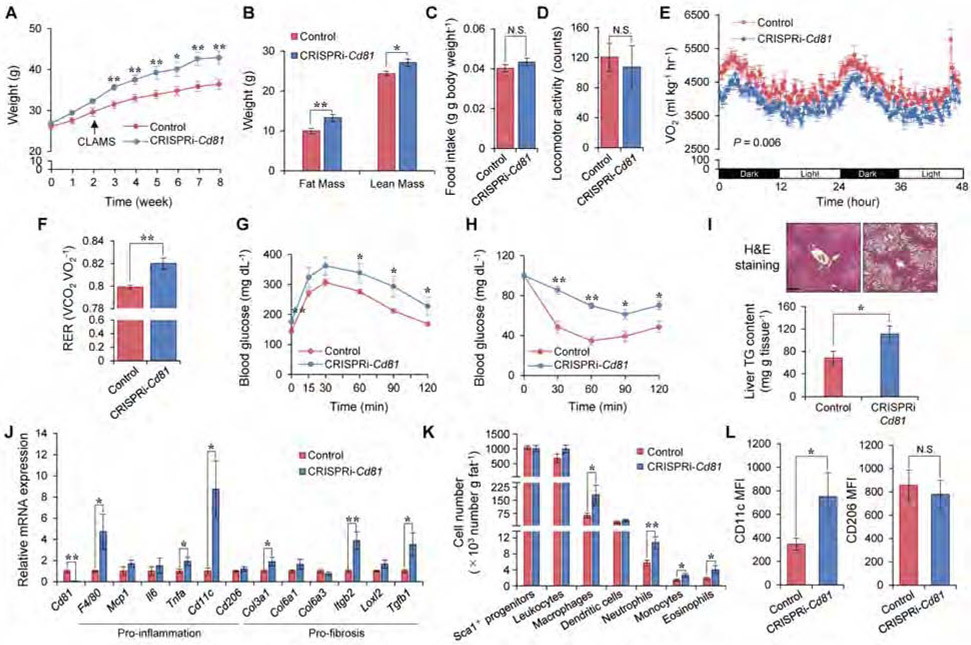
The above data lead us to hypothesize that CD81 would be an important determinant of whole-body energy homeostasis. Of note, several transmembrane tetraspanin proteins, including CD81, are expressed in the exosome membranes, and are often used as exosome markers (Colombo et al., 2014); however, we found that CD81 was dispensable for the formation and release of exosomes (Figure S7A). Thus, the contribution of possible changes in exosome secretion per se to the metabolic phenotype of CRISPRi-Cd81 mice is most likely negligible. To test how CD81 deficiency affects whole-body energy metabolism, 8 weeks old CRISPRi-Cd81 mice and littermate controls were put on a high-fat diet (HFD, 60 % fat) for 8 weeks. We found that CRISPRi-Cd81 mice gained significantly more body-weight than controls starting at 3 weeks of HFD and thereafter (Figure 6A). The greater body-weight gain in CRISPRi-Cd81 mice was due to increased lean mass and fat mass (Figure 6B), although neither food intake nor locomotor activity was different between the genotypes (Figure 6C, D). By contrast, CRISPRi-Cd81 mice had significantly lower whole-body energy expenditure (VO2) than did control mice: it is important to note that the reduced VO2 in CRISPRi-Cd81 mice was detected at 2 weeks of HFD when the differences in body-weight had not yet manifest (Figure 6E). Moreover, CRISPRi-Cd81 mice had a higher respiratory exchange ratio (RER, VCO2/VO2) than did control mice, suggesting that CD81 loss causes a fuel switch toward carbohydrate oxidation (Figure 6F). Furthermore, CRISPRi-Cd81 mice were glucose intolerant relative to control mice after 8 weeks of consuming a HFD (Figure 6G). This phenotype was not due to a metabolic consequence of obesity because glucose intolerance in CRISPRi-Cd81 mice already emerged within 2 weeks of HFD, a time at which the body weights between the two groups had not yet diverged (Figure S7B). CRISPRi-Cd81 mice were also insulin resistant relative to control mice (Figure 6H). In addition, the liver of CRISPRi-Cd81 mice contained significantly more triglycerides than did those of control mice (Figure 6I, Figure S7C).
Figure 6. CD81 loss causes obesity, glucose intolerance, and adipose tissue inflammation.
A) Body-weight of indicated mice on HFD. Control, n=9; CRISPRi-Cd81, n=6.
B) Fat mass and lean mass of mice on 8 weeks of HFD. Control, n=9; CRISPRi-Cd81, n=6.
C) Food intake of mice on HFD. Control, n=8; CRISPRi-Cd81, n=6.
D) Locomotor activity of mice in (C). (C, D) N.S., not significant by unpaired Student’s t-test.
E) Whole-body energy expenditure (VO2, ml kg−1 hr−1) of mice on 2 weeks of HFD. Control, n=8; CRISPRi-Cd81, n=6. P value by two-way repeated measures ANOVA.
F) RER of mice in (E).
G) GTT in mice on 8 weeks of HFD. Control, n=9; CRISPRi-Cd81, n=6.
H) ITT in mice in (G). (A, G, H) *P < 0.05, **P < 0.01 by two-way repeated measures ANOVA with post hoc test by unpaired Student’s t-test.
I) Top: H&E staining in the liver of mice on 10 weeks of HFD. Sale bar = 200 μm. Bottom: TG contents. Control, n=9; CRISPRi-Cd81, n=6.
J) mRNA expression of pro-inflammatory and pro-fibrosis genes in the Ing WAT of mice on 10 weeks of HFD. Control, n=7; CRISPRi-Cd81, n=6.
K) The number of total non-leukocyte stromal cells (CD45−: Sca1+), leukocytes (CD45+), macrophages (CD45+: CD64+), dendritic cells (CD45+: CD64−: CD11c+), neutrophils (CD45+: CD64−: Ly6G+), monocytes (CD45+: CD64−: Ly6C+), eosinophils (CD45+: CD64−: SiglecF+) per gram of the Ing WAT of mice on 10 weeks of HFD. Control, n=8-9; CRISPRi-Cd81, n=5-6.
L) The expression of M1-like macrophage marker (CD11c) and M2-like macrophage marker (CD206) among total macrophages in the Ing WAT from mice on 10 weeks of HFD. Control, n=9; CRISPRi-Cd81, n=6. (B, F, I, J, K, L) *P < 0.05, **P < 0.01 by unpaired Student’s t-test.
Data are represented as mean ± SEM.
We next examined the extent to which CD81 loss alters adipose tissue inflammation. Although macrophages express detectable levels of CD81, the capacity of bone marrow cells to differentiate into macrophages (BMDM) and the ability of these BMDM to express pro-inflammatory markers were not altered by the presence or absence of CD81 (Figure S7D-G). The metabolic phenotype of CRISPRi-Cd81 mice is thus unlikely to be due to an intrinsic hyperactivation of macrophages. However, we found that, in association with their relative obesity, the Ing WAT of CRISPRi-Cd81 mice expressed significantly higher levels of pro-inflammatory and pro-fibrotic genes than did control mice (Figure 6J). Indeed, analyzing Ing WAT-resident immune cells by FACS showed that obese CRISPRi-Cd81 mice had significantly more macrophages, neutrophils, monocytes, and eosinophils in the Ing WAT than did control mice fed the same HFD diet, even though the number of total Lin− Sca1+ cells and other immune cells (B cells, T cells, NK cells) in Ing WAT were similar between the two groups (Figure 6K, Figure S7H). The expression of CD11c, a marker of pro-inflammatory (M1-like) macrophages, was significantly higher in Ing WAT-resident macrophages from CRISPRi-Cd81 mice than that in control mice, whereas the expression of CD206, an M2-macrophage marker, was not different between the genotypes (Figure 6L). This pro-inflammatory macrophage phenotype was selective to the Ing WAT, as saw no such differences when examining the Epi WAT (Figure S7I, J).
CD81+ APC enrichment in human subcutaneous fat correlates negatively with metabolic health.
Given the results in mice, we aimed to determine whether the number of CD81+ APCs is associated with any metabolic traits in humans. We first isolated CD81+ APCs from the subcutaneous fat in a cohort of 7 healthy men (age 49 ± 8 years (mean ± SD), BMI 29 ± 6 kg/m2) with no history of diabetes. For the isolation of CD81+ APC, we used PDGFRα instead of Sca1 (Lin−: PDGFRα+: CD81+) because humans lack Sca1, and both PDGFRα and Sca1 mark the same CD81+ APC in the Ing WAT of mice (Figure 1E). Consistent with our findings in mice, human CD81+ cells (Lin−: PDGFRα+: CD81+) from the subcutaneous fat of human subjects included a significantly higher number of Ki67+ cells than did corresponding CD81− cells (Lin−: PDGFRα+: CD81−), suggesting that human CD81+ APCs in the subcutaneous fat are also a proliferative population (Figure S7K).
Next, we examined human subcutaneous fat from 28 individuals across a wide BMI range who were recruited from a well-described multiethnic cohort of adults assembled in the San Francisco Bay Area (Alba et al., 2018). In analyzing the correlation between the number of CD81+ APCs within the subcutaneous fat and metabolic profiles of these individuals (Table S2), we found that the percentage of CD81+APC significantly correlated inversely with HOMA-IR, fasting blood glucose levels, and diastolic blood pressure (Table 1). Furthermore, we found a significant inverse correlation between the number of subcutaneous fat CD81+ APC and both absolute visceral fat mass (kg), as measured by dual-energy X-ray absorptiometry (DXA), and visceral adiposity (% VAT /fat mass). There was a trend towards an inverse correlation between CD81+ APC enrichment and both fasting insulin and triglyceride levels, and a trend towards a positive correlation between CD81+ APC enrichment and HDL cholesterol levels, although these did not reach statistical significance. By contrast, there was no correlation with BMI, total fat mass, or total lean mass. It is important to note that no correlations were seen with these metabolic parameters when only Lin− PDGFRα+ cells or Lin− cells were used in the analyses. Taken together, these data point to reduced numbers of CD81+ APCs within the subcutaneous fat as an indicator of metabolic risk in people, in alignment with the predictions that emanate from our accumulated mouse data.
Table 1.
Correlations between % CD81+ cells and clinical parameters.
| Parameter | % CD81+: PDGFRα+: Lin− cells |
% PDGFRα+: Lin− cells |
% Lin− cells | |||
|---|---|---|---|---|---|---|
| Rho | P-value | Rho | P-value | Rho | P-value | |
| BMI (kg/m2) | −0.132 | N.S. | −0.070 | N.S. | 0.014 | N.S. |
| Fat mass (kg) | 0.162 | N.S. | 0.043 | N.S. | 0.232 | N.S. |
| Lean mass (kg) | 0.010 | N.S. | 0.313 | N.S. | 0.341 | 0.076 |
| HOMA-IRa | −0.433* | 0.024 | −0.337 | 0.086 | −0.250 | N.S. |
| Fasting glucose (mg/dL)a | −0.477* | 0.012 | −0.177 | N.S. | −0.139 | N.S. |
| Fasting insulin (mIU/L)a | −0.376 | 0.053 | −0.375 | 0.054 | −0.230 | N.S. |
| DBP (mmHg) | −0.411* | 0.030 | 0.135 | N.S. | −0.223 | N.S. |
| SBP (mmHg) | 0.238 | N.S. | −0.007 | N.S. | 0.179 | N.S. |
| VAT mass (kg) | −0.393* | 0.038 | −0.063 | N.S. | −0.140 | N.S. |
| %VAT/FM | −0.384* | 0.044 | −0.088 | N.S. | −0.248 | N.S. |
| Triglycerides (mg/dL) | −0.352 | 0.066 | 0.107 | N.S. | −0.175 | N.S. |
| Total cholesterol (mg/dL) | −0.048 | N.S. | 0.092 | N.S. | 0.090 | N.S. |
| LDL cholesterol (mg/dL) | 0.002 | N.S. | −0.050 | N.S. | 0.073 | N.S. |
| HDL cholesterol (mg/dL) | 0.326 | 0.090 | −0.120 | N.S. | 0.139 | N.S. |
BMI: body mass index; DBP: diastolic blood pressure; SBP: systolic blood pressure; VAT: visceral adipose tissue; FM: fat mass. HOMA-IR = Fasting insulin (mIU/L) x [fasting blood glucose (mg/dL)/405]. Values are presented as Spearman’s coefficients of correlation.
P-values are statistically significant (<0.05). n=28 subjects included in the analysis. N.S., not significant.
One subject on insulin was excluded from the analysis.
DISCUSSION
PDGFRα and Sca1 (in mice) are known APC markers, but they are also expressed in many other stromal cells; therefore, what distinguishing beige APC among other PDGFRα+/Sca1+ stromal cells remained unknown. The present study identified CD81 as a cell surface molecule that marks a unique subset of PDGFRα+/Sca1+ stromal cells that give rise to beige adipocytes in vivo. CD81+ APC in the Ing WAT is highly proliferative and the proliferation is further enhanced by cold exposure. This result is intriguing because previous studies did not detect an increase in EdU incorporation in response to cold exposure or β3-AR agonist treatment when all the PDGFRα+ cells were analyzed in aggregate (Lee et al., 2015; Lee et al., 2012). These results suggest that a relatively small fraction of PDGFRα+/Sca1+ stromal cells, which are marked by CD81, is proliferative and contributes to de novo beige adipogenesis. Our results that selective ablation of PPARγ in CD81+ cells attenuates beige fat biogenesis are aligned with studies that cold-induced beige fat biogenesis occurs through de novo differentiation, while some mature adipocytes reinstall the thermogenic program (Shao et al., 2019).
Besides CD81+ APC, diverse progenitor cells likely contribute to forming beige fat within the Ing WAT. For instance, Lin− PDGFRα+ MyoD+ myogenic progenitors in the Ing WAT give rise to clusters of beige adipocytes that reside near the lymph nodes and exhibit a unique metabolic profile (a.k.a., g-beige fat) (Chen et al., 2019). These g-beige progenitors are distinct from CD81+ APC and their differentiation is regulated independently from the β-AR pathway. It is possible that heterogeneous APC populations differentially respond to diverse browning stimuli (e.g., exercise, tissue injury, cachexia, and intermittent fasting), and that newly formed beige fat is composed of developmentally and functionally distinct subtypes of beige adipocytes depending on the nature of external cues (e.g., cold vs. tissue injury). A better understanding of the mechanisms that define thermogenic fat heterogeneity is a significant area of research in the field.
Importantly, CD81 is more than merely a molecular marker of beige APC. CD81 is known to form a complex with CD19/CD21-B cell receptors (BCR) and controls BCR signaling (Cherukuri et al., 2004; Mattila et al., 2013). We found that CD81 is required for beige APC proliferation by forming a complex with αV/β1 and αV/β5 integrins and controls integrin-FAK signaling in response to irisin. Furthermore, CD81 loss causes systemic glucose intolerance and insulin resistance. Consistent with these results, fat-selective loss of FAK leads to impaired glucose tolerance and insulin sensitivity (Luk et al., 2017). Because irisin increases upon exercise and acts on adipose tissues, the brain, and bone (Bostrom et al., 2012; Colaianni et al., 2015; Kim et al., 2018), it will be intriguing to determine the extent to which CD81 mediates exercise-induced metabolic changes in these organs. It is also worth noting that several hormones are known to activate integrin-FAK signaling through an interaction with integrins. For example, Angiopoietin-Like 4 (ANGPTL4) binds to β1/β5 integrins and modulates FAK signaling (Goh et al., 2010) and increased ANGPTL4 promotes beige fat biogenesis in the inguinal WAT (McQueen et al., 2017). It is thus conceivable that, besides irisin, CD81 mediates the activation of integrin-FAK signaling in response to ANGPTL4 and possibly other hormonal cues.
Selective activation of beige fat biogenesis is accompanied by improved insulin sensitivity, reduced adipose tissue inflammation, and fibrosis (Hasegawa et al., 2018; Ikeda et al., 2017; McDonald et al., 2015; Seale et al., 2011; Shinoda et al., 2015b), implying the possibility that the browning capacity of subcutaneous WAT reflects overall metabolic health. The present study found that the proliferative capacity of CD81+ APC in the inguinal WAT was higher in 129SVE mice (an obesity/diabetes-resistant strain) relative to that in BL6 mice (an obesity/diabetes-prone strain), while CD81+ APC proliferation declined in aging. Importantly, we found that the number of CD81+ APC in the subcutaneous fat of human subjects showed an inverse correlation with HOMA-IR, fasting glucose levels, diastolic blood pressure, and visceral adiposity. These results are intriguing given the recent clinical observation that mitochondrial activity in the subcutaneous fat (i.e., mitochondria-enriched adipocytes) correlates inversely with fat inflammation and hyperinsulinemia, and directly with the success of dietary-based weight-loss interventions and fat-specific glucose uptake (Heinonen et al., 2017; Jokinen et al., 2018). Thus, the number of CD81+ APCs in the subcutaneous fat of people may serve as a useful index to predict the risk of future metabolic disorders or the future success of efforts to control body weight. However, the number of subjects tested in the present study is relatively small, such that the results need to be replicated in larger cohorts under diverse dietary circumstances. Specifically, it will be interesting to explore the potential utility of CD81+ APCs as a biomarker of metabolic responsiveness in the context of weight loss interventions, including bariatric surgery, and exercise programs.
STAR Methods
LEAD CONTACT
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Shingo Kajimura (skajimur@bidmc.harvard.edu).
MATERIALS AVAILABILITY
Unique materials and reagents generated in this study are available upon request from the Lead Contact with a completed Materials Transfer Agreement.
DATA AND CODE AVAILABILITY
The RNA-seq datasets generated during this study are available at ArrayExpress (www.ebi.ac.uk) under the accession number E-MTAB-8435 and E-MTAB-8464.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All animal experiments were performed according to procedures approved by the Institutional Animal Care and Use Committee for animal care and handling at the University of California, San Francisco (UCSF). C57BL/6J wild-type mice (Stock No. 000664), NU/J mice (Stock No. 002019) and ThermoMouse (Stock No. 026690) were obtained from the Jackson Laboratory. Itgb5 null mice (Stock No. 004166) and Itgb1flox/flox mice (Stock No. 004605) available at the Jackson Laboratory were also used for the isolation of CD81+ APC. 129SVE mice (129S6/SvEvTac) were obtained from Taconic Biosciences. Male mice were maintained on a standard rodent chow at ambient temperature (22°C) under a 12hour light-dark cycle. Samples were obtained from male mice at 8-16 weeks or 60 weeks of age. For acute cold-exposure study, samples were obtained from male C57BL/6J wild-type mice (8-10 weeks old) at 4°C for 4 days, or male C57BL/6J wild-type mice (8, 24, 40, and 72 weeks old) at 8°C for 5 days using a rodent incubator (Power Scientific, Inc. RIS33SD). For chronic cold exposure, nude mice at 8-10 weeks of age were kept under 12°C for 10 days.
CRISPRi-Cd81 mice:
CRISPRi mice were generated by using a site-specific integrase-mediated approach described previously (Tasic et al., 2011). In brief, CRISPRi mice in the FVB background contain a CAG promoter within the Hipp11 (H11) locus expressing the nuclease-deficient Cas9 fused to the zinc-finger protein 10 (ZNF10) Krüppel-Associated Box (KRAB) repressor domain, together with mCherry and the puromycin resistance. CRISPRi mice were crossed with the FVB mice expressing gRNA-Cd81 on the H11 locus to generate CRISPRi-Cd81 mice. Male mice were maintained on a standard rodent chow or a 60% high-fat diet (Research Diets) at ambient temperature (22°C). For cold exposure study, CRISPRi-Cd81 mice and littermate control mice at 8-10 weeks of age were housed at 8°C for 3 days.
Lineage tracing reporter mice:
We inserted CreERT2 into the Cd81 gene locus and generated (Cd81-CreERT2 mice (Applied StemCell). Subsequently, Cd81-CreERT2 mice were crossed with Rosa26-mTmG mice (Jackson Laboratory, Stock No. 007576) to generate Cd81-lineage reporter mice (Cd81-CreER T2; Rosa26-mTmG mice). To induce Cre expression in Cd81-lineage reporter mice, tamoxifen (Sigma-Aldrich) at 3 mg in 100 μl corn oil per dose were administered intraperitoneally for 5 days. For cold exposure study, male mice (8 weeks old) were kept under 8°C for 14 days. Rosa26-mTmG mice without Cre recombinase was used as negative control. Acta2-CreERT2 mice were crossed with Rosa26-LSL-tdTomato mice to generate α-SMA+ reporter mice (Tsujino et al., 2017). To induce Cre expression in α-SMA+ reporter mice, we administered tamoxifen at 2 mg in 100 μl corn oil per dose to male mice at 10 weeks of age for 3 days. At 1 week after tamoxifen treatment, we examined tdTomato expression. Rosa26-LSL-tdTomato mice without Cre recombinase was used as negative control.
Cd81-specific Pparg KO mice:
To generate CD81+ cell-specific Pparg knockout mice (PpargCD81 KO, Cd81-CreERT2; Ppargflox/flox), Cd81-CreERT2 mice were crossed with Pparg-floxed mice (Jackson Laboratory, Stock No. 004584). To induce Cre expression, tamoxifen at 3 mg in 100 μl corn oil per dose were administered intraperitoneally for 5 days. For cold exposure study, male PpargCD81 KO mice and control mice at 8 weeks of age were housed at 8°C for 10 days.
Human Subjects
All the subjects signed consent forms to participate in the study, which was approved by the University of California San Francisco (UCSF) Institutional Review Board. Subjects were part of a multiethnic clinical cohort, termed Inflammation, Diabetes, Ethnicity, and Obesity (IDEO), consisting of 25- to 65-year-old healthy men and women living in the San Francisco Bay Area and recruited from medical and surgical clinics at the UCSF and the Zuckerberg San Francisco General Hospital, or through local public advertisements (NCT03022682). The IDEO cohort excludes those taking anti-inflammatory medications, glucocorticoids, or other medications affecting inflammation and those with a history of heart failure, liver failure, renal dysfunction, autoimmune disorders, chronic inflammatory or infectious disease, cancer, or a known history of alcohol or drug abuse. Individuals are also excluded if they have smoked or were not weight stable (>3% change over 3 months). IDEO collects demographic, medical, medication, dietary, and lifestyle data from subjects using questionnaires. The study involved a random sample of 28 individuals (16 women, 12 men) with BMI >20 kg/m2. Eleven subjects had a diagnosis of diabetes (HbA1c >6.5% or a physician diagnosis plus diabetes medication use). Of these, one individual was taking insulin. Height and weight were measured using a standard stadiometer and scale, with BMI (kg/m2) calculated from two averaged measurements. Blood pressure was measured with a standard mercury sphygmomanometer on the left arm after at least 10 min of rest. Mean values were determined from two independent measurements.
Blood samples were collected after an overnight fast and analyzed for plasma glucose, insulin, serum total cholesterol, HDL-cholesterol and triglycerides (LDL-cholesterol was estimated according to the Friedwald formula). Insulin resistance was estimated by the homeostatic model assessment of insulin resistance (HOMA-IR) index calculated from fasting glucose and insulin values (Mather et al., 2001; Muniyappa et al., 2008). Subjects taking insulin were excluded from HOMA-IR analyses. Body composition of the subjects was estimated by dual-energy X-ray absorptiometry (DXA) using a Hologic Horizon/A scanner (3-min whole-body scan, <0.1 G mGy) per manufacturer protocol. A single technologist analyzed all DXA measurements using Hologic Apex software (13.6.0.4:3) following International Society for Clinical Densitometry guidelines. Visceral adipose tissue (VAT) was measured in a 5-cm-wide region across the abdomen just above the iliac crest, coincident with the fourth lumbar vertebrae, to avoid interference from iliac crest bone pixels and matching the region commonly used to analyze VAT mass by CT scan (Bredella et al., 2013).
Subcutaneous WAT sampling:
Subcutaneous WAT samples were obtained from the subjects by 2 different methods as described previously (Alba et al., 2018; Hasegawa et al., 2018). In most cases, samples were collected by aspirational needle biopsies using a 14-16 G needle from the peri-umbilical area under local anesthesia. Some of the samples were obtained during elective abdominal or bariatric surgery. WAT samples were freed of visible connective tissue and rinsed to remove blood and clots, after which they were further washed with Krebs-Ringer bicarbonate (KRB) buffer supplemented with 1% BSA. The tissues were rinsed twice in KRB buffer containing 1% BSA and strained using 70 μm nylon filters and immediately placed in KRB buffer containing 1% BSA and 2 mg ml−1 Type 1 Collagenase (Alfa Aesar). The tissues were transferred to a 37°C incubator in 50 mL plastic tubes and incubated with agitation for 45-60 min. The digests were washed with KRB buffer containing 1% BSA and strained into fresh 50 mL plastic tubes using 70 μm filters to remove any undigested tissue, and the tubes were spun at 450 × g for 10 min at 4°C. The pellets containing the SVF cells were resuspended in RBC lysis buffer for 5-10 min to lyse the RBCs. The resulting SVF cells were further washed with KRB buffer containing 1% BSA at 4°C, and then centrifuged at 450 × g for 10 min at 4°C. The cells were resuspended in freezing medium (90% FBS/10% DMSO) at a concentration of 1.0 × 106 cells ml−1 cryopreservation medium. The SVF cells were then frozen at −80°C in an ethanol-jacketed closed container overnight, and subsequently stored in liquid nitrogen.
Cells
Mouse stromal vascular fraction (SVF) cells were obtained from male mice by collagenase digestion following the protocol that was published previously (Aune et al., 2013). For the isolation of CD81+ cells for differentiation studies, SVF cells were cultured overnight (~ 20 hour) before sorting in non-coated plates with DMEM/F12 medium (Gibco) containing 1% GlutaMAX-I (Gibco) and 10% FBS (Atlanta Biologicals). Cells were harvested using TrypLE™ Select (1X) (Gibco) after washing with PBS. Subsequently, CD81− (Lin−: Sca1+: CD81−) and CD81+ APCs (Lin−: Scal+: CD81+) were isolated using FACS Aria II and seeded onto collagen-coated plates. Cell plating was scaled according to the surface area. At 95% confluency, cells were induced to differentiate for 2 days with an adipogenic cocktail (0.5 mM IBMX, 2 μg ml−1 dexamethasone, 850 nM insulin, 1 nM T3, 125 μM indomethacin with or without 1 μM rosiglitazone) in DMEM/F12 medium containing 1% GlutaMAX-I and 10% FBS (Ohno et al., 2012). Two days after induction, cells were re-fed every 48 hours with adipocyte culture medium containing 1 nM T3, and 850 nM insulin. Cells were fully differentiated by day 6 after induction. Lipid droplets were visualized by Oil-red-O staining. To stimulate thermogenesis, differentiated cells were treated with norepinephrine (NE) at 1 or 10 μM for 4 hours. For protein expression studies, cells were incubated with forskolin at a dose of 10 μM for 6 hours.
For the analysis of integrin-FAK signaling, Itgb1 KO and Itgb5 KO CD81+ APCs and Lin−: Sca1+ cells from CRISPRi-Cd81 mice were cultured until confluency in non-coated plates in order to reduce basal FAK activity, and induced for 2 days in an adipogenic medium containing 0.5 mM IBMX, 2 μg ml−1 dexamethasone, 1 μM rosiglitazone, and 5% FBS. The medium was switched to FreeStyle293 Expression medium (Gibco) after washing with warm PBS to starve the cells. After 4 hours of starvation, cells were treated with irisin (Lake Pharma)(Kim et al., 2018) at 100 pM for indicated times. For antagonistic antibody treatment, cells were treated with specific antibodies against integrin β1 (clone: HA2/5, BD Biosciences) or integrin β5 (clone: ALULA)(Su et al., 2007) or mouse IgG as a negative control at 5 μg ml−1 for 60 min, followed by irisin treatment for 5 min.
Bone marrow-derived macrophages (BMDMs) were isolated from CRISPRi-Cd81 and control mice on a high-fat diet for 10 weeks and differentiated as described previously (Koliwad et al., 2010). In brief, male mice were euthanized with Avertin, and their tibias and femurs were flushed of bone marrow. Bone marrow was filtered and cleared of red blood cells with ACK lysis buffer. The bone marrow cells were plated, and the myeloid precursors were differentiated for 6-8 days in RPMI 1640 (Gibco) containing 10% FBS, 1% penicillin/streptomycin, 1% HEPES, and 10% previously prepared L929-conditioned media to yield BMDMs; media was changed every 2-3 days. To stimulate a pro-inflammatory response, we incubated BMDMs with a metabolic cocktail containing 30 mM glucose, 10 nM insulin, and 0.5 mM palmitic acid for 18 hours. Cells were maintained at 37°C.
METHOD DETAILS
scRNA sequencing.
Single-cell RNA extraction and mRNA amplification were performed on the C1™ Single-Cell Auto Prep Integrated Fluidic Circuit (IFC) following the methods described in the protocol (PN 100-7168, http://www.fluidigm.com/). For experiments where ERCC ExFold RNA Spike-In were used, the spikes were added to the lysis mix at a 20,000-fold dilution. The PCR thermal protocol was adapted from a publication that optimized template-switching chemistry for single-cell mRNA Seq (Fan et al., 2012) and is outlined in the C1™ Single-Cell Auto Prep System protocol. The cDNA reaction products were quantified using the Quant-iT™ PicoGreen® dsDNA Assay Kit (Invitrogen™) and subsequently diluted to a final concentration of 0.15–0.30 ng/μL using C1™ Harvest Reagent. The diluted cDNA reaction products were converted into mRNA Seq libraries using the Nextera® XT DNA Sample Preparation Kit (Illumina, FC-131-1096 and FC-131-1002, 1 kit used for 4 C1™ IFCs/384 samples) following manufacturer's instructions with minor modifications. Specifically, reactions were run at one-quarter of the recommended volume, the tagmentation step was extended to 10 min, and the extension time during the PCR step was increased from 30 seconds to 60 seconds. After the PCR step, samples were pooled, cleaned twice with 0.9X Agencourt AMPure XP SPRI beads (Beckman Coulter), eluted in C1 DNA Dilution Reagent at half of the recommended volume and quantified using a High sensitivity DNA chip (Agilent).
Transcriptome data analysis.
Sequenced tags from single-end sequenced libraries for the samples were pseudo-aligned to mouse reference transcriptome built from GENCODE Version M18. Transcript-level abundance estimates were generated using a fast RNA-seq quantification program, Kallisto (Version 0.44.0). All downstream analyses were performed using R (Version 3.5.1). Transcript-level estimates from Kallisto were imported into R and expression levels per gene were estimated using the Bioconductor package tximport (Version 1.10.0). Library size normalization was performed using edgeR (Version 3.24.0). Expression differences between samples were quantified using the limma-voom pipeline (limma Version 3.38.2). In the limma-voom pipeline used for differential gene expression analysis, raw counts were transformed into log2 counts per million, where “per million reads” were defined based on the library normalization factors computed by edgeR. A linear model was fitted to the voom normalized data, and comparison between groups, CD81+ and CD81− was obtained from the linear fitted model. Benjamini-Hochberg procedure was applied in order to control the false discovery rates. Transcriptome data of mouse APCs were analyzed from the recent scRNA sequencing data (GSE111588).
Bulk RNA-sequencing and analysis.
Total RNA was isolated from sorted CD81+ and CD81− cells from wild-type C57BL/6J mice at 10-weeks-old using RNeasy Micro Kit (QIAGEN). For transcriptome data analysis, CD81+ and CD81− cells were collected without overnight culture. cDNA synthesis and amplification were performed from 50 ng of total RNA using SMARTer® Universal Low Input RNA Kit (Clontech) according to manufacturer's instruction. Sequencing adapters were ligated, and high-throughput sequencing was performed using Illumina HiSeq 3000 instrument at the UCLA Clinical Microarray Core. To compare Fluidigm dataset and sorted bulk CD81+ transcriptome, FASTQ files from the bulk sequencing data were downsampled to 2,700,000 reads, which is the median read depth per cell for the Fluidigm dataset, using Seqtk (https://github.com/lh3/). Both Fluidigm and sorted CD81+/− FASTQ files were pseudoaligned and converted to TPM (Transcripts Per Million) by Kallisto (version 0.46.0) with default parameters. The integrated expression matrix was imported to Seurat and variable genes across the cells were identified by mean.var.plot (y.cutoff = 2,x.low.cutoff = 2). PCA and Hierarchical clustering were performed using the variable genes. The first 20 principal components (PC) were used for clustering. Biological pathway analysis was performed using Metascape (Tripathi et al., 2015).
Tissue histology and immunohistochemistry.
For hematoxylin and eosin (H&E) staining, tissues of mice were fixed in 4% paraformaldehyde (PFA) overnight at 4 °C, followed by dehydration in 70% ethanol. After the dehydration procedure, tissues were embedded in paraffin, sectioned at a thickness of 5-7 or 15 μm, and stained with H&E following the standard protocol. Images were acquired using a Revolve microscope (ECHO Laboratories). For immunostaining, paraffin-embedded tissues were deparaffinized twice in xylene and subsequently rehydrated. After incubating the slides for 20 min in boiling water, the tissues were blocked in PBS containing 10% goat or donkey serum with 0.1% Tween 20 for 60 min. After washing in PBS, slides were incubated with rabbit anti-UCP-1(1:200, ab10983, Abcam), goat anti-PERILIPIN-1 (1:100, ab61682, Abcam), rabbit anti-mCherry (1:500, ab167453, Abcam), rat anti-PDGFRα (1:100, 135908, Biolegend) and chicken anti-GFP (1:500, GFP-1020, Aves Labs) antibody overnight at 4 °C, followed by incubation with fluorescence-conjugated second antibody (1:500, Thermo Fisher) for 60 min at room temperature. Anti-mCherry antibody was used for tdTomato staining. Alexa Fluor 488 antibody was used as a second antibody for GFP, UCP1 in CD81-derived transplant. Alexa Fluor 555 antibody was used as a second antibody for mCherry. Alexa Fluor 647 antibody was used as a second antibody for UCP1 in CRISPRi-Cd81 mice, Cd81-CreERT2; Rosa26-mTmG mice, PpargCD81 KO mice study and PERILIPIN-1. After washing, the sections were stained with 4’,6-diamidino-2-phenylindole (DAPI) and mounted with mounting medium (Cytoseal 60, Thermo Scientific). Images of tissue samples were captured using the Inverted Microscope Leica DMi8 and analyzed using the Icy software (Version 2.0.0.0).
Fluorescence-activated cell sorting (FACS).
For the isolation of CD81+ and CD81− cells for differentiation studies in culture, adipose tissues from male mice at 8-16 weeks were digested using Collagenase D (1.5 U ml−1) and Dispase II (2.5 U ml−1) to isolate SVF as described previously (Aune et al., 2013), and subsequently cultured overnight (~ 20 hour) on non-coated plates. The SVF was first gated based on size and granularity. For scRNA sequencing, dead cells were removed based on SYTOX dead cell stain (1:400, Thermo Fisher). A panel of fluorescent antibodies (CD31-PE (1:300, 553373, BD Biosciences), CD45-PE (1:200, 12-0451-82, eBioscience), and TER-119-PE (1:200, 116207, Biolegend)) or MACS® Non-Adipocyte Progenitor Depletion Cocktail for mouse (130-106-639, Miltenyi Biotec) and MACS LS columns (Miltenyi Biotec) were used to deplete lineage+ (Lin+) cells. The following antibodies were used for the isolation of CD81+ APC (Lin−: Sca1+: CD81+) in mouse: Sca-1-PB (1:800, 108120, Biolegend) and CD81-APC (1:50, 104910, Biolegend) in autoMACS Rising Solution (Miltenyi Biotec) containing 0.5% BSA in the dark at 4°C for 15 min. This sorting strategy was used to isolate CD81+ APC from C57BL/6J wild-type, 129SVE mice, ThermoMouse, Itgb5 null mice, 129 Sv mice and Itgb1flox/flox mice, and Lin−: Sca1+ stromal cells from CRISPRi-Cd81 mice and control mice. CD140a-PE (1:200, 135905, Biolegend) was used to co-stain PDGFRα+ with Sca1+.
To evaluate CD81+ APC number in young vs. aged mice, stromal cells were isolated from the SVFs of Ing WAT of young mice (8-10 weeks old) and old mice (60 weeks old) without overnight culture. For intracellular staining, stromal cells were isolated from the SVFs of Ing WAT or Epi WAT without overnight culture. Cells were stained with above antibodies and fixed in 4% PFA for 15 min. Subsequently, cells were incubated in autoMACS Rising Solution containing 0.5% BSA and 0.1% saponin for 15 min and stained with Ki67-PE antibody for 30 min (1:300, 151209, Biolegend). Cell population (%) was calculated as frequency of parent. All the cells were isolated and analyzed using FACS Aria II equipped with 100 μm nozzle diameter. FlowJo software (version 10.5.3) was used for data analysis.
To identify immune cell, Ing WAT and Epi WAT were digested with Type II Collagenase (Sigma-Aldrich) at 2 mg ml−1 for 40 min at 37°C while shaking at 200 rpm. The SVF, containing immune cells and progenitors, was pelleted and separated from floating adipocytes by centrifugation at 800 × g for 8 min at 4°C. Red blood cells were lysed with ACK buffer for 3 min on ice. Cells were then filtered and blocked in TruStain FcX™ PLUS (1:200, 156604, Biolegend) before proceeding to staining for specific markers to be assessed by FACS. The following antibodies were used to identify immune cell in mouse: Sca-1-PECy7 (1:400, 122513, Biolegend), CD45-PE-Dazzle594 (1:4,000, 103145, Biolegend), F4/80-APC (1:400, 123116, Biolegend), CD64-PE (1:400, 139304, Biolegend), CD11c-BV785 (1:400, 117335, Biolegend), CD206-PerCP-Cy5.5 (1:400, 141715, Biolegend), Ly6C-BV510 (1:400, 128033, Biolegend), Ly6G-APC-Cy7 (1:400, 127623, Biolegend), SiglecF-APC (1:400, 155507, Biolegend), CD3-PE-Cy7 (1:400, 100219, Biolegend), CD4-PerCP (1:400, 100537, Biolegend), CD8a-APC (1:400, 100711, Biolegend), CD19-A700 (1:400, 115527, Biolegend), IL-23R-PE (1:400, 150903, Biolegend), NKp46-BV785 (1:400, 137637, Biolegend) in PBS containing 1% FBS and 2 mM EDTA in the dark at 4°C for 30 min. DAPI was added immediately prior to analysis to remove dead cells. The SVF was first gated based on size and granularity. This sorting strategy was used to identify immune cells from CRISPRi-Cd81 mice and control mice on a high-fat diet.
Human WAT analysis.
Prior to FACS analysis, individual cryovials of cells were rapidly thawed in a 37°C water bath (1–2 min agitation), resuspended in prewarmed animal component-free medium (Stem Cell Technologies) and seeded into separate wells of a six-well plate for 72 hours of incubation at 37°C. Cells were pooled, harvested by trypsinization with TrypLE™ Select, washed with culture medium and resuspended in autoMACS Rising Solution containing 0.5% BSA. The following antibodies were used for the isolation of CD81+ APC (Lin−: PDGFRα+: CD81+) from subcutaneous WAT in human: CD14-FITC (1:400, 301803, Biolegend), CD31-FITC (1:200, 303103, Biolegend), CD45-FITC (1:200, 304005, Biolegend), CD235a-FITC (1:500, 349103, Biolegend), PDGFRα (CD140a)-PerCP-Cy5.5 (1:200, 563575, BD Biosciences) and CD81-APC (1:500, 349510, Biolegend) in autoMACS Rising Solution containing 0.5% BSA in the dark at 4°C for 15 min. This sorting strategy was used to evaluate the correlations between CD81+ APC and clinical parameters. For intracellular staining, isolated SVF cells were stained with above antibodies without overnight culture, and immediately fixed by using Fixation/Permeabilization Solution Kit (BD Biosciences) as described protocol and stained with Ki67-PE antibody for 30 min (1:500, 350503, Biolegend). Cell population (%) was calculated as frequency of parent.
Virus construction.
We generated lentiviral shRNA constructs in pLKO.1-hygromycin (#24150, Addgene) that expressed shRNA targeting mouse Ptk2 (CCTGGCATCTTTGATATTATA) or scrambled control (CAACAAGATGAAGAGCACCAA) based on the previous study (Shibue and Weinberg, 2009). For virus production, HEK293T packaging cells were transfected with 10 μg of lentiviral plasmids and 10 μg the packaging constructs (psPAX2 and pMD2.G) using a calcium phosphate method. After 48 hours of incubation, the viral supernatant was collected and filtered.
Ptk2 KD, Itgb1 KO and Itgb5 KO cells.
For the generation of Ptk2 knockdown (KD) cells, immortalized Lin−: Sca1+: CD81+ cells from Ing WAT by using the SV40 Large T antigen as described previously (Shinoda et al., 2015a) were infected with pLKO.1-shRNA targeting Ptk2 or scrambled control, followed by hygromycin selection (150 μg ml−1). The knockdown efficiency was confirmed by immunoblotting for FAK. For the generation of Itgb1 KO cells, CD81+ APCs were isolated from the Ing WAT of Itgb1flox/flox mice after overnight culture and subsequently infected with retrovirus constructs expressing Cre (#34565, Addgene) or an empty vector control, followed by hygromycin selection. For the generation of Itgb5 KO cells, CD81+ cells were isolated from Ing WAT of Itgb5 null mice and wild-type mice (129 Sv) as control after overnight culture. By immunoblotting, we confirmed that both Itgb1 KO and Itgb5 KO cells did not express ITGB1 and ITGB5, respectively.
Cell proliferation assay.
To examine changes in APC proliferation in vivo, we administered EdU (25mg kg−1 body weight, I.P.) to C57BL/6J mice that were acclimated at 22°C or 4°C for 4 days. Two hours after EdU administration, mice were sacrificed and CD81+ APCs from the Ing WAT were isolated for analysis. EdU positive cells were detected by using Click-iT™ EdU Alexa Fluor™ 488 Flow Cytometry Assay Kit (Thermo Fisher) and FACS Aria II. For cell proliferation assay in CD81− cells and CD81+ cells, cells were isolated from Ing WAT or Epi WAT without overnight culture at the sorting day by FACS and seeded directly onto 24-well non-coated plates. Isolated cells were cultured for 5 days in DMEM/F12 medium containing 1% GlutaMAX-I and 10% FBS and re-fed every 48 hours with culture medium. Cell proliferation rate (% growth day−1) were calculated as relative values to the growth rate of CD81− cells from C57BL/6J mice at 8-10 weeks or 12 weeks of age. For cultured cell proliferation assays, Lin−: Sca1+ cells were isolated from the SVFs of Ing WAT of CRISPRi-Cd81 mice and littermate control mice at 10-12 weeks of age after overnight culture. To verify cell culture conditions, isolated Lin−: Sca1+ cells were cultured in non-coated plates or collagen-coated plates with cell culture medium. Cell proliferation rate (% growth day−1) were calculated as relative values to the growth rate of control cell on non-coated culture plates. To test the involvement of FBS concentration, isolated Lin−: Sca1+ cells were cultured in non-coated plates with cell culture medium containing FBS at indicated concentrations (0-20%) for 5 days. Cell proliferation rate (% growth day−1) were calculated as relative values to the growth rate of control cell under the condition of 10% FBS. For irisin experiments, isolated Lin−: Sca1+ cells were treated with irisin at indicated doses (1-10 ng ml−1) for 5 days in DMEM/F12 medium containing 1% GlutaMAX-I and 10% FBS. The culture medium was replaced every 48 hours. Cell proliferation rate (% growth day−1) were calculated as relative values to the growth rate of control cells without irisin treatment. For FAK inhibitor experiments in control and Ptk2 KD cell, CD81+ APCs were treated with FAK inhibitor (PF-573228, TOCRIS) ranging from 5 nM to 100 nM or vehicle (DMSO) for 4 days in DMEM (Corning) containing 5% FBS. For the FAK inhibitor experiments in CD81+ cell and Lin−: Sca1+ cells, cells were treated with FAK inhibitor at 1 μM or vehicle (DMSO) for 4 days in DMEM/F12 medium containing 1% GlutaMAX-I and 10% FBS. Culture media containing FAK inhibitor or vehicle were replaced every 24 hours. Cell proliferation rate (% growth day−1) were calculated as relative values to the growth rate of vehicle control cells. To harvest cells, cells were incubated with 0.05% trypsin-EDTA (Corning) at 37°C after washing with PBS. After neutralization with cell culture medium, cells were collected in 1.7 mL tubes. The wells were washed with cell culture medium one more time and then collected in same tube. After centrifugation at 300 × g for 5 min, cell pellets were resuspended in 100 μL of cell culture medium. Cell number was counted using a cell counter.
Fat transplantation.
CD81− and CD81+ APCs were isolated from the Ing WAT of ThermoMouse (Galmozzi et al., 2014) following overnight culture. Cells were cultured for 2 days under an adipogenic condition that contains 1 μM rosiglitazone. Subsequently, cells were gently collected using cell scraper and resuspended in a cell culture medium. Cells were implanted with 0.25% HydroMatrix™ Peptide Cell Culture Scaffold (Sigma-Aldrich). Cells (2 × 106 cells) mixed in HydroMatrix (200 μl) were injected subcutaneously into nude mice. Three days after transplantation, mice were kept under 12°C for 10 days. Mice were injected with rosiglitazone (10 mg kg−1 body weight) twice daily for 3 days from day 10 to 12. Luciferase activity was monitored at 3, 6, and 13 days after transplantation using an IVIS Spectrum Instrument (Caliper Life Sciences), as described previously (Galmozzi et al., 2014). Images were collected starting 15 min after injection of luciferin (150 mg kg−1 body weight, Goldbio). For acute stimulation, β3 adrenergic receptor agonist (CL-316, 243, Sigma-Aldrich) at 1 mg kg−1 body weight was injected intraperitoneally 5 hours before harvesting the transplanted fat pads. To examine the transcriptional profile of CD81-derived fat transplants, CD81+ APCs were isolated from the Ing WAT of C57BL/6J mice following overnight culture. Three days after transplantation, mice were kept under 12°C for 10 days. Mice were injected with rosiglitazone (10 mg kg−1 body weight) twice daily for 10 days from day 4 to 13. Transplanted fats were isolated for RNA extraction and transcriptional analyses by qRT-PCR. As comparisons, we included Ing WAT samples from wild-type mice under a thermoneutral condition (30°C) for 14 days and cold-acclimated mice at 8°C for 3 days.
Metabolic analysis in mice.
CRISPRi-Cd81 mice and littermate control mice were fed a 60% high-fat diet (HFD, Research Diets) under the ambient temperature condition. HFD feeding was started when mice were 8 weeks old. Whole-body energy expenditure, respiratory exchange ratio (RER), food intake, and locomotor activity (beam break counts) were monitored by a comprehensive lab animal monitoring system (CLAMS) (Columbus Instruments). Metabolic data were collected in mice on a HFD for 2 weeks at 22°C. Body-weight was monitored once a week. Fat mass and lean mass were measured by Body Composition Analyzer EchoMRI (Echo Medical Systems) at 8 weeks of HFD. The rectal temperature was monitored using a TH-5 thermometer (Physitemp). Electromyography (EMG) was recorded for 15 min at 30°C before cold stimulation, and for another 15 min at 8°C after the 3 days cold exposure, following our protocol (Chen et al., 2019; Yoneshiro et al., 2019). For glucose tolerance tests, mice on HFD for 2 or 8 weeks received an intraperitoneal injection of glucose (1.5 g kg−1 body weight) after 6 hours of fasting. For insulin tolerance tests, mice on HFD for 8 weeks received an intraperitoneal injection of insulin (1.0 U kg−1 body weight) after 3 hours of fasting. Blood samples were collected at several time points after injection, and glucose levels were measured using blood glucose test strips (Abbott). To measure liver triglyceride contents, liver tissues were homogenized in 350 μl ethanolic KOH (100% EtOH: 30% KOH=2:1) and incubated at 55°C overnight. Subsequently, tissue lysates were brought volume to 1 ml with 50% EtOH. After centrifugation, the supernatant was incubated with 1 M MgCl2 on ice for 10 min. Amounts of triglycerides were determined by an Infinity Triglycerides kit (Thermo Fisher). Exosome were isolated from serum at 8 weeks of HFD using exoEasy Maxi Kit (QIAGEN).
Oxygen consumption assay.
OCR in cultured adipocytes and isolated adipose tissue were measured using the Seahorse XFe Extracellular Flux Analyzer (Agilent) in a 24-well plate. CD81− and CD81+ cells were isolated from the SVFs of Ing WAT after overnight culture. Cells were differentiated as described above and maintained in XF assay medium supplemented with 1 mM sodium pyruvate, 2 mM GlutaMAX-I, and 25 mM glucose. For measurement of NE-induced respiration in cultured adipocytes, cells were treated with 10 μM NE during OCR measurement. For the measurement of uncoupled respiration in differentiated adipocytes, cells were stimulated with NE treatment and subsequently treated with 10 μM oligomycin. For tissue respiration analyses, punch-biopsy tissues of Ing WAT (1.5 mg) or iBAT (1.0 mg) were placed into XFe24 Islet Capture Microplates. OCR in Ing WAT and iBAT were measured 1 hour after incubation with assay medium supplemented with 1 mM sodium pyruvate, 2 mM GlutaMAX-I, and 25 mM glucose.
Protein interaction.
HEK293T cells were transfected with the plasmids containing gene of CD81 (#HG14244-CY, Sino Biological), Integrin αV (#27290, Addgene), Integrin β1 (#HG10587-CM, Sino Biological) or Integrin β5 (#HG10779-CM, Sino Biological) using Lipofectamine 2000 according to the manufacture’s manual. Twenty-four hours after transfection, medium was switched to Freestyle 293 expression medium and cells were incubated for 4 hours. Subsequently, cells were harvested in cold PBS and resuspended in cell lysis buffer (50 mM Tris-HCl pH7.5, 150 mM NaCl, 1 mM MgCl2, 0.1 mM CaCl2, 0.5% Brij98) containing phosphatase inhibitors (Roche) and protease inhibitors (Roche). Cell lysates were rotated for 1 hour at 4°C and then supernatants were collected after centrifugation at 17,000 × g for 10 min. The obtained lysates were rotated with protein A/G magnetic beads (Thermo Fisher) for 20 min at 4°C. After centrifugation at 1,000 × g for 2 min, the supernatants were subjected to anti-c-Myc magnetic beads (Thermo Fisher) and rotated overnight at 4°C. After washing in the buffer (50 mM Tris-HCl pH7.5, 150 mM NaCl, 1 mM MgCl2, 0.1 mM CaCl2, 0.5% Brij98) three times, precipitated proteins were eluted by boiling with 1 × Laemmli buffer. Each protein was detected by immunoblotting.
Irisin-induced FAK signaling in HEK293T cells.
Cells were treated and homogenized as reported (Chang and Finnemann, 2007; Kim et al., 2018). Briefly, HEK293T cells were transfected with the plasmids containing gene of CD81, Integrin αV (#HG11269-CM, Sino Biological) or Integrin β5 using Lipofectamine 2000 according to the manufacture’s protocol. Twenty-four hours after transfection, medium was switched to Freestyle 293 expression medium and cells were incubated for 4 hours. Subsequently, cells were treated with irisin protein for 5 min and harvested in RIPA buffer. Protein levels of phospho-FAK (Tyr397), FAK, CD81, integrin αV, integrin β5 and β-actin were visualized by immunoblot analysis.
Immunoblotting.
Cells and tissues were lysed in RIPA lysis and extraction buffer (Thermo Fisher), phosphatase inhibitors (Sigma-Aldrich) and protease inhibitors (Roche). Total protein lysates were boiled with Laemmli sample buffer containing 355 mM β-mercaptoethanol, loaded on a 4-12%, 4–15% or 8% SDS-PAGE. Subsequently, separated proteins were transferred onto PVDF membranes. PVDF membrane blots were blocked in 5% milk or 3-5% BSA in Tris-buffered saline with Tween 20 (TBS-T) for 1 hour at room temperature and incubated overnight at 4°C with rabbit anti-UCP-1 (1:1,000, ab10983, Abcam), anti-phospho-FAK (Tyr397) (1:1,000, 3283S, Cell Signaling), anti-FAK (1:1,000, 3285S, Cell Signaling), anti-Integrin αV (1:1,000, 60896S, Cell Signaling), anti-Integrin β1 (1:1,000, 34971S, Cell Signaling), anti-Integrin β5 (1:1,000, 3629S, Cell Signaling), anti-HA-tag (1:1,000, 3724, Cell Signaling), mouse anti-Myc-tag (9B11) (1:1,000, 2276S, Cell Signaling), anti-CD81 (1:100, sc-166029, Santa Cruz), anti-CD63 (1:200, sc-5275, Santa Cruz), or anti-β-actin (1:10,000, A3854, Sigma-Aldrich or ab49900, Abcam). Mitochondrial proteins were detected using MitoProfile Total OXPHOS Rodent Antibody Cocktail (1:1,000, ab110413, Abcam). Anti-rabbit IgG (711-035-152, Jackson ImmunoResearch) was used to a second antibody for UCP1, phospho-FAK (Tyr397), FAK, integrin αV, integrin β1, integrin β5, and HA-tag. Anti-mouse IgG (715-035-150, Jackson ImmunoResearch) was used as a secondary for Myc-tag, CD81, CD63, and Total OXPHOS Rodent Antibody Cocktail.
Gene expression analysis.
Total RNA was extracted from tissue or cells using TRIzol reagents (Thermo Fisher) followed by the RNeasy Mini Kit (QIAGEN) protocol. Complementary DNA was synthesized using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories) according to the provided protocol. Quantitative RT-PCR was performed using an ABI ViiA 7 PCR cycler (Applied Biosystems). Quantity of a particular gene in each sample was normalized to the TATA-binding protein, 36B4 or Hprt. Relative mRNA levels were determined by the ΔΔ Ct method and normalized to an internal calibrator specific to each gene using the formula 2−ΔΔCT, Primer sequences are provided in Table S3.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed using statistical software (SPSS 20.0, IBM). All data were represented as mean ± SEM, except where noted. Unpaired Student’s t-test was used for two-group comparisons. One-way ANOVA followed by Bonferroni's-test was used for multiple group comparisons. One-way ANOVA followed by Dunnett’s-test was used for cell proliferation studies. Unpaired Student’s t-test with Bonferroni’s correction was used for cell proliferation studies and transplantation results. Two-way repeated-measures ANOVA was used for cell proliferation assay results, cold tolerance test results, body weight results, whole-body energy expenditure results, GTT results, and ITT results. Correlations were examined with non-parametric Spearman correlation test. P < 0.05 was considered significant in all the experiments. The statistical parameters and the number of mice used per experiment are found in the figure legends.
Supplementary Material
Figure S1 (related to Figure 1). Technical validation of the single-cell RNA-seq analysis.
A) Illustration of the scRNA-seq experiment. scRNA-seq analysis in lineage− (Lin−) cells from the stromal vascular fraction (SVF) of mouse adipose tissue.
B) Sequential gating to isolate Lin− cells in the SVF from the adipose tissue.
C) Relationship between estimated RNA level (TPM: transcripts per kilobase million) in single-cell RNA-seq and External RNA Controls Consortium (ERCC) spike input. Note that estimated RNA levels are positively correlated with ERCC spike input (r = 0.953).
D) Relationship between coefficient of variation (CV) of TPM or detection rate (%) and ERCC spike input (copies per cell).
E) Meta-analysis of scRNA-seq data GSE111588. Violin plots showing mRNA levels of indicated genes (Cd81, Sca1, Acta2 and Sm22) in type 0 to 6 cell clusters. Note that Cd81, Sca1, Acta2, and Sm22 are highly enriched in Type 6 cluster.
F) Hierarchical clustering of bulk CD81+, bulk CD81− and stromal cell types identified in Fig. 1A. The horizontal distance represents similarities among each cluster. See the bulk RNA-seq transcriptomics in Fig 3.
G) Sca1 and CD81 expression levels in Lin− PDGFRα+ cells in the SVF from mouse inguinal WAT. Note that all the Lin− PDGFRα+ cells are marked by Sca1 in the SVF from the inguinal WAT.
Figure S2 (related to Figure 2). CD81+ stromal cells give rise to beige adipocytes.
A) Schematic illustration of the experiments in culture cell studies. CD81+ cells (Lin−: Sca1+: CD81+) and CD81− cells (Lin−: Sca1+: CD81−) were isolated from the SVFs of Ing WAT of BL6 mice (8-10 weeks of age) after overnight culture. Isolated cells were differentiated for 6 days under an adipogenic condition and stimulated with or without norepinephrine (NE) for 4 hours prior to harvest. During the adipogenic induction phase during day 0 to day 2, cells were cultured with rosiglitazone (Fig 2B-F) or without rosiglitazone (Fig S2B-C).
B) Relative mRNA expression of adipogenic and thermogenic genes in differentiated CD81− cells and CD81+ cells. Cells were differentiated for 6 days under an adipogenic condition without rosiglitazone. mRNA expression relative to Tbp. n=3.
C) Relative mRNA expression of Ucp1 and Pgc1a in differentiated CD81− cells and CD81+ cells. Cells were differentiated for 6 days under an adipogenic condition without rosiglitazone and stimulated with or without NE at 10 μM for 4 hours prior to harvest. mRNA expression relative to Tbp. n=3.
D) Uncoupled OCR (pmol min−1) in differentiated CD81− cells and CD81+ cells. Differentiated adipocytes were stimulated with NE at 10 μM and subsequently treated with oligomycin (10 μM). 20,000 cells were seeded onto plates. n=10. (B, C, D) *P < 0.05, **P < 0.01 by unpaired Student’s t-test.
E) Illustration of the transplantation experiments. CD81− cells and CD81+ cells were isolated from Ing WAT of ThermoMouse (UCP1-luciferase reporter mice). Cells were differentiated for 2 days under an adipogenic condition with rosiglitazone and transplanted into the subcutaneous region of nude mice. Luciferase activity of transplanted cells were monitored at day 3, 6, and 13 after transplantation. Nude mice were kept at 12°C after 3 days transplantation. Mice were treated with rosiglitazone (10 mg kg body weight−1) twice daily during day 10 to day 12.
F) Representative images of CD81-derived transplants. Arrowhead indicates a CD81-derived transplant in the subcutaneous region of nude mice.
G) Immunofluorescent staining for tdTomato and UCP1 in the transplants in (F). DAPI for counterstaining of immunofluorescent staining. Anti-mCherry antibody was used for tdTomato staining. Note that tdTomato is expressed in fat transplants originated from ThermoMouse that express a lufierase-tdTomato fusion protein. Scale bar = 100 μm.
H) Relative mRNA expression of indicated genes in CD81+ APC-derived transplants. Mice were kept at 12°C cold and rosiglitazone treatment for 10 days. Ing WAT of wild-type mice under a thermoneutral condition (TN: 30°C) and cold-acclimated mice at 8°C for 3 days were used as references. mRNA expression relative to Tbp. Ing WAT (TN), n=3; Ing WAT (Cold), n=5; Transplant, n=7. *P < 0.05, **P < 0.01 by unpaired Student’s t-test with Bonferroni’s correction.
I) Unstained signal for GFP in the Ing WAT (left) and interscapular BAT (iBAT; right) of Cd81-lineage reporter mice. Scale bar = 100 μm.
J) Immunofluorescent staining for GFP and UCP1 in the posterior region of Ing WAT of Cd81-lineage reporter mice at 8°C. Arrow indicates GFP− beige adipocytes (GFP−: UCP1+ cells). Scale bar = 50 μm.
K) Immunofluorescent staining for GFP and UCP1 at 8°C in iBAT of Cd81-lineage reporter mice. Scale bar = 100 μm.
L) Immunofluorescent staining for GFP and PDGFRα in the Ing WAT of Cd81-lineage reporter mice at 8°C. Scale bar = 50 μm. Data in (B, C, D, H) are represented as mean ± SEM.
Figure S3 (related to Figure 3). Characterization of CD81+ cells in adipose tissues.
A) The upregulated signaling pathways in CD81+ cells relative to CD81− cells from Epi WAT, and iBAT of BL6 mice (Top 5 signaling pathway). GO analysis was performed in transcriptomics data of CD81+ cells and CD81− cells by Metascape. Epi WAT,n=3; iBAT, n=2.
B) Immunoblotting for UCP1 in the Ing WAT of BL6 mice at 8 weeks old, 24 weeks old, 40 weeks old, and 72 weeks old mice. Male BL6 mice were kept at 22°C or 8°C for 5 days. β-actin as a loading control. Molecular weight (MW) is shown on the right.
C) CD81+ cell population (%) of Lin−: Sca1+ cells in the Ing WAT of young mice (8-10 weeks old) and old mice (60 weeks old). Male BL6 mice were kept at 22°C. Cells were isolated from the SVFs of above mice without overnight culture. n=12 per group. *P < 0.05 by unpaired Student’s t-test.
D) The downregulated signaling pathways in CD81+ cells relative to CD81− cells from Ing WAT (10 weeks old). Reactome gene sets analysis was performed in transcriptomics data of CD81+ cells and CD81− cells by Metascape. n=3.
Data in (C) are represented as mean ± SEM.
Figure S4 (related to Figure 4). The requirement of FAK for CD81+ APC proliferation.
A) Gene expressions related to FAK signaling pathway enriched in CD81+ cells relative to CD81− cells in the Ing WAT. The color scale shows intensity representing the mRNA levels in blue (low expression)-white-red (high expression) scheme. n=3.
B) Time-dependent effect of an ATP competitive FAK inhibitor, PF-573228, on primary CD81+ cell growth. Ing WAT-derived CD81+ cells were seeded on non-coated plates. Cells were treated with PF-573228 at a dose of 1 μM or vehicle (DMSO) for indicated time. Culture media containing FAK inhibitor or vehicle were replaced every 24 hours. n=3. **P < 0.01 by two-way repeated measures ANOVA.
C) Mice expressing dCas9-KRAB on the H11 locus (dCas9-KRAB mouse) were crossed with transgenic mice expressing gRNA targeting Cd81 on the H11 locus to generate CRISPRi-Cd81 mice.
D) Relative mRNA expression of Cd81 in Lin−: Sca1+ stromal cells from Ing WAT of CRISPRi-Cd81 mice and littermate control mice. mRNA expression relative to Tbp. n=6.
E) Relative mRNA expression of Cd81 in indicated tissues of CRISPRi-Cd81 mice and littermate control mice. mRNA expression relative to Tbp. n=3.
F) Validation of cell culture conditions to assess APC proliferation. Lin−: Sca1+ cells from the Ing WAT of CRISPRi-Cd81 and control mice were cultured on non-coated culture plates or collagen-coated plates. n=4.
G) Ing WAT-derived Lin−: Sca1+ cells from CRISPRi-Cd81 and control mice were cultured in media with indicated concentrations of FBS. n=4. (D-G) *P < 0.05, **P < 0.01 by unpaired Student’s t-test. N.S., not significant. Data in (B, D-G) are represented as mean ± SEM.
Figure S5 (related to Figure 4). The requirement of integrins for CD81-FAK signaling.
A) Transcriptional profile of integrin family members in CD81+ cells from Ing WAT of BL6 mice (10 weeks old). Isolated cells were analyzed by RNA-sequencing. n = 3.
B) Illustration of the experiment in Fig. 4F. CD81+ cells were isolated from Ing WAT of Itgb1flox/flox mice. Cells were infected with retrovirus containing Cre or an empty vector control, followed by hygromycin selection at a dose of 150 μg ml−1. Isolated cells were cultured until confluency and incubated for additional 2 days under adipogenic conditions. Subsequently, cells were cultured in FreeStyle293 Expression medium for starvation. After 4 hours of starvation, cells were treated with antagonistic antibodies against integrin β1 (ITGB1) or integrin β5 (ITGB5) or mouse IgG control at 5 μg ml−1 for 60 min. Subsequently, cells were treated with irisin at 100 pM for 5 min.
C) Illustration of the experiment in Fig. 4H. CD81+ cells were isolated from Ing WAT of Itgb5 KO mice and wild-type control (129Sv). Cells were cultured and treated with irisin as described in (B).
D) Illustration of the experiment in Fig. 4I. Ing WAT-derived Lin−: Sca1+ cells from CRISPRi-Cd81 mice and littermate control mice were cultured as described in (B).
E) Immunoblotting for FAK phosphorylation (pTyr397), total FAK, and β-actin in HEK293T cells expressing integrin αV (ITGAV) and integrin β5 (ITGB5) in the presence or absence of CD81. Cells were subsequently treated with irisin at 1 pM, 10 pM, and 100 pM for 5 min.
Figure S6 (related to Figure 5). CD81 is dispensable for brown adipocyte biogenesis in iBAT and muscle shivering.
A) Representative images of H&E staining and UCP1 immunofluorescent staining in iBAT of control mice (top) and CRISPRi-Cd81 mice (bottom) following cold exposure. Scale bar = 200 μm.
B) Relative mRNA expression of indicated genes in the iBAT of mice in (A). mRNA expression relative to Tbp. n=5.
C) OCR in the iBAT of mice after cold exposure. Punch-biopsy tissues (1.0 mg) were analyzed by the Seahorse XF Analyzer. n=4. (B, C) **P < 0.01 by unpaired Student’s t-test. N.S., not significant.
D) Representative electromyography (EMG) traces in skeletal muscle for CRISPRi-Cd81 mice and control mice at 30 °C and 8 °C.
E) Quantification of the images in (D) converted to the root mean square (RMS). n = 3. **P < 0.01 by one-way ANOVA followed by Bonferroni’s post hoc test. N.S., not significant.
F) Representative images of H&E staining in the Ing WAT of PpargCD81 KO and control mice after 8 °C cold environment for 10 days. Arrowhead indicates lymph node (LN). Scale bar = 200 μm. Data in (B, C, E) are represented as mean ± SEM.
Figure S7 (related to Figure 6). Metabolic phenotype of CRISPRi-Cd81 mice.
A) Immunoblotting for CD81 and exosome marker (CD63) in isolated exosome from the blood of CRISPRi-Cd81 mice and control mice on HFD for 8 weeks.
B) GTT in CRISPRi-Cd81 and control mice on HFD for 2 weeks. Mice were fasted for 6 hours and glucose (1.5 g kg body weight−1) was administered via i.p. Note that body-weight between CRISPRi-Cd81 mice and littermate controls was indistinguishable at 2 weeks of HFD. Control, n=8; CRISPRi-Cd81, n=6. *P < 0.05, **P < 0.01 by two-way repeated measures ANOVA with post hoc test by unpaired Student’s t-test.
C) Representative images of the liver of CRISPRi-Cd81 mice and control mice on 10 weeks of HFD.
D) Relative mRNA expression of Cd81 in the BMDM from CRISPRi-Cd81 mice and control mice, and CD81+ cells in the Ing WAT of BL6 mice. CRISPRi-Cd81 mice and control mice were on HFD for 10 weeks. mRNA expression relative to Hprt. BMDM, n=6; CD81+ cells, n=3.
E) The number of macrophages (F4/80+) in BMDM cultures differentiated from bone marrow of CRISPRi-Cd81 mice and control mice, quantified by FACS. Mice were on a high-fat diet for 10 weeks. n=6.
F) The expression of M1-like macrophage marker (CD11c) among total macrophages (F4/80+) in BMDM cultures from CRISPRi-Cd81 mice and control mice by FACS (MFI). Mice were on HFD for 10 weeks. n=6.
G) Relative mRNA expression of Tnfa in the BMDM from CRISPRi-Cd81 mice and control mice on 10 weeks of HFD. Cells were stimulated with or without metabolic cocktail (30 mM glucose, 10 nM insulin and 0.5 mM palmitic acid) for 18 hours. mRNA expression relative to Hprt. n=6. **P < 0.01 by one-way ANOVA followed by Bonferroni’s post hoc test. N.S., not significant.
H) The absolute number of leukocytes (CD45+), B cells (CD45+: CD3−: CD19+), T cells (CD45+: CD3+), CD4+ T cells (CD45+: CD3+: CD4+), CD8+ T cells (CD45+: CD3+: CD8+), Th17 cells (CD45+: CD3+: CD4+: IL-23R+), and NK cells (CD45+: CD3−: CD4−: CD8−: CD19−: NKp46+) per gram of Ing WAT of CRISPRi-Cd81 mice and control mice by FACS. Mice were on HFD for 10 weeks. Control, n=8-9; CRISPRi-Cd81, n=5-6.
I) Relative mRNA expression of indicated pro-inflammatory and pro-fibrosis genes in Epi WAT of CRISPRi-Cd81 mice and control mice. Mice were on HFD for 10 weeks. Control, n=7; CRISPRi-Cd81, n=6. mRNA expression relative to Tbp.
J) The expression (MFI) of M1-like macrophage marker (CD11c) among total macrophages (CD45+: CD64+) in Epi WAT of CRISPRi-Cd81 mice and control mice on 10 weeks of HFD. Control, n=9; CRISPRi-Cd81, n=6.
K) Ki67+ cell population (%) in CD81− (Lin−: PDGFRα+: CD81−) and CD81+ cells (Lin−: PDGFRα+: CD81+) in human subcutaneous WAT biopsies. n=7. (D-F, H-K) **P < 0.01 by unpaired Student’s t-test. N.S., not significant. Data in (B, D-K) are represented as mean ± SEM.
Table S1 (related to Figure 1D): List of cell surface markers highly expressed in Type III cell population (top 20).
Table S2 (related to Table 1): Baseline characteristics of study participants (n=28).
Table S3 (related to STAR methods): Primer sequences used for quantitative RT-PCR.
Highlights:
Beige fat progenitors are marked by cell surface proteins, PDGFRα, Sca1, and CD81.
Beige APC proliferation is regulated by temperature, genetic background, and aging.
CD81 mediates integrin-FAK signaling in response to irisin.
CD81 loss causes obesity, insulin resistance, and adipose tissue inflammation.
A subset of adipocyte progenitor cells give rise to beige fat through signaling responses to the hormone irisin through the action of specific integrins and the co-receptor CD81.
ACKNOWLEDGMENTS
We are grateful to Drs. C. Paillart for his support in the CLAMS studies, D. Scheel for histology, and Drs. Pollen, and Nowakowski for single cell analysis. This work was supported by the NIH (DK97441 and DK112268) and the Edward Mallinckrodt, Jr. Foundation to S. Kajimura, and National Center for Advancing Translational Sciences, NIH, through UCSF-CTSI grant UL1 TR000004 to S. Koliwad, the JPB Foundation to B.M. Spiegelman, and NIH (DK110098) to K. Atabai. Y. Oguri and K. Tajima were supported by the post-doctoral fellowship from the Manpei Suzuki Diabetes Foundation. Y. Oguri is supported by JSPS, K. Shinoda by NIH (DK110426 and P30DK026687), H. Kim by NIH (5T32HL007374-38), D.L. Alba by the American Diabetes Association (1-18-PMF-003), R.N. Pradhan by the Vera M. Long fellowship, T. Yoneshiro by the Uehara Memorial Foundation, and R. Datta by Larry L. Hillblom Foundation fellowship (2019-D-004-FEL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Alba DL, Farooq JA, Lin MYC, Schafer AL, Shepherd J, and Koliwad SK (2018). Subcutaneous Fat Fibrosis Links Obesity to Insulin Resistance in Chinese Americans. The Journal of clinical endocrinology and metabolism 103, 3194–3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aune UL, Ruiz L, and Kajimura S (2013). Isolation and differentiation of stromal vascular cells to beige/brite cells. Journal of visualized experiments : JoVE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berditchevski F, Zutter MM, and Hemler ME (1996). Characterization of novel complexes on the cell surface between integrins and proteins with 4 transmembrane domains (TM4 proteins). Molecular biology of the cell 7, 193–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DC, Jiang Y, Arpke RW, Close EL, Uchida A, Reading D, Berglund ED, Kyba M, and Graff JM (2017). Cellular Aging Contributes to Failure of Cold-Induced Beige Adipocyte Formation in Old Mice and Humans. Cell metabolism 25, 481. [DOI] [PubMed] [Google Scholar]
- Berry DC, Jiang Y, and Graff JM (2016). Mouse strains to study cold-inducible beige progenitors and beige adipocyte formation and function. Nature communications 7, 10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry R, and Rodeheffer MS (2013). Characterization of the adipocyte cellular lineage in vivo. Nature cell biology 15, 302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostrom P, Wu J, Jedrychowski MP, Korde A, Ye L, Lo JC, Rasbach KA, Bostrom EA, Choi JH, Long JZ, et al. (2012). A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 481, 463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredella MA, Gill CM, Keating LK, Torriani M, Anderson EJ, Punyanitya M, Wilson KE, Kelly TL, and Miller KK (2013). Assessment of abdominal fat compartments using DXA in premenopausal women from anorexia nervosa to morbid obesity. Obesity (Silver Spring, Md 21, 2458–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burl RB, Ramseyer VD, Rondini EA, Pique-Regi R, Lee YH, and Granneman JG (2018). Deconstructing Adipogenesis Induced by beta3-Adrenergic Receptor Activation with Single-Cell Expression Profiling. Cell metabolism 28, 300–309 e304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, and Finnemann SC (2007). Tetraspanin CD81 is required for the alpha v beta5-integrin-dependent particle-binding step of RPE phagocytosis. Journal of cell science 120, 3053–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Ikeda K, Yoneshiro T, Scaramozza A, Tajima K, Wang Q, Kim K, Shinoda K, Sponton CH, Brown Z, et al. (2019). Thermal stress induces glycolytic beige fat formation via a myogenic state. Nature 565, 180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherukuri A, Shoham T, Sohn HW, Levy S, Brooks S, Carter R, and Pierce SK (2004). The tetraspanin CD81 is necessary for partitioning of coligated CD19/CD21-B cell antigen receptor complexes into signaling-active lipid rafts. J Immunol 172, 370–380. [DOI] [PubMed] [Google Scholar]
- Chouchani ET, and Kajimura S (2019). Metabolic adaptation and maladaptation in adipose tissue. Nat Metab 1, 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P, Levy JD, Zhang Y, Frontini A, Kolodin DP, Svensson KJ, Lo JC, Zeng X, Ye L, Khandekar MJ, et al. (2014). Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell 156, 304–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaianni G, Cuscito C, Mongelli T, Pignataro P, Buccoliero C, Liu P, Lu P, Sartini L, Di Comite M, Mori G, et al. (2015). The myokine irisin increases cortical bone mass. Proceedings of the National Academy of Sciences of the United States of America 112, 12157–12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M, Raposo G, and Thery C (2014). Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 30, 255–289. [DOI] [PubMed] [Google Scholar]
- Fan JB, Chen J, April CS, Fisher JS, Klotzle B, Bibikova M, Kaper F, Ronaghi M, Linnarsson S, Ota T, et al. (2012). Highly parallel genome-wide expression analysis of single mammalian cells. PloS one 7, e30794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlin BS, Memetimin H, Confides AL, Kasza I, Zhu B, Vekaria HJ, Harfmann B, Jones KA, Johnson ZR, Westgate PM, et al. (2018). Human adipose beiging in response to cold and mirabegron. JCI Insight 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galmozzi A, Sonne SB, Altshuler-Keylin S, Hasegawa Y, Shinoda K, Luijten IHN, Chang JW, Sharp LZ, Cravatt BF, Saez E, et al. (2014). ThermoMouse: an in vivo model to identify modulators of UCP1 expression in brown adipose tissue. Cell reports 9, 1584–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh YY, Pal M, Chong HC, Zhu P, Tan MJ, Punugu L, Lam CR, Yau YH, Tan CK, Huang RL, et al. (2010). Angiopoietin-like 4 interacts with integrins beta1 and beta5 to modulate keratinocyte migration. Am J Pathol 177, 2791–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra C, Koza RA, Yamashita H, Walsh K, and Kozak LP (1998). Emergence of brown adipocytes in white fat in mice is under genetic control. Effects on body weight and adiposity. The Journal of clinical investigation 102, 412–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y, Ikeda K, Chen Y, Alba DL, Stifler D, Shinoda K, Hosono T, Maretich P, Yang Y, Ishigaki Y, et al. (2018). Repression of Adipose Tissue Fibrosis through a PRDM16-GTF2IRD1 Complex Improves Systemic Glucose Homeostasis. Cell metabolism 27, 180–194 e186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinonen S, Muniandy M, Buzkova J, Mardinoglu A, Rodriguez A, Fruhbeck G, Hakkarainen A, Lundbom J, Lundbom N, Kaprio J, et al. (2017). Mitochondria-related transcriptional signature is downregulated in adipocytes in obesity: a study of young healthy MZ twins. Diabetologia 60, 169–181. [DOI] [PubMed] [Google Scholar]
- Hepler C, Shan B, Zhang Q, Henry GH, Shao M, Vishvanath L, Ghaben AL, Mobley AB, Strand D, Hon GC, et al. (2018). Identification of functionally distinct fibro-inflammatory and adipogenic stromal subpopulations in visceral adipose tissue of adult mice. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Kang Q, Yoneshiro T, Camporez JP, Maki H, Homma M, Shinoda K, Chen Y, Lu X, Maretich P, et al. (2017). UCP1-independent signaling involving SERCA2b-mediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis. Nature medicine 23, 1454–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokinen R, Rinnankoski-Tuikka R, Kaye S, Saarinen L, Heinonen S, Myohanen M, Rappou E, Jukarainen S, Rissanen A, Pessia A, et al. (2018). Adipose tissue mitochondrial capacity associates with long-term weight loss success. International journal of obesity (2005) 42, 817–825. [DOI] [PubMed] [Google Scholar]
- Kajimura S, Spiegelman BM, and Seale P (2015). Brown and Beige Fat: Physiological Roles beyond Heat Generation. Cell metabolism 22, 546–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Wrann CD, Jedrychowski M, Vidoni S, Kitase Y, Nagano K, Zhou C, Chou J, Parkman VA, Novick SJ, et al. (2018). Irisin Mediates Effects on Bone and Fat via alphaV Integrin Receptors. Cell 175, 1756–1768 e1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koliwad SK, Streeper RS, Monetti M, Cornelissen I, Chan L, Terayama K, Naylor S, Rao M, Hubbard B, and Farese RV Jr. (2010). DGAT1-dependent triacylglycerol storage by macrophages protects mice from diet-induced insulin resistance and inflammation. The Journal of clinical investigation 120, 756–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Petkova AP, Konkar AA, and Granneman JG (2015). Cellular origins of cold-induced brown adipocytes in adult mice. FASEB J 29, 286–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Petkova AP, Mottillo EP, and Granneman JG (2012). In vivo identification of bipotential adipocyte progenitors recruited by beta3-adrenoceptor activation and high-fat feeding. Cell metabolism 15, 480–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidell ME, Betz MJ, Dahlqvist Leinhard O, Heglind M, Elander L, Slawik M, Mussack T, Nilsson D, Romu T, Nuutila P, et al. (2013). Evidence for two types of brown adipose tissue in humans. Nature medicine 19, 631–634. [DOI] [PubMed] [Google Scholar]
- Lin JZ, Rabhi N, and Farmer SR (2018). Myocardin-Related Transcription Factor A Promotes Recruitment of ITGA5+ Profibrotic Progenitors during Obesity-Induced Adipose Tissue Fibrosis. Cell reports 23, 1977–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JZ, Svensson KJ, Tsai L, Zeng X, Roh HC, Kong X, Rao RR, Lou J, Lokurkar I, Baur W, et al. (2014). A smooth muscle-like origin for beige adipocytes. Cell metabolism 19, 810–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk CT, Shi SY, Cai EP, Sivasubramaniyam T, Krishnamurthy M, Brunt JJ, Schroer SA, Winer DA, and Woo M (2017). FAK signalling controls insulin sensitivity through regulation of adipocyte survival. Nature communications 8, 14360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcelin G, Ferreira A, Liu Y, Atlan M, Aron-Wisnewsky J, Pelloux V, Botbol Y, Ambrosini M, Fradet M, Rouault C, et al. (2017). A PDGFRalpha-Mediated Switch toward CD9(high) Adipocyte Progenitors Controls Obesity-Induced Adipose Tissue Fibrosis. Cell metabolism 25, 673–685. [DOI] [PubMed] [Google Scholar]
- Mather KJ, Hunt AE, Steinberg HO, Paradisi G, Hook G, Katz A, Quon MJ, and Baron AD (2001). Repeatability characteristics of simple indices of insulin resistance: implications for research applications. The Journal of clinical endocrinology and metabolism 86, 5457–5464. [DOI] [PubMed] [Google Scholar]
- Mattila PK, Feest C, Depoil D, Treanor B, Montaner B, Otipoby KL, Carter R, Justement LB, Bruckbauer A, and Batista FD (2013). The actin and tetraspanin networks organize receptor nanoclusters to regulate B cell receptor-mediated signaling. Immunity 38, 461–474. [DOI] [PubMed] [Google Scholar]
- McDonald ME, Li C, Bian H, Smith BD, Layne MD, and Farmer SR (2015). Myocardin-related transcription factor A regulates conversion of progenitors to beige adipocytes. Cell 160, 105–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen AE, Kanamaluru D, Yan K, Gray NE, Wu L, Li ML, Chang A, Hasan A, Stifler D, Koliwad SK, et al. (2017). The C-terminal fibrinogen-like domain of angiopoietin-like 4 stimulates adipose tissue lipolysis and promotes energy expenditure. The Journal of biological chemistry 292, 16122–16134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick D, Sakers A, Irgebay Z, Okada C, Calvert C, Morley MP, Percec I, and Seale P (2019). Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science (New York, NY 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniyappa R, Lee S, Chen H, and Quon MJ (2008). Current approaches for assessing insulin sensitivity and resistance in vivo: advantages, limitations, and appropriate usage. American journal of physiology 294, E15–26. [DOI] [PubMed] [Google Scholar]
- Ohno H, Shinoda K, Spiegelman BM, and Kajimura S (2012). PPARgamma agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell metabolism 15, 395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren R, Takahashi S, Doss C, Levy R, and Levy S (1990). TAPA-1, the target of an antiproliferative antibody, defines a new family of transmembrane proteins. Molecular and cellular biology 10, 4007–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raajendiran A, Ooi G, Bayliss J, O'Brien PE, Schittenhelm RB, Clark AK, Taylor RA, Rodeheffer MS, Burton PR, and Watt MJ (2019). Identification of Metabolically Distinct Adipocyte Progenitor Cells in Human Adipose Tissues. Cell reports 27, 1528–1540 e1527. [DOI] [PubMed] [Google Scholar]
- Sanchez-Gurmaches J, and Guertin DA (2014). Adipocytes arise from multiple lineages that are heterogeneously and dynamically distributed. Nature communications 5, 4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TJ, Huang TL, Tran TT, Zhang H, Townsend KL, Shadrach JL, Cerletti M, McDougall LE, Giorgadze N, Tchkonia T, et al. (2011). Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proceedings of the National Academy of Sciences of the United States of America 108, 143–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalie PC, Dong H, Zachara M, Russeil J, Alpern D, Akchiche N, Caprara C, Sun W, Schlaudraff KU, Soldati G, et al. (2018). A stromal cell population that inhibits adipogenesis in mammalian fat depots. Nature 559, 103–108. [DOI] [PubMed] [Google Scholar]
- Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, Cohen P, Cinti S, and Spiegelman BM (2011). Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. The Journal of clinical investigation 121, 96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao M, Wang QA, Song A, Vishvanath L, Busbuso NC, Scherer PE, and Gupta RK (2019). Cellular Origins of Beige Fat Cells Revisited. Diabetes 68, 1874–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp LZ, Shinoda K, Ohno H, Scheel DW, Tomoda E, Ruiz L, Hu H, Wang L, Pavlova Z, Gilsanz V, et al. (2012). Human BAT possesses molecular signatures that resemble beige/brite cells. PloS one 7, e49452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibue T, and Weinberg RA (2009). Integrin beta1-focal adhesion kinase signaling directs the proliferation of metastatic cancer cells disseminated in the lungs. Proceedings of the National Academy of Sciences of the United States of America 106, 10290–10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda K, Luijten IH, Hasegawa Y, Hong H, Sonne SB, Kim M, Xue R, Chondronikola M, Cypess AM, Tseng YH, et al. (2015a). Genetic and functional characterization of clonally derived adult human brown adipocytes. Nature medicine 21, 389–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda K, Ohyama K, Hasegawa Y, Chang HY, Ogura M, Sato A, Hong H, Hosono T, Sharp LZ, Scheel DW, et al. (2015b). Phosphoproteomics Identifies CK2 as a Negative Regulator of Beige Adipocyte Thermogenesis and Energy Expenditure. Cell metabolism 22, 997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su G, Hodnett M, Wu N, Atakilit A, Kosinski C, Godzich M, Huang XZ, Kim JK, Frank JA, Matthay MA, et al. (2007). Integrin alphavbeta5 regulates lung vascular permeability and pulmonary endothelial barrier function. Am J Respir Cell Mol Biol 36, 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima K, Ikeda K, Chang HY, Chang CH, Yoneshiro T, Oguri Y, Jun H, Wu J, Ishihama Y, and Kajimura S (2019). Mitochondrial lipoylation integrates age-associated decline in brown fat thermogenesis. Nature Metabolism 1, 886–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Hippenmeyer S, Wang C, Gamboa M, Zong H, Chen-Tsai Y, and Luo L (2011). Site-specific integrase-mediated transgenesis in mice via pronuclear injection. Proceedings of the National Academy of Sciences of the United States of America 108, 7902–7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Termini CM, and Gillette JM (2017). Tetraspanins Function as Regulators of Cellular Signaling. Front Cell Dev Biol 5, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che J, Mulder LC, et al. (2015). Meta- and Orthogonal Integration of Influenza "OMICs" Data Defines a Role for UBR4 in Virus Budding. Cell host & microbe 18, 723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujino K, Li JT, Tsukui T, Ren X, Bakiri L, Wagner E, and Sheppard D (2017). Fra-2 negatively regulates postnatal alveolar septation by modulating myofibroblast function. Am J Physiol Lung Cell Mol Physiol 313, L878–L888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lans AA, Hoeks J, Brans B, Vijgen GH, Visser MG, Vosselman MJ, Hansen J, Jorgensen JA, Wu J, Mottaghy FM, et al. (2013). Cold acclimation recruits human brown fat and increases nonshivering thermogenesis. The Journal of clinical investigation 123, 3395–3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishvanath L, MacPherson KA, Hepler C, Wang QA, Shao M, Spurgin SB, Wang MY, Kusminski CM, Morley TS, and Gupta RK (2016). Pdgfrbeta+ Mural Preadipocytes Contribute to Adipocyte Hyperplasia Induced by High-Fat-Diet Feeding and Prolonged Cold Exposure in Adult Mice. Cell metabolism 23, 350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Ishibashi J, Trefely S, Shao M, Cowan AJ, Sakers A, Lim HW, O'Connor S, Doan MT, Cohen P, et al. (2019). A PRDM16-Driven Metabolic Signal from Adipocytes Regulates Precursor Cell Fate. Cell metabolism 30, 174–189 e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, Khandekar M, Virtanen KA, Nuutila P, Schaart G, et al. (2012). Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 150, 366–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneshiro T, Aita S, Matsushita M, Kayahara T, Kameya T, Kawai Y, Iwanaga T, and Saito M (2013). Recruited brown adipose tissue as an antiobesity agent in humans. The Journal of clinical investigation 123, 3404–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneshiro T, Wang Q, Tajima K, Matsushita M, Maki H, Igarashi K, Dai Z, White PJ, McGarrah RW, Ilkayeva OR, et al. (2019). BCAA catabolism in brown fat controls energy homeostasis through SLC25A44. Nature 572, 614–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P, Arch JR, and Ashwell M (1984). Brown adipose tissue in the parametrial fat pad of the mouse. FEBS letters 167, 10–14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 (related to Figure 1). Technical validation of the single-cell RNA-seq analysis.
A) Illustration of the scRNA-seq experiment. scRNA-seq analysis in lineage− (Lin−) cells from the stromal vascular fraction (SVF) of mouse adipose tissue.
B) Sequential gating to isolate Lin− cells in the SVF from the adipose tissue.
C) Relationship between estimated RNA level (TPM: transcripts per kilobase million) in single-cell RNA-seq and External RNA Controls Consortium (ERCC) spike input. Note that estimated RNA levels are positively correlated with ERCC spike input (r = 0.953).
D) Relationship between coefficient of variation (CV) of TPM or detection rate (%) and ERCC spike input (copies per cell).
E) Meta-analysis of scRNA-seq data GSE111588. Violin plots showing mRNA levels of indicated genes (Cd81, Sca1, Acta2 and Sm22) in type 0 to 6 cell clusters. Note that Cd81, Sca1, Acta2, and Sm22 are highly enriched in Type 6 cluster.
F) Hierarchical clustering of bulk CD81+, bulk CD81− and stromal cell types identified in Fig. 1A. The horizontal distance represents similarities among each cluster. See the bulk RNA-seq transcriptomics in Fig 3.
G) Sca1 and CD81 expression levels in Lin− PDGFRα+ cells in the SVF from mouse inguinal WAT. Note that all the Lin− PDGFRα+ cells are marked by Sca1 in the SVF from the inguinal WAT.
Figure S2 (related to Figure 2). CD81+ stromal cells give rise to beige adipocytes.
A) Schematic illustration of the experiments in culture cell studies. CD81+ cells (Lin−: Sca1+: CD81+) and CD81− cells (Lin−: Sca1+: CD81−) were isolated from the SVFs of Ing WAT of BL6 mice (8-10 weeks of age) after overnight culture. Isolated cells were differentiated for 6 days under an adipogenic condition and stimulated with or without norepinephrine (NE) for 4 hours prior to harvest. During the adipogenic induction phase during day 0 to day 2, cells were cultured with rosiglitazone (Fig 2B-F) or without rosiglitazone (Fig S2B-C).
B) Relative mRNA expression of adipogenic and thermogenic genes in differentiated CD81− cells and CD81+ cells. Cells were differentiated for 6 days under an adipogenic condition without rosiglitazone. mRNA expression relative to Tbp. n=3.
C) Relative mRNA expression of Ucp1 and Pgc1a in differentiated CD81− cells and CD81+ cells. Cells were differentiated for 6 days under an adipogenic condition without rosiglitazone and stimulated with or without NE at 10 μM for 4 hours prior to harvest. mRNA expression relative to Tbp. n=3.
D) Uncoupled OCR (pmol min−1) in differentiated CD81− cells and CD81+ cells. Differentiated adipocytes were stimulated with NE at 10 μM and subsequently treated with oligomycin (10 μM). 20,000 cells were seeded onto plates. n=10. (B, C, D) *P < 0.05, **P < 0.01 by unpaired Student’s t-test.
E) Illustration of the transplantation experiments. CD81− cells and CD81+ cells were isolated from Ing WAT of ThermoMouse (UCP1-luciferase reporter mice). Cells were differentiated for 2 days under an adipogenic condition with rosiglitazone and transplanted into the subcutaneous region of nude mice. Luciferase activity of transplanted cells were monitored at day 3, 6, and 13 after transplantation. Nude mice were kept at 12°C after 3 days transplantation. Mice were treated with rosiglitazone (10 mg kg body weight−1) twice daily during day 10 to day 12.
F) Representative images of CD81-derived transplants. Arrowhead indicates a CD81-derived transplant in the subcutaneous region of nude mice.
G) Immunofluorescent staining for tdTomato and UCP1 in the transplants in (F). DAPI for counterstaining of immunofluorescent staining. Anti-mCherry antibody was used for tdTomato staining. Note that tdTomato is expressed in fat transplants originated from ThermoMouse that express a lufierase-tdTomato fusion protein. Scale bar = 100 μm.
H) Relative mRNA expression of indicated genes in CD81+ APC-derived transplants. Mice were kept at 12°C cold and rosiglitazone treatment for 10 days. Ing WAT of wild-type mice under a thermoneutral condition (TN: 30°C) and cold-acclimated mice at 8°C for 3 days were used as references. mRNA expression relative to Tbp. Ing WAT (TN), n=3; Ing WAT (Cold), n=5; Transplant, n=7. *P < 0.05, **P < 0.01 by unpaired Student’s t-test with Bonferroni’s correction.
I) Unstained signal for GFP in the Ing WAT (left) and interscapular BAT (iBAT; right) of Cd81-lineage reporter mice. Scale bar = 100 μm.
J) Immunofluorescent staining for GFP and UCP1 in the posterior region of Ing WAT of Cd81-lineage reporter mice at 8°C. Arrow indicates GFP− beige adipocytes (GFP−: UCP1+ cells). Scale bar = 50 μm.
K) Immunofluorescent staining for GFP and UCP1 at 8°C in iBAT of Cd81-lineage reporter mice. Scale bar = 100 μm.
L) Immunofluorescent staining for GFP and PDGFRα in the Ing WAT of Cd81-lineage reporter mice at 8°C. Scale bar = 50 μm. Data in (B, C, D, H) are represented as mean ± SEM.
Figure S3 (related to Figure 3). Characterization of CD81+ cells in adipose tissues.
A) The upregulated signaling pathways in CD81+ cells relative to CD81− cells from Epi WAT, and iBAT of BL6 mice (Top 5 signaling pathway). GO analysis was performed in transcriptomics data of CD81+ cells and CD81− cells by Metascape. Epi WAT,n=3; iBAT, n=2.
B) Immunoblotting for UCP1 in the Ing WAT of BL6 mice at 8 weeks old, 24 weeks old, 40 weeks old, and 72 weeks old mice. Male BL6 mice were kept at 22°C or 8°C for 5 days. β-actin as a loading control. Molecular weight (MW) is shown on the right.
C) CD81+ cell population (%) of Lin−: Sca1+ cells in the Ing WAT of young mice (8-10 weeks old) and old mice (60 weeks old). Male BL6 mice were kept at 22°C. Cells were isolated from the SVFs of above mice without overnight culture. n=12 per group. *P < 0.05 by unpaired Student’s t-test.
D) The downregulated signaling pathways in CD81+ cells relative to CD81− cells from Ing WAT (10 weeks old). Reactome gene sets analysis was performed in transcriptomics data of CD81+ cells and CD81− cells by Metascape. n=3.
Data in (C) are represented as mean ± SEM.
Figure S4 (related to Figure 4). The requirement of FAK for CD81+ APC proliferation.
A) Gene expressions related to FAK signaling pathway enriched in CD81+ cells relative to CD81− cells in the Ing WAT. The color scale shows intensity representing the mRNA levels in blue (low expression)-white-red (high expression) scheme. n=3.
B) Time-dependent effect of an ATP competitive FAK inhibitor, PF-573228, on primary CD81+ cell growth. Ing WAT-derived CD81+ cells were seeded on non-coated plates. Cells were treated with PF-573228 at a dose of 1 μM or vehicle (DMSO) for indicated time. Culture media containing FAK inhibitor or vehicle were replaced every 24 hours. n=3. **P < 0.01 by two-way repeated measures ANOVA.
C) Mice expressing dCas9-KRAB on the H11 locus (dCas9-KRAB mouse) were crossed with transgenic mice expressing gRNA targeting Cd81 on the H11 locus to generate CRISPRi-Cd81 mice.
D) Relative mRNA expression of Cd81 in Lin−: Sca1+ stromal cells from Ing WAT of CRISPRi-Cd81 mice and littermate control mice. mRNA expression relative to Tbp. n=6.
E) Relative mRNA expression of Cd81 in indicated tissues of CRISPRi-Cd81 mice and littermate control mice. mRNA expression relative to Tbp. n=3.
F) Validation of cell culture conditions to assess APC proliferation. Lin−: Sca1+ cells from the Ing WAT of CRISPRi-Cd81 and control mice were cultured on non-coated culture plates or collagen-coated plates. n=4.
G) Ing WAT-derived Lin−: Sca1+ cells from CRISPRi-Cd81 and control mice were cultured in media with indicated concentrations of FBS. n=4. (D-G) *P < 0.05, **P < 0.01 by unpaired Student’s t-test. N.S., not significant. Data in (B, D-G) are represented as mean ± SEM.
Figure S5 (related to Figure 4). The requirement of integrins for CD81-FAK signaling.
A) Transcriptional profile of integrin family members in CD81+ cells from Ing WAT of BL6 mice (10 weeks old). Isolated cells were analyzed by RNA-sequencing. n = 3.
B) Illustration of the experiment in Fig. 4F. CD81+ cells were isolated from Ing WAT of Itgb1flox/flox mice. Cells were infected with retrovirus containing Cre or an empty vector control, followed by hygromycin selection at a dose of 150 μg ml−1. Isolated cells were cultured until confluency and incubated for additional 2 days under adipogenic conditions. Subsequently, cells were cultured in FreeStyle293 Expression medium for starvation. After 4 hours of starvation, cells were treated with antagonistic antibodies against integrin β1 (ITGB1) or integrin β5 (ITGB5) or mouse IgG control at 5 μg ml−1 for 60 min. Subsequently, cells were treated with irisin at 100 pM for 5 min.
C) Illustration of the experiment in Fig. 4H. CD81+ cells were isolated from Ing WAT of Itgb5 KO mice and wild-type control (129Sv). Cells were cultured and treated with irisin as described in (B).
D) Illustration of the experiment in Fig. 4I. Ing WAT-derived Lin−: Sca1+ cells from CRISPRi-Cd81 mice and littermate control mice were cultured as described in (B).
E) Immunoblotting for FAK phosphorylation (pTyr397), total FAK, and β-actin in HEK293T cells expressing integrin αV (ITGAV) and integrin β5 (ITGB5) in the presence or absence of CD81. Cells were subsequently treated with irisin at 1 pM, 10 pM, and 100 pM for 5 min.
Figure S6 (related to Figure 5). CD81 is dispensable for brown adipocyte biogenesis in iBAT and muscle shivering.
A) Representative images of H&E staining and UCP1 immunofluorescent staining in iBAT of control mice (top) and CRISPRi-Cd81 mice (bottom) following cold exposure. Scale bar = 200 μm.
B) Relative mRNA expression of indicated genes in the iBAT of mice in (A). mRNA expression relative to Tbp. n=5.
C) OCR in the iBAT of mice after cold exposure. Punch-biopsy tissues (1.0 mg) were analyzed by the Seahorse XF Analyzer. n=4. (B, C) **P < 0.01 by unpaired Student’s t-test. N.S., not significant.
D) Representative electromyography (EMG) traces in skeletal muscle for CRISPRi-Cd81 mice and control mice at 30 °C and 8 °C.
E) Quantification of the images in (D) converted to the root mean square (RMS). n = 3. **P < 0.01 by one-way ANOVA followed by Bonferroni’s post hoc test. N.S., not significant.
F) Representative images of H&E staining in the Ing WAT of PpargCD81 KO and control mice after 8 °C cold environment for 10 days. Arrowhead indicates lymph node (LN). Scale bar = 200 μm. Data in (B, C, E) are represented as mean ± SEM.
Figure S7 (related to Figure 6). Metabolic phenotype of CRISPRi-Cd81 mice.
A) Immunoblotting for CD81 and exosome marker (CD63) in isolated exosome from the blood of CRISPRi-Cd81 mice and control mice on HFD for 8 weeks.
B) GTT in CRISPRi-Cd81 and control mice on HFD for 2 weeks. Mice were fasted for 6 hours and glucose (1.5 g kg body weight−1) was administered via i.p. Note that body-weight between CRISPRi-Cd81 mice and littermate controls was indistinguishable at 2 weeks of HFD. Control, n=8; CRISPRi-Cd81, n=6. *P < 0.05, **P < 0.01 by two-way repeated measures ANOVA with post hoc test by unpaired Student’s t-test.
C) Representative images of the liver of CRISPRi-Cd81 mice and control mice on 10 weeks of HFD.
D) Relative mRNA expression of Cd81 in the BMDM from CRISPRi-Cd81 mice and control mice, and CD81+ cells in the Ing WAT of BL6 mice. CRISPRi-Cd81 mice and control mice were on HFD for 10 weeks. mRNA expression relative to Hprt. BMDM, n=6; CD81+ cells, n=3.
E) The number of macrophages (F4/80+) in BMDM cultures differentiated from bone marrow of CRISPRi-Cd81 mice and control mice, quantified by FACS. Mice were on a high-fat diet for 10 weeks. n=6.
F) The expression of M1-like macrophage marker (CD11c) among total macrophages (F4/80+) in BMDM cultures from CRISPRi-Cd81 mice and control mice by FACS (MFI). Mice were on HFD for 10 weeks. n=6.
G) Relative mRNA expression of Tnfa in the BMDM from CRISPRi-Cd81 mice and control mice on 10 weeks of HFD. Cells were stimulated with or without metabolic cocktail (30 mM glucose, 10 nM insulin and 0.5 mM palmitic acid) for 18 hours. mRNA expression relative to Hprt. n=6. **P < 0.01 by one-way ANOVA followed by Bonferroni’s post hoc test. N.S., not significant.
H) The absolute number of leukocytes (CD45+), B cells (CD45+: CD3−: CD19+), T cells (CD45+: CD3+), CD4+ T cells (CD45+: CD3+: CD4+), CD8+ T cells (CD45+: CD3+: CD8+), Th17 cells (CD45+: CD3+: CD4+: IL-23R+), and NK cells (CD45+: CD3−: CD4−: CD8−: CD19−: NKp46+) per gram of Ing WAT of CRISPRi-Cd81 mice and control mice by FACS. Mice were on HFD for 10 weeks. Control, n=8-9; CRISPRi-Cd81, n=5-6.
I) Relative mRNA expression of indicated pro-inflammatory and pro-fibrosis genes in Epi WAT of CRISPRi-Cd81 mice and control mice. Mice were on HFD for 10 weeks. Control, n=7; CRISPRi-Cd81, n=6. mRNA expression relative to Tbp.
J) The expression (MFI) of M1-like macrophage marker (CD11c) among total macrophages (CD45+: CD64+) in Epi WAT of CRISPRi-Cd81 mice and control mice on 10 weeks of HFD. Control, n=9; CRISPRi-Cd81, n=6.
K) Ki67+ cell population (%) in CD81− (Lin−: PDGFRα+: CD81−) and CD81+ cells (Lin−: PDGFRα+: CD81+) in human subcutaneous WAT biopsies. n=7. (D-F, H-K) **P < 0.01 by unpaired Student’s t-test. N.S., not significant. Data in (B, D-K) are represented as mean ± SEM.
Table S1 (related to Figure 1D): List of cell surface markers highly expressed in Type III cell population (top 20).
Table S2 (related to Table 1): Baseline characteristics of study participants (n=28).
Table S3 (related to STAR methods): Primer sequences used for quantitative RT-PCR.
Data Availability Statement
The RNA-seq datasets generated during this study are available at ArrayExpress (www.ebi.ac.uk) under the accession number E-MTAB-8435 and E-MTAB-8464.