Abstract
We assessed the gut microbiota of 90 American young adults, comparing 43 participants with major depressive disorder (MDD) and 47 healthy controls, and found that the MDD subjects had significantly different gut microbiota compared to the healthy controls at multiple taxonomic levels. At the phylum level, participants with MDD had lower levels of Firmicutes and higher levels of Bacteroidetes, with similar trends in the at the class (Clostridia and Bacteroidia) and order (Clostridiales and Bacteroidales) levels. At the genus level, the MDD group had lower levels of Faecalibacterium and other related members of the family Ruminococcaceae, which was also reduced relative to healthy controls. Additionally, the class Gammaproteobacteria and genus Flavonifractor were enriched in participants with MDD. Accordingly, predicted functional differences between the two groups include a reduced abundance of short-chain fatty acid production pathways in the MDD group. We also demonstrated that the magnitude of taxonomic changes was associated with the severity of depressive symptoms in many cases, and that most changes were present regardless of whether depressed participants were taking psychotropic medications. Overall, our results support a link between MDD and lower levels of anti-inflammatory, butyrate-producing bacteria, and may support a connection between the gut microbiota and the chronic, low-grade inflammation often observed in MDD patients.
Introduction
There is increasing recognition of the fact that the gut microbiota is associated with a wide range of health conditions and disease states in humans. Alterations to gut microbiome composition or function have been linked to gastrointestinal disorders1,2, autoimmune disorders3-8, and metabolic and cardiovascular disease8-10. Perhaps most surprisingly, given the physiological distance and the presence of the blood-brain barrier, the gut microbiota has also been implicated in psychiatric disorders or syndromes, including anxiety disorders11-13, bipolar disorder14-17, and major depressive disorder (MDD)11,18-26.
Links between the gut microbiota and the brain are likely mediated in part through the gut-brain axis (GBA), a proposed series of complex communication pathways between the gastrointestinal tract and the central nervous system27-33. The GBA includes neural, immune, endocrine, and metabolic pathways involved in the regulation of hunger and satiety, stress, immunity, and intestinal motility; the gut microbiota are believed to play a direct role in some of these. In particular, microbes or their metabolites such as short-chain fatty acids (SCFA) can stimulate afferent inputs to the vagus nerve34-36, induce enteroendocrine cells to produce neuropeptides and activate afferent nerve pathways37,38, promote normal stress responses and hypothalamic-pituitary-adrenal (HPA) axis development and function39,40, and participate in local neurotransmitter production and systemic regulation via tryptophan metabolism or direct secretion29,41-43. In the other direction, the vagus nerve can promote anti-inflammatory responses and decrease intestinal permeability34,35,44,45, stress-induced glucocorticoid induction through the HPA axis can lead to microbial changes and increased gut barrier permeability46-52, and the central nervous system can influence the gut environment through release of signalling molecules, changes to mucus secretion, and regulation of intestinal motility28,32,53-55.
In terms of depression, links to the microbiome have been established in both human and animal models56-59. In one particularly compelling case, fecal transplants from humans with MDD resulted in the development of depressive symptoms in a germ-free mouse model20. Similarly, fecal transplants from depressed human subjects into a germ-free rat model induced development of depressive symptoms, including anhedonia and anxiety, in addition to changes in tryptophan metabolism60. Additionally, certain microbes including Faecalibacterium prausnitzii, Lactobacillus spp., and Bifidobacterium spp. have been found to ameliorate the onset of anxiety and depressive symptoms that rodents develop when subjected to chronic unpredictable mild stress, maternal separation, or chemically-induced colitis61-63. Finally, recent work has demonstrated that treatment of rats bred for high-anxiety behavior with the antimicrobial minocycline altered the gut microbiota while also alleviating depressive symptoms, supporting previous work suggesting that minocycline may have merit as an adjunct treatment alongside antidepressants64-66.
In humans, studies have consistently indicated that the gut microbiota of adults with MDD are different from those of their healthy counterparts, although specific differences have varied between studies. Some studies have found that the phylum Bacteroidetes is underrepresented in subjects with depression, while Firmicutes are overrepresented20,21,24-26, although other studies have found the opposite trend18,19. Multiple studies have linked higher abundance of the genera Alistipes, Oscillibacter, and Flavonifractor and the family Enterobacteriaceae to MDD and low quality of life scores18,20-25, while Faecalibacterium, Dialister, Coprococcus, and Prevotella have been found to be lower in subjects with depression and/or low quality of life scores18,20,22,23,60. Faecalibacterium has even been found to negatively correlate with depression severity18 and this species has also been found to be negatively associated with both bipolar disorder and generalized anxiety disorder12,14.
A potential link between the gut microbiota and MDD is the low-grade, chronic inflammation that has previously been observed in a substantial proportion of depressed individuals67-69. Significant subsets of depressed subjects have been associated with higher levels of circulating inflammatory cytokines, particularly IL-6 and TNF-α70-75, in addition to hypercortisolism and dysregulation of the HPA axis76-78. Furthermore, a few studies have demonstrated that combining antidepressants with anti-inflammatory drugs improved response rates79,80, and inflammasome signalling has been linked to induction of anxiety and depressive behaviors in mice81-85. Human patients with chronic inflammatory illnesses have higher levels of depression than the general population86-90, and administration of inflammatory cytokines or immune-provoking stimuli such as lipopolysaccharide (LPS) leads to the development of “sickness behavior” and depressive symptoms in both animal models and human patients91-99. Mechanistically, inflammatory cytokines may increase blood-brain barrier permeability and in some cases cross it99-105, activate vagus nerve afferents45,104-108, impact neurotransmitter levels in the brain104,105,109-111, contribute to hyperactive dysregulation of the HPA axis112-117, and affect serotonergic neurotransmission by promoting enzymatic metabolism of the precursor tryptophan99,118,123.
The gut microbiota may contribute to such an effect through their capacity to either promote or protect against inflammation. For example, loss of bacteria that produce the anti-inflammatory, barrier-strengthening molecule butyrate, such as Faecalibacterium or Coprococcus, could lead to a loss of protection against epithelial inflammation and gut barrier disruption. Combined with increases in LPS-producing bacterial groups such as Proteobacteria or potentially pro-inflammatory species such as Flavonifractor, this could lead to increased translocation of immunogenic bacterial products and activation of low-grade systemic inflammation. In fact, studies have found that depressed subjects have increased levels of bacterial DNA in circulation and increased antibody responses to LPS124-128.
In this study, we analyzed the gut microbiota of young adults with major depressive disorder and healthy controls. We hypothesized that we would observe potential signatures of inflammation, including either reductions in protective, butyrate-producing bacterial taxa or increases in pro-inflammatory taxa. Our study differs from previous studies in terms of demographics, as most previous studies have been performed on subjects of Chinese heritage18-20,22,24-26 and a few have examined European subjects21,23,60, but to our knowledge none have so far assessed differences in American subjects. Furthermore, most previous studies have examined older age groups, with age ranges in the thirties and forties, while we utilized a sample of participants aged 18-25. Given previous work demonstrating that age, ethnic background, and geographic location can have significant impacts on the composition and function of the microbiota129-135, this study provides an important new perspective on the gut microbiome in the context of MDD. Finally, our study analyzed a large sample which included both a notable subset of MDD participants who were not taking psychotropic medications as well as MDD subjects with a range of symptom severities. This allowed us to assess the potential contributions of these characteristics to changes in the gut microbiota observed in participants with MDD. Overall, we observed that the MDD subjects exhibited lower levels of potentially protective taxa, including Faecalibacterium and Subdoligranulum, and higher levels of potentially pro-inflammatory taxa, including Flavonifractor and Gammaproteobacteria; furthermore, many of these changes track with symptom severity.
Methods
Participants and Sample Collection Procedures
Young adults were recruited from the community and local psychiatric clinics through flyers and social media advertisements between May 2018 and July 2019. All participants were recruited from the Rhode Island community and psychiatric clinics in Providence, RI. To participate in the study, prospective participants were required to be 18-25 years old, meet eligibility criteria for either the MDD group or the healthy control group, and not be subject to any exclusion criteria. To be eligible for the healthy control group, subjects were required to have a Patient-Reported Outcomes Measurement Information System (PROMIS) Depression score < 13, no lifetime history of major depression, no lifetime history of suicidal ideation, suicide attempts, or non-suicidal self-injury as assessed by the Columbia-Suicide Severity Rating Scale (C-SSRS) and Self-Injurious Thoughts and Behavior Interview (SITBI). To be eligible for the MDD group, participants needed to meet diagnostic criteria for a current major depressive episode and have PROMIS Depression scores > 21. Individuals were excluded if they had smoked cigarettes or cigars in the past 12 months, were vegan, had gastrointestinal illness in the past six months, had diarrhea in the past two weeks, used anti-diarrhea medication in the past six weeks, or used antibiotics in the past three months.
Prospective participants first completed an online screener which included the PROMIS Depression scale and questions regarding the exclusion criteria. Those who remained potentially eligible based on the online screener then completed a phone screener, in which they answered the Structured Clinical Interview for DSM-5 Disorders (SCID-V), C-SSRS, and SITBI. Those who were eligible were invited to attend a one-hour in-person assessment, at which they were re-administered the PROMIS Depression questionnaire and were asked to provide a stool sample using OMNIgene•GUT stool collection kits (DNA Genotek). Whole blood was drawn by licensed phlebotomists, allowed to clot for 15 minutes at room temperature, centrifuged for 10 minutes, and immediately aliquoted and stored at −80°C until use. Data about psychotropic medication usage was also collected.
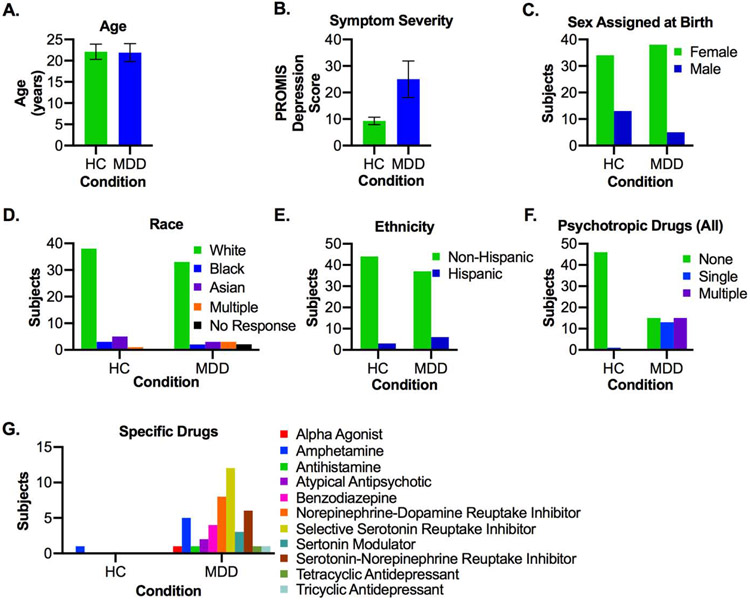
The final sample consisted of 90 participants (see Table 1 and Figure 1). The healthy control group (n=47) had a mean age of 21.7±2.1, mean PROMIS Depression scores of 9.3±1.4, and was 72.3% female, 76.7% white, and 14.0% Hispanic. The MDD group (n=43) had a mean age of 22.7±1.8, mean PROMIS Depression scores of 25.0±6.9, and was 88.4% female, 80.1% white, and 6.7% Hispanic. For later analyses, PROMIS scores were used to partition participants into mild (PROMIS<23), moderate (PROMIS 23-32), and severe (PROMIS>32) symptom groups.
Table 1: Demographic Information.
This table shows the demographic information in both the healthy control and MDD groups, including sex assigned at birth, gender identity, age, ethnicity, race, SCID score, PROMIS depression and anxiety scores, and prescription of psychoactive drugs.
| Characteristic | Specific Descriptor | Healthy Controls | Major Depressive Disorder |
|---|---|---|---|
| Sex Assigned at Birth | Female | 34 (72.3%) | 38 (88.4%) |
| Male | 13 (27.7%) | 5 (11.6%) | |
| Age (years) | Mean (SD) | 22.1 (1.8) | 21.9 (2.1) |
| Ethnicity | Hispanic | 3 (6.4%) | 6 (14.0%) |
| Non-Hispanic | 44 (93.3%) | 37 (86.0%) | |
| Race | White | 38 (80.1%) | 33 (76.7%) |
| Black | 3 (6.4%) | 2 (4.7%) | |
| Asian | 5 (10.6%) | 3 (7.0%) | |
| Multiple | 1 (2.1%) | 3 (7.0%) | |
| No Response | 0 (0%) | 2 (4.7%) | |
| PROMIS Depression Score | Mean (SD) | 9.3 (1.4) | 25.0 (6.9) |
| Prescribed Psychoactive Drugs | Single | 1 (2.1%) | 13 (30.0%) |
| Multiple | 0 (0%) | 15 (34.9%) | |
| No | 46 (97.9%) | 15 (34.9%) |
Figure 1: Demographic Characteristics of Control and MDD Groups.
A) Age. B) PROMIS Depression scores. C) Sex assigned at birth. D) Race. E) Ethnicity. F) Number of psychotropic drugs taken. G) Categories of psychotropic drugs taken. In A-B, bars represent group averages and error bars indicate standard deviation. In G, the sum of all bars in the MDD group may exceed 43, due to subjects taking multiple medications. MDD = Major Depressive Disorder
Measures for Assessing Depression and Symptoms in Participants
The PROMIS Depression – Short Form136 was created by National Institutes of Health as part of the Roadmap for Medical Research initiative to use item-response-theory methodology to develop psychometrically advanced self-report measures of health outcomes. The adult depression short form consists of an eight-item questionnaire that assesses depressive symptoms over the past seven days. Higher scores indicate greater depressive symptom severity.
The C-SSRS137 and SITBI138 are semi-structured interviews for assessing lifetime history of suicidal thoughts and behaviors and non-suicidal self-injury, respectively. All interviewers received extensive training and supervision from the first author in the administration of this interview and rating of its data. A rigorous protocol developed by the first author was implemented, with an average training period of three to four months before interviewers administered the measure independently. Interviewers conferred with the first author whenever coding questions arose.
The SCID-V139 is a semi-structured diagnostic interview of current and lifetime DSM-5 psychopathology. For the current study, only the depression module was administered. Research staff were trained by doctoral level clinicians and certified by the first author in the research procedures.
Extraction and Preparation of Fecal Samples
Upon receipt at Brown University, fecal samples were stored at −80°C until all samples had been collected. Samples were then thawed, and 300 μL of fecal suspension from each sample was transferred into two plates of the ZymoBIOMICS 96 DNA Kit (Zymo Research) to extract DNA. Samples from the two groups were randomized across the two 96-well plates. Extraction was performed according to manufacturer’s protocols, and extracted DNA was measured using the Qubit 3.0 system with Broad-Range DNA reagents (Thermo Fisher Scientific).
Sequencing
Amplicons of the V4 hypervariable regions of the 16S rRNA gene were generated according to the Earth Microbiome Protocol140. In brief, 10 μg of extracted DNA from each sample was used as template for triplicate PCR reactions utilizing individually barcoded 515F forward primers (GTGYCAGCMGCCGCGGTAA) with Illumina adapters and the 806R reverse primer (GGACTACNVGGGTWTCTAAT) with Illumina adapters. Triplicates were combined and measured using the Qubit 3.0 system with Broad-Range DNA Reagents (Thermo Fisher Scientific). Samples were pooled in equimolar concentrations and sent out for 2x250 paired-end sequencing utilizing an Illumina MiSeq system at the University of Rhode Island. We obtained a total of 3,806,054 quality-filtered sequences across all 90 samples. The average sequencing depth was 41,509 reads in the control group and 44,169 in the MDD group. Sequences can be found at the NCBI Short Read Archive under BioProject ID PRJNA591924.
Microbiome Data Analysis
Data was initially processed utilizing the QIIME2 (v2019.7) pipeline141. In brief, samples were imported using the tools plugin, demultiplexed using the demux plugin, and denoised using the dada2 plugin to obtain amplicon sequence variants (ASVs)142. Phylogenetic trees were generated using the phylogeny plugin and taxonomy was assigned using the feature-classifier plugin and the Silva (release 132) 99% identity V4 classifier. Additionally, functional potential was predicted using the picrust2 plugin143. The feature table, representative sequences, rooted phylogenetic tree, and taxonomy QIIME2 artifacts were exported, and the feature table, taxonomy, and sample metadata were merged into a unified biom file using the add-metadata function of the biom-format package144.
The exported biom file, phylogenetic tree, and representative sequences were imported into the phyloseq package (v1.28.0)145,146 in R (v3.6.1) using the import_biom function. Alpha diversity metrics were calculated using the estimate_richness function of phyloseq (Shannon’s Diversity Index, Simpson’s Diversity Index, Observed ASVs) and the estimate_pd function of the btools package (v0.0.1) (Faith’s Phylogenetic Diversity). Beta diversity (Bray Curtis Dissimilarity and both Unweighted and Weighted Unifrac Distances) was calculated using the phyloseq:distance function of the vegan package (v2.5-6)147, statistically analyzed using the adonis function, and subjected to Principal Coordinates Analysis using the ordinate function of vegan.
ASV tables were agglomerated at the phylum, class, order, family, and genus levels and exported as relative abundance tables for plotting and analysis (Data S1). The genus-level table was reformatted to conform to the requirements of the Linear Discriminant Analysis Effect Size (LEfSe) web-based tool148, and analysis to identify group biomarkers was performed according to default parameters. Similarly, the MetaCyc pathways output from picrust2 (Data S2) was formatted for and analyzed with LEfSe. All figures were generated using GraphPad Prism v8e, with the exception of Figure 3B, which is a modified LEfSe output. Statistics were performed in R for beta diversity metrics, LEfSe for differential abundance, and GraphPad Prism for alpha diversity metrics and tests for trends based on psychotropic drug usage or symptom severity. Throughout the text and figures, in cases where discriminant taxa had unclear or nonspecific names at a particular level, the next-higher taxon was included in curly brackets to indicate its provenance (for example, {Muribaculaceae} uncultured bacterium).
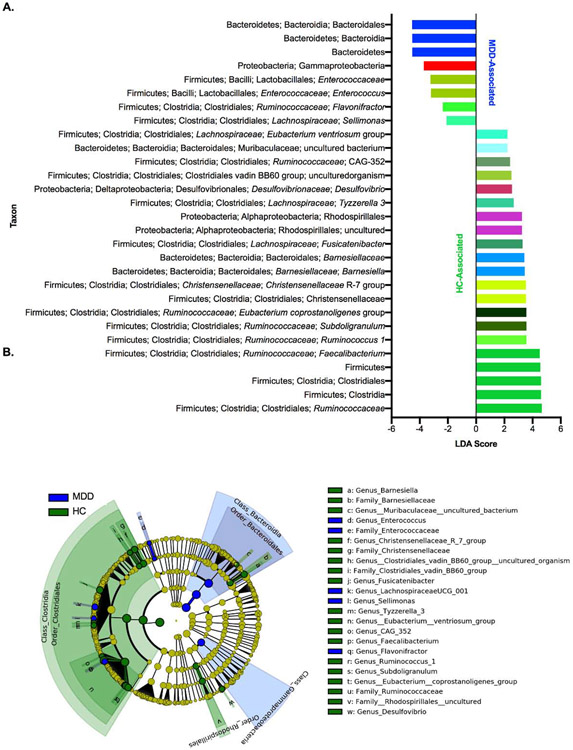
Figure 3: Differential Taxa are Associated with Control and MDD Subjects.
A) Taxa identified by LEfSe as biomarkers of samples from the control or MDD groups (cutoffs were LDA≥2 and p-value≤0.05). B) Cladogram indicating the phylogenetic relatedness of the discriminant taxa. Colors in A) correspond to the taxonomic position, matching the coloring used in Figures 4, 5, and S2. Abbreviations: LEfSe: Linear Discriminant Analysis Effect Size, MDD = Major Depressive Disorder. LEfSe is a biomarker discovery tool used to identify features (taxa, in this case) from high-dimensional genomic data that characterize differences between two or more biological conditions.
Serum Inflammatory Cytokines
We utilized the LegendPlex Human Inflammation Panel 1 kit (BioLegend, Lot B291816), which measures levels of IL-1β, IFN-α2, IFN-γ, TNF-α, MCP-1, IL-6, IL-8, IL-10, IL-12p70, IL-17A, IL-18, IL-23, and IL-33, to quantitate serum levels of pro-inflammatory cytokines. Serum was thawed, vortexed, centrifuged, heated at 70°C for ten minutes, and diluted 1:2 in Assay Buffer before measurement. Serum samples and standards were run in duplicate according to manufacturer’s instructions, and data was processed using the LegendPlex analysis software. GraphPad Prism was used to plot data and perform two-tailed t-tests with Welch’s correction, and the Bonferroni correction was applied to account for multiple hypothesis testing.
Results
Psychotropic Medication Usage
We analyzed the microbiomes of 90 participants. Demographics and range of depressive symptom severity based on PROMIS scores of the population can be found in the Materials and Methods and in Figure 1 and Table 1. In addition to demographics, we also assessed the psychotropic drug usage of the groups. In the MDD group, 15 (34.9%) were not actively taking prescribed psychotropic medications, while 30.0% were taking a single medication, and 34.9% were taking two or more medications (Table S1-2, Figure 1F). These medications were quite varied, and included a range of both anti-depressants, anxiolytics, and stimulants; in the control group, only one subject was taking a psychotropic medication, specifically an amphetamine for ADHD (Table S2, Figure 1G).
Alpha and Beta Diversity
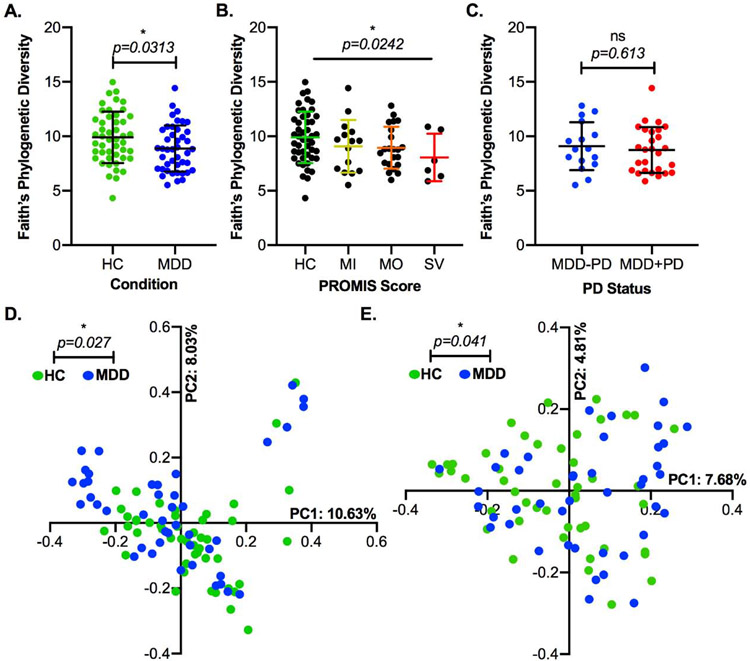
We started by analyzing the alpha diversity of the control and MDD groups. We first utilized the Observed ASVs metric, which assesses the richness of ASVs found in the samples without considering their phylogenetic relatedness, and Shannon’s Diversity Index, which reflects both the richness and evenness of the samples. On both of these measures, there was no significant difference between the two groups (Figure S1A-B). However, when using Faith’s Phylogenetic Diversity, a biodiversity metric which analyzes phylogenetic tree branch length to incorporate relatedness of taxa, we observed a slight but significant decrease in this metric in the MDD group (Figure 2A). Furthermore, this diversity metric was inversely related to the severity of depressive symptoms (Figure 2B), and was not impacted by the usage of psychotropic medications (Figure 2C).
Figure 2: Phylogenetic Diversity is Reduced in Subjects with MDD.
A) Faith’s Phylogenetic Diversity in control and MDD groups (*, p=0.0313, t-test with Welch’s correction). B) Faith’s Phylogenetic Diversity in healthy controls and depressed subjects with mild, moderate, and severe symptoms according to PROMIS depression scores (*, p=0.0242, ANOVA post-test for linear trend of column means). C) Faith’s Phylogenetic Diversity in depressed subjects taking no psychotropic medication and depressed subjects taking one or more psychotropic drugs (ns, p=0.613, t-test with Welch’s correction). D) Principal Coordinates 1 and 2 of Bray-Curtis Dissimilarity, with points colored according to their source sample’s condition group (*, p=0.027, R2=0.01, permANOVA). E) Principal Coordinates 1 and 2 of Unweighted Unifrac Distance, with points colored according to their source sample’s condition group (*, p = 0.041, R2=0.0106, permANOVA). In A-C, the central line indicates the group average and error bars indicate standard deviation. Abbreviations: HC = Healthy Control, MDD = Major Depressive Disorder, MI = Mild, MO = Moderate, SV = Severe, PD = Psychotropic Drug, PC = Principal Coordinate. Faith’s Phylogenetic Diversity is a metric of biodiversity that incorporates phylogenetic relationships. Principal Coordinates Analysis is a method of multidimensional scaling that attempts to find the main axes through a complex matrix, in this case of beta diversity values. Bray-Curtis Dissimilarity compares communities without incorporating phylogenetic relationships between taxa. Unweighted Unifrac Distance compares communities with the inclusion of phylogenetic data but does not weight by abundance.
We then utilized metrics of beta diversity to assess whether the healthy and MDD microbiomes were different at a whole-community level. First, we found that there were statistically significant differences in community composition between the two groups based on Bray-Curtis Dissimilarity, which does not consider phylogenetic relatedness of taxa in a sample. Similarly, there was a significant difference in community composition based on Unifrac Distance, which does take relatedness into account. However, the permANOVA indicates that the condition (control or MDD) explains very little of the discrimination between samples (R2=0.018 and R2=0.014, respectively); accordingly, when plotting these metrics using a Principal Coordinate Analysis (PCoA), we did not observe clear separation of the two groups (Figure 2D-E). In the case of Bray-Curtis Dissimilarity, there was a clear clustering of several samples in both groups separately from the majority, which could be attributed to the dominance of Prevotella 9 rather than Bacteroides as the predominant genus-level taxon of the phylum Bacteroidetes in those samples (Figure S1C). This pattern was not observed in the PCoA for Unifrac Distance (Figure S1D), consistent with the fact that this metric accounts for the two genera’s taxonomic relatedness. Finally, using Weighted Unifrac Distance, which accounts for both phylogenetic relatedness and abundance of taxa, there was no significant difference between the groups and no clear clustering by condition (Figure S1E).
Taxonomic Composition
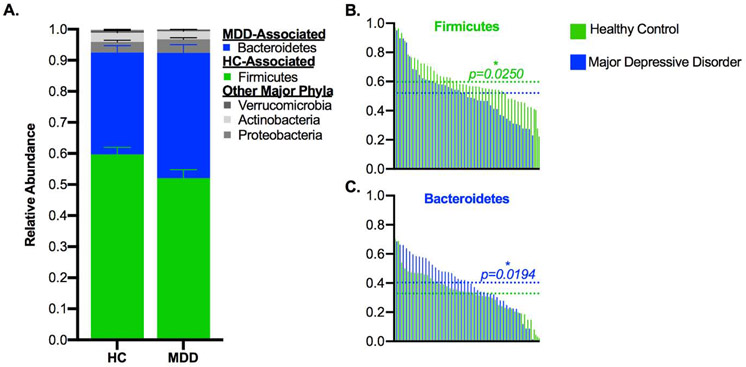
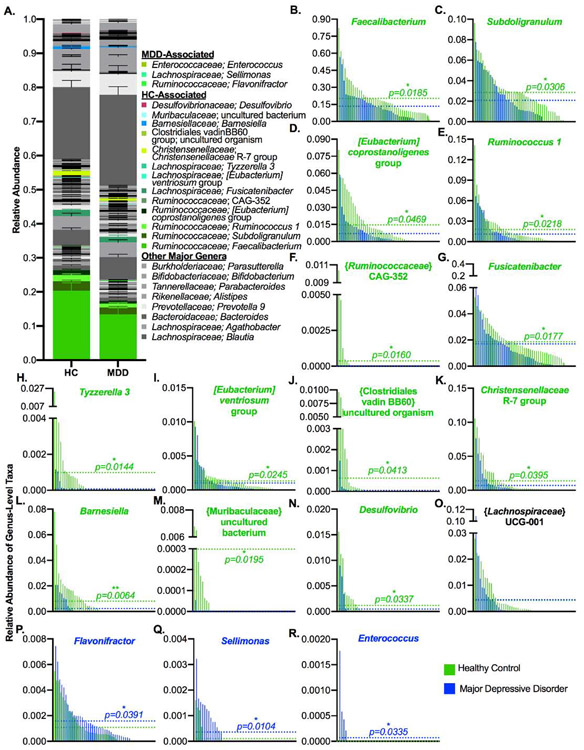
We next analyzed the composition of the samples at multiple taxonomic levels to assess whether there were differences in the communities of depressed and healthy subjects. We utilized the Linear Discriminant Analysis Effect Size (LEfSe) tool to identify taxa that were biomarkers of each group and found a range of discriminating taxa across all taxonomic levels (Figure 3, Table S4). At the phylum level, controls were enriched in Firmicutes and the MDD subjects were enriched in Bacteroidetes (Figure 4A-C, Table S4). Similar findings were obtained at the class and order levels (Figure S2A-B), with the class Clostridia and order Clostridiales of Firmicutes associated with controls while the class Bacteroidia and order Bacteroidales of Bacteroidetes were associated with depressed subjects (Figure S2C-F, Table S4). Additionally, within the phylum Proteobacteria, the order Rhodospirillales of the class Alphaproteobacteria was associated with controls, while the class Gammaproteobacteria was associated with the MDD group (Figure S2G-H). At the family level (Figure S2I, Table S4), the Clostridiales families Ruminococcaceae and Christensenellaceae were associated with the control group (Figure S2J-K), as well as the Bacteroidetes family Barnesiellaceae and an uncultured family of the order Rhodospirillales (Figure S2L-M), while the family Enterococcaceae (of the Bacilli-Lactobacillales lineage of Firmicutes) was associated with the MDD group (Figure S2N).
Figure 4: Major Phyla Are Differentially Abundant Between Healthy and MDD Subjects.
A) Stacked bar plot indicating the average relative abundance of phyla within the control and MDD groups, with discriminant phyla identified by LEfSe highlighted in color. B-C) Phyla that were discriminant of the groups, with control and MDD samples interleaved by ranked abundance of each taxon and dotted lines indicating the average relative abundance by group. In A, error bars indicate standard error of the mean. In B-C, the text color indicates the group that the phylum was associated with, and p-values are from LEfSe. Abbreviations: MDD = Major Depressive Disorder, LEfSe = Linear Discriminant Analysis Effect Size, HC = Healthy Control. LEfSe is a biomarker discovery tool used to identify features (taxa, in this case) from high-dimensional genomic data that characterize differences between two or more biological conditions.
A number of trends at the family level were also represented in their subordinate genera, although there were some contrasting patterns as well (Figure 5A, Table S4). First, the four most abundant genera within the Firmicutes family Ruminococcaceae (Faecalibacterium, Subdoligranulum, [Eubacterium] coprostanoligenes group, and Ruminococcus 1) were associated with healthy controls (Figure 5B-E), as well as the less-abundant genus-level taxon CAG-352 (Figure 5F). On the contrary, only a single Ruminococcaceae genus, Flavonifractor, was associated with MDD subjects (Figure 5P). Additionally, while the related Clostridiales family Lachnospiraceae itself was not associated with either group, a number of its member genera were associated with healthy controls, including Fusicatenibacter, Tyzzerella 3, and [Eubacterium] ventriosum group (Figure 5G-I). In contrast, the Lachnospiraceae genus Sellimonas was associated with the MDD group (Figure 5Q). The genus-level taxon {Lachnospiraceae} UCG-001 was also called as associated with the MDD group, but a closer examination reveals that this taxon is in fact more highly present and abundant in the control group and the misidentification is likely due to one extreme outlier in the MDD group (Figure 5O).
Figure 5: Numerous Genera Are Differentially Abundant Between Healthy and MDD Subjects.
A) Stacked bar plot indicating the average relative abundance of genera within the control and MDD groups, with discriminant phyla identified by LEfSe highlighted in color. B-R) Genera that were discriminant of the groups, with control and MDD samples interleaved by ranked abundance of each taxon and dotted lines indicating the average relative abundance by group. In A, stacking is done by phylogeny, so all genera are grouped by their higher taxonomic ranks. Error bars indicate standard error of the mean. In B-R the text color indicates the group that the genus was associated with, and p-values are from LEfSe. Abbreviations: MDD = Major Depressive Disorder, LEfSe = Linear Discriminant Analysis Effect Size, HC = Healthy Control. LEfSe is a biomarker discovery tool used to identify features (taxa, in this case) from high-dimensional genomic data that characterize differences between two or more biological conditions.
Other discriminatory genus-level taxa within the phylum Firmicutes included an uncultured organism of the {Clostridiales} vadin BB60 family and the R-7 group of Christensenellaceae, both associated with controls (Figure 5J-K), and Enterococcus, which was associated with MDD subjects (Figure 5R). Within the phylum Bacteroidetes, Barnesiella and an uncultured bacterium of the Muribaculaceae family were associated with controls (Figure 5L-M), while no genus-level taxa were associated with MDD subjects. Finally, within Proteobacteria, the Deltaproteobacteria genus Desulfovibrio was associated with healthy controls (Figure 5N). As considered in greater depth in the discussion, these changes generally appear to reflect a loss of protective bacteria and an increase in pro-inflammatory bacteria in the MDD group.
Impact of Psychotropic Medication
We then examined whether any of the associations of taxa with depressed subjects were driven by the consumption of psychotropic medication. We compared the taxa that were identified as significant between the healthy controls, depressed subjects taking no medication, and depressed subjects taking one or more medications. Unfortunately, as the number and types of medications varied significantly, we could not assess the impacts of specific classes or combinations of medications.
Generally, we found that the trends observed in the depressed subjects were present in both the medicated and unmedicated groups, although the effect was sometimes stronger in one group than the other (Figure S3). Specifically, the MDD-associated reductions observed in the phylum Firmicutes, the class Clostridia, the order Clostridiales, the family Ruminococcaceae, and the genus-level taxa Faecalibacterium, and [Eubacterium] coprostanoligenes group, were slightly stronger in the medicated group (Figure S3A-F). Similarly, the depression-associated increases in the phylum Bacteroidetes, the class Bacteroidia, and the order Bacteroidales, were somewhat stronger in this group (Figure S3G-I). In particular, the changes in the genera Flavonifractor and Sellimonas appeared to be driven primarily by the medicated group (Figure S3J-K). On the other hand, the MDD-associated reductions in the families Christensenellaceae and Barnesiellaceae, and genera Christensenellaceae R-7 group, Barnesiella, and an uncultured organism of the Clostridiales vadin BB60 family-level taxon appear somewhat stronger in the unmedicated group (Figure S3L-P). In the case of the genus Fusicatenibacter, reductions in the depressed group appeared to be primarily driven by the unmedicated subjects (Figure S3Q). There were also a number of cases in which there were no apparent differences between the depressed subjects based on medication (Figure S3R-AA). Finally, in the case of the family Enterococcaceae and its daughter genus Enterococcus, levels of these taxa were found only in medicated subjects, suggesting that this group may drive the effect (Figure S3AB-AC); however, as they were detected in so few samples, it is difficult to make this judgment.
Overall, the differences between depressed and healthy subjects could not be specifically attributed to psychotropic medications, as changes were typically present in both medicated and unmedicated subjects, although they may play a role in some cases. Additionally, we were unable to examine the impacts of specific classes of psychotropic drugs, given the variety and combinations of medications taken by participants.
Impact of Depression Severity
While psychotropic medication use may have had some impact on taxonomic trends, we also found that depression severity could significantly confound this interpretation. Based on PROMIS Depression scores, the proportion of subjects taking psychotropic medication increased based on their symptom severity: 46.7% of the 15 subjects with mild symptoms were taking psychotropic medication, compared with 68.2% of the 22 subjects with moderate symptoms and 83.3% of the 6 subjects with severe symptoms (Figure S4A). Accordingly, the proportions of subjects with mild symptoms was higher in the unmedicated group (53.3% vs. 28.6%), while the proportion with severe symptoms was lower (6.7% vs. 17.9%) (Figure S4B). Therefore, we decided to assess whether any of the observed trends tracked with symptom severity, which might explain differences better than whether or not depressed subjects were taking one or more of a wide range of psychotropic drugs with different mechanisms of action.
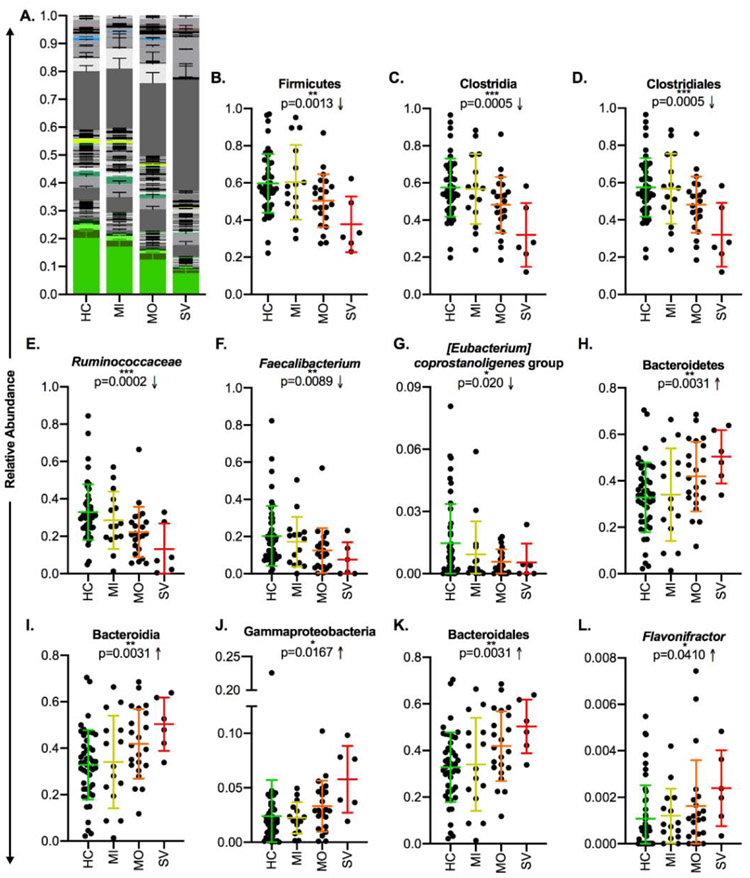
In fact, despite interindividual variation, the observed changes were more exaggerated in subjects with higher depressive symptom severity scores in the majority of taxa (Figures 6A, S5A); this trend was significant in most of these cases, although a few fell just short of statistical significance (Table S3). Specifically, the phylum Firmicutes, class Clostridia, order Clostridiales, family Ruminococcaceae and its member genera Faecalibacterium, [Eubacterium] coprostanoligenes group, Subdoligranulum, and Ruminococcus 1, and family Christensenellaceae and member genus-level taxon Christensenellaceae R-7 group were more reduced in subjects with more severe symptoms (Figures 6B-G, S5B-K). In the case of Fusicatenibacter, reductions were most pronounced in the subjects with severe symptoms, although there was not a consistent trend in the mild and moderate symptom groups (Figure S5L). In the other direction, the phylum Bacteroidetes, classes Bacteroidia and Gammaproteobacteria, order Bacteroidales, and genera Flavonifractor and Sellimonas were more abundant in subjects with more severe symptoms (Figures 6H-L, S5M-R). There were also a few cases in which such a trend was not present or where there were very few depressed samples with detectable levels of a given taxon, making it difficult to assess whether their abundance aligns with symptom severity. (Figure S5S-AD).
Figure 6: Taxonomic Differences in MDD Subjects Track Significantly with Symptom Severity.
A) Stacked bar plot indicating the average relative abundance of genera within the control subjects and MDD subjects with mild, moderate, or severe symptoms according to PROMIS depression scores, with genera that discriminated the control and MDD groups identified by LEfSe highlighted in color. B-L) Relative abundances of discriminant taxa in the control, MDD-mild symptoms, MDD-moderate symptoms, and MDD-severe symptoms groups. In A, stacking is done by phylogeny, so all genera are grouped by their higher taxonomic ranks. Error bars indicate standard error of the mean. In B-L, central lines indicate the group mean and error bars indicate standard deviation, p-values are from the ANOVA post-test for linear trend of column means, and arrows indicate the direction of the trend. Abbreviations: MDD = Major Depressive Disorder, LEfSe = Linear Discriminant Analysis Effect Size, HC = Healthy Control, MI = Mild, MO = Moderate, SV = Severe. LEfSe is a biomarker discovery tool used to identify features (taxa, in this case) from high-dimensional genomic data that characterize differences between two or more biological conditions.
In general, these results suggest that symptom severity tracks with changes in a number of discriminatory taxa. While the impact of psychotropic drug usage on this pattern cannot be ruled out due to the higher levels of medication utilization in subjects with more severe depressive symptoms (Figure S4A-B), the high degree of variability in the number and classes of drugs used (Figure 1, Tables S2-3) tends to suggest that the effect is more likely related to symptom severity.
Functional Predictions
We utilized Phylogenetic Investigation of Communities by Reconstruction of Unobserved States 2 (PICRUSt2) to predict the functional potential of the healthy and depressed microbial communities based on the 16S rRNA gene content. It should be noted that this is only a prediction based on the inference from the 16S rRNA content and cannot definitively measure functional potential or transcriptional activity. When assessing function at the MetaCyc pathway level, LEfSe detected a number of pathways that were associated with each group. Pathways associated with the MDD subjects tended to be related to vitamin (folate and thiamine) biosynthesis, LPS biosynthesis, and long-chain fatty acid biosynthesis. On the other hand, pathways associated with the healthy subjects tended to be related to fermentation to short chain fatty acids, phospholipid biosynthesis, nucleic acid metabolism, and aliphatic amino acid biosynthesis (Figure S6). Of particular note is the association with healthy controls of PWY 5676, “acetyl CoA fermentation to butanoate II”, as fermentation of acetyl-CoA is the dominant pathway by which the gut microbiota, including Faecalibacterium prausnitzii, produce the anti-inflammatory short-chain fatty acid butyrate149,150. The control group was also enriched in the PWY5100, “pyruvate fermentation to acetate and lactate II”, which produces another major microbial SCFA, acetate. Additionally, the MDD group’s association with LPS production (PWY1269 – “CMP-3-deoxy-D-manno-octulosonate biosynthesis”, NAGLIPASYN – “lipid IVA biosynthesis [E. coli]”, PWY6467 – “KDO transfer to lipid IVA III [Chlamydia]”, PWY7323 – “superpathway of GDP-mannose-derived O-antigen building blocks”), likely due to the enrichment in the Gram-negative phylum Bacteroidetes and class Gammaproteobacteria, is notable. In particular, Proteobacteria-derived LPS is known to be immunogenic and has been linked to chronic inflammation151,152.
Circulating Pro-Inflammatory Cytokines
We utilized a commercially-available kit to measure a range of important inflammation-related cytokines (IL-1β, IFN-α2, IFN-γ, TNF-α, MCP-1, IL-6, IL-8, IL-10, IL-12p70, IL-17A, IL-18, IL-23, and IL-33) present in serum collected from the participants. We did not observe any significant differences in any of the measured cytokines between the MDD and control groups (Figure S7).
Discussion
Depression has been previously found to be associated with differences in the gut microbiota, and our study adds to this body of work. However, a key concept in microbiome research is the heterogeneity of microbial populations between various groups of people; therefore, as we compare our conclusions to previous studies, it is important to consider the factors that make this study unique relative to other work. First, the majority of previous MDD-microbiome studies have been undertaken in Chinese populations, with a few others studying European subjects, while our study focused on an American population18-26,60. Given that the underlying microbiome can differ significantly by geography129-131, it is important to study the impacts of disease in a range of populations. Second, most previous studies have studied a wider and older age range than our own, which recruited only subjects between the ages of 18 and 25. As the microbiome can change through the lifespan129,131,132, recruitment of a narrow age range can limit underlying noise and increase power to detect differences between groups. Third, we were able to recruit a notable subset of MDD participants who were not taking psychotropic medication, which allowed us to compare this group to the larger group of participants who were using these drugs, which most previous studies were often unable to do. Finally, we were able to recruit MDD participants with a range of depressive symptom severities, which allowed us to assess whether the changes we observed in the MDD group were related to this metric. Based on this design, we observed a number of notable differences between MDD and control participants, and further found that many of these changes may track with symptom severity.
Despite demographic differences, many of our results do align with observations made previously in other populations. Most notably, we found that Faecalibacterium levels were reduced in subjects with MDD, supporting previous work linking lower levels of this genus to depression or bipolar disorder and lower quality of life14,18-20,22,23. We also found this pattern in a number of related genera within the family Ruminococcaceae, including Subdoligranulum, Ruminococcus 1, and [Eubacterium] coprostanoligenes group. There was a member of this family – Flavonifractor – which was instead more abundant in the MDD group, supporting previous work finding that higher levels of this genus are associated with depression, bipolar disorder, and lower quality of life15,18,23. Remarkably, we found that many of differences that we observed were exacerbated in subjects with more severe depressive symptoms, which had previously only been observed for Faecalibacterium in major depressive disorder and bipolar disorder14,18. In general, our results, particularly the negative correlation between Faecalibacterium levels and depressive symptoms and the association of MDD with increased levels of Flavonifractor align most closely with those of Jiang et al18. This may relate to the fact that this study utilized a subject population closer in age to our own, with average age in the mid-twenties, compared with other studies where the average ages ranged from mid-thirties to late forties; notably, across geographies and ethnic groups, microbiome composition tends to shift to an older phenotype by the age of forty133. Further work to examine whether microbiome alterations in depressed subjects are related to age may be warranted.
There were also a few trends that were contradictory to prior data. In particular, while several studies have found lower levels of the phylum Bacteroidetes and higher levels of the phylum Firmicutes in depressed subjects20-22,24-26, we found the opposite trend. Furthermore, while Jiang et al did find that Bacteroidetes were higher in depressed subjects, this was in fact driven by increases in the families Rikenellaceae and Porphyromonadaceae, while Bacteroidaceae was actually slightly reduced18; this contrasts with our data, in which increases in the levels of Bacteroidetes in our MDD group were largely driven by increases in Bacteroidaceae and Prevotellaceae, although these differences were not significant at the family level. Additionally, while some studies have found increases in the Gammaproteobacteria family of Enterobacteriaceae in depressed subjects18, we did not observe such a change. While we found that Gammaproteobacteria was associated with the MDD group, an examination of the data suggests that this increase was driven by the genus Parasutterella of the family Burkholderiaceae. Finally, while lower levels of Fusicatenibacter were found in a previous study to be associated with lower quality-of-life scores and potentially with depression, the relationship with depression was not found in the non-medicated subset of subjects23. This is in contrast to our results, where we instead found that low levels of this genus were primarily found in the non-medicated MDD subjects. Importantly, as noted previously, our study is fairly demographically distinct from previous studies of the gut microbiota in depression, which could be responsible for some of the disparities between our results and those of prior studies. In particular, geographic location significantly impacts lifestyle factors, including diet, that can substantially change the underlying microbial composition of a population. For example, previous work has established that the microbiomes of so-called western populations are distinct from those of non-western groups, and furthermore that the gut microbiota of non-western immigrants converges with those of native westerners over time and generations due to lifestyle and dietary changes133-135.
In light of the links between MDD and chronic inflammation, a number of microbiota differences observed in our study are notable. Perhaps most interesting is the relationship between MDD subjects and lower levels of the family Ruminococcaceae and its daughter genera Faecalibacterium, Subdoligranulum, Ruminococcus 1, and [Eubacterium] coprostanoligenes group. The genus Faecalibacterium includes only one named species, F. prausnitzii, which has been demonstrated to have anti-inflammatory properties153-156. Importantly, it produces the short-chain fatty acid butyrate157, which serves as a colonic fuel source, fosters immunoregulation, and promotes epithelial barrier integrity158-165. In fact, lower levels of this genus and species have been associated with IBD, Clostridiodes difficile colitis, autoimmune disorders, and atherosclerotic cardiovascular disease1,5,6,10,155,166-170, in addition to mental health disorders including depression12,14,18-20. Similarly, Subdoligranulum includes only a single named species, S. variable, which is also known to produce butyrate157 and has been negatively associated with IBD5,168,171. Additionally, it has previously been found to correlate negatively with depressive symptoms13. The genus-level taxon Ruminococcus 1 includes the species R. albus and R. callidus, both of which have also been negatively linked to IBD170, although they do not themselves produce butyrate157.
The relationship between [Eubacterium] coprostanoligenes group and MDD is less clear, as this genus-level taxon has not been specifically linked to depression or inflammation. However, like Subdoligranulum, its abundance has been negatively correlated with depressive symptoms13. It is named for primary component species E. coprostanoligenes, which is known for its ability to reduce cholesterol to coprostanol, which is less-well-absorbed by the host172,173. Fecal coprostanol levels or the ratio of fecal coprostanol/coprostanone to cholesterol have been found to be reduced in Crohn’s disease, ulcerative colitis, and Clostridioides difficile colitis174-178, suggesting that a reduction in E. coprostanoligenes or related species could contribute to inflammation through increased colonic cholesterol levels. Evidence suggests that this may relate to serum cholesterol as well, as administration of E. coprostanoligenes to mice and rabbits decreased serum cholesterol levels179,180 and a human study found that serum cholesterol was inversely related to fecal coprostanol:cholesterol ratios181. However, while higher levels of cholesterol have generally been thought to be negative for health, there is some evidence that very low cholesterol levels are associated with severe depression, although the evidence is mixed and may depend on gender and the type of cholesterol (HDL vs LDL)182-184.
Flavonifractor, the sole member of Ruminococcaceae that was associated with the depressed subjects, has previously been linked to lower quality of life scores and MDD18,23 as well as bipolar disorder15 and generalized anxiety disorder12. This genus, which currently includes the single named species F. plautii (formerly Clostridium orbiscindens and Eubacterium plautii)185, has also previously been linked to various autoimmune disorders3,7,186,187, chronic kidney disease188, and colorectal cancer189,190. Furthermore, F. plautii was demonstrated in vitro to have epithelial invasive potential191. Therefore, there is significant evidence to suggest that unlike its generally anti-inflammatory relatives, Flavonifractor may be associated with disease despite its ability to produce butyrate under some conditions185,192. As suggested in other work15,190, this is possibly related to the genus’ eponymous capacity for cleaving flavonoids that reach the colon, including antioxidants such as quercetin, although disentangling the impacts of flavonoids and their microbial breakdown products on inflammation in vivo is difficult185,193-198.
In addition to the Ruminococcaceae, some other members of Firmicutes were altered in the depressed group, including a few members of the family Lachnospiraceae. The genus-level taxon Tyzzerella 3 was associated with the control subjects, although it has previously been linked to generalized anxiety disorder13; however, it was also linked to healthy controls in a study of chronic kidney disease188. [Eubacterium] ventriosum group was also associated with controls, and this genus-level taxon has been found to produce butyrate and inversely correlate with the inflammatory cytokines IL-6 and IL-8199,200. Finally, Fusicatenibacter was also more abundant in the control group, and was particularly reduced in the subjects with severe symptoms; its primary member species F. saccharivorans has previously been found to be reduced in subjects with active IBD and colorectal cancer201,202. On the other hand, the genus Sellimonas was associated with the MDD subjects, and has been previously linked to rheumatoid arthritis186 and chronic kidney disease188.
Previous work has demonstrated that the genera Barnesiella and Christensenellaceae R-7 were associated with healthy controls in comparison to various inflammation-related gut diseases, including IBD, colorectal cancer, and C. difficile colitis187; however, these taxa have not previously been linked to depression. Finally, in the family Proteobacteria, the genus Desulfovibrio was associated with the healthy control subjects, although in previous studies it has been found to be associated with IBD and experimental colitis models203-205; however, in a study of generalized anxiety disorder, its source family of Desulfovibrionaceae was associated with healthy controls12. Additionally, the class Gammaproteobacteria was associated with MDD, which was largely driven by increases in the family Burkholderiaceae and genus Parasutterella, which has itself been linked to MDD18. Relatedly, we also observed a predicted enrichment in LPS biosynthesis pathways in the MDD group, which was also observed by Huang et al19; the LPS of members of Gammaproteobacteria is known to be immunogenic relative to that of some commensal Gram-negative bacteria such as Bacteroides species151,152, and depressed subjects have been found to have increased serum immunoglobulin A and M responses against the LPS of members of this class124,125.
Some limitations of this study must be acknowledged. First, while there are clear trends that significantly discriminatory taxa became increasingly divergent with increasing symptom severity, we cannot completely disentangle this phenomenon from the taking of psychotropic drugs given the strong degree of overlap between symptom severity and medication usage. Future work specifically focusing on newly-diagnosed subjects who have not previously taken psychotropic drugs would be beneficial in understanding the relationship between the microbiota, depressive symptom severity, and medication. Additionally, our study was not designed to study potential microbiome differences between subjects who respond to psychotropic treatment and non-responders – in fact, given the overlap between symptom severity and psychotropic treatment, our study potentially included many non-responders – but this would also be an important element in understanding the interplay between these factors.
Additionally, we utilized 16S rRNA sequencing, which can generally only identify taxa down to the genus level, so we may be missing important species- or strain-level differences between the communities. Furthermore, while we can use taxonomic composition to predict the gene content of the communities, full metagenomics and transcriptomics would be required to comment further on changes in the functional potential or transcriptional activity, such as potentially reduced capacity for SCFA production in depressed subjects. Moreover, while we did assay circulating cytokine levels as a potential marker of systemic inflammation, we did not observe any statistically significant different in serum cytokines. Therefore, we do not have direct evidence of increased systemic inflammation in the MDD group; however, given that increased levels of circulating pro-inflammatory cytokines have previously been observed in subsets of MDD patients, our study may not have been powered to detect these differences. Future work perhaps specifically focusing on gastrointestinal inflammation, through markers such as fecal calprotectin or lipocalin-2, may be warranted. Finally, we cannot account for all of the complex confounding variables that may also contribute to microbiome differences between the two groups, such as differential lifestyle factors such as dietary habits or non-psychotropic medication use.
Conclusions
The microbiomes of American young adults with major depressive disorder were found to be significantly different from those of healthy controls. At high taxonomic levels, depressed subjects had lower levels of the phylum Firmicutes, class Clostridia, and order Clostridiales, and correspondingly higher levels of the phylum Bacteroidetes, classes Bacteroidia and Gammaproteobacteria, and order Bacteroidales. Most notably, subjects with MDD had lower levels of the families Ruminococcaceae (including the genera Faecalibacterium, Subdoligranulum, Ruminococcus 1, and [Eubacterium] coprostanoligenes group), Christensenellaceae (including the genus-level taxon R-7 group), and Barnesiellaceae (including the genus Barnesiella). These subjects also had higher levels of the Ruminococcaceae genus Flavonifractor and the Lachnospiraceae genus Sellimonas. Additionally, we found that the majority of notable taxonomic changes in the depressed group were more pronounced in subjects with higher scores on a depressive symptom scale, although we cannot rule out the impact of psychotropic medication due to significant overlap of usage with symptom severity. Overall, our findings align with previous studies of the gut microbiota in subjects with depression, particularly that depressed subjects have lower levels of Faecalibacterium and higher levels of Flavonifractor. In general, the differences that we observed are supportive of an inflammatory state in subjects with MDD, as these subjects tended to have lower levels of butyrate-producing, anti-inflammatory bacteria such as Faecalibacterium and Subdoligranulum and higher levels of taxa previously associated with inflammatory disorders such as Flavonifractor and Gammaproteobacteria. Importantly, there was significant overlap in the proportions of the discriminant taxa between the control and MDD groups in most cases, aligning with previous observations that inflammation may play a role in the etiology of depression in a significant subset of patients but is neither necessary nor sufficient to cause its onset.
Supplementary Material
Highlights:
Microbiome characteristics were compared in young adults with and without depression
Phylogenetic diversity of the microbiome is reduced in young adults with depression
The depressed group has fewer anti-inflammatory bacteria, such as Faecalibacterium
The magnitudes of many microbiome changes align with symptom severity
Acknowledgments
We would like to acknowledge the University of Rhode Island Genomics and Sequencing Center for their sequencing expertise.
Funding
This study was funded by the National Institute of Mental Health under Award Numbers RF1 MH120830, R01MH101138, R01MH115905, and R21MH112055 (RTL), the Brown Institute for Brain Science/Norman Prince Neurosciences Institute (RTL), the National Science Foundation Graduate Research Fellowship Program under award number 1644760 (BJK), and the National Institute of General Medical Sciences Institutional Development Award P20GM121344 for the COBRE Center for Antimicrobial Resistance and Therapeutic Discovery at Brown University (PB). These funding sources had no role in the design of the study, in the collection, analysis, or interpretation or data, or in writing the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Availability
Underlying sequencing data can be found at the NCBI Short Read Archive under Project ID PRJNA591924.
Ethics
This study was approved by the Lifespan Institutional Review Board.
References
- 1.Kostic AD, Xavier RJ & Gevers D The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146, 1489–1499, doi: 10.1053/j.gastro.2014.02.009 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn J et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst 105, 1907–1911, doi: 10.1093/jnci/djt300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He Z, Shao T, Li H, Xie Z & Wen C Alterations of the gut microbiome in Chinese patients with systemic lupus erythematosus. Gut Pathog 8, 64, doi: 10.1186/s13099-016-0146-9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scher JU, Littman DR & Abramson SB Microbiome in Inflammatory Arthritis and Human Rheumatic Diseases. Arthritis Rheumatol 68, 35–45, doi: 10.1002/art.39259 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown CT et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One 6, e25792, doi: 10.1371/journal.pone.0025792 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Paiva CS et al. Altered Mucosal Microbiome Diversity and Disease Severity in Sjogren Syndrome. Sci Rep 6, 23561, doi: 10.1038/srep23561 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang S, Mao J, Zhou L, Xiong X & Deng Y The imbalance of gut microbiota and its correlation with plasma inflammatory cytokines in pemphigus vulgaris patients. Scand J Immunol 90, e12799, doi: 10.1111/sji.12799 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kasselman LJ, Vernice NA, DeLeon J & Reiss AB The gut microbiome and elevated cardiovascular risk in obesity and autoimmunity. Atherosclerosis 271, 203–213, doi: 10.1016/j.atherosclerosis.2018.02.036 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Arora T & Backhed F The gut microbiota and metabolic disease: current understanding and future perspectives. J Intern Med 280, 339–349, doi: 10.1111/joim.12508 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Jie Z et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun 8, 845, doi: 10.1038/s41467-017-00900-1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster JA & McVey Neufeld KA Gut-brain axis: how the microbiome influences anxiety and depression. Trends Neurosci 36, 305–312, doi: 10.1016/j.tins.2013.01.005 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Jiang HY et al. Altered gut microbiota profile in patients with generalized anxiety disorder. J Psychiatr Res 104, 130–136, doi: 10.1016/j.jpsychires.2018.07.007 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Chen YH et al. Association between fecal microbiota and generalized anxiety disorder: Severity and early treatment response. J Affect Disord 259, 56–66, doi: 10.1016/j.jad.2019.08.014 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Evans SJ et al. The gut microbiome composition associates with bipolar disorder and illness severity. J Psychiatr Res 87, 23–29, doi: 10.1016/j.jpsychires.2016.12.007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coello K et al. Gut microbiota composition in patients with newly diagnosed bipolar disorder and their unaffected first-degree relatives. Brain Behav Immun 75, 112–118, doi: 10.1016/j.bbi.2018.09.026 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Dickerson F, Severance E & Yolken R The microbiome, immunity, and schizophrenia and bipolar disorder. Brain Behav Immun 62, 46–52, doi: 10.1016/j.bbi.2016.12.010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen TT, Kosciolek T, Eyler LT, Knight R & Jeste DV Overview and systematic review of studies of microbiome in schizophrenia and bipolar disorder. J Psychiatr Res 99, 50–61, doi: 10.1016/j.jpsychires.2018.01.013 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang H et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav Immun 48, 186–194, doi: 10.1016/j.bbi.2015.03.016 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Huang Y et al. Possible association of Firmicutes in the gut microbiota of patients with major depressive disorder. Neuropsychiatr Dis Treat 14, 3329–3337, doi: 10.2147/NDT.S188340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng P et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host's metabolism. Mol Psychiatry 21, 786–796, doi: 10.1038/mp.2016.44 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Naseribafrouei A et al. Correlation between the human fecal microbiota and depression. Neurogastroenterol Motil 26, 1155–1162, doi: 10.1111/nmo.12378 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Chen Z et al. Comparative metaproteomics analysis shows altered fecal microbiota signatures in patients with major depressive disorder. Neuroreport 29, 417–425, doi: 10.1097/WNR.0000000000000985 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Valles-Colomer M et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat Microbiol 4, 623–632, doi: 10.1038/s41564-018-0337-x (2019). [DOI] [PubMed] [Google Scholar]
- 24.Lai WT et al. Shotgun metagenomics reveals both taxonomic and tryptophan pathway differences of gut microbiota in major depressive disorder patients. Psychol Med, 1–12, doi: 10.1017/S0033291719003027 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Rong H et al. Similarly in depression, nuances of gut microbiota: Evidences from a shotgun metagenomics sequencing study on major depressive disorder versus bipolar disorder with current major depressive episode patients. J Psychiatr Res 113, 90–99, doi: 10.1016/j.jpsychires.2019.03.017 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Lin P et al. Prevotella and Klebsiella proportions in fecal microbial communities are potential characteristic parameters for patients with major depressive disorder. J Affect Disord 207, 300–304, doi: 10.1016/j.jad.2016.09.051 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Sharon G, Sampson TR, Geschwind DH & Mazmanian SK The Central Nervous System and the Gut Microbiome. Cell 167, 915–932, doi: 10.1016/j.cell.2016.10.027 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carabotti M, Scirocco A, Maselli MA & Severi C The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann Gastroenterol 28, 203–209 (2015). [PMC free article] [PubMed] [Google Scholar]
- 29.O'Mahony SM, Clarke G, Borre YE, Dinan TG & Cryan JF Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behav Brain Res 277, 32–48, doi: 10.1016/j.bbr.2014.07.027 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Kelly JR, Clarke G, Cryan JF & Dinan TG Brain-gut-microbiota axis: challenges for translation in psychiatry. Ann Epidemiol 26, 366–372, doi: 10.1016/j.annepidem.2016.02.008 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Foster JA, Rinaman L & Cryan JF Stress & the gut-brain axis: Regulation by the microbiome. Neurobiol Stress 7, 124–136, doi: 10.1016/j.ynstr.2017.03.001 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayer EA, Tillisch K & Gupta A Gut/brain axis and the microbiota. J Clin Invest 125, 926–938, doi: 10.1172/JCI76304 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dinan TG, Stilling RM, Stanton C & Cryan JF Collective unconscious: how gut microbes shape human behavior. J Psychiatr Res 63, 1–9, doi: 10.1016/j.jpsychires.2015.02.021 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Bonaz B, Sinniger V & Pellissier S Vagus Nerve Stimulation at the Interface of Brain-Gut Interactions. Cold Spring Harb Perspect Med 9, doi: 10.1101/cshperspect.a034199 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forsythe P, Bienenstock J & Kunze WA Vagal pathways for microbiome-brain-gut axis communication. Adv Exp Med Biol 817, 115–133, doi: 10.1007/978-1-4939-0897-4_5 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Caspani G, Kennedy S, Foster JA & Swann J Gut microbial metabolites in depression: understanding the biochemical mechanisms. Microb Cell 6, 454–481, doi: 10.15698/mic2019.10.693 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raybould HE Gut chemosensing: interactions between gut endocrine cells and visceral afferents. Auton Neurosci 153, 41–46, doi: 10.1016/j.autneu.2009.07.007 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarkar A et al. Psychobiotics and the Manipulation of Bacteria-Gut-Brain Signals. Trends Neurosci 39, 763–781, doi: 10.1016/j.tins.2016.09.002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Mahony SM et al. Early life stress alters behavior, immunity, and microbiota in rats: implications for irritable bowel syndrome and psychiatric illnesses. Biol Psychiatry 65, 263–267, doi: 10.1016/j.biopsych.2008.06.026 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Sudo N et al. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J Physiol 558, 263–275, doi: 10.1113/jphysiol.2004.063388 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wikoff WR et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A 106, 3698–3703, doi: 10.1073/pnas.0812874106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yano JM et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 161, 264–276, doi: 10.1016/j.cell.2015.02.047 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barrett E, Ross RP, O'Toole PW, Fitzgerald GF & Stanton C gamma-Aminobutyric acid production by culturable bacteria from the human intestine. J Appl Microbiol 113, 411–417, doi: 10.1111/j.1365-2672.2012.05344.x (2012). [DOI] [PubMed] [Google Scholar]
- 44.Borovikova LV et al. Vagus nerve stimulation attenuates the systemic inflammatory response to endotoxin. Nature 405, 458–462, doi: 10.1038/35013070 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Johnston GR & Webster NR Cytokines and the immunomodulatory function of the vagus nerve. Br J Anaesth 102, 453–462, doi: 10.1093/bja/aep037 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Park AJ et al. Altered colonic function and microbiota profile in a mouse model of chronic depression. Neurogastroenterol Motil 25, 733–e575, doi: 10.1111/nmo.12153 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Punder K & Pruimboom L Stress induces endotoxemia and low-grade inflammation by increasing barrier permeability. Front Immunol 6, 223, doi: 10.3389/fimmu.2015.00223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vicario M et al. Chronological assessment of mast cell-mediated gut dysfunction and mucosal inflammation in a rat model of chronic psychosocial stress. Brain Behav Immun 24, 1166–1175, doi: 10.1016/j.bbi.2010.06.002 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Kelly JR et al. Breaking down the barriers: the gut microbiome, intestinal permeability and stress-related psychiatric disorders. Front Cell Neurosci 9, 392, doi: 10.3389/fncel.2015.00392 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodino-Janeiro BK et al. Role of Corticotropin-releasing Factor in Gastrointestinal Permeability. J Neurogastroenterol Motil 21, 33–50, doi: 10.5056/jnm14084 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moussaoui N et al. Changes in intestinal glucocorticoid sensitivity in early life shape the risk of epithelial barrier defect in maternal-deprived rats. PLoS One 9, e88382, doi: 10.1371/journal.pone.0088382 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soderholm JD et al. Neonatal maternal separation predisposes adult rats to colonic barrier dysfunction in response to mild stress. Am J Physiol Gastrointest Liver Physiol 283, G1257–1263, doi: 10.1152/ajpgi.00314.2002 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Rhee SH, Pothoulakis C & Mayer EA Principles and clinical implications of the brain-gut-enteric microbiota axis. Nat Rev Gastroenterol Hepatol 6, 306–314, doi: 10.1038/nrgastro.2009.35 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin CR, Osadchiy V, Kalani A & Mayer EA The Brain-Gut-Microbiome Axis. Cell Mol Gastroenterol Hepatol 6, 133–148, doi: 10.1016/j.jcmgh.2018.04.003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mayer EA Gut feelings: the emerging biology of gut-brain communication. Nat Rev Neurosci 12, 453–466, doi: 10.1038/nrn3071 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang TT et al. Current Understanding of Gut Microbiota in Mood Disorders: An Update of Human Studies. Front Genet 10, 98, doi: 10.3389/fgene.2019.00098 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dinan TG & Cryan JF Melancholic microbes: a link between gut microbiota and depression? Neurogastroenterol Motil 25, 713–719, doi: 10.1111/nmo.12198 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Evrensel A & Ceylan ME The Gut-Brain Axis: The Missing Link in Depression. Clin Psychopharmacol Neurosci 13, 239–244, doi: 10.9758/cpn.2015.13.3.239 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lach G, Schellekens H, Dinan TG & Cryan JF Anxiety, Depression, and the Microbiome: A Role for Gut Peptides. Neurotherapeutics 15, 36–59, doi: 10.1007/s13311-017-0585-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kelly JR et al. Transferring the blues: Depression-associated gut microbiota induces neurobehavioural changes in the rat. J Psychiatr Res 82, 109–118, doi: 10.1016/j.jpsychires.2016.07.019 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Hao Z, Wang W, Guo R & Liu H Faecalibacterium prausnitzii (ATCC 27766) has preventive and therapeutic effects on chronic unpredictable mild stress-induced depression-like and anxiety-like behavior in rats. Psychoneuroendocrinology 104, 132–142, doi: 10.1016/j.psyneuen.2019.02.025 (2019). [DOI] [PubMed] [Google Scholar]
- 62.Bravo JA et al. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A 108, 16050–16055, doi: 10.1073/pnas.1102999108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bercik P et al. The anxiolytic effect of Bifidobacterium longum NCC3001 involves vagal pathways for gut-brain communication. Neurogastroenterol Motil 23, 1132–1139, doi: 10.1111/j.1365-2982.2011.01796.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmidtner AK et al. Minocycline alters behavior, microglia and the gut microbiome in a trait-anxiety-dependent manner. Transl Psychiatry 9, 223, doi: 10.1038/s41398-019-0556-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Husain MI et al. Minocycline as an adjunct for treatment-resistant depressive symptoms: A pilot randomised placebo-controlled trial. J Psychopharmacol 31, 1166–1175, doi: 10.1177/0269881117724352 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Dean OM et al. Adjunctive minocycline treatment for major depressive disorder: A proof of concept trial. Aust N Z J Psychiatry 51, 829–840, doi: 10.1177/0004867417709357 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Felger JC & Lotrich FE Inflammatory cytokines in depression: neurobiological mechanisms and therapeutic implications. Neuroscience 246, 199–229, doi: 10.1016/j.neuroscience.2013.04.060 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Amodeo GT T. MA; Fagiolini A Depression and inflammation: Disentangling a clear yet complex and multifaceted link. Neuropsychiatry 7, 448–457 (2017). [Google Scholar]
- 69.Miller AH & Raison CL The role of inflammation in depression: from evolutionary imperative to modern treatment target. Nat Rev Immunol 16, 22–34, doi: 10.1038/nri.2015.5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dowlati Y et al. A meta-analysis of cytokines in major depression. Biol Psychiatry 67, 446–457, doi: 10.1016/j.biopsych.2009.09.033 (2010). [DOI] [PubMed] [Google Scholar]
- 71.Kohler CA et al. Peripheral cytokine and chemokine alterations in depression: a meta-analysis of 82 studies. Acta Psychiatr Scand 135, 373–387, doi: 10.1111/acps.12698 (2017). [DOI] [PubMed] [Google Scholar]
- 72.Haapakoski R, Mathieu J, Ebmeier KP, Alenius H & Kivimaki M Cumulative meta-analysis of interleukins 6 and 1beta, tumour necrosis factor alpha and C-reactive protein in patients with major depressive disorder. Brain Behav Immun 49, 206–215, doi: 10.1016/j.bbi.2015.06.001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu Y, Ho RC & Mak A Interleukin (IL)-6, tumour necrosis factor alpha (TNF-alpha) and soluble interleukin-2 receptors (sIL-2R) are elevated in patients with major depressive disorder: a meta-analysis and meta-regression. J Affect Disord 139, 230–239, doi: 10.1016/j.jad.2011.08.003 (2012). [DOI] [PubMed] [Google Scholar]
- 74.Goldsmith DR, Rapaport MH & Miller BJ A meta-analysis of blood cytokine network alterations in psychiatric patients: comparisons between schizophrenia, bipolar disorder and depression. Mol Psychiatry 21, 1696–1709, doi: 10.1038/mp.2016.3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Himmerich H, Patsalos O, Lichtblau N, Ibrahim MAA & Dalton B Cytokine Research in Depression: Principles, Challenges, and Open Questions. Front Psychiatry 10, 30, doi: 10.3389/fpsyt.2019.00030 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Burke HM, Davis MC, Otte C & Mohr DC Depression and cortisol responses to psychological stress: a meta-analysis. Psychoneuroendocrinology 30, 846–856, doi: 10.1016/j.psyneuen.2005.02.010 (2005). [DOI] [PubMed] [Google Scholar]
- 77.Carroll BJ et al. Pathophysiology of hypercortisolism in depression. Acta Psychiatr Scand Suppl, 90–103, doi: 10.1111/j.1600-0447.2007.00967.x (2007). [DOI] [PubMed] [Google Scholar]
- 78.Parker KJ, Schatzberg AF & Lyons DM Neuroendocrine aspects of hypercortisolism in major depression. Horm Behav 43, 60–66, doi: 10.1016/s0018-506x(02)00016-8 (2003). [DOI] [PubMed] [Google Scholar]
- 79.Mendlewicz J et al. Shortened onset of action of antidepressants in major depression using acetylsalicylic acid augmentation: a pilot open-label study. Int Clin Psychopharmacol 21, 227–231, doi: 10.1097/00004850-200607000-00005 (2006). [DOI] [PubMed] [Google Scholar]
- 80.Muller N et al. The cyclooxygenase-2 inhibitor celecoxib has therapeutic effects in major depression: results of a double-blind, randomized, placebo controlled, add-on pilot study to reboxetine. Mol Psychiatry 11, 680–684, doi: 10.1038/sj.mp.4001805 (2006). [DOI] [PubMed] [Google Scholar]
- 81.Wong ML et al. Inflammasome signaling affects anxiety- and depressive-like behavior and gut microbiome composition. Mol Psychiatry 21, 797–805, doi: 10.1038/mp.2016.46 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Y et al. NLRP3 Inflammasome Mediates Chronic Mild Stress-Induced Depression in Mice via Neuroinflammation. Int J Neuropsychopharmacol 18, doi: 10.1093/ijnp/pyv006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Alcocer-Gomez E et al. Stress-Induced Depressive Behaviors Require a Functional NLRP3 Inflammasome. Mol Neurobiol 53, 4874–4882, doi: 10.1007/s12035-015-9408-7 (2016). [DOI] [PubMed] [Google Scholar]
- 84.Iwata M, Ota KT & Duman RS The inflammasome: pathways linking psychological stress, depression, and systemic illnesses. Brain Behav Immun 31, 105–114, doi: 10.1016/j.bbi.2012.12.008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kaufmann FN et al. NLRP3 inflammasome-driven pathways in depression: Clinical and preclinical findings. Brain Behav Immun 64, 367–383, doi: 10.1016/j.bbi.2017.03.002 (2017). [DOI] [PubMed] [Google Scholar]
- 86.Barnard KD, Skinner TC & Peveler R The prevalence of co-morbid depression in adults with Type 1 diabetes: systematic literature review. Diabet Med 23, 445–448, doi: 10.1111/j.1464-5491.2006.01814.x (2006). [DOI] [PubMed] [Google Scholar]
- 87.Dickens C, McGowan L, Clark-Carter D & Creed F Depression in rheumatoid arthritis: a systematic review of the literature with meta-analysis. Psychosom Med 64, 52–60, doi: 10.1097/00006842-200201000-00008 (2002). [DOI] [PubMed] [Google Scholar]
- 88.Figueiredo-Braga M et al. Depression and anxiety in systemic lupus erythematosus: The crosstalk between immunological, clinical, and psychosocial factors. Medicine (Baltimore) 97, e11376, doi: 10.1097/MD.0000000000011376 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Byrne G et al. Prevalence of Anxiety and Depression in Patients with Inflammatory Bowel Disease. Can J Gastroenterol Hepatol 2017, 6496727, doi: 10.1155/2017/6496727 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Caneo C, Marston L, Bellon JA & King M Examining the relationship between physical illness and depression: Is there a difference between inflammatory and non inflammatory diseases? A cohort study. Gen Hosp Psychiatry 43, 71–77, doi: 10.1016/j.genhosppsych.2016.09.007 (2016). [DOI] [PubMed] [Google Scholar]
- 91.Dantzer R Cytokine-induced sickness behaviour: a neuroimmune response to activation of innate immunity. Eur J Pharmacol 500, 399–411, doi: 10.1016/j.ejphar.2004.07.040 (2004). [DOI] [PubMed] [Google Scholar]
- 92.Raison CL & Miller AH Do cytokines really sing the blues? Cerebrum 2013, 10 (2013). [PMC free article] [PubMed] [Google Scholar]
- 93.Miyaoka H et al. Depression from interferon therapy in patients with hepatitis C. Am J Psychiatry 156, 1120, doi: 10.1176/ajp.156.7.1120 (1999). [DOI] [PubMed] [Google Scholar]
- 94.O'Connor JC et al. Lipopolysaccharide-induced depressive-like behavior is mediated by indoleamine 2,3-dioxygenase activation in mice. Mol Psychiatry 14, 511–522, doi: 10.1038/sj.mp.4002148 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ohgi Y, Futamura T, Kikuchi T & Hashimoto K Effects of antidepressants on alternations in serum cytokines and depressive-like behavior in mice after lipopolysaccharide administration. Pharmacol Biochem Behav 103, 853–859, doi: 10.1016/j.pbb.2012.12.003 (2013). [DOI] [PubMed] [Google Scholar]
- 96.Zhang Y et al. Involvement of inflammasome activation in lipopolysaccharide-induced mice depressive-like behaviors. CNS Neurosci Ther 20, 119–124, doi: 10.1111/cns.12170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fu X et al. Central administration of lipopolysaccharide induces depressive-like behavior in vivo and activates brain indoleamine 2,3 dioxygenase in murine organotypic hippocampal slice cultures. J Neuroinflammation 7, 43, doi: 10.1186/1742-2094-7-43 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Anisman H, Merali Z, Poulter MO & Hayley S Cytokines as a precipitant of depressive illness: animal and human studies. Curr Pharm Des 11, 963–972, doi: 10.2174/1381612053381701 (2005). [DOI] [PubMed] [Google Scholar]
- 99.Schiepers OJ, Wichers MC & Maes M Cytokines and major depression. Prog Neuropsychopharmacol Biol Psychiatry 29, 201–217, doi: 10.1016/j.pnpbp.2004.11.003 (2005). [DOI] [PubMed] [Google Scholar]
- 100.Rochfort KD, Collins LE, Murphy RP & Cummins PM Downregulation of blood-brain barrier phenotype by proinflammatory cytokines involves NADPH oxidase-dependent ROS generation: consequences for interendothelial adherens and tight junctions. PLoS One 9, e101815, doi: 10.1371/journal.pone.0101815 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Farkas G et al. Experimental acute pancreatitis results in increased blood-brain barrier permeability in the rat: a potential role for tumor necrosis factor and interleukin 6. Neurosci Lett 242, 147–150, doi: 10.1016/s0304-3940(98)00060-3 (1998). [DOI] [PubMed] [Google Scholar]
- 102.de Vries HE et al. The influence of cytokines on the integrity of the blood-brain barrier in vitro. J Neuroimmunol 64, 37–43, doi: 10.1016/0165-5728(95)00148-4 (1996). [DOI] [PubMed] [Google Scholar]
- 103.Plotkin SR, Banks WA & Kastin AJ Comparison of saturable transport and extracellular pathways in the passage of interleukin-1 alpha across the blood-brain barrier. J Neuroimmunol 67, 41–47, doi: 10.1016/0165-5728(96)00036-7 (1996). [DOI] [PubMed] [Google Scholar]
- 104.Watkins LR, Maier SF & Goehler LE Cytokine-to-brain communication: a review & analysis of alternative mechanisms. Life Sci 57, 1011–1026, doi: 10.1016/0024-3205(95)02047-m (1995). [DOI] [PubMed] [Google Scholar]
- 105.Miller AH, Maletic V & Raison CL Inflammation and its discontents: the role of cytokines in the pathophysiology of major depression. Biol Psychiatry 65, 732–741, doi: 10.1016/j.biopsych.2008.11.029 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pavlov VA & Tracey KJ The vagus nerve and the inflammatory reflex--linking immunity and metabolism. Nat Rev Endocrinol 8, 743–754, doi: 10.1038/nrendo.2012.189 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Steinberg BE et al. Cytokine-specific Neurograms in the Sensory Vagus Nerve. Bioelectron Med 3, 7–17 (2016). [PMC free article] [PubMed] [Google Scholar]
- 108.Zanos TP et al. Identification of cytokine-specific sensory neural signals by decoding murine vagus nerve activity. Proc Natl Acad Sci U S A 115, E4843–E4852, doi: 10.1073/pnas.1719083115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Galic MA, Riazi K & Pittman QJ Cytokines and brain excitability. Front Neuroendocrinol 33, 116–125, doi: 10.1016/j.yfrne.2011.12.002 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.De Simoni MG & Imeri L Cytokine-neurotransmitter interactions in the brain. Biol Signals Recept 7, 33–44, doi: 10.1159/000014526 (1998). [DOI] [PubMed] [Google Scholar]
- 111.Miller AH, Haroon E, Raison CL & Felger JC Cytokine targets in the brain: impact on neurotransmitters and neurocircuits. Depress Anxiety 30, 297–306, doi: 10.1002/da.22084 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gadek-Michalska A T. J; Rachwalska P; Bugjaski J Cytokines, prostaglandins and nitric oxide in the regulation of stress-response systems. Pharmacological Reports 65, 1655–1662 (2013). [DOI] [PubMed] [Google Scholar]
- 113.Dantzer R, Wollman E, Vitkovic L & Yirmiya R Cytokines and depression: fortuitous or causative association? Mol Psychiatry 4, 328–332, doi: 10.1038/sj.mp.4000572 (1999). [DOI] [PubMed] [Google Scholar]
- 114.Zunszain PA, Anacker C, Cattaneo A, Carvalho LA & Pariante CM Glucocorticoids, cytokines and brain abnormalities in depression. Prog Neuropsychopharmacol Biol Psychiatry 35, 722–729, doi: 10.1016/j.pnpbp.2010.04.011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Humphreys D, Schlesinger L, Lopez M & Araya AV Interleukin-6 production and deregulation of the hypothalamic-pituitary-adrenal axis in patients with major depressive disorders. Endocrine 30, 371–376 (2006). [DOI] [PubMed] [Google Scholar]
- 116.Pace TW, Hu F & Miller AH Cytokine-effects on glucocorticoid receptor function: relevance to glucocorticoid resistance and the pathophysiology and treatment of major depression. Brain Behav Immun 21, 9–19, doi: 10.1016/j.bbi.2006.08.009 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Miller AH, Pariante CM & Pearce BD Effects of cytokines on glucocorticoid receptor expression and function. Glucocorticoid resistance and relevance to depression. Adv Exp Med Biol 461, 107–116, doi: 10.1007/978-0-585-37970-8_7 (1999). [DOI] [PubMed] [Google Scholar]
- 118.Capuron L et al. Association between decreased serum tryptophan concentrations and depressive symptoms in cancer patients undergoing cytokine therapy. Mol Psychiatry 7, 468–473, doi: 10.1038/sj.mp.4000995 (2002). [DOI] [PubMed] [Google Scholar]
- 119.Dantzer R, O'Connor JC, Lawson MA & Kelley KW Inflammation-associated depression: from serotonin to kynurenine. Psychoneuroendocrinology 36, 426–436, doi: 10.1016/j.psyneuen.2010.09.012 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Maes M et al. Serotonin-immune interactions in major depression: lower serum tryptophan as a marker of an immune-inflammatory response. Eur Arch Psychiatry Clin Neurosci 247, 154–161, doi: 10.1007/bf03033069 (1997). [DOI] [PubMed] [Google Scholar]
- 121.Myint AM & Kim YK Cytokine-serotonin interaction through IDO: a neurodegeneration hypothesis of depression. Med Hypotheses 61, 519–525, doi: 10.1016/s0306-9877(03)00207-x (2003). [DOI] [PubMed] [Google Scholar]
- 122.Riedel WJ, Klaassen T & Schmitt JA Tryptophan, mood, and cognitive function. Brain Behav Immun 16, 581–589, doi: 10.1016/s0889-1591(02)00013-2 (2002). [DOI] [PubMed] [Google Scholar]
- 123.Heyes MP et al. Quinolinic acid and kynurenine pathway metabolism in inflammatory and non-inflammatory neurological disease. Brain 115 ( Pt 5), 1249–1273, doi: 10.1093/brain/115.5.1249 (1992). [DOI] [PubMed] [Google Scholar]
- 124.Maes M, Kubera M & Leunis JC The gut-brain barrier in major depression: intestinal mucosal dysfunction with an increased translocation of LPS from gram negative enterobacteria (leaky gut) plays a role in the inflammatory pathophysiology of depression. Neuro Endocrinol Lett 29, 117–124 (2008). [PubMed] [Google Scholar]
- 125.Maes M, Kubera M, Leunis JC & Berk M Increased IgA and IgM responses against gut commensals in chronic depression: further evidence for increased bacterial translocation or leaky gut. J Affect Disord 141, 55–62, doi: 10.1016/j.jad.2012.02.023 (2012). [DOI] [PubMed] [Google Scholar]
- 126.Maes M et al. In depression, bacterial translocation may drive inflammatory responses, oxidative and nitrosative stress (O&NS), and autoimmune responses directed against O&NS-damaged neoepitopes. Acta Psychiatr Scand 127, 344–354, doi: 10.1111/j.1600-0447.2012.01908.x (2013). [DOI] [PubMed] [Google Scholar]
- 127.Keri S, Szabo C & Kelemen O Expression of Toll-Like Receptors in peripheral blood mononuclear cells and response to cognitive-behavioral therapy in major depressive disorder. Brain Behav Immun 40, 235–243, doi: 10.1016/j.bbi.2014.03.020 (2014). [DOI] [PubMed] [Google Scholar]
- 128.Berk M et al. So depression is an inflammatory disease, but where does the inflammation come from? BMC Med 11, 200, doi: 10.1186/1741-7015-11-200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Costello EK et al. Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697, doi: 10.1126/science.1177486 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gupta VK, Paul S & Dutta C Geography, Ethnicity or Subsistence-Specific Variations in Human Microbiome Composition and Diversity. Front Microbiol 8, 1162, doi: 10.3389/fmicb.2017.01162 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yatsunenko T et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227, doi: 10.1038/nature11053 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Odamaki T et al. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol 16, 90, doi: 10.1186/s12866-016-0708-5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yadav D, Ghosh TS & Mande SS Global investigation of composition and interaction networks in gut microbiomes of individuals belonging to diverse geographies and age-groups. Gut Pathog 8, 17, doi: 10.1186/s13099-016-0099-z (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nam YD, Jung MJ, Roh SW, Kim MS & Bae JW Comparative analysis of Korean human gut microbiota by barcoded pyrosequencing. PLoS One 6, e22109, doi: 10.1371/journal.pone.0022109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Vangay P et al. US Immigration Westernizes the Human Gut Microbiome. Cell 175, 962–972.e910, doi: 10.1016/j.cell.2018.10.029 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pilkonis PA et al. Item banks for measuring emotional distress from the Patient-Reported Outcomes Measurement Information System (PROMIS(R)): depression, anxiety, and anger. Assessment 18, 263–283, doi: 10.1177/1073191111411667 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Posner K et al. The Columbia-Suicide Severity Rating Scale: initial validity and internal consistency findings from three multisite studies with adolescents and adults. Am J Psychiatry 168, 1266–1277, doi: 10.1176/appi.ajp.2011.10111704 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nock MK, Holmberg EB, Photos VI & Michel BD Self-Injurious Thoughts and Behaviors Interview: development, reliability, and validity in an adolescent sample. Psychol Assess 19, 309–317, doi: 10.1037/1040-3590.19.3.309 (2007). [DOI] [PubMed] [Google Scholar]
- 139.First M W. JBW; Karg RS; Spitzer RL Structured Clinical Interview for DSM-5—Research Version (SCID-5 for DSM-5, Research Version; SCID-5-RV). (American Psychiatric Association, 2015). [Google Scholar]
- 140.Thompson LR et al. A communal catalogue reveals Earth's multiscale microbial diversity. Nature 551, 457–463, doi: 10.1038/nature24621 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bolyen E et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37, 852–857, doi: 10.1038/s41587-019-0209-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Callahan BJ et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13, 581–583, doi: 10.1038/nmeth.3869 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Douglas GM, Beiko RG & Langille MGI Predicting the Functional Potential of the Microbiome from Marker Genes Using PICRUSt. Methods Mol Biol 1849, 169–177, doi: 10.1007/978-1-4939-8728-3_11 (2018). [DOI] [PubMed] [Google Scholar]
- 144.McDonald D et al. The Biological Observation Matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. Gigascience 1, 7, doi: 10.1186/2047-217X-1-7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.McMurdie PJ & Holmes S phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8, e61217, doi: 10.1371/journal.pone.0061217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.McMurdie PJ & Holmes S Phyloseq: a bioconductor package for handling and analysis of high-throughput phylogenetic sequence data. Pac Symp Biocomput, 235–246 (2012). [PMC free article] [PubMed] [Google Scholar]
- 147.Dixon P VEGAN, a package of R functions for community ecology. J Veg Sci 14, 927–930 (2009). [Google Scholar]
- 148.Segata N et al. Metagenomic biomarker discovery and explanation. Genome Biol 12, R60, doi: 10.1186/gb-2011-12-6-r60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Vital M, Howe AC & Tiedje JM Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio 5, e00889, doi: 10.1128/mBio.00889-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Anand S, Kaur H & Mande SS Comparative In silico Analysis of Butyrate Production Pathways in Gut Commensals and Pathogens. Front Microbiol 7, 1945, doi: 10.3389/fmicb.2016.01945 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.d'Hennezel E, Abubucker S, Murphy LO & Cullen TW Total Lipopolysaccharide from the Human Gut Microbiome Silences Toll-Like Receptor Signaling. mSystems 2, doi: 10.1128/mSystems.00046-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Vatanen T et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell 165, 842–853, doi: 10.1016/j.cell.2016.04.007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Qiu X, Zhang M, Yang X, Hong N & Yu C Faecalibacterium prausnitzii upregulates regulatory T cells and anti-inflammatory cytokines in treating TNBS-induced colitis. J Crohns Colitis 7, e558–568, doi: 10.1016/j.crohns.2013.04.002 (2013). [DOI] [PubMed] [Google Scholar]
- 154.Quevrain E et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn's disease. Gut 65, 415–425, doi: 10.1136/gutjnl-2014-307649 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sokol H et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 105, 16731–16736, doi: 10.1073/pnas.0804812105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhou L et al. Faecalibacterium prausnitzii Produces Butyrate to Maintain Th17/Treg Balance and to Ameliorate Colorectal Colitis by Inhibiting Histone Deacetylase 1. Inflamm Bowel Dis, doi: 10.1093/ibd/izy182 (2018). [DOI] [PubMed] [Google Scholar]
- 157.Louis P & Flint HJ Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 19, 29–41, doi: 10.1111/1462-2920.13589 (2017). [DOI] [PubMed] [Google Scholar]
- 158.Arpaia N et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455, doi: 10.1038/nature12726 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Furusawa Y et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450, doi: 10.1038/nature12721 (2013). [DOI] [PubMed] [Google Scholar]
- 160.Hamer HM et al. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther 27, 104–119, doi: 10.1111/j.1365-2036.2007.03562.x (2008). [DOI] [PubMed] [Google Scholar]
- 161.Leonel AJ & Alvarez-Leite JI Butyrate: implications for intestinal function. Curr Opin Clin Nutr Metab Care 15, 474–479, doi: 10.1097/MCO.0b013e32835665fa (2012). [DOI] [PubMed] [Google Scholar]
- 162.Parada Venegas D et al. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front Immunol 10, 277, doi: 10.3389/fimmu.2019.00277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.VanHook AM Butyrate benefits the intestinal barrier. Science Signaling 8, ec135 (2015). [Google Scholar]
- 164.Wang HB, Wang PY, Wang X, Wan YL & Liu YC Butyrate enhances intestinal epithelial barrier function via up-regulation of tight junction protein Claudin-1 transcription. Dig Dis Sci 57, 3126–3135, doi: 10.1007/s10620-012-2259-4 (2012). [DOI] [PubMed] [Google Scholar]
- 165.Zhang M et al. Butyrate inhibits interleukin-17 and generates Tregs to ameliorate colorectal colitis in rats. BMC Gastroenterol 16, 84, doi: 10.1186/s12876-016-0500-x (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gevers D et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 15, 382–392, doi: 10.1016/j.chom.2014.02.005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kabeerdoss J, Sankaran V, Pugazhendhi S & Ramakrishna BS Clostridium leptum group bacteria abundance and diversity in the fecal microbiota of patients with inflammatory bowel disease: a case-control study in India. BMC Gastroenterol 13, 20, doi: 10.1186/1471-230X-13-20 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Quince C et al. Extensive Modulation of the Fecal Metagenome in Children With Crohn's Disease During Exclusive Enteral Nutrition. Am J Gastroenterol 110, 1718–1729; quiz 1730, doi: 10.1038/ajg.2015.357 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Takahashi K et al. Reduced Abundance of Butyrate-Producing Bacteria Species in the Fecal Microbial Community in Crohn's Disease. Digestion 93, 59–65, doi: 10.1159/000441768 (2016). [DOI] [PubMed] [Google Scholar]
- 170.Kang S et al. Dysbiosis of fecal microbiota in Crohn's disease patients as revealed by a custom phylogenetic microarray. Inflamm Bowel Dis 16, 2034–2042, doi: 10.1002/ibd.21319 (2010). [DOI] [PubMed] [Google Scholar]
- 171.Papa E et al. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One 7, e39242, doi: 10.1371/journal.pone.0039242 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Freier TA, Beitz DC, Li L & Hartman PA Characterization of Eubacterium coprostanoligenes sp. nov., a cholesterol-reducing anaerobe. Int J Syst Bacteriol 44, 137–142, doi: 10.1099/00207713-44-1-137 (1994). [DOI] [PubMed] [Google Scholar]
- 173.Ren D, Li L, Schwabacher AW, Young JW & Beitz DC Mechanism of cholesterol reduction to coprostanol by Eubacterium coprostanoligenes ATCC 51222. Steroids 61, 33–40, doi: 10.1016/0039-128x(95)00173-n (1996). [DOI] [PubMed] [Google Scholar]
- 174.Sundqvist T S. L; Tjellstrom B; Magnusson K; Midtvedt T; Norin E; Hogberg L Evidence of disturbed gut microbial metabolic activity in pediatric Crohn’s disease. Crohn’s & Colitis 360 1, otz010 (2019). [Google Scholar]
- 175.Reddy BS, Martin CW & Wynder EL Fecal bile acids and cholesterol metabolites of patients with ulcerative colitis, a high-risk group for development of colon cancer. Cancer Res 37, 1697–1701 (1977). [PubMed] [Google Scholar]
- 176.Santoru ML et al. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci Rep 7, 9523, doi: 10.1038/s41598-017-10034-5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Antharam VC et al. An Integrated Metabolomic and Microbiome Analysis Identified Specific Gut Microbiota Associated with Fecal Cholesterol and Coprostanol in Clostridium difficile Infection. PLoS One 11, e0148824, doi: 10.1371/journal.pone.0148824 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Midtvedt T et al. Increase of faecal tryptic activity relates to changes in the intestinal microbiome: analysis of Crohn's disease with a multidisciplinary platform. PLoS One 8, e66074, doi: 10.1371/journal.pone.0066074 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Li L, Batt SM, Wannemuehler M, Dispirito A & Beitz DC Effect of feeding of a cholesterol-reducing bacterium, Eubacterium coprostanoligenes, to germ-free mice. Lab Anim Sci 48, 253–255 (1998). [PubMed] [Google Scholar]
- 180.Li L, Buhman KK, Hartman PA & Beitz DC Hypocholesterolemic effect of Eubacterium coprostanoligenes ATCC 51222 in rabbits. Lett Appl Microbiol 20, 137–140, doi: 10.1111/j.1472-765x.1995.tb00410.x (1995). [DOI] [PubMed] [Google Scholar]
- 181.Sekimoto H, Shimada O, Makanishi M, Nakano T & Katayama O Interrelationship between serum and fecal sterols. Jpn J Med 22, 14–20, doi: 10.2169/internalmedicine1962.22.14 (1983). [DOI] [PubMed] [Google Scholar]
- 182.Deisenhammer EA et al. No evidence for an association between serum cholesterol and the course of depression and suicidality. Psychiatry Res 121, 253–261, doi: 10.1016/j.psychres.2003.09.007 (2004). [DOI] [PubMed] [Google Scholar]
- 183.Kim YK & Myint AM Clinical application of low serum cholesterol as an indicator for suicide risk in major depression. J Affect Disord 81, 161–166, doi: 10.1016/S0165-0327(03)00166-6 (2004). [DOI] [PubMed] [Google Scholar]
- 184.Sansone RA Cholesterol quandaries: relationship to depression and the suicidal experience. Psychiatry (Edgmont) 5, 22–34 (2008). [PMC free article] [PubMed] [Google Scholar]
- 185.Carlier JP, Bedora-Faure M, K'Ouas G, Alauzet C & Mory F Proposal to unify Clostridium orbiscindens Winter et al. 1991 and Eubacterium plautii (Seguin 1928) Hofstad and Aasjord 1982, with description of Flavonifractor plautii gen. nov., comb. nov., and reassignment of Bacteroides capillosus to Pseudoflavonifractor capillosus gen. nov., comb. nov. Int J Syst Evol Microbiol 60, 585–590, doi: 10.1099/ijs.0.016725-0 (2010). [DOI] [PubMed] [Google Scholar]
- 186.Sun Y C. Q; Lin P; Xu R; He D; Ji W; Bian Y; Shen Y; Li Q; Liu C; Dong K; Tang Y; Pei Z; Yang L; Lu H; Guo X; Xiao L Characteristics of Gut Microbiota in Patients with Rheumatoid Arthritis in Shanghai, China. Frontiers in Cellular and Infection Microbiology 9, 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Mancabelli L et al. Identification of universal gut microbial biomarkers of common human intestinal diseases by meta-analysis. FEMS Microbiol Ecol 93, doi: 10.1093/femsec/fix153 (2017). [DOI] [PubMed] [Google Scholar]
- 188.Lun H et al. Altered gut microbiota and microbial biomarkers associated with chronic kidney disease. Microbiologyopen 8, e00678, doi: 10.1002/mbo3.678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ai D et al. Using Decision Tree Aggregation with Random Forest Model to Identify Gut Microbes Associated with Colorectal Cancer. Genes (Basel) 10, doi: 10.3390/genes10020112 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Gupta A et al. Association of Flavonifractor plautii, a Flavonoid-Degrading Bacterium, with the Gut Microbiome of Colorectal Cancer Patients in India. mSystems 4, doi: 10.1128/mSystems.00438-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Armstrong H et al. Host immunoglobulin G selectively identifies pathobionts in pediatric inflammatory bowel diseases. Microbiome 7, 1, doi: 10.1186/s40168-018-0604-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Klaring K et al. Intestinimonas butyriciproducens gen. nov., sp. nov., a butyrate-producing bacterium from the mouse intestine. Int J Syst Evol Microbiol 63, 4606–4612, doi: 10.1099/ijs.0.051441-0 (2013). [DOI] [PubMed] [Google Scholar]
- 193.Winter J, Popoff MR, Grimont P & Bokkenheuser VD Clostridium orbiscindens sp. nov., a human intestinal bacterium capable of cleaving the flavonoid C-ring. Int J Syst Bacteriol 41, 355–357, doi: 10.1099/00207713-41-3-355 (1991). [DOI] [PubMed] [Google Scholar]
- 194.Schoefer L, Mohan R, Schwiertz A, Braune A & Blaut M Anaerobic degradation of flavonoids by Clostridium orbiscindens. Appl Environ Microbiol 69, 5849–5854, doi: 10.1128/aem.69.10.5849-5854.2003 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Tunon MJ, Garcia-Mediavilla MV, Sanchez-Campos S & Gonzalez-Gallego J Potential of flavonoids as anti-inflammatory agents: modulation of pro-inflammatory gene expression and signal transduction pathways. Curr Drug Metab 10, 256–271, doi: 10.2174/138920009787846369 (2009). [DOI] [PubMed] [Google Scholar]
- 196.Loke WM et al. Metabolic transformation has a profound effect on anti-inflammatory activity of flavonoids such as quercetin: lack of association between antioxidant and lipoxygenase inhibitory activity. Biochem Pharmacol 75, 1045–1053, doi: 10.1016/j.bcp.2007.11.002 (2008). [DOI] [PubMed] [Google Scholar]
- 197.Peng X et al. In vitro catabolism of quercetin by human fecal bacteria and the antioxidant capacity of its catabolites. Food Nutr Res 58, doi: 10.3402/fnr.v58.23406 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Jaganath IB, Mullen W, Lean ME, Edwards CA & Crozier A In vitro catabolism of rutin by human fecal bacteria and the antioxidant capacity of its catabolites. Free Radic Biol Med 47, 1180–1189, doi: 10.1016/j.freeradbiomed.2009.07.031 (2009). [DOI] [PubMed] [Google Scholar]
- 199.Barcenilla A et al. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol 66, 1654–1661, doi: 10.1128/aem.66.4.1654-1661.2000 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Biagi E et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One 5, e10667, doi: 10.1371/journal.pone.0010667 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Takeshita K et al. A Single Species of Clostridium Subcluster XIVa Decreased in Ulcerative Colitis Patients. Inflamm Bowel Dis 22, 2802–2810, doi: 10.1097/MIB.0000000000000972 (2016). [DOI] [PubMed] [Google Scholar]
- 202.Zhang Y et al. Changes in gut microbiota and plasma inflammatory factors across the stages of colorectal tumorigenesis: a case-control study. BMC Microbiol 18, 92, doi: 10.1186/s12866-018-1232-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Rowan F et al. Desulfovibrio bacterial species are increased in ulcerative colitis. Dis Colon Rectum 53, 1530–1536, doi: 10.1007/DCR.0b013e3181f1e620 (2010). [DOI] [PubMed] [Google Scholar]
- 204.Figliuolo VR et al. Sulfate-reducing bacteria stimulate gut immune responses and contribute to inflammation in experimental colitis. Life Sci 189, 29–38, doi: 10.1016/j.lfs.2017.09.014 (2017). [DOI] [PubMed] [Google Scholar]
- 205.Millien V, Rosen D, Hou J & Shah R Proinflammatory Sulfur-Reducing Bacteria Are More Abundant in Colonic Biopsies of Patients with Microscopic Colitis Compared to Healthy Controls. Dig Dis Sci 64, 432–438, doi: 10.1007/s10620-018-5313-z (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.