The evolution and practical breeding of crops depend on genetic variations. Conventionally, these variations result from natural mutations or physical and chemical mutagenesis, both of which occur randomly and lack direction. To accelerate crop improvement, targeted mutagenesis methods are highly desired. Clustered regularly interspaced palindromic repeat (CRISPR)‐Cas9 systems have been engineered for genome‐targeted mutagenesis in eukaryotic organisms, including plants (Chen et al., 2019). The CRISPR‐Cas9 system produces DNA double‐strand breaks (DSBs) by site‐specific cleavage, which typically results in small indels. To efficiently induce point mutations, CRISPR‐mediated base editors (BEs) have been further developed to generate targeted nucleotide substitution within editing windows. Combined with an sgRNA library, the BE platform has the potential to generate a large number of point mutations to screen pivotal amino acids (AAs) and to drive directed evolution of proteins in vivo (see review in Packer and Liu, 2015).
Acetyl‐CoA carboxylase (ACCase) catalyses the first step of fatty acid biosynthesis. Loss‐of‐function mutations in ACCase are lethal or lead to serious developmental arrest in plants (Baud et al., 2004). In field production, ACCase is the target of a major group of commercial herbicides. All known ACCase‐inhibiting herbicide‐resistant mutations occur in the carboxyltransferase (CT) domain of ACCase, which directly interacts with the herbicide (Jang et al., 2013). In rice, the OsACC gene (LOC_Os05g22940) has 35 exons and encodes a 2327‐AA protein, in which amino acids that determine herbicide resistance are largely unclear. To exploit dominant mutations endowing resistance in OsACC, 141 sgRNAs were designed in the 1653‐bp coding sequence of the CT domain located in the 34th exon for the base‐editing screen (Figure 1a). To conduct C∙G to T∙A substitutions, the eBE3 and eCDA systems, which previously had been optimized by stacking multiple copies of uracil DNA glycosylase inhibitor (UGI) (Qin et al., 2019), were used to construct the domain‐specific libraries separately. To achieve efficient A∙T‐to‐G∙C substitution, the previously reported plant adenine BE (ABE) was optimized by fusing triple copies of the nuclear localization sequence to develop an enhanced ABE (eABE) for constructing a library (Li et al., 2019; Li et al., 2018). For each sgRNA, an oligo pair of forward and reverse 20‐bp guide sequences flanked with a 4‐bp ligation adapter was synthesized and annealed following the instructions of the pHUN411 vector (Xing et al., 2014). Then, the annealed oligos were pooled and ligated into the BE vectors to replace the spectinomycin resistance gene between two BsaI sites. Approximately 1500 kanamycin‐positive and spectinomycin‐negative clones were pooled in each library. To evaluate the library quality, 96 clones of each vector were randomly selected and Sanger sequenced. As indicated in Figure 1b, more than 93.8% of clones carried correct sgRNA, indicating the high accuracy of the plasmid library. At least 64 unique sgRNAs were identified from 96 clones, and most sgRNAs were detected no more than twice. These results indicate the good evenness of sgRNA distribution of the libraries. Then, the plasmids were transformed into the Agrobacterium EHA105 strain, and ~3000 clones were pooled per library. A random sequencing assay indicated that the sgRNAs in Agrobacterium had similar accuracy and uniformity to those in E. coli (Figure 1b).
Figure 1.
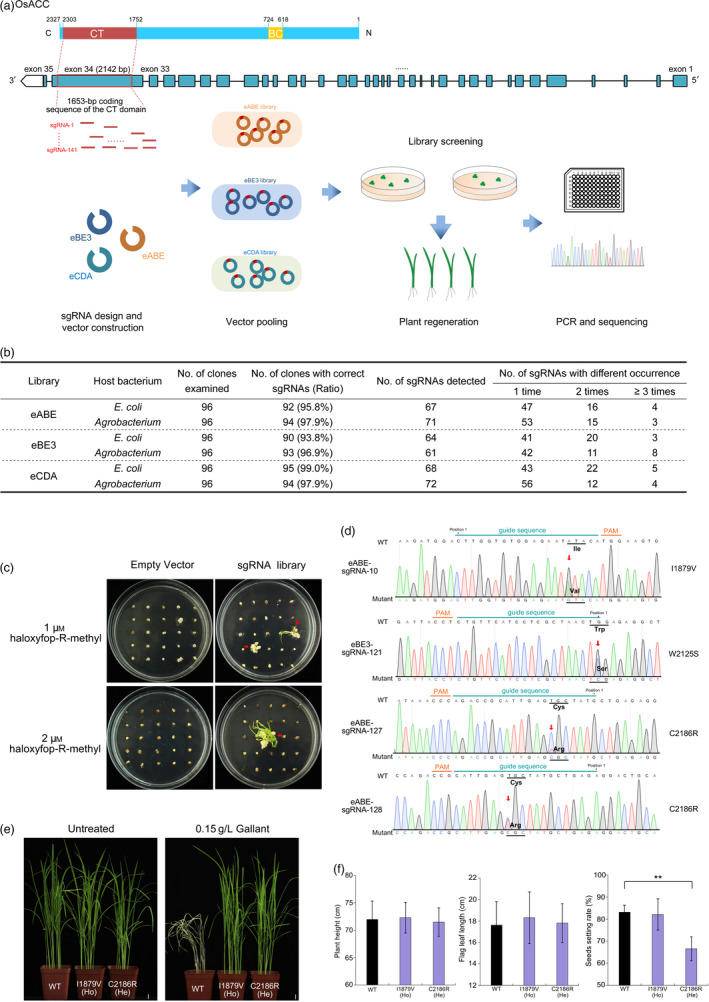
Screening amino acids of ACCase that confer herbicide resistance to rice. (a) Schematic illustration of the base‐editing screening of OsACC in rice. The protein and gene structure of OsACC is indicated in the upper panel. CT, carboxyltransferase domain; BC, biotin carboxylase domain. In the coding sequence of the CT domain, the 20‐bp sequences followed by 5'‐NGG PAM were selected as guide RNA sequences. An equimolar mixture of annealed oligos of the guide sequence was ligated into the pHUC411‐derived plant BE vectors to pool base‐editing libraries. The shoots and calli with significant herbicide resistance were collected for genotyping. (b) Clone sequencing of the sgRNA libraries in E.coli and Agrobacterium. (c) Representative selections of resistant events by screening the base‐editing library of OsACC. The transformants of empty vectors were used as controls. (d) Genotypes of the mutants screened from the base‐editing libraries. The dot indicates position 1 of the sgRNA. Red arrows indicate the base conversions. (e) Phenotypes of OsACC mutants exposed to Gallant at the recommended field concentration after 7 days. The T2 T‐DNA‐free mutants and WT plants were treated at the 3–5 leaf stage. He, heterozygous; Ho, homozygous. Scale = 1 cm. (f) Agronomic traits of OsACC mutants at the maturity stage. Data are shown as the mean ± SD (n = 12). One‐way ANOVA; **P < 0.01.
The three base‐editing libraries were stably transformed into an elite rice variety (Oryza sativa L. japonica. cv. Feigeng2020). To screen herbicide‐resistant events, an aryloxyphenoxypropionate (APP) group ACCase‐inhibited chemical, haloxyfop‐R‐methyl, was added to the regeneration medium. Approximately 5000 hygromycin‐positive calli of each library were screened under the herbicide pressure of two concentrations (1 and 2 μM), both of which completely inhibited the growth of control transformants (Figure 1c). After 4–8 weeks of selection, 15 independent shoots were recovered from the calli transformed with the eABE library. In addition, 10 independent events of eABE calli that exhibited herbicide resistance but failed in regenerating plants were also selected. Genomic DNA was isolated from resistant plants and calli to identify sgRNA by PCR sequencing. We found that 23 out of 25 resistance events, including 14 out of 15 plants and nine out of 10 calli, were transformed with sgRNA‐127. In the sgRNA‐127‐targeted genomic region, all these events carried monoallelic or biallelic T7‐to‐C conversion, leading to a C2186R mutation in the resistant cells (Figure 1d). sgRNA‐128 was observed in one resistant callus, which carried monoallelic T13‐to‐C conversion in its spacer region, leading to the same C2186R mutation. sgRNA‐10 was detected in one regenerated plant, which resulted in a monoallelic A16‐to‐G conversion and led to the I1879V mutation in OsACC. The remaining region of the CT domain in the resistant events was further screened by Sanger sequencing, and no additional mutation was found. Resistant plants and calli were also screened from the eBE3 library. Two plants and one callus, all of which harboured sgRNA‐121, were selected by haloxyfop‐R‐methyl. A monoallelic G1‐to‐C substitution was detected in all three resistance events, resulting in a W2125S mutation. No resistant event was detected from the eCDA library.
A previous study showed that the ABE‐induced C2186R substitution in OsACC conferred herbicide resistance to rice in medium (Li et al., 2018). Here, two additional single mutations, I1879V and W2125S, were produced by CRISPR‐mediated base screening to confer APP herbicide resistance in rice. Notably, the I1879V and C2186R mutations in rice correspond to the I1781V and C2088R resistance‐endowing mutations in weeds. However, the W2125S mutation is different from the conserved Cys substitution on the corresponding 2027 Trp (W2125C) in weeds, providing a new resistance allele. After transfer to greenhouse, serious growth retardation and complete sterility were observed in W2125S and homozygous C2186R plants. Furthermore, no homozygous mutant was detected in the progeny of heterozygous C2186R plants, suggesting that the homozygous C2186R mutation might be lethal to rice embryos. The growth and reproduction retardation suggest the fitness cost of these mutations. In the T2 generation, T‐DNA‐free homozygous I1879V plants and heterozygous C2186R plants were isolated and sprayed with a commercial APP herbicide Gallant to confirm resistance and evaluate the potential usage of mutations in fields. Wild‐type (WT) plants died after 7 days, while all mutants demonstrated significant resistance (Figure 1e). Compared with WT, no obvious differences were detected in T1 homozygous I1879V plants in traits of height, flag leaf length and setting rate (Figure 1f). Several reports also indicated that the I1781V mutation of ACCase had negligible fitness costs or even fitness advantages in various weeds (Jang et al., 2013). Together with our observations, the I1879V mutation may be an ideal OsACC allele to acquire herbicide resistance when breeding commercial rice varieties.
Efficient nucleotide conversion can be achieved in the editing window of different BEs, such as positions 4‐8, 3‐9 and 2‐4 of eABE, eBE3 and eCDA, respectively (Li et al., 2018; Shimatani et al., 2017; Zong et al., 2017). Accordingly, in the 1653‐bp targeted sequence, the 611‐nt (including 167 editable As and 162 editable Ts), 788‐nt (including 165 editable Cs and 192 editable Gs) and 393‐nt (including 84 editable Cs and 92 editable Gs) regions are covered by the 141 sgRNAs of the eABE, eBE3 and eCDA libraries, respectively. In grass weeds, substitutions at 7 different amino acids of the CT domain were implicated in naturally occurring resistance (Jang et al., 2013). Among them, four mutations can be generated by A∙T//G∙C conversion in rice, whereas only the nucleotide corresponding to C2186R is located inside the window of the library. Although non‐typical substitutions, such as the G‐to‐C conversion for W2125S and the mutation at position 16 of sgRNA for I1879V, were revealed, the efficiency of these unconventional substitutions should be limited using the current tools. We believe that optimized BEs with improved editing efficiency, broadened PAM recognition, expanded editing windows and more flexible conversion types would greatly facilitate in planta CRISPR‐Cas9‐mediated base‐editing screens in the near future.
Conflicts of interest
The authors declare no competing interests.
Author contributions
P.W. designed the experiments and wrote the manuscript with inputs from all authors. P.W. and D.W. supervised the project. X.L., R.Q., J.L., S.L., T.S. and R.X. performed the experiments. X.L. and R.Q. analysed the data.
Acknowledgements
This work was supported by grants from the National Transgenic Science and Technology Programs (No. 2016ZX08010‐002‐008 and No. 2019ZX08010‐003), the National Natural Science Foundation of China (No. U19A2022) and National Key Research and Development Program (No. 2017YFD0301304). We appreciate Jiangu Biotechnology (Hefei) LLC. for providing rice seeds of Feigeng2020.
Contributor Information
Dexiang Wu, Email: dexiangwu198@163.com.
Pengcheng Wei, Email: Weipengcheng@gmail.com.
References
- Baud, S. , Bellec, Y. , Miquel, M. , Bellini, C. , Caboche, M. , Lepiniec, L. , Faure, J.‐D. et al. (2004) gurke and pasticcino3 mutants affected in embryo development are impaired in acetyl‐CoA carboxylase. EMBO Rep. 5, 515–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, K. , Wang, Y. , Zhang, R. , Zhang, H. and Gao, C. (2019) CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697. [DOI] [PubMed] [Google Scholar]
- Jang, S. , Marjanovic, J. and Gornicki, P. (2013) Resistance to herbicides caused by single amino acid mutations in acetyl‐CoA carboxylase in resistant populations of grassy weeds. New Phytol. 197, 1110–1116. [DOI] [PubMed] [Google Scholar]
- Li, C. , Zong, Y. , Wang, Y. , Jin, S. , Zhang, D. , Song, Q. , Zhang, R. et al. (2018) Expanded base editing in rice and wheat using a Cas9‐adenosine deaminase fusion. Genome Biol. 19, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Qin, R. , Liu, X. , Liao, S. , Xu, R. , Yang, J. and Wei, P. (2019) CRISPR/Cas9‐mediated adenine base editing in rice genome. Rice Sci. 26, 125–128. [Google Scholar]
- Packer, M.S. and Liu, D.R. (2015) Methods for the directed evolution of proteins. Nat. Rev. Genet. 16, 379–394. [DOI] [PubMed] [Google Scholar]
- Qin, R. , Liao, S. , Li, J. , Li, H. , Liu, X. , Yang, J. and Wei, P. (2019) Increasing fidelity and efficiency by modifying cytidine base‐editing systems in rice. Crop J.. (In press). 10.1016/j.cj.2019.04.007 [DOI] [Google Scholar]
- Shimatani, Z. , Kashojiya, S. , Takayama, M. , Terada, R. , Arazoe, T. , Ishii, H. , Teramura, H. et al. (2017) Targeted base editing in rice and tomato using a CRISPR‐Cas9 cytidine deaminase fusion. Nat. Biotechnol. 35, 441–443. [DOI] [PubMed] [Google Scholar]
- Xing, H.‐L. , Dong, L. , Wang, Z.‐P. , Zhang, H.‐Y. , Han, C.‐Y. , Liu, B. , Wang, X.‐C. et al. (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong, Y. , Wang, Y. , Li, C. , Zhang, R. , Chen, K. , Ran, Y. , Qiu, J.‐L. et al. (2017) Precise base editing in rice, wheat and maize with a Cas9‐cytidine deaminase fusion. Nat. Biotechnol. 35, 438–440. [DOI] [PubMed] [Google Scholar]


