Summary
The cotton bollworm, Helicoverpa armigera, is a major insect pest for a wide range of agricultural crops. It causes significant yield loss through feeding damage and by increasing the crop’s vulnerability to bacterial and fungal infections. Although expression of Bacillus thuringiensis (Bt) toxins in transgenic crops has been very successful in protecting against insect pests, including H. armigera, field‐evolved resistance has occurred in multiple species. To manage resistant populations, new protection strategies must be continuously developed. Trans‐kingdom RNA interference (TK‐RNAi) is a promising method for controlling herbivorous pests. TK‐RNAi is based on delivering dsRNA or hairpin RNA containing essential insect gene sequences to the feeding insect. The ingested molecules are processed by the insect’s RNAi machinery and guide it to silence the target genes. Recently, TK‐RNAi delivery has been enhanced by expressing the ds‐ or hpRNAs in the chloroplast. This compartmentalizes the duplexed RNA away from the plant’s RNAi machinery, ensuring that it is delivered in an unprocessed form to the insect. Here, we report another alternative approach for delivering precursor anti‐insect RNA in plants. Insect pre‐microRNA (pre‐miR) transcripts were modified to contain artificial microRNAs (amiRs), targeting insect genes, and expressed in transgenic Nicotiana benthamiana plants. These modified pre‐miRs remained largely unprocessed in the plants, and H. armigera feeding on leaves from these plants had increased mortality, developmental abnormalities and delayed growth rates. This shows that plant‐expressed insect pre‐amiRs (plin‐amiRs) are a new strategy of protecting plants against herbivorous insects.
Keywords: trans‐kingdom RNAi, microRNA, insect control, plant protection
Introduction
Insects in the orders of Lepidoptera, Coleoptera and Hemiptera, cause feeding damage to a wide range of economically important crops such as maize, soya bean, cotton, sorghum, chickpea and pigeon pea. Global crop losses due to insect damage have been estimated to be between 13% and 18% and to cost approximately US$470 billion per year in lost productivity (Culliney, 2014). Conventional insect control relies heavily on chemical insecticides and has resulted in insects developing resistance to many commercial products (Siegwart et al., 2015). Additionally, control of resistant insects prolongs the reliance on chemical insecticides for crop protection, which not only increases selection pressure for resistance but also causes concerns over the risk of adverse effects on human health, non‐target organisms and the environment. Alternative approaches such as transgenic plants expressing Bacillus thuringiensis (Bt) toxins are being increasingly adopted as effective technologies to control insect pests, but, here too, rapid selection for field‐evolved resistant pests has occurred (Tabashnik and Carriere, 2017). The use of multiple management strategies with different modes of action aids with delaying resistance evolution; thus, there is a need to improve existing insect control strategies and develop new ones.
RNA interference (RNAi) is a multifaceted pathway in plants and insects, which produces microRNAs (miRNAs or miRs) from endogenous primary transcripts and small interfering RNAs (siRNAs) from exogenous dsRNA or hpRNAs (Carthew and Sontheimer, 2009) (Figure 1a). One of the ways it has been harnessed for insect control is by a process termed trans‐kingdom RNAi (TK‐RNAi). This entails the delivery of dsRNA into insects via transgenic plants. Over the last decade, there have been many reports on the use of this approach to protect plants (Bally et al., 2018; Gordon and Waterhouse, 2007; Huvenne and Smagghe, 2010; Mamta and Rajam, 2017) and the first commercial transgenic dsRNA product was approved by the United States Environmental Protection Agency in 2017 (US‐EPA, 2017). For TK‐RNAi to be effective in insects, the dsRNA should be at least 60 bp in length (Bolognesi et al., 2012; Li et al., 2015) and delivered in high quantity (Bally et al., 2018; Mamta and Rajam, 2017). This has been accomplished by expressing dsRNA guided by strong nuclear promoters (Baum et al., 2007; Yan et al., 2015) or from the chloroplast genome (Bally et al., 2016; Jin et al., 2015; Zhang et al., 2015). Recent studies have shown that dsRNA expressed in the chloroplast evades degradation by plant RNAi, accumulates to high levels and can confer protection against the cotton bollworm, Helicoverpa armigera, and the Colorado potato beetle, Leptinotarsa decemlineata (Bally et al., 2016; Jin et al., 2015; Zhang et al., 2015), in tobacco and potato, respectively. Although this appears to be a very effective way of protecting plants against insects, chloroplast transformation is difficult and time‐consuming for most crop species. Therefore, there is still a need for alternative TK‐RNAi delivery methods.
Figure 1.
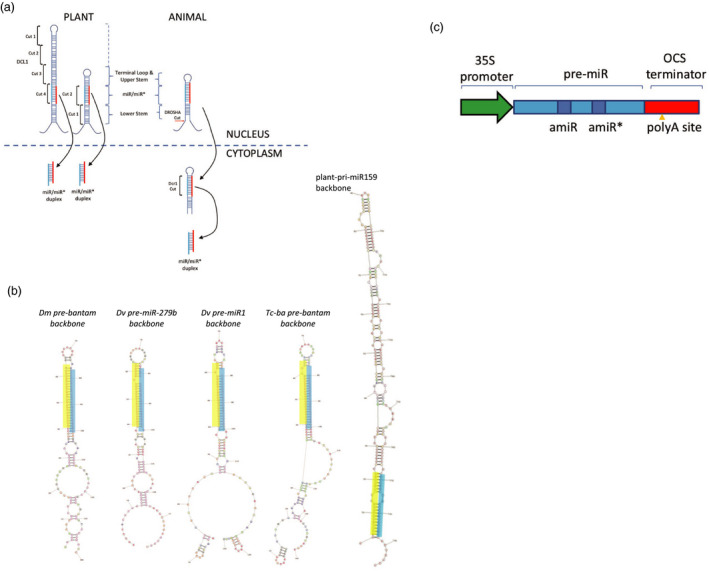
plin‐amiR design and constructs. (a) biogenesis of miRs in plants and animals. Plant pre‐miRs processed in nucleus either by top‐down or bottom‐up cleavage by Dicer‐like 1 (DCL1). Animal pri‐miR cleaved by Drosha in nucleus to form pre‐miR which is exported to cytoplasm where it is processed by Dicer 1 (DCR1). Note longer upper and lower stems in plant precursors than in animal structures. (b) Structures of plin‐amiR sequences targeting the acetylcholinesterase gene of H. armigera. Plant artificial miRNA (At‐miR159) based on miR‐159 and insect backbones: (Dm‐ba) Drosophila melanogaster bantam scaffold, (Dv‐miR279) Diabrotica virgifera virgifera mir‐279b scaffold, (Dv‐mir1) D. v. virgifera miR‐1 scaffold, (Tc‐ba) Tribolium castaneum bantam scaffold. Each insect miRNA backbone contains 40‐nucleotide flanking regions from miRNA primary transcripts listed above. The sequence that is antisense to H. armigera ACE2 is highlighted in cyan. RNAstructure version 5.5 (Mathews lab, http://rna.urmc.rochester.edu/RNAstructure.html) was used to generate the secondary structure. (c). Schematic of plin‐amiR construct.
In this study, we explored the effect of expressing modified insect pre‐miRs in plants. In insects, primary (pri) miRNA transcripts are processed in the nucleus by Drosha into ~ 100nt pre‐miR hairpin RNAs. These are transported into the cytoplasm where they are cleaved by Dicer‐1 into miRNAs. Plants do not possess a Drosha enzyme, and their pri‐miRNA transcripts are processed directly into miRs by Dicer‐like 1 in the nucleus (Figure 1a) (Bartel, 2004; Millar and Waterhouse, 2005). A comprehensive assessment of a spectrum of TK‐RNAi experimental results obtained for western corn rootworm (Bolognesi et al., 2012), using artificial diets or transgenic plants, concluded that the most important feature for successful TK‐RNAi is delivering high levels of long (>60nts) dsRNA to the insect. We wondered whether insect pre‐miRNAs would be unrecognized by the plant RNAi machinery and consequently act as effective templates for processing into miRNAs within the feeding insect. Therefore, a range of insect pre‐miR sequences were modified so that their miR and miR star (miRNA resulting from the opposite arm of the hairpin, miR*) regions were replaced with sequences targeting an important developmental gene, acetylcholinesterase (ACE2), in H. armigera. The cassettes were placed between strong plant promoter and terminator sequences and transformed into the nuclear genome of Nicotiana benthamiana. The constructs were named plant–insect chimeric artificial miRNAs (plin‐amiRs). Many of them produced RNA transcripts that were resistant to processing by the plant’s RNAi pathway and conferred resistance to H. armigera larvae by retarding growth, increasing mortality rates and causing phenotypic defects during pupation.
Results
Plant–insect artificial miRNA design
Backbones for production of artificial microRNAs (amiRs) were chosen from well‐conserved/highly abundant miRNAs in insects (Lai et al., 2003), with the hypothesis that highly abundant miRNAs are efficiently processed by Drosha and Dicer‐1. Since no insect miRNA promoter sequences or mature miRNAs were used in the artificial miRNAs, the tissue distribution or the physiological functions of miRNAs were not considered for selection. Backbones chosen were pre‐miRNAs of Drosophila melanogaster (Dm) bantam and Tribolium castaneum (Tc) bantam miRNA, a highly conserved insect miRNA that normally functions in inhibition of apoptosis (Brennecke et al., 2003). Additional potential miRNA candidates were identified in the western corn rootworm, Diabrotica virgifera virgifera (Dv), by small RNA sequencing, in combination with the transcriptome and in‐house partial genome, using an in‐house bioinformatics pipeline (Yang et al., 2019). Insect‐conserved miRNAs were identified by comparison of sequences in miRBase (www.mirbase.org). Dv‐miR‐279b and Dv‐miR1 were selected as scaffolds for testing in Dv and H. armigera. In total, five pre‐miR hairpin backbones were chosen to provide a range of predicted structures (Figure 1b and Figure S1). The backbones were named to reflect their origins, that is Dm‐ba, Tc‐ba, Dv‐miR1, Dv‐miR279 and At‐miR159 (At, Arabidopsis thaliana). The 21‐nt miR sequences in the pre‐miRs were replaced with an artificial miR (amiR) sequence targeting the H. armigera acetylcholinesterase 2 gene (ACE2); the miR* sequence was altered appropriately (Figure S1). ACE is an essential enzyme of the nervous system, having an indispensable role in terminating nerve impulse transmission at synaptic junctions of cholinergic neurons. Studies have shown that ACE2 inhibition in H. armigera results in growth inhibition of larvae, reduction in the pupal weight, malformation and mortality of the insect (Bally et al., 2016; Kumar et al., 2009). The amiR sequence (5′‐ATGGAGTTGGATGACAGGAGA‐3′) targeting ACE2 was selected using the silencing tools at www.benthgenome.com. The anti‐ACE2 amiR was not only incorporated into the four insect pre‐miRs (Figure 1a, Dm‐ba, Tc‐ba, Dv‐miR1 and Dv‐miR279) but also incorporated into a plant pre‐miR backbone (At‐miR159). These pre‐amiR sequences were cloned into pBlueGreen (Figure 1c and Figure S2), so that when transformed into plants, the expression of the RNAs was under the control of the CaMV 35S promoter and OCS terminator (Eamens et al., 2009). The pre‐amiR, including the amiR and RNA sequence contributed from the OCS terminator, is about 290 nt in length (140 nt + 150 nt) plus a poly‐A tail.
Generation and molecular characterization of plin‐amiR nuclear transformants
Multiple independent primary transgenic (T0) Nicotiana benthamiana lines were generated by Agrobacterium‐mediated transformation for each of the four plin‐amiR constructs and for the pre‐amiR159 construct. The inheritance of the T‐DNAs was followed through the T1 and T2 generations, by RT‐qPCR analysis, resulting in the selection of five independent homozygous lines for each construct. All of the transgenic plants appeared phenotypically indistinguishable from wild‐type plants, and analysis of their genomic DNA confirmed that, with the exception of some Dm‐ba lines, each line has arisen from an independent transformation event. Three of the five independent lines per construct, which were homozygous for the transgene, were used for subsequent feeding experiments.
Plin‐amiR processing in the transformed lines
To examine the processing of the plin‐amiRs in the plant, RNA was extracted from each of the transgenic lines and analysed by northern blotting (Figure 2). This revealed that the amiR was not excised from the Dm or Tc bantam pre‐miR backbones nor from the Dv pre‐miR1 backbone. As expected, the amiR was efficiently released from the At pre‐miR159 backbone. The amiR in the Dv pre‐miR279 appeared to be inefficiently excised, with the blot revealing both a larger band of the size expected for the unprocessed pre‐amiR and a ~ 21nt amiR band (Figure 2). Interestingly, the RNAs detected from the Dm and Tc pre‐bantam constructs were shorter than expected, presumably reflecting partial processing by the plant’s miRNA machinery. Overall, the analysis indicates that all four insect pre‐amiRs accumulate in N. benthamiana cells and that three of them were not processed into amiRs in the plant. The degree of processing by the plant’s machinery appears to be: most resistant Dv‐miR1> Dv‐miR279> Tc‐ba/Dm‐ba>>At‐miR159.
Figure 2.

Northern blot analysis of plant amiR and insect pre‐amiR transcripts in stably transformed N. benthamiana. Black arrow indicates 21‐nt amiRs. Red arrow indicates intact insect pre‐amiR transcripts. The probe used corresponds to the complementary sequence of mature miR‐ACE (top panel). The miR‐159 probe was used for normalization of expression levels in the different samples (bottom panel).
Bioassay and insect protection of plin‐amiR transformants
To assess the effect of the plin‐amiRs, first‐instar H. armigera larvae were fed continuously for up to 21 days (until H. armigera pupation) on seedlings from independent homozygous T2 N. benthamiana transgenic lines expressing plin‐amiRs Dv‐miR1, Dv‐miR279, Tc‐ba, Dm‐ba, At‐miR159 and wild‐type plants. The growth, mortality and development of the larvae were measured and recorded over this period. After four days, H. armigera larvae feeding on the transgenic lines had lower rates of weight gain than larvae feeding on wild‐type plants (Figure 3a). This difference became more pronounced over the next three time points (nine, twelve and fifteen days; Figure 3a), and the degree of loss of weight gain generally aligned with the degree of plin‐amiR processing by the plant (see Figure 2 and Figure 3a). The ranking for effect on weight gain was as follows: greatest inhibition Dv‐miR279> Dv‐miR1> Tc‐ba> Dm‐ba> At‐miR159>>wild‐type. Curiously, even the amiR delivered in the Arabidopsis pre‐miR backbone had some effect on inhibiting larval weight gain, but to a lesser extent than the amiR delivered in insect pre‐miR backbones (Figure 3a).
Figure 3.
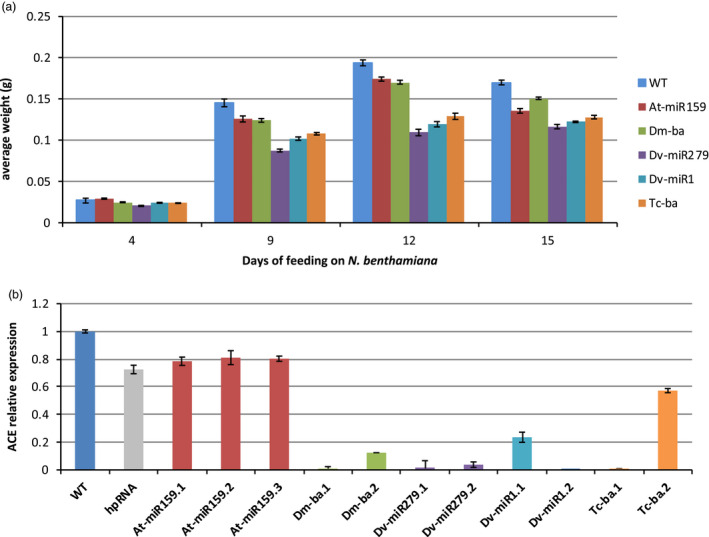
Helicoverpa armigera feeding assay on N. benthamiana expressing ACE plin‐amiRs. (a) Representation of the average larval weight (in grams), y‐axis, stable T2 N. benthamiana events used in the bioassay at 4, 9, 12, 15 and 17 days of feeding, x‐axis. (b) RT‐qPCR ACE2 transcripts level in H. armigera fed on plin‐amiR transformed lines. Error bars indicate standard error of the mean from three biological replicates.
The effect of the plin‐amiRs on acetylcholinesterase expression in the insect was evaluated using reverse transcription–polymerase chain reaction (RT‐qPCR). In larvae that fed on At‐miR159 seedlings (Figure 3b; red), the levels of ACE2 mRNA were lower, but not significantly (p> 0.5), than those of larvae fed on wild‐type seedlings. In contrast, the ACE2 mRNA levels in larvae fed on plin‐amiR transgenic seedlings were significantly reduced (P < 0.001, from 42% level reduction for Tc‐ba to 95% for Dv‐miR279, Figure 3b). Again, the degree of ACE2 inhibition generally correlated with the plin‐amiR’s resistance to processing in the plant (see Figure 2). The ranking of the degree of inhibition of ACE2 in the H. armigera larvae was as follows: Dv‐miR279> Dm‐ba> Dv‐miR1> Tc‐ba>>At‐miR159> wild‐type.
Details of the growth and behaviour of the larvae were recorded throughout the bioassay period, including data on mortality, pupal formation and moth emergence. There were significant differences in larval mortality and pupal formation between H. armigera fed on wild‐type and those fed on transgenic lines. Larvae fed on wild‐type plants had low mortality rates (22%) (Figure 4a,b, Table S1), whereas larvae fed on At‐miR159 (30%), Tc‐ba (30%), Dm‐ba (50%), Dv‐miR279 (50%) and Dv‐miR1 (60%) had higher mortality rates (Figures 4a,b, Table S1). The higher mortality was observed for 4th‐instar larvae independently of the treatment. After 21 days of feeding on wild‐type N. benthamiana, all the remaining larvae had developed normally through the six instar stages and undergone pupation. In contrast, most of the larvae that had fed on the plin‐amiR plants could not exuviate the larval cuticle, failed to pupate and died; only a few of the insects (≤than 20%) developed into normal pupae (Figure 4b, Table S1). It took 1–5 days longer for these neonates to develop into pupae than those that had fed on wild‐type plants, and those fed on Tc‐ba were the most delayed. Unexpectedly, moths emerging from pupae of larvae fed on plin‐amiR plants exhibited severe developmental defects, especially those whose larvae had fed on Tc‐ba; more than half of the moths showed developmental deformity such as smaller size body, wing deformations, antenna malformations, or only partially shed their pupal cuticle and died without completing adult emergence (Figure 4c and Table S2). No deformed phenotypes were observed in moths pupated from larvae fed on wild‐type plants.
Figure 4.

Helicoverpa armigera feeding assay on N. benthamiana expressing ACE plin‐amiRs. (a) Phenotype of dead H. armigera after 21 days of feeding on N. benthamiana expressing ACE2 plin‐amiRs showing the difference in size of the larvae. (b) Representation (%) of developmental and mortality rates of H. armigera larvae until pupation stage after feeding on N. benthamiana expressing ACE2 plin‐amiRs. (c) Developmental adult and pupal defects observed after larval feeding on N. benthamiana expressing ACE2 plin‐amiR.
Discussion
Many earlier studies have demonstrated that RNAi technologies have the potential to generate new ways of controlling agricultural insect pest populations (Bally et al., 2018; Gordon and Waterhouse, 2007; Huvenne and Smagghe, 2010; Mamta and Rajam, 2017). The first report of Lepidopteran control by TK‐RNAi was in tobacco and by targeting the cytochrome P450 monooxygenase gene, CYP6AE14, of H. armigera (Mao et al., 2007). Caterpillars feeding on transgenic cotton plants were unable to detoxify gossypol, and these larvae exhibited retarded growth. Many other examples of TK‐RNAi affecting the viability of feeding insects have been reported, but there have also been reports of variable efficacy (Huvenne and Smagghe, 2010). A comprehensive assessment of a spectrum of TK‐RNAi experimental results obtained on western corn rootworm (Bolognesi et al., 2012), using artificial diets or transgenic plants, concluded that the most important feature for successful TK‐RNAi is delivering high levels of long (>60 nts) dsRNA to insects. dsRNA and hpRNA expressed from nuclear transgenes in plants are rapidly processed by the RNAi machinery into 21‐24 nt siRNAs (Fusaro et al., 2006), and to produce an effective steady‐state pool of long dsRNA, for uptake by feeding insects, requires either transgene transcription rates that exceed those of RNAi processing or expressing the dsRNA in an RNAi‐free compartment such as the chloroplast (Bally et al., 2018; Bally et al., 2016; Jin et al., 2015; Zhang et al., 2015). Although this latter approach is very promising, it has one significant limitation: chloroplast transformation is routine in very few species, mainly members of the Solanaceae family such as N. benthamiana, N. tabacum, petunia, eggplant, potato or tomato (Ahmad et al., 2016).
Transgenic expression of amiRNAs is an effective way to silence endogenous plant or animal genes (McHale et al., 2013; Schwab et al., 2006; Ylla et al., 2016), and there are reports that an amiRNA can be more efficient than a hpRNA in silencing genes through artificial diets, such as in aphids (Guo et al., 2014). We reasoned that the upper and lower stem structures of an insect’s pre‐miRNA are shorter than those in plant pre‐miRs (Figure 1a) and may not be recognized by the plant’s Dicer complexes, which processes plant pre‐miRs either from the terminal loop (e.g. miR159) or from the stem base (e.g. miR 390) (Bologna et al., 2009; Cuperus et al., 2011), and that this unprocessed pre‐amiR may be a good template for delivering TK‐RNAi. Examining the plin‐amiR transcripts accumulating in transgenic plants revealed that a detectable level of 21 nt amiR was produced from only one of the four insect scaffolds that we tested, and at a much lower level than from a native plant pre‐miR scaffold transcript. The other three insect pre‐miR scaffolds appeared to be either unprocessed or inaccurately processed by the plant RNAi machinery. The estimated sizes of the transcripts, detected using short amiR‐specific probes, suggest that these partially processed pre‐amiRs retain> 80% of their original size and contain the amiR sequence.
Helicoverpa armigera larvae that had fed on plants expressing any one of the four plin‐amiRs, but not on plants expressing the plant‐based pre‐amiR, had reduced levels of acetylcholinesterase expression compared to larvae fed on wild‐type plants (Figure 3b). The decrease in larval acetylcholinesterase expression correlated with increased mortality rate and developmental defects, and decreased growth rate, of the larvae. Larvae fed on plants expressing the plant amiR scaffold had unaltered acetylcholinesterase expression levels but showed a modest increase in mortality rate and a pronounced increase in developmental defects and failure to successfully pupate (Figure 4 and Table S1). The amiRs generated in these transgenic plants were possibly in a less potent form, for example associated with a plant Argonaute, when taken up by the insect or were more readily degraded in transit than pre‐miRs. This strong effect on pupal development, but not on mortality of the ingesting larvae, has parallels with the observation that siRNAs can have a more pronounced effect on the production of viable young than on the viability of ingesting adult insects (Abdellatef et al., 2015; Bucher et al., 2002; Coleman et al., 2015).
Conclusion
Expressing amiRNAs embedded within insect pre‐miRs confers resistance to processing by the plant’s silencing machinery and produces strong insecticidal and phenotypic effects in herbivorous H. armigera larvae. To our knowledge, this is the first report of such an approach. To date, conventional TK‐RNAi has been less effective against Lepidopterans than against other insect orders, such as Coleopterans (Baum and Roberts, 2014; Terenius et al., 2011). This suggests that plin‐amiRs may be highly potent against agronomically important coleopteran pests, such as the Colorado potato beetle, and other insect Orders. Plin‐amiRs rely on recognizing a 21‐nt target sequence in an important insect gene and can be designed to be very specific to a target species. This facilitates the avoidance of unwanted effects against non‐target, beneficial organisms. Many strategies for further improvements to plin‐miRNA efficacy and scope can be envisioned such as alternative gene targets, concatemerization of pre‐amiRNAs and use of natural or synthetic multi‐miR‐bearing pre‐miRs. An immediate application would be to combine plin‐amiRs with Bt toxin constructs to prolong and enhance this globally adopted crop protection strategy.
Materials and methods
Plant and insect culture
Nicotiana benthamiana plants were grown at 23 °C under a 16:8 photoperiod in an environmentally controlled glasshouse. H. armigera larvae were initially procured from ABA Biologicals (Glenvale, QLD, Australia) and raised on an artificial diet (Ward’s Science, supplied by Cider House Tech, Australia) and under growth condition of 26 ± 1°C, 60% RH and 16:8 photoperiod. The eggs were placed on artificial diet and allowed to hatch in a growth chamber set at 26 °C, 60% RH and 16:8 photoperiod.
Artificial microRNA designs an N. benthamiana stable transformation
A number of potential microRNA sequences were obtained using the silencing tools at the benth genome website (www.benthgenome.com) with the acetylcholinesterase gene of H. armigera (Ace2; NCBI accession AF369793) as an input sequence. ACE2 codes for an enzyme essential for H. armigera central nervous system and is the target of many organophosphorus and carbamate insecticides (Kumar et al., 2009). Moreover, this gene was successfully used in a previous study for trans‐kingdom RNAi using hpRNA constructs expressed in N. benthamiana chloroplasts (Bally et al., 2016). The 21‐nucleotide target sequence (5′‐ ATGGAGTTGGATGACAGGAGA‐3′) of Ace2 transcript from H. armigera (Bally et al., 2016) was selected.
The mature miRNA regions within D. melanogaster (miRBase:MI0000116) and T. castaneum (miRBase:MI0008892) bantam sequences were annotated based on www.mirbase.org. The identification of mature mRNA within recently identified D. v. virgifera miRNA transcript/genomic sequence (Yang et al., 2019) was performed using sequence homology and structure to orthologous D. melanogaster and T. castaneum miRNAs in miRBase. Additional 40 nucleotides of primary transcript sequence were used as a backbone. The chimeric sequences were designed by replacing the mature insect miRNA sequences with ACE2 target sequence (Figure 1). Thus, the backbone comprised the native miRNA loop region and 40‐nt extensions from pri‐miRNA.
For the plant backbone miRNA, the target sequence was directly incorporated into the pBlueGreen artificial microRNA construct (Eamens et al., 2009), using primers pAMIR‐ACE‐F and pAMIR‐ACE‐R. For the plant–insect backbone miRNA, best potential insect miRNAs were identified and the target sequence was incorporated into the insect miRNA backbone and directly amplified as g‐blocks (Thermo Fisher Scientific Inc., Waltham, MA). The chimeric insect miRNA sequences were ordered directly as g‐blocks fragments and incorporated into the pBlueGreen vector. The resulting constructs were electroporated into Agrobacterium tumefaciens (GV3101) and transformed into N. benthamiana as described previously (Bally et al., 2016). Briefly, Agrobacterium GV3101 carrying the constructs was incubated overnight at 28 °C with shaking at 220 rpm to an OD600 of 0.4–0.6. Sterile N. benthamiana leaves were cut into pieces, co‐cultured with the Agrobacterium suspension for 30 min in 20 mL of liquid MS medium (Sigma‐Aldrich, St Louis, MO), and then placed on a piece of sterile filter paper and cultured on non‐selective MS media for three days. After three days, the infected explants were transferred to a fresh shoot induction medium (SIM) supplemented with the appropriate glufosinate‐ammonium (GFA) selection antibiotic. Healthy shoots were excised and transferred into larger pots containing the selective rooting MS medium. Putative transformants were screened for the presence of transgene by RT‐qPCR using two different sets of primer combinations (Table S3).
DNA and RNA purification
Larvae and plant tissues were ground to a fine powder under liquid nitrogen. Total DNA was extracted from young leaves of soil‐grown plants using the CTAB method (Clarke, 2009). Total RNA was extracted from 0.2 mg, both for plant and for larva samples, using TRIzol® reagent (Thermo Fisher Scientific Inc., Waltham, MA), and purified using the RNeasy Plant Mini Kit (Qiagen, Valencia, CA) following the protocols supplied by the manufacturers. Total DNA and total RNA quality was assessed with a NanoDrop 2000 UV‐VIS Spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA).
Northern blotting and Southern blotting
A 17% PAGE gel was loaded with approximately 20 μg of total RNA per lane and run at 200 V for 4 h. After staining with ethidium bromide to confirm the relative loadings and appropriate size separation, the RNA was electro‐transferred at 45 V for 1 h onto Hybond‐N+ (Amersham, Buckinghamshire, UK) membrane and cross‐linked to it by UV treatment. The membrane was incubated at 42 °C with a 32P‐labelled probe transcribed from the hp arm sequence and exposed to FujiFilm X‐100 for 16 h.
Insect feeding bioassays
Assays were performed with newly hatched larvae (<10 h old). Each larva was transferred using a moistened soft paintbrush to the appropriate cup containing N. benthamiana seedlings (1‐ to 2‐week‐old plants). For each assay, a minimum of 10 larvae were used and the experiment was monitored by assessing leaf damage and larva weight after 4 days, 7 days, 14 days and 21 days. Three replicates of each of the generated transgenic lines, along with a line expressing an hpRNA in the chloroplast (Bally et al., 2016) used as a positive control, were tested for insecticidal activity. Observations such as larval weight gain and mortality were recorded. Transgenic N. benthamiana lines showing consistently higher level of resistance in short duration bioassay were taken for long‐duration bioassay and followed until pupation and adult emergence. Fresh leaves of the respective plin‐amiR transgenic and hpRNA control N. benthamiana lines were replenished throughout the period of bioassay at regular intervals. After sixth instar, larvae pupate. The pupae were collected and kept separately until adult emergence. Observations such as growth delay, mortality and developmental defects were recorded during the bioassay.
Real‐time quantitative PCR (RT‐qPCR)
Larvae from the same generation were collected after seven days of feeding on WT and transformed plants, and pooled by groups of ten. Acetylcholinesterase transcript levels of each group were analysed by quantitative real‐time reverse transcription–polymerase chain reaction (RT‐qPCR) with a PTC‐200 Thermal Cycler (MJ Research/Bio‐Rad). In brief, synthesis of cDNA for RT‐qPCR was performed from RNA using SuperScript III Reverse Transcriptase (Life Technologies, Thermo Fisher Scientific, Inc., Waltham, MA) following the manufacturer’s instructions. RT‐PCR was performed using Brilliant III SYBR MM according to the Agilent Technologies protocol. At least two lines for each generated transformant, along with a line expressing an hpRNA in the chloroplast (Bally et al., 2016) used as a positive control, were tested. Reactions were set up in 96‐well Microseal PCR plates (Bio‐Rad) in triplicate using GAPDH (endogenous control; reference gene) and ACE2 primers (Table S3) simultaneously for normalization. Relative quantification of the transcripts was calculated using the comparative Ct method (2‐ΔΔCt).
Statistical analysis
Statistical analyses giving the means and standard errors of means (SEM) of growth, mortality, per cent hatch, phenotype and relative transcripts expression were performed by one‐way ANOVA using SAS/STAT software (SAS Institute Inc., Cary NC, 2004). For individual comparisons within a group, mean values were separated using Student’s t test at P < 0.05 significance levels as applied in previous studies (Bally et al., 2016).
Conflict of interest
The authors declare no conflict of interest.
Author contributions
JB, EF, KN and PW conceived and designed the study. JB, RL, KL and SB performed the experiments. JB, EF, MG, KN and PW contributed to the data analysis. JB, EF, KN and PW wrote the manuscript.
Supporting information
Figure S1 Pre‐miRNA backbone structures.
Figure S2 pBlueGreen vector backbone used for construction of Plin‐amiR.
Table S1 Developmental and mortality rates of H. armigera until pupation stage after feeding on N. benthamiana expressing plin‐miRNA.
Table S2 Developmental and mortality rates of H. armigera from pupation stage after feeding on N. benthamiana expressing plin‐miRNA.
Table S3 RT‐qPCR primer sequences used in this study.
Contributor Information
Julia Bally, Email: Julia.bally@qut.edu.au.
Peter M. Waterhouse, Email: peter.waterhouse@qut.edu.au.
References
- Abdellatef, E. , Will, T. , Koch, A. , Imani, J. , Vilcinskas, A. and Kogel, K.H. (2015) Silencing the expression of the salivary sheath protein causes transgenerational feeding suppression in the aphid Sitobion avenae . Plant Biotechnol. J. 13, 849–857. [DOI] [PubMed] [Google Scholar]
- Ahmad, N. , Michoux, F. , Lossl, A.G. and Nixon, P.J. (2016) Challenges and perspectives in commercializing plastid transformation technology. J. Exp. Bot. 67, 5945–5960. [DOI] [PubMed] [Google Scholar]
- Bally, J. , McIntyre, G.J. , Doran, R.L. , Lee, K. , Perez, A. , Jung, H. , Naim, F. et al. (2016) In‐plant protection against Helicoverpa armigera by production of long hpRNA in chloroplasts. Front. Plant Sci. 7, 1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bally, J. , Fishilevich, E. , Bowling, A.J. , Pence, H.E. , Narva, K.E. and Waterhouse, P.M. (2018) Improved insect‐proofing: expressing double‐stranded RNA in chloroplasts. Pest Manag. Sci. 74, 1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116, 281–297. [DOI] [PubMed] [Google Scholar]
- Baum, J.A. and Roberts, J.K. (2014) Progress towards RNAi‐mediated insect pest management. In: Advances in Insect Physiology ( Tarlochan, S. and Dhadialla, S.S.G. eds), pp. 249‐295. Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- Baum, J.A. , Bogaert, T. , Clinton, W. , Heck, G.R. , Feldmann, P. , Ilagan, O. , Johnson, S. et al. (2007) Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25, 1322–1326. [DOI] [PubMed] [Google Scholar]
- Bologna, N.G. , Mateos, J.L. , Bresso, E.G. and Palatnik, J.F. (2009) A loop‐to‐base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J. 28, 3646–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognesi, R. , Ramaseshadri, P. , Anderson, J. , Bachman, P. , Clinton, W. , Flannagan, R. , Ilagan, O. et al. (2012) Characterizing the mechanism of action of double‐stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte). PLoS ONE, 7, e47534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke, J. , Hipfner, D.R. , Stark, A. , Russell, R.B. and Cohen, S.M. (2003) bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila . Cell 113, 25–36. [DOI] [PubMed] [Google Scholar]
- Bucher, G. , Scholten, J. and Klingler, M. (2002) Parental RNAi in Tribolium (Coleoptera). Curr. Biol. 12, R85–R86. [DOI] [PubMed] [Google Scholar]
- Carthew, R.W. and Sontheimer, E.J. (2009) Origins and mechanisms of miRNAs and siRNAs. Cell, 136, 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, J.D. (2009) Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for Plant DNA isolation. Cold Spring Harbor Protocols, 2009, pdb.prot5177–pdb.prot5177. [DOI] [PubMed] [Google Scholar]
- Coleman, A.D. , Wouters, R.H. , Mugford, S.T. and Hogenhout, S.A. (2015) Persistence and transgenerational effect of plant‐mediated RNAi in aphids. J. Exp. Bot. 66, 541–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culliney, T.W. (2014) Crop losses to arthropods. In Integrated Pest Management: Pesticide Problems( Pimentel, D. and Peshin, R. , eds), pp. 201–225. Dordrecht, The Netherlands: Springer. [Google Scholar]
- Cuperus, J.T. , Fahlgren, N. and Carrington, J.C. (2011) Evolution and functional diversification of MIRNA genes. Plant Cell, 23, 431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eamens, A.L. , Smith, N.A. , Curtin, S.J. , Wang, M.B. and Waterhouse, P.M. (2009) The Arabidopsis thaliana double‐stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA, 15, 2219–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaro, A.F. , Matthew, L. , Smith, N.A. , Curtin, S.J. , Dedic‐Hagan, J. , Ellacott, G.A. , Watson, J.M. et al. (2006) RNA interference‐inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 7, 1168–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, K.H. and Waterhouse, P.M. (2007) RNAi for insect‐proof plants. Nat. Biotechnol. 25, 1231–1232. [DOI] [PubMed] [Google Scholar]
- Guo, H. , Song, X. , Wang, G. , Yang, K. , Wang, Y. , Niu, L. , Chen, X. et al. (2014) Plant‐generated artificial small RNAs mediated aphid resistance. PLoS ONE 9, e97410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huvenne, H. and Smagghe, G. (2010) Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J. Insect Physiol. 56, 227–235. [DOI] [PubMed] [Google Scholar]
- Jin, S. , Singh, N.D. , Li, L. , Zhang, X. and Daniell, H. (2015) Engineered chloroplast dsRNA silences cytochrome p450 monooxygenase, V‐ATPase and chitin synthase genes in the insect gut and disrupts Helicoverpa armigera larval development and pupation. Plant Biotechnol. J. 13, 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, M. , Gupta, G.P. and Rajam, M.V. (2009) Silencing of acetylcholinesterase gene of Helicoverpa armigera by siRNA affects larval growth and its life cycle. J. Insect Physiol. 55, 273–278. [DOI] [PubMed] [Google Scholar]
- Lai, E.C. , Tomancak, P. , Williams, R.W. and Rubin, G.M. (2003) Computational identification of Drosophila microRNA genes. Genome Biol. 4, R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Khajuria, C. , Rangasamy, M. , Gandra, P. , Fitter, M. , Geng, C. , Woosely, A. et al. (2015) Long dsRNA but not siRNA initiates RNAi in western corn rootworm larvae and adults. J. Appl. Entomol. 139, 432–445. [Google Scholar]
- Mamta, B. and Rajam, M.V. (2017) RNAi technology: a new platform for crop pest control. Physiol. Mol. Biol. Plants 23, 487–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, Y.B. , Cai, W.J. , Wang, J.W. , Hong, G.J. , Tao, X.Y. , Wang, L.J. , Huang, Y.P. et al. (2007) Silencing a cotton bollworm P450 monooxygenase gene by plant‐mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 25, 1307–1313. [DOI] [PubMed] [Google Scholar]
- McHale, M. , Eamens, A.L. , Finnegan, E.J. and Waterhouse, P.M. (2013) A 22‐nt artificial microRNA mediates widespread RNA silencing in Arabidopsis . Plant J. 76, 519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.A. and Waterhouse, P.M. (2005) Plant and animal microRNAs: similarities and differences. Funct. Integr. Genomics 5, 129–135. [DOI] [PubMed] [Google Scholar]
- Schwab, R. , Ossowski, S. , Riester, M. , Warthmann, N. and Weigel, D. (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis . Plant Cell, 18, 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegwart, M. , Graillot, B. , Blachere Lopez, C. , Besse, S. , Bardin, M. , Nicot, P.C. and Lopez‐Ferber, M. (2015) Resistance to bio‐insecticides or how to enhance their sustainability: a review. Front. Plant Sci. 6, 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabashnik, B.E. and Carriere, Y. (2017) Surge in insect resistance to transgenic crops and prospects for sustainability. Nat. Biotechnol. 35, 926–935. [DOI] [PubMed] [Google Scholar]
- Terenius, O. , Papanicolaou, A. , Garbutt, J.S. , Eleftherianos, I. , Huvenne, H. , Kanginakudru, S. , Albrechtsen, M. et al. (2011) RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J. Insect. Physiol. 57, 231–245. [DOI] [PubMed] [Google Scholar]
- US‐EPA (2017) Notice of pesticide registration. In: EPA Reg. No. 524–618, MON 87411. OPP Decision No. 533309 (Agency, U.S.E.P. eds). Washington, D.C.: Environmental Protection Agency EPA. [Google Scholar]
- Yan, Y. , Xue, L. , Miller, J.B. , Zhou, K. , Kos, P. , Elkassih, S. , Liu, L. et al. (2015) One‐pot synthesis of functional poly(amino ester sulfide)s and utility in delivering pDNA and siRNA. Polymer (Guildf) 72, 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Fishilevich, E. , German, M.A. , Gandra, P. , McEwan, R.E. , Billion, A. , Knorr, E. et al. (2019) Elucidation of the microRNA transcriptome in western corn rootworm reveals its dynamic and evolutionary complexity. Genom. Proteom. Bioinformat. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylla, G. , Fromm, B. , Piulachs, M.D. and Belles, X. (2016) The microRNA toolkit of insects. Sci. Rep. 6, 37736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Khan, S.A. , Hasse, C. , Ruf, S. , Heckel, D.G. and Bock, R. (2015) Full crop protection from an insect pest by expression of long double‐stranded RNAs in plastids. Science 347, 991–994. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Pre‐miRNA backbone structures.
Figure S2 pBlueGreen vector backbone used for construction of Plin‐amiR.
Table S1 Developmental and mortality rates of H. armigera until pupation stage after feeding on N. benthamiana expressing plin‐miRNA.
Table S2 Developmental and mortality rates of H. armigera from pupation stage after feeding on N. benthamiana expressing plin‐miRNA.
Table S3 RT‐qPCR primer sequences used in this study.


