Summary
Successful cell division involves highly regulated transcriptional and post-transcriptional control. The RNA poly(A) tail represents an important layer of RNA post-transcriptional regulation. Previous TAIL-seq analysis of S phase and M phase poly(A) tail information showed that only a small number of genes showed more than 2-fold change in their poly(A) tail length between the two cell cycle stages. In addition, the changes in poly(A) tail length between these two stages showed minimal impact on the translation of the genes as long as the poly(A) tails were longer than 20 nt. Therefore, the significance of poly(A) tail dynamics during the cell cycle remains largely unknown. Here, by re-analyzing the S phase and M phase TAIL-seq data, we uncovered an interesting global dynamics of RNA poly(A) tails in terms of their terminal modifications, implying global RNA regulation between mitotic cell cycles through poly(A) tail terminal modifications.
Subject Areas: Molecular Biology, Transcriptomics
Graphical Abstract

Highlights
-
•
Global RNA 3′ uridylation associated RNA degradation takes place in S phase
-
•
Global RNA 3′ guanylation accumulates in M phase
Molecular Biology; Transcriptomics
Introduction
The RNA poly(A) tail is an essential component for most mRNA required for their stability, nuclear export, and translation efficiency (Eckmann et al., 2011; Groisman et al., 2001, 2002; Nicholson and Pasquinelli, 2019; Richter, 1999; Villalba et al., 2011; Weill et al., 2012). The global dynamics and function of mRNA poly(A) tail-mediated post-transcriptional regulation is highly underestimated owing to technical difficulties in sequencing long homopolymeric sequences on standard second-generation sequencing platforms. TAIL-seq and PAL-seq (poly(A)-tail length profiling by sequencing) methods with customized base-calling algorithm or sequencing chemistry on Illumina platform were developed to estimate the lengths of poly(A) tails (Chang et al., 2014; Subtelny et al., 2014). In addition, TAIL-seq revealed that the 3′ ends of mRNA poly(A) tails can be modified by other bases (Chang et al., 2014; Lim et al., 2014, 2018). Short RNA poly(A) tails can be terminally U modified by TUT4/7, which can promote the degradation of the marked mRNA (Lim et al., 2014). In contrast, mixed tailing of RNA poly(A) tails by TENT4A/B can introduce G modifications that attenuate the degradation of the mRNA poly(A) tails, resulting in stabilized RNA with G modifications at the end of the tails (Lim et al., 2018).
Interestingly, studying the meiotic cell cycle of oocytes at multiple stages in multiple species, including Drosophila, Xenopus, and mouse, revealed drastic dynamics of global RNA poly(A) tails during the meiotic cell cycle with a clear link that longer poly(A) tails were generally associated with increased efficiency of mRNA translation (Eichhorn et al., 2016; Lim et al., 2016; Luong et al., 2020; Morgan et al., 2017; Subtelny et al., 2014; Yang et al., 2020). Some of the cell cycle-related genes are periodically expressed, whereas most of the genes show relatively stable steady-state transcript level throughout the mitotic cell cycle (Bertoli et al., 2013; Park et al., 2016). However, little is known about the dynamics and roles of RNA poly(A) tails during mitotic cell cycle. Combination of poly(A) tail analysis by TAIL-seq and translation efficiency by ribosome profiling revealed that less than 2% genes showed more than 2-fold change in their poly(A) tail length (Chang et al., 2014; Park et al., 2016). In addition, the length change of a poly(A) tail minimally affects mRNA translation efficiency in somatic cell cycle as long as the tails are longer than 20 nt (Park et al., 2016). Therefore, the significance of poly(A) tail dynamics during somatic cell cycle awaits further exploring. Here, we re-analyzed the S phase and M phase TAIL-seq data and found that the terminal modifications of RNAs were globally dynamic during somatic cell cycle.
Results
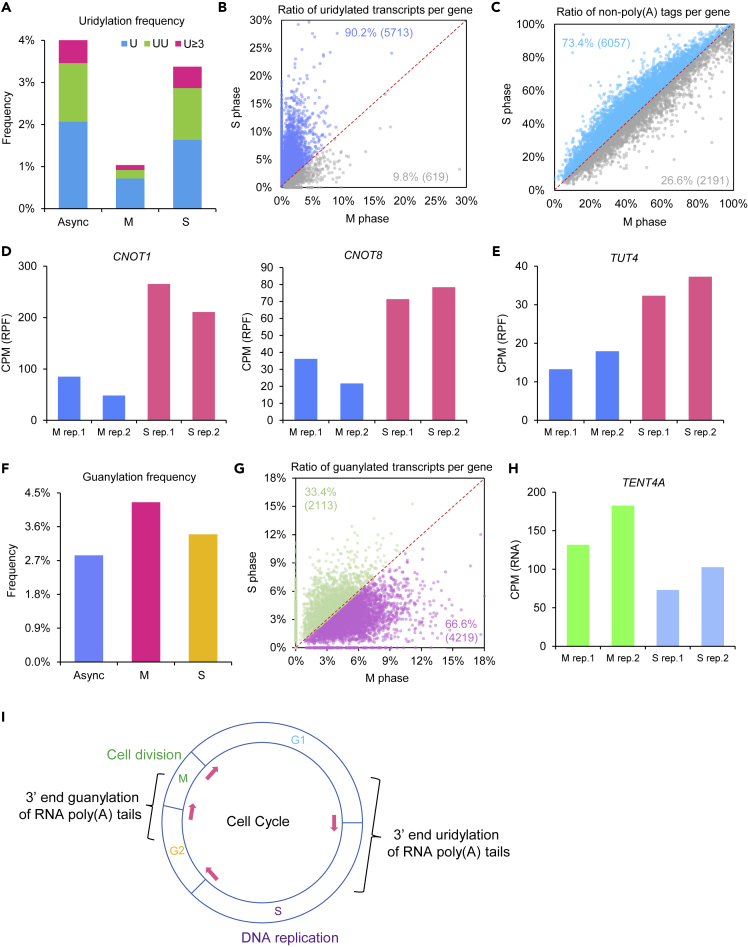
RNA poly(A) tail 3′ end uridylation can mark the RNA for rapid degradation (Lim et al., 2014; Morgan et al., 2017; Zuber et al., 2016). First, we looked into the uridylation frequency of RNA poly(A) tails during different stages of mitotic cell cycle. Very interestingly, the M phase RNA poly(A) tails showed much lower frequency of uridylation at their 3′ ends, especially the oligo-U, compared with that from the S phase cells as well as the asynchronized cells (Figure 1A). Consistently, when looking into individual genes, we can see more than 90% of genes showed lower terminal uridylation frequency in M phase compared with that in S phase (Figure 1B). We reason that the high level of uridylation in S phase may be correlated with increased global RNA degradation. TAIL-seq can capture RNA both with or without poly(A) tails (Chang et al., 2014; Park et al., 2016). As poly(A) tail deadenylation is one of the most important steps in RNA degradation, we looked into the amount of RNA with deadenylated tails to estimate the RNA degradation. Interestingly, 73% of genes showed higher percentage of deadenylated poly(A) tails in S phase compared with that in M phase (Figure 1C), confirming that higher global RNA degradation associated with uridylation in S phase. In supporting the global higher RNA degradation in S phase, we could see higher ribosome engagement of transcripts encoding core components of RNA deadenylation CCR4-NOT complexes, such as CNOT1 and CNOT8, in S phase than that in M phase (Figure 1D). Consistent with uridylation of poly(A) tails deadenylated by the CCR4-NOT complex (Yi et al., 2018), TUT4, encoding the terminal uridylation enzyme in mammals is also highly translated in S phase (Figure 1E). These findings indicate that compared with M phase there is global RNA uridylation-associated RNA degradation taking place in S phase.
Figure 1.
Poly(A) Tail Terminal Uridylation and Guanylation Cycle during the Mitotic Cell Cycle
(A) Frequency of uridylation is significantly lower at M phase compared with S phase. Frequency (y axis) is the fraction of detected reads with uridylation among all reads with poly(A) tails. Blue refers to mono-U, green refers to di-U, and red refers to three or more consecutive U. Async refers to asynchronized cells, M refers to M phase cells, and S refers to S phase cells.
(B) The fraction of transcripts with uridylation for each of the genes is significantly lower at M phase compared with S phase (p = 0, one-tailed Mann-Whitney U test). Genes with at least 30 reads with poly(A) tails in both samples are included in the analysis.
(C) The fraction of transcripts without poly(A) tails for each of the genes is significantly lower at M phase compared with S phase (p = 7.87 × 10−36, one-tailed Mann-Whitney U test). Genes with at least 30 total reads in both samples are included in the analysis.
(D) Ribosome protected fragment (RPF) counts for CNOT1 and CNOT8 are higher in S phase than those in M phase.
(E) Ribosome protected fragment (RPF) count for TUT4 is higher in S phase than that in M phase.
(F) Frequency of guanylation is significantly higher at M phase compared with S phase. Frequency (y axis) is the fraction of detected reads with guanylation among all reads with poly(A) tails.
(G) The fraction of transcripts with guanylation for each of the genes is significantly higher at M phase compared with S phase (p = 4.36 × 10−139, one-tailed Mann-Whitney U test). Genes with at least 30 reads with poly(A) tails in both samples are included in the analysis.
(H) RNA-seq quantification of transcript level for TENT4A is higher in M phase than that in S phase.
(I) A model for the transgenerational RNA cycle during mitosis.
RNA poly(A) tail 3′ end guanylation catalyzed by TENT4A/B can shield the tails from active deadenylation (Lim et al., 2018), which is opposite to the poly(A) tail uridylation. Interestingly, we could see that the guanylation in M phase cells is higher than that in S phase cells or that in asynchronized cells (Figure 1F). When looking into individual genes, two-thirds of the genes showed lower terminal guanylation frequency in M phase compared with that in S phase (Figure 1G). This increased guanylation phenomenon in M phase may be associated with the higher RNA level of TENT4A (Figure 1H). These findings indicate that compared with S phase RNA guanylation may prevent the deadenylation of the RNA in M phase.
Discussion
By reanalyzing the RNA poly(A) tail information from the S phase and M phase of the mitotic cell cycle, we revealed global RNA poly(A) tail terminal modification reconfiguration during the cell cycle (Figure 1I). The accumulation of terminal uridylation and high deadenylated transcripts at S phase suggest that there is global RNA decay happening around S phase. In turn, the accumulation of terminal guanylation and reduced deadenylated transcripts at M phase indicate that the majority of the transcriptome is protected from active deadenylation during M phase. Taken together, these data reveal that the RNA terminal guanylation and uridylation, two modifications of opposite molecular function in regulating RNA stability, are of complementary dynamics during mitotic cell cycle. Together with a recent report of m6A cell cycle dynamics (Fei et al., 2020), the findings here highlight the functional importance of RNA modifications in orchestrating the mitotic cell cycle.
Limitation of the Study
Here, we reanalyzed the previously published datasets and observed the cell cycle-associated global dynamics of RNA poly(A) tail terminal modifications. The function and regulation of this interesting global RNA terminal modification cell cycle dynamics needs further investigation in the future. Moreover, it will be interesting to see if the dynamics is differentially regulated in two daughter cells with distinct cell fates during asymmetric cell division. In addition, the TAIL-seq method used to generate the datasets analyzed here cannot measure the non-A residue modifications within the body of RNA poly(A) tails (Chang et al., 2014); new methods such as PAIso-seq and FLAM-seq may be able to reveal more information about the dynamics of poly(A) tail internal modifications during cell cycle (Legnini et al., 2019; Liu et al., 2019). Nevertheless, our findings here reveal a new layer of global post-transcriptional regulation during cell cycle, suggesting a general principle of RNA inheritance and clearance between somatic mother and daughter cells.
Resource Availability
Lead Contact
Further information and requests should be directed to and will be fulfilled by the Lead Contact, Falong Lu, (fllu@genetics.ac.cn).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
This study did not generate new datasets or code. The accession number of the published datasets analyzed in this study can be found in the Transparent Methods.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2018YFA0107001), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA24020203), and the China Postdoctoral Science Foundation (2020M670516).
Author Contributions
Y.L. performed the data analysis with input from H.N.; Y.L. and F.L. interpreted the data and wrote the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: August 21, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101402.
Supplemental Information
References
- Bertoli C., Skotheim J.M., de Bruin R.A.M. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013;14:518–528. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H., Lim J., Ha M., Kim V.N. TAIL-seq: genome-wide determination of poly(A) tail length and 3' end modifications. Mol. Cell. 2014;53:1044–1052. doi: 10.1016/j.molcel.2014.02.007. [DOI] [PubMed] [Google Scholar]
- Eckmann C.R., Rammelt C., Wahle E. Control of poly(A) tail length. Wiley Interdiscip. Rev. RNA. 2011;2:348–361. doi: 10.1002/wrna.56. [DOI] [PubMed] [Google Scholar]
- Eichhorn S.W., Subtelny A.O., Kronja I., Kwasnieski J.C., Orr-Weaver T.L., Bartel D.P. mRNA poly(A)-tail changes specified by deadenylation broadly reshape translation in Drosophila oocytes and early embryos. Elife. 2016;5:e16955. doi: 10.7554/eLife.16955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Q., Zou Z., Roundtree I.A., Sun H.L., He C. YTHDF2 promotes mitotic entry and is regulated by cell cycle mediators. Plos Biol. 2020;18:e3000664. doi: 10.1371/journal.pbio.3000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman I., Huang Y.S., Mendez R., Cao Q., Richter J.D. Translational control of embryonic cell division by CPEB and maskin. Cold Spring Harb. Symp. Quant. Biol. 2001;66:345–351. doi: 10.1101/sqb.2001.66.345. [DOI] [PubMed] [Google Scholar]
- Groisman I., Jung M.Y., Sarkissian M., Cao Q., Richter J.D. Translational control of the embryonic cell cycle. Cell. 2002;109:473–483. doi: 10.1016/s0092-8674(02)00733-x. [DOI] [PubMed] [Google Scholar]
- Legnini I., Alles J., Karaiskos N., Ayoub S., Rajewsky N. FLAM-seq: full-length mRNA sequencing reveals principles of poly(A) tail length control. Nat. Methods. 2019;16:879–886. doi: 10.1038/s41592-019-0503-y. [DOI] [PubMed] [Google Scholar]
- Lim J., Ha M., Chang H., Kwon S.C., Simanshu D.K., Patel D.J., Kim V.N. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell. 2014;159:1365–1376. doi: 10.1016/j.cell.2014.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J., Kim D., Lee Y.S., Ha M., Lee M., Yeo J., Chang H., Song J., Ahn K., Kim V.N. Mixed tailing by TENT4A and TENT4B shields mRNA from rapid deadenylation. Science. 2018;361:701–704. doi: 10.1126/science.aam5794. [DOI] [PubMed] [Google Scholar]
- Lim J., Lee M., Son A., Chang H., Kim V.N. mTAIL-seq reveals dynamic poly(A) tail regulation in oocyte-to-embryo development. Genes Dev. 2016;30:1671–1682. doi: 10.1101/gad.284802.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Nie H., Liu H., Lu F. Poly(A) inclusive RNA isoform sequencing (PAIso-seq) reveals wide-spread non-adenosine residues within RNA poly(A) tails. Nat. Commun. 2019;10:5292. doi: 10.1038/s41467-019-13228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong X.G., Daldello E.M., Rajkovic G., Yang C.R., Conti M. Genome-wide analysis reveals a switch in the translational program upon oocyte meiotic resumption. Nucleic Acids Res. 2020;48:3257–3276. doi: 10.1093/nar/gkaa010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M., Much C., DiGiacomo M., Azzi C., Ivanova I., Vitsios D.M., Pistolic J., Collier P., Moreira P.N., Benes V. mRNA 3' uridylation and poly(A) tail length sculpt the mammalian maternal transcriptome. Nature. 2017;548:347–351. doi: 10.1038/nature23318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson A.L., Pasquinelli A.E. Tales of detailed poly(A) tails. Trends Cell Biol. 2019;29:191–200. doi: 10.1016/j.tcb.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.E., Yi H., Kim Y., Chang H., Kim V.N. Regulation of poly(A) tail and translation during the somatic cell cycle. Mol. Cell. 2016;62:462–471. doi: 10.1016/j.molcel.2016.04.007. [DOI] [PubMed] [Google Scholar]
- Richter J.D. Cytoplasmic polyadenylation in development and beyond. Microbiol. Mol. Biol. Rev. 1999;63:446–456. doi: 10.1128/mmbr.63.2.446-456.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtelny A.O., Eichhorn S.W., Chen G.R., Sive H., Bartel D.P. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalba A., Coll O., Gebauer F. Cytoplasmic polyadenylation and translational control. Curr. Opin. Genet. Dev. 2011;21:452–457. doi: 10.1016/j.gde.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Weill L., Belloc E., Bava F.A., Mendez R. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat. Struct. Mol. Biol. 2012;19:577–585. doi: 10.1038/nsmb.2311. [DOI] [PubMed] [Google Scholar]
- Yang F., Wang W., Cetinbas M., Sadreyev R.I., Blower M.D. Genome-wide analysis identifies cis-acting elements regulating mRNA polyadenylation and translation during vertebrate oocyte maturation. RNA. 2020;26:324–344. doi: 10.1261/rna.073247.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H., Park J., Ha M., Lim J., Chang H., Kim V.N. PABP cooperates with the CCR4-NOT complex to promote mRNA deadenylation and block precocious decay. Mol. Cell. 2018;70:1081–1088 e1085. doi: 10.1016/j.molcel.2018.05.009. [DOI] [PubMed] [Google Scholar]
- Zuber H., Scheer H., Ferrier E., Sement F.M., Mercier P., Stupfler B., Gagliardi D. Uridylation and PABP cooperate to repair mRNA deadenylated ends in Arabidopsis. Cell Rep. 2016;14:2707–2717. doi: 10.1016/j.celrep.2016.02.060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate new datasets or code. The accession number of the published datasets analyzed in this study can be found in the Transparent Methods.