Abstract
Circular RNAs (circRNAs) are evolutionarily conserved RNA species that are formed when exons “back-splice” to each other. Current computational algorithms to detect these back-splicing junctions produce divergent results, and hence there is a need for a method to distinguish true-positive circRNAs. To this end, we developed Assembly based CircRNA Validator (ACValidator) for in silico verification of circRNAs. ACValidator extracts reads from a user-defined window on either side of a circRNA junction and assembles them to generate contigs. These contigs are aligned against the circRNA sequence to find contigs spanning the back-spliced junction. When evaluated on simulated datasets, ACValidator achieved over ∼80% sensitivity on datasets with an average of 10 circRNA-supporting reads and with read lengths of at least 100 bp. In experimental datasets, ACValidator produced higher verification percentages for samples treated with ribonuclease R compared to nontreated samples. Our workflow is applicable to non-polyA-selected RNAseq datasets and can also be used as a candidate selection strategy for prioritizing experimental validations. All workflow scripts are freely accessible on our GitHub page https://github.com/tgen/ACValidator along with detailed instructions to set up and run ACValidator.
Keywords: CircRNAs, RNAseq, RNA expression
Introduction
Circular RNAs (circRNAs) represent a large class of ubiquitously expressed noncoding RNAs that are formed when exons “back-splice” to each other. The advent of high-throughput RNA sequencing (RNAseq) technologies and bioinformatics algorithms has facilitated the identification of thousands of circRNAs in multiple cell and tissue types [1–4]. These studies have found that circRNAs are highly abundant and evolutionarily conserved, as well as exhibit cell type- and developmental stage-specific expression. CircRNAs are also more stable than linear RNAs since they are covalently closed loops without 5′/3′ termini or a polyadenylated tail. Furthermore, studies investigating their functional relevance have revealed that circRNAs can act asmicroRNA (miRNA) regulators [3, 5–7], decoys to RNA binding proteins [8], and regulators of parental gene transcription [9].
Several computational tools have been developed to identify these back-splicing events in RNAseq data. Strategies employed by these computational tools to identify circRNAs include: (i) a pseudo-reference-based strategy, which is used by the known and novel isoform explorer (KNIFE) tool [10] and (ii) a fragment-based strategy, which is used by find_circ [3], CIRCexplorer [11], Mapsplice [12], and the detect circRNAs from chimeric reads (DCC) tool [13]. While KNIFE constructs a pseudo-reference of all possible out-of-order exons to align reads against, fragment-based strategies detect circRNAs based on the mapping information of a split read’s alignment to the reference genome [14]. When segments of a split read align to the reference in a non-linear order, they are marked as potential circRNA candidates. Apart from these strategies, CircRNA Identifier(CIRI) uses concise idiosyncratic gapped alignment report (CIGAR) signatures in the alignment file to identify circRNAs [15].
Tool comparison studies have revealed that existing circRNA detection algorithms produce divergent results due to the use of different aligners, heuristics, and filtering criteria [16, 17]. Hence, there is a need for an in silico approach that can distinguish true versus false-positive circRNAs identified using these algorithms. To this end, we developed Assembly-based Circular RNA Validator (ACValidator), which can be used as an in silico verification strategy, as well as a candidate selection tool for experimental validation. While existing approaches focus on detection of circRNAs, our approach performs in silico verification of circRNAs detected using these existing approaches. ACValidator first extracts reads from a fixed window on either side of the circRNA junction of interest from the alignment file and assembles them to generate contigs. These contigs are then evaluated for alignment against the circRNA junction sequence. We defined four different stringency criteria, ranging from 10 to 60 base pairs (bps) overlap across the junction. When evaluated on simulated as well as experimental datasets, ACValidator achieves better performance in datasets with higher circRNA coverage compared to ones with lower coverage.
Materials and methods
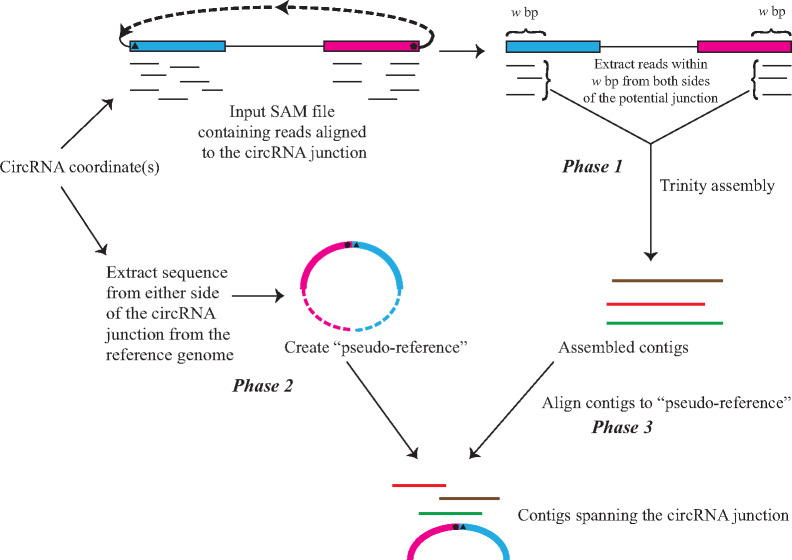
ACValidator takes as input a sequence alignment mapping (SAM) file and the circRNA coordinate(s) to be validated (Fig. 1). ACValidator operates in three phases: (i) extraction and assembly of reads from the SAM file to generate contigs; (ii) generation of a pseudo-reference file; and (iii) alignment of contigs from Phase 1 against the pseudo-reference from Phase 2. First, reads are extracted from a user-defined window size w on either side of the given SAM file [(“start-coordinate” + w) and (“end-coordinate” − w); where the start-coordinate is the splice acceptor and the end-coordinate is the splice donor of the circRNA junction]. Our datasets were run using two different window sizes, where w = insert size or w = 2* insert size, in order to understand the effect of window size on the results. We chose the above-mentioned window sizes in order to capture as many reads overlapping the circRNA junction as possible, given our use of paired-end sequencing data. However, a range of values between insert size and 2* insert size can be used, and users can adjust this parameter based on their library insert size. The tool thus extracts aligned reads within w bp on either side of the junction from the SAM file using SAMtools [18]. The extracted reads are then converted intoFASTQs and assembled using the Trinity assembler [19] to generate contigs (FASTA file). In the second phase, a pseudo-reference of the sequence surrounding the circRNA junction of interest is generated. This is performed by also extracting w bp from the end and start of the circRNA junction from the genome reference FASTA file (GRCh37; reference FASTA files of the genome of choice can be downloaded from https://genome.ucsc.edu/ or http://www.ensembl.org/) and concatenating the two sequences from end to end to capture the sequence on either side of the circRNA junction. Lastly, the assembled contigs from Phase 1 are aligned to the pseudo-reference from Phase 2 using the widely useed BWA-MEM [20] aligner. Each resulting alignment record is then examined to check whether it overlaps with the circRNA junction sequence using four different stringency criteria. The criteria require the following minimum lengths of alignment on both sides of the circRNA junction: high stringency—30 bp (total 60 bp overlap); medium stringency—20 bp (total 40 bp overlap); low stringency—10 bp (total 20 bp overlap); and very low stringency—5 bp (total 10 bp overlap). These stringency cut-offs were defined in order to capture as many tests as possible while still accounting for the extent of overlap between the contig and the circRNA junction sequence, as well as to assess whether we observe differences in sensitivity measurements across these cut-offs.
Figure 1:
ACValidator workflow. ACValidator takes as input the SAM file and the circRNA junction coordinate(s) to be validated. In Phase 1, reads from either side of the junction within a user-defined window (w) are extracted and assembled using Trinity. A pseudo-reference containing the circRNA sequence of interest is generated from the reference genome in Phase 2. The pseudo-reference consists of w bp from either side of the circRNA junction of interest (solid blue and pink blocks in Phase 2). The broken blue and pink segments represent the remaining portions of the exons that constitute the circRNA but that are not a part of the pseudo-reference. Lastly, in Phase 3, the assembled contigs are aligned to this pseudo-reference and checked for overlap with the sequence of the junction to be validated.
Datasets used for evaluation
Simulated datasets
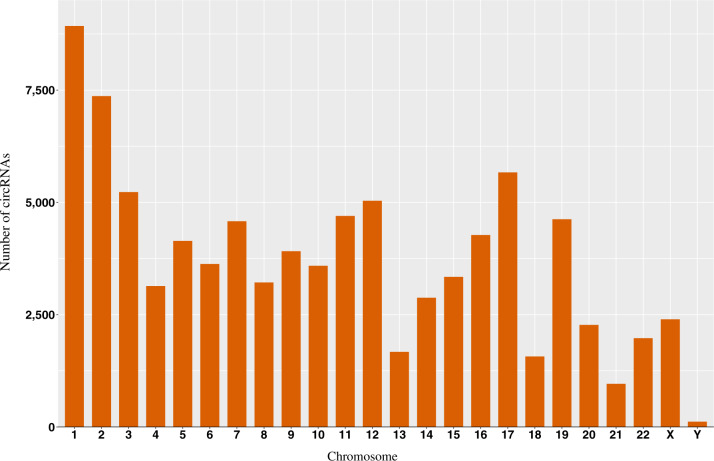
We used CIRI-simulator [15] to generate 18 synthetic RNAseq datasets that had variable average number of supporting reads for circular and linear RNAs (2–80), as well as three different read lengths (50, 100, and 150 bp) to evaluate the performance of our workflow (Table 1). CIRI-simulator takes a FASTA-formatted reference file and a GTF annotation file as input, and generates circular and linear RNA sequences. Recently, Zeng et al. [17] re-designed this tool to generate synthetic reads for circRNAs deposited in circBase [21]. We generated simulated datasets with minimum circRNA size of at least 50 bp and insert size of 300 bp. Overall, an average of 89,293 circRNAs were generated across these simulated datasets. CIRI-simulator ensures this circRNAs map to locations distributed across the entire genome, thereby eliminating any bias associated with genomic location (Fig. 2). The generated true-positive simulation datasets are named using the convention pos_<circRNA_coverage>_<linearRNA_coverage>_ <read_length> (pos: positive; Table 1). Users can reproduce all simulated datasets by using the parameters described in Table 1 in CIRI-simulator.
Table 1.
Simulation dataset parameters
| Simulation set | Simulation dataset name | Average no. of circRNA supporting reads | Range of no. of circRNA supporting reads | Average no. of linear RNA supporting reads | Read length (bp) | No. of reads generated | Window lengths tested (bp) | Overlapping base thresholds tested (bp) | No. of candidates passing top 2% expression cut-off | No. of candidates below bottom 1% expression cut-off |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | pos_2_80_150 | 2 | 2–11 | 80 | 150 | 30 926 238 | 300, 600 | 60, 40, 20, 10 | 1196 | 13 176 |
| 2 | pos_2_80_100 | 2 | 2–10 | 80 | 100 | 46 264 524 | 300, 600 | 60, 40, 20, 10 | 1616 | 12 829 |
| 3 | pos_2_80_50 | 2 | 2–13 | 80 | 50 | 92 271 226 | 300, 600 | 60, 40, 20, 10 | 620 | 12 670 |
| 4 | pos_5_40_150 | 5 | 2–17 | 40 | 150 | 22 725 820 | 300, 600 | 60, 40, 20, 10 | 712 | 5261 |
| 5 | pos_5_40_100 | 5 | 2–18 | 40 | 100 | 33 962 378 | 300, 600 | 60, 40, 20, 10 | 944 | 3887 |
| 6 | pos_5_40_50 | 5 | 2–17 | 40 | 50 | 67 667 118 | 300, 600 | 60, 40, 20, 10 | 1296 | 4122 |
| 7 | pos_10_20_150 | 10 | 3–28 | 20 | 150 | 24 867 386 | 300, 600 | 60, 40, 20, 10 | 1238 | 1391 |
| 8 | pos_10_20_100 | 10 | 2–27 | 20 | 100 | 37 173 692 | 300, 600 | 60, 40, 20, 10 | 1628 | 1802 |
| 9 | pos_10_20_50 | 10 | 2–26 | 20 | 50 | 74 092 068 | 300, 600 | 60, 40, 20, 10 | 1069 | 1641 |
| 10 | pos_20_10_150 | 20 | 5–40 | 10 | 150 | 39 314 956 | 300, 600 | 60, 40, 20, 10 | 1456 | 1304 |
| 11 | pos_20_10_100 | 20 | 4–42 | 10 | 100 | 58 844 010 | 300, 600 | 60, 40, 20, 10 | 1020 | 1039 |
| 12 | pos_20_10_50 | 20 | 4–41 | 10 | 50 | 117 432 918 | 300, 600 | 60, 40, 20, 10 | 1405 | 1047 |
| 13 | pos_40_5_150 | 40 | 8–65 | 5 | 150 | 73 285 586 | 300, 600 | 60, 40, 20, 10 | 1413 | 1303 |
| 14 | pos_40_5_100 | 40 | 11–68 | 5 | 100 | 109 808 870 | 300, 600 | 60, 40, 20, 10 | 1775 | 1246 |
| 15 | pos_40_5_50 | 40 | 10–74 | 5 | 50 | 219 344 762 | 300, 600 | 60, 40, 20, 10 | 1480 | 895 |
| 16 | pos_80_2_150 | 80 | 15–119 | 2 | 150 | 143 598 648 | 300, 600 | 60, 40, 20, 10 | 1507 | 1115 |
| 17 | pos_80_2_100 | 80 | 22–120 | 2 | 100 | 215 278 690 | 300, 600 | 60, 40, 20, 10 | 1746 | 897 |
| 18 | pos_80_2_50 | 80 | 27–115 | 2 | 50 | 430 284 334 | 300, 600 | 60, 40, 20, 10 | 1493 | 960 |
All simulation datasets are based on data generated from human cerebellum and diencephalon, SH-SY5Y cells, Hs68 cells, HeLa cells, and HEK293 cells.
Columns C–F are user-defined parameters supplied to CIRI-simulator. Minimum circRNA size used in simulation: 50 bp, insert size used in simulation: 300 bp.
Figure 2:
Chromosomal distribution of simulation datasets. Simulation datasets were generated using CIRI-simulator. The distribution of generated circRNAs across the different chromosomes is similar to the chromosomal size distribution.
Experimental datasets
All subjects were enrolled in the Banner Sun Health Research Institute (BSHRI) Brain and Body Donation Program (BBDP) in Sun City, Arizona, and written informed consent for all aspects of the program, including tissue sharing, was obtained either from the subjects themselves prior to death or from their legally appointed representative. The protocol and consent for the BBDP were approved by the Western Institutional Review Board (Puyallap, Washington).
To test ACValidator on experimental data, we analyzed six pairs of ribonuclease R (RNase R)-treated and nontreated samples (N = 12; Table 2). RNase R is an exoribonuclease that selectively digests linear RNA but leaves behind circular structures, and it is hence widely used for circRNA enrichment. Three of these sample pairs were downloaded from the sequence read archive (SRA) [22] and were generated from HeLa and Hs68 cell lines treated or not treated with RNase R. The remaining three sample pairs were generated in-house from total RNA extracted from the middle temporal gyrus (MG) of three human postmortem healthy elderly control brains.
Table 2.
Summary of experimental, non-simulated datasets used in this study
| Data source | Dataset | Cell line/tissue type | RNase R treated? | Number of reads | Number of mapped reads | Median |
|---|---|---|---|---|---|---|
| insert size | ||||||
| SRA | SRR1636985 | HeLa | Yes | 26 619 490 | 24 370 337 | 200 |
| SRR1637089 | HeLa | No | 89 866 900 | 63 842 205 | 140 | |
| SRR1636986 | HeLa | Yes | 47 011 426 | 42 027 801 | 200 | |
| SRR1637090 | HeLa | No | 71 370 620 | 53 957 685 | 130 | |
| SRR444974 | Hs68 | Yes | 316 611 710 | 271 345 091 | 160 | |
| SRR444655 | Hs68 | No | 314 106 316 | 109 706 923 | 150 | |
| In-house | MG_1 | MTG | Yes | 107 609 934 | 96 300 242 | 150 |
| generated | MG_5 | MTG | No | 96 215 516 | 86 315 844 | 150 |
| MG_2 | MTG | Yes | 96 840 790 | 86 560 619 | 150 | |
| MG_6 | MTG | No | 101 609 754 | 90 750 108 | 150 | |
| MG_3 | MTG | Yes | 111 576 344 | 100 264 691 | 150 | |
| MG_7 | MTG | No | 111 314 114 | 98 894 498 | 150 |
Polymerase chain reaction validation
Polymerase chain reaction (PCR) was performed to experimentally validate the presence of selected circRNA candidates that were in silico validated by ACValidator. cDNA was synthesized from RNA isolated from the MG of the three tissue samples of interest using SuperScript II reverse transcriptase (ThermoFisher Scientific, Waltham, MA, USA). PCR was performed using 100 ng cDNA of each sample, 12.5 µl of Kapa HiFi polymerase (2X), 1 µl of the forward primer (5 µM), 1 µl of the reverse primer (5 µM), and 9.5 µl of molecular-grade water with the following thermocycler program: denaturation—60 s at 95°C, amplification—15 s at 95°C, 15 s at 55–63°C, 30 s at 72°C (cycle 35–40X), extension—60 s at 72°C. Primers were designed using Primer version 3 (http://bioinfo.ut.ee/primer3-0.4.0) and product sizes were assessed on a TapeStation 4200 instrument (Agilent Technologies, Santa Clara, CA, USA).
Software requirements/dependencies
ACValidator can be implemented on a Linux-based high-performance computing cluster and has minimal requirements and dependencies. These requirements include the following: (i) Trinity version 2.3.1 or above; (ii) Python version 2.7.13 or higher with the pysam package installed; (iii) Bowtie2 version 2.3.0 [23] or above (required by Trinity); (iv) SAMtools version 1.4 or above and (v) BWA version 0.7.12 or above and (vi) BEDTools version 2.26 or above [24].
Results and discussion
Performance evaluation of ACValidator using simulated data
To evaluate ACValidator, we generated 18 simulation datasets with varying circular, linear RNA coverages, and read lengths (Table 1). We selected circRNA candidates above/below an expression cut-off as true positives/true negatives, respectively. For true-positive candidate set, we selected the top 2% of circRNAs based on supporting read counts (expression level), while for the true-negative candidate set, we selected the bottom 1% of circRNAs, also based on the supporting read counts (Supplementary Data, Tables S1 and S2). We replicated our analysis using two different window sizes: (i) w = insert size (300 bp) and (ii) w = 2 * insert size (600 bp) (Materials and methods section).
We observed that simulations with higher circRNA coverages and longer read lengths achieved higher sensitivity (Supplementary Data, Fig. S1). Specifically, when using w = 300 and overlap cut-off of 10 bp between the contig and pseudo-reference, simulations 7, 10, 13, and 16, which have average circRNA coverages >10 and read length of 150 bp, achieved over 81% sensitivity. For simulations 8, 11, 14, and 17, for which the average circRNA coverage was >10 but the read length was reduced to 100 bp, we observed a slight reduction in sensitivity by approximately 1–3%. However, when further reducing the read length to 50 bp, the sensitivity reduced to <=49% for simulations 9, 12, 15, and 18 (average circRNA coverage >10). When the average circRNA coverage was decreased to 5 or below (simulations 1–6), we observe an overall reduction in sensitivity while datasets with longer read lengths demonstrated improved performance compared to ones with shorter read lengths (20–26% sensitivity for simulations 3 and 6 and 57–79% for simulations 1 and 4, respectively). In datasets with lower circRNA coverage and/or shorter read length, this reduction in sensitivity was because Trinity did not find sufficient reads to assemble across these regions and hence was not able to generate contigs. We detect a similar pattern when using w = 600 and did not observe a drastic difference in sensitivity between the two window sizes (Supplementary Data, Table S1). Furthermore, we calculated the F1 score [F1 = (2*Precision* Sensitivity)/(Precision + Sensitivity)], a measure of accuracy, which indicates how well a tool achieves sensitivity and precision simultaneously (Supplementary Data, Fig. S1). We observed that for datasets with read length of 100 bp, the F1 score increases with coverage, while for datasets with read length of 150 bp, saturation is attained for datasets with average circRNA coverage >20. Overall, results from our simulation data indicate that higher circRNA coverage coupled with longer read length yields better performance of our approach.
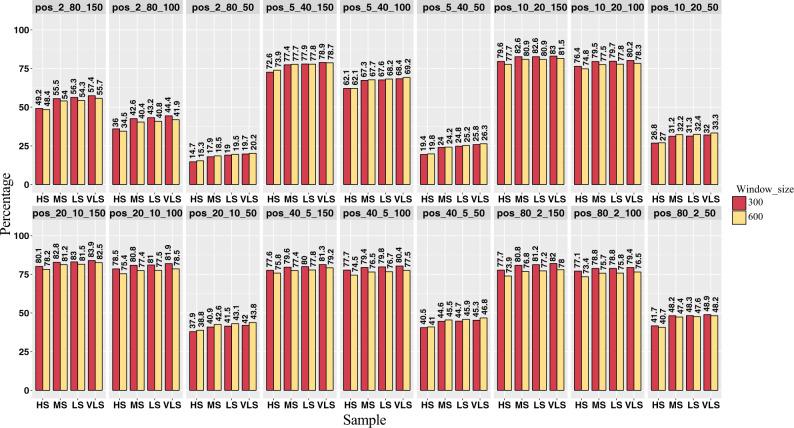
Additionally, we evaluated sensitivity using four stringency thresholds for the number of overlapping bases (30, 20, 10, and 5 bp; Materials and methods section). We observed that for datasets with average circRNA coverage >10 and read lengths of 100 or 150 bp, there was no notable difference in the sensitivity across the different thresholds for the number of overlapping bases, with an average of 79–81% sensitivity, respectively (Fig. 3). However, for simulations with circRNA coverage <10 and read length of 50 bp, the sensitivity increases by ∼4–8% (10–34%) when reducing the overlap stringency from high to very low (Fig. 3). Thus, users can choose a less stringent overlap threshold such as 10 bp when running ACValidator on low coverage/short read length datasets but for higher circRNA coverage and longer read lengths, this threshold can be increased to 40 or 60 bp.
Figure 3:
ACValidator performance across different overlap stringency thresholds in simulated datasets. Sensitivity measurements based on the top 2% of true-positive candidates were evaluated across four different overlap stringency thresholds (60, 40, 20, and 10 bp), as well as two different window sizes (300 and 600 bp). HS, high stringency; LS, low stringency; VLS, very low stringency.
Performance evaluation of ACValidator using experimental data
We next evaluated ACValidator using experimental, nonsimulated datasets generated from human tissues or cell lines (Table 2). Since we do not know the true-positive circRNAs for these datasets, we ran ACValidator on circRNAs that were called in both the RNase R-treated and nontreated datasets using six existing circRNA detection algorithms, find_circ version 1, CIRI version 2, Mapsplice version 2, KNIFE version 1.4, DCC version 0.4.4, and CIRCexplorer version 1.1.0. Each tool was run using RNAseq aligners and parameter settings as recommended by the respective developers. Co-ordinates returned by CIRI, MapSplice and DCC were converted from 1-based to 0-based coordinates to make them consistent with the other three detection tools. We considered those candidates that were called by at least three of the six tools (requiring exact co-ordinates to be detected by each tool) in both the treated and nontreated samples, and not depleted following RNase R enrichment, as true circRNAs (Supplementary Data, Table S3). A circRNA candidate is determined to be not depleted if the number of spliced reads per billion mappings [SRPBM; calculated as (number of circRNA supporting reads/total mapped reads) × 109] [2] does not decrease following enrichment. We thus ran ACValidator on these nondepleted potential true circRNA candidates for evaluation.
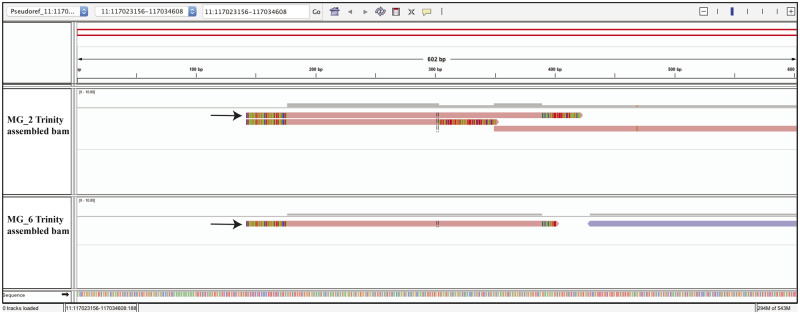
Overall, except for the SRR1636986–SRR1637090 pair, over 89% of the candidates that were called in both the treated and nontreated pairs were not depleted (i.e. SRPBM after RNase R treatment > SRPBM prior to treatment). Among these nondepleted candidates, ACValidator was able to construct contigs spanning the circRNA junction for >75% of them for the RNase R-treated samples and 47–57% of them for the nontreated samples using the medium-stringency criteria and both the window sizes (Table 3; Supplementary Data, Table S3 and Fig. S2). This increased verification rate for the treated samples is expected since RNase R treatment enriches for circRNAs and hence a higher number of back-splice junction supporting reads was observed. Furthermore, higher numbers of validated circRNAs were detected using lower stringency cut-offs for alignment overlap between contigs and junction sequences. As observed in the simulation datasets, the different window sizes did not notably affect the number of verifications among these experimental datasets (Table 3). Figure 4 shows an example of a circRNA [2, 25] that was validated by ACValidator in an MG-treated and nontreated pair (reads from this sample that align to the reference are shown in Supplementary Data, Fig. S3).
Table 3.
Summary of ACValidator results on experimental datasets
| Sample | No. overlap with pair | No. not depleted | Percent validated when w = insert size |
Percent validated when w = 2* insert size |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HS | MS | LS | VLS | HS | MS | LS | VLS | |||
| SRR1636985 | 1356 | 1243 | 73.53 | 78.52 | 79 | 79.57 | 73.45 | 78.84 | 79.57 | 80.13 |
| SRR1637089 | 1356 | 1243 | 46.34 | 49.56 | 50.2 | 51.25 | 44.97 | 49.88 | 50.52 | 51.65 |
| SRR1636986 | 780 | 594 | 79.46 | 84.68 | 85.35 | 86.03 | 80.81 | 86.36 | 87.21 | 87.71 |
| SRR1637090 | 780 | 594 | 48.48 | 54.21 | 55.39 | 55.56 | 48.32 | 55.22 | 56.23 | 56.57 |
| SRR444974 | 953 | 864 | 91.55 | 93.63 | 93.98 | 94.33 | 89.93 | 92.01 | 92.48 | 92.82 |
| SRR444655 | 953 | 864 | 51.74 | 54.98 | 55.21 | 56.6 | 53.36 | 56.94 | 57.18 | 58.91 |
| MG_1 | 1806 | 1691 | 72.8 | 77.05 | 77.94 | 78.83 | 71.67 | 77.35 | 78 | 78.71 |
| MG_5 | 1806 | 1691 | 41.4 | 48.14 | 48.97 | 49.56 | 41.34 | 48.79 | 49.67 | 50.38 |
| MG_2 | 1430 | 1331 | 71.9 | 76.86 | 77.46 | 78.14 | 69.72 | 76.11 | 76.63 | 77.31 |
| MG_6 | 1430 | 1331 | 40.5 | 47.71 | 48.31 | 49.29 | 40.35 | 47.03 | 47.63 | 48.76 |
| MG_3 | 1292 | 1148 | 71.95 | 76.74 | 77.53 | 78.48 | 70.38 | 75.87 | 76.83 | 77.7 |
| MG_7 | 1,292 | 1148 | 44.08 | 51.48 | 52.26 | 53.4 | 44.51 | 51.39 | 52.18 | 53.57 |
Figure 4:
Integrated Genomics Viewer (IGV) screen shot of a circRNA candidate (chr11:117 023 156–117 034 608) Assembled contigs generated by ACValidator on RNase R-treated (top panel) and nontreated (bottom panel) human MG samples, aligned to the corresponding pseudo-reference. This circRNA was detected in both the treated and nontreated samples by at least three of the six existing circRNA prediction algorithms, and was not depleted following RNase R treatment. The circRNA junction of interest is at 300 bp, and pink bars that span over this junction represent the contigs that validate the junction (colored segments at the ends of contigs represent soft-clipped bases; arrows indicate the generated contigs that overlap with the circRNA junction). Reads from these samples aligned to the reference are shown in Supplementary Figure 3.
In order to further evaluate the utility of our approach, we next compared the results from each individual tool to those from ACValidator. For this analysis, we used the top 100 most highly expressed candidates from among those we considered as true positives for these experimental datasets (called by at least three of six tools, in both treated and nontreated samples, and SRPBM after RNase R treatment > SRPBM before treatment; Supplementary Data, Table S3). Among the in-house treated MG samples, we observed that except for CIRCexplorer and Mapsplice, ACValidator was able to in silico validate a higher number of circRNAs, using the medium stringency (MS) criteria (20 bp overlap on either side of circRNA junction), than was detected individually by the other tools (Table 4). Among the SRA samples, however, our approach validated a fewer number of circRNAs than were detected by the individual tools except find_circ. Thus, results may vary depending on which individual tool is used for circRNA detection. Notably, the goal of ACValidator is to narrow down a list of potential high-confidence circRNAs and not for comprehensive and de novo detection of circRNAs.
Table 4.
ACValidator and other tools' results on top 100 candidates from experimental datasets.
| Sample | CIRI_count | CIRCexplorer_count | findCirc_count | Mapsplice_count | KNIFE_count | DCC_count | HS_count | MS_count | LS_count | VLS_count |
|---|---|---|---|---|---|---|---|---|---|---|
| SRR1636985 | 99 | 93 | 89 | 97 | 92 | 92 | 94 | 97 | 97 | 97 |
| SRR1637089 | 99 | 92 | 87 | 97 | 92 | 92 | 74 | 78 | 79 | 79 |
| SRR1636986 | 99 | 95 | 84 | 96 | 95 | 92 | 91 | 91 | 93 | 94 |
| SRR1637090 | 95 | 88 | 57 | 85 | 95 | 83 | 71 | 77 | 78 | 78 |
| SRR444974 | 99 | 97 | 91 | 97 | 95 | 95 | 97 | 98 | 98 | 98 |
| SRR444655 | 99 | 97 | 76 | 38 | 95 | 90 | 84 | 84 | 84 | 84 |
| MG_1 | 40 | 96 | 85 | 99 | 91 | 88 | 91 | 94 | 94 | 94 |
| MG_5 | 55 | 93 | 72 | 98 | 91 | 80 | 77 | 83 | 83 | 83 |
| MG_2 | 35 | 96 | 90 | 100 | 90 | 90 | 89 | 93 | 93 | 93 |
| MG_6 | 50 | 92 | 68 | 98 | 90 | 82 | 75 | 81 | 81 | 81 |
| MG_3 | 44 | 96 | 83 | 99 | 88 | 88 | 90 | 93 | 94 | 94 |
| MG_7 | 40 | 94 | 79 | 99 | 88 | 85 | 84 | 88 | 91 | 91 |
The top 100 most highly expressed candidates were selected from the list of circRNAs called by at least three of six tools, in both treated and nontreated samples, and having SRPBM after RNase R treatment > SRPBM before treatment.
Each <tool_name>_count column lists the number of circRNAs among the top 100 detected by the tool. Similarly, HS, MS, LS, and VLS_count columns list the number of circRNAs among the top 100 that were validated by ACValidator using those stringency thresholds.
Experimental validation of identified circRNAs
We performed experimental validations for six highly expressed circRNA candidates (average SRPBM > 650) that were in silico validated by ACValidator and that were detected by all six tools in each sample. Since we were interested in validating the presence of the circRNA and not their abundance, we performed PCR validations on these selected candidates. Each of our treated- and nontreated pair is generated from the same donor and hence, we ran validations on the non-RNase R-treated cDNA from each donor. For three of the candidates, ACValidator results were experimentally validated (Supplementary Data, Fig. S4A). For the candidate circRNA at chr10:116 879 948–116 931 050, ACValidator validated the circRNA junction in two of three samples, while chr9:113 734 352–113 735 838, chr5:38 523 520–38 530 768, and chr8:37 623 043–37 623 873 were validated in all three samples using all stringency cut-offs. For the remaining two candidates, we observed evidence of validation but because differently sized PCR products were generated, we could not determine the exact product size although it is possible that multiple circRNA species may be present. Additional PCR validations were performed on four circRNAs candidates demonstrating medium expression (average SRPBM > 300 and <600), that were validated by ACValidator, and that were called by at least three of six tools (Supplementary Data, Fig. S4B). These candidates include chr5:10 415 599–10 417 516, chr7:8 257 934–8 275 635, chr5:64 084 777–64 100 213, and chr4:56 277 780–56 284 152. All junctions were validated across the three untreated MG samples. PCR validations were also performed on two circRNAs candidates demonstrating low expression (average SRPBM < 90), that were validated by ACValidator, and that were called by three of six tools (Supplementary Data, Fig. S4C). These candidates include chr15:93 540 186–93 545 547 and chr3:3 178 943–3 186 394, and were validated across the three untreated MG samples. Sanger sequencing was also performed on the seven circRNAs candidates demonstrating medium or low expression, and although high background was observed for a large proportion of the reactions, three of the seven candidates were validated in at least one sample. These include junction 6 (for samples MG_1_5 and MG_3_7), junction 8 (for samples MG_2_6 and MG_3_7), and junction 9 (for sample MG_3_7).
Computational cost overview
We ran our evaluations on a Linux-based high-performance computing cluster running CentOS version 7. As expected, the computational cost of our approach directly correlates with the number of reads in the input sample. The only rate-limiting step in using ACValidator is read alignment to generate the SAM file, which is performed prior to starting the workflow. The python script following this step requires <2 min of runtime for an input SAM file of ∼8 GB, thus making our approach highly computationally efficient.
Future directions
We present ACValidator, a novel bioinformatics workflow, which can be used to validate circRNA candidates of interest in silico and thus helps to identify true-positive candidates. This workflow is applicable to non-polyA-selected RNAseq datasets and can be used to validate circRNAs from various sample types and diseases. When different circRNA detection algorithms identify different circRNA candidates, ACValidator can be used as a complementary/orthogonal strategy to narrow down specific candidates of interest and thus serve as a circRNA candidate prioritization tool for experimental validations or functional studies.
When evaluated on simulated datasets, ACValidator demonstrates improved performance when higher numbers of circRNA supporting reads are available along with longer read lengths of the sequencing library. Thus, coverage as well as read length are important factors contributing to the performance of ACValidator, since a higher number of reads as well as longer read lengths result in improved assembly. When tested on circRNA candidates that were not depleted between RNase R-treated and nontreated sample pairs, we observed a higher verification rate in treated samples, as expected, since those samples are enriched for circRNAs. Window size, the region from where reads are extracted for assembly, is an important parameter for our approach. Through testing of two different window sizes, one equal to and one twice the insert size, we did not observe notable differences in the number of verifications. Additionally, we applied different stringency thresholds based on the extent of contig alignment, but we did not observe notable differences in the number of validated candidates across the different thresholds for highly expressed candidates.
Although our workflow provides a novel approach for in silico verification, we are limited by a few caveats. Primarily, since our assembly analysis relies on reads that extend across circRNA junctions, we are limited in our ability to in silico validate circRNAs whose expression may be low, especially in samples that are not enriched for circRNAs. Second, since we are limited by the lack of a gold standard circRNA reference dataset, we rely on simulation datasets for evaluation of our approach, which is based on informatically predicted circRNAs detected by various studies and deposited in circBase. Further, since we do not know the true-positive events in our experimental datasets, we evaluated candidates that are not depleted by RNase R. However, it is still not known whether RNase R treatment introduces any bias in circRNA detection, especially since some circRNAs are sensitive to RNase R [2, 11, 26, 27].
Future versions of ACValidator will include contig alignment visualization options built into the workflow, as well as alternative strategies to generate contigs for circRNAs with lower expression levels. As circRNAs continue to gain attention as an interesting class of noncoding RNAs, development of novel approaches, including implementation of statistical tests to estimate false discovery rates in circRNA detection, is needed. Continued progress in improving our understanding of the biology of circRNAs will be necessary for such algorithmic development. These findings will be crucial not only for functional analysis, but also for the development of more accurate circRNA detection algorithms.
Data and tool availability
All data generated through this study are accessible through the European Genome Archive (EGA; accession EGAS00001003128). ACValidator is freely available on our GitHub page: https://github.com/tgen/ACValidator, along with detailed instructions to set up and run the tool (also available as Supplementary Text 1). ACV_launcher.sh, a wrapper/launcher script, is also included to enable verification of multiple co-ordinates.
Supplementary Material
Acknowledgements
We also thank Dr. Jonathan Keats, Dr. Sara Nasser, Ryan Richholt (TGen), and Dr. David Craig (USC) for their valuable insights and guidance, Dr. Nancy Linford (Linford Biomedical Communications) for editing assistance, and Andrea Schmitt (Banner Research) and Cynthia Lechuga (TGen) foradministrative support.
Conflict of interest statement. None declared.
Funding
Research reported in this publication was supported by the Arizona Alzheimer’s Research Center (AARC) with support from the Department of Health Services (DHS) and the State of Arizona (ADHS grant number ADHS14-052688) and the National Institute on Aging of the National Institutes of Health under Award Number P30AG019610 (Alzheimer’s Disease Core Center; ADCC). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We are also grateful to BSHRIBBDP of Sun City, Arizona for the provision of human brain tissues. The BBDP has been supported by the National Institute of Neurological Disorders and Stroke, the National Institute on Aging, the Arizona Department of Health Services, the Arizona Biomedical Research Commission and the Michael J. Fox Foundation for Parkinson’s Research [28].
References
- 1. Salzman J, Gawad C, Wang PL.. et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012;7:e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jeck WR, Sorrentino JA, Wang K.. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA (New York, NY) 2013;19:141–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Memczak S, Jens M, Elefsinioti A.. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333–8 [DOI] [PubMed] [Google Scholar]
- 4. Memczak S, Papavasileiou P, Peters O. et al. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One 2015;10:e0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Capel B, Swain A, Nicolis S.. et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993;73:1019–30 [DOI] [PubMed] [Google Scholar]
- 6. Hansen TB, Wiklund ED, Bramsen JB. et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J 2011;30:4414–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hansen TB, Jensen TI, Clausen BH. et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384–8 [DOI] [PubMed] [Google Scholar]
- 8. Du WW, Yang W, Chen Y. et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J 2016;38:1402–12 [DOI] [PubMed] [Google Scholar]
- 9. Li Z, Huang C, Bao C. et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015;22:256–64 [DOI] [PubMed] [Google Scholar]
- 10. Szabo L, Morey R, Palpant NJ. et al. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol 2015;16:126–015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang XO, Wang HB, Zhang Y.. et al. Complementary sequence-mediated exon circularization. Cell 2014;159:134–47. [DOI] [PubMed] [Google Scholar]
- 12. Wang K, Singh D, Zeng Z.. et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res 2010;38:e178–e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheng J, Metge F, Dieterich C.. Specific identification and quantification of circular RNAs from sequencing data. Bioinformatics 2016;32:1094–6 PubMed PMID: 26556385. [DOI] [PubMed] [Google Scholar]
- 14. Chen I, Chen CY, Chuang TJ.. Biogenesis, identification, and function of exonic circular RNAs. Wires RNA 2015;6:563–79 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gao Y, Wang J, Zhao F.. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol 2015;16:4–014 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hansen TB, Venø MT, Damgaard CK. et al. Comparison of circular RNA prediction tools. Nucleic Acids Res 2016;44:e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zeng X, Lin W, Guo M. et al. A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput Biol 2017;13:e1005420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li H, Handsaker B, Wysoker A. et al. ; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haas BJ, Papanicolaou A, Yassour M. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 2013;8:1494–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv 2013; 13033997
- 21. Glazar P, Papavasileiou P, Rajewsky N.. circBase: a database for circular RNAs. RNA 2014;11:1666–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leinonen R, Sugawara H, Shumway M. et al. ; On behalf of the international nucleotide sequence database Collaboration. the sequence read archive. Nucleic Acids Res 2011;39:D19–D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Langmead B, Salzberg SL.. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Quinlan AR, Hall IM.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics (Oxford, England) 2010;26:841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Salzman J, Chen RE, Olsen MN. et al. Cell-type specific features of circular RNA expression. PLoS Genet 2013;9:e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Westholm JO, Miura P, Olson S. et al. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Reports 2014;9:1966–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Szabo L, Salzman J.. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet 2016;17:679–92. PubMed PMID: 27739534; PubMed Central PMCID: PMCPMC5565156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Beach TG, Adler CH, Sue LI. et al. Arizona study of aging and neurodegenerative disorders and brain and body donation program. Neuropathology 2015;35:354–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.