Summary
Patient-derived cancer organoids hold great potential to accurately model and predict therapeutic responses. Efficient organoid isolation methods that minimize post-collection manipulation of tissues would improve adaptability, accuracy, and applicability to both experimental and real-time clinical settings. Here we present a simple and minimally invasive fine-needle aspiration (FNA)-based organoid culture technique using a variety of tumor types including gastrointestinal, thyroid, melanoma, and kidney. This method isolates organoids directly from patients at the bedside or from resected tissues, requiring minimal tissue processing while preserving the histologic growth patterns and infiltrating immune cells. Finally, we illustrate diverse downstream applications of this technique including in vitro high-throughput chemotherapeutic screens, in situ immune cell characterization, and in vivo patient-derived xenografts. Thus, routine clinical FNA-based collection techniques represent an unappreciated substantial source of material that can be exploited to generate tumor organoids from a variety of tumor types for both discovery and clinical applications.
Subject Areas: Clinical Medicine, Tissue Engineering, Cancer
Graphical Abstract
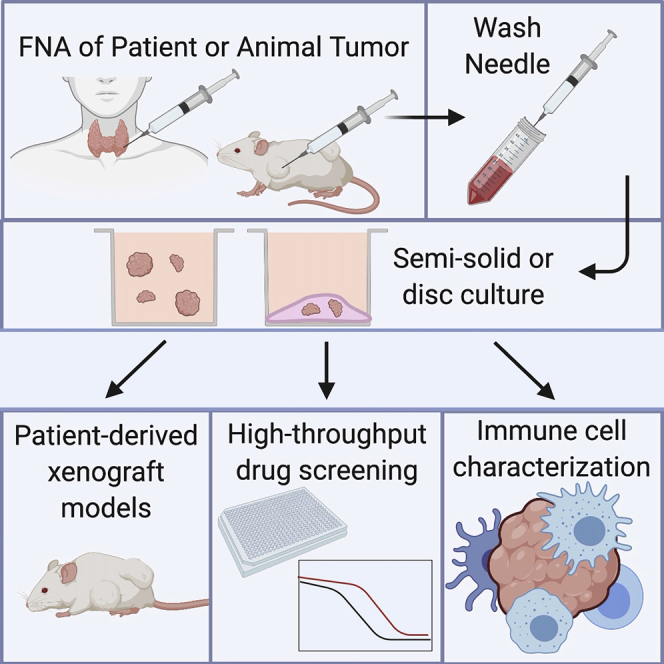
Highlights
-
•
Fine-needle aspiration (FNA) is safe, minimally invasive, and widely used clinically
-
•
FNA is a source of material for organoid culture and personalized medicine
-
•
This technique requires minimal processing, preserving histology, and immune cells
-
•
Downstream applications: high-throughput screens, immune analysis, and xenografts
Clinical Medicine; Tissue Engineering; Cancer
Introduction
Patient-derived tumor organoids hold great promise for biomedical science and clinical applications including personalized medicine (Clevers, 2016; Neal et al., 2018; Saito et al., 2019; Vlachogiannis et al., 2018). However, current methods that isolate tumor organoids from patients require tissue(s) of at least 1 cm3, which are collected from surgical resection specimens or core needle biopsies. After collection, tissues must be mechanically or enzymatically dissociated for successful isolation and plating of intact tumor organoids into Matrigel discs. Like 2D cultures, many post-isolation organoid culturing methods also rely on trypsinization of intact organoids into single cells for passaging and plating into downstream experimental formats. As such, organoid isolation is restricted to patients with surgically accessible tumors and current digestion methods disrupt native 3D growth architectures with loss of tumor infiltrating immune cells.
Fine-needle aspiration (FNA) is a minimally invasive technique that is widely used in both inpatient and outpatient settings as a first-line procedure for sampling and diagnosing tumor tissues. It is often clinically preferred over core needle biopsy because of its ability to rapidly and gently extract and separate tumor from stroma, unlike core needle biopsies, while simultaneously preserving growth architectures for gold standard pathologic diagnosis (Austin et al., 2005; Baatenburg de Jong et al., 1991; Eddyani et al., 2009; Jain et al., 2018; Lieu, 1997; Pitman et al., 2008). For palpable lesions, the sample can be obtained from patients at the bedside with minimal discomfort and limited, if any, use of local anesthetic. For deeper lesions, the technique can be combined with ultrasound (Conrad et al., 2018) or performed under computed tomography (CT) guidance to precisely target the lesion of interest. FNA can also be performed directly on resected or biopsied tumor tissues immediately after surgery. Applying this technique to isolate tumor organoids could allow for a simple, minimally invasive approach that is especially suited for preservation of intact tumor organoids and their corresponding tumor infiltrating immune cells.
FNA is distinct from larger core needle biopsies, the latter of which have been widely applied to organoid isolation (Boj et al., 2015; Choi et al., 2019; Mazzucchelli et al., 2019; Tiriac et al., 2018a, 2018b; Tsai et al., 2018). Unlike core needle biopsies, which remove intact tumor tissue including fibrotic stroma, FNA utilizes a fine needle with a bevel that is particularly adept at extracting both single cells and cell clusters out of the surrounding stroma by simply applying a rapid gentle cutting or sawing motion. FNA therefore preferentially enriches for both cohesive units of cells (such as epithelial glands) and discohesive cells (such as immune cells), while leaving behind much of the sclerotic stromal matrix that must be removed for successful downstream applications including isolation of organoids (Goldhoff et al., 2019). The gentle mechanical dislodging of cells with FNA requires no tissue digestion or processing, and important 3D architectures such as solid or glandular tumor growth patterns are often faithfully preserved. The gentle FNA extraction method and lack of further processing also greatly improves cell viability (compared with core biopsy), and FNA material can also be plated into culture or submitted for ancillary studies such as flow cytometry directly out of the needle (Boyd et al., 2015). As such, FNA biopsies provide a simple, low-risk clinical procedure for tumor cell isolation that is ideally suited for downstream research applications at minimal time and reagent costs.
Because of the ease and extraction advantages, we hypothesized that FNA could be harnessed as an efficient organoid isolation procedure that would improve take rates while simplifying the organoid collection and culturing process as a whole. In the current study, we provide evidence that FNA can be used in a patient-derived organoid culture method that is rapid and efficient and closely recapitulates the wide array of native histologic architecture and biological properties of the patient's original tissue. We demonstrate the effective use of this culture technique for high-throughput drug testing, patient-derived organoid xenograft models, and analysis of the tumor immune microenvironment.
Results
Fine-Needle Aspiration-Based Patient-Derived Organoid Culture Technique
FNAs utilize beveled 25-gauge needles with a 10-mL syringe and syringe holder to provide gentle aspiration using 1–3 needle passes for collection (see Transparent Methods and Video S1). It is well documented that this technique, with its fine beveled needle and rapid cutting motion, enriches for tumor cell clusters and discohesive cells with minimal stromal extraction (Ernst and Rimm, 2002) (Figure S1, Video S1). FNA biopsy material from patients yields intact epithelial cell aggregates and single cells that are largely free of stromal contamination, as compared with core biopsies that retain acellular matrix (illustrated in Figure S1). As such, FNA contents were plated into organoid culture directly out of the needle without any digestion or processing steps. Because FNA enriches for intact cell clusters without digestion or trypsinization, performing cell counts after FNA at the outset of the procedure is not possible for routine isolations. To approximate yields, cell counts were acquired for a representative subset of tumors to estimate the minimal tumor cellularity obtained (averaging ∼1 million cells per needle pass) by FNA (Table S1). As expected, the highest numbers of tumor cells were isolated from melanomas, which typically grow as solid sheets of discohesive cells. Renal cell carcinomas (RCCs) yielded nearly an order of magnitude fewer carcinoma cells, perhaps because tumor cells are embedded within a richly vascularized stroma. Despite the lower numbers, the cellularity was still more than sufficient for organoid culture. We note that these counts likely underestimate the true cellularity following FNA. Although not included in this study, lymph nodes likely represent the upper limits of cellularity for FNA sampling with yields up to 9.6 million cells per needle pass (Cajulis and Sneige, 1993).
Video Demonstration of FNA Procedure on Tissue Samples for Researchers.
To assess a range of tumor types for this study, FNA was performed ex vivo on surgical specimens from both human and xenografted tumors including melanomas, papillary and anaplastic thyroid carcinomas, clear cell renal carcinomas, colorectal adenocarcinomas, gastric signet ring adenocarcinomas, pancreatic adenocarcinomas, a cholangiocarcinoma, and a low-grade appendiceal mucinous tumor (Figure 1). FNA-based Patient-Derived Organoids (FNA-PDOs) were collected and cultured with equal success on human and xenograft mouse tumors. Unsampled tissue from the intact tumors was also processed for histopathology in parallel for in situ histomorphologic comparison with the FNA-PDO cultures. FNA-PDOs were then plated directly out of the needle into either semisolid or disc Matrigel formats in order to assess the suitability of both downstream culture conditions for growth and passaging (Figures 1A and 1B). Following FNA needle passes, the needles were rinsed in DMEM and centrifuged. Cell pellets were then resuspended in simplified DMEM media and Matrigel without commonly used supplements (for example, noggin, gastrin, R-spondin, Wnt3A) (Clevers, 2016; Neal et al., 2018; Saito et al., 2019; Vlachogiannis et al., 2018) in order to grow organoids and ensure selection of malignant cells over resident non-neoplastic epithelial cells. The initial take-rate was 98% (44/45) for thyroid cancer FNA-PDOs; 79% (19/24) for melanoma xenograft FNA-PDOs; 94% (16/17) for gastrointestinal FNA-PDOs including 8 colorectal carcinomas, 5 pancreatic carcinomas, 1 cholangiocarcinoma, 2 gastric carcinomas, and 1 appendiceal carcinoma; and 80% (12/15) for RCC FNA-PDOs (Table S2). This initial take-rate included cancer organoid growth in culture over 3 weeks and included both primary and metastatic lesions. Metastases, aggressive poorly differentiated primary tumors, and well-differentiated primary tumors all formed organoids with similar take rates. Successful long-term propagation for ≥5 passages following cryopreservation was achieved with a rate of 54% (13/24) for melanoma xenograft FNA-PDOs, 100% (12/12) for thyroid cancer FNA-PDOs, and 100% (4/4) for GI FNA-PDOs that have been evaluated. Renal cell carcinoma FNA-PDOs and some of the other samples were used only for short-term studies and were not cryopreserved for evaluation of long-term propagation. Despite the lack of supplements, most cancer organoid types grew well in the simplified media without overgrowth of non-malignant cells (as confirmed by two experienced pathologists, V.L.W. and O.G.M.) and the cost of culture and subsequent experimental studies was significantly decreased. The lone exception was RCC, which required enriched media containing noggin, R-spondin, and Wnt3A. Organoids from normal renal epithelium could also be successfully grown in this enriched media with an initial take rate of 80% (4/5 tissues). Normal epithelium from thyroid, skin, and liver FNAs failed to grow in simplified media (data not shown, see Transparent Methods for media details).
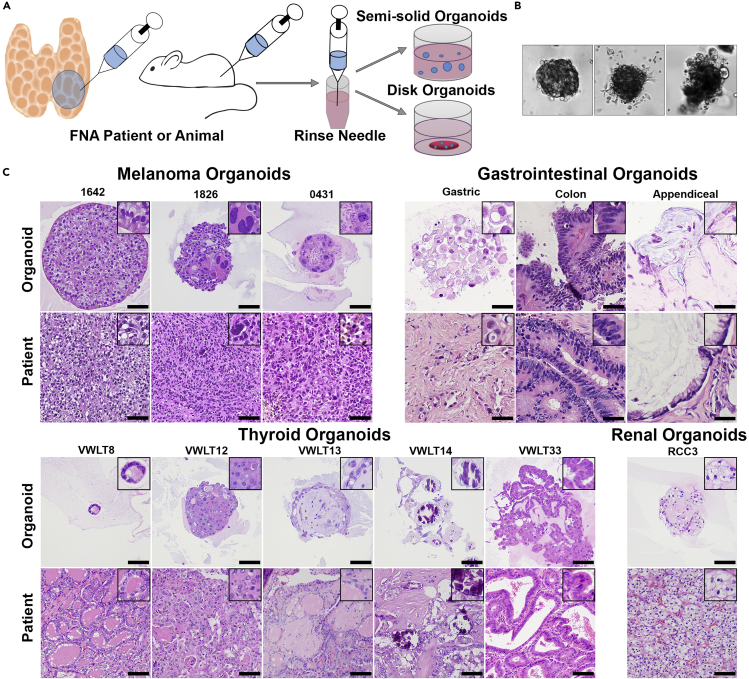
Figure 1.
FNA-Based Patient-Derived Organoid Model
(A) Fine-needle aspiration can be performed on patients, surgical specimens, or animals. One to three needle passes are typically collected, and the needle is rinsed in RPMI 1640 or DMEM. Following this rinse, the cells can be directly plated in either a semisolid or disc organoid format.
(B) Organoids begin to form within 1 week of plating and have morphologies unique to each individual patient. Three melanoma organoids shown have distinct morphology on 20× bright-field imaging.
(C) Organoids can also be embedded in paraffin blocks for morphologic evaluation using H&E staining, images taken at 20× magnification (scale bar, 50 μm). Organoid morphology, as seen on H&E stain, closely recapitulates the primary patient tumor morphology across multiple tumor types including melanoma, gastrointestinal carcinomas, thyroid carcinomas, and renal cell carcinoma.
Organoids Recapitulate Patient Tumor Morphology and Immunophenotype in Culture
A representative subset of FNA-PDOs from both disc and semisolid cultures were fixed and embedded in paraffin to assess tumor morphology (by H&E) side by side with the formalin-fixed tumor tissues collected in parallel from the patients. In all cases assessed, FNA-PDO morphology faithfully replicated the tumor morphology observed in the matched patient tissues across a range of tumor histologies (Figure 1C). This included spindled melanomas with vacuoles (patient 1642), pleomorphic melanomas with bizarre giant cells (patient 1826) or necrosis (patient 0431), signet ring gastric carcinomas with discohesive cells and intracytoplasmic mucin droplets (patient V50), colorectal adenocarcinomas with papillary glands and “dirty” necrosis (patient V80), appendiceal mucinous tumors with small clusters of malignant cells embedded in mucin (patient V12A), thyroid carcinomas with microfollicular (patient VWLT8), solid (patient VWLT12), psammomatous (patient VWLT14), and papillary architectures (patients VWLT13 and VWLT33), and clear cell RCCs with the classical clear cell morphology and nested growth patterns (Figure 1C). In addition to the morphologies observed by H&E stains, characteristic immunohistochemical staining (IHC) was also preserved across a panel of organoids (Figures 2A–2C). Melanoma FNA-PDOs strongly expressed S-100 protein with Ki-67 labeling (a proliferation marker), as seen in the matched tissue samples. Carcinomas retained their characteristic strong cytokeratin (AE1/AE3) expression. Nuclear TTF-1 expression was retained in thyroid FNA-PDOs. Thus, malignant cells isolated by FNA-PDO retained key histologic and immunophenotypic properties of the patient tumors from which they were derived.
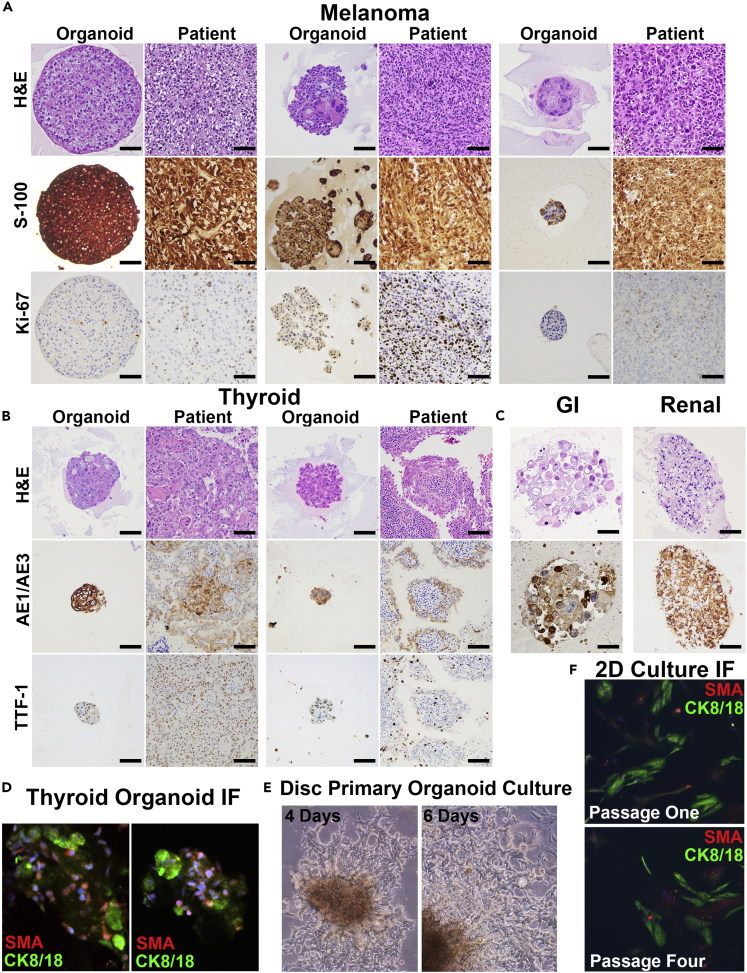
Figure 2.
Patient-Derived Organoids Maintain Identical Patient Morphology and Immunophenotype in Culture
(A) Three distinct FNA patient-derived organoids (FNA-PDOs) for melanoma. These organoids also maintain S-100 and Ki-67 immunohistochemical (IHC) staining similar to the patient tumor (20× magnification; scale bar, 50 μm).
(B) Two distinct FNA-PDOs for thyroid cancer maintain expression of markers indicative of thyroid differentiation, AE1/AE3 and TTF-1, on IHC (20× magnification; scale bar, 50 μm).
(C) Gastrointestinal and renal FNA-PDOs demonstrate preserved AE1/AE3 expression (20× magnification; scale bar, 50 μm).
(D) Immunofluorescence staining of the two thyroid cancer FNA-PDOs (shown in B) highlights both a population of tumor cells (green, CK8/18 positive staining) as well as a supportive population of cancer-associated fibroblasts (CAFs, red, SMA; DNA in blue; 20× magnification; scale bar, 100 μm).
(E) Disc organoid cultures for numerous tumor types tested demonstrate 2D cultures that extend from organoids and grow along the bottom of the plate. Bright-field images of melanoma FNA-PDO taken at 4 and 6 days of culture showing rapid growth of the 2D population (10× magnification; scale bar, 100 μm).
(F) Immunofluorescence of these 2D cultures (thyroid FNA-PDO shown) demonstrates both cancer epithelial cells (CK8/18, green) and cancer-associated fibroblasts (CAFs, SMA, red; 10× magnification; scale bar, 100 μm).
Resident stromal cells were also isolated and maintained in the 3D FNA-PDO cultures in many cases. For example, fibroblasts were clearly identified in two thyroid FNA-PDOs, as illustrated by immunofluorescence labeling of carcinoma cells (green, CK8/18 positive) surrounded by supporting cancer-associated fibroblasts (red, SMA positive, Figure 2D). In particular, the disc plating method was most conducive to maintaining cancer-associated fibroblasts. In these formats, a population of spindled fibroblasts often migrated out of the discs onto the plastic during the first few rounds of passaging, typically within 1–2 weeks of initial plating. These cells then grew exclusively as 2D cultures, as illustrated by a melanoma FNA-PDO at days 4 and 6 of culture (Figure 2E). These 2D fibroblast cultures propagated out of most FNA-PDO melanomas and thyroid carcinomas, with more variability in gastrointestinal carcinomas and RCCs. The percentage of fibroblasts migrating into 2D cultures varied between tumors and increased over time with each subsequent passage (tumor cells labeled in green with CK8/18 and fibroblasts labeled in red with SMA, Figure 2F). 2D fibroblast cultures were not observed in semisolid organoid culture formats, presumably due to the ultra-low attachment surfaces of the plates. However, fibroblasts were identified in semisolid culture within the organoids surrounding the malignant cells.
FNA-PDOs Model Therapeutic Responses
We next tested whether our FNA-PDO culture method could be applied to downstream in vitro therapeutic modeling applications (Figure 3). We first tested drug-responsive organoid growth within the semisolid format. Thyroid FNA-PDOs were treated with the BRAF inhibitor dabrafenib at clinically relevant concentrations (30 nM) for 20 days in semisolid culture. Unlike control (wild-type) thyroid carcinomas, which were insensitive to dabrafenib as expected, the BRAFV600E-mutant FNA-PDO demonstrated a partial response to therapy (Figure 3A). This is similar to the partial response rate that is observed in patients with BRAF-mutant thyroid carcinoma (Cabanillas et al., 2015).
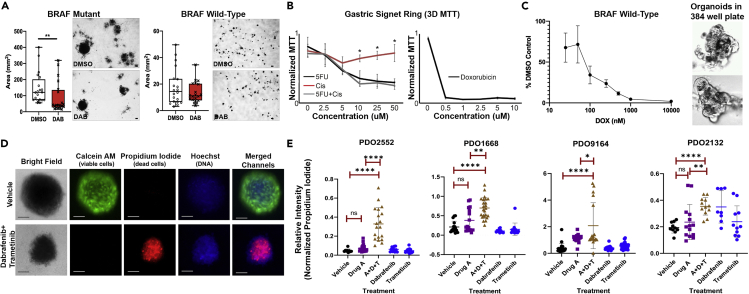
Figure 3.
FNA-Based Patient-Derived Organoids are Readily Adapted to High-throughput Screening
(A) FNAs directly plated in semisolid organoid culture can be tested for drug sensitivity. Using 24-well plates, BRAFV600E-mutant (left) and BRAF-wild-type (right) thyroid cancer organoids were treated with dabrafenib, a BRAF inhibitor (30 nM), for 20 days. BRAFV600E-mutant organoids show decreased organoid size and number following treatment. As expected, BRAF-wild-type organoids show no observable response. Images representative of three replicates. ∗∗p < 0.05, Mann Whitney test.
(B) FNA-based patient-derived gastric signet ring organoids were assayed for drug sensitivity using disc-organoid MTT assay in 96-well plates. Images representative of four replicates; error bars represent standard deviation. ∗ = p < 0.05 (Student’s t test). The x axis indicates the concentrations of each drug (alone or in combination); the y axes are MTT signals normalized to the highest absorbance value within each experiment.
(C) FNA-PDOs from aggressive anaplastic thyroid cancer were assessed for viability following doxorubicin treatment using an automated high-throughput 384-well assay. The thyroid cancer FNA-PDOs showed response to doxorubicin. Automated high content imaging also confirms organoid growth in the 384-well format, as seen in these untreated melanoma FNA-PDOs. Images representative of three replicates; error bars represent standard error of the mean (SEM).
(D) Using a high-throughput fluorescent assay, patient-derived melanoma organoids were labeled with Calcein AM (green fluorescence, labels live cells) and propidium iodide (red fluorescence, labels dead cells). To visualize both live and dead cells, a cell-permeable DNA stain Hoechst 33342 was used. Although the overall organoid structure remained intact, the organoid decreased in size and cells in the organoids were killed following 3 days of treatment with MEK inhibitor, trametinib (0.1 μM), and BRAF inhibitor, dabrafenib (10 μm). This is in agreement with clinical data showing that dabrafenib and trametinib combination is effective for treatment of BRAF V600E melanoma.31 Images representative of three replicates; scale bar, 100 μm.
(E) Organoids from indicated human melanoma tumors were generated using FNA of corresponding PDX tumors grown in BALB/c nu/nu mice. Organoids were plated in semisolid media into 96-well low-attachment plates and treated with 5 μm AMG-232, 1 μm debrafenib, 0.1 μm trametinib, or combination of three drugs for 3 days. Calcein M/Propidium Iodide viability assay was performed as shown in (C). The ratio of the intensity of red (dead cells) and green (live cells) fluorescent signal in individual organoids was plotted. Each dot represents an individual organoid. We analyzed 9–23 organoids per group for PDO2552, 11–24 for PDO1668, 17–43 for PDO 9164, and 8–14 for PDO 2132. Unpaired t test was used to determine the significance of differences between indicated treatment groups. Error bars represent standard deviation.
We next tested drug responses of organoids grown in discs plated within standard 96-well plate formats, using conventional MTT signals as a readout of organoid viability. Following 2 weeks of growth to allow mature organoid formation, a gastric signet ring FNA-PDO was treated with various chemotherapies over a subsequent period of 2 weeks. Standard-of-care palliative combination chemotherapy (5-fluorouracil [5FU] and cisplatin) demonstrated therapeutic efficacy in a concentration-dependent manner. We note that 5-FU alone was as efficacious against the FNA-PDOs as 5FU combined with cisplatin and that FNA-PDOs also responded to doxorubicin monotherapy by these assays (Figure 3B).
Results presented above indicate that the intact organoids obtained by FNA retain intrinsic chemotherapeutic sensitivities in vitro. Because other techniques trypsinize organoids into single cells during passaging and plating, we evaluated whether FNA-PDOs could be successfully trypsinized into single cells and regrown into organoids for drug sensitivity testing in 384-well plates. To this end, FNA-PDOs were dissociated into single cells with trypsin, plated into semisolid formats, incubated for 24–72 h to allow organoids to reform, and assessed for drug sensitivity with both ATP measurement (CellTiter-Glo 3D) and automated high-content imaging. As shown in Figure 3C, organoids formed efficiently in 384-well plates and concentration-response curves were generated during treatment of anaplastic thyroid carcinoma FNA-PDOs with doxorubicin.
FNA-PDOs were further tested for drug response using high-throughput fluorescence-based viability assays. Melanoma FNA-PDOs obtained from xenograft mice were treated with combination MEK inhibitor (0.1 μM trametinib) and BRAF inhibitor (10 μM dabrafenib), which are standard-of-care therapeutics for patients with BRAF-mutant melanoma. Viability was assessed by staining with Calcein AM (stains live cells) and propidium iodide (stains dead cells). Bisbenzimide (Hoechst 33342), a cell-permeable DNA stain, was used to visualize cell nuclei. Although trametinib/dabrafenib-treated FNA-organoids remained largely intact with smaller size, fluorescent imaging revealed widespread cell death after 72 h of trametinib plus dabrafenib exposure (Figure 3D). Additional studies on four distinct melanoma FNA-PDOs generated from PDX tumors using 5μm AMG-232, 1μm debrafenib, 0.1μm trametinib, or combination of three drugs for 3 days demonstrates the differential response and utility of the fluorescence-based viability assay. The response of the FNA-PDOs closely mirrors previously published work for these PDX tumors in drug-treated mice (Shattuck-Brandt et al., 2020).
Xenograft Studies Using FNA-PDOs Show Aggressive Tumor Growth
We next tested whether FNA-PDOs could efficiently xenograft into immunodeficient mice. To this end, FNA-based patient-derived organoid xenograft models (FNA-PDOX) were successfully generated by either subcutaneous implantation of FNA-PDO cultured within discs or direct injection of FNA-PDOs harvested from semisolid cultures (Figures S2A and S2B; see Transparent Methods for experimental details). Although both approaches generated FNA-PDOXs, disc implantation was more preferred for aggressive tumor types, especially melanomas. Palpable flank tumors formed between 10 and 50 days of implantation using either method and the histomorphology of the FNA-PDOX tumor was identical to both the in vitro FNA-PDO cultures and in vivo patient tumor tissues (Figure S2C). As expected, FNA-PDOX growth rates varied across tumor types and individual patient samples. However, FNA-PDOX growth closely matched the growth of conventional PDXs derived from the same patient tumor(s). Despite prior in vitro passaging and generally lower cellularity of implanted FNA-PDO discs, some FNA-PDOXs also grew unexpectedly robustly, reaching 1,000 mm3 within 35 days of implantation (Figure S2D). FNA-PDOXs were also generated in a similar time using less aggressive tumors (such as thyroid tumors) harvested from semisolid cultures (data not shown). These data demonstrate two methods for generating robust growth of FNA-PDOs in immune compromised mice.
Immune Cell Capture from FNA-PDOs
The use of human organoid models for the study of tumor immunology has been a promising area of research (Bar-Ephraim et al., 2019; Chakrabarti et al., 2018; Dijkstra et al., 2018; Finnberg et al., 2017; Jenkins et al., 2018; Neal et al., 2018; Schnalzger et al., 2019) that requires the successful co-culture of both tumor epithelial cells and infiltrating immune cells. We asked if tumor-infiltrating immune cells could be isolated by FNA and co-cultured with tumor cells in our FNA-PDO model, since FNA is a highly efficient method for extracting discohesive cell populations. Because RCCs are highly vascular tumors that are often enriched with well-defined populations of immune cells, we more thoroughly examined the immune capture efficiency of FNA-PDO using RCC as a model system (Figure 4A). Overall, viability and initial lymphocyte and myeloid subtype yields of the captured immune cells appeared equivalent between FNA and conventional digestion methods (Figures 4B–4D). As expected, absolute immune cell counts (CD45+ cells) at collection were higher for large tumor fragments as compared with FNA, approximately 8 ×105 for FNA (nine needle passes) versus 6 × 106 for whole tumor digestion (1 cm3, Figure 4E). However, immune cell survival was significantly higher in FNA-PDOs, likely due to the gentler extraction technique. After plating the same number of cells per organoid disc (100,000 total cells per 300 μL organoid disc), the immune cell survival in FNA-PDOs was significantly higher than digestion-based PDOs at 2 weeks of culture for both complete media and Wnt-enriched media (p < 0.05, Figure 4F). The culture protocols for immune-containing organoids in the literature are quite complex and often recommend Wnt3A-containing or cytokine supplemented media (Chakrabarti et al., 2018; Dijkstra et al., 2018; Finnberg et al., 2017; Jenkins et al., 2018; Long et al., 2014; Schnalzger et al., 2019; Seino et al., 2018). We next assessed whether IL-2 supplementation and media enriched with Wnt3A, noggin, and R-spondin enhanced immune subset survival within our FNA-PDO model. Enriched media enhanced CD45+ immune cell survival with significant enhancement of CD3+ T cell subset and CD14+CD11b+ myeloid cell survival at 2 weeks in FNA-PDO culture (Figures 4F–4I). IL-2 supplementation further enhanced survival of CD3+ T cell subsets at 2 weeks in FNA-PDO culture (Figure 4H). Further studies will be required to investigate various tumor types and long-term culture beyond the proof-of-principle experiments reported here. However, based on these studies, we conclude that FNA-PDO is superior to standard digestion-based PDOs in capturing resident immune cell populations from the sampled tumor microenvironment, as well as providing improved immune cell survival.
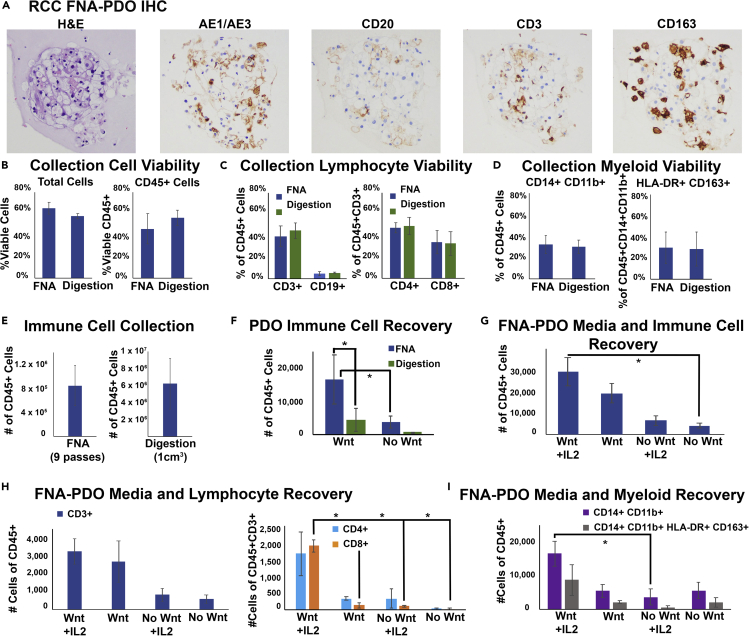
Figure 4.
FNA-Based Patient-Derived RCC Organoids Contain Immune Cells from the Tumor Microenvironment
(A) Immune cells are extracted and plated during FNA-PDO collection and generation. This representative RCC FNA-PDO demonstrates classic morphology by H&E with positive cytokeratin staining (AE1/AE3) and infiltration of B cells (CD20), T cells (CD3), and macrophages (CD163) by immunohistochemistry (IHC).
(B) Cell viability at collection was assessed using flow cytometry on fresh FNA material collected from RCCs versus fresh material following tumor digestion of RCCs, showing similar overall viability and CD45+ viability between FNA and digestion extraction methods (n = 4 tumors). Error bars represent standard error of the mean (SEM).
(C) Both FNA and digestion methods have similar lymphoid subset viability at collection (n = 4 tumors, error bars represent SEM).
(D) Both FNA and digestion methods have similar myeloid subset viability at collection (n = 4 tumors, error bars represent SEM).
(E) Absolute immune cell counts at collection for FNA (nine needle passes per tumor) and digestion (1 cm3 tissue per tumor completely digested, n = 4 tumors, error bars represent SEM).
(F) RCC FNA-PDO and digestion-PDO immune cell recovery counts at 2 weeks of disc culture demonstrate significantly improved survival of CD45+ cells from FNA-PDOs as compared with digestion-PDOs in both Wnt3A-enriched and non-Wnt-enriched complete media (n = 4 tumors, error bars represent SEM, ∗p < 0.05).
(G) FNA-PDO CD45+ cell recovery counts at 2 weeks in culture demonstrates improved CD45+ cell survival with both Wnt-enriched media and 100 IU/mL IL-2 supplementation (n = 2 tumors, error bars represent SEM, ∗p < 0.05).
(H) FNA-PDO lymphocyte recovery counts at 2 weeks in culture demonstrates CD3+ lymphocyte survival in all conditions and improved CD8+ lymphocyte survival with Wnt-enriched media and IL-2 supplementation (n = 2 tumors, error bars represent SEM, ∗p < 0.05).
(I) FNA-PDO myeloid recovery counts at 2 weeks in culture demonstrates improved survival of CD14+CD11b+ myeloid cells when cultured with Wnt-enriched media (n = 2 tumors, error bars represent SEM, ∗p=<0.05).
Discussion
FNA-PDO is a simple method that can be used to isolate tumor organoids directly from patients at the bedside or from harvested tumor tissues, including biopsied tissues as small as 2 mm3 (in our experience). Our methodology requires minimal sample processing without digestion of tissues or trypsinization of cell clusters and faithfully preserves native tumor histology. It enriches for cellular aspects of the tumor microenvironment at the expense of acellular elements, even for cancers with a highly fibrotic stroma. FNA-PDOs can be cultured and passaged without trypsinization in solid or semisolid formats, and either format is amenable to in vitro experiments including drug screens and in vivo experiments including xenograft studies. Of note, we found that the standard media used to collect FNA samples at the bedside clinically (RPMI 1640 or DMEM) can be used here for organoid culture. Finally, FNA-PDOs allow co-isolation of tumor-infiltrating immune cell populations with enhanced immune cell survival as compared with standard digestion methods. Because of its simplicity, wide applicability, and preservation of the cellular aspects of the native tumor, FNA-PDO represents an important technical advance over conventional organoid isolation methods. With the widespread clinical use of FNA, this technique can be extensively utilized across medical centers to create organoid cultures and organoid tissue banks for precision medicine and research.
We speculate that FNA-based organoid culture holds broad potential for clinical care and basic research. Like conventional organoids, FNA-PDOs could be particularly valuable for personalized therapy, since drug responses can be assessed in vitro from the same cancer over time, including before and after treatment or surgery. FNA-PDOs could be used in combination with other assays (for example, flow cytometry) to monitor patient immune responses, tumor-immune infiltration, PD-L1 expression, tumor differentiation and viability, and drug sensitivity over the course of disease and therapy. Because FNA is widely used in the clinic and FNA-PDOs require no additional processing, we anticipate that this technique could be efficiently incorporated into current clinical workflows. Generation of FNA-PDOs requires only 1–3 weeks on average, indicating that drug sensitivity testing or other ancillary studies could be completed on a similar timeline as diagnostic molecular testing. In the experimental setting, FNA-PDOs could be especially useful for serial samplings of tumors from xenografts or genetically engineered mice to determine treatment responses or immune infiltration patterns over time. Finally, FNA-PDOs can be used to isolate tumor organoids and immune cells from patients or surgically remove patient specimens for general experimental purposes.
Our current studies suggest that FNA-PDO represents a powerful new tool for use in personalized therapeutic drug testing, high-throughput drug discovery, xenograft generation, and tumor-immune microenvironment studies. When widely adapted, we anticipate that FNA-PDOs will provide the scientific community with a simple and cost-effective approach to directly interrogate key aspects of the tumor microenvironment in vitro more efficiently than traditional organoid cultures and more rapidly than traditional mouse models.
Limitations of the Study
The main limitation of the study is that this is a proof-of-principle study performed mostly on ex vivo tumor samples, without formal demonstration of the technique for in situ tumors sampled directly from patients. Although melanoma organoids were generated by in vivo sampling of PDX tumors, the mice were immunocompromised and evaluation of the immune component could not be performed. We also present the utility of this methodology for discohesive and epithelial tumors. Although we show the excellent performance of this technique on fibrotic pancreatic neoplasms and spindled anaplastic thyroid carcinomas, a formal evaluation of FNA-PDOs from soft tissue sarcomas was not performed.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Vivian Weiss (Vivian.l.weiss@vumc.org).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
This study did not generate datasets or code.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
The authors would like to acknowledge the Translational Pathology Shared Resource and the High-throughput Screening Shared Resource, supported by NCI/NIH Cancer Center Support Grant 5P30 CA68485-19 and the Vanderbilt Mouse Metabolic Phenotyping Center Grant 2 U24 DK059637-16, and the Shared Instrumental Grant S10 OD023475-01A1 for the Leica Bond RX. The project described was also supported by NIH R37 CA233770-01 (to A.V.), BCRF (IIDRP-16-001 to A.V.), NCI Research Specialist Award (R50 CA211206 to J.A.B.), NIH R35GM122516 (to E.L.), NIH CA116021 (A.R.), VA SRCS Award (A.R.), NIH R01 CA222594 (O.G.M.), ACS IRG (#IRG-58-009-58 to V.L.W.), ATA (2019-0000000090 to V.L.W.), K12 Vanderbilt Clinical Oncology Research Career Development Program (NCI K12CA090625 to V.L.W., PI W.K.R.), AACR KuRE-IT award (W.K.R.), V Foundation Scholar Award (to V.L.W.), CCRF (to V.L.W.), and ACS (133934-CSDG-19-216-01-TBG to V.L.W.).
Author Contributions
A.V. designed experiments and wrote the paper; K.B., M.W., R.S.-B., V.B., A.B., C.J., C.P., M.L., C.L., B.R., and R.H. conducted experiments; K.B. designed histology methodology; J.N., S.R., K.I., and N.B. isolated patient tissue and consented patients; A.R., W.K.R., E.L., and O.G.M. designed experiments and edited the manuscript; V.L.W. designed experiments and wrote the paper.
Declaration of Interests
The authors declare no competing interests.
Published: August 21, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101408.
Supplemental Information
References
- Austin R.M., Birdsong G.G., Sidawy M.K., Kaminsky D.B. Fine needle aspiration is a feasible and accurate technique in the diagnosis of lymphoma. J. Clin. Oncol. 2005;23:9029–9030. doi: 10.1200/JCO.2005.01.5248. author reply 9030-9021. [DOI] [PubMed] [Google Scholar]
- Baatenburg de Jong R.J., Rongen R.J., Verwoerd C.D., van Overhagen H., Lameris J.S., Knegt P. Ultrasound-guided fine-needle aspiration biopsy of neck nodes. Arch. Otolaryngol. Head Neck Surg. 1991;117:402–404. doi: 10.1001/archotol.1991.01870160056008. [DOI] [PubMed] [Google Scholar]
- Bar-Ephraim Y.E., Kretzschmar K., Clevers H. Organoids in immunological research. Nat. Rev. Immunol. 2019;20:279–293. doi: 10.1038/s41577-019-0248-y. [DOI] [PubMed] [Google Scholar]
- Boj S.F., Hwang C.I., Baker L.A., Chio, Engle D.D., Corbo V., Jager M., Ponz-Sarvise M., Tiriac H., Spector M.S. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd J.D., Smith G.D., Hong H., Mageau R., Juskevicius R. Fine-needle aspiration is superior to needle core biopsy as a sample acquisition method for flow cytometric analysis in suspected hematologic neoplasms. Cytometry B Clin. Cytom. 2015;88:64–68. doi: 10.1002/cyto.b.21183. [DOI] [PubMed] [Google Scholar]
- Cabanillas M.E., Patel A., Danysh B.P., Dadu R., Kopetz S., Falchook G. BRAF inhibitors: experience in thyroid cancer and general review of toxicity. Horm. Cancer. 2015;6:21–36. doi: 10.1007/s12672-014-0207-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajulis R.S., Sneige N. Objective comparison of cellular yield in fine-needle biopsy of lymph nodes with and without aspiration. Diagn. Cytopathol. 1993;9:43–45. doi: 10.1002/dc.2840090109. [DOI] [PubMed] [Google Scholar]
- Chakrabarti J., Holokai L., Syu L., Steele N., Chang J., Dlugosz A., Zavros Y. Mouse-derived gastric organoid and immune cell Co-culture for the study of the tumor microenvironment. Methods Mol. Biol. 2018;1817:157–168. doi: 10.1007/978-1-4939-8600-2_16. [DOI] [PubMed] [Google Scholar]
- Choi S.I., Jeon A.R., Kim M.K., Lee Y.S., Im J.E., Koh J.W., Han S.S., Kong S.Y., Yoon K.A., Koh Y.H. Development of patient-derived preclinical platform for metastatic pancreatic cancer: PDOX and a subsequent organoid model system using percutaneous biopsy samples. Front. Oncol. 2019;9:875. doi: 10.3389/fonc.2019.00875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- Conrad R., Yang S.E., Chang S., Bhasin M., Sullivan P.S., Moatamed N.A., Lu D.Y. Comparison of cytopathologist-performed ultrasound-guided fine-needle aspiration with cytopathologist-performed palpation-guided fine-needle aspiration: a single institutional experience. Arch. Pathol. Lab Med. 2018;142:1260–1267. doi: 10.5858/arpa.2017-0123-OA. [DOI] [PubMed] [Google Scholar]
- Dijkstra K.K., Cattaneo C.M., Weeber F., Chalabi M., van de Haar J., Fanchi L.F., Slagter M., van der Velden D.L., Kaing S., Kelderman S. Generation of tumor-reactive T cells by Co-culture of peripheral blood lymphocytes and tumor organoids. Cell. 2018;174:1586–1598.e12. doi: 10.1016/j.cell.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddyani M., Fraga A.G., Schmitt F., Uwizeye C., Fissette K., Johnson C., Aguiar J., Sopoh G., Barogui Y., Meyers W.M. Fine-needle aspiration, an efficient sampling technique for bacteriological diagnosis of nonulcerative Buruli ulcer. J. Clin. Microbiol. 2009;47:1700–1704. doi: 10.1128/JCM.00197-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst L.M., Rimm D.L. Quantitative examination of mechanophysical tumor cell enrichment in fine-needle aspiration specimens. Cancer. 2002;96:275–279. doi: 10.1002/cncr.10746. [DOI] [PubMed] [Google Scholar]
- Finnberg N.K., Gokare P., Lev A., Grivennikov S.I., MacFarlane A.W.t., Campbell K.S., Winters R.M., Kaputa K., Farma J.M., Abbas A.E. Application of 3D tumoroid systems to define immune and cytotoxic therapeutic responses based on tumoroid and tissue slice culture molecular signatures. Oncotarget. 2017;8:66747–66757. doi: 10.18632/oncotarget.19965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldhoff P.E., Vohra P., Kolli K.P., Ljung B.M. Fine-needle aspiration biopsy of liver lesions yields higher tumor fraction for molecular studies: a direct comparison with concurrent core needle biopsy. J. Natl. Compr. Canc Netw. 2019;17:1075–1081. doi: 10.6004/jnccn.2019.7300. [DOI] [PubMed] [Google Scholar]
- Jain D., Allen T.C., Aisner D.L., Beasley M.B., Cagle P.T., Capelozzi V.L., Hariri L.P., Lantuejoul S., Miller R., Mino-Kenudson M. Rapid on-site evaluation of endobronchial ultrasound-guided transbronchial needle aspirations for the diagnosis of lung cancer: a perspective from members of the pulmonary Pathology society. Arch. Pathol. Lab Med. 2018;142:253–262. doi: 10.5858/arpa.2017-0114-SA. [DOI] [PubMed] [Google Scholar]
- Jenkins R.W., Aref A.R., Lizotte P.H., Ivanova E., Stinson S., Zhou C.W., Bowden M., Deng J., Liu H., Miao D. Ex vivo profiling of PD-1 blockade using organotypic tumor spheroids. Cancer Discov. 2018;8:196–215. doi: 10.1158/2159-8290.CD-17-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieu D. Fine-needle aspiration: technique and smear preparation. Am. Fam. Physician. 1997;55:839–846. 853-834. [PubMed] [Google Scholar]
- Long G.V., Stroyakovskiy D., Gogas H., Levchenko E., de Braud F., Larkin J., Garbe C., Jouary T., Hauschild A., Grob J. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N. Engl. J. Med. 2014;371:1877–1888. doi: 10.1056/NEJMoa1406037. [DOI] [PubMed] [Google Scholar]
- Mazzucchelli S., Piccotti F., Allevi R., Truffi M., Sorrentino L., Russo L., Agozzino M., Signati L., Bonizzi A., Villani L. Establishment and morphological characterization of patient-derived organoids from breast cancer. Biol. Proced. Online. 2019;21:12. doi: 10.1186/s12575-019-0099-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal J.T., Li X., Zhu J., Giangarra V., Grzeskowiak C.L., Ju J., Liu I.H., Chiou S.H., Salahudeen A.A., Smith A.R. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175:1972–1988.e1916. doi: 10.1016/j.cell.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman M.B., Abele J., Ali S.Z., Duick D., Elsheikh T.M., Jeffrey R.B., Powers C.N., Randolph G., Renshaw A., Scoutt L. Techniques for thyroid FNA: a synopsis of the National Cancer Institute thyroid fine-needle aspiration state of the science conference. Diagn. Cytopathol. 2008;36:407–424. doi: 10.1002/dc.20829. [DOI] [PubMed] [Google Scholar]
- Saito Y., Muramatsu T., Kanai Y., Ojima H., Sukeda A., Hiraoka N., Arai E., Sugiyama Y., Matsuzaki J., Uchida R. Establishment of patient-derived organoids and drug screening for biliary tract carcinoma. Cell Rep. 2019;27:1265–1276.e1264. doi: 10.1016/j.celrep.2019.03.088. [DOI] [PubMed] [Google Scholar]
- Schnalzger T.E., de Groot M.H., Zhang C., Mosa M.H., Michels B.E., Roder J., Darvishi T., Wels W.S., Farin H.F. 3D model for CAR-mediated cytotoxicity using patient-derived colorectal cancer organoids. EMBO J. 2019;38:e100928. doi: 10.15252/embj.2018100928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seino T., Kawasaki S., Shimokawa M., Tamagawa H., Toshimitsu K., Fujii M., Ohta Y., Matano M., Nanki K., Kawasaki K. Human Pancreatic Tumor Organoids Reveal Loss of Stem Cell Niche Factor Dependence during Disease Progression. Cell Stem Cell. 2018;22:454–467.e6. doi: 10.1016/j.stem.2017.12.009. [DOI] [PubMed] [Google Scholar]
- Shattuck-Brandt R.L., Chen S.C., Murray E., Johnson C.A., Crandall H., O'Neal J.F., Al-Rohil R.N., Nebhan C.A., Bharti V., Dahlman K.B. Metastatic melanoma patient-derived xenografts respond to MDM2 inhibition as a single agent or in combination with BRAF/MEK inhibition. Clin. Cancer Res. 2020;26:3803–3818. doi: 10.1158/1078-0432.CCR-19-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiriac H., Belleau P., Engle D.D., Plenker D., Deschenes A., Somerville T.D.D., Froeling F.E.M., Burkhart R.A., Denroche R.E., Jang G.H. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov. 2018;8:1112–1129. doi: 10.1158/2159-8290.CD-18-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiriac H., Bucobo J.C., Tzimas D., Grewel S., Lacomb J.F., Rowehl L.M., Nagula S., Wu M., Kim J., Sasson A. Successful creation of pancreatic cancer organoids by means of EUS-guided fine-needle biopsy sampling for personalized cancer treatment. Gastrointest. Endosc. 2018;87:1474–1480. doi: 10.1016/j.gie.2017.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S., McOlash L., Palen K., Johnson B., Duris C., Yang Q., Dwinell M.B., Hunt B., Evans D.B., Gershan J. Development of primary human pancreatic cancer organoids, matched stromal and immune cells and 3D tumor microenvironment models. BMC Cancer. 2018;18:335. doi: 10.1186/s12885-018-4238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachogiannis G., Hedayat S., Vatsiou A., Jamin Y., Fernandez-Mateos J., Khan K., Lampis A., Eason K., Huntingford I., Burke R. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920–926. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video Demonstration of FNA Procedure on Tissue Samples for Researchers.
Data Availability Statement
This study did not generate datasets or code.