Abstract
The integrin family of transmembrane adhesion receptors coordinates complex signaling networks that control the ability of cells to sense and communicate with the extracellular environment. Kindlin proteins are a central cytoplasmic component of these networks, directly binding integrin cytoplasmic domains and mediating interactions with cytoskeletal and signaling proteins. The physiological importance of kindlins is well established, but how the scaffolding functions of kindlins are regulated at the molecular level is still unclear. Here, using a combination of GFP nanotrap association assays, pulldown and integrin-binding assays, and live-cell imaging, we demonstrate that full-length kindlins can oligomerize (self-associate) in mammalian cells, and we propose that this self-association inhibits integrin binding and kindlin localization to focal adhesions. We show that both kindlin-2 and kindlin-3 can self-associate and that kindlin-3 self-association is more robust. Using chimeric mapping, we demonstrate that the F2PH and F3 subdomains are important for kindlin self-association. Through comparative sequence analysis of kindlin-2 and kindlin-3, we identify kindlin-3 point mutations that decrease self-association and enhance integrin binding, affording mutant kindlin-3 the ability to localize to focal adhesions. Our results support the notion that kindlin self-association negatively regulates integrin binding.
Keywords: kindlin, integrin, cell adhesion, oligomerization, focal adhesion, protein-protein interaction, scaffold protein, cell signaling, self-association, cytoskeleton, adhesion
The phenotypes of kindlin knockout or depletion in mouse, Drosophila, and Caenorhabditis elegans clearly establish the general importance of kindlins for integrin function (1–6). Furthermore, loss-of-function mutations in two of the three mammalian kindlins (kindlin-1 and kindlin-3) result in human diseases associated with altered cell adhesion (7–9), whereas mutations in kindlin-2, or changes in its expression level, are associated with cancer (10, 11). However, while kindlins clearly bind integrin cytoplasmic tails and act as adaptor proteins that regulate integrin adhesive function and connect integrins to cytoskeletal and signaling networks (12, 13), how kindlin activity is regulated is only partially understood.
Kindlins share a conserved 4.1-ezrin-radixin-moesin (FERM) domain architecture that is distinguished from other FERM domains by the presence of an F0 subdomain, the inclusion of a long flexible loop in the F1 subdomain, and the insertion of a pleckstrin homology (PH) domain into the F2 subdomain (14–17). It is believed that the importance of kindlins in integrin-mediated signaling stems from their role as scaffolds for mediating protein–protein interactions (13, 18), and considerable efforts have been directed toward identifying kindlin binding partners (1, 12, 19). Among the best-characterized kindlin interactions is the F3 subdomain-mediated binding to integrin β tails (1, 14, 20). This interaction, which has been structurally characterized (14), is essential for normal integrin activation and for correct targeting of kindlins to focal adhesions (14, 20, 21). Kindlins also bind several other focal adhesion proteins (12, 13, 18) including integrin-linked kinase (ILK), a pseudokinase important for linking integrins to the actin cytoskeleton. The kindlin–ILK interaction, which is mediated by the kindlin F2PH subdomain and the ILK pseudokinase domain, also contributes to recruitment and retention of kindlin and ILK at focal adhesions (22–24).
It has recently been proposed that in addition to other protein–protein interactions, kindlins may also self-associate (form kindlin-kindlin interactions) (14), and that this may be important in kindlin-mediated integrin signaling. Whereas crystal structures of full-length kindlins are not yet available, the structure of a truncated kindlin-2 lacking the large F1 loop and the PH domain revealed a domain-swapped dimer (14). However, whether this dimeric form is physiologically relevant remains unclear, especially because the reported spontaneous monomer-dimer transition in solution is unfavorable, occurring on the order of days in vitro (14). Despite this, kindlin self-association has emerged as an attractive mechanistic model for how kindlins may promote integrin signaling (14); indeed, kindlins have been implicated in driving integrin clustering (25). However, it is currently unclear if kindlin self-association occurs in full-length kindlins in mammalian cells, whether or not other kindlin isoforms besides kindlin-2 can self-associate, and the impact of self-association on integrin binding and focal adhesion localization.
Despite sharing a conserved domain organization, the three mammalian kindlins exhibit specific functions and interactions (18, 23, 26, 27). Previous studies from our laboratory and others have demonstrated that the widely expressed kindlin-2 and hematopoietic-specific kindlin-3 exhibit functional differences that go beyond expression patterns. For example, when expressed in Chinese hamster ovary (CHO) cells or bovine aortic endothelial cells (BAEC) plated on fibronectin, kindlin-2 localizes to focal adhesions, whereas kindlin-3 does not (23, 28). These functional disparities can be traced, in part, to differential binding to β1 integrins and to ILK and the ILK-PINCH-parvin (IPP) complex (23, 28). Here, we extend these studies to include comparison of kindlin self-association. We report that full-length kindlin-2 and kindlin-3 can each self-associate in mammalian cells but that kindlin-3 self-associates to a greater extent than kindlin-2. We identify domains important for mediating self-association and, through analysis of kindlin-3 mutants, present evidence suggesting that kindlin self-association impairs integrin binding and inhibits focal adhesion localization.
Results
kindlin-2 and kindlin-3 can each self-associate in mammalian cells, but kindlin-3 self-association is more robust
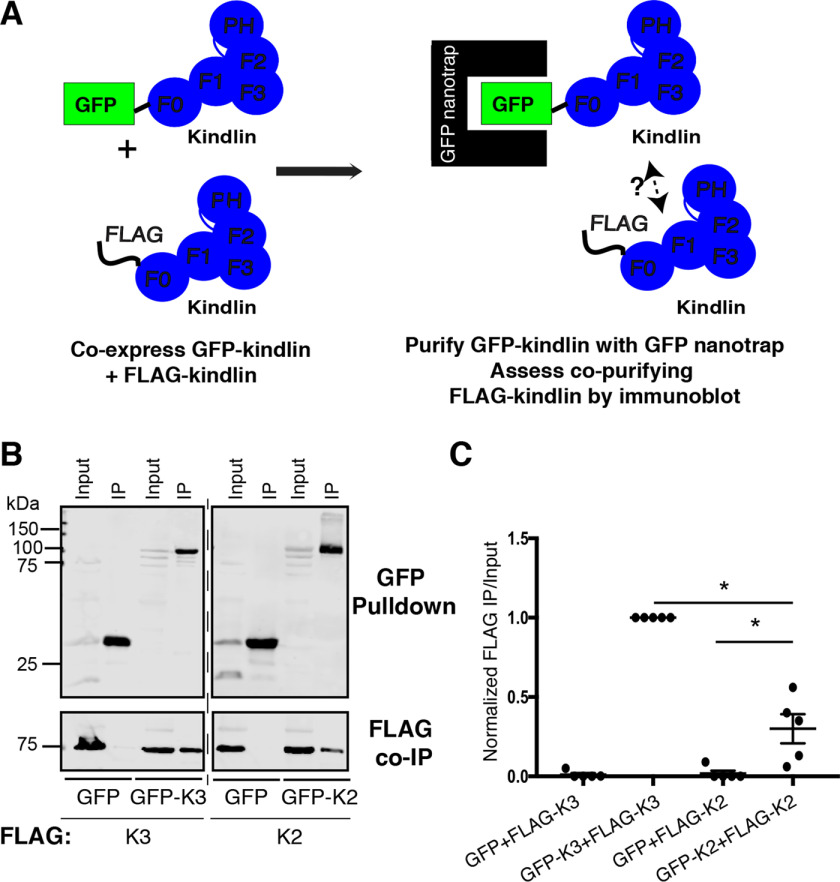
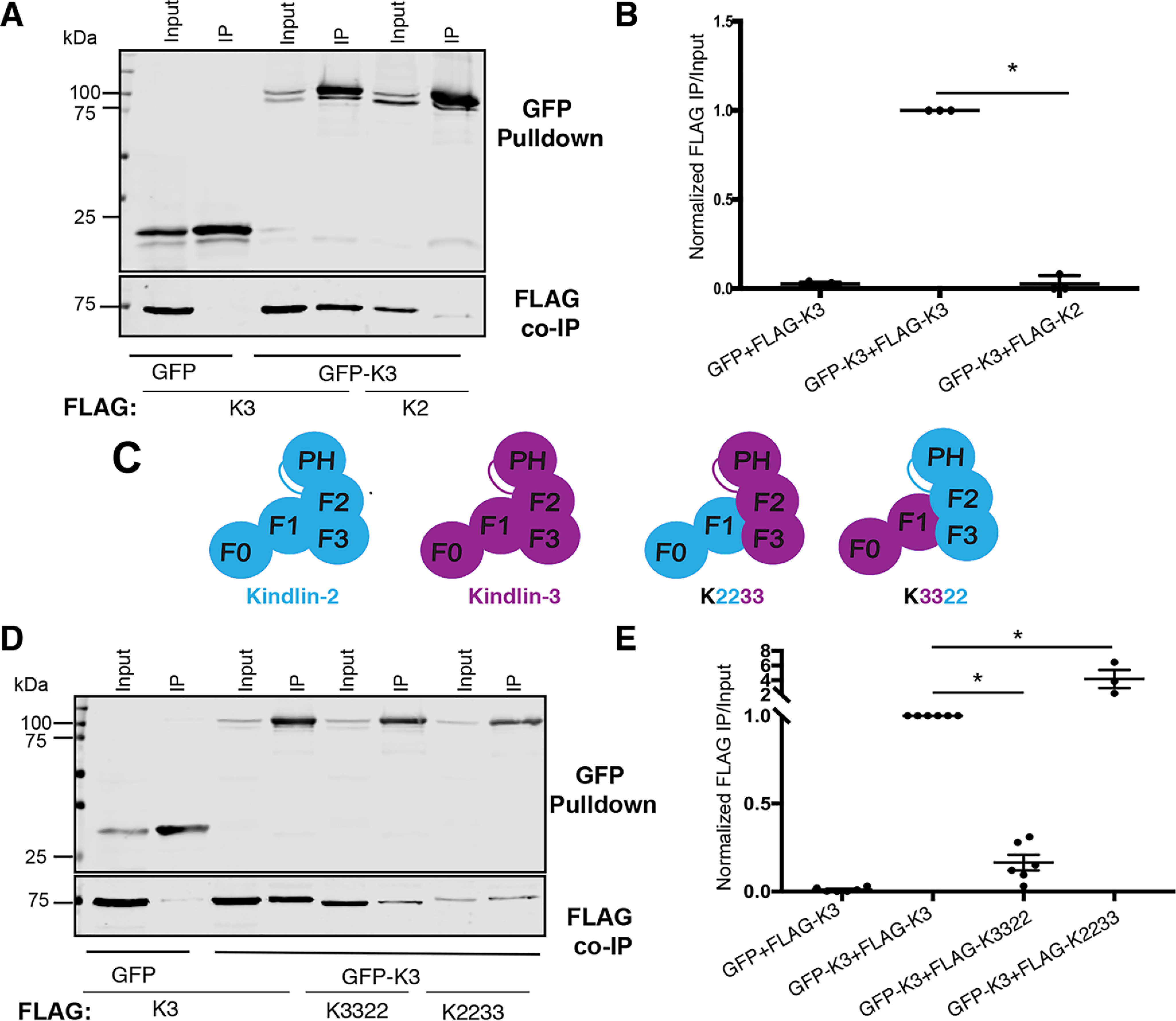
Although the crystal structure of kindlin-2 lacking the PH domain and a portion of the F1 subdomain (kindlin-2ΔPHΔF1loop) suggested that kindlin-2ΔPHΔF1loop dimerizes (14), it was unclear whether full-length kindlin-2 or the related kindlin-3 could self-associate in mammalian cells. To test this, we employed a coimmunoprecipitation (co-IP) assay using GFP-nanotrap beads and coexpressed GFP-tagged and FLAG-tagged kindlins (22, 29). First, to assess the ability of kindlin-2 to self-associate, FLAG-kindlin-2 was coexpressed with either GFP-kindlin-2 or GFP (as a negative control) in HEK293T cells (Fig. 1, A and B). In parallel, kindlin-3 self-association was assessed by coexpressing FLAG-kindlin-3 with either GFP-kindlin-3 or GFP (as a negative control). Cells were lysed and incubated with GFP-nanotrap beads to purify the GFP-tagged protein, and the amount of associating FLAG-tagged protein was assessed by immunoblotting (Fig. 1A). Whereas negligible amounts of FLAG-kindlin-3 copurified with GFP, readily detectable levels of FLAG-kindlin-3 copurified with GFP-kindlin-3 (Fig. 1, B and C). Similarly, FLAG-kindlin-2 copurified with GFP-kindlin-2 but consistently less FLAG-kindlin-2 associated with GFP-kindlin-2 than in the case of kindlin-3 (Fig. 1, B and C). This suggests that both kindlin-2 and kindlin-3 can self-associate in mammalian cells but that kindlin-3 self-associates to a greater extent than kindlin-2 (Fig. 1, B and C). We note that while these experiments clearly show that kindlins can assemble into larger complexes, they cannot provide information on the stoichiometry of the complex or whether kindlin–kindlin interactions are direct.
Figure 1.
kindlin-2 and kindlin-3 each self-associate. A, cartoon schematic illustrating GFP nanobody self-association assay. B and C, self-association of GFP or GFP-kindlin (kindlin-2 [K2] or kindlin-3 [K3]) with FLAG-kindlin was assessed by GFP nanobody pulldown followed by immunoblotting and compared with a 5% input sample (B) and quantified by normalization to GFP-kindlin-3 + FLAG-kindlin-3 (GFP-K3+FLAG-K3) self-association (C); mean ± S.E.; n = 5; *, p ≤ 0.02 (Student's t test).
The kindlin-3 F2PH domain is central to self-association
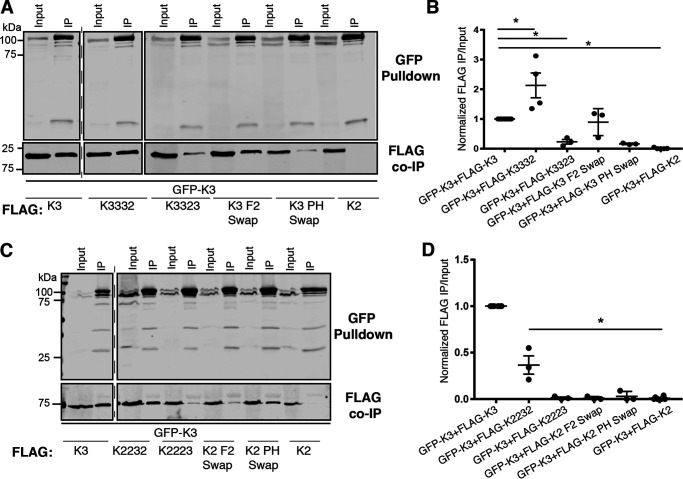
Kindlin-3 shares the same conserved domain architecture and 67% amino acid sequence similarity with kindlin-2 (20, 23). Therefore, we tested whether kindlin-3 could associate with kindlin-2. Notably, despite their similarity, GFP-kindlin-3 could associate with FLAG-kindlin-3 but not with FLAG-kindlin-2 (Fig. 2, A and B). This allowed us to map the subdomains of kindlin-3 that are important for self-association by evaluating the ability of FLAG-kindlin chimeras to restore or disturb association with GFP-kindlin-3. Two sets of chimeric FLAG-kindlins were tested: loss-of-function chimeras consisting of a FLAG-kindlin-3 backbone with inserted subdomain(s) of kindlin-2 intended to disturb association with GFP-kindlin-3 and gain-of-function chimeras composed of a FLAG-kindlin-2 backbone with inserted subdomain(s) of kindlin-3 intended to restore association with GFP-kindlin-3. Chimeric kindlins were named using four characters to denote the F0, F1, F2PH, and F3 regions, respectively, with each number denoting the kindlin isoform from which each subdomain originates (23), and chimeric kindlins were functionally characterized as previously described (23).
Figure 2.
The kindlin-3 C-terminal subdomains are central to self-association. A, association of GFP or GFP-kindlin-3 (GFP-K3) with FLAG-kindlin was assessed by GFP pulldown and immunoblotting and compared with 5% input. B, binding was quantified by normalization to GFP-kindlin-3 + FLAG-kindlin-3 (GFP-K3+FLAG-K3) association; mean ± S.E.; n = 3; *, p ≤ 0.0001 (Student's t test). C, cartoon schematic of kindlin FERM domain architecture and kindlin chimeras. D and E, association of GFP or GFP-kindlin-3 (GFP-K3) with FLAG-kindlin-3 (FLAG-K3) or FLAG-kindlin chimeras was assessed by GFP pulldown (D) and quantified by normalization to GFP-kindlin-3 + FLAG-kindlin-3 association (GFP-K3+FLAG-K3) (E); mean ± S.E.; n ≥ 3; *, p ≤ 0.007 (Student's t test).
We first tested the ability of the FLAG-K2233 and FLAG-K3322 chimeras to associate with GFP-kindlin-3 to pinpoint whether the N- or C-terminal subdomains of kindlin-3 are most important for association with GFP-kindlin-3 (Fig. 2C). In the GFP nanobody co-IP assay, FLAG-K3322 was severely impaired in association with GFP-kindlin-3 (Fig. 2, D and E), suggesting that replacement of the F2PH and F3 subdomains of kindlin-3 is detrimental to the ability of kindlin-3 to associate. In contrast, despite consistently expressing less well than FLAG-kindlin-3, when corrected for input material, FLAG-K2233 was enhanced in association with GFP-kindlin-3 (Fig. 2, D and E). We note that these chimeric kindlins are likely to be correctly folded, as we have previously shown that they bind β1 integrin tails and ILK (23). Furthermore, K3322, which exhibits reduced association with kindlin-3, shows robust targeting to focal adhesions (23). Together, these results suggest that the C-terminal region of kindlin-3 (F2PH and F3 subdomains) is important for association of FLAG-kindlin-3 with GFP-kindlin-3: replacing this region with that from kindlin-2 (K3322) impairs the ability to associate with GFP-kindlin-3, whereas inserting this region into kindlin-2 (K2233) restores and enhances association.
To determine which subdomain(s) in the C-terminal region of kindlin-3 are important for self-association, association between GFP-kindlin-3 and a series of FLAG-tagged loss-of-function chimeras was examined (Fig. 3, A and B). Four FLAG-tagged loss-of-function chimeras were tested: K3332, K3323, kindlin-3 with the F2 subdomain of kindlin-3 (K3 F2 swap), and kindlin-3 with the PH subdomain of kindlin-2 (K3 PH swap). The FLAG-K3332 chimera associated with GFP-kindlin-3; in contrast, FLAG-K3323 was severely impaired in association with GFP-kindlin-3 (Fig. 3, A and B). We consider this unlikely to be because of misfolding, as K3323 can bind β1 integrins and ILK and targets to focal adhesions (23). Swapping only the PH domain (K3 PH swap) also strongly impaired association with GFP-kindlin-3 (Fig. 3, A and B). Like kindlin-3, the PH swap construct fails to target to focal adhesions, but when expressed as a GFP fusion protein it does not form aggregates, suggesting that it is not misfolded (23). Thus, the F2PH subdomain, and particularly the PH domain, is important for the ability of kindlin-3 to self-associate in mammalian cells, as replacement of the kindlin-3 F2PH with the F2PH of kindlin-2 cannot fully support self-association.
Figure 3.
The F2PH subdomain is important for kindlin-3 self-association. A, association of GFP-kindlin-3 (GFP-K3) with FLAG-kindlin chimeras was assessed by GFP pulldown. 5% input was loaded for comparison. B, binding was quantified by normalization to GFP-kindlin-3 + FLAG-kindlin-3 association; mean ± S.E.; n ≥ 3; *, p ≤ 0.009 (Student's t test). C, association of GFP-kindlin-3 (GFP-K3) with FLAG-kindlin was assessed by GFP pulldown. Input lane is 5%. D, binding was quantified by normalization to GFP-kindlin-3 + FLAG-kindlin-3 association (GFP-K3+FLAG-K3); mean ± S.E.; n ≥ 3; *, p ≤ 0.001 (Student's t test).
Further support for the importance of the F2PH subdomain for kindlin-3 self-association was provided when we evaluated association between GFP-kindlin-3 and a series of FLAG-tagged gain-of-function chimeras (Fig. 3, C and D). We observed that FLAG-K2232 associated with GFP-kindlin-3; however, we note that this chimera only partially restored association (Fig. 3, C and D). Meanwhile, FLAG-K2223 did not associate with GFP-kindlin-3 (Fig. 3, C and D). These chimeras bind β1 integrins and ILK, suggesting that they are correctly folded (23). In addition, neither the FLAG K2 F2 swap nor PH swap chimeras associated with GFP-kindlin-3 (Fig. 3, C and D). Although the loss-of-function K3 PH swap chimera suggested that the PH domain of kindlin-3 is required, it is possible that the gain-of-function PH swap chimera does not restore association, because the kindlin-3 F2 also makes interactions or because the kindlin-3 PH domain must be presented in the context of a complete kindlin-3 F2PH. The K2PH swap is likely correctly folded, as we have shown that it targets to focal adhesions at levels comparable with those of kindlin-2 (23).
Notably, in the crystalized domain-swapped kindlin-2ΔPHΔF1loop dimer (14), the F2 subdomain is central to dimerization, and F2 subdomain point mutations that perturb dimerization have been reported, namely, both L327S328 to G327P328 (LS/GP) and A547Q548 to G547P548 (AQ/GP) (14). The LS and AQ motifs are conserved in kindlin-3 (L304S305 and A528Q529), but introduction of the equivalent LS/GP or AQ/GP mutation had no effect on kindlin-3 self-association in mammalian cells, as assessed by our nanobody IP assay (Fig. S1, A and B). Similarly, we could not observe any effect of the LS/GP and AQ/GP mutations on self-association of full-length kindlin-2 in mammalian cells (Fig. S1, C and D). Thus, while we conclude that the kindlin F2PH is important, we suggest that the mode of full-length kindlin self-association in mammalian cells likely differs from that of the kindlin-2ΔPHΔF1loop dimerization observed crystallographically (14).
Loss of the F3 subdomain enhances kindlin self-association
Our mapping of domains important for kindlin self-association consistently highlights an important role for the F2PH subdomain (Fig. 2 and 3). However, the FLAG-K2232 chimera does not fully restore association with GFP-kindlin-3 (Fig. 3D), suggesting that the kindlin-3 F2PH does not completely recapitulate binding when inserted in the context of flanking kindlin-2 subdomains, or that the kindlin-2 F3 subdomain hinders association. The latter idea is supported by our observation that whereas FLAG-K2232 only partially restored association with kindlin-3 (Fig. 3, C and D), the K2233 chimera enhanced association beyond kindlin-3 levels (Fig. 2, D and E).
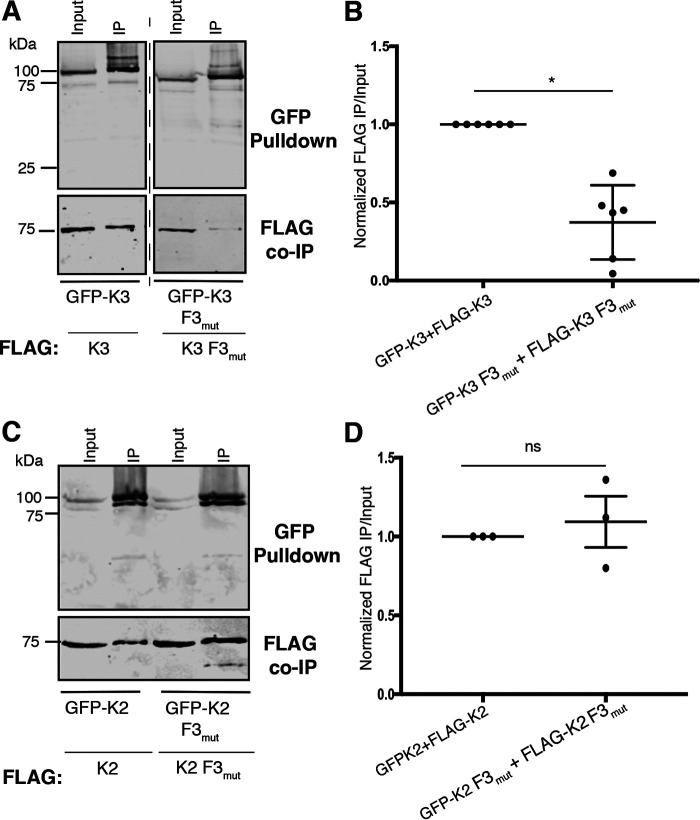
To examine the possibility that the kindlin-2 F3 domain inhibits self-association, we generated a kindlin-2ΔF3 mutant (consisting of kindlin-2 residues 1–564). FLAG-kindlin-2ΔF3 was several hundred-fold enhanced in association with GFP-kindlin-2 compared with FLAG-kindlin-2 (Fig. 4, A and B). To ensure that removal of the F3 subdomain did not alter protein integrity, we assessed the ability of GFP-kindlin-2ΔF3 to bind to the recombinant GST-tagged pseudokinase domain of ILK (GST-ILK-pKD), which our laboratory and others have shown to bind to the kindlin F2PH subdomain (23, 24). Removal of the F3 subdomain did not impair binding to ILK-pKD in complex with CH2 of α-parvin; moreover, binding was specific and was severely impaired for a K423D mutant of the ILK-pKD that we previously identified as perturbing kindlin binding (22) (Fig. 4, C and D). When we assessed the ability of kindlin-3ΔF3 (consisting of kindlin-3 residues 1–550) to self-associate, we found that FLAG-kindlin-3ΔF3 was also markedly enhanced in association with GFP-kindlin-3 (Fig. 4, E and F), suggesting that the kindlin-3 F3 subdomain also is inhibitory to association. Therefore, the F3 subdomains of both kindlin-2 and kindlin-3 are inhibitory to association, as deletion of this subdomain enhances the ability of both kindlin isoforms to self-associate.
Figure 4.
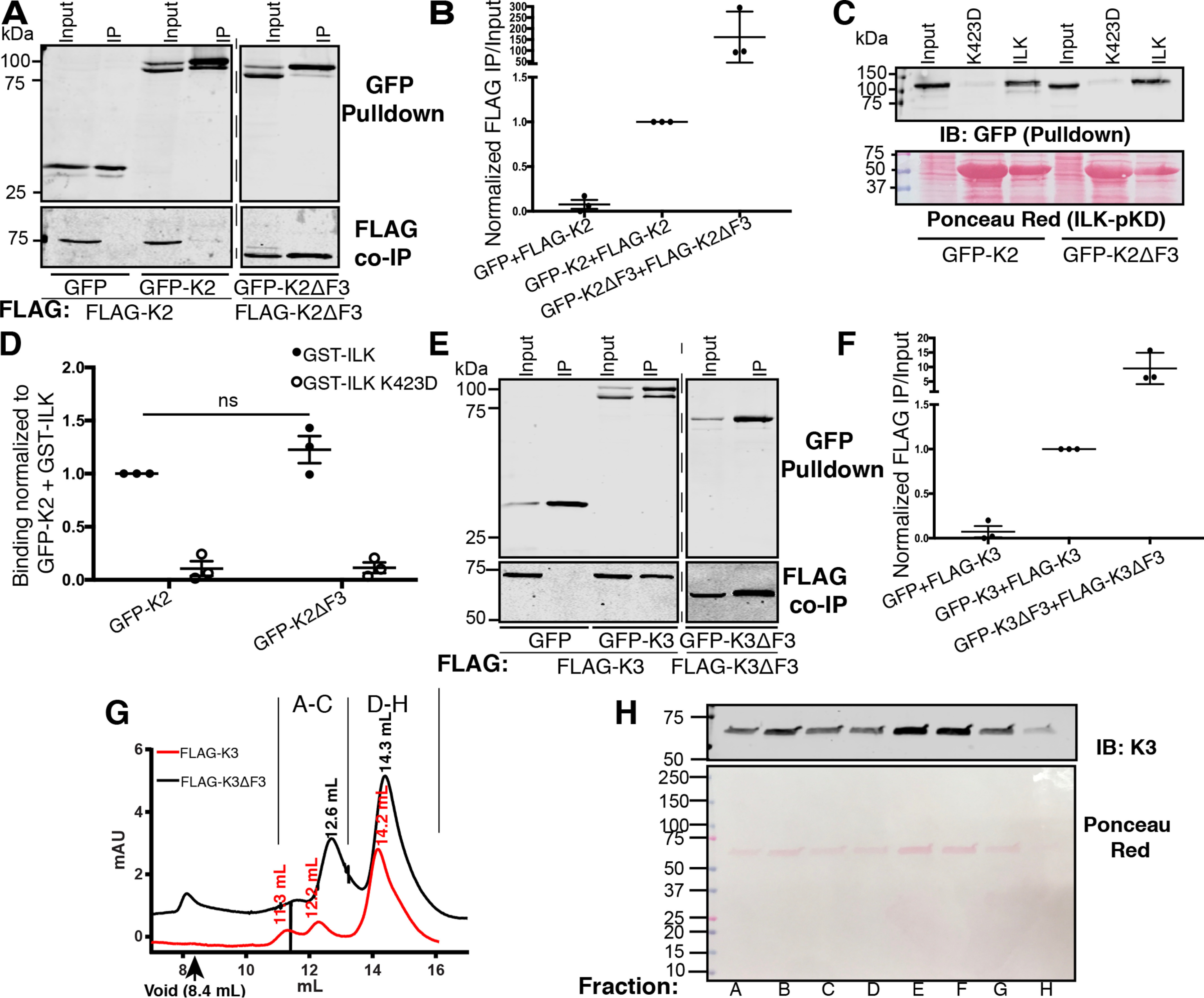
The F3 subdomain inhibits kindlin self-association. A, association of GFP or GFP-kindlin-2 (GFP-K2) with FLAG-kindlin-2 (FLAG-K2) or FLAG-kindlin-2 mutants was assessed by GFP pulldown and immunoblotting. Input lane represents 5% of input. B, binding was quantified by normalization to GFP-kindlin-2 + FLAG-kindlin-2 self-association; mean ± S.E.; n = 3. C and D, pulldown of GFP-kindlin-2 (GFP-K2) or GFP-kindlin-2ΔF3 (GFP-K2ΔF3) by immobilized GST-ILK pKD or GST-ILK K423D pKD (negative control) assessed by immunoblotting relative to a 3% input (A) and quantified by normalization to GFP-kindlin-2 (GFP-K2) + GST-ILK (B). GST-ILK pKD loading was assessed by Ponceau red staining; mean ± S.E.; n = 5; ns, not statistically significant, p > 0.05. E and F, association of GFP or GFP-kindlin-3 (GFP-K3) with FLAG-kindlin or FLAG-kindlin-2 mutants was assessed by GFP pulldown. Input lane represents 5% of input. F, binding was quantified by normalization to GFP-kindlin-3 + FLAG-kindlin-3 association (GFP-K3+FLAG-K3). G, size-exclusion chromatograms for purified FLAG-K3 (red) and FLAG K3ΔF3 (black) with peak retention volumes indicated. H, immunoblot for kindlin-3 (top) and Ponceau red staining (bottom) of selected fractions of FLAG K3ΔF3 as indicated by letters in panel E.
Self-associated kindlin-3 may consist of two to four kindlin molecules
To understand the nature of self-associated kindlin-3 multimers in mammalian cells, we purified FLAG-kindlin-3 from HEK293T cells using anti-FLAG beads to capture the FLAG-tagged protein and recombinant triple-FLAG peptide to elute the bound material. When the purified FLAG-kindlin-3 was subject to analytical size-exclusion chromatography (SEC), most FLAG-kindlin-3 eluted at a retention volume of 14.3 ml (Fig. 4E). Based on fitting against protein standards (Fig. S2, A and B), this corresponds to a species of ∼72 kDa, very close to the expected molecular weight (MW) of the FLAG-kindlin-3 monomer (expected MW, ∼78 kDa) (Fig. 4E). However, we also consistently observed material eluting at lower retention volumes of 12.2 ml and 11.3 ml, corresponding to species of ∼186 and 314 kDa, respectively (Fig. 4G and Fig. S3, A and B). As kindlin-3ΔF3 exhibits increased self-association, we also purified FLAG-kindlin-3ΔF3. Its SEC profile consisted of a major peak at 14.3 ml corresponding to a calculated MW of ∼67 kDa (Fig. 4G and Fig. S3, A and B), presumably representing monomeric FLAG-kindlin-3ΔF3 (expected MW, ∼64 kDa), but it also showed a sizeable fraction of the material eluting at a retention volume of 12.6 ml, which corresponds to a species of ∼160 kDa (Fig. 4G and Fig. S3, A and B). Importantly, based on Ponceau red staining, FLAG-kindlin3ΔF3 was the only detectable protein in the eluted fractions (Fig. 4H), suggesting that the self-association occurs by direct interactions and is not mediated via intermediary proteins. Based on the estimated molecular weights, the observed kindlin multimers may correspond to a complex of two to four kindlin molecules. Therefore, self-associated FLAG-kindlin-3 and self-associated FLAG-kindlin-3ΔF3 may be comprised of two to four FLAG-kindlin molecules.
Point mutations that enhance the ability of kindlin-3 to bind β1 integrin impair self-association
The data we have presented thus far suggest that the F2PH subdomain is important for self-association but that the F3 subdomain modulates this association. As kindlins bind directly to integrin cytoplasmic tails through the F3 subdomain (20, 23), we assessed the impact of a W/A point mutation in F3 that strongly inhibits integrin binding (20, 21) on the ability of kindlin-3 to self-associate. However, the introduction of the W596A mutation into kindlin-3 had no impact on the ability of kindlin-3 to self-associate (Fig. S3, A and B), suggesting that blocking kindlin-3 binding to integrin does not detectably alter self-association.
To test the alternative idea that kindlin-3 self-association alters binding to β1 integrin, we sought to examine the effect of enhancing the ability of kindlin-3 to bind β1 integrin on self-association. This required that we first identify kindlin-3 point mutations that enhance integrin binding. Prior studies from our laboratory have demonstrated that β1 cytoplasmic tails pull down more kindlin-2 than kindlin-3 and that this differential binding is attributed to the F3 subdomains (23). However, the specific residue(s) in the F3 subdomains that account for these differences, and which might be mutated to enhance kindlin-3 binding to β1 integrin, are unknown. To identify these residues, we analyzed the protein sequence of the kindlin-2 and kindlin-3 F3 subdomains across multiple species to identify conserved differences (Fig. 5A). As many of the key integrin-binding residues within the binding cleft are well conserved between kindlin-2 and kindlin-3 across multiple species (21, 23) (Fig. 5A), we included residues throughout the F3 domain. We selected a total of seven residues at three sites: kindlin-3 residue R594, which is K613 in kindlin-2, D601, which is E620 in kindlin-2, and the short stretch 641ERARG645, which is 660AKDQN664 in kindlin-2 (Fig. 5A).
Figure 5.
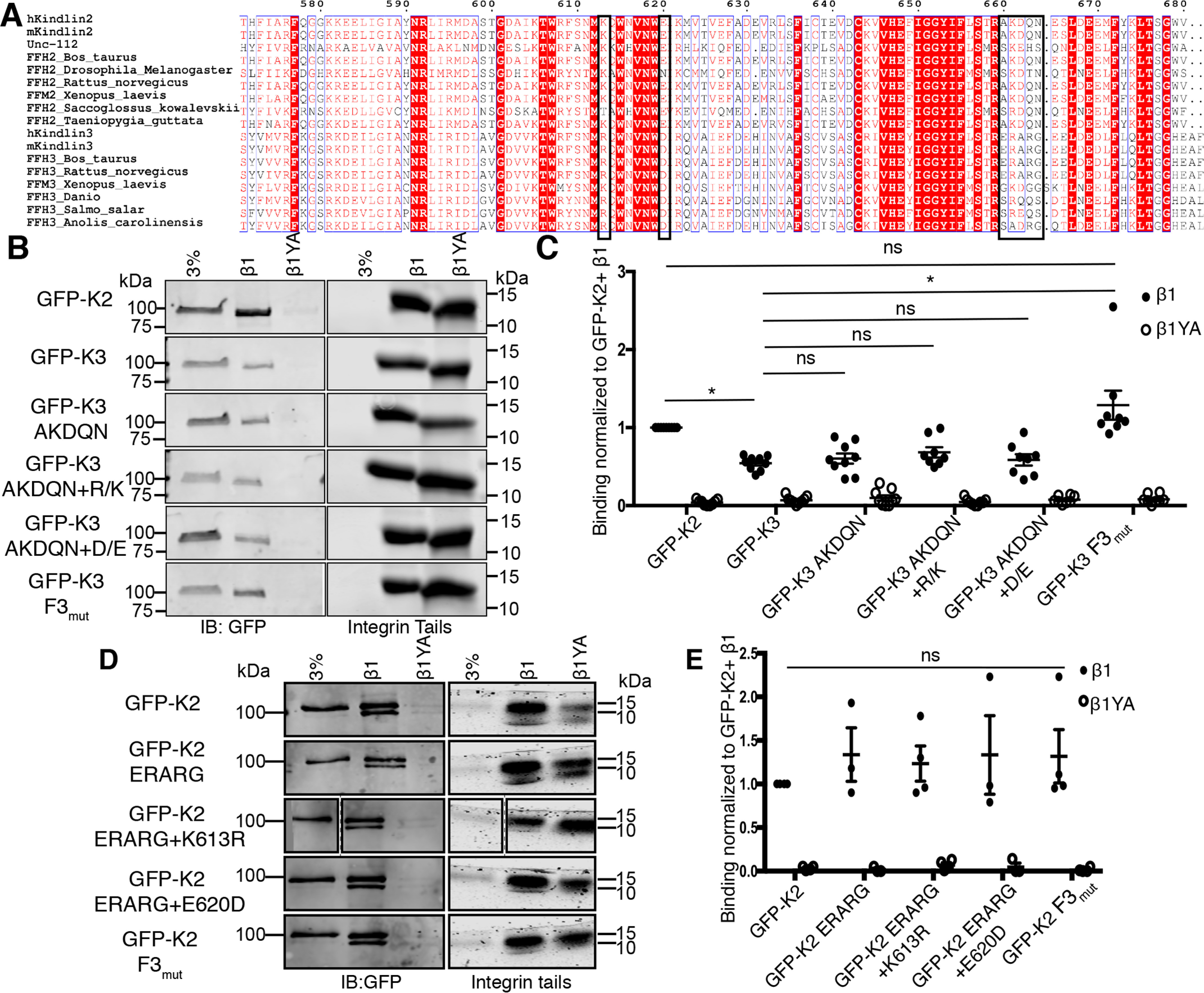
Identification of kindlin-3 mutants with enhanced integrin binding. A, multiple-sequence alignment of kindlin isoforms from different species generated with ESPript (43). Sequences boxed in black represent regions selected for mutagenesis. B and C, pulldown of GFP-kindlins by immobilized β1 or β1YA (negative control) integrin tails was assessed by immunoblotting relative to a 3% input (B) and quantified (C) by normalization to the GFP-kindlin-2 (GFP-K2) + β1 condition; mean ± S.E.; n ≥ 3; *, p ≤ 0.0008 (Student's t test) or ns (not statistically significant, p > 0.05). Integrin tail loading was assessed by Coomassie blue staining. D and E, pulldown of GFP-kindlins by immobilized β1 or β1YA (negative control) integrin tails assessed by immunoblotting relative to a 3% input (D) and quantified (E) by normalization to the GFP-kindlin-2 (GFP-K2) + β1 condition; mean ± S.E.; n ≥ 3; ns, not statistically significant, p > 0.05. Integrin tail loading was assessed by Coomassie blue staining.
We generated individual or compound GFP-kindlin-3 mutations, switching these kindlin-3 residues to the corresponding kindlin-2 residues at each of the sites and assessed binding to integrin β1 tails. As shown in Fig. 5, B and C, pulldown assays with immobilized recombinant β1 integrin tails confirmed that β1 tails pull down approximately twice as much GFP-kindlin-2 as GFP-kindlin-3, and that binding was specific, as it was abrogated by the previously characterized Y795A (Y/A) mutation in the β1 tail (14, 20, 23). Analysis of kindlin-3 mutants showed that only the compound mutations of all three sites (GFP-K3 F3mutx3) enhanced kindlin-3 binding to β1 integrin tails to the level of kindlin-2 (Fig. 5, B and C). Binding remained specific, as the mutant kindlin-3 did not bind to β1 Y/A mutant tails (Fig. 5, B and C).
Our results show that binding of kindlin-3 to β1 integrin can be enhanced by the mutation of seven amino acids in the F3 subdomain to the corresponding residues found in kindlin-2. Surprisingly, when we mutate the corresponding residues in kindlin-2 to those found in kindlin-3, we observed no significant impact on binding to integrin β1 tails (Fig. 5, D and E). This, together with analysis indicating that many of these residues are unlikely to make direct contacts with the integrin tail, raises the possibility that the enhanced ability of GFP-kindlin-3 F3mutx3 to bind to β1 tails in pulldowns is an indirect effect.
To understand the nature of potential indirect effects, we evaluated the ability of kindlin-3 F3mutx3 to self-associate. Compared with kindlin-3, kindlin-3 F3mutx3 was impaired in self-association (Fig. 6, A and B). In contrast, the analogous kindlin-2 compound mutant, kindlin-2 F3mutx3, which bound at wild type levels to β1 integrin tails, self-associated similarly to wild-type kindlin-2 (Fig. 6, C and D). This correlation between impaired self-association and enhanced integrin binding is consistent with the notion that enhanced self-association impairs binding to β1 integrin tails.
Figure 6.
A kindlin-3 mutant with enhanced integrin binding is impaired in self-association. A and B, association of GFP-kindlin-3 (GFP-K3) or GFP-kindlin-3 F3mutx3 (GFP-K3 F3mutx3) with FLAG-kindlin was assessed by GFP pulldown and immunoblotting (A) and quantified by normalization to GFP-kindlin-3 + FLAG-kindlin-3 self-association (GFP-K3+FLAG-K3) (B); mean ± S.E.; n = 6. *, p ≤ 0.03 (Student's t test). C, association of GFP-kindlin-2 (GFP-K2) or GFP-kindlin-2 F3mutx3 (GFP-K2 F3mutx3) with FLAG-kindlin was assessed and compared with a 5% input. D, binding was quantified and normalized to GFP-kindlin-3 + FLAG-kindlin-3 self-association (GFP-K3+FLAG-K3); mean ± S.E.; n = 3; ns, not statistically significant, p > 0.05.
Mutations that impair kindlin-3 self-association and enhance β1 integrin binding facilitate localization of kindlin-3 to focal adhesions
When expressed in adherent cells, GFP-kindlin-2 robustly targets to focal adhesions, but GFP-kindlin-3 is not observed in focal adhesions; this differential localization has been attributed to differences in both ILK and integrin binding (23). Because GFP-kindlin-3 F3mutx3 binds to β1 integrin at kindlin-2 levels in pulldown assays, we examined the ability of GFP-kindlin-3 F3mutx3 to localize to focal adhesions in live CHO cells stably expressing mCherry-Paxillin as a marker for focal adhesions (Fig. 7). Twenty-four hours after plating on fibronectin-coated glass, GFP-kindlin-2 localized to mCherry-Paxillin containing focal adhesions by both total internal reflection fluorescence (TIRF) and epifluorescence microscopy, while GFP-kindlin-3 localization was very similar to that of GFP alone and could not be reliably detected in focal adhesions in any of the cells visualized (Fig. 7). On the contrary, GFP-kindlin-3 F3mutx3 was observed localizing to focal adhesions by TIRF microscopy, although focal adhesion localization of this mutant was not as clearly visualized as the GFP-kindlin-2 condition (Fig. 7). This suggests that impairing self-association and increasing binding to β1 integrin permits localization to focal adhesions.
Figure 7.
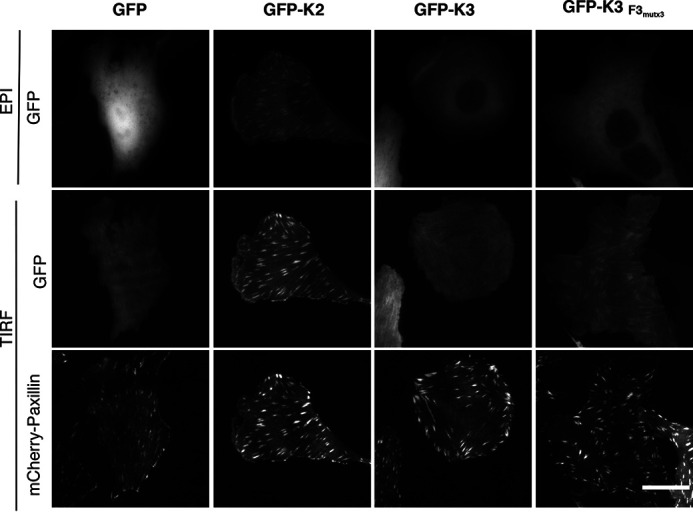
A kindlin-3 mutant with impaired self-association and enhanced integrin binding targets to focal adhesions. CHO cells stably expressing mCherry-paxillin were transiently transfected with GFP, GFP-kindlin-2 or GFP-kindlin-3 (GFP-K2 or GFP-K3), or GFP-kindlin-3 F3mutx3 (GFP-K3 F3mutx3). 18 h after replating on fibronectin-coated glass-bottom dishes, live cells were imaged by epifluorescence (EPI) and/or TIRF microscopy as indicated. Images in each channel were linearly, uniformly adjusted, and cropped for clarity. Scale bar = 20 μm.
Discussion
Kindlins are essential for normal integrin-mediated cell adhesion and signaling (1, 18). They are believed to act primarily as scaffold proteins, binding directly to integrin β subunit cytoplasmic tails and interacting with other focal adhesion and cytoskeletal proteins, such as ILK (22, 23), paxillin (30–33), and F-actin (34). Here, we show that, in mammalian cells, kindlins can also self-associate with other kindlin molecules, forming dimers or higher-order assemblies. We find that whereas both kindlin-2 and kindlin-3 can each self-associate, kindlin-3 self-associates to a greater extent, and we use this information to identify a key role for the kindlin F2PH subdomain in kindlin self-association. We further propose that the integrin-binding F3 subdomain inhibits self-association. Notably, mutations in the F3 subdomain of kindlin-3 that impair self-association of kindlin-3 enhance binding to β1 integrin cytoplasmic tails and permit localization of kindlin-3 to focal adhesions, suggesting that kindlin dimerization or oligomerization regulates integrin binding and localization.
An association between kindlin domains was first proposed for N- and C-terminal portions of C. elegans kindlin (UNC-112), but this was envisaged to be an intramolecular interaction involved in conformational regulation of kindlin interactions with ILK (PAT-4) and integrin β1 (PAT-3) (35). The first report that kindlins can self-associate (intermolecular interaction) was based on the crystal structure of a mouse kindlin-2 construct lacking the PH domain and a portion of the F1 subdomain (kindlin-2ΔPHΔF1loop) (14), which revealed an F2 subdomain-swapped dimer. The primary sequence of the kindlin F2 domain is interrupted by the PH domain, resulting in the PH domain likely being inserted into a split F2 domain, and in the crystalized dimer the F2 subdomain is assembled from two separate kindlin molecules. It was reported that purified kindlin-2ΔPHΔF1loop can dimerize in solution, albeit on a timescale of days (14), and that mutations near the observed dimerization interface could inhibit dimerization (14); whether or not this form of dimer occurs in vivo is currently unclear. However, in our assays in mammalian cells with full-length kindlins, these mutations did not impair self-association. Thus, the mode of kindlin self-association that we report with full-length kindlins in mammalian cells likely differs from that reported in the crystal structure. The different mode of self-association is consistent with our finding that the PH domain, which is missing in kindlin-2ΔPHΔF1loop, is important for kindlin self-association.
In the absence of structures of self-associated full-length kindlin dimers, trimers, or tetramers, it is difficult to predict the detailed molecular mechanism of kindlin self-association. However, our observation that deletion of the F3 subdomain enhances self-association suggests that the F3 subdomain inhibits kindlin self-association. Of note, it was recently reported that the kindlin-3–leupaxin interaction is sterically occluded by the F3 subdomain of kindlin-3, as kindlin-3ΔF3 is enhanced in leupaxin binding (33). It is possible that the F3 subdomains of kindlin-2 and kindlin-3 also sterically occlude self-association. Confirmation of this hypothesis and an understanding of how structural differences between the F3 subdomains between kindlin-2 and kindlin-3 contribute to differential self-association between the two kindlin isoforms will require high-resolution structural information.
Prior studies have demonstrated that β1 integrin tails bind kindlin-2 better than kindlin-3 in pulldown assays and that kindlin-2, but not kindlin-3, localizes to focal adhesions (23, 28). We find that kindlin-3 self-associates to a greater extent than kindlin-2, raising the possibility that kindlin-self-association is inversely correlated with integrin binding and focal adhesion localization. However, we were unable to test the consequences of increased self-association in the F3 deleted kindlins, as the F3 subdomain contains the integrin binding site (14). Instead, we turned to kindlins containing point mutations in the F3 subdomain. By performing comparative sequence analysis of the kindlin-2 and kindlin-3 F3 subdomains and testing a series of compound mutants, we identified a kindlin-3 variant with 7 substitutions in the F3 domain that resulted in enhanced binding to β1 integrin tails. Although high-resolution structural information on the kindlin-3–β1 integrin interaction is currently unavailable, the high sequence similarity between the F3 subdomains, especially among integrin-binding residues (14, 20, 21), suggests that the F3 subdomain domain architecture is highly conserved between kindlin-2 and kindlin-3. In the structure of the β1 tail bound to the F3 subdomain of kindlin-2ΔPHΔF1loop (14), kindlin-2 residues D620 and K613, which correspond to D601 and R594 in kindlin-3, are located at either end of the integrin-binding groove. A direct polar contact between the main chain of K613 and N792 in the β1 tail is visible in the structure, and K613 also packs against Y795 in the β1 tail, but no contacts are visible between D620 and the β1 tail, because the N-terminal boundary of the β1 tail fusion construct does not extend far enough. The 660AKDQN664 motif corresponding to 641ERARG645 in kindlin-3 lies in the linker between α1F3 and α2F3, although only A660 and K661 are visible in the structure. Taken together, it is not immediately clear how to rationalize the increased binding of the compound kindlin-3 mutant to integrin tails, raising the possibility that the effect is indirect. Consistent with this idea, we found that kindlin-3 F3mutx3 is impaired in self-association. This suggests that it is the impairment in self-association that enhances integrin binding and increases targeting to focal adhesions of the compound kindlin-3 mutant. Its focal adhesion targeting does not appear as robust as that for kindlin-2, presumably because its binding to ILK, which is mediated by a linker region in the F2PH domain and also contributes to differences in focal adhesion localization between kindlin isoforms, is weak (23). Notably, corresponding substitutions in kindlin-2 did not alter integrin binding or self-association, possibly because self-association is already weak in kindlin-2 and any further disruption has no measurable impact on integrin binding.
We propose that the inverse correlation between kindlin self-association and integrin binding implies that kindlin self-association impairs integrin binding. The alternative possibility, that integrin binding impairs self-association, is less likely, as a W/A mutation in kindlin-3, which has been previously shown to specifically impair the interaction between kindlin and integrin (20, 21), had no effect on self-association. Taken together, our data suggest that self-association is a regulatory mechanism controlling kindlin. If so, it will be important to determine the molecular mechanisms modulating kindlin self-association (e.g. posttranslational modifications, binding proteins, or lipids) and their consequences on other kindlin interactions. However, we note that our studies were conducted in cells that lack endogenous kindlin-3. Therefore, future studies should focus on establishing the functional roles of kindlin-3 self-association in hematopoietic cells, endothelial cells, and breast cancer cells that express endogenous kindlin-3 (36–38). In light of our findings that kindlin self-association inversely correlates with integrin binding, it will be important to test the consequences of selectively modulating kindlin-3 self-association on cell adhesion and spreading and integrin activation and signaling in these cells.
Experimental procedures
Antibodies
Primary antibodies against GFP (Rockland, catalog no. 601-101-215; Limerick, PA, USA), FLAG (Sigma, catalog no. F1804; St. Louis, MO, USA), as well as IRDye-conjugated secondary antibodies (Li-Cor; Lincoln, NE, USA), were purchased from commercial sources.
Constructs
Vectors encoding N-terminally GFP-tagged human kindlin-2, kindlin-3, GFP alone, and FLAG-tagged human kindlin-2 and kindlin-3 were as previously described (22, 23, 39). Chimeras were generated as previously described (23). Point mutations were generated by QuikChange site-directed mutagenesis by following the manufacturer's instructions (Stratagene, La Jolla, CA, USA).
Cell culture and transfection
HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 9% fetal bovine serum (FBS), sodium pyruvate, and nonessential amino acids (NEAA) (Gibco Laboratories; Gaithersburg, MD, USA) at 37°C in 5% CO2 and were obtained from colleagues at Yale University. CHO cells, which have been engineered to stably express αIIbβ3 and were described previously (40), were cultured in DMEM with 9% FBS, sodium pyruvate, and NEAA at 37°C in 5% CO2. HEK293T and CHO cells were transiently transfected with PEI (linear polyethylenimine; MW, 25,000) (Polysciences, Inc., Warrington, PA, USA). Cells were regularly tested for mycoplasma using the MycoAlert mycoplasma detection kit (Lonza, Basel, Switzerland).
GFP-nanotrap purification
GFP binding protein (GFP nanotrap), derived from a llama single-chain antibody (29), was produced in BL21 RIPL competent Escherichia coli (Millipore Sigma, St. Louis, MO, USA) as described previously (22). To summarize, bacterial pellets were lysed, and the His-tagged GFP nanotrap was purified using Ni-NTA affinity chromatography. Bound protein was eluted with imidazole, fractionated by size exclusion chromatography, and covalently coupled to NHS-activated FastFlow Sepharose (GE Healthcare, Chicago, IL USA). Beads were stored in a 50% slurry in PBS with 0.04% NaN3 and cOmplete EDTA-free protease inhibitor (Roche, Indianapolis, IN, USA) at 4°C.
GFP-nanotrap association assays
GFP- and FLAG-tagged proteins were coexpressed in HEK293T cells by transient transfection with PEI. Approximately 24 h following transfection, cells were lysed in buffer X lysis buffer (1 mm NaVO4, 50 mm NaF, 40 mm sodium pyrophosphate, 50 mm NaCl, 150 mm sucrose, 10 mm Pipes, pH 6.8) containing 0.5% Triton X-100, 0.2% deoxycholic acid, and cOmplete EDTA-free protease inhibitor mixture (Roche, Indianapolis, IN, USA) for 15 min at 4°C. After clarification by centrifugation, lysate was incubated with GFP-nanotrap beads with rocking for 2 h at 4°C. Beads were then washed three times with buffer X-T, and bound proteins were fractionated by reducing SDS-PAGE and analyzed by immunoblotting. Immunoblots were imaged on an Odyssey IR imaging system (Li-Cor, Lincoln, NE, USA) and analyzed using Image Studio Lite (Li-Cor, Lincoln, NE, USA). Ponceau stains were imaged using a 12-megapixel camera (Apple, Cupertino, CA, USA). For quantification of binding from immunoblots, the fluorescence intensity of the band corresponding to immunoprecipitated (IP) material was quantified as a fraction of the fluorescence of the 5% input material band for each condition. The binding of the internal positive control (e.g. GFP-kindlin-3 coassociation with FLAG-kindlin-3) was set to 1, and the binding under all other conditions was expressed relatively within each experiment. A GFP-kindlin/FLAG-kindlin wild type reference control was included in each experiment, where multiple constructs were tested at once.
Purification of FLAG-kindlin from HEK293T cells
FLAG-tagged kindlins were expressed in HEK293T cells by transient transfection with PEI. Approximately 24 h following transfection, cells were lysed in buffer X-G lysis buffer containing buffer X-G (1 mm NaVO4, 50 mm NaF, 40 mm sodium pyrophosphate, 50 mm NaCl, 2.5% glycerol, 10 mm Pipes, pH 6.8) containing 0.5% Triton X-100, 0.2% deoxycholic acid, and cOmplete EDTA-free protease inhibitor mixture (Roche, Indianapolis, IN, USA) for 30 min at 4°C. After clarification by centrifugation, cell lysate was incubated with anti-FLAG M2 affinity gel equilibrated with lysis buffer (Sigma, St. Louis, MO, USA) with rotation for 2 h at 4°C. Anti-FLAG beads were washed on-column with lysis buffer and then with buffer X-G. Captured protein was eluted with buffer X-G supplemented with 150 ng/μl 3× FLAG-peptide (Sigma, St. Louis, MO, USA) and concentrated using Amicon Ultra centrifugal filter units (Sigma, St. Louis, MO, USA). The concentrated material was clarified by centrifugation and analyzed on a HiLoad Superdex S200 analytical-grade size-exclusion chromatography column (GE Healthcare, Chicago, IL, USA).
Recombinant ILK-pKD production and purification
GST-ILK-pKD in complex with His-flag-α-parvin-CH2 was produced in BL21(De3) E. coli cells (Millipore Sigma, St. Louis, MO, USA) as previously described (22, 23). The complex was copurified by GSH affinity chromatography using GSH-Sepharose 4b (GE healthcare; Chicago, IL, USA).
GST-ILK pulldown binding experiments
Pulldown experiments from cell lysate performed with GST-ILK were as previously described (22, 23). GFP-kindlin was overexpressed in HEK293T or CHO cells by transient transfection with PEI. Cells were lysed in buffer X (1 mm NaVO4, 50 mm NaF, 40 mm sodium pyrophosphate, 50 mm NaCl, 150 mm sucrose, 10 mm Pipes, pH 6.8) containing 0.5% Triton X-100, 0.2% deoxycholic acid, and cOmplete EDTA-free protease inhibitor mixture (Roche, Indianapolis, IN, USA) for 15 min at 4°C. After clarification by centrifugation, lysate was diluted in buffer X-T (buffer X with 0.05% Triton X-100) and incubated with GST-ILK coupled to GSH-Sepharose 4b. Incubations were performed for 2 h with rocking at 4°C. Beads were then washed three times with buffer X-T, and bound proteins were fractionated by reducing SDS-PAGE and analyzed by immunoblotting. Immunoblots were imaged on an Odyssey IR imaging system (Li-Cor, Lincoln, NE, USA) and analyzed using Image Studio Lite (Li-Cor, Lincoln, NE, USA). Ponceau stains were imaged using a 12-megapixel cellular phone camera (Apple, Cupertino, CA, USA). For quantification of binding from immunoblots, the fluorescence intensity of the band corresponding to bound material was quantified as a fraction of the fluorescence of the 3% input material band for each condition. The binding of the internal positive control (e.g. GFP-kindlin-2 binding to GST-ILK) was set to 1, and the binding under all other conditions was expressed relatively within each experiment. Bead loading was verified by staining the nitrocellulose membrane with Ponceau S.
Integrin binding assays
For pulldown experiments from cell lysate performed with recombinant integrin tails, GFP-kindlin was overexpressed in HEK293T or CHO cells by transient transfection with PEI. Cells were lysed in buffer X (1 mm NaVO4, 50 mm NaF, 40 mm sodium pyrophosphate, 50 mm NaCl, 150 mm sucrose, 10 mm Pipes, pH 6.8) containing 0.5% Triton X-100, 0.2% deoxycholic acid, and cOmplete EDTA-free protease inhibitor mixture (Roche, Indianapolis, IN, USA) for 15 min at 4°C. After clarification by centrifugation, lysate was diluted in buffer X-T (buffer X with 0.05% Triton X-100) and incubated with His-tagged integrin tails coupled to Ni-NTA beads as previously described (20, 41). Incubations were performed for 2 h with rocking at 4°C. Beads were then washed three times with buffer X-T, and bound proteins were fractionated by reducing SDS-PAGE and analyzed by immunoblotting. Immunoblots were imaged on an Odyssey IR imaging system (Li-Cor; Lincoln, NE, USA) and analyzed using Image Studio Lite (Li-Cor; Lincoln, NE, USA). For quantification of binding from immunoblots, the fluorescence intensity of the band corresponding to bound material was quantified as a fraction of the fluorescence of the 3% input material band for each condition. The binding of the internal positive control (e.g. GFP-kindlin-2 binding to β1) was set to 1, and the binding under all other conditions was expressed relatively. Bead loading was verified by staining the relevant section of the SDS-PAGE gel with Coomassie brilliant blue.
TIRF microscopy
Live-cell imaging by TIRF microscopy was performed as previously described (22, 42). 35-mm glass-bottom microwell dishes with a 14-mm microwell diameter (MatTek Corporation, Ashland, MA, USA) were coated with 5 µg·ml−1 bovine plasma fibronectin (Sigma, St. Louis, MO, USA) for 18 h at 37°C. Cells (CHO cells stably expressing mCherry-paxillin and transiently transfected with GFP–kindlin-2 or GFP-2 mutant) were plated on the coated glass-bottom dishes in DMEM containing no glutamine and no phenol red (Gibco Laboratories, Gaithersburg, MD, USA) supplemented with 9% FBS, sodium pyruvate, NEAA, and GlutaMAX supplement (Gibco Laboratories, Gaithersburg, MD, USA). 24 h later, cells were imaged live in a temperature- and CO2-controlled environment chamber (OkoLab, Burlingame, CA, USA) mounted onto a Nikon Ti-2 Eclipse microscope (Nikon, Tokyo, Japan, USA) equipped with a motorized Ti-LA-HTIRF module with LUN4 488- and 561-nm lasers (15 mW), using a CFI Plan Apo Lambda 100× oil TIRF objective and a Prime95B RoHS cMOS camera (pixel size, 110 nm) (Photometrics, Tuscon, AZ, USA). Images were acquired and processed with the NIS-Elements AR software and ImageJ.
Statistics
Statistical tests to calculate p values were performed using Prism software. Two-tailed Student's t tests were performed as indicated in the figure legends.
Data availability
Data described in this manuscript are all contained within the manuscript. Should further information be required, please contact the corresponding author.
Supplementary Material
Acknowledgments
We thank members of the Calderwood laboratory for helpful discussions and insights.
This article contains supporting information.
Author contributions—Y. A. K. and D. A. C. conceptualization; Y. A. K., E. M. M., and C. H.-C. investigation; Y. A. K. writing-original draft; Y. A. K., C. H.-C., and D. A. C. writing-review and editing; D. A. C. supervision; D. A. C. funding acquisition; D. A. C. project administration.
Funding and additional information—This work was supported by the National Institutes of Health grants R01-NS093704 (to D. A. C.). Y. A. K. was supported by a National Science Foundation Graduate Research Fellowship (DGE1122492). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- FERM
- 4.1-ezrin-radixin-moesin
- PH
- pleckstrin homology
- ILK
- integrin-linked kinase
- co-IP
- coimmunoprecipitation
- SEC
- size-exclusion chromatography
- MW
- molecular weight
- TIRF
- total internal reflection fluorescence
- NEAA
- nonessential amino acids.
References
- 1. Montanez E., Ussar S., Schifferer M., Bosl M., Zent R., Moser M., and Fassler R. (2008) Kindlin-2 controls bidirectional signaling of integrins. Genes Dev. 22, 1325–1330 10.1101/gad.469408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moser M., Nieswandt B., Ussar S., Pozgajova M., and Fässler R. (2008) Kindlin-3 is essential for integrin activation and platelet aggregation. Nat. Med. 14, 325–330 10.1038/nm1722 [DOI] [PubMed] [Google Scholar]
- 3. Ussar S., Moser M., Widmaier M., Rognoni E., Harrer C., Genzel-Boroviczeny O., and Fässler R. (2008) Loss of kindlin-1 causes skin atrophy and lethal neonatal intestinal epithelial dysfunction. PLoS Genet. 4, e1000289 10.1371/journal.pgen.1000289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Catterson J. H., Heck M. M. S., and Hartley P. S. (2013) Fermitins, the orthologs of mammalian kindlins, regulate the development of a functional cardiac syncytium in Drosophila melanogaster. PLoS ONE 8, e62958 10.1371/journal.pone.0062958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bai J., Binari R., Ni J. Q., Vijayakanthan M., Li H. S., and Perrimon N. (2008) RNA interference screening in Drosophila primary cells for genes involved in muscle assembly and maintenance. Development 135, 1439–1449 10.1242/dev.012849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rogalski T. M., Mullen G. P., Gilbert M. M., Williams B. D., and Moerman D. G. (2000) The UNC-112 gene in Caenorhabditis elegans encodes a novel component of cell-matrix adhesion structures required for integrin localization in the muscle cell membrane. J. Cell Biol. 150, 253–264 10.1083/jcb.150.1.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Siegel D. H., Ashton G. H. S., Penagos H. G., Lee J. V., Feiler H. S., Wilhelmsen K. C., South A. P., Smith F. J. D., Prescott A. R., Wessagowit V., Oyama N., Akiyama M., Al Aboud D., Al Aboud K., Al Githami A., et al. (2003) Loss of kindlin-1, a human homolog of the Caenorhabditis elegans actin-extracellular-matrix linker protein UNC-112, causes Kindler syndrome. Am. J. Hum. Genet. 73, 174–187 10.1086/376609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mory A., Feigelson S. W., Yarali N., Kilic S. S., Bayhan G. I., Gershoni-Baruch R., Etzioni A., and Alon R. (2008) Kindlin-3: a new gene involved in the pathogenesis of LAD-III. Blood 112, 2591 10.1182/blood-2008-06-163162 [DOI] [PubMed] [Google Scholar]
- 9. Malinin N. L., Zhang L., Choi J., Ciocea A., Razorenova O., Ma Y.-Q., Podrez E. A., Tosi M., Lennon D. P., Caplan A. I., Shurin S. B., Plow E. F., and Byzova T. V. (2009) A point mutation in KINDLIN3 ablates activation of three integrin subfamilies in humans. Nat. Med. 15, 313–318 10.1038/nm.1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao T., Guan L., Yu Y., Pei X., Zhan J., Han L., Tang Y., Li F., Fang W., and Zhang H. (2013) Kindlin-2 promotes genome instability in breast cancer cells. Cancer Lett. 330, 208–216 10.1016/j.canlet.2012.11.043 [DOI] [PubMed] [Google Scholar]
- 11. Shen Z., Ye Y., Kauttu T., Seppänen H., Vainionpää S., Wang S., Mustonen H., and Puolakkainen P. (2013) Novel focal adhesion protein kindlin-2 promotes the invasion of gastric cancer cells through phosphorylation of integrin β1 and β3. J. Surg. Oncol. 108, 106–112 10.1002/jso.23353 [DOI] [PubMed] [Google Scholar]
- 12. Kadry Y. A., and Calderwood D. A. (2020) Chapter 22: Structural and signaling functions of integrins. Biochim. Biophys. Acta 1862, 183206 10.1016/j.bbamem.2020.183206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Calderwood D. A., Campbell I. D., and Critchley D. R. (2013) Talins and kindlins: Partners in integrin-mediated adhesion. Nat. Rev. Mol. Cell Biol. 14, 503–517 10.1038/nrm3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li H., Deng Y., Sun K., Yang H., Liu J., Wang M., Zhang Z., Lin J., Wu C., Wei Z., and Yu C. (2017) Structural basis of kindlin-mediated integrin recognition and activation. Proc. Natl. Acad. Sci. U S A 114, 9349–9354 10.1073/pnas.1703064114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meves A., Stremmel C., Gottschalk K., and Fässler R. (2009) The Kindlin protein family: new members to the club of focal adhesion proteins. Trends Cell Biol. 19, 504–513 10.1016/j.tcb.2009.07.006 [DOI] [PubMed] [Google Scholar]
- 16. Bouaouina M., Goult B. T., Huet-Calderwood C., Bate N., Brahme N. N., Barsukov I. L., Critchley D. R., and Calderwood D. A. (2012) A conserved lipid-binding loop in the kindlin FERM F1 domain is required for kindlin-mediated αIIbβ3 integrin coactivation. J. Biol. Chem. 287, 6979–6990 10.1074/jbc.M111.330845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yates L. A., Füzéry A. K., Bonet R., Campbell I. D., and Gilbert R. J. C. (2012) Biophysical analysis of kindlin-3 reveals an elongated conformation and maps integrin binding to the membrane-distal β-subunit NPXY motif. J. Biol. Chem. 287, 37715–37731 10.1074/jbc.M112.415208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rognoni E., Ruppert R., and Fassler R. (2016) The kindlin family: functions, signaling properties and implications for human disease. J. Cell Sci. 129, 17–27 10.1242/jcs.161190 [DOI] [PubMed] [Google Scholar]
- 19. Dong J. M., Tay F. P. L., Swa H. L. F., Gunaratne J., Leung T., Burke B., and Manser E. (2016) Proximity biotinylation provides insight into the molecular composition of focal adhesions at the nanometer scale. Sci. Signal. 9, 1–14 10.1126/scisignal.aaf3572 [DOI] [PubMed] [Google Scholar]
- 20. Harburger D. S., Bouaouina M., and Calderwood D. A. (2009) Kindlin-1 and -2 directly bind the C-terminal region of β integrin cytoplasmic tails and exert integrin-specific activation effects. J. Biol. Chem. 284, 11485–11497 10.1074/jbc.M809233200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma Y. Q., Qin J., Wu C., and Plow E. F. (2008) Kindlin-2 (Mig-2): a co-activator of β3 integrins. J. Cell Biol. 181, 439–446 10.1083/jcb.200710196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kadry Y. A., Huet-Calderwood C., Simon B., and Calderwood D. A. (2018) Kindlin-2 interacts with a highly conserved surface of ILK to regulate focal adhesion localization and cell spreading. J. Cell Sci. 131, jcs221184 10.1242/jcs.221184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huet-Calderwood C., Brahme N. N., Kumar N., Stiegler A. L., Raghavan S., Boggon T. J., and Calderwood D. A. (2014) Differences in binding to the ILK complex determines kindlin isoform adhesion localization and integrin activation. J. Cell Sci. 127, 4308–4321 10.1242/jcs.155879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fukuda K., Bledzka K., Yang J., Perera H. D., Plow E. F., and Qin J. (2014) Molecular basis of kindlin-2 binding to integrin-linked kinase pseudokinase for regulating cell adhesion. J. Biol. Chem. 289, 28363–28375 10.1074/jbc.M114.596692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ye F., Petrich B. G., Anekal P., Lefort C. T., Kasirer-Friede A., Shattil S. J., Ruppert R., Moser M., Fässler R., and Ginsberg M. H. (2013) The mechanism of kindlin-mediated activation of integrin αIIbβ3. Curr. Biol. 23, 2288–2295 10.1016/j.cub.2013.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Plow E. F., and Qin J. (2019) The kindlin family of adapter proteins: a past, present, and future prospectus. Circ. Res. 124, 202–204 10.1161/CIRCRESAHA.118.314362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bandyopadhyay A., Rothschild G., Kim S., Calderwood D. A., and Raghavan S. (2012) Functional differences between kindlin-1 and kindlin-2 in keratinocytes. J. Cell Sci. 125, 2172–2184 10.1242/jcs.096214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meller J., Rogozin I. B., Poliakov E., Meller N., Bedanov-Pack M., Plow E. F., Qin J., Podrez E. A., and Byzova T. V. (2015) Emergence and subsequent functional specialization of kindlins during evolution of cell adhesiveness. Mol. Biol. Cell 26, 786–796 10.1091/mbc.E14-08-1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rothbauer U., Zolghadr K., Muyldermans S., Schepers A., Cardoso M. C., and Leonhardt H. (2008) A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics 7, 282–289 10.1074/mcp.M700342-MCP200 [DOI] [PubMed] [Google Scholar]
- 30. Böttcher R. T., Veelders M., Rombaut P., Faix J., Theodosiou M., Stradal T. E., Rottner K., Zent R., Herzog F., and Fässler R. (2017) Kindlin-2 recruits paxillin and Arp2/3 to promote membrane protrusions during initial cell spreading. J. Cell Biol. 216, 3785–3798 10.1083/jcb.201701176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gao J., Huang M., Lai J., Mao K., Sun P., Cao Z., Hu Y., Zhang Y., Schulte M. L., Jin C., Wang J., White G. C., Xu Z., and Ma Y. Q. (2017) Kindlin supports platelet integrin αIIbβ3 activation by interacting with paxillin. J. Cell Sci. 130, 3764–3775 10.1242/jcs.205641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Theodosiou M., Widmaier M., Böttcher R. T., Rognoni E., Veelders M., Bharadwaj M., Lambacher A., Austen K., Müller D. J., Zent R., and Fässler R. (2016) Kindlin-2 cooperates with talin to activate integrins and induces cell spreading by directly binding paxillin. Elife 5, 1–24 10.7554/eLife.10130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Klapproth S., Bromberger T., Türk C., Krüger M., and Moser M. (2019) A kindlin-3–leupaxin–paxillin signaling pathway regulates podosome stability. J. Cell Biol. 218, 3436–3454 10.1083/jcb.201903109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bledzka K., Bialkowska K., Sossey-Alaoui K., Vaynberg J., Pluskota E., Qin J., and Plow E. F. (2016) Kindlin-2 directly binds actin and regulates integrin outside-in signaling. J. Cell Biol. 213, 97–108 10.1083/jcb.201501006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Qadota H., Moerman D. G., and Benian G. M. (2012) A molecular mechanism for the requirement of PAT-4 (integrin-linked kinase (ILK)) for the localization of UNC-112 (kindlin) to integrin adhesion sites. J. Biol. Chem. 287, 28537–28551 10.1074/jbc.M112.354852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bialkowska K., Ma Y. Q., Bledzka K., Sossey-Alaoui K., Izem L., Zhang X., Malinin N., Qin J., Byzova T., and Plow E. F. (2010) The integrin co-activator kindlin-3 is expressed and functional in a non-hematopoietic cell, the endothelial cell. J. Biol. Chem. 285, 18640–18649 10.1074/jbc.M109.085746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ussar S., Wang H. V., Linder S., Fässler R., and Moser M. (2006) The kindlins: subcellular localization and expression during murine development. Exp. Cell Res. 312, 3142–3151 10.1016/j.yexcr.2006.06.030 [DOI] [PubMed] [Google Scholar]
- 38. Azorin P., Bonin F., Moukachar A., Ponceau A., Vacher S., Bièche I., Marangoni E., Fuhrmann L., Vincent-Salomon A., Lidereau R., and Driouch K. (2018) Distinct expression profiles and functions of Kindlins in breast cancer. J. Exp. Clin. Cancer Res. 37, 281 10.1186/s13046-018-0955-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stiegler A. L., Grant T. D., Luft J. R., Calderwood D. A., Snell E. H., and Boggon T. J. (2013) Purification and SAXS analysis of the integrin linked kinase, PINCH, Parvin (IPP) heterotrimeric complex. PLoS ONE 8, e55591 10.1371/journal.pone.0055591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. O'Toole T. E., Katagiri Y., Faull R. J., Peter K., Tamura R., Quaranta V., Loftus J. C., Shattil S. J., and Ginsberg M. H. (1994) Integrin cytoplasmic domains mediate inside-out signal transduction. J. Cell Biol. 124, 1047–1059 10.1083/jcb.124.6.1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lad Y., Harburger D. S., and Calderwood D. A. (2007) Integrin cytoskeletal interactions. Methods Enzymol. 426, 69–84 10.1016/S0076-6879(07)26004-5 10.1016/S0076-6879(07)26004-5 [DOI] [PubMed] [Google Scholar]
- 42. Huet-Calderwood C., Rivera-Molina F., Iwamoto D. V., Kromann E. B., Toomre D., and Calderwood D. A. (2017) Novel ecto-tagged integrins reveal their trafficking in live cells. Nat. Commun. 8, 570 10.1038/s41467-017-00646-w 10.1038/s41467-017-00646-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Robert X., and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42:W320–W324. 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data described in this manuscript are all contained within the manuscript. Should further information be required, please contact the corresponding author.