Abstract
The DNA replication protein DnaA in Escherichia coli constructs higher-order complexes on the origin, oriC, to unwind this region. DnaB helicase is loaded onto unwound oriC via interactions with the DnaC loader and the DnaA complex. The DnaB–DnaC complex is recruited to the DnaA complex via stable binding of DnaB to DnaA domain I. The DnaB–DnaC complex is then directed to unwound oriC via a weak interaction between DnaB and DnaA domain III. Previously, we showed that Phe46 in DnaA domain I binds to DnaB. Here, we searched for the DnaA domain I–binding site in DnaB. The DnaB L160A variant was impaired in binding to DnaA complex on oriC but retained its DnaC-binding and helicase activities. DnaC binding moderately stimulated DnaA binding of DnaB L160A, and loading of DnaB L160A onto oriC was consistently and moderately inhibited. In a helicase assay with partly single-stranded DNA bearing a DnaA-binding site, DnaA stimulated DnaB loading, which was strongly inhibited in DnaB L160A even in the presence of DnaC. DnaB L160A was functionally impaired in vivo. On the basis of these findings, we propose that DnaB Leu160 interacts with DnaA domain I Phe46. DnaB Leu160 is exposed on the lateral surface of the N-terminal domain, which can explain unobstructed interactions of DnaA domain I–bound DnaB with DnaC, DnaG primase, and DnaA domain III. We propose a probable structure for the DnaA–DnaB–DnaC complex, which could be relevant to the process of DnaB loading onto oriC.
Keywords: DNA helicase, DNA replication, protein complex, protein dynamics, protein–protein interaction, oriC, origin DNA, DnaA, DnaB, DNA helicase
Loading of replicative helicases onto single-stranded (ss) DNA, as well as unwinding of origin DNA, is the critical step in initiating replication of the chromosome (1, 2). In Escherichia coli, the chromosomal origin oriC consists of the DNA-unwinding element (DUE) and the DnaA initiator-oligomerization region (DOR), which contains multiple DnaA-binding sites (DnaA boxes) and a single binding site for the DNA-bending factor IHF (Fig. 1A) (3–6). ATP–DnaA is the active form for initiation, whereas ADP–DnaA is inactive. Pentamers of ATP–DnaA (but not ADP–DnaA) form efficiently on the left and right DORs, and DnaA pentamer bound to the left DOR promotes unwinding of the DUE dependently on IHF (5–9). Recent studies indicate that DUE unwinding is stabilized by binding of the T-rich (upper) strand of the DUE to DnaA molecules bound to the left DOR (Fig. 1A), which is called the ssDUE recruitment mechanism and is a prerequisite for the successive loading of the replicative helicase DnaB onto the ssDNA region (1, 8–12). The DnaC loader is an AAA+ ATPase that forms a stable complex with DnaB and promotes the ssDNA interaction and conformational change of DnaB, making it competent for loading onto ssDNA (4, 13–17).
Figure 1.
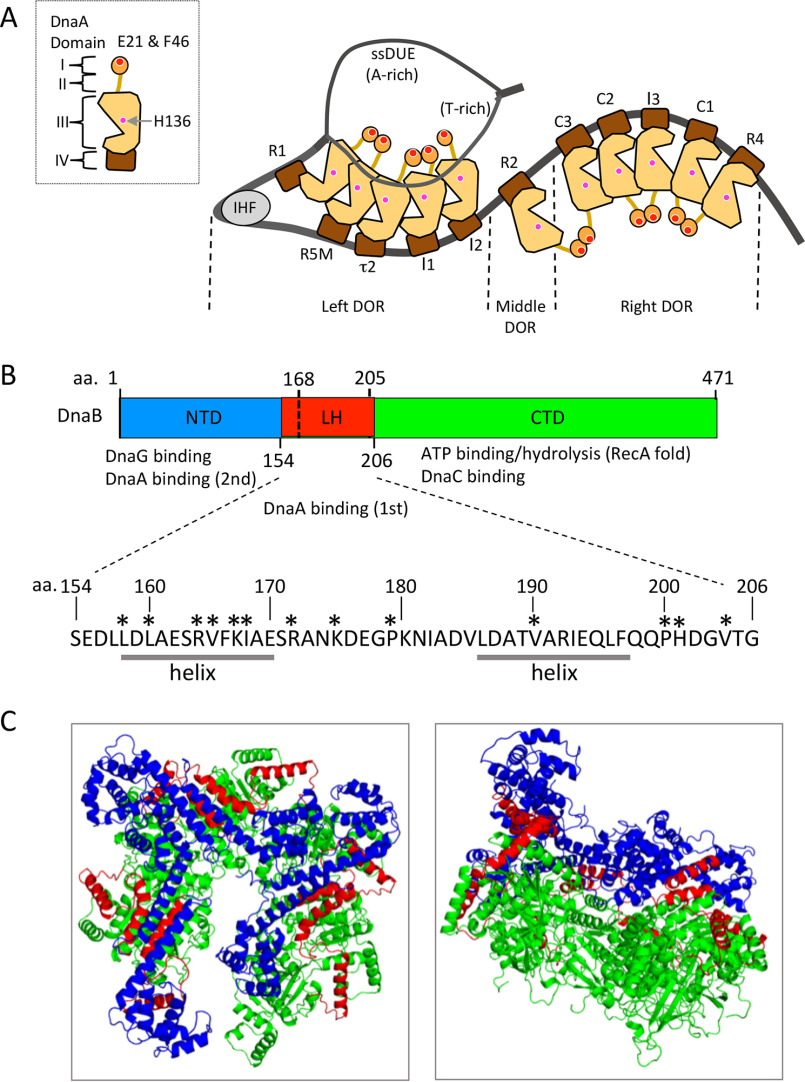
Structures of DnaA and DnaB. A, domain structure of DnaA and model of the open complex consisting of oriC, DnaA, and IHF. DnaA structure with four domains is shown (left). The DnaB-binding sites of DnaA domain I (Glu21/Phe46) and domain III (His136) are indicated by red and pink balls. DnaA boxes (R1, R2, R4, R5M, I1, I2, C1–3, and τ2) are also indicated. ATP–DnaA efficiently forms pentamers on the left and right DORs. When unwound, the T-rich (upper) strand of the DUE binds to specific sites of domain III of DnaA molecules bound to the left DOR (in particular, R1 and R5M boxes), promoting DnaB helicase loading onto the ssDNA region. B, domain structure of E. coli DnaB. The NTD, LH, CTD, and region containing Ser154–Gly206 are shown. The Ser154–Gly206 region is shown in red, and the rest of the NTD and CTD are shown in blue and light green. Specific functions of the NTD and CTD are indicated. In addition, the amino acid (aa) sequence of the Ser154–Gly206 region is shown. Residues in this study are indicated by asterisks. This region contains flexible linkers and α-helices as indicated (helix). C, tertiary structure of E. coli DnaB hexamer. DnaB within the DnaB–DnaC complex (PDB code 6QEL) is shown using a ribbon model. Substructures are colored as shown in B. Top (left) and side (right) views are shown.
DnaB within the DnaB–DnaC complex interacts with the DnaA complex on oriC, and this binding is prerequisite for loading of DnaB onto ssDNA (8, 9, 12, 18–25). The ATP-DnaA–IHF–left oriC complex promotes the loading of DnaB at the basal level; this process is stimulated by the ATP-DnaA–right oriC complex in a manner dependent upon the spacing between and relative orientation of the two complexes (9, 12). Two DnaB molecules bind to the oriC–DnaA subcomplexes, one each on the left and right (8, 9, 12).
DnaA consists of four domains (4, 6). Domain I (amino acid residues 1–84) contains the first specific binding site for DnaB, which constitutes a patch containing Glu21 and Phe46, as well as a hydrophobic patch that stimulates domain I–domain I interaction between DnaA molecules (Fig. 1A) (22, 23, 26). Domain II (amino acid residues 85–135) is a flexible linker (22, 27). Domain III (amino acid residues 136–374) contains the AAA+ family motifs that specifically interact with ATP/ADP and support domain III–domain III interactions between multiple DnaA molecules (8, 11, 12, 21, 28–33). In addition, this domain contains the ssDUE-specific binding sites Val211 and Arg245, collectively termed the H/B motifs (11). Val211 is a residue of the initiator-specific motif, an AAA+ motif shared by proteins of the initiator clade (28). In addition, the N terminus of domain III contains His136, the second specific binding site for DnaB (19, 20, 25). Even in DnaA pentamers, His136 is thought to be exposed on the surface (25). Domain IV (amino acid residues 375–457) contains a helix–turn–helix motif that specifically binds to the DnaA box (34).
DnaB, which forms a stable homohexamer with a ring or spiral configuration, consists of three domains (Fig. 1, B and C) (13, 16, 35–37). The N-terminal domain (NTD) constructs a trimer of dimers and contains DnaG primase-binding regions at the dimer–dimer interfaces (38–40). The C-terminal domain (CTD) contains a RecA ATPase fold and a DnaC helicase loader binding region (13, 16, 35–37); these domains are connected by the linker helix (LH) region. DnaC monomers bind DnaB, yielding a stable complex that facilitates a conformational change in DnaB that enables it to encircle ssDNA, thereby activating the loading of DnaB onto ssDNA (13, 14, 16, 17). When DnaB is loaded onto ssDNA, DnaC is dissociated, a process that is stimulated by DnaG binding (14, 15, 40). Loaded DnaB progresses on the ssDNA strand in a 5′-to-3′ direction and unwinds the forward dsDNA (35, 36).
The DnaA–DnaB interaction is essential for DnaB loading onto oriC (19–25). First, the DnaB–DnaC complex is recruited to the DnaA–IHF–oriC complex by binding of DnaB to multiple domain I molecules of DnaA oligomers on oriC (22, 23). Cooperative binding of DnaA domain I molecules to DnaB, which is a homohexamer, is key to increasing the affinity between the two proteins. Second, the interaction between recruited DnaB and His136, located in the domain III N terminus of oriC-bound DnaA molecules, plays an essential role in loading of DnaB onto the unwound region of oriC (25). Although this interaction is weak, it is thought to efficiently direct DnaB to the unwound region of oriC, which contains limited space.
Regardless of the importance of the DnaA–DnaB interaction, the amino acid residues within DnaB that play crucial roles in DnaA binding remain elusive, preventing a clearer understanding of the DnaA–DnaB complex structure and the mechanism of DnaA-directed loading of DnaB onto oriC. However, a previous study using various truncated forms of DnaB and DnaA proteins suggested that the region of DnaB from Ser154 to Gly206 contains a site that interacts with DnaA domain I; overproduction of this truncated DnaB region, DnaB(154–206), rescued growth inhibition in cells expressing a DnaA mutant that overinitiated replication; in addition, DnaB(154–206) bound to DnaA domain I in crude protein extracts (20). Previously, we performed NMR and mutant analyses of DnaA domain I, which showed that a patch containing Glu21 and Phe46 exposed on the surface of DnaA domain I specifically binds to DnaB and that this interaction is essential for DnaB loading onto oriC (22, 23).
In the present study, based on the above studies, we searched for the amino acids within DnaB that serve as interaction counterparts to the DnaA Glu21 and Phe46 residues. First, using the structural information of DnaB and DnaA domain I, we selected candidate residues within the DnaB Ser154–Gly206 region. Second, we functionally analyzed DnaB proteins harboring specific Ala substitutions at these residues both in vivo and in vitro. The results revealed that DnaB Leu160, located in the C terminus of DnaB NTD, was specifically important for DnaA binding and DnaB loading onto oriC. Based on our findings, we suggest novel models for the structure of the DnaA–DnaB complex and propose that DnaB loading onto oriC occurs via a specific interaction between Phe46 in DnaA domain I and Leu160 in the DnaB NTD.
Results
Searching for functional residues in the DnaB region
To identify residues within the DnaB Ser154–Gly206 region that are important for the specific interaction with DnaA, we first analyzed available tertiary structures of a DnaB homolog using homology modeling (39). Later, the results of this analysis were confirmed using a recently revealed structure of the E. coli DnaB hexamer within the DnaB–DnaC complex in the absence of ssDNA (16) (Fig. 1, B and C). The DnaB Ser154–Gly206 region corresponds to the C terminus of the NTD, the LH region, and the N terminus of the CTD (Fig. 1B). Based on the chemical nature of the Phe46 and Glu21 residues of the DnaB-binding site in DnaA domain I and structural models of the DnaB hexamer (14, 16, 22, 23), we paid special attention to hydrophobic or basic residues in this region that were likely to be exposed on the DnaB protein surface. According to these criteria, we selected 13 residues as candidates (Fig. 1B).
Next, we constructed plasmids harboring the DnaB mutants with alanine substitutions of the selected residues and performed a complementation test using the dnaB43 (Ts) mutant as a host strain (Table 1). The plasmid vector (pBAD18) contains the araB promoter and araC repressor, but even in the absence of arabinose, some leaky expression can be detected (10, 29). The dnaB43 allele contains a single A130V substitution that renders the ATPase and helicase activities highly sensitive to elevated temperature (42 °C) (24, 41).
Table 1.
Plasmid complementation test
Plasmids were introduced into ME5491 (dnaB43) cells. Transformants were incubated at 30 °C for 19 h or 42 °C for 14 h on LB-agar plates containing 0.1% arabinose and 50 µg/ml ampicillin, and then colonies were counted. Transformation efficiency (colony-forming units/µg of DNA) at each temperature and ratios of efficiencies are shown. Efficiency of pBAD/His-DnaB was defined as 1. a, only tiny colonies with a diameter of <0.5 mm were formed.
| Plasmid | dnaB allele | Transformation efficiency (× 105 transformants/μg of DNA) |
Ratio (42 °C/30 °C) | |
|---|---|---|---|---|
| 30 °C | 42 °C | |||
| pBAD18 (vector) | None | 2.0 | <1.0 × 10−2 | <7.4 × 10−3 |
| pBAD/His-DnaB | WT | 8.9 | 6.1 | [1.0] |
| pMS17 | L158A | 5.9 | <1.0 × 10−2 | <2.5 × 10−3 |
| pMS02 | L160A | 9.4 | 2.1a | 0.33 |
| pMS03 | R164A | 6.4 | 2.7a | 0.62 |
| pMS18 | V165A | 9.3 | <1.0 × 10−2 | <1.6 × 10−3 |
| pMS04 | K167A | 14 | 7.6 | 0.80 |
| pMS19 | I168A | 8.2 | <1.0 × 10−2 | <1.8 × 10−3 |
| pMS10 | R172A | 2.0 | 0.20 | 0.15 |
| pMS11 | K175A | 2.2 | 0.64 | 0.43 |
| pMS12 | P179A | 2.6 | 1.1 | 0.62 |
| pMS07 | V190A | 7.4 | 7.4a | 0.57 |
| pMS13 | P200A | 2.0 | 0.76 | 0.56 |
| pMS14 | H201A | 2.6 | 0.21 | 0.12 |
| pMS15 | V204A | 2.4 | 0.56 | 0.34 |
a Tiny colonies (<0.5 mm).
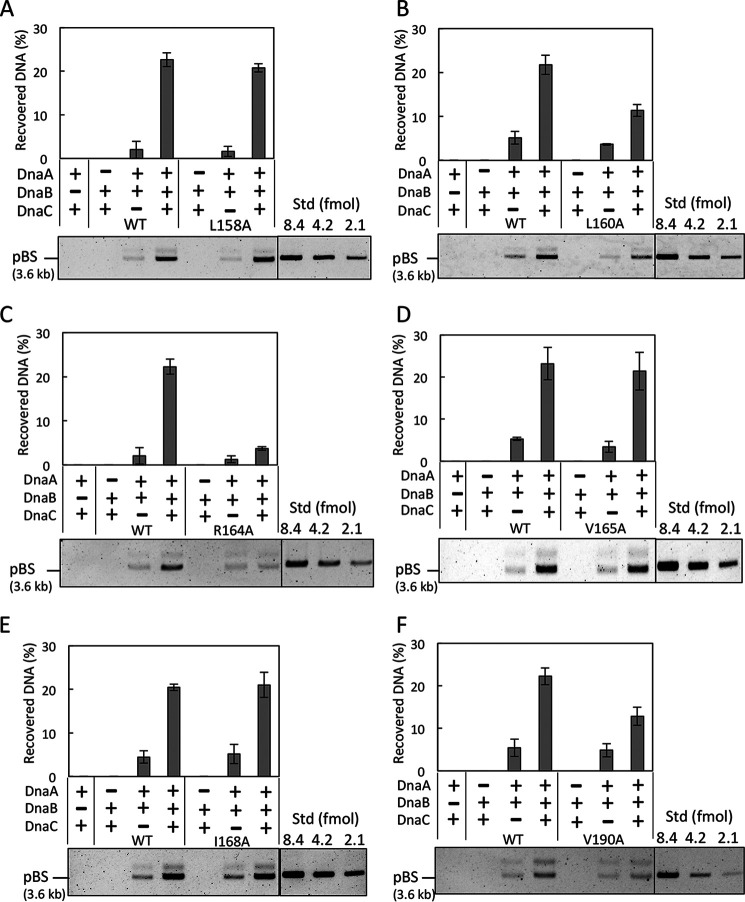
Introduction of WT (wild-type) dnaB on the plasmid (pBAD/His-DnaB) enabled colony formation at 42 °C, whereas dnaB alleles bearing the L158A, V165A, or I168A mutation (pMS17, pMS18, or pMS19, respectively) did not support growth (Table 1). Some dnaB alleles enabled only retarded growth at 42 °C: L160A, R164A, and V190A (pMS02, pMS03, and pMS07, respectively). Growth was delayed at 42 °C even in LB medium containing 0.1% arabinose: dnaB43 cells bearing pBAD/His-DnaB (WT dnaB) had a doubling time of ∼60 min, whereas cells bearing pMS03 (dnaB R164A) or pMS07 (dnaB V190A) grew much more slowly, with doubling times of ∼110 min. Cells bearing pMS02 (dnaB L160A) also grew slowly, with doubling times of ∼135 min, and when the optical density of the culture reached ∼0.4, growth ceased entirely. The levels of the mutant DnaB proteins were sustained even at 42 °C (Fig. S1). Hence, we further analyzed these six residues using purified proteins.
DnaB L160A, R164A, and V190A are impaired in DnaB loading onto unwound oriC
His-tagged forms of the six mutant DnaB proteins were overproduced and purified as described previously (17, 23, 25). We assessed the loading activity of DnaB after co-incubation with an oriC plasmid and IHF in the presence or absence of DnaA and DnaC, as described previously (17, 25). When DnaB is loaded onto unwound oriC by encircling the ssDNA in a DUE/DnaA/DnaC-dependent manner, the resultant DnaB–oriC complexes are so stable that oriC plasmid can be co-recovered with His-DnaB in pulldown experiments, even under high-salt conditions.
When WT DnaB was used, a considerable amount of oriC plasmid was recovered in a DnaA/DnaC-dependent manner (Fig. 2A). The slight recovery in the absence of DnaC might be the result of direct DnaB binding to DnaA molecules on oriC, an interaction that is labile under high-salt conditions. These results are consistent with our previous data (17, 25). Notably, recoveries of oriC plasmid by DnaB L160A, DnaB R164A, and DnaB V190A were moderately or dramatically reduced (Fig. 2, B, C, and F). By contrast, recovery by DnaB L158A, DnaB V165A, and DnaB I168A was comparable with the level achieved by the WT protein (Fig. 2, A, D, and E).
Figure 2.
DnaB pulldown assay for oriC loading activity. IHF (0.5 pmol, 61 nm) and oriC plasmid pBSoriC (17 fmol, 2.1 nm) were incubated at 37 °C for 15 min with (+) or without (−) ATP–DnaA (1 pmol, 120 nm), DnaC (2 pmol, 240 nm), and His-DnaB (2 pmol, 240 nm, as monomer). His-DnaB–bound pBSoriC was collected with Co2+-conjugated magnetic beads, washed with 100 mm NaCl, and analyzed by 1% agarose gel electrophoresis. Quantitative standards (Std) of pBSoriC were included in the same gel. The percentages of recovered DNA relative to the input DNA are indicated as Recovered DNA (%) (means ± S.D. (error bars); n = 2). Representative gel images are also shown. The position of pBSoriC (pBS; supercoiled form) is indicated. His-DnaB WT and the indicated mutant derivatives, L158A (A), L160A (B), R164A (C), V165A (D), I168A (E), and V190A (F), were used for this experiment.
DnaB L160A is specifically impaired in DnaA binding
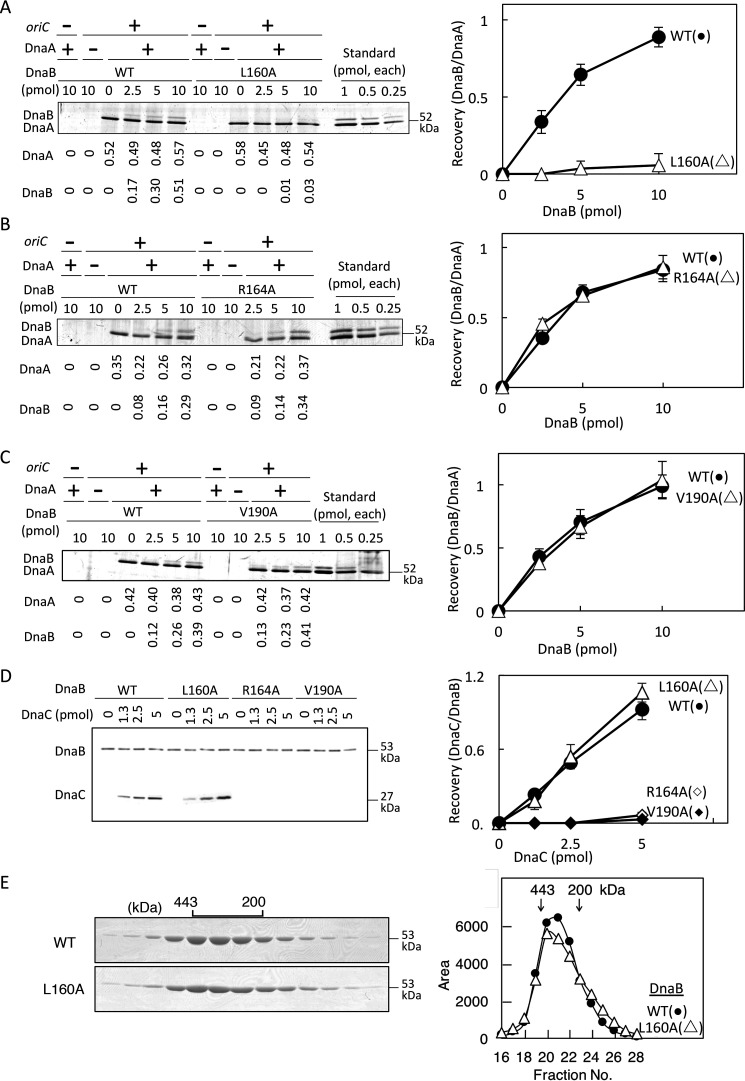
To identify a functional residue specific for DnaA binding, we analyzed DnaB L160A, DnaB R164A, and DnaB V190A in oriC pulldown experiments using a biotin-tagged oriC fragment (bio-oriC) bearing DnaA complexes. Although the affinity of DnaA monomers for DnaB is low (i.e. Kd is ∼2 μm) (19), DnaB binding of DnaA oligomers is drastically enhanced, likely by cooperative binding of multiple DnaA molecules to DnaB homohexamer (12, 22–24). Consistent with these results, WT DnaB was recovered in a manner dependent upon DnaA and bio-oriC (Fig. 3A). Unlike WT DnaB, DnaB L160A was barely recovered even in the presence of DnaA–oriC complexes (Fig. 3A), suggesting a crucial role for this residue in direct DnaA binding. By contrast, recovery of DnaB R164A and DnaB V190A was comparable with that of the WT protein (Fig. 3, B and C).
Figure 3.
DnaA and DnaC binding activities of DnaB. A–C, oriC pulldown assay for analysis of DnaA binding. Bio-oriC (0.1 pmol, 10 nm) was incubated on ice for 10 min with (+) or without (–) DnaA (5 pmol, 500 nm) and the indicated amounts (0–10 pmol or 1.0 μm, as monomer) of His-DnaB (WT or the indicated mutant), and bound materials were recovered with streptavidin-coated beads. Recovered DnaA and His-DnaB were analyzed by SDS-PAGE and silver staining (left panels). The recovered amounts (pmol, as monomers) were deduced using quantitative standards and are shown below the gel image. Quantitative standards included the same amounts (indicated as monomers) of each DnaA and His-DnaB (Standard). Based on these results, ratios of DnaB (as monomers) and DnaA were calculated (right panels). Experiments were performed in duplicate, and representative gel images and means ± S.D. (error bars) are shown. D, DnaC-binding activity of DnaB was analyzed by pulldown assay. His-DnaB (5 pmol, 500 nm, as monomer) was incubated on ice for 10 min with DnaC (0–5 pmol), and His-DnaB and bound materials were collected with Co2+-conjugated magnetic beads and analyzed by SDS-PAGE and silver staining (left). The numbers of recovered DnaB and DnaC molecules were deduced using quantitative standards, and ratios of DnaB (as monomers) and DnaC were calculated (right). E, gel filtration analysis was performed. His-DnaB (WT or L160A) (20 µg) was analyzed using a Superose 6 gel filtration column. Eluted fractions (5 μl) were analyzed by SDS-PAGE and Coomassie Brilliant Blue staining (left). Elution positions of marker proteins are indicated (443 and 200 kDa). Intensities of the DnaB bands are plotted (right).
Next, we analyzed the activities of DnaB L160A in DnaC binding and hexamer formation. To this end, we co-incubated His-DnaB proteins on ice with DnaC and performed pulldown experiments. DnaB L160A bound to DnaC at levels similar to WT DnaB (Fig. 3D). By contrast, DnaB R164A and DnaB V190A hardly bound DnaC (Fig. 3D). These results for DnaB V190A are consistent with a recent report that the α-helix containing Val190 binds to the DnaC N-terminal region (17). Also, these results for DnaB R164A are basically consistent with those of the DnaB-loading assay (Fig. 2C). At 37 °C, DnaB V190A may have some residual activities in interaction with DnaC, allowing loading to oriC at a moderate level (Fig. 2F). Gel filtration experiments suggested that like WT DnaB, DnaB L160A formed a stable homohexamer (Fig. 3E).
DnaB–DnaA interactions in the presence of DnaC
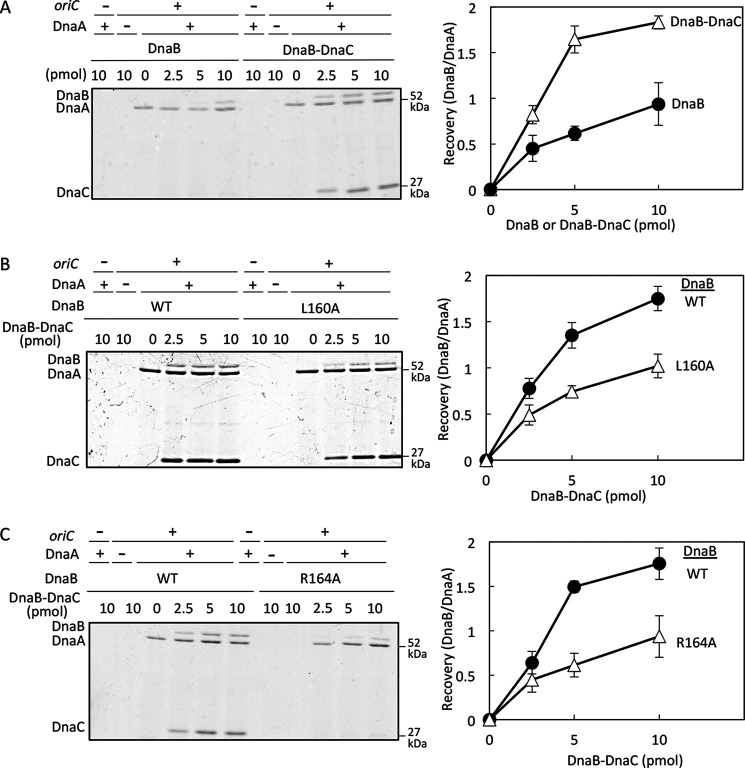
In the presence of DnaC, the affinity of DnaB for DnaA is moderately elevated, potentially due to structural changes in DnaB that increase the stability of the DnaA–DnaB interaction (12, 23, 25). We also analyzed the DnaA binding of DnaB L160A in the presence of DnaC. To this end, DnaB was incubated on ice with DnaA and bio-oriC in the presence or absence of DnaC, and bio-oriC–bound materials were recovered. Recovery of DnaB was moderately elevated in a DnaC-dependent manner (Fig. 4A), consistent with previous results (12, 23, 25). When WT DnaB and DnaB L160A were analyzed in the presence of DnaC, recovery of DnaB L160A was moderately reduced relative to WT DnaB (Fig. 4B). These results suggest that DnaB Leu160 plays an important role in binding to DnaA even in the presence of DnaC, although DnaC partly stimulates DnaB L160A binding to DnaA. Also, these findings are consistent with the moderate inhibition of DnaB L160A in oriC loading and with the results of the in vivo complementation test (Fig. 3A and Table 1).
Figure 4.
oriC pulldown assay for DnaA-binding activity of DnaB in the presence of DnaC. A, experiments were performed as described in the legend of Fig. 3 using His-DnaB WT in the presence or absence of DnaC. When present, the molecular numbers (as monomer) of DnaC input were the same as those of DnaB, and those as DnaB–DnaC complex are shown. B and C, similar experiments were performed using His-DnaB WT or mutant forms in the presence of DnaC. The molecular numbers (as monomer) of DnaC input were the same as those of DnaB and those as DnaB–DnaC complex are shown. Error bars, S.D.
Similar experiments were performed for DnaB R164A, which retained the basic activities of DnaA binding (Fig. 3B). In the presence of DnaC, DnaB R164A bound DnaA at lower levels than WT DnaB (Fig. 4C), consistent with the inability to bind DnaC (Fig. 3D).
DnaB L160A is active as a helicase but is impaired in DnaA-dependent stimulation of helicase activity
We further analyzed DnaB L160A in terms of its helicase activity and stimulation of ssDNA loading by DnaA. Helicase activity was assessed using a Y-shaped DNA in which single strands (30 nucleotides) are conjugated to dsDNA (49 bp) containing a DnaA box consensus sequence (Fig. 5A). The length between the end of the dsDNA and the DnaA box was identical to that between DUE and DnaA box R1 in oriC. In the absence of DnaA, DnaB is loaded onto the 5′-end ssDNA strand and migrates as a helicase to unwind the forward dsDNA region. DnaB L160A had intact activity in this helicase reaction (Fig. 5B), supporting the idea that DnaB L160A preserves the functional structure of the helicase.
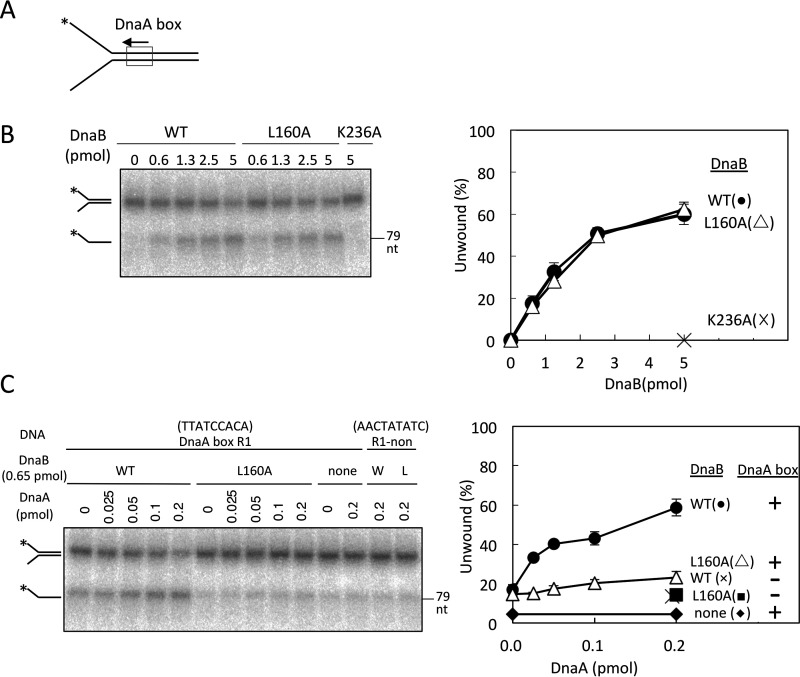
Figure 5.
DnaB helicase activity on fork DNAs. A, structure of fork DNA bearing DnaA box R1; direction of the box sequence (5′-TTATCCACA) is indicated. *, 5′-end 32P label. B, fork DNA (10 fmol, 1.1 nm) with a 32P-labeled 5′-end (*) was incubated at 37 °C for 30 min with DnaB (0–5 pmol or 560 nm; WT, L160A, or K236A). DNA was extracted and analyzed by PAGE. The proportion of ssDNA (as a fraction of total DNA) was plotted. Experiments were performed in duplicate, and a representative gel image and means ± S.D. (error bars) are shown. DnaB K236A, an ATPase-deficient mutant (17), was used as a negative control. C, fork DNAs (10 fmol, 1.1 nm) with DnaA box R1 sequence (R1) or a nonsense sequence (non) were incubated at 37 °C for 30 min with DnaB (0.65 pmol, 72 nm; WT or L160A) and DnaA (0–0.2 pmol or 22 nm). DNA was analyzed as above. DnaA box R1 and nonsense sequences are TTATCCACA and AACTATATC, respectively. W, WT; L, L160A.
In the presence of DnaA, the helicase activity of WT DnaB increased with the DnaA dose (Fig. 5C). Consistent with this, DnaA-dependent stimulation depended on the DnaA box sequence of the substrate DNA. These findings can be explained by the idea that even though the affinity is weak, the interaction of DnaB and a single DnaA molecule bound to the DnaA box can stimulate recruitment and loading of DnaB to the ssDNA region. In DnaB L160A, this DnaA-dependent stimulation was severely inhibited (Fig. 5C), supporting the idea that DnaB Leu160-dependent interaction with DnaA is functionally important in the stimulation of DnaB loading on the ssDNA.
Helicase loading to unwound DUE-mimic DNA is severely inhibited by DnaB L160A
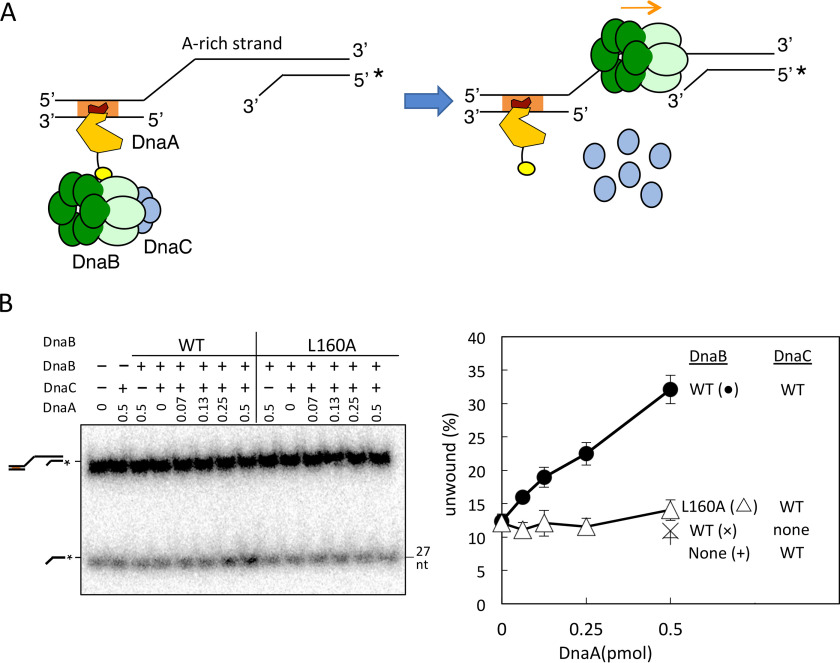
We further analyzed the DnaA-dependent stimulation of DnaB loading using a DNA fragment that partly mimicked the DUE-unwound structure. The DNA fragment contained an ssDNA region (28 nucleotides) that was conjugated at the 5′-site to dsDNA bearing a DnaA box and at the 3′-site to dsDNA with a 3′-ssDNA tail (Fig. 6A). The length of the ssDNA region (28 nucleotides) between the dsDNAs was determined based on the fact that an oriC DUE region, including two 13-mer sequences (M and R), is initially unwound in in vitro reconstituted systems, and a similar ssDNA region stably binds to DnaA oligomers constructed on oriC–DnaA box clusters (Fig. 1A) (10–12, 42). In this DNA structure, DnaB should be loaded onto the ssDNA region located between the dsDNA regions in a manner dependent upon DnaC and DnaA, and loaded DnaB should migrate by unwinding the dsDNA region with the 3′-ssDNA tail. This mechanism is consistent with the conclusion of a previous study that used various forms of gapped DNA with a single DnaA box; the results suggested that DnaB bound to DnaA box R1 should be predominantly loaded onto the A-rich (lower) strand of ssDUE (43). Consistent with these ideas, WT DnaB unwound the 3′-tailed dsDNA region in a DnaA dose–dependent manner in the presence of DnaC (Fig. 6B). However, such DnaA-dependent unwinding was severely inhibited in DnaB L160A (Fig. 6B). Only slight residual activity was observed for DnaB L160A, which is consistent with the data in Fig. 5.
Figure 6.
DnaB helicase activity for an unwound oriC-mimic gapped DNA. A, structure of the unwound oriC-mimic gapped DNA and a conceivable mechanism for DnaB helicase loading. Orange box, DnaA box R1; *, the 32P-labeled site. DnaA is shown as illustrated in Fig. 1A. The sequence of the gapped DNA is the same as that of the region spanning from DnaA box R1 to the DUE, except for the deletion of the DUE 28-mer sequence in the upper strand (see the conceivable structure of the left-half oriC open complex in Fig. 1A). DnaA binds to the DnaA box on the gapped DNA and recruits the DnaB–DnaC complex via DnaA domain I, which promotes DnaB loading onto the ssDNA region and stimulates helicase function. B, gapped DNA (10 fmol, 1.1 nm) was incubated on ice for 5 min with DnaA (0–0.5 pmol or 55 nm), followed by further incubation at 37 °C for 8 min in the presence or absence of DnaB (WT or L160A) and DnaC that had been incubated on ice for 10 min. DNA was analyzed as described in the legend to Fig. 5B. Error bars, S.D.
Construction of a structural model of the DnaA–DnaB–DnaC complex
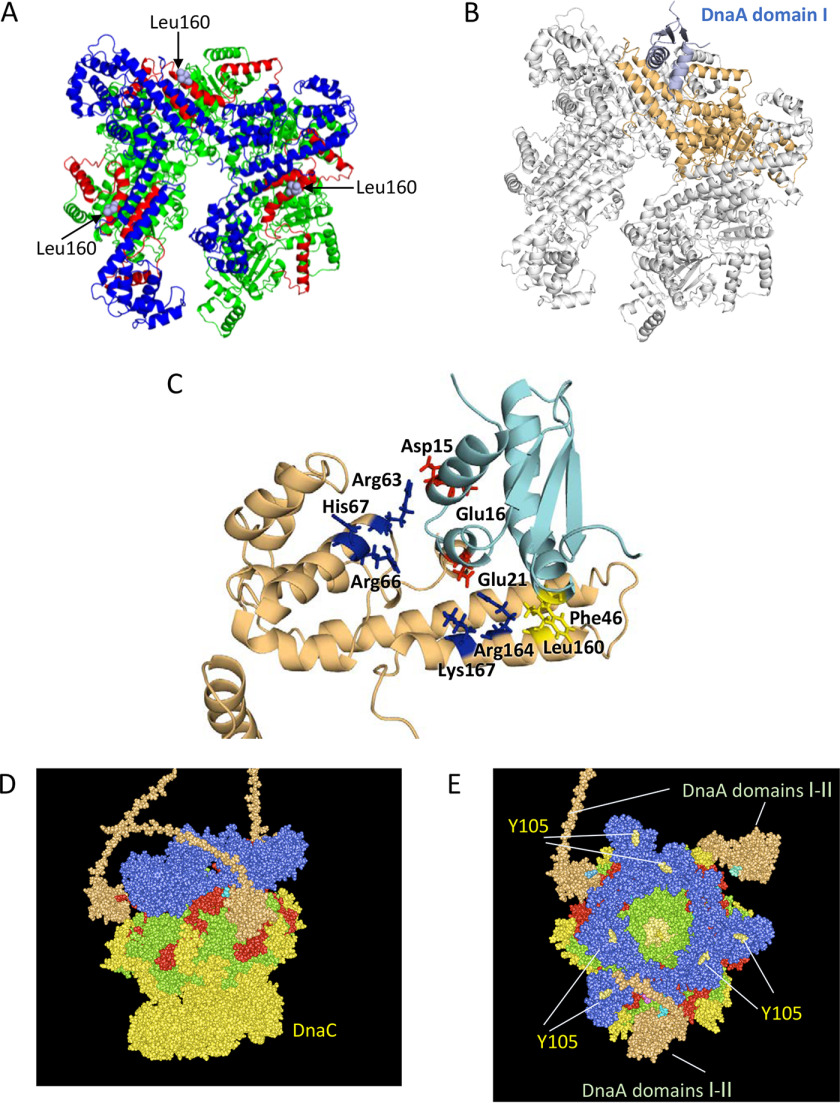
Based on the tertiary structures of DnaA domain I and the DnaB–DnaC complexes (16, 22), we constructed a structural model of DnaA domain I–DnaB complex, in which DnaB Leu160 interacts with DnaA Phe46 (Fig. 7). The Leu160 residues of three protomers of the DnaB hexamer are exposed on the surface (Fig. 7, A–C), which is reasonable in that DnaB NTD forms a trimer of dimers in which pairs of NTDs bind each other in an essentially symmetrical manner. In addition, we noted that the side chains of DnaB Arg164 and Lys167 are exposed on the surface at a position flanking Leu160 and closely adjoin each other (Fig. 7C). Structurally, these seem like the most probable sites for the counterpart of DnaA Glu21. The interaction between DnaA Glu21 and either Arg164 or Lys167 could be sufficient to support a functional DnaA–DnaB interaction. Based on these findings, we assessed the constructed complex using the Rosetta server to finely optimize structural rationality and the energy level of the complex (44); the lowest-score model is shown (Fig. 7, B and C). This model is consistent with the idea of hydrophobic interactions between DnaB Leu160 and DnaA Phe46 (Fig. S2A) and electrostatic interactions between DnaB Arg164/Lys167 and DnaA Glu21 (Fig. S2B). In addition, the lowest-score docking model suggested an interaction between a cluster of basic residues in DnaB, Arg63/Arg66/His67 and a cluster of acidic residues, DnaA Asp15/Glu16 (Fig. 7C). In a previous study, we observed that the DnaA E16A mutant protein had moderately inhibited activities in oriC plasmid replication in a system reconstituted with purified proteins (22).
Figure 7.
Structural model for DnaB helicase in complex with DnaA domains I–II. A, ribbon model of a DnaB hexamer structure (PDB code 6QEL) and the positions of Leu160 residues exposed on the surface are shown. DnaB regions are colored as shown in Fig. 1. B, docking model of DnaB and DnaA domain I is shown using a ribbon model. The overall structure with only one DnaA domain I molecule was calculated using the Rosetta server and optimized. DnaA is shown in light blue. DnaB is shown in gray, except for the DnaA-bound protomer, which is shown in brown. C, close-up view of the interface between DnaB and DnaA domains I and II. A part of the docking model (A) is enlarged. DnaB Leu160 and DnaA Phe46 (yellow), DnaA Asp15, Glu16, and Glu21 (red), and DnaB Arg63, Arg66, His67, Arg164, and Lys167 (blue) are shown as stick models. D and E, a possible structure of the DnaA–DnaB–DnaC complex is shown using a space-filling mode. Based on the above and the DnaB–DnaC complex structure (PDB code 6QEM), overall structure of the DnaB–DnaC complex bound to three molecules of DnaA domains I and II (orange) is shown. The DnaB hexamer, which is colored as shown in A, binds six molecules of DnaC (yellow) and three molecules of DnaA domains I and II (orange). An essential DnaG binding site in DnaB, Tyr105, is also shown.
In the proposed model, three DnaA domain I molecules can bind to a DnaB hexamer without physical obstruction (Fig. 7D), consistent with previous studies that predicted functional binding of multiple DnaA molecules to a single DnaB hexamer (22, 23). Also, the binding of DnaA domain I to the middle lateral regions of a DnaB hexamer allows free space around the NTD and CTD of DnaB, which are important for the efficient interactions of DnaG primase and DnaC with DnaB (Fig. 7, D and E). DnaB Tyr105 is an essential binding site of DnaG that resides on the top surface of DnaB (Fig. 7E) (38, 39, 45). The DnaB Arg164/Lys167–DnaA Glu21 interaction determines the orientation of DnaA domain I, placing DnaA domain II in the direction of the DnaB NTD. This overall orientation of DnaA domain I and DnaB hexamer may be important for the efficient interaction of the DnaB–DnaC complex, with unwound DNA strands generated at oriC initiation complexes (see “Discussion”).
Discussion
In this study, we showed that DnaB Leu160 in the C-terminal helix of the NTD is essential in DnaA binding. Like DnaA F46A and DnaA E21A, but unlike DnaA H136A, DnaB L160A does not form stable complexes with DnaA oligomers on oriC. This observation is consistent with previous structural studies of DnaA and DnaB. DnaB Leu160 is the counterpart of DnaA Phe46, as both can engage in hydrophobic interactions. DnaC binding and fundamental helicase activities were retained in DnaB L160A, supporting the idea that Leu160 plays a specific role in DnaA binding.
In the presence of DnaC, DnaB–DnaA binding is moderately stimulated, and DnaB L160A exhibited residual DnaA-binding activity, perhaps because DnaC binding affects the structure of the DnaA-binding site. Recently reported structures of the DnaB–DnaC interface revealed that DnaC interacts with the α-helix (Leu186–Phe197) in the LH region, causing a conformational change in the DnaB hexamer (16, 17). The α-helix (Leu186–Phe197) is flanked by a linker connecting to the DnaA-binding helix with Leu160 (Fig. 1, B and C). Consistent with this, DnaC might indirectly affect the structure of the DnaA-binding helix through spatial rearrangement of the α-helix (Leu186–Phe197) and the flanking linker. We infer that the DnaC-dependent changes in the affinity of DnaB for DnaA might be functionally significant in the formation and dissociation of the DnaA–DnaB–DnaC complex during the overall process of replication initiation. When DnaC is dissociated from DnaB, which is loaded onto unwound oriC, the affinity of DnaB for DnaA should be reduced. This is reasonable, in that DnaB must dissociate from DnaA to migrate along ssDNA strands. In the presence of DnaC, DnaB L160A exhibited only slight residual activity for loading onto the oriC-mimic gapped DNA, unlike the case for loading onto oriC plasmid (Figs. 2B and 6). Because the gapped DNA contained only a single DnaA box, interaction of the DnaB L160A–DnaC complex with DNA-bound DnaA was likely too unstable to substantially stimulate DnaB loading. Accordingly, we propose that the DnaB Leu160–mediated interaction with DnaA plays a predominant role in the process of DnaB loading.
In the tertiary structure of the DnaB hexamer, the DnaA domain I binding site containing Leu160 is located on the lateral surface of the C terminus of the NTD (Fig. 7). Because the N-terminal tier of the DnaB hexamer consists of a trimer of NTD dimers, three Leu160 sites are exposed on the outside of the DnaB hexamer. Structural modeling suggested that three DnaA domain I molecules could bind those sites without structural conflict (Fig. 7, D and E), and this is consistent with biochemical analyses showing that DnaB binds to multiple DnaA molecules on oriC. In addition, the model supports the idea that binding of DnaA domain I to the DnaB NTD–lateral sites does not structurally interfere with DnaC binding to the CTD (Fig. 7D). Also, the specific binding region of DnaG primase, which resides on the top surface of the N-terminal tier (38, 39, 45), does not overlap with the DnaA domain I–binding site (Fig. 7E). In addition, DnaB hexamers could form a homodimer for bidirectional replication via top-to-top interaction of NTDs during loading onto ssDNA regions of oriC (46). Even in that case, binding of DnaA domain I to the DnaB NTD–lateral sites would not cause structural conflicts. These observations indicate that the binding positions of multiple proteins to DnaB are arranged in a reasonable manner.
Glu21 in DnaA domain I, in addition to Phe46, plays an important role in DnaB binding (22). We inferred that DnaB Leu160 is the counterpart of DnaA Phe46, based on the hydrophobic nature of these two residues. Notably, the structure around DnaB Leu160 suggests that DnaB Arg164 and Lys167 basic residues, which adjoin each other on the surface and are located near Leu160, are candidate counterparts of DnaA Glu21, an acidic residue (Fig. 7C). Both DnaB Arg164 and Lys167 could participate in DnaA interactions, and either residue could be sufficient to functionally support the interactions (Fig. 3B). DnaB R164A is inactive in DnaC binding, suggesting that this mutant causes allosteric effects. The helix containing Arg164 connects via a linker to the helix containing Val190, which is important for DnaC binding. In addition, the R164A mutation stimulates ATPase activity, indicating that this mutation also has pleiotropic effects (47). Furthermore, we revealed possible interactions between DnaA Asp15/Glu16 and DnaB Arg63/Arg66/His67 (Fig. 7C). Our previous data, showing that DnaA E16A is moderately impaired in oriC plasmid replication in an in vitro reconstituted system, are consistent with this idea (22). Therefore, future studies should more closely examine DnaB Arg164/Lys167 residues and the possible interactions between DnaA Asp15/Glu16 and DnaB Arg63/Arg66/His67. In contrast to the stable binding between DnaA domain I and DiaA, a DnaA assembly–stimulating factor (23), effective hydrophobic interactions would not be formed except between DnaA Phe46 and DnaB Leu160 (Fig. S2A), which might explain the relatively weak interaction between DnaA and DnaB.
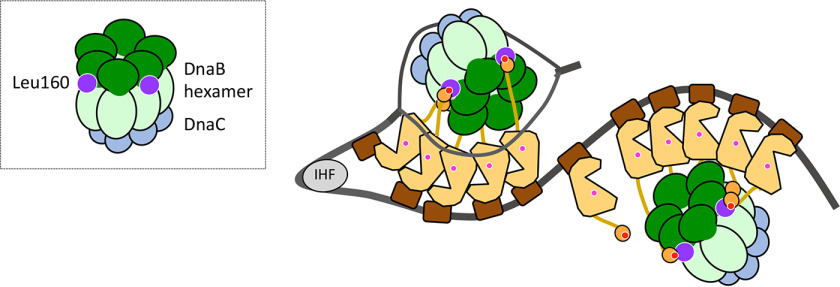
In the proposed model (Fig. 7), the relative orientation of DnaA domain I–II in the complex is determined by two sets of interactions: DnaB Leu160–DnaA Phe46 and DnaB Arg164/Lys167–DnaA Glu21. As a result, DnaB NTD is oriented toward DnaA domain III, whereas DnaB CTD, to which the DnaC helicase loader binds, is oriented in the opposite fashion (Fig. 7D). This orientation is reasonable: in the ssDUE recruitment mechanism, DnaA pentamers on oriC DOR bind the T-rich ssDUE region, and DnaC interacts with the DnaA-free A-rich ssDUE region (Fig. 8). The proposed orientation of the DnaB–DnaC complex bound to domain I of DnaA pentamers fits well with the requirement for the DnaC–ssDNA interaction, which triggers DnaB loading onto the unwound oriC region. After the first interaction via the DnaA domain I–DnaB NTD C terminus, the second interaction of DnaB with DnaA domain III His136 stimulates precise delivery of the DnaB–DnaC complex to the unwound DNA of the oriC region, as we previously proposed (25) (Fig. 8). However, we cannot exclude other possibilities regarding the relative direction of DnaB bound to DnaA, and these should be investigated in future studies.
Figure 8.
Model of DnaB loading via interaction between DnaB Leu160 and DnaA Phe46. A model of the structure of the open complex is shown as in Fig. 1. In addition, DnaB (NTD (dark green) and LH-CTD (pale green)), DnaC (cyan), DnaB Leu160 site (purple), and DnaA Phe46 site (dark red) are shown. Two or three protomers of the DnaB hexamer bind to domain I in DnaA pentamers on oriC, which recruits two DnaB hexamers and tethers them in a particular orientation. This conformation underlies the functional interaction of DnaB–DnaC complexes with the DnaA domain III site containing His136 and the unwound region of oriC, promoting DnaB loading onto the oriC region.
Self-dimerization of DnaA domain I is thought to stimulate DnaB loading onto oriC (19). DnaA domain I has a hydrophobic patch almost on the opposite side from the DnaB-binding site (22), resulting in a low-affinity domain I–domain I interaction that can be detected by chemical cross-linking (26). Regardless of its weak affinity, the domain I–domain I interaction via flexible linker domain II effectively stimulates the interaction between two DnaA molecules bound to a DNA region at a certain relative distance. Although the overall structure of domain I dimer has not been revealed, it is conceivable that only one protomer of the domain I dimer binds to the site containing DnaB Leu160, whereas the other protomer causes a secondary interaction with DnaB, increasing the affinity between DnaA domain I and DnaB. Otherwise, dimerization of domain I restricts the orientation of domain I relative to the linker domain II to stimulate efficient interaction between the site containing Phe46 in domain I and the site containing Leu160 in DnaB. Further analyses are necessary to test these possibilities.
Experimental procedures
Bacterial strains, plasmids, and the fork and gapped DNAs
For plasmid complementation tests, strains ME6299 (HfrP4X8) and ME5491 (ME6299 dnaB43 (Ts)) were acquired from the National Bioresource Project (E. coli) of the Institute of National Genetics, Japan (24, 41).
N-terminally His6-tagged DnaB was overproduced using pBAD/His-DnaB, which was constructed by inserting the WT dnaB into the pBAD/His-B arabinose-inducible vector using NheI–EcoRI sites (17). Ala-substitution mutations were introduced to the dnaB gene in pBAD/His-DnaB by PCR using mutagenic primers, and the mutations were confirmed by nucleotide sequencing. The resultant plasmids are listed in Table 1. Purifications of His-DnaB and its derivatives were performed as described previously (17, 23). Purified His-DnaB retained the oriC loading and helicase activities of the intact protein (23).
The fork and gapped DNAs were constructed by annealing of synthetic DNA strands (Table S1). Annealed products were resolved by PAGE and then purified.
Pulldown assays using His-DnaB
DnaB loading onto oriC plasmid was assayed as described previously (17, 25). Briefly, DnaA, His-DnaB, DnaC, IHF, pBSoriC oriC plasmid, and ATP were incubated at 30 °C for 15 min, followed by incubation at 4 °C for 15 min with Co2+-conjugated magnetic beads (Dynabeads, Thermo Fisher Scientific). Bead-bound materials were collected by magnetic force and washed in buffer containing 100 mm NaCl. Recovered plasmid was eluted in standard SDS sample buffer and analyzed by 1% agarose gel electrophoresis.
The DnaB–DnaC binding assay was performed as described previously (17). Briefly, His-DnaB and DnaC were incubated on ice for 10 min, followed by pulldown with nickel-conjugated magnetic beads. After washing, recovered proteins were eluted in standard SDS sample buffer and analyzed by SDS-10% PAGE and silver staining.
Biotin-tagged oriC pulldown assay
The assay was performed as described previously (8, 12, 23–25). Briefly, bio-oriC (419 bp), including the entire oriC, was incubated on ice for 10 min with DnaA, DnaB, or DnaC, followed by pulldown using streptavidin-coated beads. Recovered proteins were analyzed by SDS-10% PAGE and silver staining.
Helicase activity assay
The upper strands of fork DNAs were labeled at the 5′-end with 32P using T4 polynucleotide kinase and annealed with the lower strands, followed by isolation using spin columns and 6% polyacrylamide gel. DnaB helicase activity was assessed using fork DNAs basically as described previously (43, 46). Briefly, fork DNAs were incubated at 37 °C for the indicated times in buffer containing 20 mm HEPES-KOH (pH 7.6), 1 mm EDTA, 4 mm DTT, 5 mm magnesium acetate, 10% glycerol, 0.1% Triton X-100, 40 µg/ml BSA, 5 mm ATP, 5 mm creatine phosphate, 20 µg/ml creatine kinase, and DnaB in the presence or absence of DnaA. DNA was extracted using phenol/chloroform and analyzed by 6% PAGE; bands were visualized on a BAS2500 PhosphorImager.
Gel filtration experiments
His-DnaB was analyzed using Superose 6 PC 3.2/30 (2.4-ml column volume), equilibrated with buffer containing 20 mm HEPES-KOH (pH 7.6), 1 mm EDTA, 4 mm DTT, 5 mm magnesium acetate, 10% glycerol, 0.1% Triton X-100, 40 mm ammonium sulfate, and 20 mm NaCl; flow rate was 30 μl/min. Eluted fractions (5 μl) were analyzed by SDS-10% PAGE and Coomassie Brilliant Blue staining.
Structural model building
The docking model of DnaA domain I and DnaB was calculated by the Rosetta server using the docking2 protocol (RRID:SCR_015701) (44). For this calculation, we used the structures of DnaB from the DnaB–DnaC complex structure (PDB code 6QEL) and the DnaA NTDs (PDB code 2E0G). Hydrophobicity is indicated by color as in a previous paper (48). Electrostatic potentials were calculated using PyMOL (Schrödinger, LLC)) with APBS Tools (49). The DnaA–DnaB–DnaC complex structure model was constructed using MolFeat (FiatLux, Tokyo, Japan) and PyMOL.
Data availability
All data are contained within the article.
Supplementary Material
Acknowledgments
We thank Naoki Ochi for the construction of pBAD/His-DnaB (pHDB01); Dr. Asako Furukohri and Dr. Hisaji Maki for anti-DnaB antiserum; and the National Bioresource Project (E. coli) of the Institute of National Genetics, Japan, for strains.
This article contains supporting information.
Author contributions—C. H., E. M., and T. K. data curation; C. H., E. M., S. O., Y. A., and T. K. formal analysis; C. H., E. M., S. O., Y. A., and T. K. validation; C. H., E. M., S. O., Y. A., and T. K. investigation; C. H., E. M., Y. A., and T. K. visualization; C. H., E. M., S. O., Y. A., and T. K .writing-original draft; S. O., Y. A., and T. K. conceptualization; S. O. and T. K. supervision; S. O., Y. A., and T. K. writing-review and editing; T. K. resources; T. K. funding acquisition.
Funding and additional information—This study was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grants JP17H03656 and JP20H03212.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- DUE
- the DNA-unwinding element
- DOR
- the DnaA oligomerization region
- AAA
- ATPases associated with various cellular activities
- NTD
- N-terminal domain
- LH
- linker helix
- CTD
- C-terminal domain
- PDB
- Protein Data Bank
- LB
- Luria-Bertani
- WT
- wild-type.
References
- 1. Soultanas P. (2012) Loading mechanisms of ring helicases at replication origins. Mol. Microbiol. 84, 6–16 10.1111/j.1365-2958.2012.08012.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bell S. P., and Kaguni J. M. (2013) Helicase loading at chromosomal origins of replication. Cold Spring Harb. Perspect. Biol. 5, a010124 10.1101/cshperspect.a010124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Katayama T., Ozaki S., Keyamura K., and Fujimitsu K. (2010) Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 8, 163–170 10.1038/nrmicro2314 [DOI] [PubMed] [Google Scholar]
- 4. Kaguni J. M. (2011) Replication initiation at the Escherichia coli chromosomal origin. Curr. Opin. Chem. Biol. 15, 606–613 10.1016/j.cbpa.2011.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Leonard A. C., and Grimwade J. E. (2015) The orisome: structure and function. Front. Microbiol. 6, 545 10.3389/fmicb.2015.00545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Katayama T., Kasho K., and Kawakami H. (2017) The DnaA cycle in Escherichia coli: activation, function and inactivation of the initiator protein. Front. Microbiol. 8, 2496 10.3389/fmicb.2017.02496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rozgaja T. A., Grimwade J. E., Iqbal M., Czerwonka C., Vora M., and Leonard A. C. (2011) Two oppositely oriented arrays of low-affinity recognition sites in oriC guide progressive binding of DnaA during Escherichia coli pre-RC assembly. Mol. Microbiol. 82, 475–488 10.1111/j.1365-2958.2011.07827.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ozaki S., Noguchi Y., Hayashi Y., Miyazaki E., and Katayama T. (2012) Differentiation of the DnaA-oriC subcomplex for DNA unwinding in a replication initiation complex. J. Biol. Chem. 287, 37458–37471 10.1074/jbc.M112.372052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shimizu M., Noguchi Y., Sakiyama Y., Kawakami H., Katayama T., and Takada S. (2016) Near-atomic structural model for bacterial DNA replication initiation complex and its functional insights. Proc. Natl. Acad. Sci. U. S. A. 113, E8021–E8030 10.1073/pnas.1609649113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sakiyama Y., Kasho K., Noguchi Y., Kawakami H., and Katayama T. (2017) Regulatory dynamics in the ternary DnaA complex for initiation of chromosomal replication in Escherichia coli. Nucleic Acids Res. 45, 12354–12373 10.1093/nar/gkx914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ozaki S., Kawakami H., Nakamura K., Fujikawa N., Kagawa W., Park S.-Y., Yokoyama S., Kurumizaka H., and Katayama T. (2008) A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J. Biol. Chem. 283, 8351–8362 10.1074/jbc.M708684200 [DOI] [PubMed] [Google Scholar]
- 12. Ozaki S., and Katayama T. (2012) Highly organized DnaA-oriC complexes recruit the single-stranded DNA for replication initiation. Nucleic Acids Res. 40, 1648–1665 10.1093/nar/gkr832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arias-Palomo E., O'Shea V. L., Hood I. V., and Berger J. M. (2013) The bacterial DnaC helicase loader is a DnaB ring breaker. Cell 153, 438–448 10.1016/j.cell.2013.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chodavarapu S., Jones A. D., Feig M., and Kaguni J. M. (2016) DnaC traps DnaB as an open ring and remodels the domain that binds primase. Nucleic Acids Res. 44, 210–220 10.1093/nar/gkv961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Felczak M. M., Chodavarapu S., and Kaguni J. M. (2017) DnaC, the indispensable companion of DnaB helicase, controls the accessibility of DnaB helicase by primase. J. Biol. Chem. 292, 20871–20882 10.1074/jbc.M117.807644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arias-Palomo E., Puri N., O'Shea Murray V. L., Yan Q., and Berger J. M. (2019) Physical basis for the loading of a bacterial article physical basis for the loading of a bacterial replicative helicase onto DNA. Mol. Cell 74, 173–184 10.1016/j.molcel.2019.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nagata K., Okada A., Ohtsuka J., Ohkuri T., Akama Y., Sakiyama Y., Miyazaki E., Horita S., Katayama T., Ueda T., and Tanokura M. (2020) Crystal structure of the complex of the interaction domains of Escherichia coli DnaB helicase and DnaC helicase loader: structural basis implying a distortion-accumulation mechanism for the DnaB ring opening caused by DnaC binding. J. Biochem. 167, 1–14 10.1093/jb/mvz087 [DOI] [PubMed] [Google Scholar]
- 18. Marszalek J., and Kaguni J. M. (1994) DnaA protein directs the binding of DnaB protein in initiation of DNA replication in Escherichia coli. J. Biol. Chem. 269, 4883–4890 [PubMed] [Google Scholar]
- 19. Sutton M. D., Carr K. M., Vicente M., and Kaguni J. M. (1998) Escherichia coli DnaA protein: the N-terminal domain and loading of DnaB helicase at the E. coli chromosomal origin. J. Biol. Chem. 273, 34255–34262 10.1074/jbc.273.51.34255 [DOI] [PubMed] [Google Scholar]
- 20. Seitz H., Weigel C., and Messer W. (2000) The interaction domains of the DnaA and DnaB replication proteins of Escherichia coli. Mol. Microbiol. 37, 1270–1279 10.1046/j.1365-2958.2000.02096.x [DOI] [PubMed] [Google Scholar]
- 21. Felczak M. M., and Kaguni J. M. (2004) The box VII motif of Escherichia coli DnaA protein is required for DnaA oligomerization at the E. coli replication origin. J. Biol. Chem. 279, 51156–51162 10.1074/jbc.M409695200 [DOI] [PubMed] [Google Scholar]
- 22. Abe Y., Jo T., Matsuda Y., Matsunaga C., Katayama T., and Ueda T. (2007) Structure and function of DnaA N-terminal domains: specific sites and mechanisms in inter-DnaA interaction and in DnaB helicase loading on oriC. J. Biol. Chem. 282, 17816–17827 10.1074/jbc.M701841200 [DOI] [PubMed] [Google Scholar]
- 23. Keyamura K., Abe Y., Higashi M., Ueda T., and Katayama T. (2009) DiaA dynamics are coupled with changes in initial origin complexes leading to helicase loading. J. Biol. Chem. 284, 25038–25050 10.1074/jbc.M109.002717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Noguchi Y., and Katayama T. (2016) The Escherichia coli cryptic prophage protein YfdR binds to DnaA and initiation of chromosomal replication is inhibited by overexpression of the gene cluster yfdQ-yfdR-yfdS-yfdT. Front. Microbiol. 7, 239 10.3389/fmicb.2016.00239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sakiyama Y., Nishimura M., Hayashi C., Akama Y., Ozaki S., and Katayama T. (2018) The DnaA AAA+ domain His136 residue directs DnaB replicative helicase to the unwound region of the replication origin, oriC. Front. Microbiol. 9, 2017 10.3389/fmicb.2018.02017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Felczak M. M., Simmons L. A., and Kaguni J. M. (2005) An essential tryptophan of Escherichia coli DnaA protein functions in oligomerization at the E. coli replication origin. J. Biol. Chem. 280, 24627–24633 10.1074/jbc.M503684200 [DOI] [PubMed] [Google Scholar]
- 27. Nozaki S., and Ogawa T. (2008) Determination of the minimum domain II size of Escherichia coli DnaA protein essential for cell viability. Microbiology 154, 3379–3384 10.1099/mic.0.2008/019745-0 [DOI] [PubMed] [Google Scholar]
- 28. Iyer L. M., Leipe D. D., Koonin E. V., and Aravind L. (2004) Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 146, 11–31 10.1016/j.jsb.2003.10.010 [DOI] [PubMed] [Google Scholar]
- 29. Kawakami H., Keyamura K., and Katayama T. (2005) Formation of an ATP-DnaA-specific initiation complex requires DnaA arginine 285, a conserved motif in the AAA+ protein family. J. Biol. Chem. 280, 27420–27430 10.1074/jbc.M502764200 [DOI] [PubMed] [Google Scholar]
- 30. Erzberger J. P., Mott M. L., and Berger J. M. (2006) Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 13, 676–683 10.1038/nsmb1115 [DOI] [PubMed] [Google Scholar]
- 31. Duderstadt K. E., Chuang K., and Berger J. M. (2011) DNA stretching by bacterial initiators promotes replication origin opening. Nature 478, 209–213 10.1038/nature10455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Noguchi Y., Sakiyama Y., Kawakami H., and Katayama T. (2015) The Arg fingers of key DnaA protomers are oriented inward within the replication origin oriC and stimulate DnaA subcomplexes in the initiation complex. J. Biol. Chem. 290, 20295–20312 10.1074/jbc.M115.662601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Saxena R., Stanley C. B., Kumar P., Cuneo M. J., Patil D., Jha J., Weiss K. L., Chattoraj D. K., and Crooke E. (2020) A nucleotide-dependent oligomerization of the Escherichia coli replication initiator DnaA requires residue His136 for remodeling of the chromosomal origin. Nucleic Acids Res. 48, 200–211 10.1093/nar/gkz939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fujikawa N., Kurumizaka H., Nureki O., Terada T., Shirouzu M., Katayama T., and Yokoyama S. (2003) Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 31, 2077–2086 10.1093/nar/gkg309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. O'Donnell M. E., and Li H. (2018) The ring-shaped hexameric helicases that function at DNA replication forks. Nat. Struct. Mol. Biol. 25, 122–130 10.1038/s41594-018-0024-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oakley A. J. (2019) A structural view of bacterial DNA replication. Protein Sci. 28, 990–1004 10.1002/pro.3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Itsathitphaisarn O., Wing R. A., Eliason W. K., Wang J., and Steitz T. A. (2012) The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell 151, 267–277 10.1016/j.cell.2012.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thirlway J., Turner I. J., Gibson C. T., Gardiner L., Brady K., Allen S., Roberts C. J., and Soultanas P. (2004) DnaG interacts with a linker region that joins the N- and C-domains of DnaB and induces the formation of 3-fold symmetric rings. Nucleic Acids Res. 32, 2977–2986 10.1093/nar/gkh628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bailey S., Eliason W. K., and Steitz T. A. (2007) Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science 318, 459–463 10.1126/science.1147353 [DOI] [PubMed] [Google Scholar]
- 40. Makowska-Grzyska M., and Kaguni J. M. (2010) Primase directs the release of DnaC from DnaB. Mol. Cell 37, 90–101 10.1016/j.molcel.2009.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Saluja D., and Godson G. N. (1995) Biochemical characterization of Escherichia coli temperature-sensitive dnaB mutants dnaB8, dnaB252, dnaB70, dnaB43, and dnaB454. J. Bacteriol. 177, 1104–1111 10.1128/jb.177.4.1104-1111.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bramhill D., and Kornberg A. (1988) Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell 52, 743–755 10.1016/0092-8674(88)90412-6 [DOI] [PubMed] [Google Scholar]
- 43. Weigel C., and Seitz H. (2002) Strand-specific loading of DnaB helicase by DnaA to a substrate mimicking unwound oriC. Mol. Microbiol. 46, 1149–1156 10.1046/j.1365-2958.2002.03232.x [DOI] [PubMed] [Google Scholar]
- 44. Lyskov S., and Gray J. J. (2008) The RosettaDock server for local protein-protein docking. Nucleic Acids Res. 36, W233–W238 10.1093/nar/gkn216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chang P., and Marians K. J. (2000) Identification of a region of Escherichia coli DnaB required for functional interaction with DnaG at the replication fork. J. Biol. Chem. 275, 26187–26195 10.1074/jbc.M001800200 [DOI] [PubMed] [Google Scholar]
- 46. Kaplan D. L., and O'Donnell M. (2004) Twin DNA pumps of a hexameric helicase provide power to simultaneously melt two duplexes. Mol. Cell 15, 453–465 10.1016/j.molcel.2004.06.039 [DOI] [PubMed] [Google Scholar]
- 47. Carney S. M., Gomathinayagam S., Leuba S. H., and Trakselis M. A. (2017) Bacterial DnaB helicase interacts with the excluded strand to regulate unwinding. J. Biol. Chem. 292, 19001–19012 10.1074/jbc.M117.814178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Eisenberg D., Schwarz E., Komaromy M., and Wall R. (1984) Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 179, 125–142 10.1016/0022-2836(84)90309-7 [DOI] [PubMed] [Google Scholar]
- 49. Baker N. A., Sept D., Joseph S., Holst M. J., and McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A 98, 10037–10041 10.1073/pnas.181342398 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.