Abstract
The Arf GTPase-activating protein (Arf GAP) with SH3 domain, ankyrin repeat and PH domain 1 (ASAP1) establishes a connection between the cell membrane and the cortical actin cytoskeleton. The formation, maintenance, and turnover of actin filaments and bundles in the actin cortex are important for cell adhesion, invasion, and migration. Here, using actin cosedimentation, polymerization, and depolymerization assays, along with total internal reflection fluorescence (TIRF), confocal, and EM analyses, we show that the N-terminal N-BAR domain of ASAP1 directly binds to F-actin. We found that ASAP1 homodimerization aligns F-actin in predominantly unipolar bundles and stabilizes them against depolymerization. Furthermore, the ASAP1 N-BAR domain moderately reduced the spontaneous polymerization of G-actin. The overexpression of the ASAP1 BAR–PH tandem domain in fibroblasts induced the formation of actin-filled projections more effectively than did full-length ASAP1. An ASAP1 construct that lacked the N-BAR domain failed to induce cellular projections. Our results suggest that ASAP1 regulates the dynamics and the formation of higher-order actin structures, possibly through direct binding to F-actin via its N-BAR domain. We propose that ASAP1 is a hub protein for dynamic protein–protein interactions in mechanosensitive structures, such as focal adhesions, invadopodia, and podosomes, that are directly implicated in oncogenic events. The effect of ASAP1 on actin dynamics puts a spotlight on its function as a central signaling molecule that regulates the dynamics of the actin cytoskeleton by transmitting signals from the plasma membrane.
Keywords: actin, actin-binding protein, Arf GAP with SH3 domain ankyrin repeat and PH domain 1 (ASAP1), N-BAR domain, cytoskeleton, circular dorsal ruffles, focal adhesion, membrane dynamics, invadopodia, mechanosensing, enzyme mechanism, protein–protein interaction, protein complex
The spatiotemporal reorganization of the actin cytoskeleton is often coupled with the deformation of membranes in cellular processes including migration, adhesion, and division (1–3). The actin cytoskeleton receives, integrates, and responds to intra- and extracellular signaling events by changing its organization. Changes in actin organization are driven by the regulated assembly of globular actin (G-actin) into filamentous actin (F-actin), the formation of distinct higher-order F-actin structures, such as bundles, and the coordinated disassembly of F-actin to G-actin (4). The cortical actin cytoskeleton is highly dynamic, and interactions between the actin cytoskeleton and the juxtapositioned plasma membrane are often directly mediated by actin-binding motifs in membrane-associated molecules and lipid-dependent activation mechanisms (1, 5–8).
ASAP1, an Arf GTPase-activating protein (Arf GAP), is a direct mediator of the membrane-associated actin cytoskeleton and is critical for the regulation of circular dorsal ruffles, focal adhesions, invadopodia, and podosomes (5, 9–12). Unifying features of these cytoskeletal structures are thick actin bundles that assemble and disassemble in a strictly controlled manner. How ASAP1 regulates the actin cytoskeleton is not fully understood. ASAP1 is a multifunctional scaffolding and signaling protein (Fig. 1A) that interacts through its C-terminal Src homology 3 (SH3) domain with actin cytoskeleton effectors, such as focal adhesion kinase (13), and through its proline-rich domain with Src-kinases and CrkL (5, 10, 14). The N-terminal N-BAR domain mediates ASAP1 dimerization and interacts with the actin-based molecular motor nonmuscle myosin-2A (11). The central pleckstrin homology (PH) domain regulates the catalytic GAP domain, which converts Arf·GTP to Arf·GDP (5). PI(4,5)P2 binding to the PH domain activates ASAP1 GAP activity (15). Overexpression of ASAP1 is reported in several mammalian cancers and associated with enhanced cellular migration or invasion rates that promote metastasis (16–18).
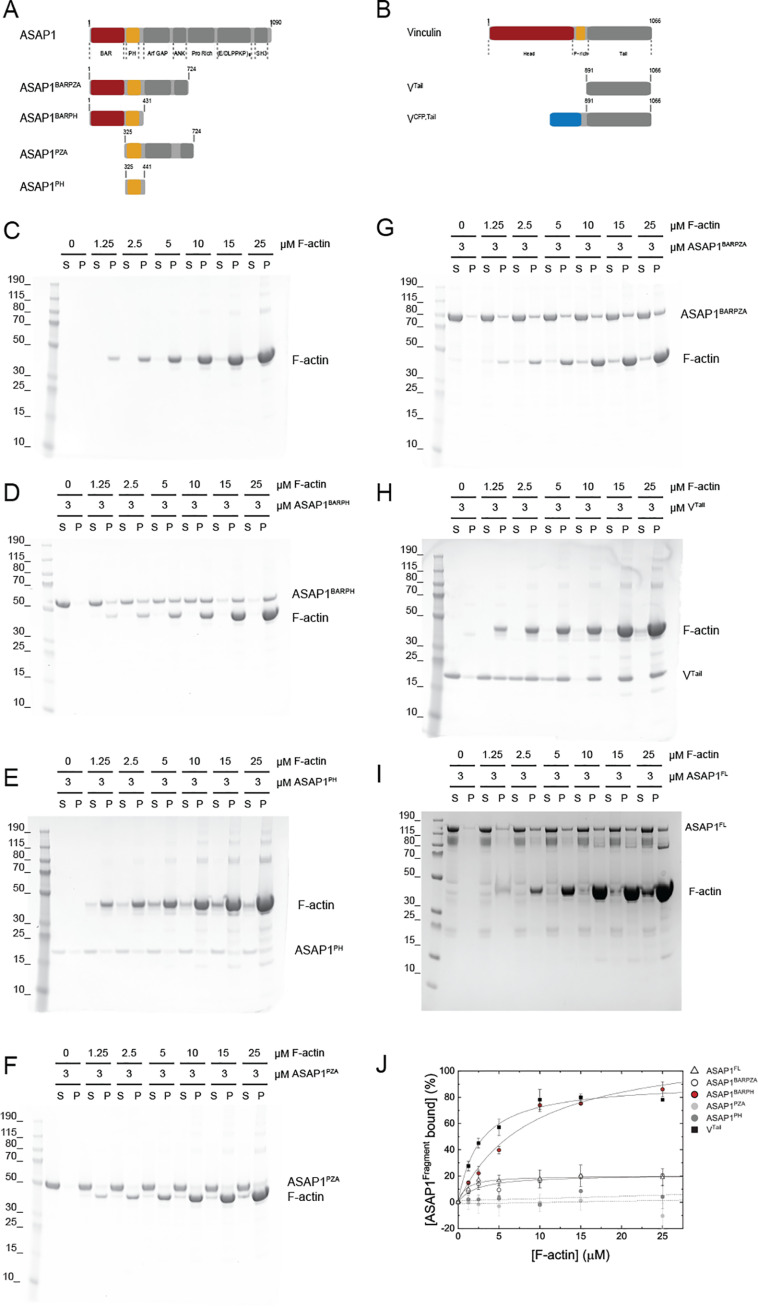
Figure 1.
The ASAP1 N-BAR domain directly binds to F-actin. A, domain organization of ASAP1 and ASAP1 fragments used in this study. B, domain organization of vinculin and vinculin fragments used in this study. C, control sedimentation of increasing concentrations of F-actin. The abbreviations S and P refer to supernatant and pellet, respectively. D, the soluble ASAP1BARPH cosediments with F-actin. E, ASAP1PH remains soluble and does not cosediment with F-actin in the concentration range tested. F, ASAP1PZA does not cosediment with F-actin, whereas the longer fragment ASAP1BARPZA partially cosediments with F-actin (G) but to a reduced extent compared with that of ASAP1BARPH (D). H, VTail cosediments with F-actin to an extent similar to that of ASAP1BARPH. ASAP1FL (I) cosediments with F-actin to an extent similar to that of ASAP1BARPZA (G). J, Quantification (mean ± S.D.) of the sedimentation behavior of ASAP1 fragments and VTail with F-actin (n = 2–3).
We investigated the possibility that ASAP1 functions not only as a scaffolding hub protein for well-described regulators of the actin cytoskeleton at the cytoskeleton-membrane interface but also as an F-actin binding protein. The data presented here reveal the direct interaction between ASAP1 and F-actin: the ASAP1 N-BAR domain directly binds F-actin and aligns it into predominantly unipolar bundles in vitro. Binding of ASAP1 to F-actin protects actin bundles from depolymerization and reduces the spontaneous polymerization of G-actin in vitro. In vivo, overexpression of ASAP1BARPH induces long cellular projections in fibroblasts, underlining its essential role at the cytoskeleton–plasma membrane interface. In summary, ASAP1BARPH inhibits actin turnover likely by stabilizing F-actin bundles, as they are found in focal adhesions and invadopodia, structures that are of importance in cancer cell migration and progression (19).
Results
Considering the pivotal role of ASAP1 in F-actin-rich structures, such as circular dorsal ruffles, focal adhesions, invadopodia, and podosomes, we investigated whether ASAP1 could directly interact with F-actin.
ASAP1BARPH directly binds to F-actin
To test for a potential interaction between ASAP1 and F-actin, we used high-speed centrifugation assays in which soluble binding proteins (Fig. 1, A–B and D–H) will cosediment with F-actin (Fig. 1C). As a control for a positive binding event, we used the well-characterized tail domain of vinculin (VTail) that binds to F-actin under the assay conditions (Fig. 1H; ∼78% at 25 μm F-actin).
We first addressed whether ASAP1FL would interact and cosediment with F-actin. Increasing concentrations of F-actin resulted in the sedimentation of ∼20% ASAP1FL at 25 μm F-actin (Fig. 1I). Next, we tested ASAP1BARPZA, which contains the three domains that support regulated catalysis and the N-BAR domain, in actin cosedimentation assays. Similar to ASAP1FL, ∼20% ASAP1BARPZA sedimented at 25 μm F-actin (Fig. 1G). To test whether this low extent of binding would be indicative for weak F-actin binding properties of ASAP1FL and ASAP1BARPZA or may be caused by autoinhibition (20), we tested a fragment of ASAP1 lacking the N-BAR domain but containing all domains necessary for ASAP1 GTPase-activating (GAP) activity, ASAP1PZA. ASAP1PZA did not sediment with F-actin to an appreciable extent compared with ASAP1BARPZA (Fig. 1F). This observation led us to test ASAP1BARPH, a short and catalytically inactive ASAP1 fragment. In contrast to ASAP1BARPZA and ASAP1PZA, ASAP1BARPH strongly bound to F-actin and showed saturated binding at substoichiometric concentrations (Fig. 1D, ∼86% at 25 μm F-actin). ASAP1PH, which lacks the N-BAR domain, did not bind F-actin to an appreciable extent under the same assay conditions (Fig. 1E). The isolated ASAP1 N-BAR domain is refractory to purification and could not be tested (11). The densitometric analysis of the sedimentation data is shown in Fig. 1 and reveals the extent of binding in the following order: ASAP1BARPH > ASAP1FL ≈ ASAP1BARPZA > ASAP1PH ≈ ASAP1PZA.
To investigate the specificity and reversibility of the interaction between ASAP1BARPH and F-actin, we performed cosedimentation assays as a function of the ionic strength. Increasing [NaCl] led to a decreased extent of complex formation between ASAP1BARPH and F-actin (Fig. S1A). A similar behavior was observed for the interaction between F-actin and VCFP,Tail (Fig. S1B). ASAP1BARPH and VCFP,Tail did not show a salt dependence in their sedimentation behavior and remained soluble in the absence of F-actin (Fig. S1, C–D). Taken together, the cosedimentation data reveal a direct interaction between ASAP1 and F-actin that is mediated by the amino-terminal N-BAR domain of ASAP1.
ASAP1BARPH aligns F-actin into thick bundles
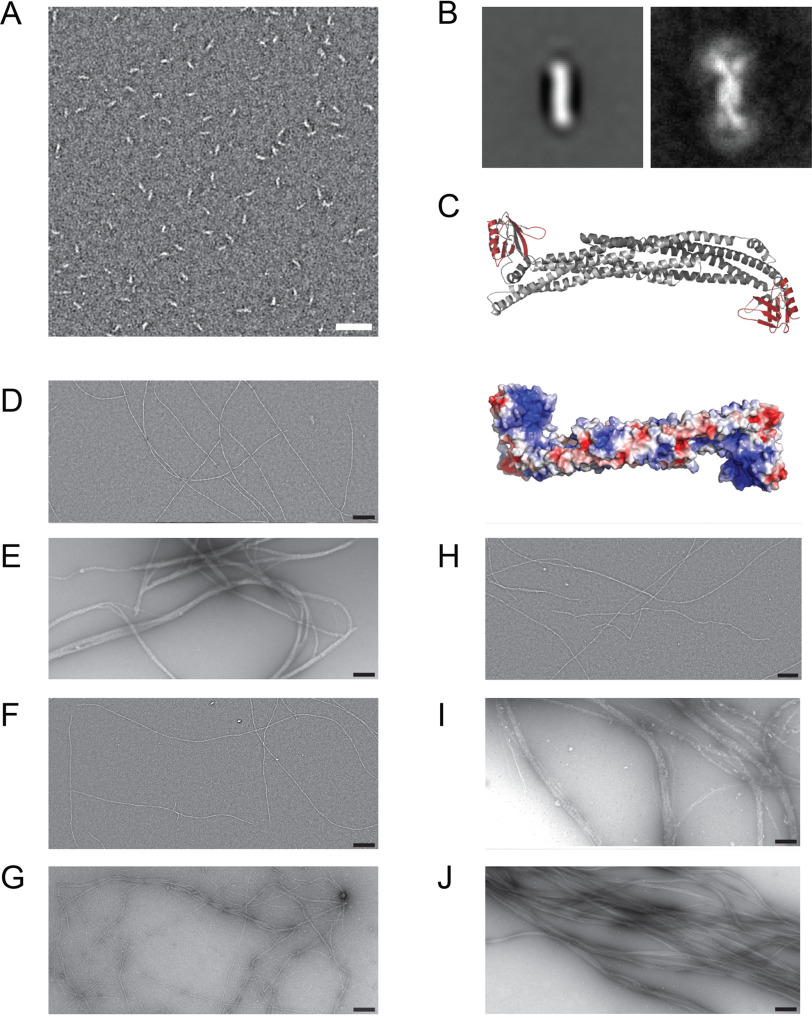
Our previous biochemical characterization showed that the ASAP1 N-BAR domain forms antiparallel homodimers (21). Concomitantly, a crescent-shaped structure was observed for ASAP1BARPH in negative-stain EM (Fig. 2A). Class averages revealed that the central BAR dimer is 4–5 nm wide and 15–16 nm long (Fig. 2B). The global average and variance images shown in Fig. 2B indicate that the PH domain is flexibly linked to the N-BAR domain. The length of the molecule extended to 26–27 nm in cases where the PH domain was directly in line with the N-BAR domains. A more systematic classification of the ASAP1BARPH fragment is shown in Fig. S2. Homology modeling of ASAP1BARPH (Fig. 2C) and superimposition with the obtained electron micrographs showed that the ASAP1PH domains extend from both ends of the molecule. Homology modeling and sequence analysis of ASAP1BARPH further indicated that the concave surface formed by the N-BAR domains contains clusters of positively charged amino acids (Fig. 2C, bottom). Based on the salt dependence of the actoASAP1BARPH interaction (Fig. S1A) and the results from previous studies on the F-actin binding properties of the F-BAR protein pacsin2 and PICK1 (22, 23), we hypothesize that electrostatic interactions between positively charged clusters on the concave surface of the ASAP1 N-BAR domain mediate binding to the negatively charged surface of F-actin.
Figure 2.
Electron micrographs of ASAP1BARPH and reconstituted F-actin:ASAP1 fragment complexes. A, EM micrograph of 100 nm ASAP1BARPH. Scale bar, 50 nm. B, Global average (left) and variance (right) images of ASAP1BARPH showing the curved structure of the tandem dimer and the flexibility of the PH domain at both ends of the molecule. The PH domain is not seen in the global average due to its variable position relative to the N-BAR domain. In the variance image, the position of the PH domain can be seen as a white arc (high variability) at each end of the molecule. Window size 36.8 nm by 36.8 nm. In the most extended conformation, the molecule is 4–5 nm wide and 26–27 nm long. C, top, homology model of ASAP1BARPH. The N-BAR domain dimer is colored gray and the PH domains are red. The long axis of the N-BAR domain is ∼14.4 nm long. Bottom, the ASAP1BARPH surface is colored according to the electrostatic potential distribution (red, −5 KbT/ec; blue, −5 KbT/ec). D–I, electron micrographs of 1000 nm F-actin (D) incubated with 1000 nm ASAP1BARPH (E), 1000 nm ASAP1PH (F), 1000 nm ASAP1FL (G), 1000 nm ASAP1PZA (H), 1000 nm ASAP1BARPZA (I), or 1000 nm VTail as a control (J). Scale bar, 200 nm.
Homodimerization of ASAP1BARPH by means of its N-terminal BAR domain creates a protein complex with potentially two F-actin binding sites that may cross-link F-actin into bundles. The cosedimentation assays shown in Fig. 1, D–H do not distinguish between F-actin binding and bundling activity, as both single actin filaments and bundles sediment at high speeds. To test the hypothesis that ASAP1BARPH bundles F-actin, we incubated F-actin with a series of ASAP1 fragments and used EM to visualize the protein complexes. The electron micrograph in Fig. 2D shows the filamentous structure of actin alone as a reference. In contrast, the reconstituted actoASAP1BARPH complex formed straight, axially aligned dense bundles (Fig. 2E) that resemble actoVTail bundles in their ultrastructure (Fig. 2J). F-actin bundling was also observed for ASAP1FL (Fig. 2G) and ASAP1BARPZA (Fig. 2I) but to a reduced extent compared with that of ASAP1BARPH (Fig. 2E), in agreement with the reduced binding properties observed in F-actin cosedimentation assays (Fig. 1, I and G). Unlike ASAP1BARPH and ASAP1BARPZA (Fig. 2, E and I), ASAP1PH (Fig. 2F) and ASAP1PZA (Fig. 2H) did not significantly bind and bundle F-actin. Notably, the dense, three-dimensional higher-order F-actin bundles did not allow the determination of the binding mode of ASAP1 fragments in the bundle.
ASAP1BARPH preferentially aligns F-actin into unipolar bundles
To explore the actoASAP1BARPH bundles and their polarity in more detail, we used total internal reflection fluorescence (TIRF) microscopy. First, we observed the ASAP1BARPH-induced bundling of F-actin in an endpoint assay in which ATTO488-labeled G-actin was polymerized in the presence of increasing concentrations of ASAP1BARPH. The resulting actin structures were analyzed after 10 min (Fig. 3, A and B). As shown in Fig. 3A, increasing concentrations of ASAP1BARPH aligned actin filaments in dense bundles that contain up to a maximum ∼6 actin filaments under the experimental conditions (Fig. 3B).
Figure 3.
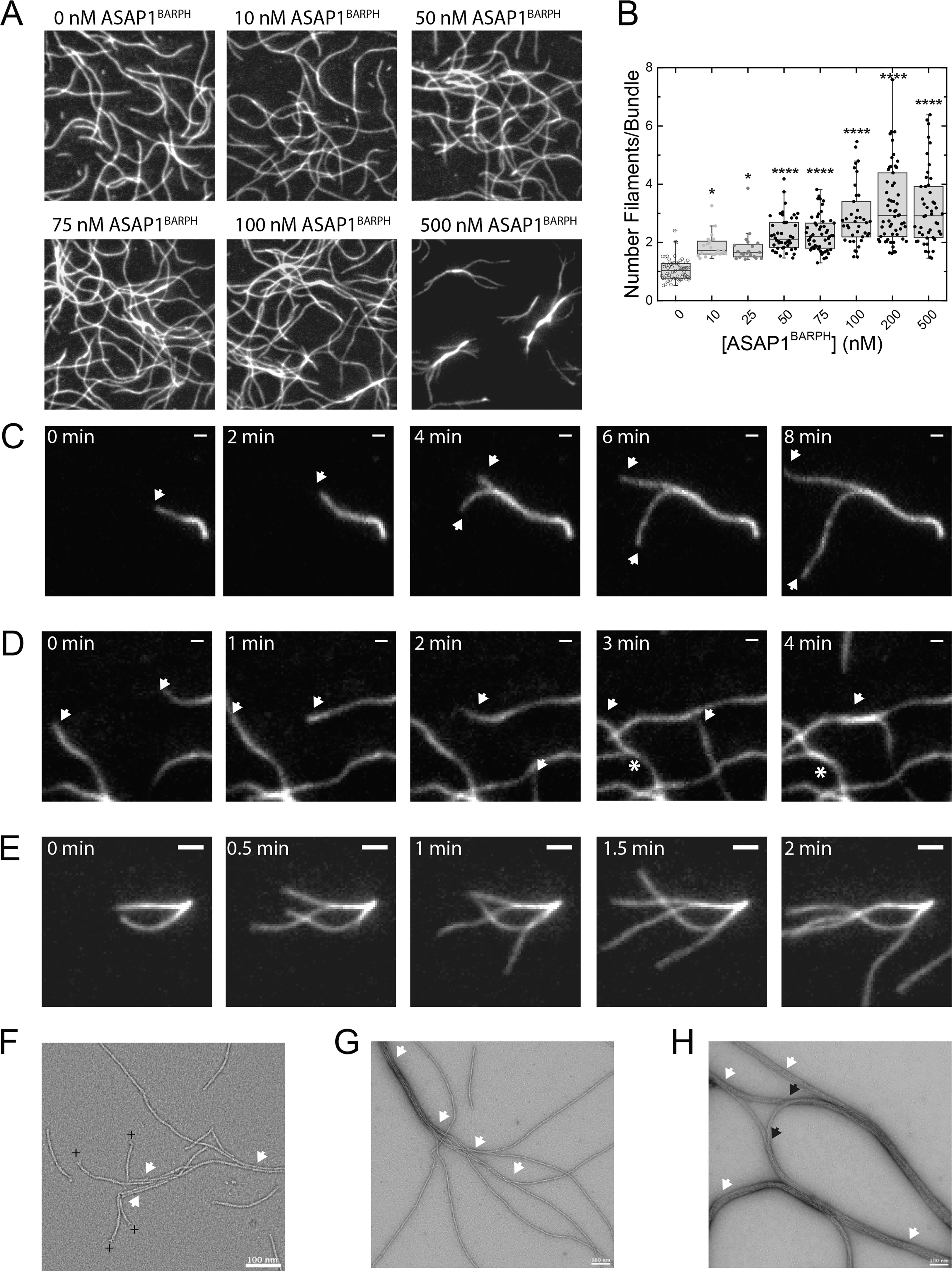
ASAP1BARPH aligns F-actin in predominantly unipolar bundles. A, actin polymerization endpoint assay. 500 nm ATTO488-labeled G-actin was polymerized in the absence or presence of increasing concentrations ASAP1BARPH and the resulting actin structures imaged after 10 min. The dimensions of the panels are 30 μm by 30 μm. B, increasing concentrations of ASAP1BARPH increase the number of actin filaments per bundle (n = 31–63). ****, p < 0.0001, and *, p < 0.05, using one-way ANOVA with Dunnett's multiple-comparison test. C–D, montage of images showing the time-dependent elongation of individual actin filaments in the presence of ASAP1BARPH. White arrows indicate parallel growth/bundling, and white asterisks indicate antiparallel growth/bundling events. C, Polymerization of 500 nm ATTO488-labeled G-actin in the presence of 500 nm ASAP1BARPH. Time interval, 2 min; scale bar, 1 μm. D, polymerization of 500 nm ATTO488-labeled G-actin in the presence of 50 nm ASAP1BARPH. Time interval, 1 min; scale bar, 1 μm. E, polymerization of 500 nm ATTO488-labeled G-actin in the presence of 25 nm ASAP1BARPH. Time interval, 30 s; scale bar, 2 μm. F–H, electron micrographs of 1000 nm F-actin capped with 20 nm gelsolin and incubated with 80 nm ASAP1BARPH (F), 80 nm ASAP1BARPH (G), or 800 nm ASAP1BARPH (H). The white arrows indicate parallel, the black arrows indicate likely antiparallel F-actin bundles. The plus sign indicates the barbed end of the actin filament. Scale bar, 100 nm.
To determine whether the actoASAP1BARPH bundles are of mixed or uniform polarity, the polymerization of ATTO488-labeled G-actin into filaments in the presence of ASAP1BARPH was observed with TIRF microscopy. Bundling events of two polymerizing filaments were identified and the polarity of the bundle determined. As shown in Fig. 3, C and D, unipolar and bipolar fusion events were observed. We also observed that in the presence of ASAP1BARPH, polymerizing actin filaments often originated from small bundles with unipolar polarity (Fig. 3E), leading to the hypothesis that ASAP1BARPH preferentially aligns actin filaments into unipolar bundles.
The same trend was observed in EM, where the orientation of individual actin filaments in short, actoASAP1BARPH bundles was determined by labeling the actin barbed end with a gelsolin cap prior to bundle formation (Fig. 3F).
Gelsolin is an F-actin-severing protein that remains attached to the barbed end of the actin filament as a cap after severing (24). The presence of a gelsolin cap was used to determine the polarity of the shortened actin filaments. Quantification of n = 77 filaments showed that 92.2% (n = 71) of the filaments in actin bundles are parallel and 7.8% antiparallel (n = 6). The observed, predominantly unipolar polarity of actoASAP1BARPH bundles agrees with the polarity of actin found in focal adhesions and cellular structures where ASAP1 and F-actin colocalize (25). In the absence of gelsolin, ASAP1BARPH paired actin filaments and promoted the formation of thick bundles. The presence of fully bundled and individual actin filaments indicates a cooperative binding mode (Fig. 3G) that agrees with the small actin bundles observed in TIRF microscopy (Fig. 3, A and E). Under saturating conditions, ASAP1BARPH completely aligned actin into thick unipolar bundles that can associate laterally into higher-order bipolar arrangements (Fig. 3H).
ASAP1BARPH stabilizes F-actin bundles against depolymerization
ASAP1 localizes to thick F-actin bundles in circular dorsal ruffles and focal adhesions (11) in vivo and induces F-actin bundling in vitro (Fig. 2E). To test whether ASAP1BARPH would stabilize F-actin within these bundles, we investigated its effect on the dilution-induced depolymerization of F-actin. Increasing concentrations of ASAP1BARPH inhibited the depolymerization of F-actin in a concentration-dependent manner (Fig. 4A). The same behavior was observed for the VTail but not the isolated ASAP1PH domain (Fig. 4, B and C), which were used as positive and negative controls, respectively. In summary, the data show that substoichiometric concentrations of ASAP1BARPH stabilize F-actin bundles against depolymerization in vitro.
Figure 4.
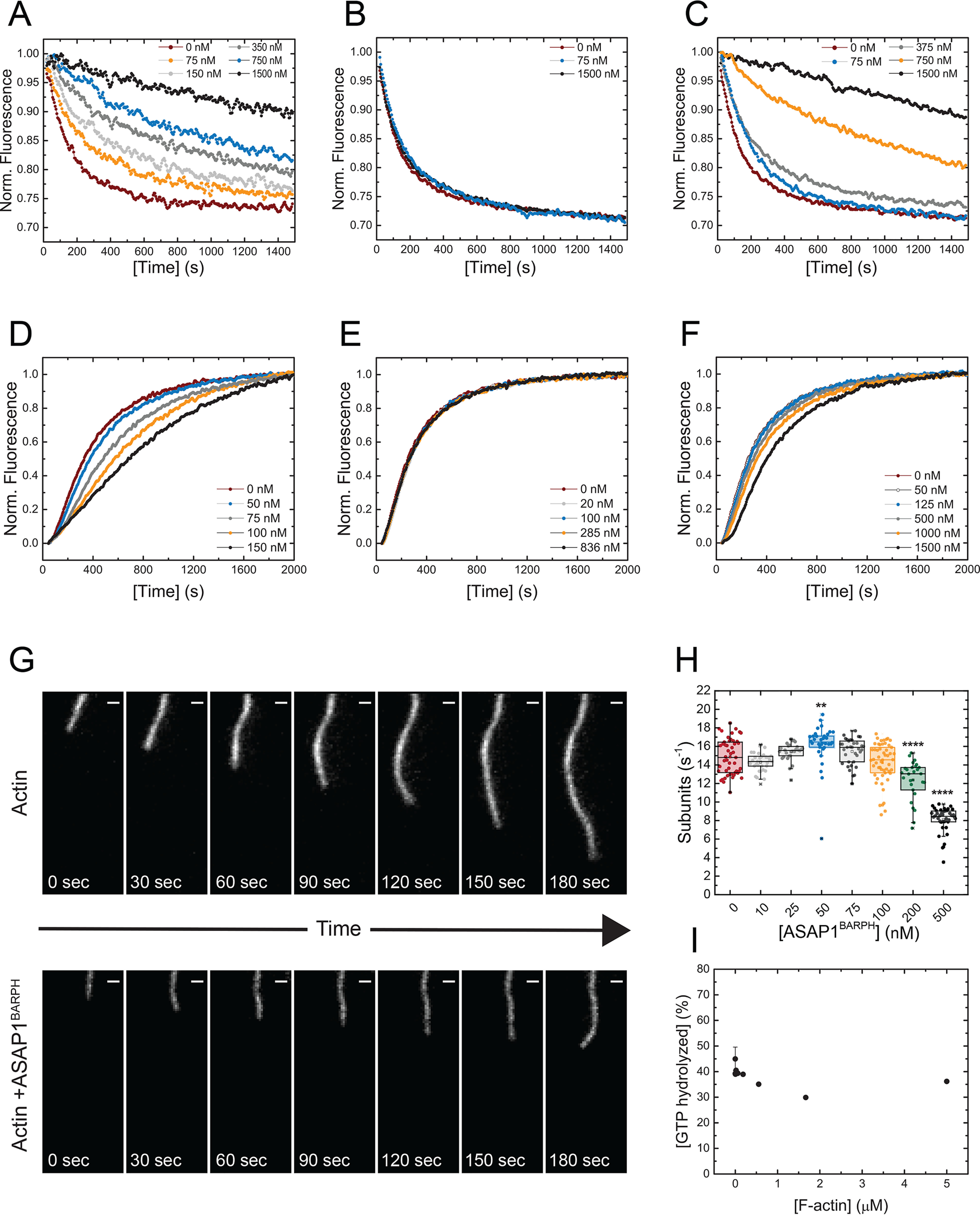
ASAP1BARPH stabilizes F-actin bundles and decreases the spontaneous polymerization of G-actin to F-actin. A, increasing concentrations of ASAP1BARPH protect F-actin bundles against depolymerization. ASAP1PH (B) and VTail (C) were used as negative and positive controls, respectively. A premixing concentration of 4 μm F-actin and the indicated concentrations of ASAP1BARPH, ASAP1PH, or VTail was used. The representative plotted fluorescence intensities are normalized to 1 for the initial fluorescence at t = 0 of each sample shown in panels A–C. D, ASAP1BARPH decreases the spontaneous polymerization of 3.5 μm G-actin (10% pyrene labeled) to F-actin. ASAP1PH does not affect actin polymerization (E), whereas VTail slightly decreases the actin polymerization rate under the same experimental conditions (F). Representative data are shown. G, montage of images showing the elongation of individual actin filaments alone (top) or in the presence of 500 nm ASAP1BARPH (bottom). Time interval, 30 s; scale bar, 1 μm. (H) Actin elongation rates as a function of ASAP1BARPH concentration (n = 28–55). ****, p < 0.0001, and **, p < 0.01, using one-way ANOVA with Dunnett's multiple-comparison test. I, F-actin does not influence the enzymatic GAP activity of ASAP1BARPZA in vitro (n = 2).
ASAP1BARPH reduces the spontaneous polymerization of G-actin
Several F-actin binding and bundling proteins alter actin polymerization (26). To test whether ASAP1BARPH affects the polymerization of G-actin to F-actin, we studied its influence on the spontaneous polymerization of G-actin in vitro. As shown in Fig. 4D, increasing concentrations of ASAP1BARPH decreased the polymerization kinetics of pyrene-labeled G-actin in a concentration-dependent manner. No significant effect on G-actin polymerization was observed for ASAP1PH, whereas the presence of VTail slightly decreased the actin polymerization kinetics (Fig. 4, E and F). In agreement with the bulk kinetic assays, increasing concentrations of ASAP1BARPH also decreased actin polymerization rates in TIRF microscopy assays in which the elongation of actin filaments was observed in real time (Fig. 4G). The presence of 500 nm ASAP1BARPH decreased the actin elongation rate around 50% compared with the control (Fig. 4H). We also observed that individual polymerizing actin filaments are straighter and less dynamic in the presence of ASAP1BARPH, indicating a stabilizing effect on the actin filament.
F-actin does not alter the enzymatic activity of ASAP1BARPZA
After we examined the effect of ASAP1BARPH on actin dynamics in vitro, we tested whether F-actin regulates the ASAP1 GAP activity, which is important for the hydrolysis of GTP bound to the small GTPase Arf1 during cytoskeletal reorganization (27). The N-BAR domain of ASAP1 has been shown to autoinhibit its GAP activity, whereas other N-BAR domain-binding proteins, such as FIP3 and nonmuscle myosin-2A, stimulate the GAP activity of recombinant ASAP1 proteins containing the N-BAR domain by 10- to 20-fold (11, 28). Unlike FIP3 and nonmuscle myosin-2A, F-actin had no detectable effect on the GAP activity of ASAP1BARPZA (Fig. 4I).
ASAP1BARPH induces the formation of cellular projections in fibroblasts
Some actin-binding proteins and BAR domain-containing proteins were previously shown to induce the formation of cellular projections (29–32).
To examine the effect of ASAP1 on actin structures in cells, we transfected NIH3T3 fibroblasts with various Flag-tagged ASAP1 constructs. Overexpression of ASAP1FL, ASAP1BARPH, or ASAP1BARPZA was sufficient to induce substantial actin remodeling in cells. The most noticeable change was the formation of long actin-filled cellular projections (Fig. 5A). In contrast, ASAP1ΔBARPH, a construct that lacks the BAR-PH tandem, failed to induce these structures (Fig. 5, A and B). We quantified the potency of ASAP1FL, ASAP1BARPZA, ASAP1ΔBARPH, and ASAP1BARPH to induce the formation of actin-filled projections by scoring the percentage of cells exhibiting at least 10 actin projections longer than 5 μm (Fig. 5B). We found that the extent to which these projections were induced correlates with the extent to which the ASAP1 fragments bind F-actin in vitro (Fig. 1J). This observation suggests that ASAP1 affects actin remodeling in cells, possibly through a direct interaction between the N-BAR domain and F-actin.
Figure 5.
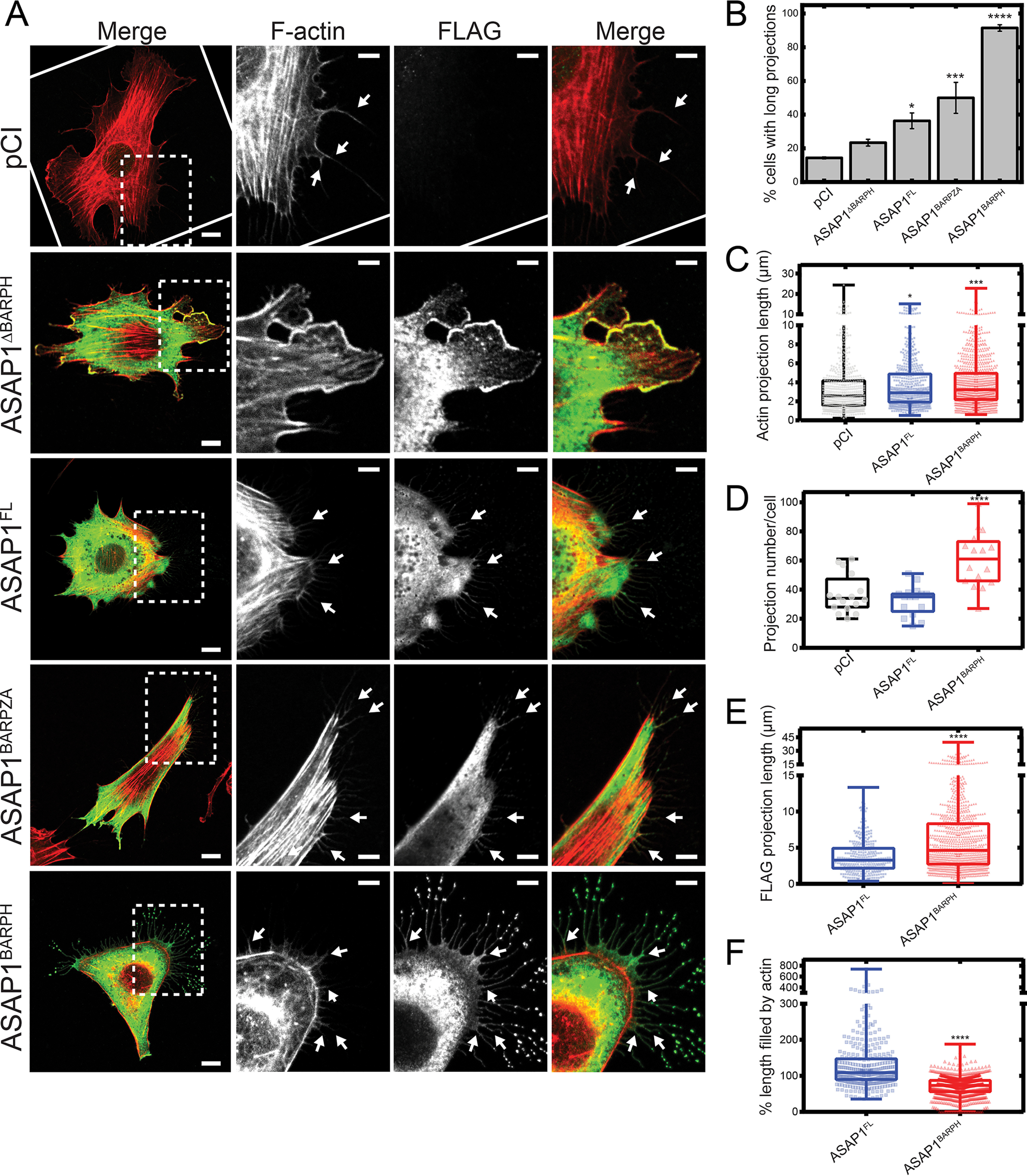
ASAP1BARPH induces the formation of cellular projections in NIH3T3 fibroblasts. Transient overexpression of ASAP1BARPH induces the formation of long projections on the surface of NIH3T3 fibroblasts. NIH3T3 fibroblasts were transfected with different Flag-tagged ASAP1 constructs and costained for F-actin and Flag-ASAP1 fragments using rhodamine-phalloidin and anti-Flag antibody, respectively. A, F-actin is shown in red and Flag-ASAP1 proteins in green in the merged images. Scale bars throughout represent 10 μm and 5 μm in whole-cell and inset images, respectively. White arrows indicate examples of actin-containing projections. B, quantification (mean ± S.E.M.) of cells with long projections (at least 10 actin projections of >5 μm) per ASAP1 construct from 3–4 experiments. An average of 60∼100 cells positive for Flag immunostaining for each construct was scored blindly for exhibiting long projections in each experiment. C, quantification of the projection length by F-actin staining (n = 589 [pCI], n = 522 [ASAP1FL], n = 967 [ASAP1BARPH]). D, quantification of the total numbers of actin projection of >0.2 μm in cells transfected with different ASAP1 constructs from 3 experiments (n = 16 [pCI], n = 16 [ASAP1FL], n = 16 [ASAP1BARPH]). E, Quantification of the projection length by anti-Flag immunostaining (n = 324 [ASAP1FL], n = 993 [ASAP1BARPH]). F, quantification of the percentage of projection length filled by actin (n = 324 [ASAP1FL], n = 993 [ASAP1BARPH]). Statistics are based on the indicated number of projections (n) from 16 cells and three experiments. Of note, ASAP1FL did not fill the entire length of the projections in ∼60% of the analyzed cases. This results in values of >100% for the projection length filled by actin. In contrast, ASAP1BARPH did not fill the entire length of the projections in ∼10% of the analyzed cases. ****, p < 0.0001, ***, p < 0.001, and *, p < 0.05, using one-way ANOVA with Dunnett's multiple-comparison test (B, C, and D) and Student's t test (E and F).
We further characterized the cellular projections induced by the overexpression of ASAP1FL and ASAP1BARPH. We found that the overexpression of ASAP1FL and ASAP1BARPH induced the formation of longer actin-filled projections than a vector control (Fig. 5C). Further, cells overexpressing ASAP1BARPH had a higher total number of projections than cells overexpressing ASAP1FL or a vector control (Fig. 5D). We noticed that the projections induced by the overexpression of ASAP1BARPH displayed vesicular structures at the tip where ASAP1BARPH was concentrated. In contrast, vesicular structures were less frequent in projections induced by the overexpression of ASAP1FL (Fig. 5A). We further measured the length of the projection induced by ASAP1FL or ASAP1BARPH by anti-Flag immunostaining and found that projections induced by ASAP1BARPH were longer than those induced by ASAP1FL (Fig. 5E). As a result, the extent to which actin filled these projections was lower in ASAP1BARPH than ASAP1FL projections (Fig. 5F).
Taken together, the data suggest that the N-BAR domain of ASAP1 is involved in the organization of the actin cytoskeleton in fibroblasts.
Discussion
The results presented here collectively demonstrate that the ASAP1 N-BAR domain directly interacts with F-actin. Dimerization of ASAP1BARPH creates a molecule that connects and aligns actin filaments into emerging higher-order, tightly packed bundles with predominantly unipolar polarity (Fig. 2E and 3, E and G). This feature qualifies ASAP1 as an actin-binding protein that organizes the eukaryotic actin cytoskeleton in a spatiotemporal manner. Although the F-actin binding properties of several BAR domain proteins have been explored in vitro, only pacsin2, PICK1, Bain1, Bin1, Gas7, PSTPIP2, MIM, and IRSp53 were described to interact with F-actin both in vitro and in vivo via their BAR domains (22, 23, 33–37). Moreover, ASAP1 and the recently described Bin1 (37) are the only N-BAR domain proteins reported to directly interact with F-actin so far.
The data presented in this study favor a model in which the ASAP1BARPH dimer binds along the lateral surface of actin filament. The putative actin-binding site or interface is solvent exposed and presumably located in the concave surface of the ASAP1BARPH dimer. Based on our experimental finding that ASAP1PH does not bind and bundle F-actin in vitro (Fig. 1E and 2F), we conclude the actin-binding site/interface lies within the N-BAR domain of ASAP1BARPH. The maximum distance between the two putative actin-binding sites of the ASAP1BARPH dimer is ∼15–16 nm (Fig. 2 and Fig. S2), spanning ∼5–6 actin subunits (2.7 nm) in an actin filament. The ASAP1 dimer may not only stabilize the filament but also cross-link actin filaments into predominantly unipolar bundles as they are found in focal adhesions and filopodia (25, 38). Bundling may also be facilitated by BAR domain tetramerization, as has been shown for the actin-bundling protein pacsin1 and PICK1 (39, 40). Intra- and interfilamental ASAP1BARPH cross-links effectively protect actin filaments from the dilution-induced depolymerization (Fig. 4A). The concentration dependence of the reaction indicates that several actin subunits can depolymerize at normal rates at low ASAP1BARPH concentrations until the subsequent cross-linked subunit is reached, which delays depolymerization. Therefore, increasing ASAP1BARPH concentrations would increase the number of cross-linked F-actin bundles and delay depolymerization (41, 42). We detected a small decrease in actin polymerization kinetics at low substoichiometric molar ratios of ASAP1BARPH (Fig. 4D), as they are likely found in eukaryotic cells (43). Higher actin:ASAP1BARPH ratios result in an ∼50% decrease in actin polymerization rates, likely caused by actin bundling under the in vitro conditions. In a cellular context, we expect that the effect of ASAP1 on actin polymerization is minor compared with its effects on actin bundling and the protection of bundles from depolymerization.
The observation that the lipid-binding ASAP1PH domain, the catalytic GAP domain, the proline-rich domain, and the SH3 domain are not required for F-actin binding in vitro suggests that ASAP1 interacts with phospholipids, its substrate Arf, and other known binding partners of these domains simultaneously with F-actin. Consistent with this idea, we found that F-actin does not significantly change the enzymatic GAP activity of ASAP1BARPZA (Fig. 4I), underlining that F-actin binding and other biochemical functions of ASAP1 are not mutually exclusive.
While this work was under revision, an independent study was published that described the interaction between ASAP1 and F-actin (44). Corroborating our results, this study found that the N-BAR domain of ASAP1 directly binds to and bundles actin filaments in vitro. This study showed a correlation between ASAP1 expression and F-actin levels in cells. Further, it showed that reduced ASAP1 expression results in the depletion of ventral stress fibers, whereas increased ASAP1 expression results in more pronounced stress fibers and the appearance of cellular projections. Our cellular studies focus on cellular projections extending from the cell periphery in more detail, as the effect of ASAP1 expression on this type of structure has not been characterized extensively (5, 12). Moreover, the results from both studies suggest that the ASAP1 N-BAR domain is under autoinhibitory regulation, as has previously been described for other BAR domain proteins, such as PICK1, syndapin, and FCHSD2 (45, 46). Both studies show that the region C terminal to the BAR-PH tandem of ASAP1 negatively affects its F-actin binding properties in vitro and the induction of actin-filled projections in cells. Future studies will test if protein–protein interactions and/or posttranslational modifications in the C-terminal region of ASAP1 relieve the autoinhibition to regulate the N-BAR–F-actin interaction in cells.
The mechanism by which ASAP1 drives the formation of actin-filled cellular projections remains elusive. Our in vitro and in vivo results, together with the known biophysical properties of ASAP1 (20), suggest that ASAP1 induces the formation of actin-filled cellular projections by the actin-binding and -bundling activity of its N-BAR domain. In contrast, other BAR domain proteins, including IRSp53, IRTKS, FCHSD2, and srGAPs, likely induce the formation of cellular projections, such as filopodia, microvilli, or stereocilia, by interacting via their BAR domain with the membrane and via their C terminus with regulators of the actin cytoskeleton, including WASP/N-WASP, EPS8, or FMNL1 (34, 47–53).
Only IRSp53 and IRTKS directly bind and bundle actin filaments in vitro, although there is controversy in the literature (34, 53, 54). For IRSp53, which binds to lipids and actin via its I-BAR domain, membrane binding rather than actin binding has been shown to drive filopodia formation (51). In contrast to the actin-free cellular projections induced by the overexpression of the IRSp53 I-BAR domain (51, 55, 56), we show that the overexpression of ASAP1BARPH promotes the formation of actin-filled cellular projections (Fig. 5). This suggests that ASAP1BARPH is sufficient to couple actin to the membrane within these projections. ASAP1 contains an N-BAR domain, which binds to and induces convex membrane curvature (20, 21). Therefore, unlike the I-BAR-containing proteins IRSp53 and IRTKS, which bind to concave membrane curvature, or the F-BAR proteins FCHSDs and srGAPs, which bind to flatter membrane surfaces, it is unlikely that ASAP1BARPH induces cellular projections solely by membrane deformation. This does not exclude that membrane binding or bending can contribute to the formation of ASAP1-dependent cellular projections, which can be facilitated by the PH domain and Arf GAP domain, which binds to membrane-associated Arf·GTP, or simultaneous binding of lipids and F-actin by the N-BAR domain. To this end, it remains elusive whether and how the ASAP1 N-BAR domain discriminates between actin and membrane binding in cells. We speculate that the interaction with binding partners and changes in local concentration of actin and lipids plays a role in this process.
ASAP1 localizes to several distinct actin structures, including focal adhesions, invadopodia, podosomes, and circular dorsal ruffles (12). Based on the data presented in this study, we suggest that ASAP1 bundles and transiently stabilizes actin filaments in these structures. In this context, ASAP1 may control the assembly and/or resolution of the cellular structures by linking the actin cytoskeleton to the plasma membrane and functioning as a scaffolding hub for regulators of the actin cytoskeleton, such as FAK, Src, and the molecular motor nonmuscle myosin-2A (11). Consistent with this idea, the N-BAR and SH3 domains and Src-mediated phosphorylation of ASAP1 contribute to the formation of invadopodia and podosomes (57).
Moreover, the finding that ASAP1 preferentially aligns F-actin into unipolar bundles is in line with the known unipolarity of actin filaments in invadopodia and podosomes and at the ends of stress fibers (19, 58). ASAP1 colocalizes with F-actin only at the junction between stress fibers and focal adhesions instead of the whole length of the stress fibers (11). The ends of stress fibers emanating from focal adhesions contain unipolar actin bundles, whereas the central region of stress fibers contains bipolar actin filaments.
The stabilizing effect of ASAP1 on F-actin bundles and turnover may also be of importance in cancer cell migration and invasion from the signaling perspective. The actin cytoskeleton supports cellular signaling events, and altered actin-bundling dynamics modulate signaling pathways that result in cancer initiation, progression, and metastasis (19), processes that are associated with ASAP1 overexpression (16–18). Therefore, the actin-bundling and stabilizing properties of ASAP1 might be part of its function in cancer cell migration and metastasis that are often associated with the alteration in the activities of F-actin bundling proteins (19).
Experimental procedures
Protein overproduction and preparation
Flag-tagged full-length mouse ASAP1b (accession number AAC98350.1), ASAP1FL (amino acids 1–1090), was expressed in the baculovirus/Sf9 insect cell system, and polyhistidine-tagged mouse ASAP1b fragments ASAP1BARPZA (amino acids 1–724), ASAP1PZA (amino acids 325–724), ASAP1BARPH (amino acids 1–431), ASAP1PH (amino acids 325–441), the tail domain of human vinculin (VTail; amino acids 891–1066, accession number AAH39174.1), and CFP-tagged VTail (VCFP,Tail, amino acids 891–1066) were recombinantly overproduced in Escherichia coli and purified to electrophoretic homogeneity as described previously (7, 20, 59). G-actin was prepared from rabbit skeletal muscle acetone powder (Pel-Freez Biologicals) (60) or purchased as ATTO488- or ATTO565-labeled G-actin (Hypermol). Pyrene-labeled G-actin was from Hypermol or Cytoskeleton. N-ethylmaleimide (NEM)-modified rabbit skeletal muscle myosin-2 was from Hypermol, and human gelsolin was purchased from Cytoskeleton.
F-actin cosedimentation assay
For cosedimentation assays shown in Fig. 1, 3 μm ASAP1, ASAP1 fragment, or VTail was incubated with increasing concentrations of F-actin (0–25 μm) in PBS supplemented with 1 mm MgCl2, incubated for 10 min at 22 °C, and sedimented for 15 min at 100,000 × g at 4 °C in a TLA-100 rotor in a Beckman Ultima MAX-XP ultracentrifuge. Following sedimentation, the supernatant was removed and the pellet resuspended in an equal volume of assay buffer. Supernatant and pellet fractions were resolved on a 4–12% Bis-Tris gel (Thermo Fisher Scientific). The gel was stained with PageBlue (Thermo Fisher Scientific), destained with water, and documented with an Odyssey system (Li-Cor Biosciences) or a ChemiDoc MP (Bio-Rad) imaging system. Fiji was used to quantify the band intensity by densitometry and corrected for the amount of ASAP1 fragment or VTail that sediments in the absence of F-actin (61). Data were plotted with Origin (OriginLab). The salt dependence of the interaction of ASAP1BARPH/VCFP,Tail and F-actin was accessed by centrifugation of 3 μm ASAP1BARPH/VCFP,Tail alone or in the presence of 15 μm F-actin in buffer containing 51.25–532.5 mm NaCl. Samples were processed and analyzed as described above.
Actin polymerization and depolymerization assays
Polymerization of 3.5 μm MgATP–G-actin (10% pyrene labeled) was induced by the addition of 1/10 10× KMEI (500 mm KCl, 100 mm imidazole, pH 7.0, 10 mm MgCl2, 10 mm EGTA) in the presence and absence of various concentrations (0.075–1.5 μm) of ASAP1BARPH, ASAP1PH, or VTail. The time-dependent change in pyrene fluorescence was recorded in a QuantaMaster fluorescence spectrophotometer (Photon Technology International; excitation wavelength, 365 nm; emission wavelength, 407 nm) at 20 °C. For polymerization data analysis, the dead time was added back to the data, the baseline fluorescence at the start of the reaction was subtracted, and the data were normalized by division by the plateau fluorescence (62). For dilution-induced F-actin depolymerization assays, 10 µl of 4 μm F-actin (25% pyrene labeled) was incubated with an equal volume of substoichiometric concentrations (0.075–1.5 μm) of ASAP1BARPH, ASAP1PH, and VTail for 30 min and diluted 20-fold with 1× KMEI buffer (50 mm KCl, 1 mm MgCl2, 1 mm EGTA, 10 mm imidazole, pH 7.0). The time-dependent decrease in pyrene fluorescence was measured in a QuantaMaster fluorescence spectrophotometer (Photon Technology International; excitation wavelength, 365 nm; emission wavelength, 407 nm) at 20 °C. For actin polymerization and depolymerization assays, the storage buffer of ASAP1 and vinculin fragments was exchanged to 1× KMEI with a Zeba Spin desalting column (Thermo Fisher Scientific) or dialysis prior to the assay.
TIRF microscopy
15–20-µl flow cells were constructed with double-sided tape. The chamber was coated with 5% BSA in 20 mm imidazole, pH 7.0, 300 mm KCl, 0.5 mm DTT and functionalized with 0.1 mg/ml NEM. The time-dependent polymerization of 0.5 μm ATTO488-labeled (degree of labeling, ∼0.3) G- to F-actin was monitored in the presence or absence of ASAP1 fragments in TIRF buffer (1× KMEI buffer supplemented with 0.17% methylcellulose, 0.2 mm ATP, 3 mg/ml glucose, 0.1 mg/ml glucose oxidase, 0.2 mg/ml catalase, and 10 mm DTT) on a Nikon Ti-Eclipse H-TIRF microscope with a 100× 1.49 numeric aperture (NA) objective at ambient temperature over 10 min. The time-dependent growth of the actin barbed end was manually tracked after a 30-pixel rolling ball radius background subtraction, image registration, and brightness adjustment, if necessary, in Fiji (61). The polarity of F-actin bundles was determined as described previously (63). To determine the number of actin filaments in a bundle, images were processed as described above, and the maximum fluorescence across an individual filament was measured for 15 filaments per flow cell and the average intensity calculated. The number of actin filaments in a bundle was determined by dividing the fluorescence intensity of the bundle by the average intensity of a single actin filament from the same experiment. Data corresponding to the mean ± S.D. of a single filament was not included in the analysis of bundles (Fig. 3B). Data analysis was performed with Fiji (61). Data were plotted with Origin (OriginLab), and statistical analysis was performed with Prism (GraphPad).
EM and image processing
Proteins were diluted to 100 nm in buffer containing 10 mm MOPS, pH 7.0, 150 mm NaCl, 0.1 mm EGTA, 2 mm MgCl2. In the presence of gelsolin, the buffer was supplemented with 0.2 mm CaCl2. Samples were applied to the grid immediately after dilution and stained with 1% uranyl acetate. Micrographs were recorded on a JEOL 1200EX II microscope using an AMT XR-60 charge-coupled device camera, operating at 80.0 kV at room temperature. Catalase crystals were used as a size calibration standard. Image processing was carried out using SPIDER as described previously (64). Initially, 4723 particles were aligned and classified into 200 classes using a variance threshold mask that incorporated the whole molecule. Classes predominated by misaligned particles were discarded, resulting in a data set with 4441 particles. This data set was classified into 5 distinct N-BAR classes using a mask drawn manually around the central N-BAR domain dimer region. The 4441-particle data set was also classified into 5 PH classes based on a mask drawn manually around one of the PH domain regions at the end of the molecule (PHtop). This process was repeated for the opposite PH domain (PHbottom). Therefore, each particle was assigned to 1 of 5 N-BAR classes (Fig. S2, B and C), 1 of 5 PHtop classes, and 1 of 5 PHbottom classes. This resulted in 125 distinct classes. All particles within a given class were recombined and averaged, resulting in the matrix shown in Fig. S2A. To quantify parallel and antiparallel interactions from EM images, the polarity of actin filaments running into paired or sparsely bundled regions was determined using the presence of gelsolin caps at the barbed ends as well as visual inspection of the actin. Only regions in which actin filaments were tightly connected over more than 50 nm were quantified.
Homology modeling
The structure of the N-BAR–PH tandem domain (amino acids 49–431) of mouse ASAP1 was modeled using the template structures of the human BAR domain of APPL1 (PDB entry 2Q13), the human BAR domain of ACAP1 (PDB entry 4NSW), and the human PH domain of ASAP1 (PDB entry 5C79) using Modeller 9v19 (65). The final N-BAR–PH model was subjected to the steepest descent energy minimization method using the molecular modeling toolkit in the Chimera molecular visualization package (66).
Immunofluorescence staining, confocal microscopy, and image analysis
NIH3T3 fibroblasts (ATCC) were grown at 37 °C in Dulbecco's modified Eagle's medium supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% fetal bovine serum (FBS) (Thermo Fisher Scientific). Fibroblasts were transiently transfected with plasmids pCI, pCI-Flag-ASAP1FL (amino acids 1–1090) (10), pCI-Flag-ASAP1BARPH (amino acids 1–431), pCI-Flag-ASAP1ΔBARPH (amino acids 431–1090), or pCI-Flag-ASAP1BARPZA (amino acids 1–724) using Lipofectamine 2000 reagent (Invitrogen). The latter plasmids were generated by PCR amplification and subsequent cloning into the pCI vector backbone. 24 h after transfection, cells were plated on 10-μg/ml fibronectin-coated coverslips and fixed in 4% paraformaldehyde after 5.5 h of adhesion. Fixed cells were incubated in 15 mm glycine for 10 min, 50 mm NH4Cl twice for 10 min each time and then permeabilized and blocked with 0.2% saponin, 0.5% BSA, and 1% FBS in PBS for 20 min. Cells were stained with anti-Flag antibody (Cell Signaling Technology) and rhodamine-phalloidin (Invitrogen) for 1 h, followed by secondary antibodies for 1 h, and mounted in DakoCytomation fluorescent mounting medium (Dako). Images for fixed cells were taken at the focal plane where cells adhere to the substrate on a Zeiss LSM 510 attached to a Zeiss Axiovert 100M with a 63×, 1.4 NA Plan Neofluar oil immersion lens (Carl Zeiss) or on a Nikon Ti2 inverted microscope and A1R live-cell resonant dual-scanner confocal system using a 60×, 1.49 NA CFI60 Apochromat TIRF oil immersion objective. Statistical analysis was performed with Prism (GraphPad).
Cellular projections were quantified by using the following criteria. Cells were scored as cells with long projections when they had at least 10 actin-containing projections longer than 5 μm. For the total number of actin projections per cell, all actin projections (stained by rhodamine-phalloidin) extending ≥0.2 μm from the cell periphery were counted. The projection length was measured by manually line-tracking projections based on rhodamine-phalloidin or anti-Flag antibody staining in Image J. The percent length filled with actin was calculated by dividing the actin projection length by the Flag projection length for each projection.
GAP assay
The GAP activity of ASAP1BARPZA was determined, as described earlier (11, 59), in the presence of up to 5 μm F-actin and myrArf1·GTP as substrate. The reaction was stopped after 3 min and protein-bound nucleotide trapped on nitrocellulose, eluted with formic acid, and analyzed by TLC.
Data availability
All data are contained within the article.
Supplementary Material
Acknowledgments
We thank Paul A. Randazzo, James R. Sellers, and Laurent Blanchoin for valuable feedback on this work. We further thank the Biophysics Core Facility and the Electron Microscopy Core Facility of the National Heart, Lung, and Blood Institute (NHLBI) for support, advice, and use of facilities.
This article contains supporting information.
Author contributions—P.-W. C., N. B., B. Y. M., J. A. S., K. C., and S. M. H. formal analysis; P.-W. C., N. B., K. C., and S. M. H. visualization; P.-W. C., N. B., K. C., and S. M. H. methodology; P.-W. C., N. B., K. C., and S. M. H. writing-review and editing; B. Y. M. and J. A. S. investigation; S. M. H. conceptualization; S. M. H. supervision; S. M. H. writing-original draft.
Conflict of interest—The authors declare that they have no conflict of interest with the contents of this work.
- G-actin
- globular actin
- GAP
- GTPase-activating protein
- BAR
- Bin/Amphiphysin/Rvs
- PH
- pleckstrin homology
- TIRF
- total internal reflection fluorescence
- F-actin
- filamentous actin
- NEM
- N-ethylmaleimide
- NA
- numeric aperture.
References
- 1. Zhao H., Pykalainen A., and Lappalainen P. (2011) I-BAR domain proteins: linking actin and plasma membrane dynamics. Curr. Opin. Cell Biol. 23, 14–21 10.1016/j.ceb.2010.10.005 [DOI] [PubMed] [Google Scholar]
- 2. Pollard T. D., and Cooper J. A. (2009) Actin, a central player in cell shape and movement. Science 326, 1208–1212 10.1126/science.1175862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Saarikangas J., Zhao H., and Lappalainen P. (2010) Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol. Rev. 90, 259–289 10.1152/physrev.00036.2009 [DOI] [PubMed] [Google Scholar]
- 4. Pollard T. D. (2016) Actin and actin-binding proteins. Cold Spring Harb. Perspect. Biol. 8, a018226 10.1101/cshperspect.a018226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Randazzo P. A., Inoue H., and Bharti S. (2007) Arf GAPs as regulators of the actin cytoskeleton. Biol. Cell 99, 583–600 10.1042/bc20070034 [DOI] [PubMed] [Google Scholar]
- 6. Chinthalapudi K., Rangarajan E. S., Brown D. T., and Izard T. (2016) Differential lipid binding of vinculin isoforms promotes quasi-equivalent dimerization. Proc. Natl. Acad. Sci. U S A 113, 9539–9544 10.1073/pnas.1600702113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chinthalapudi K., Rangarajan E. S., Patil D. N., George E. M., Brown D. T., and Izard T. (2014) Lipid binding promotes oligomerization and focal adhesion activity of vinculin. J. Cell Biol. 207, 643–656 10.1083/jcb.201404128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Koster D. V., and Mayor S. (2016) Cortical actin and the plasma membrane: inextricably intertwined. Curr. Opin. Cell Biol. 38, 81–89 10.1016/j.ceb.2016.02.021 [DOI] [PubMed] [Google Scholar]
- 9. Araki N., Hatae T., Furukawa A., and Swanson J. A. (2003) Phosphoinositide-3-kinase-independent contractile activities associated with Fcgamma-receptor-mediated phagocytosis and macropinocytosis in macrophages. J. Cell Sci. 116, 247–257 10.1242/jcs.00235 [DOI] [PubMed] [Google Scholar]
- 10. Brown M. T., Andrade J., Radhakrishna H., Donaldson J. G., Cooper J. A., and Randazzo P. A. (1998) ASAP1, a phospholipid-dependent arf GTPase-activating protein that associates with and is phosphorylated by Src. Mol. Cell. Biol. 18, 7038–7051 10.1128/mcb.18.12.7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen P. W., Jian X., Heissler S. M., Le K., Luo R., Jenkins L. M., Nagy A., Moss J., Sellers J. R., and Randazzo P. A. (2016) The Arf GTPase-activating protein, ASAP1, binds nonmuscle myosin 2A to control remodeling of the actomyosin network. J. Biol. Chem. 291, 7517–7526 10.1074/jbc.M115.701292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tanna C. E., Goss L. B., Ludwig C. G., and Chen P. W. (2019) Arf GAPs as regulators of the actin cytoskeleton-an update. Int. J. Mol. Sci 20, 442 10.3390/ijms20020442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu Y., Loijens J. C., Martin K. H., Karginov A. V., and Parsons J. T. (2002) The association of ASAP1, an ADP ribosylation factor-GTPase activating protein, with focal adhesion kinase contributes to the process of focal adhesion assembly. Mol. Biol. Cell 13, 2147–2156 10.1091/mbc.e02-01-0018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oda A., Wada I., Miura K., Okawa K., Kadoya T., Kato T., Nishihara H., Maeda M., Tanaka S., Nagashima K., Nishitani C., Matsuno K., Ishino M., Machesky L. M., Fujita H., et al. (2003) CrkL directs ASAP1 to peripheral focal adhesions. J. Biol. Chem. 278, 6456–6460 10.1074/jbc.M210817200 [DOI] [PubMed] [Google Scholar]
- 15. Jian X., Tang W. K., Zhai P., Roy N. S., Luo R., Gruschus J. M., Yohe M. E., Chen P. W., Li Y., Byrd R. A., Xia D., and Randazzo P. A. (2015) Molecular basis for cooperative binding of anionic phospholipids to the PH domain of the Arf GAP ASAP1. Structure 23, 1977–1988 10.1016/j.str.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hou T., Yang C., Tong C., Zhang H., Xiao J., and Li J. (2014) Overexpression of ASAP1 is associated with poor prognosis in epithelial ovarian cancer. Int. J. Clin. Exp. Pathol. 7, 280–287 [PMC free article] [PubMed] [Google Scholar]
- 17. Lin D., Watahiki A., Bayani J., Zhang F., Liu L., Ling V., Sadar M. D., English J., Fazli L., So A., Gout P. W., Gleave M., Squire J. A., and Wang Y. Z. (2008) ASAP1, a gene at 8q24, is associated with prostate cancer metastasis. Cancer Res. 68, 4352–4359 10.1158/0008-5472.CAN-07-5237 [DOI] [PubMed] [Google Scholar]
- 18. Sato H., Hatanaka K. C., Hatanaka Y., Hatakeyama H., Hashimoto A., Matsuno Y., Fukuda S., and Sabe H. (2014) High level expression of AMAP1 protein correlates with poor prognosis and survival after surgery of head and neck squamous cell carcinoma patients. Cell Commun. Signal. 12, 17 10.1186/1478-811X-12-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stevenson R. P., Veltman D., and Machesky L. M. (2012) Actin-bundling proteins in cancer progression at a glance. J. Cell Sci. 125, 1073–1079 10.1242/jcs.093799 [DOI] [PubMed] [Google Scholar]
- 20. Jian X., Brown P., Schuck P., Gruschus J. M., Balbo A., Hinshaw J. E., and Randazzo P. A. (2009) Autoinhibition of Arf GTPase-activating protein activity by the BAR domain in ASAP1. J. Biol. Chem. 284, 1652–1663 10.1074/jbc.M804218200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nie Z., Hirsch D. S., Luo R., Jian X., Stauffer S., Cremesti A., Andrade J., Lebowitz J., Marino M., Ahvazi B., Hinshaw J. E., and Randazzo P. A. (2006) A BAR domain in the N terminus of the Arf GAP ASAP1 affects membrane structure and trafficking of epidermal growth factor receptor. Curr. Biol. 16, 130–139 10.1016/j.cub.2005.11.069 [DOI] [PubMed] [Google Scholar]
- 22. Kostan J., Salzer U., Orlova A., Toro I., Hodnik V., Senju Y., Zou J., Schreiner C., Steiner J., Merilainen J., Nikki M., Virtanen I., Carugo O., Rappsilber J., Lappalainen P., et al. (2014) Direct interaction of actin filaments with F-BAR protein pacsin2. EMBO Rep. 15, 1154–1162 10.15252/embr.201439267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rocca D. L., Martin S., Jenkins E. L., and Hanley J. G. (2008) Inhibition of Arp2/3-mediated actin polymerization by PICK1 regulates neuronal morphology and AMPA receptor endocytosis. Nat. Cell Biol. 10, 259–271 10.1038/ncb1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sun H. Q., Yamamoto M., Mejillano M., and Yin H. L. (1999) Gelsolin, a multifunctional actin regulatory protein. J. Biol. Chem. 274, 33179–33182 10.1074/jbc.274.47.33179 [DOI] [PubMed] [Google Scholar]
- 25. Hotulainen P., and Lappalainen P. (2006) Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J. Cell Biol. 173, 383–394 10.1083/jcb.200511093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. dos Remedios C. G., Chhabra D., Kekic M., Dedova I. V., Tsubakihara M., Berry D. A., and Nosworthy N. J. (2003) Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol. Rev. 83, 433–473 10.1152/physrev.00026.2002 [DOI] [PubMed] [Google Scholar]
- 27. Randazzo P. A., Nie Z., Miura K., and Hsu V. W. (2000) Molecular aspects of the cellular activities of ADP-ribosylation factors. Sci. STKE 2000, re1 10.1126/stke.2000.59.re1 [DOI] [PubMed] [Google Scholar]
- 28. Inoue H., Ha V. L., Prekeris R., and Randazzo P. A. (2008) Arf GTPase-activating protein ASAP1 interacts with Rab11 effector FIP3 and regulates pericentrosomal localization of transferrin receptor-positive recycling endosome. Mol. Biol. Cell 19, 4224–4237 10.1091/mbc.e08-03-0290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mesarec L., Góźdź W., Iglič V. K., Kralj S., and Iglič A. (2016) Closed membrane shapes with attached BAR domains subject to external force of actin filaments. Colloids Surf. B Biointerfaces 141, 132–140 10.1016/j.colsurfb.2016.01.010 [DOI] [PubMed] [Google Scholar]
- 30. Yamagishi A., Masuda M., Ohki T., Onishi H., and Mochizuki N. (2004) A novel actin bundling/filopodium-forming domain conserved in insulin receptor tyrosine kinase substrate p53 and missing in metastasis protein. J. Biol. Chem. 279, 14929–14936 10.1074/jbc.M309408200 [DOI] [PubMed] [Google Scholar]
- 31. Loomis P. A., Zheng L., Sekerkova G., Changyaleket B., Mugnaini E., and Bartles J. R. (2003) Espin cross-links cause the elongation of microvillus-type parallel actin bundles in vivo. J. Cell Biol. 163, 1045–1055 10.1083/jcb.200309093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Suetsugu S., and Gautreau A. (2012) Synergistic BAR-NPF interactions in actin-driven membrane remodeling. Trends Cell Biol. 22, 141–150 10.1016/j.tcb.2012.01.001 [DOI] [PubMed] [Google Scholar]
- 33. Madasu Y., Yang C., Boczkowska M., Bethoney K. A., Zwolak A., Rebowski G., Svitkina T., and Dominguez R. (2015) PICK1 is implicated in organelle motility in an Arp2/3 complex-independent manner. Mol. Biol. Cell 26, 1308–1322 10.1091/mbc.E14-10-1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Millard T. H., Bompard G., Heung M. Y., Dafforn T. R., Scott D. J., Machesky L. M., and Futterer K. (2005) Structural basis of filopodia formation induced by the IRSp53/MIM homology domain of human IRSp53. EMBO J. 24, 240–250 10.1038/sj.emboj.7600535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Salzer U., Kostan J., and Djinovic-Carugo K. (2017) Deciphering the BAR code of membrane modulators. Cell Mol. Life Sci 74, 2413–2438 10.1007/s00018-017-2478-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. She B. R., Liou G. G., and Lin-Chao S. (2002) Association of the growth-arrest-specific protein Gas7 with F-actin induces reorganization of microfilaments and promotes membrane outgrowth. Exp. Cell Res. 273, 34–44 10.1006/excr.2001.5435 [DOI] [PubMed] [Google Scholar]
- 37. Drager N. M., Nachman E., Winterhoff M., Bruhmann S., Shah P., Katsinelos T., Boulant S., Teleman A. A., Faix J., and Jahn T. R. (2017) Bin1 directly remodels actin dynamics through its BAR domain. EMBO Rep. 18, 2051–2066 10.15252/embr.201744137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Skau C. T., and Waterman C. M. (2015) Specification of architecture and function of actin structures by actin nucleation factors. Annu. Rev. Biophys. 44, 285–310 10.1146/annurev-biophys-060414-034308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Karlsen M. L., Thorsen T. S., Johner N., Ammendrup-Johnsen I., Erlendsson S., Tian X., Simonsen J. B., Hoiberg-Nielsen R., Christensen N. M., Khelashvili G., Streicher W., Teilum K., Vestergaard B., Weinstein H., Gether U., et al. (2015) Structure of dimeric and tetrameric complexes of the BAR domain protein PICK1 determined by small-angle X-ray scattering. Structure 23, 1258–1270 10.1016/j.str.2015.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Halbach A., Morgelin M., Baumgarten M., Milbrandt M., Paulsson M., and Plomann M. (2007) PACSIN 1 forms tetramers via its N-terminal F-BAR domain. FEBS J 274, 773–782 10.1111/j.1742-4658.2006.05622.x [DOI] [PubMed] [Google Scholar]
- 41. Schmoller K. M., Semmrich C., and Bausch A. R. (2011) Slow down of actin depolymerization by cross-linking molecules. J. Struct. Biol. 173, 350–357 10.1016/j.jsb.2010.09.003 [DOI] [PubMed] [Google Scholar]
- 42. Breitsprecher D., Koestler S. A., Chizhov I., Nemethova M., Mueller J., Goode B. L., Small J. V., Rottner K., and Faix J. (2011) Cofilin cooperates with fascin to disassemble filopodial actin filaments. J. Cell Sci. 124, 3305–3318 10.1242/jcs.086934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu J. Q., and Pollard T. D. (2005) Counting cytokinesis proteins globally and locally in fission yeast. Science 310, 310–314 10.1126/science.1113230 [DOI] [PubMed] [Google Scholar]
- 44. Gasilina A., Vitali T., Luo R., Jian X., and Randazzo P. A. (2019) The ArfGAP ASAP1 controls actin stress fiber organization via its N-BAR domain. iScience 22, 166–180 10.1016/j.isci.2019.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Carman P. J., and Dominguez R. (2018) BAR domain proteins-a linkage between cellular membranes, signaling pathways, and the actin cytoskeleton. Biophys. Rev. 10, 1587–1604 10.1007/s12551-018-0467-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kessels M. M., and Qualmann B. (2015) Different functional modes of BAR domain proteins in formation and plasticity of mammalian postsynapses. J. Cell Sci. 128, 3177–3185 10.1242/jcs.174193 [DOI] [PubMed] [Google Scholar]
- 47. Almeida-Souza L., Frank R. A. W., Garcia-Nafria J., Colussi A., Gunawardana N., Johnson C. M., Yu M., Howard G., Andrews B., Vallis Y., and McMahon H. T. (2018) A flat BAR protein promotes actin polymerization at the base of clathrin-coated pits. Cell 174, 325–337 e314 10.1016/j.cell.2018.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cao H., Yin X., Cao Y., Jin Y., Wang S., Kong Y., Chen Y., Gao J., Heller S., and Xu Z. (2013) FCHSD1 and FCHSD2 are expressed in hair cell stereocilia and cuticular plate and regulate actin polymerization in vitro. PLoS ONE 8, e56516 10.1371/journal.pone.0056516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Coutinho-Budd J., Ghukasyan V., Zylka M. J., and Polleux F. (2012) The F-BAR domains from srGAP1, srGAP2 and srGAP3 regulate membrane deformation differently. J. Cell Sci. 125, 3390–3401 10.1242/jcs.098962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Guerrier S., Coutinho-Budd J., Sassa T., Gresset A., Jordan N. V., Chen K., Jin W. L., Frost A., and Polleux F. (2009) The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell 138, 990–1004 10.1016/j.cell.2009.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lim K. B., Bu W., Goh W. I., Koh E., Ong S. H., Pawson T., Sudhaharan T., and Ahmed S. (2008) The Cdc42 effector IRSp53 generates filopodia by coupling membrane protrusion with actin dynamics. J. Biol. Chem. 283, 20454–20472 10.1074/jbc.M710185200 [DOI] [PubMed] [Google Scholar]
- 52. Mason F. M., Heimsath E. G., Higgs H. N., and Soderling S. H. (2011) Bi-modal regulation of a formin by srGAP2. J. Biol. Chem. 286, 6577–6586 10.1074/jbc.M110.190397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Postema M. M., Grega-Larson N. E., Neininger A. C., and Tyska M. J. (2018) IRTKS (BAIAP2L1) elongates epithelial microvilli using EPS8-dependent and independent mechanisms. Curr. Biol. 28, 2876–2888.e4 10.1016/j.cub.2018.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee S. H., Kerff F., Chereau D., Ferron F., Klug A., and Dominguez R. (2007) Structural basis for the actin-binding function of missing-in-metastasis. Structure 15, 145–155 10.1016/j.str.2006.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ahmed S., Goh W. I., and Bu W. (2010) I-BAR domains, IRSp53 and filopodium formation. Semin. Cell Dev. Biol. 21, 350–356 10.1016/j.semcdb.2009.11.008 [DOI] [PubMed] [Google Scholar]
- 56. Govind S., Kozma R., Monfries C., Lim L., and Ahmed S. (2001) Cdc42Hs facilitates cytoskeletal reorganization and neurite outgrowth by localizing the 58-kD insulin receptor substrate to filamentous actin. J. Cell Biol. 152, 579–594 10.1083/jcb.152.3.579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bharti S., Inoue H., Bharti K., Hirsch D. S., Nie Z., Yoon H. Y., Artym V., Yamada K. M., Mueller S. C., Barr V. A., and Randazzo P. A. (2007) Src-dependent phosphorylation of ASAP1 regulates podosomes. Mol. Cell. Biol. 27, 8271–8283 10.1128/MCB.01781-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Livne A., and Geiger B. (2016) The inner workings of stress fibers—from contractile machinery to focal adhesions and back. J. Cell Sci. 129, 1293–1304 10.1242/jcs.180927 [DOI] [PubMed] [Google Scholar]
- 59. Chen P. W., Jian X., Luo R., and Randazzo P. A. (2012) Approaches to studying Arf GAPs in cells: in vitro assay with isolated focal adhesions. Curr. Protoc. Cell Biol. Chapter17, Unit17.13 10.1002/0471143030.cb1713s55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lehrer S. S., and Kerwar G. (1972) Intrinsic fluorescence of actin. Biochemistry 11, 1211–1217 10.1021/bi00757a015 [DOI] [PubMed] [Google Scholar]
- 61. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hansen S. D., Zuchero J. B., and Mullins R. D. (2013) Cytoplasmic actin: purification and single molecule assembly assays. Methods Mol. Biol. 1046, 145–170 10.1007/978-1-62703-538-5_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chacon-Martinez C. A., Kiessling N., Winterhoff M., Faix J., Muller-Reichert T., and Jessberger R. (2013) The switch-associated protein 70 (SWAP-70) bundles actin filaments and contributes to the regulation of F-actin dynamics. J. Biol. Chem. 288, 28687–28703 10.1074/jbc.M113.461277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Burgess S. A., Walker M. L., Thirumurugan K., Trinick J., and Knight P. J. (2004) Use of negative stain and single-particle image processing to explore dynamic properties of flexible macromolecules. J. Struct. Biol. 147, 247–258 10.1016/j.jsb.2004.04.004 [DOI] [PubMed] [Google Scholar]
- 65. Webb B., and Sali A. (2016) Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 54, 5.6.1–5.6.37 10.1002/cpbi.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.