Abstract
Rhizobia are soil bacteria that form important symbiotic associations with legumes, and rhizobial surface polysaccharides, such as K-antigen polysaccharide (KPS) and lipopolysaccharide (LPS), might be important for symbiosis. Previously, we obtained a mutant of Sinorhizobium fredii HH103, rkpA, that does not produce KPS, a homopolysaccharide of a pseudaminic acid derivative, but whose LPS electrophoretic profile was indistinguishable from that of the WT strain. We also previously demonstrated that the HH103 rkpLMNOPQ operon is responsible for 5-acetamido-3,5,7,9-tetradeoxy-7-(3-hydroxybutyramido)-l-glycero-l-manno-nonulosonic acid [Pse5NAc7(3OHBu)] production and is involved in HH103 KPS and LPS biosynthesis and that an HH103 rkpM mutant cannot produce KPS and displays an altered LPS structure. Here, we analyzed the LPS structure of HH103 rkpA, focusing on the carbohydrate portion, and found that it contains a highly heterogeneous lipid A and a peculiar core oligosaccharide composed of an unusually high number of hexuronic acids containing β-configured Pse5NAc7(3OHBu). This pseudaminic acid derivative, in its α-configuration, was the only structural component of the S. fredii HH103 KPS and, to the best of our knowledge, has never been reported from any other rhizobial LPS. We also show that Pse5NAc7(3OHBu) is the complete or partial epitope for a mAb, NB6-228.22, that can recognize the HH103 LPS, but not those of most of the S. fredii strains tested here. We also show that the LPS from HH103 rkpM is identical to that of HH103 rkpA but devoid of any Pse5NAc7(3OHBu) residues. Notably, this rkpM mutant was severely impaired in symbiosis with its host, Macroptilium atropurpureum.
Keywords: bacteria, Sinorhizobium fredii, symbiosis, lipopolysaccharide (LPS), pseudaminic acid, carbohydrate structure, antibody, soybean, Macroptilium, Cajanus, microbiology
Rhizobia are soil bacteria able to establish a symbiotic association with legumes in which a complex interchange of molecular signals between the prokaryotic and the eukaryotic partners takes place. This interchange of molecular signals culminates in the formation of specialized plant structures, called nodules, on the roots or stems of the plant host (1). The molecular dialogue between the bacteria and the plant is initiated by flavonoids exuded by the legume root, which activate the transcription of bacterial nodulation genes. Some of these genes encode proteins involved in the biosynthesis and secretion of molecular signals called Nod factors. These bacterial signals mediate species specificity and induce the development of root nodules in the plant (1–3).
In addition to Nod factors, different rhizobial surface polysaccharides (RSP) are commonly necessary for successful nodulation. Exopolysaccharides (EPS), lipopolysaccharides (LPS), capsular polysaccharides (KPS or K-antigen polysaccharides), and cyclic glucans (CG) are the main rhizobial polysaccharides involved in symbiosis. For each specific pair of rhizobium-legume symbionts, one or more RSP could act as signal molecules promoting bacterial infection and/or preventing plant defense reactions (4–7).
Sinorhizobium fredii HH103 (8) is a fast-growing rhizobial strain that nodulates Glycine max (soybean) and many other determinate- and indeterminate-nodule forming legumes (9). This bacterium produces at least five different RSP: EPS, LPS, CG, and two types of KPS. The chemical structure of these RSP, except for the LPS, have been previously described (10–13). Interestingly, two KPS are produced by S. fredii HH103, one is a homopolysaccharide built up of a pseudaminic acid derivative, i.e. α-configured 5-acetamido-3,5,7,9-tetradeoxy-7-(3-hydroxybutyramido)-l-glycero-l-manno-nonulosonic acid [α-Pse5NAc7(3OHBu)] (12), whereas the other is a homopolymer of 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo) (13). To our knowledge, S. fredii HH103 mutants affected in the production of the poly-Kdo KPS have not been described. An S. fredii RifR exoA mutant, unable to produce EPS, did not show any detectable reduction in the symbiotic capacity with soybean (10). On the contrary, soybean plants inoculated with HH103 mutants unable to produce CG only formed pseudonodules that did not fix nitrogen (11). Interestingly, S. fredii HH103 RifR mutants in genes located in the so-called rkp-1 region (such as HH103 rkpA) showed a severe reduction of nodulation with soybeans (14–16). HH103 rkpA failed to produce the poly-α-Pse5NAc7(3OHBu) KPS, but the other RSP did not show any detectable change. Even more importantly, S. fredii HH103 RifR mutants in genes located in the so-called rkp-3 region (such as rkpM, rkpP, or rkpQ) did not produce the α-Pse5NAc7(3OHBu) KPS as well, showed altered LPS electrophoretic profiles, and only induced the formation of soybean pseudonodules unable to fix nitrogen (17).
LPS are the predominant constituents in the external leaflet of Gram-negative outer membrane and can be conceptually divided in three different structural regions: a glycolipid component called lipid A that is linked to a core oligosaccharide (core OS) that is attached to an outer polysaccharide, termed the O-chain (18). On the basis of the nature of the saccharide portion, an LPS can be classified as smooth, semirough, or rough: smooth-type LPS (S-LPS) contains all three abovementioned domains, whereas a semirough-type LPS (sR-LPS) has a subunit (complete or truncated) of the O-chain, and a rough-type LPS (R-LPS) does not express any O-chain moiety (18).
To the best of our knowledge, the complete LPS structure has only been described for five different rhizobial strains: Rhizobium etli CE3, R. leguminosarum biovar viciae 3841, R. leguminosarum biovar viciae 128C53 (19), and Bradyrhizobium sp. strain BTAi1 and ORS285 (20–22); in addition, only partial structures or the chemical composition of different LPS parts of various Sinorhizobium strains have been reported (23). Nevertheless, it is widely known that rhizobial LPSs appear to play very relevant roles at different stages of the symbiotic interaction with legumes, including recognition and infection, invasion of root cortical cells, bacterial penetration into nodule cells, and formation and persistence of functional symbiosomes (1, 19, 24–31). Indeed, it has been demonstrated that alterations in the LPS structure can potently affect the symbiotic properties of the bacterium; in particular, O-chain rhizobial mutants were reported to be drastically affected in symbiosis, either in the infection event, nodule development, or bacteroid differentiation, and this phenomenon has been observed with both determinate and indeterminate nodule-forming legumes (32–36).
In this work, we focused attention on the LPS isolated from two S. fredii HH103 RifR mutants: HH103 rkpA (16) and HH103 rkpM (17). HH103 rkpA was chosen because it does not produce KPS, which could interfere with LPS purification, and its LPS silver- and immune-staining profiles were equal to those of S. fredii HH103 RifR, indicating that the LPS structure of both strains should be highly similar or identical. In parallel, HH103 rkpM was investigated because it does not produce KPS as well, and its LPS silver-staining profile was shown to be only slightly altered with respect to S. fredii HH103 RifR. Structural investigation showed that HH103 rkpA possesses an LPS containing β-Pse5NAc7(3OHBu), which was never described before as a component of this polysaccharide in rhizobia. Interestingly, we also show that S. fredii HH103 rkpM produces an LPS devoid of such pseudaminic acid derivatives, and that this mutant is severely impaired in symbiosis with the HH103 host legume Macroptilium atropurpureum.
Results
Silver staining and antibody recognition of the LPS produced by different S. fredii HH103 mutants affected in LPS and/or KPS biosynthesis
In previous works (17, 31, 37), we have studied different S. fredii HH103 mutants (in the rkpM, lpsB, lpsE, rkpK, and lpsL genes) that were affected in LPS production, as revealed by alterations in their LPS electrophoretic profiles. Relevant characteristics of these mutants are summarized in Table 1. The lpsB and lpsE genes belong to the greA lpsB lpsCDE cluster and are predicted to code for type 1 glycosyltransferases that transfer activated sugars to a variety of substrates, including glycogen and LPS (31). The rkpK and lpsL genes constitute the so-called rkp-2 region, and their encoded products are responsible for the synthesis of glucuronic and galacturonic acid, respectively (37). Finally, the rkpM gene belongs to the rkpLMNOPQ operon (rkp-3 region), which is involved in the production of the pseudaminic acid derivative that constitutes the HH103 KPS (17). In this work, we have compared their LPS profiles in a single polyacrylamide gel (Fig. 1A) to classify them according to the mobility of the silver-stained LPS bands. We hypothesized that any increase of the electrophoretic mobility of the LPS bands would be directly related to the size of the truncated LPS forms produced by each mutant. Therefore, mutants showing faster migrating LPS bands might correspond to mutants producing smaller truncated LPS core OS forms. WT S. fredii HH103 RifR and its rkpA mutant derivative were also included as controls, because this mutant does not produce KPS but its LPS shows the same electrophoretic profile of HH103 RifR (16).
Table 1.
Previously studied Sinorhizobium fredii HH103 mutants affected in LPS
| Mutated gene | Predicted function of the encoded product | RSP affecteda | Symbiotic phenotype with soybean | Reference |
|---|---|---|---|---|
| lpsB | Glycosyl transferase | LPS* | High reduction in the number of Fix+ nodules, pseudonodules | 31 |
| lpsE | Glycosyl transferase | LPS* | ||
| lpsL | UDP-glucuronate 4-epimerase | LPS* | Pseudonodules, Fix– | 37 |
| rkpK | UDP-glucose 6-dehydrogenase | LPS* EPS− | Nod+ Fix+ (as WT) | |
| rkpM | Pyridoxal phosphate (PLP)-dependent aminotransferase | LPS* KPS− | Pseudonodules, Fix– | 17 |
a LPS* refers to alterations in the electrophoretic profile and lack of recognition by the mAb NB6-228.22.
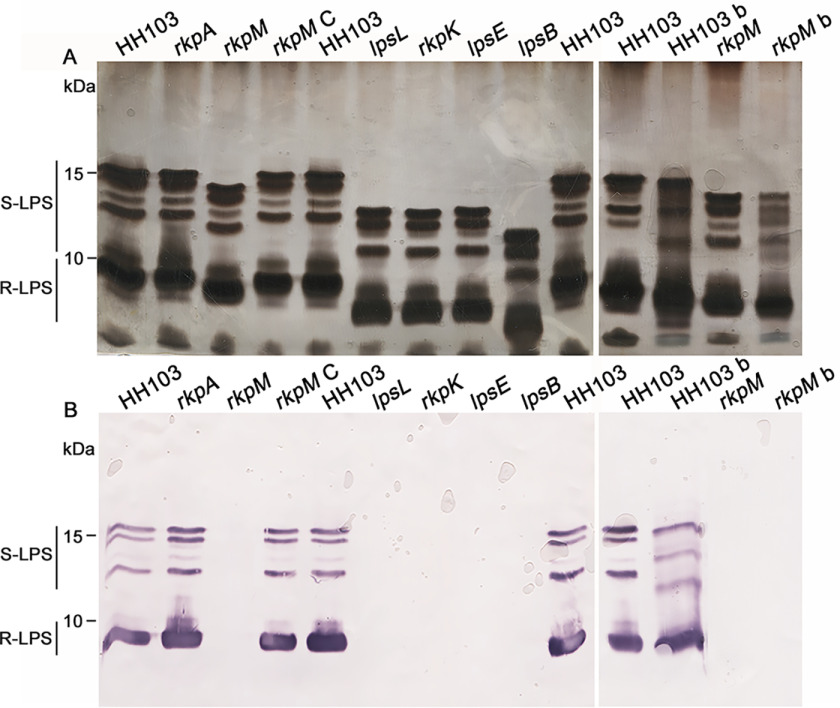
Figure 1.
Electrophoretic profile and immunodetection of the LPS produced by different S. fredii HH103 RifR mutants affected in LPS and/or KPS production. A, SDS-PAGE after silver staining of LPS crude extracts. B, immunostaining using the mAb NB6-228.22 of LPS crude extracts. In both panels A and B, the mutated gene is indicated on the top of the lane. HH103 denotes HH103 RifR, and rkpM C denotes the complemented rkpM mutant. All of the samples were extracted from free-living cells, with the only exception being samples HH103 b and rkpM b, which correspond to samples extracted from bacteroids from Glycyrrhiza uralensis nodules induced by HH103 RifR and its rkpM mutant, respectively.
Figure 1A allowed us to compare the electrophoretic alterations exhibited by the different mutants. As shown in this figure, the LPS profile of S. fredii HH103 RifR is composed of four silver-stained bands of slower mobility and two thick bands of faster mobility. This observation suggested that bands in the upper part of the gel would correspond to LPS molecules made up of lipid A, core OS, and O-chain (or part of it), whereas in the lower part of the gel a smaller LPS, migrating as a dispersed thick region, was assumed to contain the lipid A and core OS with no O-chain subunits. The biological nature of the tiny smears visualized in the upper part of the gel remains unknown. At the bottom of each lane appears a diffuse band that is not always visualized in SDS-PAGE gels carried out with HH103 LPS samples (16, 17) and whose nature is unknown.
As reported before (17, 31, 37), the electrophoretic mobility of the bands of the five HH103 mutants affected in LPS was clearly faster than that of HH103 RifR or its rkpA derivative. However, there were also differences among the different LPS mutants (Fig. 1A): mutants in lpsL, rkpK, and lpsE showed similar LPS profiles with faster electrophoretic mobility than that of HH103 rkpM but slower than that of HH103 lpsB.
Following these observations, we employed NB6-228.22, a mAb able to recognize all the silver-stained LPS bands of S. fredii HH103 RifR (38). As shown in Fig. 1B, the pattern of LPS bands recognized by NB6-228.22 in S. fredii HH103 RifR was equal to that in HH103 rkpA. Neither the tiny smears at the upper part nor the diffuse bands at the bottom of the gels reacted with the antibody. None of the S. fredii HH103 RifR mutants showing alterations in their LPS electrophoretic profiles (rkpM, lpsL, rkpK, lpsE, and lpsB) were recognized by NB6-228.22, indicating that in all these mutants the LPS epitope recognized by NB6-228.22 was missing or has become unavailable for antibody recognition. Introduction in HH103 rkpM of a cosmid containing the HH103 rkp-3 region (strain HH103 rkpM C) restored the WT LPS profile (Fig. 1A) and recognition by the mAb NB6-228.22 (Fig. 1B).
LPS profiles of S. fredii HH103 RifR free-living cells is undistinguishable from HH103 RifR bacteroids isolated from G. max or C. cajan (39). However, S. fredii HH103 RifR bacteroids isolated from Glycyrrhiza uralensis showed altered LPS profiles compared with those of S. fredii HH103 free-living cells (39). Given these premises, the LPS from bacteroids of G. uralensis nodules induced by inoculation with HH103 RifR or HH103 rkpM were also analyzed by SDS-PAGE in the present study (Fig. 1A). The LPS profile of HH103 rkpM bacteroids was slightly different from that of free-living HH103 rkpM cells and clearly altered with respect to that of HH103 bacteroids. This demonstrated that HH103 rkpM LPS is altered not only in free-living cells but also in G. uralensis bacteroids compared with those of the WT strain. The LPS of HH103 rkpM bacteroids isolated from Glycyrrhiza nodules was not recognized by the mAb NB6-228.22 (Fig. 1B).
Taking into account all the above results, we started the structural characterization of the LPS of HH103 mutants rkpA and rkpM.
Structural determination of the LPS from S. fredii HH103 rkpA and HH103 rkpM mutants
To define the detailed structure of S. fredii HH103 rkpA and HH103 rkpM LPS, the dried bacterial pellets were extracted by the hot phenol-water procedure (40), checked by SDS-PAGE, and extensively purified. To define the nature of the monosaccharides composing the isolated LPSs, a set of chemical analyses (41) on pure water phase extracts was performed. The merging of data from compositional analysis, through acetylated methyl glycosides derivatization, and absolute configuration, through octyl glycoside derivatization, revealed the presence in both LPS of d-galacturonic (d-GalA) and d-glucuronic (d-GlcA) acids, d-glucose (d-Glc), d-galactose (d-Gal), 2-amino-2-deoxy-d-glucose (d-GlcN), and Kdo. Linkage analyses (41) proved the pyranose form for all sugar residues and allowed the identification of the same branching points for the monosaccharides composing both the LPS, except for terminal-d-GalpA, which was found only in the case of HH103 rkpM, and 2-substituted-d-GalpA, which was found instead only in the case of HH103 rkpA; on the other hand, 4-substituted d-GalpA, terminal and 2-substituted d-GlcpA, 2-substituted and 3-substituted d-Glcp, 3,6-disubstituted d-Galp, 6-substituted-d-GlcpN, and terminal and 4,5-disubstituted Kdo were detected in both LPS. Importantly, as described further in the text, an additional sugar constituent, Pse5NAc7(3OHBu) (Pse), was identified solely in HH103 rkpA and only after an in-depth investigation of the isolated LPS saccharide component using NMR spectroscopy and MALDI-TOF MS.
To define the primary structure of the saccharide portion of both HH103 rkpA and HH103 rkpM LPS, an aliquot of each sample underwent a mild acid hydrolysis to selectively cleave the linkage between the Kdo, which is the first sugar component of the core OS, and the lipid A moiety. After several steps of purification comprising size-exclusion low-pressure and HPLC, the pure saccharide moieties (OS1rkpA and OS1rkpM) were analyzed via 1D and 2D NMR spectroscopy, including double-quantum filtered phase-sensitive correlation spectroscopy (DQF-COSY), total correlation spectroscopy (TOCSY), rotating frame Overhauser enhancement spectroscopy (ROESY), heteronuclear single quantum coherence (1H-13C HSQC), 1H-13C HSQC-TOCSY, and heteronuclear multiple bound correlation (1H-13C HMBC).
All sugar units were present as pyranose rings according to the 13C chemical shift values and in full accordance with chemical analyses. The analysis of both the 1H NMR (Fig. 2) and HSQC spectra (Fig. 3) of the pure OS1rkpA clearly showed the occurrence of eight anomeric signals attributed to eight different spin systems (A–H, Table 2), whereas the presence of a Kdo unit was attested by the presence of the diasterotopic H-3 methylene proton signals at δH 1.94/1.75 ppm (K, δC 33.9 ppm, Fig. 2 and 3). Furthermore, the 1H NMR spectrum (Fig. 2) showed the presence of four intense methyl signals at δH 1.05, 1.09, 1.11, and 1.13 ppm (Table 2) and of another intense methyl signal at 1.90 ppm accounting for two N-acetyl groups (NAc). Finally, the presence of additional methylene proton signals at 1.55/2.42 ppm (PseI H-3ax/H-3eq, δC 36.0 ppm) and 1.50/2.47 ppm (PseII H-3ax/H-3eq, δC 36.1 ppm) were also identified. Starting from the above methylene signals, the analysis of HSQC and HMBC spectra, supported by the correlations visible in COSY, TOCSY, and ROESY spectra, allowed the complete structural elucidation of two Pse residues (here indicated as PseI and PseII). Briefly, the diasterotopic methylene protons H-3 PseI showed long-range correlations with an oxymethine carbon atom (C-4 PseI, δC 66.3 ppm), a quaternary carbon (C-2 PseI, δC 103.2 ppm), and a carboxylic group (C-1 PseI, δC 172.8 ppm) (Fig. 4). A less intense HMBC correlation between H-3ax PseI and a nitrogen-bearing carbon atom resonating at 48.0 ppm (C-5 PseI) was also detected (Fig. 4) and identified as the site of N-acetylation, as proven by the NOE correlation between H-5 PseI (4.05 ppm) and the methyl proton at 1.90 ppm (not shown). Starting from H-5 PseI, a long-range correlation with an oxymethine carbon atom at 73.0 ppm, assigned as C-6 PseI, was visible in the HMBC spectrum (Fig. 4); further on, the methyl proton signal at 1.09 ppm correlated with an oxymethine carbon atom (C-8 PseI, 68.3 ppm) and a nitrogen-bearing carbon (C-7 PseI, 53.7 ppm), with the latter, in turn, correlated to H-6 PseI (3.77 ppm) (Fig. 4). Finally, a 3-hydroxybutyryl moiety (HBu, made up of the CH3-C group at δC 22.0 ppm, the C-CHOH-C group at δC 69.8 ppm, the C-CH2-C at δC 45.0 ppm, and the CO group at 173.4 ppm) was found to be connected by amide linkage at position 7 of PseI; the location of the HBu moiety was attained by the observation of a long-range correlation between C-1′ (173.4 ppm) and H-7 PseI (3.94 ppm) in the HMBC spectrum (Fig. 4). Because of the relatively large JH-3ax,H-4 (∼12.9 Hz) and the small JH-4,H-5 and JH-5,H-6 (<5 Hz) coupling constants, it was possible to deduce the axial orientation of H-4 PseI and the equatorial orientation of H-5 PseI that combined with the JH-6,H-7 coupling constant of ∼10 Hz, suggested the l-glycero-l-manno configuration of unit PseI (42). This was also corroborated by the observation of the chemical shift for the C-9 signal at 16.2 ppm (Table 2), which is typical of the l-epimer of the glycero-l-manno isomers; in contrast, chemical shifts at around 20 ppm are observed in the case of the d-epimers (42). Interestingly, the large difference between the H-3ax and H-3eq chemical shifts (0.88 ppm) indicated the axial orientation of the carboxyl group, which is the β configuration of PseI at the anomeric center (42). Finally, the downfield displacement of the chemical shift of C-3′ (69.8 ppm) of unit PseI suggested a site of glycosylation at such a position. Similarly, a second unit of β-Pse5NAc7(3OHBu) has been assigned (PseII), but it turned out to be a terminal residue, linked to position 3′ of PseI, as proven by the long-range correlation between the C-2 of PseII and the oxymethine proton H-3′ (4.13 ppm, Table 2 and Fig. 4) of PseI. The absolute configuration of the HBu substituent remains to be defined.
Figure 2.
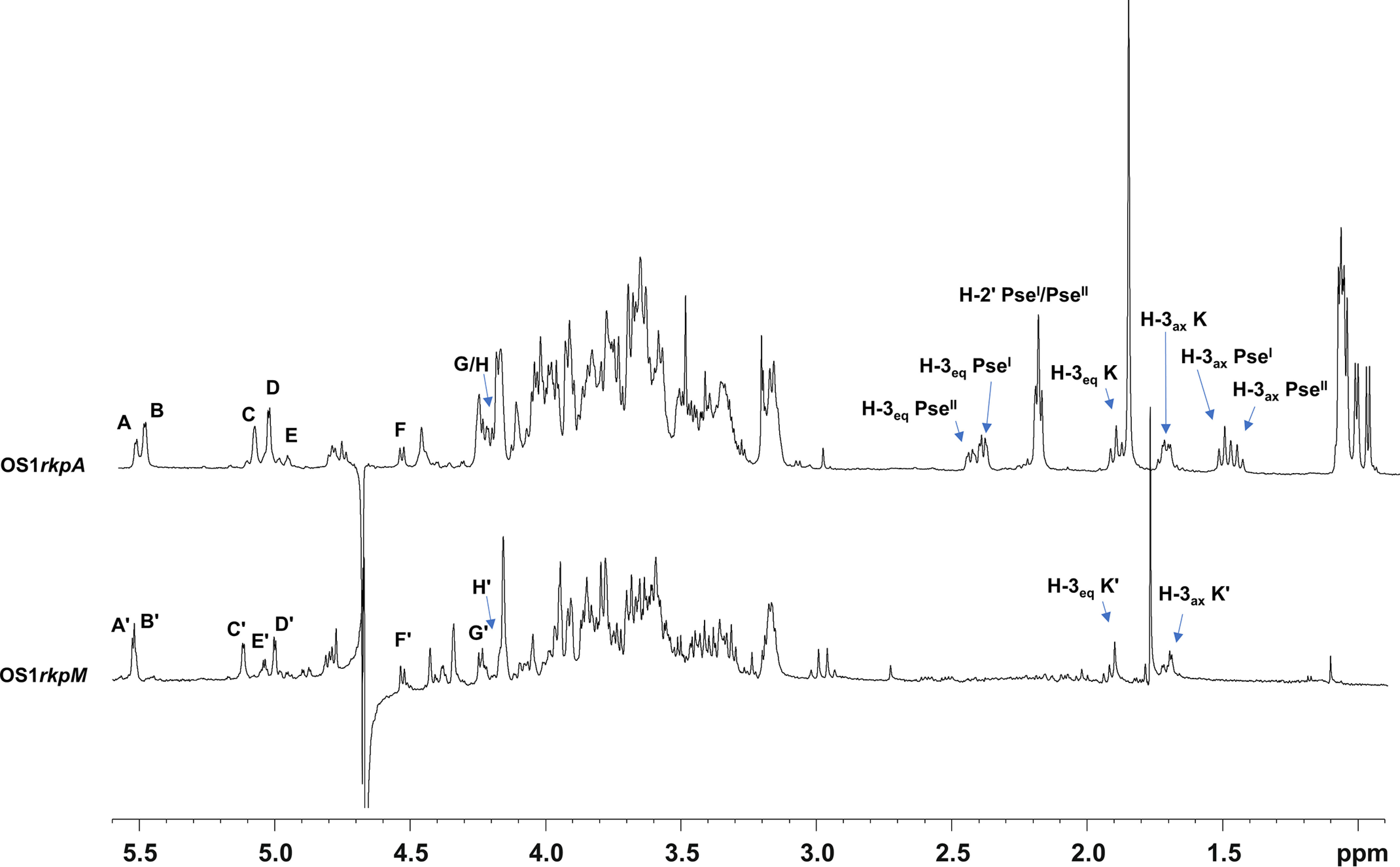
1H NMR spectra of Sinorhizobium fredii HH103 rkpA (top) and rkpM (bottom). Anomeric signals of spin systems are designated as described in Table 2. Diastereotopic methylene proton signals of PseI, PseII, and Kdo residues (K/K') are also indicated.
Figure 3.
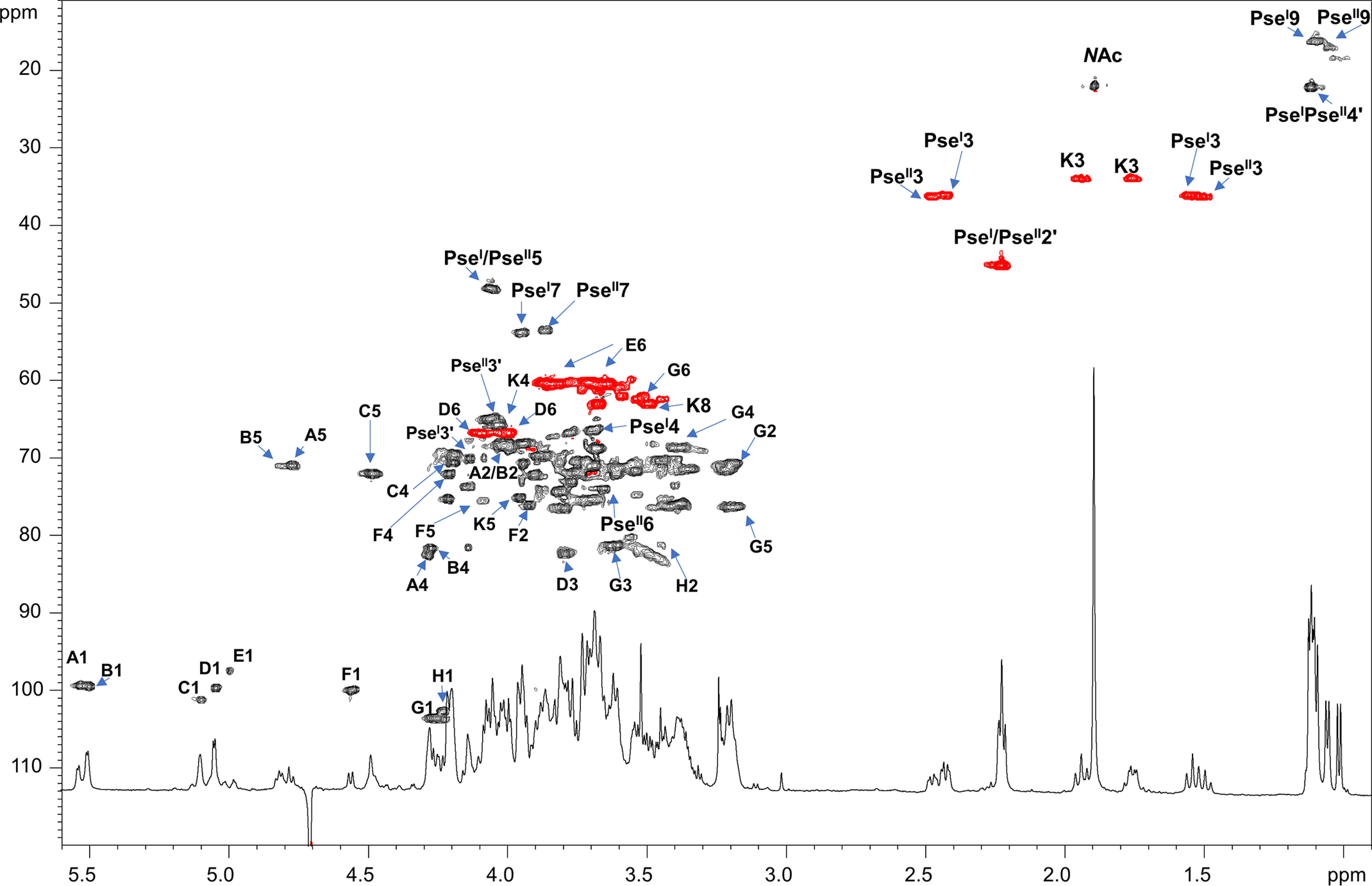
Zoom of the 1H-13C HSQC spectrum of OS1rkpA product obtained after mild acid hydrolysis of the LPS from S. fredii HH103 rkpA. Most of the heteronuclear correlations are indicated. Spin system labels are indicated as described in Table 2. The color of the cross-peaks indicates the phase: black indicates CH and CH3 signals, whereas red indicates CH2 signals.
Table 2.
Proton and carbon chemical shifts of the OS1rkpA (A-H, K, PseI and PseII) and OS2rkpA (E2, K2, J, X, and Y) products derived from mild acid hydrolysis and complete deacylation, respectively, of LPS from S. fredii HH103 rkpA
| Product | Chemical shift | Value at positiona: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1/1′ | 2/2′ | 3/3′ | 4/4′ | 5 | 6 | 7 | 8 | 9 | ||
| A | 1H | 5.53 | 4.03 | 3.69 | 4.28 | 4.77 | ||||
| 4-α-d-GalA | 13C | 99.4 | 68.3 | 71.0 | 82.5 | 71.1 | 175.3 | |||
| B | 1H | 5.50 | 3.99 | 3.68 | 4.27 | 4.81 | ||||
| 4-α-d-GalA | 13C | 99.5 | 68.4 | 71.0 | 81.6 | 71.0 | 175.1 | |||
| C | 1H | 5.09 | 3.68 | 3.88 | 4.19 | 4.49 | ||||
| t-α-d-GlcA | 13C | 101.2 | 68.7 | 69.5 | 70.5 | 71.9 | 175.5 | |||
| D | 1H | 5.04 | 3.53 | 3.79 | 4.19 | 3.91 | 4.09/3.99 | |||
| 3,6-α-d-Gal | 13C | 99.7 | 71.6 | 82.3 | 69.5 | 70.4 | 66.7 | |||
| E | 1H | 5.00 | 3.45 | 3.75 | 3.38 | 3.95 | 3.84/3.64 | |||
| t-α-d-Glc | 13C | 97.4 | 70.3 | 70.5 | 70.3 | 72.9 | 60.4 | |||
| F | 1H | 4.55 | 3.92 | 3.88 | 4.20 | 4.07 | ||||
| 2-β-d-GalA | 13C | 99.9 | 76.2 | 74.0 | 72.1 | 75.4 | 175.4 | |||
| G | 1H | 4.26 | 3.18 | 3.62 | 3.38 | 3.19 | 3.52 | |||
| 3-β-d-Glc | 13C | 103.5 | 70.7 | 81.1 | 68.5 | 76.2 | 62.2 | |||
| H | 1H | 4.23 | 3.44 | 3.40 | 3.31 | 3.42 | ||||
| 2-β-d-GlcA | 13C | 102.6 | 81.3 | 76.1 | 68.9 | 76.0 | 174.2 | |||
| K | 1H | 1.94/1.75 | 4.03 | 3.95 | 3.69 | 3.87 | 3.67/3.49 | |||
| 4,5-α-d-Kdo | 13C | ND | 96.2 | 33.9 | 65.5 | 75.0 | 70.9 | 69.7 | 63.0 | |
| PseI | 1H | 2.42/1.55 | 3.68 | 4.05 | 3.77 | 3.94 | 4.01 | 1.09 | ||
| 3′-β-l,l-Pse | 13C | 172.8 | 103.2 | 36.0 | 66.3 | 48.0 | 73.0 | 53.7 | 68.3 | 16.2 |
| HBu | 1H | 2.24 | 4.13 | 1.13 | ||||||
| 13C | 173.4 | 45.0 | 69.8 | 22.0 | ||||||
| PseII | 1H | 2.47/1.50 | 3.77 | 4.05 | 3.65 | 3.86 | 3.99 | 1.05 | ||
| t-β-l,l-Pse | 13C | 172.1 | 102.3 | 36.1 | 66.6 | 48.0 | 73.9 | 53.5 | 68.8 | 16.9 |
| HBu | 1H | 2.22 | 4.06 | 1.11 | ||||||
| 13C | 173.3 | 44.9 | 64.8 | 21.9 | ||||||
| NAc | 1.90 | |||||||||
| 21.8 | ||||||||||
| X | 1H | 5.55 | 3.30 | 3.80 | 3.47 | 4.06 | 4.16/3.68 | |||
| 6-α-d-GlcN | 13C | 90.4 | 54.1 | 69.5 | 70.1 | 72.4 | 69.2 | |||
| Y | 1H | 4.78 | 2.95 | 3.71 | 3.40 | 3.67 | 3.44 | |||
| 6-β-d-GlcN | 13C | 99.3 | 55.5 | 71.9 | 69.6 | 75.0 | 61.0 | |||
| E2 | 1H | 5.08 | 3.68 | 3.68 | 3.26 | 4.06 | 3.43 | |||
| 2-α-d-Glc | 13C | 97.3 | 75.4 | 71.9 | 69.8 | 72.0 | 61.0 | |||
| K2 | 1H | 1.74/1.99 | 4.05 | 4.20 | 3.62 | 3.92 | 3.82/3.55 | |||
| 4,5-α-d-Kdo | 13C | ND | ND | 34.3 | 70.4 | 72.2 | 72.2 | 70.3 | 62.8 | |
| J | 1H | 1.86/2.11 | 3.83 | 3.91 | 3.93 | 3.87 | 3.82/3.49 | |||
| t-α-d-Kdo | 13C | ND | ND | 33.9 | 69.4 | 70.1 | 71.4 | 69.6 | 63.0 | |
a 1′, 2′, 3′, and 4′ indicate the proton and the carbon positions of the HBu group. The proton chemical shift of the NH of the NAc group for both PseI and PseII was at 7.51 ppm, whereas that at position 7 of both PseI and PseII was at 7.18 ppm.
Figure 4.
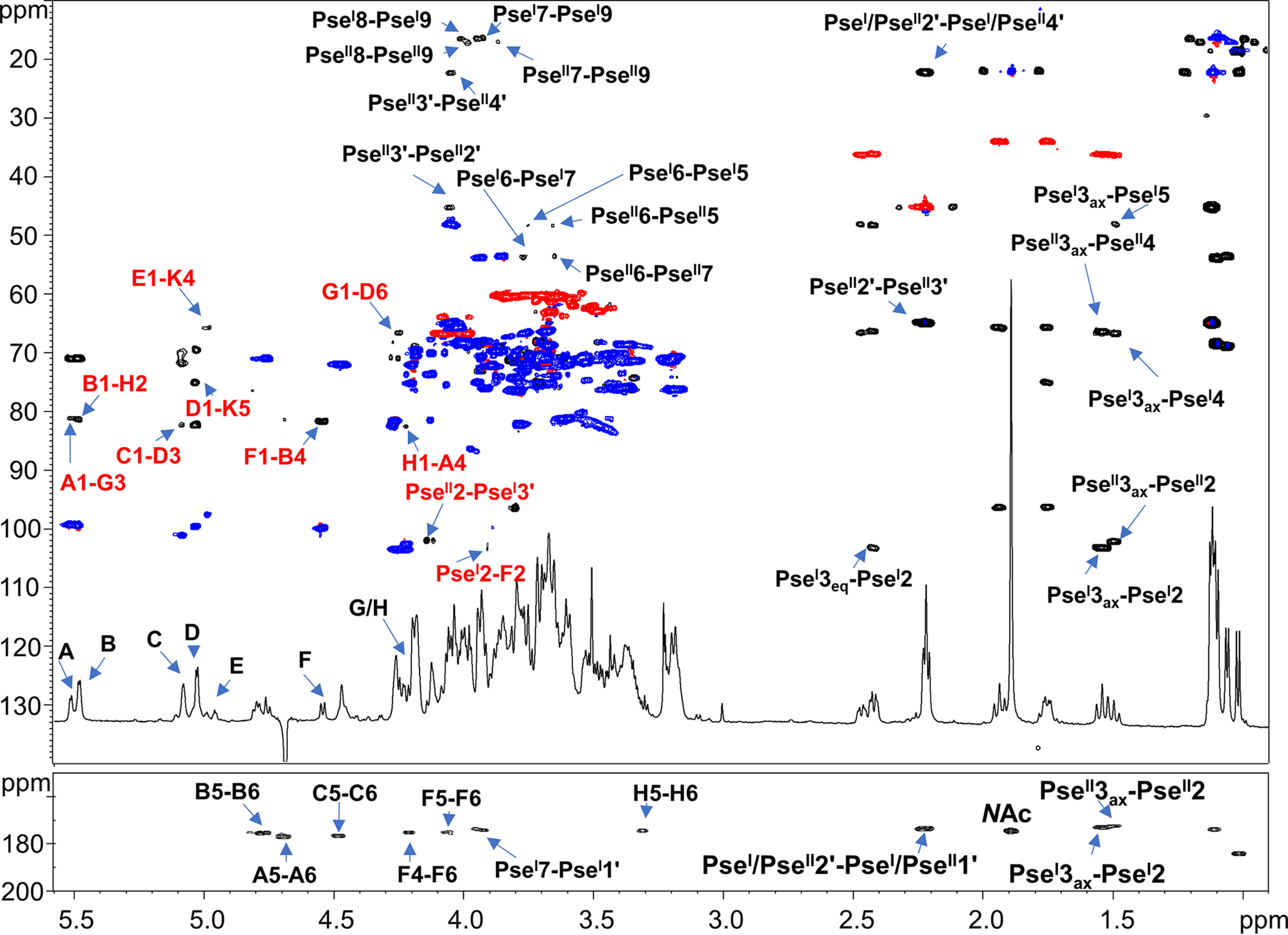
Superimposition of 1H-13C HSQC (blue/red) with 1H-13C HMBC (black) of OS1rkpA product. Densities are labeled with the same letters listed in Table 2. The most relevant interresidue long-range correlations involving sugar moieties are indicated in red, whereas in black are reported some important intraresidue long-range correlations.
As for the other spin systems, A (H-1 at 5.53 ppm, Table 2), B (H-1 at 5.50 ppm), C (H-1 at 5.09 ppm), F (H-1 at 4.55 ppm), and H (H-1 at 4.23 ppm) were identified as hexuronic acid residues because of the correlation of their H-5 signals with carboxyl groups resonating at 175.3, 175.1, 175.5, 174.9, and 174.2 ppm, respectively. The 3JH-3,H-4 and 3JH-4,H-5 coupling constant values (3 and 1 Hz, respectively) measured for A, B, and F were diagnostic of a galacto configuration, whereas the large ring 3JH,H coupling constants observed for C and H led to their identification as gluco-configured sugar units. The α-anomeric configuration for A, B, and C was attributed on the basis of the intraresidual NOE contact of H-1 with H-2 and the 3JH1,H2 coupling constant values, whereas the β-anomeric configuration for residues F and H was inferred by the large 3JH-1,H-2 values and the NOE contacts of H-1 with H-3 and H-5. Therefore, A and B were identified as α-GalA units, C as an α-GlcA, F as a β-GalA, and H as a β-GlcA. Residue D (H-1 at 5.04 ppm, Table 2) was identified as α-Gal because of the small 3JH,H values for H-3/H-4 and H-4/H-5, whereas residues E (H-1 at 5.00 ppm) and G (H-1 at 4.26 ppm) were assigned to α- and β-Glc, respectively.
Low-field shifted carbon signals (Table 2 and Fig. 3) were useful to identify substitution at O-4 of residues A and B, at O-2 of F and H, at O-3 and O-6 of D, at O-3 of G, at O-4 and O-5 of K, and, as described above, at O-3′ of PseI; on the other hand, residues C, E, and PseII were ascribed to terminal sugar units. The sequence of the monosaccharide residues was deduced by using both NOE correlations attained from the ROESY spectrum and the long-range correlations of the HMBC spectrum. In detail, the scalar interresidue long-range connections H-1 D/C-5 K, H-1 E/C-4 K, H-1 C/C-3 D, H-1 G/C-6 D, H-1 A/C-3 G, H-1 H/C-4 A, H-1 B/C-2 H, H-1 F/C-4 B, H-2 F/C-2 PseI, and H-3′ PseI/C-2 PseII (Fig. 4) were identified and led to the structural assessment of the OS1rkpA product (Fig. 5).
Figure 5.
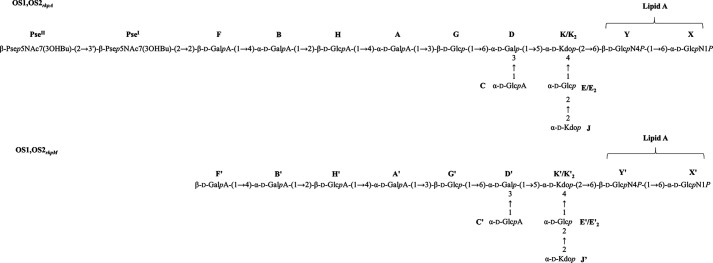
Structural assessments of the OS1rkpA, OS2rkpA, OS1rkpM, and OS2rkpM products. The complete structure of the core OS from S. fredii HH103 rkpA and rkpM was achieved by combination of data attained from the NMR investigation of the LPS after mild acid hydrolysis (yielding OS1rkpA and OS1rkpM) and full deacylation (yielding OS2rkpA and OS2rkpM) procedures. The mild acid hydrolysis selectively cleaved the linkage between the Kdo and the nonreducing glucosamine of the lipid A portion, enabling the isolation of the core OS moieties (sugar units labeled A, A'-H, H', K, K', PseI, and PseII) from the lipid A domain. The complete deacylation procedure allowed the isolation of the core OS plus the glucosamine disaccharide backbone (labeled X, X', Y, and Y') of the lipid A. The occurrence of an additional Kdo unit (J, J') was deduced by analysis of the fully deacylated products which, consequently, allowed the identification of the spin system E2, E2' (i.e. the α-d-Glcp substituted at position 2 by the Kdo unit J, J'). The core OS structures are labeled using letters in Table 2 and Table S2. p indicates the pyranose form of sugar residues, whereas P indicates the phosphate group.
To gain further structural information about HH103 rkpA LPS, an aliquot of sample underwent a complete deacylation step by hydrazinolysis followed by strong alkaline treatment. This approach provides the complete oligosaccharide moiety comprising the disaccharide lipid A backbone and allows us to evaluate the presence of substituents that could have been lost during the mild acid hydrolysis procedure. Indeed, despite the great heterogeneity observed in the product because of the harsh alkaline treatment and the intrinsic nature of the oligosaccharide, the NMR analysis of the obtained product (OS2rkpA) clearly disclosed, in addition to the signals relative to the α-GlcN (X) and β-GlcN (Y) of the lipid A (Table 2, Fig. S1), the occurrence of a further Kdo unit (J) whose methylene proton signals resonated at 1.86/2.11 ppm (δC 33.9 ppm). Finally, the observation of the low-field position of the signal for C-2 (75.4 ppm) of the α-Glc (E2) linked to the 4,5-α-Kdo (K2), and the occurrence of an NOE correlation between H-3 J and H-2 of E2 led to the determination of the sequence J-(2→2)-E2-(1→4)-K2, with the latter Kdo unit as the one linked to β-GlcN (Y) of lipid A. Therefore, summarizing all results arising from mild acid hydrolysis and full deacylation procedures, we found that HH103 rkpA LPS possesses a novel carbohydrate backbone made up of a disaccharide of β-Pse5NAc7(3OHBu), two Kdo units, one α-d-Glcp, one d-β-Glcp, one d-α-Galp, two d-α-GalpA, one d-β-GalpA, one d-α-GlcpA and one d-β-GlcpA, as sketched in Fig. 5.
A similar approach was used to elucidate the structure of OS1rkpM (Table S1). The comparison of the NMR spectra of OS1rkpM and OS1rkpA (overlapped HSQC spectra are reported in Fig. S2) revealed an identical structure apart from the absence of any Pse residue in OS1rkpM. Indeed, as shown by both the 1H and the HSQC NMR spectra (Fig. 2 and Fig. S2), no signals relative to the methylene and methyl groups of Pse residues were visible. Consequently, the β-GalA unit F' of OS1rkpM (Table S1), corresponding to residue F in the rkpA mutant, was identified as a terminal sugar because of the lack of glycosylation at position O-2 by the Pse moiety. Therefore, the structure of the saccharide backbone of rpkM mutant LPS is as reported in Fig. 5. In the case of HH103 rkpM, the NMR investigation (not shown) of the fully deacylated product (OS2rkpM) confirmed the occurrence of a terminal Kdo unit, lost during the mild acid hydrolysis treatment, as also proven by MALDI-TOF MS analysis executed on intact HH103 rkpM LPS (see below).
MALDI-TOF MS of intact LPS from HH103 rkpA and rkpM mutants
An aliquot of pure LPS isolated from HH103 rkpA and HH103 rkpM has been investigated by MALDI-TOF MS. The MS spectra, recorded in negative polarity, are reported in Fig. 6. The linear MALDI-TOF MS spectrum of HH103 rkpA LPS (Fig. 6A) showed at higher molecular masses (between m/z 3500 and 4800) a series of [M–H]– ions relative to the native LPS mixture, composed of species differing by the composition of both the acyl and the sugar moieties. More importantly, ion fragments arising from the cleavage of the labile glycosidic bond between Kdo and the lipid A portion were also identified at lower mass ranges (m/z 1500–2100). This LPS fragmentation (β-elimination) yielding both oligosaccharide ions and lipid A ions furnished important details about the structure of HH103 rkpA LPS. In particular, the MALDI-TOF MS investigation gave us the chance to appreciate the heterogeneity of both the lipid A and the core OS portion of such LPS. Indeed, the main ion fragments in Fig. 6A at m/z 1770.2 corresponded to an OS composed of four hexuronic acids (HexA), three hexoses (Hex), one Kdo, and one Pse (OSA), namely, an OS species matching with the one identified in the NMR analysis but lacking one Pse and one HexA unit. More interestingly, the ion fragment at m/z 2165.7 was attributed to an OS made up of five HexA, three Hex, two Kdo, and one Pse (OSB), confirming the occurrence of a second unit of Kdo, as observed in the NMR investigation of the fully deacylated product. Finally, the main LPS species, as stated above, were consistent with a combination of the OS ion peaks and differently acylated lipid A species.
Figure 6.
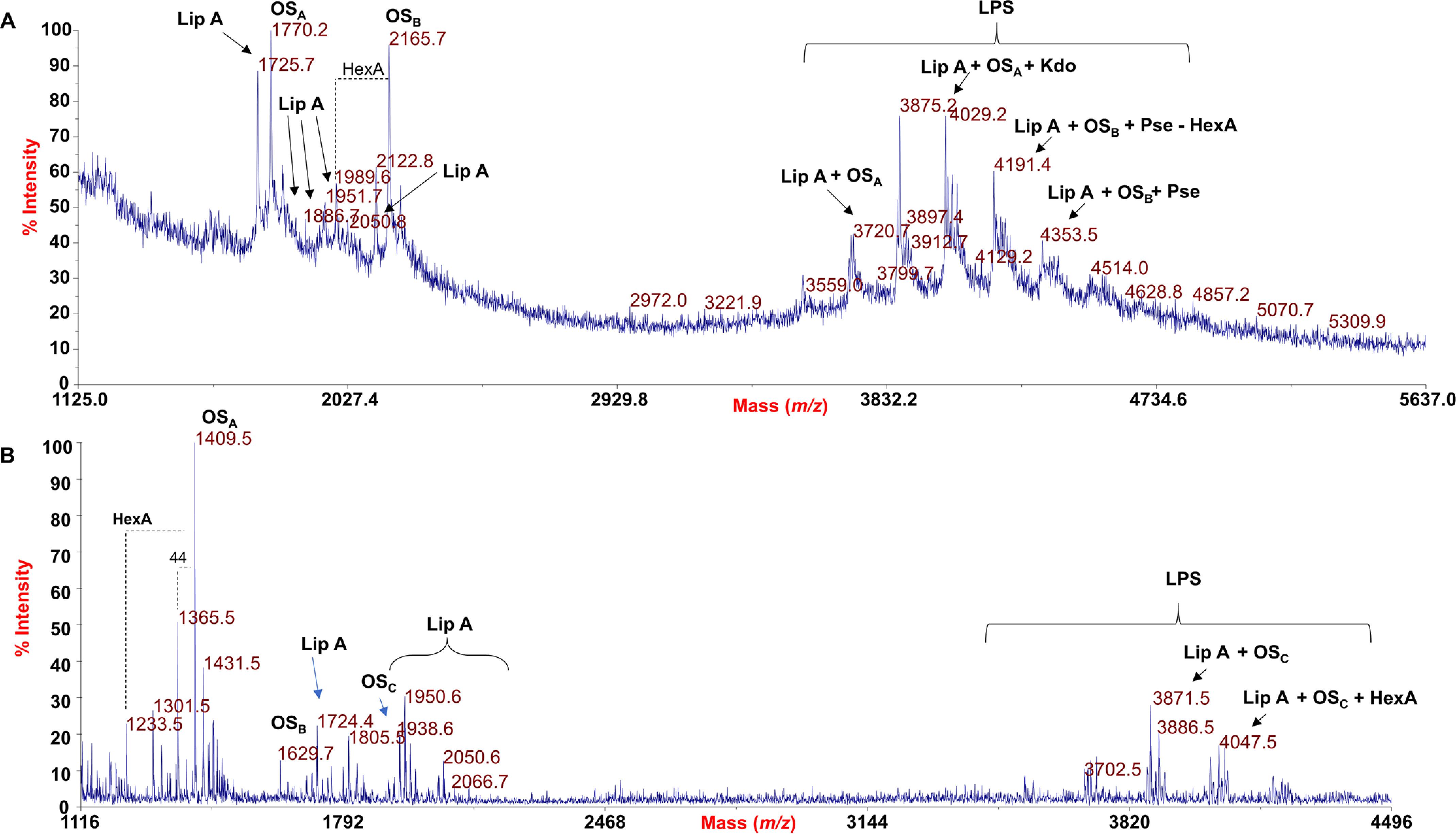
A, negative-ion MALDI mass spectrum of intact LPS from S. fredii HH103 rkpA recorded in linear mode. Both LPS molecular ions and their ion fragments, attributable to the core OS (OSA, OSB) and lipid A (LipA) species, are visible and indicated in the spectrum. B, negative-ion MALDI mass spectrum of LPS of S. fredii HH103 mutant rkpM recorded in reflectron mode. The lipid A (LipA) and core OS (OSA, OSB, OSC) species are also indicated in the spectrum. Proposed structural assessment of some LPS-related ions has been reported.
Similarly, the reflectron MALDI-TOF MS spectrum of intact LPS from the HH103 rkpM mutant revealed the occurrence of a main fragment ion at m/z 1409.5 relative to an OS built up of four HexA, three Hex, and one Kdo (OSA); OS fragments composed by a further Kdo unit (m/z 1629.7, OSB) and HexA (m/z 1805.5, OSC) have also been identified. Therefore, by MALDI-TOF MS analysis it was possible to further confirm that HH103 rkpM expresses an LPS devoid of Pse, according to NMR data. As described for the rkpA mutant, in the higher mass region, LPS species composed of the above OS species and differently acylated lipid A forms have been detected.
Fatty acid compositional analysis and MALDI-TOF MS of lipid A from HH103 rkpM and rkpA LPS
Fatty acid compositional analysis revealed the occurrence, in both HH103 mutants, of 12:0(3-OH), 13:0(3-OH), 14:0(3-OH), 15:0(3-OH), 16:0(3-OH), 17:0(3-OH), 18:0(3-OH), 19:0(3-OH), 18:1(3-OH), 18:2(3-OH), 19:1(3-OH), 20:1(3-OH), and the very-long-chain fatty acids (VLCFA) 28:0(27-OH), 30:0(29-OH), 28:0(27-OHBu), and 30:0(29-OHBu), which is consistent with the lipid A fatty acid composition from the genus Sinorhizobium (23). To unveil the structure of the glycolipid portion of the LPS from both mutants, an aliquot of the mild acid hydrolysis-precipitated product, containing lipid A, has been analyzed by negative-ion MALDI-TOF MS (the spectrum is reported in Fig. S3, A and B). The MALDI mass spectra of the isolated lipid A fractions were identical for both mutants and further confirmed, and expanded on, the high heterogeneity of such LPS moieties and the occurrence of hydroxybutyrate as an acyloxyacyl substituent carried on some of the VLCFAs. The main ion peaks and the proposed interpretation of the substituting fatty acids on the lipid A backbone are reported in Table S2. As an example, in the higher mass regions, the ion peak at m/z 2066.0 was attributed to a bis-phosphorylated penta-acylated lipid A species carrying two 14:0(3-OH), one 18:0(3-OH), and one 19:0(3-OH) as primary fatty acids and one 30:0(29-OHBu) as the secondary acyl moiety; further on, the ion peak at m/z 1952.0 corresponded to a penta-acylated lipid A species decorated by two phosphates and carrying two 14:0(3-OH), one 18:0(3-OH), one 19:0(3-OH), and one 28:0(27-OH), whose relative species missing one 14:0(3-OH) was assigned to a peak at m/z 1725.9. At m/z 2050.0, a further penta-acylated lipid A form decorated by two phosphates, two 14:0(3-OH), one 18:0(3-OH), one 18:1(3-OH), and one 30:0(29-OHBu) was also detected (Fig. S3C); the related species carrying only one phosphate group and lacking one 14:0(3-OH) moiety was identified at m/z 1743.9. The more intense ion peak at m/z 1870.0 was assigned to bis-phosphorylated lipid A forms composed of two 14:0(3-OH), one 13:0(3-OH), one 12:0(3-OH), and one 28:0(27-OHBu), whose relative species lacking one 14:0(3-OH) was identified at m/z 1643.9.
The mAb NB6-228.22 recognizes S. fredii HH103 RifR LPS but fails in the recognition of KPS
Structural investigation demonstrated that a β-configured Pse5NAc7(3OHBu) is present in the LPS of rkpA but absent from rkpM. Because NB6-228.22 binds to the LPS produced by rkpA but fails in the recognition of rkpM LPS, we investigated whether this mAb was also able to recognize the KPS produced by HH103 RifR, which is a homopolymer of α-Pse5NAc7(3OHBu) (12).
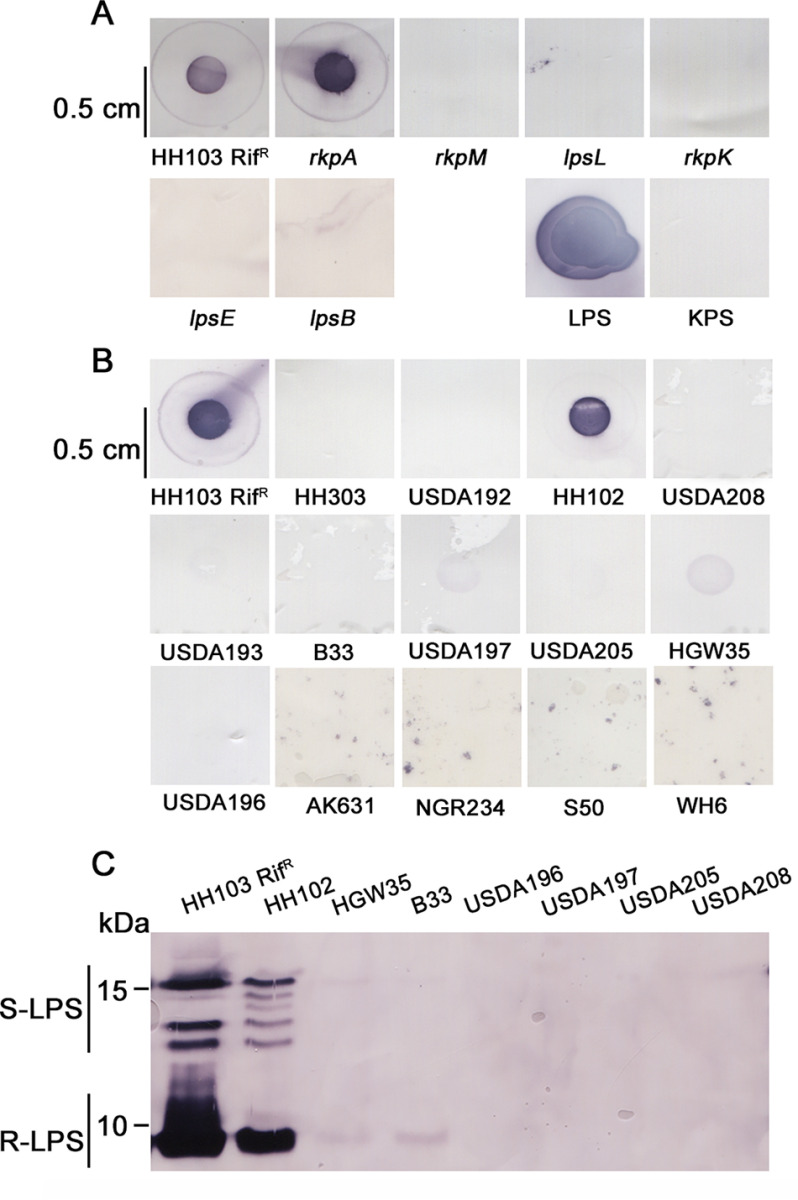
For this purpose, intact cells of S. fredii HH103 RifR and its derivatives mutated in the rkpA, rkpM, rkpK, lpsL, lpsB, and lpsE genes were transferred to a nylon membrane and were deposited on a positively charged nylon transfer membrane and immunostained with NB6-228.22. In this experiment, purified LPS and KPS samples from HH103 were also analyzed. As shown in Fig. 7A, only cells from the two strains not affected in their LPS (HH103 and its rkpA derivative) as well as the purified LPS sample showed positive recognition by NB6-228.22. These results clearly indicate that NB6-228.22 does not react with HH103 KPS.
Figure 7.
A, immunostaining of intact cells of S. fredii HH103 RifR and HH103 mutants in the rkpA, rkpM, rkpK, lpsL, lpsB, and lpsE genes using the mAb NB6-228.22. LPS, sample containing HH103 RifR crude extract preparations used for PAGE electrophoresis (they contain KPS and LPS). KPS, sample containing HH103 lpsB KPS. B, immunostaining of intact cells of S. fredii strains isolated from diverse geographical areas and S. meliloti AK631. C, immunostaining of LPS electrophoretic patterns of diverse S. fredii strains.
The epitope recognized by the mAb NB6-228.22 is not common in S. fredii strains
Both the LPS and KPS structures may vary at the strain level (1, 19). For this reason, we decided to investigate the reactivity of other S. fredii strains against NB6-228.22. Intact cells of 25 S. fredii strains (including HH103) that have been isolated in previous works from soybean plants growing in soils of different Chinese provinces were included in this study (Table S3, Fig. 7B). Only two of these strains (HH103 and HH102), which were isolated from nodules of soybean plants inoculated with soil from Hubei province (43), were recognized by the antibody. The remaining tested strains, isolated and reported in the period 1982–2003 (44), from soils of Chinese East-Central provinces (Shanghai, Hubei, Henan, and Shandong), as well as from the Xingjian Autonomous Region (Western China), were not recognized by NB6-228.22. S. fredii NGR234 and S. meliloti AK631 were included in this study, because their KPS contain two different derivatives of the pseudaminic acid (45). Intact cells of NGR234 and AK631 were not recognized by NB6-228.22 (Fig. 7B). Immunostaining of PAGE-separated LPS samples from several S. fredii strains confirmed that NB6-228.22 recognized the different LPS bands from HH102 but failed to recognize LPS from strains HGW35, B33, USDA196, USDA197, USDA205, and USDA208 (Fig. 7C).
Because the rkp-3 region is involved in the production of Pse5NAc7(3OHBu) (17), all the S. fredii strains tested with NB6-228.22 were also investigated by PCR for the presence of the rkpL, rkpM, rkpP, and rkpQ genes using genomic DNA (gDNA) as the template (Tables S3 and S4). Strain NGR234 was not included in these experiments because its rkp-3 region has been sequenced (46). Only the gDNA of HH103 and HH102 led to the amplification of the expected fragments for all the genes investigated (Table S3), whereas when gDNA of strain S50 was used, the internal fragments of rkpM and rkpP, but not those of rkpL and rkpQ, could be amplified. All amplified fragments showed an identity of 98–100% with the corresponding rkp gene of S. fredii HH103 RifR (Table S4).
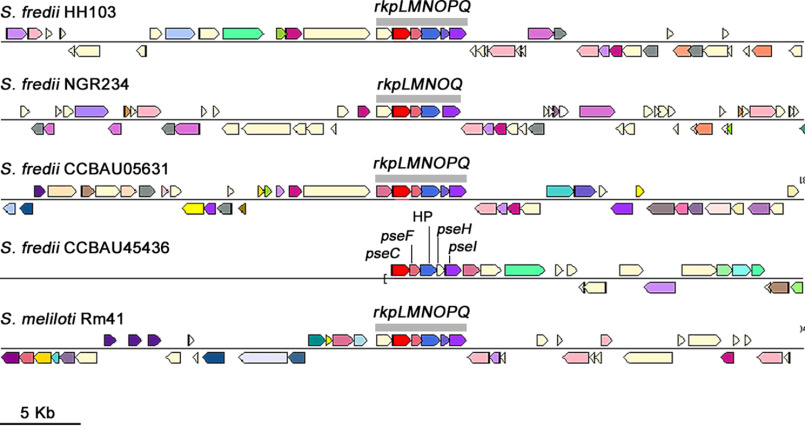
Because S. fredii S50 appeared to have only part of the rkpLMNOPQ operon, we investigated the presence of these genes in sinorhizobial strains for which sequences homologous (>85.0 identity at amino acid level) to RkpM were found in silico. The complete operon was present in S. fredii CCBAU05631, Sinorhizobium sp. strain BJ1, S. meliloti Rm41, S. medicae Str9, and S. americanum CCGM7 (Table S5). In two other strains, S. fredii NGR234 and S. saheli LMG7837, the rkpP genes were missing. S. fredii strain CCBAU45436 lacks rkpO, and the rest of the predicted proteins showed identities lower than 55%. Figure 8 shows the organization of the rkp-3 region in S. fredii strains as well as in S. meliloti Rm41.
Figure 8.
The rkp-3 region organization in the different S. fredii strains analyzed and S. meliloti Rm41 as a reference. Genes in red correspond to the rkpM gene or, in the case of CCBAU45436, to an orthologue with similar function. The rkp-3 genes shown have different colors depending on their function.
The symbiotic impairment of the S. fredii HH103 rkpM mutant is higher with Cajanus cajan than with Macroptilium atropurpureum
The symbiotic relevance of LPS and KPS in rhizobium-legume interactions has been extensively documented (1–5). In this work, we have analyzed the symbiotic phenotype of HH103 rkpM and its complemented version, HH103 rkpM C, with two HH103 host legumes, M. atropupureum and C. cajan, which form determinate and indeterminate nodules, respectively (17, 38).
In the case of M. atropurpureum (Table 3 and Fig. 9), HH103 rkpA, affected in KPS but not in LPS production, was also included in these studies. The number and fresh weight of nodules of M. atropurpureum plants inoculated with HH103 rkpA or HH103 rkpM were similar and significantly lower than those formed on plants inoculated with HH103 RifR, although nodules induced by HH103 rkpM were white. Plants inoculated with these mutants were stunted with yellow leaves, and their plant-top dry weight was not significantly different from that of the uninoculated control. In contrast, plants inoculated with HH103 RifR or HH103 rkpM C produced green leaves and plants did not show symptoms of nitrogen starvation. Nitrogenase activity (nmol ethylene/3 plants × hour, determined by acetylene reduction assays) was similar in plants growing in the three jars inoculated with HH103 RifR (136 ± 43) or HH103 rkpM C (154 ± 63) and lower in the three jars inoculated with HH103 rkpA (36 ± 14). When HH103 rkpM was used as the inoculant, a very low nitrogenase activity could be detected and only in two out of the three jars analyzed (4.7 and 15.1). Microscopy analyses showed dispersed patches of nodule cells invaded by HH103 rkpM (Fig. 10A2), which contained less bacteroids than those infected by S. fredii HH103 RifR or HH103 rkpM C (Fig. 10A1 and A3). Nodules induced by HH103 rkpA (Fig. 10A4) showed an infection level that could be considered an intermediate stage between those induced by HH103 RifR and HH103 rkpM.
Table 3.
Macroptilium atropurpureum and Cajanus cajan responses to inoculation with S. fredii HH103 RifR, its rkpM mutant, and its complemented derivativea (HH103 rkpM C)
| Legume and inoculant | No. of nodules | Fresh wt of nodules (mg) | Plant-top dry wt (mg) |
|---|---|---|---|
| Macroptilium atropurpureum | |||
| HH103 RifR | 39.00 ± 4.77 A | 50.53 ± 4.8 A | 127.61 ± 10.03 A |
| HH103 rkpM | 4.33 ± 0.47 B | 13.84 ± 2.39 B | 41.43 ± 4.56 B |
| HH103 rkpM C | 30.00 ± 2.22 A | 48.78 ± 4.68 A | 110.15 ± 11.02 A |
| HH103 rkpA | 8.45 ± 0.81 B | 17.53 ± 3.37 B | 63.45 ± 6.23 B |
| Uninoculated | None | 48.83 ± 2.78 B | |
| Cajanus cajan | |||
| HH103 RifR | 30.58 ± 1.15 A | 273.84 ± 33.84 A | 621.22 ± 59.35 A |
| HH103 rkpM | Pseudonodules | 78.68 ± 5.04 B | |
| HH103 rkpM C | 31.58 ± 3.17 A | 353.95 ± 54.42 A | 722.41 ± 89.13 A |
| Uninoculated | None | 101.88 ± 10.72 B |
aNumbers are mean (±S.E.) values per plant. M. atropurpureum roots inoculated with HH103 rkpM also developed pseudonodules. Twelve plants were analyzed for each legume/inoculant combination. For each legume, data in the same column with the same letter were not significantly different (one-way analysis of variance, α = 5%). For number of nodules, only those treatments showing nodules were compared.
Figure 9.
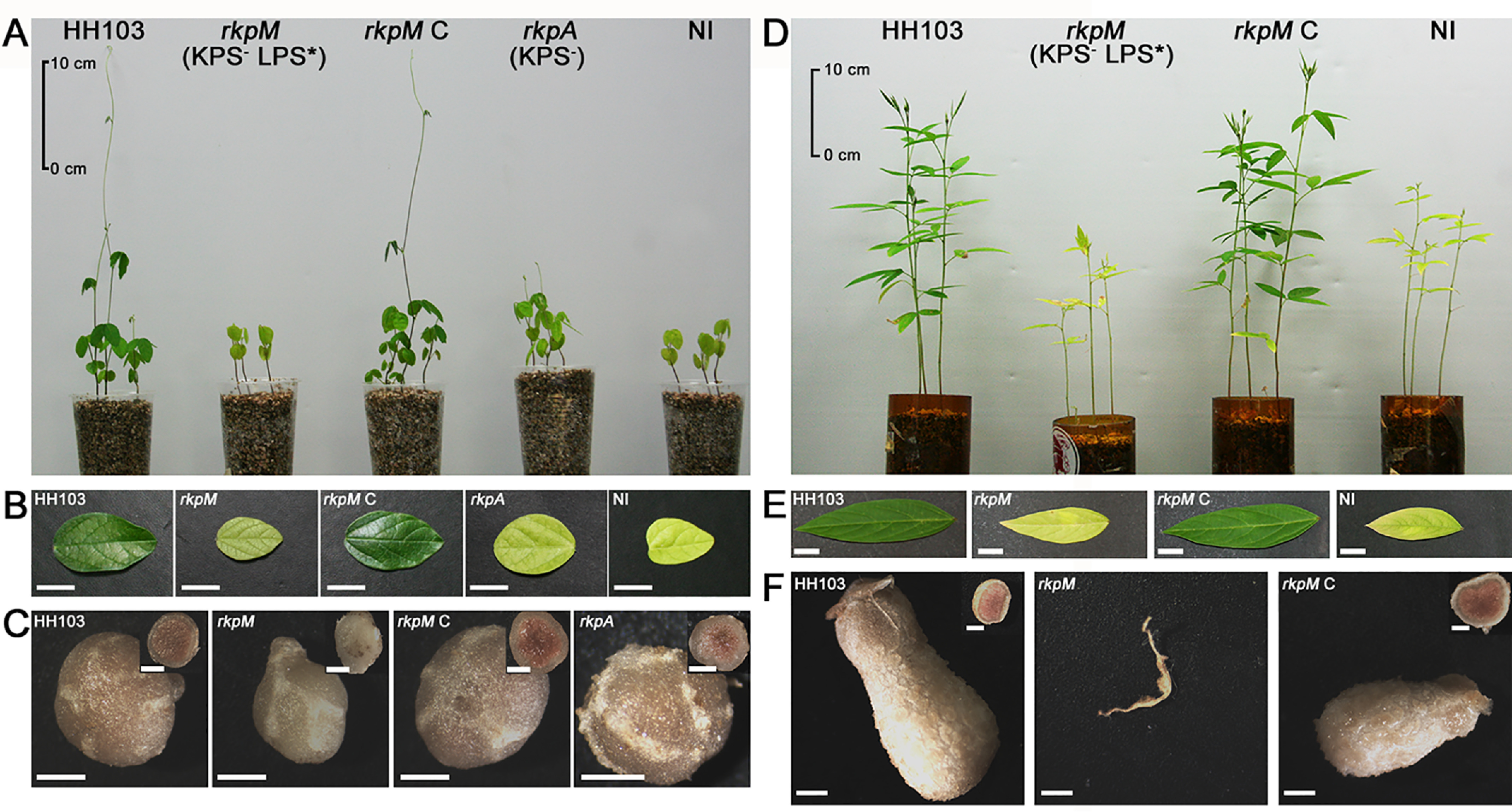
Symbiotic performance of HH103 rkpM and its complemented version (HH103 rkpM C) with Macroptilium atropurpureum (A–C) and Cajanus cajan (D–F). The HH103 rkpA mutant was also included in the studies with M. atropurpureum. The WT strain is denoted as HH103. NI stands for noninoculated plants. The phenotype regarding surface polysaccharide production of mutants rkpM and rkpA is shown is panels A and D. LPS* indicates production of an altered LPS. A and D, aerial part of the plants at the end of the assay. B and E, plant leaves. C and F, complete and transversal sections of nodules (or macroscopic root outgrowths) induced by inoculation with the different strains. The bars in B and E correspond to 1 cm and in C and F to 1 mm.
Figure 10.
Structure of Macroptilium atropurpureum and Cajanus cajan nodules elicited by inoculation with S. fredii HH103, its rkpM and rkpA mutants, and the rkpM complemented strain (HH103 rkpM C). M. atropurpureum nodules induced by HH103 RifR (A1), HH103 rkpM (A2), HH103 rkpM complemented (HH103 rkpM C, A3), and HH103 rkpA (A4). C. cajan nodules or pseudonodules were elicited by HH103 RifR (B1), HH103 rkpM (B2), and HH103 rkpM C (B3). Nodules were analyzed seven (M. atropurpureum) or eight (C. cajan) weeks after inoculation. The bar size represents 50 μm.
The symbiotic impairment of another rkpM mutant (called SVQ582) with C. cajan was previously reported (17), although microscopy of the pseudonodules produced was not carried out. Because infected cells were found in M. atropurpureum nodules, we investigated whether C. cajan pseudonodules contained cells invaded by the HH103 rkpM mutant investigated in the present work. Plant-top dry weight of C. cajan plants inoculated with HH103 rkpM was not significantly different from that shown by uninoculated controls (78 and 101 mg/plant, respectively). Plants inoculated with HH103 rkpM only formed pseudonodules and macroscopic swellings (Table 3, Fig. 9). Microscopy images showed that pseudonodules induced by this mutant were devoid of bacteroids (Fig. 10B2). C. cajan plants inoculated with S. fredii HH103 RifR or HH103 rkpM C formed nitrogen-fixing nodules, and plants were green and did not show symptoms of nitrogen starvation (Fig. 9).
Discussion
In this work, we report the characterization of the full structure of the LPS from S. fredii HH103 rkpA. We selected this mutant because its LPS shows the same electrophoretic profile and reactivity with the mAb NB6-228.22 as the WT strain LPS does. Thus, the LPS structure of HH103 rkpA is assumed to be equal to that of the WT strain. In addition, HH103 rkpA is devoid of KPS, a polysaccharide that can interfere with LPS purification procedures. Our work reveals a novel core OS with a high content in hexuronic acids and showing the presence of β-Pse5NAc7(3OHBu) located at the outermost part of the LPS. This residue was found to be linked to another one of the same nature, which might constitute an extremely short O-chain moiety (Fig. 5). In previous works, we obtained different HH103 mutants affected in LPS structure, as reflected in their altered electrophoretic profiles (Table 1) (17, 31, 37). In this work, we have also carried out the structural analysis of the LPS produced by HH103 rkpM, because its LPS electrophoretic profile was less altered than that of HH103 LPS, among the different HH103 mutants available. Our work highlights that the core OS of HH103 rkpM lacks Pse5NAc7(3OHBu) (Fig. 5). This result is in full accordance with the fact that the rkpLMNOPQ operon is involved in the biosynthesis of Pse derivatives in S. meliloti 41 (47) and S. fredii HH103 (17). Finally, the lipid A moieties of both rkpA and rkpM were found to be identical and displayed a high degree of heterogeneity in the acyl chains, which were characterized by the nonstoichiometric substitution by hydroxybutyrate on the VLCFA and by the occurrence of unsaturated chains, as previously reported for other Sinorhizobium strains (23). The lack of function of RkpM also affects the alterations that the S. fredii HH103 LPS undergoes during bacteroid development in nodules of Glycyrrhiza uralensis.
The HH103 lpsB, lpsE, lpsL, and rkpK mutants are also presumably affected in the core structure of their LPS. LpsB and LpsE are predicted glycosyl transferases (31), whereas LpsL and RkpK are responsible for the synthesis of GalA and GlcA, respectively, (37), and, as demonstrated in the present work, the HH103 LPS core contains hexoses and these two uronic acids. We planned to analyze the LPS structure of these four HH103 LPS mutants in the future.
The mAb NB6-228.22 recognizes all of the LPS silver-stained bands of WT HH103 and its rkpA mutant derivative (unable to produce KPS) as well as entire cells of these two strains and a purified HH103 LPS sample. However, it failed to perceive the LPS of HH103 rkpM, which strongly suggested that the Pse5NAc7(3OHBu) residue present in the LPS core OS of HH103 and HH103 rkpA should be the epitope (or part of it) recognized by this antibody. Interestingly, antibody NB6-228.22 failed to perceive a purified sample of the S. fredii HH103 KPS, even though Pse5NAc7(3OHBu) is the only component of this polysaccharide, and failed in the recognition of intact cells and LPS profiles of HH103 lpsL, lpsB, lpsE, and rkpK mutants (altered LPS, WT KPS). Thus, it was possible to conclude that the mAb NB6-228.22 recognizes the HH103 LPS but not its KPS. There are various possibilities that could explain why the same monosaccharide, Pse5NAc7(3OHBu), is recognized in the LPS but not in the KPS. One very likely explanation is that Pse5NAc7(3OHBu) residues are α-linked in the KPS (12) but β-linked in the LPS. Alternatively, the epitope perceived by the antibody might not be restricted to only Pse5NAc7(3OHBu), which is the case of the HH103 RifR KPS but not of the LPS in which Pse5NAc7(3OHBu) is linked to a d-GalA residue (Fig. 5). In any case, the epitope recognized by the antibody NB6-228.22 is infrequent in S. fredii strains, as demonstrated by the fact that only 2 of 25 strains analyzed (including HH103) reacted with this antibody. This fact is not unexpected, because the LPS core and O-antigen regions are known to vary at the strain level (1, 19).
PCR experiments and in silico analyses carried out in this work showed that the rkpLMNOPQ operon involved in Pse5NAc7(3OHBu) biosynthesis can be found in diverse Sinorhizobium species, but its presence might not be frequent at the strain level. In addition, some of the Sinorhizobium strains containing the region lack some of these genes. S. fredii S50 amplified the rkpM and rkpP genes but failed in the amplification of rkpQ and rkpL. The amplified internal sequences of S50 rkpM and rkpP are nearly identical to those of HH103 (98–100% identity), which suggests that the failure to amplify rkpQ and rkpL is because of the absence of these genes in S50 rather than the unsuitable nucleotide sequences of the primers used. S. fredii NGR234 and S. saheli LMG7837 are devoid of orthologues of rkpP, and S. fredii CCBAU45436 lacks rkpO. The absence of rkpP does not prevent the biosynthesis of the Pse derivative present in the NGR234 KPS (46).
In previous studies, we showed that two plants forming determinate nodules, soybean and cowpea (Vigna unguiculata), when inoculated with HH103 rkpM only formed pseudonodules that did not fix nitrogen and were devoid of intracellular bacteria, whereas this mutant was able to induce the formation of Fix+ nodules with Lotus burttii (determinate nodules) and Glycyrrhiza uralensis (indeterminate nodules) (17, 48). In this work, we have included another determinate-nodule forming legume, Macroptilium atropurpureum, and showed that nodulation of plants inoculated with HH103 rkpM was severely reduced and nitrogen fixation almost abolished compared with those inoculated with HH103 (Table 3).
In contrast to HH103 rkpM, S. fredii HH103 lpsB and lpsE mutants (presenting KPS but with an altered LPS) as well as S. fredii HH103 mutants in genes of the rkp-1 region (unable to produce KPS but with a normal LPS) are still able to induce the formation of Fix+ nodules in both soybean and cowpea plants, albeit nodulation is reduced and plants show symptoms of nitrogen starvation (14–17). Considering all these results, we can conclude that in S. fredii HH103 the combination of the absence of KPS production and an alteration of the LPS, consisting of the absence of Pse5NAc7(3OHBu), causes the most severe symbiotic impairment with these two legumes. An S. fredii HH103 RifR double (rkpH and exoA) mutant unable to produce KPS and EPS (LPS electrophoretic profile apparently unaffected) was still able to induce the formation of some nitrogen-fixing nodules with G. max cvs. Williams and Peking (14). Thus, the loss of EPS production in an S. fredii HH103 unable to produce KPS is not so harmful for the bacterial symbiotic capacity with soybeans than the simultaneous absence of KPS production and Pse5NAc7(3OHBu) residues in the LPS structure.
Thus, two types of plant responses to inoculation with HH103 rkpM can be observed in both determinate and indeterminate nodules forming legumes: G. max, V. unguiculata, and C. cajan formed uninfected pseudonodules, whereas L. burttii, M. atropurpureum, and G. uralensis were still able to form a variable number of nitrogen-fixing nodules. All these results indicate that the symbiotic impairment of mutations affecting S. fredii HH103 surface polysaccharides vary among the different legumes tested, regardless of whether they form indeterminate or determinate nodules, and that the simultaneous alteration of two polysaccharides can produce unpredictable impacts on the bacterial symbiotic capacity.
RSP, such as LPS and KPS, also can be important for stress tolerance both under free-living conditions and during the infection process and bacteroid persistence (29, 31, 49). Here, we have studied the effect of the rkpA and rkpM mutations on the final growth (expressed as the optical density at 600 nm [OD600] scored 24, 48, or 120 h after inoculation) of S. fredii HH103 in liquid minimal medium supplemented with high concentrations of either sodium chloride (100 or 150 mm) or the detergent SDS (0.35 or 0.52 mm) (Fig. S4). No differences among the different strains were found in the presence of 100 or 150 mm NaCl or 0.35 mm SDS. However, 5 days after inoculation, the growth of S. fredii HH103 RifR and HH103 rkpM C in liquid MM supplemented with 0.52 mm SDS was higher (OD600, >0.8) than that observed for HH103 rkpM (OD600 of about 0.3) or HH103 rkpA (OD600 of about 0.5) (Fig. S4). These results suggest that both KPS and LPS provide protection against SDS. Whether these phenotypes are related to the symbiotic impairment exhibited by HH103 rkpM requires further investigation.
In conclusion, with this work a novel LPS structure has been identified revealing the occurrence of an unprecedented component for rhizobial LPS, a β-configured Pse5NAc7(3OHBu), whose presence might be important for the establishment of an effective symbiosis of S. fredii HH103 with some of its host plants. This crucial point will be the object of future structure-to-function studies focused on the determination of the molecular motifs of S. fredii HH103 LPS that may be crucial for bacterium–plant cell interactions.
Experimental procedures
Extraction, purification, and SDS-PAGE analysis of rkpA and rkpM LPS
Dried bacterial cells of HH103 rkpA and rkpM were washed several times with distilled water, ethanol, acetone, and ethylic ether to remove growth culture contaminants. After lyophilization, the bacterial pellets were extracted by the phenol/water method (40). After extensive dialyses against distilled water, the extracted phases were suspended in 40 ml of digestion buffer (100 mm Tris, 50 mm NaCl, 10 mm MgCl2, pH 7.5) to which RNase (4 mg, R5503-Merck), DNase (4 mg, DN25-Merck), and proteases (4 mg) were added to remove nucleic acids and protein contaminants. Both the water and phenol fractions were checked by SDS-PAGE after silver staining; the LPS material, in both HH103 rkpA and rkpM, was found exclusively in the water phases. To further purify the samples, the LPSs were ultracentrifuged (4 °C, 208,000 × g, 16 h) and then subjected to size-exclusion chromatography on a Sephacryl S-300 (GE Healthcare, 1.5 by 90 cm, eluent of 50 mm NH4HCO3).
LPS extraction for PAGE and immune-staining analyses, as well as the separation on SDS-PAGE gels, silver staining, and immunostaining procedures with the mAb NB6-228.22 (class IgG 2b), were performed as described previously (38). Production of the mAb NB6-228.22 (class IgG 2b) was described by Buendía-Clavería and collaborators (38). Briefly, soybean nodule-derived bacteroids of HH103 were used to immunize male rats (line LOU/IAP), and spleen cells derived from these rats were fused to the myeloma line IR983F. NB6-228.22 recognizes all the LPS bands from HH103 free-living cells or HH103 bacteroids isolated from soybean nodules. NB6-228.22 was purified from the hybridoma culture supernatant.
Binding assays of intact S. fredii cells to NB6-228.22 were carried out as follows. A loop of S. fredii cells grown on solid TY for 4 days was suspended in 200 μl of sterile water. 10-μl drops of bacterial suspensions then were deposited on positively charged Nylon transfer membranes (Amersham Biosciences, UK). Intact bacterial samples absorbed to the filters were immunostained with NB6-288.22, using as the secondary antibody a goat anti-rat IgG conjugated to alkaline phosphatase (Sigma).
Chemical analyses of HH103 rkpA and rkpM LPS
Monosaccharides were identified as acetylated O-methyl glycoside derivatives obtained by methanolysis (1.25 M HCl/MeOH, 85 °C, 16 h) followed by acetylation with acetic anhydride in pyridine (80 °C, 30 min) (41). To define the sugar linkage pattern, including uronic acids and Kdo, an aliquot of each sample was methylated with iodomethane, treated with sodium tetradeuteroborate (NaBD4) to transform the methyl ester functions of acidic sugars in a hydroxymethyl group with two deuterium atoms, and then hydrolyzed with TFA (2 m, 100 °C, 2 h), carbonyl-reduced by employment again of NaBD4, acetylated with acetic anhydride and pyridine, and analyzed by gas LC-MS (GLC-MS) (41, 50). The absolute configuration of sugar residues has been determined by GLC-MS analysis of the acetylated O-(+)-2-octyl glycoside derivatives and comparison with authentic standards (41, 51).
The total fatty acid content was established by treating each LPS with 4 M HCl (100 °C, 4 h) and followed by a treatment with 5 M NaOH (100 °C, 30 min). The pH was adjusted to reach slight acidity. Fatty acids, after extraction in CHCl3, were methylated with diazomethane and analyzed by GLC-MS. The ester-bound fatty acids were selectively released by base-catalyzed hydrolysis with aqueous 0.5 m NaOH, MeOH (1:1, v/v, 85 °C, 2 h), and then the product was acidified, extracted in chloroform, methylated with diazomethane, and analyzed by GLC-MS. Moreover, an aliquot of each LPS fraction (0.5 mg) was also methanolized with 1.25 M HCl/CH3OH (80 °C for 16 h). The mixture underwent three extractions with hexane. The hexane layer, containing the fatty acids as methyl ester derivatives, was then analyzed by GLC-MS.
All the above derivatives were analyzed on Agilent Technologies gas chromatograph 6850A interfaced with mass spectrometry and a selective mass detector 5973N equipped with a SPB-5 capillary column (Supelco, 30 m by 0.25 mm inner diameter, flow rate of 0.8 ml min−1 using He as gas carrier). Electron impact mass spectra were recorded with an ionization energy of 70 eV and an ionizing current of 0.2 mA.
Isolation of the lipid A and the oligosaccharides OS1rkpA and OS1rkpM
The OS1 fractions of the two HH103 mutants investigated were obtained by mild acid hydrolysis. Purified LPS (∼10 mg) was dissolved in acetate buffer (1 ml, pH 4.4). SDS (1 mg ml−1) was added and hydrolysis proceeded (100 °C, 2 h). The solutions were extracted three times with CHCl3/MeOH/H2O (100:100:30, v/v/v) and centrifuged (4 °C, 8,800 × g, 20 min). The organic phases containing the lipid A moiety were further purified by several washes with distilled water, lyophilized, and then analyzed by MALDI-TOF MS. The water phases, containing the OS1 fractions, were recovered and lyophilized. These OS1 fractions were then purified by gel filtration chromatography on a Bio-Gel P-4 column (Bio-Rad, 1.5 by 100 cm, eluent of distilled water) and then, once dissolved in 50 mm NH4HCO3, further purified via HPLC by using a TSKgel G5000PWXL column (Supelco-Sigma, run in 50 mm NH4HCO3 at 0.8 ml min−1, mounted on an Agilent 1100 system equipped with a refractive index and a UV detector, this last set at 206 nm).
Isolation of the oligosaccharides OS2rkpA and OS2rkpM
To remove O-linked acyl chains, an aliquot of each sample was treated with anhydrous hydrazine, stirred at 37 °C for 60 min, cooled, poured into ice-cold acetone, and allowed to precipitate. The precipitate was centrifuged (1,200 × g, 30 min), washed with cold acetone in an ice bath, dried, dissolved in water, and lyophilized (52). The O-deacylated product was then N-deacylated with KOH (4 M, 120 °C, 16 h). After adjustment of the pH, free fatty acids were removed by several extractions with CHCl3 followed by centrifugation (1,200 × g, 30 min). The aqueous phase of each sample was then lyophilized, and the produced salts were removed by size-exclusion chromatography on a Sephadex G-10 column (Pharmacia, 50 by 1.5 cm, eluent of distilled water) to yield OS2rkpA and OS2rkpM fractions.
NMR spectroscopy of the isolated OS1 and OS2
1D and 2D 1H NMR spectra were recorded on a Bruker 600 DRX equipped with a cryoprobe. The solvent employed was D2O, the temperature was 298 K, and pD was 7. To obtain information on the NH chemical shifts, the experiments were recorded in an H2O-D2O mixture (53). Spectra calibration was performed with internal acetone (δH 2.225 ppm, δC 31.45 ppm). The double-quantum filtered phase-sensitive correlation spectroscopy (DQF-COSY) experiment was carried out by using data sets of 4096 × 256 points. Total correlation spectroscopy (TOCSY) experiments were executed with spinlock times of 100 ms, using data sets (t1 × t2) of 4096 × 256 points. Rotating frame Overhauser enhancement spectroscopy (ROESY) and NOESY experiments were recorded by using data sets (t1 × t2) of 4096 × 256 points and by using mixing times between 100 and 400 ms. In all homonuclear experiments, the data matrix was zero-filled in both dimensions to give a matrix of 4 K × 2 K points and was resolution enhanced in both dimensions by a cosine-bell function before Fourier transformation. The determination of coupling constants was obtained by 2D phase-sensitive DQF-COSY (54, 55). Heteronuclear single quantum coherence (1H-13C HSQC) and heteronuclear multiple bound correlation (1H-13C HMBC) experiments were recorded in 1H-detection mode by single-quantum coherence with proton decoupling in the 13C domain using data sets of 2048 × 256 points. 1H-13C HSQC was executed using sensitivity improvement and in the phase-sensitive mode using Echo/Antiecho gradient selection, with multiplicity editing during the selection step (56). The 1H-13C HMBC experiment was optimized on long-range coupling constants with a low-pass J filter to suppress one-bound connectivity, using gradient pulses for selection. A delay of 60 ms was employed for the evolution of long-range correlations. It was used a long-range coupling constant value of 6 Hz. The data matrix in both heteronuclear experiments was extended to 2048 × 1024 points using forward linear prediction (57).
MALDI-TOF MS of intact LPS from HH103 rkpA and rkpM and their isolated lipid A fractions
MALDI-TOF mass spectra of intact LPS from HH103 rkpA were recorded in linear mode and negative ion polarity on an Applied Biosystems MDS SCIEX 4800 MALDI TOF/TOF™ analyzer, equipped with delayed extraction technology. Ions formed by a pulsed UV laser (Nd:YAG laser, λ = 355 nm) were accelerated by 24 kV. The MS structural investigation of HH103 rkpM LPS was performed in reflectron mode and negative ion polarity on an ABSCIEX TOF/TOFTM 5800 Applied Biosystems mass spectrometer equipped with an Nd:YAG laser (λ = 349 nm), with a 3-ns pulse width and a repetition rate of up to 1000 Hz. In both cases, LPS MALDI preparations were executed as previously reported (50, 58, 59). Reflectron MALDI-TOF MS spectra of lipid A isolated from both HH103 mutant strains analyzed in this work were recorded on the above-described AB SCIEX TOF/TOFTM 5800 Applied Biosystems mass spectrometer. Lipid A fractions were dissolved in CHCl3/CH3OH (50:50, v/v). The matrix solution was 2,4,6-trihydroxyacetophenone in CH3OH/0.1% TFA/CH3CN (7:2:1, v/v) at a concentration of 75 mg ml−1 (60). 0.5 μl of the sample and 0.5 μl of the matrix solution were deposited on a stainless steel plate and left to dry at room temperature.
Molecular and microbiological techniques
Bacterial strains and plasmids used in this work are listed in Table S6. S. fredii strains were grown at 28 °C on complete TY medium (61), liquid YMB (62), or liquid MM (63). YMA medium corresponds to YMB solidified with agar (20 g/liter). When necessary, the media were supplemented with antibiotics (concentrations in µg ml−1): rifampicin (Rif), 25; tetracycline (Tc), 2–4 (10 for Escherichia coli); gentamycin (Gm), 5 (10 for E. coli); spectinomycin (Spc), 50 or 100.
Assays to determine bacterial tolerance to SDS were carried out in MM containing 0.35 or 0.52 mm SDS. A 50 mm SDS stock solution in water was sterilized by filtration (0.2-μm pore size) and added to sterile MM to reach final concentrations of 0.35 or 0.52 mm SDS. Aliquots (50 μl) of 10-fold dilutions of early-stationary bacterial cultures (OD600 of 1.0) were used to inoculate tubes containing 5 ml of MM supplemented with the detergent. Bacterial growth was spectrophotometrically monitored at 1, 2, and 5 days after inoculation. Similarly, assays to determine salt tolerance were carried out in MM supplemented with 100 or 150 mm NaCl.
PCR amplifications were carried out as previously described (64). Primers for PCR amplifications are listed in Table S4.
Nodulation tests
Plant tests were carried out on Glycyrrhiza uralensis (L), Macroptilium atropurpureum (Moc. & Sessé ex DC) Urb., and Cajanus cajan (L.) Millsp. Inoculated G. uralensis nodules were used as a source of bacteroids for the visualization of LPS electrophoretic profiles. M. atropurpureum and C. cajan plants were used for standard nodulation assays in which parameters of symbiotic significance were determined. Nodule structures by optic and electronic microscopy of nodules induced by the HH103 rkpM mutant were also investigated.
M. atropurpureum and G. uralensis seeds were surface sterilized by sequential exposure to sulfuric acid for 20 min and sodium hypochlorite 12% (w/s) for 5 min, followed by five washing steps with sterile distilled water. C. cajan seeds were sterilized by sequential exposure to 96% (v/v) ethanol for 30 s and sodium hypochlorite 12% (w/s) for 5 min.
Two germinated seeds of G. uralensis or C. cajan were transferred to individual Leonard jars containing 800 ml of sterilized vermiculite supplemented with Fåhraeus nutrient solution (64). Three M. atropurpureum germinated seeds were placed in 250-ml Leonard jars. Each seedling was inoculated with 1 ml of a particular bacterial culture grown in YMB (OD600 of 0.4–0.5). Inoculated plants were grown for seven (M. atropurpureum) or eight (C. cajan) weeks with a 16-h photoperiod at 25 °C in the light and 18 °C in the dark. Nitrogenase activity of M. atropurpureum nodules was detected by acetylene reduction assays as described previously (65). Briefly, M. atropurpureum roots were cut off and placed in a 10-ml sealed test tube or a 250-ml sealed flask, respectively. The reaction was started with an air gas phase containing 10% acetylene. After incubating for 1 h, ethylene was measured with a gas chromatograph (5890A Hewlett Packard) using a Porapack Q column and nitrogen carrier gas at 80 °C. Plant tops were dried at 80 °C for 48 h and weighed.
Bacteria were isolated from surface-sterilized nodules as previously reported (38). Bacteria isolated (70 colonies) from 7 nodules induced for each strain on M. atropurpureum or C. cajan roots showed the expected antibiotic resistance markers: RifR for HH103, RifR GmR for HH103 rkpM and rkpA mutants, and RifR GmR TcR for HH103 rkpM C.
Microscopy of nodules
Optical microscopy studies were carried out as previously described (66). Briefly, M. atropurpureum plants were inoculated and grown for seven weeks in a plant growth chamber. Small fragments of M. atropurpureum nodules then were immediately fixed in 4% (v/v) glutaraldehyde in 0.1 m cacodylate buffer, pH 7.2, for 2 h at 4 °C after collection. Samples were washed several times in 0.1 m cacodylate buffer, pH 7.2, dehydrated in acetone at progressively higher concentrations, and embedded in Spurr resine (low-viscosity embedding kit, by Dr. Spurr). The semithin sections (1 μm) were obtained on a Leica EM UC7 ultramicrotome, stained with Toluidine blue, and viewed in an Olympus BX61 light microscope.
Extraction of bacteroid cells from nodules
Bacteroids from G. uralensis nodules were extracted as previously described (67). Nodules were surface sterilized for 1 min in 100% bleach and then washed three times in water. Nodules were crushed in MMS [40 mm 3-(4-morpholino)-propane sulfonic acid, 20 mm KOH, 2 mm MgSO4, 0.3 M sucrose, pH 7.0] buffer, and the liquid containing bacteroids was removed with a pipette. Large particles were removed by passing through a 40-μm cell strainer cap (BD Falcon, BD Biosciences, San Jose, CA, USA). Samples were centrifuged once at 70 × g for 5 min and posteriorly twice at 2200 × g for 5 min to resuspend the resulting pellet fraction in MMS medium. LPS extraction from isolated bacteroids was carried out by following the same protocol as that used for the extraction of LPS from free-living cultures (38).
Data availability
All data described are contained within the manuscript.
Supplementary Material
Acknowledgments
We thank the Centro de Investigación, Tecnología e Innovación (CITIUS), of the University of Seville for light microscopy facilities.
This article contains supporting information.
Author contributions—F. D. L., I. S., A. S., S. A.-J., M.-R.-C., M. S. D., A. P., D. G., J.-E. R.-S., A. M., and J.-M. V. conceptualization; F. D. L., I. S., A. S., S. A.-J., M.-R.-C., M. S. D., A. P., D. G., J.-E. R.-S., A. M., and J.-M. V. formal analysis; F. D. L., I. S., A. S., S. A.-J., M.-R.-C., M. S. D., A. P., D. G., J.-E. R.-S., A. M., and J.-M. V. funding acquisition; F. D. L., I. S., A. S., C. A.-V., S. A.-J., M.-R.-C., M. S. D., A. P., D. G., J.-E. R.-S., and J.-M. V. investigation; F. D. L., I. S., A. S., S. A.-J., M.-R.-C., M. S. D., A. P., D. G., J.-E. R.-S., A. M., and J.-M. V. writing-original draft; F. D. L., I. S., A. S., S. A.-J., M.-R.-C., M. S. D., A. P., D. G., J.-E. R.-S., A. M., and J.-M. V. writing-review and editing.
Funding and additional information—This work was funded by grants P11-CVI-7500 from the Andalusian Government, and BIO2016-78409-R from the Ministry of Economy and Competitiveness (Spanish Government). M. S. D. is a Career Member of CONICET and worked thanks to grant CONICET PIP 112-201501-00232.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- KPS
- K-antigen polysaccharide
- RSP
- rhizobial surface polysaccharides
- EPS
- exopolysaccharides
- LPS
- lipopolysaccharides
- CG
- cyclic glucans
- OS
- oligosaccharide
- Pse5NAc7(3OHBu)
- 5-acetamido-3,5,7,9-tetradeoxy-7-(3-hydroxybutyramido)-l-glycero-l-manno-nonulosonic acid
- DQF-COSY
- double-quantum filtered phase-sensitive correlation spectroscopy
- TOCSY
- total correlation spectroscopy
- ROESY
- rotating frame Overhauser enhancement spectroscopy
- 1H-13C HSQC
- heteronuclear single quantum coherence
- 1H-13C HMBC
- heteronuclear multiple bound correlation
- NAc
- N-acetyl groups
- VLCFA
- very-long-chain fatty acids
- OD600
- optical density at 600 nm
- GLC-MS
- gas LC-MS.
References
- 1. López-Baena F. J., Ruiz-Sainz J. E., Rodríguez-Carvajal M. A., and Vinardell J. M. (2016) Bacterial molecular signals in the Sinorhizobium fredii-soybean symbiosis. Int. J. Mol. Sci. 17, 755 10.3390/ijms17050755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gage D. J. (2004) Infection and invasion of roots by symbiotic, nitrogen fixing rhizobia during nodulation of temperate legumes. Mol. Biol. Rev. 68, 280–300 10.1128/MMBR.68.2.280-300.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jones K. M., Kobayashi H., Davies B. W., Taga M. E., and Walker G. C. (2007) How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat. Rev. Microbiol. 5, 619–633 10.1038/nrmicro1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Krol E., and Becker A. (2009) Surface polysaccharides as fitness factors of rhizospheric nitrogen-fixing bacteria. in Bacterial Polysaccharides: Current Innovations and Future Trends, (Ullrich M., ed), pp. 187–211, Caister Academic Press, London [Google Scholar]
- 5. Janczarek M., Rachwał K., Marzec A., Grzadziel J., and Palusińska- Szysz M. (2015) Signal molecules and cell-surface components involved in early stages of the legume–rhizobium interactions. Appl. Soil Ecol. 85, 94–113 10.1016/j.apsoil.2014.08.010 [DOI] [Google Scholar]
- 6. Kawaharada Y., Kelly S., Nielsen M. W., Hjuler C. T., Gysel K., Muszyński A., Carlson R. W., Thygesen M. B., Sandal N., Asmussen M. H., Vinther M., Andersen S. U., Krusell L., Thirup S., Jensen K. J., et al. (2015) Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 523, 308–312 10.1038/nature14611 [DOI] [PubMed] [Google Scholar]
- 7. Kawaharada Y., Nielsen M. W., Kelly S., James E. K., Andersen K. R., Rasmussen S. R., Füchtbauer W., Madsen L. H., Heckmann A. B., Radutoiu S., and Stougaard J. (2017) Differential regulation of the Epr3 receptor coordinates membrane-restricted rhizobial colonization of root nodule primordia. Nat. Commun. 8, 14534 10.1038/ncomms14534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vinardell J.-M., Acosta-Jurado S., Zehner S., Göttfert M., Becker A., Baena I., Blom J., Crespo-Rivas J. C., Goesmann A., Jaenicke S., Krol E., McIntosh M., Margaret I., Pérez-Montaño F., Schneiker-Bekel S., et al. (2015) The Sinorhizobium fredii HH103 genome: a comparative analysis with S. fredii strains differing in their symbiotic behaviour with soybean. Mol. Plant Microbe Interact. 28, 811–824 10.1094/MPMI-12-14-0397-FI [DOI] [PubMed] [Google Scholar]
- 9. Margaret I., Becker A., Blom J., Bonilla I., Goesmann A., Göttfert M., Lloret J., Mittard-Runte V., Rückert C., Ruiz-Sainz J. E., Vinardell J. M., and Weidner S. (2011) Symbiotic properties and first analyses of the genomic sequence of the fast growing model strain Sinorhizobium fredii HH103 nodulating soybean. J. Biotechnol. 155, 11–19 10.1016/j.jbiotec.2011.03.016 [DOI] [PubMed] [Google Scholar]
- 10. Rodríguez-Navarro D. N., Rodríguez-Carvajal M. A., Acosta-Jurado S., Soto M. J., Margaret I., Crespo-Rivas J. C., Sanjuan J., Temprano F., Gil-Serrano A., Ruiz-Sainz J. E., and Vinardell J. M. (2014) Structure and biological roles of Sinorhizobium fredii HH103 exopolysaccharide. PLoS ONE 9, e115391 10.1371/journal.pone.0115391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Crespo-Rivas J. C., Margaret I., Hidalgo A., Buendía-Clavería A. M., Ollero F. J., López-Baena J., Murdoch P., Rodríguez-Carvajal M. A., Soria-Díaz M. E., Reguera M., Lloret J., Sumpton D. P., Mosely J. A., Thomas-Oates J. E., van Brussel A. A. N., et al. (2009) Sinorhizobium fredii HH103 cgs mutants are unable to nodulate determinate- and indeterminate-nodule forming legumes and overproduce an altered EPS. Mol. Plant Microbe Interact. 22, 575–588 10.1094/MPMI-22-5-0575 [DOI] [PubMed] [Google Scholar]
- 12. Gil-Serrano A. M., Rodríguez-Carvajal M. A., Tejero-Mateo P., Espartero J. L., Menéndez M., Corzo J., Ruiz-Sainz J. E., and Buendía-Clavería A. M. (1999) Structural determination of a 5-acetamido-3,5,7,9-tetradeoxy-7-(3-hydroxybutyramido)-l-glycero-l-manno-nonusolonic acid-containing homopolysaccharide isolated from Sinorhizobium fredii HH103. Biochem. J. 342, 527–535 10.1042/0264-6021:3420527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chataigné G., Couderc F., and Poinsot V. (2008) Polysaccharides analysis of sinorhizobial capside by on-line anion exchange chromatography with pulse amperiometric detection and mass spectrometry coupling. J. Chromatogr. A 1185, 241–250 10.1016/j.chroma.2008.01.065 [DOI] [PubMed] [Google Scholar]
- 14. Parada M., Vinardell J. M., Ollero F. J., Hidalgo A., Gutiérrez R., Buendía Clavería A., Lei W., Margaret I., López-Baena F. J., Gil-Serrano A., Rodríguez Carvajal M. A., Moreno J., and Ruiz-Sainz J. E. (2006) Sinorhizobium fredii HH103 mutants affected in capsular polysaccharide (KPS) are impaired for nodulation with soybean and Cajanus cajans. Mol. Plant Microbe Interact. 19, 43–52 10.1094/MPMI-19-0043 [DOI] [PubMed] [Google Scholar]
- 15. Hidalgo A., Margaret I., Crespo-Rivas J. C., Parada M., Murdoch P. S., López A., Buendía-Clavería A. M., Moreno J., Albareda M., Gil-Serrano A. M., Rodríguez-Carvajal M. A., Palacios J. M., Ruiz-Sainz J. E., and Vinardell J. M. (2010) The rkpU gene of Sinorhizobium fredii HH103 is required for bacterial K-antigen polysaccharide production and for efficient nodulation with soybean but not with cowpea. Microbiology 156, 3398–3411 10.1099/mic.0.042499-0 [DOI] [PubMed] [Google Scholar]
- 16. Margaret-Oliver I., Lei W., Parada M., Rodríguez-Carvajal M. A., Crespo-Rivas J. C., Hidalgo Á., Gil-Serrano A., Moreno J., Rodríguez-Navarro D. N., Buendía-Clavería A., Ollero J., Ruiz-Sainz J. E., and Vinardell J. M. (2012) Sinorhizobium fredii HH103 does not strictly require KPS and/or EPS to nodulate Glycyrrhiza uralensis, an indeterminate nodule-forming legume. Arch. Microbiol. 194, 87–102 10.1007/s00203-011-0729-2 [DOI] [PubMed] [Google Scholar]
- 17. Margaret I., Crespo-Rivas J. C., Acosta-Jurado S., Buendía-Clavería A. M., Cubo M. T., Gil-Serrano A., Moreno J., Murdoch P. S., Rodríguez-Carvajal M. A., Rodríguez-Navarro D. N., Ruiz-Sainz J. E., Sanjuan J., Soto M. J., and Vinardell J. M. (2012) Sinorhizobium fredii HH103 rkp-3 genes are required for KPS biosynthesis, affect LPS structure and are essential for infection of legumes forming determinate nodules. Mol. Plant Microbe Interact. 25, 825–838 10.1094/MPMI-10-11-0262 [DOI] [PubMed] [Google Scholar]
- 18. Di Lorenzo F., De Castro C., Lanzetta R., Parrilli M., Silipo A., and Molinaro A. (2015) Lipopolysaccharides as microbe-associated molecular patterns: a structural perspective. in Carbohydrates in Drug Design and Discovery (Jiménez-Barbero J., Javier Canada F., and Martín-Santamaría S., eds.), pp. 38–63, Royal Society of Chemistry (RSC), London [Google Scholar]
- 19. Carlson R. W., Forsberg L. S., and Kannenberg E. L. (2010) Lipopolysaccharides in Rhizobium-legume symbioses. in Endotoxins: Structure, Function and Recognition, (Wang X. and Quinn P. J., eds), pp. 339–386, Springer, New York: [DOI] [PubMed] [Google Scholar]
- 20. Silipo A., Leone M. R., Erbs G., Lanzetta R., Parrilli M., Chang W. S., Newman M. A., and Molinaro A. (2011) A unique bicyclic monosaccharide from the Bradyrhizobium lipopolysaccharide and its role in the molecular interaction with plants. Angew. Chem. Int. Ed. Engl. 50, 12610–12612 10.1002/anie.201106548 [DOI] [PubMed] [Google Scholar]
- 21. Silipo A., Vitiello G., Gully D., Sturiale L., Chaintreuil C., Fardoux J., Gargani D., Lee H. I., Kulkarni G., Busset N., Marchetti R., Palmigiano A., Moll H., Engel R., Lanzetta R., et al. (2014) Covalently linked hopanoid-lipid A improves outer-membrane resistance of a Bradyrhizobium symbiont of legumes. Nat. Commun. 5, 5106 10.1038/ncomms6106 [DOI] [PubMed] [Google Scholar]
- 22. Di Lorenzo F., Palmigiano A., Duda K. A., Pallach M., Busset N., Sturiale L., Giraud E., Garozzo D., Molinaro A., and Silipo A. (2017) Structure of the lipopolysaccharide from the Bradyrhizobium sp. ORS285 rfaL mutant strain. ChemistryOpen 6, 541–553 10.1002/open.201700074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gudlavalleti S. K., and Forsberg L. S. (2003) Structural characterization of the lipid A component of Sinorhizobium sp. NGR234 rough and smooth form lipopolysaccharide. Demonstration that the distal amide-linked acyloxyacyl residue containing the long chain fatty acid is conserved in rhizobium and Sinorhizobium sp. J. Biol. Chem. 278, 3957–3968 10.1074/jbc.M210491200 [DOI] [PubMed] [Google Scholar]
- 24. Brewin N. J., Perotto S., Kannenberg E. L., Rae A. L., Rathburn E. A., Lucas M. M., Kardailsky I., Gunder A., Bolaños L., Donovan N., and Drøbak B. K. (1993) Mechanisms of cell and tissue invasion by Rhizobium leguminosarum: the role of cell surface interactions. in Advances in Molecular Genetics of Plant-Microbe Interactions (Nester E. W. and Verma D. P. S., eds.), pp. 369–380, Kluwer Academic Publishers, New York [Google Scholar]
- 25. Noel K. D., van den Bosch K. A., and Kulpaca B. (1986) Mutation in Rhizobium phaseoli that lead to arrested development of infection threads. J. Bacteriol. 168, 1392–1401 10.1128/jb.168.3.1392-1401.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stacey G., So J. S., Roth L. E., Lakshmi B., and Carlson R. W. (1991) A lipopolysaccharide mutant from Bradyrhizobium japonicum that uncouples plant from bacterial differentiation. Mol. Plant Microbe Interact. 4, 332–340 10.1094/mpmi-4-332 [DOI] [PubMed] [Google Scholar]
- 27. Lagares A., Caetano-Anolles G., Niehaus K., Lorenzen J., Ljunggren H. D., Pühler A., and Favelukes G. (1992) A Rhizobium meliloti lipopolysaccharide mutant altered in competitiveness for nodulation of alfalfa. J. Bacteriol. 174, 5941–5952 10.1128/jb.174.18.5941-5952.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lagares A., Hozbor D. F., Niehaus K., Otero A. J., Lorenzen J., Arnold W., and Pühler A. (2001) Genetic characterization of a Sinorhizobium meliloti chromosomal region in lipopolysaccharide biosynthesis. J. Bacteriol. 183, 1248–1258 10.1128/JB.183.4.1248-1258.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Campbell G. R., Reuhs B. L., and Walker G. C. (2002) Chronic intracellular infection of alfalfa nodules by Sinorhizobium meliloti requires correct lipopolysaccharide core. Proc. Natl. Acad. Sci. U S A 99, 3938–3943 10.1073/pnas.062425699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Campbell G. R., Sharypova L. A., Scheidle H., Jones K. M., Niehaus K., Becker A., and Walker G. C. (2003) Striking complexity of lipopolysaccharide defects in a collection of Sinorhizobium meliloti mutants. J. Bacteriol. 185, 3853–3862 10.1128/jb.185.13.3853-3862.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Margaret I., Lucas M., Acosta-Jurado S., Buendía-Clavería A. M., Fedorova E., Hidalgo A., Rodríguez-Carvajal M. A., Rodríguez-Navarro D. N., Ruiz-Sainz J. E., and Vinardell J. M. (2013) The Sinorhizobium fredii HH103 lipopolysaccharide is not only relevant at early soybean nodulation stages but also for symbiosome stability in mature nodules. PLoS ONE 8, e74717 10.1371/journal.pone.0074717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carlson R. W., Kalembasa S., Turowski D., Pachori P., and Noel K. D. (1987) Characterization of the lipopolysaccharide from a Rhizobium phaseoli mutant that is defective in infection thread development. J. Bacteriol. 169, 4923–4928 10.1128/jb.169.11.4923-4928.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cava J. R., Elias P. M., Turowski D. A., and Noel K. D. (1989) Rhizobium leguminosarum CFN42 genetic regions encoding lipopolysaccharide structures essential for complete nodule development on bean plants. J. Bacteriol. 171, 8–15 10.1128/jb.171.1.8-15.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Noel K. D., Forsberg L. S., and Carlson R. W. (2000) Varying the abundance of O-antigen in Rhizobium etli and its effect on symbiosis with Phaseolus vulgaris. J. Bacteriol. 182, 5317–5324 10.1128/jb.182.19.5317-5324.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gao M., D'Haeze W., De Rycke R., Wolucka B., and Holsters M. (2001) Knockout of an azorhizobial dTDP-L-rhamnose synthase affects lipopolysaccharide and extracellular polysaccharide production and disables symbiosis with Sesbania rostrata. Mol. Plant Microbe Interact. 14, 857–866 10.1094/MPMI.2001.14.7.857 [DOI] [PubMed] [Google Scholar]
- 36. Noh J. G., Jeon H. E., So J. S., and Chang W. S. (2015) Effects of the Bradyrhizobium japonicum waaL (rfaL) gene on hydrophobicity, motility, stress tolerance, and symbiotic relationship with soybeans. Int. J. Mol. Sci. 16, 16778–16791 10.3390/ijms160816778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Acosta-Jurado S., Navarro-Gómez P., Crespo-Rivas J.-C., Medina C., Murdoch P. D. S., Cuesta-Berrio L., Rodríguez-Carvajal M.-Á., Ruiz-Sainz J.-E., and Vinardell J.-M. (2017) The Sinorhizobium fredii HH103 rkp-2 region is involved in the biosynthesis of lipopolysaccharide and exopolysaccharide but not in K-antigen polysaccharide production. Plant Soil 417, 415–431 10.1007/s11104-017-3268-z [DOI] [Google Scholar]
- 38. Buendía-Clavería A. M., Moussaid A., Ollero F. J., Vinardell J. M., Torres A., Moreno J., Gil-Serrano A. M., Rodríguez-Carvajal M. A., Tejero-Mateo P., Peart J. L., Brewin N. J., and Ruiz Sainz J. E. (2003) A purL mutant of Sinorhizobium fredii HH103 is symbiotically defective and altered in its lipopolysaccharide. Microbiology 149, 1807–1818 10.1099/mic.0.26099-0 [DOI] [PubMed] [Google Scholar]
- 39. Crespo-Rivas J. C., Guefrachi I., Mok K. C., Villaécija-Aguilar J. A., Acosta-Jurado S., Pierre O., Ruiz-Sainz J. E., Taga M. E., Mergaert P., and Vinardell J. M. (2016) Sinorhizobium fredii HH103 bacteroids are not terminally differentiated and show altered O-antigen in nodules of the inverted repeat-lacking clade legume Glycyrrhiza uralensis. Environ. Microbiol. 18, 2392–2404 10.1111/1462-2920.13101 [DOI] [PubMed] [Google Scholar]
- 40. Westphal O., and Jann K. (1965) Bacterial lipopolysaccharides extraction with phenol-water and further applications of the procedure. Methods Carbohydr. Chem. 5, 83–91 [Google Scholar]
- 41. De Castro C., Parrilli M., Holst O., and Molinaro A. (2010) Microbe-associated molecular patterns in innate immunity: extraction and chemical analysis of gram-negative bacterial lipopolysaccharides. Methods Enzymol. 480, 89–115 10.1016/S0076-6879(10)80005-9 [DOI] [PubMed] [Google Scholar]
- 42. Knirel Y. A., Shashkov A. S., Tsvetkov Y. E., Jansson P. E., and Zãhringer U. (2003) Tetradeoxynon-2-ulosonic acids in bacterial glycopolymers: chemistry and biochemistry. Adv. Carbohydr. Chem. Biochem. 58, 371–417 10.1016/s0065-2318(03)58007-6 [DOI] [PubMed] [Google Scholar]
- 43. Dowdle S. F., and Bohlool B. B. (1985) Predominance of fast-growing Rhizobium japonicum in a soybean field in the People's Republic of China. Appl. Environ. Microbiol. 50, 1171–1176 10.1128/AEM.50.5.1171-1176.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thomas-Oates J., Bereszczak J., Edwards E., Gill A., Noreen S., Zhou J. C., Chen M. Z., Miao L. H., Xie F. L., Yang J. K., Zhou Q., Yang S. S., Li X. H., Wang L., Spaink H. P., et al. (2003) A catalogue of molecular, physiological and symbiotic properties of soybean-nodulating rhizobial strains from different soybean cropping areas of China. Syst. Appl. Microbiol. 26, 453–465 10.1078/072320203322497491 [DOI] [PubMed] [Google Scholar]
- 45. Reuhs B. L., Geller D. P., Kim J. S., Fox J. E., Kolli V. S. K., and Pueppke S. G. (1998) Sinorhizobium fredii and Sinorhizobium meliloti produce structurally conserved lipopolysaccharides and strain-specific K antigens. Appl. Environ. Microbiol. 64, 4930–4938 10.1128/AEM.64.12.4930-4938.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Le Quéré A. J. L., Deakin W. K., Schmeisser C., Carlson R. W., Streit W. R., Broughton W. J., and Forsberg L. S. (2006) Structural characterization of a K-antigen capsular polysaccharide essential for normal symbiotic infection in Rhizobium sp. NGR234. J. Biol. Chem. 281, 28981–28992 10.1074/jbc.M513639200 [DOI] [PubMed] [Google Scholar]
- 47. Kiss E., Kereszt A., Barta F., Stephens S., Reuhs B. L., Kondorosi A., and Putnoky P. (2001) The rkp-3 gene region of Sinorhizobium meliloti Rm41 contains strain-specific genes that determine K antigen structure. Mol. Plant Microbe Interact. 14, 1395–1403 10.1094/MPMI.2001.14.12.1395 [DOI] [PubMed] [Google Scholar]
- 48. Acosta-Jurado S., Rodríguez-Navarro D. N., Kawaharada Y., Perea J. F., Gil-Serrano A., Jin H., An Q., Rodríguez-Carvajal M. A., Andersen S. U., Sandal N., Stougaard J., Vinardell J. M., and Ruiz-Sainz J. E. (2016) Sinorhizobium fredii HH103 invades Lotus burttii by crack entry in a Nod factor and surface polysaccharide-dependent manner. Mol. Plant Microbe Interact. 29, 925–937 10.1094/MPMI-09-16-0195-R [DOI] [PubMed] [Google Scholar]
- 49. Bourassa D. V., Kannenberg E. L., Sherrier D. J., Buhr R. J., and Carlson R. W. (2017) The lipopolysaccharide lipid A long-chain fatty acid is important for the Rhizobium leguminosarum growth and stress adaptation in free-living and nodule environments. Mol. Plant-Microbe Interact. 30, 161–175 10.1094/MPMI-11-16-0230-R [DOI] [PubMed] [Google Scholar]
- 50. Di Lorenzo F., Sturiale L., Palmigiano A., Lembo-Fazio L., Paciello I., Coutinho C. P., Sá-Correia I., Bernardini M., Lanzetta R., Garozzo D., Silipo A., and Molinaro A. (2013) Chemistry and biology of the potent endotoxin from a Burkholderia dolosa clinical isolate from a cystic fibrosis patient. Chembiochem 14, 1105–1115 10.1002/cbic.201300062 [DOI] [PubMed] [Google Scholar]
- 51. Leontein K., Lindberg B., and Lōnngren J. (1978) Assignment of absolute configuration of sugars by glc of their acetylated glycosides formed from chiral alcohols. Methods Carbohydr. Chem. 62, 359–362 10.1016/S0008-6215(00)80882-4 [DOI] [Google Scholar]
- 52. Holst O. (2000) Deacylation of lipopolysaccharides and isolation of oligosaccharide phosphates. Methods Mol. Biol. 145, 345–353 10.1385/1-59259-052-7:345 [DOI] [PubMed] [Google Scholar]
- 53. Kasimova A. A., Shneider M. M., Arbatsky N. P., Popova A. V., Shashkov A. S., Miroshnikov K. A., Balaji V., Biswas I., and Knirel Y. A. (2017) Structure and gene cluster of the K93 capsular polysaccharide of Acinetobacter baumannii B11911 containing 5-N-acetyl-7-N-[(R)-3-hydroxybutanoyl]pseudaminic acid. Biochemistry 82, 483–489 10.1134/S0006297917040101 [DOI] [PubMed] [Google Scholar]
- 54. Piantini U., Sorensen O. W., and Ernst R. R. (1982) Multiple quantum filters for elucidating NMR coupling networks. J. Am. Chem. Soc. 104, 6800–6801 10.1021/ja00388a062 [DOI] [Google Scholar]
- 55. Rance M., Sørensen O. W., Bodenhausen G., Wagner G., Ernst R. R., and Wüthrich K. (2012) Improved spectral resolution in COSY (1)H NMR spectra of proteins via double quantum filtering. Biophys. Res. Commun. 425, 527–533 10.1016/j.bbrc.2012.08.019 [DOI] [PubMed] [Google Scholar]
- 56. States D. J., Haberkorn R. A., and Ruben D. J. (1982) A two-dimensional nuclear Overhauser experiment with pure absorption phase in four quadrants. J. Magnetic Res. 48, 286–292 10.1016/0022-2364(82)90279-7 [DOI] [Google Scholar]
- 57. Stern A. S., Li K. B., and Hoch J. C. (2002) Modern spectrum analysis in multidimensional NMR spectroscopy: comparison of linear-prediction extrapolation and maximum-entropy reconstruction. J. Am. Chem. Soc. 124, 1982–1993 10.1021/ja011669o [DOI] [PubMed] [Google Scholar]
- 58. Sturiale L., Palmigiano A., Silipo A., Knirel Y. A., Anisimov A. P., Lanzetta R., Parrilli M., Molinaro A., and Garozzo D. (2011) Reflectron MALDI TOF and MALDI TOF/TOF mass spectrometry reveal novel structural details of native lipooligosaccharides. J. Mass Spectrom. 46, 1135–1142 10.1002/jms.2000 [DOI] [PubMed] [Google Scholar]
- 59. Sturiale L., Garozzo D., Silipo A., Lanzetta R., Parrilli M., and Molinaro A. (2005) New conditions for matrix-assisted laser desorption/ionization mass spectrometry of native bacterial R-type lipopolysaccharides. Rapid Commun. Mass Spectrom. 19, 1829–1834 10.1002/rcm.1994 [DOI] [PubMed] [Google Scholar]
- 60. Di Lorenzo F., Palmigiano A., Al Bitar-Nehme S., Sturiale L., Duda K. A., Gully D., Lanzetta R., Giraud E., Garozzo D., Bernardini M. L., Molinaro A., and Silipo A. (2017) The lipid A from Rhodopseudomonas palustris strain BisA53 LPS possesses a unique structure and low immunostimulant properties. Chemistry 23, 3637–3647 10.1002/chem.201604379 [DOI] [PubMed] [Google Scholar]
- 61. Beringer J. E. (1974) R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84, 188–198 10.1099/00221287-84-1-188 [DOI] [PubMed] [Google Scholar]
- 62. Vincent J. M. (ed) (1970) The modified Fåhraeus slide technique. in Appendix III, A Manual for the Practical Study of Root Nodule Bacteria, pp 144–145, Blackwell Scientific, New York [Google Scholar]
- 63. Robertsen B. K., Aman P., Darvill A. G., McNeil M., and Albersheim P. (1981) The structure of acidic extracellular polysaccharides secreted by Rhizobium leguminosarum and Rhizobium trifolii. Plant Physiol. 67, 389–400 10.1104/pp.67.3.389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vinardell J. M., Ollero F. J., Hidalgo A., López-Baena F. J., Medina C., Ivanov-Vangelov K., Parada M., Madinabeitia N., Espuny M. R., Bellogín R. A., Camacho M., Rodríguez-Navarro D. N., Soria-Díaz M. E., Gil-Serrano A. M., and Ruiz-Sainz J. E. (2004) NolR regulates diverse symbiotic signals of Sinorhizobium fredii HH103. Mol. Plant Microbe Interact. 17, 676–685 10.1094/MPMI.2004.17.6.676 [DOI] [PubMed] [Google Scholar]
- 65. Acosta-Jurado S., Alias-Villegas C., Navarro-Gómez P., Zehner S., Murdoch P. D., Rodríguez-Carvajal M. A., Soto M. J., Ollero F. J., Ruiz-Sainz J. E., Göttfert M., and Vinardell J. M. (2016) The Sinorhizobium fredii HH103 MucR1 global regulator is connected with the nod regulon and is required for efficient symbiosis with Lotus burttii and Glycine max cv. Williams. Mol. Plant Microbe Interact. 29, 700–712 10.1094/MPMI-06-16-0116-R [DOI] [PubMed] [Google Scholar]
- 66. Acosta-Jurado S., Alias-Villegas C., Navarro-Gómez P., Almozara A., Rodríguez-Carvajal M. A., Medina C., and Vinardell J. M. (2020) Sinorhizobium fredii HH103 syrM inactivation affects the expression of a large number of genes, impairs nodulation with soybean and extends the host-range to Lotus japonicus. Environ. Microbiol. 22, 1104–1124 10.1111/1462-2920.14897 [DOI] [PubMed] [Google Scholar]
- 67. Finan T. M., McWhinnie E., Driscoll B., and Watson R. J. (1991) Complex symbiotic phenotypes result from gluconeogenic mutations in Rhizobium meliloti. Mol. Plant Microbe Interact. 4, 386–389 10.1094/MPMI-4-386 [DOI] [Google Scholar]
- 68. Keyser H. H., Hu T. S., Bohlool B. B., and Weber D. F. (1982) Fast growing rhizobia isolated from root nodules of soybean. Science 215, 1631–1632 10.1126/science.215.4540.1631 [DOI] [PubMed] [Google Scholar]
- 69. Lamrabet Y., Bellogín R. A., Cubo T., Espuny M. R., Gil A., Krishnan H. B., Megias M., Ollero F. J., Pueppke S., Ruiz-Sainz J. E., Spaink H. P., Tejero-Mateo P., Thomas-Oates J., and Vinardell J. M. (1999) Mutation in GDP-fucose synthesis genes of Sinorhizobium fredii alters Nod factors and significantly decreases competitiveness to nodulate soybeans. Mol. Plant Microbe Interact. 12, 207–217 10.1094/MPMI.1999.12.3.207 [DOI] [PubMed] [Google Scholar]
- 70. Madinabeitia N., Bellogín R. A., Buendía-Clavería A. M., Camacho M., Cubo T., Espuny M. R., Gil-Serrano A., de Lyra M. C. C. P., Moussaid A., Ollero F. J., Soria Díaz M. E., Vinardell J. M., Zeng J., and Ruiz-Sainz J. E. (2002) Sinorhizobium fredii HH103 has a truncated nolO gene due to a –1 frameshift mutation that is conserved among other geographically distant S. fredii strains. Mol. Plant Microbe Interact. 15, 150–159 10.1094/MPMI.2002.15.2.150 [DOI] [PubMed] [Google Scholar]
- 71. Trinick M. J. (1980) Relationships amongst the fast-growing Rhizobium of Lablab purpureus, Leucaena leucocephala, Mimosa sp., Acacia farnesiana, and Sesbania grandiflora and their affinities with other Rhizobium groups. J. Appl. Bacteriol. 49, 39–53 10.1111/j.1365-2672.1980.tb01042.x [DOI] [Google Scholar]
- 72. Casse F., Boucher C., Julliot J. S., Michel M., and Denarié J. (1979) Identification and characterization of large plasmids in Rhizobium meliloti using agarose-gel electrophoresis. J. Gen. Microbiol. 113, 229–242 10.1099/00221287-113-2-229 [DOI] [Google Scholar]
- 73. Kereszt A., Kiss E., Reuhs B. L., Carlson R. W., Kondorosi A., and Putnoky P. (1998) Novel rkp gene clusters of Sinorhizobium meliloti involved in capsular polysaccharide production and invasion of the symbiotic nodule: the rkpK gene codes a UDP-glucose dehydrogenase. J. Bacteriol. 180, 5426–5431 10.1128/JB.180.20.5426-5431.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Perotto S., Brewin N. J., and Kannenberg E. L. (1994) Cytological evidence for a host defense response that reduces cell and tissue invasion in pea nodules by lipopolysaccharide-defective mutants of Rhizobium leguminosarum strain 3841. Mol. Plant Microbe Interact. 7, 99–112 10.1094/MPMI-7-0099 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data described are contained within the manuscript.