Highlights
-
•
Differential distribution of ACE I/D polymorphism observed in Asian countries.
-
•
Allele D of ACE I/D polymorphism was associated with susceptibility to SARS-CoV-2 infection.
-
•
The minor allele of ACE I/D was linked with COVID-19 mortality in the Asian population.
Keywords: SARS-CoV-2, COVID-19, ACE, Asian population
Abstract
Background
Severe acute respiratory syndrome corona virus-2 (SARS-CoV-2) is believed to have emerged from Wuhan, China, and spreads over 215 countries worldwide. The spike protein of SARS-CoV-2 binds to angiotensin-converting enzyme-2 (ACE-2) receptors and enter the host cells. Several reports have been highlighted the importance of ACE-2 on the pathogenesis of COVID-19. In the present study, we hypothesize that a functional insertion/deletion polymorphism in the ACE gene could be associated with SARS-CoV-2 infection and mortality.
Materials and methods
PubMed and Google scholar search engines were used to obtained data on the prevalence of ACE I/D polymorphism in different countries of the Asia continent. Data on COVID-19 infection rate (per million), mortality/million, and percentage of recovery were acquired form worldometer website. The Spearman rank correlation test performed to investigate the correlation of allele ‘D’ with SARS-CoV-2 infection, mortality rate, and recovery percentage.
Results
Epidemiological investigation revealed a significant positive correlation of D allele of ACE polymorphism with SARS-CoV-2 infection (r = 0.502, p = 0.008, n = 26) and mortality rate (r = 0.620, p = 0.002, n = 22) in Asian population. However, no significant role of ACE I/D polymorphism was observed with recovery rate of patients from SARS-CoV-2 infection (r = −0.208, p = 0.352, n = 22).
Conclusions
Allele D of ACE insertion/deletion polymorphism is associated with the rate of infection and mortality in the Asian population.
1. Introduction
The coronavirus disease 2019 (COVID-19), is caused by severe acute respiratory syndrome corona virus-2 (SARS-CoV-2), has emerged from Wuhan, China, in December 2019 [1] and spread rapidly across worldwide. On 11th March 2020, the World Health Organization (WHO) declared COVID-19 as a pandemic (https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020). Till date (27th July 2020), 17.17 million of subjects were infected with SARS-CoV-2 in 215 countries, and about 0.66 million of death has been reported throughout the globe (https://www.worldometers.info/coronavirus/#countries). The recovery rate of SARS-CoV-2 infected subjects remained at 62.23% (https://www.worldometers.info/coronavirus/#countries).
The spike protein of SARS-CoV-2 efficiently binds to the angiotensin-converting enzyme 2 (ACE2) receptors and facilitate the invading of the virus into the host cell [2]. ACE2 receptor is a type I transmembrane glycoprotein consisting of 805 amino acid length [3]. ACE2 receptors also mediate the cellular entry of SARS-CoV and NL63 [4]. SARS-CoV shares about 76% similarity in amino acid sequence with SARS-CoV-2 [5]. Various reports in the mouse model have deciphered importance of ACE2 in susceptibility and pathogenesis of SARS-CoV infections: the virus entry rate is enhanced with overexpression of ACE-2 receptors [6], antibodies to ACE-2 receptors significantly blocks entry of SARS-CoV [4] and ACE2 knockout mice had lower pulmonary lesion compared to the wildtype [7]. These observations collectively indicate an essential role ACE2 receptor in the pathogenesis of SARS-CoV-2.
Angiotensin-converting enzyme (ACE) share about 40% of sequence homology with ACE-2 [8], and several lines of evidence have demonstrated a counter-regulatory relationship between these two molecules [9]. The administration of ACE inhibitors significantly increased the expression levels of ACE-2 [10]. As levels of ACE and ACE2 are interwoven, and ACE2 plays a vital role in SARS-CoV-2 infection and pathogenesis, we hypothesized that differential levels of ACE could be associated with SARS-CoV-2 infections and mortality rate. ACE gene is located at the long arm (q23.3) of the 17th chromosome and consists of 26 exons and 25 introns [11]. A functional insertion-deletion (I/D) polymorphism of 286 bp has been reported in the intron 16 of the ACE gene [11]. The deletion (D) and insertion allele (I) are associated with an elevated and diminished ACE level and enzyme activity, respectively [12], [13]. Previous reports demonstrated the association of ACE D allele with the incidence of pneumonia in SARS patients [14] and the death of subjects with acute respiratory distress syndrome [15]. These observations tempted us to investigate the importance of ACE I/D polymorphism in susceptibility to SARS-CoV-2 infection and related mortality.
In the present epidemiological investigation, we tested the possible association between ACE I/D polymorphism with SARS-CoV-2 infection, mortality rate, and percentage of recovery in the Asian population.
2. Materials and methods
2.1. Data
For COVID-19 related information in the Asian continent, we explored the worldometer site (https://www.worldometers.info/coronavirus/) and various data such as country name, number cases per million, number of death per million, number of patients recovered from SARS-CoV-2 infections were obtained (assessed on 27th July 2020). The recovery rate in the percentage of SARS-CoV-2 patients was calculated by using the total number of infected cases and the number of patients who recovered after treatment. Total expenditure on health as percentage of gross domestic product (GDP) of various countries (www.who.int/countries/en/), doctors, and nurse density per 10,000 individuals (https://www.who.int/data/gho/data/indicators/indicator-details/GHO/medical-doctors-(per-10-000-population) were acquired from the World Health Organisation database.
2.2. Prevalence of ACE I/D polymorphism
Literature search database PubMed and google scholar were screened for the prevalence of ACE I/D polymorphism in different countries of Asia where SARS-CoV-2 infection has been reported. All publications were inspected, and authors' details, country name, ACE I/D genotypes number or frequency, allele number, or frequency were extracted. For countries with more than one report, genotype or allele data were pooled, and allele frequency was calculated.
2.3. Exclusion criteria
The genotypes distributions of ACE I/D polymorphism in all reports were screened for Hardy-Weinberg equilibrium (HWE), and the studies deviated from the HWE were excluded from the present investigation. As the health care facility depends upon the economic condition of the country, for mortality analysis, countries with expenditure on health sector lower than 3% of their GDP were not included.
2.4. Statistical analysis
The prevalence of ACE I/D genotypes and alleles frequency were calculated by manual counting. All statistical analysis were performed by Graphpad Prism 8.3.0. Correlation of D allele with SARS-CoV-2 infection rate, mortality rate, and recovery percentage was investigated by Spearman rank coefficient analysis, where a P-value < 0.05 was considered significant.
3. Results
A data search in worldometer website (on 27th July 2020) revealed a total of 38 Asian countries are affected with SARS-CoV-2 with a total of 4.12 million cases, about 94 thousand death, and about 72.63 percent of infected patients were recovered from this infection. Out of the 38 countries, where the presence of SARS-CoV-2 infection has been reported in the Asian population, data on the prevalence of ACE I/D genotypes or alleles were available for 28 countries namely Azerbaijan, Bahrain, China, India, Iran, Israel, Japan, Jordan, Kuwait, Kyrgyzstan, Lebanon, Malaysia, Mongolia, Nepal, Oman, Philippines, Saudi Arabia, Singapore, South Korea, Taiwan, Thailand, Turkey, United Arab Emirates, Uzbekistan, Bangladesh, Hong Kong, Pakistan, and Palestine. Distribution of ACE I/D genotypes in Jordan and Thailand population were not following HWE, thus excluded from the present study. Furthermore, ACE I/D polymorphism data of 27 reports from different countries (China: 06, India: 09, Japan: 02, Lebanon: 01, Malaysia: 02, Mongolia: 01, Pakistan: 03, South Korea: 01, Turkey: 01, and Uzbekistan: 01) were excluded from the present study, as the genotypes distributions did not obey HWE equilibrium. Data from 26 countries comprising of 148 publications were considered for the current investigation (Table 1 ). Majority of the reports on the frequency of ACE I/D genotype or allele were from the Chinese population (n = 51), and details of publications included in the present report are shown in the supplementary file. The frequency of allele D ranges from 32.08 to 74.27% (Table 1).
Table 1.
SARS-CoV-2 data and details information of earlier reports enrolled in the present study.
| Country Name | SARS-CoV-2 infected Cases per million of population | SARS-CoV-2 related Deaths per million of population | Recovery Rate (%) | Number of Paper considered for prevalence of genotype investigation* | Total healthy controls (n) | I/I genotype (n) | I/D genotype (n) | D/D genotype (n) | I allele (n) | D allele (n) | Frequency of allele D (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Azerbaijan | 2962 | 41 | 75.48 | 2 | 173 | 32 | 75 | 66 | 139 | 207 | 59.82 |
| Bahrain | 22,952 | 82 | 91.20 | 1 | 560 | 87 | 248 | 225 | 422 | 698 | 62.32 |
| Bangladesh | 1356 | 18 | 55.43 | 1 | 59 | 19 | 26 | 14 | 64 | 54 | 45.76 |
| China | 58 | 3 | 94.07 | 51 | 7980 | 3068 | 3691 | 1221 | 9827 | 6133 | 38.42 |
| Hong Kong | 351 | 2 | 56.75 | 1 | 326 | 43 | 133 | 150 | 219 | 433 | 66.41 |
| India | 1040 | 24 | 63.97 | 8 | 1621 | 558 | 796 | 267 | 1912 | 1330 | 41.02 |
| Iran | 3464 | 187 | 86.96 | 10 | 1655 | 350 | 815 | 490 | 1515 | 1795 | 54.22 |
| Israel | 6636 | 51 | 43.61 | 4 | 389 | 43 | 168 | 178 | 254 | 524 | 67.35 |
| Japan | 232 | 8 | 74.06 | 19 | 5399 | 2132 | 2496 | 771 | 6760 | 4038 | 37.39 |
| Kuwait | 14,919 | 101 | 85.26 | 2 | 283 | 34 | 118 | 131 | 186 | 380 | 67.13 |
| Kyrgyzstan | 5024 | 196 | 62.13 | 2 | 210 | 78 | 98 | 34 | 254 | 166 | 39.52 |
| Lebanon | 550 | 7 | 45.12 | 4 | 757 | 75 | 310 | 372 | 460 | 1054 | 69.61 |
| Malaysia | 275 | 4 | 96.66 | 1 | 72 | 18 | 35 | 19 | 71 | 73 | 50.69 |
| Mongolia | 88 | 0 | 75.69 | 1 | 100 | 42 | 50 | 8 | 134 | 66 | 33 |
| Nepal | 638 | 2 | 70.53 | 2 | 239 | 105 | 104 | 30 | 314 | 164 | 34.30 |
| Oman | 14,861 | 75 | 72.75 | 1 | 124 | 8 | 55 | 61 | 71 | 177 | 71.37 |
| Pakistan | 1235 | 26 | 86.93 | 4 | 448 | 130 | 225 | 93 | 485 | 411 | 45.87 |
| Palestine | 2049 | 15 | 35.83 | 3 | 243 | 16 | 93 | 134 | 125 | 361 | 74.27 |
| Philippines | 733 | 18 | 32.45 | 1 | 95 | 28 | 49 | 18 | 105 | 85 | 44.73 |
| Saudi Arabia | 7660 | 78 | 82.53 | 3 | 530 | 61 | 219 | 250 | 341 | 719 | 67.83 |
| Singapore | 8605 | 5 | 90.37 | 2 | 642 | 287 | 298 | 57 | 872 | 412 | 32.08 |
| South Korea | 276 | 6 | 91.04 | 8 | 2344 | 823 | 1129 | 392 | 2775 | 1913 | 40.80 |
| Taiwan | 19 | 0.3 | 96.06 | 3 | 713 | 300 | 309 | 104 | 909 | 517 | 36.25 |
| Turkey | 2679 | 67 | 92.65 | 11 | 1090 | 240 | 503 | 347 | 983 | 1197 | 54.90 |
| United Arab Emirates | 5952 | 35 | 88.57 | 2 | 275 | 39 | 127 | 109 | 205 | 345 | 62.72 |
| Uzbekistan | 613 | 3 | 54.08 | 1 | 60 | 12 | 32 | 16 | 56 | 64 | 53.33 |
Note: Data of SARS-CoV-2 infected cases, related death and recovery rate were obtained from https://www.worldometers.info/coronavirus/ assessed on 27.07.2020. Details of papers included in the study for analysis.
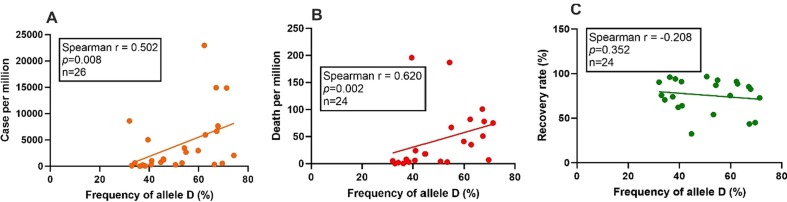
Spearman rank correlation analysis between SARS-CoV-2 infection rate per million of population and ACE I/D polymorphism revealed a positive correlation of D allele with the rate of infection (r = 0.502, p = 0.008, n = 26) (Fig. 1 A).
Fig. 1.
Correlation of ACE allele D with SARS-CoV-2 infection, death, and recovery rate in the Asian population. SARS-CoV-2 cases, mortality, and recovery from infection data were obtained from worldometer site (assessed on 27.07.2020). The prevalence of ACE ‘D’ allele in healthy subjects of Asian countries was obtained from earlier published literature. Spearman rank coefficient analysis was performed to investigate the correlation of allele ‘D’ with SARS-CoV-2 infection/million (A: r = 0.502, p = 0.008, n = 26), the mortality rate per million (B: r = 0.620, p = 0.002, n = 22) and patients recovery rate (C: r = −0.208, p = 0.352, n = 22). A p-value of less than 0.05 was taken as significant.
As the mortality from a disease is associated with the health facility of the country, in the present study countries those spends less than 3% of their GDP in the health sector, were excluded (Bangladesh, Pakistan, Hong Kong, and Palestine) and a total of 22 countries were included for mortality related analysis. No significant correlation was observed between SARS-CoV-2 related mortality rate with doctors (r = 0.070, p = 0.762), nursing staff (r = −0.089, p = 0.699) intensity per 10,000 population of the countries or the percentage of GDP spend in the health sector (r = 0.023, p = 0.917). Interestingly, the D allele of the ACE I/D polymorphism was positively correlated with the SARS-CoV-2 mortality rate per million (r = 0.620, p = 0.002, n = 22) (Fig. 1 B). However, no significant correlation was observed between allele D with the rate of recovery of SARS-CoV-2 patients in the Asian population (r = −0.208, p = 0.352, n = 22) (Fig. 1 C).
4. Discussion
ACE2 receptor plays a crucial role in cellular invading of SARS-CoV-2 as well as various pathophysiological conditions of the COVID-19 [16]. The spike protein of the SARS-CoV-2 binds to ACE2 receptors and facilitates the internalization of the virus [2]. The dominant allele ‘D’ of ACE I/D polymorphism has been associated with elevated levels of ACE [12], [13]. In the present study, we observed a significant positive correlation between the frequency of D allele and the number of SARS-CoV-2 infected cases/million in the Asian populations. The mechanism of how major allele (D) of ACE I/D polymorphism is associated with a higher infection rate of SARS-CoV-2 is not known. Since the levels of ACE and ACE2 are a counter complement to each other[17], lower levels of ACE2 could be expected in subject those harboring the major allele D of ACE I/D polymorphism. Furthermore, the soluble form of ACE2 protein which is believed to counteract SARS-CoV-2 [18] is scarcely present in the circulation than the membrane-bound ACE2 receptor [16]; thus the probability of interaction between SARS-CoV-2 and ACE2 receptor is higher, and that may facilitate the invasion of the host cell up to a greater extent.
We observed a significant positive correlation between allele D and mortality rate in the Asian populations indicating higher levels of ACE are deleterious in SARS-CoV-2 infected subjects. Mortality related to SARS-CoV-2 infections has been linked to various comorbid conditions such as hypertension, diabetes, hyperlipidemia, coronary artery disease, and renal disease [19]. High producer of ACE, the DD genotype has been associated with susceptibility to hypertension [20], type 2 diabetes [21], coronary artery disease [22] and hyperlipidemia [22] indicating a possible role of ACE I/D polymorphism with SARS-CoV-2 related mortality.
An earlier study in thirty-three countries, including 25 European, three north-African, and five from middle East continents, highlighted a negative correlation between deletion allele frequency of ACE I/D polymorphism with SARS-CoV-2 infection and mortality rate [23]. However, in the present report, we observed a positive correlation of allele D with infection an mortality rate of COVID-19. The possible reasons for the discrepancy would be the differential distributions of the allele among populations. Further, SARS-CoV-2 infection and mortality of subjects are dependent on a wide range of complex phenomena, and genetic mutation is one among them [24], [25].
Although the present analysis elegantly demonstrated the association of ACE I/D polymorphism with susceptibility to SARS-CoV-2 infection and related mortality, it has some limitations. Other genetic polymorphisms in the ACE gene were not considered in the present study. Furthermore, various essential factors were also not considered in the current investigation, such as the testing capacity of countries, inter-country movement of subjects, local health policy of the government, age group of the infected cases, population density, other comorbidities condition of the infected patients. In summary, we conclude ACE I/D polymorphism is associated with infection frequency and disease outcome of SARS-CoV-2 infection. However, hospital-based case controls studies in different populations are required to validate our findings.
CRediT authorship contribution statement
Abhijit Pati: Investigation, Formal analysis. Harishankar Mahto: Investigation, Formal analysis. Sunali Padhi: Writing - original draft. Aditya K Panda: Conceptualization, Supervision, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
Authors would like to thank all faculty members of the Department of Bioscience and Bioinformatics, Khallikote University, for encouragement and supports.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cca.2020.08.008.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Lu H., Stratton C.W., Tang Y.W. Outbreak of pneumonia of unknown etiology in Wuhan, China: The mystery and the miracle. J. Med. Virol. 2020;92(4):401–402. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tipnis S.R., Hooper N.M., Hyde R., Karran E., Christie G., Turner A.J. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol Chem. 2000;275(43):33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 4.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Science China. Life sciences. 2020;63(3):457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang X.H., Deng W., Tong Z., Liu Y.X., Zhang L.F., Zhu H., Gao H., Huang L., Liu Y.L., Ma C.M., Xu Y.F., Ding M.X., Deng H.K., Qin C. Mice transgenic for human angiotensin-converting enzyme 2 provide a model for SARS coronavirus infection. Comp. Med. 2007;57(5):450–459. [PubMed] [Google Scholar]
- 7.Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A.S., Liu D., Qin C., Jiang C., Penninger J.M. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11(8):875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lv Y., Li Y., Yi Y., Zhang L., Shi Q., Yang J. A Genomic Survey of Angiotensin-Converting Enzymes Provides Novel Insights into Their Molecular Evolution in Vertebrates. Molecules. 2018;23(11):2923. doi: 10.3390/molecules23112923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaddam R.R., Chambers S., Bhatia M. ACE and ACE2 in inflammation: a tale of two enzymes. Inflamm. Allergy Drug Targets. 2014;13(4):224–234. doi: 10.2174/1871528113666140713164506. [DOI] [PubMed] [Google Scholar]
- 10.Li X.C., Zhang J., Zhuo J.L. The vasoprotective axes of the renin-angiotensin system: Physiological relevance and therapeutic implications in cardiovascular, hypertensive and kidney diseases. Pharmacol. Res. 2017;125(Pt A):21–38. doi: 10.1016/j.phrs.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villard E., Soubrier F. Molecular biology and genetics of the angiotensin-I-converting enzyme: potential implications in cardiovascular diseases. Cardiovasc. Res. 1996;32(6):999–1007. doi: 10.1016/s0008-6363(96)00170-8. [DOI] [PubMed] [Google Scholar]
- 12.Kostadinova E.S., Miteva L.D., Stanilova S.A. ACE serum level and I/D gene polymorphism in children with obstructive uropathies and other congenital anomalies of the kidney and urinary tract. Nephrology (Carlton, Vic.) 2017;22:609–616. doi: 10.1111/nep.12824. [DOI] [PubMed] [Google Scholar]
- 13.Jeong K.H., Lee T.W., Ihm C.G., Lee S.H., Moon J.Y. Polymorphisms in two genes, IL-1B and ACE, are associated with erythropoietin resistance in Korean patients on maintenance hemodialysis. Exp. Mol. Med. 2008;40(2):161–166. doi: 10.3858/emm.2008.40.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itoyama S., Keicho N., Quy T., Phi N.C., Long H.T., Ha L.D., Ban V.V., Ohashi J., Hijikata M., Matsushita I., Kawana A., Yanai H., Kirikae T., Kuratsuji T., Sasazuki T. ACE1 polymorphism and progression of SARS. Biochem. Biophys. Res. Commun. 2004;323(3):1124–1129. doi: 10.1016/j.bbrc.2004.08.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adamzik M., Frey U., Sixt S., Knemeyer L., Beiderlinden M., Peters J., Siffert W. ACE I/D but not AGT (-6)A/G polymorphism is a risk factor for mortality in ARDS. Eur. Resp. J. 2007;29(3):482–488. doi: 10.1183/09031936.00046106. [DOI] [PubMed] [Google Scholar]
- 16.Verdecchia P., Cavallini C., Spanevello A., Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur. J. Int. Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarke N.E., Turner A.J. Angiotensin-Converting Enzyme 2: The First Decade. Int. J. Hyperten. 2012;2012 doi: 10.1155/2012/307315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciaglia E., Vecchione C., Puca A.A. COVID-19 Infection and Circulating ACE2 Levels: Protective Role in Women and Children. Front. Pediatr. 2020;8(206) doi: 10.3389/fped.2020.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanyaolu A., Okorie C., Marinkovic A., Patidar R., Younis K., Desai P., Hosein Z., Padda I., Mangat J., Altaf M. Comorbidity and its Impact on Patients with COVID-19. SN Compr. Clin. Med. 2020:1–8. doi: 10.1007/s42399-020-00363-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krishnan R., Sekar D., Karunanithy S., Subramanium S. Association of angiotensin converting enzyme gene insertion/deletion polymorphism with essential hypertension in south Indian population. Genes Dis. 2016;3(2):159–163. doi: 10.1016/j.gendis.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng Y., Niu T., Xu X., Chen C., Li Q., Qian R., Wang G., Xu X. Insertion/deletion polymorphism of the ACE gene is associated with type 2 diabetes. Diabetes. 2002;51(6):1986–1988. doi: 10.2337/diabetes.51.6.1986. [DOI] [PubMed] [Google Scholar]
- 22.Amara A., Mrad M., Sayeh A., Lahideb D., Layouni S., Haggui A., Fekih-Mrissa N., Haouala H., Nsiri B. The Effect of ACE I/D Polymorphisms Alone and With Concomitant Risk Factors on Coronary Artery Disease. Clin. Appl. Thromb./Hemost. Off. J. Int. Acad. Clin. Appl. Thromb./Hemost. 2018;24(1):157–163. doi: 10.1177/1076029616679505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delanghe J.R., Speeckaert M.M., De Buyzere M.L. COVID-19 infections are also affected by human ACE1 D/I polymorphism. Clin. Chem. Lab. Med. 2020;58(7):1125–1126. doi: 10.1515/cclm-2020-0425. [DOI] [PubMed] [Google Scholar]
- 24.Hirschhorn J.N., Lohmueller K., Byrne E., Hirschhorn K. A comprehensive review of genetic association studies, Genetics in medicine : official journal of the American College of Medical. Genetics. 2002;4(2):45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 25.Huang T., Shu Y., Cai Y.D. Genetic differences among ethnic groups. BMC Genom. 2015;16:1093. doi: 10.1186/s12864-015-2328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.