Abstract
The transition from acetate production by a microorganism in its early growth phase to acetate re-uptake in its late growth phase has been termed acetate switch. It has been observed in several heterotrophic prokaryotes, but not in an autotroph. Furthermore, all reports hitherto have involved the tricarboxylic acid cycle. This study reports the first observation of acetate switch in a methanogenic autotroph Methanococcus maripaludis S2, which uses the Wolfe cycle for its anaerobic respiration. When grown in minimal medium with carbon dioxide as the sole carbon source, and either ammonium or dinitrogen as the sole nitrogen source, M. maripaludis S2 dissimilated acetate in the early growth phase and assimilated it back in the late growth phase. The acetate switch was more pronounced in the dinitrogen-grown cultures. We postulate that the acetate dissimilation in M. maripaludis S2 may serve as a metabolic outlet for the carbon overflow in the early growth phase, and the assimilation in the late growth phase may be due to the scarcity of the carbon source. Based on the primary and secondary protein structures, we propose that MMP0253 may function as the adenosine diphosphate (ADP)-forming acetyl-CoA synthetase to catalyse acetate formation from acetyl-CoA. To verify this, we produced MMP0253 via the ligation-independent cloning technique in Escherichia coli strain Rosetta (DE3) using pNIC28-Bsa4 as the vector. The recombinant protein showed catalytic activity, when added into a mixture of acetyl-CoA, ADP, and inorganic phosphate (Pi). The concentration profile of acetate, together with the enzymatic activity of MMP0253, shows that M. maripaludis S2 can produce acetate and exhibit an acetate switch.
Keywords: acetate switch, acetate dissimilation, acetate assimilation, Methanococcus maripaludis, ADP-forming acetyl-CoA synthetase, Acd
Introduction
Many microorganisms alternate their metabolic pathways to adapt to the nutrients in their immediate environments. They prioritise growth when nutrients are abundant and survival when they are scarce. One such behaviour is the ‘acetate switch’, which has been studied extensively in Escherichia coli.1 Acetate switch involves the transition from acetate dissimilation (production and secretion) to assimilation (uptake and utilisation). In the early growth phase, E. coli dissimilates acetate when the carbon flux into the cells exceeds the capacity of its central pathways or when its tricarboxylic acid (TCA) cycle does not operate fully due to limited oxygen.1 This dissimilation allows E. coli to generate adenosine triphosphate (ATP) and reduce acetyl-CoA (Ac-CoA) accumulation.1 In the later growth phase, when the metabolic bottleneck allows the uptake of acetate due to respiratory capacity, E. coli switches to acetate assimilation. The acetate switch is defined as the point where acetate dissimilation equals assimilation. Figure 1 illustrates a typical acetate switch profile in E. coli. Besides E. coli, acetate switch has been observed in several other bacteria2-4 and 3 halophilic archaea.5 To date, all reported cases have involved heterotrophs and the TCA cycle.
Figure 1.
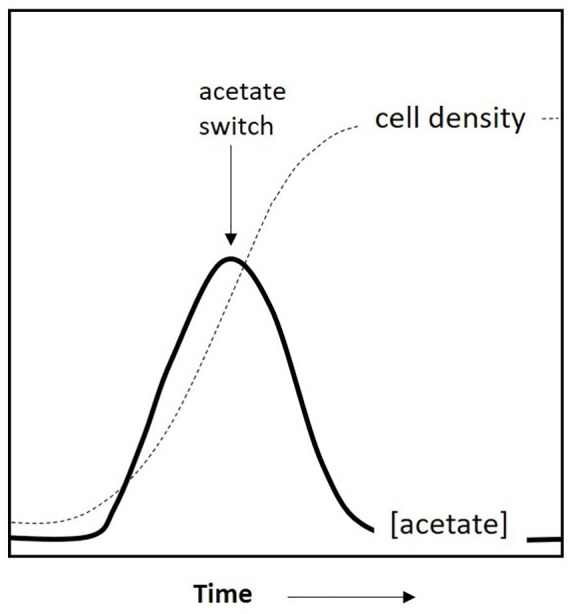
A schematic of acetate switch profile. Acetate concentration increases to a maximum in the early growth phase and decreases thereafter. Acetate concentration reaches zero as the cells enter the stationary phase.
This study reports the first observation of an acetate switch in a methanogenic autotroph, specifically Methanococcus maripaludis S2. This archaeon is mesophilic, hydrogenotrophic,6 and can thrive on minimal nutrients, carbon dioxide (CO2) as the sole carbon source, and either ammonium or dinitrogen (N2) as the sole nitrogen source. Acetate assimilation by adenosine monophosphate (AMP)-forming acetate-CoA ligase (Acs) is a known phenomenon7 in M. maripaludis S2. However, the only record of acetate dissimilation by M. maripaludis S2 is the work of Abdel Azim et al,8 who reported an acetate concentration of about 1 mmol/L after incubating M. maripaludis for 150 hours in a minimal medium supplemented with propionate. Other methanogens such as Methanosphaera stadtmanae,9 Methanococcus voltae,10 Methanospirillum hungatei,11 and Methanothrix sp.12 are known to assimilate acetate, but have not been reported to produce acetate. On the other hand, Methanococcus jannaschii13 can dissimilate acetate, but is unable to assimilate it. Notably, Methanosarcina acetivorans14 and the non-methanogenic Pyrobaculum islandicum15 can both dissimilate and assimilate acetate, but under different growth conditions. No methanogen or autotroph has been reported to display an acetate switch.
In this study, batch culture experiments of M. maripaludis S2 were performed in a minimal medium (void of propionate) to investigate its acetate switch. CO2 and either ammonium or N2 were used as the sole carbon and nitrogen sources, respectively. As adenosine diphosphate (ADP)-forming Ac-CoA synthetase (Acd) is known16 to catalyse reversible formation of acetate from Ac-CoA in some other archaea, we propose MMP0253 (GenBank accession no. CAF29809) as the putative Acd in M. maripaludis. We produced it and confirmed its enzymatic activity.
Materials and Methods
Gases and chemicals
Pure H2, CO2, N2, and CH4 gases were purchased from AIR Liquide, Singapore. Mass flow controllers (Red-Y Compact 2 Series, Vögtlin Instruments, Basel-Landschaft, Switzerland) were used to prepare gas mixtures (H2/CO2 and H2/CO2/N2). K2HPO4, MnSO4·H2O, and NH4Cl were obtained from VWR, Singapore. Nitrilotriacetic acid and Na2S·9H2O were purchased from Thermo Fisher Scientific, New Jersey, United States. Other chemicals, if not stated otherwise, were purchased from Sigma-Aldrich, Singapore.
Strain and medium preparation
Methanococcus maripaludis S2 (DSM 14266) was purchased from Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures. Ammonium samples were prepared as reported.17 No carbon source was added into the medium and vitamins were also omitted. In N2 samples, NH4Cl was omitted in the minimal medium and Fe(NH4)2(SO4)2·6H2O was replaced by 140 µL of FeSO4·7H2O solution (0.01% w/v). About 460 mL of the minimal medium was dispensed into 1 L serum bottles (Chemglass Life Sciences LLC, New Jersey, USA). To create an anaerobic environment, the bottles were flushed with 80/20 v/v H2/CO2 gas mixture at 2 bars for 30 minutes. Subsequently, the bottles were sterilised by autoclaving at 121°C for 21 minutes. After the bottles had cooled down to room temperature, 2.0 mL of Na2S stock solution (12.5% w/v) was injected into each bottle.
Batch cultivation
The inoculum was pre-cultured for 2 days in the respective minimal medium. At the start of the experiment, 40 mL of the pre-cultured inoculum was transferred to each serum bottle containing fresh medium. H2/CO2 gas mixture (74%-85% of H2 by volume, balance was CO2) was used to pressurise the ammonium-grown cultures to 259-265 kPa. H2/CO2/N2 gas mixture (68%-73% H2% and 10%-17% CO2 by volume, balance was N2) was used to pressurise the N2-grown cultures to 304-313 kPa. The cultures were incubated at 37°C and 180 rpm (Orbital Shaking Incubator LM-570RD, Yihder, China). Minimal medium without inoculum was used as a negative control. At the start of each day, the bottles were de-pressurised, flushed, and re-pressurised with H2/CO2 and H2/CO2/N2, respectively. At the end of each day, the cultures were kept at 20°C without shaking, until the following morning. All growth experiments were performed in duplicates.
Analytical procedures
About 2 mL of liquid sample was withdrawn hourly using a disposable hypodermic needle (25G × 1”, Terumo Corporation, USA) attached to a sterile disposable syringe (3 mL, Nipro Medical Corporation, USA) and deposited in a quartz cell (Agilent Technologies, USA). The optical densities of the samples were measured at 600 nm using a double-beam UV/Vis spectrophotometer (Agilent Technologies, Carry, USA). The concentration of acetate in the liquid sample was determined by a high-performance liquid chromatography (HPLC) (1260 Infinity I, Agilent Technologies, USA) equipped with Aminex HPX-87 H column and G7165A multiple wavelength detector (190-240 nm). About 5 mM of H2SO4 solution at 0.5 mL/min was used as the mobile phase. The column oven was set at 60°C. The liquid samples were centrifuged at 20 g for 5 minutes before analysis. Acetate was calibrated using standard solutions at 0.01, 0.05, 0.1, 0.25, 0.5, and 0.75 mM. Two additional methods were used to confirm the presence of acetate: gas chromatography (GC) (PerkinElmer Clarus 580, USA) and 500 MHz proton nuclear magnetic resonance (NMR) spectroscopy (Bruker, USA). A control sample with minimal medium and no inoculum was also tested with all 3 methods to confirm our detection and measurement of acetate.
Identification of Acd in M. maripaludis S2 and comparison against other known Acd
The genome18 of M. maripaludis S2 was studied using Basic Local Alignment Search Tool (BLAST)19 with Acd in M. jannaschii (GenBank accession no. WP_010870094) as a query. MMP0253 was identified as a potential Acd. Its primary and secondary structures were compared against the known16,20 Acd enzymes in M. jannaschii, Haloarcula marismortui (GenBank accession no. AAV45866) and Archaeoglobus fulgidus (GenBank accession no. WP_048095590) using BLAST and HHpred.21
Production of recombinant MMP0253
MMP0253 was cloned using the ligation-independent cloning (LIC) technique with pNIC28-Bsa4 as the vector.22 Briefly, the vector was linearised by BsaI and overhangs were created by treating the linear vector with T4 DNA polymerase and deoxyguanosine triphosphate (dGTP). The acd insert was amplified by the polymerase chain reaction (PCR) technique with Phusion DNA polymerase and primers. Subsequently, the purified PCR product was treated with T4 DNA polymerase and deoxycytidine triphosphate (dCTP) to create overhangs. The vector and insert were then annealed, and E. coli strain Rosetta (DE3) was transformed. The E. coli with the recombinant plasmid was cultivated in Terrific Broth (TB) medium at 37°C and 200 rpm. The protein expression was induced by isopropyl β-D-1-thiogalactopyranoside (IPTG) at 18°C. The recombinant protein was purified by immobilised metal ion affinity chromatography (IMAC) followed by gel filtration.
Test for enzymatic activity
The activity of recombinant MMP0253 was tested at 37°C under oxic conditions. Each assay mixture (0.1 mL) contained 200 mM Tris-HCl (pH 7.7), 5 mM MgCl2, 0.1 mM 5,5'-dithiobis-(2-nitrobenzoic acid) (Ellman’s reagent), 0.1 mM Ac-CoA, 0.5 mM ADP, and 5 mM K2HPO4. The absorbance at 412 nm was measured by Infinite 200 PRO (Tecan, Switzerland) before the addition of MMP0253 and 2 min after its addition. Each assay condition was tested in triplicates.
Results
Acetate concentration profiles
All 3 methods (HPLC, GC, and proton NMR spectroscopy) indicated the presence of acetate in our samples. Thus, in spite of its low concentration, there was a high confidence in the acetate detection. Figure 2 shows the acetate concentration profiles in both ammonium and N2-grown cultures over multiple days. The 500 MHz proton NMR spectroscopy showed a signal at 1.839 ppm (see Supplementary Material 1), which matches the signal of pure acetate. No acetate was detected in the control sample.
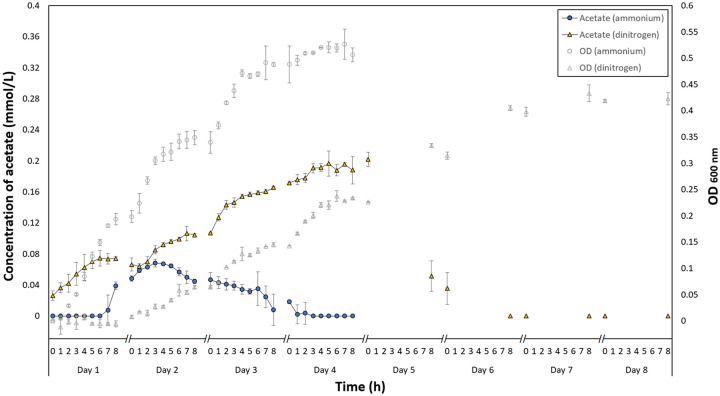
Figure 2.
Concentration of acetate (in mmol/L) in ammonium-grown cultures (filled circle, n = 2) and dinitrogen-grown cultures (filled triangle, n = 2). The optical density (OD) measured at 600 nm of ammonium (empty circle) and dinitrogen (empty triangle) grown cultures were also shown. The data were collected hourly over an 8-hour period in the first 4 days. For subsequent days, only dinitrogen-grown cultures were monitored, and the data were collected at time t = 0 and t = 8 h each day. The symbol // indicates a break in data collection, during which the cultures were kept at 20°C without any shaking. Error bars indicate one standard deviation of the sample data. Some error bars are smaller than the symbols. OD indicates optical density.
In the ammonium-grown cultures, acetate reached a peak concentration of 0.069 ± 0.005 mmol/L midway through the growth phase on the second day and declined thereafter. In the N2-grown cultures, the acetate concentration was non-zero at the very beginning, which is probably due to the dissimilated acetate in the N2-acclimated inoculum. The daily concentration profile exhibited a repeated trend during the first 4 days. It increased at a slowing rate and plateaued near the day’s end. The acetate concentration reached a peak of 0.202 ± 0.009 mmol/L on the fourth day, midway through the growth phase. In both ammonium and N2-grown cultures, the acetate concentration reached zero as the cells entered the stationary phase.
The overall profiles in both cultures follow an initial rising and subsequent falling trend with a distinct peak characteristic of the acetate switch. In other words, the growth of M. maripaludis S2 exhibited 2 phases: an initial acetogenic phase followed by an acetotrophic phase. Acetate assimilation and dissimilation are likely to be competitive and balance each other at the acetate peaks. Both profiles of ammonium and N2-grown cultures are markedly similar to the acetate switch profile in E. coli.1 There have been no reports of this for an autotroph in the literature thus far.
MMP0253 showed similarities to other known Acd
Our amino acid sequence analysis via BLAST shows that MMP0253 is 35% (e-value: 5.6e-116) similar to Acd in H. marismortui, 44% (e-value: 2.4e-110) similar to A. fulgidus, and 52% (e-value: 9.5e-120) similar to M. jannaschii. These seemingly low percentages do not discredit the claim of MMP0253 as an Acd, because the characterised Acd in H. marismortui, A. fulgidus, and M. jannaschii are also 36% to 42% similar to each other. Our analysis of the 4 secondary protein structures via HHpred showed marked resemblance (see Supplementary Material 2). All 4 have 2 conserved histidine residues (His-242 and His-538 in M. maripaludis), which may be analogous to the active sites for the transient phosphorylation in acetate formation.23 Residues associated with the formation of Ac-CoA binding site and signature motifs of Acd24 can also be found in MMP0253. In essence, the structural evidence at both levels suggests that MMP0253 in M. maripaludis is homologous to Acd in the other 3 archaea. Abdel Azim et al,8 Goyal et al,25 and Richards et al26 have also annotated MMP0253 as Acd.
MMP0253 catalysed acetate formation from Ac-CoA
After recombinant MMP0253 was added into the reagents mixture, a rapid yellow colourisation was observed. Acetate formation from Ac-CoA released coenzyme A (HS-CoA), which reacted with Ellman’s reagent to give yellow-coloured 2-nitro-5-thiobenzoate dianions. Therefore, the observed colourisation (absorbance change of approximately 0.4) confirms the catalytic activity of MMP0253 (Table 1). In the absence of MMP0253, the absorbance change was negligible even after 30 minutes. When Ac-CoA, ADP, or K2HPO4 was not added into the reagents mixture, the absorbance change was about 0.1. However, this is due to the interaction between MMP0253 and Ellman’s reagent as verified by adding MMP0253 into a mixture of Tris-HCl (pH 7.7), MgCl2, and Ellman’s reagent only. This suggests that the acetate formation from Ac-CoA catalysed by MMP0253 requires all 3 reagents, Ac-CoA, ADP, and inorganic phosphate (Pi).
Table 1.
Test for the enzymatic activity of MMP0253 at 37°C under oxic conditions.
| Reagents and enzymes |
Change in absorbance at 412 nm | |||
|---|---|---|---|---|
| Acetyl-CoA | ADP | K2HPO4 | MMP0253 | |
| X | X | X | X | 0.3695 ± 0.0278 |
| X | X | X | 0.1164 ± 0.0081 | |
| X | X | X | 0.1155 ± 0.0139 | |
| X | X | X | 0.0772 ± 0.0028 | |
| X | 0.0899 ± 0.0255 | |||
| X | X | X | 0.0038 ± 0.0004 | |
Abbreviations: ADP, adenosine diphosphate; X, present in the assay mixture.
Unless indicated otherwise, each assay mixture (0.1 mL) consisted of 200 mM Tris-HCl (pH 7.7), 5 mM MgCl2, 0.1 mM 5,5'-dithiobis-(2-nitrobenzoic acid) (Ellman’s reagent), 0.1 mM acetyl-CoA, 0.5 mM ADP, and 5 mM K2HPO4. Absorbance readings at 412 nm were obtained 2 minutes after the addition of MMP0253. For the tests in which MMP0253 was not added, absorbance readings were obtained after 30 minutes. Each assay was done in triplicates.
Discussion
While acetate assimilation by M. maripaludis S2 has been reported in the literature,7 its dissimilation has not been conclusively established prior to this study. Although one record8 of acetate dissimilation does exist for this methanogen, it was limited to the growth with propionate. Our experimental results confirm that M. maripaludis S2 can produce acetate when grown in a minimal medium (void of propionate) with either ammonium or N2 as the nitrogen source. Our preliminary enzymatic study also shows that MMP0253 may function as Acd, which is known16 to catalyse the reversible formation of acetate from Ac-CoA and generate ATP via substrate-level phosphorylation: Ac-CoA + ADP + Pi ↔ Acetate + ATP + HS-CoA.
Having established acetate dissimilation in M. maripaludis S2, we observe that our acetate concentration profiles have a marked resemblance to the acetate switch profile in E. coli. Acetate switch, a phenomenon that involves the transition from an acetogenic phase to an acetotrophic phase, has only been observed in heterotrophs (Table 2).1 Furthermore, all reported microorganisms use the TCA cycle.1 In contrast, M. maripaludis S2 is an autotroph, which uses the Wolfe cycle for methanogenesis (Figure 3).
Table 2.
A comparison of the characteristics of the bacteria and archaea with acetate switch.
| Microorganism | Domain | Type | Acetate dissimilation enzymes | Acetate assimilation enzymes | References |
|---|---|---|---|---|---|
| Escherichia coli | Bacteria | Heterotroph | Pta-Ack | Acs Reverse Pta-Ack |
Brown et al,27 Kumari et al28 |
| Salmonella enterica | Bacteria | Heterotroph | EutD-ACK | Acs Reverse Pta-Ack Reverse EutD-Ack |
Starai et al4,29 |
| Bacillus subtilis | Bacteria | Heterotroph | Pta-Ack | Acs | Grundy et al,2 Presecan-Siedel et al30 |
| Corynebacterium glutamicum | Bacteria | Heterotroph | No data | Reverse Pta-Ack | Dominguez et al,31 Reinscheid et al,32 Gerstmeir et al33 |
| Vibrio fischeri | Bacteria | Heterotroph | No data | Acs | Studer et al34 |
| Staphylococcus aureus | Bacteria | Heterotroph | Pta-Ack | Acs | DeMars and Bose35 |
| Campylobacter jejuni | Bacteria | Heterotroph | Pta-Ack | Acs | Wright et al36 |
| Mycobacterium tuberculosis | Bacteria | Heterotroph | Pta-Ack | Acs Reverse Pta-Ack |
Rucker et al3 |
|
Haloarcula marismortui
Haloferax volcanii Halobacterium saccharovorum |
Archaea | Heterotroph | Acd | Acs | Brasen and Schonheit5 |
| Methanococcus maripaludis | Archaea | Autotroph | Acd | Acs | This study |
Abbreviations: Acd, ADP-forming acetyl-CoA synthetase; Ack, acetate kinase; Acs, AMP-forming acetate-CoA ligase; EutD, eut operon-encoded phosphotransacetylase; Pta, phosphotransacetylase.
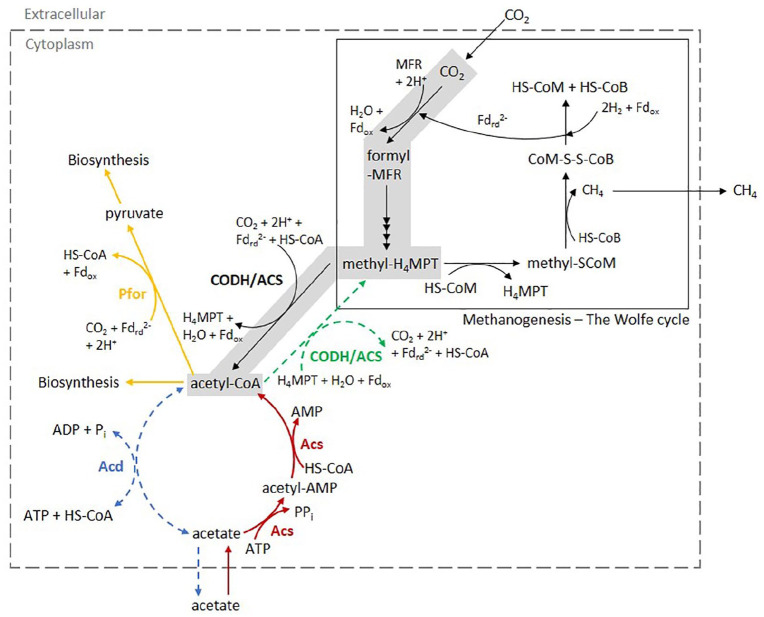
Figure 3.
Simplistic view of methanogenesis and biochemical reactions involved acetate and acetyl-CoA. Methanogenesis is shown in the rectangle with solid line border. Grey background indicates the reductive acetyl-CoA pathway. Acetate dissimilation and assimilation were shown. Hypothetical pathways were indicated as dotted lines. Enzymes involved in the Wolfe cycle were not included. Metabolites: acetyl-CoA indicates acetyl coenzyme A; acetyl-AMP, acetyladenylate; ADP, adenosine diphosphate; AMP, adenosine monophosphate; ATP, adenosine triphosphate; Fdrd2-, reduced ferredoxin; Fdox, oxidised ferredoxin; MFR, methanofuran; H4MPT, tetrahydromethanopterin; HS-CoA, coenzyme A; HS-CoB, coenzyme B; HS-CoM, coenzyme M; Pi, inorganic phosphate; PPi, inorganic pyrophosphate. Enzymes (bolded): Acd, ADP-forming acetyl-CoA synthetase; Acs, AMP-forming acetate-CoA ligase; CODH/ACS, carbon monoxide dehydrogenase/acetyl-CoA synthase complex; Pfor, pyruvate: ferredoxin oxidoreductase.
While we do not have a solid evidence to explain the above acetate profile in this archaeon, we put forth a hypothesis based on the literature on other microorganisms. It is established1 that acetate dissimilation in E. coli and other heterotrophs serves as a metabolic outlet for carbon overflow, which occurs when their TCA cycles are saturated or inhibited. The dissimilation also helps to reduce Ac-CoA accumulation and recycle HS-CoA. In the late growth phase when the carbon source depletes, these microorganisms assimilate acetate as an additional carbon source. In addition, as acetate accumulates in the growth medium, it diffuses back across the cell membrane. This acidifies the cytoplasm and interferes with other biosynthetic processes.3 The cell removes acetate by converting it into Ac-CoA via a single-step reaction.
Based on the above explanation of the acetate switch in other microorganisms, we can postulate that a similar hypothesis may hold for M. maripaludis S2. When the carbon flux into the Wolfe cycle exceeds its capacity, M. maripaludis must find a metabolic outlet for the overflow. While the Wolfe cycle has various exits leading to the formations of purines, thymidylate, and methionine, the formation of Ac-CoA from methyl-tetrahydromethanopterin (methyl-H4MPT) is the largest.37 This may lead to an accumulation of Ac-CoA and diminution of HS-CoA pools, which the cell may avoid via biosynthesis or acetate dissimilation. However, biosynthesis needs ATP, and its primary supply (Wolfe cycle) is already saturated. Hence, acetate dissimilation seems to be the only metabolic outlet to relieve the carbon overflow. While no concrete evidence exists for this hypothesis at the present, we observed that it is consistent with the daily acetate profiles (Figure 2). As our study involved batch culture and CO2 was replenished daily, the CO2 supply (flux or overflow) in the liquid medium gradually decreased during each day. As carbon supply decreased over time, carbon overflow decreased, and hence dissimilation decreased and eventually plateaued off. When CO2 was replenished the following morning, carbon overflow occurred at the start and dissimilation continued at a higher rate than the previous evening.
The hypothesis can also explain the greater acetate dissimilation in N2-grown cultures. Diazotrophy is an ATP-demanding process.38 Hence, the Wolfe cycle in the N2 samples will saturate earlier for the same flux of CO2 (mmol/gDCW/h). Furthermore, the biosynthesis level with N2 is known39 to be lower. Therefore, the N2-grown cultures will experience a higher carbon overflow and hence greater acetate dissimilation. Abdel Azim et al8 reported an acetate dissimilation when incubating M. maripaludis in a minimal medium supplemented with propionate. While they hypothesised that propionate might have reduced the electrochemical proton gradient across the cell membrane, forcing the cell to restore it via the ATP-consuming proton extrusion process, they did not explain why acetate was produced. This ATP-consuming process may have an effect similar to diazotrophy and thus caused acetate dissimilation to occur.
In the late growth phase, M. maripaludis transitioned to acetate assimilation. This is possibly due to carbon deficiency, similar to the phenomenon in other microorganisms as discussed earlier. The acidification of the cytoplasm by acetic acid may play a role in inducing M. maripaludis to convert it to Ac-CoA; however, further study is needed to verify this.
Supplemental Material
Supplemental material, Supplementary_material_1_xyz4053208aba416 for First Observation of an Acetate Switch in a Methanogenic Autotroph (Methanococcus maripaludis S2) by Chi Hung Vo, Nishu Goyal, Iftekhar A Karimi and Markus Kraft in Microbiology Insights
Supplemental material, Supplementary_material_2_xyz40532449c19c2 for First Observation of an Acetate Switch in a Methanogenic Autotroph (Methanococcus maripaludis S2) by Chi Hung Vo, Nishu Goyal, Iftekhar A Karimi and Markus Kraft in Microbiology Insights
Supplemental material, Supplementary_material_3_xyz4053215cc5e01 for First Observation of an Acetate Switch in a Methanogenic Autotroph (Methanococcus maripaludis S2) by Chi Hung Vo, Nishu Goyal, Iftekhar A Karimi and Markus Kraft in Microbiology Insights
Supplemental material, Supplementary_material_4_xyz40532c5298d3e for First Observation of an Acetate Switch in a Methanogenic Autotroph (Methanococcus maripaludis S2) by Chi Hung Vo, Nishu Goyal, Iftekhar A Karimi and Markus Kraft in Microbiology Insights
Acknowledgments
We thank Professors Zhou Zhi (Purdue), Zhou Kang (NUS), Zhao Dan (NUS), Yan Ning (NUS), He Jianzhong (NUS), and Dr Karthika Nagarajan (NUS) for their invaluable help, guidance, and suggestions during the course of this work. Similarly, we thank Matthew James Rogers (NUS), Prerna Goyal (NUS), Lim Ee Yang (NUS), and Dr Hugo Gerald Schmidt (CARES Ltd). We are indebted to Professor Tong Yen Wah (NUS) and Energy and Environmental Sustainability for Megacities (E2S2) for lending us equipment, which enhanced and expedited our work. We thank Dr Dai Chencheng (CARES Ltd) for his assistance with high-performance liquid chromatography. We thank the NTU Protein Production Platform (www.proteins.sg) for the cloning, expression tests, and purification of protein construct Acd.
Footnotes
Declaration of Conflicting Interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research is supported by the National Research Foundation, Prime Minister’s Office, Singapore, under its CREATE programme. Concomitantly, this work is supported by the National University of Singapore through its Graduate Research Scholarship. This work also received partial financial support under a Tier 1 grant R-279-000-541-114 from the Ministry of Education, Singapore.
Author Contributions: All authors designed the experiment. CHV performed the experiment. All authors wrote and edited the manuscript. All authors read and approved the final manuscript.
Availability of Data and Material: The main datasets generated and/or analysed during the current study are attached as supplementary materials. Any other datasets are available from the corresponding author on reasonable request.
ORCID iDs: Chi Hung Vo  https://orcid.org/0000-0002-8242-8494
https://orcid.org/0000-0002-8242-8494
Markus Kraft  https://orcid.org/0000-0002-4293-8924
https://orcid.org/0000-0002-4293-8924
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Wolfe AJ. The acetate switch. Microbiol Mol Biol Rev. 2005;69:12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Grundy FJ, Waters DA, Takova TY, Henkin TM. Identification of genes involved in utilization of acetate and acetoin in Bacillus subtilis. Mol Microbiol. 1993;10:259-271. [DOI] [PubMed] [Google Scholar]
- 3. Rucker N, Billig S, Bucker R, Jahn D, Wittmann C, Bange FC. Acetate dissimilation and assimilation in Mycobacterium tuberculosis depend on carbon availability. J Bacteriol. 2015;197:3182-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Starai VJ, Escalante-Semerena JC. Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J Mol Biol. 2004;340:1005-1012. [DOI] [PubMed] [Google Scholar]
- 5. Brasen C, Schonheit P. Mechanisms of acetate formation and acetate activation in halophilic archaea. Arch Microbiol. 2001;175:360-368. [DOI] [PubMed] [Google Scholar]
- 6. Jones WJ, Paynter MJB, Gupta R. Characterization of Methanococcus maripaludis sp. nov., a new methanogen isolated from salt marsh sediment. Arch Microbiol. 1983;135:91-97. [Google Scholar]
- 7. Shieh JS, Whitman WB. Pathway of acetate assimilation in autotrophic and heterotrophic Methanococci. J Bacteriol. 1987;169:5327-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abdel Azim A, Rittmann SKMR, Fino D, Bochmann G. The physiological effect of heavy metals and volatile fatty acids on Methanococcus maripaludis S2. Biotechnol Biofuels. 2018;11:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fricke WF, Seedorf H, Henne A, et al. The genome sequence of Methanosphaera stadtmanae reveals why this human intestinal archaeon is restricted to methanol and H2 for methane formation and ATP synthesis. J Bacteriol. 2006;188:642-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Whitman WB, Ankwanda E, Wolfe RS. Nutrition and carbon metabolism of Methanococcus voltae. J Bacteriol. 1982;149:852-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ekiel I, Smith IC, Sprott GD. Biosynthetic pathways in Methanospirillum hungatei as determined by 13C nuclear magnetic resonance. J Bacteriol. 1983;156:316-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Westermann P, Ahring BK, Mah RA. Threshold acetate concentrations for acetate catabolism by aceticlastic methanogenic bacteria. Appl Environ Microbiol. 1989;55:514-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sprott GD, Ekiel I, Patel GB. Metabolic pathways in Methanococcus jannaschii and other methanogenic bacteria. Appl Environ Microbiol. 1993;59:1092-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rother M, Metcalf WW. Anaerobic growth of Methanosarcina acetivorans C2A on carbon monoxide: an unusual way of life for a methanogenic archaeon. Proc Natl Acad Sci USA. 2004;101:16929-16934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hu YJ, Holden JF. Citric acid cycle in the hyperthermophilic archaeon Pyrobaculum islandicum grown autotrophically, heterotrophically, and mixotrophically with acetate. J Bacteriol. 2006;188:4350-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Musfeldt M, Schonheit P. Novel type of ADP-forming acetyl coenzyme A synthetase in hyperthermophilic Archaea: heterologous expression and characterization of isoenzymes from the sulfate reducer Archaeoglobus fulgidus and the methanogen Methanococcus jannaschii. J Bacteriol. 2002;184:636-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goyal N, Padhiary M, Karimi IA, Zhou Z. Flux measurements and maintenance energy for carbon dioxide utilization by Methanococcus maripaludis. Microb Cell Fact. 2015;14:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hendrickson EL, Kaul R, Zhou Y, et al. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J Bacteriol. 2004;186:6956-6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brasen C, Schonheit P. Unusual ADP-forming acetyl coenzyme A synthetases from the mesophilic halophilic euryarchaeon Haloarcula marismortui and from the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Arch Microbiol. 2004;182:277-287. [DOI] [PubMed] [Google Scholar]
- 21. Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244-W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keates T, Cooper CD, Savitsky P, et al. Expressing the human proteome for affinity proteomics: optimising expression of soluble protein domains and in vivo biotinylation. N Biotechnol. 2012;29:515-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brasen C, Schmidt M, Grotzinger J, Schonheit P. Reaction mechanism and structural model of ADP-forming acetyl-CoA synthetase from the hyperthermophilic archaeon Pyrococcus furiosus: evidence for a second active site histidine residue. J Biol Chem. 2008;283:15409-15418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sanchez LB, Galperin MY, Muller M. Acetyl-CoA synthetase from the amitochondriate eukaryote Giardia lamblia belongs to the newly recognized superfamily of acyl-CoA synthetases (nucleoside diphosphate-forming). J Biol Chem. 2000;275:5794-5803. [DOI] [PubMed] [Google Scholar]
- 25. Goyal N, Zhou Z, Karimi IA. Metabolic processes of Methanococcus maripaludis and potential applications. Microb Cell Fact. 2016;15:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Richards MA, Lie TJ, Zhang J, Ragsdale SW, Leigh JA, Price ND. Exploring hydrogenotrophic methanogenesis: a genome scale metabolic reconstruction of Methanococcus maripaludis. J Bacteriol. 2016;198:3379-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brown TD, Jones-Mortimer MC, Kornberg HL. The enzymic interconversion of acetate and acetyl coenzyme A in Escherichia coli. J Gen Microbiol. 1977;102:327-336. [DOI] [PubMed] [Google Scholar]
- 28. Kumari S, Beatty CM, Browning DF, et al. Regulation of acetyl coenzyme A synthetase in Escherichia coli. J Bacteriol. 2000;182:4173-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Starai VJ, Garrity J, Escalante-Semerena JC. Acetate excretion during growth of Salmonella enterica on ethanolamine requires phosphotransacetylase (EutD) activity, and acetate recapture requires acetyl-CoA synthetase (Acs) and phosphotransacetylase (Pta) activities. Microbiology. 2005;151:3793-3801. [DOI] [PubMed] [Google Scholar]
- 30. Presecan-Siedel E, Galinier A, Longin R, et al. Catabolite regulation of the pta gene as part of carbon flow pathways in Bacillus subtilis. J Bacteriol. 1999;181:6889-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dominguez H, Nezondet C, Lindley ND, Cocaign M. Modified carbon flux during oxygen limited growth of Corynebacterium glutamicum and the consequences for amino acid overproduction. Biotechnol Lett. 1993;15:449-454. [Google Scholar]
- 32. Reinscheid DJ, Schnicke S, Rittmann D, Zahnow U, Sahm H, Eikmanns BJ. Cloning, sequence analysis, expression and inactivation of the Corynebacterium glutamicum pta-ack operon encoding phosphotransacetylase and acetate kinase. Microbiology. 1999;145:503-513. [DOI] [PubMed] [Google Scholar]
- 33. Gerstmeir R, Wendisch VF, Schnicke S, et al. Acetate metabolism and its regulation in Corynebacterium glutamicum. J Biotechnol. 2003;104:99-122. [DOI] [PubMed] [Google Scholar]
- 34. Studer SV, Mandel MJ, Ruby EG. AinS quorum sensing regulates the Vibrio fischeri acetate switch. J Bacteriol. 2008;190:5915-5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. DeMars Z, Bose JL. Redirection of metabolism in response to fatty acid kinase in Staphylococcus aureus. J Bacteriol. 2018;200:e00345-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wright JA, Grant AJ, Hurd D, et al. Metabolite and transcriptome analysis of Campylobacter jejuni in vitro growth reveals a stationary-phase physiological switch. Microbiology. 2009;155:80-94. [DOI] [PubMed] [Google Scholar]
- 37. Thauer RK. The Wolfe cycle comes full circle. Proc Natl Acad Sci USA. 2012;109:15084-15085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hoffman BM, Lukoyanov D, Yang ZY, Dean DR, Seefeldt LC. Mechanism of nitrogen fixation by nitrogenase: the next stage. Chem Rev. 2014;114:4041-4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kessler PS, Daniel C, Leigh JA. Ammonia switch-off of nitrogen fixation in the methanogenic archaeon Methanococcus maripaludis: mechanistic features and requirement for the novel GlnB homologues, NifI(1) and NifI(2). J Bacteriol. 2001;183:882-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplementary_material_1_xyz4053208aba416 for First Observation of an Acetate Switch in a Methanogenic Autotroph (Methanococcus maripaludis S2) by Chi Hung Vo, Nishu Goyal, Iftekhar A Karimi and Markus Kraft in Microbiology Insights
Supplemental material, Supplementary_material_2_xyz40532449c19c2 for First Observation of an Acetate Switch in a Methanogenic Autotroph (Methanococcus maripaludis S2) by Chi Hung Vo, Nishu Goyal, Iftekhar A Karimi and Markus Kraft in Microbiology Insights
Supplemental material, Supplementary_material_3_xyz4053215cc5e01 for First Observation of an Acetate Switch in a Methanogenic Autotroph (Methanococcus maripaludis S2) by Chi Hung Vo, Nishu Goyal, Iftekhar A Karimi and Markus Kraft in Microbiology Insights
Supplemental material, Supplementary_material_4_xyz40532c5298d3e for First Observation of an Acetate Switch in a Methanogenic Autotroph (Methanococcus maripaludis S2) by Chi Hung Vo, Nishu Goyal, Iftekhar A Karimi and Markus Kraft in Microbiology Insights