Abstract
Nowadays, the emergence of multidrug-resistant bacterial strains initiates the urgent need for the elucidation of the new drug targets for the discovery of antimicrobial drugs. Filamenting temperature-sensitive mutant Z (FtsZ), a eukaryotic tubulin homolog, is a GTP-dependent prokaryotic cytoskeletal protein and is conserved among most bacterial strains. In vitro studies revealed that FtsZ self-assembles into dynamic protofilaments or bundles and forms a dynamic Z-ring at the center of the cell in vivo, leading to septation and consequent cell division. Speculations on the ability of FtsZ in the blockage of cell division make FtsZ a highly attractive target for developing novel antibiotics. Researchers have been working on synthetic molecules and natural products as inhibitors of FtsZ. Accumulating data suggest that FtsZ may provide the platform for the development of novel antibiotics. In this review, we summarize recent advances in the properties of FtsZ protein and bacterial cell division, as well as in the development of FtsZ inhibitors.
Keywords: FtsZ, multidrug resistance, cell division, antimicrobial, inhibitors, natural antimicrobial compounds
INTRODUCTION
The antibacterial resistance phenomenon has seen an increase to dangerous, unprecedented levels in practically all parts of the world over the past 20 years. While antibiotic resistance was once generally restricted to the hospitals and long-term care facilities, it has been emerging now in community settings, becoming one of the world’s most pressing concerns associated with the topic of public health [1]. In 2017, the Center for Disease Control (CDC) estimated 19,832 methicillin-resistant Staphylococcus aureus (MRSA) related deaths in the United States [2]. Based on the extent of threat, CDC categorizes these threats into three levels: “concerning” indicates the lowest level of threat, “serious” classifies as optimum level, while “urgent” falls in the highest level of resistance. As fatal diseases that rank as “serious” on the threat level, immediate action is required to ensure that the threats of MRSA and extensively drug-resistant and multidrug-resistant Mycobacterium tuberculosis (XDR/MDR-TB) do not upgrade to the highest level. According to the CDC, the urgent threats include carbapenem-resistant Acinetobacter, Candida auris, Clostridioides difficile, carbapenem-resistant Enterobacteriaceae (CRE), and drug-resistant Neisseria gonorrhoeae [3]. Not only pathogenic strains of bacteria have increased the multidrug resistance to antibiotic, but there is also a lack of advancement in the discovery of new antibiotics agents; hence, they have punctuated the need for novel chemotherapeutic strategies [4]. Conventionally, the cellular processes of transcription, folate biosynthesis, translation, cytoplasmic membrane, peptidoglycan biosynthesis, DNA replication, and bacterial structures among others were targeted by all known clinical antibiotics [5]. Now, urgency is placed on exploring new molecular targets and new strategies for antibiotic development; one of the most promising targets to develop novel antibiotics is bacterial cell division.
Cell division, or cytokinesis, is the essential process for the survival of all bacterial cells [6,7]. It is a three-step process in rod-shaped bacteria, comprising first of cell elongation, then septum formation, and finally cell division into two identical daughter cells. Filamenting temperature-sensitive mutant Z (FtsZ) is a key cytoskeletal cell division protein in most bacteria. At the division site, during the process of cell division, FtsZ forms ring-like structure (Z-ring), takes on the role as a scaffold for the recruitment of multiprotein complex (known as divisome), and may also generate the force that is vital for the viability of the cell [8-10]. The Z-ring, besides being very dynamic in vivo [11,12] as well as consisting of mainly FtsZ and dozens of other proteins, is also known to take part in the localization and stability of divisional proteins through a considerable number of protein-protein interactions [13]. Over the past 30 years, the bacterial cell division has been a substantial topic of discussion for the development of novel antimicrobials compounds [14,15].
The current review aims to discuss the biochemical properties of FtsZ proteins and assess what makes them an appropriate target for antibacterial drugs. Furthermore, we discuss the example of compounds that have been identified to interrupt FtsZ assembly and its interaction with other proteins. Correspondingly, we deliberate the significant challenges in developing drugs against this novel target.
PATHWAY OF BACTERIAL CYTOKINESIS
The process of cytokinesis has been widely studied in Bacillus subtilis and Escherichia coli, as well as in S. aureus [16-20], Streptococcus pneumonia [21-24], and M. tuberculosis [25-32]. In these bacterial cells, the cell envelope layer is invaginated by the divisome which, in turn, forms a septum between replicated chromosomes at the middle of the cell. Dozens of proteins recruited to the site of divisome at the cell center are involved in the process of septation. FtsZ, a bacterial tubulin homologue, is the center of the divisome (contractile ring, Z-ring). Many of these proteins in E. coli and B. subtilis, named filamentous temperature-sensitive (Fts), have been determined by screening for a temperature-sensitive mutant that grows normally and divides at the permissive temperature, but at the restrictive temperature, it continuously elongates without the division of cell. However, some proteins are also used, such as Div that stands for the division in B. subtilis, and are recognized through other methods [33]. More than two dozen proteins have been found as components or regulators of the divisome, and the number is still increasing. There might be different associated proteins in different bacteria. In E. coli, an abundance of proteins is essential for cell division, including FtsZ, FtsA, ZipA, FtsE, FtsX, FtsK, FtsB, FtsQ, FtsL, FtsW, FtsN, and FtsI. Studies revealed that E. coli has a consecutive, almost straight assembly of the protein’s pathway in the following direction: FtsZ [FtsA, ZapA, ZipA] (FtsE, FtsX), FtsK, FtsQ, (Fts B, FtsL), FtsW, FtsI, FtsN, AmiC, and EnvC [34-39].
At the early stage of the Z-ring assembly, FtsZ polymers anchor to the membrane by FtsA and ZipA at the future division site in E. coli. However, in B. subtilis, FtsA, ZapA, SepF, and perhaps EzrA are first localized to Z-ring of FtsZ proteins. Moreover, these are dependent on FtsZ for their localization to the site of cell division. The downstream proteins such as DivIC (FtsB from E. coli), DivIB (FtsQ from E. coli), FtsL, PBP2B (FtsI from E. coli), and possibly FtsW are recruited next for the localization of septum [31]. Although new studies of E. coli have shown that instead of arising as a sequence of binary interactions, the divisome assembly occurs as a complex network of protein-protein interactions [40], it is proposed that in these two organisms the assembly of the divisome complex could potentially be more parallel than once supposed.
FtsZ STRUCTURE AND FUNCTION
FtsZ is a structural homolog of eukaryotic cytoskeletal protein tubulin [41,42]. The sequence similarity of FtsZ in the majority of archaeal and bacterial species is about 40–50% [43]. Although the sequence resemblance between FtsZ and tubulin is <20%, they have a high structural identity [44]. In a single E. coli, there are approximately 5000 copies of FtsZ [45]. FtsZ polymerizes into protofilaments, and at the center of the dividing cell, they are aligned to form the Z-ring [10].
TREADMILLING DYNAMICS
Like eukaryotic tubulin, FtsZ binds and hydrolyzes guanosine-5’-triphosphate (GTP); on the other hand, in the presence of GTP, it forms a single-stranded protofilament or bundles instead of forming hollow microtubules. FtsZ from E. coli assembles into mostly single protofilaments that are highly dynamic, with an average of 200 nm in the presence of GTP in vitro [46]. The filaments will disassemble after GTP hydrolysis with a halftime of the subunits exchange of around 5–7 seconds [46-48], exhibiting a dynamic treadmilling action [49]. It is very consistent with the halftime of the exchange rate of the Z-ring in vivo, which is around 8–10 seconds [12]. Two studies in 2017 showed that FtsZ protofilaments demonstrate polarity and undergo treadmilling in both B. subtilis and E. coli cells [50,51]. During treadmilling process, subunits are selectively bound to one end and detached from the other end, and as a result, single FtsZ protofilaments or bundles travel as a unit around the cell circumference, following the irregular path of the ring. Although each treadmilling filament moment is processive, in the same cell, different FtsZ protofilaments can move in the opposite direction. Thus, it appears that each unit of protofilament has its own directionality [7,52]. Occurring inside protofilaments or by treadmilling at the ends of protofilaments, the turnover in the FtsZ subunit, regulated by FtsZ GTPase activity, is proportional to treadmilling [53]. It remains uncertain how single-stranded FtsZ protofilaments treadmill as they have innate polarity, and why one end gains while the other loses subunits. Wagstaff et al. suggested a possible mechanism; by examining the crystal structures of FtsZ, a correlation was identified between various FtsZ conformational states and its abilities to form polymers [18]. As treadmilling and polymerization of FtsZ are crucial for normal septal morphology, the inhibition of such activities should inhibit the process of cell division.
Recently, Monteiro et al. studied the effect of the compound PC190723, an FtsZ inhibitor, and DMPI (3-{1-[(2,3-dimethylphenyl) methyl]piperidin-4-yl}-1-methyl-2-pyridin-4-yl-1H-indole), the lipid II flippase MurJ inhibitor, on the Z-ring of S. aureus cells [54]. It has been demonstrated that the PC190723 compound inhibits FtsZ treadmilling and blocks the constriction of Z-ring only at initial stages, whereas the MurJ inhibitor DMPI has been shown to block the ring constriction at all stages. Furthermore, many small molecules and peptides, either synthetic or natural, target FtsZ polymerization, some having the potential to initiate antimicrobial activities. Based on the research that treadmilling motion of the FtsZ protein is essential for cell division, it may be possible to develop a novel drug that precisely prevents this motion, similar to how the chemotherapy drug taxol suppresses the moment of the cytoskeleton in malignant growth cells [55].
FTSZ AS A NOVEL ANTIBACTERIAL DRUG TARGET
Antibiotics are one of the most prominent weapons to use against microbial infections [56]. However, currently, the misuse of antibiotics creates selective pressure for the existence of resistant bacterial strains, and subsequently, several clinically used antibiotics, such as aminoglycosides, β-lactams, sulfonamides, tetracyclines, are becoming less effective against antibiotic-resistant bacteria [57]. Therefore, to control these problems, finding novel medicines is a necessity. Bacterial cell division proteins have several features; for this reason, they are suggested to be the right candidate for the antibacterial goal. First, FtsZ is a key protein for the viability of bacterial cells [58-60]. In addition, the cell division proteins in most of the bacteria are highly conserved but mainly lack or have limited sequence similarity in eukaryotic cells. The third benefit is that FtsZ is absent in higher eukaryotes, which asserts that inhibitors of FtsZ will not be toxic to human cells [41,61,62]. During the last few decades, the biological and biochemical properties of FtsZ caught the attention of scientists. A biological molecule that inhibits the GTPase activity or assembly of FtsZ can establish an active antibacterial agent.
Due to the structural similarity between FtsZ and tubulin, the tubulin inhibitors and their derivation could be used to screen anti-FtsZ compounds. Scientists at the Southern Research Institute screened the library of 200 2-alkoxycarbonylamin-o-pyridine compounds, which are inhibitors of tubulin polymerization, to inhibit FtsZ and control M. tuberculosis growth. With this approach, they found many compounds that have desirable properties [63]. Colchicine, a well-recognized tubulin inhibitor, along with the compound 2-alkoxycarbonylamin-o-pyridine SRI-3072 and SRI-7614, inhibited the polymerization and GTP hydrolysis of M. tuberculosis FtsZ in a dependent manner. Both components (SRI-3072 and SRI-7614) were effective against both drug-resistant and susceptible strain of M. tuberculosis. SRI-7614 inhibited both tubulin and FtsZ, while SRI-3072 only inhibited FtsZ polymerization. In addition, they showed that SRI-3072 could inhibit M. tuberculosis growth in the infected bone marrow macrophages of mice and was able to effectively enter intracellular parasite and even the host cell [29,63].
Even though FtsZ and tubulin show high structural similarity, there are some structural features of FtsZ that are different from tubulin [62]. First, the primary sequence of FtsZ differs from that of tubulin by more than 80% [53]. Second, all the subunits of FtsZ are similar, while the microtubules consist of two types of tubulin subunits (α and β). In addition, the lateral interaction of FtsZ protofilament changes from the longitudinal association of tubulin protofilaments, which leads to the development of an arch-shaped Z-ring [64]. These evidences are useful for the development of new antibacterial agents that will ultimately target the FtsZ but with few side effects. Actually, most of the tubulin inhibitors such as albendazole, nocodazole, thiabendazole, 3-methoxybenzamide, or paclitaxel show little inhibition of FtsZ polymerization or GTPase activity in vitro [65]. It is demonstrated that such an inhibitor can be determined to discriminate between FtsZ and tubulin.
NATURAL-PRODUCT SCREENING FOR FtsZ
The natural sources of medication have a key role in the treatment of human diseases. These traditional medicines have become an integral part of the primary healthcare organization [66]. It is reported that herbal plants have tremendous healing potentials and play a crucial role in traditional medicines [67]. Here, we describe natural products and their derivatives, which showed significant inhibition on FtsZ and divisome.
Alkaloids
Sanguinarine, known as the bloodroots, is a benzophenanthridine alkaloid derived from the rhizomes of Sanguinaria canadensis, having a wide variety of antimicrobial activities. In both E. coli and B. subtilis, it causes filamentation, which indicates a cell division defect. It is proposed that sanguinarine is not limited to inhibiting FtsZ assembly; it can also reduce FtsZ bundling by directly binding FtsZ. The cytotoxic ability of sanguinarine can be inferred due to this compound’s ability to depolymerizing microtubules [68,69].
Berberine is a planted alkaloid extracted from many species of Berberis. It has been reported that berberine inhibits E. coli FtsZ assembly and its GTPase activity in vitro [70]. It is predicted that the berberine binding spot overlaps with the GTP binding pocket in E. coli FtsZ. Given the availability of crystal structure of S. aureus FtsZ, a series of 9-phenoxyalkyl berberine derivatives were designed that could attach to the S. aureus FtsZ interdomain cleft. The value of minimum inhibitory concentration (MIC) was 2–8 µg/mL against S. aureus and 4–16 µg/mL against Enterococcus faecalis. Furthermore, the berberine analogs prevent the growth of Gram-negative organisms such as Klebsiella pneumoniae and E. coli with 32–128 µg/mL minimum inhibitory concentration (MIC) values [57]. Therefore, further study is needed to ascertain the antimicrobial action of berberine.
Phenylpropanoids and terpenoids
Phenylpropanoids are secondary metabolites of the plant that protect plants from predators and pathogens. Many of these compounds have antibiotic activities and display the inhibition of FtsZ activity [71]. Hemaiswarya et al. reported eight phenylpropanoids against E. coli FtsZ, demonstrating them as the inhibitors of FtsZ polymerization and GTPase activity. The light scattering technique showed a dose-dependent reduction in the FtsZ polymerization [72]. Phenylacrylamide possesses antibacterial activity against Streptococcus pyogenes and S. aureus and prevents their cell division [73], and Vanillin derivatives 3a and 4u were individually tested against M. tuberculosis FtsZ [74,75]. Scopoletin, an analog of coumarin related to quercetin and esculetin, inhibits B. subtilis FtsZ polymerization and its GTPase activity [76]. The cinnamaldehyde reduces E. coli FtsZ polymerization, inhibits its GTPase activity, and is non-lethal to red blood cells [77].
Curcumin is a natural compound extracted from Curcuma longa; for ages, it is known for both being used as a source of color as well as a flavoring agent in India and South Asia and retaining several biological properties, such as anticancer, antioxidant, and antibacterial [78]. It has been explored that curcumin binds to tubulin and has antiproliferative activity by disrupting microtubules [79]. With the help of computational docking program, cavity depth analysis, and molecular electrostatic potential (MEP), Kaur et al. recognized possible curcumin attaching sites in E. coli FtsZ and B. subtilis FtsZ, further verified through mutagenesis studies [80]. Curcumin raises GTPase activity and disrupts FtsZ polymerization, therefore, decreasing the steady-state length of polymer assembly [81]. Colchicine was extremely active against tubulin polymerization but demonstrated no effect on M. tuberculosis FtsZ polymerization [63]. Sulfoalkylresorcinol prevents the GTPase activity of FtsZ protein in vitro and shows antimicrobial activity against several bacterial organisms [82]. A synthetic derivative, n-undecyl gallate, was reported to disturb ZapA localization and possibly Z-ring development in Xanthomonas citri subsp. citri [83]. Totarol is a diterpenoid phenol isolated from Podocarpus totara that impedes the growth of many Gram-positive bacteria, including M. tuberculosis, which leads to the inhibition of FtsZ polymerization and prevention of its GTPase activity. It causes filamentation in B. subtilis and demonstrates a mislocalized Z-ring [84]. Initially, it was described that totarol has no activity against human tubulin; recently, it has been verified that totarol attaches to proteins and inhibits FtsZ by forming clumps [85]. Germacrene D-4-ol and germacrene D extracted from the oil of pine needles belong to the terpenoids family. They show antimicrobial activity toward many bacterial species. A docking model expects a binding position of the germacrene family to be a hydrophobic pocket in the FtsZ protein [86].
Polyphenols
In vitro, a high throughput screening assay was used, examining from a library of over 100,000 extracts of plants and microbial fermented broths, to discover a novel inhibitor of FtsZ; they found viriditoxin as an FtsZ inhibitor. Viriditoxin not only exhibits the inhibition of both FtsZ polymerization and GTPase activity in vivo and in vitro but also shows broad-spectrum activity against Gram-positive antibiotic-resistant and sensitive strains. Moreover, it is non-toxic to human cells, which makes it a fascinating compound for the uncovering of the drug [65]. Plumbagin is a naphthoquinone derivative of Plumbago zeylanica, being produced as a secondary metabolite in the roots [87]. It has been shown that plumbagin prevents FtsZ polymerization in a dose-dependent manner. The computational study revealed that the binding site of plumbagin is close to the C-terminal of B. subtilis FtsZ. Although it does not display any effect on the E. coli FtsZ, plumbagin obviously hinders the polymerization of B. subtilis FtsZ. This designates that there is a considerable variation in the structure of FtsZ proteins from different bacteria [88]. SA-011 was manufactured as an analog of plumbagin and prevented the GTPase activity of Bacillus anthracis FtsZ a bit stronger in comparison to berberine [89]. Resveratrol could inhibit Z-ring development and also suppress the FtsZ mRNA expression [90].
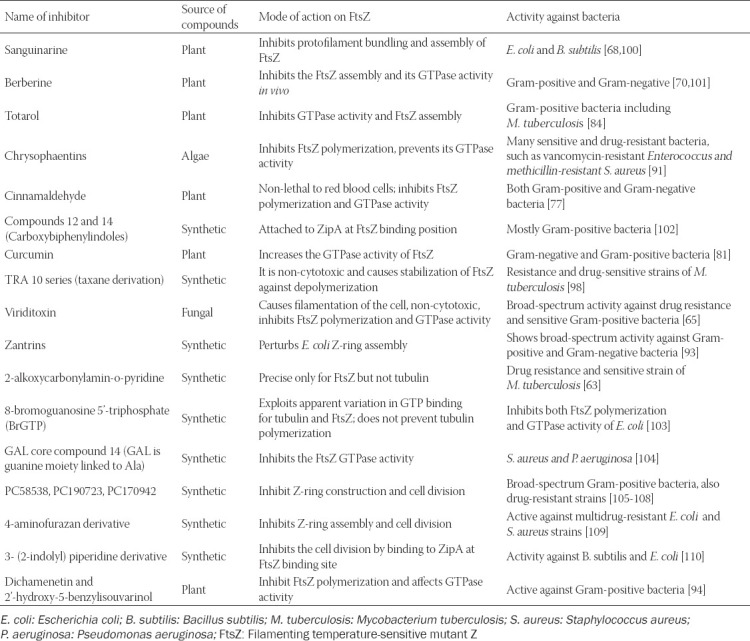
Chrysophaentins are polyoxygenated, polyhalogenated, and bisdiarylbutene ether macrocycle isolated from algae Chrysophaeumtaylori. Plaza et al. suggested that chrysophaentins inhibit both the GTPase activity and the polymerization of FtsZ, which results in activation against various bacteria strains such as Enterococcus faecium and S. aureus [91]. In vitro and in vivo, the compound was explored to be a competitive inhibitor of the FtsZ by attaching to the GTP binding site of the protein. Molecular docking experiments demonstrated that the chrysophaentins block a significant part of the GTP binding site of the proteins [91,92]. Five polyphenols known as Zantrins have been recognized. These compounds inhibit the GTPase activity of E. coli FtsZ [93], prevent the FtsZ polymerization, and disrupt E. coli Z-ring assembly. Zantrins have broad-spectrum antibacterial activity against both Gram-positive and Gram-negative pathogenic bacteria. Two more polyphenols extracted from natural sources have a parallel structure of Zantrin Z1, and likewise, the same antibacterial activity [94] (summary of the FtsZ targeting compounds is listed in Table 1).
TABLE 1.
Summary of the cell division protein inhibitors
Taxane derivative
Paclitaxel, a stabilizing agent of the microtubule that is originally isolated from the bark of the Pacific yew tree, is recognized as a very significant cancer chemotherapy drug [95]. To treat human tumors by targeting tubulin, the taxane derivative taxol (paclitaxel) was used [96]. About 120 taxane derivatives and many compounds with antituberculosis activities were recognized. Optimization of the particular compounds led to the detection of the taxane group, C-seco-taxane multidrug-resistant reversal agents (C-seco-TRAs): they are non-cytotoxic but active against both drug-resistant and sensitive strain of M. tuberculosis, having MIC99 values in the micromolar range [97,98]. Recently, for the inhibition of the vital mycobacterium FtsZ, a diverse range of commercial analogs and a novel series of sulindac derivatives were screened. Demonstrating studies recommend that these analogs attach to a precise area of M. tuberculosis FtsZ polymer which differs from eukaryotic tubulin [99]. However, due to the low structural similarity and sequence identity between the FtsZ and tubulin, recognition for this exact class of compound for M. tuberculosis FtsZ may be recommended.
CONCLUSION
Several bacterial cell division proteins with similar or novel functions have recently been explored to possess a key role in antibiotic discovery. The inhibition of their functions and ability to assemble as part of the division machinery consequently leads to the loss of viability in several bacterial species. Although the core cell division proteins are the same in most bacterial species examined thus far, various additional proteins exist that are sole to their genus; in this way, their inhibition may contribute to a species-specific antimicrobial agent. Our knowledge of the bacterial cell division proteins has recently prompted the exploration of a major player, FtsZ, as a novel antibacterial drug target, but this is still not enough. Therefore, novel or known antibacterial drug targets need to be further elucidated for the development of antibiotics.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (Grant No. 31970050) to Y.C. and Opening Foundation of Key Laboratory of Resource Biology and Biotechnology in Western China (Northwest University), Ministry of Education (ZSK2019004) to Y.C.
Footnotes
Conflict of interest statement: The authors declare no conflict of interests
REFERENCES
- 1.Martelli G, Giacomini D. Antibacterial and antioxidant activities for natural and synthetic dual-active compounds. Eur J Med Chem. 2018;158:91–105. doi: 10.1016/j.ejmech.2018.09.009. https://doi.org/10.1016/j.ejmech.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Kourtis AP, Hatfield K, Baggs J, Mu Y, See I, Epson E, et al. Vital signs: Epidemiology and recent trends in methicillin-resistant and in methicillin-susceptible Staphylococcus aureus bloodstream infections United States. MMWR Morb Mortal Wkly Rep. 2019;68(9):214–9. doi: 10.15585/mmwr.mm6809e1. https://doi.org/10.15585/mmwr.mm6809e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States. Atlanta, GA: U.S. Department of Health and Human Services, CDC; 2019. Available from: www.cdc.gov/DrugResistance/Biggest-Threats.html. http://dx.doi.org/10.15620/cdc:82532 . [Google Scholar]
- 4.Wright GD. Antibiotic resistance in the environment: A link to the clinic? Curr Opin Microbiol. 2010;13(5):589–94. doi: 10.1016/j.mib.2010.08.005. https://doi.org/10.1016/j.mib.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Walsh CT, Wencewicz TA. Prospects for new antibiotics: A molecule-centered perspective. J Antibiot (Tokyo) 2014;67(1):7–22. doi: 10.1038/ja.2013.49. https://doi.org/10.1038/ja.2013.49. [DOI] [PubMed] [Google Scholar]
- 6.Amos LA, Löwe J. Overview of the diverse roles of bacterial and archaeal cytoskeletons. Subcell Biochem. 2017;84:1–26. doi: 10.1007/978-3-319-53047-5_1. https://doi.org/10.1007/978-3-319-53047-5_1. [DOI] [PubMed] [Google Scholar]
- 7.Krupka M, Margolin W. Unite to divide: Oligomerization of tubulin and actin homologs regulates initiation of bacterial cell division. F1000Res. 2018;7:235. doi: 10.12688/f1000research.13504.1. https://doi.org/10.12688/f1000research.13504.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osawa M, Anderson DE, Erickson HP. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320(5877):792–4. doi: 10.1126/science.1154520. https://doi.org/10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osawa M, Anderson DE, Erickson HP. Curved FtsZ protofilaments generate bending forces on liposome membranes. EMBO J. 2009;28(22):3476–84. doi: 10.1038/emboj.2009.277. https://doi.org/10.1038/emboj.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erickson HP, Osawa M. FtsZ constriction force curved protofilaments bending membranes. Subcell Biochem. 2017;84:139–60. doi: 10.1007/978-3-319-53047-5_5. https://doi.org/10.1007/978-3-319-53047-5_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stricker J, Maddox P, Salmon ED, Erickson HP. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc Natl Acad Sci U S A. 2002;99(5):3171–5. doi: 10.1073/pnas.052595099. https://doi.org/10.1073/pnas.052595099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson DE, Gueiros-Filho FJ, Erickson HP. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J Bacteriol. 2004;186(17):5775–81. doi: 10.1128/JB.186.17.5775-5781.2004. https://doi.org/10.1128/jb.186.17.5775-5781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weiss DS. Last but not least: New insights into how FtsN triggers constriction during Escherichia coli cell division. Mol Microbiol. 2015;95(6):903–9. doi: 10.1111/mmi.12925. https://doi.org/10.1111/mmi.12925. [DOI] [PubMed] [Google Scholar]
- 14.Hurley KA, Santos TM, Nepomuceno GM, Huynh V, Shaw JT, Weibel DB. Targeting the bacterial division protein FtsZ. J Med Chem. 2016;59(15):6975–98. doi: 10.1021/acs.jmedchem.5b01098. https://doi.org/10.1021/acs.jmedchem.5b01098. [DOI] [PubMed] [Google Scholar]
- 15.Lock RL, Harry EJ. Cell-division inhibitors: New insights for future antibiotics. Nat Rev Drug Discov. 2008;7(4):324–38. doi: 10.1038/nrd2510. https://doi.org/10.1038/nrd2510. [DOI] [PubMed] [Google Scholar]
- 16.Eswara PJ, Brzozowski RS, Viola MG, Graham G, Spanoudis C, Trebino C, et al. An essential Staphylococcus aureus cell division protein directly regulates FtsZ dynamics. Elife. 2018;7:e38856. doi: 10.7554/eLife.38856. https://doi.org/10.7554/elife.38856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster TJ. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol Rev. 2017;41(3):430–49. doi: 10.1093/femsre/fux007. https://doi.org/10.1093/femsre/fux007. [DOI] [PubMed] [Google Scholar]
- 18.Wagstaff JM, Tsim M, Oliva MA, García-Sanchez A, Kureisaite-Ciziene D, Andreu JM, et al. A polymerization-associated structural switch in FtsZ that enables treadmilling of model filaments. mBio. 2017;8(3):e00254–17. doi: 10.1128/mBio.00254-17. https://doi.org/10.1128/mbio.00254-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujita J, Maeda Y, Nagao C, Tsuchiya Y, Miyazaki Y, Hirose M, et al. Crystal structure of FtsA from Staphylococcus aureus. FEBS Lett. 2014;588(10):1879–85. doi: 10.1016/j.febslet.2014.04.008. https://doi.org/10.1016/j.febslet.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 20.Matsui T, Han X, Yu J, Yao M, Tanaka I. Structural change in FtsZ induced by intermolecular interactions between bound GTP and the T7 loop. J Biol Chem. 2014;289(6):3501–9. doi: 10.1074/jbc.M113.514901. https://doi.org/10.1074/jbc.m113.514901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng Z, Zhang J, Xu D, Jiang YL, Zhou CZ, Chen Y. Multi-functional regulator MapZ controls both positioning and timing of FtsZ polymerization. Biochem J. 2019;476(10):1433–44. doi: 10.1042/BCJ20190138. https://doi.org/10.1042/bcj20190138. [DOI] [PubMed] [Google Scholar]
- 22.Perez AJ, Cesbron Y, Shaw SL, Bazan Villicana J, Tsui HT, Boersma MJ, et al. Movement dynamics of divisome proteins and PBP2x: FtsW in cells of Streptococcus pneumoniae. Proc Natl Acad Sci U S A. 2019;116(8):3211–20. doi: 10.1073/pnas.1816018116. https://doi.org/10.1073/pnas.1816018116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fleurie A, Lesterlin C, Manuse S, Zhao C, Cluzel C, Lavergne JP, et al. MapZ marks the division sites and positions FtsZ rings in Streptococcus pneumoniae. Nature. 2014;516(7530):259–62. doi: 10.1038/nature13966. https://doi.org/10.1038/nature13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salvarelli E, Krupka M, Rivas G, Mingorance J, Gómez-Puertas P, Alfonso C, et al. The cell division protein FtsZ from Streptococcus pneumoniae exhibits a GTPase activity delay. J Biol Chem. 2015;290(41):25081–9. doi: 10.1074/jbc.M115.650077. https://doi.org/10.1074/jbc.m115.650077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Hsin J, Zhao L, Cheng Y, Shang W, Huang KC, et al. FtsZ protofilaments use a hinge-opening mechanism for constrictive force generation. Science. 2013;341(6144):392–5. doi: 10.1126/science.1239248. https://doi.org/10.1126/science.1239248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das N, Dai J, Hung I, Rajagopalan MR, Zhou HX, Cross TA. Structure of CrgA, a cell division structural and regulatory protein from Mycobacterium tuberculosis in lipid bilayers. Proc Natl Acad Sci U S A. 2015;112(2):E119–26. doi: 10.1073/pnas.1415908112. https://doi.org/10.1073/pnas.1415908112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Popp D, Iwasa M, Erickson HP, Narita A, Maéda Y, Robinson RC. Suprastructures and dynamic properties of Mycobacterium tuberculosis FtsZ. J Biol Chem. 2010;285(15):11281–9. doi: 10.1074/jbc.M109.084079. https://doi.org/10.1074/jbc.m109.084079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y, Anderson DE, Rajagopalan M, Erickson HP. Assembly dynamics of Mycobacterium tuberculosis FtsZ. J Biol Chem. 2007;282(38):27736–43. doi: 10.1074/jbc.M703788200. https://doi.org/10.1074/jbc.M703788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White EL, Ross LJ, Reynolds RC, Seitz LE, Moore GD, Borhani DW. Slow polymerization of Mycobacterium tuberculosis FtsZ. J Bacteriol. 2000;182(14):4028–34. doi: 10.1128/jb.182.14.4028-4034.2000. https://doi.org/10.1128/jb.182.14.4028-4034.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kraxner KJ, Polen T, Baumgart M, Bott M. The conserved actinobacterial transcriptional regulator FtsR controls expression of FtsZ and further target genes and influences growth and cell division in Corynebacterium glutamicum. BMC Microbiol. 2019;19(1):179. doi: 10.1186/s12866-019-1553-0. https://doi.org/10.1186/s12866-019-1553-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Errington J, Daniel RA, Scheffers DJ. Cytokinesis in bacteria. Microbiol Mol Biol Rev. 2003;67(1):52–65. doi: 10.1128/MMBR.67.1.52-65.2003. https://doi.org/10.1128/mmbr.67.1.52-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Misra HS, Maurya GK, Chaudhary R, Misra CS. Interdependence of bacterial cell division and genome segregation and its potential in drug development. Microbiol Res. 2018;208:12–24. doi: 10.1016/j.micres.2017.12.013. https://doi.org/10.1016/j.micres.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 33.Harry E, Monahan L, Thompson L. Bacterial cell division: The mechanism and its precison. Int Rev Cytol. 2006;253:27–94. doi: 10.1016/S0074-7696(06)53002-5. https://doi.org/10.1016/s0074-7696(06)53002-5. [DOI] [PubMed] [Google Scholar]
- 34.Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354(6349):161–4. doi: 10.1038/354161a0. https://doi.org/10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 35.Lutkenhaus J, Pichoff S, Du S. Bacterial cytokinesis: From Z ring to divisome. Cytoskeleton (Hoboken) 2012;69(10):778–90. doi: 10.1002/cm.21054. https://doi.org/10.1002/cm.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Erickson HP, Anderson DE, Osawa M. FtsZ in bacterial cytokinesis: Cytoskeleton and force generator all in one. Microbiol Mol Biol Rev. 2010;74(4):504–28. doi: 10.1128/MMBR.00021-10. https://doi.org/10.1128/mmbr.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukherjee A, Dai K, Lutkenhaus J. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc Natl Acad Sci U S A. 1993;90(3):1053–7. doi: 10.1073/pnas.90.3.1053. https://doi.org/10.1073/pnas.90.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Boer P, Crossley R, Rothfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature. 1992;359(6392):254–6. doi: 10.1038/359254a0. https://doi.org/10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- 39.Du S, Lutkenhaus J. Assembly and activation of the Escherichia coli divisome. Mol Microbiol. 2017;105(2):177–87. doi: 10.1111/mmi.13696. https://doi.org/10.1111/mmi.13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goehring NW, Gonzalez MD, Beckwith J. Premature targeting of cell division proteins to midcell reveals hierarchies of protein interactions involved in divisome assembly. Mol Microbiol. 2006;61(1):33–45. doi: 10.1111/j.1365-2958.2006.05206.x. https://doi.org/10.1111/j.1365-2958.2006.05206.x. [DOI] [PubMed] [Google Scholar]
- 41.Löwe J, Amos LA. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391(6663):203–6. doi: 10.1038/34472. https://doi.org/10.1038/34472. [DOI] [PubMed] [Google Scholar]
- 42.Nogales E, Downing KH, Amos LA, Löwe J. Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol. 1998;5(6):451–8. doi: 10.1038/nsb0698-451. https://doi.org/10.1038/nsb0698-451. [DOI] [PubMed] [Google Scholar]
- 43.Vaughan S, Wickstead B, Gull K, Addinall SG. Molecular evolution of FtsZ protein sequences encoded within the genomes of Archaea Bacteria and Eukaryota . J Mol Evol. 2004;58(1):19–29. doi: 10.1007/s00239-003-2523-5. https://doi.org/10.1007/s00239-003-2523-5. [DOI] [PubMed] [Google Scholar]
- 44.Burns R. Synchronized division proteins. Nature. 1998;391:121–3. doi: 10.1038/34286. https://doi.org/10.1038/34286. [DOI] [PubMed] [Google Scholar]
- 45.Li GW, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157(3):624–35. doi: 10.1016/j.cell.2014.02.033. https://doi.org/10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y, Bjornson K, Redick SD, Erickson HP. A rapid fluorescence assay for FtsZ assembly indicates cooperative assembly with a dimer nucleus. Biophys J. 2005;88(1):505–14. doi: 10.1529/biophysj.104.044149. https://doi.org/10.1529/biophysj.104.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Y, Erickson HP. Rapid in vitro assembly dynamics and subunit turnover of FtsZ demonstrated by fluorescence resonance energy transfer. J Biol Chem. 2005;280(23):22549–54. doi: 10.1074/jbc.M500895200. https://doi.org/10.1074/jbc.m500895200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Y, Erickson HP. FtsZ filament dynamics at steady state: Subunit exchange with and without nucleotide hydrolysis. Biochemistry. 2009;48(28):6664–73. doi: 10.1021/bi8022653. https://doi.org/10.1021/bi8022653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loose M, Mitchison TJ. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat Cell Biol. 2014;16(1):38–46. doi: 10.1038/ncb2885. https://doi.org/10.1038/ncb2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bisson-Filho AW, Hsu YP, Squyres GR, Kuru E, Wu F, Jukes C, et al. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science. 2017;355(6326):739–43. doi: 10.1126/science.aak9973. https://doi.org/10.1126/science.aak9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang X, Lyu Z, Miguel A, McQuillen R, Huang KC, Xiao J. GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science. 2017;355(6326):744–7. doi: 10.1126/science.aak9995. https://doi.org/10.1126/science.aak9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schoenemann KM, Margolin W. Bacterial division: FtsZ treadmills to build a beautiful wall. Curr Biol. 2017;27(8):R301–3. doi: 10.1016/j.cub.2017.03.019. https://doi.org/10.1016/j.cub.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 53.Mukherjee A, Lutkenhaus J. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 1998;17(2):462–9. doi: 10.1093/emboj/17.2.462. https://doi.org/10.1093/emboj/17.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Monteiro JM, Pereira AR, Reichmann NT, Saraiva BM, Fernandes PB, Veiga H, et al. Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. Nature. 2018;554(7693):528–32. doi: 10.1038/nature25506. https://doi.org/10.1038/nature25506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yvon AM, Wadsworth P, Jordan MA. Taxol suppresses dynamics of individual microtubules in living human tumor cells. Mol Biol Cell. 1999;10(4):947–59. doi: 10.1091/mbc.10.4.947. https://doi.org/10.1091/mbc.10.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hemaiswarya S, Kruthiventi AK, Doble M. Synergism between natural products and antibiotics against infectious diseases. Phytomedicine. 2008;15(8):639–52. doi: 10.1016/j.phymed.2008.06.008. https://doi.org/10.1016/j.phymed.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 57.Sun N, Chan FY, Lu YJ, Neves MA, Lui HK, Wang Y, et al. Rational design of berberine-based FtsZ inhibitors with broad-spectrum antibacterial activity. PLoS One. 2014;9(5):e97514. doi: 10.1371/journal.pone.0097514. https://doi.org/10.1371/journal.pone.0097514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beall B, Lutkenhaus J. FtsZ in Bacillus subtilis is required for vegetative septation and for asymmetric septation during sporulation. Genes Dev. 1991;5(3):447–55. doi: 10.1101/gad.5.3.447. https://doi.org/10.1101/gad.5.3.447. [DOI] [PubMed] [Google Scholar]
- 59.Dai K, Lutkenhaus J. FtsZ is an essential cell division gene in Escherichia coli . J Bacteriol. 1991;173(11):3500–6. doi: 10.1128/jb.173.11.3500-3506.1991. https://doi.org/10.1128/jb.173.11.3500-3506.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pinho MG, Errington J. Dispersed mode of Staphylococcus aureus cell wall synthesis in the absence of the division machinery. Mol Microbiol. 2003;50(3):871–81. doi: 10.1046/j.1365-2958.2003.03719.x. https://doi.org/10.1046/j.1365-2958.2003.03719.x. [DOI] [PubMed] [Google Scholar]
- 61.Downing KH. Structural basis for the interaction of tubulin with proteins and drugs that affect microtubule dynamics. Annu Rev Cell Dev Biol. 2000;16:89–111. doi: 10.1146/annurev.cellbio.16.1.89. https://doi.org/10.1146/annurev.cellbio.16.1.89. [DOI] [PubMed] [Google Scholar]
- 62.Nogales E, Wolf SG, Downing KH. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 1998;391(6663):199–203. doi: 10.1038/34465. https://doi.org/10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 63.White EL, Suling WJ, Ross LJ, Seitz LE, Reynolds RC. 2-Alkoxycarbonylaminopyridines: Inhibitors of Mycobacterium tuberculosis FtsZ. J Antimicrob Chemother. 2002;50(1):111–4. doi: 10.1093/jac/dkf075. https://doi.org/10.1093/jac/dkf075. [DOI] [PubMed] [Google Scholar]
- 64.Leung AK, Lucile White E, Ross LJ, Reynolds RC, DeVito JA, Borhani DW. Structure of Mycobacterium tuberculosis FtsZ reveals unexpected, G protein-like conformational switches. J Mol Biol. 2004;342(3):953–70. doi: 10.1016/j.jmb.2004.07.061. https://doi.org/10.1016/j.jmb.2004.07.061. [DOI] [PubMed] [Google Scholar]
- 65.Wang J, Galgoci A, Kodali S, Herath KB, Jayasuriya H, Dorso K, et al. Discovery of a small molecule that inhibits cell division by blocking FtsZ, a novel therapeutic target of antibiotics. J Biol Chem. 2003;278(45):44424–8. doi: 10.1074/jbc.M307625200. https://doi.org/10.1074/jbc.m307625200. [DOI] [PubMed] [Google Scholar]
- 66.Verma S, Singh S. Current and future status of herbal medicines. Vet World. 2008;1(11):347. https://doi.org/10.5455/vetworld.2008.347-350. [Google Scholar]
- 67.Dubey N, Kumar R, Tripathi P. Global promotion of herbal medicine: India’s opportunity. Curr Sci. 2004;86(1):37–41. [Google Scholar]
- 68.Beuria TK, Santra MK, Panda D. Sanguinarine blocks cytokinesis in bacteria by inhibiting FtsZ assembly and bundling. Biochemistry. 2005;44(50):16584–93. doi: 10.1021/bi050767+. https://doi.org/10.1021/bi050767+ [DOI] [PubMed] [Google Scholar]
- 69.Lopus M, Panda D. The benzophenanthridine alkaloid sanguinarine perturbs microtubule assembly dynamics through tubulin binding. A possible mechanism for its antiproliferative activity. FEBS J. 2006;273(10):2139–50. doi: 10.1111/j.1742-4658.2006.05227.x. https://doi.org/10.1111/j.1742-4658.2006.05227.x. [DOI] [PubMed] [Google Scholar]
- 70.Domadia PN, Bhunia A, Sivaraman J, Swarup S, Dasgupta D. Berberine targets assembly of Escherichia coli cell division protein FtsZ. Biochemistry. 2008;47(10):3225–34. doi: 10.1021/bi7018546. https://doi.org/10.1021/bi7018546. [DOI] [PubMed] [Google Scholar]
- 71.Puupponen-Pimiä R, Nohynek L, Meier C, Kähkönen M, Heinonen M, Hopia A, et al. Antimicrobial properties of phenolic compounds from berries. J Appl Microbiol. 2001;90(4):494–507. doi: 10.1046/j.1365-2672.2001.01271.x. https://doi.org/10.1046/j.1365-2672.2001.01271.x. [DOI] [PubMed] [Google Scholar]
- 72.Hemaiswarya S, Soudaminikkutty R, Narasumani ML, Doble M. Phenylpropanoids inhibit protofilament formation of Escherichia coli cell division protein FtsZ. J Med Microbiol. 2011;60(Pt 9):1317–25. doi: 10.1099/jmm.0.030536-0. https://doi.org/10.1099/jmm.0.030536-0. [DOI] [PubMed] [Google Scholar]
- 73.Li X, Sheng J, Song D, Guo L, Ma S. Phenylacrylamides as novel FtsZ-targeted potential antimicrobials. Lett Drug Des Discov. 2015;12(3):234–40. https://doi.org/10.2174/1570180811666141009235409. [Google Scholar]
- 74.Sun J, Yin Y, Sheng GH, Yang ZB, Zhu HL. Synthesis, molecular modeling and structural characterization of vanillin derivatives as antimicrobial agents. J Mol Struct. 2013;1039(8):214–8. https://doi.org/10.1016/j.molstruc.2013.01.071. [Google Scholar]
- 75.Sun J, Li MH, Wang XY, Zhang Y, Yuan RJ, Liu HY, et al. Vanillin derivatives as the selective small molecule inhibitors of FtsZ. Med Chem Res. 2014;23:2985–94. https://doi.org/10.1007/s00044-013-0886-8. [Google Scholar]
- 76.Duggirala S, Nankar RP, Rajendran S, Doble M. Phytochemicals as inhibitors of bacterial cell division protein FtsZ: Coumarins are promising candidates. Appl Biochem Biotechnol. 2014;174(1):283–96. doi: 10.1007/s12010-014-1056-2. https://doi.org/10.1007/s12010-014-1056-2. [DOI] [PubMed] [Google Scholar]
- 77.Domadia P, Swarup S, Bhunia A, Sivaraman J, Dasgupta D. Inhibition of bacterial cell division protein FtsZ by cinnamaldehyde. Biochem Pharmacol. 2007;74(6):831–40. doi: 10.1016/j.bcp.2007.06.029. https://doi.org/10.1016/j.bcp.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 78.Kocaadam B, Şanlier N. Curcumin, an active component of turmeric (Curcuma longa), and its effects on health. Crit Rev Food Sci Nutr. 2017;57(13):2889–95. doi: 10.1080/10408398.2015.1077195. https://doi.org/10.1080/10408398.2015.1077195. [DOI] [PubMed] [Google Scholar]
- 79.Gupta KK, Bharne SS, Rathinasamy K, Naik NR, Panda D. Dietary antioxidant curcumin inhibits microtubule assembly through tubulin binding. FEBS J. 2006;273(23):5320–32. doi: 10.1111/j.1742-4658.2006.05525.x. https://doi.org/10.1111/j.1742-4658.2006.05525.x. [DOI] [PubMed] [Google Scholar]
- 80.Kaur S, Modi NH, Panda D, Roy N. Probing the binding site of curcumin in Escherichia coli and Bacillus subtilis FtsZ a structural insight to unveil antibacterial activity of curcumin. Eur J Med Chem. 2010;45(9):4209–14. doi: 10.1016/j.ejmech.2010.06.015. https://doi.org/10.1016/j.ejmech.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 81.Rai D, Singh JK, Roy N, Panda D. Curcumin inhibits FtsZ assembly: An attractive mechanism for its antibacterial activity. Biochem J. 2008;410(1):147–55. doi: 10.1042/BJ20070891. https://doi.org/10.1042/bj20070891. [DOI] [PubMed] [Google Scholar]
- 82.Kanoh K, Adachi K, Matsuda S, Shizuri Y, Yasumoto K, Kusumi T, et al. New sulfoalkylresorcinol from marine-derived fungus Zygosporium sp. KNC52. J Antibiot (Tokyo) 2008;61(3):192–4. doi: 10.1038/ja.2008.29. https://doi.org/10.1002/chin.200839206. [DOI] [PubMed] [Google Scholar]
- 83.Silva IC, Regasini LO, Petrônio MS, Silva DH, Bolzani VS, Belasque J, Jr, et al. Antibacterial activity of alkyl gallates against Xanthomonas citri subsp citri . J Bacteriol. 2013;195(1):85–94. doi: 10.1128/JB.01442-12. https://doi.org/10.1128/jb.01442-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jaiswal R, Beuria TK, Mohan R, Mahajan SK, Panda D. Totarol inhibits bacterial cytokinesis by perturbing the assembly dynamics of FtsZ. Biochemistry. 2007;46(14):4211–20. doi: 10.1021/bi602573e. https://doi.org/10.1021/bi602573e. [DOI] [PubMed] [Google Scholar]
- 85.Anderson DE, Kim MB, Moore JT, O’Brien TE, Sorto NA, Grove CI, et al. Comparison of small molecule inhibitors of the bacterial cell division protein FtsZ and identification of a reliable cross-species inhibitor. ACS Chem Biol. 2012;7(11):1918–28. doi: 10.1021/cb300340j. https://doi.org/10.1021/cb300340j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sarac Z, Matejić JS, Stojanović-Radić ZZ, Veselinović JB, Džamić AM, Bojović S, et al. Biological activity of Pinus nigra terpenes - evaluation of FtsZ inhibition by selected compounds as contribution to their antimicrobial activity. Comput Biol Med. 2014;54:72–8. doi: 10.1016/j.compbiomed.2014.08.022. https://doi.org/10.1016/j.compbiomed.2014.08.022. [DOI] [PubMed] [Google Scholar]
- 87.Jamal MS, Parveen S, Beg MA, Suhail M, Chaudhary AG, Damanhouri GA, et al. Anticancer compound plumbagin and its molecular targets: A structural insight into the inhibitory mechanisms using computational approaches. PLoS One. 2014;9(2):e87309. doi: 10.1371/journal.pone.0087309. https://doi.org/10.1371/journal.pone.0087309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bhattacharya A, Jindal B, Singh P, Datta A, Panda D. Plumbagin inhibits cytokinesis in Bacillus subtilis by inhibiting FtsZ assembly a mechanistic study of its antibacterial activity. FEBS J. 2013;280(18):4585–99. doi: 10.1111/febs.12429. https://doi.org/10.1111/febs.12429. [DOI] [PubMed] [Google Scholar]
- 89.Park HC, Gedi V, Cho JH, Hyun JW, Lee KJ, Kang J, et al. Characterization and in vitro inhibition studies of Bacillus anthracis FtsZ: A potential antibacterial target. Appl Biochem Biotechnol. 2014;172(6):3263–70. doi: 10.1007/s12010-014-0752-2. https://doi.org/10.1007/s12010-014-0752-2. [DOI] [PubMed] [Google Scholar]
- 90.Hwang D, Lim YH. Resveratrol antibacterial activity against Escherichia coli is mediated by Z-ring formation inhibition via suppression of FtsZ expression. Sci Rep. 2015;5:10029. doi: 10.1038/srep10029. https://doi.org/10.1038/srep10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Plaza A, Keffer JL, Bifulco G, Lloyd JR, Bewley CA. Chrysophaentins A-H, antibacterial bisdiarylbutene macrocycles that inhibit the bacterial cell division protein FtsZ. J Am Chem Soc. 2010;132(26):9069–77. doi: 10.1021/ja102100h. https://doi.org/10.1002/chin.201052182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Keffer JL, Huecas S, Hammill JT, Wipf P, Andreu JM, Bewley CA. Chrysophaentins are competitive inhibitors of FtsZ and inhibit Z-ring formation in live bacteria. Bioorg Med Chem. 2013;21(18):5673–8. doi: 10.1016/j.bmc.2013.07.033. https://doi.org/10.1016/j.bmc.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Margalit DN, Romberg L, Mets RB, Hebert AM, Mitchison TJ, Kirschner MW, et al. Targeting cell division: Small-molecule inhibitors of FtsZ GTPase perturb cytokinetic ring assembly and induce bacterial lethality. Proc Natl Acad Sci U S A. 2004;101(32):11821–6. doi: 10.1073/pnas.0404439101. https://doi.org/10.1073/pnas.0404439101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Urgaonkar S, La Pierre HS, Meir I, Lund H, RayChaudhuri D, Shaw JT. Synthesis of antimicrobial natural products targeting FtsZ: (+/-)-dichamanetin and (+/-)-2’ “-hydroxy-5”-benzylisouvarinol-B. Org Lett. 2005;7(25):5609–12. doi: 10.1021/ol052269z. https://doi.org/10.1021/ol052269z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Khanna C, Rosenberg M, Vail DM. A review of paclitaxel and novel formulations including those suitable for use in dogs. J Vet Intern Med. 2015;29(4):1006–12. doi: 10.1111/jvim.12596. https://doi.org/10.1111/jvim.12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.St George M, Ayoub AT, Banerjee A, Churchill CD, Winter P, Klobukowski M, et al. Designing and testing of novel taxanes to probe the highly complex mechanisms by which taxanes bind to microtubules and cause cytotoxicity to cancer cells. PLoS One. 2015;10(6):e0129168. doi: 10.1371/journal.pone.0129168. https://doi.org/10.1371/journal.pone.0129168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huang Q, Tonge PJ, Slayden RA, Kirikae T, Ojima I, Fts Z. A novel target for tuberculosis drug discovery. Curr Top Med Chem. 2007;7(5):527–43. doi: 10.2174/156802607780059790. https://doi.org/10.2174/156802607780059790. [DOI] [PubMed] [Google Scholar]
- 98.Huang Q, Kirikae F, Kirikae T, Pepe A, Amin A, Respicio L, et al. Targeting FtsZ for antituberculosis drug discovery: Noncytotoxic taxanes as novel antituberculosis agents. J Med Chem. 2006;49(2):463–6. doi: 10.1021/jm050920y. https://doi.org/10.1021/jm050920y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mathew B, Hobrath JV, Ross L, Connelly MC, Lofton H, Rajagopalan M, et al. Screening and development of new inhibitors of FtsZ from M. tuberculosis . PLoS One. 2016;11(10):e0164100. doi: 10.1371/journal.pone.0164100. https://doi.org/10.1371/journal.pone.0164100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Godowski KC. Antimicrobial action of sanguinarine. J Clin Dent. 1989;1(4):96–101. [PubMed] [Google Scholar]
- 101.Boberek JM, Stach J, Good L. Genetic evidence for inhibition of bacterial division protein FtsZ by berberine. PLoS One. 2010;5(10):e13745. doi: 10.1371/journal.pone.0013745. https://doi.org/10.1371/journal.pone.0013745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sutherland AG, Alvarez J, Ding W, Foreman KW, Kenny CH, Labthavikul P, et al. Structure-based design of carboxybiphenylindole inhibitors of the ZipA-FtsZ interaction. Org Biomol Chem. 2003;1(23):4138–40. doi: 10.1039/b312016c. https://doi.org/10.1039/b312016c. [DOI] [PubMed] [Google Scholar]
- 103.Läppchen T, Hartog AF, Pinas VA, Koomen GJ, den Blaauwen T. GTP analogue inhibits polymerization and GTPase activity of the bacterial protein FtsZ without affecting its eukaryotic homologue tubulin. Biochemistry. 2005;44(21):7879–84. doi: 10.1021/bi047297o. https://doi.org/10.1021/bi047297o. [DOI] [PubMed] [Google Scholar]
- 104.Paradis-Bleau C, Beaumont M, Sanschagrin F, Voyer N, Levesque RC. Parallel solid synthesis of inhibitors of the essential cell division FtsZ enzyme as a new potential class of antibacterials. Bioorg Med Chem. 2007;15(3):1330–40. doi: 10.1016/j.bmc.2006.11.015. https://doi.org/10.1016/j.bmc.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 105.Stokes NR, Sievers J, Barker S, Bennett JM, Brown DR, Collins I, et al. Novel inhibitors of bacterial cytokinesis identified by a cell-based antibiotic screening assay. J Biol Chem. 2005;280(48):39709–15. doi: 10.1074/jbc.M506741200. https://doi.org/10.1074/jbc.m506741200. [DOI] [PubMed] [Google Scholar]
- 106.Haydon DJ, Stokes NR, Ure R, Galbraith G, Bennett JM, Brown DR, et al. An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science. 2008;321(5896):1673–5. doi: 10.1126/science.1159961. https://doi.org/10.1126/science.1159961. [DOI] [PubMed] [Google Scholar]
- 107.Haydon DJ, Bennett JM, Brown D, Collins I, Galbraith G, Lancett P, et al. Creating an antibacterial with in vivo efficacy: Synthesis and characterization of potent inhibitors of the bacterial cell division protein FtsZ with improved pharmaceutical properties. J Med Chem. 2010;53(10):3927–36. doi: 10.1021/jm9016366. https://doi.org/10.1021/jm9016366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Adams DW, Wu LJ, Czaplewski LG, Errington J. Multiple effects of benzamide antibiotics on FtsZ function. Mol Microbiol. 2011;80(1):68–84. doi: 10.1111/j.1365-2958.2011.07559.x. https://doi.org/10.1111/j.1365-2958.2011.07559.x. [DOI] [PubMed] [Google Scholar]
- 109.Ito H, Ura A, Oyamada Y, Tanitame A, Yoshida H, Yamada S, et al. A 4-aminofurazan derivative-A189-inhibits assembly of bacterial cell division protein FtsZ in vitro and in vivo . Microbiol Immunol. 2006;50(10):759–64. doi: 10.1111/j.1348-0421.2006.tb03851.x. https://doi.org/10.1111/j.1348-0421.2006.tb03851.x. [DOI] [PubMed] [Google Scholar]
- 110.Jennings LD, Foreman KW, Rush TS, 3rd, Tsao DH, Mosyak L, Li Y, et al. Design and synthesis of indolo[2,3-a]quinolizin-7-one inhibitors of the ZipA-FtsZ interaction. Bioorg Med Chem Lett. 2004;14(6):1427–31. doi: 10.1016/j.bmcl.2004.01.028. https://doi.org/10.1016/j.bmcl.2004.01.028. [DOI] [PubMed] [Google Scholar]


