Abstract
A 5-day in vivo rat model was evaluated as an approach to estimate chemical exposures that may pose minimal risk by comparing benchmark dose (BMD) values for transcriptional changes in the liver and kidney to BMD values for toxicological endpoints from traditional toxicity studies. Eighteen chemicals, most having been tested by the National Toxicology Program in 2-year bioassays, were evaluated. Some of these chemicals are potent hepatotoxicants (eg, DE71, PFOA, and furan) in rodents, some exhibit toxicity but have minimal hepatic effects (eg, acrylamide and α,β-thujone), and some exhibit little overt toxicity (eg, ginseng and milk thistle extract) based on traditional toxicological evaluations. Male Sprague Dawley rats were exposed once daily for 5 consecutive days by oral gavage to 8–10 dose levels for each chemical. Liver and kidney were collected 24 h after the final exposure and total RNA was assayed using high-throughput transcriptomics (HTT) with the rat S1500+ platform. HTT data were analyzed using BMD Express 2 to determine transcriptional gene set BMD values. BMDS was used to determine BMD values for histopathological effects from chronic or subchronic toxicity studies. For many of the chemicals, the lowest transcriptional BMDs from the 5-day assays were within a factor of 5 of the lowest histopathological BMDs from the toxicity studies. These data suggest that using HTT in a 5-day in vivo rat model provides reasonable estimates of BMD values for traditional apical endpoints. This approach may be useful to prioritize chemicals for further testing while providing actionable data in a timely and cost-effective manner.
Keywords: 5-day HTT model, high-throughput transcriptomics, benchmark dose, apical toxicity, hepatotoxicity
The National Toxicology Program (NTP) generates toxicological information on a broad range of substances with poorly characterized hazards. Traditionally, potential hazards have been evaluated in 28- or 90-day subchronic toxicity studies, 2-year chronic toxicity and carcinogenicity studies, or developmental toxicity studies in rats and mice (https://ntp.niehs.nih.gov/publications/index.html; last accessed June 17, 2020). Risk assessors use data from these rodent toxicity studies to estimate chemical exposure levels that may pose minimal human risk. However, these studies typically are expensive and require a lengthy time period before the reporting phase. Since its founding in 1978, the NTP has evaluated more than 2800 environmental substances, of which more than 600 have been tested in chronic toxicity and carcinogenicity studies. However, this number is small compared with the more than 50 000 chemicals submitted to the U.S. EPA in its Toxic Substances Control Act (TSCA) program since 1979 (https://www.epa.gov/reviewing-new-chemicals-under-toxic-substances-control-act-tsca/statistics-new-chemicals-review#stats; last accessed June 17, 2020). For toxicological evaluations to keep pace with new chemical entities submitted to the U.S. EPA and other federal and state agencies, alternative approaches, in the form of shorter in vivo and/or in vitro models, are needed to characterize the dose-response relationships for chemical-induced apical effects.
The intent of the Tox21 program has been to address the challenge of limited throughput with traditional toxicity testing by providing high-throughput biological response information focusing on assays evaluating single pathways, such as estrogen receptor activation (Judson et al., 2015; Kleinstreuer et al., 2017). Although this approach provided high chemical throughput, the biological space probed (eg, range of biological responses of cells and tissues) was limited. High-throughput transcriptomics (HTT) has been proposed as an alternative timely and cost-effective screening approach that covers a large biological response space to chemical exposures (Ramaiahgari et al., 2019). The NTP is considering the use of 5-day exposure studies, which include HTT as an alternative data stream for understanding a quantitative estimate of hazard, as a bioactivity-based bridge between traditional apical endpoints and HTT data generated from in vitro assays. In these 5-day assays, animals are exposed to test articles via exposure routes relevant to human exposure for 5 consecutive days and humanely euthanized approximately 24 h after the last exposure. Select tissues are removed and isolated total RNA is evaluated using HTT to test whether short-term in vivo exposures can provide a rapid estimate of the benchmark dose (BMD) values for traditional apical endpoints, as well as provide a broad screen for interpretable biological activity. Previous studies using a 5-day assay in rats have shown that the lowest transcriptional BMD values in specific target tissues (bladder, liver, and thyroid) correlated well with the lowest noncancer apical BMD values within the same target tissues (Thomas et al., 2013).
The objective of this study was to further evaluate the 5-day HTT model in rats as a predictive tool using a set of 18 well-studied chemicals. The chemicals selected all have established apical toxicity data in mice and/or rats (mostly from historical NTP chronic or subchronic toxicity studies). The results presented herein are a proof of concept study to determine whether the transcriptional BMD values for liver and kidney (as “sentinel” tissues) after 5 days of oral chemical exposure in male rats can estimate the lowest apical (histopathological) BMD values determined from chronic or subchronic toxicity studies.
Previous in vivo studies assessing transcriptional BMD values and their relationship to apical BMD values were performed such that the in-life portion of the studies tightly mirrored the corresponding guideline toxicity studies (ie, by matching dose levels, vehicle, exposure route, and strain). Furthermore, the studies evaluated transcriptional changes in the specific target organs as identified in the guideline studies. However, in a real-world scenario where a tiered testing approach is being employed, such information (eg, target organ) will not be available. Also, due to the short-term nature of the studies, many chemicals will likely not be formulated in feed or water due to potential palatability issues, hence the likely default administration route will be oral gavage. With such a scenario in mind, we wanted to test the 5-day approach to determine if transcriptional BMD values exhibited forward predictability with regards to apical (histopathological) BMD values. More specifically, (1) test the hypothesis that liver and kidney, being common target organs for toxicity with high levels of chemical exposure and systemic integration, would provide a transcriptional gene set (ie, a set of genes that function in a coordinated way to carry out specific molecular biological processes) BMD value similar to the apical BMD value independent of the target organ; and (2) test the hypothesis that male rat liver and kidney transcriptional gene set BMD values could estimate apical BMD values from other sex and species (female rats, male, and female mice).
MATERIALS AND METHODS
Rationale for chemical selection for the 5-day assays
The chemicals (Table 1) and chemical concentrations and vehicles (Table 2) used in this study were selected based on the following criteria. (1) The chemical was previously evaluated in repeated dosing studies. All selected chemicals were tested in 90-day subchronic or 2-year chronic studies except FEN (Table 3). (2) The chemical was administered by the oral route (gavage, drinking water, or feed). (3) Chemical-related increased incidences of non-neoplastic/neoplastic histopathological effects were observed in liver and/or other tissues in the historical chronic or subchronic toxicity studies. Liver and kidney histopathology were a focus as these organs represent “sentinel” tissues often affected by chemical exposures. The 18 chemicals selected for the 5-day assays in male rats are shown in Tables 1 and 2 and were further divided into 3 categories based on the presence/absence of liver toxicity in male rats (since this was the sex/species used for the 5-day assays) as previously observed in the chronic or subchronic toxicity studies (Table 3). (1) DE71 (NTP, 589, 2016), FUR (Von Tungeln et al., 2017), MET (NTP, 491, 2000), COU (NTP, 422, 1993a), TCAB (NTP, 558, 2010a), PUL (NTP, 563, 2011d), DEHP (NTP, 217, 1982), PFOA (NTP, 598, 2020), FEN, and HCB (Arnold et al., 1985) are hepatotoxic (most are carcinogenic) in male rats. TCPP is also a liver toxicant but data shown from the NTP 90-day subchronic study are only in mice (due to perinatal exposure only in rats). (2) THU (NTP, 570, 2011a), ACR (NTP, 575, 2012), BDCA (NTP, 583, 2015), and EE2 (NTP, 548, 2010b) exhibit minimal/no hepatotoxicity (only basophilic and/or eosinophilic foci) in male rats but produce adverse effects in other tissues. TBBPA was not toxic in male rats but caused tumors and non-neoplastic effects in the uterus of female rats in the NTP 2-year chronic study (NTP, 587, 2014). (3) GIN (NTP, 567, 2011b) and MTE (NTP, 565, 2011c) exhibit little overt toxicity to the liver or other tissues in male rats and were used as negative controls.
Table 1.
Chemicals Selected for 5-day Assays in Male Sprague Dawley Rats
| Chemical (Abbreviation) | CAS Number | Supplier | Lot Number | % Purity |
|---|---|---|---|---|
| Acrylamide (ACR) | 79-06-1 | Sigma-Aldrich (St Louis, Missouri) | BCBR0859V | 99 |
| Bromodichloroacetic acid (BDCA) | 71133-14-7 | Chemfinet (Tarrytown, New York) | NJ 87-90/9/2005 | 93.6 |
| Coumarin (COU) | 91-64-5 | Sigma-Aldrich | MKBX9839V | 100 |
| Di(2-ethylhexyl) phthalate (DEHP) | 117-81-7 | Sigma-Aldrich | 01514TH | 99.7 |
| Pentabromodiphenyl ether mixture (DE71) | 32534-81-9 | Great Lakes Chemical Corp (West Lafayette, Indiana) | 2550OA30A | 101.8 |
| Ethinyl estradiol (EE2) | 57-63-6 | Toronto Research Chemicals, Inc (North York, Ontario) | 16-XJZ-61-1 | 99 |
| Fenofibrate (FEN) | 49562-28-9 | Gojira Fine Chemicals LLC (Bedford Heights, Ohio) | 091722 | 100.3 |
| Furan (FUR) | 110-00-9 | Sigma-Aldrich | SHBG4510V | 100 |
| Ginseng (GIN) | 50647-08-0 | Plus Pharma, Inc (Vista, California) | 3031978 | NA |
| Hexachlorobenzene (HCB) | 118-74-1 | Sigma-Aldrich | 03915CU | 99 |
| Methyl eugenol (MET) | 93-15-2 | Sigma-Aldrich | MKBX2654V | 98.3 |
| Milk thistle extract (MTE) | 84604-20-6 | Indena USA, Inc (Seattle, Washington) | 27691/M6 | NA |
| Perfluorooctanoic acid (PFOA) | 335-67-1 | Sigma-Aldrich | 03427TH | 93.7 |
| Pulegone (PUL) | 89-82-7 | TCI America (Portland, Oregon) | OGI01 | 96 |
| Tetrabromobisphenol A (TBBPA) | 79-94-7 | Albemarle Corporation (Baton Rouge, Louisiana) | M03207KA | 99 |
| 3,3′,4,4′-Tetrachloroazobenzene (TCAB) | 14047-09-7 | AccuStandard (New Haven, Connecticut) | 10009-52-01 | 99.8 |
| Tris(chloropropyl) phosphate (TCPP) | 13674-84-5 | Albemarle Corporation | M072911NP | 97 |
| α,β-Thujone (THU) | 76231-76-0 | TCI America | 5J7FG | 79.9 |
Abbreviation: NA, not applicable.
Table 2.
Chemical Concentrations and Vehicles Selected for 5-day Assays in Male Sprague Dawley Rats
| Chemical | Gavage Vehicle | Concentrations Testeda,b |
|---|---|---|
| ACR | Deionized water | 0, 0.078, 0.156, 0.3125, 0.625, 1.25, 2.5, 5, 10 |
| BDCA | Deionized water | 0, 1.25, 2.5, 5, 10, 20, 40, 80, 160 |
| COU | Corn oil | 0, 3.125, 6.25, 12.5, 25, 50, 100, 200, 400 |
| DEHP | Corn oil | 0, 8, 16, 31.25, 62.5, 125, 250, 500, 1000 |
| DE71 | Corn oil | 0, 0.38, 0.75, 1.5, 3, 15, 50, 100, 200, 500 |
| EE2 | Corn oil | 0, 0.02, 0.067, 0.2, 0.6, 1.8, 5.4, 16.2, 48.6 |
| FEN | 0.5% aqueous methylcellulosec | 0, 8, 16, 31.25, 62.5, 125, 250, 500, 1000 |
| FUR | Corn oil | 0, 0.125, 0.25, 0.5, 1, 2, 4, 8, 16 |
| GIN | Deionized water | 0, 39.1, 78.125, 156.25, 312.5, 625, 1250, 2500, 5000 |
| HCB | Corn oil | 0, 0.004, 0.015, 0.0625, 0.25, 1, 4, 16, 64 |
| MET | 0.5% aqueous methylcellulose | 0, 4.625, 9.25, 18.5, 37, 75, 150, 300, 600 |
| MTE | Corn oil | 0, 39.1, 78.125, 156.25, 312.5, 625, 937.5, 1250, 1750 |
| PFOA | 2% Tween 80c | 0, 0.156, 0.3125, 0.625, 1.25, 2.5, 5, 10, 20 |
| PUL | Corn oil | 0, 2.4, 4.7, 9.4, 18.75, 37.5, 75, 150, 300 |
| TBBPA | Corn oil | 0, 4, 8, 16, 31.25, 62.5, 125, 250, 500, 1000, 2000 |
| TCAB | Corn oil: acetone (99:1) | 0, 0.1, 0.3, 1, 3, 10, 30, 100, 200, 400 |
| TCPP | 0.5% aqueous methylcellulose | 0, 18.75, 37.5, 75, 150, 300, 600, 1000, 2000 |
| THU | 0.5% aqueous methylcellulose | 0, 1.5, 3, 6.25, 12.5, 25, 50, 100, 200 |
mg/kg Except ethinyl estradiol (µg/kg).
All formulations were analyzed using qualified or validated analytical methods and, in general, were within 15% of the target concentration.
In deionized water.
Table 3.
Chronic or Subchronic Toxicity Studies for Selected Chemicals
| Chemical | Study Type | Hepatotoxic | Rat Strain | Route/Vehicle | Concentrations Tested a |
|---|---|---|---|---|---|
| ACR | 2-year (NTP TR 575) | No (otherb) | F344/N | Drinking water | 0, 0.33, 0.66, 1.32, 2.71 |
| BDCA | 2-year (NTP TR 583) | No (other) | F344/NTac | Drinking water | 0, 11, 21, 43 |
| COU | 2-year (NTP TR 422) | Yes | F344/N | Gavage/corn oil | 0, 25, 50, 100 |
| DEHP | 2-year (NTP TR 217) | Yes | F344 | Feed/NA | 0, 322, 674 |
| DE71 | 2-year (NTP TR 589) | Yes | Wistar Han | Gavage/corn oil | 0, 3, 15, 50 |
| EE2 | 2-year (NTP TR 548) | No (other) | NCTR SD (F1C) | Feed/NA | 0, 0.15, 0.6, 3.3 |
| FEN | 29-day (TG-GATEs)c | Yes | Sprague Dawley | Gavage/0.5% aqueous methylcellulosed | 0, 10, 100, 1000 |
| FUR | 2-year (Von Tungeln et al., 2017) | Yes | F344/N | Gavage/corn oil | 0, 0.02, 0.044, 0.092, 0.2, 0.44, 0.92, 2 |
| 2-year (NTP TR 402)e | Yes | F344/N | Gavage/corn oil | 0, 2, 4, 8 | |
| GIN | 2-year (NTP TR 567) | No | F344/N | Gavage/deionized water | 0, 1250, 2500, 5000 |
| HCB | 2-year (Arnold et al., 1985) | Yes | Sprague Dawley | Feed/NA | 0, 0.02, 0.09, 0.47, 2.35 |
| MET | 2-year (NTP TR 491) | Yes | F344/N | Gavage/0.5% aqueous methylcellulose | 0, 37, 75, 150, 300 |
| MTE | 2-year (NTP TR 565) | No | F344/N | Feed/NA | 0, 570, 1180, 2520 |
| PFOA | 2-year (NTP TR 598) | Yes | Sprague Dawley | Feed/NA | 0, 2.2, 4.4, 8.8 |
| PUL | 2-year (NTP TR 563) | Yes | F344/N | Gavage/corn oil | 0, 18.75, 37.5, 75 |
| TBBPA | 2-year (NTP TR 587) | No (other) | Wistar Han | Gavage/corn oil | 0, 250, 500, 1000 |
| TCAB | 2-year (NTP TR 558) | Yes | Sprague Dawley | Gavage/corn oil: acetone (99:1) | 0, 10, 30, 100 |
| TCPP | 90-day (NTP TOX)f | Yes | B6C3F1g | Feed/NA | 0, 250, 500, 1000, 2000, 4000 |
| THU | 2-year (NTP TR 570) | No (other) | F344/N | Gavage/0.5% aqueous methylcellulose | 0, 12.5, 25, 50 |
mg/kg Except ethinyl estradiol (µg/kg).
Not hepatotoxic but toxic to other tissues.
TG-GATES (https://toxico.nibiohn.go.jp/english).
In deionized water.
Apical data used only for female rats and male and female mice.
Report in-progress (unpublished data).
Data shown for mice only.
Abbreviation: NA, not applicable.
Animal exposures
Studies were approved by the Battelle Animal Care and Use Committee and conducted in accordance with all relevant NIH and NTP animal care and use policies. Male Sprague Dawley (Hsd: Sprague Dawley SD) rats were received at Battelle from Envigo (Haslett, Michigan), acclimated for approximately 2 weeks, and then randomized based on body weights. The rats were provided food (irradiated NTP-2000 diet) and water ad libitum and were 8–10 weeks of age on Day 0. Eight to 10 concentrations plus a vehicle control for each of the 18 selected chemicals (Table 2) were administered by oral gavage (5 ml/kg) once per day for 5 consecutive days (Days 0–4) with n = 4 rats per exposure concentration and vehicle control. The body weight for each rat was recorded prior to dose administration on Day 0. Each rat was observed twice daily over the course of the study for moribundity and mortality. Exposure concentrations for each chemical were selected based on those used in the chronic or subchronic toxicity study and included concentrations that were higher and lower than those previously tested (Table 3). Whenever possible, the gavage vehicle was matched to the vehicle used in the chronic or subchronic study (Table 3); however, multiple chemicals were orally administered in feed (MTE, DEHP, PFOA, EE2, HCB, and TCPP) or drinking water (BDCA and ACR) in the chronic or subchronic study. On Day 5, the rats were weighed and humanely terminated via exsanguination whereas under 100% CO2 or 70% CO2:30% O2 anesthesia. The liver and left and right kidneys were rapidly collected and weighed. The left liver lobe and right kidney were transferred into RNAlater (Qiagen, Valencia, California) in accordance with the manufacturer’s instructions (ie, storage at 4°C overnight to allow the RNAlater to penetrate the tissues, followed by removal of the RNAlater supernatant and frozen storage of the tissues until RNA extraction).
RNA extraction and HTT using the rat S1500+
Total RNA was extracted from liver and kidney using the RNeasy Mini Kit (Qiagen) with a DNA digestion step. RNA purity and quality were determined by the absorbance ratio (260/280) and calculation of the RNA integrity number, respectively. RNA was sent frozen to BioSpyder (Carlsbad, California) for HTT analysis using the rat S1500+ TempO-Seq platform (Mav et al., 2018, https://ntp.niehs.nih.gov/results/tox21/s1500-gene-set-consensus-strategy-index.html; last accessed June 17, 2020). The S1500+ measures approximately 3000 transcripts covering nearly 90% of all well-curated gene sets. Briefly, mRNA targets were hybridized with a detector oligo (DO) mix (2 DOs per transcript) in 96-well plates, followed by nuclease digestion of excess oligos, ligation, and elution to generate a pool of amplification templates that share common PCR primer-binding sites. During product amplification, each sample well was assigned a specific, barcoded primer pair, allowing for proper identification and matching of mRNAs and samples following sequencing. Sample amplicons were pooled and cleaned up using a PCR clean-up kit. Libraries were then sequenced using a HiSeq 2500 Ultra-High-Throughput Sequencing System (Illumina, San Diego, California). Sequencing readouts were demultiplexed to generate FASTQ files.
Rat S1500+ data quality control and normalization
An initial quality control (QC) of the sequence data (FASTQ files) was performed using FASTQC. Sequence data were then aligned to the rat S1500+ probe sequences using Bowtie version 1.2.2. For the alignment, 3 mismatches were allowed and only reads that aligned uniquely to a probe were retained. Sequence data were evaluated based on the number of sequenced reads, alignment rate, number of aligned reads, and the distribution of the reads across the S1500+ probes. Samples meeting any of the following criteria were flagged: sequencing read depth < 500 K, alignment rate < 40%, aligned reads < 500 K, or if > 50% of the probes had less than 5 reads. Furthermore, principal component analysis (PCA), hierarchical clustering, and correlation plots were generated for each chemical/dose group and evaluated to determine which samples did not clearly cluster with other replicates within the same group. Samples flags from the QC and visual inspection of the PCA/hierarchical clustering/correlation plots were used to determine which samples were outliers and should be removed.
Counts per million (CPM) reads were used to adjust for DO read count discrepancies. This normalization first determined the total number of reads in a sample. The read count for each gene in the sample was then divided by the total number of reads. Finally, each DO count/total DO read ratio was multiplied by 1 × 106. Log2(CPM + 1) transformation was performed to achieve a normal distribution and to limit problems of heteroscedasticity during the dose-response modeling. The normalized gene expression signal of log2(CPM + 1) was used for dose-response analysis.
Transcriptional BMD analysis
To avoid analytical bias, we applied a predefined analysis process that balances reproducibility with noise elimination which was previously peer-reviewed (NTP, 2018). The authors acknowledge that there are a diverse set of parameter options in the analysis pipeline that have the potential to influence the transcriptional gene set BMD analysis and the subsequent comparisons with apical endpoints. However, our goal here was not to explore the impact of parameter settings on transcriptional gene set BMD estimates, but to understand how effectively 5-day transcriptional gene set BMDs derived from a standardized analysis pipeline compared with apical BMDs.
Dose-response analyses of log2(CPM + 1)-normalized DO counts from the rat S1500+ platform were performed using BMD Express 2.2. Data were prefiltered (1 data set per chemical) using Williams Trend Test (p < .05, 1 × 104 dose-level permutations) in combination with a fold change cut-off of > ǀ1.5ǀ to remove DOs that did not demonstrate a response to chemical treatment from BMD analysis. DOs that passed the prefilter were fit to multiple continuous models (linear, exponential 2–5, polynomial of degree 2–4, and power). An assumption of constant variance across dose groups was made due to the log transformation of the data. To be considered further, model fits had to demonstrate convergent BMD, BMDL, and BMDU values. For the models with convergent values, the one with the lowest Akaike information criterion (AIC) was chosen as the best fit model for each DO. A benchmark response (BMR) of 1 SD from the modeled response at control for each DO was used to identify a BMD along with 95% upper and lower bound values. Prior to analysis, DOs that mapped to >1 gene, had global goodness of fit p values of <.1, had a BMD > the top dose level or BMDU/BMDL ratios (between upper and lower 95% confidence limits) of >40 were removed. All DOs which passed these selection criteria were converted into their corresponding NCBI Entrez Gene ID and then parsed into the Gene Ontology Biological Process (GO BP) gene sets. GO BP gene sets that contained at least 3 genes and had BMDs for at least 5% of the genes in the set (based on the total annotated gene number) were declared active and a BMD was determined by calculating the median BMD for that gene set (BMDs were not extrapolated outside the evaluated concentration range). The approach used to analyze the data was peer reviewed by an NTP expert panel (https://www.ncbi.nlm.nih.gov/books/NBK535511/; last accessed June 17, 2020) and deemed to be acceptable for use as it avoids spurious derivation of BMD values from very small gene sets (count threshold) and gene sets that are very large (percentage threshold). Importantly, it has become common practice when performing genomic dose-response modeling to avoid use of statistical tests that are impacted by the number of probes coming through the analysis. By using such thresholds, this approach avoids the situation where gene sets can be declared active in 1 study and inactive in another, hence facilitating comparability/scalability of results. For this study, “transcriptional BMD” is defined as the median gene set (GO BP) BMD. The 3 lowest/most sensitive transcriptional BMDs are shown for liver (Supplementary Table 2) and kidney (Supplementary Table 3) and the single lowest value was used for subsequent analyses.
Apical BMD analysis
Apical BMD analysis was performed on chemical-related increased incidences of non-neoplastic and neoplastic histopathological effects in all tissues in male and female rats and mice observed in the historical chronic or subchronic toxicity studies (Table 3) using BMDS Wizard or Version 3. The chronic and subchronic study data were modeled using the multistage 1 and 2 models. If both models gave an acceptable fit to the data, the BMD for the model with the lowest AIC was used. Often, both models gave the same result, because the parameter fit can give a value of 0 for the quadratic term in the multistage 2 model, which makes the multistage 1 and 2 models the same. The apical BMD was calculated as the BMD10 (ie, the BMD based on 10% extra risk with a 95% lower confidence limit [BMDL]). To identify the most sensitive apical endpoints, an expert review of the data was performed, and the 3 apical effects estimated to produce the lowest BMD values were selected for modeling. After modeling the data, the BMD values were ranked, and the lowest value was chosen for comparison with the transcriptional BMD values. The 3 lowest/most sensitive apical BMDs are shown for non-neoplastic and neoplastic lesions (Supplementary Tables 5–12).
For 1 chemical, fenofibrate, incidence data that could be modeled was only available from a 28-day subchronic study included in the TG-GATEs data set. In this study, there was 1 histopathological effect (“degeneration, granular, eosinophilic, hepatocyte”) which showed 100% incidence at all nonzero dose levels (5 animals/dose group) making it difficult to determine an apical BMD using only the incidence data. Therefore, histopathological severity scores were evaluated using categorical regression analysis using EPA’s CatReg software (https://www.epa.gov/bmds/catreg; last accessed June 17, 2020). Severity scores were analyzed using the cumulative odds model with logit link function, log10 dose scaling, and 0 background response. The apical BMD for FEN was calculated as the BMD10 using a 10% extra risk of a level 1 severity score as the BMR.
BMD values for chemical-induced changes in organ weights for liver and kidney were calculated using BMDS Version 3 with continuous models, nonconstant variance, and a BMR = 0.1 * control response and are reported as the BMD10. Some of the chemicals did not induce sufficient changes in organ weights within the selected dose range to determine a BMD.
Statistical analyses of terminal body and organ weights
Statistical analyses of changes in body and organ (liver and kidney) weights were performed using both the Jonckheere (trend) test and Williams or Dunnett (pairwise) test (vs vehicle control).
RESULTS
Survival, Terminal Body, and Organ Weights
Male rats were exposed to chemical or vehicle control (Table 2) for 5 consecutive days as described in the Materials and Methods section. All treated rats survived until scheduled termination except those exposed to the highest doses of COU (n “surviving” = 1 at 400 mg/kg), MET (n = 2 at 600 mg/kg), PUL (n = 1 at 300 mg/kg), TCPP (n = 3 at 1000 mg/kg), and THU (n = 3 at 200 mg/kg) which were found dead or euthanized due to moribund condition. Terminal body weights were significantly decreased for rats exposed to PFOA (20 mg/kg) and PUL (75 and 150 mg/kg) compared with vehicle controls (data not shown). Rats exposed to COU, FEN, HCB, PFOA, and PUL exhibited significant weight loss based on trend tests (data not shown). Absolute liver weights and liver weight/terminal body weight ratios were significantly increased in rats exposed to FUR (8 and 16 mg/kg), PFOA (2.5 mg/kg and higher), DEHP (125 mg/kg and higher), FEN (16 mg/kg and higher), TCPP (1000 and 2000 mg/kg), HCB (16 and 64 mg/kg), TCAB (100 mg/kg and higher), TBBPA (500 mg/kg and higher), DE71 (15 mg/kg and higher), PUL (75 and 150 mg/kg), and GIN (312.5 mg/kg) compared with vehicle controls (data not shown). Rats exposed to FUR, PFOA, DEHP, FEN, TCPP, HCB, TCAB, TBBPA, DE71, MTE, and MET exhibited significant increases in absolute liver weights and liver weight/terminal body weight ratios based on trend tests (data not shown). Absolute left and right kidney weights and kidney weight/terminal body weight ratios were significantly increased only in rats exposed to DEHP (1000 mg/kg) compared with vehicle control (data not shown). Rats exposed to TCPP and GIN exhibited significant increases in absolute right and left kidney weights, respectively, and kidney weight/terminal body weight ratios based on trend tests (data not shown). All survival data and terminal body and organ weights for all chemicals can be found in the NTP Chemical Effects in Biological Systems (CEBS) database https://doi.org/10.22427/NTP-DATA-002-00058-0002-0000-7 (last accessed June 17, 2020). All data showing body and organ weight changes with a dose-related trend were subject to BMD analysis. Data for organ weight BMDs are reported in Supplementary Table 4.
Liver and Kidney Transcriptional BMDs in Male Rats
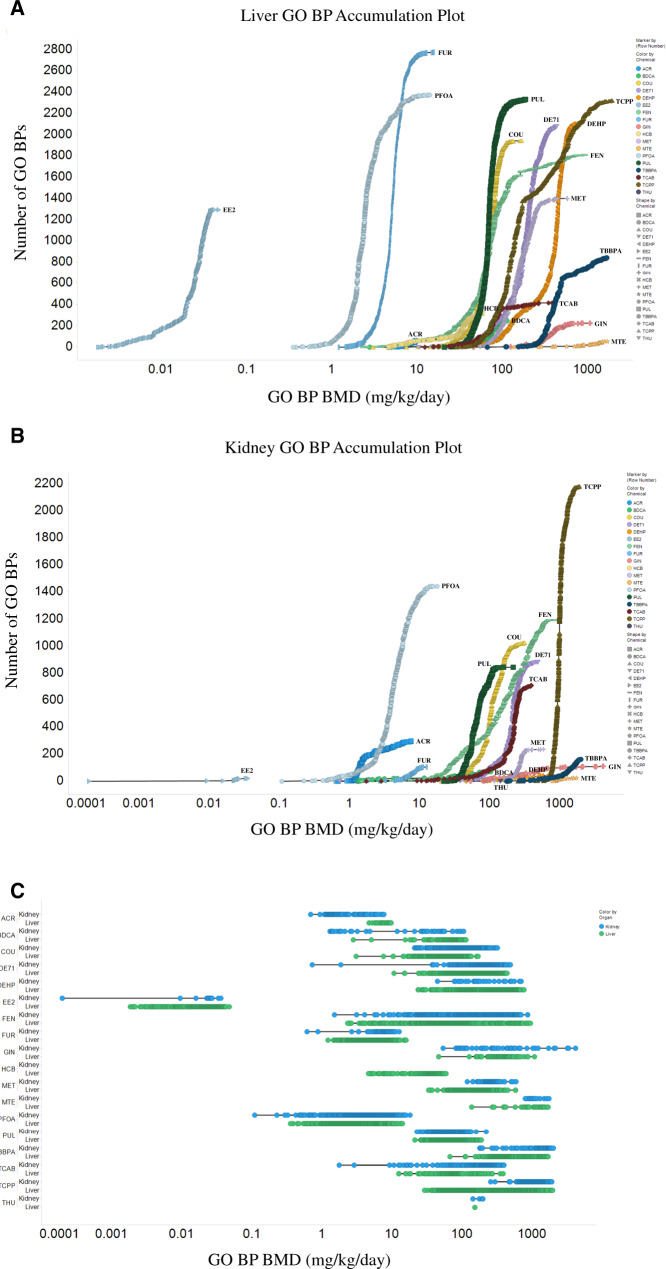
HTT dose-response (S1500+) data for 5-day liver and kidney from male rats were analyzed using BMD Express 2.2 to determine transcriptional BMDs as described in the Materials and Methods section. The number of active gene sets (GO BPs) in liver and kidney (Figure 1) varied dramatically across chemicals. The chemical with the lowest number of active gene sets (GO BPs) was THU (only 1 in liver and 7 in kidney). The chemical with the highest number of active gene sets (GO BPs) was TCPP (4504 total; 2322 in liver and 2182 in kidney). The number of active GO BPs and genes that passed the prefilters is shown in Supplementary Table 1.
Figure 1.
Gene set (Gene Ontology Biological Process, GO BP) accumulation plots for liver (A) and kidney (B) from the 5-day assays in male rats (THU cannot be seen on the plot in A due to only 1 active GO BP for liver, and HCB is not on the plot in B due to no active GO BPs for kidney). Line plots for gene sets (GO BPs) in liver and kidney are shown in C. Each point represents a single active GO BP. All benchmark doses (BMDs) are in mg/kg/day.
Supplementary Table 2 shows the 3 lowest/most sensitive transcriptional (GO BP) BMDs (mg/kg) for liver for each of the 18 chemicals evaluated in male rats. For THU, only 1 transcriptional BMD was identified for liver. Supplementary Table 3 shows the 3 lowest/most sensitive transcriptional (GO BP) BMDs (mg/kg) for kidney for each chemical evaluated in male rats. For HCB, no transcriptional BMDs were identified for kidney. Chemical exposure induced alterations in transcripts from a wide variety of biological pathways in both liver and kidney. For some of the BMD values, there were more than 1 GO term at the same dose level (eg, for PUL, there were 3 GO terms for the BMD value of 25.25 mg/kg). All transcriptional (liver and kidney GO BP) BMD values (which were not extrapolated outside the evaluated concentration range) were above the lowest concentration tested (Table 2) for all chemicals except FEN. All active GO BPs (and associated BMDs) for liver and kidney for all chemicals (as well as all of the rat S1500+ gene expression data) can be found in the NTP CEBS database (https://doi.org/10.22427/NTP-DATA-002-00058-0002-0000-7). For all chemicals shown in Supplementary Table 4, the lowest transcriptional (GO BP) BMD for kidney and/or liver was lower (more sensitive) than the respective organ weight BMD.
Chronic or Subchronic Apical BMDs in Male Rats
Apical BMD analysis was performed on chemical-related increased incidences of non-neoplastic and neoplastic histopathological effects in all tissues in male rats observed in the chronic or subchronic studies using BMDS as described in the Materials and Methods section. Supplementary Table 5 shows the 3 lowest/most sensitive apical BMDs and BMDLs for non-neoplastic histopathological lesions in male rats for each of the chemicals. For GIN, the NTP 2-year chronic study showed no treatment-related histopathological findings in rats or mice. For TBBPA and MTE, the NTP 2-year chronic studies showed no treatment-related histopathological findings in male rats. For FUR, apical data from the most recent 2-year bioassay (Von Tungeln et al., 2017) were used for male rats. For TCPP, data in rats from the NTP 90-day subchronic study were not used because rats were only exposed perinatally. There were only 2 non-neoplastic lesions for DEHP and THU, and just 1 for FEN. Supplementary Table 6 shows the 3 lowest/most sensitive apical BMDs and BMDLs for neoplastic histopathological lesions in male rats for each of the chemicals. For PUL, EE2, and FEN, treatment-related neoplastic lesions were not observed in male rats. There were only 2 neoplastic lesions for DEHP, THU, and HCB and just 1 for COU. For FUR, PFOA, COU, DE71, PUL, MET, TCAB, and DEHP, which are all hepatotoxic chemicals based on the historical chronic toxicity studies (Table 3), the lowest 3 apical BMDs/BMDLs included histopathological (non-neoplastic or neoplastic) effects in liver.
Comparison of Transcriptional and Apical BMDs in Male Rats
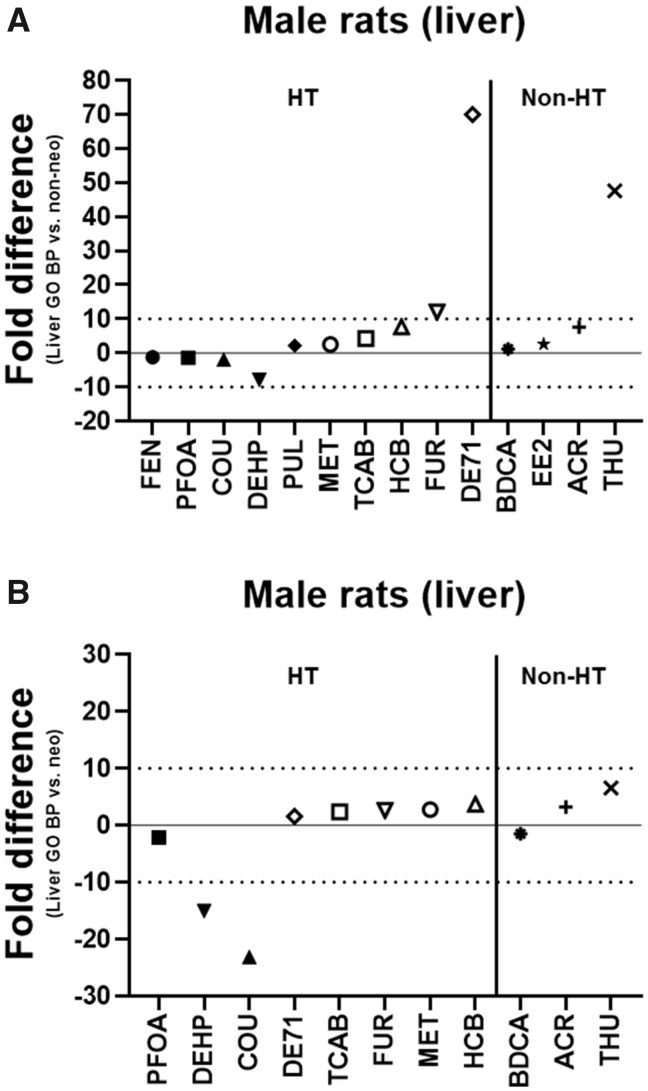
The lowest transcriptional (GO BP) BMDs for liver and kidney in male rats were compared with the lowest apical BMDs for non-neoplastic and neoplastic lesions in male rats to determine how accurately the 5-day transcriptional BMDs estimated the apical (histopathological) BMDs from the historical chronic or subchronic toxicity studies. The lowest liver transcriptional (GO BP) BMD was within 10-fold of the lowest apical (non-neoplastic) BMD in male rats for all chemicals except FUR (12-fold), THU (48-fold), and DE71 (70-fold) and within 5-fold for BDCA, COU, EE2, MET, PFOA, PUL, FEN, and TCAB (Figure 2A). The liver transcriptional BMD was lower (more sensitive) than the apical (non-neoplastic) BMD for COU, DEHP, PFOA, and FEN and higher (less sensitive) for the other chemicals. The lowest liver transcriptional (GO BP) BMD was within 10-fold of the lowest apical (neoplastic) BMD in male rats for all chemicals that induced neoplastic lesions except DEHP (15-fold) and COU (23-fold) and within 5-fold for BDCA, ACR, FUR, MET, PFOA, DE71, HCB, and TCAB (Figure 2B). The liver transcriptional BMD was lower (more sensitive) than the apical (neoplastic) BMD for BDCA, COU, DEHP, and PFOA and higher (less sensitive) for the other chemicals. For both DEHP and COU, despite the difference being > 10-fold, the lowest liver transcriptional BMD was below the lowest apical (neoplastic) BMD.
Figure 2.
Lowest liver transcriptional versus apical benchmark doses (BMDs) in male rats. The fold difference of the lowest liver transcriptional Gene Ontology Biological Process (GO BP) BMD and the lowest apical (non-neoplastic [A] or neoplastic [B]) BMD is shown in male rats. A negative fold difference indicates that the liver GO BP BMD is less than the apical (non-neoplastic or neoplastic) BMD. A positive fold difference indicates that the liver GO BP BMD is greater than the apical (non-neoplastic or neoplastic) BMD. The solid line represents no difference, whereas the dotted lines represent a 10-fold difference between the liver GO BP BMD and the apical (non-neoplastic or neoplastic) BMD. HT indicates hepatotoxic chemicals. Non-HT indicates chemicals that exhibit minimal/no hepatotoxicity but produce adverse effects in other tissues.
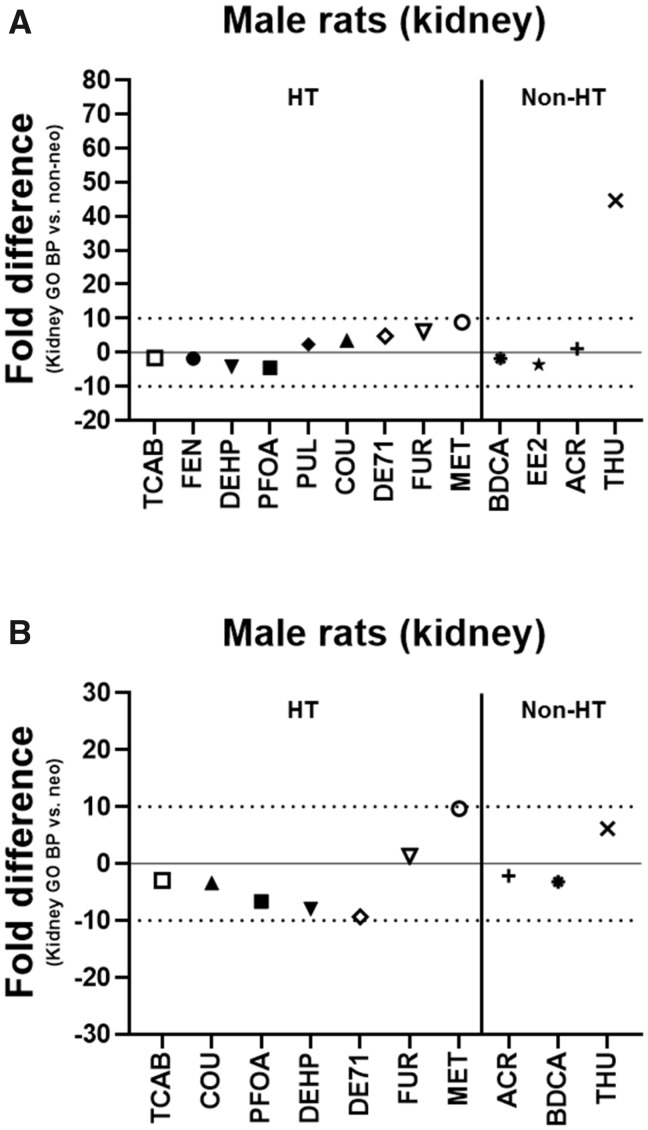
The lowest kidney transcriptional (GO BP) BMD was within 10-fold of the lowest apical (non-neoplastic) BMD in male rats for all chemicals except THU (45-fold) and within 5-fold for 10/13 of the chemicals (BDCA, ACR, COU, DEHP, EE2, PFOA, PUL, DE71, FEN, and TCAB) (Figure 3A). The kidney transcriptional BMD was lower (more sensitive) than the apical (non-neoplastic) BMD for BDCA, DEHP, EE2, PFOA, FEN, and TCAB and higher (less sensitive) for the other chemicals. The lowest kidney transcriptional (GO BP) BMD was within 10-fold of the lowest apical (neoplastic) BMD in male rats for all chemicals that induced neoplastic lesions and within 5-fold for BDCA, ACR, COU, FUR, and TCAB (Figure 3B). The kidney transcriptional BMD was lower (more sensitive) than the apical (neoplastic) BMD for all chemicals except FUR, MET, and THU.
Figure 3.
Lowest kidney transcriptional versus apical benchmark doses (BMDs) in male rats. The fold difference of the lowest kidney transcriptional Gene Ontology Biological Process (GO BP) BMD and the lowest apical (non-neoplastic [A] or neoplastic [B]) BMD is shown in male rats. A negative fold difference indicates that the kidney GO BP BMD is less than the apical (non-neoplastic or neoplastic) BMD. A positive fold difference indicates that the kidney GO BP BMD is greater than the apical (non-neoplastic or neoplastic) BMD. The solid line represents no difference, whereas the dotted lines represent a 10-fold difference between the kidney GO BP BMD and the apical (non-neoplastic or neoplastic) BMD. HT indicates hepatotoxic chemicals. Non-HT indicates chemicals that exhibit minimal/no hepatotoxicity but produce adverse effects in other tissues.
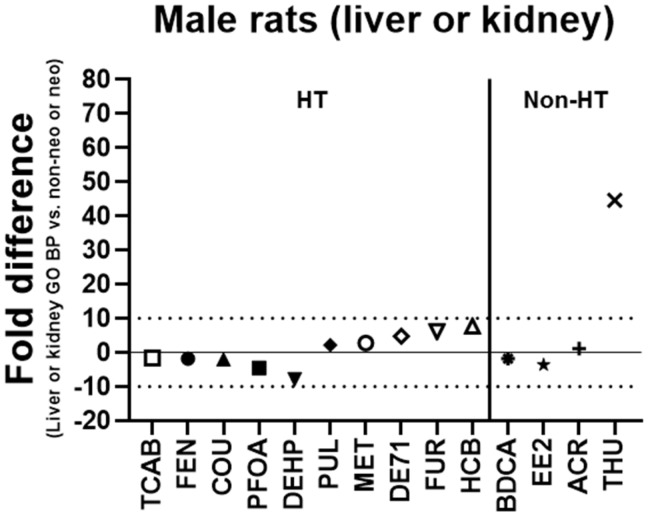
The lowest transcriptional (GO BP) BMD (liver or kidney) was also compared with the lowest apical (non-neoplastic or neoplastic) BMD in male rats. The lowest transcriptional (GO BP) BMD was within 10-fold of the lowest apical (histopathological) BMD in male rats for 13/14 (93%) of the chemicals with the exception being THU (45-fold) and within 5-fold for 10/14 (71%) of the chemicals (BDCA, ACR, COU, EE2, MET, PFOA, PUL, DE71, FEN, and TCAB) (Figure 4). The transcriptional BMD was lower (more sensitive) than the apical BMD for BDCA, COU, DEHP, EE2, PFOA, FEN, and TCAB and higher (less sensitive) for the other chemicals. The lowest kidney GO BP BMD was less than the lowest liver GO BP BMD for 9 of the tested chemicals (ACR, BDCA, DE71, EE2, FEN, FUR, PFOA, TCAB, and THU).
Figure 4.
Lowest transcriptional (liver or kidney) versus apical (non-neoplastic or neoplastic) benchmark doses (BMDs) in male rats. The fold difference of the lowest transcriptional Gene Ontology Biological Process (GO BP) BMD (liver or kidney) and the lowest apical (non-neoplastic or neoplastic) BMD is shown in male rats. A negative fold difference indicates that the liver or kidney GO BP BMD is less than the apical (non-neoplastic or neoplastic) BMD. A positive fold difference indicates that the liver or kidney GO BP BMD is greater than the apical (non-neoplastic or neoplastic) BMD. The solid line represents no difference, whereas the dotted lines represent a 10-fold difference between the liver or kidney GO BP BMD and the apical (non-neoplastic or neoplastic) BMD. HT indicates hepatotoxic chemicals. Non-HT indicates chemicals that exhibit minimal/no hepatotoxicity but produce adverse effects in other tissues.
Comparison of Transcriptional and Apical BMDs in Female Rats and Male and Female Mice
Apical BMD analysis was similarly performed on increased incidences of histopathological effects in all tissues in female rats as well as in male and female mice observed in the chronic or subchronic studies to determine if the transcriptional BMDs derived from the 5-day assays in male rats estimated apical (histopathological) BMDs independent of sex and species. Supplementary Table 7 shows the 3 lowest/most sensitive apical BMDs and BMDLs for non-neoplastic histopathological lesions in female rats for each of the chemicals. For FUR, apical data from the original 2-year bioassay (NTP, 402, 1993b) were used for female rats. FEN was not tested in female rats in the TG-GATEs 29-day subchronic study. For PFOA, all non-neoplastic lesions with treatment-related effects had decreasing incidences in female rats. There were only 2 non-neoplastic lesions for FUR, TBBPA, THU, and MTE. Supplementary Table 8 shows the 3 lowest/most sensitive apical BMDs and BMDLs for neoplastic histopathological lesions in female rats for each of the chemicals. For COU, EE2, THU, MTE, and HCB, neoplastic lesions were not observed in female rats. For DEHP in female rats, there were only neoplastic histopathological effects, and there were only 2 neoplastic lesions for BDCA, PFOA, and PUL.
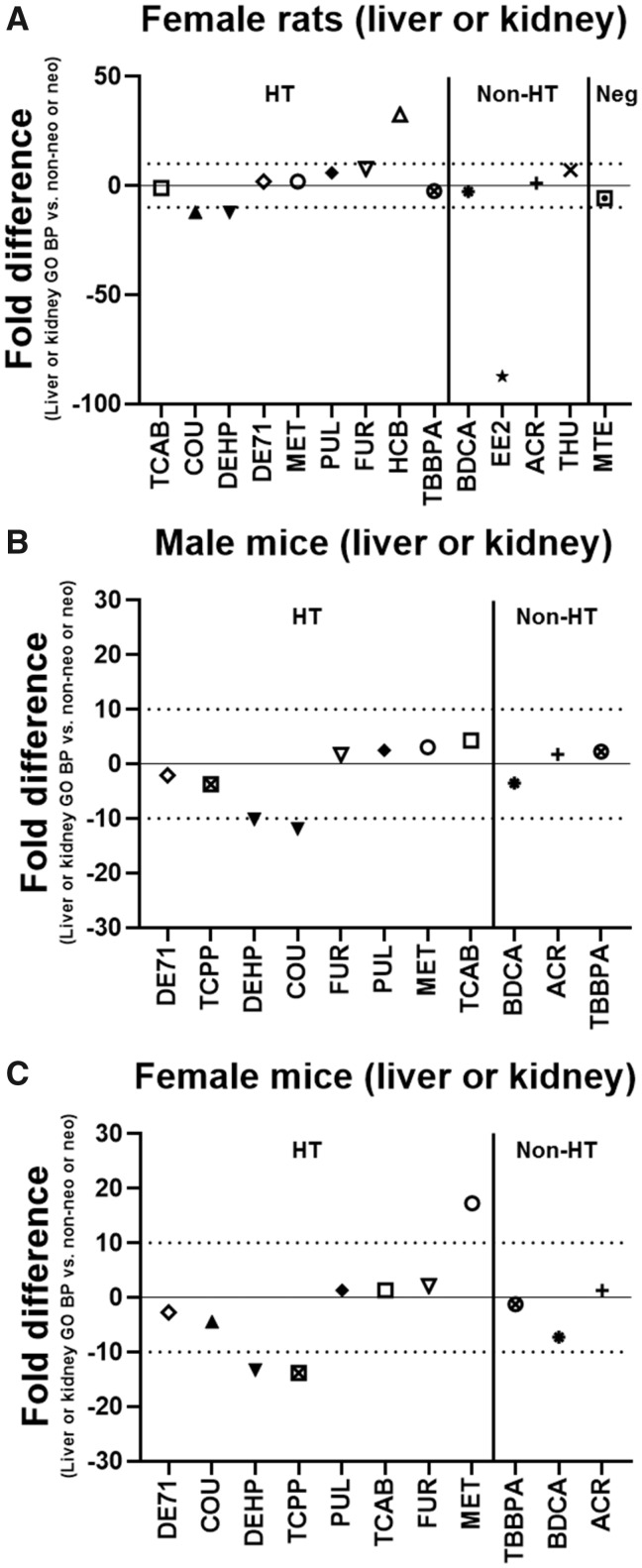
The lowest transcriptional (GO BP) BMD (liver or kidney) in male rats was compared with the lowest apical (non-neoplastic or neoplastic) BMD in female rats. The lowest transcriptional (GO BP) BMD in male rats was within 10-fold of the lowest apical (histopathological) BMD in female rats for all chemicals except DEHP (12-fold), COU (12-fold), HCB (33-fold), EE2 (88-fold), and PFOA (353-fold) and within 5-fold for BDCA, ACR, MET, DE71, TCAB, and TBBPA (Figure 5A). The transcriptional BMD was lower (more sensitive) than the apical BMD for BDCA, EE2, TCAB, COU, DEHP, TBBPA, PFOA, and MTE and higher (less sensitive) for the other chemicals. For EE2, COU, PFOA, and DEHP, despite the difference being > 10-fold, the lowest transcriptional BMD was below the lowest apical BMD.
Figure 5.
Lowest transcriptional (liver or kidney) versus apical (non-neoplastic or neoplastic) benchmark doses (BMDs) for different sex and species. The fold difference of the lowest transcriptional Gene Ontology Biological Process (GO BP) BMD (liver or kidney) in male rats and the lowest apical (non-neoplastic or neoplastic) BMD in female rats (A), male mice (B), and female mice (C) is shown. A negative fold difference indicates that the liver or kidney GO BP BMD is less than the apical (non-neoplastic or neoplastic) BMD. A positive fold difference indicates that the liver or kidney GO BP BMD is greater than the apical (non-neoplastic or neoplastic) BMD. The solid line represents no difference, whereas the dotted lines represent a 10-fold difference between the liver or kidney GO BP BMD and the apical (non-neoplastic or neoplastic) BMD. HT indicates hepatotoxic chemicals. Non-HT indicates chemicals that exhibit minimal/no hepatotoxicity but produce adverse effects in other tissues. Neg indicates chemicals that exhibit little overt toxicity to the liver or other tissues. PFOA (−353-fold) is not shown on the graph in A for female rats due to scaling.
Supplementary Tables 9 and 11 show the 3 lowest/most sensitive apical BMDs and BMDLs for non-neoplastic histopathological lesions in male and female mice, respectively, for each of the chemicals. For FUR, apical data from the original 2-year bioassay (NTP, 402, 1993b) were used for male and female mice. For MTE, GIN, and THU, the NTP 2-year chronic studies showed no treatment-related histopathological findings in male or female mice. PFOA, EE2, HCB, and FEN were not tested in male or female mice in the corresponding chronic or subchronic studies. There were only 2 non-neoplastic lesions for TCPP and just 1 for DEHP in male mice. There were only 2 non-neoplastic lesions for BDCA and just 1 for TCPP in female mice. Supplementary Tables 10 and 12 show the 3 lowest/most sensitive apical BMDs and BMDLs for neoplastic histopathological lesions in male and female mice, respectively, for each of the chemicals. For TCPP, there were no treatment-related neoplastic lesions observed in male or female mice. For DEHP in female mice, there were only neoplastic histopathological effects. For TBBPA, there were no treatment-related neoplastic lesions observed in female mice and just 1 in male mice. There were only 2 neoplastic lesions for COU, PUL, TCAB, and DEHP in male mice. There were only 2 neoplastic lesions for DEHP and just 1 for PUL in female mice.
The lowest transcriptional (GO BP) BMD (liver or kidney) in male rats was compared with the lowest apical (non-neoplastic or neoplastic) BMD in male and female mice. The lowest transcriptional (GO BP) BMD in male rats was within 10-fold of the lowest apical (histopathological) BMD in male mice for all chemicals except COU (12-fold) and within 5-fold for 9/11 of the chemicals (BDCA, ACR, MET, PUL, TCPP, DE71, TCAB, FUR, and TBBPA) (Figure 5B). The transcriptional BMD was lower (more sensitive) than the apical BMD for BDCA, DE71, COU, DEHP, and TCPP and higher (less sensitive) for the other chemicals. For COU, despite the difference being > 10-fold, the lowest transcriptional BMD was below the lowest apical BMD. The lowest transcriptional (GO BP) BMD in male rats was within 10-fold of the lowest apical (histopathological) BMD in female mice for all chemicals except DEHP (13-fold), TCPP (14-fold), and MET (17-fold) and within 5-fold for ACR, COU, PUL, DE71, TCAB, FUR, and TBBPA (Figure 5C). The transcriptional BMD was lower (more sensitive) than the apical BMD for BDCA, DE71, COU, DEHP, TCPP, and TBBPA and higher (less sensitive) for the other chemicals. For DEHP and TCPP, despite the difference being > 10-fold, the lowest transcriptional BMD was below the lowest apical BMD.
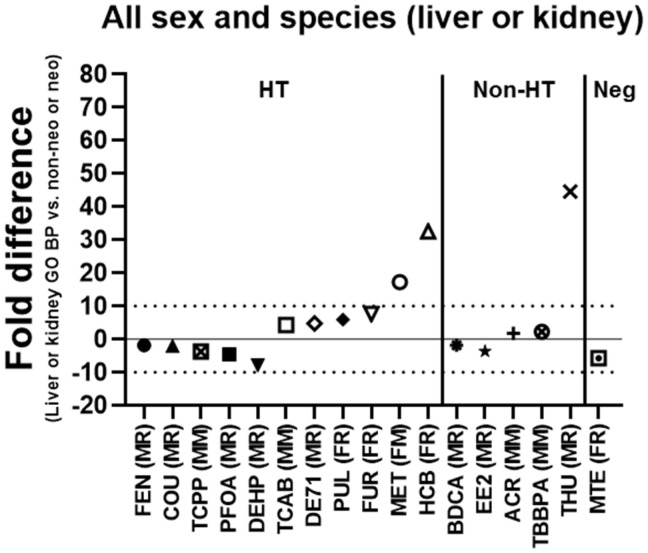
In addition, the lowest transcriptional (GO BP) BMD (liver or kidney) in male rats was compared with the overall lowest apical (non-neoplastic or neoplastic) BMD in all sex and species (male or female rats or mice). The lowest transcriptional (GO BP) BMD in male rats was within 10-fold of the overall lowest apical (histopathological) BMD for 14/17 (82%) of the chemicals with the exceptions being MET (17-fold), HCB (33-fold), and THU (45-fold) and within 5-fold for DE71, TCAB, BDCA, ACR, COU, EE2, PFOA, TCPP, FEN, and TBBPA (Figure 6). The transcriptional BMD was lower (more sensitive) than the apical BMD for PFOA, EE2, COU, BDCA, FEN, DEHP, TCPP, and MTE and higher (less sensitive) for the other chemicals.
Figure 6.
Lowest transcriptional (liver or kidney) versus apical (non-neoplastic or neoplastic) benchmark doses (BMDs) independent of sex and species. The fold difference of the lowest transcriptional Gene Ontology Biological Process (GO BP) BMD (liver or kidney) in male rats and the overall lowest apical (non-neoplastic or neoplastic) BMD in male or female rats or mice is shown. A negative fold difference indicates that the liver or kidney GO BP BMD is less than the apical (non-neoplastic or neoplastic) BMD. A positive fold difference indicates that the liver or kidney GO BP BMD is greater than the apical (non-neoplastic or neoplastic) BMD. The solid line represents no difference, whereas the dotted lines represent a 10-fold difference between the liver or kidney GO BP BMD and the apical (non-neoplastic or neoplastic) BMD. HT indicates hepatotoxic chemicals. Non-HT indicates chemicals that exhibit minimal/no hepatotoxicity but produce adverse effects in other tissues. Neg indicates chemicals that exhibit little overt toxicity to the liver or other tissues. Male rats (MR), female rats (FR), male mice (MM), or female mice (FM) indicate which sex and species the overall lowest apical (non-neoplastic or neoplastic) BMD was observed in for each of the chemicals shown.
DISCUSSION
The National Research Council (NRC) report (NRC, 2007) proposed the use of in vitro toxicity pathway perturbations in place of toxicity testing in animals. Toxicity pathways were defined as normal biological pathways that, when sufficiently altered by chemical exposure, mediate toxic responses. Identification of these pathways and defining “sufficiently altered” are some of the challenges in adopting this approach. In this study, we attempted to evaluate pathway-based toxicity testing approaches in male rats exposed to test chemicals for 5 consecutive days. While the NRC proposed evaluating human cells in vitro, the present approach used male rats as a bridge between in vitro approaches and traditional guideline toxicity studies. A 5-day exposure paradigm in male rats was chosen to build on the efforts of Thomas et al. (2013) and to link to the large transcriptional data available for chemical-treated male rats in the TG-GATEs and Drug Matrix databases. Toxicity pathways were evaluated agnostically using HTT and GO BPs.
The lowest transcriptional gene set (GO BP) BMD values for liver and/or kidney from the 5-day assays in male rats provided estimates, within a factor of 10 for most of the chemicals, of the lowest apical (histopathological) BMD values in male rats from the corresponding historical chronic or subchronic toxicity studies. In fact, 13/14 (93%) of the lowest liver or kidney GO BP BMDs were within 10-fold, and 10/14 (71%) within 5-fold, of the lowest non-neoplastic or neoplastic BMDs. These data suggest that liver or kidney GO BP BMD values in a short-term, in vivo assay are generally predictive of BMD values for apical (histopathological) effects in long-term chronic studies. Reasons for this are unclear and outside the scope of this study but may be due to specific “early” molecular initiating events in the 5-day assay that may mediate the development and progression over time of the toxic response.
These data indicate that liver and kidney can act as “sentinel” tissues to estimate apical BMD values for histopathological effects in liver and kidney as well as other target tissues. Furthermore, this indication was also true for apical (histopathological) data in male and female rats and mice from the chronic or subchronic studies because 14/17 (82%) of the lowest liver or kidney GO BP BMDs were within 10-fold of the lowest non-neoplastic or neoplastic BMDs across all sex and species; therefore, the 5-day model appears to be applicable in a manner independent of sex and species. In contrast, the 5-day approach used by Thomas et al. (2013) only compared apical effects to transcriptional analyses within the same target tissues in rats. Whether the liver or kidney transcriptional BMD value better estimated the apical (histopathological) BMD value depended on the chemical and type of lesion (non-neoplastic or neoplastic). This observation highlights the advantage of using 2 tissues as “sentinels” with an agnostic assay platform. Although, in male rats, the kidney appeared to provide better estimates of apical BMDs overall than the liver. When considering non-neoplastic and neoplastic BMDs in male rats, the lowest kidney GO BP BMD was less than the lowest apical BMD (and therefore protective) for 13 of the chemicals and within 10-fold for all but one of the chemicals (THU). For comparison, the lowest liver GO BP BMD was less than the lowest apical BMD in male rats for 8 of the chemicals and outside 10-fold for 5 of the chemicals. These findings suggest that the use of a single sentinel tissue (such as kidney) may be sufficient to provide reasonable estimates of apical BMDs in the 5-day assay, but this needs to be explored further.
THU was the chemical most consistently outside of the 10-fold “cut-off” with regards to the difference between the transcriptional and apical (histopathological) BMD values. THU is a potent neurotoxicant that only altered a single transcriptional GO BP in liver. In this infrequent example, perhaps evaluating only liver and kidney as sentinel tissues is a potential limitation of this 5-day model when considering highly neuroactive compounds such as THU, which was found to cause seizures resulting in early death or termination of the animals in the 2-year chronic study. GIN, which was considered a negative control (along with MTE), caused no treatment-related histopathological effects in the 2-year chronic bioassay in rats or mice but did exhibit some transcriptional activity in the 5-day assay in male rats. There was an approximately 10% weight loss (vs vehicle control) in female rats at 5000 mg/kg in the chronic study (data not shown), but this exposure concentration (indicative of a dose causing biological effects) was much higher than the liver and kidney transcriptional (GO BP) BMD values.
The BMD predictions in all sex and species (Figure 6) based on HTT ranged from a −8-fold underestimation for DEHP (over-sensitive) to a +45-fold overestimation for THU (under-sensitive). Such a wide error range is a current limitation of the 5-day assay that needs to be taken into consideration when estimating BMDs for new chemicals with unknown biological effects. THU, MTE, and GIN highlight some of the uncertainties that exist within the 5-day assay. For MTE and GIN, which were relatively nontoxic chemicals in the NTP chronic bioassays, several gene sets (GO BPs) were still active in liver and kidney in the 5-day assays, although the assumption of relative nontoxicity held because the lowest liver GO BP BMDs were below the BMDs for biological effects (minor histopathological effects in liver for MTE and body weight loss for GIN). However, the chemical (THU) with specific pharmacological activity (neurotoxicity) was not effectively identified in the 5-day assay because very few GO BPs were active in liver and kidney, although 1 of the rats administered the highest dose of THU exhibited seizures (data not shown) in the 5-day assay (as in the chronic bioassay) which would serve as a warning. Several important points can be taken from the observations with THU. (1) Chemical agents that are highly selective in their toxic effects may be underestimated in their potency, particularly if they occur in systems not sampled by transcriptomics, or they lead to secondary systemic disturbances that manifest later in life. (2) The value of serendipitous discovery is highlighted when applying in vivo systems and illustrates the need to keep employing these systems in complex toxicological phenomena until we have an adequate understanding at the molecular level to capture proximal effects in biological systems. (3) In addition, it highlights the need to identify toxicological effects which are inert or weak perturbagens at the transcriptional level. THU and many pharmaceutical agents cover privileged chemical space. Assuming there is no prior knowledge of their molecular target, the diversity of in vitro systems that may be needed to detect their toxic effects at potency levels reflective of their in vivo potency will be quite broad.
The 5-day assay was able to estimate apical BMD values for both hepatotoxicants and nonhepatotoxicants (ACR, BDCA, TBBPA, and EE2) as well as chemicals that induced little toxicity (eg, MTE), although further validation is needed using more chemicals. It is important to emphasize that the transcriptional and apical (histopathological) BMD values do not necessarily need to be similar for 5-day HTT to be a useful predictive model. For chemicals with transcriptional BMDs < apical BMDs (HTT is more sensitive) the transcriptional BMD value provides a more conservative estimate of the apical BMD value. These data showed that some of the chemicals had GO BP BMD values that were below the apical BMD values, and were therefore protective, but this was not consistent across all chemicals. Future studies should attempt to address how to better achieve more conservative estimates of apical BMDs in the 5-day assay.
The 5-day HTT model could potentially provide estimates of apical BMD values for poorly characterized chemicals of concern in the environment (eg, at Superfund sites) to assess the margin of exposure (MOE) in humans. The 5-day HTT model estimated apical (histopathological) BMD values but did not predict-specific apical toxicities (ie, it did not distinguish between toxic and nontoxic chemicals based on the number or type of GO BPs activated). It is likely that a physiological response to any chemical will occur given sufficient exposure, and alterations in the most sensitive transcriptional pathways are indicative of biological activity and do not necessarily represent mechanisms of toxicity. If the MOE is high (ie, the BMD is much greater than the estimated dose of human exposure), the 5-day HTT model will be health protective whether the chemical is considered toxic or not. If the MOE estimated for a chemical using the 5-day HTT model is of concern, the chemical can be prioritized for further testing using more traditional approaches. Thus, the use of the 5-day HTT model can reduce the extensive time and effort involved in determining the specific apical toxicity of chemicals. If further testing of a chemical is necessary, these transcriptional data can provide, not only a dose range-finding study, but also an opportunity to sort through GO BPs that tend to be associated with apical toxicity and recommend specific guideline studies. If this information can be collected, it may be possible to identify a limited set of gene expression signatures that could better predict certain apical toxicities.
In summary, the DNTP is currently evaluating HTT as an approach to provide estimates of chemical exposure that produce minimal biological activity. HTT was evaluated in a 5-day in vivo rat model using 18 chemicals with the objective to determine if minimum biological effect levels as defined by gene set BMD values for liver and kidney expression data provided accurate estimates of BMD values for the most sensitive apical toxicological effects derived from comprehensive guideline toxicity and cancer assessments. For many of the chemicals tested, the lowest gene set BMD value for liver and/or kidney from the 5-day assays in male rats was within a factor of 5 of the lowest histopathological BMD value for not only male rats, but also female rats and male and female mice. These data suggest that the most sensitive liver and/or kidney gene set BMD values derived from short-term (repeated dosing) toxicity studies provide reasonable estimates of the most sensitive BMD values derived from histopathological effects captured across a variety of target tissues in much more resource-intensive guideline studies. These findings support the use of in vivo gene set BMD values for MOE-based prioritization for hazard characterization and as a rapid approach for estimating a BMD value when guideline studies are not available.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
FUNDING
National Toxicology Program, National Institute of Environmental Health Sciences (NIEHS); National Institutes of Health (NIH). This article may be the work product of an employee or group of employees of the National Institute of Environmental Health Sciences (NIEHS), National Institutes of Health (NIH); however, the statements, opinions, or conclusions contained therein do not necessarily represent the statements, opinions, or conclusions of NIEHS, NIH, or the United States government. NIH IRP (project ES103318-04) (2019).
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Supplementary Material
Disclaimer: Biomolecular Screening and Alternative Approaches for the National Toxicology Program https://intramural.nih.gov/search/searchview.taf?ipid=110658&ts=1575385967 (last accessed June 17, 2020).
REFERENCES
- Arnold D. L., Moodie C. A., Charbonneau S. M., Grice H. C., McGuire P. F., Bryce F. R., Collins B. T., Zawidzka Z. Z., Krewski D. R., Nera E. A., et al. (1985). Long-term toxicity of hexachlorobenzene in the rat and the effect of dietary vitamin A. Food Chem. Toxicol. 23, 779–793. [DOI] [PubMed] [Google Scholar]
- Judson R. S., Magpantay F. M., Chickarmane V., Haskell C., Tania N., Taylor J., Xia M., Huang R., Rotroff D. M., Filer D. L., et al. (2015). Integrated model of chemical perturbations of a biological pathway using 18 in vitro high-throughput screening assays for the estrogen receptor. Toxicol. Sci. 148, 137–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstreuer N. C., Ceger P., Watt E. D., Martin M., Houck K., Browne P., Thomas R. S., Casey W. M., Dix D. J., Allen D., et al. (2017). Development and validation of a computational model for androgen receptor activity. Chem. Res. Toxicol. 30, 946–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mav D., Shah R. R., Howard B. E., Auerbach S. S., Bushel P. R., Collins J. B., Gerhold D. L., Judson R. S., Karmaus A. L., Maull E. A., et al. (2018). A hybrid gene selection approach to create the S1500+ targeted gene sets for use in high-throughput transcriptomics. PLoS One 13, e0191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council. (2007). Toxicity Testing in the 21st Century: A Vision and a Strategy. The National Academies Press, Washington, DC. [Google Scholar]
- National Toxicology Program. (2018). NTP Research Report on National Toxicology Program Approach to Genomic Dose-response Modeling: Research Report 5 [Internet]. National Toxicology Program. [PubMed]
- Ramaiahgari S. C., Auerbach S. S., Saddler T. O., Rice J. R., Dunlap P. E., Sipes N. S., DeVito M. J., Shah R. R., Bushel P. R., Merrick B. A., et al. (2019). The power of resolution: Contextualized understanding of biological responses to liver injury chemicals using high-throughput transcriptomics and benchmark concentration modeling. Toxicol. Sci. 169, 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R. S., Wesselkamper S. C., Wang N. C., Zhao Q. J., Petersen D. D., Lambert J. C., Cote I., Yang L., Healy E., Black M. B., et al. (2013). Temporal concordance between apical and transcriptional points of departure for chemical risk assessment. Toxicol. Sci. 134, 180–194. [DOI] [PubMed] [Google Scholar]
- National Toxicology Program. (1982). Carcinogenesis bioassay of di(2-ethylhexyl)phthalate (CAS No. 117-81-7) in F344 rats and B6C3F1 mice (feed studies). Natl. Toxicol. Program Tech. Rep. Ser. 217, 1–127. [PubMed] [Google Scholar]
- National Toxicology Program. (1993. a ). NTP toxicology and carcinogenesis studies of coumarin (CAS No. 91-64-5) in F344/N rats and B6C3F1 mice (gavage studies). Natl. Toxicol. Program Tech. Rep. Ser. 422, 1–340. [PubMed] [Google Scholar]
- National Toxicology Program. (1993. b ). Toxicology and carcinogenesis studies of furan (CASRN 110-00-9) in F344/N rats and B6C3F1 mice (gavage studies). Natl. Toxicol. Program Tech. Rep. Ser. 402, 1–286. [PubMed] [Google Scholar]
- National Toxicology Program. (2000). NTP toxicology and carcinogenesis studies of methyleugenol (CAS No. 93-15-2) in F344/N rats and B6C3F1 mice (gavage studies). Natl. Toxicol. Program Tech. Rep. Ser. 491, 1–412. [PubMed] [Google Scholar]
- National Toxicology Program. (2010. a ). Toxicology and carcinogenesis studies of 3,3′,4,4′-tetrachloroazobenzene (TCAB) (CAS No. 14047-09-7) in Harlan Sprague-Dawley rats and B6C3F1 mice (gavage studies). Natl. Toxicol. Program Tech. Rep. Ser. 558, 1–206. [PubMed] [Google Scholar]
- National Toxicology Program. (2010. b ). Toxicology and carcinogenesis study of ethinyl estradiol (CAS No. 57-63-6) in Sprague-Dawley rats (feed study). Natl. Toxicol. Program Tech. Rep. Ser. 548, 1–210. [PubMed] [Google Scholar]
- National Toxicology Program. (2011. a ). Toxicology and carcinogenesis studies of alpha, beta-thujone (CAS No. 76231-76-0) in F344/N rats and B6C3F1 mice (gavage studies). Natl. Toxicol. Program Tech. Rep. Ser. 570, 1–260. [PubMed] [Google Scholar]
- National Toxicology Program. (2011. b ). Toxicology and carcinogenesis studies of ginseng (CAS No. 50647-08-0) in F344/N rats and B6C3F1 mice (gavage studies). Natl. Toxicol. Program Tech. Rep. Ser. 567, 1–149. [PubMed] [Google Scholar]
- National Toxicology Program. (2011. c ). Toxicology and carcinogenesis studies of milk thistle extract (CAS No. 84604-20-6) in F344/N rats and B6C3F1 mice (feed studies). Natl. Toxicol. Program Tech. Rep. Ser. 565, 1–177. [PubMed] [Google Scholar]
- National Toxicology Program. (2011. d ). Toxicology and carcinogenesis studies of pulegone (CAS No. 89-82-7) in F344/N rats and B6C3F1 mice (gavage studies). Natl. Toxicol. Program Tech. Rep. Ser. 563, 1–201. [PubMed] [Google Scholar]
- National Toxicology Program. (2012). Toxicology and carcinogenesis studies of acrylamide (CASRN 79-06-1) in F344/N rats and B6C3F1 mice (feed and drinking water studies). Natl. Toxicol. Program Tech. Rep. Ser. 575, 1–234. [PubMed] [Google Scholar]
- National Toxicology Program. (2014). Toxicology studies of tetrabromobisphenol A (CASRN 79-94-7) in F344/NTac rats and B6C3F1/N mice and toxicology and carcinogenesis studies of tetrabromobisphenol A in Wistar Han [Crl: WI(Han)] rats and B6C3F1/N mice (gavage studies). Natl. Toxicol. Program Tech. Rep. Ser. 587, 1–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Toxicology Program. (2015). Toxicology studies of bromodichloroacetic acid (CASRN 71133-14-7) in F344/N rats and B6C3F1/N mice and toxicology and carcinogenesis studies of bromodichloroacetic acid in F344/NTac rats and B6C3F1/N mice (drinking water studies). Natl. Toxicol. Program Tech. Rep. Ser. 583, 1–100. [Google Scholar]
- National Toxicology Program. (2016). Toxicology studies of a pentabromodiphenyl ether mixture [DE-71 (Technical Grade)] (CASRN 32534-81-9) in F344/N rats and B6C3F1/N mice and toxicology and carcinogenesis studies of a pentabromodiphenyl ether mixture [DE-71 (Technical Grade)] in Wistar Han [Crl: WI(Han)] rats and B6C3F1/N mice (gavage studies). Natl. Toxicol. Program Tech. Rep. Ser. 589, 1–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Toxicology Program. (2020). Toxicology and carcinogenesis studies of perfluorooctanoic acid (CASRN 335-67-1) administered in feed to Sprague Dawley (Hsd: Sprague Dawley SD) rats. Natl. Toxicol. Program Tech. Rep. Ser. 598, 1–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Tungeln L. S., Walker N. J., Olson G. R., Mendoza M. C., Felton R. P., Thorn B. T., Marques M. M., Pogribny I. P., Doerge D. R., Beland F. A. (2017). Low dose assessment of the carcinogenicity of furan in male F344/N Nctr rats in a 2-year gavage study. Food Chem. Toxicol. 99, 170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.