Abstract
Lead (Pb) exposure is ubiquitous with permanent neurodevelopmental effects. The hippocampus brain region is involved in learning and memory with heterogeneous cellular composition. The hippocampus cell type-specific responses to Pb are unknown. The objective of this study is to examine perinatal Pb treatment effects on adult hippocampus gene expression, at the level of individual cells. In mice perinatally exposed to control water or a human physiologically relevant level (32 ppm in maternal drinking water) of Pb, 2 weeks prior to mating through weaning, we tested for hippocampus gene expression and cellular differences at 5 months of age. We sequenced RNA from 5258 hippocampal cells to (1) test for treatment gene expression differences averaged across all cells, (2) compare cell cluster composition by treatment, and (3) test for treatment gene expression and pathway differences within cell clusters. Gene expression patterns revealed 12 hippocampus cell clusters, mapping to major expected cell types (eg, microglia, astrocytes, neurons, and oligodendrocytes). Perinatal Pb treatment was associated with 12.4% more oligodendrocytes (p = 4.4 × 10−21) in adult mice. Across all cells, Pb treatment was associated with expression of cell cluster marker genes. Within cell clusters, Pb treatment (q < 0.05) caused differential gene expression in endothelial, microglial, pericyte, and astrocyte cells. Pb treatment upregulated protein folding pathways in microglia (p = 3.4 × 10−9) and stress response in oligodendrocytes (p = 3.2 × 10−5). Bulk tissue analysis may be influenced by changes in cell type composition, obscuring effects within vulnerable cell types. This study serves as a biological reference for future single-cell toxicant studies, to ultimately characterize molecular effects on cognition and behavior.
Keywords: metals, gene expression/regulation, glia, neurotoxicity, developmental, hippocampus, single cell, oligodendrocyte, Pb, perinatal
Lead (Pb) exposure remains ubiquitous in many municipalities and rural areas. The removal of Pb from paint and gasoline has been a major public health success (Needleman et al., 1990), though Pb’s persistence in soil, dust, and historic house paint makes abatement from our lives and environments difficult (Dissanayake and Erickson, 2012). The Centers for Disease Control and Prevention estimate that approximately 500 000 children ages 1–5 in the United States have blood Pb levels above the reference level (≥5 μg/dl) (Raymond and Brown, 2017), and no safe level of Pb has been identified. Developmental Pb exposure has profound permanent effects on behavior and cognition (Bellinger et al., 1984; Needleman et al., 1990; Sanders et al., 2009). Young children are particularly susceptible to Pb’s effects due to (1) 4–5 times higher Pb absorption than nonpregnant adults (WHO, 2010), (2) higher Pb intake per unit body weight, (3) incomplete blood-brain barrier development, and (4) neurological effects which occur at all levels of exposure (WHO, 2007). Early life Pb exposure results in permanent cognitive and behavioral changes (Bellinger et al., 1984; Needleman et al., 1990; Sanders et al., 2009).
Many brain regions are influenced by Pb exposure (Sanders et al., 2009). In particular, the hippocampus, a brain region in the temporal lobe that is central to long-term memory formation and an important part of the limbic system (responsible for emotional regulation), is a key target of Pb’s effects. Magnetic resonance spectroscopy of the hippocampus of men with high cumulative Pb exposure revealed higher myoinositol-to-creatinine ratios with exposure (Weisskopf et al., 2007), suggesting that the hippocampus may have cellular changes in response to Pb. Toxicology studies have shown RNA expression differences in the bulk hippocampus with both acute and chronic Pb exposures (An et al., 2014; Schneider et al., 2012). For example, in mice, 3 days of intraperitoneal Pb exposure (15 mg/kg body weight) starting at postnatal day 12 was associated with hippocampal microglial activation and inflammatory cytokine generation (Liu et al., 2015). Multiple brain regions are influenced by Pb exposures, and the hippocampus has emerged as a brain region where consistent Pb-driven changes were observed.
Bulk brain region studies are important first steps to characterize relevant exposure levels and disease endpoints (Bakulski et al., 2012; Malloy et al., 2019). The next challenge in developmental toxicology studies is distinguishing whether the persistent molecular effects identified are due to global changes, altered cellular proportions, or cell type-specific changes (Jaffe and Irizarry, 2014). The hippocampus is a highly heterogeneous structure, containing many cell types such as neurons, astrocytes, oligodendrocytes, and microglial cells. Recently, single-cell RNA profiling through droplet-based techniques has allowed for the deep unbiased profiling of thousands of cells in the hippocampus. Use of this technique showed that the hippocampus has even more cellular heterogeneity than was initially thought (Ståhl et al., 2016). As all of the previous in-depth molecular profiling of Pb-exposed brain tissues utilized bulk tissues analyses, the majority of our knowledge comes from averaging across this complex heterogeneity. Identifying cell type-specific effects of developmental Pb exposure has potential long-term clinical benefits.
Understanding the cell type-specific impacts of Pb exposure is critical to elucidate mechanisms of its effects and reveal possible avenues of prevention and treatment of adverse outcomes due to Pb exposure. Here, we apply single-cell RNA profiling to comprehensively characterize the effects of perinatal Pb exposure on the adult hippocampus in a cell-specific manner in mice. To anchor these novel analyses, we use a human relevant perinatal Pb dose (32 ppm in drinking water) and a collection time point (5 months) where we have well-documented toxic effects in our model system, including persistent metabolic effects and epigenetic reprograming (Faulk et al., 2013, 2014a,b; Montrose et al., 2017; Svoboda et al., 2019), including neuron-specific DNA methylation effects (Dou et al., 2019). Here, our results reveal global RNA changes after Pb exposure, consistent with prior research. Critically, we also identify oligodendrocytes and microglia as particularly vulnerable cell types to Pb exposure. This work demonstrates cell type-specific RNA changes and an exposure-dependent shift in cell type representation.
MATERIALS AND METHODS
Animal treatment paradigm
The mice for this experiment were “wild-type” a/a nonagouti mice derived from a colony of the viable yellow agouti (Avy) strain maintained for over 230 generations. This results in forced heterozygosity on an invariant genetic background which is approximately 93% identical to the C57BL/6J strain (Waterland and Jirtle, 2003; Weinhouse et al., 2014). Postpubertal virgin a/a females (∼5 weeks old) were randomized into exposure groups: control or drinking water containing 32 ppm Pb-acetate. Pb-acetate water was made following our previously described protocol (Faulk et al., 2014c). Throughout the experiment, mice were maintained on a phytoestrogen-free modified AIN-93G diet (TD.95092, 7% Corn Oil Diet, Harlan Teklad). This dose and route of exposure was designed to be relevant to human perinatal exposure. Using this exposure paradigm previously, in dams at weaning, we collected blood by cardiac puncture and assessed maternal blood Pb levels by inductively coupled plasma mass spectrometry at the Michigan Department of Community Health (Faulk et al., 2013). In our previous study, we observed that treatment with water containing 32 ppm Pb-acetate resulted in blood Pb levels of 32.1 (±11.4) µg/dl in the pregnant dams. These levels are within the range of those observed in the US population during the 1970s—in 1971, the “normal” range of blood Pb in children was stated to be between 15 and 40 µg/dl (Steinfeld, 1971).
After an initial 2 weeks of Pb treatment, a/a females were mated with a/a males. Drinking water treatment continued through gestation and lactation. After weaning, 1 male and 1 female wild-type (a/a) offspring per litter were maintained on Pb-free drinking water. At 5 months, a timepoint at which are “mature adults” with fully developed brains, mice were sacrificed for experimental analyses following the NIEHS Toxicant Exposures and Responses by Genomic and Epigenomic Regulators of Transcription (TaRGET) II Consortium protocol (Wang et al., 2018). All mice analyzed were derived from separate litters. All animals had access to food and water ad libitum throughout the experiment, were housed in polycarbonate-free cages, and were maintained in accordance with the guidelines set by the Institute of Laboratory Animal Resources. The study protocol was approved by the University of Michigan Institutional Animal Care and Use Committee (PRO00008852).
Hippocampus isolation and dissociation
Immediately following euthanasia with CO2, mice underwent whole-body perfusion with cell-culture grade saline (0.9%, Sigma). The hippocampal region of the brain was isolated, and then dissociated into a viable single-cell suspension using the Adult Mouse and Rat Brain Dissociation kit (Miltenyi) using a gentleMACS Octo Dissociator with Heaters automated tissue dissociation instrument. This process incorporates both enzymatic and mechanical digestion to remove viable cells from the extracellular matrix and removes debris through gradient centrifugation, taking a total of approximately 3 h. Cells were cryopreserved using Recovery Cell Culture Freezing Medium (Thermo Fisher). Viability and cell concentrations were quantified upon thawing for single-cell analysis using a Luna FL Automated Cell Counter (Logos) by coquantification of fluorescence from acridine orange and propidium iodide dyes. We profiled 8 total samples: 4 Pb treated and 4 control, with biological replicates from 2 male and 2 female animals in each treatment group. Cells were resuspended in a PBS + 0.02% Bovine serum albumin solution at a concentration of approximately 1000 cells/mL for single-cell transcriptomic analysis processing.
Single-cell RNA-sequencing
Single cells were processed for high throughput RNA-sequencing using the Chromium (10× Genomics) instrument at the University of Michigan Advanced Genomics Core Facility, targeting approximately 3000–5000 cells per sample. Cells from the 8 hippocampus samples were processed. The Chromium device partitions cells into individual oil droplets containing gel beads which are hybridized to oligonucleotides containing a partial Illumina sequencing primer, a unique molecular identifier (UMI), a poly-dT primer sequence for the capture of mRNA, and a cell-specific 10× barcode. Chromium uses a pool of approximately 750 000 different 10× barcodes to index each cells transcriptome separately, with each gel bead having a single 10× barcode. Cells are lysed, and reverse transcription reactions conducted within the oil droplet result in cDNAs which incorporate a cell-specific oligo barcode. The oil beads were broken and cDNAs from all cells are pooled together, the cDNA pool is amplified by qPCR, and Illumina P5 and P7 sequencing primers added during Sample Index pPCR. Prepared libraries for all 8 samples were pooled and sequenced on a NovaSeq 6000 (Illumina) using 2× 96 paired-end reads to capture sample index, cell barcode, and transcriptional information. Data are available on the Gene Expression Omnibus (GEO at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE151723 with accession number GSE151723).
Data processing
Raw single-cell RNA-sequencing data were processed using the CellRanger (10× Genomics) analysis pipeline. mkfastq was used to demultiplex the raw Illumina BCL files into fastq format. count was used to align the reads to the mouse reference genome (mm10), filter reads, count barcodes, and count UMIs. The output from count yields digital gene expression (DGE) matrices for each sample that contain the UMI counts per gene, per cell barcode. DGEs were loaded into R statistical software for use with the single-cell transcriptomic data analysis suite Seurat (Butler et al., 2018), then normalized using the “LogNormalize” method in Seurat. Code to produce all R analyses in this manuscript is available (https://github.com/bakulskilab).
Single-cell data analysis
We tested for differences by treatment group in cell viability prior to processing on the 10× instrument using a t test. For data quality control, we applied multiple filter criteria (Supplementary Figure 1). We excluded droplets with fewer than 1000 expressed genes and we excluded genes that were measured in fewer than 3 cells, leaving a total of 5258 cells and 17 143 genes expressed across the 8 samples. We calculated the proportion of mitochondrial genes expressed per cell. We tested for differences in quality control metrics by treatment using t tests. Specifically, we tested for differences in the number of droplets sequenced, the proportion of cells passing quality control and following filtering, and the mean proportion of mitochondrial genes expressed.
Dimension reduction was performed on scaled DGEs using principal component analysis. For data visualization, t-Distributed Stochastic Neighbor Embedding (tSNE) (Maaten and Hinton, 2008) was conducted on the top 12 principal components. The number of principal components was chosen heuristically based on an elbow plot (Supplementary Figure 2), where the variance explained by each principal component began to level out. Cell clusters were predicted based on the top 12 principal components using the FindClusters function, which first uses a k-nearest neighbors graph based on Euclidean distance in principal component analysis space, followed by clustering by the Louvain algorithm. Key gene markers which define each cluster were calculated using the FindAllMarkers function, which uses the nonparametric Wilcoxon rank sum test to identify the top expression markers which are upregulated in each cell cluster. We assessed cluster identity using known markers of the various cell populations in the hippocampus as well as predicting each cluster’s identity based on the expression of cell type-specific marker genes identified previously in single-cell RNA-seq profiling of the mouse hippocampus (Zeisel et al., 2015).
To test for individual genes differentially expressed with Pb treatment across all cells, we used the edgeR package. We used glmQLFit with a nested design to adjust for each individual mouse within the 2 treatment groups and also included the logit of each cell’s detection rate (proportion of nonzeros) as a recommended covariate (Soneson and Robinson, 2018). To test for differential expression in Pb treatment, we used glmQLFTest with a contrast to compare the average effects of the mice in the Pb group with those in the control group. We then calculated false discovery rate (FDR) q values to account for multiple comparisons. We tested for differential expression by Pb across all cells.
To assess the human relevance of these findings, we tested genes differentially expressed by Pb for enrichment with human psychiatric disorder genetic loci. The Psychiatric Genomics Consortium conducted a psychiatric cross-disorder genome-wide association study (GWAS) in 33 332 cases and 27 888 controls of European ancestry (Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013). The 5 disorders considered as cases were autism spectrum disorder, attention deficit-hyperactivity disorder, bipolar disorder, major depressive disorder, and schizophrenia. Among our Pb response genes from all hippocampus cells, we annotated human genes using biomaRt (Durinck et al., 2009). In total, 93.8% of these mouse genes had a corresponding human ortholog, which we matched to the corresponding psychiatric cross-disorder GWAS test statistics. We tested for enrichment of Pb response genes with psychiatric cross-disorder genes at p < .01 in both studies using a Fishers exact test. As a follow-up analysis, we tested each of GWAS summary statistics (p < .001) for the 5 disorders individually for enrichment with Pb response genes. We used individual GWAS for autism spectrum disorder (Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium, 2017), attention deficit-hyperactivity disorder (Demontis et al., 2019), bipolar disorder (Stahl et al., 2019), major depressive disorder (Howard et al., 2019), and schizophrenia (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014).
To test for cluster-specific individual genes differentially expressed with Pb treatment, we next conducted cell cluster-specific analyses. We subset cells from each individual cluster, then tested for differential expression by Pb as described above. We conducted a post hoc differential gene expression power calculation using 4 samples per treatment group and varying numbers of cells, ranging from all cells in the study (n = 5258) in the case of simulated analysis across all clusters down to the number of cells in cluster 4 (n = 181). To investigate whether bulk analysis with all cells together reflects cell proportion changes, we tested the overlap of top differentially expressed genes and cluster markers.
To test for biological processes enriched with Pb treatment, we performed gene set enrichment analysis using the RNA-Enrich option of LRpath (Lee et al., 2016). p values effect estimates and log counts per million from the edgeR differential expression results were used as input for LRpath. For enrichment analysis, we did directional tests in LRpath, querying the gene ontology (GO) Biological Processes database. First, enrichment analysis was performed on differentially expressed genes in the analysis across all clusters. Cluster-specific enrichment analysis was done on cluster-specific differentially expressed genes.
To test for differences in cell proportion by Pb treatment, we summarized the proportion of cells assigned to each cell cluster by animal. We collapsed cluster counts by our identified cell types, and for each cell type, we tested for differences in the proportion of cells by treatment (Pb vs control) using beta regression.
Assessing effects of Pb on neural stem cell differentiation in vitro
To validate our findings, we reanalyzed data from a human cell culture Pb treatment experiment (GEO accession: GSE84712). In this study, human embryonic stem cell-derived neural progenitor cells were cultured into neural rosettes and aggregates, which allows for differentiation of neural and glial cells in a mixed culture (Jiang et al., 2017). Cells were exposed to 0 µM, 3 µM (0.625 ppm), or 30 µM (6.25 ppm) Pb-acetate in a time course experiment. From Pb treatment days 0–26, gene expression in cells was analyzed daily using RNA-sequencing (Jiang et al., 2017). To estimate the cell proportions present in the Pb treatment neural and glial cell culture, using the RNA-sequencing data, we applied Multisubject Single Cell (MuSiC) deconvolution (Wang et al., 2019b). We used an independent mouse hippocampus single-cell reference panel (GSE95315) to inform MuSiC cell types (Hochgerner et al., 2018), predicting proportions of oligodendrocytes, oligodendrocyte progenitor cells, neuronal immediate precursors, and neuroblasts in the bulk RNA expression data. We plotted the cell type proportions estimated in each of the 3 treatment groups by treatment day. In addition, we plotted the gene expression levels of prioritized genes by treatment day.
RESULTS
Sequencing of Hippocampal Cells
To identify effects of perinatal Pb treatment in the hippocampus on a cell-specific basis, we profiled eight 5-month old dissociated hippocampal samples (N = 4 Pb exposed and N = 4 controls; 50% male). The observed number of droplets sequenced ranged from 376 198 to 455 785 per animal (Supplementary Table 1). Following quality control filtering for number of genes expressed and percent mitochondrial genes expressed, we obtained results from 5258 hippocampal cells across all animals. Per sample, the geometric mean percent mitochondrial genes expressed ranged from 3.0% to 4.7% and was on average 0.79% lower with Pb treatment (p value = .046) (Supplementary Figure 3). Given the proportion of mitochondrial genes were relatively low, we did not exclude cells on the basis of mitochondrial gene expression. Neither mean UMI nor mean genes expressed per cell differed significantly between control and exposed mice (UMI difference: 515.4, p value = .09; genes per cell difference: 100.5, p value = .15), although both trended higher in controls.
Hippocampus Cell Cluster Identification
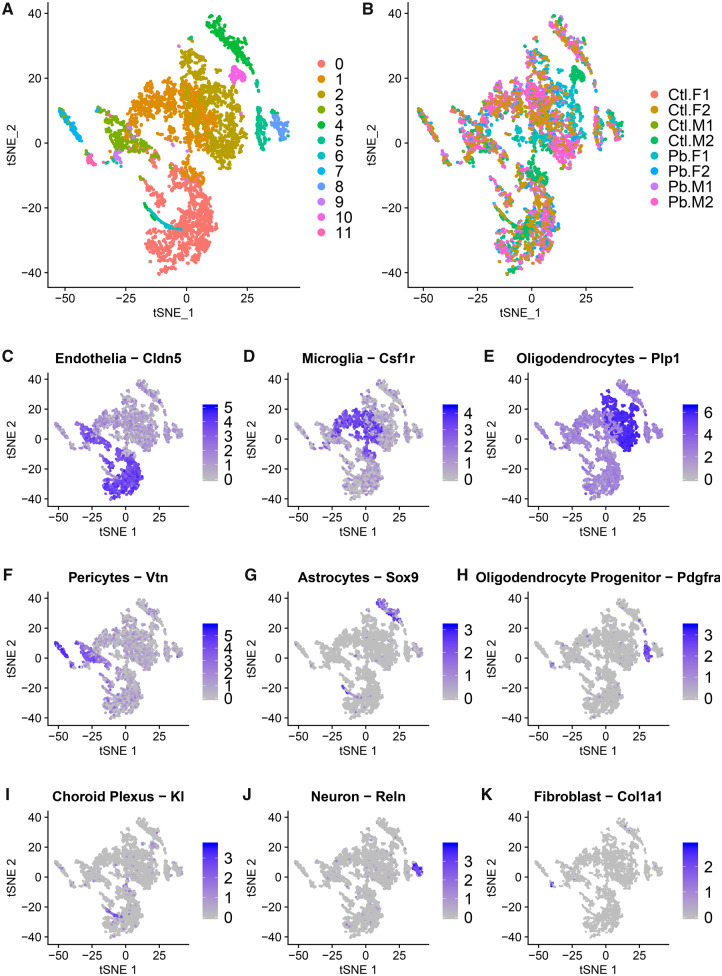
Unsupervised cell clustering resulted in 12 cell clusters (Figure 1A). Clusters 0–5 and 7–9 had cells represented from each sample. Cluster 6 had no cells from 2 control samples and 1 Pb sample. Cluster 11 had no cells from 1 control sample. Cluster 10 cells were derived from 1 control sample (88 of 93 cells) and 1 Pb-exposed sample (Figure 1B). Cell clusters ranged in size from 44 to 1329 cells (Table 1).
Figure 1.
t-Distributed Stochastic Neighbor Embedding (tSNE) by cell clusters 0–11 (A) and sample (B). Samples are labeled as control samples (Ctl) and Pb treated (Pb), the sex of the sample is specified (male [M] and female [F]) and biological replicates are indicated 1–2. Figures are painted by known markers for cell types: Cldn5 for endothelial cells (C), Csf1r for microglial cells (D), Plp1 for oligodendrocytes (E), Vtn for pericytes (F), Sox9 for astrocytes (G), Pdgfra for oligodendrocyte precursors (H), Kl for choroid plexus (I), Reln for neurons (J), and Col1a1 for fibroblasts (K).
Table 1.
Number of Cells Observed per Cluster and the Predicted Cell Type Identity of That Cluster
| Cluster | Number of Cells | Predicted Identity | Top Marker Genes | Ctrl % | Pb % | p Value |
|---|---|---|---|---|---|---|
| 0 | 1329 | Endothelial | Itm2a, Cldn5, Slco1a4 | 26.85 | 24.56 | .67 |
| 1 | 1303 | Microglia | Hexb, C1qc, Ctss | 26.61 | 23.95 | .15a |
| 2 | 1192 | Oligodendrocytes | Plp1, Aplp1, Cldn11 | 14.58 | 26.36 | .0002a |
| 3 | 356 | Pericytes | Myl9, Rgs5, Tagln | 7.23 | 6.56 | .66a |
| 4 | 306 | Astrocytes | Aldoc, Slc1a3, Slc1a2 | 5.59 | 5.92 | .85 |
| 5 | 181 | Oligodendrocyte progenitor | Pdgfra, Clql1, Matn4 | 2.49 | 3.88 | .04 |
| 6 | 129 | Choroid plexus | Kl, Igfbp2, 1500015O10Rik | 4.19 | 1.66 | .82 |
| 7 | 115 | Pericytes | Ndufa4l2, Ifitm1, Rgs4 | 1.88 | 2.33 | a |
| 8 | 109 | Neurons | Reln, Snhg11, Ndnf | 2.19 | 2.02 | .96 |
| 9 | 101 | Microglia | H2-Eb1, H2-Aa, H2-Ab1 | 2.25 | 1.77 | a |
| 10 | 93 | Oligodendrocytes | Ptprd, Ttll7, Ermn | 5.35 | 0.14 | a |
| 11 | 44 | Fibroblasts | Dcn, Spp1, Slc7a11 | 0.79 | 0.86 | .14 |
Microglia (1 and 9), oligodendrocyte (2 and 10), and pericyte (3 and 7) clusters were collapsed when testing for differences in proportions by Pb exposure.
We identified likely cell types for each cluster using published marker genes (Supplementary Table 2). Endothelial cells were the most abundant cells, identified in cluster 0 by their expression of Claudin 5 (Cldn5) (Figure 1C). Microglial cells were the next most abundant cells, identified in clusters 1 and 9 by their expression of colony stimulating factor 1 receptor (Csf1r) (Figure 1D). Oligodendrocytes can then be observed in clusters 2 and 10 by expression of Proteolipid protein 1 (Plp1) (Figure 1E). Pericytes were identified in clusters 3 and 7 by their expression of vitronectin (Vtn) (Figure 1F). Astrocytes were identified in cluster 4 by their expression of SRY-box 9 (Sox9) (Figure 1G). Cluster 5 is likely oligodendrocyte progenitor by their expression of platelet-derived growth factor receptor (Pdgfra) (Figure 1H). Klotho (Kl) expression marked choroid plexus cells in cluster 6 (Figure 1I). Neurons were identified in cluster 8 by their expression of Reelin (Reln) (Figure 1J). Lastly, fibroblasts were the least abundant cells, identified in cluster 11 by their expression of collagen type I alpha 1 (Col1a1) (Figure 1K).
The geometric mean number of genes expressed per cell varied by cell cluster (mean = 2537.2, range = 1658.3–3619.4) (Supplementary Figure 4). Cell types with the highest number of genes expressed were choroid plexus, pericytes, oligodendrocyte progenitor cells, and neurons (clusters 3, 5, 6, and 8). Cell types with the lowest number of expressed genes were microglia, astrocytes, and pericytes (clusters 1, 4, 7, and 9).
Overall Differences in Gene Expression by Pb Treatment
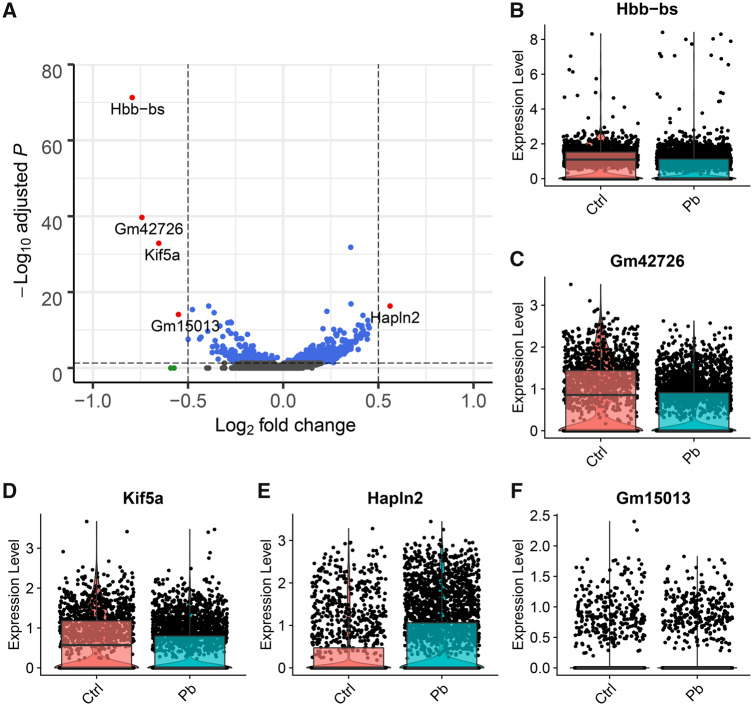
Across all cells (ncells = 5258) and all genes (ngenes = 17 143), we tested for differences in gene expression by Pb treatment (Supplementary Table 3). We observed 1230 genes that differed by Pb treatment (q value < 0.05), and of these, 5 had absolute log fold change (FC) > 0.5 (Figure 2A). Among these genes, only one (Hapln2) had higher expression with Pb treatment, whereas 4 had lower expression with Pb treatment. Hemoglobin, beta adult s chain (Hbb-bs) was 0.74 log-fold lower expressed in Pb-treated animals (q value = 5.0 × 10−72). A predicted gene on chromosome 5, Gm42726, had 0.74 log-fold lower expression in Pb-exposed animals than controls (q value = 2.0 × 10−40) (Figure 2B). Kinesin family member 5A (Kif5a) was 0.65 log-fold lower expressed in Pb-exposed animals than controls (q value = 1.3 × 10−33) (Figure 2C). Hyaluronan and proteoglycan link protein 2 (Hapln2) was 0.56 log-fold higher expressed in Pb-exposed animals than controls (q value = 4.7 × 10−17) (Figure 2D). Predicted gene Gm15013 on the X chromosome was 0.55 log-fold lower expressed in Pb-exposed animals than controls (q value = 7.8 × 10−15) (Figure 2E). These genes are not uniformly expressed by all cell types (Supplementary Figure 5).
Figure 2.
Across all cells measured, genes differentially expressed in the mouse hippocampus at 5 months of age following perinatal Pb exposure. Volcano plot of the log(fold change) difference in expression between Pb exposed and control versus the −log10(FDR q value) for the test statistic. Each point represents a gene. Genes reaching q value < 0.05 are blue, genes reaching log(fold change) > ± 0.5 are green, and genes reaching both thresholds are red (A). Violin plots showing per cell gene expression in Pb-exposed animals versus controls for Hbb-bs (B) Gm42726 (C), Kif5a (D), Hapln2 (E), and Gm15013 (F). FDR: false discovery rate.
To assess the contribution of cell cluster proportions to the overall Pb differences, we tested the overall Pb differentially expressed gene list for enrichment in cell cluster marker genes. Specifically, the top 10 markers for oligodendrocytes in cluster 2 (Plp1, Aplp1, Cldn11, Cnp, Ptgds, Car2, Mag, Cryab, Mog, and Mal) and the top 10 markers for oligodendrocyte progenitor cells in cluster 5 (Tnr, Lhfpl3, Cacng4, Cdo1, Gpr17, Neu4, 3110035E14Rik, Opcml, Pcdh15, and Vcan) were among the Pb treatment bulk analysis 1230 differentially expressed genes with q value < 0.05 (Fisher p value for overlap = 3.5 × 10−12). Pb treatment bulk analysis contained several of the top 10 cluster markers for oligodendrocytes in cluster 10 (Ptprd, Pcdh9, Kcna1, Edil3, Agpat4, Ttll7, Cntn2). Overall Pb treatment genes were significantly enriched for cluster 10 marker genes (Fisher p value for overlap = 9.5 × 10−7).
Human Psychiatric Disorder Gene Enrichment Testing
We compared overall Pb treatment genes that mapped to the human genome (N = 1517, p value < .01) with human psychiatric cross-disorder genes (n = 3676, p value < .01). Overall Pb treatment genes were highly enriched for human psychiatric cross-disorder genes (p value = .002, OR: 1.25, 95% CI: 1.08, 1.44). The top overlapping genes (p value < 10−3) in both datasets were SEZ6L2, HSPA1B, PEX5L, PTPRD, KAZN, CKB, DSCAM, NRXN3, DDN, NRXN2, F2, NALCN, DPP10, PPP1R16B, NREP, CSMD1, CACNA1S, PUF60, TOP1, HLA-DMB, CNTN6, RPEL1, PAK6, PCDH15, SEC14L5, SPON1, AGAP1, SUFU, LRMP, ZDHHC2, NSG1, and PTPRO. We compared Pb treatment genes with each of the 5 individual disorders (p value < 10−3) comprising the cross-disorder analysis. Pb treatment genes were most highly enriched in major depressive disorder genes (p value = .003, OR: 1.36, 95% CI: 1.11, 1.65). Pb treatment genes were marginally enriched for attention deficit-hyperactivity disorder genes (p value = .03, OR: 1.33, 95% CI: 1.01, 1.73) and autism spectrum disorder genes (p value = .05, OR: 1.42, 95% CI: 0.98, 2.02). Pb treatment genes were not enriched for bipolar disorder (p value = .10, OR: 1.18, 95% CI: 0.96, 1.44) or schizophrenia genes (p value = .51, OR: 1.06, 95% CI: 0.88, 1.28).
Pb Treatment-Related Differences Within Cell Clusters
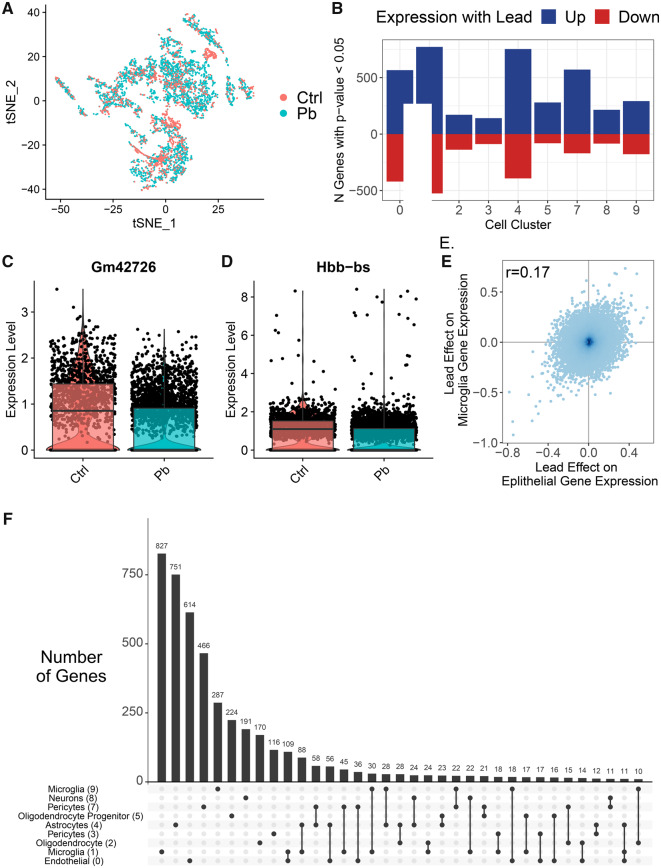
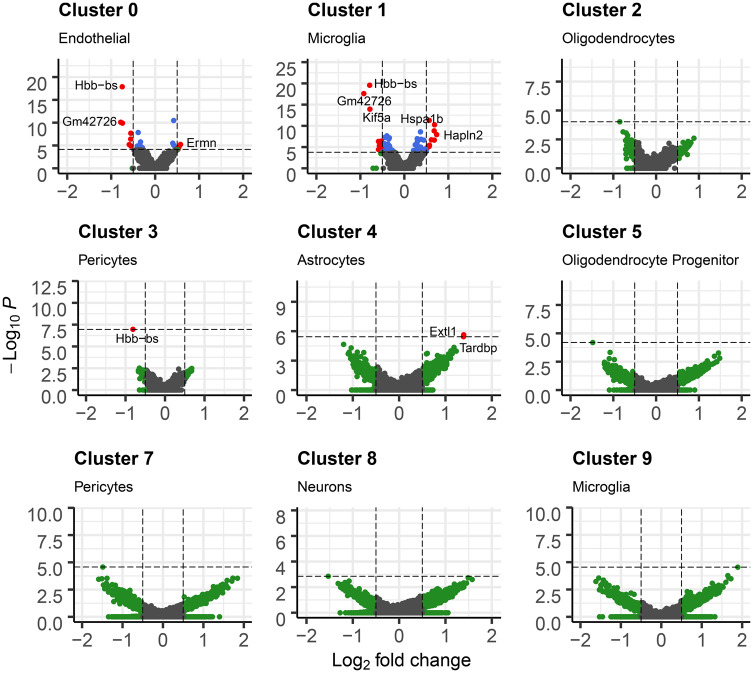
We tested for differences in gene expression by Pb treatment on a cluster-specific basis (Supplementary Table 4). Due to zero cell counts for some samples in clusters 6, 10, and 11, we were unable to run tests in those clusters. Among the remaining clusters, we observed varying numbers of nominally associated genes (p value < .05) in each cluster, with more nominal genes in clusters 0, 1, 4, and 7. At more stringent levels, we found genes with differential expression by Pb treatment (q value < 0.05 and absolute log[FC] > 0.5) in 4 clusters: clusters 0, 1, 3, and 4 (Figure 3). Microglia (cluster 1) had 22 genes differentially expressed with Pb. The top genes were Gm42726, which had 0.92 lower log(FC) expression by Pb (Figure 4C) and Hbb-bs which had 0.79 lower log(FC) expression by Pb (Figure 4D). Endothelial cells (cluster 0) had 9 genes differentially expressed by Pb. Pericytes (cluster 3) had 1 such gene, and astrocytes (cluster 4) had 2 genes meeting those criteria.
Figure 3.
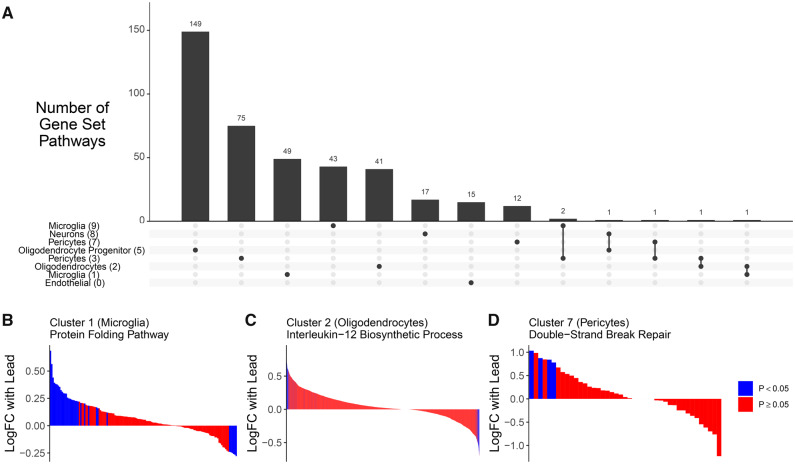
Cluster-specific differences in gene expression by Pb exposure. tSNE by Pb exposure (A). Barchart of number of nominal (p value < .05) differentially expressed genes by Pb exposure per cluster (B). Violin plots of gene expression levels in cluster 1 cells of Gm42726 (C) and Hbb-bs (D). Scatter plot of log(fold change) estimates of lead effect on gene expression in epithelial cluster and microglia cluster (E). Upset plot of genes different/common across clusters with nominal p value (<.05). Each column corresponds to a set of clusters, shown by filled in dots. The upper bar chart shows number of nominal genes in that set of clusters. Additional intersections with < 10 genes not shown in plot (F).
Figure 4.
Multipanel volcano plot for each cell cluster representing the associations between Pb treatment and gene expression. Cutoff for log fold change is 0.05. p value cutoffs were set at maximum p value if no genes had FDR < 0.05, or at largest p value that had FDR < 0.05. Points are gray if genes do not meet log fold change or p value cutoff, green if they only meet p value cutoff, blue if genes only meet log fold change cutoff, and red if genes meet both log fold change and p value cutoff.
To assess the consistency of Pb’s effects across cell types, we tested the pairwise Spearman correlations of gene-specific effect estimates. Log(FC) by Pb treatment were most correlated between endothelial (cluster 0) and microglia (cluster 1), with Spearman r = 0.17 (Figure 4E). Pairwise log(FC) estimate relationships among other clusters had correlations r < 0.1, except for endothelial (cluster 0) versus pericytes (cluster 3) (r = 0.16) and microglia (cluster 1) versus pericytes (cluster 3) (r = 0.13) (Supplementary Figure 6). Most genes differentially expressed by Pb treatment within clusters were cluster specific, with comparatively few genes overlapping between clusters (Figure 4F). A total of 166 genes were nominally differentially expressed in 3 or more clusters. One gene, Hbb-bs, was nominal in all clusters, except in neurons (cluster 8).
We conducted a post hoc differential gene expression power analysis using a negative binomial model, 4 samples per treatment group, and varying numbers of cells, ranging from all cells in the study (n = 5258) in the case of simulated analysis across all clusters down to the number of cells in cluster 4 (n = 181). Assuming a scenario with 1000 Pb treatment truly significantly associated genes, we calculated the proportion of genes detected at various magnitudes (FC) of association with Pb. The range of FCs examined is within those observed in this study. For the all clusters analysis and the larger cell clusters, we are reasonably well powered to detect associations (up to 100%), particularly for the larger FC genes (Supplementary Table 5). Even for the smaller cell clusters, we would be able to identify at least 5% of true associations. These analyses emphasize the utility of pioneering these methods for toxicology applications.
Biological Processes Gene Set Enrichment
Gene set enrichment by Pb treatment was performed on an overall and cluster-specific basis. In our overall analysis (similar to a bulk RNA analysis), the top pathways in Pb treatment were upregulation of oligodendrocyte differentiation and ensheathment of neurons, and downregulation of lymphocyte activation (Supplementary Table 6).
The majority of genes implicated in cluster-specific pathway analyses were unique to a single cluster (Figure 5A). The top pathways in cluster-specific analysis include enrichment in the following gene sets: downregulation of mRNA poly(A) tail shortening (endothelial cells in cluster 0), upregulation of protein folding (microglia in cluster 1, Figure 5B), upregulation of interleukin-12 biosynthetic process (oligodendrocytes in cluster 2, Figure 5C), down regulation of regulation of tyrosine phosphorylation of Stat5 protein (pericytes in cluster 3), upregulation of regulation of plasma lipoprotein particle levels pathway (astrocytes in cluster 4), upregulation of positive regulation of stem cell differentiation pathway (oligodendrocyte progenitor cells in cluster 5), downregulation of double-strand break repair via nonhomologous end joining (pericytes in cluster 7), downregulation of phospholipase C-activating G-protein coupled receptor signaling pathway (neurons in cluster 8), and upregulation negative regulation of protein oligomerization (microglia in cluster 9, Figure 5D) (Supplementary Table 7).
Figure 5.
Gene set enrichment analysis on cluster-specific gene expression differences with Pb exposure in the hippocampus. In clusters 0–3, 5, and 7–9, gene ontologies were identified with FDR < 0.05 and these ontologies contained genes that were largely unique (A). In panels (B) and (C), genes within a cluster are shown on the x-axis, ranked by their magnitude of association with Pb exposure and painted by the level of significance of their association with Pb exposure. In cluster 1, the protein folding pathway was upregulated (GO:0006457) (B). In cluster 2, the interleukin 12 biosynthetic process pathway was upregulated (GO:0006950) (C). In cluster 7, double-strand break repair via nonhomologous end joining pathway was downregulated (GO:0006303) (D).
Pb Treatment Differences in Cell Cluster Proportions
We observed Pb-treated cells were nonrandomly distributed across cell clusters (Figure 3A). We tested for differences in the proportions of cells annotated to cell types by treatment group (Supplementary Figure 7) with beta regression. The proportion of oligodendrocyte cells was higher on average with Pb treatment (p value = .0002), and the proportion of oligodendrocyte progenitor cells was marginally higher with Pb treatment on average (p value = .04) (Table 1). These findings suggest that developmental Pb exposure may alter differentiation trajectories in the brain toward an oligodendrocyte lineage.
Assessing the Effects of Pb on Oligodendrocyte Lineage Differentiation In Vitro
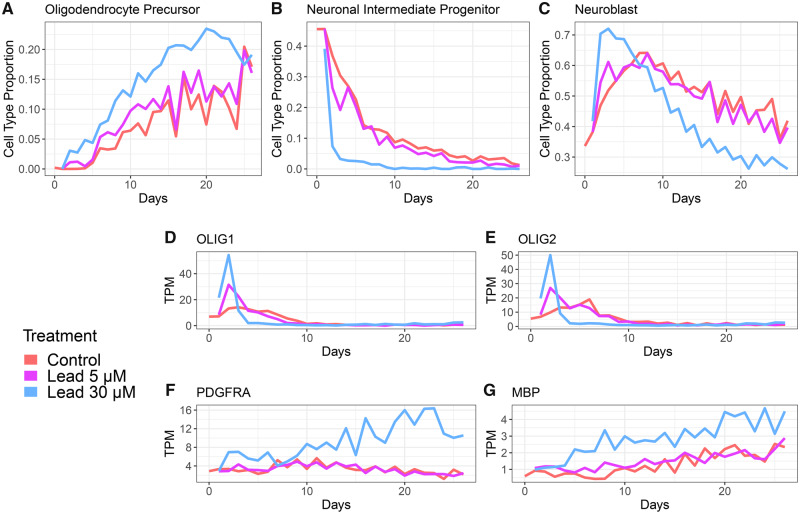
To further assess the impact of developmental Pb exposure on oligodendrocyte differentiation in a different model system, we reanalyzed data from a time course experiment in which human embryonic stem cell-derived neural progenitor cells were Pb exposed in vitro. Neural progenitor cell cultures were exposed to 3 or 30 µM Pb-acetate or control conditions and gene expression was measured daily for 26 days by bulk RNA-seq. Through bioinformatic deconvolution of cell type proportions from these bulk RNA-seq data, we observed a Pb dose-dependent increase in the estimated proportion of oligodendrocyte progenitor cells across time, with the highest estimates consistently appearing in the 30 µM lead treatment (Figure 6A). There was also an early drop in proportion of neuronal intermediate progenitors with 30 µM lead treatment (Figure 6B), along with an early spike in neuroblast proportion, which then decreased relative to control in the later time points (Figure 6C). The proportion of fully differentiated oligodendrocyte cells was predicted as 0 by MuSiC across all treatments and timepoints, suggesting that these cells are not found in the neuronal progenitor cell-derived cultures (Supplementary Figure 8). We further validated these findings through quantification of expression of 2 transcription factors which are essential for driving oligodendrocyte differentiation from neuronal stem cells: OLIG1 and OLIG2 (Dai et al., 2015; Lu et al., 2001; Yu et al., 2013). We observed a dose-dependent increase in the expression of OLIG1 and OLIG2 at the earliest time points (Figs. 6D and 6E). Moreover, at the 30 µM Pb dose, the expression of the oligodendrocyte progenitor marker gene PDGFRA was elevated relative to the control and 3 µM Pb doses (Figure 6F). Expression of an additional oligodendrocyte marker gene, MBP, was also consistently higher throughout the time period in the highest exposed group relative to the low and control groups (Figure 6G).
Figure 6.
Human neural progenitor cells treated with control (red), 3 µM (green), or 30 µM (blue) lead acetate (GSE84712). Daily RNA-sequencing analysis was performed until 26 days. Estimation of cell proportions (dataset used as reference panel: GSE95315) of oligodendrocyte precursors (A), neuronal intermediate progenitors (B), and neuroblasts (C). Expression levels in transcripts per million (TPM) of key oligodendrocyte genes OLIG1 (D), OLIG2 (E), OLIG3 (F), and MBP (G) are shown.
DISCUSSION
Toxicological and epidemiological evidence shows that early life exposure to Pb has profound impacts on the developing brain, with no safe level of Pb exposure identified. The mechanisms and effects of Pb’s effects in the brain, however, remain incompletely understood. Here, we used single-cell RNA-sequencing to compare gene expression patterns in adult mouse hippocampus following perinatal Pb treatment. These methods allow for unparalleled resolution of cell type-specific effects of Pb treatment. Through unbiased clustering and marker gene analyses, we identified and separated multiple known cell types in the brain, including oligodendrocytes, endothelial cells, microglia, neurons, pericytes, astrocytes, and fibroblasts. We found that in utero Pb treatment shifts the proportions of these cells and these findings were validated using data from an in vitro human neural progenitor Pb exposure experiment. In mice, we observed in utero Pb treatment also causes cell type-specific transcriptomic alterations which persist into adulthood. Many of these changes were in genes and pathways associated with psychiatric and neurological diseases, as well as in some novel genes and pathways not previously associated with Pb treatment. Overall, these findings highlight the power of single-cell analyses for the assessment of the cell type-specific effects of toxicant exposure and provide new mechanistic insights into the lifelong effects of developmental Pb treatment.
We identified significant shifts in adult hippocampal cell type proportions following perinatal Pb treatment. Specifically, Pb-exposed animals had significantly higher proportions of oligodendrocytes and oligodendrocyte progenitor cells relative to control animals. Oligodendrocyte proliferation is a known response to white matter damage (Armstrong et al., 2016). Our findings of oligodendrocytes being a particularly Pb-susceptible cell population corroborate others’ prior work in this area. In a second species (human vs mouse) and model system (cell culture vs animal model), we consistently observed that Pb treatment was associated with greater proportion of oligodendrocytes. In vitro treatment of oligodendrocyte progenitor cells with Pb caused decreased differentiation capacity, decreased myelin basic protein (MBP) expression, and decreased cell branching (Ma et al., 2015). Wistar rats exposed to 1% Pb in their drinking water for 3–6 months had significant hypomyelination and demyelination in their nerve fibers (Coria et al., 1984), reflecting aberrant oligodendrocyte function. In vitro studies show that oligodendrocyte progenitor cells are more susceptible to the effects of Pb than differentiated oligodendrocytes (Deng et al., 2001), providing further mechanistic evidence that link developmental Pb exposures to altered neurological function. In agreement with these previous studies, we also identified an increase in oligodendrocyte progenitor cells in Pb-exposed animals and human neuronal cell cultures. Both stress and glucocorticoid signaling have been linked to increased oligodendrocyte differentiation from hippocampal neural stem cells in a mouse model (Chetty et al., 2014). Our findings suggest that developmental exposure to a chemical stressor, Pb, can generate a similar phenotype. Further research is necessary to determine both the mechanism by which Pb may promote differentiation toward the oligodendrocyte lineage as well as the mechanisms by which Pb exposure impacts mature oligodendrocyte function.
Of relevance to interpreting results from bulk tissue molecular profiling studies in toxicology, we also simulated the standard approach in the field for differential expression analysis by comparing gene expression across bulk tissues in treated and control animals. In these analyses, we identified overall gene expression differences with Pb treatment that were driven by differences in cell proportions, specifically greater numbers of oligodendrocytes with Pb treatment. Indeed, the top cell type markers for multiple clusters, including oligodendrocytes and oligodendrocyte progenitor cells, predominated in the list of differentially expressed genes. These results highlight the importance of considering cell type heterogeneity and alterations in cell type proportions as major drivers of differences in bulk tissues molecular profiles between treated and control animals. Moreover, bulk tissue studies can be prone to contamination from surrounding tissue. In only one of our samples, we identified a gene expression signature of the choroid plexus, a brain region adjacent to the hippocampus. We suspect that these additional cells were captured during the dissection process of this sample. Previous studies of differences in hippocampal gene expression by treatment have identified known markers of the choroid plexus, including Ttr, Igfbp2, and Kl as the top differentially expressed genes (Sárvári et al., 2015). Our results suggest that these findings could actually reflect differential hippocampus contamination with choroid plexus tissue in the experimental groups. The precision to be able to detect and isolate this contamination is a significant advantage of single-cell RNA-seq analyses in toxicology studies.
Many of our findings of genes and pathways differentially expressed with in utero Pb treatment align with recent literature linking altered gene expression, genetic variants, or known biological mechanisms associated with psychiatric and neurological diseases. We observed Pb treatment response genes were enriched in psychiatric cross-disorder genes, annotated from prior genome-wide association studies. In addition, we identified that protein folding pathways were significantly downregulated in microglial cells with Pb treatment. Dysregulation of protein folding is a major underlying etiologic factor for the development of neurodegenerative disease (Selkoe, 2003; Stocker et al., 2020). One of our top downregulated genes overall, and within individual clusters, was Hbb-bs. A genetic mouse model for Alzheimer’s disease, the Tg2576 mouse has lower levels of Hbb-bs in their hippocampus relative to wild-type mice (Wu et al., 2019), suggesting that this gene may play a role in Alzheimer’s pathology in the hippocampus. We also identified significant downregulation of Kif5a, a kinesin gene involved in organelle transport. KIF5A is downregulated in postmortem temporal lobe samples from Alzheimer’s patients (Wang et al., 2019a), which impairs the axonal transport of mitochondria and mitochondrial trafficking in the neuron. KIF5A was also identified in a recent GWAS of amyotrophic lateral sclerosis (Nicolas et al., 2018). Alzheimer’s like pathology has been identified in aged nonhuman primates which were exposed to Pb starting at birth until 400 days of age (Wu et al., 2008). In addition, we observed upregulated plasma lipoprotein pathways in astrocytes. Lipoprotein levels and polymorphisms in Apolipoprotein E are hallmarks of Alzheimer’s disease risk (Liu et al., 2013). DNA damage is another factor in neurological disease, where high levels of double-strand breaks are observed in Alzheimer’s patients (Adamec et al., 1999; Shanbhag et al., 2019). This is consistent with elevated double-strand break repair pathways in pericytes in our study. Surprisingly, we were able to detect these psychiatric and neurological associated molecular alterations at only 5 months of age in mice. Our results suggest that molecular alterations due to the effects of Pb exposure persist into early adulthood and may precede histopathological alterations later in life. This work identifies in utero Pb treatment with overlapping gene signatures to psychiatric and neurological diseases, suggesting exposure to neurotoxicants early in life may relate to or mediate later life psychiatric and neurological disease outcomes.
Our results should be interpreted in light of a number of study limitations. Although we profiled the transcriptomes of thousands of cells, due to the nature of single-cell transcriptomic profiling experimental platforms, we were only able to assess effects in a small number of biological replicates. Future applications of single-cell RNA-seq in toxicology can take advantage of emerging methods, such as cell hashing with barcoded antibodies to allow for sample multiplexing during cell capture (Stoeckius et al., 2018), as well as decreasing high throughput sequencing costs to increase the number of biological replicates profiled. Single-cell transcriptomic studies are also conducted on dissociated tissue samples, which lose the spatial architecture of the tissue. Moreover, droplet-based platforms require viable cells and have differential capture rates based on cellular characteristics, such as shape and size, or genes expressed (Ye et al., 2019). For example, here we captured an overabundance of glial cells relative to neurons. Future single-cell studies of the neuron-specific effects of toxicant exposure could first capture neurons or neuronal nuclei using a fluorescence activated or magnetic bead-based sorting method to ensure a population of enriched cells for downstream analysis. Emerging literature also identifies background contamination of highly expressed transcripts (a so-called “soup” effect) that is detectable across all droplets (Young and Behjati, 2018), likely due to dying cells releasing RNA into the buffer solution being run through the Chromium instrument, which are then detectable in all droplets containing that buffer. Ongoing improvements in experimental technology and bioinformatic analysis methods will likely dramatically increase the utility of single-cell transcriptomics for toxicology studies. Future studies could also integrate orthogonal validation techniques, such as immunohistochemistry analysis or RNA in situ hybridization approaches in intact tissues. These analyses were not available for the mice in this study, which were part of the TaRGET II Consortium. All tissues were earmarked for high throughput omics analyses, rather than pathologic assessment. Ongoing analyses by the consortium, including whole transcriptome and methylome profiling of other brain tissues, are current underway for integration with these single-cell data.
Our study also has a number of strengths. To the best of our knowledge, this is the first single-cell analysis of the effects of a neurotoxicant on the brain. This approach allowed us to simultaneously assess the effects of Pb on the cell type proportions of the developing brain as well as identify cell type-specific effects. Deconvoluting the effects of a toxicant is essential for identifying mechanisms of action and biomarkers of exposure, for example, identifying whether an exposure during development alters the cell type proportions in the adult organ or alters gene expression patterns of individual cell types in a tissue. Our study also used an established exposure paradigm to understand the effects of perinatal toxicant exposure conducted in the context of a large national consortium. In our model system, the human relevant perinatal Pb treatment dose tested (32 ppm) and sample collection time point (5 months) have well-documented toxic effects (Faulk et al., 2013, 2014a,b; Montrose et al., 2017; Svoboda et al., 2019), including DNA methylation effects in the cortex (Dou et al., 2019). Further, we validated our finding of increased proportion of oligodendrocyte lineage cells using data from an in vitro study of Pb-exposed human neural progenitor cells. Overall, these results inform our understanding of the molecular effects of early life Pb exposure as well as the interpretation of data from bulk tissue RNA-seq data in toxicology experiments.
Pb exposure remains a persistent public health challenge in the United States and worldwide. Our results here use single-cell transcriptomic profiling to help to elucidate the molecular effects of early life exposure to Pb. We identify that glial cell populations, particularly cellular proportions of oligodendrocytes and gene expression in microglia, appear to be the most susceptible to the effects of Pb, and that these effects persist into adulthood. Neurons were underrepresented in our sample. We also identified a number of important genes and pathways simultaneously associated with neurodegenerative diseases and Pb exposure. Future work should characterize the mechanism by which these alterations lead to frank pathology in the brain through aging, whether through epigenetic alterations, oxidative stress, or persistently activated inflammatory pathways. Future work should also consider sex-stratification as the effects of perinatal Pb exposure may vary by sex. Cutting edge techniques, including single-cell RNA-seq and spatial transcriptomic profiling, will continue to provide new insights to these biological processes at unprecedented resolution.
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
NIEHS TaRGET II Consortium award to the University of Michigan (U01 ES026697) for the mouse exposure study; NIEHS Michigan Center on Lifestage Environmental Exposures and Disease (M-LEEaD; P30 ES017885) for the sequencing experiment; NIH (grants R01 ES025531, R01 ES025574, R01 AG055406, R01 MD013299, UG3 OD023285, UH3 OD023285, and P30 AG053760 to J.F.D. and K.M.B.); NIH (R01 ES028802 and UL1TR002240 to J.A.C.); NIEHS (T32 ES007062 to B.P.U.P.); NIEHS (T32 ES007062 to K.N.); and NICHD (T32 HD079342 to K.N.).
Supplementary Material
ACKNOWLEDGMENTS
Sequencing data are available on the Gene Expression Omnibus (GEO at https://www.ncbi.nlm.nih.gov/geo/ with accession number pending). Code to produce all R analyses in this manuscript is available (https://github.com/bakulskilab).
REFERENCES
- Adamec E., Vonsattel J. P., Nixon R. A. (1999). DNA strand breaks in Alzheimer’s disease. Brain Res. 849, 67–77. [DOI] [PubMed] [Google Scholar]
- An J., Cai T., Che H., Yu T., Cao Z., Liu X., Zhao F., Jing J., Shen X., Liu M., et al. (2014). The changes of miRNA expression in rat hippocampus following chronic lead exposure. Toxicol. Lett. 229, 158–166. [DOI] [PubMed] [Google Scholar]
- Armstrong R. C., Mierzwa A. J., Sullivan G. M., Sanchez M. A. (2016). Myelin and oligodendrocyte lineage cells in white matter pathology and plasticity after traumatic brain injury. Neuropharmacology 110, 654–659. [DOI] [PubMed] [Google Scholar]
- Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium. (2017). Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol. Autism 8, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakulski K. M., Dolinoy D. C., Sartor M. A., Paulson H. L., Konen J. R., Lieberman A. P., Albin R. L., Hu H., Rozek L. S. (2012). Genome-wide DNA methylation differences between late-onset Alzheimer’s disease and cognitively normal controls in human frontal cortex. J. Alzheimers Dis. 29, 571–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellinger D. C., Needleman H. L., Leviton A., Waternaux C., Rabinowitz M. B., Nichols M. L. (1984). Early sensory-motor development and prenatal exposure to lead. Neurobehav. Toxicol. Teratol. 6, 387–402. [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetty S., Friedman A. R., Taravosh-Lahn K., Kirby E. D., Mirescu C., Guo F., Krupik D., Nicholas A., Geraghty A. C., Krishnamurthy A., et al. (2014). Stress and glucocorticoids promote oligodendrogenesis in the adult hippocampus. Mol. Psychiatry 19, 1275–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coria F., Berciano M. T., Berciano J., Lafarga M. (1984). Axon membrane remodeling in the lead-induced demyelinating neuropathy of the rat. Brain Res. 291, 369–372. [DOI] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. (2013). Identification of risk loci with shared effects on five major psychiatric disorders: A genome-wide analysis. Lancet 381, 1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., Bercury K. K., Ahrendsen J. T., Macklin W. B. (2015). Olig1 function is required for oligodendrocyte differentiation in the mouse brain. J. Neurosci. 35, 4386–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis D., Walters R. K., Martin J., Mattheisen M., Als T. D., Agerbo E., Baldursson G., Belliveau R., Bybjerg-Grauholm J., Bækvad-Hansen M., et al. (2019). Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 51, 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., McKinnon R. D., Poretz R. D. (2001). Lead exposure delays the differentiation of oligodendroglial progenitors in vitro. Toxicol. Appl. Pharmacol. 174, 235–244. [DOI] [PubMed] [Google Scholar]
- Dissanayake V., Erickson T. B. (2012). Ball and chain: The global burden of lead poisoning. Clin. Toxicol. (Phila). 50, 528–531. [DOI] [PubMed] [Google Scholar]
- Dou J. F., Farooqui Z., Faulk C. D., Barks A. K., Jones T., Dolinoy D. C., Bakulski K. M. (2019). Perinatal lead (Pb) exposure and cortical neuron-specific DNA methylation in male mice. Genes (Basel) 10, 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S., Spellman P. T., Birney E., Huber W. (2009). Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulk C., Barks A., Liu K., Goodrich J. M., Dolinoy D. C. (2013). Early-life lead exposure results in dose- and sex-specific effects on weight and epigenetic gene regulation in weanling mice. Epigenomics 5, 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulk C., Barks A., Sánchez B. N., Zhang Z., Anderson O. S., Peterson K. E., Dolinoy D. C. (2014. a). Perinatal lead (Pb) exposure results in sex-specific effects on food intake, fat, weight, and insulin response across the murine life-course. PLoS One 9, e104273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulk C., Liu K., Barks A., Goodrich J. M., Dolinoy D. C. (2014. b). Longitudinal epigenetic drift in mice perinatally exposed to lead. Epigenetics 9, 934–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulk C., Liu K., Barks A., Goodrich J. M., Dolinoy D. C. (2014. c). Longitudinal epigenetic drift in mice perinatally exposed to lead. Epigenetics 9, 934–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochgerner H., Zeisel A., Lönnerberg P., Linnarsson S. (2018). Conserved properties of dentate gyrus neurogenesis across postnatal development revealed by single-cell RNA sequencing. Nat. Neurosci. 21, 290–299. [DOI] [PubMed] [Google Scholar]
- Howard D. M., Adams M. J., Clarke T.-K., Hafferty J. D., Gibson J., Shirali M., Coleman J. R. I., Hagenaars S. P., Ward J., Wigmore E. M., et al. (2019). Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci. 22, 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A. E., Irizarry R. A. (2014). Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 15, R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P., Hou Z., Bolin J. M., Thomson J. A., Stewart R. (2017). RNA-seq of human neural progenitor cells exposed to lead (Pb) reveals transcriptome dynamics, splicing alterations and disease risk associations. Toxicol. Sci. 159, 251–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Patil S., Sartor M. A. (2016). RNA-enrich: A cut-off free functional enrichment testing method for RNA-seq with improved detection power. Bioinformatics 32, 1100–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. C., Liu C. C., Kanekiyo T., Xu H., Bu G. (2013). Apolipoprotein e and Alzheimer disease: Risk, mechanisms and therapy. Nat. Rev. Neurol. 9, 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. T., Chen B. Y., Zhang J. Q., Kuang F., Chen L. W. (2015). Lead exposure induced microgliosis and astrogliosis in hippocampus of young mice potentially by triggering TLR4-MyD88-NFκB signaling cascades. Toxicol. Lett. 239, 97–107. [DOI] [PubMed] [Google Scholar]
- Lu Q. R., Cai L., Rowitch D., Cepko C. L., Stiles C. D. (2001). Ectopic expression of olig1 promotes oligodendrocyte formation and reduces neuronal survival in developing mouse cortex. Nat. Neurosci. 4, 973–974. [DOI] [PubMed] [Google Scholar]
- Ma T., Wu X., Cai Q., Wang Y., Xiao L., Tian Y., Li H. (2015). Lead poisoning disturbs oligodendrocytes differentiation involved in decreased expression of NCX3 inducing intracellular calcium overload. Int. J. Mol. Sci. 16, 19096–19110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maaten L., Hinton G. (2008). Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605. [Google Scholar]
- Malloy M. A., Kochmanski J. J., Jones T. R., Colacino J. A., Goodrich J. M., Dolinoy D. C., Svoboda L. K. (2019). Perinatal bisphenol A exposure and reprogramming of imprinted gene expression in the adult mouse brain. Front. Genet. 10, 951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montrose L., Faulk C., Francis J., Dolinoy D. C. (2017). Perinatal lead (Pb) exposure results in sex and tissue-dependent adult DNA methylation alterations in murine IAP transposons. Environ. Mol. Mutagen. 58, 540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman H. L., Schell A., Bellinger D., Leviton A., Allred E. N. (1990). The long-term effects of exposure to low doses of lead in childhood. An 11-year follow-up report. N. Engl. J. Med. 322, 83–88. [DOI] [PubMed] [Google Scholar]
- Nicolas A., Kenna K. P., Renton A. E., Ticozzi N., Faghri F., Chia R., Dominov J. A., Kenna B. J., Nalls M. A., Keagle P., et al. (2018). Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron 97, 1268–1283.e1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond J., Brown M. J. (2017). Childhood blood lead levels in children aged <5 years—United States, 2009–2014. MMWR Surveill. Summ. 66, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders T., Liu Y., Buchner V., Tchounwou P. B. (2009). Neurotoxic effects and biomarkers of lead exposure: A review. Rev. Environ. Health 24, 15–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sárvári M., Kalló I., Hrabovszky E., Solymosi N., Rodolosse A., Vastagh C., Auer H., Liposits Z. (2015). Hippocampal gene expression is highly responsive to estradiol replacement in middle-aged female rats. Endocrinology 156, 2632–2645. [DOI] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J. S., Anderson D. W., Talsania K., Mettil W., Vadigepalli R. (2012). Effects of developmental lead exposure on the hippocampal transcriptome: influences of sex, developmental period, and lead exposure level. Toxicol. Sci. 129, 108–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe D. J. (2003). Folding proteins in fatal ways. Nature 426, 900–904. [DOI] [PubMed] [Google Scholar]
- Shanbhag N. M., Evans M. D., Mao W., Nana A. L., Seeley W. W., Adame A., Rissman R. A., Masliah E., Mucke L. (2019). Early neuronal accumulation of DNA double strand breaks in Alzheimer’s disease. Acta Neuropathol. Commun. 7, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C., Robinson M. D. (2018). Bias, robustness and scalability in single-cell differential expression analysis. Nat. Methods 15, 255–261. [DOI] [PubMed] [Google Scholar]
- Stahl E. A., Breen G., Forstner A. J., McQuillin A., Ripke S., Trubetskoy V., Mattheisen M., Wang Y., Coleman J. R. I., Gaspar H. A., et al. (2019). Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 51, 793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ståhl P. L., Salmén F., Vickovic S., Lundmark A., Navarro J. F., Magnusson J., Giacomello S., Asp M., Westholm J. O., Huss M., et al. (2016). Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82. [DOI] [PubMed] [Google Scholar]
- Steinfeld J. L. (1971). Medical aspects of childhood lead poisoning. Pediatrics 48, 464–468. [PubMed] [Google Scholar]
- Stocker H., Nabers A., Perna L., Mollers T., Rujescu D., Hartmann A., Holleczek B., Schöttker B., Gerwert K., Brenner H. (2020). Prediction of Alzheimer’s disease diagnosis within 14 years through Aβ misfolding in blood plasma compared to APOE4 status, and other risk factors. Alzheimers Dement. 16, 283–291. [DOI] [PubMed] [Google Scholar]
- Stoeckius M., Zheng S., Houck-Loomis B., Hao S., Yeung B. Z., Mauck W. M., Smibert P., Satija R. 3rd (2018). Cell hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 19, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda L., Neier K., Cavalcante R., Tsai Z., Jones T., Liu S., et al. (2019). Perinatal exposure to lead results in altered DNA methylation in adult mouse liver and blood: implications for target versus surrogate tissue use in environmental epigenetics. bioRxiv:783209.
- Wang Q., Tian J., Chen H., Du H., Guo L. (2019. a). Amyloid beta-mediated KIF5A deficiency disrupts anterograde axonal mitochondrial movement. Neurobiol. Dis. 127, 410–418. [DOI] [PubMed] [Google Scholar]
- Wang T., Pehrsson E. C., Purushotham D., Li D., Zhuo X., Zhang B., Lawson H. A., Province M. A., Krapp C., Lan Y., et al. (2018). The NIEHS TaRGET II Consortium and environmental epigenomics. Nat. Biotechnol. 36, 225–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Park J., Susztak K., Zhang N. R., Li M. (2019. b). Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat. Commun. 10, 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland R. A., Jirtle R. L. (2003). Transposable elements: Targets for early nutritional effects on epigenetic gene regulation. Mol. Cell. Biol. 23, 5293–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhouse C., Anderson O. S., Bergin I. L., Vandenbergh D. J., Gyekis J. P., Dingman M. A., Yang J., Dolinoy D. C. (2014). Dose-dependent incidence of hepatic tumors in adult mice following perinatal exposure to bisphenol A. Environ. Health Perspect. 122, 485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisskopf M. G., Hu H., Sparrow D., Lenkinski R. E., Wright R. O. (2007). Proton magnetic resonance spectroscopic evidence of glial effects of cumulative lead exposure in the adult human hippocampus. Environ. Health Perspect. 115, 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. 2007. Blood Lead Levels in Children. World Health Organization, Regional Office for Europe, European Environment and Health Information System (Fact Sheet No. 45). Copenhagen.
- WHO. 2010. Exposure to Lead: A Major Public Health Concern. World Health Organization, Preventing Disease Through Healthy Environments.
- Wu J., Basha M. R., Brock B., Cox D. P., Cardozo-Pelaez F., McPherson C. A., Harry J., Rice D. C., Maloney B., Chen D., et al. (2008). Alzheimer’s disease (AD)-like pathology in aged monkeys after infantile exposure to environmental metal lead (Pb): Evidence for a developmental origin and environmental link for AD. J. Neurosci. 28, 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Fang K., Wang W., Lin W., Guo L., Wang J. (2019). Identification of key genes and pathways for Alzheimer’s disease via combined analysis of genome-wide expression profiling in the hippocampus. Biophysics. Rep. 5, 98–109. [Google Scholar]
- Ye C., Speed T. P., Salim A. (2019). Decent: Differential expression with capture efficiency adjustment for single-cell RNA-seq data. Bioinformatics 35, 5155–5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. D., Behjati S. (2018). SoupX removes ambient RNA contamination from droplet based single cell RNA sequencing data. bioRxiv:303727. [DOI] [PMC free article] [PubMed]
- Yu Y., Chen Y., Kim B., Wang H., Zhao C., He X., Liu L., Liu W., Wu L. M. N., Mao M., et al. (2013). Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell 152, 248–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A., Munoz-Manchado A. B., Codeluppi S., Lonnerberg P., La Manno G., Jureus A., Marques S., Munguba H., He L., Betsholtz C., et al. (2015). Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.