Abstract
Arsenic exposure via drinking water is a serious environmental health concern. Epidemiological studies suggest a strong association between prenatal arsenic exposure and subsequent childhood respiratory infections, as well as morbidity from respiratory diseases in adulthood, long after systemic clearance of arsenic. We investigated the impact of exclusive prenatal arsenic exposure on the inflammatory immune response and respiratory health after an adult influenza A virus (IAV) lung infection. C57BL/6J mice were exposed to 100 ppb sodium arsenite in utero, and subsequently infected with IAV (H1N1) after maturation to adulthood. Assessment of lung tissue and bronchoalveolar lavage fluid at various time points post-IAV infection reveals greater lung damage and inflammation in arsenic-exposed mice versus control mice. Single-cell RNA sequencing analysis of immune cells harvested from IAV-infected lungs suggests that the enhanced inflammatory response is mediated by dysregulation of innate immune function of monocyte-derived macrophages, neutrophils, natural killer cells, and alveolar macrophages. Our results suggest that prenatal arsenic exposure results in lasting effects on the adult host innate immune response to IAV infection, long after exposure to arsenic, leading to greater immunopathology. This study provides the first direct evidence that exclusive prenatal exposure to arsenic in drinking water causes predisposition to a hyperinflammatory response to IAV infection in adult mice, which is associated with significant lung damage.
Keywords: arsenic, influenza, immunotoxicology, single-cell RNA-seq
It is estimated that over 200 million people worldwide are exposed to elevated levels of arsenic in their drinking water that exceed the 10 ppb limit established in 2001 as the maximum municipal drinking water concentration by the U.S. Environmental Protection Agency (Ayotte et al., 2017). Approximately 44 million individuals in the United States rely on unregulated private wells for their water, and of those it is estimated that approximately 2.5 million are exposed to >10 ppb of arsenic in their drinking water (Klein et al., 2012). Arsenic concentrations in the United States typically fall in the moderate range, but higher levels of contamination (> 100 ppb) have been documented (Ayotte et al., 2017; Karagas et al., 2002). Arsenic is an established carcinogen and is also associated with numerous noncancerous negative health outcomes (Chen and Ahsan, 2004; Naujokas et al., 2013). For example, multiple epidemiological studies have shown an association between arsenic ingestion and endocrine dysfunction, and increased respiratory infections (Davey et al., 2008; Farzan et al., 2016; Smith et al., 2006).
Recent epidemiological studies suggest that negative respiratory health effects of arsenic exposure are more prominent in individuals that are exposed to arsenic prenatally (Smith et al., 2006). Results from retrospective studies of arsenic exposure in Chile have linked perinatal arsenic exposure with bronchiectasis, a disease characterized by chronic airway infection, inflammation, and significant morbidity resulting from progressive airway damage, in adulthood, decades after the cessation of arsenic exposure (Roh et al., 2018). Additional clinical correlates were observed in a study of a well-characterized U.S. cohort of 412 infants born to mothers drinking well water during pregnancy (Farzan et al., 2015, 2016; Nadeau et al., 2014). In this study, maternal urinary arsenic concentrations were positively correlated with respiratory infections in infants requiring a physician visit (Farzan et al., 2016). Associations were also observed with reported respiratory symptoms and respiratory infections clinically significant enough (as judged by a primary care provider) to be treated with prescription medications (Farzan et al., 2016). However, investigating effects of early life exposures on morbidity in adulthood is challenging in human populations, and no studies to date have examined effects of prenatal arsenic exposure in a U.S. population on immune function in adulthood.
Previously, our group utilized a novel murine model to demonstrate that exposure of adult mice to 100 ppb of arsenic in drinking water resulted in dysregulated immune responses to a respiratory virus infection (H1N1 IAV) with enhanced inflammation and immunopathologic lung damage (Kozul et al., 2009a). Ramsey et al. (2013a,b), examined the effects of continuous prenatal with postnatal arsenic exposure in mice, and reported evidence of enhanced inflammatory responses in exposed animals infected with influenza A virus (IAV) at 1 week of life. However, there are no studies examining the effects of exclusive prenatal exposure of arsenic on the immune response to IAV infection in adult mice, months after exposure to arsenic. Thus, we conducted studies to test the hypothesis that prenatal arsenic exposure results in long-term effects on the adult host immune response to IAV infection, long after a finite exposure, leading to greater pulmonary immunopathology. In this study, mice that were exposed to 100 ppb sodium arsenite during gestation, via maternal drinking water, were allowed to mature immunologically to adulthood without arsenic and were then infected intranasally with a sublethal inoculum of IAV. We demonstrate that arsenic exposure enhanced lung inflammation and immunopathologic lung damage, with impairment of respiratory integrity in mice infected with IAV. We coupled our phenotypic studies with single-cell RNA sequencing (scRNA-seq) of immune cells early in the course of infection to obtain high resolution information about impacts of arsenic on individual cell types that presage the development of clinically significant lung damage. With our scRNA-seq analysis of immune cells in the lung, we demonstrate that the enhanced inflammatory response is likely mediated by dysregulation of innate immune function of monocyte-derived macrophages, neutrophils, alveolar macrophages (AM), and natural killer (NK) cells. To our knowledge this is the first scRNA-seq study to examine the effect of arsenic on IAV lung infection.
MATERIALS AND METHODS
Animal husbandry
All animal studies were conducted in accordance with guidelines approved by the Association for Assessment and Accreditation of Laboratory Animal Care using a protocol approved by the Institutional Animal Care and Use Committee (IACUC) at the Geisel School of Medicine at Dartmouth. In addition, studies were in compliance with the Guiding Principles in the Use of Animals in Toxicology. All animals were treated humanely and with regard for alleviation of suffering. Six-week-old C57BL/6J mice (Jackson Laboratories, Bar Harbor, Maine) were housed on Carefresh bedding (International Absorbents, Inc, Ferndale, Washington) in autoclaved cages and fed TEK-LAD AIN-76A diet (Envigo, Madison, Wisconsin) ad libitum. Background total arsenic concentration in the chow was < 20 ppb, only a small fraction of which is bioavailable inorganic species (Kozul et al., 2008). At the start of each experiment, F0 mice were placed on this diet for 2 weeks. Subsequently, mice were placed into breeding pairs and when female mice showed cervical plug formation, those in the arsenic treatment group were given drinking water with 100 ppb sodium arsenite ad libitum. The concentration of arsenic in drinking water was verified through ICP-MS, and arsenic was removed from drinking water immediately after pups were born. Thus, the F1 offspring were exposed to arsenic exclusively in utero and for the entirety of gestation. Average litter size observed for arsenic-exposed dams (13 dams: 6.42 ± 0.63 [SD] pups/litter) and -unexposed dams (12 dams: 6.95 ± 0.76 [SD] pups/litter) was not significantly different. F1 pups were randomized during weaning at 3 weeks of age within respective exposure and sex groups. F1 mice were allowed to mature to 7 and 8 weeks of age prior to experimental infection. Because studies have shown that IAV infection may have sex-dependent effects, the effects of prenatal arsenic exposure on host responses to IAV infection were examined in both male and female mice (Klein et al., 2012).
Influenza virus infection
Mice that were at least 7 and 8 weeks old, and matched for body weight, were anesthetized with ketamine/xylazine and inoculated intranasally with 0.5 LD50 of influenza A/PR8/34 as previously described (DeBerge et al., 2013). Morbidity was monitored over the course of infection by daily measurement of weight loss. No animals required sacrifice at this inoculum because of unacceptable morbidity (weight loss >20%).
Lung sample collection
Bronchoalveolar lavage fluid (BALF) was collected with an 18-gauge catheter inserted into the trachea after anesthesia. Lungs were lavaged in situ with 1 ml phosphate-buffered saline (PBS). Remaining cells were collected with 4 sequential aliquots. BALF albumin concentration was assessed on the first ml of fluid by ELISA (Bethyl Labs, Montgomery, Texas). Cytospin preparations of BALF were stained for immune cell enumeration using the Protocol Hema 3 stain set (Fisher, Houston, Texas). The concentration of 32 immunoregulatory cytokines (Eotaxin, G-CSF, GM-CSF, IFN-gamma, IL-10, IL-12 [p40], IL-12 [p70], IL-13, IL-15, IL-17, IL-1alpha, IL-1β, IL-2, IL-2, IL-4, IL-5, IL-6, IL-7, IL-9, IP-10, KC, LIF, LIX, M-CSF, MCP-1, MIG, MIP-1α, MIP-1β, MIP-2, RANTES, TNFα, VEGF) in BALF and whole-lung homogenate was measured by multiplex ELISA (Millipore, Danvers, Massachusetts). IL-1β in lung homogenate was validated with ELISA (BioLegend, San Diego, California).
Virus titers
Whole lungs from infected mice were snap frozen and stored at −80°C. Influenza virus titers were determined as TCID50 (50% tissue culture infective dose) of whole-lung homogenates on Madin-Darby canine kidney (MDCK) cells monolayers; 10x serial dilutions of lung whole-lung homogenates were added in triplicate to MDCK cells in 96-well plates and incubated at 37°C for 5 days. Points of limiting dilution were identified by chicken red blood cell hemagglutination (LaBarre and Lowy, 2001).
Statistical analysis
For logistical reasons, infections were conducted in batches on multiple days, always including arsenic exposed and control groups. Albumin, BALF cell count, viral titer, and cytokine data were analyzed using linear models with arsenic exposure and batch factors. Males and females were analyzed separately because sex and batch factors were not independent. Litter could not be accounted for in the statistical analyses, as would have been preferred. However, each experiment was composed of mice from multiple litters, and the number of mice and litters for all experiment are provided in Supplementary Table 1. Details of the statistical analyses are provided in the figure legends.
Whole-lung homogenate preparation for scRNA-seq
Lungs were perfused with PBS via the right ventricle, harvested, and mechanically disassociated prior to straining through 70- and 30-µm filters to obtain a single-cell suspension. Dead cells were removed (annexin V EasySep kit, StemCell Technologies, Vancouver, Canada), and samples were enriched for cells of hematopoietic origin by magnetic separation using anti-CD45-conjugated microbeads (Miltenyi, Auburn, California). Single-cell suspensions of 6 samples were loaded on a Chromium Single Cell system (10X Genomics) to generate barcoded single-cell gel beads in emulsion, and scRNA-seq libraries were prepared using Single Cell 3′ Version 2 chemistry. Libraries were multiplexed and sequenced on 4 lanes of a Nextseq 500 sequencer (Illumina) with 3 sequencing runs. Demultiplexing and barcode processing of raw sequencing data was conducted using Cell Ranger v. 3.0.1 (10X Genomics; Dartmouth Genomics Shared Resource Core). Reads were aligned to mouse (GRCm38) and IAV (A/PR8/34, genome build GCF_000865725.1) genomes to generate unique molecular index (UMI) count matrices. Gene expression data have been deposited in the NCBI GEO database and are available at accession No. GSE142047.
Flow cytometry
We collected whole lungs perfused with PBS via the right ventricle, and mechanically disassociated cells prior to straining through 70- µm and 30-µm filters to obtain a single-cell suspension. A780 CD45, APC CD11b, PerCP-Cy5.5 F4/80, PE NK1.1, PerCP-Cy5.5 CD3, PE-Cy7 Klrg1, and APC CD107a antibodies were purchased from BioLegend. FITC HA (hemagglutinin) was purchased from Virostat (Westbrook, Maine). Samples were run on Gallios 10 Color (Beckman Coulter, Indianapolis, Indiana), and all data were analyzed with Kaluza Analysis 2.1 software (Beckman Coulter).
Preprocessing of scRNA-seq data
Count matrices produced using Cell Ranger were analyzed in the R statistical working environment (version 3.6.1). Preliminary visualization and quality analysis were conducted using scran (v 1.14.3, Lun et al., 2016) and Scater (v. 1.14.1, McCarthy et al., 2017) to identify thresholds for cell quality and feature filtering. Sample matrices were imported into Seurat (v. 3.1.1, Stuart et al., 2019) and the percentage of mitochondrial, hemoglobin, and influenza A viral transcripts calculated per cell. Cells with < 1000 or >20 000 unique molecular identifiers (UMIs: low quality and doublets), fewer than 300 features (low quality), greater than 10% of reads mapped to mitochondrial genes (dying) or greater than 1% of reads mapped to hemoglobin genes (red blood cells) were filtered from further analysis. Total cells per sample after filtering ranged from 1895 to 2482, no significant difference in the number of cells was observed in arsenic versus control. Data were then normalized using SCTransform (Hafemeister and Satija, 2019) and variable features identified for each sample. Integration anchors between samples were identified using canonical correlation analysis and mutual nearest neighbors, as implemented in Seurat V3 (Stuart et al., 2019) and used to integrate samples into a shared space for further comparison. This process enables identification of shared populations of cells between samples, even in the presence of technical or biological differences, while also allowing for nonoverlapping populations that are unique to individual samples.
Clustering and reference-based cell identity labeling of single immune cells from IAV-infected lung with scRNA-seq
Principal components were identified from the integrated dataset and were used for Uniform Manifold Approximation and Projection (UMAP) visualization of the data in 2D space. A shared nearest neighbor graph was constructed using default parameters, and clusters identified using the SLM algorithm in Seurat at a range of resolutions (0.2–2). The first 30 principal components were used to identify 22 cell clusters ranging in size from 25 to 2310 cells. Gene markers for clusters were identified with the findMarkers function in scran. To label individual cells with cell type identities, we used the singleR package (v. 3.1.1) to compare gene expression profiles of individual cells with expression data from curated, FACS-sorted leukocyte samples in the Immgen compendium (Aran et al., 2019; Heng et al., 2008). We manually updated the Immgen reference annotation with 263 sample group labels for fine-grain analysis and 25 CD45+ cell type identities based on markers used to sort Immgen samples (Guilliams et al., 2014). The reference annotation is provided in Supplementary Table 2, cells that were not labeled confidently after label pruning were assigned “Unknown.”
Viral transcript detection in single immune cells by scRNA-seq
For each cell, reads that aligned to the influenza A genome segments (H1N1/PR8) were calculated both as a count and as a percentage of total reads from the cell. Infected cells were classified as cells with >0.05% viral reads, which Steuerman et al. (2018), proposed as a reasonable threshold for identification of infected cells in mouse lung with single cell sequencing. We then compared the percentage of infected cells per sample between arsenic and control groups, at both the sample-wide and cell-type-specific levels using linear models in R. We also examined the percentage of cells in which viral transcripts were detected (viral read count >0) and obtained similar results.
Differential abundance analysis of immune cell populations by scRNA-seq
To assess the effect of prenatal arsenic exposure on the distribution of host cells within cell types or clusters, we tested for differences in the proportion of cells in each cluster using EdgeR as described by Amezquita et al. (2019). Briefly, tables of cell type counts per sample across cell types were imported into EdgeR, the NB, and QL dispersions for each cluster were estimated, and differences in abundance were identified with the glmQLFTest. Differences in abundance with a false discovery rate (FDR)-corrected p value < .05 were considered significant.
Differential gene expression by immune cells
Differential gene expression within individual cell types was performed by pooling raw count data from cells of each cell type on a per-sample basis to create a pseudo-bulk count table for each cell type. Differential expression (DE) analysis was only performed on cell types that were sufficiently represented (> 10 cells) in each sample. In droplet-based scRNA-seq, ambient RNA from lysed cells is incorporated into droplets, and can result in spurious identification of these genes in cell types where they are not actually expressed. We therefore used a method developed by Young and Behjati (2018) to estimate the contribution of ambient RNA for each gene and identified genes in each cell type that were estimated to be >25% ambient-derived. These genes were excluded from analysis in a cell-type-specific manner. Genes expressed in less than 5% of cells were also excluded from analysis. DE analysis was then performed in Limma (limma-voom with quality weights) following a standard protocol for bulk RNA-seq (Law et al., 2014). Significant genes were identified using MA/QC criteria of p < .05, log2FC >1.
Analysis of arsenic effect on immune cell gene expression by scRNA-seq
Sample-wide effects of arsenic on gene expression were identified by pooling raw count data from all cells per sample to create a count table for pseudo-bulk gene expression analysis. Genes with less than 50 counts in any sample, or less than 150 total counts were excluded from analysis. DE analysis was performed using limma-voom as described above.
Gene set analysis of immune cells by scRNA-seq
DE of biological pathways in a sample-wide and cell-type-specific manner was performed using the Camera function in limma using Hallmark gene sets from the molecular signatures database (Liberzon et al., 2015). Pathways with FDR < .1 were considered significant. To compare pathway enrichment across cell types, we compiled a list of all pathways significant in at least one cell type and obtained FDR-corrected p values and direction of regulation in all cell clusters. Visualizations of these data were created using ggplot2 (v. 3.2.1).
RESULTS
Prenatal Arsenic Exposure and Predisposition to Lung Damage After IAV Infection
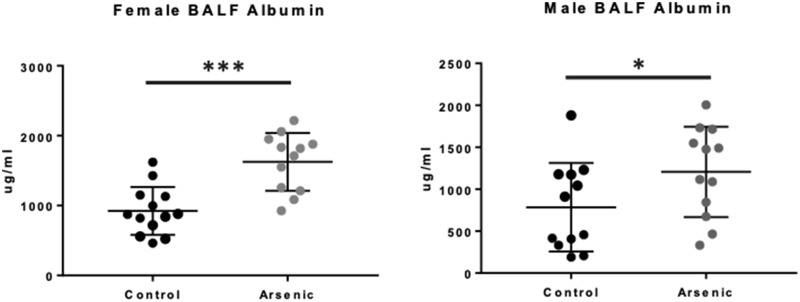
Weight loss was measured over the course of infection, and we observed no significant differences in weight loss in exposed versus unexposed mice (of either sex) over the course of 7 days post-IAV infection (Supplementary Figure 1). To assess the net impact of prenatal arsenic exposure on the structural and functional integrity of the infected lung, we measured the pathological leak of a serum protein otherwise prevented by the intact alveolar capillary membrane. Measurement of BALF albumin as a marker of its disrupted barrier function is a well-characterized indicator of lung damage (Dauber et al., 1985), from both direct viral cytopathic effects and host immune-mediated epithelial injury. We observed significantly greater BALF albumin concentrations on day 7 after IAV infection in arsenic-exposed versus -unexposed F1 mice, from both males and females (Figure 1).
Figure 1.
Effect of prenatal arsenic on immunopathology from adult influenza A virus (IAV) infection. Albumin concentration was measured by ELISA on bronchoalveolar lavage fluid (BALF) obtained from arsenic-exposed versus -unexposed mice at day 0 (n = 3) and day 7 IAV after infection (n = 10–13). Results from day 7 IAV infection are from 2 independent experiments pooled. Data are presented as means ± SD. Data were analyzed using linear models that accounted for arsenic exposure and batch. *p < .05, ***p < .001.
Impact of Prenatal Arsenic Exposure on Subsequent Respiratory Virus Replication
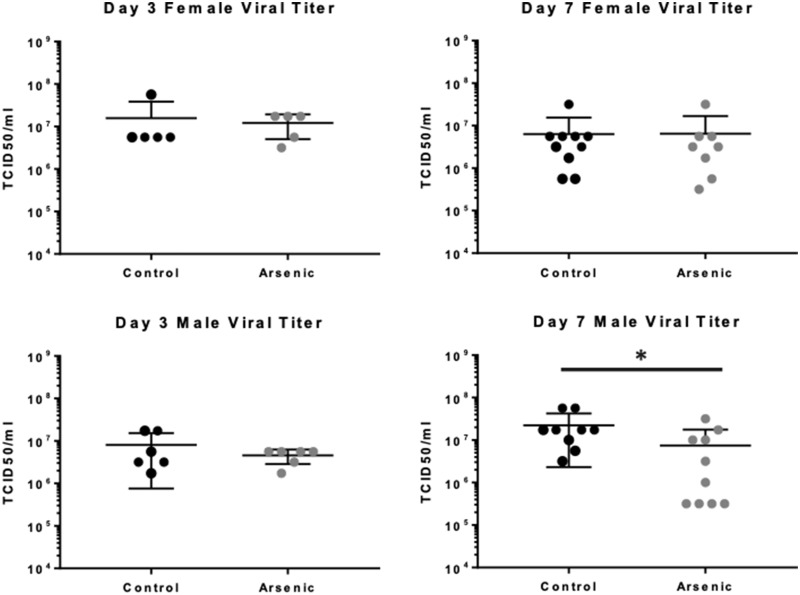
The kinetics of antiviral host responses may have a significant impact on lung damage in IAV infection, and previous work has shown that chronic and ongoing arsenic exposure of IAV-infected adult mice can result in impaired clearance of IAV (Kozul et al., 2009a). To determine if prenatal arsenic exposure predisposed adult mice to alterations in clearance after infection, we measured virus titers in whole-lung homogenates at days 3 and 7 after IAV infection. Titers were similar at both time points measured in exposed versus unexposed female mice, but we observed a slightly lower titer in exposed males at day 7 after infection (Figure 2), suggesting that prenatal arsenic may predispose to later alterations in virus clearance after subsequent infection.
Figure 2.
Effect of prenatal arsenic exposure on viral clearance from adult influenza A virus infection. TCID50 was assessed on Madin-Darby canine kidney cell cultures inoculated with serial dilutions of lung homogenates harvested on day 3 (n = 5–6) and day 7 (n = 8–10) after infection. Results are from 2 independent experiments pooled. Data are presented as means ± SD. Data were analyzed using linear models that accounted for arsenic exposure and batch. *p < .05.
Host Inflammatory Responses
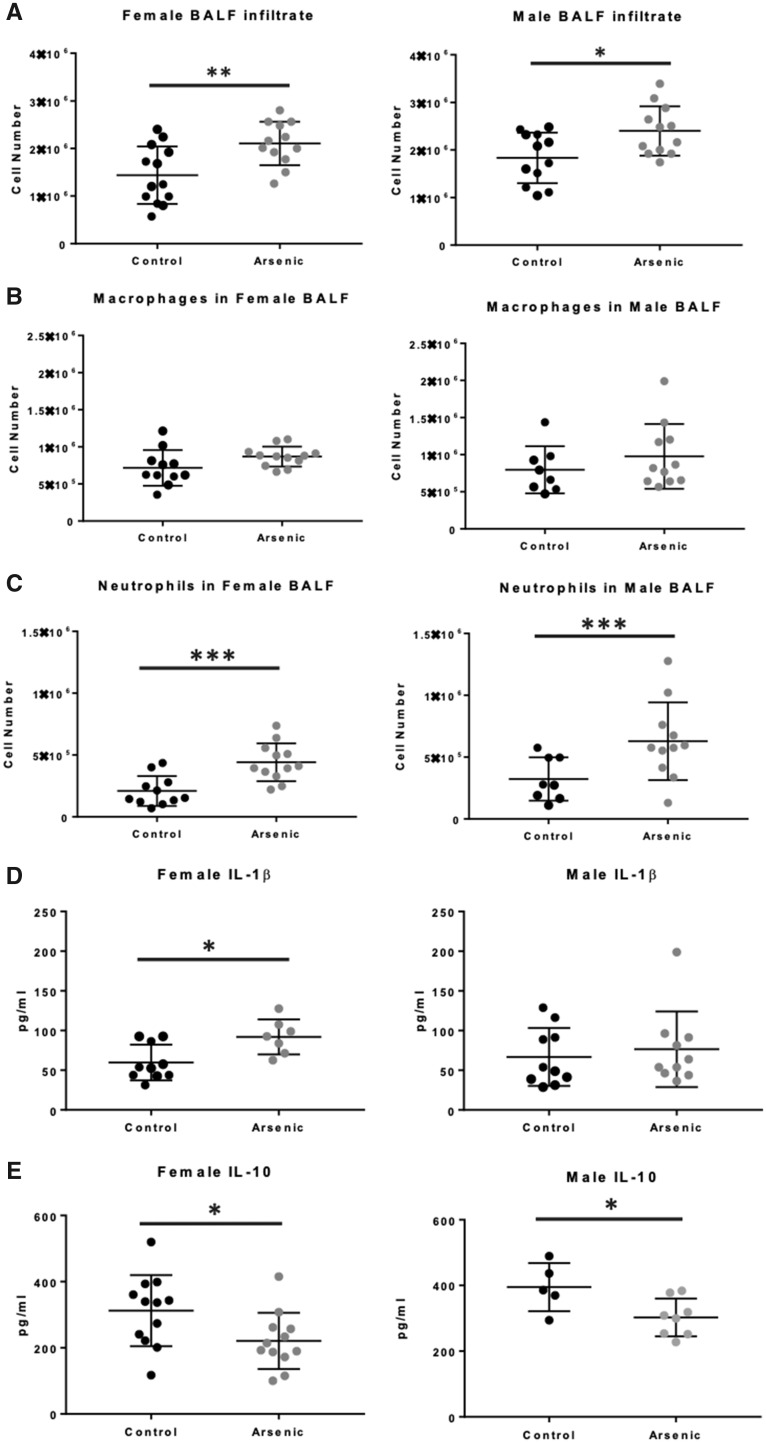
Previous studies have revealed that a significant component of damage to the airways in IAV infection is immunopathologic, ie, mediated by increased infiltration and effector activities of antiviral immune cells (Duan and Thomas, 2016). Prenatal arsenic exposure predisposed to an increase in immune cell infiltration, evident in the day 7 BALF from both male and female IAV-infected mice (Figure 3A). Although there was a small increase in the number of BALF macrophages in arsenic-exposed versus control-infected mice, the difference did not reach statistical significance (Figure 3B). However, in utero arsenic exposure predisposed to a significant increase in the neutrophil content of day 7 BALF of both male and female IAV-infected mice (Figure 3C).
Figure 3.
Effect of prenatal arsenic on adult inflammatory immune cell recruitment in influenza A virus (IAV) infection. A, Total immune cells in bronchoalveolar lavage fluid (BALF) at day 7 IAV infection were counted after trypan blue exclusion (n = 12–13 mice). B and C, Discrete leukocyte populations were counted by morphology on cytospin preparations (n = 8–12 mice). D and E, IL-1β and IL-10 expression from whole-lung homogenate at day 7 IAV infection from ELISA (n = 8–11 mice). Data are presented as means ± SD. IL-10 expression in BALF at day 7 IAV infection from multiplex. Females (n = 4–5). Males (n = 5–8, mice). Data were analyzed using linear models that accounted for arsenic exposure and batch. *p < .05, **p < .01, ***p < .001.
Studies were also conducted to examine the effect of arsenic exposure on a large panel of immunoregulatory cytokines in lung homogenate and in BALF after IAV infection. As expected, IAV infection elicited robust host immune responses by day 7 in all animal infected (Supplementary Figure 2). Two important mediators were notably impacted by prenatal arsenic, IL-1β, and IL-10. Arsenic exposure significantly decreased the concentration of the anti-inflammatory cytokine IL-10 in BALF from both male and female mice (Figure 3D). Though arsenic exposure resulted in higher concentrations of the canonical proinflammatory IL-1β at day 7 after IAV infection, only in female homogenates did the differences reach statistical significance (Figure 3E). Taken together, the data in Figures 1–3 suggest that prenatal arsenic exposure predisposes to enhanced lung injury by day 7 after IAV infection, likely due to increased cellular infiltration, as well as alterations in the proinflammatory/anti-inflammatory cytokine milieu. Additional studies employing scRNA-seq technology, described below, were conducted at an earlier time point in infection to elucidate in greater detail the effects of prenatal arsenic exposure on specific immune cells and their function in subsequently infected adult mice.
scRNA-seq Analysis: Prenatal Arsenic Exposure Does Not Alter the Detailed Immune Cell Landscape
To develop a detailed picture of the early immune cell populations in the lung after IAV infection, we performed a high-resolution molecular analysis of infiltrating immune cells and their individual transcriptomes on day 3 after infection. We chose to focus our analysis on cells harvested from IAV-infected females because the difference in magnitude of lung inflammation and injury specifically attributable to arsenic exposure was greatest in the females. We hypothesized that the transcriptional profiles analyzed on day 3 would begin to provide mechanistic insights into the distinctive phenotypes we observed at a later, clinically significant, time point (ie, day 7). A provocative recent study of gene expression by lung infiltrating cells in IAV-infected female mice demonstrated the potential power of single-cell transcriptional analysis in generating novel hypotheses in the complex responses to virus infection (Steuerman et al., 2018). However, no studies have used scRNA-seq to perform high resolution examination of the impact of arsenic exposure on the observed dysregulation of host antiviral responses. Infiltrating cells were isolated from dissociated whole lungs of adult female mice, previously exposed to arsenic in utero, on day 3 after IAV infection. We obtained 1.21 ± 0.26 × 106 viable CD45+ cells from whole-lung homogenates and observed no significant difference (p = .79) in the total number of cells recovered from exposed versus unexposed mice. scRNA-seq provides the opportunity to examine both the effects of arsenic on the landscape of cell subtypes in the lung in a highly detailed manner, as well as gene expression within individual cells, and cell subtypes, that may not be evident in a bulk RNA-seq analysis. These two lines of analysis, differential abundance (DA), and DE are inseparable, as both depend on the resolution of cell type classification. At the coarse end of the spectrum, as in a bulk RNA-seq analysis, differences in gene expression may reflect differences in subpopulation structure. We began with a coarse-grained DA analysis of major cell types detected by scRNA-seq, and then examined cell populations at progressively detailed levels of resolution to identify key biological processes altered by prenatal arsenic exposure.
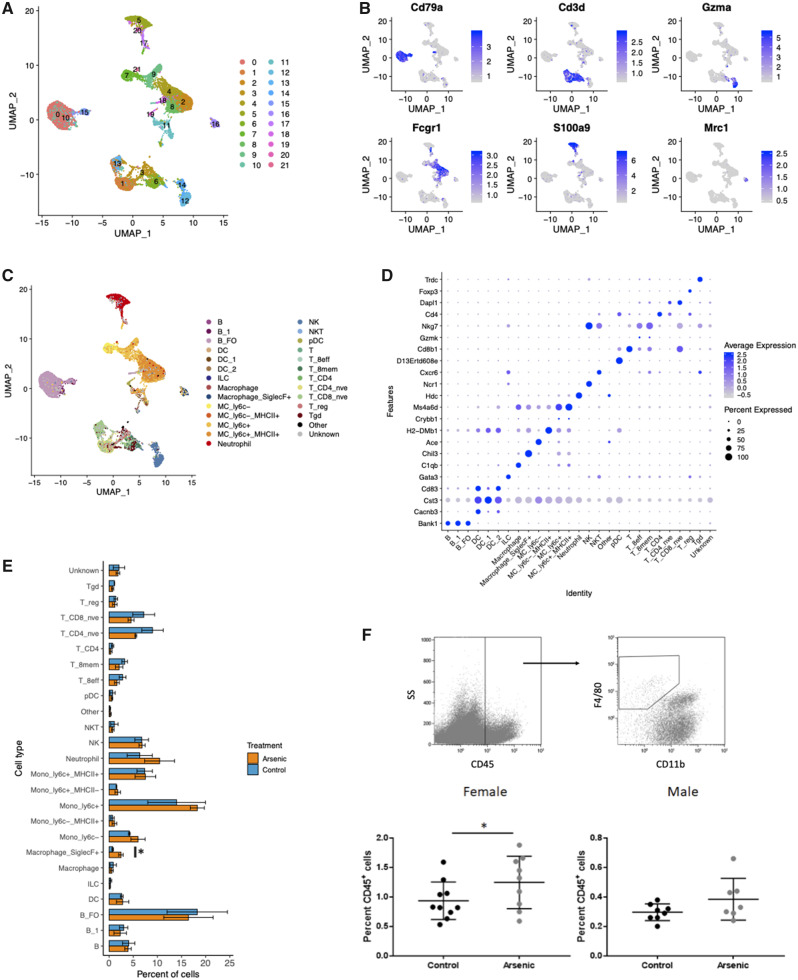
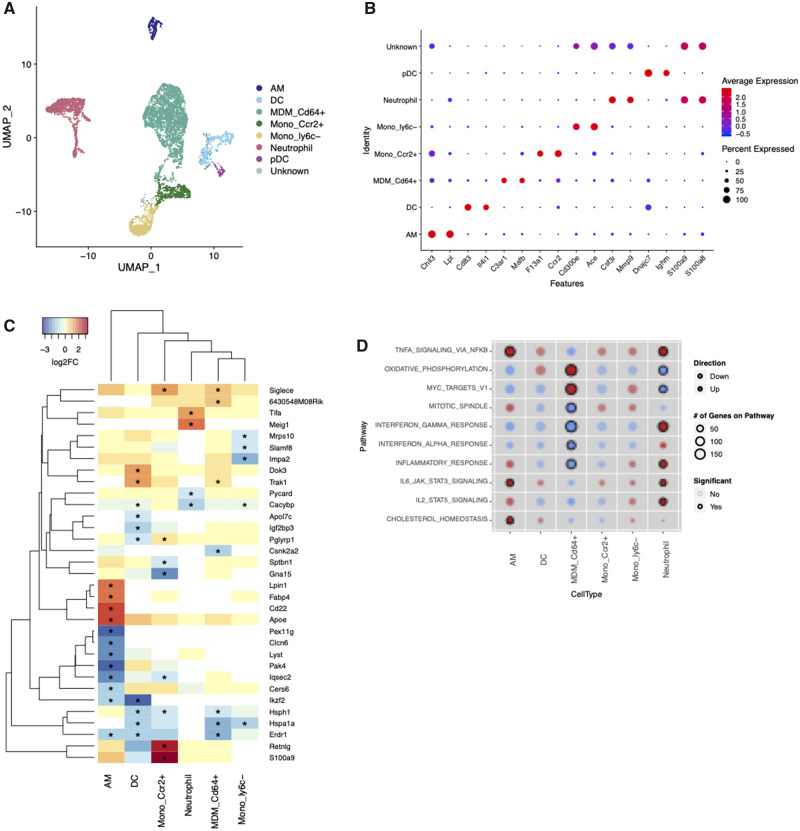
We first integrated scRNA-seq data from all samples and used independent methods to identify cell clusters (ie, cells with similar gene expression profiles) and cell type and subtype identities (utilizing a reference compendium) for analysis of DA of immune cell subtypes in the IAV-infected lung. Our clustering approach identified 22 cell clusters, which we visualized in 2D space using a UMAP (Figure 4A). We examined differentially expressed genes in each cluster, compared with all other cell clusters, and found that many highly expressed genes represent canonical cell type markers. Visualization of top marker gene expression confirmed that cells were clustering by cell type, rather than by sample or treatment group (Figure 4B). We additionally visualized clustering of cells from individual samples in 2D space and observed notable consistency across 6 samples (Supplementary Figure 3). This observation indicates that prenatal arsenic exposure did not predispose to gross changes in immune cell populations on day 3 after IAV infection. In our parallel cell labeling approach, 98% of cells (12 989 cells) were mapped to 1 of 263 sample groups in the Immgen compendium. Cells assigned to CD45− populations accounted for 0.2% of cells in the dataset, confirming that the CD45+ immune cell isolation protocol was specific for cells of hematopoietic origin (Figure 4C). The UMAP visualization of all recovered cells allowed clear separation of B, T, NK, dendritic cell (DC), neutrophil, Siglec-F+ macrophage, and monocyte-derived cell populations (Figure 4C). To further validate the reference-based cell type labeling approach, we identified marker genes expressed in each cell type at significantly higher levels than 90% of other cell types. This confirmed, eg, that Bank1 (B Cell Scaffold Protein with Ankyrin Repeats 1) was highly expressed exclusively in cells labeled as B cells, and Foxp3 (Forkhead box 3) was expressed only in cells labeled as regulatory T cells (Figure 4D). Cells labeled as Siglec-F+ macrophages (resident AMs) uniquely expressed multiple canonical AM markers, but the most highly significant gene marker was Chil3 (Figure 4D). Taken together, the data presented in Figure 4 demonstrate that our scRNA-seq protocol permitted clear identification of the major immune cell populations and subtypes at day 3 post-IAV infection, and that prenatal arsenic exposure did not result in elimination of a specific immune cell subtype or recruitment of a unique subtype to the lungs after IAV infection. Furthermore, enumeration of the DA of immune cell types and their subpopulations revealed that the only population that was significantly more abundant in arsenic-exposed mice at this early time point was Siglec-F+ (resident alveolar) macrophages (3.2-fold increase, Figure 4E). This finding was validated with flow cytometric quantitation of cells harvested from lungs of IAV-infected F1 mice, and similar increases in the proportion of AMs were observed in arsenic exposed infected F1 female mice compared with unexposed (Figure 4F).
Figure 4.
Identification of leukocyte populations in mouse lung with single-cell RNA sequencing. Cell capture in droplets, library preparation, sequencing, and QA/QC of cells resulted in RNA expression data from 2207 ± 255 cells per sample. All cells from all 6 samples were normalized and integrated in Seurat, and clusters identified using the shared nearest neighbor method on the first 30 principal components. A, Uniform Manifold Approximation and Projection (UMAP) projection of cells from all samples colored by cluster. All 22 clusters are represented in each sample. B, UMAP projection of cells from all samples colored by cell type identity label from derived from singleR reference-based annotation score (Spearman’s rank correlation). The 258 cells that did not meet singleR score cutoffs were assigned “Unknown.” Cells assigned with CD45− cell types are labeled as “Other.” C, Expression of select canonical cell type markers is displayed on UMAP projections of all cells in the experiment. Cd3e, T cells; Cd79b, B cells; Gzma, natural killer cells; Fcgr1(Cd64), monocyte-derived cells; Retnlg, neutrophils; Ear1, alveolar macrophages, eosinophils. D, Dot plot representing the expression of top gene markers (identified with scran findMarkers function) across all cell type identities. Red represents high mean expression level compared with other cell types; blue represents low expression. Size of dot represents the percent of cells within the cell type expressing the gene. E, Comparison of the percent of cells within each cell type in arsenic-exposed versus control mice. Bars represent means ± SD, N = 3 samples per group. Statistical analysis was conducted in EdgeR, *FDR < .1. F, Flow cytometry of alveolar macrophages from lung of day 3 IAV-infected mice. Data were analyzed using linear models that accounted for arsenic exposure and batch (Female n = 9–10; Male n = 7–8) *p < .05.
Prenatal Arsenic Exposure Results in Cell-type-specific Differential Gene Expression After IAV Infection
We hypothesized that differences in transcriptional activity of specific cell types at the day 3 early time point could provide insight into drivers of later lung pathology. We first focused on mononuclear phagocyte system (MPS) cells and granulocytes, which are critical to the early response to IAV infection. Though these populations were clearly distinct from lymphocyte lineage cells in our initial clustering analysis (Figure 4), the boundaries between the various monocyte-derived cell populations were more difficult to distinguish. To more clearly define cell groups within these heterogeneous populations, we created an MPS and granulocyte subset of the scRNA-seq dataset and reclustered the cells to identify distinct subpopulations (Figure 5A). Clusters were labeled based on the reference-based identity of the majority of cells, as well as top cluster-specific marker genes (Figure 5B). Cell populations with sufficient numbers for DE analysis (at least 10 cells per sample) were resident AMs, neutrophils, DCs, CD64+ inflammatory monocyte/macrophages (MDM_CD64+), nonclassical monocytes (Mono_ly6c−), and blood monocytes (Mono_Ccr2+). We identified significantly differentially expressed genes (p < .05, log2FC >1) within each cell type (Log2Fold Change, nominal p values, and FDR-corrected p values for all genes detected in these cell types are available in Supplementary Tables 3–5, respectively). Resident AMs had the largest number of genes that met these significance criteria (208), whereas monocyte-derived macrophages had the smallest number of DE genes (10). We compiled a list of all genes that showed significant DE and examined the arsenic effect on expression of these genes in each cell type (Figure 5C, genes with p < .001). For the majority of these genes, the effect of previous arsenic exposure was cell-type specific. To explore whether cell-type specificity of arsenic effects generalized beyond the genes that reached high statistical significance, we examined transcriptional differences at the pathway level using gene set enrichment analysis. Significant pathway enrichment (FDR < .05) was detected only in CD64+ MDMs, neutrophils and resident AMs, and the impact of prenatal arsenic on the profiles of these cell types differed considerably (Figure 5D). AMs from arsenic-exposed mice had significant upregulation of TNFα signaling (via NFκB), IL-6-JAK-Stat3 signaling, and cholesterol homeostasis gene sets. Neutrophils had significant upregulation of genes in the JAK-Stat5 signaling pathway, as well as the NFκB and JAK-Stat3 response pathways (Figure 5D). At the same time, monocyte-derived macrophages from arsenic-exposed mice showed significantly decreased type I interferon responses, whereas oxidative phosphorylation pathway genes were significantly upregulated in these cells.
Figure 5.
Cell-type-specific effects of arsenic in granulocyte and mononuclear phagocyte system (MPS) cells. A subset of the dataset containing MPS cells and granulocytes was reclustered in Seurat to identify distinct clusters for gene expression analysis. A, Uniform Manifold Approximation and Projection (UMAP) projection of MPS cells and granulocytes from all samples colored by cluster. DC, dendritic cell; MDM_CD64+, monocyte-derived macrophage; Mono_Ccr2+, monocyte; Mono_ly6c+, nonclassical monocyte; pDC, plasmacytoid dendritic cell. B, Dot plot of top gene markers (identified with scran findMarkers function) expression across all cell type identities. Red represents high mean expression level compared with other cell types; blue represents low expression. Size of dot represents the percent of cells within the cell type expressing the gene. C, Expression of top significantly differentially expressed genes (p < .005, logFC > 2 in at least one cell type) across all cell clusters. Red, increased expression; blue, decreased expression. Yellow, no expression change with arsenic. White, below detection * significantly differentially expressed p < .05. D, Comparison of gene set enrichment across cell types. Color of dots represents overall direction of regulation, red = up, blue = down, and size of dot represents the number of genes on the pathway in each cell type. Dots are shaded by p value, darker color = smaller p value. Significant enrichment (FDR < .1) is represented by black circles.
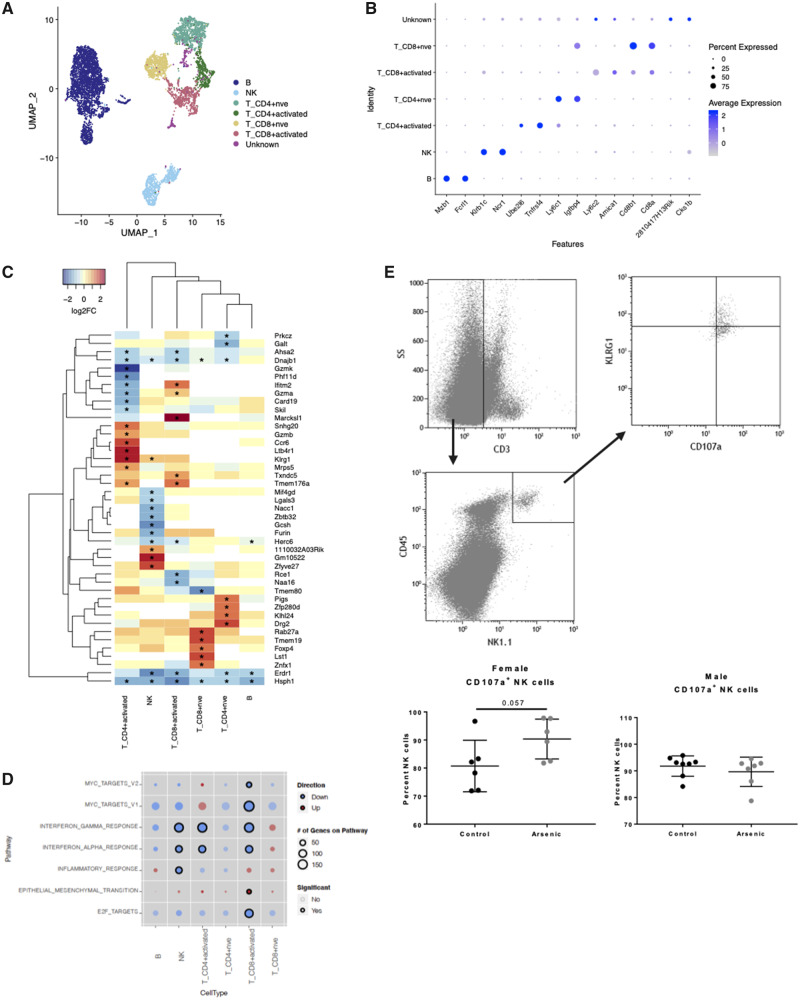
We used the same approach to identify cell-type-specific effects of arsenic exposure on lymphoid lineage cells. Cell clusters were labeled based on the reference-based identity of the majority of cells, as well as top cluster-specific marker genes. NK, B, naïve CD4+, and naïve CD8+ cells localized to distinct clusters with sufficient cells for DE analysis (Figure 6). Other T cells localized to 2 clusters that were comprised predominantly of activated CD4+ and activated CD8+ cells, but also contained gamma delta T cells and NK T cells, reflecting the heterogeneity of this cell population. We found that gene expression was affected by arsenic in NK cells and activated T-cell populations to a greater extent than in naïve B and T cells (Figure 6 and Supplementary Tables 6–8). Two genes, Hsph1 and Erd1, were downregulated in all lymphocyte populations, whereas the majority of significant genes were cell-type specific (Figure 6C). Arsenic-exposed mice had significantly higher expression of genes related to NK cell maturation and effector function including Zeb2 (1.5-fold), Klrg1 (1.5-fold), and Tbx21 (1.3-fold) (Supplementary Table 6). Examination of differential gene expression at the pathway level revealed significant downregulation of interferon-dependent gene sets in NK and activated T cells (Figure 6D). There was a trend of decreased type I and II interferon signaling in most other lymphoid populations, but effects were not significant (FDR < .1).
Figure 6.
Cell-type-specific effects of arsenic in lymphoid cells. A, Distinct subclusters were defined for differential gene expression analysis. B, B cell; NK, natural killer cell; T_CD8+ activated, CD8+ effector and memory T cells; T_CD4+ activated, regulatory T cells, CD4+ memory T cells, and effector T cells; T_CD4nve, naïve CD4+ T cells; T_CD8nve, naïve CD8+ T cells; T_CD8; Other CD8+ T cells. B, Dot plot of top gene markers (identified with scran findMarkers function) expression across all cell type identities. Blue represents high mean expression level compared with other cell types; blue represents low expression. Size of dot represents the percent of cells within the cell type expressing the gene. C, Expression of top significantly differentially expressed genes (p < .005, logFC > 2 in at least one cell type) across all cell clusters. Red, increased expression; blue, decreased expression. Yellow, no expression change with arsenic. White, below detection * significantly differentially expressed p < .05. D, Comparison of gene set enrichment across cell types. Color of dots represents overall direction of regulation, red = up, blue = down, and size of dot represents the number of genes on the pathway in each cell type. Dots are shaded by p value, darker color = smaller p value. Significant enrichment (FDR < .1) is represented by black circles. E, Flow cytometry of NK cells from lung of day 3 IAV-infected mice (Female n = 6; Male n = 7–8). Data were analyzed using linear models that accounted for arsenic exposure and batch.
Because NK cells have been suggested to play a significant negative role in IAV prognosis (Abdul-Careem et al., 2012), and our scRNA seq gene expression data suggested enhanced activation and effector potential, we analyzed cell surface expression of CD107a, a preformed protein found in cytotoxic granules (along with perforin and granzymes), which is a sensitive marker of recent granule exocytosis when detected on the cell surface (Alter et al., 2004). Functional confirmation of NK cell activity suggested that arsenic-exposed female mice had a strong trend to exhibit greater cytolytic activity, potentially participating in further lung epithelial injury (Figure 6E). Together, our cell-type-specific DE analysis revealed, for the first time, that prenatal arsenic exposure had significant, and cell-type specific, effects on gene expression particularly in AMs, CD64+ monocyte-derived cells, neutrophils, and NK cells at 3 days post-IAV infection.
IAV Infection Rate of Infiltrating Host Cells Is Not Altered in Mice Exposed to Arsenic
Though the bulk of IAV infection and replication occurs in respiratory epithelial cells, it has become increasingly evident that nearly all cells are susceptible to IAV infection (Hufford et al., 2012; Manicassamy et al., 2010). The quantitative contribution of nonepithelial replication to total viral load as well as the impact of infection on immune cell transcriptional activity and functional consequences are poorly understood. Steuerman et al. (2018) characterized transcriptomic profiles of uninfected versus infected cells in the mouse lung and identified a large signature gene expression response to infection that was conserved across all cell types. They additionally used detection of viral transcripts in individual cells as a proxy for infection, and identified cell-type-specific infection rates, as well as transcriptional responses to intracellular infection versus exposure to the infectious/inflammatory lung environment (Steuerman et al., 2018). Using a similar approach with our scRNA-seq dataset, we examined infection rates in infiltrating cells and associated transcriptional responses to investigate whether differential rates of infection could account for the effects we observed of previous arsenic exposure on gene expression. Though we observed cell-type-specific infection rates similar to Steuerman et al. (low rates of infection in T cells, relatively high rates of infection in AMs), we observed no significant differences in the total number of IAV-infected cells, or the cell-type-specific viral transcript levels, in arsenic exposed versus control mice. These results were validated with detection of a key influenza protein (IAV hemagglutinin) by flow cytometry performed on multiple cell types (Supplementary Figure 4).
Cell-type-specific Effects of Prenatal Arsenic Exposure Are Masked in Composite Immune Cell Gene Expression Data
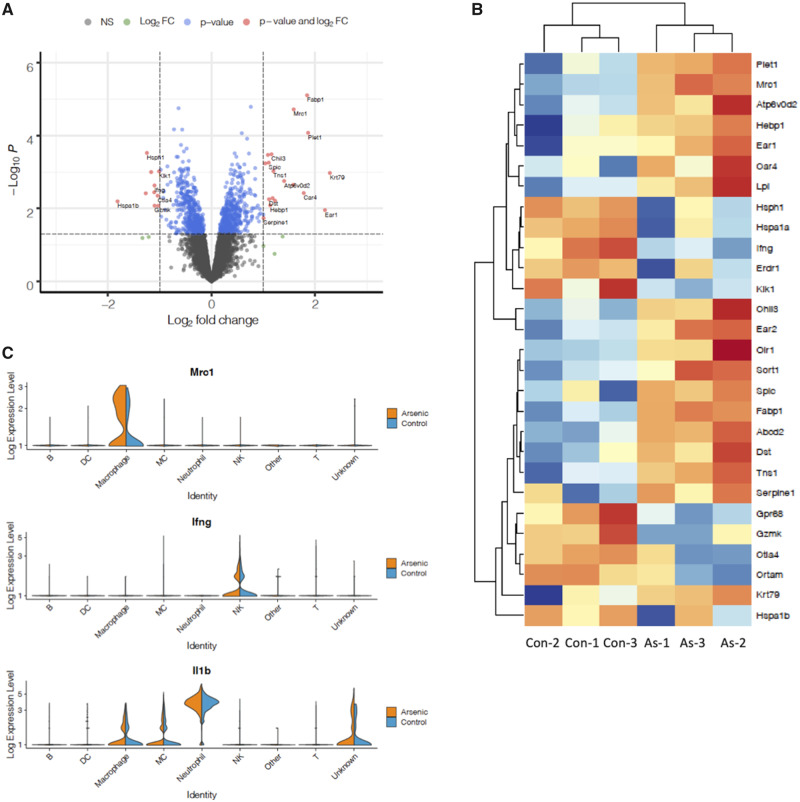
Our scRNA-seq results revealed significant changes in gene expression within individual cell types at 3 days postinfection (dpi), and a significant change in abundance of AMs. To investigate the combined effects of cell-type-specific DE and DA of cell populations in immune cells in the lung, we pooled all cells per sample to create a pseudo-bulk RNA-seq dataset and compared the differentially expressed genes identified in this whole-lung dataset with our cell-type-specific findings. Only 2 genes, Fabp1 and Mrc1, were differentially expressed with an FDR-corrected p value < .05. Using MA/QC criteria of a nominal p value < .05 and log2FC > 1, we identified 28 significantly differentially expressed genes (Figs. 7A and 7B, Supplementary Table 9). Upon examining the expression patterns of the most significantly upregulated genes in the bulk analysis, we found that the majority were highly expressed in the AM population (eg, Mrc1, Figure 7C). It is important to note that no effect of arsenic was observed on these genes when arsenic versus unexposed AMs were compared in the cell-type-specific analysis (Figure 5C and Supplementary Tables 3–5). This suggests that in a bulk tissue analysis a significant difference in abundance of even a small cell population can have a large impact on the overall gene expression profile. In contrast to the upregulated genes, many significantly downregulated genes were evident in all cell types (eg, Hsph1, Hspa1a, Hspa1b) and exhibited small but consistent decreases in expression. We observed DE of several immunologically important genes, such as IFNγ, which had expression limited to a few cell types and decreased in the pseudo-bulk analysis as well as in the cell-type-specific analysis. Notably absent from the significantly differentially expressed gene lists at 3 dpi were IL-1β and IL-10, which we previously showed changed at the protein level at 7 dpi. We found that IL-1β was expressed at high levels in neutrophils (Figure 7C) and more moderate levels in monocyte-derived cells and macrophages but did not change significantly in individual populations or the pseudo-bulk analysis. Gene expression levels of IL-10 were too low to allow for single cell analysis.
Figure 7.
Overall effect of exclusively prenatal arsenic exposure on leukocyte gene expression in the lung at 3 days postinfection. A, Volcano plot comparing significance (−log p value) versus log2 fold change of arsenic effect for all genes. All cells from each sample were pooled to create pseudo-bulk RNA-seq samples and analyzed in EdgeR, N = 3 per treatment group. B, Expression of significant genes (p < .05, log2FC > 1) in each sample is displayed in the heatmap, with expression values normalized to the mean per row. Red, increased expression; blue, decreased expression. C, Violin plots comparing expression of 3 genes of interest in arsenic versus control samples across cell types. Width of peaks represents percent of cells within each type at a given expression level.
We examined transcriptional differences in the pseudo-bulk dataset at the pathway level using gene set enrichment analysis, and found that inflammation-related (TNFα signaling via NFκB, Il-6-JAK-STAT3 signaling), metabolic (cholesterol homeostasis and adipogenesis) and xenobiotic metabolism gene sets were significantly upregulated in cells from mice exposed to prenatal arsenic, compared with controls (Table 1). Several gene sets related to cell cycle control and carcinogenesis (MYC targets, G2M checkpoint) were downregulated in cells from arsenic-exposed mice. Overall, we found substantial overlap in pathways identified in the pseudo-bulk RNA-seq analysis and scRNA-seq analysis, and this was particularly the case for pathways affected by arsenic in AM and neutrophil populations. However, the pseudo-bulk analysis failed to identify pathways specifically altered in CD64+ MDMs and NK cells, of particular note is that decreased interferon alpha (IFNα) signaling was masked in the bulk analysis.
Table 1.
Overall Arsenic Effect on Pathway-level Gene Expression
| Hallmark Gene Set | No. of Genes in Dataset | Direction | FDR |
|---|---|---|---|
| E2F_TARGETS | 140 | Down | 5.31E−05 |
| CHOLESTEROL_HOMEOSTASIS | 54 | Up | 0.000326084 |
| MYC_TARGETS_V1 | 193 | Down | 0.008222994 |
| IL6_JAK_STAT3_SIGNALING | 71 | Up | 0.009576791 |
| XENOBIOTIC_METABOLISM | 111 | Up | 0.009576791 |
| TNFA_SIGNALING_VIA_NFKB | 168 | Up | 0.009576791 |
| COAGULATION | 63 | Up | 0.013944503 |
| ADIPOGENESIS | 147 | Up | 0.014601948 |
| ANGIOGENESIS | 18 | Up | 0.025714191 |
| HYPOXIA | 124 | Up | 0.029940812 |
| SPERMATOGENESIS | 43 | Down | 0.037158419 |
| BILE_ACID_METABOLISM | 46 | Up | 0.037158419 |
| MYC_TARGETS_V2 | 52 | Down | 0.037158419 |
| G2M_CHECKPOINT | 133 | Down | 0.037158419 |
| EPITHELIAL_MESENCHYMAL_TRANSITION | 66 | Up | 0.037927805 |
| MYOGENESIS | 64 | Up | 0.041120419 |
| PROTEIN_SECRETION | 84 | Up | 0.050861297 |
Hallmark gene sets that were differentially expressed (FDR-corrected p < .1) are listed, along with the number of genes on the pathway in the dataset, the overall direction of regulation, and the FDR-corrected p value.
Abbreviation: FDR, false discovery rate.
Together, our scRNA-seq analysis of immune cells in the lung produced several novel observations regarding the effect of prenatal arsenic exposure on the response of lung immune cells at day 3 of IAV infection. First, we did not identify any subpopulations of cells unique to either the arsenic group or the control group. Second, mice exposed to prenatal arsenic had significantly more AMs (Siglec-F+ macrophages), but not monocyte-derived macrophages, in the lungs at day 3 than vehicle-treated animals. This observation highlights the power of the scRNA-seq approach to provide detailed information about cell type abundance, which not only provided new phenotypic information, but was critical to our further investigation of how prenatal arsenic exposure affected gene expression. Finally, cell-type-specific gene expression analysis provided insight into early (3 days) changes in cell type activity that may explain the pathology observed later in infection by revealing a deficiency in type I interferon signaling in CD64+ monocytes, enhanced cytolytic transcriptional profile of NK cells, and an upregulation of several proinflammatory pathways (eg, NFkB, and IL-6-JAK-STAT-mediated signaling cascades) in AM and neutrophil populations.
DISCUSSION
In this study, we demonstrate for the first time the long-term impact of exclusive prenatal arsenic exposure on the antiviral immune responses in adult mice, after maturation, and well after the toxicant and its metabolites have cleared to the point of undetectability (Hughes et al., 2003; Twaddle et al., 2018). We demonstrate that arsenic exposure enhanced capillary leak, indicating significant lung damage, and show that increased damage was accompanied by an increase in inflammatory immune cell infiltrates in BALF, an increase in the proinflammatory cytokine IL-1β, and a reduction in the anti-inflammatory cytokine IL-10 (Dinarello, 1997; Rojas et al., 2017). In a detailed examination of lung immune cells using scRNA-seq, we identified increased expression of pathways in arsenic-exposed mice that drive inflammation early in infection. We further leveraged cell-type-specific gene expression analysis to identify AMs, neutrophils, CD64+ MDMs, and NK cells as cells that mediate lung damage in mice exposed to arsenic exclusively in utero.
Previous studies have shown impaired viral clearance and dysregulated immune responses after IAV infection in chronic arsenic exposure in mature animals (Kozul et al., 2009a,b), as well as in neonates infected with IAV (Ramsey et al., 2013b). Although these studies assessed the impact of arsenic exposure extending throughout the course of the infection, our study demonstrated effects of exclusively prenatal arsenic exposure 7 and 8 weeks after birth, long after arsenic was cleared from the body. In contrast to the study by Kozul et al. (2009a), we observed no difference in weight loss over the course of infection or viral titer in mice exposed to arsenic exclusively in utero, despite observing a similarly enhanced inflammatory response. A potential cause of this difference may be the dose of IAV that was used. For this study, we optimized a sublethal LD50 dose that would cause significant observable injury by 7 dpi (prior to unacceptable morbidity or mortality). This inoculum caused mice to lose a maximum of 20% weight by day 7 IAV infection. Because of this weight loss, we were unable to assess immune responses later in IAV infection. As shown in previous work, a consequence of such a high dose of IAV is that the kinetics of clearance are delayed compared with a lower dose (Vogel et al., 2014).
An additional potential reason for differences between our results and previous work is the consideration of sex as a factor. Results from a previous study investigating prenatal arsenic exposure and IAV infection in neonates pooled data from females and males (Ramsey et al., 2013b). Kozul et al. (2009a) only assessed adult male mice exposed to chronic arsenic exposure and acute IAV infection. In this study, we evaluated males and females separately and observed a significant increase in IL-1β expression in females exposed to arsenic, whereas no significant increase in IL-1β was observed in males (Figure 3D). Future studies investigating prenatal arsenic exposure impact on host responses to IAV infection should account for sex-dependent effects.
The increase in inflammation and injury in our study is consistent with previous epidemiological observations of increased morbidity and mortality from lung disease in humans exposed to arsenic-contaminated water during early life (Smith et al., 2006). These phenotypic findings are consistent with research demonstrating epigenetic effects of arsenic exposure, including changes in DNA methylation, in human populations (Bailey and Fry, 2014; Winterbottom et al., 2019). Although additional research, beyond the scope of this study, will be required to elucidate how prenatal arsenic exposure selectively regulates immune gene expression 7 and 8 weeks after cessation of exposure, our scRNA-seq analysis of cellular and molecular events early in infection highlights affected cell populations that should be targets of further mechanistic investigation.
Our scRNA-seq transcriptional analysis of immune cells identified signals consistent with our studies on immune cells and cytokines in BALF, and identified MDM’s, neutrophils, AMs, and possibly NK cells as likely drivers of enhanced inflammation leading to lung damage in arsenic-exposed animals. At 7 dpi, we observed an increase in IL-1β protein which has been linked to increased lung injury (Dinarello, 1997; Ganter et al., 2008). We found that signaling pathways, including IL-6-JAK-Stat and NFκB, which ultimately participate in regulating pro-IL-1β transcription, were upregulated specifically in AMs and neutrophils. IFNα production is a crucial early immune response to IAV infection and has well-described anti-inflammatory properties. Ablation of IFNα signaling in mice leads to severe inflammation and morbidity after IAV infection, without alterations in virus titer (Arimori et al., 2013), and IFNα signaling plays a key role in limiting inflammation by restricting IL-1β and promoting IL-10 signaling (Arimori et al., 2013; Guarda et al., 2011). Our scRNA-seq analysis identified a significant decrease in IFNα response signaling in CD64+ MDMs, as well as multiple lymphocyte populations at day 3 postinfection. Whether the deficiency in IFNα response of CD64+ MDMs is the result of alterations of gene regulation in the monocytes themselves or is a response to secondary paracrine loops upon recruitment into the lung is uncertain.
In addition to observing significant changes to type I interferon response pathways and enhanced inflammatory pathways in certain leukocytes at day 3 IAV infection, we also found changes in gene expression that further contribute to inflammation and adverse health outcomes. We observed that prenatal arsenic exposure increased Fabp4 expression in macrophages. Previous work has shown that Fabp4 expression is important in proper expression of CXCL1 and proper recruitment of neutrophils (Liang et al., 2019). We also observed significant changes to heat shock protein expression (Hsph1, Hspa1a, Hspa1b, and Dnaja1) in leukocytes. It has been shown in previous studies that arsenic exposure leads to abnormal expression of various heat shock proteins across different tissues contributing to a variety of diseases (Han et al., 2005; Ngalame et al., 2013). Future work should further elucidate how changes in the expression of these candidate genes contribute to greater inflammation and injury in this model.
We also observed changes in NK cell effector function. We observed trends in greater CD107a+ NK cells from female mice exposed to prenatal arsenic. CD107a is a surface marker indicative of degranulation from CD8+ T cells and NK cells. Depletion of NK cells lead to greater survival in mice administered lethal doses of IAV, and that adoptive transfer of NK cells exposed to IAV into an IAV-infected mouse leads to greater mortality (Zhou et al., 2013). Our results suggest that prenatal arsenic exposure may increase cytotoxicity from the NK cell compartment, and this may be a contributing factor to the enhanced immunopathology. Analysis of NK-specific gene expression changes in female mice identified significant upregulation of Zeb2, Klrg1, and Tbx21 that may be promoting this effect. Zeb2 is heavily involved in NK cell differentiation and promoting proper effector function (van Helden et al., 2015). Klrg1 is considered an inhibitory mechanism for NK effector function, however it is also considered a maturation marker for NK cells (Huntington et al., 2007). We performed flow cytometry to assess the number of Klrg1hi NK cells and saw no significant differences. Lastly, we observed a significant increase in expression of Tbx21, which encodes the transcription factor T-bet. T-bet plays an important role in NK cell cytotoxicity in the development of cytotoxic molecule like perforin and granzyme B (Simonetta et al., 2016).
Although scRNA-seq enables high-resolution gene expression analysis that promises to increase precision of toxicological investigations (Zhang et al., 2019), limitations of this approach should also be considered in the interpretation of our results. The platform we used to generate single-cell sequencing libraries in droplets (10X Genomics Chromium system) requires live cells in a single-cell suspension, and cell-type selection bias occurs during both sample preparation (a result of tissue disruption, digestion) and the cell capture step (a result of cell characteristics such as size and RNA quantity). Cell type enrichment is often required to obtain sufficient numbers of low abundant populations of interest and must also be considered in the interpretation of results. To obtain sufficient numbers of cell types hypothesized to be mediating the phenotype observed in this study, we specifically targeted the scRNA-seq analysis to cells of hematopoietic origin. Future studies, however, should also examine effects of prenatal arsenic exposure on nonhematopoietic cell types. For example, the majority of IAV infection and replication occurs in respiratory epithelial cells, which we and others have shown to be affected by low dose arsenic exposure (Goodale et al., 2017; Manicassamy et al., 2010). The low proportion of neutrophils observed in both our scRNA-seq dataset and the Steuerman et al. dataset (Steuerman et al., 2018) suggests that neutrophils are prone to under-sampling in scRNA-seq, likely because they are fragile and removed during the dead cell removal process (Glasser and Fiederlein, 1990). Cell isolation method is important to consider when examining relative proportions of cell types. Meanwhile, methods such as single-nucleus RNA sequencing, which is compatible with freezing (Wu et al., 2019), and spatial transcriptomics, which enables high-resolution RNA capture from intact tissue (Burgess, 2019), hold promise for sequencing difficult-to-obtain cell types and maintaining the spatial information of cells within tissues in future studies. We additionally focused our scRNA-seq study on females, which had a more robust phenotype. Specific interrogation of sex differences in the effects of prenatal arsenic exposure will be an interesting direction for future studies, particularly in light of a recent study that reported sexual dimorphism in transcriptional programs of macrophages (Gal-Oz et al., 2019). Various feasibility limitations of single-cell sequencing technology (Bacher and Kendziorski, 2016) have likely resulted in the relatively small sample sizes employed in scRNA-seq studies to date. We believe that biological replication was a strength of this study, and indeed critical to the investigation of arsenic effects on DA and gene expression. Advances in single-cell sequencing technology will hopefully allow future studies to increase biological replication and power to detect more subtle effects, as well as investigate additional experimental factors such as sex and time point during the course of infection.
Together, our scRNA-seq analysis suggests that prenatal arsenic exposure may predispose to enhanced IL-1β and suppressed IL-10 signaling via both direct regulation in macrophages and neutrophils, and via early suppression of IFNα signaling. In addition, our analysis suggests that enhanced cytolytic activity from NK cells may contribute to greater observed immunopathology in our model. These data demonstrate the power of single cell sequencing by revealing major tissue levels effects of toxicant exposure, as well as heterogeneous responses within individual cells and cell populations that ultimately lead to aberrant inflammation and lung damage after respiratory virus infection.
In recent studies of children of the New Hampshire Birth Cohort, Farzan et al. reported increased respiratory infection in children, correlated with maternal urinary arsenic levels during pregnancy. We speculate that this finding may actually reflect the impact of enhanced inflammatory responses in childhood respiratory infection, which lead to the clinical manifestations and recognition of infection (rather than infection rate per se). Based on our studies in a mouse model presented here, we hypothesize that this predisposition in children to the more evident clinical manifestations of respiratory infections may result, at least in part, from long-term effects of prenatal arsenic exposure. Importantly, our data show for the first time a direct link between exclusive prenatal arsenic exposure and the predisposition to aberrant immune responses to respiratory virus infection, and persistence of this effect long after cessation of exposure.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr Fred Kolling for his scientific advice on single-cell sequencing.
FUNDING
National Heart, Lung, and Blood Institute of the National Institutes of Health (HL122585 to R.I.E.); National Institute of Environmental Health Sciences of the National Institutes of Health (P42ES007373 to B.A.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Flow cytometry on Gallios 10 Color and multiplexed cytokine results from whole-lung homogenate were carried out in DartLab, the Immune Monitoring and Flow Cytometry Shared Resource, supported by a National Cancer Institute Cancer Center Support Grant to the Norris Cotton Cancer Center (P30CA023108-37). Validation of arsenic levels in mouse drinking water was carried out at the Dartmouth Trace Element Core Facility, which was established by grants from the National Institute of Health (NIH) and National Institute of Environmental Health Sciences (NIEHS) Superfund Research Program (P42ES007373) and the Norris Cotton Cancer Center at Dartmouth Hitchcock Medical Center. Single cell capture, sequencing, and raw data processing were conducted through the Dartmouth Center for Quantitative Biology, which is supported by National Institute of General Medical Sciences (P20GM130454) and NIH S10 (S10OD025235) awards, in collaboration with the Dartmouth Genomics and Molecular Biology Shared Resource, which was established by equipment grants from the NIH and NSF and is supported in part by a Cancer Center Core Grant (P30CA023108) from the National Cancer Institute.
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
REFERENCES
- Abdul-Careem M. F., Mian M. F., Yue G., Gillgrass A., Chenoweth M. J., Barra N. G., Chew M. V., Chan T., Al-Garawi A. A., Jordana M., et al. (2012). Critical role of natural killer cells in lung immunopathology during influenza infection in mice. J. Infect. Dis. 206, 167–177. [DOI] [PubMed] [Google Scholar]
- Alter G., Malenfant J. M., Altfeld M. (2004). CD107a as a functional marker for the identification of natural killer cell activity. J. Immunol. Methods 294, 15–22. [DOI] [PubMed] [Google Scholar]
- Amezquita R. A., Carey V. J., Carpp L. N., Geistlinger L., Lun A. T. L., Marini F., Rue-Albrecht K., Risso D., Soneson C., Waldron L., et al. (2019). Orchestrating single-cell analysis with bioconductor. BioRxiv 590562. [Google Scholar]
- Aran D., Looney A. P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R. P., Wolters P. J., Abate A. R., et al. (2019). Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimori Y., Nakamura R., Yamada H., Shibata K., Maeda N., Kase T., Yoshikai Y. (2013). Type I interferon limits influenza virus-induced acute lung injury by regulation of excessive inflammation in mice. Antivir. Res. 99, 230–237. [DOI] [PubMed] [Google Scholar]
- Ayotte J. D., Medalie L., Qi S. L., Backer L. C., Nolan B. T. (2017). Estimating the high-arsenic domestic-well population in the conterminous United States. Environ. Sci. Technol. 51, 12443–12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher R., Kendziorski C. (2016). Design and computational analysis of single-cell RNA-sequencing experiments. Genome Biol. 17, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey K., Fry R. C. (2014). Long-term health consequences of prenatal arsenic exposure: Links to the genome and the epigenome. Rev. Environ. Health 29, 9–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess D. J. (2019). Spatial transcriptomics coming of age. Nat. Rev. Genet. 20, 317. [DOI] [PubMed] [Google Scholar]
- Chen Y., Ahsan H. (2004). Cancer burden from arsenic in drinking water in Bangladesh. Am. J. Public Health 94, 741–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauber I. M., Pluss W. T., VanGrondelle A., Trow R. S., Weil J. V. (1985). Specificity and sensitivity of noninvasive measurement of pulmonary vascular protein leak. J. Appl. Physiol. 59, 564–574. [DOI] [PubMed] [Google Scholar]
- Davey J. C., Nomikos A. P., Wungjiranirun M., Sherman J. R., Ingram L., Batki C., Lariviere J. P., Hamilton J. W. (2008). Arsenic as an endocrine disruptor: Arsenic disrupts retinoic acid receptor-and thyroid hormone receptor-mediated gene regulation and thyroid hormone-mediated amphibian tail metamorphosis. Environ. Health Perspect. 116, 165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerge M. P., Ely K. H., Cheng G.-S., Enelow R. I. (2013). ADAM17-mediated processing of TNF-α expressed by antiviral effector CD8+ T cells is required for severe T-cell-mediated lung injury. PLoS One 8, e79340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello C. A. (1997). Interleukin-1. Cytokine Growth Factor Rev. 8, 253–265. [DOI] [PubMed] [Google Scholar]
- Duan S., Thomas P. G. (2016). Balancing immune protection and immune pathology by CD8(+) T-cell responses to influenza infection. Front. Immunol. 7, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzan S. F., Chen Y., Wu F., Jiang J., Liu M., Baker E., Korrick S. A., Karagas M. R. (2015). Blood pressure changes in relation to arsenic exposure in a U.S. pregnancy cohort. Environ. Health Perspect. 123, 999–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzan S. F., Li Z., Korrick S. A., Spiegelman D., Enelow R., Nadeau K., Baker E., Karagas M. R. (2016). Infant infections and respiratory symptoms in relation to in utero arsenic exposure in a U.S. cohort. Environ. Health Perspect. 124, 840–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Oz S. T., Maier B., Yoshida H., Seddu K., Elbaz N., Czysz C., Zuk O., Stranger B. E., Ner-Gaon H., Shay T. (2019). ImmGen report: Sexual dimorphism in the immune system transcriptome. Nat. Commun. 10, 4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganter M. T., Roux J., Miyazawa B., Howard M., Frank J. A., Su G., Sheppard D., Violette S. M., Weinreb P. H., Horan G. S., et al. (2008). Interleukin-1β causes acute lung injury via αvβ5 and αvβ6 integrin-dependent mechanisms. Circ. Res. 102, 804–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser L., Fiederlein R. L. (1990). The effect of various cell separation procedures on assays of neutrophil function. A critical appraisal. Am. J. Clin. Pathol. 93, 662–669. [DOI] [PubMed] [Google Scholar]
- Goodale B. C., Rayack E. J., Stanton B. A. (2017). Arsenic alters transcriptional responses to Pseudomonas aeruginosa infection and decreases antimicrobial defense of human airway epithelial cells. Toxicol. Appl. Pharmacol. 331, 154–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarda G., Braun M., Staehli F., Tardivel A., Mattmann C., Förster I., Farlik M., Decker T., Du Pasquier R. A., Romero P., et al. (2011). Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity 34, 213–223. [DOI] [PubMed] [Google Scholar]
- Guilliams M., Ginhoux F., Jakubzick C., Naik S. H., Onai N., Schraml B. U., Segura E., Tussiwand R., Yona S. (2014). Dendritic cells, monocytes and macrophages: A unified nomenclature based on ontogeny. Nat. Rev. Immunol. 14, 571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister C., Satija R. (2019). Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. BioRxiv 576827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S. G., Castranova V., Vallyathan V. (2005). Heat shock protein 70 as an indicator of early lung injury caused by exposure to arsenic. Mol. Cell. Biochem. 277, 153–164. [DOI] [PubMed] [Google Scholar]
- Heng T. S. P., Painter M. W., Elpek K., Lukacs-Kornek V., Mauermann N., Turley S. J., Koller D., Kim F. S., Wagers A. J., Asinovski N., et al. (2008). The Immunological Genome Project: Networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Hufford M. M., Richardson G., Zhou H., Manicassamy B., García-Sastre A., Enelow R. I., Braciale T. J. (2012). Influenza-infected neutrophils within the infected lungs act as antigen presenting cells for anti-viral CD8(+) T cells. PLoS One 7, e46581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M. F., Kenyon E. M., Edwards B. C., Mitchell C. T., Razo L. M. D., Thomas D. J. (2003). Accumulation and metabolism of arsenic in mice after repeated oral administration of arsenate. Toxicol. Appl. Pharmacol. 191, 202–210. [DOI] [PubMed] [Google Scholar]
- Huntington N. D., Tabarias H., Fairfax K., Brady J., Hayakawa Y., Degli-Esposti M. A., Smyth M. J., Tarlinton D. M., Nutt S. L. (2007). NK cell maturation and peripheral homeostasis is associated with KLRG1 up-regulation. J. Immunol. 178, 4764–4770. [DOI] [PubMed] [Google Scholar]
- Karagas M. R., Stukel T. A., Tosteson T. D. (2002). Assessment of cancer risk and environmental levels of arsenic in New Hampshire. Int. J. Hyg. Environ. Health 205, 85–94. [DOI] [PubMed] [Google Scholar]
- Klein S. L., Hodgson A., Robinson D. P. (2012). Mechanisms of sex disparities in influenza pathogenesis. J. Leukoc. Biol. 92, 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozul C. D., Ely K. H., Enelow R. I., Hamilton J. W. (2009. a). Low-dose arsenic compromises the immune response to influenza A infection in vivo. Environ. Health Perspect. 117, 1441–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozul C. D., Hampton T. H., Davey J. C., Gosse J. A., Nomikos A. P., Eisenhauer P. L., Weiss D. J., Thorpe J. E., Ihnat M. A., Hamilton J. W. (2009. b). Chronic exposure to arsenic in the drinking water alters the expression of immune response genes in mouse lung. Environ. Health Perspect. 117, 1108–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozul C. D., Nomikos A. P., Hampton T. H., Warnke L. A., Gosse J. A., Davey J. C., Thorpe J. E., Jackson B. P., Ihnat M. A., Hamilton J. W. (2008). Laboratory diet profoundly alters gene expression and confounds genomic analysis in mouse liver and lung. Chem. Biol. Interact. 173, 129–140. [DOI] [PubMed] [Google Scholar]
- LaBarre D. D., Lowy R. J. (2001). Improvements in methods for calculating virus titer estimates from TCID50 and plaque assays. J. Virol. Methods 96, 107–126. [DOI] [PubMed] [Google Scholar]
- Law C. W., Chen Y., Shi W., Smyth G. K. (2014). voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Gupta K., Quintero J. R., Cernadas M., Kobzik L., Christou H., Pier G. B., Owen C. A., Çataltepe S. (2019). Macrophage FABP4 is required for neutrophil recruitment and bacterial clearance in Pseudomonas aeruginosa pneumonia. FASEB J. 33, 3562–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J. P., Tamayo P. (2015). The molecular signatures database hallmark gene set collection. Cell Syst. 1, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun A. T. L., McCarthy D. J., Marioni J. C. (2016). A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res 5, 2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manicassamy B., Manicassamy S., Belicha-Villanueva A., Pisanelli G., Pulendran B., García-Sastre A. (2010). Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc. Natl. Acad. Sci. U.S.A. 107, 11531–11536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy D. J., Campbell K. R., Lun A. T. L., Wills Q. F. (2017). Scater: Pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau K. C., Li Z., Farzan S., Koestler D., Robbins D., Fei D. L., Malipatlolla M., Maecker H., Enelow R., Korrick S., et al. (2014). In utero arsenic exposure and fetal immune repertoire in a US pregnancy cohort. Clin. Immunol. 155, 188–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naujokas M. F., Anderson B., Ahsan H., Aposhian H. V., Graziano J. H., Thompson C., Suk W. A. (2013). The broad scope of health effects from chronic arsenic exposure: Update on a worldwide public health problem. Environ. Health Perspect. 121, 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngalame N. N. O., Micciche A. F., Feil M. E., States J. C. (2013). Delayed temporal increase of hepatic Hsp70 in ApoE knockout mice after prenatal arsenic exposure. Toxicol. Sci. 131, 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey K. A., Bosco A., McKenna K. L., Carter K. W., Elliot J. G., Berry L. J., Sly P. D., Larcombe A. N., Zosky G. R. (2013. a). In utero exposure to arsenic alters lung development and genes related to immune and mucociliary function in mice. Environ. Health Perspect. 121, 244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey K. A., Foong R. E., Sly P. D., Larcombe A. N., Zosky G. R. (2013. b). Early life arsenic exposure and acute and long-term responses to influenza A infection in mice. Environ. Health Perspect. 121, 1187–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh T., Steinmaus C., Marshall G., Ferreccio C., Liaw J., Smith A. H. (2018). Age at exposure to arsenic in water and mortality 30–40 years after exposure cessation. Am. J. Epidemiol. 187, 2297–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas J. M., Avia M., Martín V., Sevilla N. (2017). IL-10: A multifunctional cytokine in viral infections. J. Immunol. Res. 2017, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetta F., Pradier A., Roosnek E. (2016). T-bet and Eomesodermin in NK cell development, maturation, and function. Front. Immunol. 7, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. H., Marshall G., Yuan Y., Ferreccio C., Liaw J., von Ehrenstein O., Steinmaus C., Bates M. N., Selvin S. (2006). Increased mortality from lung cancer and bronchiectasis in young adults after exposure to arsenic in utero and in early childhood. Environ. Health Perspect. 114, 1293–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuerman Y., Cohen M., Peshes-Yaloz N., Valadarsky L., Cohn O., David E., Frishberg A., Mayo L., Bacharach E., Amit I., et al. (2018). Dissection of influenza infection in vivo by single-cell RNA sequencing. Cell Syst. 6, 679–691.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M., Hao Y., Stoeckius M., Smibert P., Satija R. (2019). Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twaddle N. C., Vanlandingham M., Fisher J. W., Doerge D. R. (2018). Metabolism and disposition of arsenic species from controlled dosing with sodium arsenite in adult female CD-1 mice. III. Toxicokinetic studies following oral and intravenous administration. Food Chem. Toxicol. 121, 676–686. [DOI] [PubMed] [Google Scholar]
- van Helden M. J., Goossens S., Daussy C., Mathieu A.-L., Faure F., Marçais A., Vandamme N., Farla N., Mayol K., Viel S., et al. (2015). Terminal NK cell maturation is controlled by concerted actions of T-bet and Zeb2 and is essential for melanoma rejection. J. Exp. Med. 212, 2015–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel A. J., Harris S., Marsteller N., Condon S. A., Brown D. M. (2014). Early cytokine dysregulation and viral replication are associated with mortality during lethal influenza infection. Viral Immunol. 27, 214–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterbottom E. F., Moroishi Y., Halchenko Y., Armstrong D. A., Beach P. J., Nguyen Q. P., Capobianco A. J., Ayad N. G., Marsit C. J., Li Z., et al. (2019). Prenatal arsenic exposure alters the placental expression of multiple epigenetic regulators in a sex-dependent manner. Environ. Health 18, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Kirita Y., Donnelly E. L., Humphreys B. D. (2019). Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: Rare cell types and novel cell states revealed in fibrosis. J. Am. Soc. Nephrol. 30, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. D., Behjati S. (2018). SoupX removes ambient RNA contamination from droplet based single cell RNA sequencing data. BioRxiv 303727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Caudle W. M., Pi J., Bhattacharya S., Andersen M. E., Kaminski N. E., Conolly R. B. (2019). Embracing systems toxicology at single-cell resolution. Curr. Opin. Toxicol. 16, 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G., Juang S. W. W., Kane K. P. (2013). NK cells exacerbate the pathology of influenza virus infection in mice. Eur. J. Immunol. 43, 929–938. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.