Abstract
A series of 2-chloro-5-[(4-chlorophenyl)sulfamoyl]-N-(alkyl/aryl)-4-nitrobenzamide derivatives (5a–5v) has been synthesized and confirmed by physicochemical(Rf, melting point) and spectral means (IR, 1HNMR, 13CNMR). The results of in vitro antidiabetic study against α-glucosidase indicated that compound 5o bearing 2-CH3-5-NO2 substituent on phenyl ring was found to be the most active compound against both enzymes. The electron donating (CH3) group and electron withdrawing (NO2) group on a phenyl ring highly favoured the inhibitory activity against these enzymes. The docking simulations study revealed that these synthesized compounds displayed hydrogen bonding, electrostatic and hydrophobic interactions with active site residues. The structure activity relationship studies of these compounds were also corroborated with the help of molecular modeling studies. Molecular dynamic simulations have been done for top most active compound for validating its α-glucosidase and α-amylase inhibitory potential, RMSD analysis of ligand protein complex suggested the stability of top most active compound 5o in binding site of target proteins. In silico ADMET results showed that synthesized compounds were found to have negligible toxicity, good solubility and absorption profile as the synthesized compounds fulfilled Lipinski’s rule of 5 and Veber’s rule.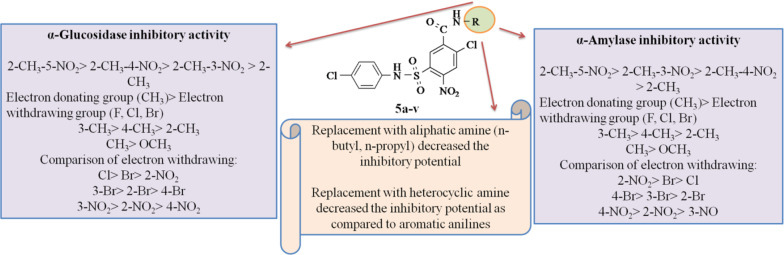
Keywords: α-Glucosidase, α-Amylase, Molecular docking, Molecular dynamic simulations, ADMET
Introduction
Diabetes mellitus (DM) is a complex metabolic disorder resulting either due to relative or absolute deficiency of pancreatic insulin secretion or insensitivity to insulin action, ensuing in postprandial hyperglycemia and assorted diabetic complications [1, 2]. According to World Health Organization reports, at present around 250 million peoples are living with diabetes and this number is expected to be more than 366 million by 2030 [3] and these statistics are predicted to reach 592 million by 2035 of which 46% may still remain undiagnosed. The reduction of postprandial hyperglycemia by inhibiting carbohydrate hydrolyzing enzymes in gastrointestinal tract is one of the promising approaches for management of diabetes [4, 5]. α-Amylase is involved in hydrolyzing long chain of starch and α-glucosidase release glucose into the small intestine by breaking down oligosaccharides and disaccharides [2, 6]. α-Glucosidase and α-amylase inhibitors reduced postprandial blood glucose level by delaying the hydrolysis of carbohydrate by inhibiting the digestive enzymes [7]. Acarbose, Miglitol and Voglibose are currently available drugs used as α-glucosidase and α-amylase inhibitors, but due to their deleterious side effects such as abdominal distention, diarrhoea and bloating, flatulence [8–10] there is need to explore and synthesize new drug candidates for the management of type-II diabetes mellitus with no or low risk of side effects.
The sulphonamide moiety (–SO2NH2) is an effective pharmacophore revealing the clinical and medicinal importance of sulphonamide drugs in the field of drug discovery [11]. The lead molecules bearing sulphonamide structure exhibited diverse biological properties viz. antibacterial [12, 13], diuretics, carbonic anhydrase (CA) inhibitors [14], antithyroid, antidiabetic [11, 15, 16], anticancer [17], antitubercular [18], selective Cox II inhibitors [19], anti-inflammatory [20], aldose reductase inhibitor [21], anti-oxidant [22], and anticancer [20] etc. Benzamides are the carbonic acid amide of benzoic acid and have also been described for exhibiting various biological activities i.e. antimicrobial [23, 24], anti-inflammatory [25], anticancer [26, 27], antidiabetic [28], antidepressant, antitubercular [29], anticonvulsant [30] and analgesic [31] etc. 2,4-Dichlorobenzoic acid derivatives have also been reported for their antidiabetic potential exhibiting α-glucosidase and α-amylase inhibitory activity, as described in our previous studies [32, 33]. Singh et al., reported the benzamides as glucokinase activators possessing hypoglycaemic activity [34]. Thiazole-2-yland N-pyridin-2-yl benzamides from benzoic acids showed glucokinase activation and possessed good antidiabetic potential in animal rat model [35, 36]. A series of sulfamoyl benzamide derivatives have also been reported by Grewal et al., having glucokinase activation potential for the treatment of type 2 diabetes [37]. In view of the vital importance of benzamides in management of type 2 diabetes, we have synthesized a series of 2-chloro-5-[(4-chlorophenyl)sulfamoyl]-N-(alkyl/aryl)-4-nitrobenzamides and evaluated its antidiabetic potential in the current report.
Results and discussion
Chemistry
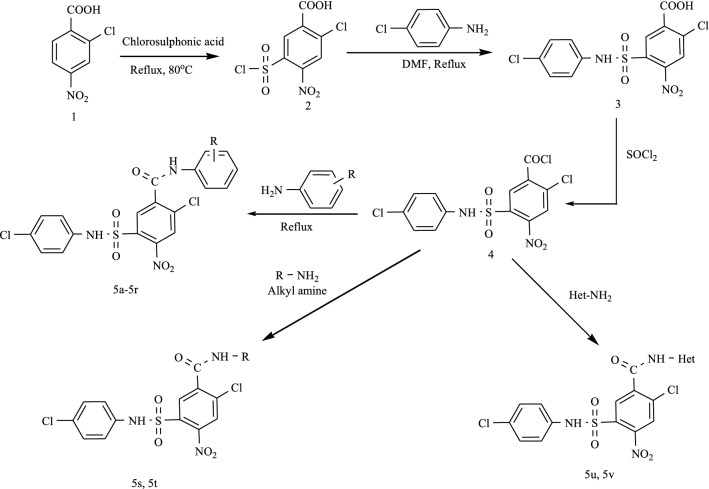
The 2-chloro-5-(chlorosulfonyl)-4-nitrobenzoic acid (2) was prepared from 2-chloro-4-nitro benzoic acid according to our previously reported procedure [32]. The reaction of commercially available para chloro substituted aniline with compound 2 in DMF yielded 2-chloro-5-[(4-chlorophenyl)sulfamoyl]-4-nitrobenzoic acid in appropriate amount. The treatment of compound 3 with excess of thionyl chloride in presence of DMF as a catalyst afforded intermediate 4, which was further refluxed with aromatic/aliphatic/heterocyclic amines in DMF to provide the target compounds 5a–5v (Table 1, Scheme 1).
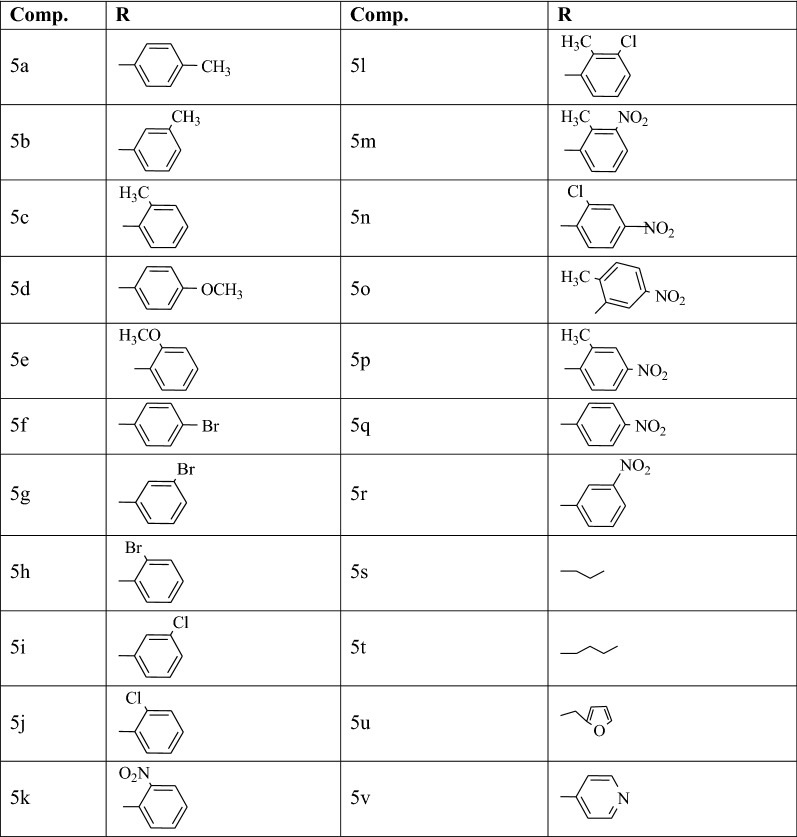
Table 1.
List of synthesized 2-chloro-5-[(4-chlorophenyl)sulfamoyl]-N-(alkyl/aryl)-4-nitrobenzamide compounds
Scheme 1.
General scheme for synthesis of 2-chloro-5-[(4-chlorophenyl)sulfamoyl]-N-(alkyl/aryl)-4-nitrobenzamide derivatives
The structure of 2-chloro-5-[(4-chlorophenyl)sulfamoyl]-N-(alkyl/aryl)-4-nitrobenzamide compounds was elucidated by IR, 1H NMR and 13C NMR spectral analysis. The stretching frequency due to NH and carbonyl of amide bond were obtained at 3294–3524 cm−1 and 1614–1692 cm−1 respectively. The bands around 1302–1398 cm−1 and 1127–1183 cm−1 were assigned to asymmetric and symmetric stretching of SO2 of sulfonamide group respectively. The IR spectrum of synthesized compounds exhibits a band around 1506–1587 cm−1 to 1302–1378 cm−1 assignable to asymmetric and symmetric stretching of NO2. In the 1H NMR spectra of compound, singlet for NH protons of SO2NH and CONH appeared at δ 3.37–4.08 ppm and δ 10.19–10.81 ppm, respectively. The two aromatic protons of 2-chloro-4-nitro benzoic acid appeared around at δ 8.50 ppm and δ 7.50 ppm. The aromatic protons showed the chemical shift values in region of δ 6.58–8.58 ppm based on their chemical structure. In 13C NMR, signals for various carbons appeared in the region of δ 17.72 to 168.51 ppm.
In vitro antidiabetic evaluation
α-Glucosidase inhibitory activity
All the synthesized compounds were tested for their in vitro α-glucosidase inhibitory activity and revealed their varying degree of inhibitory potential with IC50 values of 10.75 ± 0.52 to 130.90 ± 2.42 μM (Table 2) as compared to reference acarbose (IC50 = 39.48 ± 0.80 μM). The compound 5o (R = 2-CH3-5-NO2) was found to be most active among this series of synthesized compounds. Most of the compounds exhibited good inhibitory potential with significant IC50 as compared to positive reference.
Table 2.
α-Glucosidase and α-amylase inhibitory activity (IC50) of synthesized derivatives (5a–5v) and their docking affinity with α-glucosidase (modeled protein) and α-amylase (PDB-1qho)
| Comp. | IC50 α-glucosidase (µM) | Binding score (α-glucosidase: modeled protein) | IC50 α-amylase (µM) | Binding score (α-amylase: 1qho) |
|---|---|---|---|---|
| 5a | 31.39 ± 1.66 | − 9.4 | 7.40 ± 0.15 | − 8.9 |
| 5b | 24.78 ± 2.69 | − 9.7 | 5.30 ± 1.23 | − 9.0 |
| 5c | 26.77 ± 1.13 | − 9.3 | 8.00 ± 0.71 | − 9.8 |
| 5d | 38.57 ± 0.01 | − 9.2 | 38.00 ± 0.51 | − 9.1 |
| 5e | 41.75 ± 1.08 | − 9.3 | 50.30 ± 0.21 | − 8.7 |
| 5f | 50.24 ± 0.89 | − 9.4 | 16.00 ± 0.33 | − 9.7 |
| 5g | 35.92 ± 0.60 | − 9.6 | 16.70 ± 0.41 | − 9.8 |
| 5h | 40.64 ± 1.49 | − 9.6 | 19.30 ± 0.63 | − 9.7 |
| 5i | 14.02 ± 0.93 | − 9.5 | 27.12 ± 0.51 | − 8.5 |
| 5j | 15.75 ± 0.90 | − 9.6 | 20.90 ± 1.24 | − 9.5 |
| 5k | 36.93 ± 1.30 | − 9.7 | 12.50 ± 0.91 | − 8.8 |
| 5l | 29.01 ± 0.86 | − 9.3 | 6.30 ± 0.42 | − 9.3 |
| 5m | 24.47 ± 1.23 | − 9.4 | 1.52 ± 0.84 | − 9.7 |
| 5n | 29.54 ± 1.53 | − 9.4 | 35.30 ± 0.45 | − 9.6 |
| 5o | 10.75 ± 0.52 | − 9.3 | 00.90 ± 0.31 | − 9.2 |
| 5p | 19.51 ± 0.43 | − 9.4 | 02.10 ± 0.52 | − 9.1 |
| 5q | 43.88 ± 1.18 | − 9.4 | 11.20 ± 0.67 | − 9.0 |
| 5r | 34.36 ± 0.62 | − 9.7 | 15.30 ± 1.24 | − 9.4 |
| 5s | 106.23 ± 0.61 | − 8.2 | 48.05 ± 0.23 | − 7.9 |
| 5t | 130.90 ± 2.42 | − 8.6 | 55.14 ± 0.71 | − 8.3 |
| 5u | 89.04 ± 1.76 | − 8.9 | 38.20 ± 0.34 | − 8.6 |
| 5v | 52.37 ± 1.92 | − 9.0 | 40.40 ± 0.87 | − 8.9 |
| Acarbose | 39.48 ± 0.88 | − 8.0 | 5.60 ± 0.30 | − 8.4 |
α-Amylase inhibitory activity
All the compounds were also evaluated for α-amylase inhibitory activity and the inhibition potential with IC50 values were found in range of 0.90 ± 0.31 μM to 55.14 ± 0.71 μM (Table 2). The compound 5o showed excellent inhibitory potential against α-amylase with IC50 value of 0.90 ± 0.31 μM. Compounds 5b, 5m, 5p showed most significant inhibitory potential against α-amylase with IC50 values of 5.30 ± 1.23, 1.52 ± 0.84 and 2.10 ± 0.52 μM, respectively, when compared to acarbose, used as reference compound (IC50 = 5.60 ± 0.30 μM).
Structure activity relationship
The compound 5o (R = 2-CH3-5-NO2) was the most active compound (IC50 = 10.75 ± 0.52 μM; 0.90 ± 0.31 μM) which may be due to the presence of electron withdrawing and electron donating group which generate an uniform electron flow, leading the compound to be more active and potent inhibitor against both enzymes. This fact is supported by the similar results of Adegboye et al. [38]. In compounds 5m (R = 2-CH3-3-NO2) and 5p (R = 2-CH3-4-NO2) difference in inhibitory potential was mainly affected by position of NO2 substituent.
However the inhibitory activity increased when the phenyl ring was substituted with CH3 at meta position, as observed in compound 5b (IC50 = 24.78 ± 2.69 μM; 5.30 ± 1.23 μM) in comparison to compounds 5a and 5c having CH3 substitution at para and ortho positions. Further a decrease in inhibitory activity was observed for compounds 5d (IC50 = 38.57 ± 0.01 μM; 38.00 ± 0.51 μM) and 5e (IC50 = 41.75 ± 1.08 μM; 50.30 ± 0.21 μM) bearing OCH3 substituted phenyl ring instead of compounds having CH3 substituted phenyl ring. The compounds 5f–5k, 5q and 5r bearing electron withdrawing groups were found to have considerable inhibitory potential. The results illustrated that compounds 5g (R = 3-Br), 5i (R = 3-Cl), 5r (R = 3-NO2), substitution at meta position of phenyl ring was found to be most favored for the α-glucosidase inhibitory activity while compounds 5f and 5q bearing electron withdrawing groups at para position were found to be most favorable for α-amylase inhibitory activity. This fact is supported by Taha et al. [39]. The compounds 5u (IC50 = 89.04 ± 1.76 μM, 38.20 ± 0.34 μM) and 5v (IC50 = 52.37 ± 1.92 μM, 40.40 ± 0.87 μM) substituted with heterocyclic amine displayed reduced inhibitory activities compared to aryl amines. This fact is supported by similar results of Kumar et al. [40] and Charaya et al. [35]. Substituting the compounds with n-propyl amine and butyl amine resulted in diminished activity as in compounds 5s (IC50 = 106.23 ± 0.61 μM, 48.05 ± 0.23 μM) and 5t (IC50 = 130.90 ± 2.42 μM, 55.14 ± 0.71 μM). This fact is supported by the similar study on benzamide derivatives by Charaya et al. [35].
Molecular docking
In silico molecular docking study was performed to investigate binding interactions and to explore binding modes of synthesized compounds with their respective targets. The binding affinities of all the synthesized compounds are reported in Table 2.
α-Glucosidase enzyme
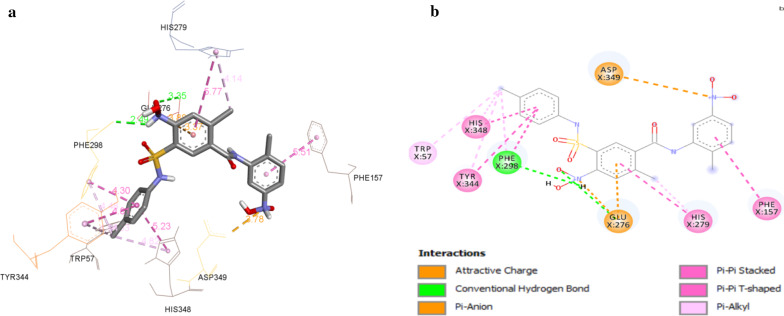
The docking results revealed that all the synthesized compounds displayed binding energy ranging from − 9.7 to − 8.0 kcal/mol and depicted various types of significant binding interactions like hydrogen bonding, electrostatic and hydrophobic interactions with the amino acid residues of active site of enzyme. The binding mode of most active compound 5o and modeled protein is presented in Fig. 1. The oxygen of 2-Cl-4-NO2 established hydrogen bonding interaction with Glu:276 amino acid residue at a distance of 3.35 Å whereas Phe:298 amino acid was found to engage in hydrogen bond interactions with both protonated nitrogen of NO2 of same with bond lengths of 2.49 Å. The nitrogen of 2-CH3-5-NO2 substituted compound displayed charge–charge interaction with Asp:349 amino acid residue (3.78 Å) while the nitrogen of 2-Cl-4-NO2 presented charge–charge interaction with Glu:276 amino acid residue (3.80 Å). The 2-Cl-4-NO2 substituted phenyl ring created pi-anion interaction with residue Glu:276 of modeled protein at a distance of 3.37 Å. It was noticed that Phe:157 residue (5.51 Å) formed pi–pi T shaped interaction with 2-CH3-5-NO2 substituted phenyl ring and para chloro substituted phenyl ring displayed two pi–pi T shaped and one pi–pi stacked interaction with His:348, Tyr:344 and Phe:298 amino acid residues. In addition 2-Cl-4-NO2 substituted phenyl ring created pi–pi stacked and pi–pi T shaped interaction with His:279 amino acid residues with bond length of 5.77 Å. Pi-alkyl interactions were established by chlorine of 2-Cl-4-NO2 substituted phenyl ring with His:279 residues at a distance of 4.14 Å. The chlorine of para chlorosubstituted phenyl ring was found to engage in forming pi–alkyl interactions with Tyr:344, His:348, Phe:298, Trp:57 amino acid residues of modeled protein. The involvement of 2-CH3-5-NO2 substituted phenyl ring in forming more hydrophobic interactions i.e. pi–pi interactions may be contributing to better activity of compound 5o as compared to compounds 5n (R = 2-CH3-3-NO2) and 5p (2-CH3-4-NO2). Comparison of compound 5c (R = 2-CH3) with 5n (R = 2-CH3-3-NO2), 5o (R = 2-CH3-5-NO2), 5p (R = 2-CH3-4-NO2), 5n, 5o, 5p displayed more hydrophobic interactions with Phe:177, Arg:312, Val:108, His:279, Phe:157, His:348, Tyr:344, Phe:298 amino acid residues of modeled protein which may have resulted in their higher inhibitory potential. The binding interaction between compounds 5c (R = 2-CH3) and residues of modeled protein was nearly same as 5a (R = 4-CH3) and 5b (R = 3-CH3). The difference was that ortho methyl substituted phenyl ring maintained pi–pi stacked, pi–alkyl and pi–pi T interactions (hydrophobic interactions) with Try:344, His:348, Phe:298, Phe:177, Phe:158, Tyr:344 amino acid residue that made 5c more active than 5a and 5b.
Fig. 1.
a 3D Binding confirmation of compound 5o with active site residues of α-glucosidase. b 2D binding confirmation of compound 5o with amino acid residue of nearby active site
The compound 5e (R = 2-OCH3) formed less number of hydrogen bonding, electrostatic and hydrophobic interactions as compared to compound 5d (R = 4-OCH3), resulting in decreased inhibitory potential of compound 5e. The binding of compound 5i (R = 3-Cl) facilitated one more pi–alkyl interaction with other hydrogen bonding, hydrophobic and electrostatic interactions same as that of compound 5j (R = 2-Cl), which may be contributing to better potential of compound 5i. Considering the moderately active compound 5r (R = 3-NO2), additional hydrophobic interaction such as pi–pi interactions with amino acid residues were observed as compared to compounds 5k (R = 2-NO2) and 5q (R = 4-NO2). In comparison to compounds bearing aromatic anilines, a decrease in inhibitory potential was observed in compounds 5s (R = n-propyl), and 5t (R = n-butyl), due to less pi–pi interactions between the inhibitory compounds and amino acid residues. The binding interactions of compound 5u (R = C4H3O-CH2 (2-furfuryl)) with residues of modeled protein were nearly same as that of 5v (R = C5H5N-(pyridine-2-yl)) but the difference was that 2-furfuryl ring exhibited pi–pi T shaped interaction with Trp:177 residue and four hydrogen bond interaction with Asp:329, Arg:376, His:90, Trp:93 residues of α-glucosidase with other interactions while compound 5v formed three hydrogen bond interactions, which made 5u more active than 5v against α-glucosidase enzyme.
α-Amylase enzyme
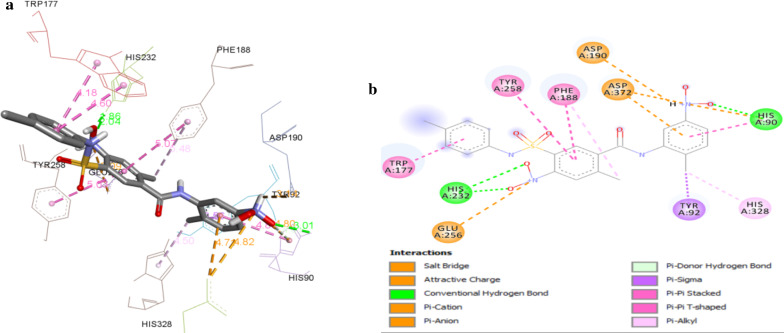
The docking results revealed that all the synthesized compounds displayed binding energy ranging from − 9.8 to − 7.9 kcal/mol. The binding mode of most active compound 5o and 1qho is presented in Fig. 2. The oxygen of 2-CH3-5-NO2 established hydrogen bonding interaction with His:90 amino acid residue at a distance of 3.01 Å whereas His:232 amino acid was found to engage in hydrogen bond interactions with both oxygen of NO2 of 2-Cl-4-NO2 substituted phenyl ring with bond lengths of 2.04 Å and 1.86 Å. The nitrogen of 2-CH3-5-NO2 displayed charge–charge interaction with Asp:372 amino acid residue (4.82 Å) while the protonated nitrogen of 2-CH3-5-NO2 presented salt bridge charge–charge interaction with Asp:190 amino acid residue (3.14 Å). The charge–charge interaction was also found between the nitrogen of 2-Cl-4-NO2 substituted phenyl ring and Glu:256 amino acid residue with bond length of 5.09 Å. The 2-CH3-5-NO2 substituted phenyl ring created pi-anion interaction with residue Asp:372 of α-amylase while nitrogen of 2-CH3-5-NO2 substituted phenyl ring formed pi-cation interaction with His:90 residue. It was shown that His:90 residue (4.88 Å) formed pi–pi T shaped interaction with 2-CH3-5-NO2 substituted phenyl ring and para chloro substituted phenyl ring displayed pi–pi stacked interaction with Trp:177 residue (4.60 Å). In addition 2-Cl-4-NO2 substituted phenyl ring created pi–pi stacked and pi–pi T shaped interaction with Tyr:258, Phe:188 amino acid residues with bond lengths of 5.3 Å and 5.07 Å, respectively. The pi–alkyl interactions were established by chlorine of 2-Cl-4-NO2 substituted phenyl ring and methyl of 2-CH3-5-NO2 substituted phenyl ring with Phe:188 and His:328 residues at a distance of 5.48 Å and 4.50 Å, respectively. The CH3 of 2-CH3-5-NO2 substituted phenyl ring was found to engage in forming pi-sigma interaction with Tyr:92 residue (3.56 Å) while oxygen of NO2 created pi-donor hydrogen bond with His:90 residue at a distance of 4.01 Å.
Fig. 2.
a 3D Binding confirmation of compound 5o with active site residues of α-amylase. b 2D binding confirmation of compound 5o with amino acid residue of nearby active site
The involvement of 2-CH3-5-NO2 substituted phenyl ring in forming more hydrophobic interactions may be contributing to better activity of compound 5o as compared to compounds 5n (R = 2-CH3-3-NO2) and 5p (2-CH3-4-NO2).The comparison of compound 5c (R =CH3) with 5n (R = 2-CH3-3-NO2), 5o (R = 2-CH3-5-NO2) and 5p (R = 2-CH3-4-NO2), 5n, 5o, 5p displayed more electrostatic and hydrophobic interactions with Asp:372, Asp:190, Glu:256, His:90, Trp:177, Tyr:258, Phe:188, His:328 and Tyr:92 residues of α-amylase enzyme, which may have resulted in increase in inhibitory potential. The binding interactions between compound 5b (R = 3-CH3) and residues of α-amylase were nearly same as 5a (R = 4-CH3) and 5c (R = 2–CH3). The difference was that meta methyl substituted phenyl ring and methyl group maintained pi–pi stacked, pi-alkyl and pi-sigma interactions (hydrophobic interactions) with Trp:177 amino acid residue which made 5b more active than 5a and 5c. The compound 5e (R = 2-OCH3) formed less number of hydrogen bonding, electrostatic and hydrophobic interactions as compared to compound 5d (R = 4-OCH3), resulting in decrease in inhibitory potential of compound 5e. The binding of compound 5f (R = 4-Br) facilitated two pi–pi T shaped and one pi–pi stacked interaction of para bromo substituted phenyl ring with Tyr:258, Phe:188, Trp:177 amino acid residues and two pi–pi stacked interactions of para chloro substituted phenyl ring with phe:188, Tyr:92 amino acid residues with other interactions, which made compound 5f more active than compounds 5g (R = 3-Br) and 5h (R = 2-Br). Considering the moderate active compound 5q (R = 4-NO2), additional hydrophobic interaction such as pi–pi T shaped, pi–pi stacked interactions with Tyr:258 and Trp:177 residues were observed as compared to compounds 5k (R = 2-NO2) and 5r (R = 3-NO2). In comparison to compounds bearing aromatic anilines, a decrease in inhibitory potential was observed in compounds 5s (R = n-propyl), and 5t (R = n-butyl), due to less pi–pi interactions between the inhibitory compounds and amino acid residues. The binding interaction of compound 5u (R = C4H3O-CH2 (2-furfuryl)) with residues of α-amylase was nearly same as 5v(R = C5H5N- (pyridine-2-yl))but the difference was that 2-furfuryl ring exhibited pi–pi T shaped interaction with Trp:177 residue and four hydrogen bond interactions with Asp:329, Arg:376, His:90, Trp:93 residues of α-amylase with other interactions while compound 5v formed three hydrogen bond interactions, which made 5u more active than 5v against α-amylase.
Molecular dynamics study
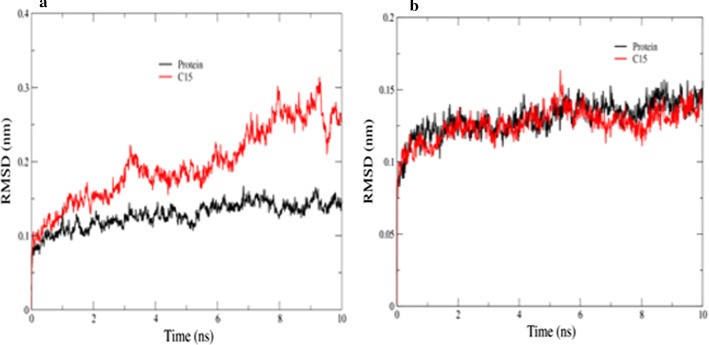
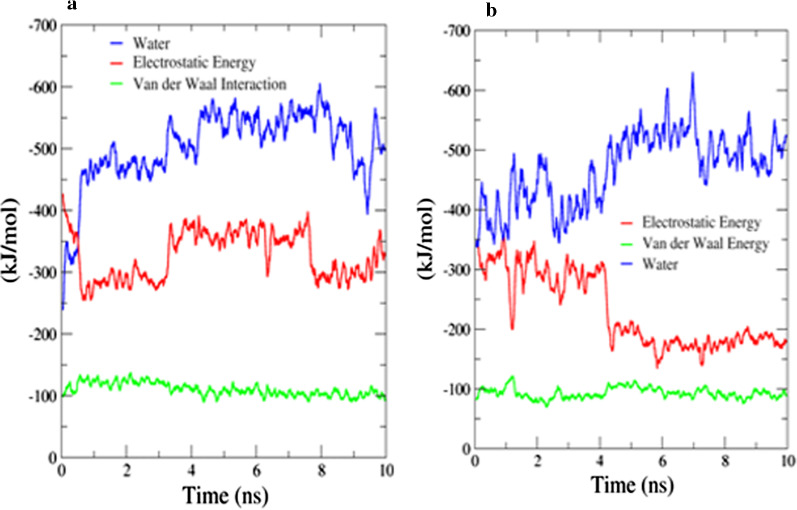
A stable protein backbone atoms RMSD vs time is an indication of the near-equilibrium system. As shown in Fig. 3a and b, the protein backbone in both systems attains a constant phase after an initial surge. Whereas, due to the extensive involvement of water molecules (− 500 kJ/mol) the ligand-bound protein backbone has higher RMSD fluctuations compared to the naked protein as represented in Fig. 3a. Figure 4 represent that electrostatic interactions are dominated between the ligand 5o and protein.
Fig. 3.
Plot of protein backbone RMSD against time where red color line represent 5o bound modeled protein and black line represent naked protein. a For 3aj7 modeled protein with 5o. b For 1qho with 5o
Fig. 4.
Plot of binding energy (electrostatic and Van der Waal) vs time: a For 3aj7 modeled protein with 5o. b For 1qho with 5o
The results obtained from the MD simulations demonstrated that water molecules are predominately involved in ligand–protein interactions (Fig. 5). As shown in the figure, the fall in electrostatic energy that corresponds to the ligand–protein interactions is compensated by the water molecules.
Fig. 5.
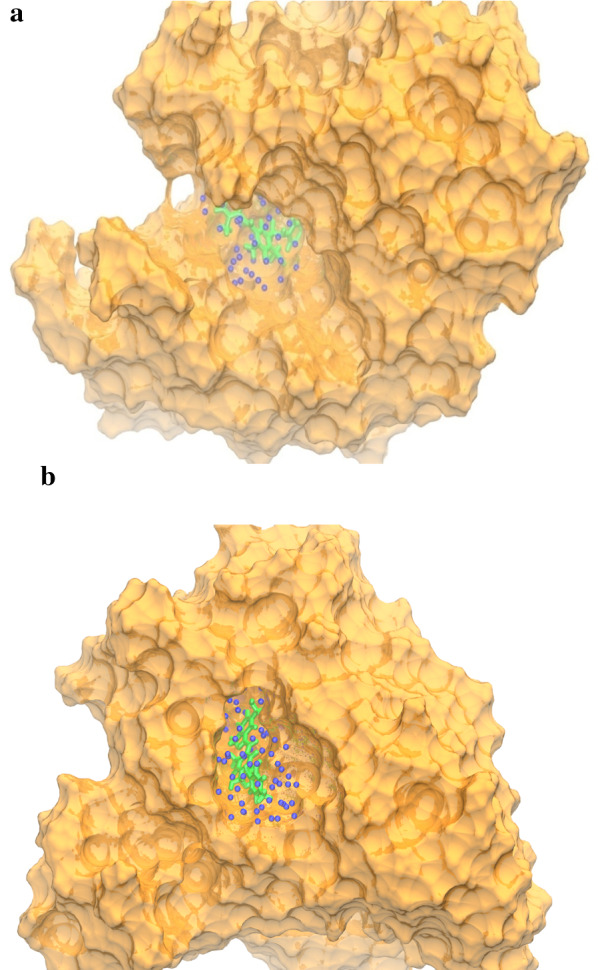
a Active site of modeled protein of S. cerevisiae α-glucosidase (Golden color) with ligand 5o shown in green color, oxygen atom of water molecule as sphere in blue color. b Active site of α-amylase (Golden color) with ligand 5o shown in green color, oxygen atom of water molecule as sphere in blue color
In silico ADMET properties prediction
Lipinski’s rule of five, topological polar surface area, aqueous solubility and number of rotatable bonds, these calculated parameters are presented in Additional file 1: Table S1. The human intestinal absorption values were found in range of 93.10 to 95.93% which established the moderate to good absorption capacity of synthesized compounds and supported their interaction with target cell.
The in vitro Caco-2 cell permeable property in the range of 0.36–0.55 nm/s, in vitro MDCK cell permeability in range of 0.01–0.97 nm/s designated low permeability of target compounds with the concerned cell line. The synthesized compounds displayed values in range of 95.75–100% confirmed their strong binding capacity with proteins. The in vivo blood brain barrier penetration ranges from 0.01 to 0.32 supported their low to moderate distribution in vivo with medium to good penetration capacity (Additional file 1: Table S2). Bioactivity and toxicity risk values of synthesized compounds are illustrated in Additional file 1: Table S3.
Conclusion
A series of 2-chloro-5-[(4-chlorophenyl)sulfamoyl]-N-(alkyl/aryl)-4-nitrobenzamide derivatives (5a-5v) has been synthesized and all the compounds were found to possess potent to moderate inhibitory potential against α-glucosidase and α-amylase. Compound 5o (2-chloro-5-[(4-chlorophenyl) sulfamoyl]-N-(2-methyl-5-nitrophenyl)-4-nitrobenzamide) was found to be highly active having fourfold inhibitory potential against α-glucosidase and around six times inhibitory activity against α-amylase in comparison to standard drug acarbose. Molecular docking results of antidiabetic study showed reasonable dock score and binding interactions of synthesized molecules with their respective targets. Analysis of RMSD of ligand protein complex during molecular dynamic simulations suggested stability of the most active compounds 5o in binding site of respective target proteins i.e. α-glucosidase and α-amylase enzymes. Prediction of computational drug like properties showed that most of synthesized compounds are safe with acceptable ADMET and druggable properties.
Materials and methods
Chemicals
The analytical grade chemicals and reagents were used as such in experiments without any purification. Decibel melting point apparatus was used for checking the melting point of the synthesized compounds and are reported as uncorrected. The silica gel-precoated aluminum sheets for thin-layer chromatography (TLC) were employed to keep a vigil of the reaction progress. FT-IR (Diffuse Reflectance Method (DRS) -8000A, Shimadzu, Japan) spectrophotometer was utilized for recording infrared spectra and the Bruker Avance III, 400 MHz NMR spectrometer was employed for nuclear magnetic resonance spectra (1H NMR, 13C NMR; Chemical shift δ values- ppm). α-Glucosidase from Saccharomyces cerevisiae (EC 3.2.1.20, Sigma Aldrich) and α-amylase from malt (232-588-1, HiMedia) have been used for in vitro studies.
General procedure for synthesis of 2-chloro-5-[(4-chlorophenyl)sulfamoyl]-N-(alkyl/aryl)-4-nitrobenzamide (5a–5v)
Synthesis of 2-chloro-5-(chlorosulfonyl)-4-nitro benzoic acid (2)
Compound 2 was synthesized from 2-chloro-4-nitro benzoic acid (1) as previously reported method in literature [32].
Synthesis of 2-chloro-5-[(4-chlorophenyl)sulfamoyl]-4-nitrobenzoic acid (3)
2-Chloro-5-(chlorosulfonyl)-4-nitrobenzoic acid (1 g, 0.003 mol) was refluxed with p-nitro aniline (0.003 mol) using dimethyl formamide as solvent, till the completion of reaction [34]. The reaction progress was monitored by TLC. The reaction mixture was cooled and yielded precipitates were washed and recrystallized.
Synthesis of 2-chloro-5-[(4-chlorophenyl)sulfamoyl]-N-(alkyl/aryl)-4-nitrobenzamide (5a–5o)
Compound 3 (0.5 g, 0.0012 mol) was further treated with excess of thionyl chloride in presence of catalytic amount of DMF with calcium chloride (CaCl2) guard tube to get 2-chloro-5-[(4-chlorophenyl)sulfamoyl]-4-nitrobenzoylchloride (4). Compound 4 was dissolved in DMF and refluxed with anilines/amines/heterocyclic amines to get the desired products in appropriate yield [41]. After refluxing, mixture was cooled and poured on crushed ice, separated product was filtered and washed with dilute HCl and dried.
Physicochemical and spectral characterization
2-Chloro-5-[(4-chlorophenyl)sulfamoyl]-N-(4-methylphenyl)-4-nitrobenzamide (5a)
% Yield: 37.70; m.p.: 90–92 °C; Rf: 0.81 (Chloroform); FTIR (KBr): νmax (cm−1): 3502.79 (N–H str.), 3171.70 (C–H str., Ar), 2977.30, 2889.01 (C–H str., Aliphatic), 1641.45 (C=O), 1600.41 (N–H bend), 1586.48 (asym. NO2 str.), 1349.25 (sym. NO2 str.), 1315.47 (asym. SO2 str.), 1157.31 (sym. SO2 str.), 733.44 (C–Cl);1HNMR (300 MHz, DMSO-d6), δ ppm: 2.15 (s, 3H, CH3), 4.01 (s, 1H, NH), 6.78–6.80 (d, 2H, CH of C3, C5 of –CONH–C6H5CH3–), 7.11–7.13 (d, 2H, CH of C2, C6 of –CONH–C6H5CH3–), 7.60 (s, 1H, CH of C6 of ClNO2C6H2CONH–), 7.88–7.89 (d, 2H, CH of C2 and C6 of ClC6H4NH), 8.23 (s, 1H, CH of C3 of ClNO2C6H2CONH–), 8.42–8.43 (d, 2H, CH of C3 and C5 of ClC6H4NH), 10.69 (s, 1H, NH); 13CNMR (300 MHz, DMSO-d6), δ ppm: 163.59 (C=O), 148.75 (C–S), 142.92 (C–NO2), 136.44 (C–NH), 133.79 (C–Cl), 131.62, 130.79, 130.34, 129.71, 125.76, 123.26, 120.15, 119.55, 117.91, 21.00.
2-Chloro-5-[(4-chlorophenyl)sulfamoyl]-N-(3-methylphenyl)-4-nitrobenzamide (5b)
% Yield: 50.81; m.p.: 102–104 °C; Rf: 0.82 (H:E— 8:2); FTIR (KBr): νmax (cm−1): 3502.97 (N–H str.), 3117.98 (C–H str., Ar), 2982.94, 2882.00 (C–H str., Aliphatic), 1621.03 (C=O), 1602.20 (N–H bend), 1544.11 (asym. NO2 str.), 1370.45 (asym. SO2 str.), 1340.55 (sym. NO2 str.), 1170.07 (sym. SO2 str.), 766.16 (C–Cl); 1HNMR (300 MHz, DMSO-d6), δ ppm: 2.35 (s, 3H, CH3), 3.92 (s, 1H, NH), 5.48 (s, 1H, CH of C2 –CONH–C6H5CH3–), 6.88 (s, 1H, CH of C6 of ClNO2C6H2CONH–), 7.10–7.12 (d, 2H, CH of C2 and C6 of ClC6H4NH), 7.19 (s, 1H, CH of C3 of ClNO2C6H2CONH–), 7.23–7.25 (d, 1H, CH of C6 of –CONH–C6H5CH3–), 7.35–7.40 (m, 2H, CH of C4, C5 –CONH–C6H5CH3–), 7.51–7.53 (d, 2H, CH of C3 and C5 of ClC6H4NH), 10.31 (s, 1H, NH); 13CNMR (300 MHz, DMSO-d6), δ ppm: 168.51 (C=O), 151.75 (C–S), 143.40 (C–NO2), 139.92 (C–NH), 138.59 (C–Cl), 132.07, 130.56, 130.11, 129.23, 128.68, 124.09, 120.89, 119.21, 115.21, 21.41.
2-Chloro-5-[(4-chlorophenyl)sulfamoyl]-N-(2-methylphenyl)-4-nitrobenzamide (5c)
% Yield: 84.91; m.p.: 88–90 °C; Rf: 0.75 (Chloroform); FTIR (KBr): νmax (cm−1): 3354.96 (N–H str.), 3197.07 (C–H str., Ar), 2915.46, 2837.25 (C–H str., Aliphatic), 1602.25 (C=O), 1578.76 (N–H bend), 1511.25 (asym. NO2 str.), 1374.26 (asym. SO2 str.),1348.27 (sym. NO2 str.), 1146.44 (sym. SO2 str.), 753.00 (C–Cl); 1HNMR (300 MHz, DMSO-d6), δ ppm: 2.35 (s, 3H, CH3), 3.45 (s, 1H, NH), 6.58–6.60 (d, 1H, CH of C3 –CONH–C6H5CH3–), 7.26–7.33 (t, 2H, CH of C4, C5 of –CONH–C6H5CH3–), 7.40–7.41 (d, H, CH of C6 –CONH–C6H5CH3–), 7.81 (s, 1H, CH of C6 of ClNO2C6H2CONH–), 8.00–8.02 (d, 2H, CH of C2 and C6 of ClC6H4NH), 8.36–8.37 (d, 2H, CH of C3 and C5 of ClC6H4NH), 8.51 (s, 1H, CH of C3 of ClNO2C6H2CONH–), 10.31 (s, 1H, NH); 13CNMR (300 MHz, DMSO-d6), δ ppm: 162.32 (C=O), 153.39 (C–S), 142.32 (C–NO2), 136.24 (C–NH), 133.93 (C–Cl), 131.57, 129.24, 127.88, 125.93, 123.22, 120.56, 117.54, 115.14, 17.72.
2-Chloro-5-[(4-chlorophenyl)sulfamoyl]-N-(4-methoxyphenyl)-4-nitrobenzamide (5d)
% Yield: 88.69; m.p.: 150–152 °C; Rf: 0.23 (Chloroform); FTIR (KBr): νmax (cm−1): 3438.05 (N–H str.), 3083.26 (C–H str., Ar), 1614.01 (C=O), 1594.19 (N–H bend), 1555.62 (asym. NO2 str.), 1352.32 (sym. NO2 str.), 1304.87 (asym. SO2 str.), 1261.51 (C–O–C str.), 1175.58 (sym. SO2 str.), 727.73 (C–Cl);1HNMR (300 MHz, DMSO-d6), δ: 3.38 (s, 3H, OCH3 of –CONH–C6H5OCH3–), 3.89 (s, 1H, NH), 7.02–7.03 (d, 2H, CH of C3, C5 of –CONH–C6H5OCH3–), 7.34–7.37 (d, 2H, CH of C2, C6 of –CONH– C6H5OCH3–), 7.52 (s, 1H, CH of C6 of ClNO2C6H2CONH–), 7.81–7.83 (d, 2H, CH of C2 and C6 of ClC6H4NH), 8.09–8.11 (d, 2H, CH of C3 and C5 of ClC6H4NH), 8.50 (s, 1H, CH of C3 of ClNO2C6H2CONH–), 10.32 (s, 1H, NH); 13CNMR (300 MHz, DMSO-d6), δ ppm: 164.63 (C=O), 154.30 (C–S), 144.57 (C–NO2), 138.48 (C–NH), 136.24 (C–Cl), 131.58, 129.95, 128.12, 126.09, 124.46, 123.24, 120.20, 32.90.
2-Chloro-5-[(4-chlorophenyl)sulfamoyl]-N-(2-methoxyphenyl)-4-nitrobenzamide (5e)
% Yield: 85.30; m.p.: 164–166 °C; Rf: 0.53 (B:EA–7:3); FTIR (KBr): νmax (cm−1): 3447.82 (N–H str.), 3012.86 (C–H str., Ar), 1682.45 (C=O), 1597.64 (N–H bend), 1530.54 (asym. NO2 str.), 1378.16 (sym. NO2 str.), 1307.76 (asym. SO2 str.), 1252.68 (C–O–C str.), 1174.67 (sym. SO2 str.), 752.65 (C–Cl); 1HNMR (300 MHz, DMSO-d6), δ ppm: 3.38 (s, 3H, OCH3), 3.89 (s, 1H, NH), 7.07–7.09 (d, 1H, CH of C3 –CONH–C6H5OCH3–), 7.27.7.33 (m, 3H, CH of C4, C5, C6 of –CONH–C6H5 OCH3–), 7.52 (s, 1H, CH of C6 of ClNO2C6H2CONH–), 7.81–7.82 (d, 2H, CH of C2 and C6 of ClC6H4NH), 8.28–8.30 (d, 2H, CH of C3 and C5 of ClC6H4NH), 8.49 (s, 1H, CH of C3 of ClNO2C6H2CONH–), 10.48 (s, 1H, NH); 13CNMR (300 MHz, DMSO-d6), δ ppm: 164.21 (C=O), 157.32 (C–S), 145.07 (C–NO2), 139.07 (C–NH), 136.16 (C–Cl), 131.34, 129.45, 128.24, 127.14, 125.18, 121.68, 121.51, 120.55, 35.60.
N-(4-Bromophenyl)-2-chloro-5-[(4-chlorophenyl)sulfamoyl]-4-nitrobenzamide (5f)
% Yield: 91.30; m.p.: 200–202 °C; Rf: 0.56 (Chloroform); FTIR (KBr): νmax (cm−1): 3392.85 (N–H str.), 3012.55 (C–H str., Ar), 1614.45 (C=O), 1591.36 (N–H bend), 1562.37 (asym. NO2 str.), 1353.50 (sym. NO2 str.), 1308.72 (asym. SO2 str.), 1145.63 (sym. SO2 str.), 732.07 (C–Cl), 691.49 (C–Br);1HNMR (300 MHz, DMSO-d6), δ ppm: 3.60 (s, 1H, NH), 7.46 (s, 1H, CH of C6 of ClNO2C6H2CONH–), 7.54–7.55 (d, 2H, CH of C2 and C6 of ClC6H4NH), 8.00–8.02 (d, 2H, CH of C2, C6 of –CONH–C6H5Br–), 8.24–8.26 (d, 2H, CH of C3 and C5 of ClC6H4NH), 8.36–8.37 (d, 2H, CH of C3, C5 –CONH–C6H5CH3–), 8.51 (s, 1H, CH of C3 of ClNO2C6H2CONH–), 10.55 (s, 1H, NH);13CNMR (300 MHz, DMSO-d6), δ ppm: 166.34 (C=O), 158.33 (C–S), 144.35 (C–NO2), 140.21 (C–NH), 135.49 (C–Cl), 132.90, 130.58, 129.89, 129.21, 128.24, 127.08, 120.18.
N-(3-Bromophenyl)-2-chloro-5-[(4-chlorophenyl)sulfamoyl]-4-nitrobenzamide (5g)
% Yield: 44.92; m.p.: 181–183 °C; Rf: 0.73 (Chloroform); FTIR (KBr): νmax (cm−1): 3502.79 (N–H str.), 3058.19 (C–H str., Ar), 1614.45 (C=O), 1588.41 (N–H bend), 1566.61 (asym. NO2 str.), 1372.38 (asym. SO2 str.), 1302.95 (sym. NO2 str.), 1175.63 (sym. SO2 str.), 778.76 (C–Cl), 675.80 (C–Br); 1HNMR (300 MHz, DMSO-d6), δ ppm: 3.71 (s, 1H, NH), 7.37 (s, 1H, CH of C6 of ClNO2C6H2CONH–), 7.51 (s, 1H, CH of –CONH–C6H5Br–), 7.71–7.80 (d, 2H, CH of C2 and C6 of ClC6H4NH–), 7.93–7.98 (m, 3H, CH C4, C5 and C6 of –CONH–C6H5Br–), 8.21–8.23 (d, 2H, CH of C3 and C5 of ClC6H4NH), 8.48 (s, 1H, CH of C3 of ClNO2C6H2CONH–), 10.40 (s, 1H, NH); 13CNMR (300 MHz, DMSO-d6), δ ppm: 165.33 (C=O), 157.35 (C–S), 143.24 (C–NO2), 139.51 (C–NH), 136.54 (C–Cl), 133.05, 130.77, 129.10, 128.39, 128.17, 127.63, 126.71, 126.12, 121.49.
N-(2-Bromophenyl)-2-chloro-5-[(4-chlorophenyl)sulfamoyl]-4-nitrobenzamide (5h)
% Yield: 55.07; m.p.: 180–182 °C; Rf: 0.70 (Chloroform); FTIR (KBr): νmax (cm−1): 3392.85 (N–H str.), 3028.29 (C–H str., Ar), 1614.62 (C=O), 1594.19 (N–H bend), 1565.96 (asym. NO2 str.), 1349.23 (sym. NO2 str.), 1377.20 (asym. SO2 str.), 1152.15 (sym. SO2 str.), 754.34 (C–Cl), 661.15 (C–Br);1HNMR (300 MHz, DMSO-d6), δ ppm: 3.83 (s, 1H, NH), 6.57–6.61 (t, 1H, CH of C5 of –CONH–C6H5Br–), 6.90–6.92 (d, 1H, CH of C6 –CONH–C6H5Br–), 7.12–7.14 (t, 1H, CH of C4 of –CONH–C6H5Br–), 7.32–7.40 (d, 2H, CH of C2 and C6 of ClC6H4NH), 7.51–7.52 (d, 1H, CH of C3 of –CONH–C6H5Br–), 7.82 (s, 1H, CH of C6 of ClNO2C6H2CONH–), 8.16–8.18 (d, 2H, CH of C3 and C5 of ClC6H4NH), 8.50 (s, 1H, CH of C3 of ClNO2C6H2CONH–), 10.39 (s, 1H, NH); 13CNMR (300 MHz, DMSO-d6), δ ppm: 167.04 (C=O), 156.56 (C–S), 143.06 (C–NO2), 138.20 (C–NH), 135.47 (C–Cl), 135.07, 130.23, 129.04, 128.93, 126.13, 125.34, 122.21.
2-Chloro-N-(3-chlorophenyl)-5-[(4-chlorophenyl)sulfamoyl]-4-nitrobenzamide (5i)
% Yield: 93.25; m.p.: 220–222 °C; Rf: 0.72 (B:EA– 7:3); FTIR (KBr): νmax (cm−1): 3455.53 (N–H str.), 3056.96 (C–H str., Ar), 1687.80 (C=O), 1592.15 (N–H bend), 1542.11 (asym. NO2 str.), 1366.59 (sym. NO2 str.), 1308.72 (asym. SO2 str.), 1165.99 (sym. SO2 str.), 782.92 (C–Cl); 1HNMR (300 MHz, DMSO-d6), δ ppm: 3.39 (s, 1H, NH), 7.51 (s, 1H, CH of C6 of ClNO2C6H2CONH–), 7.80–7.83 (d, 2H, CH of C2 and C6 of ClC6H4NH), 8.01–8.04 (m, 3H, CH C4, C5 and C6 of –CONH–C6H5Cl–), 8.23 (s, 1H, CH of –CONH–C6H5Cl–), 8.30–8.32 (d, 2H, CH of C3 and C5 of ClC6H4NH), 8.49 (s, 1H, CH of C3 of ClNO2C6H2CONH–), 10.20 (s, 1H, NH); 13CNMR (300 MHz, DMSO-d6), δ ppm: 167.04 (C=O), 155.28 (C–S), 145.02 (C–NO2), 139.44 (C–NH), 136.10 (C–Cl), 132.12, 130.16, 129.12, 128.14, 127.33, 127.17, 126.39, 124.17.
2-Chloro-N-(2-chlorophenyl)-5-[(4-chlorophenyl)sulfamoyl]-4-nitrobenzamide (5j)
% Yield: 87.71; m.p.: 176–178 °C; Rf: 0.5 (B:EA– 7:3); FTIR (KBr): νmax (cm−1): 3337.42 (N–H str.), 3060.18 (C–H str., Ar), 1665.56 (C=O), 1592.43 (N–H bend), 1533.43 (asym. NO2 str.), 1377.20 (sym. NO2 str.), 1392.63 (asym. SO2 str.), 1173.70 (sym. SO2 str.), 755.22 (C–Cl); 1HNMR (300 MHz, DMSO-d6), δ ppm: 3.89 (s, 1H, NH), 7.44 (s, 1H, CH of C6 of ClNO2C6H2CONH–), 7.85–7.90 (m, 3H, CH of C4,C5 and C6 –CONH–C6H5Cl–), 7.98–8.00 (d, 2H, CH of C2 and C6 of ClC6H4NH), 8.21–8.22 (d, 2H, CH of C3 and C5 of ClC6H4NH), 8.31–8.33 (d, 1H, CH of C3 of –CONH–C6H5Cl–), 8.52 (s, 1H, CH of C3 of ClNO2C6H2CONH–), 10.19 (s, 1H, NH);13CNMR (300 MHz, DMSO-d6), δ ppm: 166.34 (C=O), 157.02 (C–S), 146.93 (C–NO2), 139.24 (C–NH), 135.56 (C–Cl), 131.57, 131.25, 129.53, 128.55, 127.21, 125.24, 121.86.
2-Chloro-5-[(4-chlorophenyl)sulfamoyl]-4-nitro-N-(2-nitrophenyl)benzamide (5k)
% Yield: 44.61; m.p.: 150–152 °C; Rf: 0.8 (B:EA– 7:3); FTIR (KBr): νmax (cm−1): 3524.01 (N–H str.), 3198.03 (C–H str., Ar), 1614.45 (C=O), 1593.50 (N–H bend), 1563.33 (asym. NO2 str.), 1376.23 (asym. SO2 str.), 1347.30 (sym. NO2 str.), 1146.26 (sym. SO2 str.), 746.55 (C–Cl);1HNMR (300 MHz, DMSO-d6), δ ppm: 3.64 (s, 1H, NH), 7.26–7.28 (d, 1H, CH of C6 of –CONH–C6H5NO2–), 7.46–7.48 (d, 1H, CH of C3 of –CONH–C6H5NO2–), 7.65 (s, 1H, CH of C6 of ClNO2C6H2CONH–), 7.81–7.82 (d, 2H, CH of C2 and C6 of ClC6H4NH), 7.90–7.96 (m, 2H, CH of C4,C5 of –CONH–C6H5NO2–), 8.16–8.19 (d, 2H, CH of C3 and C5 of ClC6H4NH), 8.50 (s, 1H, CH of C3 of ClNO2C6H2CONH–), 10.30 (s, 1H, NH);13CNMR (300 MHz, DMSO-d6), δ ppm: 167.03 (C=O), 156.04 (C–S), 149.28 (C–NO2), 141.14 (C–NH), 137.43 (C–Cl), 135.29, 133.38, 132.21, 130.67, 130.22, 129.18, 128.15, 126.02.
2-Chloro-N-(3-chloro-2-methylphenyl)-5-[(4-chlorophenyl)sulfamoyl]-4-nitrobenzamide (5l)
% Yield: 57.62; m.p.: 212–214 °C; Rf: 0.65 (B:EA– 7:3); FTIR (KBr): νmax (cm−1): 3447.82 (N–H str.), 3096.77 (C–H str., Ar), 2947.05, 2885.31 (C–H str., Aliphatic), 1692.29 (C=O), 1592.79 (N–H bend), 1531.51 (asym. NO2 str.), 1380.09 (asym. SO2 str.), 1306.80 (sym. NO2 str.), 1174.67 (sym. SO2 str.), 772.81 (C–Cl); 1HNMR (300 MHz, DMSO-d6), δ ppm: 2.26 (s, H, CH3), 3.45 (s, 1H, NH), 7.24–7.30 (m, 2H, CH C4, C5 andC6 of –CONH–C6H5CH3Cl–), 7.63 (s, 1H, CH of C6 of ClNO2C6H2CONH–), 7.81–7.82 (d, 2H, CH of C2 and C6 of ClC6H4NH), 8.28–8.30 (d, 2H, CH of C3 and C5 of ClC6H4NH), 8.58 (s, 1H, CH of C3 of ClNO2C6H2CONH–), 10.59 (s, 1H, NH);13CNMR (300 MHz, DMSO-d6), δ ppm: 165.37 (C=O), 158.36 (C–S), 147.85 (C–NO2), 140.10 (C–NH), 138.55 (C–Cl), 137.21, 135.46, 130.80, 129.61, 129.04, 125.15, 121.47, 118.28, 23.57.
2-Chloro-5-[(4-chlorophenyl)sulfamoyl]-N-(2-methyl-3-nitrophenyl)-4-nitrobenzamide (5m)
% Yield: 38.29; m.p.: 170–172 °C; Rf: 0.61 (C:T– 9:1); FTIR (KBr): νmax (cm−1): 3469.45 (N–H str.), 3095.80 (C–H str., Ar), 2882.55 (C–H str., Aliphatic), 1692.32 (C=O), 1598.62 (N–H bend), 1530.06 (asym. NO2 str.), 1351.59 (sym. NO2 str.), 1302.76 (asym. SO2 str.), 1177.56 (sym. SO2 str.), 734.45 (C–Cl);1HNMR (300 MHz, DMSO-d6), δ ppm: 2.14 (s, H, CH3 of –CONH–C6H5CH3NO2–), 3.37 (s, 1H, NH), 7.45 (s, 1H, CH of C6 of ClNO2C6H2CONH–), 7.94–7.95 (d, 2H, CH of C2 and C6 of ClC6H4NH), 8.09–8.13 (m, 2H, CH C4, C5 andC6 of –CONH–C6H5CH3NO2–), 8.46–8.47 (d, 2H, CH of C3 and C5 of ClC6H4NH), 8.72 (s, 1H, CH of C3 of ClNO2C6H2CONH–), 10.50 (s, 1H, NH);13CNMR (300 MHz, DMSO-d6), δ ppm: 166.48 (C=O), 158.10 (C–S), 146.02 (C–NO2), 138.52 (C–NH), 137.46 (C–Cl), 136.78, 136.15, 131.20, 129.89, 129.25, 128.19, 126.71, 126.26, 117.19, 22.76.
2-Chloro-N-(2-chloro-4-nitrophenyl)-5-[(4-chlorophenyl)sulfamoyl]-4-nitrobenzamide (5n)
% Yield: 79.16; m.p.: 214–216 °C; Rf: 0.34 (B:EA– 7:3); FTIR (KBr): νmax (cm−1): 3461.39 (N–H str.), 3187.49 (C–H str., Ar), 1626.02 (C=O), 1587.77 (asym. NO2 str.), 1378.16 (asym. SO2 str.), 1320.40 (sym. NO2 str.), 1127.06 (sym. SO2 str.), 747.47 (C–Cl); 1HNMR (300 MHz, DMSO-d6), δ ppm: 3.37 (s, 1H, NH), 7.53 (s, 1H, CH of C6 of ClNO2C6H2CONH–), 7.80–7.81 (d, 2H, CH of C2 and C6 of ClC6H4NH–), 8.11–8.13 (d, 2H, CH of C3 and C5 of NO2C6H4NH), 8.25–8.26 (d, H, CH of C6 of –CONH–C6H5ClNO2), 8.36–8.38 (d, H, CH of C5 of –CONH–C6H5ClNO2), 8.51 (s, 1H, CH of C3 of ClNO2C6H2CONH–), 8.61 (s, H, CH of C3 of –CONH–C6H5ClNO2), 10.53 (s, 1H, NH);13CNMR (300 MHz, DMSO-d6), δ ppm: 168.41 (C=O), 160.22 (C–S), 145.06 (C–NO2), 138.02 (C–NH), 136.61 (C–Cl), 132.42, 130.59, 128.62, 128.41, 126.57, 124.78, 122.38, 118.76, 26.08.
2-Chloro-5-[(4-chlorophenyl)sulfamoyl]-N-(2-methyl-5-nitrophenyl)-4-nitrobenzamide (5o)
% Yield: 93.61; m.p.: 203–205 °C; Rf: 0.30 (B:EA– 7:3); FTIR (KBr): νmax (cm−1): 3488.67 (N–H str.), 3082.35 (C–H str., Ar), 2979.35, 2899.74 (C–H str., Aliphatic), 1630.01 (C=O), 1511.16 (asym. NO2 str.), 1382.02 (asym. SO2 str.), 1345.60 (sym. NO2 str.), 1138.02 (sym. SO2 str.), 737.34 (C–Cl);1HNMR (300 MHz, DMSO-d6), δ ppm: 2.14 (s, H, CH3 of –CONH–C6H5CH3NO2–), 3.37 (s, 1H, NH), 7.24–7.29 (m, 2H, CH C3, C4 of –CONH–C6H5CH3NO2–), 7.62 (s, 1H, CH of C6 of ClNO2C6H2CONH–), 7.84–7.87 (d, 2H, CH of C2 and C6 of ClC6H4NH), 8.32 (d, 2H, CH of C3 and C5 of ClC6H4NH), 8.41 (d, H, CH of C6, of –CONH–C6H5CH3NO2–), 8.29–8.58 (s, 1H, CH of C3 of ClNO2C6H2CONH–), 10.67 (s, 1H, NH);13CNMR (300 MHz, DMSO-d6), δ ppm: 163.88 (C=O), 153.37 (C–S), 147.61(C–NO2), 139.52 (C–NH), 137.36 (C–Cl), 130.91, 129.21, 128.35, 127.24, 124.82, 122.63, 118.04, 110.49, 18.53.
2-Chloro-5-[(4-chlorophenyl)sulfamoyl]-N-(2-methyl-4-nitrophenyl)-4-nitrobenzamide (5p)
% Yield: 56.66; m.p.: 104–106 °C; Rf: 0.50 (B:EA– 7:3); FTIR (KBr): νmax (cm−1): 3473.73 (N–H str.), 3090.98 (C–H str., Ar), 2838.62 (C–H str., Aliphatic), 1640.49 (C=O), 1586.11 (N–H bend), 1529.58 (asym. NO2 str.), 1396.49 (asym. SO2 str.), 1350.19 (sym. NO2 str.), 1154.21 (sym. SO2 str.), 740.69 (C–Cl);1HNMR (300 MHz, DMSO-d6), δ ppm: 2.12 (s, H, CH3 of –CONH–C6H5CH3NO2–), 3.46 (s, 1H, NH), 7.51 (s, H, CH of C3 of –CONH–C6H5CH3NO2–), 7.67 (s, 1H, CH of C6 of ClNO2C6H2CONH–), 7.84–7.87 (d, 2H, CH of C2 and C6 of ClC6H4NH–), 8.21–8.25 (m, 2H, CH C5, C6 of –CONH–C6H5CH3NO2–), 8.47–8.48 (d, 2H, CH of C3 and C5 of ClC6H4NH–), 8.64 (s, 1H, CH of C3 of ClNO2C6H2CONH–), 10.70 (s, 1H, NH);13CNMR (300 MHz, DMSO–d6), δ ppm: 167.72 (C=O), 154.62 (C–S), 148.40 (C–NO2), 141.67 (C–NH), 135.94 (C–Cl), 130.20, 128.44, 126.61, 124.87, 124.39, 120.86, 112.81, 17.59.
2-Chloro-N-(4-chlorophenyl)-5-[(4-chlorophenyl)sulfamoyl]-4-nitrobenzamide (5q)
% Yield: 95.24; m.p.: 177–179 °C; Rf: 0.65 (B:EA— 7:3); FTIR (KBr): νmax (cm−1): 3483.02 (N–H str.), 3108.34 (C–H str., Ar), 1633.74 (C=O), 1599.82 (N–H bend), 1530.54 (asym. NO2 str.), 1396.49 (asym. SO2 str.), 1353.09 (sym. NO2 str.), 1183.49 (sym. SO2 str.), 754.34 (C–Cl);1HNMR (300 MHz, DMSO-d6), δ ppm: 3.67 (s, 1H, NH), 7.44–7.46 (d, 2H, CH of C2 and C6 of –ClC6H4NH–), 7.60 (s, 1H, CH of C6 of ClNO2C6H2CONH–), 7.95–7.96 (d, 2H, CH of C2 and C6 of –CONHC6H5Cl–), 8.00–8.02 (d, 2H, CH of C3 and C5 of –CONH–C6H5Cl–), 8.24–8.26 (d, 2H, CH of C3 and C5 of –ClC6H4NH–), 8.36 (s, 1H, CH of C3 of ClNO2C6H2CONH–), 10.63 (s, 1H, NH);13CNMR (300 MHz, DMSO-d6), δ ppm: 166.17 (C=O), 156.21 (C–S), 149.28 (C–NO2), 138.40 (C–NH), 138.06 (C–Cl), 136.11, 131.94, 132.52, 129.89, 126.84, 125.71, 123.20, 112.90.
2-Chloro-5-[(4-chlorophenyl)sulfamoyl]-4-nitro-N-(3-nitrophenyl)benzamide (5r)
% Yield: 84.37; m.p.: 135-137 °C; Rf: 0.59 (B:EA- 7:3); FTIR (KBr): νmax (cm−1): 3294.47 (N–H str.), 3102.55 (C–H str., Ar), 1618.98 (C=O), 1569.12 (N–H bend), 1549.18 (asym. NO2 str.), 1398.42 (asym. SO2 str.), 1353.37 (sym. NO2 str.), 1133.20 (sym. SO2 str.), 732.87 (C–Cl);1HNMR (300 MHz, DMSO-d6), δ ppm: 4.08 (s, 1H, NH), 7.05 (s, 1H, CH of C6 of ClNO2C6H2CONH–), 7.34–7.49 (t, 1H, CH of C5 of –CONH–C6H5NO2–), 7.91–7.93 (d, 2H, CH of C2 and C6 of ClC6H4NH), 8.00–8.02 (d, 1H, CH of C6 of –CONH–C6H5NO2–), 8.24–8.26 (d, 2H, CH of C3 and C5 of ClC6H4NH), 8.36–8.39 (d, 1H, CH of C4 of –CONH–C6H5NO2–), 8.64 (s, 1H, CH of C3 of ClNO2C6H2CONH–), 8.95 (s, 1H, CH of C2of –CONH–C6H5NO2–), 10.81 (s, 1H, NH);13CNMR (300 MHz, DMSO-d6), δ ppm: 165.98 (C=O), 161.31 (C–S), 149.25 (C–NO2), 139.61 (C–NH), 132.26 (C–Cl), 130.24, 125.93, 124.56, 123.59, 118.85, 113.87.
2-Chloro-5-[(4-chlorophenyl)sulfamoyl]-4-nitro-N-propylbenzamide (5s)
% Yield: 57.89; m.p.: 116–118 °C; Rf: 0.66 (Chloroform); FTIR (KBr): νmax (cm−1): 3524.01 (N–H str.), 3089.05 (C–H str., Ar), 2967.82, 2874.42 (C–H str., Aliphatic), 1650.13 (C=O), 1598.78 (N–H bend), 1533.43 (asym. NO2 str.), 1372.38 (sym. SO2 str.), 1309.34 (sym. NO2 str.), 1168.88 (sym. SO2 str.), 755.90 (C–Cl). 1HNMR (300 MHz, DMSO-d6), δ ppm: 1.82–1.90 (m, 7H, –CONH–C3H7), 3.39 (s, 1H, NH), 7.50 (s, 1H, CH of C6 of ClNO2C6H2CONH–), 7.80–7.83 (d, 2H, CH of C2 and C6 of ClC6H4NH), 8.21–8.23 (d, 2H, CH of C3 and C5 of ClC6H4NH), 8.51 (s, 1H, CH of C3 of ClNO2C6H2CONH–), 10.32 (s, 1H, NH); 13CNMR (300 MHz, DMSO-d6), δ ppm: 162.67 (C=O), 158.75 (C–S), 146.08 (C–NO2), 139.51 (C–NH), 135.49 (C–Cl), 132.72, 128.73, 128.06, 126.60, 31.15, 26.04, 21.41.
N-Butyl-2-chloro-5-[(4-chlorophenyl)sulfamoyl]-4-nitrobenzamide (5t)
% Yield: 84.54; m.p.: 111–113 °C; Rf: 0.54 (Chloroform); FTIR (KBr): νmax (cm−1): 3446.85 (N–H str.), 3186.46 (C–H str., Ar), 2959.53, 2871.74 (C–H str., Aliphatic), 1658.64 (C=O), 1597.82 (N–H bend), 1531.51 (asym. NO2 str.), 1372.38 (asym. SO2 str.), 1309.80 (sym. NO2 str.), 1174.67 (sym. SO2 str.), 750.37 (C–Cl);1HNMR (300 MHz, DMSO-d6), δ ppm: 2.08–2.18 (m, 9H, –CONH–C4H9), 3.71 (s, 1H, NH), 7.46 (s, 1H, CH of C6 of ClNO2C6H2CONH–), 7.79–7.80 (d, 2H, CH of C2 and C6 of ClC6H4NH), 8.22–8.24 (d, 2H, CH of C3 and C5 of NO2C6H4NH), 8.50 (s, 1H, CH of C3 of ClNO2C6H2CONH–), 10.51 (s, 1H, NH);13CNMR (300 MHz, DMSO-d6), δ ppm: 160.69 (C=O), 155.52 (C–S), 144.08 (C–NO2), 138.84 (C–NH), 136.60 (C–Cl), 133.53, 129.57, 128.48, 127.57, 32.15, 27.34, 18.99.
2-Chloro-5-[(4-chlorophenyl)sulfamoyl]-N-[(furan-2-yl)methyl]-4-nitrobenzamide (5u)
% Yield: 67.48; m.p.: 191–193 °C; Rf: 0.40 (B:EA– 7:3); FTIR (KBr): νmax (cm−1): 3503.75 (N–H str.), 3056.03 (C–H str., Ar), 2981.34 (C–H str., Aliphatic), 1665.16 (C=O), 1596.15 (N–H bend), 1506.43 (asym. NO2 str.), 1396.80 (asym. SO2 str.), 1376.23 (sym. NO2 str.), 1149.59 (sym. SO2 str.), 743.60 (C–Cl);1HNMR (300 MHz, DMSO-d6), δ: 2.59 (s, 2H, CH of CONH–CH2–C4H3O), 3.71 (s, 1H, NH), 7.61 (s, 1H, CH of C6 of ClNO2C6H2CONH–), 7.79–7.87 (m, 3H, CH of C2, C3 and C4 of CONH–CH2–C4H3O), 7.92–7.93 (d, 2H, CH of C2 and C6 of ClC6H4NH), 8.22–8.24 (d, 2H, CH of C3 and C5 of ClC6H4NH), 8.50 (s, 1H, CH of C3 of ClNO2C6H2CONH–), 10.58 (s, 1H, NH); 13CNMR (300 MHz, DMSO–d6), δ ppm: 165.97 (C=O), 157.03 (C–S), 145.82 (C–NO2), 142.54 (C–NH), 136.72 (C–Cl), 130.46, 129.18, 125.49, 121.85, 120.80, 113.64, 32.59.
2-Chloro-5-[(4-chlorophenyl)sulfamoyl]-4-nitro-N-(pyridin-4-yl)benzamide (5v)
% Yield: 71.32; m.p.: 197-199 °C; Rf: 0.31 (B:EA- 7:3); FTIR (KBr): νmax (cm−1): 3503.75 (N–H str.), 3113.16 (C–H str., Ar), 1665.56 (C=O), 1598.05 (N–H bend), 1548.50 (asym. NO2 str.), 1371.07 (asym. SO2 str.), 1316.44 (sym. NO2 str.), 1170.81 (sym. SO2 str.), 755.14 (C–Cl);1HNMR (300 MHz, DMSO-d6), δ ppm: 3.71 (s, 1H, NH), 7.57 (s, 1H, CH of C6 of ClNO2C6H2CONH-), 7.80–7.83 (d, 2H, CH of C2 and C6 of ClC6H4NH–), 7.93–7.96 (d, 2H, CH of C2 and C6 of –CONH–C5H4NH–), 8.24–8.26 (d, 2H, CH of C3 and C5 of ClC6H4NH–), 8.36–8.39 (d, 2H, C3 and C5 CH of –CONH–C5H4NH), 8.50 (s, 1H, CH of C3 of ClNO2C6H2CONH–), 10.56 (s, 1H, NH); 13CNMR (300 MHz, DMSO–d6), δ ppm: 164.70 (C=O), 156.22 (C–S), 148.17 (C–NO2), 144.89 (C–NH), 138.96 (C–Cl), 130.34, 128.50, 125.55, 120.17.
In vitro antidiabetic studies
α-Glucosidase inhibitory assay
The method adopted for performing α-glucosidase inhibitory assay was similar to our prevenient study, Thakral and Singh [32]. Graph Pad Prism program, version 5 was employed for calculation of the 50% inhibitory concentration (IC50) of all compound [32, 42, 43].
α-Amylase inhibitory assay
Xiao et al., and Yoshikawa et al., illustrated a method, with little modification this method has been adopted for measuring the activity [32, 44].
Homology modeling
The 3D model for α-glucosidase is developed by comparative homology modeling technique using SWISS-MODEL web server (https://swissmodel.expasy.org/) [45] and then the quality of modeled structure was validated by Ramachandran plot (RAMPAGE) (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php). The details are available in our previous report [32].
Molecular docking
Ligand molecules were prepared as per reported method [32] using MarvinSketch and AutoDock tools. The crystal structures of α-amylase, 1qho [32, 46] from Bacillus sterothermophilus, maltose/acarbose complex downloaded from the protein data bank (http://www.rcsb.org) and α-glucosidase modeled structure [32] was used for docking in antidiabetic evaluation. Docking studies were carried out as reported in our previous study and literature using AutoDock Vina program [32, 47].
Molecular dynamic simulations
The respective structures placed in the center of the cubic box, the remaining volume of the box was filled by SPCe [48] water molecules. The whole box is then neutralized by adding the respective number of positive and negative ions using GROMACS 5.4 [49] by replacing the equal number of water molecules. Further energy minimization followed by 10 ns equilibration performed by using OPLS [50] force fields integrated into GROMACS 5.4 package to represent the potential energy of the system.
Computation of drug like parameters and ADMET profiling
Molinspiration (http://www.molinspiration.com/) online tool kit and OSIRIS property explorer was used for computing drug like characteristics from 2D chemical structures of aforementioned compounds [51–54]. Pre-ADMET online server (https://preadmet.bmdrc.kr/) was used for calculating pharmacokinetic parameters like adsorption, distribution, metabolism and excretion and some of the computed properties are human intestinal absorption (HIA %), Caco-2 cell permeability (nm/s), MDCK (Medin-Darbey Canine Kidney Epithelial Cells) cell permeability (nm/s), plasma protein binding (%)¸ blood brain barrier penetration (C. brain/C. blood) and Pgp inhibition [55]. Bioactivity of synthesized compounds was predicted by Molinspiration (http://www.molinspiration.com/) online tool kit [56] and toxicity parameters like mutagenicity, tumorigenicity irritating effects and reproductive effects were computed by OSIRIS property explorer [57].
Supplementary information
Additional file 1: Table S1. Topological polar surface area, aqueous solubility, number of rotatable bonds, and calculated Lipinski’s rule of five for the synthesized 2-chloro-5-[(4-chlorophenyl)sulfamoyl]-N-(alkyl/aryl)-4-nitrobenzamide derivatives; Table S2. ADME property values of synthesized 2-chloro-5-[(4-chlorophenyl) sulfamoyl]-N-(alkyl/aryl)-4-nitrobenzamide derivatives using Pre-ADMET online server; Table S3. Bioactivity and toxicity risk of synthesized 2-chloro-5-[(4-chlorophenyl)sulfa-moyl]-N-(alkyl/aryl)-4-nitrobenzamide derivatives
Acknowledgements
The authors are thankful to Chairman, Department of Pharmaceutical Sciences, G. J. U. S. and T., Hisar for providing necessary facilities to carry out this research work and Prof. Neeraj Dilbaghi, Dept of Bio &Nano Technology, G. J. U. S. and T., Hisar for providing lab facility for in vitro studies. The authors are also thankful to Amit Singh, Discipline of Chemistry, Indian Institute of Technology, Gandhinagar for facilitation in computational studies.
Authors’ contributions
The authors (ST, RN, MK and VS) have done synthetic work, in vitro and in silico evaluation. All authors have read and approved the manuscript.
Funding
The author (VS) gratefully acknowledges the financial support (CIL/2017/356) as minor project for purchase of chemicals and Junior Research Fellow (JRF) award to Ms. Samridhi Thakral by Dr. A. P. J. Abdul Kalam Central Instrumentation laboratory, G. J. U. S. and T., Hisar under DST-PURSE program.
Availability of data and materials
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13065-020-00703-4.
References
- 1.Kar K, Krithika U, Mithuna BP, Kumar SS, Reji A, Kumar BRP. Design, synthesis and glucose uptake activity of some novel glitazones. Bioorg Chem. 2014;56:27–33. doi: 10.1016/j.bioorg.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Li K, Yao F, Xue Q, Fan H, Yang L, Li X, Sun L, Liu Y. Inhibitory effects against α-glucosidase and α-amylase of the flavonoids-rich extract from Scutellaria baicalensis shoots and interpretation of structure–activity relationship of its eight flavonoids by a refined assign-score method. Chem Cent J. 2015;12(1):82. doi: 10.1186/s13065-018-0445-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chinthala Y, Thakur S, Tirunagari S, Chinde S, Kumar A, Domatti AK, Arigari NK, Srinivas KVNS, Alam S, Jonalla K, Khan F, Tiwari A, Grover P. Synthesis, docking and ADMET studies of novel chalcone triazoles for anti-cancer and anti-diabetic activity. Eur J Med Chem. 2015;93:564–573. doi: 10.1016/j.ejmech.2015.02.027. [DOI] [PubMed] [Google Scholar]
- 4.Kim KY, Nguyen TH, Kurihara H, Kim SM. α-Glucosidase inhibitory activity of bromophenol purified from the red alga Polyopes lancifolia. J Food Sci. 2010;75(5):145–150. doi: 10.1111/j.1750-3841.2010.01629.x. [DOI] [PubMed] [Google Scholar]
- 5.Wang G, Li X, Wang J, Xie Z, Li L, Chen M, Chen S, Peng Y. Synthesis, molecular docking and α-glucosidase inhibition of 2-((5, 6-diphenyl-1, 2, 4-triazin-3-yl) thio)-N-arylacetamides. Bioorg Med Chem Lett. 2017;27(5):1115–1118. doi: 10.1016/j.bmcl.2017.01.094. [DOI] [PubMed] [Google Scholar]
- 6.Lopéz D, Cherigo L, Mejia LC, Loza-Mejía MA, Martínez-Luis S. α-Glucosidase inhibitors from a mangrove associated fungus, Zasmidium sp. strain EM5-10. BMC Chem. 2019;13(1):22. doi: 10.1186/s13065-019-0540-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abuelizz HA, Iwana NA, Ahmad R, Anouar EH, Marzouk M, Al-Salahi R. Synthesis, biological activity and molecular docking of new tricyclic series as α-glucosidase inhibitors. BMC Chem. 2019;13(1):52. doi: 10.1186/s13065-019-0560-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barakat A, Ali M, Al-Majid AM, Yousuf S, Choudhary MI, Khalil R, Ul-Haq Z. Synthesis of thiobarbituric acid derivatives: in vitro α-glucosidase inhibition and molecular docking studies. Bioorg Chem. 2017;75:99–105. doi: 10.1016/j.bioorg.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Gollapalli M, Taha M, Javid MT, Almandil NB, Rahim F, Wadood A, Mosaddik A, Ibrahim M, Alqahtani MA, Bamarouf YA. Synthesis of benzothiazole derivatives as a potent α-glucosidase inhibitor. Bioorg Chem. 2019;85:33–48. doi: 10.1016/j.bioorg.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 10.Imran S, Taha M, Ismail NH, Kashif SM, Rahim F, Jamil W, Hariono M, Yusuf M, Wahab H. Synthesis of novel flavone hydrazones: in vitro evaluation of α-glucosidase inhibition, QSAR analysis and docking studies. Eur J Med Chem. 2015;105:156–170. doi: 10.1016/j.ejmech.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 11.El-Karim SSA, Anwar MM, Syam YM, Nael MA, Ali HF, Motaleb MA. Rational design and synthesis of new tetralin-sulfonamide derivatives as potent anti-diabetics and DPP-4 inhibitors: 2D & 3D QSAR, in vivo radiolabeling and bio distribution studies. Bioorg Chem. 2018;81:481–493. doi: 10.1016/j.bioorg.2018.09.021. [DOI] [PubMed] [Google Scholar]
- 12.Naaz F, Srivastava R, Singh A, Singh N, Verma R, Singh VK, Singh RK. Molecular modeling, synthesis, antibacterial and cytotoxicity evaluation of sulfonamide derivatives of benzimidazole, indazole, benzothiazole and thiazole. Bioorgan Med Chem. 2018;26(12):3414–3428. doi: 10.1016/j.bmc.2018.05.015. [DOI] [PubMed] [Google Scholar]
- 13.Krishnaiah M, de Almeida NR, Udumula V, Song Z, Chhonker YS, Abdelmoaty MM, do Nascimento VA, Murry DJ, Conda-Sheridan M. Synthesis, biological evaluation, and metabolic stability of phenazine derivatives as antibacterial agents. Eur J Med Chem. 2018;143:936–947. doi: 10.1016/j.ejmech.2017.11.026. [DOI] [PubMed] [Google Scholar]
- 14.Ahmed A, Channar PA, Saeed A, Kalesse M, Kazi MA, Larik FA, Abbas Q, Hassan M, Raza H, Seo SY. Synthesis of sulfonamide, amide and amine hybrid pharmacophore, an entry of new class of carbonic anhydrase II inhibitors and evaluation of chemo-informatics and binding analysis. Bioorg Chem. 2019;86:624–630. doi: 10.1016/j.bioorg.2019.01.060. [DOI] [PubMed] [Google Scholar]
- 15.Navarrete-Vázquez G, Morales-Vilchis MG, Estrada-Soto S, Ramírez-Espinosa JJ, Hidalgo-Figueroa S, Nava-Zuazo C, Tlahuext H, Leon-Rivera I, Medina-Franco JL, López-Vallejo F, Webster SP. Synthesis of 2-{2-[(α/β-naphthalen-1-ylsulfonyl) amino]-1, 3-thiazol-4-yl} acetamides with 11β-hydroxysteroid dehydrogenase inhibition and in combo antidiabetic activities. Eur J Med Chem. 2014;74:179–186. doi: 10.1016/j.ejmech.2013.12.042. [DOI] [PubMed] [Google Scholar]
- 16.Ghareb N, El-Sayed NM, Abdelhameed R, Yamada K, Elgawish MS. Toward a treatment of diabesity: rational design, synthesis and biological evaluation of benzene-sulfonamide derivatives as a new class of PTP-1B inhibitors. Bioorg Chem. 2019;86:322–338. doi: 10.1016/j.bioorg.2019.01.052. [DOI] [PubMed] [Google Scholar]
- 17.Durgapal SD, Soman SS. Evaluation of novel coumarin-proline sulfonamide hybrids as anticancer and antidiabetic agents. Synth Commun. 2019 doi: 10.1080/00397911.2019.1647439. [DOI] [Google Scholar]
- 18.Singh V, Pacitto A, Donini S, Ferraris DM, Boros S, Illyés E, Szokol B, Rizzi M, Blundell TL, Ascher DB, Pato J. Synthesis and structure–activity relationship of 1-(5-isoquinolinesulfonyl) piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH. Eur J Med Chem. 2019;174:309–329. doi: 10.1016/j.ejmech.2019.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ugwu DI, Okoro UC, Ahmad H. New carboxamide derivatives bearing benzenesulphonamide as a selective COX-II inhibitor: design, synthesis and structure-activity relationship. PLoS ONE. 2017;12(9):e0183807. doi: 10.1371/journal.pone.0183807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banuppriya G, Sribalan R, Padmini V. Synthesis and characterization of curcumin-sulfonamide hybrids: biological evaluation and molecular docking studies. J Mol Struct. 2018;1155:90–100. [Google Scholar]
- 21.Ji Y, Chen X, Chen H, Zhang X, Fan Z, Xie L, Ma B, Zhu C. Designing of acyl sulphonamide based quinoxalinones as multifunctional aldose reductase inhibitors. Bioorg Med Chem. 2019;27(8):1658–1669. doi: 10.1016/j.bmc.2019.03.015. [DOI] [PubMed] [Google Scholar]
- 22.Abbas A, Murtaza S, Tahir MN, Shamim S, Sirajuddin M, Rana UA, Naseem K, Rafique H. Synthesis, antioxidant, enzyme inhibition and DNA binding studies of novel N-benzylated derivatives of sulfonamide. J Mol Struct. 2016;1117(5):269–275. [Google Scholar]
- 23.Gatadi S, Gour J, Shukla M, Kaul G, Dasgupta A, Madhavi YV, Chopra S, Nanduri S. Synthesis and evaluation of new 4-oxoquinazolin-3 (4H)-yl) benzoic acid and benzamide derivatives as potent antibacterial agents effective against multidrug resistant Staphylococcus aureus. Bioorg Chem. 2019;83:569–579. doi: 10.1016/j.bioorg.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Dev J, Poornachandra Y, Kumar N, Ravikumar N, Ranjithreddy P, Kumar S, Nanubolu JB, Kumar G, Narsaiah B. Synthesis of novel pyrazolo [3, 4-b] quinolinyl acetamide analogs, their evaluation for antimicrobial and anticancer activities, validation by molecular modeling and CoMFA analysis. EurJ Med Chem. 2017;130:223–239. doi: 10.1016/j.ejmech.2017.02.052. [DOI] [PubMed] [Google Scholar]
- 25.Caliendo G, Santagada V, Perissutti E, Severino B, Fiorino F, Warner TD, Wallace JL, Ifa DR, Antunes E, Cirino G, de Nucci G. Synthesis of substituted benzamides as anti-inflammatory agents that inhibit preferentially cyclooxygenase 1 but do not cause gastric damage. Eur J Med Chem. 2001;36(6):517–530. doi: 10.1016/s0223-5234(01)01251-x. [DOI] [PubMed] [Google Scholar]
- 26.Tian Y, Zhang T, Long L, Li Z, Wan S, Wang G, Yu Y, Hou J, Wu X, Zhang J. Design, synthesis, biological evaluation and molecular modeling of novel 2-amino-4-(1-phenylethoxy) pyridine derivatives as potential ROS1 inhibitors. Eur J Med Chem. 2018;143:182–199. doi: 10.1016/j.ejmech.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Guo J, Zhu M, Wu T, Hao C, Wang K, Yan Z, Huang W, Wang J, Zhao D, Cheng M. Discovery of indolin-2-one derivatives as potent PAK4 inhibitors: structure-activity relationship analysis, biological evaluation and molecular docking study. Bioorg Med Chem. 2017;25(13):3500–3511. doi: 10.1016/j.bmc.2017.04.047. [DOI] [PubMed] [Google Scholar]
- 28.Avalakki AS, Jadhav SB, Bandawane DD, Bhalekar PA. Synthesis and antidiabetic evaluation of some novel compounds. Indian J Chem. 2019;58:849–854. [Google Scholar]
- 29.Giacobbo BC, Pissinate K, Rodrigues-Junior V, Villela AD, Grams ES, Abbadi BL, Subtil FT, Sperotto N, Trindade RV, Back DF, Campos MM. New insights into the SAR and drug combination synergy of 2-(quinolin-4-yloxy) acetamides against Mycobacterium tuberculosis. Eur J Med Chem. 2017;126:491–501. doi: 10.1016/j.ejmech.2016.11.048. [DOI] [PubMed] [Google Scholar]
- 30.Foster JE, Nicholson JM, Butcher R, Stables JP, Edafiogho IO, Goodwin AM, Henson MC, Smith CA, Scott KR. Synthesis, characterization and anticonvulsant activity of enaminones Part 6: Synthesis of substituted vinylic benzamides as potential anticonvulsants. Bioorg Med Chem. 1999;7(11):2415–2425. doi: 10.1016/s0968-0896(99)00185-6. [DOI] [PubMed] [Google Scholar]
- 31.Carson JR, Coats SJ, Codd EE, Dax SL, Lee J, Martinez RP, Neilson LA, Pitis PM, Zhang SP. N, N-Dialkyl-4-[(8-azabicyclo [32 1]-oct-3-ylidene) phenylmethyl] benzamides, potent, selective δ opioid agonists. Bioorg Med Chem Lett. 2004;14(9):2109–2112. doi: 10.1016/j.bmcl.2004.02.051. [DOI] [PubMed] [Google Scholar]
- 32.Thakral S, Singh V. 2, 4-Dichloro-5-[(N-aryl/alkyl) sulfamoyl] benzoic acid derivatives: in vitro antidiabetic activity, molecular modeling and in silico ADMET screening. Med Chem. 2019;15(2):186–195. doi: 10.2174/1573406414666180924164327. [DOI] [PubMed] [Google Scholar]
- 33.Thakral S, Singh V. Synthesis, biological evaluation, QSAR, molecular docking and ADMET studies of N-aryl/N,N-dimethyl substituted sulphonamide derivatives. Anti-Infect Agents. 2019 doi: 10.2174/2211352517666190902130014. [DOI] [Google Scholar]
- 34.Singh R, Lather V, Pandita D, Judge V, Arumugam KN, Grewal AS. Synthesis, docking and antidiabetic activity of some newer benzamide derivatives as potential glucokinase activators. Lett Drug Des Discov. 2017;14(5):540–553. [Google Scholar]
- 35.Charaya N, Pandita D, Grewal AS, Lather V. Design, synthesis and biological evaluation of novel thiazol-2-yl benzamide derivatives as glucokinase activators. Comput Biol Chem. 2018;73:221–229. doi: 10.1016/j.compbiolchem.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 36.Grewal AS, Kharb R, Prasad DN, Dua JS, Lather V. N-pyridin-2-yl benzamide analogues as allosteric activators of glucokinase: design, synthesis, in vitro, in silico and in vivo evaluation. Chem Biol Drug Des. 2019;93(3):364–372. doi: 10.1111/cbdd.13423. [DOI] [PubMed] [Google Scholar]
- 37.Grewal AS, Sharma K, Singh S, Singh V, Pandita D, Lather V. Design, synthesis and antidiabetic activity of novel sulfamoyl benzamide derivatives as glucokinase activators. J Pharm Tech Res Manag. 2018;6(2):113–122. [Google Scholar]
- 38.Adegboye AA, Khan KM, Salar U, Aboaba SA, Chigurupati S, Fatima I, Taha M, Wadood A, Mohammad JI, Khan H, Perveen S. 2-Aryl benzimidazoles: synthesis, in vitro α-amylase inhibitory activity, and molecular docking study. Eur J Med Chem. 2018;150:248–260. doi: 10.1016/j.ejmech.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 39.Taha M, Irshad M, Imran S, Chigurupati S, Selvaraj M, Rahim F, Ismail NH, Nawaz F, Khan KM. Synthesis of piperazine sulfonamide analogs as diabetic-II inhibitors and their molecular docking study. Eur J Med Chem. 2017;141:530–537. doi: 10.1016/j.ejmech.2017.10.028. [DOI] [PubMed] [Google Scholar]
- 40.Kumar CA, Veeresh B, Ramesha KC, Raj CA, Mahadevaiah KM, Prasad SB, Naveen S, Madaiah M, Rangappa KS. Antidiabetic studies of 1-benzhydryl-piperazine sulfonamide and carboxamide derivatives. J Applicable Chem. 2017;6(2):232–240. [Google Scholar]
- 41.Lad NP, Manohar Y, Mascarenhas M, Pandit YB, Kulkarni MR, Sharma R, Salkar K, Suthar A, Pandit SS. Methylsulfonyl benzothiazoles (MSBT) derivatives: search for new potential antimicrobial and anticancer agents. Bioorg Med Chem Lett. 2017;27(5):1319–1324. doi: 10.1016/j.bmcl.2016.08.032. [DOI] [PubMed] [Google Scholar]
- 42.Kim KY, Nguyen TH, Kurihara H, Kim SM. α-Glucosidase inhibitory activity of bromophenol purified from the red alga Polyopes lancifolia. J Food Sci. 2010;75:145–150. doi: 10.1111/j.1750-3841.2010.01629.x. [DOI] [PubMed] [Google Scholar]
- 43.Nguyen TH, Kim SM. α-Glucosidase inhibitory activities of fatty acids purified from the internal organ of sea cucumber Stichopus japonicas. J Food Sci. 2015;80:841–847. doi: 10.1111/1750-3841.12810. [DOI] [PubMed] [Google Scholar]
- 44.Rani N, Sharma SK, Vasudeva N. Assessment of antiobesity potential of Achyranthes aspera Linn. seed. Evid Based Complement Alternat Med. 2012 doi: 10.1155/2012/715912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T. SWISS-MODEL: modeling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dauter Z, Dauter M, Brzozowski AM, Christensen S, Borchert TV, Beier L, Wilson KS, Davies GJ. X-ray structure of novamyl, the five-domain “maltogenic” α-amylase from Bacillus stearothermophilus: maltose and acarbose complexes at 1.7 Å resolution. Biochemistry. 1999;38(26):8385–8392. doi: 10.1021/bi990256l. [DOI] [PubMed] [Google Scholar]
- 47.Trott O, Olson JA. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berendsen HJC, Grigera JR, Straatsma TP. The missing term in effective pair potentials. J Phys Chem. 1987;91(24):6269–6271. [Google Scholar]
- 49.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 50.Jorgensen WL, Tirado-Rives J. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J Am Chem Soc. 1988;110(6):1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 51.El-Gohary NS, Shaaban MI. Antimicrobial and antiquorum-sensing studies. Part 3: synthesis and biological evaluation of new series of [1, 3, 4] thiadiazoles and fused [1, 3, 4] thiadiazoles. Arch Pharm. 2015;348:283–297. doi: 10.1002/ardp.201400381. [DOI] [PubMed] [Google Scholar]
- 52.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 53.Cardoso MF, Rodrigues PC, Oliveira MEI, Gama IL, da Silva IM, Santos IO, Rocha DR, Pinho RT, Ferreira VF, de Souza MCB, da Silva FDC. Synthesis and evaluation of the cytotoxic activity of 1, 2-furanonaphthoquinones tethered to 1, 2, 3-1H-triazoles in myeloid and lymphoid leukemia cell lines. Eur J Med Chem. 2014;84:708–717. doi: 10.1016/j.ejmech.2014.07.079. [DOI] [PubMed] [Google Scholar]
- 54.Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 55.Murugavel S, Kannan D, Bakthadoss M. Experimental and computational approaches of a novel methyl (2E)-2-{[N-(2-formylphenyl)(4-methylbenzene) sulfonamido] methyl}-3-(4-chlorophenyl) prop-2-enoate: a potential antimicrobial agent and an inhibition of penicillin-binding protein. J Mol Struct. 2016;1115:33–54. [Google Scholar]
- 56.Balam SK, Krishnammagari SK, Harinath JS, Sthanikam SP, Chereddy SS, Pasupuleti VR, Yellapu NK, Peddiahgari VGR, Cirandur SR. Synthesis of N-(3-picolyl)-based 1, 3, 2λ5-benzoxazaphosphinamides as potential 11β-HSD1 enzyme inhibitors. Med Chem Res. 2015;24:1119–1135. [Google Scholar]
- 57.de Oliveira KN, Souza MM, Sathler PC, Magalhaes UO, Rodrigues CR, Castro HC, Palm PR, Sarda M, Perotto PE, Cezar S, de Brito MA. Sulphonamide and sulphonyl-hydrazone cyclic imide derivatives: antinociceptive activity, molecular modeling and in silico ADMET screening. Arch Pharm Res. 2012;35:1713–1722. doi: 10.1007/s12272-012-1002-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Topological polar surface area, aqueous solubility, number of rotatable bonds, and calculated Lipinski’s rule of five for the synthesized 2-chloro-5-[(4-chlorophenyl)sulfamoyl]-N-(alkyl/aryl)-4-nitrobenzamide derivatives; Table S2. ADME property values of synthesized 2-chloro-5-[(4-chlorophenyl) sulfamoyl]-N-(alkyl/aryl)-4-nitrobenzamide derivatives using Pre-ADMET online server; Table S3. Bioactivity and toxicity risk of synthesized 2-chloro-5-[(4-chlorophenyl)sulfa-moyl]-N-(alkyl/aryl)-4-nitrobenzamide derivatives
Data Availability Statement
Not applicable.