Abstract
Most KRAS G12C mutant non-small cell lung cancer (NSCLC) patients experience clinical benefit from selective KRAS G12C inhibition, while patients with colorectal cancer (CRC) bearing the same mutation rarely respond. To investigate the cause of the limited efficacy of KRAS G12C inhibitors in CRC, we examined the effects of AMG510 in KRAS G12C CRC cell lines. Unlike NSCLC cell lines, KRAS G12C CRC models have high basal receptor tyrosine kinase (RTK) activation and are responsive to growth factor stimulation. In CRC lines, KRAS G12C inhibition induces higher phospho-ERK rebound than in NSCLC cells. Although upstream activation of several RTKs interferes with KRAS G12C blockade, we identify EGFR signaling as the dominant mechanism of CRC resistance to KRAS G12C inhibitors. The combinatorial targeting of EGFR and KRAS G12C is highly effective in CRC cells, patient-derived organoids and xenografts, suggesting a novel therapeutic strategy to treat KRAS G12C CRC patients.
Introduction
The small GTPase KRAS is mutated and constitutively active in 15% of all human tumors(1). This protein acts as a molecular switch, transitioning from an active (GTP-bound) to an inactive state (GDP-bound). This transition is highly regulated and physiologic feedback loops limit the duration of KRAS activation. Guanine exchange factors (GEFs), such as SOS1, and GTPase activating proteins (GAPs), like NF1, directly mediate KRAS activation(2, 3). When mutated, KRAS loses the ability to interact with GAPs and therefore becomes constitutively activated, driving downstream MAPK and PI3K-AKT signaling pathways and enhancing cancer cell proliferation and survival(4). Different mutations in KRAS may modify the intrinsic activation (GTP-bound state) of the mutant protein and its interaction with downstream effectors, such as RAF proteins(5, 6). Moreover, KRAS signaling is also regulated by negative feedback mechanisms through the transcriptional induction of protein phosphatases (DUSPs and Sproutys) and SPREDs(2). The complexity of the signaling network and the heterogeneous features of the multiple KRAS mutant alleles have contributed to the difficulty in developing molecularly targeted therapies against mutant KRAS, a target considered undruggable for decades.
However, in recent years, the unique characteristics of one of the KRAS mutant alleles, KRAS G12C, have been exploited for the design of covalent inhibitors that can specifically bind the cysteine on the mutant residue(7). These inhibitors can bind G12C mutant KRAS beneath the effector binding switch-II domain when KRAS is in the GDP-bound (inactive) state trapping KRAS in its inactive form and taking advantage of the retained, intrinsic cycling of the G12C mutant from GTP-bound to GDP-bound states(7, 8). Given this particular feature of the KRAS G12C mutant protein, the efficacy of these compounds can be increased by concomitant receptor tyrosine kinase (RTK) inhibition that further maintain KRAS G12C in a GDP-bound conformation, facilitating the binding of the covalent inhibitor(7–9).
Besides RTK inhibition, several other combinatorial strategies have already been proposed, with the goal of potentiating the effects of these inhibitors(10–13). Importantly, the unique feature of KRAS G12C covalent inhibitors, which only bind the mutant allele, renders these drugs a potentially ideal partner for combinatorial strategies, as toxicities should be minimized and limited to the combinatorial partner.
Several clinical trials testing the efficacy of KRAS G12C targeted drugs are currently ongoing and early results from the first two trials using AMG510 and MRTX849 have documented promising outcomes in the clinic. However, clinical studies unexpectedly reported that the response rate to these drugs is high in non-small cell lung cancers (NSCLC) patients but limited in colorectal cancer (CRC) patients(14, 15).
In this work, we sought to understand the mechanisms underlying this lineage difference. We hypothesized that NSCLC bearing KRAS G12C could be intrinsically different from KRAS G12C mutant CRC, despite harboring the same oncogenic mutation. We and others previously reported that BRAF V600E CRC have limited sensitivity to single agent RAF inhibitors, and this is a result of an adaptive response selectively triggered in CRC(16–18).
Therefore, we examined the adaptive response to KRAS G12C inhibition and its kinetics in KRAS G12C mutant NSCLC and CRC cell models. We show that KRAS G12C mutant CRCs maintain sensitivity to upstream RTK signaling, particularly EGFR. Using biochemical and pharmacological approaches, we show that EGFR reactivation restricts the efficacy of KRAS G12C inhibition in CRC. We find that KRAS G12C and EGFR blockade is effective to overcome this adaptive resistance, both in cell lines and in patient-derived models. These findings have immediate relevance for clinical strategies to overcome unresponsiveness to KRAS G12C blockade in CRC patients.
Results
ERK inhibition is not sustained in CRC cell lines treated with AMG510
To understand the mechanistic basis for differential clinical responses to treatment with the selective KRAS G12C inhibitor AMG510 in CRC and NSCLC, we analyzed the effect of this drug in a panel of KRAS G12C mutant cell lines. These included two NSCLC (NCIH358 and LU65) and seven CRC cell lines (C106, RW7213, SW837, SNU1411, JVE015 and LIM2099). Response to AMG510 was measured in a dose-response proliferation assay. Sensitivity to single agent was comparable among the models, showing no clear difference between CRC and NSCLC cell lines (Figure 1A). Therefore, to explore the quality, duration, and adaptive signaling response to drug treatment, we compared the NSCLC cell lines to four CRC models, consisting of two representative models with low IC50 to AMG510 (C106, RW7213) and two representatives with high IC50 values (SW837 and SNU1411). Cells were subjected to increasing concentrations of AMG510 for 1 hour and 24 hours (Figure 1B). After one hour of drug treatment, KRAS G12C inhibition led to downregulation of phosphorylated ERK in a dose-dependent manner in all cell models, regardless of tumor lineage. However, after 24 hours of drug exposure, we observed that both NSCLC cell lines further downregulated ERK phosphorylation, while all the CRC cell lines showed a rebound in phospho-ERK levels (Figure 1C–D). We then examined the impact of AMG510 on the signaling network over time in NSCLC and CRC cell lines. At baseline, the CRC cell line C106 expressed lower levels of phosphorylated MEK and ERK with similar total protein relative to the NSCLC line NCIH358. Treatment with AMG510 decreased levels of active RAS-GTP, but the suppression was more pronounced in the NCIH358 compared to C106. In C106, residual phospho-MEK and ERK were measurable already after one hour of treatment while the NSCLC cell line (NCIH358) showed signs of rebound in phospho-MEK and phospho-ERK only at 72 hours after treatment (Supplementary Figure 1A–B–C). Altogether, these data show that MAPK pathway inhibition upon AMG510 treatment is transient and less profound in CRC than lung models and suggest that this may be due to greater feedback suppression of MAPK signaling in CRC that is then released with KRAS G12C inhibition.
Figure 1: Response to KRAS G12C inhibition in CRC cell lines.
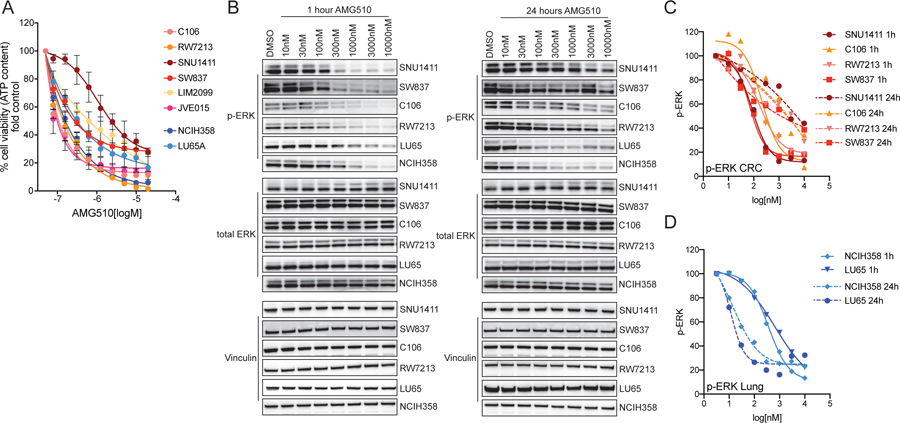
(A) Short term proliferation assay of CRC cell lines (shades of red) and NSCLC cell lines (shades of blue). Cells were treated for 120 hours with increasing concentration of AMG510 and then ATP content was measured using CellTiterGlo. Data represents the average and standard deviation of 3 biological replicates. (B) Western blot analysis of ERK activation upon AMG510 dose-response treatment after 1 hour and 24 hours treatment. Vinculin is used as loading control. (C) Densitometry analysis of CRC cell lines in western blot in (B). p-ERK values are normalized on both total ERK and Vinculin and on DMSO control. (D) Densitometry analysis of NSCLC cell lines western blot in (B). p-ERK values are normalized on both total ERK and Vinculin and on DMSO control.
Colorectal cancer cells bearing the KRAS G12C mutation display high basal RTK activation
KRAS mutation acts downstream of RTK signaling and leads to constitutive ERK activation, in turn, high ERK levels feedback inhibit receptor signaling(19). KRAS mutant proteins are thought to be constitutively active, therefore it would be expected that KRAS mutant tumors do not rely on RTK activation. To characterize the basal level of RTKs in patient tumors we assembled a series of CRC and NSCLC clinical samples carrying the KRAS G12C mutation. We analyzed eight CRC and five NSCLC patient specimens (Supplementary table 1), all collected before any targeted therapy treatment, and evaluated the expression of 72 different proteins, including RTKs, using an SRM-mass spectrometry assay (see methods). Detectable levels of EGFR, HER2, and MET were present in both lineages without clear differences, while HER3 expression was more prevalent in CRC patient samples (Figure 2A). We did not detect ALK, ROS1, RET, FGFR, or AXL expression in any of the samples. Two CRC patients and two NSCLC patients later went on the phase 1 trial of AMG510 (Supplementary table 1). Despite similar basal total RTK levels in the tumors from trial enrolled patients, both NSCLC cases achieved clinical benefit, while both CRC patients progressed upon AMG510 treatment. These data indicate that even in the presence of KRAS G12C mutation, tumors often express upstream RTKs, predominantly consisting of HER family and MET receptors. However, the levels of total RTK expression do not appear to underlie lineage specific differential response to KRAS G12C inhibition between CRC and NSCLC patients.
Figure 2: RTK component and responsiveness in CRC cell lines.
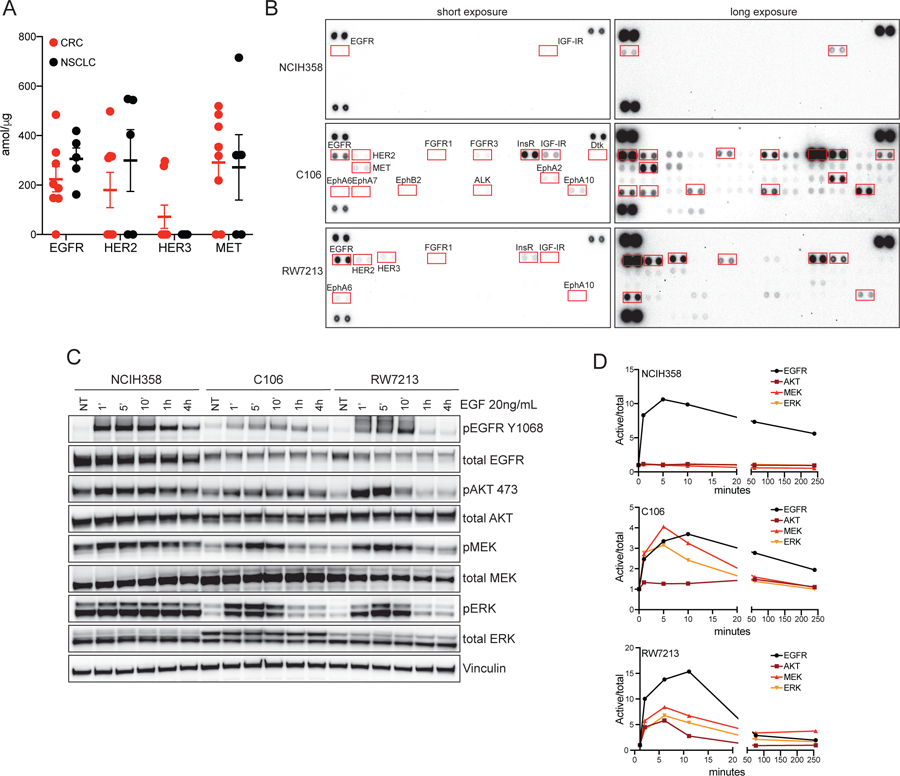
(A) Mass spectrometry analysis of RTK in CRC and NSCLC patients-derived biopsies. (B) Phospho-RTK array of NCIH358 (NSCLC), C106 and RW7213 (CRC) cell lines. Cell were FBS starved for 24h and stimulated with 20% FBS for ten minutes before harvesting. (C) Western blot analysis of KRAS downstream effectors in NCIH358 (NSCLC), C106 and RW7213 (CRC) cell lines upon time course EGF treatment. Vinculin is used as loading control. (D) Densitometry analysis of p-AKT, p-MEK and p-ERK of western blot in (C). Phospho-proteins values are normalized on total proteins and Vinculin and on not treated (NT) control.
While we were able to detect total RTK expression in the clinical samples, we could not assess activated RTK levels or evaluate the functional consequences of detectable RTK expression in these tumors. To test this, we measured the levels of active RTKs in CRC and NSCLC cell lines using phospho-RTK arrays. The NSCLC cell line NCIH358 had no detectable basal RTK activation and low levels of the p-EGFR and p-IGF-IR could be detected only with signal enhancement through long exposure. In contrast, both CRC cell lines, C106 and RW7213, showed a number of detectable phosphorylated RTKs (Figure 2B). Marked phospho-EGFR signal was present in both CRC models and was accompanied by detectable levels of other phosphorylated receptors. Co-immunoprecipitation assays using cetuximab as a capture antibody confirmed that EGFR is bound to the other HER receptors MET and FGFR suggesting hetero-dimerization processes (Supplementary Figure 2A–B). Several Ephrin receptors were also found activated in CRC cell lines. Interestingly EphR activation has been reported as mechanism of resistance to cetuximab in CRC and head and neck squamous cell carcinoma and CRC, suggesting that the cross talk between these two classes of RTKs can potentially amplify EGFR signaling(20).
We next investigated the functional effect of the high levels of basal RTK activation in the CRC cell lines by evaluating the signaling consequences of EGF stimulation over time. At baseline, signaling downstream of RAS (phospho-AKT, phospho-MEK, phospho-ERK) was lower in the colon cancer cell lines compared to the NSCLC line NCIH358. EGF treatment led to induction of phospho-EGFR in all cell models (Figure 2C). However, activation of downstream effectors showed lineage-specific differences. The NSCLC cell line NCIH358 did not show any modulation of AKT, MEK or ERK phosphorylation upon EGF treatment, suggesting that KRAS G12C does not get (further) activated by EGFR stimulation and can sustain downstream signaling without upstream inputs (Figure 2C–D). In contrast, in the CRC cell lines C106 and RW7213, downstream effectors followed the same kinetics of EGFR activation, showing that KRAS G12C can be activated by upstream signaling components and that basal activation of RAS downstream effectors may be lower due to negative feedback loops (Figure 2C–D). Altogether, these data indicate that even in the presence of the same KRAS activating mutation, lineage-specific differences exist in the degree of RTK activation, contribution of upstream receptors to ERK activation (and thus rate of cycling of KRAS between the GTP- and GDP-bound forms), and feedback mechanisms that would be released from suppression by selective KRAS inhibitors-mediated ERK inhibition.
Cetuximab sensitizes KRAS G12C CRC cell lines to AMG510 and concomitant EGFR and KRAS G12C blockade reverts secondary resistance to anti EGFR antibodies
Based on the high levels of basally active EGFR, the contribution of EGFR to KRAS G12C signaling outputs, and the known relevance of EGFR signaling in CRC, we next evaluated whether KRAS G12C-EGFR combined inhibition could effectively suppress signaling and growth in the CRC models. Biochemical analysis and densitometric quantitation of phosphoprotein levels showed that concomitant KRAS G12C and EGFR inhibition, with the anti-EGFR antibody cetuximab, led to sustained inhibition of phospho-MEK and phospho-ERK, as well as the ERK downstream targets DUSP6 and CyclinD1, indicating inhibition of ERK output (Figure 3A and Supplementary Figure 3A–B). CRC cell lines were also tested in short-term proliferation assays. ATP content measurement showed that concurrent treatment with the EGFR inhibitor cetuximab synergized with AMG510 in all the cell lines tested (Figure 3B and Supplementary Figure 4A). Growth suppression was durable as similar effects were also seen in long-term proliferation assays stained with crystal violet (Figure 3C).
Figure 3: EGFR inhibition sensitizes CRC cell lines to AMG510.
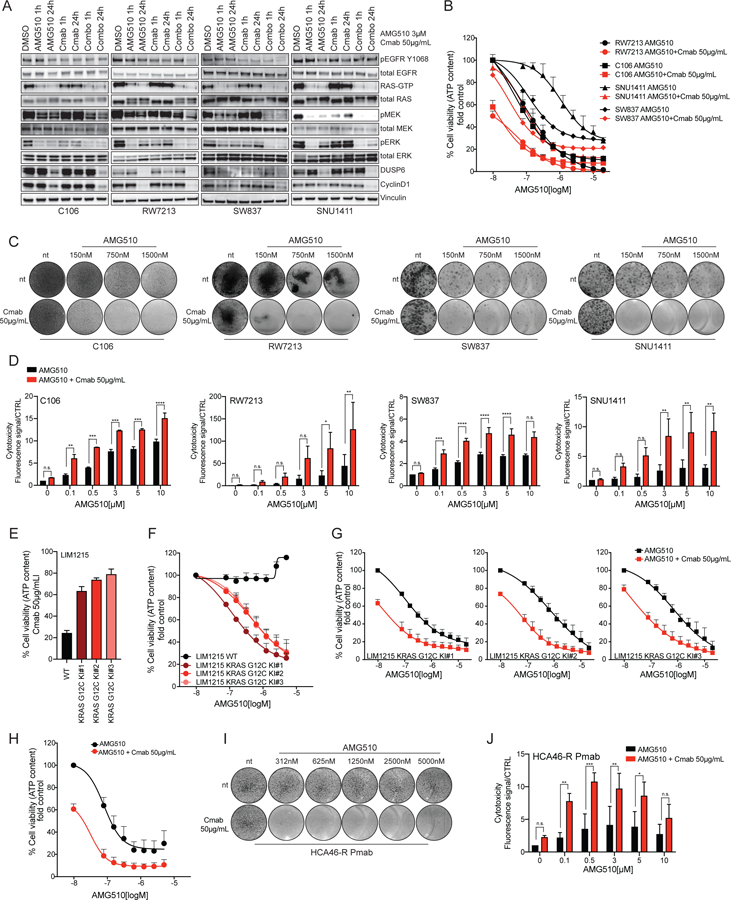
(A) Western blot analysis of cetuximab plus AMG510 combinatorial treatment in time course in C106, RW7213, SNU1411 and SW837 CRC cell lines. RAS-GTP pull down assay is included, and Vinculin is used as loading control. (B) Short-term proliferation assay of C106, RW7213, SNU1411 and SW837 treated with increasing concentration of AMG510 with or without 50ug/mL cetuximab. First data points of the combination curves represent the response to cetuximab alone. Cells were treated for 120 hours with increasing concentration of AMG510 and then ATP content was measured using CellTiterGlo. Data represents the average and standard deviation of 3 biological replicates. The AMG510 single agent curves used in this graph, are the same used in fig 1A. (C) Long-term drug screening proliferation assay of C106, RW7213, SNU1411 and SW837 treated with increasing concentration of AMG510 with or without 50μg/mL cetuximab. Cetuximab only treated cells are shown in the first lower circles. Cells were treated for 9 to 13 days according to the time when untreated controls reached confluence. (D) CellTox assay of C106, RW7213, SNU1411 and SW837 treated with increasing concentration of AMG510 with or without 50μg/mL cetuximab. Cetuximab only treated cells are shown as the first red bar at 0μM AMG510. Cells were treated for 120 hours. Data represents the average and standard deviation of 3 biological replicates. Statistical significance was calculated using one-way ANOVA with Bonferroni’s correction. Asterisks indicates *=p<0.05, **=p<0.01, ***=p<0.001, ****=p<0.0001, n.s.= not significant. (E) LIM1215 KRAS G12C Knock-In (KI) clones response to cetuximab; LIM1215 parental cells were included as control. (F) LIM1215 KRAS G12C KI clones response to AMG510 in short-term proliferation assay. Cells were treated for 120 hours with increasing concentration of AMG510 and then ATP content was measured using CellTiterGlo. Data represents the average and standard deviation of 3 biological replicates. (G) LIM1215 KRAS G12C KI clones response to AMG510+cetuximab in short-term proliferation assay. First data points of the combination curves represent the response to cetuximab alone. Cells were treated for 120 hours with increasing concentration of AMG510 and then ATP content was measured using CellTiterGlo. Data represents the average and standard deviation of 3 biological replicates. (H) HCA46-R Pmab cell line response to AMG510+cetuximab in short-term proliferation assay. First data point of the combination curve represents the response to cetuximab alone. Cells were treated for 120 hours with increasing concentration of AMG510 and then ATP content was measured using CellTiterGlo. Data represents the average and standard deviation of 3 biological replicates. (I) Long-term drug screening proliferation assay of HCA46-R Panitumumab treated with increasing concentration of AMG510 with or without 50ug/mL cetuximab. Cetuximab only treated cells are shown in the first lower circles. (J) CellTox assay of HCA46-R Pmab treated with increasing concentration of AMG510 with or without 50ug/mL cetuximab. Cetuximab only treated cells are shown as the first red bar at 0μM AMG510. Data represents the average and standard deviation of 3 biological replicates. Statistical significance was calculated using one-way ANOVA with Bonferroni’s correction. Asterisks indicates *=p<0.05, **=p<0.01, ***=p<0.001, ****=p<0.0001, n.s.= not significant.
We next sought to determine whether combined KRAS and EGFR inhibition could lead to cell death together with cell proliferation arrest. We found that the combination treatment induced a pronounced cytotoxic effect, as measured by detection of DNA of cells with impaired membrane integrity, in all the models tested and across dose levels of AMG510 (Figure 3D). These data indicate that combined KRAS G12C and EGFR inhibition leads to increase cell death rate.
To further assess the role of EGFR signaling in AMG510 resistance, we used the KRAS wild type, cetuximab sensitive, CRC cell line LIM1215 to generate isogenic models carrying the KRAS G12C allele using an AAV mediated knock-in approach(21). As expected, all KRAS G12C mutant clones (KI#1, KI#2 and KI#3) showed resistance to cetuximab as a single agent (Figure 3E). KRAS G12C mutant clones were more sensitive to AMG510 than their wild type counterpart (Figure 3F). The addition of cetuximab in combination with KRAS G12C inhibition was synergistic compared to the single agent alone (Figure 3G and Supplementary Figure 4B). These results provide further support that EGFR and KRAS G12C should be concomitantly inhibited to overcome resistance to KRAS G12C inhibition in CRC.
We and others previously reported that KRAS acquired mutations are responsible for secondary resistance to anti-EGFR therapies in CRC(22, 23). We reasoned that KRAS inhibitors could have clinical applications in patients who develop KRAS mutations at relapse after EGFR blockade. To directly test this possibility, we used a CRC cell line (HCA46-R Pmab), in which prolonged treatment with panitumumab lead to the emergence of a KRAS G12C allele(24). We examined whether AMG510 could be effective in this setting. While single agent AMG510 treatment was partially effective, its combination with cetuximab induced both cell proliferation arrest and cell death (Figure 3H–J and Supplementary Figure 4C). These data suggest that also in the secondary resistance setting CRC cells could retain upstream signaling dependency.
CRC patient-derived models are sensitive to combinatorial EGFR-KRAS G12C inhibition
To test whether and to what extent these findings were applicable to clinically relevant models we collected biopsies and exploited established models from CRC patients generated in our institutions (Supplementary tables 2–3).
Taking advantage of our molecularly and therapeutically annotated CRC patient-derived bio-bank (25–27), in a cohort of more than one hundred patient-derived organoids (PDO), we selected three different KRAS G12C mutant PDOs and one KRAS G12D mutant PDO (as negative control) and tested the effects of single agents AMG510, cetuximab and their combination.
In the KRAS G12C PDOs, single agent AMG510 or cetuximab did not or poorly inhibited growth, while the combination suppressed proliferation with high synergy scores (Figure 4A–B–C).
Figure 4: AMG510+cetuximab combination is effective in KRAS G12C mutant patient-derived models.
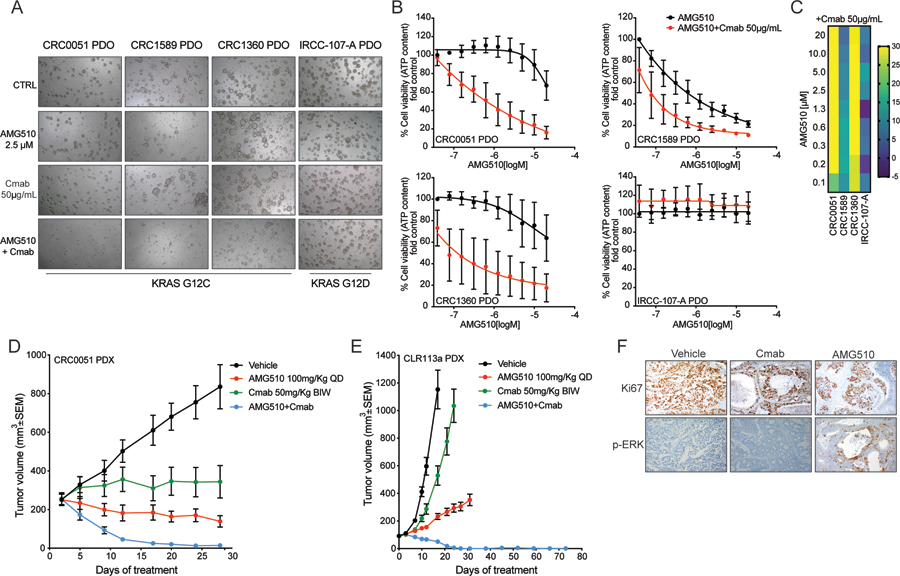
(A) Bright-field microscopy images of patient-derived organoid (PDO) treated with vehicle, AMG510, cetuximab and the combination 7 days after treatment. (B) ATP content quantification with CellTiterGlo assay. (C) Synergy score. Heatmap of excess over the Bliss (Bliss score) for AMG510-Cetuximab combination. (D) CRC0051 KRAS G12C mutant CRC patient-derived xenografts (PDXs) were treated with vehicle alone, AMG510 100mg/kg oral BID, cetuximab 50 mg/kg intraperitoneal twice a week (BIW), or the combination of the 2 drugs at the same doses. Error bars represent SEM (5 animals per group) (E) CLR113a KRAS G12C mutant CRC patient-derived xenografts (PDXs) were treated with vehicle alone, AMG510 100mg/kg oral BID, cetuximab 50 mg/kg intraperitoneal twice a week (BIW), or the combination of the 2 drugs at the same doses. Error bars represent SEM (5 animals per group). (F) Ki67 and phospho-ERK immunohistochemical staining of the CLR113a PDX samples collected at the end of treatment from vehicle, cetuximab and AMG510 treated arms. Ki67 IHC intensity of 97% with no necrosis in vehicle treated mice, 92% with necrosis in cetuximab treated mice, and 83% with necrosis in AMG510 treated mice.
In parallel, we generated patient-derived xenografts (PDXs) from two patients with KRAS G12C mutant CRC and tested AMG510 and cetuximab as single agents and their combination in vivo. Both models exhibited slowed but persistent growth when mice were treated with either cetuximab or AMG510 single agent treatment. Combination treatment induced massive or even complete tumor shrinkage in the treated animals (Figure 4D–E). No tumor was visible after 10 weeks of treatment and no samples could be collected at the end of the PDX experiment (Figure 4E). Interestingly, IHC staining for the proliferation marker Ki67 was highly positive in tumor samples derived from vehicle, cetuximab and AMG510 treated arms (Ki67 IHC intensity of 97% with no necrosis in vehicle treated mice, 92% with necrosis in cetuximab treated mice, and 83% with necrosis in AMG510 treated mice) but strong phospho-ERK staining was observed only in the AMG510 treated samples, suggesting that strong MAPK pathway rebound occurs in patient-derived samples as well (Figure 4F).
Discussion
Recent clinical data have shown promising activity of agents targeting mutant (G12C) KRAS, a heretofore-undruggable target. These trials, however, show substantial differences in the response rate between NSCLC and CRC patients(14, 15).
We found that KRAS G12C inhibitors produce less profound and more transient inhibition of KRAS downstream signaling in CRC versus NSCLC models. Our data indicate that, despite harboring the same mutation, there are intrinsic differences in signaling between CRC and NSCLC. Our data suggest that in CRC the combination of (1) signaling rebound upon KRAS G12C inhibition, (2) the higher extent of RTK activation and (3) the stronger response to EGFR direct stimulation can be used to define the intrinsic dependency of bowel tumors on EGFR-MAPK pathway. These results, together with the emerging clinical experience with single agent KRAS G12C inhibitors, indicate that combinatorial treatment is necessary to effectively treat KRAS G12C mutant CRC.
This is, to our knowledge, the first study aimed at specifically characterizing the effects of KRAS G12C inhibition in CRC. Indeed, the main goal of this work was to understand and exploit CRC lineage-specific features to develop more effective pharmacological strategies.
It is known that RTK inhibition in NSCLC preclinical models can potentiate KRAS G12C covalent inhibitors, by slowing down exchange factors favoring a GDP-bound status of KRAS(8). However this combinatorial strategy is considered only marginal in this setting as other combinatorial partners including PI3K, mTOR, and Aurora Kinase inhibitors have been shown to have higher efficacy than EGFR inhibition in combination with KRAS G12C inhibition in lung cancer(10, 12, 13).
We found that expression of EGFR and its dimerization partners is common in KRAS G12C CRC and NSCLC clinical samples. Basal EGFR expression, as measured by immunohistochemistry or mass spectrometry(28, 29), has not been associated with EGFR inhibitor sensitivity in CRC, and indeed we find no clear difference between receptor expression levels by lineage or in the subset of CRC and NSCLC patients treated with AMG510. In cell line models, however, we are able to assess activated RTK levels and we find that the degree of this dependency on RTK signaling across KRAS G12C mutant tumors can be very different. Specifically, we show that KRAS G12C mutant CRC cell lines still retain their dependency on EGFR for downstream signaling.
The clinical and signaling effects of AMG510 in CRC is reminiscent of the clinical experience in BRAF V600E CRC, where the concomitant inhibition of upstream signaling reactivation results in improved clinical efficacy (30), which has led to the recent FDA approval of the BRAF inhibitor encorafenib in combination with cetuximab (BEACON CRC; NCT02928224).
Clinical trials results have shown limited efficacy of KRAS G12C covalent inhibitors in CRC; only one CRC patient treated with MRTX849 had a 47% tumor reduction by RECIST criteria(15). Our results show that in CRC cell models and in PDX, treatment with AMG510 single agent induces strong reactivation of MAPK pathway. These data indicate that even in the best scenario, upfront combination treatment is the best option for KRAS G12C mutant CRC patients, as this approach would enhance the efficacy of KRAS G12C inhibitors and would also limit the emergence of acquired resistance.
We showed that KRAS G12C mutant CRC still retains EGFR-MAPK dependency, as demonstrated by the signaling rebound upon KRAS G12C inhibition, the more pronounced activation of RTKs and the high response to EGFR direct stimulation. We can speculate that also other KRAS mutant alleles in CRC could retain these characteristics suggesting that also other KRAS mutant CRC could potentially benefit from EGFR-based combinations. Therefore, direct or indirect KRAS blockade-based combinations could be considered as an option in these specific settings.
Combined inhibition of KRAS G12C with SHP2, a phosphatase that serves as a common mediator of RTK signaling to RAS(31), has been proposed as another combination to overcome the feedback reactivation in KRAS G12C tumors(32).
Available clinical data with SHP2 inhibitors is currently limited and it is not yet known if these inhibitors can be administered to patients in sufficient doses to effectively inhibit RTK signaling. Further, preclinical models suggest growth suppression but not complete tumor regression with SHP2 and KRAS G12C inhibitors, in contrast to the complete, durable tumor inhibition achieved in a patient-derived xenograft treated with EGFR and KRAS G12C inhibitors.
Since RTK signaling in CRC is mostly dominated by wild-type EGFR, we propose to combine KRAS G12C inhibitors with anti-EGFR monoclonal antibodies to block receptor signaling rather than inhibiting EGFR kinase activity. Moreover, the differential activity of EGFR inhibitors in CRC versus NSCLC further indicate signaling differences in these two lineages(33). The anti-EGFR antibodies cetuximab and panitumumab are approved for the treatment of RAS/RAF wild type CRC(34, 35), while EGFR tyrosine kinase inhibitors (TKI) are approved for the treatment of EGFR mutant lung cancer(36). EGFR-targeting TKI have not been effective in CRC(37), probably because EGFR genetic alterations are rare in CRC and these drugs are more efficient in EGFR-mutant setting. As release of wild type receptor signaling from feedback inhibition is thought to mediate adaptive resistance to targeted therapies, the role of wild type EGFR in activating signaling in CRC could render CRCs more sensitive to this feedback release than NSCLC.
Taken together, our results demonstrate that biochemical characterization of response to KRAS G12C inhibitors in CRC provides a mechanistic explanation for their limited activity in the clinic and, at the same time, provides a strong rationale to combine EGFR blockade with these agents in CRC patients.
Methods
Cell lines and Compounds
The cell lines NCIH358, LU65, SNU1411, JVE015, LIM2099, LIM1215, LIM1215 KRAS G12C KI cell lines and RW7213 were cultured in RPMI (Lonza). C106 cell line was cultured in Iscove’s modified medium (Lonza). SW837 cell line was cultured in DMEM/F12 (Lonza). HCA46-R Pmab cell line was cultured in DMEM (Lonza). Each media was supplemented with 10% FBS, 2mM L-glutamine, 100U/ml penicillin and 100mg/mL streptomycin. SW837 and NCIH358 cell lines were purchased from ATCC; C106 and HCA46 cells were purchased from ECACC, SNU1411 cells were purchased from KCLB; LU65 were purchased from AcceGen Biotech; LIM2099, LIM1215 were kindly provided from Dr. Whitehead (Australia); JVE015 were provided from Prof. Tom Van Wezel (LUMC); RW7213 were provided by Dr. Diego Arando.
All the cell lines were determined to be mycoplasma free using the Venor® GeM Classic kit (Minerva biolabs) and tested by Short Tandem Repeats profiling at 10 different loci.
AMG510 was purchased from ChemGood (C-1499) and cetuximab was either gifted from Merck Serono under an academic material transfer agreement or purchased from the Pharmacy at Memorial Sloan Kettering Cancer Center.
Patient-Derived Organoids
All the procedures performed with patient specimens were conducted under the approval of the local Ethical Committee of the institutions, after the written informed consent of the patients. CRC1589, CRC1360 and CRC0051 patient-derived organoids (PDOs) were derived from PDXs established from three distinct KRAS G12C CRC patients while IRCC107A patient-derived organoid (PDO) was established directly from tissue biopsy obtained at the time of surgery. To generate PDX-derived organoids, PDX samples were first dissociated mechanically with gentle MACS Dissociator (Milteny Biotec) and then underwent to enzymatically digestion of extracellular matrix using Human Tumor Dissociation Kit (Miltenyi Biotec). The derived single cell suspension was centrifuged at 400g for 5 minutes and the resuspendend in the organoids’ Basal medium. Cell suspension was filtered with 70-uM cell strainer in order to avoid cell clustering formation. The flow-through was centrifuged and the pellet was resuspended in Matrigel (Corning) and dispensed as 100ul droplet in the center of 37°C prewarmed 12-well plate. After an incubation of 15 minutes at 37°C for matrigel solidification, organoids were added with ENAS medium and incubated at 5%CO2 and 37°C. ENAS medium is refreshed twice per week.
Differently, to generate PDOs directly from patient tissue biopsy, tumor tissue was first smashed in small pieces and incubated in PBS with collagenase A (0.5 mg/mL; Roche), hyaluronidase (20 mg/mL; Sigma), and 10 mmol/L ROCK inhibitor for 30 minutes at 37°C with shaking. After incubation, 5% FBS was added and the mixture was centrifuged at 400 g for 5 minutes. The pellet was washed three times in PBS to remove debris and collagenase. At the end of the washing phase, the pellet was resuspended in Matrigel (Corning) and 50 uL of organoids-Matrigel suspension were dispensed into the center of each well of a 37°C prewarmed 24-well plate. Different densities of tumor cells were plated and left to solidify for 10 to 30 minutes at 37°C before ENAS medium was added and the cells were incubated at 37°C, 5% CO2. Fresh medium was replaced every 2 to 3 days. Outgrowing organoids were passaged every 10 to 15 days after mechanical and enzymatic disruption. Organoid drug treatment was performed in each organoid-specific culture medium at 37°C and 5% CO2.
The organoid Basal Medium is composed of Advanced DMEM/F12 medium (Thermo Fisher) containing 100 U/mL penicillin, 100µg/mL streptomycin, 2 mM GlutaMAX (Gibco), 10 mM Hepes, 50 ug/mL Primocin, (Invitrogen). ENAS medium is prepared by adding 1 × B27 supplement (Invitrogen), 1 × N2 supplement (Invitrogen), 1.25 mM N-acetyl-cysteine (Sigma Aldrich), 10 mM Nicotinamide (Sigma Aldrich), 10 nM gastrin (Sigma), 50 ng/mL recombinant mouse EGF (Life Technologies), 100 ng/mL Noggin (PeproTech), 500 nM TGFb type I receptor inhibitor A83–01 (Tocris) and 10 μM p38 MAPK inhibitor SB202190 (Sigma Aldrich) to the organoid Basal Medium.
Antibodies and Western blotting
After seeding and drug treatments, cells were washed with cold PBS and lysed in RIPA buffer (Pierce #89901) plus phosphatase and protease inhibitors (Thermo Scientific #1861277, #1861278). Lysates were cleared by centrifugation at 14000rpm at 4°C and quantified using BCA method (Pierce #23224).
Samples were prepared using LDS+Reducing agent Novex buffers (Invitrogen #NP0008, #NP0009). 10 to 20μg of lysates were loaded and run on NuPageTM 4–12% Bis-Tris gels (ThermoFisher #NP0321BOX) followed by transfer to nitrocellulose membranes (Biorad #1620233). Membranes were incubated over night with the indicated antibodies, washed and incubated again for 45 minutes with anti-rabbit or anti-mouse secondary antibodies. Detection was performed using Immobilion Western (Millipore #WBKLS0500).
Primary antibodies were obtained from Cell Signaling Technology and were used at a concentration of 1:1000: p-Akt S473 (#9271), p-MEK1/2 S217/221 (#9154), p-p44/42 MAPK T202/204 (#9101), Total MEK1/2 (#9122), Total ERK1/2 (#9102), Total AKT (#9272), Total EGFR (#2232S), phospho-EGFR Tyr1068 (#3777S) and Vinculin (#13901S). DUSP6 antibody was purchased from Abcam (#ab54940) and used at 1:500 dilution. CyclinD1 antibody was purchased from Santa Cruz (#sc-718) and used at 1:1000 dilution. Densitometry analyses were performed using ImageJ software.
EGFR Immunoprecipitation
For Immunoprecipitation (IP) assays, cells were seeded and starved for 24h and then stimulated with 20% FBS for 10 minutes. Cells were then washed in cold PBS and lysed using NP-40 buffer (150 mM NaCl, 10 mM Tris pH 8, 1% NP-40, 10% glycerol). 20 μg of proteins were used as total lysates. 1500μg of protein lysates were incubated rotating at 4°C for 1,5 hours in the presence of 10 μg/mL cetuximab, Agarose protein G beads (Invitrogen #15920–010). Beads were centrifuged at 3000 rpm for 1 min and supernatant was removed. Similarly, beads were washed three times using NP-40 buffer and once using nuclease-free sterile water. Finally, immunoprecipitates were eluted using 1X sample buffer (NuPAGE™ LDS Sample Buffer, Thermo Fisher Scientific #NP0008 and NuPAGE Sample Reducing Agent, Thermo Fisher Scientific #NP0009) at 95°C for 5 min
RAS-GTP pull-down assay
RAS-GTP pull –down assay was performed according to manufacturer’s protocol (Thermo Scientific #16117). Briefly, 500μg of lysates were loaded into columns together with agarose beads and RAS-RBD bait and incubated for 1 hour at 4°C. After the incubation, beads were washed three times and resuspended in LDS+Reducing agent Novex buffers (Invitrogen #NP0008, #NP0009). A fraction of lysates was used to measure total RAS amount. Pull-Down and total lysates were subjected to western blotting procedure as described above. The kit provided primary antibody against pan-RAS.
Phospho-RTK array
Human Phospho-RTK arrays (R&D Systems, ARY001B) were used to detect activated RTKs according to manufacturer’s instructions. Cells were washed with cold PBS and lysed using the provided lysis buffer plus phosphatase and protease inhibitors. 200 μg lysates were incubated on membranes overnight. Membranes were subsequently washed and exposed to chemiluminescent reagent.
Mass Spectrometry
Selected reaction monitoring-mass spectrometry (SRM-MS) of 72 biomarkers of the tissue sections from FFPE blocks was conducted as previously described(38, 39). Briefly, two tissue section sections (10µM each) from FFPE blocks were placed on DIRECTOR slides, deparaffinized, and stained with hematoxylin. Tumor areas were marked by board-certified pathologist, then was micro-dissected and solubilized to tryptic fragments as per manufacturer instructions (Expression Pathology, Rockville, MD). Protein concentration of the tryptic peptides was calculated using microBCA. Stable heavy isotope-labeled internal standard peptides for 72 biomarkers was added to the solution and injected into the mass spectrometer (TSQ Quantiva, ThermoFisher Scientific, San Jose, CA). On-column injection resulted in 5fmol of isotopically labeled internal standard peptide and 1µg of total tumor protein. Data analysis of the 72 biomarkers was conducted using Pinnacle software (Optys Tech, Boston, MA). All patients whose tumors were analyzed signed written informed consent for molecular profiling of their tumors (IRB 12–245) and approval was obtained for the mass spectrometry assay.
Drug proliferation assays
For short-term proliferation assays, 2,000 cells were seeded in 96-well culture plates in medium containing 10% FBS. After 24 hours, serial dilutions of AMG510 (alone or in combination with Cetuximab) were added to cell in 10% FBS medium (ratio 1:1). Cell viability was determined at day 5 after the start of the treatment by measuring ATP content using Cell Titer-Glo® Luminescent Cell Viability assay (Promega). DMSO-only treated cells were used as control. Excess over the Bliss (Bliss score) was calculated using effect of the combo (observed) versus what was predicted/expected based on the effect of each single agent expressed as percentage. Negative values on values close to zero show no/very little synergy. Higher percentages indicate higher synergy.
For long-term proliferation assays, cells were seeded (5000 per well for SNU1411, SW837 and HCA46R, 80000 cells per well for RW7213) in 24-well plates or in 6-well plates (C106 80000 cells per well) in 10% FBS medium. The following day, plates were treated with different concentrations of drugs as indicated. Cells were fixed with 4% paraformaldehyde and stained with 10% Crystal Violet in a Methanol/Water solution (Sigma Aldrich) 9 to 13 days after the start of treatment, according to the confluence reached by the untreated control. Every 7 days, media and drug were refreshed. In all the experiments, plates were incubated at 37°C in 5% CO2.
Short-term assays were performed at least three times. All long-term assays were performed independently at least three times. Analyses and graphs were performed with GraphPad Prism Software using the function log (inhibitor) versus response-variable slope (4 parameters).
Drug cytotoxicity assay
For short term cytotoxicity assay, 2,000 cell/well were seeded in 96-well plates and treated after 24 hours with AMG510 alone or in combination with cetuximab as indicated. After 5 days from the start of treatment, cell cytotoxicity was determined measuring the fluorescence of death cells using CellTox Green cytotoxicity assay (Promega), according to the kit instruction. The CellTox green results were normalized on results of Cell Titer-Glo® Luminescent Cell Viability assay conducted on the same plates. In all the experiments, plates were incubated at 37°C in 5% CO2. All assays were performed independently at least three times
In vivo studies
The CLR113 PDX was derived from a liver metastasis under written consensus. Tumor tissue was transplanted orthotopically into NSG mice to establish the PDX (IRB protocols 06–107, 14–091). Once a tumor became visible in the first mouse, it was transplanted and expanded to other animals. Tumor tissue was implanted subcutaneously in the flank of 4–6-week-old NSG female mice and treatment of the mice began when tumor reached approximately 100mm3 in size. Mice were randomized (n = 5 mice per group) to receive drug treatments or vehicle as control. Studies were performed in compliance with institutional guidelines under an IACUC approved protocol. The animals were immediately euthanized as soon as the tumors reached the IACUC set limitations.
The CRC0051 PDX was derived from a colorectal cancer patient metastasis, under written consensus. Patient tissue was transplanted into a NOD-SCID mice to establish PDX and then transplanted and expanded to other animals. Tumor was implanted subcutaneously in the flank of 11–13-week-old NODSCID mice and treatment of the mice began when tumor reached approximately 250 mm3. Mice were randomized (n=7 per arm) and received drug treatment or vehicle control.
AMG 510 was given 100mg/kg daily by gavage. Cetuximab was administered 50mg/kg twice a week, by intra-peritoneal injections.
The entire experiment was performed in compliance with institutional guidelines under the Ethical committee of Candiolo Cancer Institute consensus and under the Italian Ministry of Health approved protocol. The animals were sacrificed on the base of the rules defined in the approved protocol.
Supplementary Material
Statement of significance.
The efficacy of KRAS G12C inhibitors in NSCLC and CRC is lineage-specific. RTK dependency and signaling rebound kinetics are responsible for sensitivity or resistance to KRAS G12C inhibition in CRC. EGFR and KRAS G12C should be concomitantly inhibited to overcome resistance to KRAS G12C blockade in colorectal tumors.
Acknowledgments
The authors thank Dr. Maurizio Scaltriti and Dr. David Solit for critical reading of the manuscript. Authors also thank the Molecular Cytology Core Facility at MSKCC for technical support. This work is supported by AIRC, Associazione Italiana per la Ricerca sul Cancro, Investigator Grants 20697 (to A.Be.), 21407 (to F.D.N.) and 22802 (to L.T.). AIRC IG 2018-ID. 21923 project-PI Bardelli Alberto. AIRC under 5 per Mille 2018-ID 21091 program - P.I. Bardelli Alberto, G.L. Bertotti Andrea, G.L. Di Nicolantonio, G.L. Marsoni Silvia, G.L. Siena Salvatore, G.L. Trusolino Livio (to A.B., A. Be. F.D.N., S.Ma., S.S. and L.T). European Research Council Consolidator Grant 724748 – BEAT (to A.Be.). H2020 grant agreement no. 754923 COLOSSUS (to L.T.). H2020 INFRAIA grant agreement no. 731105 EDIReX (to A.Be.). AIRC/CRUK/FC AECC Accelerator Award 22795 (to A.B. and L.T.). Fondazione Piemontese per la Ricerca sul Cancro-ONLUS, 5×1000 Ministero della Salute 2015 Project “STRATEGY” (F.D.N.) Fondazione Piemontese per la Ricerca sul Cancro-ONLUS, 5×1000 Ministero della Salute 2015 Project “IMMUNOGENOMICA” (to A.B. and L.T.), 2014 and 2016 (to L.T.). A.Be. and L.T. are members of the EurOPDX Consortium. AIRC MFAG 2017 ID 20236 (to S.A.). This work is also supported by the National Institutes of Health R01 CA233736 (R.Y.) and Cancer Center Core Grant P30 CA008748. This research is the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Disclosure of potential
R.Y. has received research funding from Array BioPharma, Novartis and Boehringer Ingelheim and has consulted for Array BioPharma. Y.R.M.G. has received support for travel and expenses from AstraZeneca and has received training via a Clinical and Translational Science Award (CTSA) grant (UL1TR00457). NV received honoraria or travel accommodation from Merck Serono, Pfizer, Bayer, Eli-Lilly and Menarini Silicon Biosystems. S.T., W-L.L. and A.B. are mProbe Inc. employees. L.T. receives research grants from Symphogen, Servier, Pfizer, Merus and Menarini and is a paid consultant for Eli Lilly, AstraZeneca, and Merck KGaA. B.T.L. has served as a consultant/advisory board member for Roche, Biosceptre International, Thermo Fisher Scientific, Mersana Therapeutics, Hengrui Therapeutics, Guardant Health and has received research funds from Genentech (Inst), Daiichi Sankyo (Inst), Hengrui Therapeutics (Inst), Illumina (Inst), BioMed Valley Discoveries (Inst), AstraZeneca (Inst), GRAIL (Inst). S. S is an employee/paid consultant for Amgen, Bayer, Bristol-Myers Squibb, CheckMab, Clovis, Daiichi Sankyo, Incyte, Merck, Novartis, Roche-Genentech, and Seattle Genetics. N.R. is on the SAB and receives research funding from Chugai, on the SAB and owns equity in Beigene, and Fortress. N.R. is also on the SAB of Daiichi-Sankyo, Astra-Zeneca-MedImmune, and F-Prime, and is a past SAB member of Millenium-Takeda, Kadmon, Kura, and Araxes. N.R. is a consultant to Novartis, Boehringer Ingelheim, Tarveda, and Foresight and consulted in the last three years with Eli Lilly, Merrimack, Kura Oncology, Araxes, and Kadman. N.R. owns equity in ZaiLab, Kura Oncology, Araxes, and Kadman. N.R. also collaborates with Plexxikon. A. B. reports receiving commercial research grants from Neophore; is an advisory board member/unpaid consultant for Roche, Illumina, Guardant, and Third Rock; holds ownership interest (including patents) in Neophore and Phoremost; and is an advisory board member/unpaid consultant for Horizon Discovery, Biocartis, and Neophore. S.M. consulted for Boehringer-Ingelheim.
Footnotes
conflicts of interest
All the other authors declare no conflicts of interests.
References
- 1.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nature medicine. 2017;23:703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simanshu DK, Nissley DV, McCormick F. RAS Proteins and Their Regulators in Human Disease. Cell. 2017;170:17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–77. [DOI] [PubMed] [Google Scholar]
- 4.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nature reviews Cancer. 2007;7:295–308. [DOI] [PubMed] [Google Scholar]
- 5.Yuan TL, Amzallag A, Bagni R, Yi M, Afghani S, Burgan W, et al. Differential Effector Engagement by Oncogenic KRAS. Cell reports. 2018;22:1889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunter JC, Manandhar A, Carrasco MA, Gurbani D, Gondi S, Westover KD. Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations. Molecular cancer research : MCR. 2015;13:1325–35. [DOI] [PubMed] [Google Scholar]
- 7.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lito P, Solomon M, Li LS, Hansen R, Rosen N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science. 2016;351:604–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patricelli MP, Janes MR, Li LS, Hansen R, Peters U, Kessler LV, et al. Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State. Cancer discovery. 2016;6:316–29. [DOI] [PubMed] [Google Scholar]
- 10.Misale S, Fatherree JP, Cortez E, Li C, Bilton S, Timonina D, et al. KRAS G12C NSCLC Models Are Sensitive to Direct Targeting of KRAS in Combination with PI3K Inhibition. Clinical cancer research : an official journal of the American Association for Cancer Research. 2019;25:796–807. [DOI] [PubMed] [Google Scholar]
- 11.Lou K, Steri V, Ge AY, Hwang YC, Yogodzinski CH, Shkedi AR, et al. KRAS(G12C) inhibition produces a driver-limited state revealing collateral dependencies. Science signaling. 2019;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molina-Arcas M, Moore C, Rana S, van Maldegem F, Mugarza E, Romero-Clavijo P, et al. Development of combination therapies to maximize the impact of KRAS-G12C inhibitors in lung cancer. Science translational medicine. 2019;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue JY, Zhao Y, Aronowitz J, Mai TT, Vides A, Qeriqi B, et al. Rapid non-uniform adaptation to conformation-specific KRAS(G12C) inhibition. Nature. 2020. January;577(7790):421–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575:217–23. [DOI] [PubMed] [Google Scholar]
- 15.Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, et al. The KRAS(G12C) Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discovery. 2020. January;10(1):54–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prahallad A, Heynen GJ, Germano G, Willems SM, Evers B, Vecchione L, et al. PTPN11 Is a Central Node in Intrinsic and Acquired Resistance to Targeted Cancer Drugs. Cell reports. 2015;12:1978–85. [DOI] [PubMed] [Google Scholar]
- 17.Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer discovery. 2012;2:227–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yaeger R, Yao Z, Hyman DM, Hechtman JF, Vakiani E, Zhao H, et al. Mechanisms of Acquired Resistance to BRAF V600E Inhibition in Colon Cancers Converge on RAF Dimerization and Are Sensitive to Its Inhibition. Cancer research. 2017;77:6513–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lake D, Correa SA, Muller J. Negative feedback regulation of the ERK1/2 MAPK pathway. Cell Mol Life Sci. 2016;73:4397–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Robertis M, Loiacono L, Fusilli C, Poeta ML, Mazza T, Sanchez M, et al. Dysregulation of EGFR Pathway in EphA2 Cell Subpopulation Significantly Associates with Poor Prognosis in Colorectal Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2017;23:159–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Nicolantonio F, Arena S, Gallicchio M, Zecchin D, Martini M, Flonta SE, et al. Replacement of normal with mutant alleles in the genome of normal human cells unveils mutation-specific drug responses. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20864–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diaz LA Jr., Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Misale S, Arena S, Lamba S, Siravegna G, Lallo A, Hobor S, et al. Blockade of EGFR and MEK intercepts heterogeneous mechanisms of acquired resistance to anti-EGFR therapies in colorectal cancer. Science translational medicine. 2014;6:224ra26. [DOI] [PubMed] [Google Scholar]
- 25.Bertotti A, Migliardi G, Galimi F, Sassi F, Torti D, Isella C, et al. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer discovery. 2011;1:508–23. [DOI] [PubMed] [Google Scholar]
- 26.Lazzari L, Corti G, Picco G, Isella C, Montone M, Arcella P, et al. Patient-Derived Xenografts and Matched Cell Lines Identify Pharmacogenomic Vulnerabilities in Colorectal Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2019;25:6243–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertotti A, Papp E, Jones S, Adleff V, Anagnostou V, Lupo B, et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature. 2015;526:263–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serna G, Ruiz-Pace F, Cecchi F, Fasani R, Jimenez J, Thyparambil S, et al. Targeted multiplex proteomics for molecular prescreening and biomarker discovery in metastatic colorectal cancer. Scientific reports. 2019;9:13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung KY, Shia J, Kemeny NE, Shah M, Schwartz GK, Tse A, et al. Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23:1803–10. [DOI] [PubMed] [Google Scholar]
- 30.Kopetz S, Grothey A, Yaeger R, Van Cutsem E, Desai J, Yoshino T, et al. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E-Mutated Colorectal Cancer. The New England journal of medicine. 2019;381:1632–43. [DOI] [PubMed] [Google Scholar]
- 31.Grossmann KS, Rosario M, Birchmeier C, Birchmeier W. The tyrosine phosphatase Shp2 in development and cancer. Adv Cancer Res. 2010;106:53–89. [DOI] [PubMed] [Google Scholar]
- 32.Ryan MB, Fece de la Cruz F, Phat S, Myers DT, Wong E, Shahzade HA, et al. Vertical pathway inhibition overcomes adaptive feedback resistance to KRASG12C inhibition. Clinical Cancer Research. 2020. April 1;26(7):1633–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Troiani T, Napolitano S, Della Corte CM, Martini G, Martinelli E, Morgillo F, et al. Therapeutic value of EGFR inhibition in CRC and NSCLC: 15 years of clinical evidence. ESMO Open. 2016;1:e000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saltz LB, Meropol NJ, Loehrer PJ Sr., Needle MN, Kopit J, Mayer RJ. Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2004;22:1201–8. [DOI] [PubMed] [Google Scholar]
- 35.Van Cutsem E, Peeters M, Siena S, Humblet Y, Hendlisz A, Neyns B, et al. Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2007;25:1658–64. [DOI] [PubMed] [Google Scholar]
- 36.Diaz-Serrano A, Gella P, Jimenez E, Zugazagoitia J, Paz-Ares Rodriguez L. Targeting EGFR in Lung Cancer: Current Standards and Developments. Drugs. 2018;78:893–911. [DOI] [PubMed] [Google Scholar]
- 37.Kuo T, Fisher GA. Current status of small-molecule tyrosine kinase inhibitors targeting epidermal growth factor receptor in colorectal cancer. Clin Colorectal Cancer. 2005;5 Suppl 2:S62–70. [DOI] [PubMed] [Google Scholar]
- 38.Hembrough T, Thyparambil S, Liao WL, Darfler MM, Abdo J, Bengali KM, et al. Application of selected reaction monitoring for multiplex quantification of clinically validated biomarkers in formalin-fixed, paraffin-embedded tumor tissue. J Mol Diagn. 2013;15:454–65. [DOI] [PubMed] [Google Scholar]
- 39.Catenacci DV, Liao WL, Thyparambil S, Henderson L, Xu P, Zhao L, et al. Absolute quantitation of Met using mass spectrometry for clinical application: assay precision, stability, and correlation with MET gene amplification in FFPE tumor tissue. PloS one. 2014;9:e100586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


