Abstract
Single-cell CRISPR screens enable the exploration of mammalian gene function and genetic regulatory networks. However, use of this technology has been limited by reliance on indirect indexing of single-guide RNAs (sgRNAs). Here we present direct-capture Perturb-seq, a versatile screening approach in which expressed sgRNAs are sequenced alongside single-cell transcriptomes. Direct-capture Perturb-seq enables detection of multiple distinct sgRNA sequences from individual cells and thus allows pooled single-cell CRISPR screens to be easily paired with combinatorial perturbation libraries that contain dual-guide expression vectors. We demonstrate the utility of this approach for high-throughput investigations of genetic interactions and, leveraging this ability, dissect epistatic interactions between cholesterol biogenesis and DNA repair. Using direct capture Perturb-seq, we also show that targeting individual genes with multiple sgRNAs per cell improves the efficacy of CRISPR interference and activation, facilitating the use of compact, highly active CRISPR libraries for single-cell screens. Last, we show that hybridization-based target enrichment permits sensitive, specific sequencing of informative transcripts from single-cell RNA-seq experiments.
CRISPR-based genetic tools have recently been paired with high-resolution phenotypic profiling to enable genetic screens with information rich readouts1–3. These efforts have dramatically expanded our ability to investigate genetic control over complex cellular processes. One such approach, independently implemented as Perturb-seq4,5, CRISP-seq6, Mosaic-seq7, and CROP-seq8 and herein referred to as single-cell CRISPR screening, combines pooled CRISPR screens with single-cell RNA-sequencing (scRNA-seq) readouts to facilitate unbiased exploration of gene function and systematic delineation of genetic regulatory networks. However, current implementations face technical and practical limitations that unnecessarily restrict their use. Here, we present advances that address these limitations, specifically poor scalability, dependence on specialized vector systems and high cost9–12, and by doing so, we enable facile and scalable single-cell analysis of both single and combinatorial genetic perturbations. In particular, we establish a method for interrogating programmed pairs of CRISPR sgRNAs by scRNA-seq, thus enabling efforts to study redundant gene isoforms or paralogs, investigate cis-regulatory genome architecture13, evade knockout rescue14, generate precise genetic edits15,16, or map genetic interactions (GIs)17.
The technological crux of all single-cell CRISPR screens is the assignment of perturbation identities to single-cell phenotypes. To achieve this, scRNA-seq screening platforms typically rely on polyadenylated indexes. These indexes are co-expressed with non-polyadenylated sgRNAs, but unlike the sgRNAs, they can be recorded on standard scRNA-seq platforms that capture only polyadenylated RNAs (Supplementary Fig. 1a,b). However, recombination of indexed sgRNA libraries during lentiviral delivery can uncouple indexes from their assigned sgRNAs9–12. This means that such platforms are limited to arrayed use and restricted scale9,11. Notably, one method, CROP-seq, has minimized this problem8. CROP-seq uses a clever vector system to deliver sgRNAs to cells. This vector duplicates the sequence of a single encoded sgRNA during lentiviral transduction to produce two expression cassettes on the same construct: one that expresses a functional sgRNA and another that expresses a polyadenylated transcript carrying the sgRNA sequence at the 3’ end. In this way, CROP-seq ensures delivery of pooled guide libraries to cells with faithful pairing of sgRNAs and polyadenylated “indexes”. However, due to constraints on cassette size, CROP-seq is thought to be incompatible with delivery of multiple sgRNAs.
To establish tools for more versatile single-cell CRISPR screens, we sought to directly sequence sgRNAs alongside single-cell transcriptomes in a method we refer to as “direct capture Perturb-seq”. Breifly, droplet-based scRNA-seq uses molecular barcoding to identify transcripts from individual cells. This barcoding occurs during reverse transcription (RT), when both unique molecular identifiers (UMIs) and cell barcodes (CBCs) are added to the 3’ or 5’ ends of mRNA sequences (Supplementary Fig. 1a,b)18–20. For direct capture Perturb-seq, we extended this barcoding to non-polyadenylated sgRNAs by addition of guide-specific primers during RT (Fig. 1a,b). To maximize flexibility, we designed platforms for direct capture with both 5’ and 3’ scRNA-seq. For 5’ scRNA-seq, this required the simple addition of an unbarcoded guide-specific RT primer to standard protocols (Fig. 1a and Supplementary Fig. 1b), an approach also reported by Mimitou et al. while this work was under review21. For 3’ scRNA-seq, the RT configuration necessitated that we implement an entirely new scRNA-seq platform (Fig. 1b). This platform concurrently delivers target-specific, barcoded primers to single-cell reactions alongside barcoded oligo-dT (Fig. 1b and Supplementary Fig. 1a). These target-specific primers anneal to capture sequences (cs1 and cs2) in modified sgRNA constant regions and thus enable RT of sgRNAs and efficient recording of sgRNA sequences (Supplementary Fig. 1c,d,e,f, Supplementary Note 1, and Supplementary Table 1). We selected the capture sequences for our platform carefully, to ensure for example, that their incorporation into an optimized sgRNA constant region (CR1) would not compromise guide activity. However, these capture sequences are not guide-specific, and thus, in principle, will enable multiplexed capture of additional features, such as antibodies22,23 and other oligo-tagged markers24. Herein, we refer to guides with cs1 incorporated in a stem loop of our standard Streptococcus pyogenes Cas9 sgRNAs as sgRNA-CR1cs1 and guides with cs2 incorporated at the 3’ end as sgRNA-CR1cs2. We note that an alternate configuration with incorporation of cs1 at the 3’ end compromises activity and therefore is not recommended (Supplementary Fig. 1f).
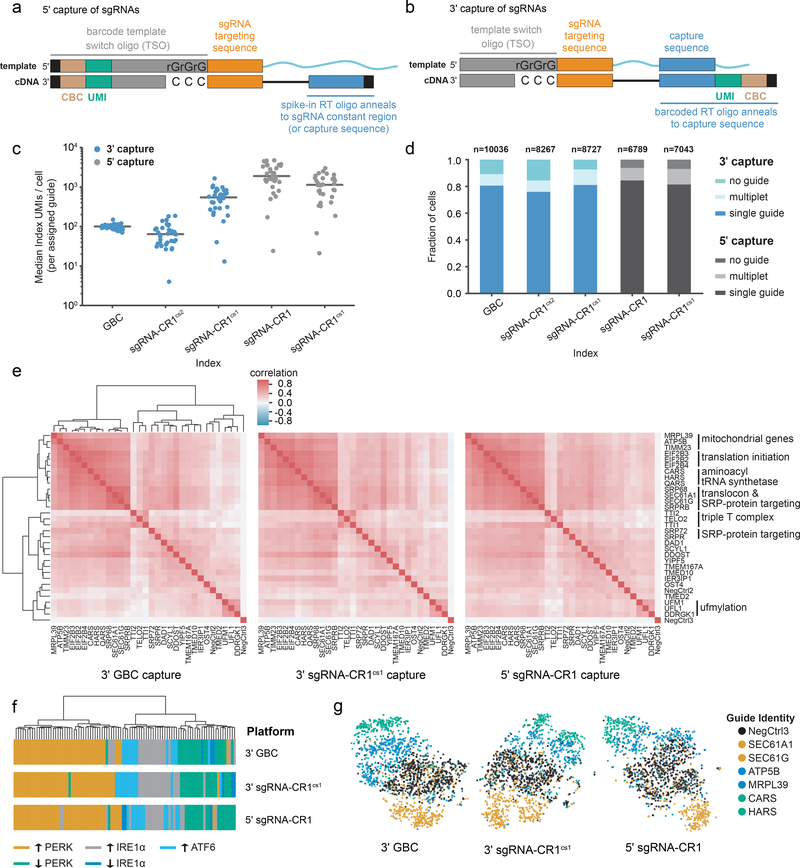
Figure 1: Design and validation of direct capture Perturb-seq for 3’ and 5’ single-cell RNA-sequencing.
a) Schematic of sgRNA capture during 5’ scRNA-seq. An sgRNA containing a standard constant region (top) anneals to a guide-specific RT oligo. Indexing of reverse transcribed cDNA (bottom) occurs after template switch. This strategy is compatible with unmodified sgRNAs (shown) or with sgRNAs with an integrated capture sequence. b) Schematic of sgRNA capture via an integrated capture sequence by 3’ scRNA-seq. A capture sequence within the constant region of the sgRNA (top) anneals to a barcoded, target-specific RT primer. Indexed cDNA (bottom) is produced by reverse transcription. c) Index (GBC or guide) capture rates per cell across experiments conducted with GBC Perturb-seq and direct capture Perturb-seq. Data represent median index UMI counts per cell for cells bearing each of n=32 sgRNAs across platforms. Grey lines indicate median values. “sgRNA-CR1” indicates 5’ capture of standard sgRNAs without a capture sequence. d) Index (GBC or guide) assignment rates across experiments conducted with GBC Perturb-seq and direct capture Perturb-seq. The total number of cells per experiment as well as the fractions of cells assigned no guide, a single guide, or more than one guide are indicated. “sgRNA-CR1” indicates 5’ capture of standard sgRNAs without a capture sequence. e) Clustering of perturbations from UPR Perturb-seq experiments conducted with GBC Perturb-seq and direct capture Perturb-seq. Heatmaps represent Spearman’s rank correlations between pseudo-bulk expression profiles for each of n=32 perturbations. For visual comparison, the rows and columns of all three heatmaps are ordered identically based on the hierarchical clustering of GBC Perturb-seq data. Functional annotations are indicated. f) Hierarchical clustering of UPR-regulated genes based on co-expression in each of the indicated Perturb-seq experiments. Colors indicate membership in different UPR-regulated groups as determined by Adamson et al.4 g) Single-cell projections are based on t-sne visualization of 10 independent components (n=1795 cells for 3’ GBC Perturb-seq, n=1595 cells for 3’ sgRNA-CR1cs1 Perturb-seq, and n=1424 cells for 5’ sgRNA-CR1 Perturb-seq). Colors indicate functional similarities among targeted genes.
To test the performance of guide capture, we next performed 5 parallel CRISPRi-based25,26 screens in K562 cells designed to compare 3’ direct capture and 5’ direct capture to indexing by a polyadenylated barcode transcript (hereafter referred to as guide barcode or GBC Perturb-seq). On each platform, we screened one or more sgRNA libraries containing the same 32 targeting sequences (against 30 genes whose depletion leads to activation of the unfolded protein response, UPR,4 and including 2 non-targeting controls4) (Supplementary Note 2 and Supplementary Table 2). To enable comparison, we prepared each of these libraries using arrayed cloning and lentiviral packaging, and after performing our screens, used custom protocols to amplify index molecules (GBCs or guides) for deep sequencing alongside mRNA sequences (Supplementary Fig. 1g and Supplementary Note 3; see Methods). At a constant sequencing depth, screens using both direct capture platforms gave higher index capture than the GBC-based method (4.1-fold higher for 3’ sgRNA-CR1cs1 capture; 15.5-fold higher for 5’ sgRNA-CR1cs1 capture; 7.8-fold higher for 5’ sgRNA-CR1 capture), with the exception of 3’ capture of sgRNA-CR1cs2, which had modestly lower capture (0.56-fold) (Supplementary Fig. 2a). To assign guide identities to cells, we then fit a two-component Poisson-Gaussian mixture model to the log2-transformed guide UMIs per cell for each guide (Supplementary Fig. 2b; see Methods). This approach aims to separate true guide-expressing cells from “background cells” which arise from spurious cell barcode-sgRNA pairing (potentially due to PCR chimeras or capture of ambient guides). Unlike capture of GBCs, we found that guide capture was sequence-dependent with capture rates varying across guides by targeting sequence (Fig. 1c and Supplementary Fig. 2c,d). Importantly, this variation was correlated across screens and was related to the nucleotides at the 5’ ends of guide RNAs but not to overall GC content (Supplementary Fig. 2e–g). Nevertheless, our assignment procedure robustly assigned guide identities to 84–94% of cells (compared to 89% for GBC Perturb-seq) with roughly expected guide distributions across all platforms (Fig. 1d and Supplementary Fig. 2h). Moreover, indicative of robust assignment, we found strong (and comparable) target depletion across platforms (median knockdown: 90% for GBC capture, 94% for 3’ sgRNA-CR1cs1 capture, 93% for 3’ sgRNA-CR1cs2 capture, 95% for 5’ sgRNA-CR1 capture, 93% for 5’ sgRNA-CR1cs1 capture) (Supplementary Fig. 3a).
We then sought to benchmark the performance of direct capture Perturb-seq for the study of genes and genetic networks. High-content Perturb-seq phenotypes should enable (1) functional clustering of target genes, (2) identification of transcriptional phenotypes caused by individual perturbations, (3) delineation of gene expression regulons, and (4) identification of cell-to-cell heterogeneities. We therefore asked how phenotypes from our direct capture screens with the highest guide assignment rates (3’ capture of sgRNA-CR1cs1 and 5’ capture of standard CR1 sgRNAs) performed on each of these tasks (compared to GBC Perturb-seq). First, we hierarchically clustered target genes based on their pseudo-bulk expression profiles (Fig. 1e). This recapitulated known functional and physical interactions and, when compared to results generated with GBC Perturb-seq, produced highly similar relationships (cophenetic correlation with GBC Perturb-seq: r=0.95 for 3’ sgRNA-CR1cs1; r=0.95 for 5’ sgRNA-CR1). Next, we evaluated transcriptional responses and found good agreement across screens / platforms (for the top 100 differentially expressed genes, r = 0.88 for 3’ sgRNA-CR1cs1 capture compared to GBC Perturb-seq and r=0.87 for 5’ sgRNA-CR1 capture compared to GBC Perturb-seq) with especially high correlations for perturbations that led to differential expression of >100 genes (Supplementary Fig. 3b,c). This result confirms our ability to accurately assign guide identities and suggests that our guide-specific RT primers do not globally alter single-cell gene expression profiles. Next, we tested the utility of direct capture Perturb-seq for the discovery of genetic networks. For this, we relied on our prior empirical classification of genes regulated by the three separate signaling branches of the UPR4. Examining the covariance of these genes across single cells in our current data, we found gene expression modules that were conserved across platforms (cophenetic correlation with GBC Perturb-seq: r=0.93 for 3’ sgRNA-CR1cs1 capture; r=0.95 for 5’ sgRNA-CR1 capture), with modules tending to cluster functionally based on their regulation by the three UPR branches (Fig. 1f). Lastly, we quantitatively evaluated the single-cell performance of our platforms by training a random forest classifier to classify perturbed and unperturbed (control) cells for each targeting guide. Despite the intrinsic noise of scRNA-seq data, prediction accuracies were highly similar across platforms (correlation with GBC Perturb-seq: r=0.91 for 3’ sgRNA-CR1cs1 capture, r=0.90 for 5’ sgRNA-CR1 capture) (Fig. 1g and Supplementary Figure 3d).
To demonstrate the versatility of direct capture Perturb-seq, we next performed a 3’ direct capture Perturb-seq experiment in induced pluripotent stem cells (iPSCs) with Cas927, now using pooled lentiviral packaging and transduction of 40 sgRNAs (Supplementary Table 3). In iPSCs, we again found high guide capture rates (mean capture of 999 UMIs/cell; Supplementary Fig. 4a,b) and transcriptional phenotypes that were correlated for guides targeting the same gene (Supplementary Fig. 4c,d).
Recently, we showed that GBC Perturb-seq can be coupled with epistasis analysis to provide mechanistic insights into how genes interact17. However, GBC Perturb-seq is not scalable. Motivated by this limitation, we next explored the use of direct capture Perturb-seq to study genetic interactions, specifically a complex set of GIs we recently identified between genes that control cholesterol biosynthesis (e.g. FDPS, MVD, and IDI1) and genes that facilitate DNA repair (e.g. ATR and genes encoding components of the 9–1-1 complex)28. For this, we cloned a CRISPRi library of 92 programmed sgRNA pairs targeting 41 genes and 81 gene pairs using a strategy for pooled cloning of dual-guide vectors (Fig. 2a, Supplementary Fig. 5a, and Supplementary Note 4). Notably, we made and tested this library in two configurations, using two combinations of guide constant region sequences (CR3cs1/CR1cs1 and CR2cs2/CR1cs1) (Supplementary Notes 1,4 and Supplementary Table 4). Screening this library in K562s by 3’ direct capture Perturb-seq revealed adequate capture of guides from both vector positions (position A, median of 776 UMIs/cell; position B, median of 511 UMIs/cell; Supplementary Fig. 5b,c), and after mapping these guides to cells, we observed >90% guide assignment with >67% of guide-bearing cells expressing two sgRNAs, as expected given multiple infections, doublets from cell loading, and imperfect guide calling (Fig. 2b; see Methods). Importantly, consistent with similar dual-guide expression systems4,17,28, we also observed comparable knockdown between the two positions in our dual-guide vector. Specifically, for three guides in our library (sgHUS1, sgFDPS, and sgTOPBP1) encoded in both positions paired with a non-targeting guide, we achieved target knockdown of 84% and 84% (position A and position B), 81% and 73% (position A and position B), 70% and 74% (position A and position B), respectively. Lastly, we note that the design of our dual-guide expression system minimizes intramolecular recombination between linked sgRNA sequences by using distinct U6 promoters and sgRNA constant regions, as previously demonstrated by Adamson et al.4; however, it does not prevent paired sgRNA pairs from shuffling due to intermolecular recombination events. Nevertheless, because direct capture allows us to assign sgRNAs to cells in an unbiased way, we were able to identify novel sgRNA pairs and excluded them from downstream analysis.
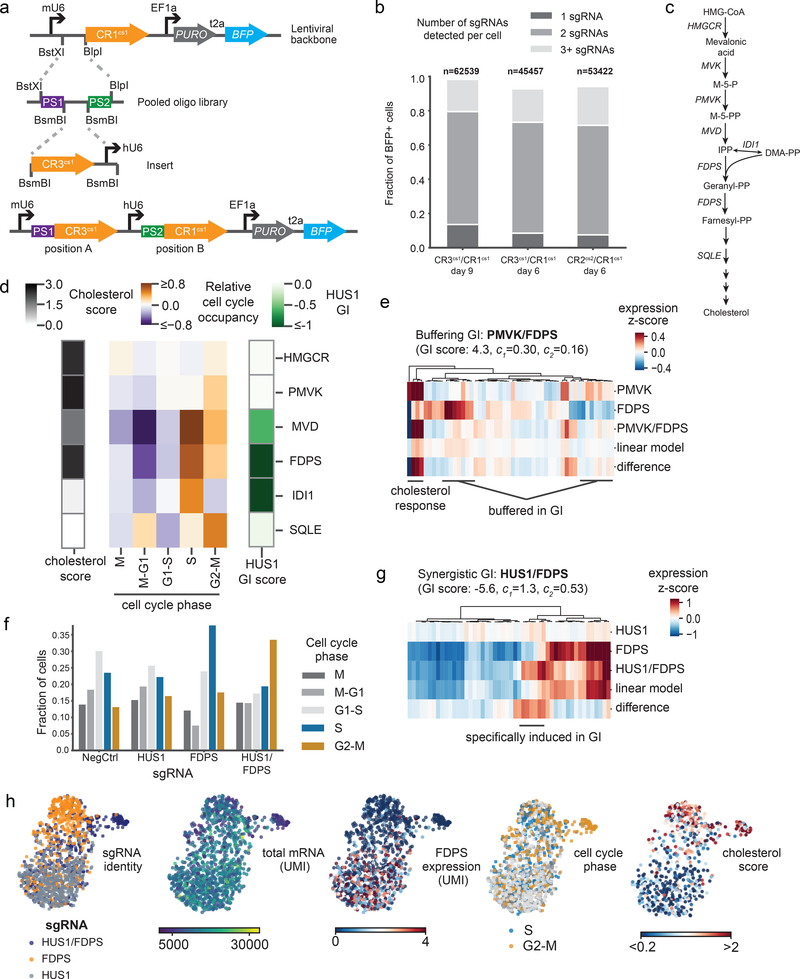
Figure 2: Direct capture Perturb-seq and pooled dual-guide cloning allows systematic dissection of genetic interactions between cholesterol biosynthesis and DNA repair genes.
a) Schematic of programmed dual-guide library cloning strategy. Paired sgRNA targeting regions are synthesized on a single oligo and cloned into a direct capture Perturb-seq vector by ligation. Then, an sgRNA constant region and hU6 promoter are inserted between the sgRNA targeting regions to generate a dual-guide array in a lentiviral backbone. This example shows a CR3cs1/CR1cs1 library design. b) Guide assignment rates for dual-guide direct capture Perturb-seq experiments. The fraction of cells carrying sgRNAs (marked by BFP) varied due to strong CRISPRi growth defects; the total number of cells were therefore first scaled by BFP positivity. The total number of cells and fraction of cells assigned a single guide, two guides, or more than two guides are indicated. c) Schematic of the cholesterol biosynthesis pathway. d) Heatmap of cell cycle and cholesterol phenotypes for cells with depletion of enzymes in the cholesterol biosynthesis pathway. Cell cycle occupancy for each perturbation depicted indicates the relative enrichment or depletion of cells in each phase relative to unperturbed cells. The cholesterol score is the mean z-scored expression of enzymes in the cholesterol biosynthesis pathway. The “HUS1 GI” is a metric of the growth defect caused by an genetic perturbations paired with HUS1 knockdown relative to the genetic perturbation alone as determined by Horlbeck et al.28 All genes were significantly depleted by CRISPRi (percent knockdown: HMGCR 94%; PMVK 92%; MVD 83%; FDPS 78%; IDI1 82%; SQLE 84%). Number of cells per perturbation: non-targeting control n=527, HMGCR n=608, PMVK n=389, MVD n=184, FDPS n=439, IDI1 n=131, SQLE n=255. e) Heatmap of gene expression for the 50 most differentially expressed genes between cells carrying each indicated perturbation. Expression values are the z-scored expression relative to unperturbed cells (n=389 PMVK cells, n=1921 FDPS cells, and n=517 PMVK/FDPS cells). Cells were combined to generate the expression signatures. Knockdown was consistent between single-gene and dual-gene targeting (FDPS knockdown 73% alone vs. 82% paired; PMVK knockdown 92% alone vs. 86% paired). The indicated GI score was previously determined by Horlbeck et al.28, where GI scores >3 are considered strongly buffering interactions. f) Fraction of cells in each cell cycle phase across cells with the indicated perturbations. Number of cells per perturbation: non-targeting control n=780, HUS1 n=905, FDPS n=439, HUS1/FDPS n=831. g) Heatmap of gene expression for the 50 most differentially expressed genes between cells carrying each indicated perturbation. Expression values are the z-scored expression relative to unperturbed cells (n=905 HUS1 cells, n=439 FDPS cells, and n=831 HUS1/FDPS cells). Cells were combined to generate the expression signatures. Knockdown was consistent between single-gene and dual-gene targeting (FDPS knockdown 78% alone vs. 72% paired; HUS1 knockdown 95% alone vs. 85% paired) The indicated GI score was previously determined by Horlbeck et al.28, where GI scores <−3 are considered strongly synergistic interactions. h) Single-cell UMAP projections with informative cell features highlighted (n=2175 cells).
We previously proposed a model for GIs between cholesterol biosynthesis and DNA repair genes wherein repression of the former leads to the buildup of toxic metabolic intermediates in cells, which then cause replicative stress and genotoxin-activated cell cycle arrest28. This model emerged from a set of low-content and low-throughout biochemical and functional experiments that primarily investigated the relationship between two genes: FDPS and a key mediator of the replication checkpoint machinery, HUS1. By contrast, direct capture Perturb-seq allows simultaneous interrogation of many single and double genetic perturbations with information rich phenotypes. The method therefore enabled us to thoroughly examine how cells respond to depletion of several enzymes in the cholesterol biosynthesis pathway (Fig. 2c). From this, we made three clear observations. First, as expected, perturbation of early pathway steps led to feedback marked by upregulation of cholesterol biosynthesis genes (Fig. 2d). Second, repression of intermediate pathway genes (FDPS, MVD, and IDI1), which are synthetic lethal with DNA repair genes, led to an accumulation of cells in S-phase of the cell cycle (Fig. 2d). Notably, the difference in phenotypes among genes within this linear biosynthetic pathway supports the idea that buildup of toxic intermediates, rather than depletion of cholesterol itself, leads to S-phase arrest. Lastly, as predicted by our model, we observed a buffering relationship between genes that regulate the early and intermediate steps in cholesterol biosynthesis. Specifically, when we fit a regression model to decompose dual-gene perturbations into linear combinations of single-gene effects, we observed that PMVK repression suppressed the FDPS-specific transcriptional response, while maintaining the cholesterol feedback response shared by both perturbations (Fig. 2e). This further suggests that S-phase arrest is caused by loss of the enzymatic activity of the intermediate genes, not from a loss of cholesterol itself.
While repression of HUS1 causes modest cell-cycle aberration alone, in combination with FDPS knockdown, we observed a substantial bypass of the S-phase checkpoint and an accumulation of cells in G2/M (Fig. 2f, Supplementary Fig. 5d). Additionally, we found that these cells (with perturbation of both HUS1 and FDPS) demonstrate a neomorphic phenotype characterized by a transcriptional response not induced by either perturbation alone (Fig. 2g). At the single-cell level, this generated a population of G2/M-arrested cells with notably decreased total mRNA content, likely representing dying cells (Fig. 2h). Based on this, we propose an updated model where synthetic lethality in these cells is caused specifically by failure to detect replication stress in HUS1-depleted cells, resulting in inappropriate cell cycle progression and mitotic catastrophe from unresolved damage. Broadly, this example highlights the power of high-resolution, single-cell phenotypes for the mechanistic dissection of GIs and demonstrates how direct capture Perturb-seq can be used to understand GIs in a comprehensive, unbiased fashion without the need for specific hypotheses.
To further enable single-cell CRISPR screening efforts, we next tested the idea that genetic perturbation libraries that co-deliver multiple sgRNAs per gene to the same cell could increase screening efficiency by requiring fewer constructs per gene. To test this, we selected pairs of CRISPRi and CRISPR activation (CRISPRa) sgRNAs with high predicted activity29 against individual genes that span a range of biological functions and expression levels (87 for CRISPRi; 49 for CRISPRa; Supplementary Table 5 and Supplementary Table 6). Then, using our pooled library cloning strategy and direct capture Perturb-seq in K562 cells, we compared the activity (knockdown or activation) of the guide pairs expressed from a dual-guide vector to single sgRNAs (expressed from the same dual-guide vector paired with non-targeting control sgRNAs). For both CRISPRi and CRISPRa, the multiplexed sgRNAs nearly doubled CRISPR activity over what was achieved with the best single guide (CRISPRi: sgRNAs 1+control, median relative target expression=0.20; sgRNAs 1+2, median relative target expression=0.11; Wilcoxon signed-rank two-sided test n=87 genes, W=378, p=8e-11; CRISPRa: sgRNAs 1+control, median fold-activation=2.9; sgRNAs 1+2, median fold-activation=4.7; Wilcoxon signed-rank two-sided test n=49 genes, W=162, p=7e-6; Fig. 3a,b and Supplementary Fig. 6a,b). Moreover, in both cases, the multiplexed sgRNAs appeared to perform better than expected based on a dominant model of guide activity, suggesting some degree of synergy between multiplexed sgRNAs (CRISPRi Wilcoxon signed-rank two-sided test: n=87 genes, W=698, p=3e-7; CRISPRa Wilcoxon signed-rank two-sided test: n=49 genes, W=233, p=0.0002; Supplementary Fig. 6c,d,e,f) which is consistent with previous reports30,31. These results show that compact, highly active libraries (expressing multiple sgRNAs per gene) can be used to scale single-cell experiments with direct capture Perturb-seq, interrogating more genes while minimizing false negatives due to insufficient expression modulation.
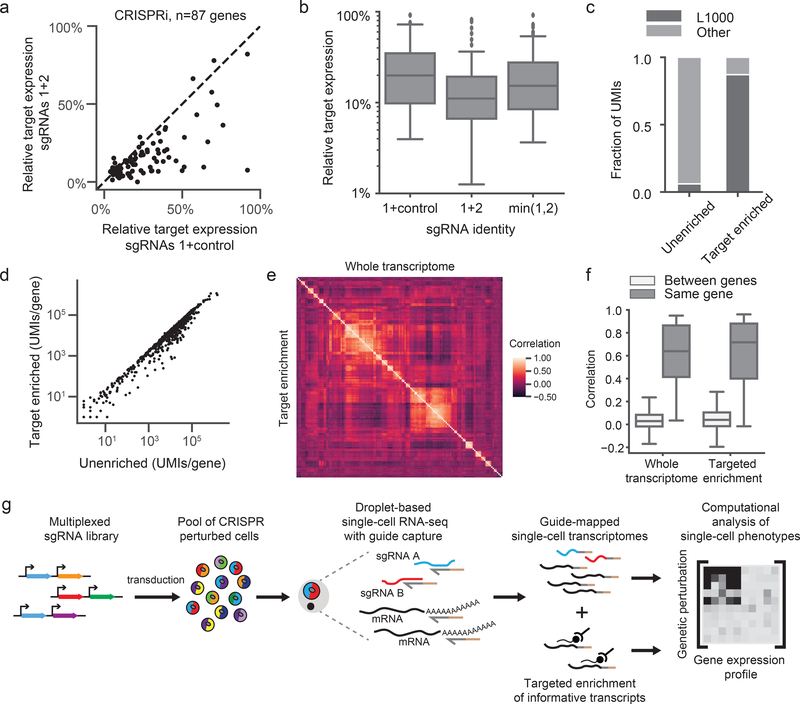
Figure 3: Multiplexed CRISPRi/CRISPRa and hybridization-based target enrichment enable scalable and versatile single-cell CRISPR screens.
a) Scatterplot of the relative target expression per gene comparing CRISPRi knockdown with a single sgRNA (expressed from a dual-guide vector paired with a non-targeting control) versus multiplexed sgRNAs. Multiplexed sgRNAs significantly improve knockdown (sgRNAs 1+control median relative target expression=0.20; sgRNAs 1+2 median relative target expression=0.11; Wilcoxon signed-rank two-sided test n=87 genes, W=378, p=8e-11). sgRNA 1, best predicted sgRNA for each gene. sgRNA 2, second best predicted sgRNA for each gene. b) Box plots of the relative target expression per gene in the multiplexed CRISPRi experiment denoting quartile ranges (box), median (center mark), and 1.5 × interquartile range (whiskers). “min(1,2)” indicates the minimum remaining target expression between sgRNA 1 (paired with negative control) and sgRNA 2 (paired with negative control), ie. the predicted multiplexed sgRNA knockdown based on a dominant model of knockdown. The multiplexed sgRNAs performed better than the dominant model (Wilcoxon signed-rank two-sided test n=87 genes, W=698, p=3e-7). c) The fraction of total UMIs for L1000 genes (n=978) versus other genes, before and after target enrichment. d) Scatterplot of the total number of UMIs for each gene, before and after target enrichment (n=978 genes). The Pearson correlation of log10 normalized UMIs is r=0.98. e) Heatmap depicts clustering of guides in our multiplexed CRISPRi experiment. Heatmap represents Spearman’s rank correlations between pseudo-bulk expression profiles of well-expressed genes (>1 UMI/cell). Data from all perturbations with >10 differentially expressed genes compared to controls are included (n=145 genes). The upper triangle (correlation matrix) was calculated on the whole transcriptome while the lower triangle (correlation matrix) was calculated on the target-enriched transcriptome. Both triangles were identically ordered based on hierarchical clustering of the whole transcriptome correlation matrix. f) Pearson correlations of pseudo-bulk differential expression profiles of well-expressed genes (>1 UMI/cell) caused by sgRNAs targeting the same gene (for n=39 genes whose knockdown led to differential gene expression) versus sgRNAs targeting different genes (n=111592 pairs). sgRNAs targeting the same gene had significantly more similar profiles than sgRNAs targeting different genes, both before and after target enrichment (unenriched median r=0.64, Mann-Whitney U two-sided test U=117224, p=1.4e-24; enriched median r=0.72, Mann-Whitney U two-sided test U=259898, p=1.7e-21). Box plots denote quartile ranges (box), median (center mark), and 1.5 × interquartile range (whiskers). g) Schematic overview of direct capture Perturb-seq workflow.
Lastly, we addressed the fact that current droplet-based scRNA-seq implementations are constrained by sequencing the whole transcriptome for phenotyping, which can be prohibitively expensive. This requirement is compounded by the fact that the distribution of gene expression is skewed (i.e. 2% of expressed genes consume >50% of sequencing reads; Supplementary Fig. 7a) and biased across gene functions. Indeed, genes with important biological functions (e.g., transcription factors, cell-surface receptors, kinases) are often lowly-expressed and difficult to measure (Supplementary Fig. 7b). However, given a suitable method for targeted enrichment of scRNA-seq libraries, many transcriptional states could be faithfully inferred from a subset of gene expression measurements32,33. Diverse approaches exist for enriching transcripts using multiplexed PCR34, custom RT beads35, or linear amplification36; however, each of these is limited by number of target genes, quality, and/or a priori gene selection. Instead, we hypothesized that we could use hybridization-based target enrichment to specifically sequence thousands of select transcripts, thereby limiting sequencing while maintaining high-content phenotypes.
To test target enrichment, we therefore empirically-designed hybridization baits for 978 genes, the L1000 landmark genes33 (Supplementary Note 5 and Supplementary Table 7). We chose these genes because they can serve as a reduced representation of the whole transcriptome and their expression levels span four orders of magnitude, providing ample range to examine potential biases introduced by hybridization capture. In our test, we performed a pulldown on 3’ scRNA-seq library and deeply sequenced recovered molecules. Hybridization capture increased the percentage of mRNA molecules aligning to target genes by >14-fold, from 6% in an unenriched control to 87% after target enrichment (Fig. 3c). Thus, at only ~0.1x sequencing depth of the original library, the enriched library contains more UMIs per cell for most targeted genes (Supplementary Fig. 7c). Enriched gene expression profiles were highly correlated with unenriched profiles at the global (r=0.98), single-cell (median r=0.93), and single-gene (median r=0.75) levels (Fig. 3d, Supplementary Fig. 7d,e,f, and Supplementary Note 6), and perturbation-dependent differential gene expression was highly similar before and after enrichment (median r=0.71; Supplementary Fig. 7g). Given these results, we next tested the ability of our reduced transcriptome subset to functionally cluster genes. Hybridization capture on our multiplexed CRISPRi Perturb-seq libraries revealed that L1000-targeted gene expression profiles can recapitulate relationships between genetic perturbations established by sequencing the entire transcriptome (cophenetic correlation r=0.95; Fig. 3e,f and Supplementary Fig. 7h). Altogether, these results demonstrate that hybridization capture is a simple and sensitive procedure for informative enrichment of tailored gene sets from scRNA-seq libraries. Moreover, because our target enrichment procedure is performed on final libraries, target genes do not need to be selected a priori and can be iteratively refined for a single experiment. This technology motivates the future optimization of gene sets that maximize biological information while minimizing sequencing requirements (analogous to the L1000 landmark genes for hybridization-based fluorescent assays33).
Since its inception, single-cell CRISPR screening has made it possible to simultaneously examine high-dimensional genotypic and phenotypic landscapes. Here, we described an improved Perturb-seq approach that substantially expands the scale and flexibility of this technology (Fig. 3g and Supplementary Table 8). Importantly, our 5’ and 3’ direct capture Perturb-seq platforms (now commercially available from 10x Genomics) have crucial advantages under different circumstances. For example, 5’ direct capture is compatible with standard sgRNAs, has higher guide capture rates, and allows for V(D)J clonotype analysis, whereas 3’ direct capture is compatible with many molecular recording and lineage tracing approaches37. We specifically demonstrated the value of direct capture for the mechanistic dissection of GIs, which is a laborious undertaking with other methods, and for generating compact, highly-active CRISPR libraries. Additionally, to decrease the cost of Perturb-seq experiments, we implemented hybridization-based target enrichment. With our target enrichment strategy, biologically meaningful gene panels (e.g., immune, developmental, metabolic, tumor suppressors/oncogenes, etc.) can be probed without unnecessary sequencing of housekeeping genes. Taken together, direct capture Perturb-seq and target enrichment greatly expand the accessibility, scalability, and flexibility of single-cell CRISPR screens.
Online Methods
Cell culture and viral production
RPMI-1640 with 25mM HEPES, 2.0 g/L NaHCO3, 0.3 g/L L-Glutamine supplemented with 10% FBS, 2 mM glutamine, 100 units/mL penicillin and 100 μg/mL streptomycin was used to grow K562 cells. HEK293T cells, used for packaging lentivirus, were grown in Dulbecco’s modified eagle medium (DMEM) in 10% FBS, 100 units/mL penicillin and 100 μg/mL streptomycin. Induced pluripotent stem cells (iPSCs) expressing Cas9 (WTC CRISPRn Gen1C27) were maintained under feeder-free conditions on growth factor-reduced Matrigel (Corning) in mTeSR medium (STEMCELL Technologies). Accutase (STEMCELL Technologies) was used to enzymatically dissociate iPSCs into single cells to passage with 10 μM p160-Rho-associated coiled-coil kinase (ROCK) inhibitor Y-27632 (Selleckchem) added to promote cell survival. Lentivirus was produced by co-transfecting HEK293T cells with transfer plasmids and standard packaging vectors using TransIT®-LTI Transfection Reagent (Mirus, MIR 2306).
Plasmid construction and development of sgRNA capture sequences
Direct capture guide RNAs were designed by appending non-random capture sequences to the 3’ end of standard guide sequences or by inserting these sequences into the loop region of the so-called “stem loop 2” (Supplementary Fig. 1c and Supplementary Table 1). Expression vectors encoding these guides are available at Addgene. To test the activity of modified guides and guide expression vectors, expression constructs carrying GFP-targeting guides with variant constant regions were transduced into GFP+ K562 dCas9-KRAB cells4 with centrifugation (2 hours at 1000 x g at 33°C). Cells were analyzed by flow cytometry on an LSR II flow cytometer (BD Biosciences). Data in Supplementary Figure 1d were processed as follows: Measurements of median GFP were recorded from GFP+ K562 dCas9-KRAB cells transduced with the indicated guides. These measurements were adjusted by subtracting background fluorescence (collected from control cells that do not express GFP) and then divided by measurements of median GFP (also background-subtracted) recorded from cells without a GFP-targeting guide (untransduced cells grown in the same wells). These “GFP remaining” ratios were then normalized to those derived from cells transduced with a positive control guide, our standard sgRNA-CR1 (on plate control) and are reported as averages of triplicates from separate infections. Data in Supplementary Figure 5a are shown as the Gaussian kernel density estimates of normalized flow-cytometry measurements representing GFP expression of all cells with the indicated guide RNAs. We chose final guide designs based on these GFP depletion results (Supplementary Note 1). The reverse complements of our final capture sequences (cs1 5’-GCTTTAAGGCCGGTCCTAGCAA-3’ and cs2 5’-GCTCACCTATTAGCGGCTAAGG-3’) were incorporated into gel beads in the Chromium Single Cell 3ʹ Reagent Kits v3 with Feature Barcoding technology.
Pilot UPR direct capture Perturb-seq
For these experiments, we constructed three CRISPRi libraries (the UPR GBC, UPR sgRNA-CR1cs1, and UPR sgRNA-CR1cs2 libraries) by arrayed cloning (Supplementary Note 2 and Supplementary Table 2). Each of these libraries encodes guide RNAs programmed with 32 unique guide RNA targeting regions: 30 which target genes whose depletion was previously shown to activate the unfolded protein response (UPR) by GBC Perturb-seq4 and 2 non-targeting controls (sgNegCtrl2 and sgNegCtrl3). Our sequence-verified libraries were then packaged into lentiviruses by arrayed transfection of individual vectors, and lentiviral preparations from each library were pooled for co-transduction into K562 dCas9-KRAB cells25 (spinfected 2 hours at 1000 x g at 33 C). To ensure representation of guides at the time of scRNA-seq (7 days after transduction), lentiviral pooling was performed in a manner that accounted for both packaging variability and guide effects on cell growth after transduction (as determined by individual test infections). Pooling ratios were designed to ensure even representation among targeting guides and delivery of sgNegCtrl2 and sgNegCtrl3 at 4-fold excess. Three days post infection, we measured BFP expression (a marker for guide transduction) on an LSR II flow cytometer (BD Biosciences) and calculated the multiplicity of infection (MOI) for each library (0.04 for the GBC library, 0.05 for the sgRNA-CR1cs1 library, and 0.05 for the sgRNA-CR1cs2 library). Transduced cells were sorted to near purity (FACSAria2, BD Biosciences). Up to this point, cell viability for all three libraries remained >87%.
Seven days post infection, cells were separated into droplet emulsions using the Chromium Controller (10x Genomics) across 5 lanes (cell pools >94% BFP+). Cells transduced with the UPR GBC library were loaded on two lanes. On one lane, cell capture was performed with Chromium Single Cell 3’ Gel Beads v2 (GBC Perturb-seq), while on the other, cell capture was performed with Chromium Single Cell 5’ Gel Beads and a spike-in of 5 pmols of oJR160 (5’-AAGCAGTGGTATCAACGCAGAGTACCAAGTTGATAACGGACTAGCC-3’) to the RT Master Mix (5’ direct capture Perturb-seq using standard CR1 sgRNAs). Similarly, cells transduced with the UPR sgRNA-CR1cs1 library were loaded onto two lanes. On one lane, cell capture was performed with Chromium Single Cell 3’ Gel Beads v3 (GBC Perturb-seq), while on the other, cell capture was performed with Chromium Single Cell 5’ Gel Beads and a 5 pmol spike-in of oJR161 (5’- AAGCAGTGGTATCAACGCAGAGTACTTGCTAGGACCGGCCTTAAAGC-3’) to the RT Master Mix (5’ direct capture Perturb-seq using sgRNA-CR1cs1). Finally, cells transduced with UPR sgRNA-CR1cs2 were loaded onto a single lane with Chromium Single Cell 3’ Gel Beads v3 (3’ direct capture Perturb-seq using sgRNA-CR1cs2). For all lanes, cells were loaded to recover ~10,000 cells (~260 cells per guide). Approximately 100 pmols of 10x RT oligo (poly-dT RT Primer PN 2000007) are added to each 10x RT reaction based on quantification by NanoDrop spectrophotometer (Thermo Scientific). Therefore, for 5’ direct capture Perturb-seq, we chose to add our guide capture oligos at ~5%. The recovered cells and subsamples thereof are analyzed in Figure 1c–g and Supplementary Figures 2 and 3.
iPSC 3’ direct capture Perturb-seq
To test direct capture Perturb-seq with iPSC cells, we constructed a sequence-verified library of 40 guides using the sgRNA-CR1cs1 design by arrayed cloning (Supplementary Note 2 and Supplementary Table 3). This “iPSC sgRNA-CR1cs1” library was then packaged into lentivirus (pooled format), and transduced into iPSCs carrying inducible Cas929 at an MOI of 10%. iPSCs were treated daily with 2 μM doxycycline (Sigma) to drive Cas9 expression, and after two days, BFP+ cells were enriched on a BD FACS Aria2 (BFP is a marker of guide vector transduction). Seven days post-infection, cells were separated into droplet emulsions using the Chromium Controller (10x Genomics) with Chromium Single Cell 3’ Gel Beads v3. The recovered cells and subsamples thereof are analyzed in Supplementary Figure 4.
Dual-guide 3’ direct capture Perturb-seq to evaluate genetic interactions
To dissect the interaction between cholesterol biosynthesis and DNA repair, we constructed two sequence-verified dual-guide libraries of manually curated guide pairs by pooled cloning (CR3cs1/CR1cs1 and CR2cs2/CR1cs1; Supplementary Note 4 and Supplementary Table 4). For each library, lentivirus was prepared in a pooled format and transduced into K562 dCas9-KRAB cells (spinfection for 2 hours at 1000 x g). Three days post infection, we calculated MOIs (using BFP expression) and sorted transduced cells to near purity (LSR II and FACSAria2, BD Biosciences). To maximize the probability of observing interpretable transcriptional responses, we sampled cells at two time points (day 6 and day 9 post-transduction). At 6 days post infection, we separated cells transduced with CR3cs1/ CR1cs1 (MOI=0.15, 89% BFP+) and CR2cs2/ CR1cs1 (MOI=0.18, 93% BFP+) libraries into droplet emulsions using the Chromium Controller with Chromium Single Cell 3’ Gel Beads v3. At 9 days post infection, we did the same for a second population of cells transduced with the CR3cs1/ CR1cs1 library (MOI=0.1, 83% BFP+), both times aiming to recover 15,000 cells per lane.
Multiplexed CRISPRi and CRISPRa 3’ direct capture Perturb-seq
To determine whether guide multiplexing can be used to construct compact CRISPRi and CRISPRa libraries, we built and analyzed dual-guide libraries wherein vectors contain either one targeting guide (paired with a negative control guide) or two guides targeting a single gene (Supplementary Note 4). For these libraries, we manually chose gene targets representing a broad range of biological functions and expression levels (87 for CRISPRi, 49 for CRISPRa). We selected guide RNA targeting sequences predicted to be highly active (the top two by rank in hCRISPRi v2.1 and hCRISPRa v230) (Supplementary Tables 5, 6). In this manuscript, we refer to guides containing the top ranked targeting sequence as “sgRNA 1” and the next best as “sgRNA 2”. Of note, when the selected targeting sequence pairs targeted genomic sequence <80 bp apart, we also included the next best-ranked guide RNA spaced >80 bp away from the first. We refer to guides containing these targeting sequences as “sgRNA 3”. Additionally, for genes with two annotated transcription start sites (TSSs), we paired the top sgRNAs targeting each TSS. We cloned these libraries in pooled format (Supplementary Note 3). We then packaged each library into lentivirus (pooled format) and transduced K562 dCas9-KRAB cells25 (CRISPRi) and K562 dCas9-SunTag/scFV-VP64 cells25 (CRISPRa) with the appropriate library. Three days post infection, we calculated MOIs (by BFP expression) of 0.1 and 0.045, respectively, and sorted transduced cells to near purity (LSR II and FACSAria2, BD Biosciences). Then, 8 days post infection, we separated cells (CRISPRi at 90% BFP+ and CRISPRa at 88% BFP+) into droplet emulsions using the Chromium Controller with Chromium Single Cell 3’ Gel Beads v3, aiming to recover 15,000 cells per lane.
Sequencing library preparation
GBC Perturb-seq sequencing libraries were prepared according to the Chromium Single Cell 3’ Reagent Kits v2 User Guide (10x Genomics CG00052) with 11 cycles of PCR during cDNA amplification and 11 cycles of Sample Index PCR. Library molecules containing guide barcodes (GBCs) were specifically amplified using KAPA HiFi ReadyMix with 30 ng of the final library as template, 0.6 mM 052-P5 (5’-AATGATACGGCGACCACCGAGATCTACAC-3’), and 0.6 mM of i7 barcoded 055-N708 (5’-CAAGCAGAAGACGGCATACGAGATCCTCTCTGGTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACCTCCCTAGCAAACTGGGGCACAAG-3’). PCR cycling was performed according to the following protocol: (1) 95 C for 3 min, (2) 14 cycles of 98 C for 15 s, then 70 C for 10 s, (3) 72 C for 1 min. The resulting GBC sequencing library was purified via a 0.8X SPRI selection.
Our 3’ direct capture Perturb-seq libraries were prepared using a protocol modified from the Chromium Single Cell 3’ Reagent Kits v3 User Guide (10x Genomics, CG000184) (Supplementary Fig. 1b). This protocol can now be found at https://support.10xgenomics.com/single-cell-gene-expression/library-prep/doc/user-guide-chromium-single-cell-3-reagent-kits-user-guide-v3-chemistry-with-feature-barcoding-technology-for-crispr-screening. Briefly, following 11 cycles of cDNA amplification, library amplicons were size separated into two fractions (Supplementary Fig. 1g): one enriched for amplicons containing guide sequences (by performing a 0.6X-1.2X double-sided SPRI), and the other (eluted from the 0.6X left-sided SPRI) containing larger cDNA amplicons. We processed the latter into gene expression sequencing libraries according to the Chromium Single Cell 3’ Reagent Kits v3 User Guide (in this case, using 10 cycles of Sample Index PCR). In parallel, we used the guide-enriched cDNA amplicons to make perturbation index sequencing libraries. For this, guide-enriched cDNAs amplicons were purified by an additional 1X SPRI selection (30 uL elution). The eluted material (5 uL) was then used as template in the following nested PCR strategy: PCR1 with 50 uL Amp Mix (10x Genomics, PN#2000047), 45 uL Feature SI Primers 1 (10x Genomics, PN#2000098), and cycling by (1) 98 C for 45 s, (2) 12 cycles of 98 C for 20 s, then 60 C for 5 s, then 72 for 5 s, (3) 72 C for 1 min. PCR2 with the products of the first PCRs (5 uL after cleanup using a 1X SPRI selection and elution in 30 uL), 50 uL of Amp Mix (10x Genomics, PN#2000047), 35 uL of Feature SI Primers 2 (10x Genomics, PN#2000098), and cycling by (1) 98 C for 45 s, (2) 5 cycles of 98 C for 20 s, then 54 C for 30 s, then 72 for 20 s, (3) 72 C for 1 min. Finally, the resulting guide sequencing libraries were cleaned up via a double-sided 0.7X-1.0X SPRI selection.
Our 5’ direct capture Perturb-seq sequencing libraries were prepared using a protocol modified from the Chromium Single Cell V(D)J Reagent Kits User Guide (10x Genomics CG000086) (Supplementary Fig. 1g and Supplementary Note 3). For this, we used two direct capture spike-in oligos oJR160 and oJR161, each with an adapter identical to the adapter sequence contained on the Poly-dT RT Primer from 10x Genomics (PN-2000007). This adapter serves as a primer binding site for the Non-Poly(dT) primer (10x Genomics, PN-220106) during cDNA amplification, and thus allows amplification of reverse transcribed guides to occur concurrently with standard cDNA amplification. Following 11 cycles of amplification, cDNAs amplicons were size separated into two fractions as described immediately above and in Supplementary Note 3 (step 3). Following this, the fractions were processed into gene expression libraries (according to the Chromium Single Cell V(D)J Reagent Kits User Guide with 14 cycles of Sample Index PCR) and index sequencing libraries (as described in Supplementary Note 3, steps 4 and 5). For our 5’ sgRNA-CR1cs1 experiment, guide molecules were amplified using 0.6 mM oJR163 and oJR166 (5’- CAAGCAGAAGACGGCATACGAGATCATGCCTAGTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGTACTTGCTAGGACCGGCCTTAAAGC-3’). Because the resulting index library had a contaminating low-molecular weight species (suspected primer dimers) an additional selection for 248–302 bp fragments was performed using a BluePippin (Sage Science) prior to sequencing.
Hybridization-based target enrichment
To enrich select transcripts from single-cell gene expression libraries for deep sequencing (so-called “target enrichment”), we developed a hybridization capture protocol. Briefly, using 120 nt biotinylated oligos, which we generated according to specific design criteria outlined in Supplementary Note 5, we performed streptavidin pulldowns of target sequencing amplicons from indexed 10x Genomics gene expression libraries. We tested our enrichment approach in two scenarios: (1) a single library from our dual-guide 3’ direct capture Perturb-seq experiment to evaluate genetic interactions, and (2) all 16 libraries (in two pools of 8) from our multiplexed CRISPRi 3’ direct capture Perturb-seq experiment. Using the former, we compared deep sequencing of both the unenriched and enriched libraries, and using the latter, we tested the functional utility of L1000 transcriptomes. For pooling libraries, we mixed 187.5 ng from each to obtain a total mass of 1500 ng and dried the library with a SpeedVac. We then followed steps 4 to 7 of the published Twist Biosciences protocol for hybrid capture available at: https://www.twistbioscience.com/sites/default/files/resources/2019–01/DOC-001031%20Twist%20Protocol%20Custom%20Panels%20REV%201.0.pdf. This protocol consists of a 16-hour probe hybridization at 70°C, pulldown of hybridized probes using streptavidin beads, and 5-cycles of post-capture PCR prior to sequencing.
Sequencing
For our UPR direct capture Perturb-seq experiments, both the gene expression and index sequencing libraries were sequenced using a NovaSeq 6000 S2 Reagent kit (Illumina) and a custom sequencing strategy (26 bp Read 1, 125 bp Read 2, and 8 bp Index Read 1) where the extended Read 2 was used to sequence guide RNA targeting regions in our 5’ guide sequencing libraries. For all other libraries, we sequenced using the standard format for scRNA-seq from 10x Genomics (28 bp Read 1, 98 bp Read 2, and 8 bp Index Read 1) on a NovaSeq 6000 System (Illumina) with NovaSeq 6000 S4 Reagent kits (Illumina).
Data processing, statistics, and analysis
We used Cell Ranger 3.0 software (10x Genomics) for alignment of scRNA-seq reads, collapsing reads to unique molecular identifier (UMI) counts, cell calling, and depth normalization of transcriptome libraries. Index reads were aligned to expected sequences using bowtie for GBC Perturb-seq and bowtie2 for direct capture Perturb-seq. We observed index alignment rates of 0.82 for GBCs, 0.35 for 3’ sgRNA-CR1cs2, 0.62 for 3’ sgRNA-CR1cs1, 0.71 for 5’ sgRNA-CR1, and 0.62 for 5’ sgRNA-CR1cs1 in our UPR direct capture Perturb-seq experiments. Downstream analyses were performed in Python, using a combination of Numpy, Scipy, Pandas, scikit-learn, pomegranate, polo, and seaborn libraries.
Tests for differences in distributions (for example, of capture rates or correlations of guides within and between gene targets) were conducted with a two-sided Mann-Whitney U test (scipy.stats mannwhitneyu with use_continuity=True, alternative=‘two-sided’). Tests for differences in distributions for paired samples (for example, knockdown by single versus multiplexed guides) were carried out with a two-sided Wilcoxon signed rank test (scipy.stats wilcoxon with zero_method=‘wilcox’, correction=False, alternative=‘two-sided’). Tests for differential gene expression were performed with a two-sample, two-sided Kolmogorov–Smirnov test and corrected for multiple-hypothesis testing at an FDR of 0.01 using the Benjamini–Yekutieli procedure. As indicated in the text, differentially expressed genes were also identified by random forest classifiers (scikit-learn extremely randomized trees with 1000 trees in the forest to predict perturbation status). The advantage of this approach is that we assess the similarity of average expression profiles across platforms regardless of the strength of the perturbation because we do not employ a strict cutoff. Correlations reported are Pearson correlation coefficients unless otherwise indicated. Sample sizes used to calculate statistics are provided in the figures and legends. Additional information is available in the Nature Research Life Sciences Reporting Summary linked to this article, and details specific to individual data analyses can be found in Supplementary Note 6.
Perturbation identity mapping
Within our sequencing data, we found evidence of perturbation index reads containing spurious cell barcode (CBC) / index pairs. We attribute these to the droplet encapsulation of ambient indexes (GBC transcripts or guides) and PCR chimeras. Therefore, to accurately assign guide identities to cells, true CBC/index pairs had to be determined. For GBC Perturb-seq, we did this using a threshold that separates the bimodal distribution of GBC coverage (reads per UMI) as previously described4. However, for direct capture Perturb-seq, we found that coverage distributions were not bimodal, at least not at the downsampled sequencing depth we used to compare libraries in our UPR experiments (25 million aligned indexing reads per GBC or guide sequencing library). At this sequencing depth, saturation of the index libraries is 0.75 for GBC, 0.96 for 3’ sgRNA-CR1cs2, 0.71 for 3’ sgRNA-CR1cs1, 0.28 for 5’ sgRNA-CR1, and 0.60 for 5’ sgRNA-CR1cs1. Instead, we found that each guide had a bimodal distribution of the number of UMIs per CBC (capture rates) and that these rates vary across guide RNA targeting regions (perhaps influenced by targeting region-dependent variability in guide stability, Cas9 binding, and RT efficiency) (Supplementary Fig. 2e,f).
Given targeting region-variable capture rates, to assign guide identities to cells, we fit a two-component mixture model, consisting of a Poisson (lower) and Gaussian (upper) distribution, to the log2 transformed capture rates (UMIs per CBC) for each guide RNA targeting region, as exemplified in Supplementary Figure 2b. These mixture models enabled us to separate the upper modes (representing transduced cells) from the lower modes (representing background) and thus assign guides to cells. Each cell with a posterior probability >0.5 of belonging to an upper mode component was assigned a given guide identity. This procedure produced a coherent proportion of cells assigned to each guide identity (Supplementary Fig. 2h) and a coherent multiplet rate across platforms–within 1–1.6 fold of expectations based on library transduction (assuming Poisson infection distribution) and published multiple encapsulation rates (Fig. 1d).
In our UPR experiments, only cells with a single assigned guide were considered for downstream analysis; however, for dual-guide experiments, cells with two assigned guides were used. Across all dual-guide direct capture Perturb-seq experiments, we observed that >67% of cells contained exactly two sgRNAs. We attribute many of the cells with less than two assigned guides to stringent guide assignment cutoffs. For example, given ~90% assignment rate in single-guide experiments (on par with GBC Perturb-seq and CROP-seq), we expect only ~81% of dual-guide cells to be assigned exactly 2 guides. Yet our mapping strategy is clearly overly conservative as it assumes that guides are independently paired. To increase assignments rates in future applications, our mapping framework could be extended to fit a multivariate mixture model that jointly calls guides by leveraging shared information. Cells with more than two guides, on the other hand, may arise from either multiple infection events or double loading into droplets. Our loading scheme (designed to recover ~15,000 cells per lane) increased these doublets but notably also minimized reagent cost per recovered cell. Lastly, with direct capture Perturb-seq, we can identify cells bearing undesired sgRNA pairs (generated from intermolecular lentiviral recombination between programmed pairs9–12) and computationally exclude them from downstream analysis. In our data, we observed rates of novel pairs varying from 0.09–0.15 across experiments, which is roughly consistent with a previous report28.
Expression normalization, average expression profiles, and target knockdown or activation
We normalized for differences in capture and sequencing coverage across cells by rescaling each cell to have the same total gene expression UMIs (i.e., each row of the raw expression matrix is rescaled to have the same sum). We then z-normalized expression of each gene with respect to the mean and standard deviation of that gene in the control cell population. We generated pseudo-bulk RNA-seq phenotypes for individual guides or guide pairs by averaging the normalized expression profiles of well-expressed genes–excluding genes with a mean expression <1 UMI per cell (Fig. 1e and Fig. 3e) and <0.5 UMI per cell (Supplementary Figure 3b) across all cells assigned that guide or guide pair (and excluding multiplets). We computed on-target gene knockdown as the ratio of the mean number of target UMIs in perturbed cells versus the mean number of target UMIs in control cells (bearing non-targeting sgRNAs), and we computed on-target gene activation as the ratio of the mean number of target UMIs in perturbed cells vs the mean number of target UMIs in controls.
Data availability
Raw and processed sequencing data are available at Gene Expression Omnibus under accession code GSE146194.
Code availability
Cell Ranger 3.0 is available from 10x Genomics (https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest). Our previously published analytic framework for Perturb-seq analysis17 is available at https://github.com/thomasmaxwellnorman/Perturbseq_GI. Python scripts and Jupyter notebooks for direct capture guide identity assignment are available at https://github.com/josephreplogle/guide_calling. Python Jupyter notebooks for the design of hybridization capture probes are available at https://github.com/josephreplogle/target_enrichment.
Supplementary Material
Acknowledgements
We thank Sara E. Vazquez, Alina Guna, Marco Jost, Dian Yang, Reuben Saunders, Xiaojie Qiu, Eric Chow, Rene Sit, and all members of the Weissman and Adamson labs and 10x Genomics for helpful discussions. This work was funded by National Institutes of Health Grants P50 GM102706, U01 CA168370, R01 DA036858, and RM1HG009490 (all to J.S.W.), the Defense Advanced Research Projects Agency (DARPA) (HR0011-19-2-0007), the Chan Zuckerberg Initiative, and Princeton University. J.S.W. is a Howard Hughes Medical Institute Investigator. J.M.R. is an NIH/NINDS Ruth L. Kirschstein National Research Service Award fellow (F31 NS115380). T.M.N. is a fellow of the Damon Runyon Cancer Research Foundation (DRG-[2211-15]). J.A.H. is the Rebecca Ridley Kry Fellow of the Damon Runyon Cancer Research Foundation (DRG-2262-16). J.C. is funded by the Jane Coffin Childs Memorial Fund for Medical Research and the NIH K99/R00 Pathway to Independence Award (GM134154).
Competing Interests
10x Genomics was involved in producing this work. I.T.F., J.G.A., L.J.A., K.A.P., E.J.M., J.M.T., D.P.R., N.S., and T.S.M. are employees of 10x Genomics. The Regents of the University of California with T.M.N., J.S.W., and B.A. as inventors have filed patent applications related to CRISPRi/a screening, Perturb-seq, and GI mapping. J.S.W. consults for and holds equity in KSQ Therapeutics, Maze Therapeutics, and Tenaya Therapeutics, is a venture partner at 5AM Ventures, and is a member of the Amgen Scientific Advisory Board. J.M.R. and T.M.N. consult for Maze Therapeutics. B.A. is a member of a ThinkLab Advisory Board for and holds equity in Celsius Therapeutics.
References
- 1.Packer J & Trapnell C Single-Cell Multi-omics: An Engine for New Quantitative Models of Gene Regulation. Trends Genet 34, 653–665 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feldman D et al. Optical Pooled Screens in Human Cells. Cell 179, 787–799.e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubin AJ et al. Coupled Single-Cell CRISPR Screening and Epigenomic Profiling Reveals Causal Gene Regulatory Networks. Cell 176, 361–376.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adamson B et al. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell 167, 1867–1882.e21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dixit A et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 167, 1853–1866.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaitin D et al. Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell 167, 1883–1896.e15 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Xie S, Duan J, Li B, Zhou P & Hon GC Multiplexed Engineering and Analysis of Combinatorial Enhancer Activity in Single Cells. Mol Cell 66, 285–299.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Datlinger P et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat Methods 14, 297–301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hill AJ et al. On the design of CRISPR-based single-cell molecular screens. Nat Methods 15, 271 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adamson B, Norman TM, Jost M & Weissman JS Approaches to maximize sgRNA-barcode coupling in Perturb-seq screens. Biorxiv 298349 (2018). doi: 10.1101/298349 [DOI] [Google Scholar]
- 11.Xie S, Cooley A, Armendariz D, Zhou P & Hon GC Frequent sgRNA-barcode recombination in single-cell perturbation assays. Plos One 13, e0198635 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feldman D, Singh A, Garrity AJ & Blainey PC Lentiviral co-packaging mitigates the effects of intermolecular recombination and multiple integrations in pooled genetic screens. Bioirxiv 262121 (2018). doi: 10.1101/262121 [DOI] [Google Scholar]
- 13.Gasperini M et al. A Genome-wide Framework for Mapping Gene Regulation via Cellular Genetic Screens. Cell 176, 377–390.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smits AH et al. Biological plasticity rescues target activity in CRISPR knock outs. Nat Methods 16, 1087–1093 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Anzalone AV et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature (2019). doi: 10.1038/s41586-019-1711-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ran AF et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norman TM et al. Exploring genetic interaction manifolds constructed from rich single-cell phenotypes. Science 365, 786–793 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macosko EZ et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein AM et al. Droplet Barcoding for Single-Cell Transcriptomics Applied to Embryonic Stem Cells. Cell 161, 1187–1201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng GX et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun 8, ncomms14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mimitou EP et al. Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat Methods 16, 409–412 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoeckius M et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 14, 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peterson VM et al. Multiplexed quantification of proteins and transcripts in single cells. Nat Biotechnol 35, 936–939 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Zhang S-Q et al. High-throughput determination of the antigen specificities of T cell receptors in single cells. Nat Biotechnol 36, 1156–1159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilbert LA et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 159, 647–61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilbert LA et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell 154, 442–51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandegar MA et al. CRISPR Interference Efficiently Induces Specific and Reversible Gene Silencing in Human iPSCs. Cell Stem Cell 18, 541–553 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horlbeck MA et al. Mapping the Genetic Landscape of Human Cells. Cell 174, 953–967.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horlbeck MA et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. Elife 5, e19760 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreno AM et al. In Situ Gene Therapy via AAV-CRISPR-Cas9-Mediated Targeted Gene Regulation. Mol Ther 26, 1818–1827 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savell KE et al. A Neuron-Optimized CRISPR/dCas9 Activation System for Robust and Specific Gene Regulation. Eneuro 6, ENEURO.0495–18.2019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cleary B, Cong L, Cheung A, Lander ES & Regev A Efficient Generation of Transcriptomic Profiles by Random Composite Measurements. Cell 171, 1424–1436.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian A et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 171, 1437–1452.e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salomon R et al. Droplet-based single cell RNAseq tools: a practical guide. Lab Chip 19, 1706–1727 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Saikia M et al. Simultaneous multiplexed amplicon sequencing and transcriptome profiling in single cells. Nat Methods 16, 59–62 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vallejo AF et al. Resolving cellular systems by ultra-sensitive and economical single-cell transcriptome filtering. Biorxiv 800631 (2019). doi: 10.1101/800631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan MM et al. Molecular recording of mammalian embryogenesis. Nature 570, 77–82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed sequencing data are available at Gene Expression Omnibus under accession code GSE146194.