Abstract
Tyrosine hydroxylase (Th) encodes the rate-limiting enzyme in catecholamine biosynthesis, and the regulation of its transcription is critical for the specification and maintenance of catecholaminergic neuronal phenotypes. For many genes, regulatory genomic DNA sequences that are upstream of the proximal promoter control expression levels as well as region-specific expression patterns. The regulatory architecture of the genomic DNA upstream of the Th proximal promoter, however, is poorly understood. In this study, we examined the 11kb upstream nucleotide sequence of Th from nine mammalian species and identified five highly conserved regions. Using cultured human cells and mouse olfactory bulb tissue, chromatin immunoprecipitation (ChIP) assays show that these conserved regions recruit transcription factors that are established regulators of Th transcription (such as NURR1, PITX3, FOXA2, MEIS2 and PAX6). This analysis also identified a conserved binding site for CTCF, and functional studies in cultured human cells and ChIP assays with mouse tissue show that CTCF is a novel regulator of Th transcription in the forebrain. Together, the findings in this study provide key insights into the upstream regulatory genomic architecture and regulatory mechanisms controlling mammalian Th gene transcription.
Keywords: transcription, evolution, genomic, dopamine, catecholamine
INTRODUCTION
Tyrosine hydroxylase (Th) encodes the rate-limiting enzyme for catecholamine neurotransmitter biosynthesis. Homozygous loss of Th in mice is embryonic lethal due to disruption of cardiac and/or cardiovascular development [1,2]. Heterozygous Th mutant mice are viable and fertile with a normal physical appearance, but they have reduced noradrenaline levels in multiple brain regions that are linked to impaired associative and latent learning [3]. In humans, individuals with mutations in the Th coding region on both Th alleles can develop Th deficiency, which encompasses a spectrum of movement disorders that typically first manifests in infants [4].
The spatial organization of catecholaminergic neurons is conserved in the mammalian brain [5], which indicates that there are evolutionarily conserved regulatory mechanisms controlling the expression of genes required for catecholaminergic phenotypes. Studies examining molecular mechanisms regulating Th transcription have concentrated on the human and rodent Th proximal promoter region (<1kb upstream), and these studies have identified several promoter cis-regulatory elements that modulate gene expression [6–10]. Although the proximal promoter is necessary for Th expression, it is not sufficient to drive reporter gene expression in vivo [11–13] and regulatory regions outside the proximal promoter are required to activate Th expression. Studies with the human and rat Th locus have shown that the 11kb and 9kb upstream regions, respectively, can drive reporter gene expression in adult catecholaminergic regions with minimal ectopic expression [14,6,15,12]. Despite their importance, a systematic and thorough examination of evolutionary conservation within these upstream sequences is lacking. Previous studies have compared human and rodent upstream sequences, but these studies concentrated on consensus transcription factor binding sites and short conserved motifs, and did not define potential upstream enhancer or repressor territories [6,8]. Given the conservation in the spatial organization of mammalian catecholaminergic neurons as well as the extensive use of rodents and other mammals to understand the function of these neurons in humans, there is a high priority on identifying conserved upstream regulatory regions that control Th expression in the mammalian nervous system. Moreover, identifying conserved upstream regions will also provide novel insight into the molecular mechanisms controlling Th expression necessary for the specification and maintenance of catecholaminergic neuronal phenotypes.
To establish whether there are conserved territories upstream of the mammalian Th promoter, this study aligned 11kb genomic sequences upstream of Th from nine mammalian species. The conserved regions identified by this alignment were tested for their ability to recruit established transcription factors that regulate Th expression in the midbrain and olfactory bulb (OB), which contain the two largest groups of dopaminergic neurons in the brain.
MATERIALS AND METHODS
Nucleotide sequence alignment
Upstream genomic DNA sequences for Th were downloaded from Ensembl (http://www.ensembl.org). The species used for the alignments were: Baboon (Papio anubis), Cow (Bos taurus), Dog (Canis lupus familiaris), Dolphin (Tursiops truncatus), Human (Homo sapiens); Mouse (Mus musculus), Panda (Aliuropoda melanoleuca), Rat (Rattus norvegicus), and Vervet (Chlorocebus sabaeus). Sequence alignments and visualization were performed using Multi-LAGAN and mVista web-based services (http://genome.lbl.gov/vista/mvista/submit.shtml)[16,17]. All sequence alignments and comparisons were made relative to the human Th 11kb upstream region.
ChIP assays
For ChIP experiments to detect NURR1 occupancy, human SH-SY5Y cells were grown in 60mm culture dishes to ~80% confluence and then transfected with p3XFlag-CMV-mNurr1 (a gift from Dr. Kaoru Saijo, UC Berkeley) using Lipofectamine LTX (Life Technologies). Twenty-four hours after transfection, cells were washed with PBS and then cross-linked with 1% formaldehyde in PBS at room temperature for 9min. Fixation was terminated by addition of 125mM glycine, and cells were washed twice with PBS before being pelleted by centrifugation at 4°C for 5min and resuspended with SDS Lysis Buffer (Millipore). The chromatin was sheared with a Bioruptor sonicator (Diagenode) and immunoprecipitation of cross-linked protein-DNA complexes used the Magna ChIP Protein-A/G kit (Millipore) following the manufacturer’s instructions. Immunoprecipitation reactions used 4μg of either mouse anti-FLAG M2 antibody (Sigma) or normal mouse IgG (Santa Cruz Biotechnology). Reverse cross-linking was done overnight in the presence of Proteinase K at 62°C with shaking.
For ChIP assays to detect binding by PITX3 and FOXA2, lysates from non-transfected SH-SY5Y cells were prepared as described above. Immunoprecipitation reactions used either 2.5μg of mouse anti-PITX3 antibody (Thermo, 38-2850), 1.5μg of rabbit anti-FOXA2 antibody (Abcam, ab108422) or an equivalent amount of normal IgG (Santa Cruz Biotech).
For ChIP experiments to detect CTCF, MEIS2, PBX1/2/3 and PAX6 occupancy in vivo, adult C57BL6 mice (aged 2–5 months) of both sexes were used for tissue from the OB, cortex, ventral midbrain and liver. The tissue was washed with ice cold PBS and then cross-linked with 1% formaldehyde in PBS on ice for 15 min. Fixation was terminated with 125mM glycine, and the tissue was washed twice with PBS before addition of SDS Lysis Buffer (Millipore). Immunoprecipitation of protein/chromatin complexes was performed as described above with SH-SY5Y cells and used either 6μg of goat anti-CTCF antibody (Santa Cruz), 10μg of goat anti-MEIS2 antibody (Santa Cruz Biotech, sc-10600), 2μg of mouse anti-PBX1/2/3 antibody (Santa Cruz Biotech, sc-28313), 5μg of rabbit anti-PAX6 antibody (Abcam, ab5790-100), or an equivalent amount of normal IgG (Santa Cruz Biotech).
For ChIP experiments with neurospheres, cultures were generated from dissociated anterior subventricular zone tissue of adult mice (aged 3 months) grown in DMEM/F12 media supplemented with B27 (Life Technologies), bFGF (20ng/mL; BD Bioscience) and EGF (20ng/mL; BD Bioscience). Initial cultures were expanded for 7–8 days, and then dissociated and used to reseed new cultures. The passaged cultured were expanded for 7 days, after which the neurospheres were pelleted and washed with PBS before being cross-linked with 1% formaldehyde in PBS at room temperature for 9min. Fixation reactions were terminated by the addition of 125mM glycine, and the tissue was washed twice with PBS before addition of SDS Lysis Buffer (Millipore). Immunoprecipitation of protein/chromatin complexes was performed as described above with SH-SY5Y cells and used 6μg of either goat anti-CTCF antibody (Santa Cruz) or normal goat IgG (Santa Cruz).
All immunoprecipitated Th promoter genomic DNA fragments were amplified and quantified using a 7600 Fast Real-time PCR System (Applied Biosystems) using SYBR Green PCR master mix (Applied Biosystems). Primer sequences used for amplifying were:
mouse Th E1 forward strand 5′-GGGATTTGCAGGAGCTTGCTCA-3′
mouse Th E1 reverse strand 5′-CTTGGACTCTCAGGAGCCAACT-3′
mouse Th E2 forward strand 5′-TTCCATGAAAGCACAACTGGC-3′
mouse Th E2 reverse strand 5′-CAGGGTCGGCTGCTGAGGAT-3′
mouse Th E3 forward strand 5′-TGGTCTGACTTTCAGCTGCCCAAT-3′
mouse Th E3 reverse strand 5′-CAATACCACTCACTGACCTCACTG-3′
mouse Th E4 forward strand 5′-GTGACCACCACTCACGGGCT-3′
mouse Th E4 reverse strand 5′-CCTGTGCACCAGTGAGTCACATAA-3′
mouse Th E5 forward strand 5′-TCCAGGAGAACAGACGCCAGC-3′
mouse Th E5 reverse strand 5′-GCCAGGCTGAAGGCAAGCACA-3′
mouse Th negative control forward strand 5′-TGCCTCAGCAGAGCCTGAGT-3′
mouse Th negative control reverse strand 5′-AAGCTCCCCGTGACTGTGTG-3′
human Th E1 forward strand 5′-CCAAATCCTTCTGGGCCAGGA-3′
human Th E1 reverse strand 5′-CCGTTCTCTCTTCAACAATAGCC-3′
human Th E2 forward strand 5′-TTCCATGAAAGCACAACTGGC-3′
human Th E2 reverse strand 5′-CAGGGTCGGCTGCTGAGGAT-3′
human Th E3 forward strand 5′-TCGCTCTGGGCCTGACTTCC-3′
human Th E3 reverse strand 5′-AACACAGGACAGAATCCGCCGT-3′
human Th E5 forward strand 5′-TTGGAGCAAAGCGGACAAGCTCA-3′
human Th E5 reverse strand 5′-GCGCATTCACTTCAGGTACCTC-3′
human Th negative control forward strand 5′-AGGCTGAGGCCTCTCCTTCCA-3′
human Th negative control reverse strand 5′-GAACTCCACCGTGAACCAGTACA-3′
All ChIP experiments were conducted as three independent assays and the mean relative enrichment of the target region is reported with error bars representing the standard error of the mean. Statistical significance was assessed using either two-tailed Student’s t-test or ANOVA with appropriate post-hoc tests.
CTCF over-expression and knock-down
To measure Th promoter activity when Ctcf was over-expressed, SH-SY5Y cultures were seeded in Primaria-coated 6-well plates (Corning) at 3x105 cells/well and incubated at 37°C for 24 hours before using Lipofectamine LTX reagent (ThermoFisher Scientific) to co-transfect 4μg of pCMV-Myc-CTCF (a gift from Dr. Mary Donohoe, Burke Medical Research Instiute), 2μg of pGL4.20 reporter plasmid containing Firefly luciferase under control of the rat Th 4.5kb promoter (generated in our laboratory), and 1μg of pRL-CMV (Promega), which constitutively expresses Renilla luciferase in order to control for variations in transfection efficiency. After 24 hours, cells were harvested and Firefly and Renilla luciferase activity levels were measured using the Dual-Glo Luciferase Assay System (Promega) with a LMaxII luminometer (Molecular Devices). Luciferase activities are reported as the mean of at least three independent measurements with error bars representing the standard error of the mean.
To measure Th promoter activity when Ctcf was knocked-down, SH-SY5Y cultures were seeded in Primaria-coated 6-well plates (Corning) at 3x105 cells/well and incubated at 37°C for 24 hours before being transfected with siRNA (Dharmacon Accell siRNA) according to the manufacture’s instruction. Control cultures were treated with Accell Delivery Media (Dharmacon). All cultures were maintained at 37°C for 48 hours following transfection before were divided to conduct either Western blot or qRT-PCR analyses.
Knock-down of CTCF in cultures was confirmed by Western blots with goat anti-CTCF (Santa Cruz Biotech., sc-15914X at 1:1000 dilution) and mouse anti-beta-Actin (Sigma, A5316, at 1:6000 dilution). Blots were imaged with an Odyssey Imaging System (Li-Cor Biosciences) and intensities were quantified using ImageJ software (National Institutes of Health). Protein band intensities are reported as the mean of three individual trials with error bars representing the standard error of mean.
Th expression levels in transfected cells were measured by qRT-PCR. Total RNA from cells was collected using the GenCatch Total RNA Extraction System (Epoch Life Science). First-strand cDNA syntheses were generated with SuperScript III reverse transcriptase (Invitrogen). Quantitative PCR reactions were performed with TaqMan assays (Applied Biosystems) for human Th (Hs00165941_m1) and beta-Actin (Hs3044422). All reactions were carried out on a 7500 Fast Real-Time PCR System (Applied Biosystems). All samples were run in triplicate and expression levels for Th were normalized to beta-Actin. The mean relative expression levels are reported with error bars representing the standard error of the mean.
RESULTS
Genomic sequence alignments and identification of conserved upstream regions
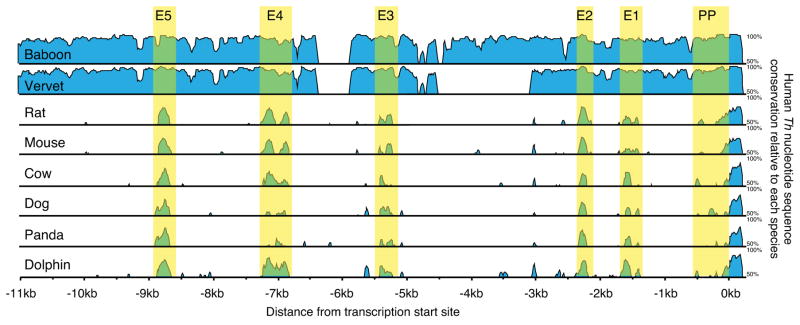
To identify regions of evolutionary conservation, 11kb genomic DNA sequences upstream of the Th proximal promoter from nine mammalian species were aligned. This analysis identified five regions of high homology based on the criteria of being a minimum length of 75 nucleotides length, having more than 50% homology to human, and being conserved in all nine species. These conserved regions were named E1–E5, going from the proximal to distal end of the Th upstream region (Fig 1–3). All five of the regions were found in only placental mammals and were absent in alignments with Th upstream regions from avians, reptiles, amphibians and fish (data not shown).
Figure 1.
Conservation of 11kb genomic sequences upstream of mammalian Th. Graphic representation of human nucleotide sequence conservation as a percentage relative to each of the other mammalian species examined. The five upstream regions of high conservation (E1–E5) and proximal promoter (PP) are highlighted in yellow.
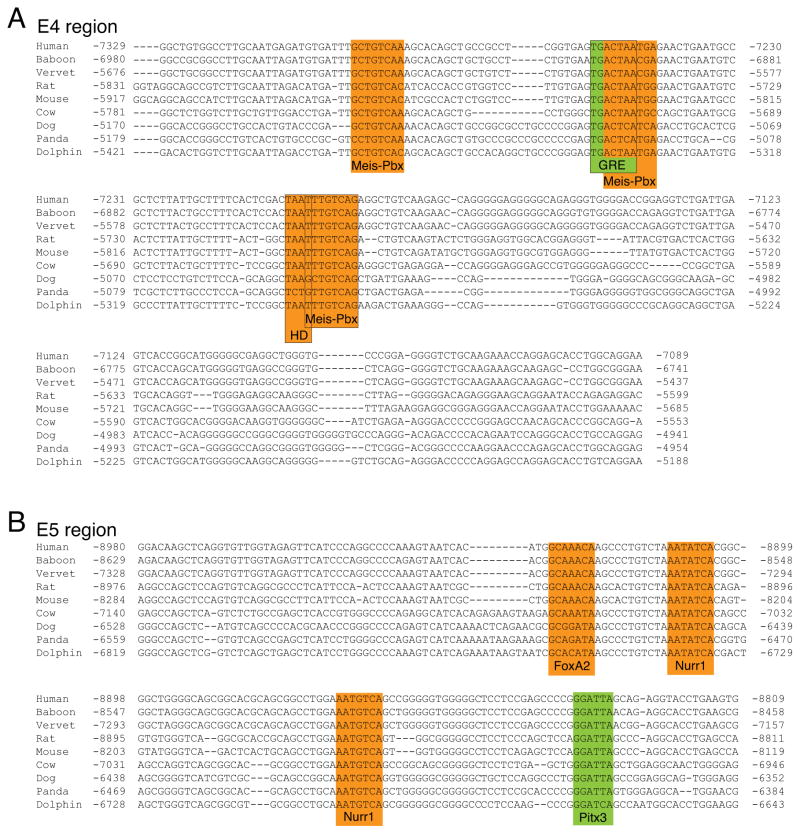
Figure 3.
Genomic sequence alignment and transcription factor binding sites for the E4 and E5 upstream regions. A, alignment of the E4 region reveals three novel and conserved binding site motifs and MEIS-PBX heterodimers [61](MEIS-PBX; highlighted in orange). One of the MEIS-PBX sites overlaps a previously identified AP-1 binding site reported to mediate the induction of Th expression in response to glucocorticoid receptor activation [53](GRE; highlighted in green). Another MEIS-PBX site overlaps with partially conserved core homeodomain binding site motif (HD; highlighted in orange). B, alignment of the E5 region shows that a previously identified PITX3 binding site (highlighted in green) is conserved in mammals [24]. Analysis of this region also identified previously unreported and conserved binding site motifs for FOXA2 and NURR1 (highlighted in orange).
Recruitment of established regulators of Th transcription
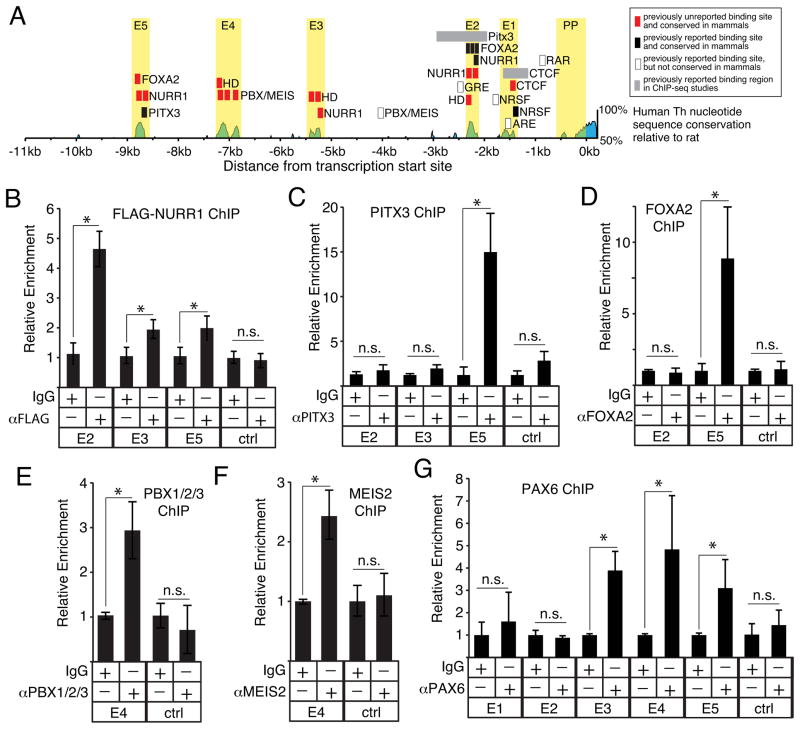
Several individual transcription factor binding sites upstream of the Th proximal promoter have been reported, and many of these sites overlap with the conserved regions identified by our alignment (Fig 4A). NURR1 (NR4A2) is a transcription factor necessary for Th expression in the midbrain [18,19]. NURR1 can also drive dopaminergic differentiation in the OB [20]. The E2 region identified in this study contains a previously reported NURR1 binding site [21,22] and our analysis shows that this site is highly conserved in mammals (Fig 2B, Fig 4A). Our analysis also identified potential novel NURR1 binding sites in the E3 and E5 regions (Fig 2C, Fig 3B, Fig 4A). To test whether these unreported binding sites recruit NURR1, chromatin immunoprecipitation (ChIP) assays were performed in SH-SY5Y cells transfected with a FLAG-tagged NURR1 expression plasmid. These ChIP assays showed that NURR1 is recruited to the E2, E3 and E5 regions, but not to a negative control region in intron 10–11 (Fig 4B). Together, the ChIP assays indicate that NURR1 regulates Th transcription by targeting multiple upstream regulatory regions.
Figure 4.
Recruitment of established regulators of Th transcription to conserved upstream regions. A, graphic representation of transcription factor binding sites previously reported and identified in the current study. B, ChIP assays testing NURR1 occupancy on the E2, E3 and E5 regions within SH-SY5Y cells over-expressing FLAG-NURR1 (n=3). C–D, ChIP assays testing PITX3 and FOXA2 occupancy, respectively, on the E2, E3 and E5 regions within SH-SY5Y cells (n=3 for each). E–F, ChIP assays testing PBX1/2/3 and MEIS2 occupancy, respectively, on the E4 region within OB tissue (n=6 and n=5 for PBX and MEIS, respectively). G, ChIP assays testing PAX6 occupancy on the E2, E3 and E4 regions within OB tissue (n=4). For all ChIP assays, negative control regions are indicated by “crtl”, asterisks indicate a significant difference (p ≤ 0.01) and “n.s.” indicates non-significant enrichment.
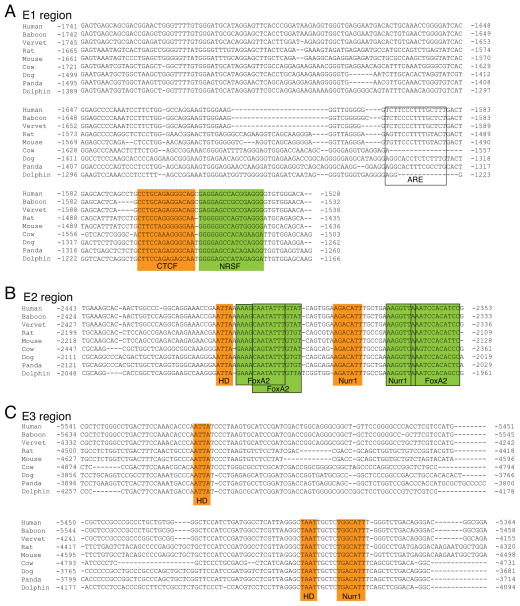
Figure 2.
Genomic sequence alignment and transcription factor binding sites for the E1, E2 and E3 upstream regions. A, alignment of the E1 region reveals a novel and conserved binding site motif for CTCF (highlighted in orange). The alignment shows that a previously reported NRSF binding site [35] is partially conserved in mammals (highlighted in green). The E1 region also contains an aryl-hydrocarbon response element (ARE) that was previously identified in mice [60]; highlighted with open box). This mouse ARE, however, is only partially conserved in mammals and may be a species-specific regulatory element. B, alignment of the E2 region shows that previously reported binding sites for FoxA2 and Nurr1 [21,22] within this region are conserved in mammals (sites are highlighted in green). This alignment also reveals an additional, novel and conserved binding site for Nurr1 and a core homeodomain (HD) binding motif (highlighted in orange). C, alignment of the E3 region reveals a novel and conserved Nurr1 binding site as well as two core homeodomain (HD) motifs (highlighted in orange).
The specification and maintenance of the midbrain dopaminergic phenotype is regulated by the interaction of NURR1 with other transcription factors, such as Paired-like homeodomain 3 (PITX3) and Forkhead Box A2 (FOXA2). Both PITX3 and FOXA2 physically interact with NURR1 and function as co-activators [23,22]. In the E5 region, our analysis found that a previously reported PITX3 binding site [24] is conserved in mammals (Fig 3B, Fig 4A) and also identified a previously unreported FOXA2 core binding motif (5′-AAAYA-3′)[25,26] that is conserved in rodents and primates (Fig 2B). In addition, the E2 region contains three conserved FOXA2 binding sites (Figu 2B) that were previously identified in embryonic midbrain progenitors [22]. A previous ChIP-seq study also indicated PITX3 targets a region that overlaps E2 [23], but a consensus PITX3 binding site motif (5′-TAATCC-3′) was not identified within this region. The analysis of the E2 region, however, did find a core homeodomain sequence (5′-TAAT-3′) 15bp upstream from the NURR1 sites (Fig 2B, Fig 4A). Inspection of the E3 region also found a core homeodomain motif 5bp upstream from a Nurr1 site (Fig 3A, Fig 4A).
ChIP assays in SH-SY5Y cells showed that endogenous PITX3 and FOXA2 are recruited to the E5, but not E2 or E3 regions (Fig 4C,D). These findings indicate that NURR1, PITX3 and FOXA2 collectively target the E5 region to regulate Th expression. By contrast, the binding site motifs identified in the E2 and E3 regions were not sufficient to recruit either PITX3 or FOXA2 under these conditions used in these studies. Previous work with SH-SY5Y cells also detected PITX3 binding to the E5, but not E2, region [24]. By contrast, studies with mouse embryonic midbrain cells and cultured MN9D cells reported PITX3 and FOXA2 binding to the E2 region [23,22]. Given the importance of combinatorial interactions between transcription factors in regulating DNA binding site specificity and the formation of transcription regulatory protein complexes [27,28], the differential recruitment of PITX3 and FOXA2 to the E2 region may reflect cell type differences in either post-translational modifications or the co-expression of other proteins.
In OB dopaminergic neurons, the heterodimeric transcription factors Pre B-cell Leukemia homeodomain 1 (PBX1) and Myeloid ecotropic viral integration site 2 (MEIS2) promote specification of the OB dopaminergic phenotype by directly binding to a site upstream of the mouse Th proximal promoter [29,30]. Our analysis, however, found that this target site, which is between the E2 and E3 regions, was not conserved throughout mammals (Fig 4A). By contrast, inspection of the E4 region identified three MEIS-PBX consensus binding motifs that are well conserved (Fig 3A, Fig 4A), and ChIP assays with mouse OB tissue showed that both MEIS2 and PBX1/2/3 associate with the E4 region (Fig 4E,F). The high conservation of the MEIS-PBX binding motifs within E4 suggests that this region mediates the conserved regulation of Th expression by MEIS and PBX proteins in the mammalian OB, whereas the previously identified MEIS-PBX site may be an important co-regulator of Th expression specifically in rodents.
MEIS2 also regulates Th transcription in the OB by physically interacting with the Paired Box 6 (PAX6) transcription factor [29]. PAX6 is necessary for OB dopaminergic neuron development and survival [31–33], but whether it targets a specific Th cis-regulatory region has not been established. PAX6 contains two functional DNA-binding domains, a paired domain and homeodomain, that each bind to distinct recognition sequences [34]. None of the upstream regions identified in this study contain consensus binding sites for the paired domain, but the E2, E3 and E4 regions contain core homeodomain (5′-TAAT-3′) binding site motifs (Fig 2B,C, Fig 3A, Fig 4A). ChIP assays with mouse OB tissue showed that PAX6 associates with the E3, E4 and E5 regions, but not with either E1, E2 or a negative control region (Fig 4G). Together, these studies indicate that MEIS2, PBX1 and PAX6 regulate Th transcription in the OB, in part, by collectively targeting the E4 upstream region.
The Neuron-Restrictive Silencer Factor/RE1-Silencing Transcription factor (NRSF/REST) mediates epigenetic regulation of Th transcription in cultured cell lines and binds to a site found within the E1 region (Fig 2A and Fig 4A)[35–37]. This NRSF binding site shows conservation, but the in vivo relevance of this site is unclear. NRSF expression levels in the adult brain under physiological conditions are very low and there are several alternatively-spliced NRSF isoforms that have different DNA binding affinities [38,39]. Furthermore, NRSF is expressed at high levels during embryonic development, but the ENCODE database shows that NRSF does not occupy either the E1 region or the Th promoter in human embryonic stem cells [40]. Thus, NRSF may not regulate Th expression either during development or in the normal adult brain. Further studies are required, however, to establish whether NRSF regulates Th expression by targeting the E1 region under pathological conditions associated with elevated NRSF expression levels, such as MPTP-mediated neurodegeneration [36].
CTCF is a novel, direct regulator of Th promoter activity in the forebrain
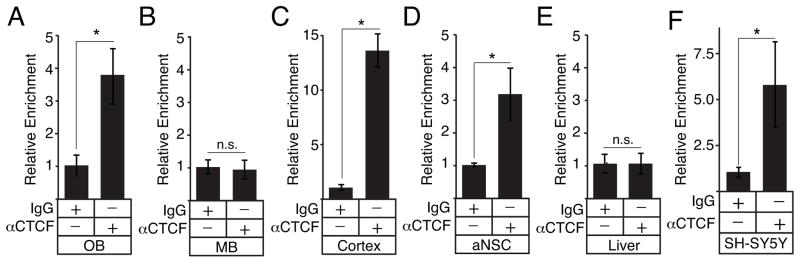
Analysis of the E1 region identified an unreported and conserved CCCTC-binding factor (CTCF) consensus site adjacent to the previously reported NRSF site (Fig 2A, Fig 4A). CTCF is broadly expressed in many tissues and is a bifunctional regulator of transcription [41]. CTCF has not been previously reported to regulate Th expression, but ChIP-seq data from over 45 different cell lines in the ENCODE database show that CTCF associates with a region that overlaps with E1 (Fig 4A). To establish whether CTCF targets the E1 region in vivo, ChIP assays were performed with tissue from several mouse brain regions. These assays showed that CTCF occupied the E1 region in the OB, but not in the midbrain (Fig 5A, B), suggesting that CTCF is a region-specific regulator of Th expression. In a small subset of cortical neurons, Th is transcribed, but not translated into protein [42]. ChIP assays showed that CTCF also occupied the E1 region in cortical tissue (Fig 5C), suggesting that CTCF is a forebrain-specific regulator of Th. ChIP assays were also conducted with forebrain progenitors in neurospheres derived from the adult subventricular zone. These assays also found CTCF occupancy in the E1 region, revealing that CTCF regulation of Th is not limited to only mature neurons (Fig 5D). To establish whether CTCF targeted the Th promoter outside of the nervous system, ChIP assays were conducted with liver tissue. These assays showed no significant occupancy of E1 (Fig 5E) and indicate that CTCF is not required for maintaining Th repression in non-neural tissues.
Figure 5.
Tissue-specific recruitment of CTCF to the E1 upstream region. A–F, relative enrichment of the E1 region in ChIP assays for CTCF with mouse olfactory bulb (OB), midbrain (MB), cortex, adult neurosphere cultures (aNSC), liver tissue, and SH-SY5Y neuroblastoma cells, respectively (n=3 for each).
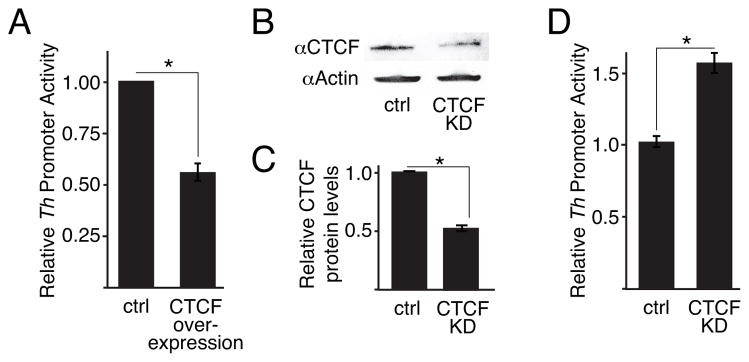
To address whether modifying CTCF expression levels could alter Th promoter activity, luciferase transcription assays were conducted with SH-SY5Y cells since we had shown that CTCF is recruited to the E1 region in this cell line (Fig 5F). These assays showed that co-transfection of a Ctcf expression plasmid reduced Th promoter activity (Fig 6B), whereas siRNA-mediated partial knock-down of CTCF in these cells increased Th promoter activity (Fig 6C–E). These findings indicate that CTCF functions as a repressor of Th transcription in these cells. Moreover, together with the ChIP assays, these findings show that CTCF is a forebrain-specific and direct regulator of Th transcription.
Figure 6.
CTCF regulates Th promoter activity. A, luciferase reporter activity with the 4.5kb rat Th promoter in SH-SY5Y cells with over-expressing CTCF (n=3). B and C, western blots and quantitation of protein levels, respectively, for SH-SY5Y cells with partial Ctcf expression knock down by RNAi (n=3 for each). D, luciferase reporter activity with the 4.5kb rat Th promoter in SH-SY5Y cells with Ctcf expression knocked down by RNAi (n=3). For all studies, asterisks indicate a significant difference (p ≤ 0.01) and “n.s.” indicates non-significant difference.
DISCUSSION
Our analysis identified five highly conserved regions in the genomic DNA upstream of the Th proximal promoter in placental mammals. The restricted conservation of these regions to placental mammals likely reflects the presence of a Th gene paralogue (Th2) in non-placental mammals and other vertebrates (avians, reptiles, amphibians and fish) that may have arisen from whole-genome duplication in early vertebrate evolution [43]. In situ hybridization studies in both developing and adult fish showed that the two Th paralogues have largely complementary expression patterns [43–45], suggesting that the cis-regulatory mechanisms controlling the single placental mammalian Th gene are distributed between the two paralogs in other vertebrates.
Our identification of highly conserved territories within the mammalian Th upstream region ranging in length between ~100bp to ~200bp significantly advances our understanding of genomic control of Th expression. These findings contrast with a previous analysis of humans, rats and mice that only identified five short conserved sequences (18–60bp in length)[6]. Four of these five previously reported sequences overlap with the regions identified in our alignments, but the limited number of species analyzed together with an apparent emphasis on contiguous sequences with near perfect homology likely prevented this previous analysis from identifying the larger territories of high conservation described in the current study.
The five upstream regions defined in the present study contain both previously and newly identified binding sites for transcription factors that regulate Th transcription during development and homeostasis. NURR1 is a key regulator of Th expression in several brain regions, and our study showed that NURR1 is recruited to the E2, E3 and E5 regions. NURR1 is a bifunctional regulator of Th transcription, and whether it either activates or represses transcription is influenced by its interaction with other transcription factors [23,21]. Thus, the recruitment of NURR1 to E2, E3 and E5 regions is expected to make the regulatory role of these regions bifunctional as well, which suggests these regions can act as either distal enhancers or repressors depending on specific transcription factor expression profile of the cell.
NURR1 is also reported to bind the Th proximal promoter [46,47], but a previous analysis of nucleotide conservation within the Th proximal promoter found that this site was poorly conserved outside of rodents [10]. In addition, the position of this putative site in the promoter potentially conflicts with the binding of general transcription factors, such as TBP. In light of these observations, the findings from the present study suggest that the targeting of the E2, E3 and E5 regions is the conserved mechanism by which NURR1 regulates Th transcription.
The findings in this study suggest that the E5 region is important for regulating Th transcription in midbrain neurons. This region can recruit the transcription factors (NURR1, PITX3 and FOXA2; this study) and co-activator proteins (DJ1 and MTA1; [24]) that are established regulators of the midbrain dopaminergic phenotype. Other studies have also indicated that the sequences within the E2 regions are also important for driving Th expression in the developing midbrain by also recruiting NURR1, PITX3 and FOXA2 [23,21,22], which suggests that that regulatory protein complexes assembled on the E5 region may work coordinately with those assembled on the E2 region. The role of PITX3 or FOXA2 on these regulatory regions within the midbrain, however, is limited to the substantia nigra since the loss of either PITX3 or FOXA2 only disrupts development of dopaminergic neurons in the substantia nigra [48–50,25,51]. By contrast, NURR1 is critical for all ventral midbrain dopaminergic neurons [18,19], and thus, other transcription factors may interact with NURR1 on the E2, E3 and E5 regions in midbrain regions outside the substantia nigra to regulate Th transcription. Further studies are required to establish whether these conserved upstream regions are targeted by other transcription factors that regulate specification and maintenance of midbrain dopaminergic phenotypes, such as LMX1A/B, EN1, NGN2 and OTX2 (reviewed in [52].
In the OB, our studies indicate that MEIS2, PBX1 and PAX6 regulate Th transcription, in part, by collectively targeting the E4 upstream region. Our findings also suggest that the E3 and E5 regions may also contribute to regulation of Th in the OB by recruiting Pax6. The presence of a homeodomain motif in E3 suggest that PAX6 could directly bind this region, but the absence of either a paired domain or homeodomain recognition motif in E5 suggests that PAX6 associates with this region indirectly through interactions with other DNA-binding proteins. An important goal for future studies is to establish whether the association of PAX6 to the Th upstream regions is mediated by direct binding to the Th genomic DNA or through protein-protein interactions with factors, like MEIS2, that are also recruited to these regions. The high conservation of the MEIS-PBX binding motifs within E4 suggests that this region mediates the conserved regulation of Th expression by MEIS and PBX proteins in the mammalian OB, whereas the previously identified MEIS-PBX site may be an important co-regulator of Th expression specifically in rodents. It should also be noted that the one of the conserved MEIS-PBX sites overlap with AP-1 binding site that is responsible for induction of Th expression in response to glucocorticoid receptor activation [53]. AP-1 also has a highly conserved site in the Th proximal promoter that is an important modulator of stimulus-induced transcription [10,54,55]. This further suggests that the E4 region is an important regulator of Th expression in the olfactory system, and future studies will address with MEIS, PBX or PAX6 proteins bound to E4 interact with AP-1 bound to either E4 or the proximal promoter to drive Th transcription.
The present study identified CTCF as a novel and region-specific regulator of Th transcription. The CTCF recognition sequence in the E1 region is strongly conserved and the ChIP assays showed that CTCF occupied this region preferentially in the forebrain. Since CTCF can mediate interactions between distant genomic regions [41], CTCF may be important for bringing E1 into the proximity of other Th distal regulatory regions. CTCF is a bifunctional regulator of transcription and its ability to either repress or enhance transcription is context-dependent [41]. Our functional studies indicate that CTCF is a repressor of Th expression, and this property may be important for restricting Th transcription to specific neuronal subpopulations in the forebrain [42,56]. Since CTCF is broadly expressed in the brain and other tissues [57], further studies are required to establish how brain region-specific recruitment to the E1 region is achieved.
The conserved upstream regions identified in this study significantly advance our understanding of the genomic DNA regulatory architecture for the mammalian Th gene. Moreover, our findings that these regions recruit transcription factors that are established regulators of Th expression provides insight into conserved mechanisms that regulate specification and maintenance of catecholaminergic neuronal phenotypes in the mammalian brain. These mechanisms, however, also likely include contributions from regulatory regions downstream downstream of the transcription start site. Previous studies with transgenic mice showed that downstream regions can augment reporter expression levels [58,59]. The first 2kb downstream of the human Th gene was found to be sufficient for activating transcription in specific brain regions, but it did not suppress ectopic reporter gene expression in non-catecholaminergic regions [58]. This suggests that the combined input from upstream and downstream regulatory regions is required to drive Th expression at high levels specifically in catecholaminergic neurons.
Acknowledgments
We thank Dr. Kaoru Saijo (Univ. California, Berkeley) for the NURR1 expression plasmid and Dr. Mary Donohoe for the CTCF expression plasmid.
Footnotes
Conflicts of Interest:
The authors declare that they have no conflicts of interest.
References
- 1.Kobayashi K, Morita S, Sawada H, Mizuguchi T, Yamada K, Nagatsu I, Hata T, Watanabe Y, Fujita K, Nagatsu T. Targeted disruption of the tyrosine hydroxylase locus results in severe catecholamine depletion and perinatal lethality in mice. The Journal of biological chemistry. 1995;270(45):27235–27243. doi: 10.1074/jbc.270.45.27235. [DOI] [PubMed] [Google Scholar]
- 2.Zhou QY, Quaife CJ, Palmiter RD. Targeted disruption of the tyrosine hydroxylase gene reveals that catecholamines are required for mouse fetal development. Nature. 1995;374(6523):640–643. doi: 10.1038/374640a0. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi K, Noda Y, Matsushita N, Nishii K, Sawada H, Nagatsu T, Nakahara D, Fukabori R, Yasoshima Y, Yamamoto T, Miura M, Kano M, Mamiya T, Miyamoto Y, Nabeshima T. Modest neuropsychological deficits caused by reduced noradrenaline metabolism in mice heterozygous for a mutated tyrosine hydroxylase gene. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2000;20(6):2418–2426. doi: 10.1523/JNEUROSCI.20-06-02418.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willemsen MA, Verbeek MM, Kamsteeg EJ, de Rijk-van Andel JF, Aeby A, Blau N, Burlina A, Donati MA, Geurtz B, Grattan-Smith PJ, Haeussler M, Hoffmann GF, Jung H, de Klerk JB, van der Knaap MS, Kok F, Leuzzi V, de Lonlay P, Megarbane A, Monaghan H, Renier WO, Rondot P, Ryan MM, Seeger J, Smeitink JA, Steenbergen-Spanjers GC, Wassmer E, Weschke B, Wijburg FA, Wilcken B, Zafeiriou DI, Wevers RA. Tyrosine hydroxylase deficiency: a treatable disorder of brain catecholamine biosynthesis. Brain: a journal of neurology. 2010;133(Pt 6):1810–1822. doi: 10.1093/brain/awq087. [DOI] [PubMed] [Google Scholar]
- 5.Smeets WJ, Gonzalez A. Catecholamine systems in the brain of vertebrates: new perspectives through a comparative approach. Brain Res Brain Res Rev. 2000;33(2–3):308–379. doi: 10.1016/s0165-0173(00)00034-5. [DOI] [PubMed] [Google Scholar]
- 6.Kessler MA, Yang M, Gollomp KL, Jin H, Iacovitti L. The human tyrosine hydroxylase gene promoter. Brain research Molecular brain research. 2003;112(1–2):8–23. doi: 10.1016/s0169-328x(02)00694-0. [DOI] [PubMed] [Google Scholar]
- 7.Kim TE, Park MJ, Choi EJ, Lee HS, Lee SH, Yoon SH, Oh CK, Lee BJ, Kim SU, Lee YS, Lee MA. Cloning and cell type-specific regulation of the human tyrosine hydroxylase gene promoter. Biochemical and biophysical research communications. 2003;312(4):1123–1131. doi: 10.1016/j.bbrc.2003.11.029. [DOI] [PubMed] [Google Scholar]
- 8.Schimmel JJ, Crews L, Roffler-Tarlov S, Chikaraishi DM. 4.5 kb of the rat tyrosine hydroxylase 5′ flanking sequence directs tissue specific expression during development and contains consensus sites for multiple transcription factors. Brain research Molecular brain research. 1999;74(1–2):1–14. doi: 10.1016/s0169-328x(99)00234-x. [DOI] [PubMed] [Google Scholar]
- 9.Yang C, Kim HS, Seo H, Kim KS. Identification and characterization of potential cis-regulatory elements governing transcriptional activation of the rat tyrosine hydroxylase gene. Journal of neurochemistry. 1998;71(4):1358–1368. doi: 10.1046/j.1471-4159.1998.71041358.x. [DOI] [PubMed] [Google Scholar]
- 10.Wang M, Banerjee K, Baker H, Cave JW. Nucleotide sequence conservation of novel and established -regulatory sites within the tyrosine hydroxylase gene promoter. Front Biol (Beijing) 2015;10(1):74–90. doi: 10.1007/s11515-014-1341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Merlie JP, Todd RD, O’Malley KL. Identification of cell type-specific promoter elements associated with the rat tyrosine hydroxylase gene using transgenic founder analysis. Brain research Molecular brain research. 1997;50(1–2):33–42. doi: 10.1016/s0169-328x(97)00163-0. [DOI] [PubMed] [Google Scholar]
- 12.Min N, Joh TH, Kim KS, Peng C, Son JH. 5′ upstream DNA sequence of the rat tyrosine hydroxylase gene directs high-level and tissue-specific expression to catecholaminergic neurons in the central nervous system of transgenic mice. Brain research Molecular brain research. 1994;27(2):281–289. doi: 10.1016/0169-328x(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 13.Sasaoka T, Kobayashi K, Nagatsu I, Takahashi R, Kimura M, Yokoyama M, Nomura T, Katsuki M, Nagatsu T. Analysis of the human tyrosine hydroxylase promoter-chloramphenicol acetyltransferase chimeric gene expression in transgenic mice. Brain research Molecular brain research. 1992;16(3–4):274–286. doi: 10.1016/0169-328x(92)90236-5. [DOI] [PubMed] [Google Scholar]
- 14.Iacovitti L, Wei X, Cai J, Kostuk EW, Lin R, Gorodinsky A, Roman P, Kusek G, Das SS, Dufour A, Martinez TN, Dave KD. The hTH-GFP reporter rat model for the study of Parkinson’s disease. PLoS One. 2014;9(12):e113151. doi: 10.1371/journal.pone.0113151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsushita N, Okada H, Yasoshima Y, Takahashi K, Kiuchi K, Kobayashi K. Dynamics of tyrosine hydroxylase promoter activity during midbrain dopaminergic neuron development. Journal of neurochemistry. 2002;82(2):295–304. doi: 10.1046/j.1471-4159.2002.00972.x. [DOI] [PubMed] [Google Scholar]
- 16.Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, Sidow A, Batzoglou S. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome research. 2003;13(4):721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic acids research. 2004;32(Web Server issue):W273–279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saucedo-Cardenas O, Quintana-Hau JD, Le WD, Smidt MP, Cox JJ, De Mayo F, Burbach JP, Conneely OM. Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc Natl Acad Sci U S A. 1998;95(7):4013–4018. doi: 10.1073/pnas.95.7.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zetterstrom RH, Solomin L, Jansson L, Hoffer BJ, Olson L, Perlmann T. Dopamine neuron agenesis in Nurr1-deficient mice. Science. 1997;276(5310):248–250. doi: 10.1126/science.276.5310.248. [DOI] [PubMed] [Google Scholar]
- 20.Vergano-Vera E, Diaz-Guerra E, Rodriguez-Traver E, Mendez-Gomez HR, Solis O, Pignatelli J, Pickel J, Lee SH, Moratalla R, Vicario-Abejon C. Nurr1 blocks the mitogenic effect of FGF-2 and EGF, inducing olfactory bulb neural stem cells to adopt dopaminergic and dopaminergic-GABAergic neuronal phenotypes. Dev Neurobiol. 2015;75(8):823–841. doi: 10.1002/dneu.22251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim TE, Seo JS, Yang JW, Kim MW, Kausar R, Joe E, Kim BY, Lee MA. Nurr1 represses tyrosine hydroxylase expression via SIRT1 in human neural stem cells. PLoS One. 2013;8(8):e71469. doi: 10.1371/journal.pone.0071469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi SH, He XB, Rhee YH, Park CH, Takizawa T, Nakashima K, Lee SH. Foxa2 acts as a co-activator potentiating expression of the Nurr1-induced DA phenotype via epigenetic regulation. Development. 2014;141(4):761–772. doi: 10.1242/dev.095802. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs FM, van Erp S, van der Linden AJ, von Oerthel L, Burbach JP, Smidt MP. Pitx3 potentiates Nurr1 in dopamine neuron terminal differentiation through release of SMRT-mediated repression. Development. 2009;136(4):531–540. doi: 10.1242/dev.029769. [DOI] [PubMed] [Google Scholar]
- 24.Reddy SD, Rayala SK, Ohshiro K, Pakala SB, Kobori N, Dash P, Yun S, Qin J, O’Malley BW, Kumar R. Multiple coregulatory control of tyrosine hydroxylase gene transcription. Proc Natl Acad Sci U S A. 2011;108(10):4200–4205. doi: 10.1073/pnas.1101193108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferri AL, Lin W, Mavromatakis YE, Wang JC, Sasaki H, Whitsett JA, Ang SL. Foxa1 and Foxa2 regulate multiple phases of midbrain dopaminergic neuron development in a dosage-dependent manner. Development. 2007;134(15):2761–2769. doi: 10.1242/dev.000141. [DOI] [PubMed] [Google Scholar]
- 26.Wederell ED, Bilenky M, Cullum R, Thiessen N, Dagpinar M, Delaney A, Varhol R, Zhao Y, Zeng T, Bernier B, Ingham M, Hirst M, Robertson G, Marra MA, Jones S, Hoodless PA. Global analysis of in vivo Foxa2-binding sites in mouse adult liver using massively parallel sequencing. Nucleic acids research. 2008;36(14):4549–4564. doi: 10.1093/nar/gkn382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merika M, Thanos D. Enhanceosomes. Curr Opin Genet Dev. 2001;11(2):205–208. doi: 10.1016/s0959-437x(00)00180-5. [DOI] [PubMed] [Google Scholar]
- 28.Ptashne M, Gann A. Genes & signals. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2002. [Google Scholar]
- 29.Agoston Z, Heine P, Brill MS, Grebbin BM, Hau AC, Kallenborn-Gerhardt W, Schramm J, Gotz M, Schulte D. Meis2 is a Pax6 co-factor in neurogenesis and dopaminergic periglomerular fate specification in the adult olfactory bulb. Development. 2014;141(1):28–38. doi: 10.1242/dev.097295. [DOI] [PubMed] [Google Scholar]
- 30.Grebbin BM, Hau AC, Gross A, Anders-Maurer M, Schramm J, Koss M, Wille C, Mittelbronn M, Selleri L, Schulte D. Pbx1 is required for adult subventricular zone neurogenesis. Development. 2016;143(13):2281–2291. doi: 10.1242/dev.128033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohwi M, Osumi N, Rubenstein JL, Alvarez-Buylla A. Pax6 is required for making specific subpopulations of granule and periglomerular neurons in the olfactory bulb. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2005;25(30):6997–7003. doi: 10.1523/JNEUROSCI.1435-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dellovade TL, Pfaff DW, Schwanzel-Fukuda M. Olfactory bulb development is altered in small-eye (Sey) mice. The Journal of comparative neurology. 1998;402(3):402–418. [PubMed] [Google Scholar]
- 33.Ninkovic J, Pinto L, Petricca S, Lepier A, Sun J, Rieger MA, Schroeder T, Cvekl A, Favor J, Gotz M. The transcription factor Pax6 regulates survival of dopaminergic olfactory bulb neurons via crystallin alphaA. Neuron. 2010;68(4):682–694. doi: 10.1016/j.neuron.2010.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun J, Rockowitz S, Xie Q, Ashery-Padan R, Zheng D, Cvekl A. Identification of in vivo DNA-binding mechanisms of Pax6 and reconstruction of Pax6-dependent gene regulatory networks during forebrain and lens development. Nucleic acids research. 2015;43(14):6827–6846. doi: 10.1093/nar/gkv589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim SM, Yang JW, Park MJ, Lee JK, Kim SU, Lee YS, Lee MA. Regulation of human tyrosine hydroxylase gene by neuron-restrictive silencer factor. Biochemical and biophysical research communications. 2006;346(2):426–435. doi: 10.1016/j.bbrc.2006.05.142. [DOI] [PubMed] [Google Scholar]
- 36.Suo H, Wang P, Tong J, Cai L, Liu J, Huang D, Huang L, Wang Z, Huang Y, Xu J, Ma Y, Yu M, Fei J, Huang F. NRSF is an essential mediator for the neuroprotection of trichostatin A in the MPTP mouse model of Parkinson’s disease. Neuropharmacology. 2015;99:67–78. doi: 10.1016/j.neuropharm.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 37.Yang JW, Choi EY, Park MJ, Lee MA. Expression of tyrosine hydroxylase is epigenetically regulated in neural stem cells. Biochemical and biophysical research communications. 2011;414(4):712–718. doi: 10.1016/j.bbrc.2011.09.141. [DOI] [PubMed] [Google Scholar]
- 38.Palm K, Belluardo N, Metsis M, Timmusk T. Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene. The Journal of neuroscience: the official journal of the Society for Neuroscience. 1998;18(4):1280–1296. doi: 10.1523/JNEUROSCI.18-04-01280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mori N, Mizuno T, Murai K, Nakano I, Yamashita H. Effect of age on the gene expression of neural-restrictive silencing factor NRSF/REST. Neurobiol Aging. 2002;23(2):255–262. doi: 10.1016/s0197-4580(01)00286-x. [DOI] [PubMed] [Google Scholar]
- 40.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ong CT, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15(4):234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baker H, Kobayashi K, Okano H, Saino-Saito S. Cortical and striatal expression of tyrosine hydroxylase mRNA in neonatal and adult mice. Cell Mol Neurobiol. 2003;23(4–5):507–518. doi: 10.1023/A:1025015928129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamamoto K, Ruuskanen JO, Wullimann MF, Vernier P. Two tyrosine hydroxylase genes in vertebrates New dopaminergic territories revealed in the zebrafish brain. Molecular and cellular neurosciences. 2010;43(4):394–402. doi: 10.1016/j.mcn.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 44.Filippi A, Mahler J, Schweitzer J, Driever W. Expression of the paralogous tyrosine hydroxylase encoding genes th1 and th2 reveals the full complement of dopaminergic and noradrenergic neurons in zebrafish larval and juvenile brain. The Journal of comparative neurology. 2010;518(4):423–438. doi: 10.1002/cne.22213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen YC, Priyadarshini M, Panula P. Complementary developmental expression of the two tyrosine hydroxylase transcripts in zebrafish. Histochemistry and cell biology. 2009;132(4):375–381. doi: 10.1007/s00418-009-0619-8. [DOI] [PubMed] [Google Scholar]
- 46.Iwawaki T, Kohno K, Kobayashi K. Identification of a potential nurr1 response element that activates the tyrosine hydroxylase gene promoter in cultured cells. Biochemical and biophysical research communications. 2000;274(3):590–595. doi: 10.1006/bbrc.2000.3204. [DOI] [PubMed] [Google Scholar]
- 47.Kim KS, Kim CH, Hwang DY, Seo H, Chung S, Hong SJ, Lim JK, Anderson T, Isacson O. Orphan nuclear receptor Nurr1 directly transactivates the promoter activity of the tyrosine hydroxylase gene in a cell-specific manner. Journal of neurochemistry. 2003;85(3):622–634. doi: 10.1046/j.1471-4159.2003.01671.x. [DOI] [PubMed] [Google Scholar]
- 48.Smidt MP, Smits SM, Bouwmeester H, Hamers FP, van der Linden AJ, Hellemons AJ, Graw J, Burbach JP. Early developmental failure of substantia nigra dopamine neurons in mice lacking the homeodomain gene Pitx3. Development. 2004;131(5):1145–1155. doi: 10.1242/dev.01022. [DOI] [PubMed] [Google Scholar]
- 49.Hwang DY, Ardayfio P, Kang UJ, Semina EV, Kim KS. Selective loss of dopaminergic neurons in the substantia nigra of Pitx3-deficient aphakia mice. Brain research Molecular brain research. 2003;114(2):123–131. doi: 10.1016/s0169-328x(03)00162-1. [DOI] [PubMed] [Google Scholar]
- 50.van den Munckhof P, Luk KC, Ste-Marie L, Montgomery J, Blanchet PJ, Sadikot AF, Drouin J. Pitx3 is required for motor activity and for survival of a subset of midbrain dopaminergic neurons. Development. 2003;130(11):2535–2542. doi: 10.1242/dev.00464. [DOI] [PubMed] [Google Scholar]
- 51.Stott SR, Metzakopian E, Lin W, Kaestner KH, Hen R, Ang SL. Foxa1 and foxa2 are required for the maintenance of dopaminergic properties in ventral midbrain neurons at late embryonic stages. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2013;33(18):8022–8034. doi: 10.1523/JNEUROSCI.4774-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arenas E, Denham M, Villaescusa JC. How to make a midbrain dopaminergic neuron. Development. 2015;142(11):1918–1936. doi: 10.1242/dev.097394. [DOI] [PubMed] [Google Scholar]
- 53.Sheela Rani CS, Soto-Pina A, Iacovitti L, Strong R. Evolutionary conservation of an atypical glucocorticoid-responsive element in the human tyrosine hydroxylase gene. Journal of neurochemistry. 2013;126(1):19–28. doi: 10.1111/jnc.12294. [DOI] [PubMed] [Google Scholar]
- 54.Liu N, Cigola E, Tinti C, Jin BK, Conti B, Volpe BT, Baker H. Unique regulation of immediate early gene and tyrosine hydroxylase expression in the odor-deprived mouse olfactory bulb. The Journal of biological chemistry. 1999;274(5):3042–3047. doi: 10.1074/jbc.274.5.3042. [DOI] [PubMed] [Google Scholar]
- 55.Nagamoto-Combs K, Piech KM, Best JA, Sun B, Tank AW. Tyrosine hydroxylase gene promoter activity is regulated by both cyclic AMP-responsive element and AP1 sites following calcium influx. Evidence for cyclic amp-responsive element binding protein-independent regulation. The Journal of biological chemistry. 1997;272(9):6051–6058. doi: 10.1074/jbc.272.9.6051. [DOI] [PubMed] [Google Scholar]
- 56.Saino-Saito S, Sasaki H, Volpe BT, Kobayashi K, Berlin R, Baker H. Differentiation of the dopaminergic phenotype in the olfactory system of neonatal and adult mice. The Journal of comparative neurology. 2004;479(4):389–398. doi: 10.1002/cne.20320. [DOI] [PubMed] [Google Scholar]
- 57.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137(7):1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Choi EY, Yang JW, Park MS, Sun W, Kim H, Kim SU, Lee MA. Transgenic mice expressing yellow fluorescent protein under control of the human tyrosine hydroxylase promoter. J Neurosci Res. 2012;90(10):1949–1959. doi: 10.1002/jnr.23085. [DOI] [PubMed] [Google Scholar]
- 59.Kaneda N, Sasaoka T, Kobayashi K, Kiuchi K, Nagatsu I, Kurosawa Y, Fujita K, Yokoyama M, Nomura T, Katsuki M, et al. Tissue-specific and high-level expression of the human tyrosine hydroxylase gene in transgenic mice. Neuron. 1991;6(4):583–594. doi: 10.1016/0896-6273(91)90061-4. [DOI] [PubMed] [Google Scholar]
- 60.Akahoshi E, Yoshimura S, Uruno S, Ishihara-Sugano M. Effect of dioxins on regulation of tyrosine hydroxylase gene expression by aryl hydrocarbon receptor: a neurotoxicology study. Environ Health. 2009;8:24. doi: 10.1186/1476-069X-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Penkov D, Mateos San Martin D, Fernandez-Diaz LC, Rossello CA, Torroja C, Sanchez-Cabo F, Warnatz HJ, Sultan M, Yaspo ML, Gabrieli A, Tkachuk V, Brendolan A, Blasi F, Torres M. Analysis of the DNA-binding profile and function of TALE homeoproteins reveals their specialization and specific interactions with Hox genes/proteins. Cell Rep. 2013;3(4):1321–1333. doi: 10.1016/j.celrep.2013.03.029. [DOI] [PubMed] [Google Scholar]