Abstract
Pseudomonas fluorescens Pf-5 is a plant commensal bacterium that inhabits the rhizosphere and produces secondary metabolites that suppress soilborne plant pathogens. The complete sequence of the 7.1-Mb Pf-5 genome was determined. We analyzed repeat sequences to identify genomic islands that, together with other approaches, suggested P. fluorescens Pf-5's recent lateral acquisitions include six secondary metabolite gene clusters, seven phage regions and a mobile genomic island. We identified various features that contribute to its commensal lifestyle on plants, including broad catabolic and transport capabilities for utilizing plant-derived compounds, the apparent ability to use a diversity of iron siderophores, detoxification systems to protect from oxidative stress, and the lack of a type III secretion system and toxins found in related pathogens. In addition to six known secondary metabolites produced by P. fluorescens Pf-5, three novel secondary metabolite biosynthesis gene clusters were also identified that may contribute to the biocontrol properties of P. fluorescens Pf-5.
Supplementary information
The online version of this article (doi:10.1038/nbt1110) contains supplementary material, which is available to authorized users.
Main
Pseudomonas spp. are ubiquitous inhabitants of soil, water and plant surfaces that belong to the Gamma subclass of Proteobacteria. Many pseudomonads live in a commensal relationship with plants, utilizing nutrients exuded from plant surfaces and surviving environmental stress by occupying protected sites provided by the plant's architecture. These commensal species can have profound effects on plants by suppressing pests, enhancing access to key nutrients, altering physiological processes or degrading environmental pollutants. Pseudomonads have an exceptional capacity to produce a wide variety of metabolites, including antibiotics that are toxic to plant pathogens1,2. Antibiotic production by plant-associated Pseudomonas spp. enhances the fitness of the producing strain3 and suppresses pathogens that would otherwise jeopardize plant health1,2,4. Certain antibiotic-producing strains of Pseudomonas spp. function as biological control agents; their capacity to protect plants from disease distinguishes them as microorganisms with immense effects on agricultural productivity.
Among the plant commensals, P. fluorescens Pf-5 is notable as a biological control organism, for its rhizosphere competence and the spectrum of antibiotics and other secondary metabolites that it produces. P. fluorescens Pf-5 inhabits the rhizosphere of many plants and suppresses plant diseases caused by soilborne plant pathogens5,6,7,8,9,10,11. P. fluorescens Pf-5 produces a suite of antibiotics including pyrrolnitrin5, pyoluteorin11 and 2,4-diacetylphloroglucinol12. It also produces hydrogen cyanide and the siderophores pyochelin and pyoverdine, which can suppress target pathogens in the rhizosphere through iron competition13,14. In this study, we report the complete genome sequence of P. fluorescens Pf-5, and highlight genes with a demonstrated or proposed role in biological control or rhizosphere colonization.
Note: Supplementary information is available on the Nature Biotechnology website.
Results
Genome features and comparative genomics
The P. fluorescens Pf-5 genome is composed of one circular chromosome of 7,074,893 bp (Fig. 1). A total of 6,144 open reading frames (ORFs) were identified within the P. fluorescens Pf-5 genome (Table 1). Among the predicted genes, 3,822 (63%) are assigned a putative function and 330 (5%) are hypothetical genes. The P. fluorescens Pf-5 genome is larger than that of the three other pseudomonads whose genomic sequences have been published: P. aeruginosa PAO115, an opportunistic animal pathogen, P. putida KT244016, a saprophytic soil bacterium, and P. syringae DC300017, a plant pathogen.
Figure 1. Circular representation of the P. fluorescens Pf-5 overall genome structure.
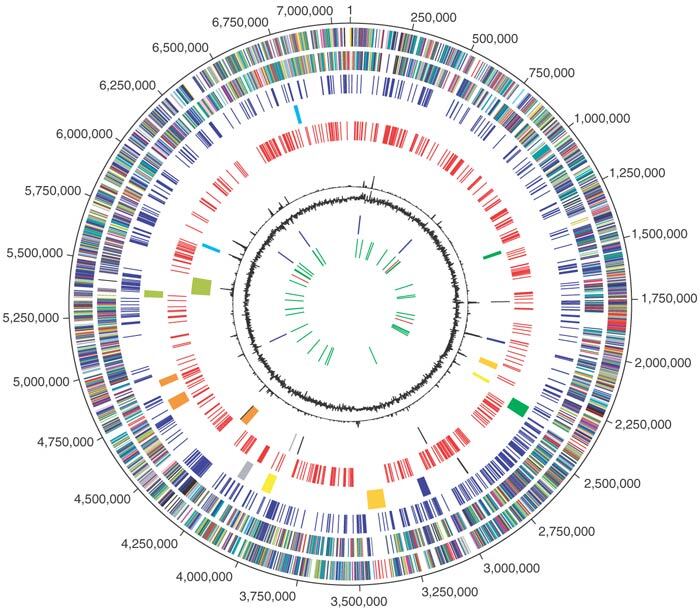
The outer scale designates coordinates in base pairs (bp). The first circle shows predicted coding regions on the plus strand color-coded by role categories: violet, amino acid biosynthesis; light blue, biosynthesis of cofactors, prosthetic groups and carriers; light green, cell envelope; red, cellular processes; brown, central intermediary metabolism; yellow, DNA metabolism; light gray, energy metabolism; magenta, fatty acid and phospholipid metabolism; pink, protein synthesis and fate; orange, purines, pyrimidines, nucleosides and nucleotides; olive, regulatory functions and signal transduction; dark green, transcription; teal, transport and binding proteins; gray, unknown function; salmon, other categories; blue, hypothetical proteins. The second circle shows predicted coding regions on the minus strand color-coded by role categories. The third circle shows the set of 656 P. fluorescens Pf-5 genes that are not conserved in any of the other three Pseudomonas genomes whose sequences have been published (see Fig. 2). The fourth circle shows nine regions encoding secondary metabolism gene clusters coded by color as follows: green, decapeptide biosynthesis; blue, pyoluteorin biosynthesis; gold, polyketide biosynthesis; yellow, pyochelin biosynthesis; gray, pyrrolnitrin biosynthesis; orange, pyoverdine biosynthesis; olive, nonribosomal peptide synthesis; cyan, 2,4-diacetylphloroglucinol synthesis. The fifth circle shows REP repeat elements. The sixth circle shows transposases in black, the predicted PFGI-1 mobile island in olive, and putative phage regions as follows: green, prophage 1; blue, prophage 2; gold, prophage 3; yellow, prophage 4; gray, prophage 5; orange, prophage 6; cyan, prophage 7. The seventh circle shows trinucleotide composition in black. The eighth circle shows percentage G+C in relation to the mean G+C in a 2,000-bp window. The ninth circle shows rRNA genes in blue and the tenth circle shows tRNA genes in green and sRNA genes in red.
Table 1.
General genome features of sequenced pseudomonads
| P. fluorescens | P. aeruginosa a | P. putida a | P. syringaea , b | |
|---|---|---|---|---|
| Size (base pairs) | 7,074,893 | 6,264,403 | 6,181,863 | 6,538,260 |
| G+C content (%) | 63.3 | 66.6 | 61.6 | 58.4 |
| Protein-coding genes | ||||
| No. similar to known proteins | 3,822 | 3,021 | 3,311 | 3,524 |
| No. similar to proteins of unknown function c | 890 | NA | 522 | 647 |
| No. of conserved hypotheticals d | 1,113 | 769 | 995 | 976 |
| No. with no database match e | 330 | 1,780 | 596 | 628 |
| Total | 6,144 | 5,570 | 5,420 | 5,763 |
| Average ORF size (bp) | 1,021 | 1,005 | 998 | 991 |
| Coding (%) | 88.8 | 89.4 | 88 | 87 |
| rRNA | 5 | 4 | 7 | 5 |
| tRNA | 71 | 63 | 74 | 63 |
aThe distribution of ORFs in the P. aeruginosa, P. putida and P. syringae chromosomes are derived from the original annotation. These numbers, particularly hypothetical and conserved hypothetical proteins, may be significantly different with updated blast searches and annotation.
bThe genome information for P. syringae represents combined data from the chromosome and two plasmids.
cUnknown function, significant sequence similarity to a named protein for which no function is currently attributed.
dConserved hypothetical protein, sequence similarity to a translation of another ORF, however no experimental evidence for protein expression exists.
eHypothetical protein, no significant similarity to any other sequenced protein.
Only a limited degree of gene synteny was observed between P. fluorescens Pf-5 and the other sequenced pseudomonads (Supplementary Fig. 1 online). Regions of limited conservation of gene order are centered in an X-pattern around the terminus of replication18, suggestive of frequent rearrangement and inversion events around the terminus of replication, as found in other genera18. A four way BLAST comparison of the sequenced pseudomonads identified a core set of over 4,000 genes conserved in all four species (Fig. 2). Based on this analysis, 656 genes unique to P. fluorescens Pf-5 were identified. Only four insertion elements were identified within the P. fluorescens Pf-5 genome, fewer than in other Pseudomonas spp. genomes.
Figure 2. Venn diagram showing the number of P. fluorescens Pf-5 predicted proteins with significant homology (P < 10−5) with the predicted proteins from the sequenced P. aeruginosa PAO1, P. putida KT2440 and P. syringae DC3000 genomes.
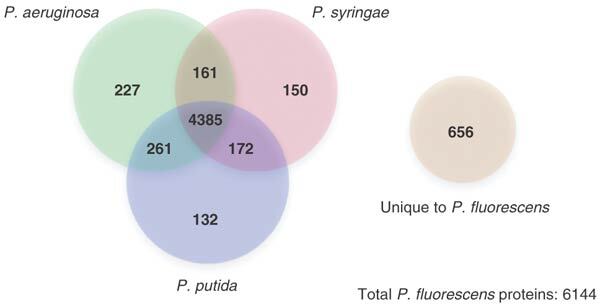
The number outside the circles (656) represents the number of P. fluorescens Pf-5 proteins that do not have significant homologs in any of the Pseudomonas species examined.
We used four independent criteria to identify unique genomic islands within the P. fluorescens Pf-5 genome that potentially reflect recent biological adaptations to the commensal lifestyle of this bacterium. The four criteria were (i) distribution of the 656 genes unique to P. fluorescens Pf-5, (ii) atypical trinucleotide composition, (iii) presence of putative integrated phage and transposable elements and (iv) distribution of a unique P. fluorescens Pf-5 REP element (see below).
Analysis of short repeats in the P. fluorescens Pf-5 genome identified a REP repeat similar to that described in the genome of P. putida19. An initial count of 14 mers revealed an imperfect palindrome in two varieties: 5′-GCCGGCTTGCCGGC-3′ (n = 379) and 5′-GCTGGCTTGCCAGC-3′ (n = 278). A hidden Markov model (HMM) trained with the palindrome and its flanking sequences identified 1,052 gap-free copies, with sequence similarity across 34 bp. The 34-bp sequences were found as a single element, as pairs of elements or in clusters of up to 11 elements with alternating orientation. The 34-bp sequences are rarely detected in the genome sequences of P. aeruginosa (n = 21) or P. syringae (n = 17), and are related to but distinctly different from the previously characterized REP element in P. putida (n = 509). The REP sequences are not uniformly distributed throughout the genome of P. fluorescens Pf-5; instead, the presence of discrete regions lacking REP sequences is striking (Fig. 1). REP sequences were absent in regions under strong selective pressure for maintenance, such as rRNA and ribosomal protein gene clusters. Regions unique to P. fluorescens Pf-5, with an atypical trinucleotide composition and in some cases including putative phage genes, correlated well with regions lacking REP repeat sequences. Hence, the analysis of the distribution of conserved repetitive elements in a genome may represent a useful approach for identifying recently-acquired genomic islands.
We identified nine secondary metabolite gene clusters in the genome of P. fluorescens Pf-5. Two clusters contain biosynthetic genes for the siderophores pyoverdine and pyochelin; these genes are conserved in other pseudomonads and do not display atypical trinucleotide composition or unusual REP distribution. One cluster that codes for the production of hydrogen cyanide is conserved in P. aeruginosa and likewise does not show unusual REP distribution or trinucleotide composition. The other six secondary metabolite gene clusters, which are unique to P. fluorescens Pf-5, essentially lack REP elements. Several of the six gene clusters have atypical trinucleotide compositions and two are located adjacent to putative prophage, which is suggestive of recent lateral acquisitions by the P. fluorescens Pf-5 genome.
Seven prophage (Supplementary Fig. 2 online) and a phage-related genomic island collectively encompass ∼268 kbp of the genome of P. fluorescens Pf-5 (Fig. 1). Particularly notable are prophage 1 with a tail gene module that is closely related to Shigella flexneri phage SfV, a pyocin-like prophage 3 with a P2-like myovirus tail region and a large lambdoid prophage 6 that is mosaic in nature and similar to putative prophage elements found in Yersinia spp., Burkholderia spp. and Ralstonia solanacearum. The remaining elements represent defective prophages that are reduced in size and/or complexity and often contain degraded phage-related genes. Six out of eight phage-like regions are associated with intact or mutated genes encoding site-specific integrases (Supplementary Fig. 2 online). In addition to typical bacteriophage functions, some prophage elements carry genes encoding for putative lytic and serotype-converting enzymes or bacteriocins. Only prophages 1 and 3 have counterparts in P. putida, P. syringae pv. tomato and P. aeruginosa (Supplementary Fig. 2 online). The 115-kbp mobile genomic island, PFGI-1, is site-specifically integrated into one of the two tRNALys genes. PFGI-1 is a hybrid genomic island that has elements of both temperate phages and conjugative plasmids and is closely related to the mobile genomic island pKLC102 from P. aeruginosa C20. The predicted phage regions correlate with the absence of REP repeat elements, often display atypical nucleotide composition and are found adjacent to SOS-regulon genes or within genes encoding for tRNA.
A number of other regions were evident that displayed an atypical trinucleotide composition and lacked REP elements, including three regions with putative polysaccharide biosynthesis genes, and one region with homologs of filamentous hemagglutinin (PFL1552, PFL1556) that may represent putative adhesins.
A type I restriction modification system (PFL2964/PFL2965) is present in P. fluorescens Pf-5, which may explain why P. fluorescens Pf-5 has proven recalcitrant to the introduction of foreign DNA via molecular biology approaches. This system is absent in other sequenced pseudomonads with the exception of one subunit that is shared with Pseudomonas putida. Genes for copper resistance, organized in pairs (copAB, copCD, and copRS), are dispersed throughout the genome of Pf-5. These genes are likely to be responsible for the moderate level of copper resistance exhibited by Pf-5 (M.D. Henkels and C.M.P., unpublished data).
Metabolism, transport and regulation
Central metabolic pathways in P. fluorescens Pf-5 are similar to those in other sequenced pseudomonads. Components of complete Entner-Doudoroff, pentose phosphate and tricarboxylic acid cycle pathways were identified. Consistent with Pseudomonas metabolism21,22, P. fluorescens Pf-5 lacks a phosphofructokinase gene and thus doesn't use the Embden-Meyerhof-Parnas pathway for hexose catabolism.
Similar to P. putida16,22, P. fluorescens Pf-5 is diverse in its catabolic capabilities. Present in its genome are several extracellular hydrolytic enzymes, including chitinases, proteases and lipases, which are involved in the degradation of polymers commonly found in soil. Also present are components for the catabolism of most amino acids. In contrast to P. aeruginosa, P. fluorescens Pf-5 shares with P. putida and/or P. syringae, a number of hydrolases that support utilization of plant-derived carbohydrates, including those that degrade sucrose, maltose, trehalose and xylans. The P. fluorescens Pf-5 genome also contains catabolic capabilities for more complex compounds, including aromatic derivatives and long-chain hydrocarbons. Consistent with the description of the species23, there are complete pathways for degradation of the aromatic compounds catechol and protocatechuate by ortho cleavage leading to the β-ketoadipate pathway. Also present are many derivative pathways, including vanillinate, benzoate, p-hydroxybenzoate, and phenylacetate, which are directed at degradation of aromatic compounds originating from plant products like lignin. Similar to P. aeruginosa15, a vast array of genes involved in β-oxidation exists within P. fluorescens Pf-5. Many of these genes are involved in degradation of hydrocarbon molecules of various chain lengths, providing capabilities to utilize long chain alkanes, fatty acids and oils commonly found in plant tissues.
P. fluorescens Pf-5 possesses an extensive set of transport genes. P. fluorescens Pf-5 is capable of transporting various sugars, presumably derived from the breakdown of plant carbohydrates. P. fluorescens Pf-5 has more than double the number of sugar phosphotransferase system transporters seen in the other pseudomonads, with predicted specificities for fructose, N-acetylglucosamine, trehalose and cellobiose. P. fluorescens Pf-5 retains broad capabilities for amino acid uptake: it has 21 amino acid/polyamine/choline family amino acid transporters, similar to the numbers found in P. putida and P. aeruginosa and more than in P. syringae. These broad uptake capabilities may contribute to spermosphere and rhizosphere colonization, since sugars and amino acids are major components of seed and root exudates24,25. P. fluorescens Pf-5 also has an expanded collection of metabolite efflux systems, with 24 Resistance to Homoserine/Threonine (RhtB) family amino acid efflux pumps, and 37 Drug/Metabolite Transporter (Dmt) family metabolite efflux pumps, possibly indicating a need to protect against toxic concentrations of metabolites or metabolic analogs.
Bacteria with large genomes and complex metabolisms typically have sophisticated regulatory networks relative to bacteria with smaller genomes15. P. fluorescens Pf-5 is no exception to this generalization with a complex array of regulatory systems including 32 predicted sigma factors, more than 300 genes encoding predicted transcriptional regulators and a variety of two-component signal transduction systems consisting of 82 histidine kinase domains and 120 response regulator domains.
Secondary metabolite biosynthesis
P. fluorescens Pf-5 produces four metabolites that are toxic to fungi or oomycetes and important in biocontrol: two well-characterized polyketides pyoluteorin11 and 2,4-diacetylphloroglucinol12, the chlorinated tryptophan derivative pyrrolnitrin5, and hydrogen cyanide26, which is formed by oxidation of glycine27 (Fig. 3a,b). The gene clusters for each of these compounds are found in the genomic sequence of P. fluorescens Pf-5 and are similar to those previously reported28,29,30,31. Three previously unknown gene clusters with characteristics of secondary metabolite biosynthetic regions were identified (Fig. 3a). Combined with gene clusters for the biosynthesis of two siderophores (pyoverdine and pyochelin), it is estimated that 5.7% of the P. fluorescens Pf-5 genome (∼400 kb) is dedicated to secondary metabolism, the largest percentage among the Pseudomonas spp. genomes sequenced to date.
Figure 3. Secondary metabolite gene clusters in P. fluorescens Pf-5.
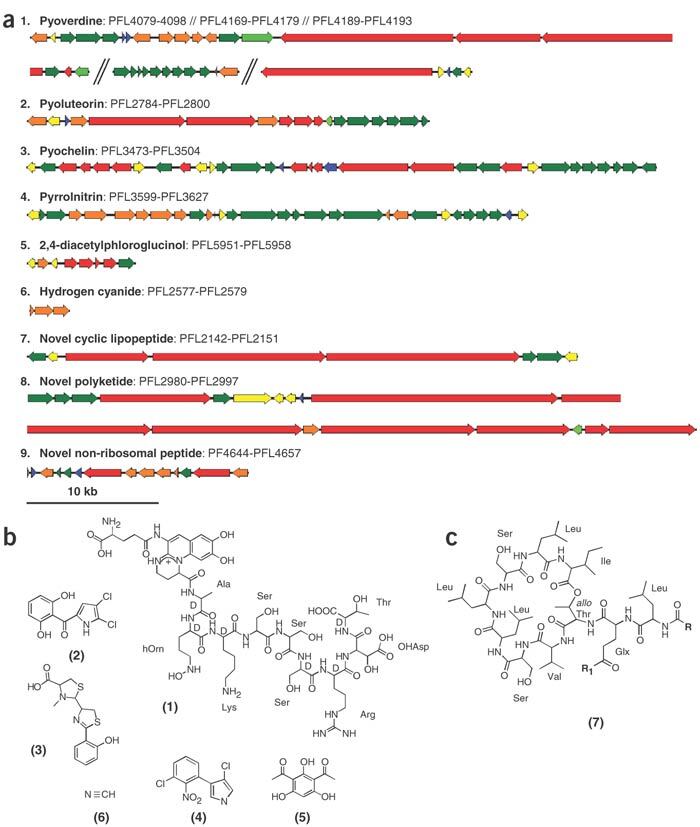
(a) Representation of each biosynthetic cluster discovered on the genome of P. fluorescens Pf-5. Red, structural genes (NRPS/PKS); yellow, regulatory genes; green, transporter/resistance genes; orange, accessory genes; and blue, hypothetical genes. (b) Chemical structure of the product of the biosynthetic gene clusters for compounds known to be produced by P. fluorescens Pf-5. (1) pyoverdine; (2) pyoluteorin; (3) pyochelin; (4) pyrrolnitrin; (5) 2,4-diacetylphloroglucinol; and (6) hydrogen cyanide. (c) Predicted structure of a novel cyclic lipopeptide. Glx, (R1= −NH2) or glutamic acid (R1= −OH). Precise chemical predictions are not available for pathways 8 and 9.
The biosyntheses of pyoluteorin and 2,4-diacetylphloroglucinol are catalyzed by polyketide synthases (PKSs) involving the sequential addition and modification of simple carboxyl acids to a growing carbon chain. A novel gene cluster of ∼102 kb was identified (Fig. 3a, cluster 8) whose predicted gene functions include four PKSs (PFL2990, PFL2991, PFL2993 and PFL2994), a hybrid nonribosomal peptide synthetase (NRPS)/PKS (PFL2989), a cytochrome p450 (PFL2992) and a methyltransferase (PFL2995). Each of the four PKSs contain two to four modules that together are predicted to synthesize a molecule composed of a starting serine residue and 14 unknown carboxyl acids.
A 42-kb gene cluster was identified that contains three NRPSs, each catalyzing the integration of two to four amino acid residues into a predicted cyclic decapeptide (Fig. 3a, cluster 7). This putative product resembles cyclic lipopeptides of the viscosin group (Fig. 3c). The C-terminus of the last NRPS (PFL2147) encodes two thioesterase domains, which could be responsible for both cyclization and addition of a lipidic side chain, a similar role to SyrC in the syringomycin biosynthesis. The first NRPS (PFL2145) lacks the characteristic loading domain, whose functions could potentially be provided by a protein containing an AMP-binding domain (PFL2162) also encoded within the cluster. Sequence data of known gene clusters for viscosin-like compounds are not available. Lipopeptides produced by certain strains of Pseudomonas spp. have surfactant and antibiotic properties that can contribute to biological control of plant disease32. P. fluorescens Pf-5 produces detectable surfactant activity and RT-PCR data indicates that the genes from this cluster are expressed under conditions where the surfactant is produced (V.O. Stockwell, M.D. Henkels and J.E.L., unpublished data).
Secondary metabolites produced by P. fluorescens have a direct role in the bacterium's capacity to suppress plant diseases, and can serve as signals influencing gene expression by coinhabiting cells in the rhizosphere. The three putative secondary metabolites whose biosynthesis is conferred by the newly discovered gene clusters are of unknown function but, in parallel with the roles of known secondary metabolites produced by P. fluorescens Pf-5, they could serve as important compounds to agriculture as well as in the ecology of the bacterium. Using the predicted structures of these novel compounds should facilitate their isolation and characterization.
Siderophore biosynthesis
The fluorescent pseudomonads are characterized by the production of pyoverdines, a diverse class of siderophores containing a chromophore, which is responsible for the UV fluorescence typifying the group, linked to a small peptide of varied length and composition that is synthesized nonribosomally33. In P. fluorescens Pf-5, genes required for pyoverdine biosynthesis and uptake are present in three gene clusters, separated by 136.4 kb and 9.8 kb (Fig. 3a). Despite some deviations in gene placement and orientation, the general organization of pyoverdine biosynthesis and uptake genes is similar to that found in other fluorescent pseudomonads34. Additionally, the gene clusters responsible for the nonribosomal biosynthesis of the siderophore pyochelin (Fig. 3a) and the iron chelator salicylic acid were identified and found linked in the genome.
Commensal lifestyle
P. fluorescens Pf-5 has the capacity to colonize plant surfaces, a characteristic shared with many bacterial plant pathogens. Therefore, characteristics contributing to epiphytic fitness on plant surfaces, such as iron acquisition and stress tolerance, are common to both plant-commensal and plant-pathogenic bacteria. Pseudomonas spp. are known to use siderophores produced by other microorganisms as sources of iron, and this capacity contributes to their fitness in iron-limited environments35. TonB-dependent, outer-membrane receptor proteins, which are responsible for the specific uptake of each ferric-siderophore complex, also play an important signaling role in the bacterial cell36. The genome of P. fluorescens Pf-5 contains 45 genes for putative TonB-dependent receptors, and sequences of 28 of the genes are similar to known receptors of ferric-siderophore complexes found in other organisms. These include six putative receptors for pyoverdines, one in the pyoverdine biosynthetic gene cluster, another linked to the pyochelin biosynthetic gene cluster, and four distributed widely throughout the genome. P. fluorescens Pf-5 also has multiple receptors for the general classes of hydroxamate siderophores and catechol siderophores, as well as many receptors whose precise specificity is not clear (Supplementary Fig. 3 online). The presence of multiple receptor genes with apparent redundancy for a given siderophore argues for the importance of iron acquisition in the ecology of the bacterium, and also may be related to a signaling role for these proteins.
Like P. fluorescens Pf-5, other Pseudomonas spp. have many genes encoding for putative TonB-dependent proteins in their genomes. A very complex evolutionary history of this gene family is apparent from a phylogenetic tree constructed from each of the four Pseudomonas spp. whose genomes have been sequenced (Supplementary Fig. 3 online). Within each node of the phylogenetic tree, there are representative proteins from each of the four species, indicating that this is an ancient class of proteins in the genus. Different Pseudomonas spp. are enriched for proteins within a given node of the phylogenetic tree. The specificities of the receptors for recognition of pyoverdines, which are a diverse class of siderophores, are not well understood, and the presence of multiple copies of pyoverdine receptors may confer on P. fluorescens Pf-5 the ability to use a broad spectrum of these compounds.
Active oxygen species such as superoxide (O2−) are produced by plant cells in response to numerous stimuli37. These active oxygen species have antimicrobial properties, and rhizosphere bacteria counter their toxicity with superoxide dismutases (SODs), which convert superoxide to H2O2, and catalases, which convert peroxide to water. P. fluorescens Pf-5 has two SODs and six catalases, more than the other sequenced pseudomonads. Other enzymes in P. fluorescens Pf-5 that may play a role in scavenging reactive oxygen species include ten peroxidases (three cytochrome C peroxidases, three glutathione peroxidases, two dyp-type peroxidases, a thiol peroxidase and an alkyl peroxidase). None of the peroxidase genes are unique to P. fluorescens Pf-5; all are shared with at least one other Pseudomonas genome. The presence of numerous copies of genes conferring tolerance to oxidative stress in the genome of P. fluorescens Pf-5 supports the proposed importance of oxidative stress tolerance to fitness in the rhizosphere38.
P. fluorescens Pf-5 lacks a number of virulence factors found in plant pathogens, consistent with its commensal lifestyle. There is no evidence in the P. fluorescens Pf-5 genome for the biosynthesis of known P. syringae phytotoxins or cellulases, pectinases, or pectin lyases associated with degradation of plant cell walls and cell wall components. Additionally, we found no evidence in P. fluorescens Pf-5 for a type III protein secretion system, frequently found in bacterial pathogens of plants and animals39. This is in contrast to the presence of a 20-kb gene cluster resembling a type III secretion system in the rhizosphere bacterium P. fluorescens SBW2540.
Discussion
The complete P. fluorescens Pf-5 genome sequence provides a variety of insights into this organism's commensal lifestyle and biocontrol capabilities. It has revealed pathways for biosynthesis of hitherto unknown secondary metabolites that may contribute to biocontrol. Its potential to use other siderophores and diverse host-derived compounds, and its antibiotic- and oxidative stress–resistance capacities likely provide a foundation for the success of P. fluorescens Pf-5 as a commensal organism in the highly competitive environment of the rhizosphere. This study has also pioneered the analysis of repeat sequences as a methodology for identifying recent lateral acquisitions. From these analyses it seems that the larger genome size of P. fluorescens Pf-5 compared with other pseudomonads is mainly attributable to the acquisition of various genomic islands, particularly phage-derived islands and secondary metabolite gene clusters. Finally, we emphasize that the complete genome sequence of P. fluorescens Pf-5 provides a framework for future studies to understand the biological basis of biocontrol.
Methods
Genome sequencing and annotation.
The complete genome sequence of P. fluorescens Pf-5 was determined using the whole-genome shotgun method41. Physical and sequencing gaps were closed using a combination of primer walking, generation and sequencing of transposon-tagged libraries of large-insert clones, and multiplex PCR42. Identification of putative protein-encoding genes and annotation of the genome were performed as previously described43. An initial set of ORFs predicted to encode proteins was identified with GLIMMER44. ORFs consisting of fewer than 30 codons and those containing overlaps were eliminated. Frame shifts and point mutations were corrected or designated 'authentic.' Functional assignment, identification of membrane-spanning domains, determination of paralogous gene families and identification of regions of unusual nucleotide composition were performed as previously described43. Sequence alignments and phylogenetic trees were generated using the methods described previously43.
Trinucleotide composition.
Distribution of all 64 trinucleotides (3 mers) was determined, and the 3-mer distribution in 2,000-bp windows that overlapped by half their length (1,000 bp) across the genome was computed. For each window, we computed the χ2 statistic on the difference between its 3-mer content and that of the whole chromosome. A large value for χ2 indicates the 3-mer composition in this window is different from the rest of the chromosome. Probability values for this analysis are based on assumptions that the DNA composition is relatively uniform throughout the genome, and that 3-mer composition is independent. Because these assumptions may be incorrect, we prefer to interpret high χ2 values as indicators of regions on the chromosome that appear unusual and demand further scrutiny.
Comparative genomics.
The P. fluorescens Pf-5 and other sequenced Pseudomonas genomes were compared at the nucleotide level by suffix tree analysis using MUMmer45. P. fluorescens Pf-5 ORFs were compared by BLAST against the complete set of ORFs from Pseudomonas genomes using an E-value cutoff of 10−5.
GenBank accession.
The complete annotated genome sequence is available in GenBank accession number CP000076.
Supplementary information
Visualization of P. fluorescens genomic synteny compared to selected Pseudomonas spp. (PDF 34742 kb)
Dot plot comparison of five largest P. fluorescens Pf-5 prophages with those from P. fluorescens Q8r1-96, P. fluorescens Pf0-1, P. putida KT2440, P. syringae pv. tomato DC3000 and P. aeruginosa PAO1. (PPT 43 kb)
Phylogenetic tree of the family of TonB-dependent receptor proteins from Pf-5, P. aeruginosa PA01, P.syringae DC3000 and P. putida KT2440. (PPT 86 kb)
Acknowledgements
This work was supported by the Initiative for Future Agriculture and Food Systems of the USDA Cooperative State Research, Education and Extension Service, grant number 2001-52100-11329. We thank Daniel Arp, the TIGR faculty, sequencing facility and informatics group for expert advice and assistance. We also thank Genevieve DeClerk for bioinformatics support and Walter Mahaffee for his helpful review of the manuscript.
Accession codes
Accessions
GenBank/EMBL/DDBJ
Competing interests
The authors declare no competing financial interests.
Contributor Information
Ian T Paulsen, Email: ipaulsen@tigr.org.
Joyce E Loper, Email: loperj@mail.science.oregonstate.edu.
References
- 1.Haas D, Keel C. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 2003;41:117–153. doi: 10.1146/annurev.phyto.41.052002.095656. [DOI] [PubMed] [Google Scholar]
- 2.Raaijmakers JM, Vlami M, de Souza JT. Antibiotic production by bacterial biocontrol agents. Antonie Van Leeuwenhoek. 2002;81:537–547. doi: 10.1023/a:1020501420831. [DOI] [PubMed] [Google Scholar]
- 3.Mazzola M, Cook RJ, Thomashow LS, Weller DM, Pierson LS., III Contribution of phenazine antibiotic biosynthesis to the ecological competence of fluorescent pseudomonads in soil habitats. Appl. Environ. Microbiol. 1992;58:2616–2624. doi: 10.1128/aem.58.8.2616-2624.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weller DM, Raaijmakers JM, Gardener BB, Thomashow LS. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 2002;40:309–348. doi: 10.1146/annurev.phyto.40.030402.110010. [DOI] [PubMed] [Google Scholar]
- 5.Howell CR, Stipanovic RD. Control of Rhizoctonia solani in cotton seedlings with Pseudomonas fluorescens and with an antibiotic produced by the bacterium. Phytopathology. 1979;69:480–482. [Google Scholar]
- 6.Kraus J, Loper JE. Lack of evidence for a role of antifungal metabolite production by Pseudomonas fluorescens Pf-5 in biological control of Pythium damping-off of cucumber. Phytopathology. 1992;82:264–271. [Google Scholar]
- 7.Pfender WF, Kraus J, Loper JE. A genomic region from Pseudomonas fluorescens Pf-5 required for pyrrolnitrin production and inhibition of Pyrenophora tritici-repentis in wheat straw. Phytopathology. 1993;83:1223–1228. [Google Scholar]
- 8.Rodriguez F, Pfender WF. Antibiosis and antagonism of Sclerotinia homoeocarpa and Drechslera poae by Pseudomonas fluorescens Pf-5 in vitro and in planta. Phytopathology. 1997;87:614–621. doi: 10.1094/PHYTO.1997.87.6.614. [DOI] [PubMed] [Google Scholar]
- 9.Sharifi-Tehrani A, Zala M, Natsch A, Moenne-Loccoz Y, Defago G. Biocontrol of soil-borne fungal plant diseases by 2,4-diacetylphloroglucinol-producing fluorescent pseudomonads with different restriction profiles of amplified 16S rDNA. Eur. J. Plant Pathol. 1998;104:631–643. [Google Scholar]
- 10.Xu GW, Gross DC. Selection of fluorescent pseudomonads antagonistic to Erwinia carotovora and suppressive of potato seed piece decay. Phytopathology. 1986;76:414–422. [Google Scholar]
- 11.Howell CR, Stipanovic RD. Suppression of Pythium ultimum induced damping-off of cotton seedlings by Pseudomonas fluorescens and its antibiotic pyoluteorin. Phytopathology. 1980;70:712–715. [Google Scholar]
- 12.Nowak-Thompson B, Gould SJ, Kraus J, Loper JE. Production of 2,4-diacetylphloroglucinol by the biocontrol agent Pseudomonas fluorescens Pf-5. Can. J. Microbiol. 1994;40:1064–1066. [Google Scholar]
- 13.Buysens S, Heungens K, Poppe J, Höfte M. Involvement of pyochelin and pyoverdin in suppression of Pythium-induced damping-off of tomato by Pseudomonas aeruginosa 7NSK2. Appl. Environ. Microbiol. 1996;62:865–871. doi: 10.1128/aem.62.3.865-871.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loper JE, Buyer JS. Siderophores in microbial interactions on plant surfaces. Mol. Plant Microbe Interact. 1991;4:5–13. [Google Scholar]
- 15.Stover CK, et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:947–948. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 16.Nelson KE, et al. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 2002;4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- 17.Buell CR, et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. 2003. Proc. Natl. Acad. Sci. USA. 2003;100:10181–10186. doi: 10.1073/pnas.1731982100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisen JA, Heidelberg JF, White O, Salzberg SL. Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol. 2000;1:0011.1–0011.9. doi: 10.1186/gb-2000-1-6-research0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aranda-Olmedo I, Tobes R, Manzanera M, Ramos JL, Marqués S. Species-specific repetitive extragenic palindromic (REP) sequences in Pseudomonas putida. Nucleic Acids Res. 2002;30:1826–1833. doi: 10.1093/nar/30.8.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klockgether J, Reva O, Larbig K, Tummler B. Sequence analysis of the mobile genome island pKCL102 of Pseudomonas aeruginosa C. J. Bacteriol. 2004;186:518–534. doi: 10.1128/JB.186.2.518-534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lessie TG, Phibbs PV., Jr. Alternative pathways of carbohydrate utilization in pseudomonads. Annu. Rev. Microbiol. 1984;38:359–387. doi: 10.1146/annurev.mi.38.100184.002043. [DOI] [PubMed] [Google Scholar]
- 22.dos Santos VAPM, Heim S, Moore ERB, Strätz M, Timmis KN. Insights into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environ. Microbiol. 2004;6:1264–1286. doi: 10.1111/j.1462-2920.2004.00734.x. [DOI] [PubMed] [Google Scholar]
- 23.Krieg NR, Holt JG. Bergey's Manual of Systematic Bacteriology. 1984. [Google Scholar]
- 24.Curl EA, Truelove B. The Rhizosphere. 1986. [Google Scholar]
- 25.Roberts DP, et al. Role of pfkA and general carbohydrate catabolism in seed colonization by Enterobacter cloacae. Appl. Environ. Microbiol. 1999;65:2513–2519. doi: 10.1128/aem.65.6.2513-2519.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whistler CA, Corbell NA, Sarniguet A, Ream W, Loper JE. The two-component regulators GacS and GacA influence accumulation of the stationary-phase sigma factor sigmaS and the stress response in Pseudomonas fluorescens Pf-5. J. Bacteriol. 1998;180:6635–6641. doi: 10.1128/jb.180.24.6635-6641.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castric PA. Glycine metabolism by Pseudomonas aeruginosa: hydrogen cyanide biosynthesis. J. Bacteriol. 1977;130:826–831. doi: 10.1128/jb.130.2.826-831.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laville J, et al. Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHA0. J. Bacteriol. 1998;180:3187–3196. doi: 10.1128/jb.180.12.3187-3196.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammer PE, Hill DS, Lam ST, Van Pee KH, Ligon JM. Four genes from Pseudomonas fluorescens that encode the biosynthesis of pyrrolnitrin. Appl. Environ. Microbiol. 1997;63:2147–2154. doi: 10.1128/aem.63.6.2147-2154.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bangera MG, Thomashow LS. Identification and characterization of a gene cluster for synthesis of the polyketide antibiotic 2,4-Diacetylphloroglucinol from Pseudomonas fluorescens Q2-87. J. Bacteriol. 1999;181:3155–3163. doi: 10.1128/jb.181.10.3155-3163.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nowak-Thompson B, Chaney N, Wing JS, Gould SJ, Loper JE. Characterization of the pyoluteorin biosynthetic gene cluster of Pseudomonas fluorescens Pf-5. J. Bacteriol. 1999;181:2166–2174. doi: 10.1128/jb.181.7.2166-2174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Souza JT, de Boer M, de Waard P, van Beek TA, Raaijmakers JM. Biochemical, genetic and zoosporicidal properties of cyclic lipopeptide surfactants produced by Pseudomonas fluorescens. Appl. Environ. Microbiol. 2003;69:7161–7172. doi: 10.1128/AEM.69.12.7161-7172.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer JM. Pyoverdines: pigments, siderophores and potential taxonomic markers of fluorescent Pseudomonas species. Arch. Microbiol. 2000;174:135–142. doi: 10.1007/s002030000188. [DOI] [PubMed] [Google Scholar]
- 34.Ravel J, Cornelis P. Genomics of pyoverdine-mediated iron uptake in pseudomonads. Trends Microbiol. 2003;11:195–200. doi: 10.1016/s0966-842x(03)00076-3. [DOI] [PubMed] [Google Scholar]
- 35.Loper JE, Henkels MD. Utilization of heterologous siderophores enhances levels of iron available to Pseudomonas putida in the rhizosphere. Appl. Environ. Microbiol. 1999;65:5357–5363. doi: 10.1128/aem.65.12.5357-5363.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clarke TE, Tari LW, Vogel HJ. Structural biology of bacterial iron uptake systems. Curr. Top. Med. Chem. 2001;1:7–30. doi: 10.2174/1568026013395623. [DOI] [PubMed] [Google Scholar]
- 37.Mori IC, Schroeder JI. Reactive oxygen species activation of plant Ca2+ channels. A signaling mechanism in polar growth, hormone transduction, stress signaling, and hypothetically mechanotransduction. Plant Physiol. 2004;135:702–708. doi: 10.1104/pp.104.042069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim YC, Miller CD, Anderson AJ. Superoxide dismutase activity in Pseudomonas putida affects utilization of sugars and growth on root surfaces. Appl. Environ. Microbiol. 2000;66:1460–1467. doi: 10.1128/aem.66.4.1460-1467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cornelis GR, Van Gijsegem F. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 2000;54:735–774. doi: 10.1146/annurev.micro.54.1.735. [DOI] [PubMed] [Google Scholar]
- 40.Preston GM, Bertrand N, Rainey PB. Type III secretion in plant growth-promoting Pseudomonas fluorescens SBW25. Mol. Microbiol. 2001;41:999–1014. doi: 10.1046/j.1365-2958.2001.02560.x. [DOI] [PubMed] [Google Scholar]
- 41.Fraser CM, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 42.Tettelin H, Radune D, Kasif S, Khouri H, Salzberg SL. Optimized multiplex PCR: efficiently closing a whole-genome shotgun sequencing project. Genomics. 1999;62:500–507. doi: 10.1006/geno.1999.6048. [DOI] [PubMed] [Google Scholar]
- 43.Paulsen IT, et al. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. USA. 2002;99:13148–13153. doi: 10.1073/pnas.192319099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salzberg SL, Delcher AL, Kasif S, White O. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delcher AL, Phillippy A, Carlton J, Salzberg SL. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Visualization of P. fluorescens genomic synteny compared to selected Pseudomonas spp. (PDF 34742 kb)
Dot plot comparison of five largest P. fluorescens Pf-5 prophages with those from P. fluorescens Q8r1-96, P. fluorescens Pf0-1, P. putida KT2440, P. syringae pv. tomato DC3000 and P. aeruginosa PAO1. (PPT 43 kb)
Phylogenetic tree of the family of TonB-dependent receptor proteins from Pf-5, P. aeruginosa PA01, P.syringae DC3000 and P. putida KT2440. (PPT 86 kb)


