Abstract
Based on a broad public database compilation, we support the hypothesis that germinal polymorphisms may regulate the expression of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) cellular target itself and proteases controlling the process of its shedding or, conversely, its internalization. Consequently, a genetic influence on individual susceptibility to coronavirus disease 2019 (COVID-19) infection is strongly suspected.
Keywords: pharmacogenetics, SNP, genetic variation, coronavirus, infection
General Background
In addition to the need for virus detection, evaluation of individual serological response [1], and biological analytical tools to manage COVID-19 on a population level, there is an urgent need to obtain objective information to identify at-risk individuals and to understand the marked variability in the severity of the disease in general, as well as in given populations. A current hypothesis is that SARS-CoV-2 clinical manifestations are governed by human genetics [2]. Thus, in this context, here we develop two complementary themes: (i) a more thorough examination of the membrane shedding of angiotensin-converting enzyme 2 (ACE2), the SARS-CoV-2 cellular target, and its potential repercussion on virus propagation; and (ii) a description of the interindividual variability of the genes (SNPs) involved in ACE2 processing and their potential impact on the risk of contracting COVID-19.
ACE2 Expression and COVID-19
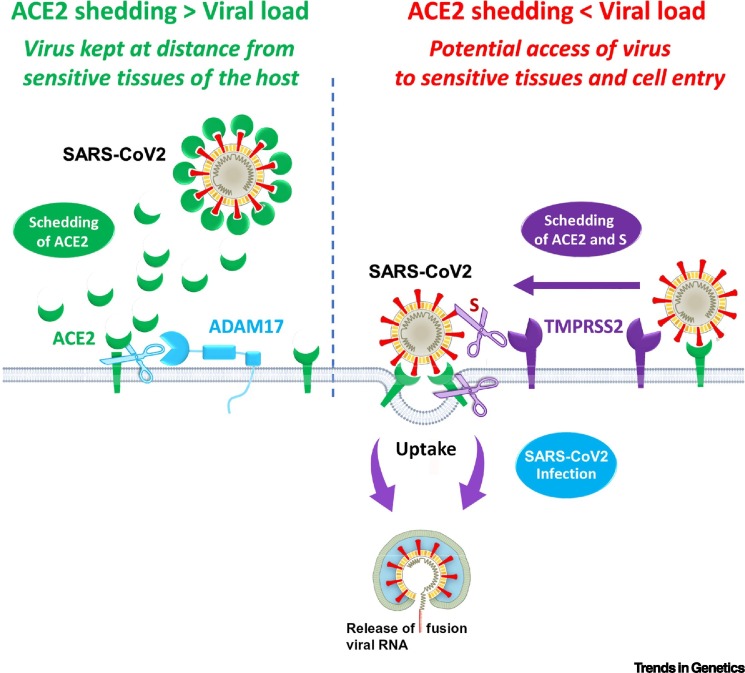
Chen et al. recently examined a large Genotype–Tissue Expression (GTEx) database and investigated the expression of ACE2 in different human tissues [3]. The authors stressed that, counterintuitively, expression of the SARS-CoV-2 target was inversely related to certain risk factors, showing higher levels in Asian females compared with Asian males and a significant decrease in patients with type 2 diabetes mellitus. Globally, at a population level, there was a negative correlation between ACE2 expression and COVID-19 severity. Recent data provide evidence that ACE2 is effectively shed from membranes, a process that is fine-tuned at different levels [4] involving two cell membrane proteases: disintegrin and metalloproteinase domain-containing protein 17 (ADAM17) and transmembrane protease serine 2 (TMPRSS2) [4]. More precisely, ADAM17 acts directly on ACE2 and leads to ACE2 shedding into the extracellular cellular space, while TMPRSS2 cleaves not only ACE2, but also the S protein of SARS-CoV-2, thus leading to membrane fusion and cellular uptake of the virus. Consequently, while ADAM17 and TMPRSS2 both act on ACE2, they may have opposite effects on net ACE2 shedding (Figure 1 ). When the respective proteolytic activities of ADAM17 and TMPRSS2 result in more ACE2 shedding than internalization, it follows that this situation may constitute a natural barrier to infection. This could be due to the interaction between soluble ACE2 with the virus at a distance from sensitive tissues. Subsequently, on this basis, one can hypothesize that, when the viral load is high, the shedding barrier effect is overwhelmed, thus facilitating subsequent infection.
Figure 1.
Graphic Support for the Hypothesis of Angiotensin-Converting Enzyme 2 (ACE2)-Related Virus Neutralization at Distance due to the Presence of Enzymatic Membrane Mechanisms Regulating ACE2 Shedding and Virus Entry.
Abbreviations: ADAM17, disintegrin and metalloproteinase domain-containing protein 17; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TMPRSS2, transmembrane protease serine 2.
ACE2, TMPRSS2, and ADAM17 Gene Polymorphisms
Cao et al. [5] compiled a database analysis of all 1700 variants in the region of the ACE2 gene located on the X chromosome. They identified 15 unique expression quantitative trait loci (eQTLs; see Glossary) variants [14 SNPs and 1 insertion/deletion (INDEL)] with higher minor allele frequencies (MAF) in the Asian population than in a European population (MAF of 0.05 versus 0.35–0.48 for the top six most common variants). Interestingly, their data showed that the 11 most common variants (MAF >0.05) were associated with increased expression of ACE2 in tissues, suggesting, according to the authors, a different sensitivity to SARS-CoV-2 infectivity. However, the functional basis of the influence of SNP on ACE2 expression remains to be established.
In this context, we performed a complementary in silico study including SNPs regulating gene expression not only for ACE2, but also for ADAM17 and TMPRSS2 (Table S1 in the supplemental information online). Overall, and based on the ACE2 expression-associated MAF between ethnic populations, it appears that Asians express a higher level of ACE2 than Caucasians, while Africans show an intermediary level of ACE2 expression. This is consistent with the findings previously reported by Cao et al . [5]. It is still debatable whether these differences should be taken into consideration in epidemiological studies on COVID-19 covering ethnic associations with disease occurrence [6]. Importantly, the diseases associated with a high level of SARS-Cov-2 infection (hypertension and diabetes) were found to be related to a lower expression of ACE2, in relation to the respective allelic distribution. This relationship concurs well with the study by Chen et al. pointing towards a negative correlation between ACE2 expression and COVID-19 severity [3].
It has been reported that subjects with rs383510/T and rs2070788/G genotypes of TMPSRSS2 located on chromosome 21q22.3 are more prone to develop a severe form of A (H1N1) influenza and acute respiratory distress syndrome [7]. Of note, males have been shown to be more likely to develop a severe form of H1N1 influenza and there is evidence that androgens are positive regulators of TMPRSS2 [8]. Importantly, the alleles at risk (T for rs383510 and G for rs2070788) are linked to increased gene expression (Table S1 in the supplemental information online), logically supporting the hypothesis of a higher level of viral cell entry. It is tempting to extrapolate this SNP influence to SARS-CoV-2 infectivity.
The ADAM17 locus on chromosome 2p25.1 presents two clusters and three unique SNPs that induce strong differences in terms of allelic profiles between Asian and European populations that are associated with hypertension [9] and/or sepsis [10]. Of note, most of these SNPs are located in the promoter region of ADAM17 and are associated with either positive or negative eQTL, depending on the SNP and the tissue (Table S1 in the supplemental information online). Therefore, there is a strong possibility that genetic polymorphisms influencing ADAM17 expression also contribute to the modulation of ACE2 shedding intensity.
Practical Consequences
Taken together, the above-discussed data advocate in favor of a multifactorial genetic impact on the risk of SARS-Cov-2 infectivity and possible disease severity. A relatively simple and easy-to-perform test, such as quantitative PCR [11] or MASSarray [12], would allow large-scale individual SNP profiling for ACE2, ADAM17, and TMPRSS2 to identify possible at-risk populations vulnerable to viral infection. On this basis, a ‘multiSNPs risk score’ could be established that would be applicable to large populations and, thus, it might then be possible to identify subjects carrying a combination of favorable alleles for ACE2, ADAM17, and TMPRSS2 conferring a lesser risk of contracting SARS-Cov-2 infection, and vice versa. Such an analytical strategy was recently developed based on patient genetic characteristics for immunogenetic profiling designed to personalized immunotherapy [12]. A similar supervised genetic approach to COVID-19 risk assessment could complement the current unsupervised GWAS investigations, which require large population studies exploring the whole patient genome for DNA variations in an attempt to explain interindividual differences in COVID-19 severity [e.g., the Howard Hughes Medical Institute (HHMI) genetic projecti and the COVID-19 Human Genetic Effort [2]). Ideally, in a final step, a multifactorial predictive index could be established incorporating SNP analysis and other, more established, risk factors.
In summary, until now, genetic influences on COVID-19 interindividual susceptibility have been largely underestimated; thus, we hope that the discussion might fill this gap and will pave the way for confirmatory investigations at experimental and clinical levels.
Acknowledgments
The authors would like to thank Jocelyn Gal, Emmanuel Chamorey, George Morgan, and Christiane Brahimi-Horn for helpful comments. Funding is acknowledged from the French Government (Agence Nationale de Recherche, ANR) through the 'Investments for the Future' LABEX SIGNALIFE (ANR-11-LABX-0028-01) and (AD-ME project R19162DD); CANC'AIR Genexposomic project, Canceropole PACA; DREAL PACA, ARS PACA, Région Sud, INSERM cancer; INCA Plan Cancer; and Children Medical Safety Research Institute (CMSRI, Vaccinophagy project R17033DJA).
Glossary
- Expression quantitative trait locus (eQTL)
a genomic locus that explains the variation in gene expression of nearby genes.
- Insertion/deletion (Indel)
an insertion or deletion of bases in the genome.
- Minor allele frequency (MAF)
frequency at which the second most common allele occurs in a given population.
Footnotes
Supplemental information associated with this article can be found online https://doi.org/10.1016/j.tig.2020.08.003.
Resources
iwww.hhmi.org/news/patients-with-severe-forms-of-coronavirus-disease-could-offer-clues-to-treatmentSupplemental Information
Supplementary material
References
- 1.Xie C. Comparison of different samples for 2019 novel coronavirus detection by nucleic acid amplification tests. Int. J. Infect. Dis. 2020;93:264–267. doi: 10.1016/j.ijid.2020.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casanova J.-L. A global effort to define the human genetics of protective immunity to SARS-CoV-2 infection. Cell. 2020;181:1194–1199. doi: 10.1016/j.cell.2020.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J. Individual variation of the SARS-CoV-2 receptor ACE2 gene expression and regulation. Aging Cell. 2020;19 doi: 10.1111/acel.13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heurich A. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2014;88:1293–1307. doi: 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao Y. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 2020;6:4–7. doi: 10.1038/s41421-020-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yi Y. COVID-19: what has been learned and to be learned about the novel coronavirus disease. Int. J. Biol. Sci. 2020;16:1753–1766. doi: 10.7150/ijbs.45134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng Z. Identification of TMPRSS2 as a susceptibility gene for severe 2009 pandemic A(H1N1) influenza and A(H7N9) influenza. J. Infect. Dis. 2015;212:1214–1221. doi: 10.1093/infdis/jiv246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin B. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. 1999;59:4180–4184. [PubMed] [Google Scholar]
- 9.Li Y. Association between ADAM17 promoter polymorphisms and ischemic stroke in a Chinese population. J. Atheroscler. Thromb. 2014;21:878–893. doi: 10.5551/jat.22400. [DOI] [PubMed] [Google Scholar]
- 10.Shao Y. Association study between promoter polymorphisms of ADAM17 and progression of sepsis. Cell. Physiol. Biochem. 2016;39:1247–1261. doi: 10.1159/000447830. [DOI] [PubMed] [Google Scholar]
- 11.Katsanis S.H., Katsanis N. Molecular genetic testing and the future of clinical genomics. Nat. Rev. Genet. 2013;14:415–426. doi: 10.1038/nrg3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Refae S. Germinal Immunogenetics predict treatment outcome for PD-1/PD-L1 checkpoint inhibitors. Investig. New Drugs. 2020;38:160–171. doi: 10.1007/s10637-019-00845-w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material