Abstract
Introduction
The emergence of a novel coronavirus, SARS-CoV-2, has highlighted the need for rapid, accurate, and point-of-care diagnostic testing. As of now, there is not enough testing capacity in the world to meet the stated testing targets, which are expected to skyrocket globally for broader testing during reopening
Aim
This review focuses on the development of lab-on-chip biosensing platforms for diagnosis of COVID-19 infection.
Results
We discuss advantages of utilizing lab-on-chip technologies in response to the current global pandemic, including their potential for low-cost, rapid sample-to-answer processing times, and ease of integration into a range of healthcare settings. We then highlight the development of magnetic, colorimetric, plasmonic, electrical, and lateral flow-based lab-on-chip technologies for the detection of SARS-CoV-2, in addition to other viruses. We focus on rapid, point-of-care technologies that can be deployed at scale, as such devices could be promising alternatives to the current gold standard of reverse transcription-polymerase chain reaction (RT-PCR) diagnostic testing.
Conclusion
This review is intended to provide an overview of the current state-of-the-field and serve as a resource for innovative development of new lab-on-chip assays for COVID-19 detection.
Keywords: Lab-on-chip, Biosensor, Coronavirus, COVID-19, Diagnostic
Introduction
As of May 29 2020, the COVID-19 pandemic was responsible for over 5.8 million diagnosed cases and over 360,000 deaths worldwide.73 Coronaviruses are a large family of viruses characterized by their spiky viral capsids, and have been responsible for a number of outbreaks including SARS and MERS. COVID-19 is caused by the SARS-CoV-2 coronavirus. The virus was first reported in December 2019 in Wuhan City in the Hubei Province of China and has since spread to over 187 countries globally.33,42 Researchers are still actively working to better characterize the biology of the virus and its epidemiology in humans23,27 to enhance our understanding of disease transmission and clinical manifestation. In addition to immediate impact on global health, the COVID-19 pandemic has had a significant social and economic impact worldwide, in part due to implemented social distancing measures and world-wide closures.10
Improved molecular and serological diagnostic testing is key to improved patient outcomes and preventing spread of infection.20,46 Testing availability has become increasingly important as new data indicate that a large proportion of infected individuals are asymptomatic, resulting in possible further spread of disease while hosts remain symtom-free.36 Furthermore, rapid diagnostic testing is critical to evaluating risk associated with reopening workplaces, educational institutions, and other social and cultural establishments. Both the public and private sector have been actively working to meet demand for diagnostic testing capacity and required reagents and consumables, such as swabs, extraction kits, and buffers.10,70 To date, almost all diagnostic testing for the virus occurs in centralized laboratories, and involves expensive laboratory equipment, lengthy assays, and trained laboratory technicians.27,70
The gold standard molecular test for the detection of SARS-CoV-2 RNA is reverse transcription-polymerase chain reaction (RT-PCR), which relies on nucleic acid amplification for viral detection.10,75 Serological assays are also used to measure the presence of target antibodies and/or antigens, and are utilized as an indicator of past infection. These assays generally take the form of a standard enzyme-linked immunosorbent assay (ELISA) or lateral flow assay (LFA).20,36,89 While these tests are robust in a clinical laboratory setting, they are extremely time intensive. Development of a point-of-care assay would allow for more timely testing, and earlier containment of infected patients.44
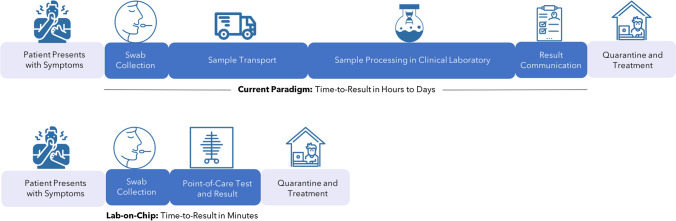
Figure 1 highlights the utility of point-of-care testing in decreasing the existing diagnostic timelines. Given the potential for rapid, point-of-care results, lab-on-chip sensors can decrease the total-analytical-time from hours/days to minutes. This could allow patients to get care sooner, reduce unknowing transmission to others, and minimize the burden on overstrained clinical labs. While a few recent studies have worked towards detection of SARS-CoV-2 using portable lab-on-chip platforms, this is still an emerging area of research with significant potential for future impact on disease surveillance, monitoring, and diagnosis.
Figure 1.
Potential impact of lab-on-chip diagnostics to patient workflow. (Top) Existing diagnostic workflows require sample collection, transport, processing, and result communication to the patient. (Bottom) Point-of-care tests enabled by lab-on-chip technologies reduce lengthy workflow and can provide a result within minutes.
Icons courtesy of the Noun Project.
In this review, we discuss on-chip biosensors, their applicability to the current pandemic, and relevant design criteria, including cost, total-analytical-time, sensitivity, portability, and limit of detection. We begin by providing a brief overview of the biology of SARS-CoV-2. We also highlight key viral biomarkers that can be employed for diagnostic testing, including RNA and surface antigens. After reviewing FDA Emergency Use Authorization (EUA)-approved clinical diagnostics, we then discuss emerging lab-on-chip systems for SARS-SoV-2 detection. We explore technology platforms such as magnetic enrichment, lateral flow devices, plasmonic devices, and electrochemical sensors. In each category, we overview recent literature directly related to COVID-19. We also discuss other lab-on-chip platforms for viral detection that could be translated to SARS-CoV-2 detection. We end with an insightful summary about how the field of lab-on-chip devices can help contribute to the point-of-care diagnosis of COVID-19. Ultimately, this review aims to serve as a useful guide to those interested in understanding existing and emerging lab-on-chip platforms for COVID-19 diagnosis at the point-of-care.
SARS-CoV-2 Biology and Biomarkers
Understanding the clinical manifestations of COVID-19 disease and the biology of SARS-CoV-2 virus is critical to the design of effective diagnostic platforms. It is important to note that our understanding of virus and the disease is rapidly changing as new peer-reviewed research is published. Coronaviruses are relatively large viruses (> 100 nm), which express a spike protein on their envelope. Expression of this protein allows the virus to enter human cells by binding to ACE receptors.17,30,84 Patients who are infected with SARS-CoV-2 typically present with a range of symptoms, including fever, shortness of breath, dry cough, fever, muscle pain, and loss of taste and/or smell.29,51
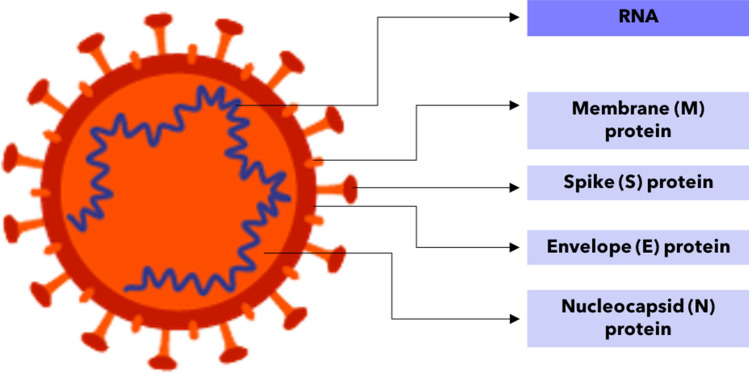
The two most commonly targeted biomarkers used for viral diagnostics are viral genetic material in the form of DNA or RNA and viral proteins found on the viral envelope.46 Antibodies to the virus can also be detected through serology tests. The following two subsections breakdown the structure of the SARS-CoV-2 virus, with a focus on nucleic acid and protein biomarkers for lab-on-chip applications. An overview of the SARS-CoV-2 viral capsid and its relevant nucleic acid and protein biomarkers is shown in Fig. 2.
Figure 2.
Structure and biomarkers of SARS-CoV-2 Virus. Viral RNA, membrane protein, spike protein, envelope protein, and nucleocapsid protein shown on the SARS-CoV-2 virus.
Adapted from Cyranoski, Nature News Feature.17
SARS-CoV-2 RNA Detection
SARS-CoV-2 is a single-stranded RNA virus that has high genetic similarity to SARS-CoV and other coronaviruses.24 Currently, the gold standard for molecular testing is reverse transcription-polymerase chain reaction (RT-PCR), which amplifies SARS-CoV-2 genetic material. Targeted genes include the ORF1b, ORF8, nucleocapsid (N), spike (S) protein, RNA-dependent RNA polymerase (RdRP), and envelope (E) genes.10 As published in an April 2020 study, 90% of 112 available molecular assays for detecting SARS-CoV-2 used PCR or RT-PCR.10
Nucleic acids can either be isolated using lengthy extraction protocols, or can be captured directly from the sample.90 Both of these methods are most often followed by PCR to amplify the viral gene of interest. Some methods of viral nucleic acid sensing without PCR include hybridization or enzymatic assays, such as a study that uses a DNAzyme to cleave target DNA and generate a signal if viral DNA is present.87 Some emerging RNA-based technologies also integrate CRISPR technology for viral nucleic acid sensing.46,89
SARS-CoV-2 Antigen Detection
Viral antigens be captured by using antibody-antigen interactions, with the capture antibody immobilized on a nanoparticle or sensor surface. There are at least four main structural proteins in SARS-Cov-2 that may be useful targets for viral detection, including the spike (S) protein, membrane (M) protein, envelope (E) protein, and nucleocapsid (N) protein. The structure of the spike protein has been thoroughly evaluated and is found to be essential for entry into the host cell, making it a promising sensor target.78 Diagnostic tests have already been commercialized for the spike (S) and nucleocapsid (N) proteins.16,19,52
Lab-on-Chip Biosensors: Overview and Fabrication
Lab-on-Chip Biosensors
A biosensor is an analytical device that detects the presence of a particular biological substance.62,67 Typically, biosensors include (1) a recognition element, which selectively captures the biological target of interest; (2) a transduction element, which converts the recognition into a measurable signal; and (3) electronics and/or an amplifier to read out the signal.4 When integrated with sample collection and processing, biosensors can be powerful platforms for the rapid quantification of biological analytes of interest for both disease diagnosis and environmental monitoring.7 Given the need for rapid information on (1) population infection status, and (2) the presence of virus in the environment, biosensors can play a critical role in disease diagnosis and surveillance in the ongoing global pandemic.
Small-scale, sample-to-answer diagnostics, often called “lab-on-a-chip” devices have been applied to a range of clinical scenarios. These platforms have grown in popularity due to their ability to automate laboratory functions and to integrate several laboratory functions onto a single chip.8,31,43,74 Through innovations in micro- and nano-scale technology, these devices afford advantages in sensitivity, total-analytical-time, portability, and ease-of-use.60,65,67
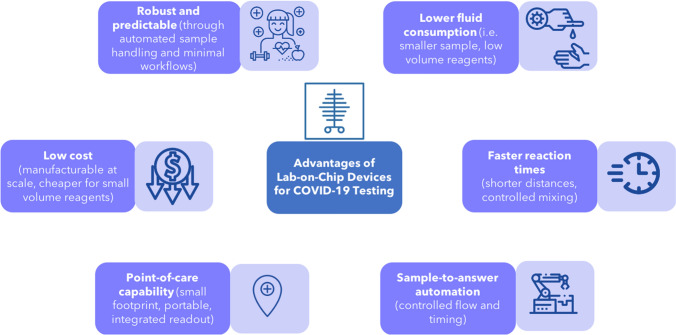
A number of advantages of lab-on-chip devices are particularly relevant to the current COVID-19 pandemic. Specifically, lab-on-chip devices are robust, rapid, sensitive, low-cost, and can provide results at the point-of-care.7,31,64,66,67,69 In the context of COVID-19, these advantages would help support crucial efforts to increase access to testing. An overview of the advantages of lab-on-chip devices in the context of COVID-19 testing can be found in Fig. 3.
Figure 3.
Advantages of lab-on-chip devices for COVID-19 testing. Advantages of lab-on-chip devices include low fluid consumption, fast reaction times, sample-to-answer automation, point-of-care capability, low cost, and robustness.
Icons courtesy of the Noun Project.
Lab-on-Chip Device Fabrication
Innovations in microfluidic and microfabrication processes have enabled the production of micro- and nano-scale lab-on-chip (LOC) devices. Microfluidic LOC devices can be classified into five different groups based on the liquid propulsion principle: capillary, pressure-driven, centrifugal, electrokinetic and acoustic-driven.40 LOC devices can be manufactured from a range of materials, including silicon, glass, and polymeric materials though various fabrication methodologies.7 Examples of commonly employed microfabrication methods include photolithography, deposition, etching, lift-off lithography, and bulk/surface micromachining.88 Additionally, soft lithography is a patterning technique that is frequently used for soft materials (e.g. polydimethylsiloxane). Soft lithography includes methods such as replica molding, microcontact printing, and micro-transfer molding.82 Recently, paper-based LOC devices have emerged as a low-cost, portable, and disposable point-of-care platforms.61 Commonly employed fabrication methods for paper-based devices include wax printing, alkyl ketene dimer (AKD) printing, flexographic printing, and layer-by-layer 3D affixing.85
Existing Clinical Diagnostics for SARS-CoV-2
Many centralized clinical diagnostics rely on nucleic acid extraction followed by RT-PCR (reverse transcriptase polymerase chain reaction).13,53 This works through first purifying nucleic acids from the collected sample through centrifugation or magnetic bead separation, followed by amplification of relevant RNA sequences. The presence of SARS-CoV-2 RNA indicates that the patient currently has the disease.36,72 A number of primers have been developed and validated to capture the sequences of RNA targeted during the assay.58 The clinical microbiology community has been assessing not only new diagnostic tests, but also the sample collection method (i.e. nasopharyngeal or oropharyngeal swabs), transport media (viral media or other), and monitoring protocol.70 Recently, a group from Rutgers University developed a validated test that uses saliva samples rather than a more invasive upper respiratory swab.3,21,56 The FDA has approved a number of molecular diagnostics under Emergency Use Authorizations (EUA) to increase the availability of technologies.75 In addition to in vitro diagnostic authorizations, the FDA has also authorized personal protective equipment, ventilators, and other medical devices. A selection of the over 75 EUA approved in vitro diagnostics (as of May 19, 2020) are shown in Table 1.
Table 1.
Selected FDA EUA approved diagnostics.
| Date EUA issued | Manufacturer | Diagnostic (letter of authorization) | Technology | Authorized settings |
|---|---|---|---|---|
| 04-02-2020 | Centers for Disease Control and Prevention’s (CDC) | CDC 2019-nCoV real-time RT-PCR diagnostic panel (CDC) | Molecular | H |
| 12-03-2020 | Roche Molecular Systems, Inc. (RMS) | cobas SARS-CoV-2 | Molecular | H, M |
| 13-03-2020 | Thermo Fisher Scientific, Inc. | TaqPath COVID-19 combo kit | Molecular | H |
| 16-03-2020 | Laboratory Corporation of America (LabCorp) | COVID-19 RT-PCR test | Molecular | H |
| 16-03-2020 | Hologic, Inc. | Panther Fusion SARS-CoV-2 assay | Molecular | H |
| 17-03-2020 | Quest Diagnostics Infectious Disease, Inc. | Quest SARS-CoV-2 rRT-PCR | Molecular | H |
| 17-03-2020 | Quidel Corporation | Lyra SARS-CoV-2 assay | Molecular | H |
| 18-03-2020 | Abbott Molecular | Abbott RealTime SARS-CoV-2 assay | Molecular | H |
| 19-03-2020 | GenMark Diagnostics, Inc. | ePlex SARS-CoV-2 test | Molecular | H, M |
| 19-03-2020 | DiaSorin Molecular LLC | Simplexa COVID-19 direct assay | Molecular | H, M |
| 20-03-2020 | Cepheid | Xpert Xpress SARS-CoV-2 test | Molecular | H, M, W |
| 23-03-2020 | BioFire Defense, LLC | BioFire COVID-19 test | Molecular | H, M |
| 27-03-2020 | Luminex Molecular Diagnostics, Inc. | NxTAG CoV extended panel assay | Molecular | H |
| 27-03-2020 | Abbott Diagnostics Scarborough, Inc. | ID NOW COVID-19 | Molecular | H, M, W |
| 30-03-2020 | QIAGEN GmbH | QIAstat-Dx respiratory SARS-CoV-2 panel | Molecular | H, M |
| 02-04-2020 | Becton, Dickinson & Company (BD) | BioGX SARS-CoV-2 reagents for BD MAX system | Molecular | H, M |
| 03-04-2020 | Co-Diagnostics, Inc. | Logix smart coronavirus disease 2019 (COVID-19) Kit | Molecular | H |
| 03-04-2020 | ScienCell Research Laboratories | ScienCell SARS-CoV-2 Coronavirus real-time RT-PCR (RT-qPCR) detection Kit | Molecular | H |
| 10-04-2020 | Atila BioSystems, Inc. | iAMP COVID-19 detection kit | Molecular | H |
| 14-04-2020 | Chembio Diagnostic System, Inc | DPP COVID-19 IgM/IgG system | Serology IgM and IgG | H, M |
| 14-04-2020 | Ortho Clinical Diagnostics, Inc. | VITROS immunodiagnostic products anti-SARS-CoV-2 total reagent pack | Serology total antibody | H, M |
| 15-04-2020 | Maccura Biotechnology (USA) LLC | SARS-CoV-2 fluorescent PCR kit | Molecular | H |
| 24-04-2020 | Ortho-Clinical Diagnostics, Inc. | VITROS immunodiagnostic products anti-SARS-CoV-2 IgG reagent pack | Serology IgG only | H, M |
| 24-04-2020 | Autobio Diagnostics Co. Ltd. | Anti-SARS-CoV-2 rapid test | Serology IgM and IgG | H, M |
| 26-04-2020 | Abbott Laboratories Inc. | SARS-CoV-2 IgG assay | Serology IgG only | H, M |
| 29-04-2020 | Bio-Rad Laboratories, Inc | Platelia SARS-CoV-2 total Ab assay | Serology total antibody | H |
| 30-04-2020 | Wadsworth Center, New York State Department of Health | New York SARS-CoV microsphere immunoassay for antibody detection | Serology total antibody | H |
| 01-05-2020 | Bio-Rad Laboratories, Inc | Bio-Rad SARS-CoV-2 ddPCR Test | Molecular | H |
| 06-05-2020 | Sherlock BioSciences, Inc. | Sherlock CRISPR SARS-CoV-2 Kit | Molecular | H |
| 06-05-2020 | BioMérieux SA | SARS-COV-2 R-GENE | Molecular | H |
| 07-05-2020 | Rutgers Clinical Genomics Laboratory at RUCDR Infinite Biologics - Rutgers | Rutgers Clinical Genomics Laboratory TaqPath SARS-CoV-2-assay | Molecular | H |
| 15-05-2020 | Everlywell, Inc. | Everlywell COVID-19 test home collection kit | Home collection kit | N/A |
A selection of molecular and serology tests approved from the over 75 total. Adapted from FDA Emergency Use Authorizations22
H high complexity tests, M medium complexity tests, W CLIA waiver
Emerging Lab-on-Chip Diagnostics for SARS-CoV-2
This section will discuss novel lab-on-chip devices for detection of SARS-CoV-2, as well as research from high-impact papers on other viruses that could be relevant to COVID-19. These viral detection methods make use of magnetic, optical, colorimetric, electrical, and lateral flow-based properties in nanotechnology to capture and transduce diagnostic signals. A brief overview of each modality will be given, followed by examples of specific studies that employ each technique to SARS-CoV-2 or other viruses. The description of how these technologies have been applied for other viruses may be useful for adapting and developing new lab-on-chip devices for COVID-19 detection. Limit of detection is an extremely important criteria for these tests. Classic RT-PCR diagnosis of infected patients is critical to constrain SARS-CoV-2 spread because due to asymptomatic infection despite high viral loads.37 Early studies of influenza viruses and community-acquired human coronaviruses showed that the viral loads in asymptomatic individuals could be relatively low.26 New research has found symptomatic children had higher initial RNA load in nasopharyngeal swab samples than asymptomatic patients, and that SARS-CoV-2 RNA can also be detected in wastewater samples.25,54 Table 2 gives an overview of the surveyed technologies.
Table 2.
Overview of surveyed technologies: Overview of magnetic, colorimetric, plasmonic, electrochemical, and lateral flow assays.11
| Methods | Biomarker | Limit of detection | Time-to-result | Sample preparation | Portability | Scale | Use cases | References |
|---|---|---|---|---|---|---|---|---|
| CDC RT-PCR diagnostic panel | SARS-CoV-2 RNA | 100 copies/µL | 3 h | Complex | Unportable | Low throughput | ID/AIS/ES | CDC Instruction Manual: CDC-006-00019, Revision: 0311 |
| Magnetic | ||||||||
| Magnetic NP capture | SARS-CoV-2 viral RNA | 10 copies | 30 min (extraction) | Simple | Unportable | Low throughput | ID/AIS/ES | Zhao et all90 (not peer reviewed) |
| Self-assemble magnetic nanoparticles | Adenovirus-5 and Herpes simplex virus-1 antigen | 5 viral particles/10 µL | <30 min | Simple | Unportable | High throughput | ID/AIS | Perez et al50 |
| Fluorescent-magnetic-catalytic nanospheres | H9N2 avian influenza virus antigen | 10 pg/mL (electrical) and 69.8 ng/mL (fluorescence) | 1–2 h | Simple | Portable | Low throughput | ID | Peng et al49 |
| Optical | ||||||||
| Catalytic colorimetric reagent | Anti-SARS-CoV-2 antibodies | N/A | Within 15 min | Simple | Portable | Low throughput | ID | Zhengtu et al35 |
| Fluorescently labeled biosensor | SARS-CoV-2 Antibody | N/A | 10 min | Simple | Portable | High throughput capable | ID | Zhenhua et al13 |
| Functionalized QD | Respiratory syncytial virus antigen | N/A | 6 days (plaques) | Complex | Unportable | Low throughput | ID | Tripp et al71 |
| Liposome-quantum dot complexes | HIV DNA | 0.1 fM | <1 hour | Complex | Unportable | Moderate throughput | ID/AIS | Zhou et al91 |
| PDA liposomes | Influenza antigen | 11 HAUs | <1 hour | Simple | Portable | Moderate throughput | ID | Riechert et al55 |
| RT-LAMP pH-based colorimetric sensor | ZIKV RNA | 1 copy/uL | 10 min | Complex | Portable | Moderate throughput | ID/AIS | Kaarj et al28 |
| Plasmonic | ||||||||
| Surface plasma | SARS-CoV-2 RNA | 0.22 pM | Within 15 min | Simple | Unportable | High throughput capable | ID | Guangyu et al53 |
| Plasmonics nanoprobe | HIV-1 DNA | 0.5 μM | 10 s (detection) | Complex | Unportable | High throughput capable | ID | Wabuyele et al76 |
| SPR SERS | HBV DNA | 50 aM | <1 h | Complex | Portable | High throughput capable | ID/AIS/ES | Li et al34 |
| Electrochemical sensor | ||||||||
| Field-effect transistor | SARS-CoV-2 antigen | 2.42 × 102 copies/mL | 30 s | Simple | Portable | Low throughput | ID/AIS/ES | Seo et al58 |
| Potentiostat sensor | SARS-CoV-2 antigen | 10 fM | 10–30 s | Simple | Portable | Moderate throughput | ID/AIS | Mahari et al38 (not peer reviewed) |
| Screen-printed carbon electrodes | SARS DNA | 2.5 pM | 20 min | Complex | Portable | Moderate throughput | ID | Martínez-Paredes et al41 |
| Nanoparticle-streptavidin conjugates | HBV DNA | 2.0 pM | N/A | Complex | Unportable | High throughput capable | ID | Wang et al 79 |
| AgNPs modified carbon electrode | Influenza antigen | sub pM | 15 min | Simple | Unportable | Moderate throughput | ID/AIS | Sepunaru et al 59 |
| Lateral flow immunoassay | ||||||||
| Lanthanide-doped nanoparticles | SARS-CoV-2 antibody | 1:1000 dilution | 10 min | Complex | Portable | Moderate throughput | ID | Chen et al13 |
| Raman scattering | Influenza A H1N1 virus and HAdV | 50 pfu/mL (HAdV)and 10 pfu/mL (H1N1) | 30 min | Simple | Unportable | High throughput capable | ID | Wang et al80 |
Technologies specific for SARS-CoV-2 in bold. Methods which can directly applied the collected sample from patients are termed “Simple”. Methods requiring extra sample processing steps are regarded as “Complex”. Devices that can only process one sample per time are defined as low throughput. Devices that have potential to process multiple samples per time, even if their ability is not mentioned in original papers, are termed “High throughput capable”
We have split the technologies into use cases based upon limit of detection requirements— ID infection diagnosis, AIS asymptomatic screening, and ES environmental surveillance
Magnetic Technologies
Magnetic nanoparticles can be used for easy extraction of target biomolecules in a complex solution, or to create supramolecular structures with readable magnetic properties. Because of their versatile properties, magnetic nanoparticles can be used for biomarker enrichment, detection, and even cell lysis.5
Recently, Zhao et al. developed magnetic nanoparticles coated with poly (amino ester)-carboxyl groups (PC) for efficient SARS-COV-2 RNA extraction, combining the lysis, extraction, and binding steps of viral genetic material into a single step for RT-PCR reactions.90 RNA molecules are absorbed onto the nanoparticles due to a strong interaction between the carboxyl groups and the nucleic acids. The extracted RNA can be directly introduced into an RT-PCR reaction without an additional elution step. This extraction method is an improvement over traditional silica-based spin column RNA extractions, where samples require pre-lysis to release the nucleic acids from viral particles and multiple centrifugation steps are required. Using magnetic nanoparticle technology, viral RNA can be purified within 20 minutes. This study demonstrated efficacy using an RT-PCR targeting two different regions, the ORF1ab and N gene, with a 10 copy sensitivity.90
In a non-Covid-19 application, Perez et al. created monodisperse magnetic nanoparticles conjugated with antibodies. These particles self-assembled in the presence of viral particles to create supramolecular structures with enhanced magnetic properties detectable by magnetic resonance methods (NMR/MRI).50 The detection of virus in solution was measured via changes in water T2 relaxation times. The research team was able to specifically detect adenovirus-5 and herpes simplex virus-1 at concentrations of 5 viral particles/10 μL. This platform could be modified to use magnetic particles to elucidate presence of COVID-19 viral particles as well.
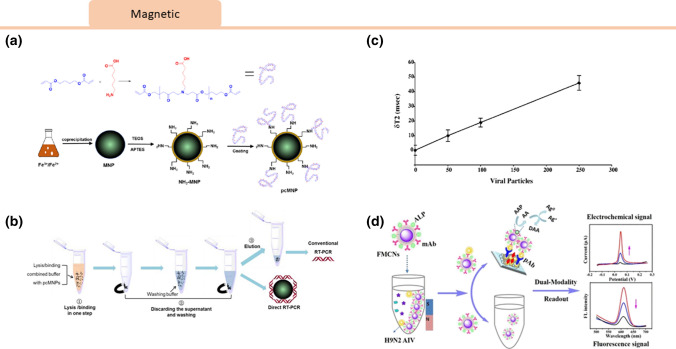
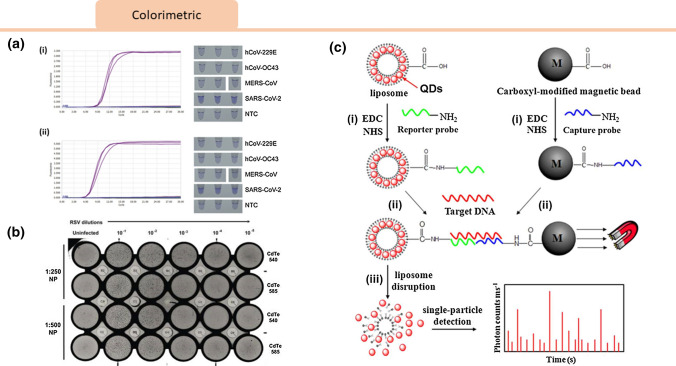
By combining both magnetic and fluorescent properties, Peng et al. introduced a dual-modality immunoassay using fluorescent-magnetic-catalytic nanospheres (FMCNs) functionalized with antibodies to capture an H9N2 avian influenza virus antigen. This sensor gave both a fluorescence and amplified electrochemical readout via alkaline phosphatase (ALP)-induced metallization.49 Due to the magnetic properties of the FMCNs, viral targets could be purified and separated from samples without pretreatment. The detection limits for the electrical and fluorescence signals were 10 pg/mL, and 69.8 ng/mL, respectively (Fig. 4).
Figure 4.
Viral magnetic lab-on-chip sensors. Magnetic nanoparticle technology for viral detection. (a) A schematic representation of the pcMNP-based SARS-CoV-2 viral RNA extraction method proposed by Zhao et al.90 A poly (amino ester) with carboxyl groups (PC) polymer was synthesized and used to coat magnetic nanoparticles to yield pcMNPs. (b) This method combines the lysis and binding steps into one step, and the pcMNPs-RNA complexes can be directly introduced into subsequent RT-PCR reactions. Permission has been requested from the author. (c) In a method proposed by Perez et al.,50 self-assembly of functionalized magnetic particles in the presence of viral particles could be measured via changes in water T2 relaxation times. Reprinted with permissions from J. Am. Chem. Soc. 2003, 125, 34, 10192–10193. Copyright 2003 American Chemical Society. (d) Peng et al.49 introduced a dual-modality immunoassay using fluorescent-magnetic-catalytic nanospheres (FMCNs) functionalized with antibodies to capture H9N2 avian influenza virus antigen, giving both a fluorescence and amplified electrochemical readout. Reprinted with permissions from ACS Appl. Mater. Interfaces 2019, 11, 44, 41148–41156. Copyright 2019 American Chemical Society.
Colorimetric and Fluorescent Sensors
Colorimetric and fluorescent sensors are a common detection method by which spectral change is often used to transduce the presence of a particular biomarker.1,2,87 Colorimetric and fluorescent components have been integrated with a number of viral molecular assays.
Isothermal amplification reactions, such as loop-mediated isothermal amplification (LAMP), can be observed in real-time by intercalating the fluorescent dye crystal violet (CV),47 which exhibits a violet color in aqueous solution. CV attaches to the major groove of dsDNA and converts into the colorless leuco type (LCV)77 in the presence of sulfite ions. As a product of this chemical reaction, the reaction solution for LAMP becomes colored only in the presence of dsDNA.45 In one study by Park et al, the authors were able to detect SARS-CoV-2 RNA with a limit of detection of 100 copies per reaction after a 30 minute amplification period.48 Specificity could be improved through different primer designs, but the high specificity of other RT-LAMP assays suggests that such methodologies are strong candidates for diagnostic use.
Promising optical detection methods have been demonstrated with other viruses, both for naked eye or fluorescence readout. Quantum dots, semiconductor nanoparticles that emit light upon excitation, are a common tool used to create optical signals. Tripp et al. showed that functionalized nanoparticles conjugated to monoclonal antibodies could be used to detect respiratory syncytial virus in vitro and in vivo by employing the fluorescent properties of CdTe quantum dots.71 Zhou et al. developed liposome-quantum dot complexes that enabled detection of attomolar HIV RNA concentrations. By sequestering quantum dots within liposomes and covalently linking that to an oligonucleotide capture sequence present on magnetic beads, a complex could be formed upon RNA hybridization. This complex could then be easily isolated for photon counting readout.91 Colorimetric sensors in the visible light range can produce a signal readable that can be seen with the naked eye without the need for instrumentation. In one study, polydiacetylene liposomes functionalized with sialic acid were used to bind and detect influenza virus, making use of the influenza hemagglutinin-sialic acid interaction to alter the color of the liposome complexes.55 Colorimetric sensors for viral genetic material can also make use of pH-based sensing. A hydrogen ion is released as a by-product of the DNA polymerase reaction. This enzyme property can be utilized in LAMP and PCR reactions with a decrease in pH indicating amplification. Kaarj et al. introduced a microfluidic assay for the Zika flavivirus that used a RT-LAMP mixture with a pH indicator to detect ZIKV RNA.28 Visible color changes were then quantified by smartphone imaging and a viral limit of detection at 1 copy/ μL was observed (Fig. 5).
Figure 5.
Viral Colorimetric Sensors: (a) Cross-reactivity tests for RT-LAMP assay targeting SARS-CoV-2 with real-time amplification fluorescence signal and end-point LCV colorimetric results for Nsp3_1-61 (i) and Nsp3_2-24 (ii) primer sets. No cross-reactivity was evident in RT-LAMP assays targeting Nsp3 to other human coronaviruses including hCoV-229E, hCoV-OC43, and MERS-CoV.48 Reprinted from The Journal of Molecular Diagnostics, Gun-Soo et al., Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assays Targeting SARS-CoV-2, Copyright (2020), with permission from Elsevier. (b) Respiratory syncytial virus-nanoparticles (RSV-NP) virus plaque assay introduced by Tripp et al. 540 nm and 585 nm CdTe quantum dots (QDs) are evaluated at days 5 or 6 pi that revealed presence of viral particles.71 Reprinted from Int J Nanomedicine. 2007;2(1):117–124, with permissions from Dove Medical Press. (c) Schematic of Liposome−QD (L/QD) complexes-based DNA detection. Prepared L/QD complexes, L/QD complex-tagged reporter probes and magnetic bead-modified capture probes (i) can form sandwich hybrids through target DNA, which is purified by magnet separation (ii). The QDs released from liposome disruption can be counted by single-particle detection.91 Reprinted with permissions from J. Am. Chem. Soc. 2013, 135, 6, 2056–2059. Copyright (2013) American Chemical Society.
Plasmonic Sensors
Plasmonic sensors harness the properties of surface plasmons – electromagnetic oscillations at the surface between a metal and dielectric that are highly sensitive to binding events.34,39,63,86 Such sensors can be used for label-free detection of nucleic acids, proteins, and even cells.7,64,68,69
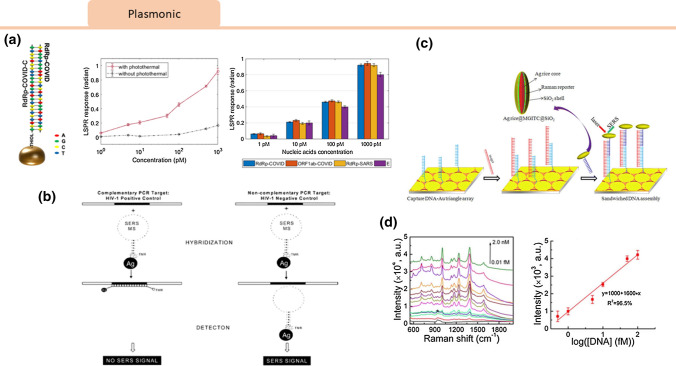
Qiu et al. demonstrated a dual-function plasmonic biosensor that combined the plasmonic photothermal effect (PPT) and localized surface plasmon resonance (LSPR) sensing transduction for SARS-CoV-2 RNA without the need for RT-PCR.53 In this method, gold nanoislands (AuNIs) were functionalized with complimentary single-stranded DNA receptors for RNA target capture.53 Due to the unique properties of plasmonic nanoparticles, heating energy is localized near the nanoparticles which can be used as a heat source for thermal processing. Hybridization occurs between the target and the conjugated probe, but a single mismatch can cause the melting temperature to decrease significantly. The PPT effect increases the hybridization rate and LSPR sensing response, providing fast and sensitive detection of nucleic acids by improving hybridization kinetics. In this study, hybridization with the target genetic material released thermoplasmonic heat as the particles were illuminated at the plasmonic resonance frequency. This elevated the in-situ hybridization temperature and allowed for accurate discrimination of two similar gene sequences, SARS and SARS-COV-2. The biosensor demonstrated high specificity and a low detection limit of SARS-COV-2 sequences down to 0.22 pM.53
Plasmonic sensors have also been demonstrated for detection of other viruses such as HIV. In a recent study, detection of HIV RNA was performed using a device consisting of a nanoparticle and stem-loop capture molecule tagged with a Raman label to detect the viral DNA. Upon hybridization with the target RNA, the stem-loop configuration is disrupted, causing the Raman label to separate from the metal nanoparticle and quench the surface-enhanced Raman spectroscopy signal.76 In another example, Li et al. coupled silver nano-rice antennae with a patterned gold triangle nanoarray chip to create spatially broadened plasmonic “hot spots” that increased the intensity and area of the surface plasmon resonance. This enhancement of the signal upon detection of HIV RNA enabled the selective detection of HIV RNA down to 50 attomolar (Fig. 6).34
Figure 6.
Viral Plasmonic Sensors: (a) Qiu et al.53 introduced a dual-functional plasmonic biosensor that combined the plasmonic photothermal effect (PPT) and localized surface plasmon resonance (LSPR). The PPT effect and LSPR sensing response improved hybridization kinetics, allowing for rapid and sensitive detection of SARS-CoV-2 RNA. Concentrations of various viral oligos were measured using the dual-functional LSPR biosensors. Reprinted with permissions from ACS Nano 2020, 14, 5, 5268–5277. https://pubs.acs.org/doi/10.1021/acsnano.0c02439. Further permissions related to the material excerpted should be directed to the ACS. (b) A plasmonics nanoprobe was designed by Wabuyele and Vo-Dinh76 consisting of a metal nanoparticle and step-loop capture DNA molecule tagged with a Raman label to detect HIV 1 RNA. Hybridization with target disrupts the stem-loop causing the Raman label to separate from the metal nanoparticle and quenching of the SERS signal. Reprinted with permissions from Anal. Chem. 2005, 77, 23, 7810–7815. Copyright 2005 American Chemical Society. (c) Li et al. 34 designed silver nanorice antennae with a patterned gold triangle nanoarray chip that created plasmonic “hot spots” which enhanced the SERS signal upon detection of HBV RNA. (d) SERS corresponded to various concentrations of the HBV target. The linear region of the Raman intensity at 1335 cm−1 plotted as a function of the logarithmic concentration of HBV concentration is highlighted. Reprinted with permissions from Anal. Chem. 2013, 85, 4, 2072–2078. Copyright 2013 American Chemical Society.
Electrical and Electrochemical Sensors
Electrochemical sensors use resistive or capacitive changes to detect binding changes of relevant analytes.9,18
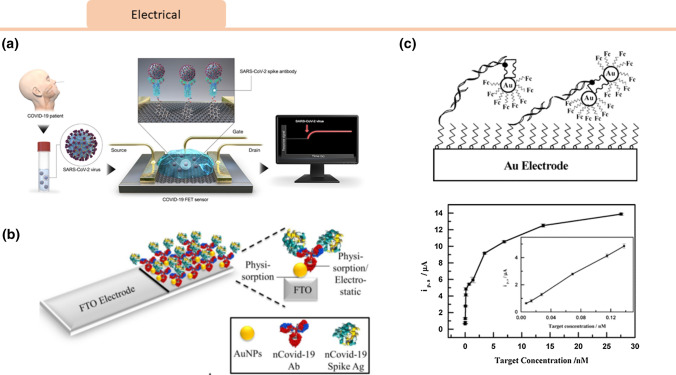
Mahari et al. built a biosensing device (eCovSens) using a screen-printed carbon electrode (SPCE) and compared it with a commercial potentiostat consisting of an fluorine doped tin oxide (FTO) electrode. They evaluated their novel device in terms of sensitivity, specificity, time of detection, sample volume, portability, and stability for nCovid-19 antigen detection.38 The eCovSens device consisted of a bio-recognition element (nCovid-19 Ab), a transducer (carbon electrode), and an electronic system (an in-house instrument) to detect changes in the voltage. Conjugated gold nanoparticles both detect the viral particles and act as a catalyst to amplify the electrochemical signal by enhancing electrical conductivity. nCovid-19 particles captured on the modified electrode led to changes in current proportional to target analyte concentration. Viral particles were successfully captured and detected using this device with a limit of detection (LOD) of 90 fM using spiked saliva samples. The eCovSens portable point-of-care device can produce results within 10–30 s.
Seo et al. developed a field-effect transistor (FET) device to detect the SARS-CoV-2 spike protein.58 FET biosensors have high sensitivity and selectivity, and are typically configured to capture analytes through biorecognition on the conducting channel. Changes in surface charge upon binding will lead to transducable differences in source-drain current measurements. Graphene sheets atop a FET were functionalized with an anti SARS-COV-2 spike protein antibody. This sensor configuration allowed for detection of SARS-COV-2 spike protein in purified antigen, cultured virus, and nasopharyngeal swab specimens with a limit of detection of 1 fg/mL in phosphate-buffered saline and 100 fg/mL in clinical transport medium.58
A wide variety of non-SARS-CoV-2 electrochemical biosensors have been developed using potentiometric or amperometric read-outs. For the detection of SARS RNA, an oligonucleotide capture monolayer was assembled onto disposable gold nanostructured screen-printed carbon electrodes. Upon hybridization there was an enzymatic amplification signal that could be measured with voltammetry with a detection limit of 2.5 pmol/L.41 In another example, nanoparticle-streptavidin conjugates covered with ferrocene caps enabled amplified voltammetric detection of viral RNA with excellent linearity for target concentrations between 6.9 and 150 pM.79 In another study, influenza virus was absorbed onto silver nanoparticles, and when the complex was exposed to a carbon electrode it underwent oxidation. The frequency of measured current spikes was linearly proportional to viral concentration, enabling viral detection to the sub-pM level (Fig. 7).59
Figure 7.
Viral Electrochemical Sensors: Electrochemical biosensors for viral detection. (a) Seo et al. 58 introduced a field-effect transistor (FET)-based biosensing device for detecting SARS-CoV-2 antigen in clinical samples by coating graphene sheets of the FET with a specific antibody against SARS-CoV-2 spike protein. Reprinted with permissions from ACS Nano 2020, 14, 4, 5135–5142. https://pubs.acs.org/doi/10.1021/acsnano.0c02823. Further permissions related to the material excerpted should be directed to the ACS. (b) Mahari et al. 38 proposed a potentiostat based sensor using a fluorine doped tin oxide electrode (FTO) with gold nanoparticles immobilized with nCovid-19Ab to measure changes in electrical conductivity upon encounter with nCovid-19 antigen.Permission has been requested from the author. (c) A voltammetric viral RNA biosensor was developed by Wang et al. 79 to detect hybridization via oxidation of ferrocene caps on gold nanoparticle/streptavidin conjugates. A plot of faradaic currents from their device against the 39-mer target concentration is shown. Reprinted with permissions from (Anal. Chem. 2003, 75, 15, 3941–3945). Copyright (2003) American Chemical Society.
Lateral Flow Assays
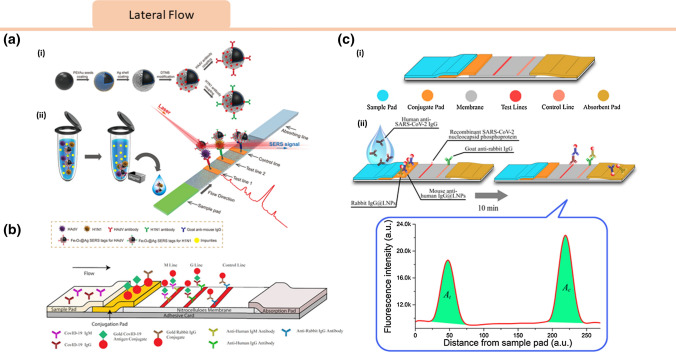
Lateral flow assays are among the most common low-cost diagnostic modalities. They are often configured as a paper substrate with wax printed channels that allow fluid flow over the testing region.60,81 Lateral flow immunoassay (LFIA) based point-of-care (POC) devices have been widely used for qualitative and quantitative analysis.57 Most LFIAs comprise of an application pad, conjugate pad, test zone, control zone, absorbent pad, and substrate pad.12 LFIA based strips allow for large-scale production and low cost due to inexpensive and off-the-shelf components, which make them promising for rapid detection of SARS-CoV-2.83
Lateral flow detect the presence of a target substance as a liquid sample runs along the surface of a functionalized pad with reactive molecules that can produce a visible readout. A lateral flow test has been developed for the detection of SARS-COV-2 antibodies. Chen et al. developed a lanthanide-doped polysterene nanoparticle (LNPs) based LFIA for screening anti-SARS-CoV-2 IgG in human serum (Fig. 8c).14 In their design, the target IgG was captured by recombinant nucleocapsid SARS-CoV-2 phosphoprotein coated on the test zone. Upon binding, a fluorescent signal from LNP-labeled mouse anti-human IgG antibody was measured, enabling quantitative detection. While this test was developed for antibody detection, lateral flow technology is also promising for SARS-COV-2 antigen detection. In another application, a surface-enhanced Raman scattering-based lateral flow immunoassay was developed to detect influenza A H1N1 virus and human adenovirus using Au/Ag-coated iron oxide magnetic nanoparticles with LOD (Fig. 8a) for H1N1 and HAdV 50 pfu/mL and 10 pfu/mL, respectively.80 The magnetic nanoparticle allowed for sample enrichment in a complex mixture and acted as a stable SERS substrate to enhance the Raman signal. Zhengtu Li et al. developed a rapid and simple LFIA device to detect SARS-CoV-2 immunoglobulin M and G (IgM and IgG) antibodies simultaneously using a blood sample within 15 minutes (Fig. 8b), with an overall testing sensitivity of 88.66% and specificity of 90.63%.35 According to their study, this LFIA strip performed well on fingerstick blood, serum, and venous blood plasma for both confirmed COVID‐19 patients and negative patients.
Figure 8.
Viral Lateral Flow Sensors: (a) Schematic of antibody-modified Fe3O4@Ag magnetic tags and magnetic SERS strip for respiratory viruses detection. (i) Fe3O4@Ag magnetic tags are modified with dye molecules (DTNBs) and capture antibodies acting as capturing and enhancing substrate while dual-labeled DTNB molecules generating SERS signals. (ii) The magnetic SERS-LFIA system components and operating procedure.80 Reprinted with permissions from ACS Appl. Mater. Interfaces 2019, 11, 21, 19495–19505. Copyright (2019) American Chemical Society. (b) Schematic of SARS-CoV-2 IgM-IgG combined antibody test. Two mouse anti-human monoclonal antibodies (anti-IgG and anti-IgM) are coated on different test lines, while surface antigen from SARS-CoV-2 is conjugated to colloidal gold nanoparticles on conjugation pads.35 This is an open access article distributes under the terms of the Creative Commons CC BY. (c) Schematic of lanthanide-doped nanoparticles-based lateral flow immunoassay (LFIA). (i) LFIA strips components. (ii) Analytical procedure: lanthanide-doped polystyrene nanoparticles (LNPs) are captured at the test and control line, where fluorescence at excitation and emission wavelengths of 365 and 615 nm is read. Their ratio determines the anti-SARS-CoV-2 IgG concentration in the sample.14This figure is reused with permission from ACS and the article can be accessed here: https://pubs.acs.org/doi/10.1021/acs.analchem.0c00784.
Summary: Insights for Development of Lab-on-Chip COVID-19 Biosensors
Scaling up the number of tests needed across the United States, based on the implied ratios from major states, means that testing facilities would have to collect and process up to 4.5 million tests per week for the United States alone. This indicates that the announced targets set out by large US states will be a challenge to meet. Development of lab-on-chip biosensors for SARS-CoV-2 and other viruses highlights their promise for rapid and sensitive viral detection. In this paper, we reviewed magnetic, plasmonic, colorimetric, electric, and lateral flow-based technologies. For immediate application of these strategies COVID-19 detection, it is important that the device be portable, rapid, and have a high sensitivity for SARS-CoV-2. The devices that we have reviewed vary in sensitivity, portability, and speed and thus have advantages and disadvantages for applications to SARS-CoV-2.
Limit of detection is of key importance due to relatively low viral concentration in a patient sample. From our survey, magnetic, plasmonic, and electrochemical devices appear to exhibit the lowest limits of detection, making them the most immediately relevant to this application. One drawback about the magnetic and plasmonic technologies, however, is that they often require specialized instrumentation both for fabrication and operation, making portability challenging. Further work to improve the portability of magnetic and plasmonic strategies could make them more portable and therefore harness their advantages in sensitivity to be applicable to the point of care.
In contrast, the colorimetric, electrochemical, and lateral flow assays are more portable, allowing for operation in the field because they do not require laboratory infrastructure or instrumentation for a result. To date, these technologies seem to focus on detection of antibodies to and antigens on the virus itself, rather than nucleic acids. These technologies show promise for as an alternative to serology tests on a compact platform. In contrast, plasmonic and magnetic technologies require a laboratory infrastructure, but have the advantage of higher throughput, allowing more samples to be processed at one. Among our surveyed technologies, the electrochemical and plasmonic assays enable fastest readout, while the colorimetric and lateral flow assays take a few minutes for development to get a result (Fig. 9).
Figure 9.
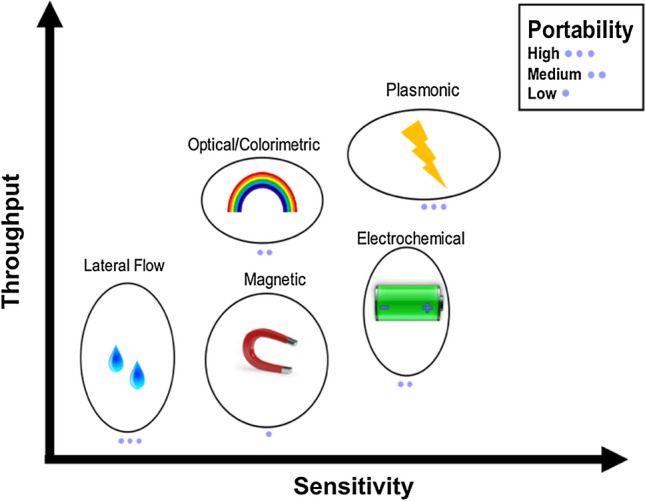
Technology Summary. Summary of nano-scale biosensing methods reviewed with comparison of throughout capability, sensitivity, and portability trends broadly estimated from literature on the axes. Images attributed from Creative Commons.
Other considerations that are relevant to technology applicability to COVID-19 are cost and integration with smartphones. Lateral flow assays are very popular in resource-limited settings due to the relatively low cost to manufacture at scale. Thus, such technologies that are integrated on paper or other low-cost substrates could have promise for deployment across the world. They also have the advantage that they can be operated by untrained users in a range of healthcare settings. Another technology that could allow this are smartphone compatible tests. Recently, a number of technologies have been developed to integrate lab-on-chip operation and readout with smartphones.6,15,32 This sort of integration could allow for data confidentiality, simple operation, and built-in optics, allowing modifications of more complex lab-on-chip formats for field use. Careful attention to cost and integration with common instrumentation such as smartphones could allow new technologies to be applied more readily to the point-of-care.
This paper reviewed current developments in the field of lab-on-chip diagnostic sensors for COVID-19. Significant progress has been made using these simple integrated formats for the detection of both SARS-CoV-2 RNA and protein biomarkers. Additional work towards detection of other viruses may become relevant as we continue to innovate and develop new platforms for COVID-19. With public and private support including new grants, expedited review, and rapid approvals, it is our hope that new technologies continue to be made available to improve our response to this pandemic.
Acknowledgments
The authors would like to acknowledge financial support from the Norris Cotton Cancer Center Developmental Funds, the PhD Innovation Program, and Agilent University Research Program.
Conflict of interest
Carly Tymm and Junhu Zhou declare that they have no conflicts of interest. Dr. Amogha Tadimety, Alison Burklund, and Dr. John X.J. Zhang are founders of nanopath diagnostics, a company working to develop in vitro diagnostic tests.
Ethical Standards
No animal studies were carried out by the authors for this article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Carly Tymm and Junhu Zhou have contributed equally to this work.
References
- 1.Ahmed, S. R., et al. In situ self-assembly of gold nanoparticles on hydrophilic and hydrophobic substrates for influenza virus-sensing platform. Sci. Rep. 7, 2017. [DOI] [PMC free article] [PubMed]
- 2.Ahn SJ, et al. Rapid and simple colorimetric detection of multiple influenza viruses infecting humans using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. BMC Infect. Dis. 2019;19:1–12. doi: 10.1186/s12879-019-4277-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apoorva, M. Just spit and wait: new coronavirus test offers advantages. New York Times 2020
- 4.Bhalla N, Jolly P, Formisano N, Estrela P. Introduction to biosensors. Essays Biochem. 2016;60:1–8. doi: 10.1042/EBC20150001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burklund A, Petryk J, Hoopes PJ, Zhang JXJ. Microfluidic enrichment of bacteria coupled to contact-free lysis on a magnetic polymer surface for downstream molecular detection. Biomicrofluidics. 2020;14(3):034115. doi: 10.1063/5.0011908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burklund A, Saturley-Hall HK, Franchina FA, Hill JE, Zhang JXJ. Printable QR code paper microfluidic colorimetric assay for screening volatile biomarkers. Biosens. Bioelectron. 2019;128:97–103. doi: 10.1016/j.bios.2018.12.026. [DOI] [PubMed] [Google Scholar]
- 7.Burklund A, Tadimety A, Nie Y, Hao N, Zhang JXJ. Advances in diagnostic microfluidics. Adv. Clin. Chem. 2019 doi: 10.1016/bs.acc.2019.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Burklund A, Zhang JXJ. Microfluidics-based organism isolation from whole blood: an emerging tool for bloodstream infection diagnosis. Ann. Biomed. Eng. 2019;47:1657–1674. doi: 10.1007/s10439-019-02256-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carregal-Romero S, et al. Multiplexed sensing and imaging with colloidal nano- and microparticles. Annu. Rev. Anal. Chem. (Palo Alto. Calif.) 2013;6:53–81. doi: 10.1146/annurev-anchem-062012-092621. [DOI] [PubMed] [Google Scholar]
- 10.Carter LJ, et al. Assay techniques and test development for COVID-19 diagnosis. ACS Cent. Sci. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.CDC Instruction Manual (CDC-006-00019 Revision 03).
- 12.Chen A, Yang S. Replacing antibodies with aptamers in lateral flow immunoassay. Biosens. Bioelectron. 2015;71:230–242. doi: 10.1016/j.bios.2015.04.041. [DOI] [PubMed] [Google Scholar]
- 13.Chen Z, et al. Rapid and sensitive detection of anti-SARS-CoV-2 IgG using lanthanide-doped nanoparticles-based lateral flow immunoassay. Anal. Chem. 2020 doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- 14.Chen Z, et al. Rapid and sensitive detection of anti-SARS-CoV-2 IgG using lanthanide-doped nanoparticles-based lateral flow immunoassay. Anal. Chem. 2020;92:7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- 15.Chin CD, et al. Microfluidics-based diagnostics of infectious diseases in the developing world. Nat. Med. 2011;17:1015–1019. doi: 10.1038/nm.2408. [DOI] [PubMed] [Google Scholar]
- 16.Covid-19. https://www.corisbio.com/Products/Human-Field/Covid-19.php.
- 17.Cyranoski D. Profile of a killer: the complex biology powering the coronavirus pandemic. Nature. 2020;581(7806):22–26. doi: 10.1038/d41586-020-01315-7. [DOI] [PubMed] [Google Scholar]
- 18.Das J, Ivanov I, Sargent EH, Kelley SO. DNA clutch probes for circulating tumor DNA analysis. J. Am. Chem. Soc. 2016;138:11009–11016. doi: 10.1021/jacs.6b05679. [DOI] [PubMed] [Google Scholar]
- 19.Diao, B., et al. Diagnosis of acute respiratory syndrome coronavirus 2 infection by detection of nucleocapsid protein running title: diagnosis of COVID-19 by N antigen detection. 2020. 10.1101/2020.03.07.20032524
- 20.Fauci AS, Lane HC, Redfield RR. Covid-19—navigating the uncharted. N. Engl. J. Med. 2020;382:1268–1269. doi: 10.1056/NEJMe2002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.FDA approves first at-home saliva collection test for coronavirus. (2020)
- 22.FDA. Emergency use authorizations. (2020)
- 23.Forster P, Forster L, Renfrew C, Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. 2020 doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Futerman, A. The cell biology of SARS-CoV-2. Interf. Rev. (2020)
- 25.Han, M. S., et al. Viral RNA load in mildly symptomatic and asymptomatic children with COVID-19. Seoul. Emerg. Infect. Dis. 26, 2020 [DOI] [PMC free article] [PubMed]
- 26.Heimdal, I., et al. Human coronavirus in hospitalized children with respiratory tract infections: A 9-year population-based study from Norway. https://pubmed.ncbi.nlm.nih.gov/30418633/. Accessed 2 Jul 2020 [DOI] [PMC free article] [PubMed]
- 27.Hoffmann M, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271-280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaarj K, Akarapipad P, Yoon JY. Simpler, faster, and sensitive Zika virus assay using smartphone detection of loop-mediated isothermal amplification on paper microfluidic chips. Sci. Rep. 2018;8:1–11. doi: 10.1038/s41598-018-30797-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaul D. An overview of coronaviruses including the SARS-2 coronavirus—molecular biology, epidemiology and clinical implications. Curr. Med. Res. Pract. 2020;10:54–64. doi: 10.1016/j.cmrp.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan S, et al. Emergence of a novel coronavirus, severe acute respiratory syndrome coronavirus 2: biology and therapeutic options. J. Clin. Microbiol. 2020;58:1–9. doi: 10.1128/JCM.00187-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kulasinghe A, Wu H, Punyadeera C, Warkiani M. The use of microfluidic technology for cancer applications and liquid biopsy. Micromachines. 2018;9:397. doi: 10.3390/mi9080397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laksanasopin T, et al. A smartphone dongle for diagnosis of infectious diseases at the point of care. Sci. Transl. Med. 2015;7:273re1. doi: 10.1126/scitranslmed.aaa0056. [DOI] [PubMed] [Google Scholar]
- 33.Lan J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020 doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 34.Li M, et al. Plasmonic nanorice antenna on triangle nanoarray for surface-enhanced Raman scattering detection of hepatitis B virus DNA. Anal. Chem. 2013;85:2072–2078. doi: 10.1021/ac303387a. [DOI] [PubMed] [Google Scholar]
- 35.Li Z, et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020 doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loeffelholz MJ, Tang Y-W. Laboratory diagnosis of emerging human coronavirus infections-the state of the art. Emerg. Microbes Infect. 2020 doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lohse, S., et al. Pooling of samples for testing for SARS-CoV-2 in asymptomatic people. Lancet Infect. Dis. 2020 [DOI] [PMC free article] [PubMed]
- 38.Mahari, S., A. Roberts, D. Shahdeo, and S. Gandhi. eCovSens-ultrasensitive novel in-house built printed circuit board based electrochemical device for rapid detection of nCovid-19. bioRxiv 2020. 10.1101/2020.04.24.059204.
- 39.Marinakos SM, Chen S, Chilkoti A. Plasmonic detection of a model analyte in serum by a gold nanorod sensor. Anal. Chem. 2007;79:5278–5283. doi: 10.1021/ac0706527. [DOI] [PubMed] [Google Scholar]
- 40.Mark D, Haeberle S, Roth G, Von Stetten F, Zengerle R. Microfluidic lab-on-a-chip platforms: requirements, characteristics and applications. Chem. Soc. Rev. 2010;39:1153–1182. doi: 10.1039/b820557b. [DOI] [PubMed] [Google Scholar]
- 41.Martínez-Paredes G, González-García MB, Costa-García A. Genosensor for SARS virus detection based on gold nanostructured screen-printed carbon electrodes. Electroanalysis. 2009;21:379–385. [Google Scholar]
- 42.Matsuyama S, et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2- expressing cells. Proc. Natl. Acad. Sci. USA. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mauk M. Miniaturized devices for point of care molecular detection of HIV. Lab Chip. 2017;17:382–394. doi: 10.1039/c6lc01239f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mboowa G. Current and emerging diagnostic tests available for the novel COVID-19 global pandemic. AAS Open Res. 2020;3:8. doi: 10.12688/aasopenres.13059.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyamoto S, Sano S, Takahashi K, Jikihara T. Method for colorimetric detection of double-stranded nucleic acid using leuco triphenylmethane dyes. Anal. Biochem. 2015;473:28–33. doi: 10.1016/j.ab.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 46.Morales-Narváez E, Dincer C. The impact of biosensing in a pandemic outbreak: COVID-19. Biosens. Bioelectron. 2020 doi: 10.1016/j.bios.2020.112274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oscorbin IP, Belousova EA, Zakabunin AI, Boyarskikh UA, Filipenko ML. Comparison of fluorescent intercalating dyes for quantitative loop-mediated isothermal amplification (qLAMP) Biotechniques. 2016;61:20–25. doi: 10.2144/000114432. [DOI] [PubMed] [Google Scholar]
- 48.Park G-S, et al. Development of reverse transcription loop-mediated isothermal amplification assays targeting SARS-CoV-2. J. Mol. Diagn. 2020 doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng X, et al. Fluorescent-magnetic-catalytic nanospheres for dual-modality detection of H9N2 avian influenza virus. ACS Appl. Mater. Interfaces. 2019;11:41148–41156. doi: 10.1021/acsami.9b16718. [DOI] [PubMed] [Google Scholar]
- 50.Perez JM, Simeone FJ, Saeki Y, Josephson L, Weissleder R. Viral-induced self-assembly of magnetic nanoparticles allows the detection of viral particles in biological media. J. Am. Chem. Soc. 2003;125:10192–10193. doi: 10.1021/ja036409g. [DOI] [PubMed] [Google Scholar]
- 51.Prevention, C. for D. C. and. Coronavirus Disease 2019: Symptoms (2020)
- 52.Products - STANDARD Q COVID-19. http://sdbiosensor.com/xe/product/7672.
- 53.Qiu G, et al. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020 doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 54.Randazzo W, et al. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181:115942. doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reichert A, Nagy JO, Spevak W, Charych D. Polydiacetylene liposomes functionalized with sialic acid bind and colorimetrically detect influenza virus. J. Am. Chem. Soc. 1995;117:829–830. [Google Scholar]
- 56.New Rutgers Saliva Test for Coronavirus gets FDA Approval. (2020)
- 57.Sajid M, Kawde AN, Daud M. Designs, formats and applications of lateral flow assay: a literature review. J. Saudi Chem. Soc. 2015;19:689–705. [Google Scholar]
- 58.Seo G, et al. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020 doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 59.Sepunaru L, Plowman BJ, Sokolov SV, Young NP, Compton RG. Rapid electrochemical detection of single influenza viruses tagged with silver nanoparticles. Chem. Sci. 2016;7:3892–3899. doi: 10.1039/c6sc00412a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharma S, Zapatero-Rodríguez J, Estrela P, O’Kennedy R. Point-of-care diagnostics in low resource settings: present status and future role of microfluidics. Biosensors. 2015;5:577–601. doi: 10.3390/bios5030577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Songjaroen T, Dungchai W, Chailapakul O, Laiwattanapaisal W. Novel, simple and low-cost alternative method for fabrication of paper-based microfluidics by wax dipping. Talanta. 2011;85:2587–2593. doi: 10.1016/j.talanta.2011.08.024. [DOI] [PubMed] [Google Scholar]
- 62.Soper SA, et al. Point-of-care biosensor systems for cancer diagnostics/prognostics. Biosens. Bioelectron. 2006;21:1932–1942. doi: 10.1016/j.bios.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 63.Sugimoto H, Fujii M. Downloaded via DARTMOUTH COLG on May 5. ACS Photonics. 2018;5:29. [Google Scholar]
- 64.Tadimety A, Zhang Y, Tsongalis GJ, Zhang XJ. Screening circulating nucleic acids of pancreatic ductal adenocarcinoma using a plasmonic nanosensor. J. Mol. Diagnostics. 2017 [Google Scholar]
- 65.Tadimety A, et al. Liquid biopsy on chip: a paradigm shift towards the understanding of cancer metastasis. Integr. Biol. 2017;9:9–29. doi: 10.1039/c6ib00202a. [DOI] [PubMed] [Google Scholar]
- 66.Tadimety A, et al. Liquid biopsy on chip: a paradigm shift towards the understanding of cancer metastasis. Integr. Biol. 2017;9:22–49. doi: 10.1039/c6ib00202a. [DOI] [PubMed] [Google Scholar]
- 67.Tadimety A, et al. Advances in liquid biopsy on-chip for cancer management: technologies, biomarkers, and clinical analysis. Crit. Rev. Clin. Lab. Sci. 2018;55:1–23. doi: 10.1080/10408363.2018.1425976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tadimety, A., et al. Plasmonic gold nanorods with sequence specific conjugation for circulating tumor DNA screening. Int. Conf. Opt. MEMS Nanophotonics2018-July, 1–5, 2018
- 69.Tadimety A, et al. Design of peptide nucleic acid probes on plasmonic gold nanorods for detection of circulating tumor DNA point mutations. Biosens. Bioelectron. 2019;130:236–244. doi: 10.1016/j.bios.2019.01.045. [DOI] [PubMed] [Google Scholar]
- 70.Tang Y-W, Schmitz JE, Persing DH, Stratton CW. The laboratory diagnosis of COVID-19 infection: current issues and challenges. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tripp RA, et al. Bioconjugated nanoparticle detection of respiratory syncytial virus infection. Int. J. Nanomed. 2007;2:117. doi: 10.2147/nano.2007.2.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Udugama B, et al. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020 doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 73.University University J. H. Johns Hopkins University Coronavirus Resource Center. Coronavirus Resource. 2020;1:2–3. [Google Scholar]
- 74.Vashist SK. Point-of-care diagnostics: recent advances and trends. Biosensors. 2017;7:62. doi: 10.3390/bios7040062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vashist SK. In vitro diagnostic assays for COVID-19: recent advances and emerging trends. Diagnostics. 2020;10:1–7. doi: 10.3390/diagnostics10040202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wabuyele MB, Vo-Dinh T. Detection of human immunodeficiency virus type 1 DNA sequence using plasmonics nanoprobes. Anal. Chem. 2005;77:7810–7815. doi: 10.1021/ac0514671. [DOI] [PubMed] [Google Scholar]
- 77.Wakelin LPG, Adams A, Hunter C, Waring MJ. Interaction of crystal violet with nucleic acids. Biochemistry. 1981;20:5779–5787. doi: 10.1021/bi00523a021. [DOI] [PubMed] [Google Scholar]
- 78.Walls AC, et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang J, et al. Amplified voltammetric detection of DNA hybridization via oxidation of ferrocene caps on gold nanoparticle/streptavidin conjugates. Anal. Chem. 2003;75:3941–3945. doi: 10.1021/ac0344079. [DOI] [PubMed] [Google Scholar]
- 80.Wang C, et al. Magnetic SERS strip for sensitive and simultaneous detection of respiratory viruses. ACS Appl. Mater. Interfaces. 2019;11:19495–19505. doi: 10.1021/acsami.9b03920. [DOI] [PubMed] [Google Scholar]
- 81.Warren AD, Kwong GA, Wood DK, Lin KY, Bhatia SN. Point-of-care diagnostics for noncommunicable diseases using synthetic urinary biomarkers and paper microfluidics. Proc. Natl. Acad. Sci. 2014;111:3671–3676. doi: 10.1073/pnas.1314651111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xia Y, Whitesides GM. Soft lithography. Annu. Rev. Mater. Sci. 1998;28:153–184. [Google Scholar]
- 83.Yager P, et al. Microfluidic diagnostic technologies for global public health. Nature. 2006;442:412–418. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- 84.Yan R, et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (80−) 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yetisen AK, Akram MS, Lowe CR. Paper-based microfluidic point-of-care diagnostic devices. Lab Chip. 2013;13:2210–2251. doi: 10.1039/c3lc50169h. [DOI] [PubMed] [Google Scholar]
- 86.Yu C, Irudayaraj J. Multiplex biosensor using gold nanorods. Anal. Chem. 2007;79:572–579. doi: 10.1021/ac061730d. [DOI] [PubMed] [Google Scholar]
- 87.Zagorovsky K, Chan WCW. A plasmonic DNAzyme strategy for point-of-care genetic detection of infectious pathogens. Angew. Chem. Int. Ed. 2013;52:3168–3171. doi: 10.1002/anie.201208715. [DOI] [PubMed] [Google Scholar]
- 88.Zhang JXJ, Hoshino K. Molecular Sensors and Nanodevices: Principles, Designs, and Applications in Biomedical Engineering. New York: Academic Press; 2018. [Google Scholar]
- 89.Zhang J, et al. Navigating the pandemic response life cycle: molecular diagnostics and immunoassays in the context of COVID-19 management. IEEE Rev. Biomed. Eng. 2020 doi: 10.1109/RBME.2020.2991444. [DOI] [PubMed] [Google Scholar]
- 90.Zhao Z, et al. A simple magnetic nanoparticles-based viral RNA extraction method for efficient detection of SARS-CoV-2. bioRxiv. 2020 [Google Scholar]
- 91.Zhou J, Wang QX, Zhang CY. Liposome-quantum dot complexes enable multiplexed detection of attomolar DNAs without target amplification. J. Am. Chem. Soc. 2013;135:2056–2059. doi: 10.1021/ja3110329. [DOI] [PubMed] [Google Scholar]