Abstract
TMPRSS2 is an important membrane-anchored serine protease involved in human prostate cancer progression and metastasis. A serine protease physiologically often comes together with a cognate inhibitor for execution of proteolytically biologic function; however, TMPRSS2’s cognate inhibitor is still elusive. To identify the cognate inhibitor of TMPRSS2, in this study, we applied co-immunoprecipitation and LC/MS/MS analysis and isolated hepatocyte growth factor activator inhibitors (HAIs) to be potential inhibitor candidates for TMPRSS2. Moreover, the recombinant HAI-2 proteins exhibited a better inhibitory effect on TMPRSS2 proteolytic activity than HAI-1, and recombinant HAI-2 proteins had a high affinity to form a complex with TMPRSS2. The immunofluorescence images further showed that TMPRSS2 was co-localized to HAI-2. Both KD1 and KD2 domain of HAI-2 showed comparable inhibitory effects on TMPRSS2 proteolytic activity. In addition, HAI-2 overexpression could suppress the induction effect of TMPRSS2 on pro-HGF activation, extracellular matrix degradation and prostate cancer cell invasion. We further determined that the expression levels of TMPRSS2 were inversely correlated with HAI-2 levels during prostate cancer progression. In orthotopic xenograft animal model, TMPRSS2 overexpression promoted prostate cancer metastasis, and HAI-2 overexpression efficiently blocked TMPRSS2-induced metastasis. In summary, the results together indicate that HAI-2 can function as a cognate inhibitor for TMPRSS2 in human prostate cancer cells and may serve as a potential factor to suppress TMPRSS2-mediated malignancy.
Subject terms: Metastasis, Prostate cancer, Proteolysis
Introduction
Prostate cancer (PCa) is one of the most frequently diagnosed malignancies worldwide, and the second leading cause of cancer death in males in USA [1]. PCa progression is a slow process, which undergoes several stages including prostatic intraepithelial neoplasia, adenocarcinoma, and metastasis [2]. Metastasis is the major cause for the morbidity and mortality of PCa patients. Currently, there is no effective therapy available for metastatic PCa patients.
For cancer metastasis, primary tumor cells need to break down the local barrier, gain a movement ability or penetrate blood vessels, and eventually spread out and grow in distant tissues or organs. There are many factors or mechanisms involved in cancer cell invasion and metastasis, including growth factors, Epithelial–Mesenchymal transition (EMT), and protease system [3]. Among them, pericellular proteases receive attention because of their pivotal roles in the degradation of extracellular matrix (ECM), and the activation of cell-surface growth factors [4]. To ensure the appropriate cleavage of a protease on its target protein(s), the proteolytic machinery requires a cognate inhibitor to regulate the enzymatic activity of the protease in a spatial and temporal manner. Once proteases get activated and cleave their substrates, their cognate inhibitors will soon be recruited to the active proteases and block their proteolytic activity. Thus, the imbalance between a protease and its cognate inhibitor in favor of proteolysis often promotes ECM degradation and pericellular growth factor activation, leading to pathogenesis and disease progression such as cancer [5].
Among pericellular proteases, membrane-anchored serine proteases are recently in the spotlight because some of them contribute to the development and progression of cancer, including TMPRSS2 and Matriptase [6]. TMPRSS2 is an androgen-regulated cell-surface serine protease [7], and plays an important role in PCa cell invasion, tumor growth and metastasis [8, 9]. Several substrates have been identified for TMPRSS2-induced PCa progression such as Matriptase, pro-HGF and extracellular matrix (Nidogen-1 and Laminin β1) [8, 9]. Moreover, TMPRSS2 can cleave and activate PAR2 receptor for calcium releasing [10], which mediates pain responses [11]. In addition, TMPRSS2 also plays an important role in the infection of influenza virus and SARS-CoV-2 by activating the Hemagglutinin (HA) of influenza virus [12] and priming the spike (S) proteins of SARS-CoV2 coronavirus [13], respectively. After the proteolytic processing of HA or S proteins, both viruses gained the ability to fuse with host membrane and promote infection [12, 13]. On the other hand, although a fusion of the 5’ untranslational region of TMPRSS2 to ETS transcription factors frequently occurs in PCa [14, 15], the increased expression and subcellular mislocalization of TMPRSS2 are also found and correlated with PCa progression [8, 9, 16, 17]. Thus, TMPRSS2 plays a critical role in PCa tumor growth, metastasis, and viral infection.
HAI-1 and HAI-2 are two membrane-anchored, Kunitz-type serine protease inhibitors, via non-covalent interactions to their target proteases. They were originally identified as inhibitors for HGFA (hepatocyte growth factor activator) [18–20]. The role of HAI-1 in the inhibition of human cancer progression has been widely reported, such as PCa, breast and ovarian cancer [20]. In addition to HGFA, HAI-1 also can inhibit the other trypsin-like proteases including matriptase [21], hepsin [22] and prostasin [23]. Matriptase, HGFA and hepsin have been demonstrated as pro-metastatic factors to promote human cancer progression [22, 24, 25]. On the other hand, HAI-2 is found to be expressed in many tissues, such as pancreas, prostate, and kidney [19]. HAI-2 can efficiently inhibit several pericellular serine proteases including plasmin, HGFA, prostasin, Matriptase and Matriptase-2 [19, 26–31]. The down-regulation of HAI-2 has been observed in many cancers due to the hyper-methylation of the promoter region of SPINT2 (gene encoded HAI-2), such as hepatocellular carcinoma, prostate cancer, ovarian and breast carcinoma [20]. Dysregulation between HAIs and their target proteases have been reported to be implicated in various cancer progression [32]. Moreover, overexpression of HAI-2 can suppress Matriptase activation, PCa cell invasion, tumor growth and metastasis [28]. Thus, HAI-2 functions as a negative regulator of pericellular proteases and human cancer progression [32].
TMPRSS2 has been shown as an oncogenic protease to promote PCa progression. In the term of the proteolytic balance between a protease and a cognate inhibitor, it is still elusive if there is a cognate inhibitor to modulate the function of TMPRSS2. In this study, we identified HAI-2 to be a novel cognate inhibitor for TMPRSS2, with an inhibitory capability to modulate TMPRSS2 proteolytic activity, PCa cell invasion and metastasis. Thus, the data together provide a new insight into the PCa treatment by reversing the imbalance between a membrane-anchored serine protease and its cognate inhibitor.
Results
TMPRSS2 promoted PCa cell invasion
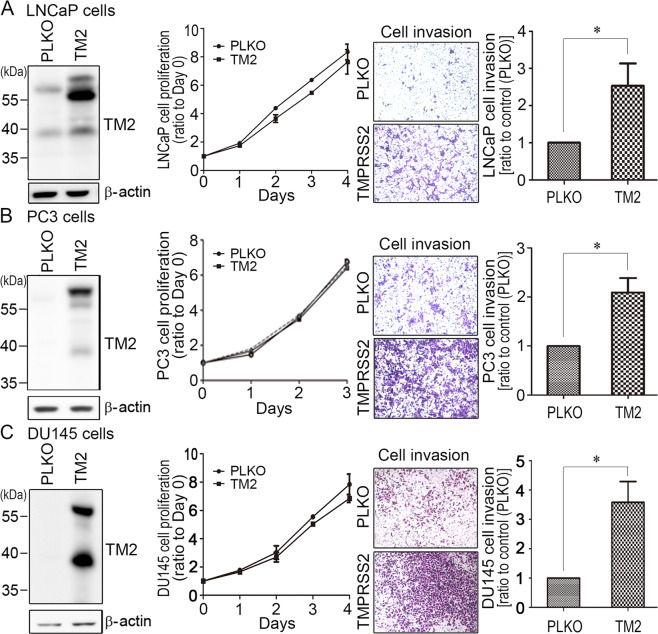
To examine whether TMPRSS2 played a role in PCa cell invasion, PCa cells (LNCaP, PC3 and DU145) were transduced with TMPRSS2 lentiviral particles. Control cells were infected with PLKO lentiviral particles. As shown in Fig. 1, TMPRSS2 overexpression had no significant effect on the cell growth (left two panels). Interestingly, TMPRSS2 overexpression could significantly increase the PCa cell invasion (Fig. 1, right two panels). Moreover, DU145 cells transfected with wild-type (WT) TMPRSS2 plasmids significantly increased the cell invasion, while protease-dead S441A mutant TMPRSS2 had no significant effect on the cell motility. It suggests that the proteolytic activity of TMPRSS2 is important for promoting PCa cell invasion (Fig. S1). The results together indicate that TMPRSS2 plays a positive role in PCa cell invasion.
Fig. 1. TMPRSS2 promoted PCa cell invasion.
TMPRSS2 was overexpressed in (a) LNCaP cells, (b) PC3 cells, and (c) DU145 cells. Left panels: Immunoblot analysis of TMPRSS2 expression in vector and TMPRSS2-overexpressing PCa cells. Middle panels: Analysis of the cell proliferation of control (PLKO) and TMPRSS2-overexpressing PCa cells using trypan blue exclusion assays and hemocytometer counting. Three independent experiments were performed for statistical calculation. Right panels: Effect of TMPRSS2 overexpression on the invasion of PCa cells. For analyzing the invasive ability of stable TMPRSS2-overexpressing PCa cells, cells were seeded at a density of 4 × 105 (LNCaP) or 5 × 104 (PC3 and DU145) per Boyden chamber with Matrigel-coating and cultured for 42 h (LNCaP) or 16 h (PC3 and DU145). Cells were fixed and stained with crystal violet. Invaded cells were imaged under a light microscope. The results were a representative of three independent experiments. The cell invasion results were statistically calculated and represented as mean ± SEM from three independent experiments.
Identification of TMPRSS2 interacting protein(s) in PCa cells
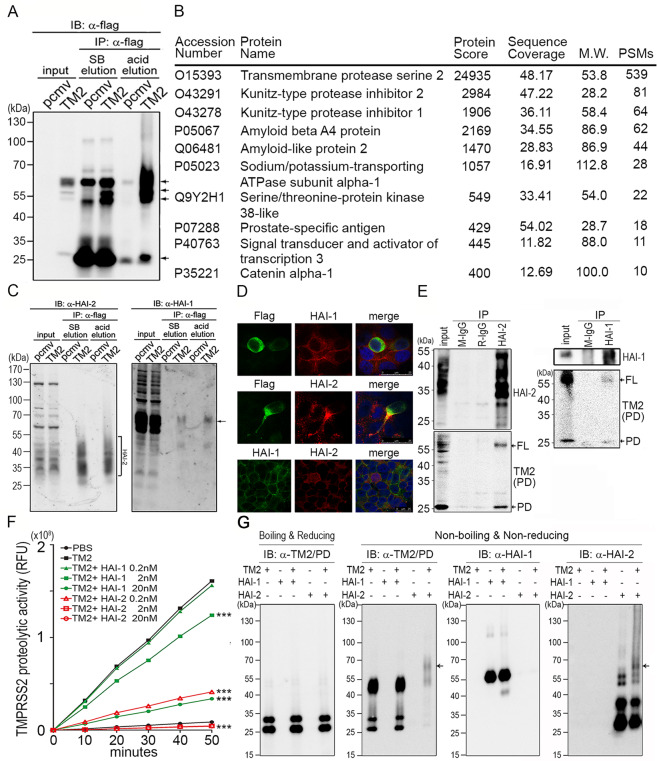
To understand the mechanisms how TMPRSS2 promoted PCa cell invasion and progression, in addition to its substrates (Matriptase, extracellular matrix and HGF) [8, 9], we further set up to identify TMPRSS2’s interacting protein(s) or endogenous inhibitor(s) in PCa cells. As shown in Fig. 2a, after pull-down using α-flag beads and immunoblotting, co-immunoprecipitated proteins were more efficiently eluted with less IgG and non-specific proteins by using the acid solution than that using SDS sample buffer. The eluent from acid elution was then collected and subjected to in-solution digestion and LC-MS/MS analysis. The results showed that more than 300 proteins were identified. Based on the subcellular localization, protein scores and coverages of the identified proteins, there were 10 candidates with high potentials as TMPRSS2-interacting proteins (Fig. 2b, a list of potential TMPRSS2-associated proteins). Among them, Kunitz-type protease inhibitor 2 and 1 (HAI-2 and HAI-1) received our attention because of their functions to be serine protease inhibitors. The results indicate that HAI-1 or HAI-2 may function as a cognate inhibitor for TMPRSS2 in PCa cells.
Fig. 2. Identification and validation of TMPRSS2-interacting proteins.
a Identification of TMPRSS2-interacting protein(s) by co-immunoprecipitation using M2 beads carrying anti-flag Abs. Arrows indicate the signals of TMPRSS2 or TMPRSS2 complexes. TMPRSS2-overexpressing LNCaP cells were lysed and the cell lysates were incubated with M2 beads at 4 °C, gentle rotation, overnight. The beads were then washed using TBST, four times and subjected to Laemmli sample buffer (SB) elution and acid elution, respectively. b List of proteins identified after Co-IP and LC/MS/MS analysis. The eluent from acid elution was subjected to LC/MS/MS analysis. The top ten protein identities with high #PSMs were listed in the table. c Interactions between TMPRSS2 and HAI-1/HAI-2 were validated by co-immunoprecipitation and immunoblot analysis in TMPRSS2-overexpressing LNCaP cells. The eluted proteins from immunoprecipitation were subjected to immunoblot analysis with anti-HAI-1 (Genetex) and anti- HAI-2 (LTK) antibodies. Square bracket, HAI-2. Arrows, HAI-1. d Analysis of the subcellular localization of HAI-1, HAI-2 and TMPRSS2 in PCa cells. Immunofluorescence staining of HAI-1, HAI-2 and TMPRSS2 was performed in PCa CWR22Rv1 cells. Cells were transiently transfected with TMPRSS2 plasmids using lipofectamine 3000 (Thermo, IL, USA). Two days after transfection, cells were fixed with 4% paraformaldehyde and blocked with 5% BSA plus 2% normal goat serum. Cells were stained with a monoclonal anti-Flag antibody (TMPRSS2, green), HAI-1 (M19, red) and HAI-2 (DC16, red). e Interaction between endogenous HAI-2 and TMPRSS2 in PCa cells. The interaction between HAI-2/HAI-1 and TMPRSS2 was examined in LNCaP cells using co-immunoprecipitation and immunoblot analysis. LNCaP cells were lysed and the cell lysates were incubated with an anti-HAI-2 antibody or an anti-HAI-1 antibody at 4 °C overnight. Protein G Sepharose beads were then added into the solution to isolate antibody-captured protein complexes. Normal mouse IgG (M-IgG) and rabbit IgG (R-IgG) were used as negative controls. The eluted proteins were subjected to immunoblot analysis with anti-HAI-2, anti-HAI-1 and anti-TMPRSS2 (AL-20) antibodies. FL full length of TMPRSS2. PD protease domain of TMPRSS2. f Analysis of the effects of the recombinant extracellular regions of HAI-1 and HAI-2 proteins (R&D) on the proteolytic activity of recombinant TMPRSS2 proteins by the measurement of the fluorescence signal of Boc-Gln-Ala-Arg-AMC substrate (ENZO). 0.2 nM of rTMPRSS2 proteins were incubated with the indicated concentrations of recombinant HAI-1 or HAI-2 proteins and then subjected to fluorescence detection in various time points. Measurements were performed in triplicate. Three independent experiments were performed, statistically calculated and presented as mean ± S.D. g TMPRSS2 formed complexes with HAI-2. Fifty nanograms (50 ng) of human rTMPRSS2 proteins were incubated with 50 ng of human rHAI-1 or rHAI-2 for 1 hour in PBS at 37 °C. The complex formation between rTMPRSS2 and rHAI-1 or rHAI-2 was analyzed using immunoblot analyses under boiling/reducing or non-boiling/non-reducing condition, using anti-TMPRSS2 (Abcam), anti-HAI-1(Genetex), as well as anti-HAI-2 (LTK) antibodies. The arrows indicated the complexes of rTMPRSS2 and rHAI-2 proteins.
Interaction of TMPRSS2 with HAI-1 and HAI-2
We then further validated the interaction between TMPRSS2 and HAI-1/2 using the co-immunoprecipitation assay. As shown in Fig. 2c, both HAI-1 and HAI-2 proteins could be pulled down by TMPRSS2. The molecular mass of HAI-1 proteins was ~65 kDa, while the molecular mass of HAI-2 was ranged from 30 to 55 kDa. The broad distribution of HAI-2 may be due to N-glycosylation [28, 33]. To further examine the subcellular localizations of TMPRSS2, HAI-1 and HAI-2 in PCa cells, immunofluorescence staining was performed in CWR22Rv1 cells. As shown in Fig. 2d, the subcellular localization of TMPRSS2, HAI-1 and HAI-2 were mainly observed on the plasma membrane. Compared with HAI-1, HAI-2 showed a high intensity of co-localization with TMPRSS2 in the invadopodia-like region of the cell. To further validate the interaction between TMPRSS2 and HAI-2, the results (Fig. 2e) from immunoprecipitation showed that TMPRSS2 proteins could be co-immunoprecipitated more effectively by HAI-2 than those by HAI-1. The data together indicates that HAI-2 has tight interaction and co-localization with TMPRSS2 in PCa cells.
HAI-2 could inhibit TMPRSS2 activity
To further examine the role of HAI-1 and HAI-2 in the proteolytic activity of TMPRSS2, the extracellular portions of recombinant TMPRSS2 (rTMPRSS2) proteins were purified and used for the proteolytic activity assays in the presence of the indicated concentrations of purified recombinant HAI-1 or HAI-2 (rHAI-1 or rHAI-2) proteins. The results (Fig. 2f) showed that rHAI-2 exhibited more effectively inhibitory effects on the proteolytic activity of TMPRSS2 than rHAI-1, although both rHAIs could inhibit TMPRSS2’s proteolytic activity in a dose-response manner. In fact, 0.2 nM of rHAI-2 had a similar effect to 20 nM of rHAI-1 on inhibiting TMPRSS2 activity, while 0.2 nM rHAI-1 showed no significant effects on TMPRSS2. To further examine whether HAI-1 or HAI-2 could form complexes with TMPRSS2, we used rTMPRSS2 and rHAIs to conduct in-tube complex formation assays. After incubation, the solutions were then subjected to immunoblot analysis under a non-boiling, non-reducing condition. As shown in Fig. 2g, rTMPRSS2 proteins formed complexes with rHAI-2 proteins in a molecular mass range of 65–70 kDa, while there was no significant complex formation between rTMPRSS2 and rHAI-1 proteins. However, there was a cleaved product (~40 kDa) of rHAI-1 proteins after the addition of rTMPRSS2 into the HAI-1 solution, indicating that HAI-1 might be able to be proteolytically processed by rTMPRSS2. Due to high glycosylation, the molecular mass of HAI-2 ranged from 25 to 55 kDa. The results together indicate that HAI-2 can interact with TMPRSS2 to inhibit the protease activity.
Identification of the functional domain of HAI-2 to inhibit TMPRSS2 proteolytic activity
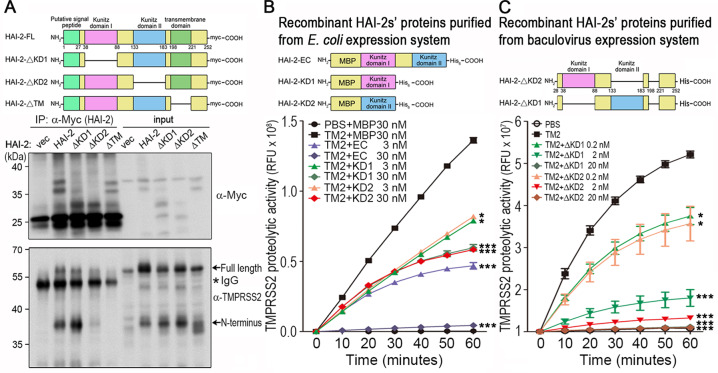
HAI-2 contains two Kunitz domains, which possess the inhibitory activity against its targets [19, 27]. In order to identify which functional domain of HAI-2 was critical for inhibiting TMPRSS2 activity, the plasmids encoding full-length HAI-2 (HAI-2-FL), KD1-deleted HAI-2 (HAI-2-∆KD1), KD2-deleted HAI-2 (HAI-2-∆KD2) and transmembrane domain-deleted HAI-2 (HAI-2-∆TM) were constructed. HEK293A cells were transfected with the mutant HAI-2 plasmids in the presence of TMPRSS2 plasmids, and lysates were subjected to immunoprecipitation and immunoblot analysis. The result (Fig. 3a) showed that HAI-2-∆KD2 and HAI-2-∆TM proteins had a decreased interaction with TMPRSS2, while HAI-2-∆KD1 had a better interaction with activated TMPRSS2 because of an increased level of the N-terminus of TMPRSS2 after immunoprecipitation. The data together indicates that the KD2 of HAI-2 is important for the interaction with TMPRSS2 in cells. The DNA fragments encoding the extracellular region (EC), KD1 and KD2 proteins of HAI-2 were then constructed into pMAL vectors. The recombinant proteins (EC, KD1 and KD2 of HAI-2) were purified from the E. coli expression system. Using proteolytic activity assays, the results (Fig. 3b) showed that the extracellular region of rHAI-2 proteins displayed more inhibitory effects on TMPRSS2 activity than KD1 or KD2 alone, although both HAI-2’s KD1 and KD2 could suppress the proteolytic activity of TMPRSS2 in a dose-response manner. Moreover, rHAI-2 proteins with the deletion of KD1 (∆KD1) and KD2 domain (∆KD2) were also purified from baculovirus expression system, and used to determine their inhibitory effects on TMPRSS2. As shown in Fig. 3c, rHAI-2 proteins with the deletion of KD1 or KD2 still could significantly suppress the proteolytic activity of TMPRSS2 in a dose-response manner. In order to support the interaction between HAI-2 and TMPRSS2, we performed the computational docking of HAI-2’s KD1 and KD2 with TMPRSS2 protease domain using the crystal structures (HAI-2’s KD1, PDB ID: 4U3274; the model of HAI-2’s KD2 and TMPRSS2 protease domain was constructed by SWISS-MODEL [34, 35]) and ClusPro web server [36–40]. The predicted docking models showed that both KD1 and KD2 of HAI-2 had close contacts with the protease domain of TMPRSS2 (Fig. S2A, B). Moreover, both active sites (R48 for KD1 and R143 for KD2) in HAI-2’s KD1 and KD2 domains deeply inserted into the protease domain and directly contacted with the catalytic residue (S441) of TMPRSS2 (Fig. S2C, D). The results of docking prediction suggest that both KD1 and KD2 of HAI-2 can interact with the protease domain of TMPRSS2 with a direct contact between KDs’ active sites and TMPRSS2’s catalytic residue. The data together indicate that HAI-2’s KD1 and KD2 domains have an inhibitory effect against TMPRSS2, and the KD2 of HAI-2 may function as an important domain to interact with TMPRSS2 in cells.
Fig. 3. Identification of the functional domain of HAI-2 for the inhibition of TMPRSS2 proteolytic activity.
a Schematic structure of wild-type and various mutants of HAI-2. Wild-type HAI-2 (HAI-2-FL), ΔKD1 HAI-2 (the deletion of kunitz domain 1), ΔKD2 HAI-2 (the deletion of kunitz domain 2) and ΔTM HAI-2 (the deletion of transmembrane domain) were constructed in pcDNA3.1 vectors with a Myc/His tag. Interaction between various HAI-2 mutants and TMPRSS2 was analyzed by co-immunoprecipitation and immunoblot analysis. HEK293 cells were transfected with wild-type HAI-2, ΔKD1 HAI-2, ΔKD2 HAI-2 and ΔTM HAI-2 (Myc tag) plasmids in the presence of TMPRSS2 (a flag tag) plasmids. Cell lysates were used for immunoprecipitation using an anti-Myc antibody. The eluted proteins were subjected to SDS-PAGE and western blotting analysis with anti-Myc or anti TMPRSS2 (Abcam) antibodies. The arrows indicated the full length or amino (N) terminus of TMPRSS2. The (*) indicated IgG. b The extracellular region (EC, a.a.38–a.a.183), kunitz domain I (KD1, a.a.38–a.a.88) and kunitz domain II (KD2, a.a.133–a.a.183) portions of HAI-2 were constructed into pMAL-c2x vectors. The plasmids were used to transform E. coli strain BL21-DE3 for the protein overexpression and purification. The effects of purified rHAI-2-EC, rHAI-2-KD1 and rHAI-2-KD2 proteins on TMPRSS2’s proteolytic activity were examined by measuring the fluorescence signals of Boc-Gln-Ala-Arg-AMC substrates. Measurements were performed in triplicate in each experiment. Three independent experiments were carried out and statistically calculated as mean ± S.D. c The HAI-2 mutants with deletion of KD1 (HAI-2-ΔKD1) or deletion of KD2 (HAI-2-ΔKD2) were constructed into Bacmid vectors for their overexpression in insect cells. The effects of purified recombinant HAI-2-ΔKD1 and HAI-2-ΔKD2 proteins on TMPRSS2’s proteolytic activity were performed by monitoring the fluorescence signals of Boc-Gln-Ala-Arg-AMC substrate. Measurements were performed in triplicate each experiment. Three independent experiments were carried out and statistically calculated as mean ± S.D.
HAI-2 inhibited TMPRSS2-induced PCa invasion and substrate procession
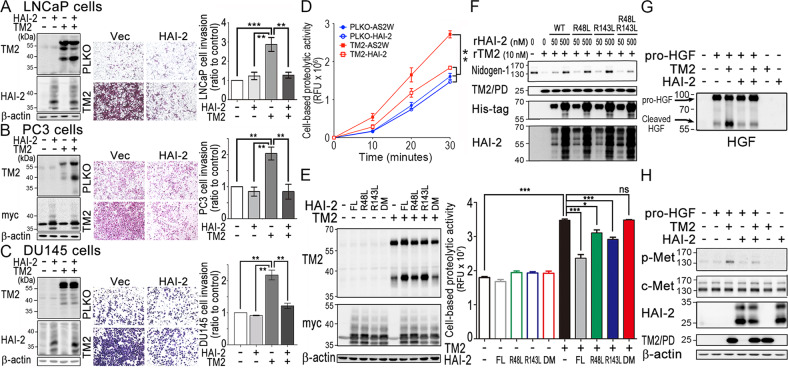
To further evaluate the importance of HAI-2 in the modulation of TMPRSS2, we co-overexpressed HAI-2 and TMPRSS2 in LNCaP, PC3 and DU145 cells, and then analyzed their invasive ability using transwell assays. The ectopic expression levels of TMPRSS2 and HAI-2 were validated by immunoblot analysis. The results showed that TMPRSS2 overexpression induced LNCaP (Fig. 4a), PC3 (Fig. 4b), and DU145 cell invasion (Fig. 4c), and HAI-2 overexpression could suppress TMPRSS2-induced PCa cell invasion, down to the basal level. To further investigate the role of HAI-2 and TMPRSS2 in PCa cells, the expression levels of TMPRSS2 and HAI-2 were analyzed by qRT-PCR. In addition, Matriptase was also examined since this protease has been identified as a substrate of TMPRSS2 [8]. As shown in Fig. S3A, compared to LNCaP cells, TMPRSS2 mRNA levels were decreased in VCaP, CWR22Rv1, PC3, and DU145 cells, while there were comparable mRNA expression levels of HAI-2 in these cells. Moreover, the western blot results (Fig. S3B) showed that the protein levels of TMPRSS2, Matriptase, and HAI-2 were consistently correlated with their gene expression levels in these cells. The protein levels of TMPRSS2 were higher in LNCaP and VCaP cells, compared to CWR22Rv1, PC3 and DU145 cells (Fig. S3B). The activated levels of Matriptase (a substrate of TMPRSS2) were also correlated with TMPRSS2 expression in LNCaP and VCaP cells (Fig. S3B). Since the expression levels of TMPRSS2, Matriptase and HAI-2 were low in DU145 cells (Fig. S3B), DU145 cells were further used to examine the role of HAI-2 in TMPRSS2-induced pericellular proteolytic activity. As shown in Fig. 4d, the overexpression of TMPRSS2 in DU145 cells enhanced the pericellular proteolytic activity and the overexpression of HAI-2 could suppress TMPRSS2-induced proteolytic activity, indicating that PCa with overexpression of TMPRSS2 or down-regulation of HAI-2 augments TMPRSS2’s proteolytic activity to cleavage TMPRSS2’s substrates. Moreover, we overexpressed TMPRSS2 and different HAI-2 proteins (wild-type, R48L, R143L, and R48L/R143L HAI-2) in CWR22Rv1 cells. As shown in Fig. 4e, HAI-2 also could inhibit the proteolytic activity of TMPRSS2 in CWR22Rv1 cells, and the HAI-2 mutants within KD1 (R48L) or KD2 (R143L) reduced their inhibitory effect on TMPRSS2 activities. Compare to wild-type HAI-2, R48L/R143L HAI-2 (double mutation on KD1 and KD2 domain) almost lost its function on suppressing TMPRSS2 activity. Since Nidogen-1 (a component of extracellular matrix) has been identified to be one of TMPRSS2’s substrates [8], we then used purified rTMPRSS2 proteins to confirm the role of TMPRSS2 in Nidogen-1 cleavage and found that TMPRSS2 could cleave Nidogen-1 in a dose-response manner (Fig. S4A). Moreover, we examined the effects of HAI-2 proteins on TMPRSS2-cleaved Nidogen-1. As shown in Fig. 4f and S4B, rHAI-2 proteins could significantly inhibit TMPRSS2 to cleave Nidogen-1, while R48L/R143L HAI-2 proteins showed reduced effects to suppress TMPRSS2-induced Nidogen-1 degradation. Purified rHAI-2 proteins alone had no effect on Nidogen-1 degradation (Fig. S4C). In order to examine the role of HAI-2 in TMPRSS2-activated c-Met signaling, DU145 cells were treated with pro-HGF which was pre-incubated with or without rTMPRSS2 in the presence or absence of rHAI-2 proteins. The results showed that rTMPRSS2 could proteolytically activate pro-HGF (Fig. 4g), leading to c-Met activation (indicated by its phosphorylation) (Fig. 4h), and rHAI-2 could suppress TMPRSS2-induced pro-HGF procession and c-Met phosphorylation (Fig. 4g, h). Thus, the results together indicate that HAI-2 can suppress TMPRSS2’s function on substrate procession and PCa cell invasion.
Fig. 4. Inhibitory role of HAI-2 in TMPRSS2-induced cell invasion and substrate cleavage.
a–c Role of HAI-2 in TMPRSS2-induced PCa cell invasion. PCa LNCaP (a), PC3 (b), and DU145 cells (c) were transfected with TMPRSS2 plasmids in the presence or absence of HAI-2 plasmids and stable pools were established after antibiotic selections. Control cells were transfected with pLKO (vector for TMPRSS2) or pcDNA 3.1 (vector for HAI-2). Cell lysates were collected, subjected to SDS-PAGE and immunoblot analysis using anti-TMPRSS2 (AL20) and anti-HAI-2 antibodies. β-actin was used as control. Left panels: Immunoblots of TMPRSS2 and HAI-2 in the transfectants. Middle and Right panels: Effect of TMPRSS2- or HAI-2-overexpression on the invasion of the PCa cells. For invasion assays, cells were seeded at a density of 4 × 105 (LNCaP) or 5 × 104 (PC3 and DU145) per Boyden chamber with Matrigel-coating and cultured for 40 h (LNCaP) or 16 h (PC3 and DU145). Cells were fixed and stained with crystal violet. Invaded cells were imaged under a light microscope. The cell invasion results were statistically calculated from three independent experiments and represented as mean ± SEM. d Analysis of the role of HAI-2 in TMPRSS2-induced pericellular proteolysis of PCa cells by monitoring the fluorescence signals of Boc-Gln-Ala-Arg-AMC substrate. DU145 transfectants with the overexpression of HAI-2, TMPRSS2 and TMPRSS2/HAI-2 were seeded, cultured one day, and starved for 16 h. Cells were then washed with PBS and incubated with 10 μM of fluorescence substrates. The conditioned media were then collected at indicated times and used for the measurement of the fluorescence signals. Results were statistically calculated from three independent experiments. e Analysis of the pericellular proteolytic activity of CWR22Rv1 cells with or without TMPRSS2 overexpression in the presence or absence of various HAI-2 mutants. CWR22Rv1 cells were transfected with wild-type HAI-2, HAI-2-R48L, HAI-2-R143L, HAI-2-R48L/R143L double mutant plasmids in the presence of TMPRSS2 plasmid or vector alone. Cell lysates were used for SDS-PAGE and immunoblotting analysis using anti-TMPRSS2 (AL20) or anti-Myc antibodies (Santa Cruz). β-actin was used as control. Immunoblots were shown in the left panel. The effects of TMPRSS2 and HAI-2 mutant overexpression on the pericellular proteolytic activity of CWR22Rv1 cells were performed by monitoring the fluorescence signal of Boc-Gln-Ala-Arg-AMC substrate (ENZO). For the pericellular proteolysis assays, transfectants were washed with PBS and incubated with 30 μM of fluorescence substrates. The conditioned media were then collected after incubation at 37 °C for 40 min and used for the measurement of fluorescence signals. f Examination of the effects of purified rHAI-2 mutant proteins (from E. coli) on the proteolytic activity of TMPRSS2 on Nidogen-1 cleavage. Three micrograms (3 μg) of Matrigel (Corning) were incubated with rTMPRSS2 proteins (10 nM) in the presence of different concentrations of purified recombinant HAI-2 mutant proteins with MBP/His tag. After incubation, the mixture was then subjected to SDS-PAGE and western blot analysis with anti-Nidogen-1 (GeneTex), anti-TMPRSS2 (TM2; Abcam), anti-His (Thermo) and anti-HAI-2 (LTK) antibodies. g, h Inhibitory effect of HAI-2 on TMPRSS2-induced pro-HGF cleavage and c-Met activation in PCa cells. Fifty nanograms (50 ng) of pro-HGF were incubated with 20 nM rTMPRSS2 or 200 nM rHAI-2 (R&D) in PBS for an hour, and some of the solutions after the proteolytic processing were subjected to the immunoblot analysis using an anti-HGF antibody (g). The remaining of the TMPRSS2-processing solutions was added to serum-starved DU145 cells for 10 min, and cell lysates were collected, followed by SDS-PAGE and immunoblot analysis using anti-HAI-2 (LTK), anti-TMPRS22 (TM2; Abcam), anti-phospho-Met (p-met, Cell Signaling), anti-c-Met (Spring), and anti-β-actin antibodies (h).
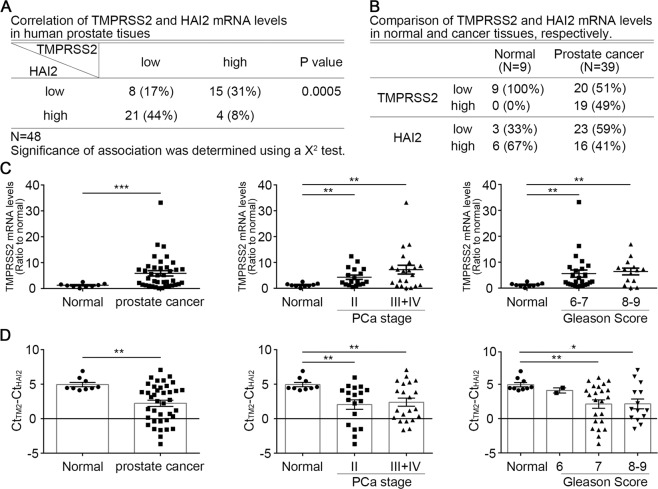
Correlation of HAI-2 and TMPRSS2 in PCa specimens
To evaluate the clinical relevance of TMPRSS2 and HAI-2 expression, we used PCa cDNA arrays to examine the levels of TMPRSS2 and HAI-2 in PCa specimens [28]. The mRNA levels of TMPRSS2 showed an inverse correlation with HAI-2 mRNA levels in the PCa specimens, compared to normal prostate tissues (Figs. 5a and S5A). The expression levels of TMPRSS2 were relatively low in normal prostate tissues and increased in PCa specimens, while the expression levels of HAI-2 displayed an inverse trend to TMPRSS2 with downregulation in PCa specimens (Fig. 5b). The levels of TMPRSS2 were correlated with the PCa progression following the tumor stages and Gleason scores (Fig. 5c). The levels of HAI-2 in PCa patients with Gleason scores 6–7 and 8–9 were significantly reduced, compared with normal prostate tissues [28]. To further analyze the correlation between HAI-2 and TMPRSS2 in PCa progression, we further analyzed the expression ratio of TMPRSS2 to HAI-2. The results showed that the ratio of TMPRSS2 to HAI-2 was increased in PCa groups, compared to normal groups. Moreover, the ratio of TMPRSS2 to HAI-2 was also significantly increased during the PCa progression which was analyzed by tumor stages and Gleason scores (Fig. 5d). Analysis of The Cancer Genome Atlas Prostate Adenocarcinoma (TCGA-PRAD) database further supported the increased ratio of TMPRSS2 to HAI-2 during PCa progression (Fig. S5B). The results indicate that an increased ratio of TMPRSS2 to HAI-2 may contribute to the dysregulation of TMPRSS2 activity and thereby PCa progression.
Fig. 5. Correlation of the expression of TMPRSS2 and HAI-2 in PCa specimens.
The expression levels of TMPRSS2 and HAI-2 mRNA in normal human prostate and PCa were analyzed using q-PCR in a PCa cDNA array including 9 normal and 39 cancerous prostate specimens. qPCR results were presented using the comparative Ct values with normalization to β-actin levels and presented as means ± SEM. a Correlation of TMPRSS2 and HAI-2 mRNA levels in archival PCa specimens. b Correlation of TMPRSS2 and HAI-2 mRNA levels at normal prostate tissues and cancer tissues of archival PCa specimens. c The expression levels of TMPRSS2 and HAI-2 mRNA in the PCa tissues with different tumor stages and Gleason scores. Data were analyzed using the comparative Ct method with normalization to β-actin levels and presented as means ± S.E.M (*p < 0.05; **p < 0.01; ***p < 0.001). d Ratio of TMPRSS2 to HAI-2 mRNA levels in PCa tissues at different tumor stages and Gleason scores. The ratios of TMPRSS2 to HAI-2 were further statistically calculated for the difference after the subtraction of HAI-2 Ct values by TMPRSS2 Ct values (TMPRSS2 Ct value minus HAI-2 Ct value) and presented as means ± S.E.M (*p < 0.05; **p < 0.01; ***p < 0.001).
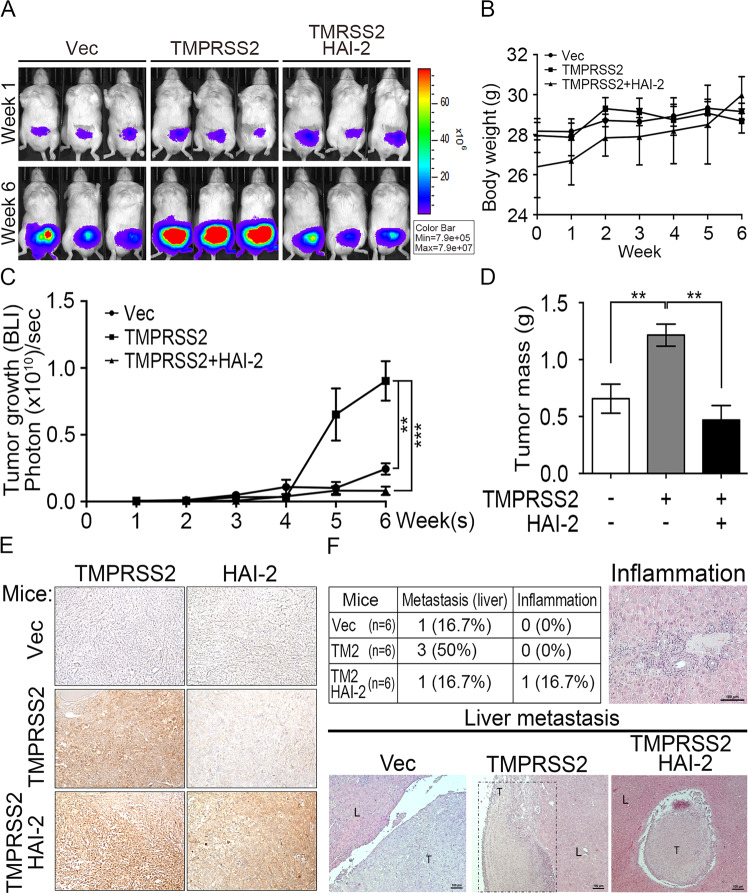
HAI-2 inhibited TMPRSS2-induced tumor growth and metastasis in PCa xenograft mouse models
In order to evaluate the significance of HAI-2 on TMPRSS2 regulation in PCa, we further examined whether HAI-2 could suppress TMPRSS2-induced PCa tumor growth and metastasis. Since DU145 cells had relatively low expression levels of HAI-2, TMPRSS2 and Matriptase among PCa cells, we then used DU145 cells as a cell model to examine the role of HAI-2 in TMPRSS2-induced PCa tumor growth and metastasis. The results from an orthotopic xenograft animal model showed that TMPRSS2 overexpression significantly increased the tumor growth (Fig. 6a, c) and tumor mass (Fig. 6d) with no significant effect on the mouse body weights (Fig. 6b). Moreover, HAI-2 overexpression suppressed TMPRSS2-induced tumor growth and masses in the xenograft animal experiment (Fig. 6a–d). The protein levels of TMPRSS2 and HAI-2 in tumor lesions were further detected using immunohistochemical staining and shown in Fig. 6e. In addition, we searched for the metastatic lesions after the animals were euthanized. The results (Fig. 6f) showed that TMPRSS2 overexpression significantly increased the liver metastatic rate of PCa DU145 cells from 16.7% to 50%, and HAI-2 overexpression inhibited TMPRSS2-induced PCa metastasis (Fig. 6f, up-left Table). The metastatic tumor tissues were further examined using H&E staining and the images showed the representative metastatic or inflammatory lesions (Fig. 6f, bottom and right). The data together indicate that HAI-2 plays a suppression role in TMPRSS2 action on prostate tumor growth and metastasis.
Fig. 6. Suppression role of HAI-2 in TMPRSS2-induced PCa tumor growth and metastasis.
DU145 cells (2 × 105 cells per mouse) were injected into the anterior prostates of mice. The tumor volumes were weekly measured as bioluminescence images using an in vivo IVIS system. Three representative images at week 1 (W1) and week 6 (W6) from each group (n = 6) were shown in a. b The body weights of mice were measured weekly. c Tumor growth was statistically calculated from the bioluminescence images at each week as mean ± SEM, and plotted. *p < 0.05; **p < 0.01. d After 6 weeks, tumor weights at each group were measured, plotted, and statistically calculated as mean ± SEM. *p < 0.05; **p < 0.01. e Immunohistochemical analysis of TMPRSS2 (left panel) and HAI-2 (right panel) in the primary tumors using anti-TMPRSS2 and anti-HAI-2 Abs. f The histological section from the livers of xenografted mice were stained with haematoxylin. and eosin, and then were subjected to the measurement of the metastatic lesions and inflammation. Representative H&E images were shown in the up right (inflammation) and bottom panels (metastatic lesions).
Discussion
Cancer metastasis is the main reason for the dead of patients. Dysregulation of androgen-regulated TMPRSS2 has been shown to increase prostate cancer (PCa) metastasis [8, 9]. It thus becomes an important issue to identify a potential inhibitor for TMPRSS2, in order to explore whether there is an imbalance between TMPRSS2 and its cognate inhibitor for TMPRSS2 dysregulation, or to provide new information for the development of a novel approach against TMPRSS2-induced PCa progression and metastasis. In this study, we used co-immunoprecipitation and LC/MS/MS analysis and isolated more than 300 TMPRSS2-associated proteins. Among them, we identified HAI-2 to be a novel cognate inhibitor for TMPRSS2 in human PCa cells and further found out HAI-2 was a critical inhibitor to suppress TMPRSS2 protease activity and TMPRSS2-induced PCa cell invasion and metastasis (Fig. 7).
Fig. 7. Schematic model for the inhibition of TMPRSS2 by HAI-2.
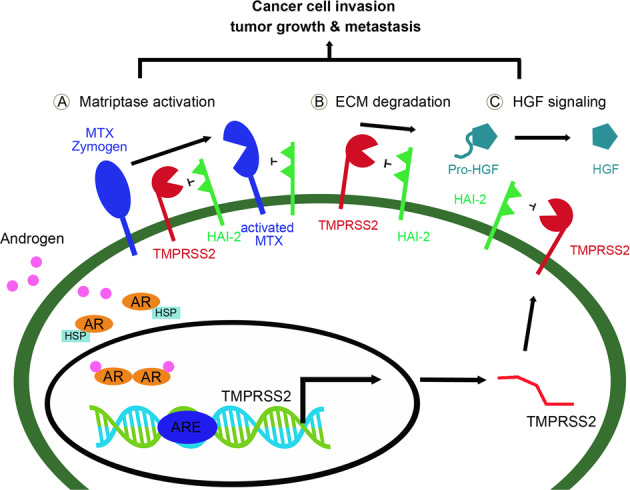
After binding to androgens, androgen receptors (ARs) are released from heat shock proteins (HSP), dimerized and moved to the nucleus. The AR dimers would bind to the ARE (androgen response elements) of TMPRSS2 and turn on the gene expression. After androgen-induced transcription/translation or its aberrant up-regulation, TMPRSS2 proteins will be translocated to the plasma membrane for its biological functions on Matriptase activation (a), ECM degradation (b), and the processing of pro-HGF to HGF (c), leading to PCa cell invasion, tumor growth and metastasis. HAI-2 can inhibit the proteolytic activity of TMPRSS2 to reduce the activation of Matriptase, the ECM degradation and the proteolytic activation of pro-HGF to HGF, and simultaneously suppresses the activated Matriptase, all contributing to down regulation of pericellular proteolysis, c-Met signaling, PCa cell motility, tumor growth and metastasis. As a result, inhibition of TMPRSS2 by HAI-2 can prevent PCa progression.
Our present study indicates TMPRSS2 is important for the invasion and metastasis of PCa cells. However, it does not show a positive correlation between TMPRSS2 levels and invasive capability among the prostate cancer cell lines no matter of androgen receptor (AR) expression. Since TMPRSS2 is an androgen-regulated transmembrane serine protease and highly expressed in AR-positive PCa LNCaP and VCaP cells, it implies that TMPRSS2 may briefly play a role in AR-positive PCa cell invasion. Indeed, TMPRSS2 has been shown to be important for androgen action on prostate cancer cell invasion and matriptase activation [8, 9] and its expression levels were related to the activated level of its substrate matriptase (Fig. S3B). Thus, the data suggests that TMPRSS2 may be mainly involved in AR-positive PCa cell motility and progression. In this study, in order to investigate the functional roles of TMPRSS2 and HAI-2 in prostate cancer cells under the less interference of androgen receptor signaling on PCa cell invasion and metastasis, prostate cancer cell lines such as PC3 and DU145 cells with little/no TMPRSS2 expression were selected to be the cell models for the study. Our results together indicate that TMPRSS2 alone also can promote prostate cancer cell invasion no matter of AR, which is ably suppressed by HAI-2.
During the past decade, several groups devoted to look for small chemical drugs to inhibit TMPRSS2, such as bromhexine hydrochloride [41], benzylsulfonyl-dArg-Pro-4-amidinobenzylamide or other synthetic inhibitors [42, 43]. However, small molecule-based targeted drugs have been estimated to be at least an average of 6.3 targets [44] suggesting that small molecules for targeted therapy exhibit more off targets and consequent unknown side effects. Instead, protein-based drugs have been demonstrated with better target-specificity and fewer side effects than chemical targeted drugs. Thus, the advantage of the protein-derived drugs strongly encourages us to identify the cognate inhibitor(s) of TMPRSS2, and the finding of TMPRSS2’s cognate inhibitor will provide an important information to develop protein-based therapeutic approaches against TMPRSS2-mediated malignancy or virus infection. Recent reports have also shown that downregulated expression of two cognate inhibitors of matriptase (HAI-1 and HAI-2) is correlated with the progression of PCa and other cancers [28, 45–47], and occurs in parallel with matriptase activation and cancer cell invasion [28, 48]. Thus, reduced levels of HAI-1 or HAI-2 contribute to matriptase activation and cancer progression. In fact, an engineered serine protease inhibitor HAI-1 has been developed with an excellent inhibitory effect on its target serine protease matriptase [49]. Thus, the results together indicate that the protease-inhibitory domains of HAI-2 and HAI-1 may exhibit some potentials for the development of therapeutic recombinant proteins after protein engineering, and will be clinically useful for TMPRSS2- or their target protease-induced prostate cancer progression, even used against viral infection.
Protein glycosylation plays a critical role in regulating protein function, structure and stability, which are also involved in human disease progression [50]. HAI-2 has been reported to be a heavily glycosylated protein [19], and HAI-2 also exhibits different patterns of glycosylation in PCa cells (Fig. S6A). N-Glycan modification on HAI-2 has been reported to be crucial for HAI-2 to inhibit matriptase in breast cancer cells [33], but our data showed that the N-glycosylation of recombinant HAI-2 proteins was not quite important for rHAI-2 proteins to suppress TMPRSS2 proteolytic activity (Fig. S6B, C). Thus, the importance of glycosylation on HAI-2 function to inhibit its target proteases may be dependent on cell types or targeted proteases and remains more investigation. Since Matriptase is one of TMPRSS2’s substrates to mediate TMPRSS2 signaling for PCa cell invasion, tumor growth and metastasis [8], in this study, we unexpectedly identified HAI-2 to be a cognate inhibitor of TMPRSS2. Moreover, our results further show that HAI-2 can suppress TMPRSS2-mediated Matriptase activation (Fig. S7). Thus, the results indicate that HAI-2 functions as a bi-specific inhibitor by simultaneous suppression on TMPRSS2 and Matriptase at this proteolytic cascade in human PCa cells. The data suggest that the application of the same protease inhibitor on a protease cascade may provide an efficient regulation on the proteolytic signal pathway.
Our present study showed that HAI-2 could suppress TMPRSS2-induced c-Met activation by inhibiting the procession of pro-HGF. Lucas et al. also showed that HGF activated by TMPRSS2 promoted c-Met signaling activation and induced a pro-invasive epithelial-to-mesenchymal transition phenotype. They further demonstrated that TMPRSS2-activated HGF promoted prostate cancer cell invasion [9]. Moreover, c-Met expression was positively correlated with the Gleason score [51], and the levels of serum HGF and urine c-Met are elevated in PCa patients with metastatic tumor compared to patients with localized tumor [52]. The results together indicate that there is a link between the ratio of TMPRSS2/HAI-2 and c-Met activity axis, and dysregulation of TMPRSS2/HAI-2 ratio in favor of proteolysis to promote PCa cancer progression may be through modulating c-Met signaling.
In addition to being a protease inhibitor, HAI-2 may also serve as a chaperone or stabilizer for TMPRSS2 proteins because the overexpression of HAI-2 could further significantly increase the protein levels of TMPRSS2 in PC3 and HEK293T cells. This observation is similar to the role of HAI-1 to Matriptase, because HAI-1 has been reported to be crucial in the biosynthesis, intracellular trafficking, activation of Matriptase, in addition to its role in the protease inhibition [53]. Thus, the data together suggest that HAI-2 may also serve as a chaperone-like inhibitor for TMPRSS2, like HAI-1 to Matriptase, to ensure their appropriate intracellular processing, proteolytic activation and subsequent inhibition.
Tumor microenvironment has been proposed to play important roles in tumor progression. In general, the paracrine or juxtacrine signaling from the tumor microenvironment (cytokines, chemokines, or growth factors) can facilitate tumor cell survival, proliferation or motility [54]. In the tumor microenvironment, cancer-associated fibroblasts (CAFs) act as a pivotal role in supplying pre-metastatic factors including pro-HGF [55]. In head and neck squamous cell carcinoma (HNSCC), pro-HGF that is secreted by CAFs increases HNSCC glycolysis and promotes HNSCC progression [56]. In hormone-refractory PCa patients, the levels of HGF in the plasma are increased and the high HGF levels are associated with poor prognosis [57]. Moreover, CAFs have been reported to promote PCa drug resistance to anti-androgen treatment [58]. Our results indicate that HAI-2 can inhibit TMPRSS2-induced pro-HGF maturation to HGF, leading to inhibition of c-MET signaling. Following the PCa progression, CAFs increase in the tumor microenvironment and secret a high abundance of pro-HGF. Once HAI-2 down-regulation or TMPRSS2 upregulation following the PCa progression will offer an increased proteolytic activity for pro-HGF maturation and lead to the activation of c-MET signaling. This molecular mechanism may provide an explanation regarding why HAI-2 downregulation or TMPRSS2 overexpression can enhance PCa cell motility and metastasis via proteolytic processing of environmental pro-HGF. Therefore, the blockade of TMPRSS2 or pro-HGF maturation may shed the light on treating refractory PCa patients.
In summary, we used co-immunoprecipitation, LC/MS/MS approaches, biochemical and image analyses, to identify HAI-2 as a cognate inhibitor for TMPRSS2. Moreover, the forced expression of HAI-2 could decrease TMPRSS2-induced PCa cell invasion, tumor growth and metastasis. Thus, the results together provide information to explain how TMPRSS2 can be modulated and inhibited by a non-covalent inhibitor HAI-2. These findings may further indicate the dysregulation of TMPRSS2 and HAI-2 in favor of pericellular proteolysis plays a role in human PCa progression, and give a hint that targeting pericellular serine proteases may be an alternative approach for developing a novel cancer therapy against PCa.
Material and methods
Due to page limits, the detailed information of cell culture, invasion assay, co-immunoprecipitation, tissue array qPCR analysis, tumor xenografts was described in Supplementary information. Detailed information for all primary antibodies used in this study was listed in Supplementary Table 1. The sequences of primers used for Quantitative Real-Time RT-PCR in this study were listed in Supplementary Table 2.
Statistical analysis
The intensities of images or bands were measured using ImageJ software, analyzed using GraphPad Prism 8.0 (GraphPad Software, CA, USA). For tumor growth, the statistical analysis was determined by a two-way ANOVA with Bonferroni correction. All other statistical analyses were performed by a two-tailed unpaired t test. P values of < 0.05 were considered significant in all studies. *p < 0.05; **p < 0.01; ***p < 0.001.
Supplementary information
Supplementary Materials, Methods and Information
Acknowledgements
This study was supported by Ministry of Science and Technology Grants MOST 104-2320-B-002-044-MY3, MOST 105-2911-I-002-521, MOST 106-2320-B-002-046-MY3 and MOST 108-2320-B-002-024-MY3, National Health Research Institutes Grant NHRI-EX106-10401BI and NHRI-EX109-10725BI, and National Taiwan University Grant NTU-CESRP-104R7602C4, NTU105R89612 and NYU107L890504 to M.S. Lee. We appreciate the service of the First Core Laboratory of National Taiwan University College of Medicine. We thank Dr. Chen-Yong Lin at the Georgetown University for his gifts of antibodies. We would like to acknowledge Dr. Chia-Jung Yu (Chang Gung University, Taoyuan, Taiwan; Chang Gung Memorial Hospital, Linkou, Taoyuan, Taiwan) and the Proteomics Core Laboratory of Chang Gung University for providing proteomics technical support.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chun-Jung Ko, Ting-Wei Hsu
Supplementary information
The online version of this article (10.1038/s41388-020-01413-w) contains supplementary material, which is available to authorized users.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Shen CA-SaMM. Molecular genetics of prostate cancer. Genes Dev. 2000;14:26. doi: 10.1101/gad.819500. [DOI] [PubMed] [Google Scholar]
- 3.Mierke CT. Physical break-down of the classical view on cancer cell invasion and metastasis. Eur J Cell Biol. 2013;92:89–104. doi: 10.1016/j.ejcb.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Lopez-Otin C, Bond JS. Proteases: multifunctional enzymes in life and disease. J Biol Chem. 2008;283:30433–7. doi: 10.1074/jbc.R800035200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sevenich L, Joyce JA. Pericellular proteolysis in cancer. Genes Dev. 2014;28:2331–47. doi: 10.1101/gad.250647.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bugge TH, Antalis TM, Wu Q. Type II transmembrane serine proteases. J Biol Chem. 2009;284:23177–81. doi: 10.1074/jbc.R109.021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin CY, Anders J, Johnson M, Dickson RB. Purification and characterization of a complex containing matriptase and a Kunitz-type serine protease inhibitor from human milk. J Biol Chem. 1999;274:18237–42. doi: 10.1074/jbc.274.26.18237. [DOI] [PubMed] [Google Scholar]
- 8.Ko CJ, Huang CC, Lin HY, Juan CP, Lan SW, Shyu HY, et al. Androgen-induced TMPRSS2 activates matriptase and promotes extracellular matrix degradation, prostate cancer cell invasion, tumor growth, and metastasis. Cancer Res. 2015;75:2949–60. doi: 10.1158/0008-5472.CAN-14-3297. [DOI] [PubMed] [Google Scholar]
- 9.Lucas JM, Heinlein C, Kim T, Hernandez SA, Malik MS, True LD, et al. The androgen-regulated protease TMPRSS2 activates a proteolytic cascade involving components of the tumor microenvironment and promotes prostate cancer metastasis. Cancer Discov. 2014;4:1310–25. doi: 10.1158/2159-8290.CD-13-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson S, Greer B, Hooper J, Zijlstra A, Walker B, Quigley J, et al. The membrane-anchored serine protease, TMPRSS2, activates PAR-2 in prostate cancer cells. Biochem J. 2005;388:967–72. doi: 10.1042/BJ20041066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lam DK, Dang D, Flynn AN, Hardt M, Schmidt BL. TMPRSS2, a novel membrane-anchored mediator in cancer pain. Pain. 2015;156:923–30. doi: 10.1097/j.pain.0000000000000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garten W, Braden C, Arendt A, Peitsch C, Baron J, Lu Y, et al. Influenza virus activating host proteases: Identification, localization and inhibitors as potential therapeutics. Eur J Cell Biol. 2015;94:375–83. doi: 10.1016/j.ejcb.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, et al. SARS-CoV-2 Cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–80. e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 15.Seth A, Watson DK. ETS transcription factors and their emerging roles in human cancer. Eur J Cancer. 2005;41:2462–78. doi: 10.1016/j.ejca.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 16.Chen YW, Lee MS, Lucht A, Chou FP, Huang W, Havighurst TC, et al. TMPRSS2, a serine protease expressed in the prostate on the apical surface of luminal epithelial cells and released into semen in prostasomes, is misregulated in prostate cancer cells. Am J Pathol. 2010;176:2986–96. doi: 10.2353/ajpath.2010.090665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucas JM, True L, Hawley S, Matsumura M, Morrissey C, Vessella R, et al. The androgen-regulated type II serine protease TMPRSS2 is differentially expressed and mislocalized in prostate adenocarcinoma. J Pathol. 2008;215:118–25. doi: 10.1002/path.2330. [DOI] [PubMed] [Google Scholar]
- 18.Shimomura T, Denda K, Kitamura A, Kawaguchi T, Kito M, Kondo J, et al. Hepatocyte growth factor activator inhibitor, a novel Kunitz-type serine protease inhibitor. J Biol Chem. 1997;272:6370–6. doi: 10.1074/jbc.272.10.6370. [DOI] [PubMed] [Google Scholar]
- 19.Kawaguchi T, Qin L, Shimomura T, Kondo J, Matsumoto K, Denda K, et al. Purification and cloning of hepatocyte growth factor activator inhibitor type 2, a Kunitz-type serine protease inhibitor. J Biol Chem. 1997;272:27558–64. doi: 10.1074/jbc.272.44.27558. [DOI] [PubMed] [Google Scholar]
- 20.Kawaguchi M, Kataoka H. Mechanisms of hepatocyte growth factor activation in cancer tissues. Cancers (Basel) 2014;6:1890–904. doi: 10.3390/cancers6041890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin CY, Anders J, Johnson M, Sang QA, Dickson RB. Molecular cloning of cDNA for matriptase, a matrix-degrading serine protease with trypsin-like activity. J Biol Chem. 1999;274:18231–6. doi: 10.1074/jbc.274.26.18231. [DOI] [PubMed] [Google Scholar]
- 22.Herter S, Piper DE, Aaron W, Gabriele T, Cutler G, Cao P, et al. Hepatocyte growth factor is a preferred in vitro substrate for human hepsin, a membrane-anchored serine protease implicated in prostate and ovarian cancers. Biochem J. 2005;390:125–36. doi: 10.1042/BJ20041955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan B, Wu TD, Li W, Kirchhofer D. Identification of hepatocyte growth factor activator inhibitor-1B as a potential physiological inhibitor of prostasin. J Biol Chem. 2005;280:34513–20. doi: 10.1074/jbc.M502119200. [DOI] [PubMed] [Google Scholar]
- 24.Lee SL, Dickson RB, Lin CY. Activation of hepatocyte growth factor and urokinase/plasminogen activator by matriptase, an epithelial membrane serine protease. J Biol Chem. 2000;275:36720–5. doi: 10.1074/jbc.M007802200. [DOI] [PubMed] [Google Scholar]
- 25.Miyazawa K. Hepatocyte growth factor activator (HGFA): a serine protease that links tissue injury to activation of hepatocyte growth factor. FEBS J. 2010;277:2208–14. doi: 10.1111/j.1742-4658.2010.07637.x. [DOI] [PubMed] [Google Scholar]
- 26.Delaria KA, Muller DK, Marlor CW, Brown JE, Das RC, Roczniak SO, et al. Characterization of placental bikunin, a novel human serine protease inhibitor. J Biol Chem. 1997;272:12209–14. doi: 10.1074/jbc.272.18.12209. [DOI] [PubMed] [Google Scholar]
- 27.Qin L, Denda K, Shimomura T, Kawaguchi T, Kitamura N. Functional characterization of Kunitz domains in hepatocyte growth factor activator inhibitor type 2. FEBS Lett. 1998;436:111–4. doi: 10.1016/s0014-5793(98)01105-3. [DOI] [PubMed] [Google Scholar]
- 28.Tsai CH, Teng CH, Tu YT, Cheng TS, Wu SR, Ko CJ, et al. HAI-2 suppresses the invasive growth and metastasis of prostate cancer through regulation of matriptase. Oncogene. 2014;33:4643–52. doi: 10.1038/onc.2013.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maurer E, Gutschow M, Stirnberg M. Hepatocyte growth factor activator inhibitor type 2 (HAI-2) modulates hepcidin expression by inhibiting the cell surface protease matriptase-2. Biochem J. 2013;450:583–93. doi: 10.1042/BJ20121518. [DOI] [PubMed] [Google Scholar]
- 30.Kirchhofer D, Peek M, Lipari MT, Billeci K, Fan B, Moran P. Hepsin activates pro-hepatocyte growth factor and is inhibited by hepatocyte growth factor activator inhibitor-1B (HAI-1B) and HAI-2. FEBS Lett. 2005;579:1945–50. doi: 10.1016/j.febslet.2005.01.085. [DOI] [PubMed] [Google Scholar]
- 31.Wu SR, Lin CH, Shih HP, Ko CJ, Lin HY, Lan SW, et al. HAI-2 as a novel inhibitor of plasmin represses lung cancer cell invasion and metastasis. Br J Cancer. 2019;120:499–511. doi: 10.1038/s41416-019-0400-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roversi FM, Olalla Saad ST, Machado-Neto JA. Serine peptidase inhibitor Kunitz type 2 (SPINT2) in cancer development and progression. Biomed Pharmacother. 2018;101:278–86. doi: 10.1016/j.biopha.2018.02.100. [DOI] [PubMed] [Google Scholar]
- 33.Lai YJ, Chang HH, Lai H, Xu Y, Shiao F, Huang N, et al. N-Glycan Branching Affects the Subcellular Distribution of and Inhibition of Matriptase by HAI-2/Placental Bikunin. PLoS ONE. 2015;10:e0132163. doi: 10.1371/journal.pone.0132163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–5. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–8. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: a fully automated algorithm for protein-protein docking. Nucleic Acids Res. 2004;32:W96–9. doi: 10.1093/nar/gkh354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics. 2004;20:45–50. doi: 10.1093/bioinformatics/btg371. [DOI] [PubMed] [Google Scholar]
- 38.Kozakov D, Brenke R, Comeau SR, Vajda S. PIPER: an FFT-based protein docking program with pairwise potentials. Proteins. 2006;65:392–406. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- 39.Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, et al. The ClusPro web server for protein-protein docking. Nat Protoc. 2017;12:255–78. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozakov D, Beglov D, Bohnuud T, Mottarella SE, Xia B, Hall DR, et al. How good is automated protein docking? Proteins. 2013;81:2159–66. doi: 10.1002/prot.24403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chou FP, Chen YW, Zhao XF, Xu-Monette ZY, Young KH, Gartenhaus RB, et al. Imbalanced matriptase pericellular proteolysis contributes to the pathogenesis of malignant B-cell lymphomas. Am J Pathol. 2013;183:1306–17. doi: 10.1016/j.ajpath.2013.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vogel LK, Saebo M, Skjelbred CF, Abell K, Pedersen ED, Vogel U, et al. The ratio of Matriptase/HAI-1 mRNA is higher in colorectal cancer adenomas and carcinomas than corresponding tissue from control individuals. BMC Cancer. 2006;6:176. doi: 10.1186/1471-2407-6-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benaud CM, Oberst M, Dickson RB, Lin CY. Deregulated activation of matriptase in breast cancer cells. Clin Exp Metastasis. 2002;19:639–49. doi: 10.1023/a:1020985632550. [DOI] [PubMed] [Google Scholar]
- 44.Mestres J, Gregori-Puigjane E, Valverde S, Sole RV. Data completeness–the Achilles heel of drug-target networks. Nat Biotechnol. 2008;26:983–4. doi: 10.1038/nbt0908-983. [DOI] [PubMed] [Google Scholar]
- 45.Pisters LL, Troncoso P, Zhau HE, Li W, von Eschenbach AC, Chung LW. c-met proto-oncogene expression in benign and malignant human prostate tissues. J Urol. 1995;154:293–8. [PubMed] [Google Scholar]
- 46.Russo AL, Jedlicka K, Wernick M, McNally D, Kirk M, Sproull M, et al. Urine analysis and protein networking identify met as a marker of metastatic prostate cancer. Clin Cancer Res. 2009;15:4292–8. doi: 10.1158/1078-0432.CCR-09-0599. [DOI] [PubMed] [Google Scholar]
- 47.Aalinkeel R, Nair MP, Sufrin G, Mahajan SD, Chadha KC, Chawda RP, et al. Gene expression of angiogenic factors correlates with metastatic potential of prostate cancer cells. Cancer Res. 2004;64:5311–21. doi: 10.1158/0008-5472.CAN-2506-2. [DOI] [PubMed] [Google Scholar]
- 48.Dozmorov MG, Hurst RE, Culkin DJ, Kropp BP, Frank MB, Osban J, et al. Unique patterns of molecular profiling between human prostate cancer LNCaP and PC-3 cells. Prostate. 2009;69:1077–90. doi: 10.1002/pros.20960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Debes JD, Tindall DJ. Mechanisms of androgen-refractory prostate cancer. N Engl. J Med (Comment) 2004;351:1488–90. doi: 10.1056/NEJMp048178. [DOI] [PubMed] [Google Scholar]
- 50.Stowell SR, Ju T, Cummings RD. Protein glycosylation in cancer. Annu Rev Pathol. 2015;10:473–510. doi: 10.1146/annurev-pathol-012414-040438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jin JS, Cheng TF, Tsai WC, Sheu LF, Chiang H, Yu CP. Expression of the serine protease, matriptase, in breast ductal carcinoma of Chinese women: correlation with clinicopathological parameters. Histol Histopathol. 2007;22:305–9. doi: 10.14670/HH-22.305. [DOI] [PubMed] [Google Scholar]
- 52.Kang JY, Dolled-Filhart M, Ocal IT, Singh B, Lin CY, Dickson RB, et al. Tissue microarray analysis of hepatocyte growth factor/Met pathway components reveals a role for Met, matriptase, and hepatocyte growth factor activator inhibitor 1 in the progression of node-negative breast cancer. Cancer Res. 2003;63:1101–5. [PubMed] [Google Scholar]
- 53.Oberst MD, Chen LY, Kiyomiya K, Williams CA, Lee MS, Johnson MD, et al. HAI-1 regulates activation and expression of matriptase, a membrane-bound serine protease. Am J Physiol Cell Physiol. 2005;289:C462–70. doi: 10.1152/ajpcell.00076.2005. [DOI] [PubMed] [Google Scholar]
- 54.Pietras K, Ostman A. Hallmarks of cancer: interactions with the tumor stroma. Exp Cell Res. 2010;316:1324–31. doi: 10.1016/j.yexcr.2010.02.045. [DOI] [PubMed] [Google Scholar]
- 55.Tao L, Huang G, Song H, Chen Y, Chen L. Cancer associated fibroblasts: An essential role in the tumor microenvironment. Oncol Lett. 2017;14:2611–20. doi: 10.3892/ol.2017.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar D, New J, Vishwakarma V, Joshi R, Enders J, Lin F et al. Cancer-associated fibroblasts drive glycolysis in a targetable signaling loop implicated in head and neck squamous cell carcinoma progression. Cancer Res. 2018;78:3769–82. [DOI] [PMC free article] [PubMed]
- 57.Humphrey PA, Halabi S, Picus J, Sanford B, Vogelzang NJ, Small EJ, et al. Prognostic significance of plasma scatter factor/hepatocyte growth factor levels in patients with metastatic hormone- refractory prostate cancer: results from cancer and leukemia group B 150005/9480. Clin Genitourin Cancer. 2006;4:269–74. doi: 10.3816/CGC.2006.n.006. [DOI] [PubMed] [Google Scholar]
- 58.Eder T, Weber A, Neuwirt H, Grunbacher G, Ploner C, Klocker H et al. Cancer-associated fibroblasts modify the response of prostate cancer cells to androgen and anti-androgens in three-dimensional spheroid culture. Int J Mol Sci 2016; 17:1458 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials, Methods and Information