Abstract
Expression of the transcription repressor Gfi-1 is required for the maintenance of murine hematopoietic stem cells. In human cells, ectopic expression of Gfi-1 inhibits and RNA interference-mediated Gfi-1 downregulation enhances proliferation and colony formation of p210BCR/ABL-expressing cells.
To investigate the molecular mechanisms that may explain the effects of perturbing Gfi-1 expression in human cells, Gfi-1-regulated genes were identified by microarray analysis in K562 cells expressing the tamoxifen-regulated Gfi-1-ER protein. STAT 5B and Mcl-1, two genes important for the proliferation and survival of hematopoietic stem cells, were identified as direct and functionally relevant Gfi-1 targets in p210BCR/ABL-transformed cells because: i) their expression and promoter activity was repressed by Gfi-1; and ii) when constitutively expressed blocked the proliferation and colony formation inhibitory effects of Gfi-1.
Consistent with these findings, genetic or pharmacological inhibition of STAT 5 and/or Mcl-1 markedly suppressed proliferation and colony formation of K562 and CD34+ CML cells.
Together, these studies suggest that the Gfi-1→ STAT 5B/Mcl-1 regulatory pathway identified here can be modulated to suppress the proliferation and survival of p210BCR/ABL-transformed cells including CD34+ CML cells.
Keywords: cell survival, oncogene, tumor suppressor gene, therapy
INTRODUCTION
The process of hematopoiesis is regulated by a network of transcription factors acting cooperatively or antagonistically on multipotent or lineage-restricted hematopoietic progenitors (1). Genetic evidence indicates that “graded” expression of some transcription factors (i.e. PU.1, c-Myb, C/EBPα) has profound consequences for the expansion of certain hematopoietic cell compartments or for lineage choice (2–5). The critical importance of transcription factor expression levels for the coordinated regulation of early (i.e., self-renewal or commitment) and late (multipotent versus lineage-restricted differentiation) stages of normal hematopoiesis is also emphasized by a large body of evidence pointing out to de-regulated transcription factor expression/activity as a mechanism which promotes leukemic transformation (6). Conversely, perturbation of transcription factor levels can suppress the proliferation and survival of leukemic cells (6), suggesting that their biological effects can be exploited for therapeutic purposes in leukemic cells.
C/EBPα exemplifies a transcription factor with distinct roles in normal and leukemic cells because it regulates the balance between differentiation and proliferation in the early stages of normal myelopoiesis (5, 7) but is functionally or genetically inactivated in many types of myeloid leukemia in which the homeostatic coordination between proliferation and differentiation is lost (8, 9).
Moreover, when ectopically expressed it induces differentiation and inhibits proliferation of myeloid leukemia lines and primary cells from patients with CML-blast crisis (10–13). These effects are important for optimal suppression of p210BCR/ABL-dependent leukemia in mice since expression of a DNA binding-deficient mutant which does not induce granulocytic differentiation (13,14) did not prolong survival of leukemic mice as effectively as expression of transcription-activation competent C/EBPα (14).
While granulocytic differentiation of p210BCR/ABL-expressing cells is only induced by DNA binding and transcription activation-competent C/EBPα proteins (14), the role of transcription activation for the anti-proliferative effects of C/EBPα is less clear. We have shown that a DNA binding-deficient C/EBPα mutant suppresses the proliferation of p210BCR/ABL- transformed cells (14) and others have demonstrated that the interaction of C/EBPα with cell cycle regulatory and chromatin remodeling proteins is sufficient for proliferation inhibition (15–18).
Nevertheless, we searched for transcriptionally-regulated biologically relevant targets of C/EBPα that may explain its anti-proliferative effects by oligonucleotide microarray hybridization with RNA of p210BCR/ABL-transformed cells expressing wild type or DNA binding-deficient C/EBPα (19). One of the genes whose expression was activated by C/EBPα in a DNA binding-dependent manner is the transcriptional repressor Gfi-1 (19) which has a dual function in hematopoietic cells: maintenance of stem cells and differentiation of late granulocytic progenitors (20–24).
We showed that Gfi-1 is a direct C/EBPα target and that its expression is required for C/EBPα-dependent inhibition of proliferation but not for induction of differentiation in K562 cells (19).
Moreover, expression of wild type Gfi-1 but not of the transcription repression defective Gfi-1 P2A mutant inhibited proliferation and colony formation of p210BCR/ABL- expressing cell lines and primary CML cells (19), suggesting that Gfi-1-regulated genes may be targeted for therapeutic purposes in p210BCR/ABL-transformed cells. We show here that the effects of Gfi-1 depend, in part, on transcriptional repression of the expression of STAT 5B and Mcl-1, two genes required for proliferation and survival of hematopoietic stem cells (25–28). Moreover, genetic or pharmacological inhibition of STAT 5 and Mcl-1 expression/activity markedly suppresses proliferation and colony formation of p210BCR/ABL-expressing cells including primary CD34+ CML cells.
MATERIALS AND METHODS
Reagents
Obatoclax (Gemin X Pharmaceuticals, Malvern, PA), Pimozide (Sigma-Aldrich, St.Louis, MO) and Stat5 inhibitor (Novagen, San Diego, CA) were dissolved in DMSO (10, 39 and 25 mM, respectively) and stored at 4°C or –20°C.
Plasmids
MigRI-Gfi-1 and MigRI-Gfi-1 P2A were the kind gift of Dr J. Zhu (Laboratory of Immunology, NIH).
MigRI-Gfi-1-ER was generated by polymerase chain reaction (PCR) as described (19). pMSCV/IRES/YFP STAT5a H299R,S771F was cloned by releasing STAT5aH299R, S711F from pDNR STAT5 1*6 (29) and cloning it at EcoRI/XhoI restriction digestion sites of pMSCV/IRES/YFP.
The FG12 Mcl-1/IRES-GFP lentivirus was kindly provided by Dr Maria S. Soengas (Spanish National Cancer Centre, Madrid, Spain).
The Mcl-1 shRNA/GFP lentivirus (30) was kindly provided by Dr M. Andreeff (MD Anderson Hospital, Houston, Tx).
The STAT-5 and Mcl-1 shRNA pLKO.1 lentiviruses were purchased from Open Biosystems (Huntsville, AL, USA).
The promoter of the human STAT 5B gene (nucleotides −3464 to −3227) was obtained by PCR using the forward 5’-TGTGCCTAGAGCTGCAAGAA-3’, and the reverse 5’- AAAATTAGCTGGGCATGGTG-3’ and cloned at the SacI/XhoI sites of the pGL3 basic vector. The Gfi-1 binding site (b.s.)MUT STAT 5B promoter/Luc plasmid was generated by mutagenesis from wild type STAT-5B/Luc plasmid.
The proximal promoter of the human Mcl-1 gene (nucleotides −661 to +255) was amplified by PCR using the forward 5’-CCTATATGTAAATAGCACCAAG-3’, and the reverse 5’-GGCCTCCTTCTCCGTAGCC-3’ and cloned at the EcoRV site of the pGL3 basic vector. The Gfi-1 binding site (b.s.)MUT Mcl-1 promoter/Luc plasmid was generated by mutagenesis from the −661+255 wild type Mcl-1/Luc plasmid.
The deletion mutant −201 to +255 Mcl-1/Luc plasmid was generated by cloning a PCR fragment obtained with the forward 5’-TTACGTAACCGGCACTCAGA-3’, and the reverse-5’GCCTCCTTCTCCGTAGCC-3’ at the SacI/XhoI sites of the basic pGL3 vector.
Cell cultures and viral infection
Ph1 K562 and LAMA-84 cells and derivative cell lines were cultured in Iscove’s modified Dulbecco medium (IMDM) supplemented with heat-inactivated FBS. Fresh leukapheresis or peripheral blood samples were obtained with written informed consent from CML-chronic phase (CP) patients (n = 11). Cord blood cells were obtained from the Labor and Delivery Unit of Thomas Jefferson University. Samples were enriched for CD34+ cells using CliniMACS (Miltenyi Biotec, Auburn, CA) according to the manufacturer’s instructions. Purity of CML CD34+ cells was more than 95% and more than 95% were Ph1 as assessed by dual-color dual-fusion fluorescence in situ hybridization.
For certain colony formation assays of Obatoclax-treated cells, CD34+ CML cells were stained with phycoerythrin-conjugated anti-CD34 (BD Bioscience) and allophycocyanin-conjugated anti-CD38 (Miltenyi Biotec) antibodies and sorted to isolate CD34+CD38- and CD34+CD38+ fractions using a MoFlo sorter (Dako Cytomation).
CML and cord blood CD34+ cells were cultured in StemSpan SFEM medium (StemCell Technologies, Vancouver, Canada) supplemented with StemSpan CC100 (20 ng/mL IL-3 and IL-6, 100 ng/mL Kit Ligand (KL), 100 ng/mL Flt-3 ligand; StemCell Technologies) and 50 ng/mL thrombopoietin (StemCell Technologies).
For retroviral infections, amphotropic Phoenix cells were transiently transfected with the indicated plasmids. The infectious supernatant was collected 48 h later and was used to infect (a 48-h procedure) K562 or LAMA-84 cells. 24 h later, infected cells were sorted (EPICS Profile Analyzer; Coulter, Hialeah, FL) for green or yellow fluorescence protein (GFP/YFP) expression.
Lentiviral stocks were prepared by transient co-transfection of 293T cells with the scramble or the Mcl1/STAT 5B shRNA vector, the pCMVΔR8.74 encoding Gag, Pol, Tat and Rev, and the pMD.G plasmid encoding VSV-G), and ProFection® Mammalian Transfection according to guidelines provided by Promega (Madison, WI). The infectious supernatant was collected 48 h later and was used to infect K562 or CD34+ CML cells.
Infected cells were selected by GFP positivity or puromycin resistance (STAT 5 shRNA, TRCN0000020356 and TRCN0000020357; Mcl1 shRNA, TRCN000005518, lentiviruses, Open Biosystem).
The K562-C/EBPα-ER and the K562-C/EBPα-ER/Gfi-1shRNA cell lines were previously described (19).
Cell proliferation and colony formation assays
Proliferation (counts of viable cells), cell cycle analysis and colony formation assays of parental and derivative K562 cells were performed as described (19).
Real time Q-PCR analysis
For real time Q-PCR, total RNA was isolated using the RNeasy Mini kit (Qiagen, Hilden, Germany), digested with DNase-RNase free (Roche, Nutley, NJ) for 1h at 37 oC and deactivated for 15 min at 65 oC. 2μg was reverse-transcribed and the resulting cDNA used as PCR template. All reactions were done in triplicate. Primer pairs designed using the ABI Primer Expression Software are: human STAT 5B FW (5’- GTAAACCATGGCTGTGTGGA-3’), RV (5’-AAATAATGCCGCACCTCAAT-3’); STAT 5A FW (5’-ACTACACTCCTGTGCTGGCTA-3’), RV (5’- GGTCGAATTCTCCATCCTGA-3), human Mcl-1 FW (5’- TAAGGACAAAACGGGACTGG-3’), RV(5’-ACCAGCTCCTACTCCAGCAA-3’); human HPRT FW(5’-AGACTTTGCTTTCCTTGGTCAGG-3’), RV(5’-GTCTGGCTTATATCCAACACTTCG-3’). Real time PCR was done using the GoTaq® qPCR Master Mix (Promega) in a My IQ thermocycler (Biorad) and quantified using MyIQ software (Bio-Rad). HPRT, a housekeeping gene with constant expression, was used as an internal control to normalize input cDNA. For relative comparison of each gene, we used the MyIQ software that analyzed the Ct value of real-time PCR signals with the ΔΔCt method.
Chromatin immunoprecipitation (ChIP) assays
ChIP assays were performed using the EZ-ChIP assay kit (Upstate Biotechnoly, Lake Placid, NY). Briefly, exponentially growing Gfi-1-ER-K562 cells (untreated or 4-HT-treated, 24 h) were cross-linked with 1% formaldehyde, incubated for 10 min at room temperature (RT), and treated with glycine (125 mM for 5 min at RT). Cells were then washed with ice-cold PBS, resuspended in lysis buffer (1 ml) in the presence of protease inhibitors and sonicated at 28% power for 12 pulses of 10 s each in a Branson Sonifer 450 (Branson Ultrasonics, Danbury, CT). Chromatin was pre-cleared with 50 μl of protein A-agarose beads for 60 min at 4 °C with rotation, and pre-cleared lysates were immunoprecipitated with the anti-estrogen receptor α antibody (12 μg; SRA-1010 Stressgen) at 4 °C overnight. Immunoprecipitations without antibody (no antibody control) and an anti-mouse IgG were included in each experiment. Immune complexes were collected with 50 μl of protein A-agarose beads for 60 min at 4 °C (except for 10 μl of supernatant of the no antibody control, saved as Input) and washed once with 1 mL each of: low salt buffer (0.1% SDS; 1% Triton X-100; 2 mM EDTA; 20 mM Tris-HCl, pH 8.0; and 150 mM NaCl), high salt buffer (0.1% SDS; 1% Triton X-100; 2 mM EDTA; 20 mM Tris-HCl, pH 8.0; and 1.5 M NaCl), LiCl buffer (250 mM LiCl; 1%
NP-40; 1% sodium deoxycholate; 1 mM EDTA; and 10 mM Tris-HCl, pH 8.0); and twice with 10 mM Tris-HCl, pH 8.0 and 1 mM EDTA. Then, complexes were eluted with elution buffer (1% SDS and 0.1 M NaHCO3). Cross-links were reversed by heating at 65°C overnight in the presence of 0.2 M NaCl. ChIP DNA (2 μl) was next used as a template for real time Q- PCR using the STAT 5B (sense 5-GGGAGGAGGCTGTAGGATGG-3’, antisense 5’ - ACCTGGAGATGCAGCAATGTC-3’ from −3926 to-3778), STAT 5B (sense 5’ – GGCATTCCCACCCACCCTTC-3’ and antisense 5-GGAGGCTGAGGCAGGAGAATC-3’, from −3438 to −3288), STAT 5B (sense 5’-TGCCACCACACCATGAGACC-3’, antisense 5’-CCAGCTACTGAGGAGGCTAAGG-3’from −2173 to −2047) or the Mcl-1 (sense 5’-CCCGTCCTCTTCCTTCAACCC-3’ and antisense 5’-CGTGGCTACCTCTGTGCTTCC-3’, from −403 to −297) primers. ChIP q- PCR signals were analyzed with the Percent Input Method.
Luciferase reporter assays
293T cells plated into 6-well plates at 35×104 cells/ well and cultured overnight were transfected the following day with 1,5 μg of wild type or mutant STAT 5B or Mcl-1 promoter-Luc plasmids, 1/50 of Renilla reporter, and the MigRI empty vector or MigRI- Gfi-1 or MigRI-Gfi-1P2A expression vector for a total of 6μg DNA/ well using the Profection Mammalian Transfection System (Promega). After 48h, cells were washed, lysed and levels of firefly and Renilla luciferase activities measured using a dual luciferase assay kit (Promega). All transfections were performed in triplicates in three independent experiments.
Microarray analysis
RNA was isolated from 4-HT- or vehicle-treated Gfi-1-ER K562 cells, purified with RNAesy Column (Qiagen, CA) and used to generate biotin labeled cRNA. Ten micrograms of cRNA were fragmented and hybridized to HGU133 2.0 Plus arrays (Affymetrix, Santa Clara, CA). Signal values were determined using Gene Chip Operating System 1.0 (GCOS, Affymetrix). A qualifier was considered to be differentially expressed if: i) was detected in at least 50% of the samples, ii) the fold change was at least 1.5, and iii) the p-value based on t-test was ≤ 0.05.
Statistical Analyses
Means were compared using the unpaired, 2-tailed Student’s t test. A p value of less than 0.05 was considered statistically significant in all calculations.
RESULTS
Wild type Gfi-1 represses STAT 5B and Mcl-1 expression in K562 cells
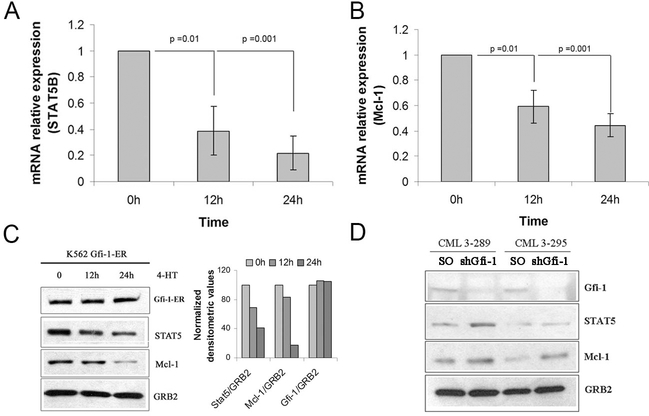
To identify transcription-regulated Gfi-1 targets which may be responsible for its biological effects, Gfi-1-regulated genes were identified by probing oligonucleotide microarrays with RNA from 4-HT-treated (12 or 24 h) Gfi-1-ER-K562 or MigRI-K562 cells. In triplicate experiments, expression of approximately 850 genes was downregulated or upregulated ≥1.5-fold upon Gfi-1 activation. Among the downregulated genes, expression of STAT 5B and Mcl-1 mRNA was reduced 1.6 and 1.5 fold, respectively (p> 0.001 for both); of interest, expression of STAT 5A was also decreased (1.2 fold; p= 0.02).
The decrease in STAT 5B and Mcl-1 transcripts in 4-HT-treated Gfi-1-ER-K562 cells was confirmed by real time PCR (Figure 1A and 1B). Of interest, expression of STAT 5A was also decreased (Supplementary Figure 1). Consistent with the lower mRNA expression, anti-STAT 5 and anti-Mcl-1 Western blotting of whole lysates of 4-HT- treated Gfi-1-ER-K562 cells demonstrated that levels of STAT 5 and Mcl-1 were also reduced (approximately 2- to 4-fold) after a 24-h Gfi-1 activation (Figure 1C).
Figure 1.
Expression of STAT 5 and Mcl-1 is downregulated by Gfi-1 in p210BCR/ABL-expressing K562 cells. Expression of STAT 5B (A) and Mcl-1 (B) mRNA measured by real time PCR in 4-HT-treated Gfi-1-ER- K562 cells; (C) Left, levels of STAT 5 and Mcl-1 protein in 4-HT-treated Gfi-1-ER-K562 cells; Right, densitometric analysis of STAT 5 and Mcl-1 expression after normalization for GRB2 expression; (D) Levels of STAT 5 and Mcl-1 in Gfi-1-silenced CD34+ CML cells.
STAT 5 and Mcl-1 were detected by anti-STAT5 rabbit polyclonal antibody (C-17; Santa Cruz Biotechnology), and anti-Mcl-1 rabbit polyclonal antibody (S-19; Santa Cruz Biotechnology, Santa Cruz, CA). Blots were normalized by anti-GRB2 (610112; BD Transduction Laboratories, Lexington, KY) Western blotting.
A decrease in STAT 5 and Mcl-1 expression was also observed upon activation of ectopically expressed Gfi-1-ER in Ph1 CML-blast crisis LAMA-84 cells (Supplementary Figure 2A); Gfi-1-ER activation also suppressed colony formation of these cells (Supplementary Figure 2B).
The functional relationship between Gfi-1 and STAT 5 or Mcl-1 expression was also assessed in primary CD34+ CML cells; CD34+ cells from two CM-chronic phase samples were transduced with a Puro-Gfi-1 shRNA lentivirus able to downregulate Gfi-1 expression (19); after selection in the presence of puromycin to kill residual non- transduced cells, lysates were prepared and subjected to western blotting to assess Gfi-1, Mcl-1 and STAT 5 expression. Gfi-1-silenced CML CD34+ cells express higher levels of STAT 5 and Mcl-1 than the scramble-transduced counterpart (Figure 1D).
STAT 5B and Mcl-1 are direct Gfi-1 targets
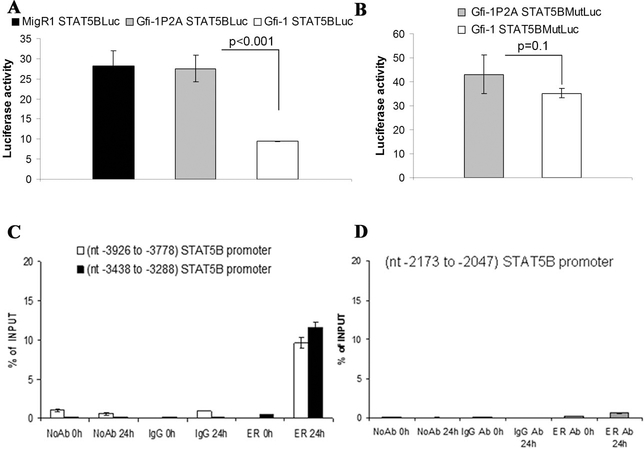
Search of the human STAT 5B promoter using transcription factor binding site databases (i.e. www.gene-regulation.com ) identified a putative Gfi-1 binding site at nucleotides - 3350 to −3344. Thus, we cloned the −3464 to −3227 promoter region in the basic pGL3- Luc plasmid and performed luciferase assays in 293T cells co-transfected with wild type Gfi-1 or the transcription repression-deficient Gfi-1P2A mutant. In three independent experiments, STAT 5B-driven luciferase activity was repressed approximately two-fold by wild type Gfi-1 (Figure 2A). Then, we performed luciferase assays using a reporter plasmid in which the putative Gfi-1 binding site (5-AAATC-3’) at nucleotides −3349– 3345 was mutated into the sequence 5’-AGTTA-3’. As shown in Figure 2B, expression of wild type Gfi-1 had minimal effects on the activity of this mutated promoter. PCR amplification of anti-ER chromatin immunoprecipitates from lysates of 4-HT-treated Gfi- 1-ER/K562 cells revealed that Gfi-1 bound segments of the STAT 5B promoter near to or including the putative Gfi-1 binding site responsible for Gfi-1-dependent repression of STAT 5B promoter activity but not a more proximal promoter segment which does not include putative Gfi-1 binding sites (Figure 2C and D).
Figure 2.
STAT 5B is a direct transcriptional target of Gfi-1. Effect of wild type Gfi-1 or the transcription repression defective Gfi-1P2A on luciferase activity driven by the (− 3464 to −3227) STAT 5B 5’ flanking region (A) or by the corresponding promoter with mutated Gfi-1 binding site (B). Values represent the mean + SD of three experiments performed in triplicate; (C and D) ChIP assays (representative of two experiments) showing the interaction of Gfi-1-ER with segments of the STAT 5B promoter.
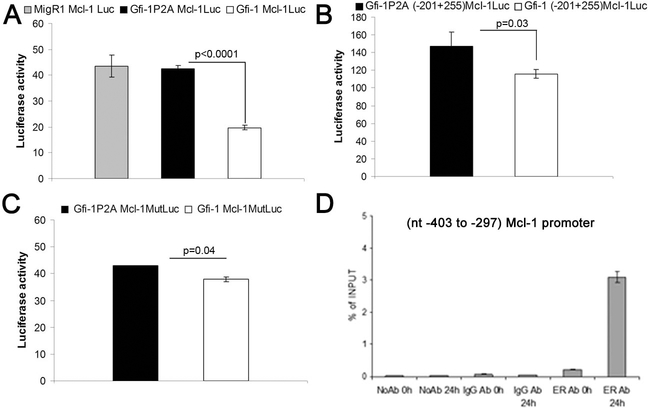
Luciferase assays were also performed on cell extracts of 293T cells co-transfected with the Mcl-1 5’ flanking sequence (−661 to +255)-Luc plasmid and the wild type or Gfi- 1P2A expression vector. Luciferase activity driven by this segment of the Mcl-1 promoter was repressed upon expression of wild type Gfi-1 but not of the transcription repression-deficient Gfi-1P2A mutant (Figure 3A). Search of this region of the Mcl-1 promoter identified a putative Gfi-1 binding site at nucleotides – 311 to – 307 (5’- AAATC-3’). Thus, we assessed the luciferase activity of a deletion mutant (−201 to + 255 Mcl-1/Luc) lacking this putative binding site after co-transfection of 293T cells with Gfi- 1 or the Gfi-1P2A mutant. Expression of wild type Gfi-1 induced only a modest (~ 20%) decrease in luciferase activity (Figure 3B), suggesting that the −661 to −202 segment of the Mcl-1 promoter contains a functional Gfi-1 binding site. Thus, we generated a reporter plasmid (−661 to +255 Mcl-1(Gfi-1 b.s. MUT)/Luc with a non functional putative Gfi-1 binding site at nucleotides −311 to −307 and assessed Gfi-1-regulated luciferase activity in 293 T cells. Wild type Gfi-1 still repressed luciferase activity driven by the mutant promoter but less than that driven by the promoter with an intact Gfi-1 binding site (Figure 3C, compare panels A and C); this suggests that additional Gfi-1 binding site(s) downstream of that at nucleotides −311 to −307 may be involved in the Gfi-1-dependent repression of the Mcl1 promoter or that Gfi-1 regulates the Mcl-1 promoter, in part, via indirect mechanisms. Since expression of STAT 5 is repressed by Gfi-1 (this study) and the Gfi-1-regulated (661 to +255) Mcl-1 promoter contains a putative STAT 5 binding site at nucleotides −231 to −226, we reasoned that the effects of Gfi-1 on the Mcl-1 promoter may depend, in part, on repression of STAT 5B expression. However, ectopic expression of STAT 5B did not enhance luciferase activity driven by the −661 to +255 Mcl-1 promoter (not shown), suggesting that Gfi-1 is unlikely to repress the Mcl-1 promoter via STAT 5. Moreover, wild type Gfi-1 was still able to repress the activity of the −201 to +255 Mcl-1 promoter which lacks the putative STAT 5 binding site (Figure 3B), suggesting that its effect is direct rather than STAT 5-dependent. Consistent with this interpretation, ChIP assays from lysates of 4-HT-treated Gfi-1-ER-K562 cells showed interaction of Gfi-1 with the Mcl-1 promoter as revealed by PCR amplification of a segment of the Mcl-1 5’ flanking region (Figure 3D).
Figure 3.
Mcl-1 is a direct transcriptional target of Gfi-1.
Effect of Gfi-1 on the luciferase activity driven by: (A), the (−661 to +255)Mcl-1 5’ flanking region; (B), the (−201 to +255) 5’ flanking region; (C) the (−661 to +255) 5’ flanking region carrying a mutated Gfi-1 b.s. at nucleotides −311 to −307. (D) ChIP assays (representative of two experiments)showing the interaction of Gfi-1-ER to a segment of the Mcl-1 promoter containing a putative Gfi-1 binding site (−311 to −307).
Ectopic expression of STAT 5B and Mcl-1 rescues the colony formation inhibitory effects of Gfi-1
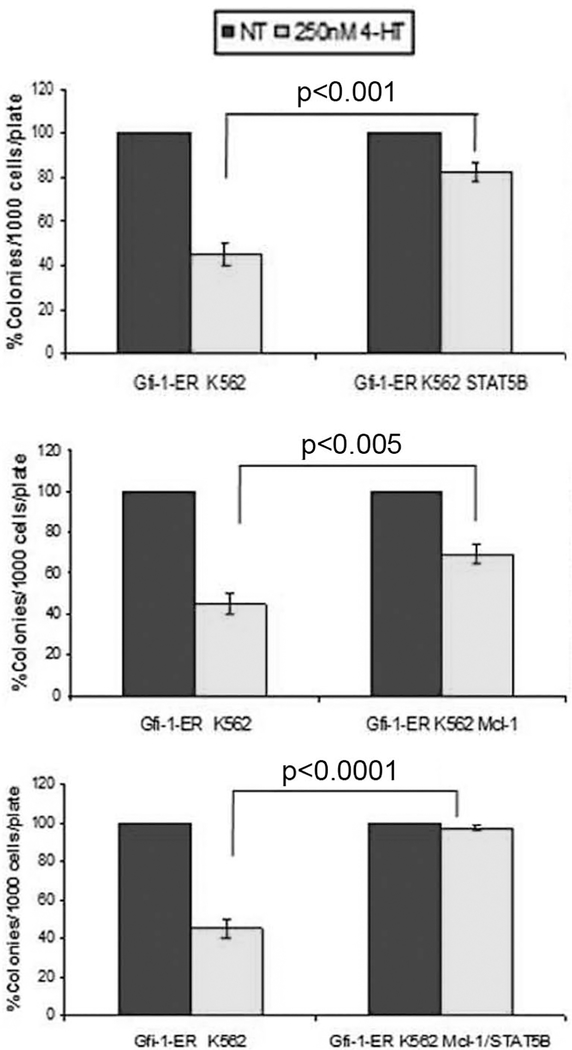
Activation of Gfi-1 markedly suppresses colony formation of 4-HT-treated Gfi-1-ER- K562 cells (ref 19; Figure 4). To assess whether the effect of Gfi-1 can be rescued by expression of STAT 5B and/or Mcl-1, we generated derivative Gfi-1-ER-K562 cell lines expressing STAT 5B, Mcl-1 or both and assessed colony formation upon treatment with 4-HT. As shown in Figure 4A and 4B, expression of STAT 5B or Mcl-1 rescued, in part, the colony formation inhibitory effect of Gfi-1; however, expression of both blocked completely the effect of Gfi-1 (Figure 4C). Thus, the effects of Gfi-1 could be mimicked by inhibition of STAT 5 and Mcl-1 expression and/or activity.
Figure 4.
Constitutive expression of STAT 5B and Mcl-1 rescues the colony formation inhibitory effect of Gfi-1 on K562 cells. Histograms show methylcellulose colony formation (mean + SD, three independent experiments) of untreated and 4-HT-treated:
(A) Gfi-1-ER-K562 or STAT 5B-expressing Gfi-1-ER-K562 cells; (B) Gfi-1-ER-K562 or Mcl-1-expressing Gfi-1-ER-K562 cells; (C) Gfi-1-ER-K562 or STAT 5B/Mcl-1- expressingGfi-1-ER-K562 cells. Colonies were counted 7 days after plating the indicated number of cells.
STAT 5 and Mcl-1 RNA interference suppresses colony formation of p210BCR/ABL-expressing cells
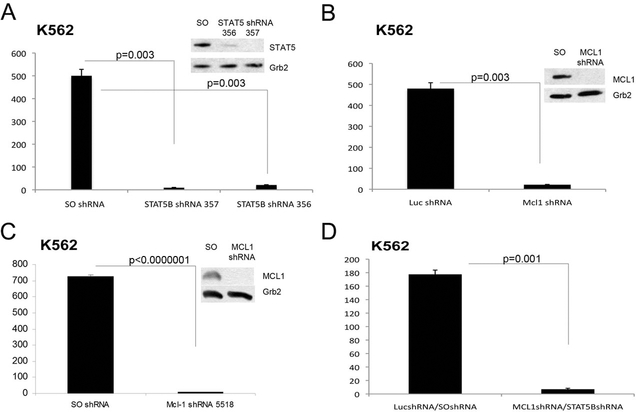
The requirement of Mcl-1 and STAT 5 expression was first assessed in K562 cells lentivally transduced with Mcl-1 and/or STAT 5 shRNAs or with scramble controls. Due to the high homology between STAT 5A and STAT 5B mRNAs, we could not identify shRNAs targeting only STAT 5B. K562 cells lentivirally transduced with the STAT 5B shRNA 356 or 357 showed an almost complete downregulation of STAT 5 expression which correlated with markedly decreased colony formation (Figure 5A). Likewise, Mcl- 1 expression was essentially undetectable in K562 cells transduced with the Mcl- 1shRNA/GFP lentiviral vector (Figure 5B) or the Mcl-1 shRNA/Puro vector (Figure 5C) and Mcl-1-silenced cells were markedly less clonogenic than the scramble-transduced counterpart (Figure 5B and 5C). As expected, colony formation of K562 cells co-infected with the Mcl-1/GFP and the STAT 5B 357/Puro shRNAs was markedly suppressed compared to that of the scramble-transduced counterpart (Figure 5D).
Figure 5.
Colony formation of K562 cells is suppressed by shRNA-mediated STAT 5 and/or Mcl-1 downregulation. Histograms represent methylcellulose colony formation (mean + SD, three experiments) of K562 cells lentivirally transduced with control or STAT 5B (A), Mcl-1 (B and C) or Mcl-1 and STAT 5B (D) shRNA and plated in methylcellulose (500 cells/plate) in the presence of puromycin (A, C and D) or after GFP sorting (B). For the experiments in panel D, co-infected cells were first GFP-sorted and then plated in methylcellulose in the presence of puromycin. Colonies were counted after 7 days. Insets show STAT 5 or Mcl-1 expression in shRNA lentivirus-transduced cells.
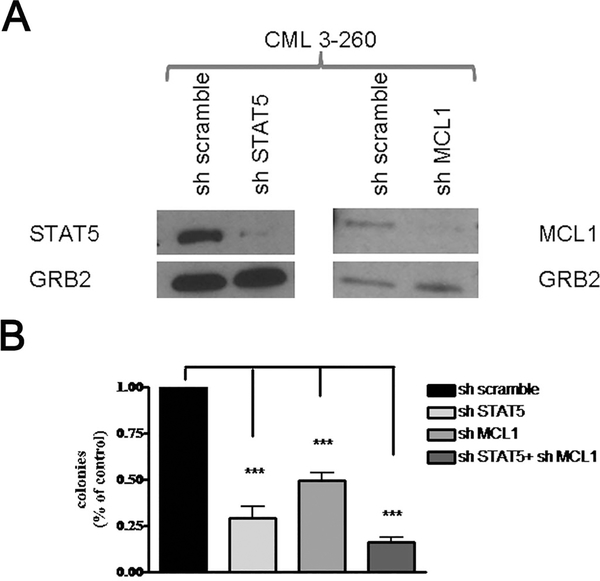
Then, we tested the requirement of STAT 5 and Mcl-1 expression in primary CML cells. CD34+ CML cells from five CP patients were lentivirally transduced with control or STAT 5B or Mcl-1 shRNAs, and plated in methylcellulose in the presence of puromycin (STAT 5B experiment) or after GFP sorting (Mcl-1 experiment). Colonies were counted after 12 days. For the experiments of Mcl-1 and STAT 5B shRNA-mediated downregulation, CD34+ cells were first transduced with the Mcl-1/GFP or the Luc/GFP shRNA lentivirus; GFP-positive cells were then transduced with the scramble or the STAT 5B/Puro shRNA lentivirus and plated in methylcellulose in the presence of puromycin. An aliquot of the lentivirally-transduced cells was maintained in liquid culture in the presence of puromycin (2 days; 2μM) and lysates were assessed for STAT 5 and Mcl-1 expression (Figure 6A). Colonies were scored after 12 days. Either shRNA suppressed colony formation of CD34+ CML cells but the combination of the STAT 5B and Mcl-1 shRNAs had a more potent inhibitory effect (Figure 6B).
Figure 6.
Colony formation of CD34+ CML cells is suppressed by shRNA-mediated STAT 5 and/or Mcl-1 downregulation. Peripheral blood CD34+ cells from CML patients (n=5) were lentivirally transduced with control shRNAs (scramble or Luc), STAT 5 or Mcl-1 shRNA and lysed to assess protein levels (A) or plated in methylcellulose (B) directly in the presence of puromycin (scramble and STAT 5 shRNA-transduced) or after sorting for GFP expression (Luc and Mcl-1 shRNA-transduced). For STAT 5 and Mcl-1 RNAi, GFP-positive cells (Luc or Mcl-1 shRNA-transduced) were re-infected with scramble or STAT 5 shRNA and plated in methylcellulose in the presence of puromycin (2 μM). Colonies were counted after 12 days. Data are presented as % colony formation inhibition by STAT 5 and/or Mcl-1 shRNA-transduced over that of scramble-transduced cells taken as 100. *** represent p value < 0.001.
Pharmacological inhibition of Mcl-1 and STAT 5 activity suppresses proliferation and colony formation of p210BCR/ABL-expressing cells
In contrast to ABT-737 which antagonizes Bcl-2 and BCL-XL (30, 31), Obatoclax is a pan-Bcl-2 inhibitor which also blocks the activity of Mcl-1 (32, 33).
Thus, we treated K562 cells with Obatoclax (500 nM) and assessed their proliferation and colony formation.
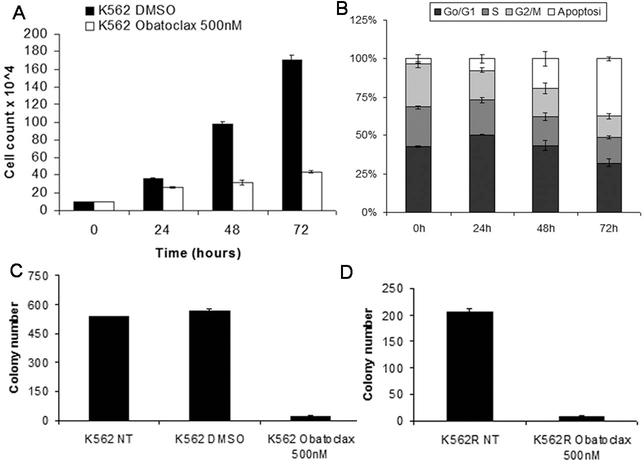
Compared to untreated cells, the number of Obatoclax-treated K562 cells was markedly reduced at each time point (Figure 7A). Obatoclax-treated cells showed a decrease in the S and G2/M phases and approximately 35% of the cells exhibited hypodiploid DNA content indicative of apoptosis (Figure 7B).
Figure 7.
Effects of the pan-Bcl-2 antagonist Obatoclax on proliferation and colony formation of K562 cells. Cell number (A), DNA content (B) and colony formation (C and D) of untreated or Obatoclax-treated parental (C) or TKI-resistant (D) K562 cells.
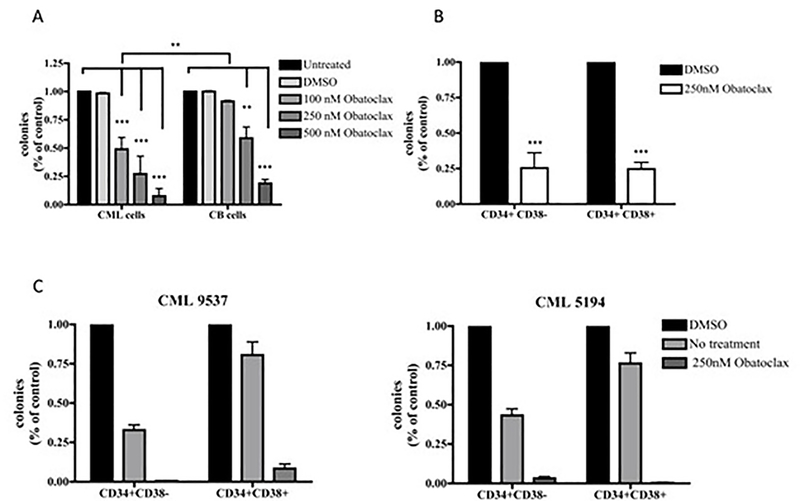
The decrease in proliferation and survival of Obatoclax-treated cells was confirmed by methylcellulose clonogenic assays. As shown in Figure 7C, clonogenic cells were less than 5% after treatment with Obatoclax. An identical inhibitory effect on colony formation was also observed upon Obatoclax treatment of the tyrosine kinase inhibitor (TKI)-resistant T315I-K562 derivative line (Figure 7D).Colony assays were also performed using Obatoclax-treated CD34+ cells from normal donors or CML chronic phase patients; as shown in Figure 8A, treatment with Obatoclax markedly decreased the number of clonogenic CD34+ CML cells whereas a larger fraction of normal cord blood CD34+ cells was spared especially upon treatment with a concentration of 100 nM. The effect of Obatoclax was also assessed on the CD34+CD38+ and the CD34+CD38-subsets purified from three CML samples; as shown in Figure 8B, treatment with Obatoclax appears to suppress with the same efficiency colony formation derived from these two progenitor subsets. To further investigate the effect of Obatoclax on these progenitor subsets, secondary colony formation assays were performed from two CML samples using cells from solubilized methylcellulose plates (Figure 8C); treatment with Obatoclax led to an almost complete suppression of colony formation from both subsets. Of interest, cells derived from the Obatoclax-treated CD34+CD38- subset and re-plated in methylcellulose without any treatment (No treatment colonies) formed fewer colonies of the CD34+CD38+ counterpart (Figure 8C), suggesting that the more primitive CML progenitors may be exquisitely sensitive to Obatoclax treatment.
Figure 8.
Effect of the pan-Bcl-2 antagonist Obatoclax on colony formation of normal and CML CD34+ cells. (A) Normal (n=3) or CML CD34+ (n=5) cells were cultured in the presence of Obatoclax (100, 250 or 500nM for 3 hours) and plated in methylcellulose (3 × 104 cells/ plate) supplemented with the same concentration of Obatoclax. Two CML samples were treated only with 500 nM Obatoclax; (B) CD34+CD38+ and CD34+CD38-cells from the same samples (n=3) were cultured in the presence of Obatoclax (250 nM, 3 hours) and plated in methylcellulose (2 × 104/plate) supplemented with the same concentration of Obatoclax; colonies were counted 12 days after plating. Results are expressed as percent inhibition (mean + SD) of colony formation from Obatoclax-treated versus untreated samples taken as 100. The difference in colony formation inhibition between normal cord blood versus CML CD34+ cells was statistically significant only at the 100 nM concentration; (C) Colonies derived from the CD34+CD38- or the CD34+CD38+ subsets [untreated (DMSO) or treated with Obatoclax] were solubilized and cells (untreated or Obatoclax-treated; 20,000 cells/plate) were re-plated in methylcellulose; colonies were counted after 12 days. Results are expressed as percent inhibition (mean +SD) of colony formation from Obatoclax-treated or untreated cells derived from primary plates of Obatoclax-treated cells (No treatment) versus untreated cells from control (DMSO) primary plates.
We also assessed the effects of N’-((4-oxo-4H-chromen-3-yl) methylene) nicotinohydrazide (Novagen) and pimozide (34), two recently described STAT 5 inhibitors; however, neither suppressed proliferation and colony formation of K562 cells as effectively as Obatoclax (Supplementary Figure 3).
Discussion
We reported previously that the transcription repressor Gfi-1 is a C/EBPα-regulated gene required for its proliferation-inhibitory effects but not for induction of myeloid differentiation in p210BCR/ABL-transformed cells (19). Moreover, silencing of Gfi-1 expression in primary CD34+ CML cells led to markedly increased primary and secondary colony formation (19), although these cells were no longer able to form colonies after the fourth or fifth re-plating (not shown). Based on these observations and the finding that ectopic expression of Gfi-1 suppressed colony formation of CD34+ CML cells (19), we reasoned that Gfi-1 may repress the expression of genes important for the proliferation and/or survival of primitive hematopoietic progenitors and that the products of these genes may be targeted by genetic or pharmacological approaches in leukemic cells. Candidate Gfi-1-regulated genes were identified in K562 cells expressing the 4- hydroxytamoxifen (4-HT)-regulated Gfi-1-ER protein. 12 hours after 4-HT treatment, almost 850 genes were downregulated or upregulated ≥ 1.5 fold; the upregulated genes were approximately 200, suggesting that some of the changes in gene expression induced by Gfi-1 may be indirect and/or Gfi-1 has hitherto overlooked transcription-enhancing effects. Among the genes whose expression was downregulated upon Gfi-1 activation, STAT 5B and Mcl-1 attracted our attention because of their involvement in proliferation and survival of hematopoietic stem cells (25–28). Both genes appear to be transcriptionally repressed by Gfi-1 directly. The Mcl-1 5’flanking region used in our luciferase assays contains a STAT 5 putative binding site and previous studies suggested that the expression of Mcl-1 may be regulated by STAT 5 transcriptionally (35,36); however, overexpression of STAT 5B did not enhance luciferase activity driven by the Mcl-1 promoter in 293 T cells. Moreover, silencing of STAT 5 expression in K562 cells only induced a modest decrease in Mcl-1 expression (not shown), suggesting that STAT 5 downregulation is not crucial for the effects of Gfi-1 on Mcl-1 expression.
In a recent study investigating the role of Gfi-1 in normal hematopoiesis (37), Gfi-1 knockout mice showed an expansion of early myeloid progenitors associated with increased expression of HoxA9, Pbx1 and Meis1 which appear to be direct targets of Gfi-1. However, expression of HoxA9 is low in K562 cells and activation of Gfi-1 did not induce a decrease in HoxA9 levels, suggesting that Gfi-1 exerts its effects through multiple downstream effectors.
The functional relationship between Gfi-1 and STAT 5B and Mcl-1 is supported by the increased expression of STAT 5 and Mcl-1 in Gfi-1-silenced primary CD34+ CML cells and by the finding that the colony formation inhibitory effect of Gfi-1 on K562 cells was rescued by expression of STAT 5B and/or Mcl-1.
Together, these studies suggest that Gfi-1 plays an intermediate role in a C/EBPα- dependent regulatory pathway whereby increased levels of C/EBPα promote a decrease in the expression of proliferation and survival regulatory genes such as STAT 5B and Mcl-1. Indeed, activation of C/ΕΒα in K562 cells induced a decrease in the expression of STAT 5 and Mcl-1 which was blocked in Gfi-1-silenced C/EBPa-ER-K562 cells (Supplementary Figure 4). The decrease in expression/activity of STAT 5 and Mcl-1 may be important not only for the ability of different levels of C/EBPα expression to influence self-renewal and commitment to granulocyte/monocyte progenitors of normal hematopoietic stem/progenitor cells (5, 7) but also for the consequences on the proliferation and survival of myeloid leukemia cells.
Although downregulation of STAT 5 and Mcl-1 expression has been implicated in the anti-leukemia effects of several compounds such as sorafenib, PKC-412, omacetaxine, and quercetin (38–41), there is no direct genetic evidence in support of an essential role of these two genes for the proliferation and survival of p210BCR/ABL-transformed cells.
As shown here, STAT 5 or Mcl-1 downregulation led to a complete eradication of K562 clonogenic cells and markedly suppressed colony formation of CD34+ CML cells.
Compared to K562 cells, the less potent inhibitory effect on colony formation of CML primary cells probably reflects the residual expression of these two proteins in primary CML cells rather than an intrinsic differential requirement for STAT 5 and/or Mcl-1 between K562 and primary CD34+ CML cells.
Although specific pharmacological inhibitors of Mcl-1 and STAT 5B are not yet available, we thought that it would be informative to test whether Obatoclax, a Bcl-2 family antagonist which also inhibits Mcl-1 (32), and Pimozide, a neuroleptic drug recently identified as an inhibitor of STAT 5-dependent transcription (34), would be as effective as STAT 5 and Mcl-1 RNA interference in suppressing proliferation and survival of p210BCR/ABL-transformed cells.
Although preliminary, our data suggest that Obatoclax is highly effective in inhibiting colony formation of CD34+ CML cells (including the more primitive CD34+CD38- subset) and could be used at concentrations that may spare a high proportion of normal progenitors; in comparison, Pimozide is considerably less effective even if used at micromolar concentrations.
In summary, Gfi-1 appears to be a crucial mediator of the proliferation and survival inhibitory effects of C/EBPα in p210BCR/ABL-transformed cells via transcription repression of STAT 5B and Mcl-1, two “usual suspects” for an important role in normal hematopoiesis and in leukemogenesis (25–28).
The functional relationship between Gfi-1 and STAT 5B and Mcl-1 expression is likely to be altered in other types of myeloid leukemia with genetic or functional inactivation of C/EBPα (8, 9); like p210BCR/ABL-transformed cells, these leukemic cells may also rely on STAT 5 and Mcl-1 expression and be exquisitely sensitive to inhibition of their activity.
Supplementary Material
Acknowledgments
We thank Dr Michael Andreeff (MD Anderson Hospital, Houston, TX) for the kind gift of the Mcl-1 shRNA/GFP lentiviral plasmid.
This work was supported by National Cancer Institute grants CA 95111 and P01 78890 (B.C). M.R.L was supported by a fellowship of the American-Italian Cancer Foundation (AICF).A.R.S. and G.F-A. were supported by a fellowship of the Associazione Italiana Ricerca sul Cancro (AIRC). S.C. was supported, in part, by a fellowship of Fondazione Cassa di Risparmio di Vignola (MO).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary information is available at Leukemia’s website.
References
- 1.Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene 26: 6816–6828, 2007. [DOI] [PubMed] [Google Scholar]
- 2.Laslo P, Spooner CJ., Warmflash A, Lancki DN, Lee HS, Sciammes R et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell 126: 755–766, 2006. [DOI] [PubMed] [Google Scholar]
- 3.Chou ST., Khandros E, Baley LC, Nichols KE, Vakoc CR, Yao Y et al. Graded expression of PU.1/Sfp1 gene transcription by GATA factors regulates hematopoietic cell fate. Blood 114:983–994, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakamoto H, Dai G, Tsujino K, Huang X, Fujimoto T,Mucenski M et al. Proper levels of c-Myb are discretely defined at distinct steps of hematopoietic cell development. Blood 108: 896–903, 2006. [DOI] [PubMed] [Google Scholar]
- 5.Zhang P, Iwasaki-Arai J, Iwasaki H, Fenyus ML., Dayaram T, Owens BM et al. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBPalpha. Immunity 21: 853–863, 2004. [DOI] [PubMed] [Google Scholar]
- 6.Pabst T and Mueller BU. Transcriptional dysregulation during myeloid transformation in AML. Oncogene 26: 6829–6837, 2007. [DOI] [PubMed] [Google Scholar]
- 7.Radomska HS., Huettner CS, Zhang P, Chen T, Scadden DT and Tenen DG CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol. Cell. Biol. 18: 4301–4314, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat. Rev. Cancer 3: 89–101, 2003. [DOI] [PubMed] [Google Scholar]
- 9.Nerlov C C/EBPα mutations in acute myeloid leukemias. Nat. Rev. Cancer 4: 394–400, 2004. [DOI] [PubMed] [Google Scholar]
- 10.Perrotti D, Cesi V, Trotta R, Guerzoni C, Santilli G, Campbell K et al. BCR- ABL suppresses C/EBPα expression through inhibitory action of hnRNP E2. Nat. Genet, 30: 48–58, 2002. [DOI] [PubMed] [Google Scholar]
- 11.Tavor S, Park DJ, Gery S Vuong PT, Gombart AF, Koeffler HP Restoration of C/EBPα exression in BCR-ABL+ cell line induces terminal granulocytic differentiation. J. Biol. Chem, 278: 52651–52659, 2003. [DOI] [PubMed] [Google Scholar]
- 12.Pabst T, Mueller BU, Harakawa N, Schoch C, Haferlach T, Behre G et al. AML1-ETO downregulates the granulocytic differentiation factor C/EBPalpha in t(8; 21) myeloid leukemia. Nat. Med, 7: 444–451, 2001. [DOI] [PubMed] [Google Scholar]
- 13.Ferrari-Amorotti G, Keeshan K, Zattoni M, Guerzoni C, Iotti G, Donato N, and Calabretta B Leukemogenesis induced by wild-type and ST1571-resistant BCR/ABL is potently suppressed by C/EBPα. Blood, 108: 1353–1362, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keeshan K, Santilli G, Corradini F, Perrotti D, and Calabretta B Transcription activation function of C/EBPα is required for induction of granulocytic differentiation. Blood 102: 1267–1275, 2003. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Iakova P, Wilde M, Welm A, Goode T, Roesler WJ, Timchenko NA. C/EBPalpha arrests cell proliferation through direct inhibition of Cdk2 and Cdk4. Mol Cell. 8:817–28, 2001. [DOI] [PubMed] [Google Scholar]
- 16.Xu X, Lindberg B, Wewer UM, Friis-Hansen L, Nerlov C. E2F repression by C/EBPalpha is required for adipogenesis and granulopoiesis in vivo. Cell 107:247–58, 2001. [DOI] [PubMed] [Google Scholar]
- 17.Müller C, Calkhoven CF, Sha X, Leutz A. The CCAAT enhancer-binding protein alpha (C/EBPalpha) requires a SWI/SNF complex for proliferation arrest. J. Biol. Chem. 279:7353–8, 2004. [DOI] [PubMed] [Google Scholar]
- 18.D’Alo’ F, Johansen LM, Nelson EA, Radomska HS, Evans EK, Zhang P et al. The amino terminal of E2F interaction domains are critical for C/EBP alpha-mediated induction of granulopoietic development of hematopoietic cells. Blood, 102: 3163–3171, 2003. [DOI] [PubMed] [Google Scholar]
- 19.Lidonnici MR., Audia A, Soliera AR, Prisco M, Ferrari-Amorotti G, waldron T et al. Expression of the transcription repressor Gfi-1 is regulated by C/EBP(alpha) and is involved in its proliferation and colony formation-inhibitory effects in p210BCR/ABL- expressing cells. Cancer Res. 70: 7949–7959, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hock H, Haniblen MJ, Rooke HM, Schindler JW, Saleque S, Fujiwara Y and Orkin SH Gfi-1 restricts proliferation and preserves functional integrity of hematopoietic stem cells. Nature, 431: 1002–1007, 2004. [DOI] [PubMed] [Google Scholar]
- 21.Zeng H, Yucel R, Kosan C, Klein-Hitpass L, and Moroy T Transcription factor Gfi-1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J 23: 4116–4125, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Elf SE, Miyata Y, Sashida G, Liu Y, et al. P53 regulates hematopoietic stem cell quiescence. Cell Stem Cell 4: 37–48, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dahl R, Iyer SR., Owens KS, Cuylear DD, and Simon MC The transcriptional repressor GFI-1 antagonizes PU.1 activity through protein-protein interaction. J. Biol. Chem. 282: 6473–6483, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hock H, Hamblen MJ, Rooke HM, Trover D, Bronson RT, Cameron S, and Orkin SH Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity, 18: 109–120, 2003. [DOI] [PubMed] [Google Scholar]
- 25.Schuringa JJ, Chung KY., Morrone G and Moore MA. Constitutive activation of STAT5A promotes human hematopoietic stem cell self-renewal and erythroid differentiation. J. Exp. Med. 200: 623–635, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wierenga ATJ., Vellenga E, and Schuringa JJ. Maximal STAT5-induced proliferation and self-renewal at intermediate STAT5 activity levels. Mol.Cell Biol. 28: 6668–6680, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Opferman JT., Iwasaki H, Ong CC, Suh H, Mizuno S, Akashi K and Korsmeyer SJ. Obligate role of anti-apoptotic MCL-1 in the survival of hematopoietic stem cells. Science 307: 1101–1104, 2005. [DOI] [PubMed] [Google Scholar]
- 28.Campbell CJV., Lee JB, Levadoux-Martin M, Wynder T, Xenocostas A, Leber B, and Bathia M The human stem cell hierarchy is defined by a functional dependence on Mcl-1 for self-renewal capacity. Blood 116: 1433–1442, 2010. [DOI] [PubMed] [Google Scholar]
- 29.Onishi M, Nosaka T, Misawa ALF., Gorman DM., McMahon M, Miyajima A, and Kitamura T Identification and characterization of a constitutively active STAT 5 mutant that promotes cell proliferation. Mol. Cell. Biol. 18: 3871–3879, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oltersdorf T, Elmore SW., Shoemaker AR., Armstrong RC., Augeri DJ, Belli BA et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 435: 677–681, 2005. [DOI] [PubMed] [Google Scholar]
- 31.Konopleva M, Contractor R, Tsao T, Samudio I, Ruvolo PP., Kitada S et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell 10: 375–388, 2006. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen M, Marcellus RC., Roulston A., Watson M, Serfass L Murthy Madirayan SR et al. Small molecule obatoclax (GX15–070) antagonizes MCL-1 and overcomes MCL-1-mediated resistance to apoptosis. Proc. Natl. Acad. Sci. USA 104: 19512–19517, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konopleva M, Watt J, Contractor R, Tsao T, Harris D, Estrov Z, et al. Mechanisms of antileukemic activity of the novel Bcl-2 homology domain-3 mimetic GX15–07 (obatoclax). Cancer Res. 68: 3413–3420, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson EA., Walker SR., Weisberg E, Bar-Natan M, Barrett R, Gashin LB., Terrel S et al. The STAT 5 inhibitor pimozide decreases survival of chronic myelogenous leukemia cells resistant to kinase inhibitors. Blood 117: 3421–3429, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerenyi MA., Grebien F, Gehart H, Schirfer M, Artaker M, Kovacic B et al. Stat5 regulates cellular iron uptake of erythroid cells via IRP-2 and TfR-1. Blood 112: 3878–3888, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aichberger KJ., Mayerhofer M, Ktauth M-T, Skvana H, Florian S, Sonneck K et al. Identification of mcl-1 as a BCR/ABL-dependent target in chronic myeloid leukemia (CML): evidence for cooperative antileukemic effects of imatinib and mcl-1 antisense oligonucleotides. Blood 105: 3303–3311, 2005. [DOI] [PubMed] [Google Scholar]
- 37.Horman SR., Velu SC, Chaubey A, Bourdeau T, Zhu J, Paul WE et al. Gfi-1 integrates progenitor versus granulocytic transcriptional programming. Blood 113: 5466–5475, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rahmani M, Nguyen TK., Dent P, and Grant S The multikinase inhibitor sorafenib induces apoptosis in highly imatinib mesylate-resistant bcf/abl+ human leukemia cells in association with signal transducer and activation of transcription 5 innhibition and myeloid cell leukemia-1 downregulation. Mol. Pharmacol. 72: 788–795, 2007. [DOI] [PubMed] [Google Scholar]
- 39.Yoshimoto G, Miyamoto T, Jabbarzadeh-Tabrizi S, Iino T, Rocnick JL., et al. FLT3-ITD upregulates MCL-1 to promote survival of stem cells in acute myeloid leukemia via FLT3-ITD-specific STAT 5 activation. Blood 114: 5024–5043, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allan EK., Holyoake TL, Craig AR, and Jorgensen HG. Omacetaxine may have a role in chronic myeloid leukaemia eradication through downregulation of Mcl-1 and induction of apoptosis in stem/progenitor cells. Leukemia 25: 985–994, 2011. [DOI] [PubMed] [Google Scholar]
- 41.Cheng S, Gao N, Zhang Z, Chen G, Budhraja A, Ke Z et al. Quercetin induces tumor-selective apoptosis through downregulation of Mcl-1 and activation of Bax. Clin. Cancer Res. 16: 5679–5691, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.